Significance
Glycosylation is often critical for the biological activity of plant natural products. Here we identify 2 enzymes required for glycosylation of the antifungal triterpene glycoside avenacin A-1 and show that 1 of these (AsUGT91G16) is a classical cytosolic uridine diphosphate-dependent glycosyltransferase, while the other (AsTG1) is a noncanonical vacuolar sugar transferase belonging to glycosyl hydrolase family 1. The genes encoding these 2 enzymes form part of the avenacin biosynthetic gene cluster. Our findings elucidate the steps required for the biosynthesis of the branched oligosaccharide chain required for antifungal activity of avenacin A-1 and thus for disease resistance in oat. They further highlight the importance of noncanonical sugar transferases in natural product glycosylation.
Keywords: natural products, glycosylation, plant defense, biosynthetic gene clusters, saponins
Abstract
Plants produce an array of natural products with important ecological functions. These compounds are often decorated with oligosaccharide groups that influence bioactivity, but the biosynthesis of such sugar chains is not well understood. Triterpene glycosides (saponins) are a large family of plant natural products that determine important agronomic traits, as exemplified by avenacins, antimicrobial defense compounds produced by oats. Avenacins have a branched trisaccharide moiety consisting of l-arabinose linked to 2 d-glucose molecules that is critical for antifungal activity. Plant natural product glycosylation is usually performed by uridine diphosphate-dependent glycosyltransferases (UGTs). We previously characterized the arabinosyltransferase that initiates the avenacin sugar chain; however, the enzymes that add the 2 remaining d-glucose molecules have remained elusive. Here we characterize the enzymes that catalyze these last 2 glucosylation steps. AsUGT91G16 is a classical cytosolic UGT that adds a 1,2-linked d-glucose molecule to l-arabinose. Unexpectedly, the enzyme that adds the final 1,4-linked d-glucose (AsTG1) is not a UGT, but rather a sugar transferase belonging to Glycosyl Hydrolase family 1 (GH1). Unlike classical UGTs, AsTG1 is vacuolar. Analysis of oat mutants reveals that AsTG1 corresponds to Sad3, a previously uncharacterized locus shown by mutation to be required for avenacin biosynthesis. AsTG1 and AsUGT91G16 form part of the avenacin biosynthetic gene cluster. Our demonstration that a vacuolar transglucosidase family member plays a critical role in triterpene biosynthesis highlights the importance of considering other classes of carbohydrate-active enzymes in addition to UGTs as candidates when elucidating pathways for the biosynthesis of glycosylated natural products in plants.
Collectively, plants synthesize an enormous array of chemicals. These compounds have important ecological functions, such as providing protection against abiotic stress, pests, and pathogens; serving as attractants for pollinators and seed dispersal agents; enabling establishment of symbiotic interactions; and shaping plant microbiomes. Metabolic diversification is therefore most likely a reflection of adaptation to survival in different ecological niches. Plant natural products are derived from a repertoire of different types of scaffolds. These scaffolds are often further modified (by, e.g., oxidation, methylation, acylation, and glycosylation) to generate a wealth of chemical diversity, with more than 1 million specialized metabolites reported from plants so far (1). Among these modifications, glycosylation has a profound impact on the solubility, cellular localization, and bioactivity of the molecules that plants produce (2).
Enzymes that build or break down the diverse glycosylated structures found in living organisms are collectively referred to as carbohydrate-active enzymes (CAZymes) (3). CAZymes include the glycosytransferases, a large enzyme superfamily that has been classified into 106 different enzyme families (http://www.cazy.org/glycosyltransferases.html). The enzymes primarily responsible for glycosylation of plant natural products belong to glycosyltransferase family 1 (GT1) (4, 5). GT1 enzymes use uridine diphosphate (UDP)-activated sugar donors to transfer sugar units onto small molecules and thus are referred to as UDP-dependent glycosyltransferases (UGTs). GT1 enzymes make major contributions to the glycodiversification of plant natural products (4, 5). They are also involved in the regulation of plant growth via modulation of phytohormone homeostasis (6), and they enable xenobiotic degradation through glucoconjugation, a step that precedes transfer of the modified xenobiotic to the vacuole (7).
Triterpene glycosides are one of the largest and most structurally diverse groups of plant natural products. These molecules are also referred to as saponins (a name derived from sapo, the Latin word for soap) because of their surfactant properties. The names of some triterpene glycoside-producing plant species—for example, soapwort (Saponaria offininalis), soapberry (Sapindus species), and soapbark (Quillaja saponaria)—reflect their original use as sources of soap. Triterpene glycosides display a tremendous range of structural diversity and biological activities. They protect plants against attack by pests and pathogens and can determine other agronomically important traits, such as flavor; for example, they are associated with bitterness in soybean. They are also of considerable interest for industrial and pharmaceutical applications (8, 9). Examples include the sweetener glycyrrhizin, from licorice, the vaccine adjuvant QS-21 from Q. saponaria, and ginsenosides, heath-beneficial triterpene glycosides from ginseng (10).
Triterpene glycosides commonly have 1 or more simple or branched sugar chains, often consisting of 3 or more sugars, attached to the triterpene scaffold. These sugars are critical for many of the bioactive properties of these compounds (2). Despite the importance of glycosylation for biological activity, the enzymes required for the biosynthesis of triterpene sugar chains are not well understood. To date, 26 triterpene UGTs have been characterized, of which 19 add sugars directly to the scaffold and 7 add a second sugar to give disaccharide moieties (11–21). The discovery of enzymes capable of the glycodiversification of triterpene scaffolds will open up new opportunities to exploit this important class of compounds for agronomic, medicinal, and industrial biotechnology applications using synthetic biology approaches.
Here we elucidate the steps required for glucosylation of antimicrobial triterpene glycosides known as avenacins, which confer disease resistance in oat (Avena species) (22). The major avenacin, A-1 (1) (Fig. 1A and SI Appendix, Table S1), is produced in the roots and provides protection against soil-borne pathogens (22). The branched trisaccharide chain attached to the triterpene scaffold is critical for antifungal activity and disease resistance (22–25). This sugar chain consists of 1 l-arabinose molecule and 2 d-glucose molecules. We recently identified the enzyme that initiates this sugar chain by adding l-arabinose to the carbon 3 position of the triterpene scaffold (AsAAT1/UGT99D1) (11); however, the enzyme(s) required for the addition of the remaining 1,2- and 1,4-linked d-glucose molecules were not previously known. Here, using a combination of transcriptomics, phylogenetics, and functional analysis, we identify and characterize these enzymes. We show that AsUGT91G16, a classical cytosolic UGT, adds a 1,2-linked d-glucose molecule to the l-arabinose. Unexpectedly, the enzyme that adds the final 1,4-linked d-glucose is not a UGT, but rather a sugar transferase belonging to glycosyl hydrolase family 1 (GH1). Unlike classical UGTs, this enzyme (AsTG1) is vacuolar rather than cytosolic. We further show that AsTG1 is synonymous with Sad3, a locus that we had previously defined in a forward mutagenesis screen as being required for avenacin biosynthesis (22, 26), and that AsUGT91G16 along with AsTG1 form part of the avenacin biosynthetic gene cluster (27, 28).
