Significance
Vertebrate photoreceptor cells contain a specialized organelle, the outer segment, which produces electrical responses to light. To maximize light capture efficiency, the outer segment is filled with hundreds of stacked membrane “discs” containing the molecular machinery responsible for performing visual signaling. Importantly, discs undergo continuous renewal throughout the lifetime of a photoreceptor. Our data suggest that new discs are formed through a mechanism analogous to the formation of lamellipodia in migrating cells. Each new disc is born as an evagination of the outer segment membrane driven by the expanding branched actin network. This work contributes to our understanding of the molecular mechanisms responsible for disc formation and guides future therapies directed at correcting defects in this process.
Keywords: photoreceptor, ectosome, actin cytoskeleton, Arp2/3, cilium
Abstract
The light-sensitive outer segment of the vertebrate photoreceptor is a highly modified primary cilium filled with disc-shaped membranes that provide a vast surface for efficient photon capture. The formation of each disc is initiated by a ciliary membrane evagination driven by an unknown molecular mechanism reportedly requiring actin polymerization. Since a distinct F-actin network resides precisely at the site of disc morphogenesis, we employed a unique proteomic approach to identify components of this network potentially driving disc morphogenesis. The only identified actin nucleator was the Arp2/3 complex, which induces the polymerization of branched actin networks. To investigate the potential involvement of Arp2/3 in the formation of new discs, we generated a conditional knockout mouse lacking its essential ArpC3 subunit in rod photoreceptors. This knockout resulted in the complete loss of the F-actin network specifically at the site of disc morphogenesis, with the time course of ArpC3 depletion correlating with the time course of F-actin loss. Without the actin network at this site, the initiation of new disc formation is completely halted, forcing all newly synthesized membrane material to be delivered to the several nascent discs whose morphogenesis had already been in progress. As a result, these discs undergo uncontrolled expansion instead of normal enclosure, which leads to formation of unusual, large membrane whorls. These data suggest a model of photoreceptor disc morphogenesis in which Arp2/3 initiates disc formation in a “lamellipodium-like” mechanism.
The outer segment is a ciliary organelle of the vertebrate photoreceptor cell responsible for generating electrical responses to light. In order to capture photons efficiently, the outer segment contains a stack of disc-shaped membranes, or “discs,” densely packed with visual pigment (1). Throughout the entire lifespan of a vertebrate animal, outer segments undergo a continuous process of membrane renewal. Each day, dozens of new discs are formed at the outer segment base while an equal number of old discs are shed from its tip (2). The formation of each disc begins with a ciliary membrane evagination at the outer segment base, followed by membrane elongation, flattening, and enclosure inside the outer segment (3–7).
The ciliary membrane evagination, which initiates the formation of a new disc, is thought to involve the actin cytoskeleton (8, 9). Ultrastructural analyses of photoreceptors have shown a high density of actin filaments specifically at this site whose appearance in development coincides with the morphogenesis of the first discs (at approximately P7 in mice) (9–12). Blocking actin filament formation by cytochalasin D treatment of frog and rabbit retinas prevented the formation of new discs while causing excessive growth of the several discs that were undergoing membrane expansion at the time of treatment (8, 13–15). It was concluded that lipids and proteins normally delivered to the newly budding discs were uncontrollably added to the nascent discs still connected to the plasma membrane.
The critical role of the actin cytoskeleton in disc morphogenesis highlights the importance of understanding the molecular mechanism controlling the formation of the actin network at this site. In principle, there are 2 mechanisms by which filamentous actin (F-actin) can be nucleated and expanded. Actin filaments can elongate in a branched or unbranched manner, resulting in web-like or parallel actin networks, respectively (16). Each type of actin network employs a distinct set of proteins regulating its initiation, expansion, and turnover. Actin filament assembly begins with a rate-limiting nucleation step in which monomeric actin molecules assemble into dimers and trimers. Branched networks, such as those found in lamellipodia, are always nucleated by the Arp2/3 complex, whereas linear networks, such as are found in filopodia, are typically nucleated by members of the formin or Spire protein families (16, 17). Thus, discerning which actin nucleator is employed in photoreceptor disc formation is a critical step for understanding how a new disc is born.
In the present study, we first conducted a proteomic analysis aiming to identify actin-related proteins potentially involved in disc morphogenesis. This proteome contained the Arp2/3 complex, along with an array of other proteins known to regulate actin dynamics. Notably, no other actin nucleators were identified. To determine whether the Arp2/3 complex is indeed involved in disc morphogenesis, we generated a conditional knockout mouse lacking functional Arp2/3 in rod photoreceptors. Once the knockout was induced, the F-actin network at the site of new disc formation disappeared and all newly synthesized membrane material was delivered to the several discs whose morphogenesis was already ongoing. As a result, these discs underwent uncontrolled expansion, leading to the formation of unusual, large membrane whorls. Conceptually, this phenotype resembles the acute effect of cytochalasin D treatment, but the degree of observed pathology was far more exaggerated. Interestingly, some of these grossly overgrown disc membranes were enclosed, indicating that the process of disc enclosure is independent of actin polymerization. Taken together, our data suggest a model in which the process of disc morphogenesis is initiated through an Arp2/3-dependent “lamellipodium-like” mechanism.
Results
An F-Actin Network Is Crucial for Initiating Disc Morphogenesis but Not Disc Enclosure.
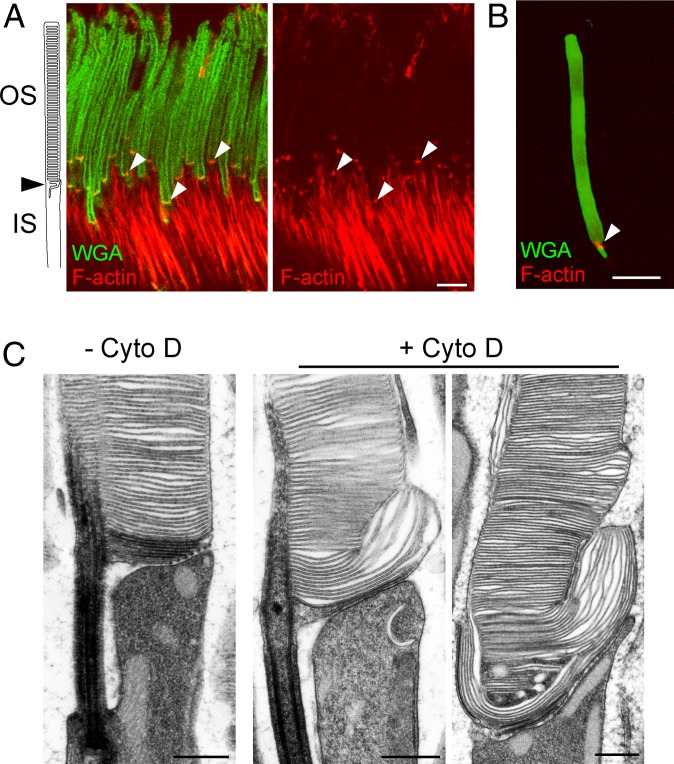
The presence of F-actin at the photoreceptor outer segment base in the mouse can be particularly well-appreciated using superresolution microscopy (Fig. 1A and Movie S1). Consistent with previous reports, we observed that photoreceptor outer segments contained an intensely labeled patch of actin located precisely at the site of new disc formation. No additional F-actin was observed in the rest of the outer segment, a result corroborated in experiments with isolated outer segments (Fig. 1B), indicating that it is efficiently retracted from nascent discs as they transition from budding to flattening. The rest of photoreceptor F-actin staining was observed in the inner segment, at a distance of at least several microns from this patch.
Fig. 1.
F-actin localizes to the base of photoreceptor outer segments and is crucial for initiating disc morphogenesis. (A) An immunofluorescence image of a WT mouse retinal cross-section spanning photoreceptor inner (IS) and outer segments (OS). Outer segments were labeled with WGA-Alexa Fluor 488 (green) and F-actin with phalloidin-Alexa Fluor 594 (red). The phalloidin staining was detected using STED superresolution imaging using a 775-nm depletion laser. Arrowheads denote the distinct F-actin puncta residing at the base of each photoreceptor outer segment. (Scale bar, 5 µm.) Movie S1 provides a STED 3D rendering of the same F-actin staining. (B) A confocal immunofluorescence image of a photoreceptor outer segment, which was isolated from an adult WT mouse retina and attached to a poly-L-lysine–coated microscope slide. The outer segment is labeled with WGA (green) and F-actin with phalloidin (red). The arrowhead highlights the actin punctum at the base of the outer segment. (Scale bar, 5 µm.) (C) Electron micrographs taken at the base of photoreceptor outer segments of WT mice either intravitreally injected (+Cyto D) or not injected (−Cyto D) with cytochalasin D. The mice were killed 6 h postinjection. Retinal sections were contrasted with tannic acid, which stains newly forming, open discs more intensely than mature, enclosed discs. (Scale bars, 500 nm.)
