Significance
Disruption of normal transcriptional splicing is a common mutational mechanism for disease-predisposing alleles. Some splice-altering mutations can be difficult to detect, and their effects difficult to characterize, because they lie deep within exons or introns. We developed cBROCA, an experimental approach that characterizes the transcriptional effects of genomic mutations anywhere in a locus. With patient RNA as template, cBROCA yields quantitative estimates of all effects of altered splicing in tumor suppressor genes. cBROCA analysis of mutations from patients with familial cancers revealed a wide variety of consequences of abnormal splicing, including whole or partial exon skipping, exonification of intronic sequence, loss or gain of exonic or intronic splicing enhancers or silencers, complete intron retention, and combinations of these alterations.
Keywords: mutation, splicing, cancer
Abstract
Mutations responsible for inherited disease may act by disrupting normal transcriptional splicing. Such mutations can be difficult to detect, and their effects difficult to characterize, because many lie deep within exons or introns where they may alter splice enhancers or silencers or introduce new splice acceptors or donors. Multiple mutation-specific and genome-wide approaches have been developed to evaluate these classes of mutations. We introduce a complementary experimental approach, cBROCA, which yields qualitative and quantitative assessments of the effects of genomic mutations on transcriptional splicing of tumor suppressor genes. cBROCA analysis is undertaken by deriving complementary DNA (cDNA) from puromycin-treated patient lymphoblasts, hybridizing the cDNA to the BROCA panel of tumor suppressor genes, and then multiplex sequencing to very high coverage. At each splice junction suggested by split sequencing reads, read depths of test and control samples are compared. Significant Z scores indicate altered transcripts, over and above naturally occurring minor transcripts, and comparisons of read depths indicate relative abundances of mutant and normal transcripts. BROCA analysis of genomic DNA suggested 120 rare mutations from 150 families with cancers of the breast, ovary, uterus, or colon, in >600 informative genotyped relatives. cBROCA analysis of their transcripts revealed a wide variety of consequences of abnormal splicing in tumor suppressor genes, including whole or partial exon skipping, exonification of intronic sequence, loss or gain of exonic and intronic splicing enhancers and silencers, complete intron retention, hypomorphic alleles, and combinations of these alterations. Combined with pedigree analysis, cBROCA sequencing contributes to understanding the clinical consequences of rare inherited mutations.
As genetic testing for inherited disease becomes increasingly widespread, rare mutations are very frequently encountered whose consequences are unknown. Some of these mutations alter splicing, with biological and clinical consequences that can range from mild to severe. Because these mutations are individually rare, comparing their frequencies between cases versus controls is generally not informative, and they must be evaluated experimentally. Multiple RNA-based approaches have been applied to this problem, including RT-PCR followed by next-generation sequencing (1), TOPO TA cloning of complementary DNA (cDNA) products (2), minigene analysis (3), genome-wide RNA sequencing approaches (4, 5), genome editing with measurement of RNA expression in cells (6), and an RNA-Seq assay targeted to splicing events in breast and ovarian cancer genes (7).
cBROCA is an experimental approach using patient cells that complements these methods. cBROCA is based on isolating RNA from patient lymphoblast cells that have been grown in the presence of puromycin so as to inhibit nonsense-mediated decay, followed by hybridization to the BROCA panel of cancer-predisposition genes (8), followed by multiplexed sequencing to >1,000-fold median depth. The approach is analogous to evaluation of genomic DNA by BROCA, but with cDNA rather than genomic DNA as template. Transcript sequences are aligned, splice junctions predicted computationally, and splicing effects evaluated by comparing transcript profiles of cases versus controls.
The purpose of this project was to apply cBROCA sequencing to cDNA of cancer patients from severely affected families in order to characterize rare mutations potentially leading to abnormal splicing. Depending on the gene, between 5% and 20% of known cancer-predisposing mutations act by altering splicing (SI Appendix, Table S1). Our goal was to develop and deploy an effective way to characterize newly discovered potential splice-altering mutations. In addition to identifying mutations at canonical splice sites, cBROCA revealed rare variants of 4 sorts: 1) variants at canonical splice sites yielding multiple abnormal transcripts, some of which were not predictable by in silico tools; 2) intronic variants occurring at some distance from splice junctions, without strong (or any) in silico support; 3) exonic variants altering exonic splicing enhancers or silencers; and 4) genomic copy number variants (CNVs) altering splicing.
cBROCA was developed in order to discover and evaluate mutations that alter transcription in genes predisposing to cancer, but the same approach could be applied to any gene panel that includes intronic and untranslated region (UTR) sequences in gene capture.