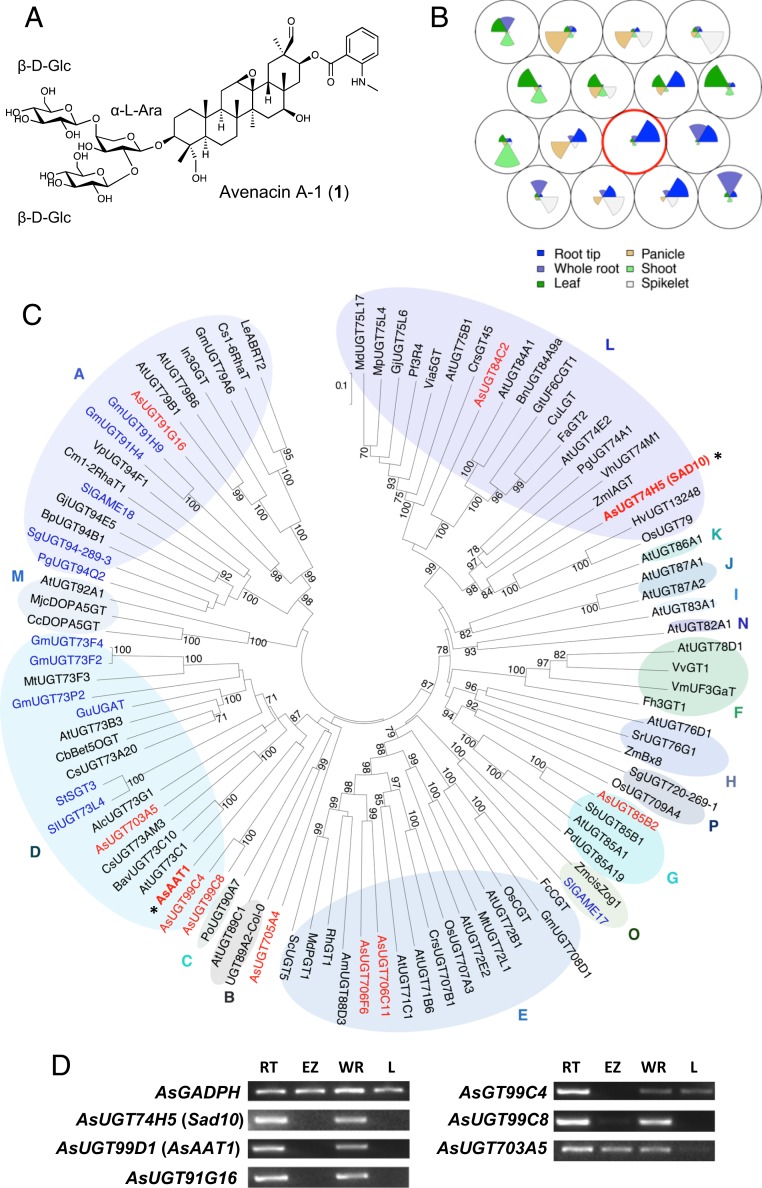
Fig. 1.
Mining for candidate avenacin glycosyltransferase genes. (A) Structure of avenacin A-1 (1), the major avenacin found in oat roots (all triterpene structures referred to in this work are numbered in parenthesis in bold, and their full names and structures are provided in SI Appendix, Table S1). (B) SOM of A. strigosa transcriptomic data for 6 tissues: root tip, whole root, leaf, panicle, shoot, and spikelet. Each circular node represents A. strigosa transcripts with similar expression profiles, with the average expression profile plotted within each node. The node containing avenacin biosynthetic genes is circled in red. (C) Phylogenetic analysis of A. strigosa UGT candidates (red) with characterized UGTs from other plant species (listed in SI Appendix, Tables S4 and S5). Functionally characterized glycoside glycosyltransferases (SI Appendix, Table S6) are indicated in blue, and the characterized avenacin biosynthetic pathway UGTs AsUGT74H5 (Sad10) (29) and AsUGT99D1 (AsAAT1) (11) are marked by asterisks. The UGT phylogenetic groups (groups A to P) are labeled as described previously (37, 38). The tree was constructed using the neighbor-joining method with 1,000 bootstrap replicates (the % support for branch points is shown). The scale bar shows 0.1 substitutions per site at the amino acid level. (D) RT-PCR expression profiles of genes for candidate UGTs belonging to groups A and D. The previously characterized avenacin biosynthesis genes AsUGT74H5 (Sad10) and AsUGT99D1 (AsAAT1) and the AsGAPDH housekeeping gene are included as controls. RNA was extracted from 3-d-old A. strigosa seedlings. RT, root tip; EZ, elongation zone; WR, whole root; L, leaf.
Results and Discussion
Glycosylation of plant natural products is usually carried out by uridine diphosphate-dependent glycosyltransferases (UGTs) belonging to the carbohydrate-active enzyme (CAZY) glycosyltransferase 1 (GT1) family (4, 5). Indeed, AsAAT1 is a member of this family (11). Thus, we hypothesized that the addition of the 1,2- and 1,4-linked d-glucose molecules to l-arabinose may be carried out by 1 or more GT1 UGTs. AsAAT1 and the other previously characterized avenacin pathway genes are expressed preferentially in the tips of oat roots, the site of accumulation of avenacin A-1 (11, 28–33). We exploited this highly tissue-specific expression pattern to identify potential UGT candidate genes in diploid oat (Avena strigosa) using a transcriptome mining approach.
RNA-seq data were generated for 6 different tissues: whole roots, root tips, leaves, panicles, shoots, and spikelets (SI Appendix, Table S2, http://db.ncgr.ac.cn/oat/RNAseq.php and Dataset S1). Gene expression profiles were clustered using a self-organizing map (34, 35). This methodology provides an unbiased and unsupervised method for clustering large datasets and has previously been used to identify genes involved in plant-specialized metabolic pathways (36). Six transcripts corresponding to the known avenacin biosynthetic pathway genes (including AsAAT1) mapped to a node of the self-organizing map that was of high quality, indicating the coexpression of these genes (Fig. 1B and SI Appendix, Figs. S1 and S2). Within the transcripts that clustered with the avenacin biosynthetic pathway genes in 100% of self-organizing map runs, 17 transcripts were identified as UGT-like, of which 9 were predicted to encode full-length sequences (SI Appendix, Tables S3 and S4 and Dataset S2).
We then carried out phylogenetic analysis of the predicted amino acid sequences of the 9 new candidate UGTs (Fig. 1C) along with characterized UGTs from other plants, including those previously reported to glycosylate triterpenoids (SI Appendix, Table S5). The phylogenetic tree approximately recapitulated the monophyletic groups A to P previously defined by Ross et al. (37) and Caputi et al. (38). Four oat UGTs from groups A and D were prioritized for further investigation, since both of these groups contain enzymes known to extend triterpene sugar chains (SI Appendix, Table S6). Gene expression analysis revealed that the genes for 2 of these (AsUGT91G16 and AsUGT99C8) had expression patterns closely resembling those of the characterized avenacin pathway genes (Fig. 1D).
AsUGT91G16 and AsUGT99C8 were expressed in Escherichia coli as N-terminal 9X histidine-tagged proteins. Protein preparations enriched for the recombinant enzymes were prepared using immobilized metal affinity chromatography and incubated with bis-deglucosyl avenacin A-1 (2) (SI Appendix, Table S1) (generated by the enzymatic hydrolysis of avenacin A-1) in the presence of UDP-glucose. AsUGT91G16 generated a more polar product with a mass consistent with addition of a hexose when analyzed by HPLC-MS (Fig. 2A), while AsUGT99C8 did not yield any detectable product (SI Appendix, Fig. S3).
Fig. 2.