As described earlier, cytochalasin D treatment of frog and rabbit retinas prevented the budding of new photoreceptor discs while redirecting membrane material to the nascent discs already present at the time of treatment (8, 13–15). We corroborated these observations in the mouse by conducting intraocular injections of cytochalasin D, which similarly resulted in uncontrolled elongation of several nascent discs and a complete lack of the most proximal short discs evaginating from the outer segment cilium (Fig. 1C). A key aspect of our experiments was the use of a recently developed methodology that distinguishes nascent discs still exposed to the extracellular space (“open discs”) from mature discs enclosed inside the outer segment (“closed discs”) (4). This distinction is achieved by contrasting membranes with tannic acid, a compound that poorly penetrates cellular membranes, thereby yielding a more intense staining of membranes exposed to the extracellular space than intracellular membranes. We found that some of the overgrown discs were lightly stained, indicating that they were enclosed at the time of tissue fixation, while the most proximal discs were darkly stained, indicating that they were open. Therefore, polymerization of actin filaments is not needed for the expansion of the disc membrane or membrane enclosure, but is essential for the initiation of new disc budding.
The Proteome of rds Ectosomes Contains Arp2/3 and other Actin-Associated Proteins Potentially Involved in Disc Morphogenesis.
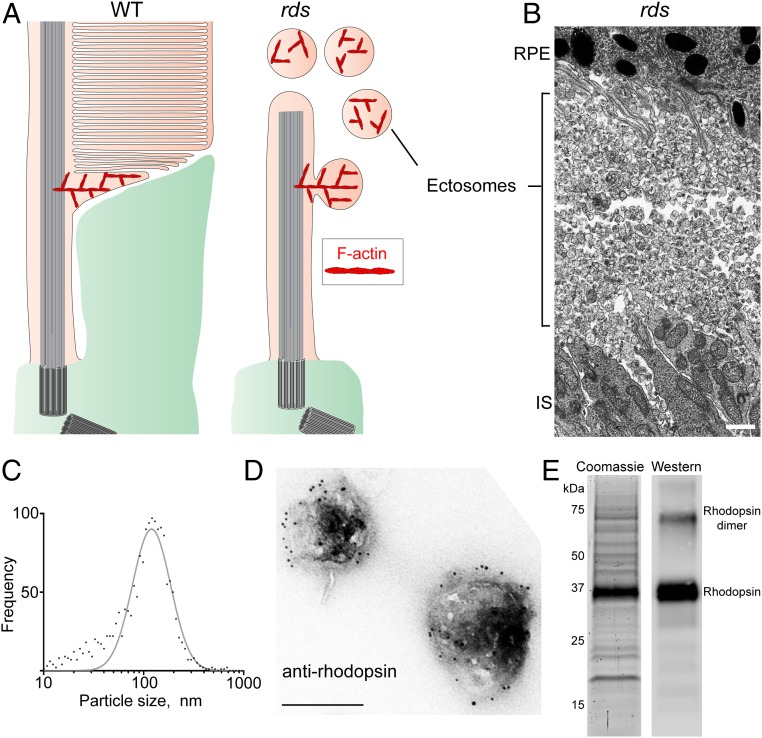
Given the importance of the F-actin network at the site of disc morphogenesis, we investigated which proteins in this network are responsible for F-actin nucleation. We began addressing this question by using mass spectrometry to identify actin-binding proteins enriched at the site of disc morphogenesis. A challenge of these experiments is that the patch of F-actin within the active zone of disc formation occupies a very small fraction of the total outer segment volume (Fig. 1 A and B) and cannot be easily isolated or enriched. As a result, proteins residing within this patch are anticipated to be underrepresented in the outer segment proteome. To circumvent this problem, we took advantage of the rds mouse, which lacks the disc-specific protein peripherin (18–20). Without peripherin, the evaginating membranes normally destined to become discs in WT mice sever from the photoreceptor cilium in the form of vesicles (21–24) recently determined to be ciliary ectosomes (25) (Fig. 2A). As a result, photoreceptors of rds mice form only a primary cilium surrounded by an enormous number of ectosomes (Fig. 2B). Importantly, these ectosomes contain actin filaments (12, 26), suggesting that the actin nucleator(s) responsible for initiating disc formation is trapped inside. Unlike the F-actin patches, which are essentially inseparable from the rest of the outer segment, ciliary ectosomes could be isolated in great purity.
Fig. 2.
Purification of ciliary ectosomes produced by rds photoreceptors. (A) A cartoon highlighting the F-actin network localized to the site of photoreceptor disc formation in WT mice and the site of photoreceptor ciliary ectosome release in rds mice. The actin filaments remain trapped inside the ectosomes after their scission from the ciliary membrane (12, 26). Note that the size of the first, actin-containing disc is exaggerated to aid the illustration of the model (see refs. 6 or 4 for to-scale depictions of its size, which is ∼0.3 µm). (B) An electron micrograph showing the massive accumulation of ciliary ectosomes in the subretinal space of a P14 rds mouse. (Scale bar, 1 µm.) IS, inner segment; RPE, retinal pigment epithelium. (C) A Gaussian histogram representing the size distribution of the purified ectosomes measured with a ZetaView particle tracking analyzer. The diameter of the purified ectosomes was 116 ± 70 nm (mean ± SD). (D) Immunogold staining of purified ectosomes with 4D2 anti-rhodopsin antibody. (Scale bar, 200 nm.) (E, Left) A Coomassie-stained SDS/PAGE gel loaded with lysate of ectosomes purified from the subretinal space of rds mice. (E, Right) Western blotting of an aliquot of the ectosome lysate using an anti-rhodopsin antibody.
To isolate these ectosomes, we gently detached the retinas of P14 rds mice from the eyecups and collected the vesicular material from the subretinal space. Ectosomes were purified following previously established protocols reported for similar microvesicles in a procedure including differential and gradient centrifugation steps (27, 28) (SI Appendix, Fig. S1). The particle size distribution in the resulting ectosome preparation, measured using a ZetaView particle size analyzer, showed that the majority of vesicles followed a Gaussian size distribution, indicative of their high purity (Fig. 2C). The identity of these vesicles as ciliary ectosomes was confirmed by their immunogold staining with an anti-rhodopsin antibody 4D2, which produced robust labeling along the membrane (Fig. 2D). Consistently, Coomassie-stained SDS/PAGE gels were dominated by an intense band identified as rhodopsin by Western blotting (Fig. 2E).
We next conducted LC-MS/MS analysis of this ectosome preparation. To reduce contributions from contaminating particulate material generated upon detaching retinas from eyecups, we repeated our membrane isolation procedure using WT mouse eyecups. The LC-MS/MS of the resulting preparation was performed alongside ectosomes obtained from rds mice. The results of these analyses are presented in SI Appendix, Table S1, whereas SI Appendix, Table S2 lists 295 confidently identified proteins enriched at least 2-fold in the ectosome preparation (calculated from the relative ion intensities of the corresponding peptides).
The resulting proteome contained all major outer segment proteins (highlighted in green in SI Appendix, Table S2) and 20 proteins directly involved in various aspects of actin dynamics (Table 1). Five of these 20 proteins were known actin nucleators, each one representing a subunit of the Arp2/3 complex. Notably, several Arp2/3 subunits were also identified in the previously reported mouse rod outer segment proteome (29). This suggests that the budding of new photoreceptor discs is driven by the expansion of a branched actin network nucleated by Arp2/3, rather than a linear network nucleated by formins or Spire proteins.
Table 1.
Proteomic identifications of actin-associated proteins in ectosomes purified from rds mice
| Description | Gene name | Confidence score | Unique peptides | rds/WT ratio |
| Actin | Actb, Actg | 2,617 | 36 | 9.0 |
| Actin nucleating | ||||
| Actin-related protein 2 | Actr2 | 231 | 9 | 18.2 |
| Actin-related protein 2/3 complex subunit 1A | Arpc1a | 106 | 2 | 14.4 |
| Actin-related protein 2/3 complex subunit 2 | Arpc2 | 111 | 4 | 6.7 |
| Actin-related protein 2/3 complex subunit 4 | Arpc4 | 70 | 3 | 41.4 |
| Actin-related protein 3 | Actr3 | 80 | 4 | 15.4 |
| Actin cross-linking | ||||
| α-actinin 1 | Actn1 | 671 | 11 | 9.6 |
| α-actinin 4 | Actn4 | 948 | 18 | 20.3 |
| Actin severing | ||||
| Cofilin 1 | Cfl1 | 201 | 4 | 26.3 |
| Destrin | Dstn | 142 | 3 | 30.1 |
| WD repeat-containing protein 1 | Wdr1 | 90 | 4 | >100 |
| Actin filament capping | ||||
| F-actin-capping protein subunit α1 | Capza1 | 158 | 5 | 42.6 |
| F-actin-capping protein subunit β | Capzb | 201 | 8 | 41.9 |
| Actin motors | ||||
| Myosin 10 | Myh10 | 674 | 11 | 19.5 |
| Myosin 9 | Myh9 | 559 | 9 | 8.3 |
| Unconventional myosin VI | Myo6 | 65 | 2 | 93.3 |
| Unconventional myosin VIIa | Myo7a | 147 | 6 | 6.5 |
| Actin signaling | ||||
| Photoreceptor cilium actin regulator | Pcare | 215 | 11 | 15.7 |
| Transforming protein RhoA | Rhoa | 165 | 6 | 10.9 |
| Wiskott–Aldrich syndrome protein family member 3 | Wasf3 | 60 | 2 | >100 |
Rod-Specific Inactivation of Arp2/3 Eliminates F-Actin at the Site of Disc Morphogenesis.