Results
Participants were cancer patients from families severely affected with cancers of the breast, ovary, endometrium, or colon, but with negative (normal) results from conventional genetic testing. For each family, cBROCA analysis was undertaken for any rare mutation in any tumor suppressor gene associated with a patient’s cancer, regardless of the exonic or intronic position of the mutation in the gene. Overall, cBROCA analysis was undertaken for 120 candidate splice mutations in 16 tumor suppressor genes in 150 families with >600 genotyped relatives: BRCA1 (26 mutations), BRCA2 (31 mutations), ATM (20 mutations), PALB2 (11 mutations), BRIP1 (6 mutations), CHEK2 (6 mutations), CDH1 (4 mutations), PTEN (4 mutations); BARD1, MLH1, RAD51C, and RAD51D (2 mutations each); and APC, GEN1, MSH2, and TP53 (1 mutation each).
cBROCA analysis of these mutations revealed a wide variety of abnormal splicing, including whole or partial exon skipping, exonification of intronic sequence, loss or gain of splicing enhancers and silencers in either exons or introns, complete intron retention, hypomorphic alleles, and combinations of these alterations.
Validation.
To validate analysis by cBROCA, 2 classes of mutations were evaluated by the approach described in Methods. The first validation class comprised genomic mutations at canonical splice sites that were consistently predicted by in silico tools to alter splicing (SI Appendix, Table S2). For these 20 mutations, splice effects revealed by cBROCA were as expected, with extremely high Z scores. cBROCA analysis clarified 1 feature of the characterizations of several of these mutations, in that proportions of mutant transcripts from the mutant allele (PM) estimated from cBROCA were generally not identical to proportions of mutant transcripts estimated from single mutation-specific methods. In particular, for many heterozygous mutations, PCR-based methods yielded substantially more than 50% mutant transcripts from patients’ diploid RNA. These high fractions likely reflect bias in PCR amplification of shorter mutant transcripts versus full-length normal transcripts. cBROCA estimates are not based on PCR amplification so estimates of proportions of mutant transcripts may be more accurate. The second validation class comprised genomic mutations in potential splice regulatory regions, either exonic or intronic, predicted consistently by in silico tools to have no effect on splicing (SI Appendix, Table S3). For these 58 mutations, all cBROCA analyses were consistent with in silico predictions, indicating no splice effects of the genomic mutations. These benign mutations were not coinherited with cancer more often than expected by chance in severely affected families. (For 3 of these mutations, consequences to protein function in the context of normal splicing have been reported, as indicated in SI Appendix, Table S3.)
We turned next to more complex cases. The examples that follow illustrate the range of splice alterations revealed by cBROCA. For each class of mutations, detailed results of all cBROCA analyses are provided in the supplementary tables.
Multiple Transcripts from Single Mutations at Canonical Splice Sites.
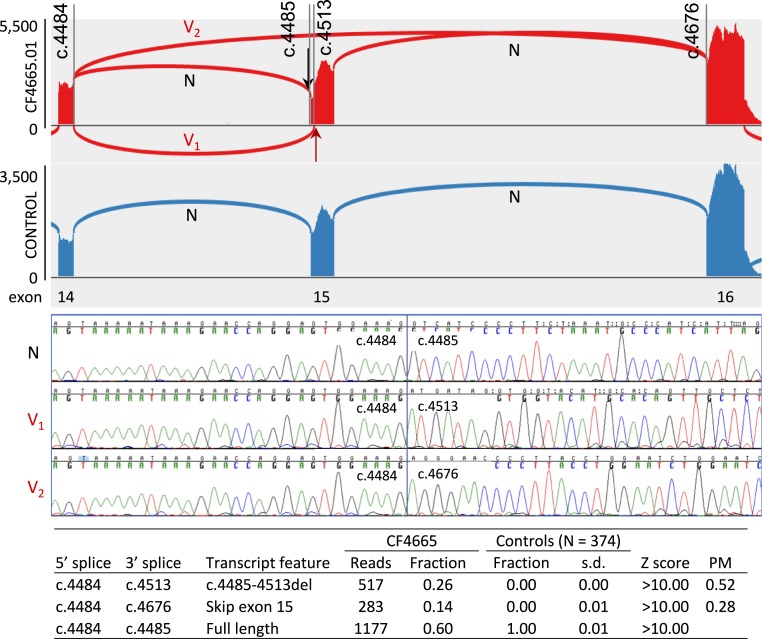