Functional analysis of AsUGT91G16. (A) Heterologous expression in E. coli. HPLC-UV-MS chromatograms following incubation of recombinant AsUGT91G16 with deglucosylated avenacin A-1 (2) and UDP-glucose are shown. Protein preparations from E. coli transformed with an expression vector containing recombinant AsUGT91G16 yielded a new product peak at 4.8 min with the expected mass of (2) with an added hexose (m/z = 931.1). Protein preparations for the control reaction were boiled at 95 °C for 10 min. The minor peak at 4.1 min is likely a degradation product that has lost the unstable C12-C13 epoxide group. Data are representative of 2 separate experiments. Absorbance was measured at 357 nm. (B) Transient expression in N. benthamiana. HPLC-CAD-MS traces for extracts from agroinfiltrated leaves are shown. Coexpression of AsbAS1/SAD1, AsCYP51H10/SAD2, and AsAAT1/UGT99D1 yields 3β-(α-l-arabinopyranosyloxy)-12,13β-epoxy, and 16β-hydroxy-β-amyrin (3). The addition of AsUGT91G16 resulted in consumption of (3) and the appearance of new peaks (4a) and (4b), which have masses consistent with being (3) with the addition of 1 hexose [m/z = 751], along with 2 other minor peaks with masses consistent with the addition of 2 hexoses [m/z = 913]. The NMR structure of the AsUGT91G16 product isolated following large-scale agroinfiltration and extraction (4b) is shown (NMR assignment shown in SI Appendix, Table S7). (4b) is most likely a degradation product of (4a) generated by loss of the epoxide. IS, internal standard (digitoxin). (C) Analysis of wild-type and mutant oat lines. HPLC-CAD analysis of methanolic root extracts is shown. Root extracts from the wild-type A. strigosa line (S75) contain the avenacins A-1 (1), A-2 (5), B-1 (6), and B-2 (7) (39). The sad1 mutant lacks all 4 avenacins. The sad4 mutant accumulates the 4 avenacins and monodeglucosylated forms of the 2 major avenacins, A-1 and A-2 (8) and (9). The sad3 mutant accumulates monodeglucosyl avenacins (8) and (9). The 4 asugt91g16 mutants (#85, #543, #1073, and #1473) lack avenacins and instead accumulate 2 major products, (10) and (11) (18.3 min and 20.5 min, respectively) that are less polar than the avenacins. An avenacin A-1 standard is also shown.
We next expressed AsUGT91G16 in Nicotiana benthamiana leaves by Agrobacterium-mediated transient expression, in combination with AsbAS1/SAD1, AsCYP51H10/SAD2, and AsAAT1/UGT99D1, which together compose the arabinosylated triterpene scaffold 3β-(α-l-arabinopyranosyloxy)-12,13β-epoxy,16β-hydroxy-β-amyrin (3) (11) (Fig. 2B). Coexpression of AsUGT91G16 together with these enzymes resulted in the disappearance of (3) and the appearance of 1 major and 3 minor polar peaks. The major peak (4a) (retention time 16.8 min) and 1 of the minor peaks (4b) (retention time 14.4 min) both had masses consistent with the addition of a single hexose to (3), and the other 2 minor peaks (retention times 13.3 and 15.3 min) had masses consistent with the addition of 2 hexoses. We speculated that (4b) is a breakdown product of (4a) formed by opening of the C12-C13 epoxide, which is known to be unstable (39) (Fig. 2B). The latter 2 minor peaks may be the result of modifications of the AsUGT91G16 product by endogenous N. benthamiana glycosyltransferases.
We then scaled up our N. benthamiana infiltration experiments using vacuum infiltration following a method that we developed previously (40), agroinfiltrated 150 plants, and extracted the leaves to purify the AsUGT91G16 product for structural determination by NMR. Under these conditions, the major extracted product was (4b) (SI Appendix, Fig. S4). NMR analysis of (4b) confirmed that AsUGT91G16 is indeed a triterpene 3-O-arabinose-β-[1,2]-glycosyltransferase (Fig. 2B and SI Appendix, Table S7). The analysis also identified the scaffold of the purified compound 4b as 12-keto,16β-hydroxy-β-amyrin, consistent with breakdown of the epoxide group of (4a) during large-scale extraction.
We previously carried out a forward genetic screen for avenacin-deficient mutants of A. strigosa following sodium azide mutagenesis and identified a collection of approximately 100 mutants, roughly one-half of which have been assigned to characterized loci (22, 26, 28, 31–33, 41). We sequenced AsUGT91G16 in 49 uncharacterized avenacin-deficient mutants and identified 4 mutants with single nucleotide polymorphisms (SNPs) in this gene (SI Appendix, Table S8). Root extracts of wild-type A. strigosa contain the major avenacin A-1 (1), along with 3 other structural variants: A-2 (5), B-1 (6), and B-2 (7) (Fig. 2C and SI Appendix, Table S1) (39). Synthesis of all 4 avenacins is abolished in sad1 mutants, which are blocked in the first step in the pathway (AsbAS1/SAD1) (30). We previously showed that mutants affected at 2 other uncharacterized loci, Sad3 and Sad4, lack avenacins and accumulate monodeglucosyl avenacins missing the 1,4-linked d-glucose, of which monodeglucosyl avenacin A-1 (8) and monodeglucosyl avenacin A-2 (9) are the most abundant (22, 26). HPLC analysis of root extracts of the 4 ugt91g16 mutants revealed that these mutants share a common metabolite profile that is distinct from that of the wild-type line and also from sad1, sad3, and sad4 mutants. The ugt91g16 mutants accumulate 2 new, less polar compounds, (10) and (11) (Fig. 2C). These 2 peaks have masses consistent with avenacins A-1 and A-2 minus a hexose and also lacking oxidation at the C-30 position (Fig. 2C and SI Appendix, Fig. S5). A lack of C-30 oxidation was also previously observed in AsAAT1 mutants, suggesting that the introduction of this modification is dependent on previous glycosylation (11). Collectively, our data indicate that AsUGT91G16 adds the 1,2-linked d-glucose molecule to the sugar moiety of avenacin A-1.
All of the ugt91g16 mutants had reduced root fluorescence and stunted roots (SI Appendix, Fig. S6). Analysis of seedlings of F2 progeny derived from crosses between the wild-type and each of 2 ugt91g16 mutant lines (#543 and #1473) confirmed that the reduced avenacin phenotype cosegregated both with the SNPs in these lines and with the short root phenotype, and that these phenotypes are likely due to recessive mutations at AsUGT91G16 (SI Appendix, Table S9). The stunted root phenotype is most likely due to accumulation of toxic pathway intermediates. Root growth defects were previously observed for sad3 and sad4 mutants and attributed to the accumulation of incompletely glycosylated avenacin pathway intermediates (26). Asugt91g16 mutants, like other characterized avenacin-deficient mutants, were compromised in resistance to the root-infecting pathogen Gaeumannomyces graminis var. tritici, which causes take-all disease of cereals (SI Appendix, Fig. S7) (22).
Neither AsUGT91G16 nor AsUGT99C8 is able to complete the sugar chain of avenacin A-1 by adding the 1,4-linked d-glucose (SI Appendix, Fig. S8). We have previously shown that 5 of the 7 characterized avenacin biosynthetic genes are located on an ∼400-kb bacterial artificial chromosome contig in A. strigosa (28) (SI Appendix, Fig. S1). Genetic analysis indicates that the other 2 characterized avenacin pathway genes (Sad6 and AsAAT1) and other loci that we have identified by mutation but not yet cloned (including Sad3) are also likely to be clustered with these 5 genes (11, 22, 26, 33). A genome sequence is not yet available for A. strigosa; however, another avenacin-producing diploid oat species, Avena atlantica, has been subjected to whole-genome shotgun sequencing, and the assembled contigs have been mapped by survey resequencing of recombinant inbred progeny derived from a cross between this accession and A. strigosa accession Cc7651 (IBERS) as described in Louveau et al. (11). Examination of these assembled contigs revealed that AsUGT91G16 is located on a short (22.9 kb) scaffold next to a gene predicted to encode a member of the GH1 family (Fig. 3A and SI Appendix, Fig. S9 and Table S10). Plant GH1 hydrolases have previously been reported to function in plant development and stress responses, for example, by modulating iron deficiency and rhizobacteria-induced systematic resistance responses and by activating glucosinolate defense compounds and plant growth hormones by hydrolysis (42–44).