To directly test whether nascent photoreceptor discs require Arp2/3 for evagination, we generated mice specifically lacking Arp2/3 in their rods. This was achieved by breeding mice containing a floxed ArpC3 allele (30) with iCre75 mice expressing Cre recombinase under control of the rhodopsin promoter (31). Previous studies demonstrated that deletion of the ArpC3 subunit abolishes the ability of the Arp2/3 complex to nucleate actin (30, 32, 33). We will refer to the resulting conditional knockout (ArpC3f/f;iCre75+) as the ArpC3−/− mouse.
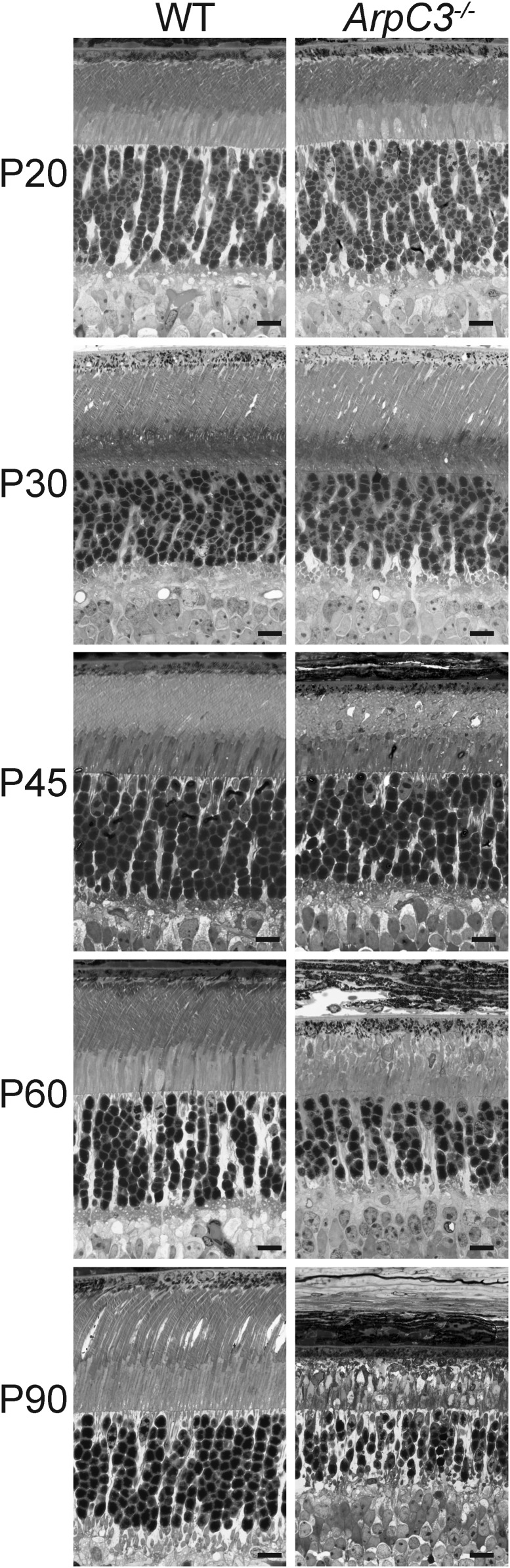
The retinal phenotype of this mouse was first analyzed in semithin plastic sections (Fig. 3). ArpC3−/− rods looked essentially normal at P20 and P30, but displayed a defect in the outer segment structure beginning at P45, which became more severe at P60 and P90 (Fig. 3 and SI Appendix, Fig. S2). We also observed a pattern of progressive photoreceptor degeneration with about one third of cells lost by P90 (Fig. 3 and SI Appendix, Fig. S3). Because Arp2/3 is known to be occasionally involved in the process of vesicular trafficking (34), we tested whether the ArpC3 knockout causes mislocalization of outer segment proteins. However, the localization of multiple outer segment-specific proteins remained normal at P60 (SI Appendix, Fig. S4) despite a prominent defect in outer segment morphology, suggesting that vesicular trafficking to the outer segment is unaffected.
Fig. 3.
Photoreceptor degeneration and disorganization of their outer segments in ArpC3−/− mice. Representative light microscopy images of 0.5-µm-thick toluidine blue-stained plastic sections through the photoreceptor layer of WT and ArpC3−/− retinas at the indicated postnatal days. (Scale bars, 10 µm.)
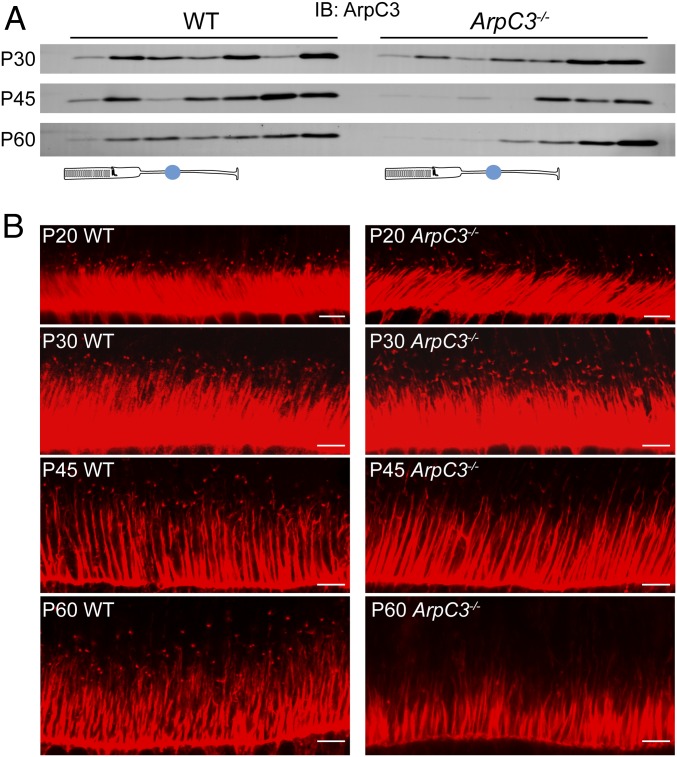
We next investigated the time course of ArpC3 depletion in ArpC3−/− rods. Unfortunately, none of the tested commercially available anti-ArpC3 antibodies produced a specific immunofluorescent signal in rods that was absent in the knockout control. Therefore, we resorted to an alternative technique combining serial tangential sectioning of the frozen flat-mounted retina with Western blotting of ArpC3 in individual sections (refs. 35 and 36 and Fig. 4A). The distribution of ArpC3 was analyzed in 25-µm-thick sections obtained from ArpC3−/− and control WT mice. In WT mice, ArpC3 was distributed throughout the entire retina. In the photoreceptor layer (represented by the top 4 or 5 sections), ArpC3 tended to be enriched in inner segments, which is consistent with its expected pattern of predominantly cytoplasmic distribution. In ArpC3−/− mice, ArpC3 was only marginally reduced in the photoreceptor layer at P30, but was nearly absent from this layer at P45 and P60, indicating that ArpC3 was eliminated from the majority of rods between P30 and P45. The remaining trace amounts of ArpC3 likely originated from cones not expressing Cre recombinase and perhaps the processes of Müller cells extending into the photoreceptor layer. These data show that the lifetime of the ArpC3 protein in ArpC3−/− rods is relatively long because Cre recombination in the iCre75 line completes by approximately P21 (31). Such a relatively long half-life of ArpC3 may be specific for rods, as the phenotype of conditional ArpC3 knockout in other neurons manifests more quickly (30).
Fig. 4.
Simultaneous loss of ArpC3 protein and F-actin puncta at the site of disc morphogenesis in ArpC3−/− mice. (A) ArpC3 immunostaining in serial tangential sections obtained from WT and ArpC3−/− retinas at the indicated postnatal days. Each lane contained material from an entire 25-µm section. The photoreceptor layer marked by a cartoon, normally occupying the top ∼100 to 125 µm of the retina, spans the first 4 to 5 sections (but only the first 3 to 4 sections in degenerating ArpC3−/− retinas at P60). (B) F-actin staining in WT and ArpC3−/− mouse photoreceptors at the indicated postnatal days. F-actin was fluorescently labeled with Alexa Fluor-conjugated phalloidin. (Scale bars, 5 µm.)