A single splice-altering genomic mutation may yield multiple mutant transcripts. Analysis by cBROCA yields quantitative estimates of these effects. BRCA1 c.4485-1G>A at chr17:41,226,539C>T (hg19) in family CF4665 is an informative example to illustrate the approach (Fig. 1). The proband of family CF4665 was diagnosed with breast cancer at age 36; her mother died of ovarian cancer. Analysis by cBROCA indicates that BRCA1 c.4485-1G>A leads to 2 mutant transcripts, in different proportions. One mutant transcript (V1) exploits a cryptic acceptor splice site in BRCA1 exon 15 at chr17:41,226,510 (BRCA1 c.4513), created by the weakened canonical splice site due to the genomic mutation. This altered splice is supported by 26% (517/1,977) of reads at this site from cDNA from patient cells and does not appear in cDNA from controls (0.00 ± 0.01 of reads in 374 controls), yielding Z > 10. The mutant transcript from the cryptic acceptor splice has a deletion of 29 base pairs (c.4485-c.4513) and a premature stop at codon 1496. The other mutant transcript skips BRCA1 exon 15 by splicing from chr17:41,228,504 of exon 14 to chr17:41,223,256 of exon 16. This altered splice is supported by 14% (283/1,977) of reads at this site from cDNA of patient cells and similarly does not appear in cDNA of controls, also yielding Z > 10. This mutant transcript has a premature stop at codon 1519. RT-PCR and Sanger sequencing validated the 3 transcripts. Given that the cells are heterozygous for mutant and normal alleles and were grown in puromycin to protect transcripts from nonsense-mediated decay, all transcripts appear at approximately the proportions in which they were generated. The proportions of altered transcripts specifically from the mutant allele (PM) are therefore ∼52% with partial deletion of exon 15, 28% with complete deletion of exon 15, and 20% normal.
Fig. 1.
An example of analysis of BRCA1 by cBROCA. At BRCA1 exons 14 to 16, BRCA1 c.4485-1G>A (black arrow) of patient CF4665.01 produces 2 mutant transcripts (V1 and V2) and normal transcript (N) (red graph). Wild-type BRCA1 of controls produces only the normal transcript (blue graph). Mutant transcript V1 introduces a frameshift by splicing from c.4484 of exon 14 to a cryptic acceptor at c.4513 of exon 15, yielding a premature stop at codon 1496. This transcript is supported by 26% of reads (517/1,977). Mutant transcript V2 skips BRCA1 exon 15 by splicing from c.4484 to c.4676 of exon 16, yielding a premature stop of codon 1509. This transcript is supported by 14% of reads (283/1,977). Sanger sequencing validated these splice effects. Estimates of proportions of transcripts from the mutant allele (PM) are therefore 52% V1, 28% V2, and 20% wild-type.
As the results for this mutation suggest, analysis by cBROCA can reveal multiple altered transcripts due to the same genomic mutation: that is, pleiotropic effects of the mutant allele. From RNA extracted directly from patient tissues, transcripts with premature stops would be subject to nonsense-mediated decay so might be detected at very low levels or missed entirely. Knowing all transcriptional effects of a splice mutation can be important because some mutations may be more severe than suggested by observing only the (disproportionately in-frame) mutant transcripts that survive nonsense-mediated decay.
Splice site mutations in 7 genes that lead to multiple mutant transcripts in severely affected families are described in SI Appendix, Table S4. Some of these mutations have been previously reported, with functional evaluation (9–14). For others, results of cBROCA analysis provide experimental support for interpreting transcriptional consequences. Two examples are shown in Fig. 2 and described here.
Fig. 2.