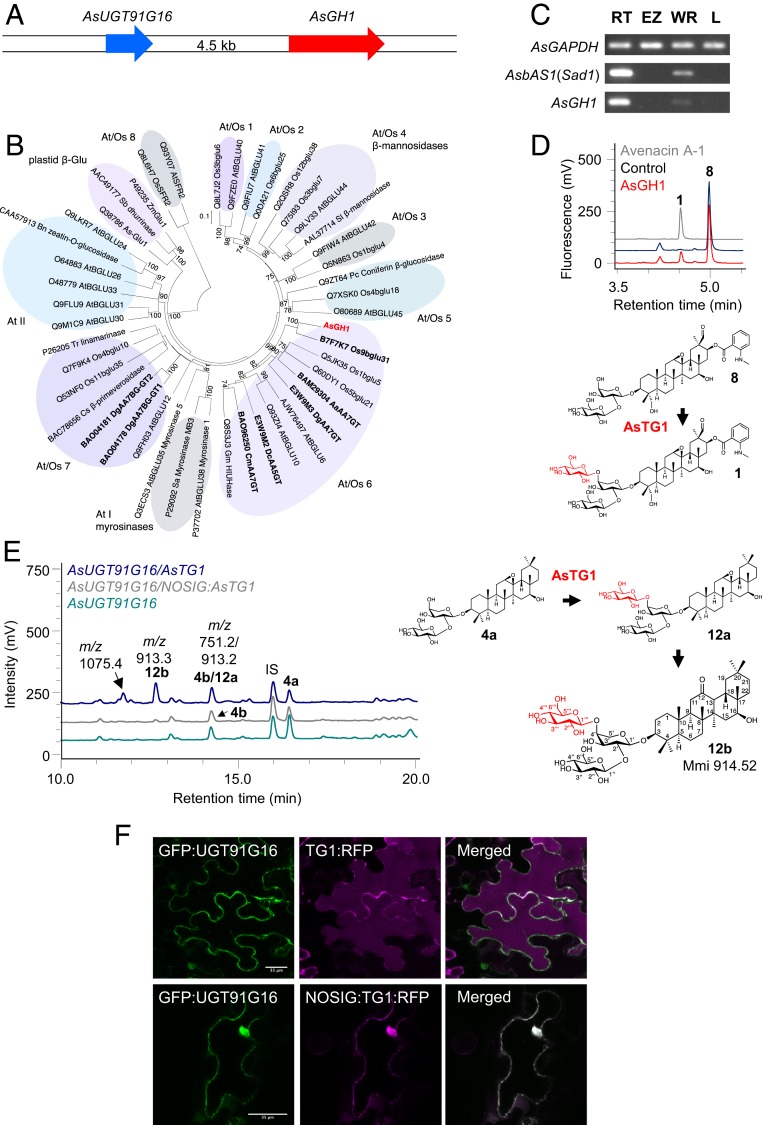
Fig. 3.
Characterization of AsTG1. (A) The AsUGT91G16 and AsGH1 genes are adjacent in the oat genome. (B) Phylogenetic tree of AsGH1 and other plant GH1 sequences. Rice (49) and Arabidopsis thaliana (63) GH1 enzymes were included in the analysis, along with characterized sequences from other plant species (SI Appendix, Table S11). The phylogenetic clusters that include the rice and Arabidopsis sequences (At/Os 1 to 8) and the Arabidopsis clusters (At I and II) are labeled as designated in Opassiri et al. (49), and the monocot plastidial β-glucosidases are labeled as in Ketudat Cairns et al. (64). Enzymes with reported transglycosidase activity (SI Appendix, Table S12) are indicated in bold type, and the A. strigosa GH1 is shown in red. The tree was constructed using the neighbor-joining method with 1,000 bootstrap replicates (% support for branch points is shown). The scale bar shows 0.1 substitutions per site at the amino acid level. (C) RT-PCR expression profile of AsGH1 in different oat tissues. AsbAS1/Sad1 and the AsGAPDH housekeeping gene were included as controls. RNA was extracted from 3-d-old A. strigosa seedlings. (D) Analysis of AsGH1 activity following heterologous expression in E. coli. HPLC-UV-MS traces are shown following incubation of protein preparations with 4-nitrophenyl β-d-glucoside and avenacin A-1 lacking the 1,4-linked d-glucose (8). A boiled protein preparation served as a control. The data are representative of 2 separate experiments. Absorbance was measured at 357 nm. (E) Transient expression in N. benthamiana. HPLC-CAD-MS traces for extracts from agroinfiltrated leaves are shown. Data are representative of 2 experiments. IS, internal standard (digitoxin). (F) Localization of AsUGT91G16 and AsTG1 in N. benthamiana leaves. Coexpression of an N-terminal AsUGT91G16 GFP fusion (GFP:AsUGT91G16) with a C-terminal mRFP fusion to AsTG1 (TG1:RFP) (Upper) and a C-terminal mRFP fusion to AsTG1 without the N-terminal signal peptide (NOSIG:TG1:RFP) (Lower). AsTG1:RFP localizes to the vacuole and the apoplast and does not colocalize with GFP:UGT91G16 in the cytoplasm. NOSIG-AsTG:RFP1 colocalizes with the GFP fusion to AsUGT91 in the cytoplasm and nucleus. GFP fusions are shown in green (Left), RFP fusions are shown in magenta (Middle), and merged images are shown in white (Right). Images were obtained at 3 d postinfiltration. (Scale bar: 35 μm.)
GH1 enzymes cleave glycosidic linkages in a 2-step mechanism, retaining the anomeric configuration of the product with respect to the sugar donor (45, 46) (www.cazy.org/glycosyltransferases.html). The first step cleaves the sugar donor to form a covalent glycosyl-enzyme intermediate. The second step transfers the sugar to an acceptor molecule, which is typically water, resulting in hydrolysis of the sugar donor. However, if the acceptor molecule is an alternative sugar acceptor, then transglycosylation can occur (47, 48).
Phylogenetic analysis indicates that AsGH1 belongs to the At/Os6 subfamily of GH1 enzymes, as defined by Opassiri et al. (49) (Fig. 3B and SI Appendix, Table S11). Intriguingly, several members of this subfamily have previously been shown to glucosylate anthocyanins using acyl sugars as sugar donors (SI Appendix, Table S12) (50–52). Therefore, we hypothesized that the AsGH1 enzyme could be the missing avenacin sugar transferase. Consistent with this idea, the AsGH1 gene was seen to be preferentially expressed in the root tips, like the other characterized avenacin biosynthetic genes (Fig. 3C). Unlike classical UDP-dependent sugar transferases, which are normally localized in the cytosol, the oat GH1 enzyme has an N-terminal 18-aa leader sequence, suggesting that it may be targeted for secretion (SI Appendix, Fig. S10).
His-tagged AsGH1 was expressed in E. coli and assayed for activity toward avenacin A-1 with the 1,4-linked d-glucose removed (8) in the presence of the model β-glucosidase sugar substrate 4-nitrophenyl β-d-glucose (4NPGlc) (53). HPLC-UV-MS analysis revealed conversion of the monodeglucosylated substrate (8) to a product with a retention time and mass consistent with that of avenacin A-1 (1) (Fig. 3D); therefore, we named this enzyme A. strigosa transglucosidase 1 (AsTG1).
We next evaluated the activity of AsTG1 by transient expression in N. benthamiana, as we had done previously for AsUGT91G16. Coexpression of AsbAS1/SAD1, AsCYP51H10/SAD2, AsAAT1/UGT99D1 and the β-[1,2]- glucosyltransferase AsUGT91G16 yielded (4a) and its breakdown product (4b) (Fig. 3E), as seen previously (Fig. 2B). Inclusion of AsTG1 led to the appearance of a new, more polar peak (12b) on HPLC-charged aerosol detector (CAD)-MS chromatography with a mass consistent with addition of a hexose to (4a/4b), a retention time of 12.7 min, and m/z = 913.3 (Fig. 3E). In addition, the peak at 14.4 min corresponding to the retention time of (4b) was slightly larger and the mass data revealed another molecular ion (m/z = 913.3), indicating coelution of a new compound (12a) with (4b). AsTG1 activity is dependent on AsUGT91G16, since when AsTG1 was coexpressed with AsbAS1/SAD1, AsCYP51H10/SAD2, and AsAAT1/UGT99D1 in the absence of AsUGT91G16, only accumulation of 3β-(α-l-arabinopyranosyloxy)-12,13β-epoxy,16β-hydroxy-β-amyrin (3) was observed (SI Appendix, Fig. S11). AsTG1 activity was also dependent on the presence of the signal sequence (Fig. 3E).