In a parallel set of experiments, we analyzed the presence of F-actin puncta at the base of ArpC3−/− rod outer segments (Fig. 4B). Once again, the actin puncta appeared normal up to P30, but the number and the intensity of puncta was drastically reduced at P45. At P60, the puncta was nearly absent, aside from a few originating from cones not expressing Cre recombinase (SI Appendix, Fig. S5).
Taken together, these data show that the depletion of ArpC3, the disappearance of F-actin puncta at the site of disc morphogenesis, and the morphological defect in outer segments tightly correlate among themselves, which suggests that Arp2/3 is responsible for actin nucleation at this site.
Rod-Specific Inactivation of Arp2/3 Disrupts Disc Morphogenesis.
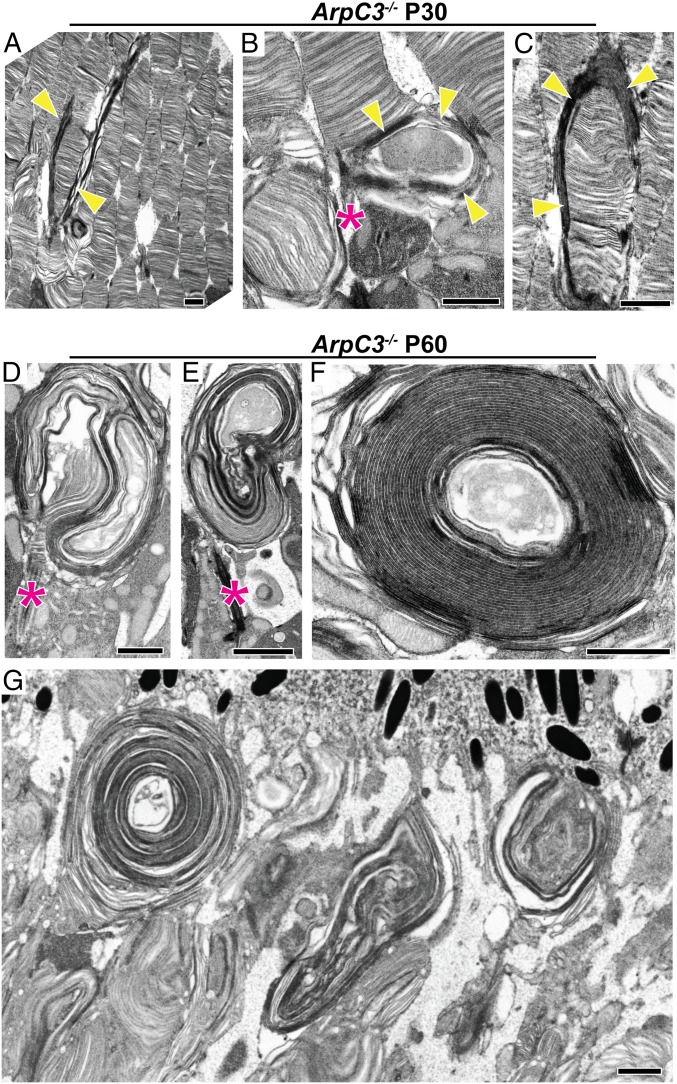
Ultrastructural analysis of ArpC3−/− rods using transmission electron microscopy (TEM) revealed that the earliest occurrence of outer segment pathology takes place in a handful of rods at approximately P30, which is even earlier than detectable by light microscopy (Fig. 5 A–C). At this age, a small fraction of rods displayed uncontrolled elongation of their nascent discs, very similar to the effect of cytochalasin D treatment except with more extended membrane outgrowth. An example of an affected rod surrounded by still normal-looking cells is shown in Fig. 5A, whereas 2 other affected rods are shown at a higher magnification in Fig. 5 B and C. Occasionally, outer segments were completely wrapped by these expanding membranes (Fig. 5C). At P45, membrane outgrowth affected the majority of cells (SI Appendix, Fig. S6), although many still had normal disc stacks at the distal outer segment end and a few outer segments appeared normal, the latter consistent with a few F-actin puncta still present at this age (Fig. 4B). Such a cell-to-cell variability in the onset of this phenotype is most likely explained by a variable onset of Cre recombination in individual rods spanning approximately 2 wk in the iCre75 mice (31). By P60, nearly all rod outer segments had grossly abnormal morphology, with the majority producing large layered membrane whorls emanating from the cilia (Fig. 5 D–G). Occasionally, outgrowing membranes formed nearly perfect spirals (Fig. 5 F and G). These whorls contained both enclosed and open membranes, as judged from the intensity of tannic acid staining.
Fig. 5.
Overgrowth of nascent photoreceptor disc membranes in ArpC3−/− mice. (A–C) Electron micrographs of photoreceptor outer segments containing overgrown nascent disc membranes from ArpC3−/− mice at P30. Retinal sections were contrasted with tannic acid to discern disc membranes exposed to the extracellular space (darkly stained membranes) from those enclosed within the outer segment (lightly stained membranes). Overgrown nascent discs are marked by yellow arrowheads. The connecting cilium in B is marked by an asterisk (magenta). (D–G) Electron micrographs of retinal sections from P60 ArpC3−/− mice showing the large overgrown membrane whorls emanating from the photoreceptor cilium. The connecting cilia are marked by asterisks. (Scale bars: 1 µm.)
The similarity between the ArpC3 knockout phenotype and the consequences of cytochalasin D treatment suggests a common pathophysiological mechanism. In both cases, the outer segments’ failure to initiate the formation of new discs leads to uncontrolled elongation of the several nascent discs, which are the only ones capable of incorporating lipids and proteins delivered to the outer segment. The difference is that the cytochalasin D experiment is acute, whereas the consequences of the ArpC3 knockout persist as long as rods remain viable. Indeed, many rods survive for weeks or even months after the onset of this phenotype, allowing essentially unlimited membrane extension.
In the final set of experiments, we investigated the ultrastructural composition of the membrane whorls in ArpC3−/− rods using 4-tilt scanning TEM (STEM) tomography, which allows the reconstruction of their 3-dimensional architecture through a depth of several micrometers. Movies S2 and S3 each capture an entire whorl from 2 different locations in the P60 ArpC3−/− retina. Interestingly, the tomogram in Movie S3 suggests that a whorl may contain intertwining membranes originating from more than 1 cell. The latter is particularly well-visualized in the segmented, 3D-rendered representation of this tomogram in Movie S4.
Discussion
The central result reported in this study is that Arp2/3, the master nucleator of branched actin networks, is required for the morphogenesis of photoreceptor discs. Combined with the results of several classical studies (8–13), our data build a strong case that Arp2/3 nucleates F-actin at the site of nascent disc formation. Whereas the ArpC3 knockout inactivates Arp2/3 in the entire rod cell, there are 4 arguments supporting direct Arp2/3 involvement at this site. First, the time course of ArpC3 loss from knockout rods correlates with both the loss of F-actin puncta at the outer segment base and the prominence of uncontrolled disc outgrowth. Second, there is no notable actin cytoskeleton disruption in ArpC3 knockout rods outside the active zone of new disc formation. Third, the Arp2/3-dependent formation of branched actin cytoskeleton is typically associated with membrane protrusions needed for cell motility, expansion, phagocytosis, or division (34). Yet, the Arp2/3 inactivation in our experiments is induced in fully matured rods, which are not known to be engaged in any of these processes. One exception is the occasional involvement of Arp2/3 in vesicular trafficking (34); however, we did not observe any mislocalization of multiple outer segment proteins. Fourth, close inspection of the classical ultrastructural images of actin filaments at the site of new disc formation (11) suggests that they are branched.