Analysis by cBROCA of mutations in BARD1 and ATM leading to multiple abnormal transcripts. (A) In family CF5058, BARD1 c.159-1C>T occurs in the proband, who was diagnosed with ovarian cancer (Ov) at age 58, her son who was diagnosed with prostate cancer (Pr) at age 52, and two other female relatives who underwent risk-reducing bilateral salpingo-oophorectomy (BSO) and remain cancer-free. Heterozygosity for the variant allele is indicated by VN and homozygosity for the nonmutant allele by NN. (B) In family CF1395, ATM c.5674+1G>T occurs in relatives who developed cancers of the breast (Br, including bilateral breast cancer [Bil Br]), ovary (Ov), prostate (Pr), pancreas (Pa), and cholangiocarcinoma/bile duct (Chlnc); and acute lymphocytic leukemia (ALL), leukemia (Leuk), non-Hodgkin lymphoma (NHL), and Hodgkin Disease (HD). Heterozygosity for the variant allele is indicated by VN and homozygosity for the nonmutant allele by NN. (C) In family CF5058, BARD1 c.159-1C>T (black arrow) yields two abnormal transcripts: V1, which skips exon 2, including critical residues Cys53, Cys66, His68, and Cys71 of the BARD1 RING domain, and V2, which skips exons 2 and 3 with an immediate stop at codon 53. Neither of these mutant transcripts appears in controls. (D) In family CF1395, ATM c.5674+1G>T yields multiple abnormal transcripts, nearly all of which (V1 cluster) extend into ATM intron 36 where they are subject to decay. Evidence of these transcripts was detected by cBROCA in RNA from cells treated with puromycin to inhibit nonsense-mediated decay. Other abnormal transcripts skip exons 37 and 38 (V2) or exon 37 (V3), both leading to premature stops.
BARD1 c.159-1(IVS2-1)C>T (CF5058) yields 2 abnormal transcripts, neither of which appears in controls. From the mutant allele, 41% of transcripts [2 × (235/1,144)] (from SI Appendix, Table S4) skip exon 2, and therefore skip critical residues Cys53, Cys66, His68, and Cys71 of the BARD1 RING domain (15), and 35% of transcripts [2 × (201/1,144)] skip exons 2 and 3, with an immediate stop at codon 53. The proband with this mutation was diagnosed with ovarian cancer at age 58, and her son was diagnosed with prostate cancer at age 52. Two other female relatives carrying the mutation are now older than age 60; both had risk-reducing bilateral salpingo-oophorectomy and remain cancer-free.
ATM c.5674+1G>T (CF1395) yields multiple abnormal transcripts, nearly all of which extend into ATM intron 36 where they are subject to decay. Evidence of these transcripts was detected by cBROCA given puromycin treatment that inhibited nonsense-mediated decay. Two other abnormal transcripts skip exon 37 or exons 37 and 38 and introduce stops. Carriers of this mutation developed an unusually wide range of cancers, all at middle age or older, several of which are not generally associated with mutations in ATM: cancers of the breast, ovary, prostate, pancreas, and bile duct, and acute lymphocytic leukemia (ALL), non-Hodgkin lymphoma, and Hodgkin disease.
Exonification and Other Effects of Intronic Mutations.
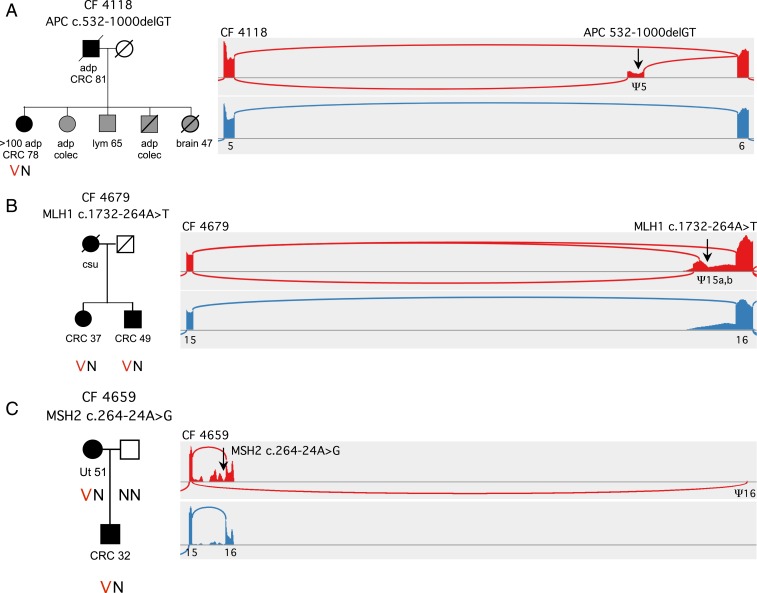
Multiple mutations in 9 different genes occurred at intronic sites other than splice junctions yet were suggested by cBROCA analysis to significantly alter splicing. Analyses of 15 such mutations are shown in SI Appendix, Table S5. For several of these, functional studies have been reported (16–20), and cBROCA results are in agreement with each of these reports. Among the previously unreported mutations, 3 altered splicing by unusual mechanisms (Fig. 3). Commercial testing had returned negative (wild-type) results for each of these families.