Vacuum infiltration was used to scale up agroinfiltration of N. benthamiana and the peak at 12.7 min (12b) purified following extraction from 150 plants. NMR analysis confirmed the scaffold of (12b) as 12-keto,16β-hydroxy-β-amyrin, and verified that the trisaccharide moiety was consistent with that of avenacin A-1 (SI Appendix, Table S13). By analogy with the AsUGT91G16 products, (12b) is likely the C12 ketone degradation product of (12a) following loss of the C12-C13 epoxide.
Localization of fluorescence-tagged proteins in N. benthamiana leaves by confocal microscopy suggests that AsUGT91G16 is cytosolic, as expected for a GT1 family UGT, and that AsTG1 is localized to the vacuole (Fig. 3F and SI Appendix, Figs. S12 to S14). The localization of AsTG1 is dependent on its signal sequence, as deletion of this sequence resulted in colocalization with AsUGT91G16 in the cytosol and nucleus (Fig. 3F).
Avenacin-deficient oat mutants affected at the uncharacterized loci, Sad3 and Sad4, accumulate monodeglycosyl avenacins missing the 1,4-linked d-glucose (26). Sad3 is genetically linked to the avenacin biosynthetic gene cluster (22), and mutations at this locus result in complete loss of avenacin production (22, 26). In contrast, Sad4 is unlinked, sad4 mutants are only partially compromised in avenacin glycosylation, and these mutants are also affected in ability to glycosylate other oat metabolites (22, 26). Like the ugt91g16 mutants, sad3 and sad4 mutants have stunted root growth (26). DNA sequence analysis revealed that all 4 sad3 mutants had an SNP in the AsTG1 gene (SI Appendix, Table S14). When F2 progeny derived from a cross between the A. strigosa wild type and a sad3 mutant (#1139) were examined, the reduced root fluorescence phenotype behaved as a single recessive Mendelian mutation (SI Appendix, Table S15) and cosegregated absolutely with the short root phenotype and the SNP, indicating that AsTG1 is synonymous with Sad3.
Conclusion
Here we have characterized the 2 enzymes required for the final glucosylation steps in biosynthesis of the antifungal defense compound, avenacin A-1. We show that 1 of these enzymes, AsUGT91G16, is a classical UDP sugar transferase belonging to family A of the GT1 family. In contrast, the enzyme that adds the final sugar (AsTG1) is an unusual vacuolar sugar transferase belonging to the glycosyl hydrolase family 1 (GH1) family of carbohydrate-active enzymes. Most of the characterized avenacin pathway enzymes are either known or likely to be endoplasmic reticulum-associated or cytosolic (11, 28, 33); however, the pathway end product avenacin A-1, like many other plant specialized metabolites, is localized in the vacuole (28). We have previously shown that AsSCPL1 (SAD7), the serine carboxypeptidase-like acyltransferase responsible for avenacin acylation, is located in the vacuole (28). Our results indicate that the partially glycosylated precursor 3β-{[β-d-glucopyranosyl-(1->2)-α-l-arabinopyranosyl]oxy}-12,13β-epoxy,16β-hydroxy-β-amyrin (4a), which is the product of AsUGT91G16, is transported from the cytosol to the vacuole, where it is then further glucosylated by AsTG1 and is also acylated (Fig. 4). This transport process is not specific to oat, since it also occurs in N. benthamiana. Several GH1 enzymes have previously been reported to glucosylate anthocyanins in plants using acyl sugars as sugar donors (50–52). Our study provides evidence of a role for GH1 enzymes in triterpene glycosylation and hints at a broader significance for this family of carbohydrate-active enzymes in glycosylation of plant natural products. Of note, it will be important to consider the alternative (non-UDP) sugar donor preferences and vacuolar localization of GH1 family sugar transferases when designing strategies for metabolic engineering of plant pathways in yeast and other microbial heterologous expression hosts. We further show that the AsUGT91G16 and AsTG1 genes are located next to each other on a short genome contig, and that AsTG1 corresponds to Sad3, a locus that we previously defined genetically as being part of the avenacin biosynthetic gene cluster in a forward screen for avenacin-deficient mutants (22, 26). Therefore, these 2 genes form part of the extended gene cluster, although the physical distance between AsUGT91G16/AsTG1 and the other characterized pathway genes (SI Appendix, Fig. S1) is not known. The generation of a high-quality genome sequence for A. strigosa will shed light on the organization of the complete avenacin biosynthetic gene cluster.
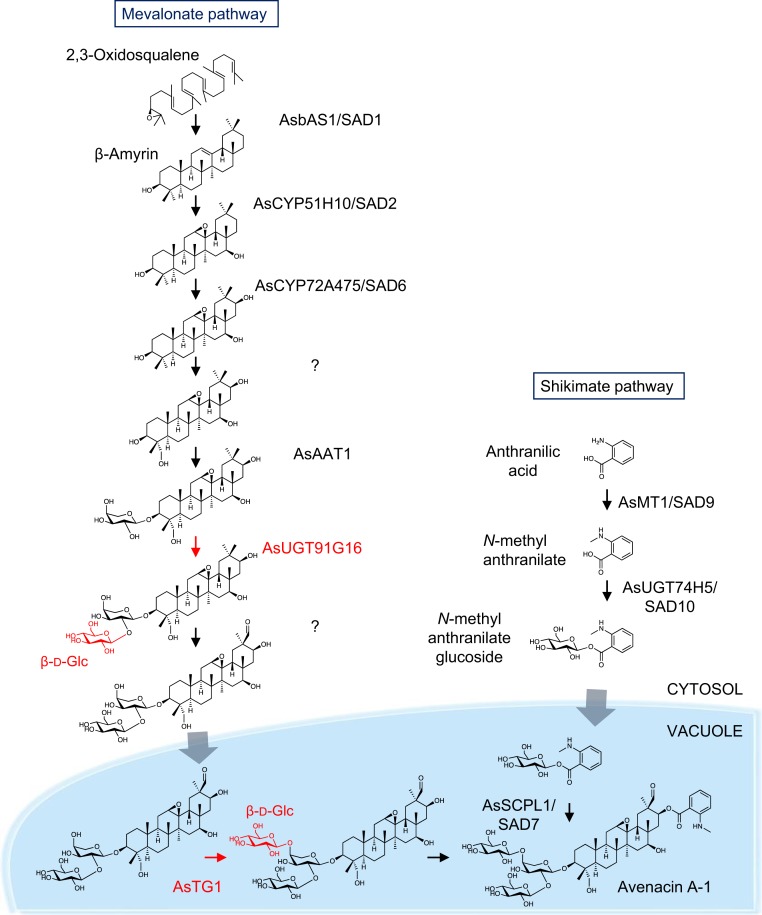
Fig. 4.
Biosynthesis of avenacin A-1 in oat. β-Amyrin is formed by the cyclization of 2,3-oxidosqualene by the β-amyrin synthase AsbAS1 (SAD1) and modified by the cytochrome P450 AsCYP51H10 (SAD2) by both epoxidation and hydroxylation (30, 31). AsCYP72A475 (SAD6) oxidizes the scaffold at the C-16 carbon position (33), and this product is then further oxidized at the C-23 position by an uncharacterized cytochrome P450 and glycosylated sequentially in the cytosol at the C-3 carbon position by an arabinosyltransferase, AsAAT1, and a glucosyltransferase, AsUGT91G16 (11). This product is then oxidized at the C-30 position by another uncharacterized cytochrome P450 and transported into the vacuole, where it is further glucosylated by the transglucosidase family member AsTG1. Anthranilic acid generated by the shikimate pathway is methylated by the methyltransferase AsMT1 (SAD9) and glucosylated in the cytoplasm by AsUGT74H5 (SAD10) to give N-methyl anthranilate (NMA) glucoside (28, 29). NMA glucoside is transported to the vacuole, where it serves as the activated acyl donor for the serine carboxypeptidase-like acyltransferase AsSCPL1 (SAD7), which acylates avenacin A-1 at the C-21 carbon position (32). Gray arrows indicate unknown transport mechanisms.