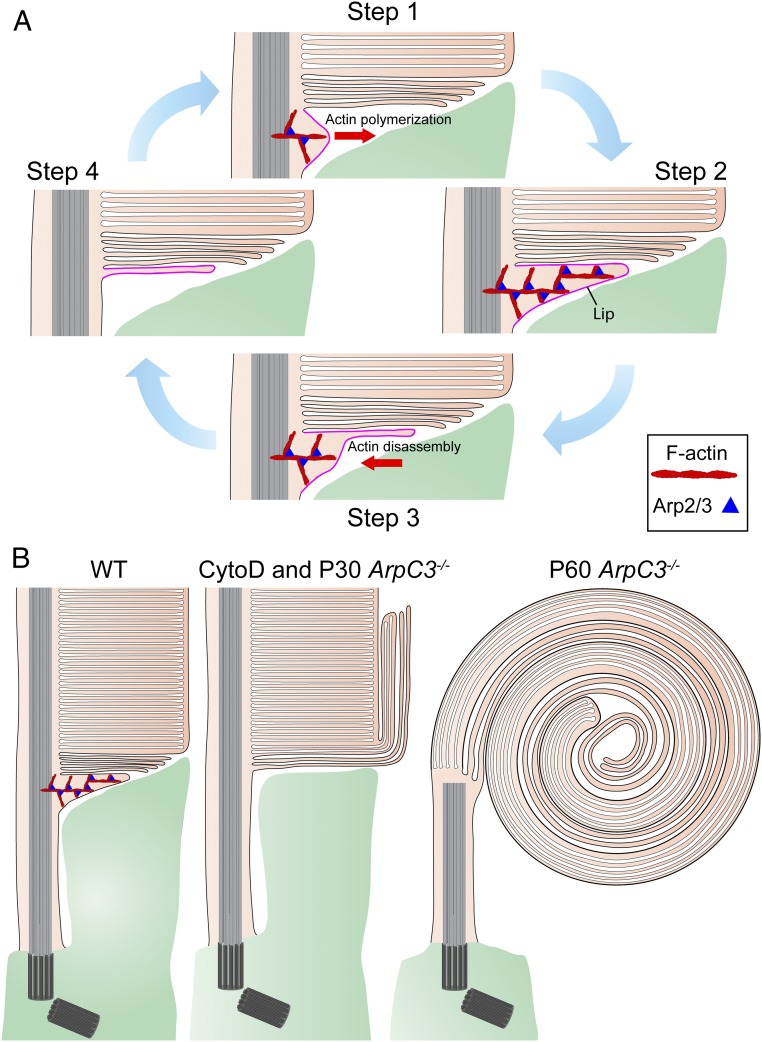
Taken together, these findings suggest a mechanistic model of disc morphogenesis summarized in Fig. 6. In this model, the formation of each new disc is initiated by Arp2/3-dependent actin polymerization, which pushes the ciliary plasma membrane away from the axoneme to form an evaginating membrane structure called a “lip” in early literature (9) (Fig. 6A, steps 1 and 2). The actin network is then disassembled but the lip is not retracted (as in the case of most other actin-mediated protrusions) and is instead flattened and transitioned into a bona fide nascent disc (Fig. 6A, steps 3 and 4). This nascent disc keeps elongating, as proteins and lipids continue to be delivered through the connecting cilium, until it reaches its final diameter and ultimately encloses inside the outer segment. Prevention of actin polymerization, either by cytochalasin D or Arp2/3 knockout, does not allow a new lip to form, and, therefore, no further discs can be produced. Yet, the influx of outer segment membrane material persists, causing uncontrolled elongation of nascent discs. The degree of membrane elongation and the shape of the resulting structures are determined by how long this defect persists (Fig. 6B).
Fig. 6.
Schematic representation of the Arp2/3-dependent mechanism of disc formation. (A) Cartoon depicting the 4 sequential steps in which the formation of a nascent disc is initiated. In step 1, Arp2/3-nucleated actin polymerization pushes the ciliary plasma membrane outward to initiate a membrane evagination. In step 2, actin polymerization continues until reaching a fully formed membrane protrusion (the “lip”). In step 3, the actin network within the lip is disassembled, allowing the membrane protrusion to flatten. In step 4, actin is completely retracted, yielding a fully flattened disc. This nascent disc subsequently elongates until it reaches the final outer segment diameter and is enclosed inside the outer segment. Once actin is retracted, the cycle repeats. (B) Cartoon illustrating the phenotypes of rod photoreceptors with actin polymerization disrupted by either cytochalasin D treatment or ArpC3 knockout. Both approaches completely halt the initiation of disc formation described in A, but permit elongation of preexisting nascent discs through the continued delivery of membrane material. In the case of cytochalasin D treatment or in young ArpC3−/− mice in which disc initiation is halted for a relatively short time, outer segments contain both normal disc stacks and outgrowth of nascent disc membranes. In older ArpC3−/− mice, the nascent disc membrane outgrowth has continued for a long time, resulting in the formation of massive membrane whorls. Note that the cartoon “lip” in A and B is exaggerated in size to aid the illustration of the model.
Another important point of this study is that our proteome contains candidates that could be implicated in nearly all stages of the lifecycle of a lip (Table 1). Perhaps most importantly, we identified Wasf3, a member of the WASP family of proteins, which are essential for the activation of the Arp2/3 complex (37). Also potentially upstream of Arp2/3 is RhoA, a key regulator of actin dynamics (17) that may contribute to determining when and where this cycle begins. Filament capping, which controls the size and shape of the expanding F-actin network, may involve CapZα/β. The disassembly of this network, allowing the transitioning from a lip to a nascent disc, may engage cofilin 1, destrin (aka actin depolymerization factor), and/or WD repeat-containing protein 1. Our proteome also contains 2 α-actinins and 4 myosins, which may be needed for actin cross-linking and actomyosin contractility, respectively. Some of these candidates have already been shown to be localized at the site of new disc formation [e.g., α-actinin (38), myosin II (39), and myosin VIIa (40)] or linked to photoreceptor defects [myosin VI (41), myosin VIIa (42), and PCARE (43)]. We should stress, however, that the involvement of each of these candidate proteins in making new discs would require direct experimental validation, and some proteins involved in this process may be missing from our proteome; for example, we do not identify nudC, a cofilin 1 interacting protein shown to regulate actin dynamics whose knockdown displays a complex phenotype including a pattern of overgrown nascent discs (44).
One additional observation obtained in our study is that some of the overgrown disc membranes, both in cytochalasin D-treated and ArpC3 knockout retinas, were enclosed and others were open, as judged from the intensity of their tannic acid staining. This result argues that initiation of disc formation and disc enclosure are performed by independent molecular mechanisms and that only the former relies on actin polymerization.
Finally, our study further bolsters the mechanistic link between disc morphogenesis and ciliary ectosome release. As illustrated in Fig. 2A, the current evolutionary model of disc morphogenesis suggests that discs evolved from ectosomes retained at the primary cilium of photoreceptor cells (25). Ciliary ectosomes are ubiquitously employed by other cell types for a range of functions, including the disposal of specific proteins from the cilia, the control of cell cycle reentry, and, likely, intercellular communication (45–49). Recent studies demonstrated that release of ciliary ectosomes is driven by a buildup of an actin network (47, 48) and that chemical inhibition of Arp2/3 prevented ectosome release (48). The degree of overlap between these 2 cellular mechanisms is an exciting subject of future investigation.
In conclusion, the results reported in this study argue that the formation of disc membranes in vertebrate photoreceptors is performed by a lamellipodium-like mechanism. Our findings provide a productive conceptual framework toward complete understanding of this complex biological process and reveal an array of molecular players likely involved at each of its critical steps.
Methods
Animals.
Handling of mice was performed in accordance with the approved protocol by the institutional animal care and use committees of Duke University (A254-16-12). WT C57BL/6J mice (stock no. 000664) and rds−/− mice (stock no. 001979) were obtained from Jackson Labs. ArpC3f/f mice (30) were provided by Scott Soderling, Duke University, Durham, NC and crossed with iCre75 mice (30) obtained from Jason Chen, Baylor College of Medicine, Houston, TX to obtain ArpC3f/f;iCre75+mice. All mice were housed under a 12/12 h diurnal light cycle.
Antibodies.
We previously described our rabbit polyclonal anti-PRCD (50) and sheep anti-Rom1 antibodies (50, 51). The following antibodies were gifts: mouse monoclonal antibodies 4D2 against the N terminus of rhodopsin and 1D1 against CNGα1 from Robert S. Molday, University of British Columbia, Vancouver, BC, Canada; rabbit polyclonal anti-peripherin antibody from Gabriel H. Travis, University of California, Los Angeles, CA; and rabbit polyclonal anti-CNGβ1 antibody from Steven Pittler, University of Alabama at Birmingham, Birmingham, AL. We tried the following commercial antibodies to ArpC3 for immunofluorescent staining of rod photoreceptors but did not observe specific labeling from any of them, which was absent in P60 ArpC3−/− rods: mouse anti-ArpC3 (SC-166630; Santa Cruz), rabbit anti-ArpC3 (NBP1-89016; Novus Biologicals), goat anti-ArpC3 (ab37916; Abcam), mouse anti-ArpC3 (612234; BD Biosciences), rabbit anti-ArpC3 (O15145; RayBiotech), and rabbit anti-ArpC3 (LS-C409316-50; LifeSpan BioSciences). For Western blotting, we used mouse anti-ArpC3 (612234; BD Biosciences), which produced the best signal out of these 6 tested antibodies.
Immunofluorescence.