Materials and Methods
Oat Material.
The wild-type A. strigosa accession S75 and avenacin-deficient mutants have been described previously (22).
RNA Extraction and Analysis.
cDNA was generated from 3-d-old seedlings of A. strigosa accession S75 for whole roots, root tips (terminal 0.2 cm of the root), root elongation zones (from 0.2 cm to the first root hair), and young leaves. Expression of candidate genes was analyzed by mRNA RT-PCR as described previously (11) and normalized relative to the transcript levels of the housekeeping gene encoding GAPDH (AsGAPDH). The gene-specific primers used for PCR amplification are listed in SI Appendix, Table S16.
RNA-seq analysis was performed using material collected from 6 different A. strigosa tissues: root tips, whole roots, shoots, leaves, panicles, and spikelets. Root tips were harvested from 3-d-old seedlings as described above. Whole roots and shoots were collected from 3-wk-old glasshouse-grown plants. Mature leaves were harvested before earing, and spikelets and panicles (large spikes) were collected when the spacing between the flag leaf and top second 2 leaves was ∼4 cm and 12 cm, respectively. For each tissue, ≥100 μg of total RNA was isolated using TRIzol reagent (Invitrogen). Poly(A)+ mRNA was purified using the DynaBeads mRNA Purification Kit (Life Technologies). Paired-end cDNA libraries were constructed using the RNA-Seq NGS Library Preparation Kit for Whole-Transcriptome Discovery (Gnomegen) with 10 cycles of PCR amplification. The resulting paired-end cDNA libraries were sequenced using the Illumina HiSeq 2500 system with a read length of 100 to 150 bp. More information on the RNA-seq analysis is provided in SI Appendix. Raw and assembled data are available at http://db.ncgr.ac.cn/oat/RNAseq.php. Self-organizing maps (SOMs) were implemented and visualized in R (54) using the Kohonen package (34, 35) (SI Appendix and Datasets S1 and S2).
Phylogenetic Analysis.
For UGT sequences, amino acid sequences were deduced from the predicted full-length coding sequences of the A. strigosa UGTs identified in the SOM analysis (SI Appendix, Table S4). Representative amino acid sequences of characterized family 1 UGTs from other plant species (SI Appendix, Table S5) were obtained from the National Center for Biotechnology Information (NCBI) database and incorporated into the phylogenetic analysis. For GH1 sequences, representative amino acid sequences of characterized plant GH1 family members (SI Appendix, Table S11) were obtained from the NCBI database. Protein sequences were aligned using MAFFT (https://mafft.cbrc.jp/alignment/software/). Unrooted trees were constructed in MEGA7 by the neighbor-joining method with 1,000 bootstrap replicates (55, 56).
Cloning and In Vitro Analysis of Candidate Glucosyltransferases.
Expression constructs were created using Gateway technology (Invitrogen). Coding sequences (CDSs), including stop codons of A. strigosa UGT91G16 and AsTG1 (AsGH1), were amplified from oat genomic DNA and root tip cDNA, respectively, with a 2-step PCR method. The first amplification step used gene-specific primers (SI Appendix, Table S16) to attach partial AttB adapters to the full-length CDS. The second amplification step attached the full AttB site to each end of the CDS. Amplified fragments were purified with the Qiagen QIAquick PCR Purification Kit. The purified CDS fragments were transferred into the pDONR207 vector using BP Clonase II Enzyme Mix (Invitrogen) according to the manufacturer’s instructions. To generate a plasmid with AsTG1 missing the predicted N-terminal signal sequence (AsTG1-NOSIG), 60 ng of the pDONR207-AsTG1 plasmid was used as a template with Fgw-nosigAsTG1 and Rgw-AsTG1 primers (SI Appendix, Table S16) by 2-step Gateway cloning. Before the PCR purification step, 1 μL of DpnI (New England BioLabs) was added to the amplified PCR product, and the mixture was then incubated at 37 °C for 1 h to digest the pDONR207-AsTG1 DNA template. Sequence-verified coding sequences were then transferred by LR clonase II into pH9GW (Invitrogen), a Gateway-compatible version of pET-28 encoding 9 N-terminal histidines (57). AsUGT99C8 (originally referred to as AsGT14h20) had previously been cloned into pH9GW as described above as part of an earlier investigation of avenacin glycosyltransferases (58). Constructs were transformed into E. coli Rosetta strain DE3 (Novagen), and recombinant partially purified enzymes were assayed for their ability to glycosylate 3β-{bisdeglucosyl-(1->2)-,(1->4)-} avenacin A-1 (2) and 3β-{deglucosyl-(1->4)-} avenacin A-1 (9) (SI Appendix).
Transient Expression in N. benthamiana.
N. benthamiana plants were grown in greenhouses maintained at 23 to 25 °C with 16 h of supplementary light per day, as described previously (59). Candidate glucosyltransferase CDSs were cloned into the pEAQ-HT-DEST1 expression vector (59) using LR clonase II (Invitrogen) according to the manufacturer’s instructions. Agroinfiltration was carried out as described previously (40, 59). In brief, expression constructs were transformed into Agrobacterium tumefaciens strain LBA4404 and the strains infiltrated into N. benthamiana leaves. To coexpress combinations of genes, A. tumefaciens strains containing different expression constructs were diluted to 0.9 OD600 and mixed in equal volumes to result in a final concentration per strain of 0.15 OD600. A strain containing a construct for expression of the feedback-insensitive form of HMG-CoA reductase, tHMGR, was included in all combinations to increase triterpene production (40). A strain with a green fluorescent protein (GFP) expression construct was used as a “filler” to maintain the same concentration of bacteria in all infiltration combinations. Details of metabolite analysis, purification, and structural determination by NMR are provided in SI Appendix.
Characterization of Oat Mutants and Segregating F2 Populations.
Oat genomic DNA was extracted based on the method described by Pallota et al. (60). The AsUGT91G16 gene was sequenced in 47 uncharacterized sodium azide-generated avenacin-deficient mutants (22, 31). The AsTG1 gene was sequenced in 4 independent sad3 mutants (26). The primers used for amplification and sequencing are listed in SI Appendix, Table S16. The purified PCR products were sequenced by GATC Biotech. To analyze segregating progeny, 3-d-old seedlings of F2 progeny from crosses between mutant lines and the S75 wild type were phenotyped for root length and fluorescence and then transferred to soil in 96-well trays under glasshouse conditions. Genomic DNA was extracted from 2-wk-old seedlings, and genotyping was performed using the amplification and sequencing primers listed in SI Appendix, Table S16.
Metabolite Analysis of Oat Root Extracts.
Roots were harvested from 3-d-old seedlings of wild-type and mutant A. strigosa lines and then flash-frozen in liquid N2. Aliquots (∼25 mg) were extracted in 0.5 mL of 80% methanol with two 3-mm tungsten beads at 1,400 rpm at 25 °C for 1 h, after which the temperature was increased to 42 °C for a further 30 min. Samples were centrifuged briefly, and 0.45 mL of supernatant was partitioned twice with 0.3 mL of hexane. Aliquots (0.2 mL) of the methanolic fractions were dried in a Genevac EZ-2 Elite centrifugal evaporator, resuspended in 100 μL of methanol, and filtered through Corning Costar Spin-X centrifuge tube filters (Sigma-Aldrich), after which 50 μL of the filtrate was combined with 50 μL of 50% methanol. Samples were analyzed by HPLC-CAD-UV-MS using a 50 × 2.1-mm, 2.6-μ Kinetex XB-C18 column (Phenomenex) with a column oven temperature of 30 °C. Detection was by CAD (Corona Ultra RS; Dionex); UV/Vis absorbance (Shimadzu SPD-M20A), collecting spectra from 200 to 500 nm; and electrospray MS (Shimadzu LC-2020 dual-source MS), collecting in positive and negative modes from m/z 50 to 1,500. The gradient was run at 0.3 mL/min with 100% water as Buffer A and 100% acetonitrile as Buffer B as follows: 20% Buffer B from 0 to 3 min, 20 to 60% Buffer B from 3 to 28 min, 60 to 100% Buffer B from 28 to 30 min, a linear gradient between 30 and 3 min, 100 to 20% Buffer B from 33 to 34 min, and held at 20% Buffer B until 35 min.
Pathogenicity Tests.
Pathogenicity tests to evaluate root infection of the A. strigosa wild-type and avenacin-deficient mutant lines with the fungal pathogen G. graminis var. tritici (isolate T5) were performed as described previously (22). Seedlings were scored for root lesions at 21 d after inoculation using a 7-point scale (11).
Subcellular Localization.
To make N- and C-terminal AsUGT91G16 fusions with GFP, AsUGT91G16 was amplified from the pH9-GW-AsUGT91G16 plasmid with primers AsUGT91-NTGW and Rgw-UGT91 or with primers Fgw-GTUGT91 and Rgw-UGT91-NOSTOP (SI Appendix, Table S16) and then cloned into pMDC45 and pMDC83, respectively (61). To make C-terminal protein fusions with GFP or RFP, AsTG1 and AsTG1-NOSIG (i.e., AsTG1 lacking the N-terminal signal sequence) were amplified from pDEST1-AsTG1 or pDEST1-AsTG1-NOSIG plasmids with primers Fgw-AsTGsig-FULL and Rgw-AsTG-NOSTOP-FULL or primers Fgw-AsTG-NOSIG-FULL and Rgw-AsTG-NOSTOP-FULL (61), respectively, and then cloned into pB7RWG2 (62) for RFP constructs and pMDC83 (61) for GFP constructs. Fluorescent fusion protein constructs were verified using RFP- or GFP-specific sequencing primers (SI Appendix, Table S16).
A. tumefaciens strain GV3101 was used for all fluorescent protein fusion assays. To assess the enzymatic activity of the fluorescent protein fusion constructs, agroinfiltrations were carried out following the method described for transient expression in N. benthamiana. Metabolites were detected by HPLC using a 50 × 2.1-mm, 2.6-μ Kinetex XB-C18 column (Phenomenex) with a column oven temperature of 25 °C. Detection was by CAD (Corona Ultra RS; Dionex). The gradient was run at 0.3 mL/min with 100% water as Buffer A and 100% acetonitrile as Buffer B and was as follows: 10% Buffer B from 0 to 1.5 min, 10 to 50% Buffer B from 1.5 to 21 min, 50 to 95% Buffer B from 21 to 21.5 min, a linear gradient between 21.5 and 23.5 min, 95 to 10% Buffer B from 23.5 to 24 min, and held at 10% until 25 min.
The subcellular localization of fusion constructs was assessed in N. benthamiana leaves. Agroinfiltration was carried out as described above but with the following changes: strains were diluted to 0.6 OD600 before mixing in equal volumes to give a final concentration of 0.2 OD600, except for combinations of GFP:UGT91G16 with AsTG1:RFP and NOSIG:AsTG1:RFP, in which strains were further diluted to achieve a final concentration of 0.04 OD600 per strain. Leaves were imaged with a Leica TCS SP8X confocal microscope after the number of days indicated.
Supplementary Material
Acknowledgments
This work was supported by the UK Biological Sciences Research Council (BBSRC) Doctoral Training Programme (A. Orme); the joint Engineering and Physical Sciences Research Council/Biotechnological and BBSRC-funded OpenPlant Synthetic Biology Research Centre Grant BB/L014130/1 (to M.J.S. and A. Osbourn); the Centre of Excellence for Plant and Microbial Sciences, established between the John Innes Centre and the Chinese Academy of Sciences and funded by the BBSRC and the Chinese Academy of Sciences; National Natural Science Foundation of China Grant 31788103, “Molecular Design for Future Crops” (to B.H.); BBSRC Strategic Programme Grant BB/P012523/1, “Molecules from Nature”; and the John Innes Foundation (A. Osbourn and R.M.).
Footnotes
Competing interest statement: This work is subject to a patent filing.
This article is a PNAS Direct Submission. W.B.F. is a guest editor invited by the Editorial Board.
Data deposition: Raw and assembled RNA-seq data can be accessed at http://db.ncgr.ac.cn/oat/RNAseq.php. Sequences have been deposited in the GenBank database (accession nos. MN396758–MN396761).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914652116/-/DCSupplemental.
References
- 1.Afendi F. M., et al. , KNApSAcK family databases: Integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol. 53, e1 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Louveau T., Osbourn A., The sweet side of plant-specialized metabolism. Cold Spring Harb. Perspect. Biol., a034744 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cantarel B. L., et al. , The Carbohydrate-Active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogt T., Jones P., Glycosyltransferases in plant natural product synthesis: Characterization of a supergene family. Trends Plant Sci. 5, 380–386 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Bowles D., Lim E. K., Poppenberger B., Vaistij F. E., Glycosyltransferases of lipophilic small molecules. Annu. Rev. Plant Biol. 57, 567–597 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Piotrowska A., Bajguz A., Conjugates of abscisic acid, brassinosteroids, ethylene, gibberellins, and jasmonates. Phytochemistry 72, 2097–2112 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Brazier-Hicks M., Gershater M., Dixon D., Edwards R., Substrate specificity and safener inducibility of the plant UDP glucose-dependent family 1 glycosyltransferase super-family. Plant Biotechnol. J. 16, 337–348 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Augustin J. M., Kuzina V., Andersen S. B., Bak S., Molecular activities, biosynthesis and evolution of triterpenoid saponins. Phytochemistry 72, 435–457 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Sawai S., Saito K., Triterpenoid biosynthesis and engineering in plants. Front. Plant Sci. 2, 25 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osbourn A., Goss R. J. M., Field R. A., The saponins: Polar isoprenoids with important and diverse biological activities. Nat. Prod. Rep. 28, 1261–1268 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Louveau T., et al. , Analysis of two new arabinosyltransferases belonging to the Carbohydrate-Active Enzyme (CAZY) Glycosyl Transferase Family1 provides insights into disease resistance and sugar donor specificity. Plant Cell 30, 3038–3057 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahimi S., et al. , Triterpenoid-biosynthetic UDP-glycosyltransferases from plants. Biotechnol. Adv., S0734-9750(19)30075-8 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Shibuya M., Nishimura K., Yasuyama N., Ebizuka Y., Identification and characterization of glycosyltransferases involved in the biosynthesis of soyasaponin I in Glycine max. FEBS Lett. 584, 2258–2264 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Sayama T., et al. , The Sg-1 glycosyltransferase locus regulates structural diversity of triterpenoid saponins of soybean. Plant Cell 24, 2123–2138 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang P., et al. , Production of bioactive ginsenosides Rh2 and Rg3 by metabolically engineered yeasts. Metab. Eng. 29, 97–105 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Yano R., et al. , Isolation and characterization of the soybean Sg-3 gene that is involved in genetic variation in sugar chain composition at the C-3 position in soyasaponins. Plant Cell Physiol. 59, 792–805 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Jung S. C., et al. , Two ginseng UDP-glycosyltransferases synthesize ginsenoside Rg3 and Rd. Plant Cell Physiol. 55, 2177–2188 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Takagi K., et al. , Genetic and functional characterization of Sg-4 glycosyltransferase involved in the formation of sugar chain structure at the C-3 position of soybean saponins. Phytochemistry 156, 96–105 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Meesapyodsuk D., Balsevich J., Reed D. W., Covello P. S., Saponin biosynthesis in Saponaria vaccaria: cDNAs encoding β-amyrin synthase and a triterpene carboxylic acid glucosyltransferase. Plant Physiol. 143, 959–969 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kita M., et al. , Molecular cloning and characterization of a novel gene encoding limonoid UDP-glucosyltransferase in Citrus. FEBS Lett. 469, 173–178 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Itkin M., et al. , The biosynthetic pathway of the nonsugar, high-intensity sweetener mogroside V from Siraitia grosvenorii. Proc. Natl. Acad. Sci. U.S.A. 113, E7619–E7628 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papadopoulou K., Melton R. E., Leggett M., Daniels M. J., Osbourn A. E., Compromised disease resistance in saponin-deficient plants. Proc. Natl. Acad. Sci. U.S.A. 96, 12923–12928 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turner E. M. C., An enzymic basis for pathogenic specificity in Ophiobolus graminis. J. Exp. Bot. 12, 169–175 (1961). [Google Scholar]