Anesthetized mice were transcardially perfused with a fixative solution, optimized for superresolution microscopy of the actin cytoskeleton (52), containing 4% paraformaldehyde in 80 mM Pipes (pH 6.8), 5 mM EGTA, and 2 mM MgCl2. The eyes were enucleated and postfixed in the same solution overnight at 4 °C. After fixation, the eyes were dissected, and eyecups were embedded in 7% low-melt agarose (A3038; Sigma-Aldrich) and cut by a vibratome (VT1200S; Leica) into 100-µm-thick slices as described previously (53). The sections were blocked in PBS containing 7% goat serum and 0.5% Triton X-100 for 1 h at 22 °C. Next, the slices were incubated with 1 µg/mL WGA conjugated with Alexa Fluor 488 (W11261; Thermo Fisher Scientific), 130 nM phalloidin conjugated with Alexa Fluor 594 (A12381; Thermo Fisher Scientific), and 10 µg/mL Hoechst (H3569; Thermo Fisher Scientific) in the same blocking buffer for 30 min at 22 °C. The sections were washed 3 times in PBS and mounted onto slides in glycerol and coverslipped. Images were taken with the Stimulated Emission Depletion (STED) Leica DMi8 motorized inverted microscope and 93×, NA 1.3 glycerol objective (Leica) using the 775-nm STED depletion laser for the 594 acquisition channel. The imaging area was strictly limited to fields of view not containing melanin granules of the retinal pigment epithelium, which were found to cause severe tissue damage after contact with the high-power STED depletion laser. Hoechst was excluded from staining for STED to avoid interference with the depletion laser. Image processing and analysis was conducted with LAS X software (Leica Microsystems).
Osmotically intact rod outer segments (ROSs) were isolated as described previously (54) with small modifications. Retinas from at least four 2-mo-old WT mice were isolated in mouse Ringer’s solution containing 130 mM NaCl, 3.6 mM KCl, 2.4 mM MgCl2, 1.2 mM CaCl2, and 10 mM Hepes (pH 7.4), adjusted to 314 mOsm. The retinas were pooled into 400 µL ice-cold OptiPrep (8%; Sigma-Aldrich) in Ringer’s solution and vortexed at maximum speed for 60 s in a 1.5-mL Eppendorf tube. The tube was then centrifuged at 200 × g for 30 s to sediment large retinal debris. A total of 350 µL of the supernatant was loaded on top of a 1.8-mL step gradient composed of 10% and 18% OptiPrep in Ringer’s solution and centrifuged for 30 min at 20,000 rpm in a swing-bucket SW-55 rotor (Beckman Coulter) at 4 °C. ROSs were carefully collected from the 10/18% OptiPrep interface and pipetted onto glass-bottom microwell dishes (P35G-1.5–14-c; MatTek) previously coated using 0.01% poly-L-lysine solution (A005C; Millipore). The dish was kept on ice for 20 min to allow ROSs to attach to the bottom before replacing the Ringer’s solution in the dish with fixative solution containing 4% paraformaldehyde in 80 mM Pipes, pH 6.8, 5 mM EGTA, and 2 mM MgCl2 and incubating overnight at 4 °C. The ROSs were washed 3 times in PBS and then stained with WGA and phalloidin as described earlier. Images of ROSs were taken with a confocal microscope (Eclipse 90i and A1 confocal scanner; Nikon) with a 60× objective (1.4 NA Plan Apochromat VC; Nikon) using Nikon NIS-Elements software. Image analysis and processing was performed with ImageJ and/or Nikon NIS-Elements software.
Histological Techniques and Transmission Electron Microscopy.
Serial tangential sectioning with Western blotting was performed as described previously (35, 36). Briefly, eyecups were dissected and the lens removed in ice cold Ringer’s solution. Using a surgical trephine, a 2-mm retinal punch was taken from the center of the eyecup while avoiding the optic nerve and transferred onto a PVDF membrane with photoreceptors facing up. The membrane and retina was sandwiched between glass slides, frozen in liquid nitrogen, and cut on a cryostat (CM3050S; Leica) into 25-µm-thick serial tangential sections, starting with the outer segment layer. The sections were collected and placed into 0.6-mL centrifuge tubes precooled on dry ice and stored at −80 °C until further use.
Fixation and processing of mouse eyes for electron microscopy was performed as described previously (4). Mice were deeply anesthetized and transcardially perfused with 2% paraformaldehyde, 2% glutaraldehyde, and 0.05% calcium chloride in 50 mM Mops (pH 7.4), resulting in exsanguination. The eyes were enucleated and fixed for an additional 2 h in the same buffer at 22 °C. To obtain thin plastic retinal sections, eyecups were cut in half through the optic nerve and embedded in Spurr’s resin (Electron Microscopy Sciences). The embedded retinal cross-sections were cut through the optic nerve in 500-nm slices and stained with toluidine blue for light microscopy as in ref. 55.
For electron microscopy, eyes were fixed and processed as described earlier. The fixed eyes were washed in PBS, and eyecups were dissected and embedded in PBS containing 5% agar (A1296; Sigma-Aldrich) and cut into 200-µm-thick slices on a vibratome (VT1200S; Leica) (4). The vibratome sections were stained with 1% tannic acid (Electron Microscopy Sciences) and 1% uranyl acetate (Electron Microscopy Sciences), gradually dehydrated with ethanol, and infiltrated and embedded in Spurr’s resin. Sections (70 nm) were cut, placed on copper grids, and counterstained with 2% uranyl acetate and 3.5% lead citrate (19314; Ted Pella). The samples were imaged on a JEM-1400 electron microscope (JEOL) at 60 kV with a digital camera (Orius; Gatan).
Three-Dimensional Electron Microscopy Tomography.
Electron tomography experiments were conducted on an FEI Titan Halo operating in the scanning transmission electron microscope (STEM) mode at 300 kV, with the possibility to resolve micrometer-thick plastic-embedded specimens down to nanoscale spatial resolution as described previously (56). Multiple serial sections, ∼800 nm thick, were cut from a resin block of the ArpC3−/− mouse retina processed as described earlier and placed in a ribbon-like shape on 50-nm Luxel film slot grids. The grids were glow-discharged, and a mix of 10-nm, 15-nm, and 30-nm gold particles was deposited on both surfaces to act as fiducial markers; the grids were carbon-coated to limit any specimen shrinkage caused by the electron beam/sample interaction. Electron tomography was performed following a 4-tilt series scheme described in ref. 57, with the specimen tilted from −60° to +60° every 0.5° at 4 evenly distributed azimuthal angle positions. To increase the effective thickness in the final 3D representation, corresponding reconstructions of the specimen originating from up to 3 consecutive sections were generated through filtered back-projection and then stitched together. This process leaves minor discontinuities in the volume density at the individual tomogram junctions. With magnification set to 10,000× and a matching pixel size of approximately 3 nm, the total accessible specimen volume was approximately 6.32 µm × 7.5 µm × 2.5 µm (Movie S3), which is large enough to contain most of the membrane whorls encountered in this study. Movie S2 represents a higher-magnification (28,500×) tomogram encompassing a large fragment of a whorl with an accessible volume of approximately 2.1 µm × 2.1 µm × 1.5 µm with pixel size of approximately 1 nm. For the image segmentation shown in Movie S4, membrane contours were manually traced as separate objects and meshed into a 3D model using IMOD software (58).
In Vivo Treatment of Mouse Retinas with Cytochalasin D.
Adult mice, anesthetized with 100 mg/kg ketamine and 10 mg/kg xylazine, were intravitreally injected at 9 AM with 0.5 µL of 250 µM cytochalasin D (C8273; Sigma-Aldrich) dissolved in PBS containing 1% DMSO. The mice were kept alive for 6 h before conducting perfusion fixation at 3 PM and processing the eyes for electron microscopy as described earlier.
Purification of Ectosomes from the Subretinal Space of rds Mice.
Two litters encompassing ∼14 mice (∼28 retinas) per genotype of rds and WT mice were killed at P14 for each experiment. Eyes were enucleated and their cornea and lens dissected and removed to form eyecups consisting of the RPE/choroid and attached retina. All eyecups were pooled into 2 mL of ice-cold Ringer’s solution containing protease inhibitor mixture (4693159001; Sigma-Aldrich) and, while immersed in this buffer, retinas were gently detached from the RPE/choroid to release the ectosomes trapped in the subretinal space. The suspended ectosomes and detached eyecups were gently agitated on ice in a 2-mL low-retention microcentrifuge tube (02-681-332; Fisher Scientific) for 10 min before centrifugation at 300 × g for 5 min to pellet the retinas and large debris. The supernatant was transferred to a clean 2-mL tube before centrifugation at 2,000 × g for 10 min to pellet large contaminating particles. This supernatant was transferred to an ultracentrifuge tube (344057; Beckman Coulter) and diluted with ice-cold Ringer’s solution to a volume of 4 mL before centrifugation at 150,000 × g in a TLA 100.3 rotor (Beckman Coulter) at 4 °C. The pellet was gently rinsed with ice-cold PBS before resuspending the pellet in 200 µL of PBS containing protease inhibitors. The resuspended pellet was layered on top of a 10-mL linear gradient of 10 to 70% sucrose in PBS made with a gravity-fed gradient maker (GM20; CBS Scientific) in an ultracentrifuge tube (344059; Beckman Coulter). The gradient was centrifuged for 5.5 h at 200,000 × g at 4 °C in a SW41 rotor (Beckman Coulter). A total of nine 1-mL fractions were collected by inserting a 21-gauge needle into the ultracentrifuge tube exactly 1 cm from the bottom and pumping the gradient at 1 mL/min through Masterflex tubing (HV96400-14–1.6; Cole-Parmer) with a peristaltic pump (EP1; Bio-Rad) into new ultracentrifuge tubes. The density of each 1-mL fraction was determined by measuring each fraction’s weight. The fractions were then diluted to 10 mL with PBS and centrifuged for 1 h at 40,000 rpm at 4 °C in a SW41 rotor. The pellet in each tube was resuspended with 100 µL of 2% SDS in 100 mM Tris, pH 8.0. A 30-µL aliquot was taken from each fraction for Western blotting, while the rest was processed for mass spectrometry as described later.