- 24.Crombie W. M. L., Crombie L., Green J. B., Lucas J. A., Pathogenicity of “take-all” fungus to oats: Its relationship to the concentration and detoxification of the four avenacins. Phytochemistry 25, 2075–2083 (1986). [Google Scholar]
- 25.Armah C. N., et al. , The membrane-permeabilizing effect of avenacin A-1 involves the reorganization of bilayer cholesterol. Biophys. J. 76, 281–290 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mylona P., et al. , Sad3 and sad4 are required for saponin biosynthesis and root development in oat. Plant Cell 20, 201–212 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi X., et al. , A gene cluster for secondary metabolism in oat: Implications for the evolution of metabolic diversity in plants. Proc. Natl. Acad. Sci. U.S.A. 101, 8233–8238 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mugford S. T., et al. , Modularity of plant metabolic gene clusters: A trio of linked genes that are collectively required for acylation of triterpenes in oat. Plant Cell 25, 1078–1092 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Owatworakit A., et al. , Glycosyltransferases from oat (Avena) implicated in the acylation of avenacins. J. Biol. Chem. 288, 3696–3704 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haralampidis K., et al. , A new class of oxidosqualene cyclases directs synthesis of antimicrobial phytoprotectants in monocots. Proc. Natl. Acad. Sci. U.S.A. 98, 13431–13436 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi X., et al. , A different function for a member of an ancient and highly conserved cytochrome P450 family: From essential sterols to plant defense. Proc. Natl. Acad. Sci. U.S.A. 103, 18848–18853 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mugford S. T., et al. , A serine carboxypeptidase-like acyltransferase is required for synthesis of antimicrobial compounds and disease resistance in oats. Plant Cell 21, 2473–2484 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leveau A., et al. , Towards take-all control: A C-21β oxidase required for acylation of triterpene defence compounds in oat. New Phytol. 221, 1544–1555 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wehrens R., Buydens L. M. C., Self- and super-organizing maps in R: The Kohonen package. J. Stat. Softw. 21, 5 (2007). [Google Scholar]
- 35.Kohonen T., Essentials of the self-organizing map. Neural Netw. 37, 52–65 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Payne R. M. E., et al. , An NPF transporter exports a central monoterpene indole alkaloid intermediate from the vacuole. Nat. Plants 3, 16208 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ross J., Li Y., Lim E., Bowles D.J.. Higher plant glycosyltransferases. Genome Biol. 2, REVIEWS3004 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caputi L., Malnoy M., Goremykin V., Nikiforova S., Martens S., A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 69, 1030–1042 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Crombie L., Crombie W. M. L., Whiting D. A., Structures of the oat root resistance factors to “take-all” disease, avenacins A-1, A-2, B-1 and B-2 and their companion substances. J. Chem. Soc., Perkin Trans. I, 1917–1922 (1986). [Google Scholar]
- 40.Reed J., et al. , A translational synthetic biology platform for rapid access to gram-scale quantities of novel drug-like molecules. Metab. Eng. 42, 185–193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qin B., et al. , High-throughput screening of mutants of oat that are defective in triterpene synthesis. Phytochemistry 71, 1245–1252 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Lee K. H., et al. , Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126, 1109–1120 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Bednarek P., et al. , A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323, 101–106 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Zamioudis C., Hanson J., Pieterse C. M., β-Glucosidase BGLU42 is a MYB72-dependent key regulator of rhizobacteria-induced systemic resistance and modulates iron deficiency responses in Arabidopsis roots. New Phytol. 204, 368–379 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Koshland D. E., Stereochemistry and the mechanism of enzymatic reactions. Biol. Rev. Camb. Philos. Soc. 28, 416–436 (1953). [Google Scholar]
- 46.Henrissat B., Davies G., Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7, 637–644 (1997). [DOI] [PubMed] [Google Scholar]
- 47.Sinnott M. L., Catalytic mechanism of enzymatic glycosyl transfer. Chem. Rev. 90, 1171–1202 (1990). [Google Scholar]
- 48.Bissaro B., Monsan P., Fauré R., O’Donohue M. J., Glycosynthesis in a waterworld: New insight into the molecular basis of transglycosylation in retaining glycoside hydrolases. Biochem. J. 467, 17–35 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Opassiri R., et al. , Analysis of rice glycosyl hydrolase family 1 and expression of Os4bglu12 β-glucosidase. BMC Plant Biol. 6, 33 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsuba Y., et al. , A novel glucosylation reaction on anthocyanins catalyzed by acyl-glucose-dependent glucosyltransferase in the petals of carnation and delphinium. Plant Cell 22, 3374–3389 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miyahara T., Takahashi M., Ozeki Y., Sasaki N., Isolation of an acyl-glucose-dependent anthocyanin 7-O-glucosyltransferase from the monocot Agapanthus africanus. J. Plant Physiol. 169, 1321–1326 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Miyahara T., et al. , Isolation of anthocyanin 7-O-glucosyltransferase from Canterbury bells (Campanula medium). Plant Biotechnol. 31, 555–559 (2014). [Google Scholar]
- 53.Luang S., et al. , Rice Os9BGlu31 is a transglucosidase with the capacity to equilibrate phenylpropanoid, flavonoid, and phytohormone glycoconjugates. J. Biol. Chem. 288, 10111–10123 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.R Core Team , R: A language and environment for statistical computing, version 3.5.2. https://www.R-project.org/. 2017. Accessed 20 December 2018.
- 55.Saitou N., Nei M., The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987). [DOI] [PubMed] [Google Scholar]
- 56.Kumar S., Stecher G., Tamura K., MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O’Maille P. E., et al. , Quantitative exploration of the catalytic landscape separating divergent plant sesquiterpene synthases. Nat. Chem. Biol. 4, 617–623 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Louveau T., “Investigation of glycosyltransferases from oat”, PhD thesis, University of East Anglia, UK (2014).
- 59.Sainsbury F., Saxena P., Geisler K., Osbourn A., Lomonossoff G. P., Using a virus-derived system to manipulate plant natural product biosynthetic pathways. Methods Enzymol. 517, 185–202 (2012). [DOI] [PubMed] [Google Scholar]
- 60.Pallotta M. A., et al. , “Marker-assisted wheat breeding in the southern region of Australia” in Proceedings of the Tenth International Wheat Genetics Symposium, Pogna N. E., Romano M., Pogna E. A., Galterio Z., Eds. (Istituto Sperimentale per la Cerealicoltura, 2003) pp. 789–791. [Google Scholar]
- 61.Curtis M. D., Grossniklaus U., A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karimi M., Inzé D., Depicker A., GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 (2002). [DOI] [PubMed] [Google Scholar]
- 63.Xu Z., et al. , Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family 1. Plant Mol. Biol. 55, 343–367 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Ketudat Cairns J. R., et al. , Protein-carbohydrate interactions leading to hydrolysis and transglycosylation in plant glycoside hydrolase family 1 enzymes. J. Appl. Glycosci. 59, 51–62 (2012). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.