Immunogold Labeling of Purified Ectosomes.
Ectosomes purified from the density gradient as described earlier were examined by TEM following a previously established protocol (27). Briefly, the ectosomes collected from 1.17- to 1.20-g/L density gradient fractions described earlier were resuspended in PBS containing 2% paraformaldehyde and deposited onto formvar-coated copper grids (01702-F; Ted Pella). The grids were washed with 50 mM glycine in PBS and then blocked in 5% donkey serum in PBS for 10 min before labeling with 4D2 mouse anti-rhodopsin antibodies in blocking buffer. Next, the grids were washed in PBS, blocked in 5% donkey serum, and labeled with donkey anti-mouse secondary conjugated with 6-nm colloidal gold particles (Jackson ImmunoResearch Laboratories). The grids were washed in PBS before fixation with 1% glutaraldehyde, rinsed with water, and contrasted negatively with uranyl-oxalate solution (2% uranyl acetate in 75 mM oxalic acid, pH 7.0) and positively with methyl cellulose uranyl acetate solution (1.8% methyl cellulose in 0.4% uranyl acetate, pH 4.0). The grids were imaged on a JEM-1400 electron microscope (JEOL) at 60 kV with a digital camera (Orius; Gatan).
ZetaView Particle Size Analysis.
Ectosomes purified at the density gradient were resuspended in 300 µL PBS and subjected to particle size analysis by a ZetaView nanoparticle tracking instrument (S/N227; Particle Metrix). The ectosome suspension was diluted at least 1,000-fold with PBS to obtain a particle concentration within the optimal range of the instrument at about ∼80,000 particles per microliter. The diluted suspension was injected into the ZetaView, and a total of 1,648 particles were imaged and traced from 7 positions in the size distribution mode. ZetaView software was used for analysis (version 8.02.30.02).
Mass Spectrometry.
Ectosomes collected from the sucrose gradient fractions with density 1.17 to 1.20 g/L were pooled into one sample before processing for mass spectrometry as described previously (59, 60). Briefly, proteins solubilized in 2% SDS, 100 mM Tris·HCl (pH 8.0) were reduced with 10 mM DTT (D0632; Sigma-Aldrich), alkylated with 25 mM iodoacetamide (I1149; Sigma-Aldrich), and subjected to tryptic hydrolysis using the HILIC beads SP3 protocol (60). The resulting peptides were analyzed with a nanoAcquity UPLC system (Waters) coupled to an Orbitrap Q Exactive HF mass spectrometer (Thermo Fisher Scientific) employing the LC-MS/MS protocol in a data-independent acquisition mode. The peptides were separated on a 75-μm × 150-mm, 1.7-μm C18 BEH column (Waters) using a 120-min gradient of 8 to 32% of acetonitrile in 0.1% formic acid at a flow rate of 0.3 mL/min at 45 °C. Eluting peptides were sprayed into the ion source of the Orbitrap Q Exactive HF at a voltage of 2.0 kV. Progenesis QI Proteomics software (Waters) was used to assign peptides to the features and generate searchable files, which were submitted to Mascot (version 2.5) for peptide identification. For peptide identification, we searched against the UniProt reviewed mouse database (September 2019 release) using carbamidomethyl at Cys as a fixed modification and Met oxidation as a variable modification. Proteins are included in SI Appendix, Table S1 based on 3 criteria: 1) at least 1 peptide is identified in each independent experiment, 2) at least 2 peptides are identified in one experiment, and 3) the identification confidence score is >95% in both experiments.
Western Blotting.
Lysates were generated by adding SDS, at a final concentration of 2%, to aliquots taken from ectosome suspensions after different steps of purification. An appropriate volume of 4× Laemmli buffer containing 400 mM DTT was added to these protein lysates before subjecting them to SDS/PAGE. The separated proteins were transferred to low-fluorescence PVDF membrane (Bio-Rad) and probed with mouse monoclonal antibody 1D4 against the C terminus of rhodopsin (ab5417; Abcam). Goat secondary antibody conjugated with Alexa Fluor 800 (Invitrogen) was used for detection on an Odyssey infrared imaging system (LiCor Bioscience).
Data Availability.
The raw mass spectrometry data were deposited in the MassIVE repository (dataset identifier MSV000084598; ref. 61) and can be accessed at ftp://massive.ucsd.edu/MSV000084598/. All other data are included in the manuscript and SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants EY012859 (V.Y.A.), EY005722 (V.Y.A.), EY025558 (W.J.S.), EY029929 (T.R.L.), GM103412 (M.H.E.), CA014236 (Duke University), and UL1TR002553 (Duke University) and an Unrestricted Award from Research to Prevent Blindness Inc. (Duke University).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The raw proteomics data reported in this article have been deposited in the MassIVE repository (dataset identifier MSV000084598).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913518117/-/DCSupplemental.
References
- 1.Arshavsky V. Y., Burns M. E., Photoreceptor signaling: Supporting vision across a wide range of light intensities. J. Biol. Chem. 287, 1620–1626 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young R. W., The renewal of photoreceptor cell outer segments. J. Cell Biol. 33, 61–72 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burgoyne T., et al. , Rod disc renewal occurs by evagination of the ciliary plasma membrane that makes cadherin-based contacts with the inner segment. Proc. Natl. Acad. Sci. U.S.A. 112, 15922–15927 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding J. D., Salinas R. Y., Arshavsky V. Y., Discs of mammalian rod photoreceptors form through the membrane evagination mechanism. J. Cell Biol. 211, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kinney M. S., Fisher S. K., The photoreceptors and pigment epithelium of the larval Xenopus retina: Morphogenesis and outer segment renewal. Proc. R. Soc. Lond. B Biol. Sci. 201, 149–167 (1978). [DOI] [PubMed] [Google Scholar]
- 6.Steinberg R. H., Fisher S. K., Anderson D. H., Disc morphogenesis in vertebrate photoreceptors. J. Comp. Neurol. 190, 501–508 (1980). [DOI] [PubMed] [Google Scholar]
- 7.Volland S., et al. , Three-dimensional organization of nascent rod outer segment disk membranes. Proc. Natl. Acad. Sci. U.S.A. 112, 14870–14875 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams D. S., Linberg K. A., Vaughan D. K., Fariss R. N., Fisher S. K., Disruption of microfilament organization and deregulation of disk membrane morphogenesis by cytochalasin D in rod and cone photoreceptors. J. Comp. Neurol. 272, 161–176 (1988). [DOI] [PubMed] [Google Scholar]
- 9.Chaitin M. H., Schneider B. G., Hall M. O., Papermaster D. S., Actin in the photoreceptor connecting cilium: Immunocytochemical localization to the site of outer segment disk formation. J. Cell Biol. 99, 239–247 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaitin M. H., Bok D., Immunoferritin localization of actin in retinal photoreceptors. Invest. Ophthalmol. Vis. Sci. 27, 1764–1767 (1986). [PubMed] [Google Scholar]
- 11.Arikawa K., Williams D. S., Organization of actin filaments and immunocolocalization of alpha-actinin in the connecting cilium of rat photoreceptors. J. Comp. Neurol. 288, 640–646 (1989). [DOI] [PubMed] [Google Scholar]
- 12.Chaitin M. H., Carlsen R. B., Samara G. J., Immunogold localization of actin in developing photoreceptor cilia of normal and rds mutant mice. Exp. Eye Res. 47, 437–446 (1988). [DOI] [PubMed] [Google Scholar]
- 13.Vaughan D. K., Fisher S. K., Cytochalasin D disrupts outer segment disc morphogenesis in situ in rabbit retina. Invest. Ophthalmol. Vis. Sci. 30, 339–342 (1989). [PubMed] [Google Scholar]
- 14.Hale I. L., Fisher S. K., Matsumoto B., The actin network in the ciliary stalk of photoreceptors functions in the generation of new outer segment discs. J. Comp. Neurol. 376, 128–142 (1996). [DOI] [PubMed] [Google Scholar]
- 15.Nemet I., Tian G., Imanishi Y., Submembrane assembly and renewal of rod photoreceptor cGMP-gated channel: Insight into the actin-dependent process of outer segment morphogenesis. J. Neurosci. 34, 8164–8174 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanchoin L., Boujemaa-Paterski R., Sykes C., Plastino J., Actin dynamics, architecture, and mechanics in cell motility. Physiol. Rev. 94, 235–263 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Ridley A. J., Life at the leading edge. Cell 145, 1012–1022 (2011). [DOI] [PubMed] [Google Scholar]
- 18.van Nie R., Iványi D., Démant P., A new H-2-linked mutation, rds, causing retinal degeneration in the mouse. Tissue Antigens 12, 106–108 (1978). [DOI] [PubMed] [Google Scholar]
- 19.Travis G. H., Sutcliffe J. G., Bok D., The retinal degeneration slow (rds) gene product is a photoreceptor disc membrane-associated glycoprotein. Neuron 6, 61–70 (1991). [DOI] [PubMed] [Google Scholar]
- 20.Connell G., et al. , Photoreceptor peripherin is the normal product of the gene responsible for retinal degeneration in the rds mouse. Proc. Natl. Acad. Sci. U.S.A. 88, 723–726 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen A. I., Some cytological and initial biochemical observations on photoreceptors in retinas of rds mice. Invest. Ophthalmol. Vis. Sci. 24, 832–843 (1983). [PubMed] [Google Scholar]
- 22.Jansen H. G., Sanyal S., Development and degeneration of retina in rds mutant mice: Electron microscopy. J. Comp. Neurol. 224, 71–84 (1984). [DOI] [PubMed] [Google Scholar]
- 23.Nir I., Papermaster D. S., Immunocytochemical localization of opsin in the inner segment and ciliary plasma membrane of photoreceptors in retinas of rds mutant mice. Invest. Ophthalmol. Vis. Sci. 27, 836–840 (1986). [PubMed] [Google Scholar]
- 24.Usukura J., Bok D., Changes in the localization and content of opsin during retinal development in the rds mutant mouse: Immunocytochemistry and immunoassay. Exp. Eye Res. 45, 501–515 (1987). [DOI] [PubMed] [Google Scholar]
- 25.Salinas R. Y., et al. , Photoreceptor discs form through peripherin-dependent suppression of ciliary ectosome release. J. Cell Biol. 216, 1489–1499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaitin M. H., Actin filaments in the photoreceptor cilium of the rds mutant mouse. Exp. Eye Res. 53, 107–113 (1991). [DOI] [PubMed] [Google Scholar]
- 27.Livshits M. A., et al. , Isolation of exosomes by differential centrifugation: Theoretical analysis of a commonly used protocol. Sci. Rep. 5, 17319 (2015). Erratum in: Sci Rep.6, 21447 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thery C., Amigorena S., Raposo G., Clayton A., Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell. Biol. Chapter 3, Unit 3.22 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Liu Q., et al. , The proteome of the mouse photoreceptor sensory cilium complex. Mol. Cell. Proteomics 6, 1299–1317 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim I. H., et al. , Disruption of Arp2/3 results in asymmetric structural plasticity of dendritic spines and progressive synaptic and behavioral abnormalities. J. Neurosci. 33, 6081–6092 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li S., et al. , Rhodopsin-iCre transgenic mouse line for Cre-mediated rod-specific gene targeting. Genesis 41, 73–80 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Suraneni P., et al. , The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J. Cell Biol. 197, 239–251 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gournier H., Goley E. D., Niederstrasser H., Trinh T., Welch M. D., Reconstitution of human Arp2/3 complex reveals critical roles of individual subunits in complex structure and activity. Mol. Cell 8, 1041–1052 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Rotty J. D., Wu C., Bear J. E., New insights into the regulation and cellular functions of the ARP2/3 complex. Nat. Rev. Mol. Cell Biol. 14, 7–12 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Sokolov M., et al. , Massive light-driven translocation of transducin between the two major compartments of rod cells: A novel mechanism of light adaptation. Neuron 34, 95–106 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Lobanova E. S., et al. , Transducin translocation in rods is triggered by saturation of the GTPase-activating complex. J. Neurosci. 27, 1151–1160 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alekhina O., Burstein E., Billadeau D. D., Cellular functions of WASP family proteins at a glance. J. Cell Sci. 130, 2235–2241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arikawa K., Williams D. S., Alpha-actinin and actin in the outer retina: A double immunoelectron microscopic study. Cell Motil. Cytoskeleton 18, 15–25 (1991). [DOI] [PubMed] [Google Scholar]
- 39.Chaitin M. H., Coelho N., Immunogold localization of myosin in the photoreceptor cilium. Invest. Ophthalmol. Vis. Sci. 33, 3103–3108 (1992). [PubMed] [Google Scholar]
- 40.Liu X., Vansant G., Udovichenko I. P., Wolfrum U., Williams D. S., Myosin VIIa, the product of the Usher 1B syndrome gene, is concentrated in the connecting cilia of photoreceptor cells. Cell Motil. Cytoskeleton 37, 240–252 (1997). [DOI] [PubMed] [Google Scholar]
- 41.Schubert T., et al. , Deletion of myosin VI causes slow retinal optic neuropathy and age-related macular degeneration (AMD)-relevant retinal phenotype. Cell. Mol. Life Sci. 72, 3953–3969 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu X., Udovichenko I. P., Brown S. D., Steel K. P., Williams D. S., Myosin VIIa participates in opsin transport through the photoreceptor cilium. J. Neurosci. 19, 6267–6274 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corral-Serrano J. C., et al. , C2orf71a/pcare1 is important for photoreceptor outer segment morphogenesis and visual function in zebrafish. Sci. Rep. 8, 9675 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boitet E. R., Reish N. J., Hubbard M. G., Gross A. K., NudC regulates photoreceptor disk morphogenesis and rhodopsin localization. FASEB J. 33, 8799–8808 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J., Barr M. M., Cell-cell communication via ciliary extracellular vesicles: Clues from model systems. Essays Biochem. 62, 205–213 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wood C. R., Huang K., Diener D. R., Rosenbaum J. L., The cilium secretes bioactive ectosomes. Curr. Biol. 23, 906–911 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phua S. C., et al. , Dynamic remodeling of membrane composition drives cell cycle through primary cilia excision. Cell 168, 264–279.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nager A. R., et al. , An actin network dispatches ciliary GPCRs into extracellular vesicles to modulate signaling. Cell 168, 252–263.e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nachury M. V., Mick D. U., Establishing and regulating the composition of cilia for signal transduction. Nat. Rev. Mol. Cell Biol. 20, 389–405 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skiba N. P., et al. , Proteomic identification of unique photoreceptor disc components reveals the presence of PRCD, a protein linked to retinal degeneration. J. Proteome Res. 12, 3010–3018 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gospe S. M., 3rd, et al. , Membrane attachment is key to protecting transducin GTPase-activating complex from intracellular proteolysis in photoreceptors. J. Neurosci. 31, 14660–14668 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leyton-Puig D., et al. , PFA fixation enables artifact-free super-resolution imaging of the actin cytoskeleton and associated proteins. Biol. Open 5, 1001–1009 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lobanova E. S., et al. , Mechanistic basis for the failure of cone transducin to translocate: Why cones are never blinded by light. J. Neurosci. 30, 6815–6824 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsang S. H., et al. , Role for the target enzyme in deactivation of photoreceptor G protein in vivo. Science 282, 117–121 (1998). [DOI] [PubMed] [Google Scholar]
- 55.Lobanova E. S., et al. , Transducin γ-subunit sets expression levels of α- and β-subunits and is crucial for rod viability. J. Neurosci. 28, 3510–3520 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sousa A. A., Azari A. A., Zhang G., Leapman R. D., Dual-axis electron tomography of biological specimens: Extending the limits of specimen thickness with bright-field STEM imaging. J. Struct. Biol. 174, 107–114 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Phan S., et al. , 3D reconstruction of biological structures: Automated procedures for alignment and reconstruction of multiple tilt series in electron tomography. Adv. Struct. Chem. Imaging 2, 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kremer J. R., Mastronarde D. N., McIntosh J. R., Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996). [DOI] [PubMed] [Google Scholar]
- 59.Spencer W. J., et al. , Progressive rod-cone degeneration (PRCD) protein requires N-terminal S-acylation and rhodopsin binding for photoreceptor outer segment localization and maintaining intracellular stability. Biochemistry 55, 5028–5037 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hughes C. S., et al. , Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol. 10, 757 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spencer W. J., Skiba N. P., Arshavsky V. Y., The proteome of ciliary ectosomes present in the subretinal space of rds mice. MassIVE. https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?accession=MSV000084598. Deposited 19 November 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw mass spectrometry data were deposited in the MassIVE repository (dataset identifier MSV000084598; ref. 61) and can be accessed at ftp://massive.ucsd.edu/MSV000084598/. All other data are included in the manuscript and SI Appendix.