Fig. 3.
Analysis by cBROCA of intronic mutations leading to exonification. (A) In family CF4118, APC c.532-1000delGT (black arrow) leads to exonification of 165 bp in APC intron 5 and an in-frame stop. Exonification is very likely due to disruption of a silencing enhancer at chr5:112,115,485-112,115,492, and thereby activation of a splice acceptor at chr5:112,115,380 and of a splice donor at chr5:112,115,548. This exon does not appear in transcripts of controls. (B) In family CF4679, MLH1 c.1732-264A>T creates 2 exons that do not appear in normal MHL1 transcripts: one, with an insertion of 141 bp in the message and a stop at codon 587; and the other, with an insertion of 85 bp in the message and a stop at codon 581. The mechanism of exonification is likely creation of a donor splice at c.1732-264, activating previously silent acceptor sites at c.1732-406 and at c.1732-350. (C) In family CF4659, MSH2 c.2635-24A>G destroys a splicing branch-point, activating multiple cryptic splice sites in MSH1 intron 15, leading to multiple unstable transcripts. The mutation also activates a cryptic acceptor splice more than 30 kb downstream, at MSH2 c.2634+31432, creating transcripts that skip exon 16 and the 3′UTR, with a stop after 922 codons. Abbreviations are adenomatous polyps (adp), prophylactic colectomy (colec), colorectal cancer (CRC), lymphoma (lym), uterine cancer (Ut), cancer of unknown primary site (csu).
APC c.532-1000delGT at chr5:112,115,487 (CF4118) is a private genomic mutation 1 kilobase (kb) from the nearest splice junction. cBROCA analysis revealed an exon at chr5:112,115,381–112,115,547, an insertion of 165 base pairs (bp) in the message with a stop at codon 199. The mechanism of exonification is likely disruption by the genomic mutation of a canonical splicing silencer motif at chr5:112,115,485–112,115,492, thereby activating a cryptic splice acceptor at chr5:112,115,380 (NNSPLICE 0.99, MaxEnt 9.75) and a cryptic splice donor at chr5:112,115,548 (NNSPLICE 0.98, MaxEnt 6.65). No transcripts in controls include this exon. cBROCA analysis indicates that 89% of transcripts [2 × (127/286)] (from SI Appendix, Table S5) from the mutant allele include this exon. The proband and multiple relatives of family CF4118 developed characteristic features of familial adenomatous polyposis.
MLH1 c.1732-264A>T at chr3:37,088,746 (CF4679) was revealed by cBROCA analysis to create 2 exons, at chr3:37,088,604–37,088,744, an insertion of 141 bp in the message with a stop at codon 587, and chr3:37,088,660–37,088,744, an insertion of 85 bp in the message with a stop at codon 581. cBROCA analysis indicated that 65% of transcripts [2 × (692+318)/3,095] (from SI Appendix, Table S5) from the mutant allele include 1 of these 2 exons. Transcripts from nonmutant alleles have background reads 5′ of exon 16, but none include the exons. The mechanism of exonification is likely creation by the genomic mutation of a new donor splice at c.1732-264 (NNSPLICE 0.99, MaxEnt 9.72 for the mutant sequence), activating previously silent acceptor sites at c.1732-406 and at c.1732-350. The proband of family CF4679 was diagnosed with colon cancer at age 31; immunohistochemistry of her tumor indicated loss of MLH1 and PMS2 proteins. Her brother was diagnosed with colon cancer at age 42, and their mother died of cancer of unknown primary site.
MSH2 c.2635-24A>G at chr2:47,709,894 (CF4659) was revealed by cBROCA analysis to destroy a splicing branch point, leading to multiple unstable transcripts. The mutation leads to activation of multiple cryptic splice sites in intron 15, with in-frame stops in each of the run-on transcripts; these represent 46% of transcripts [2 × (811/3,526)] (from SI Appendix, Table S5) from the mutant allele. The mutation also activates a cryptic acceptor splice more than 30 kb downstream, at MSH2 c.2634+31432, creating transcripts that skip exon 16 and the 3′ UTR, with a stop after 922 codons. The mechanism for this aberrant splicing is likely the destruction by the genomic mutation of the intron 15 branch point (21). The proband of family CF4659 developed endometrial cancer at age 51, and her son was diagnosed with colorectal cancer at age 32. Immunohistochemistry of tumors from both patients indicated loss of MSH2 and MSH6.
Exonic Mutations in Splicing Enhancers and Silencers.
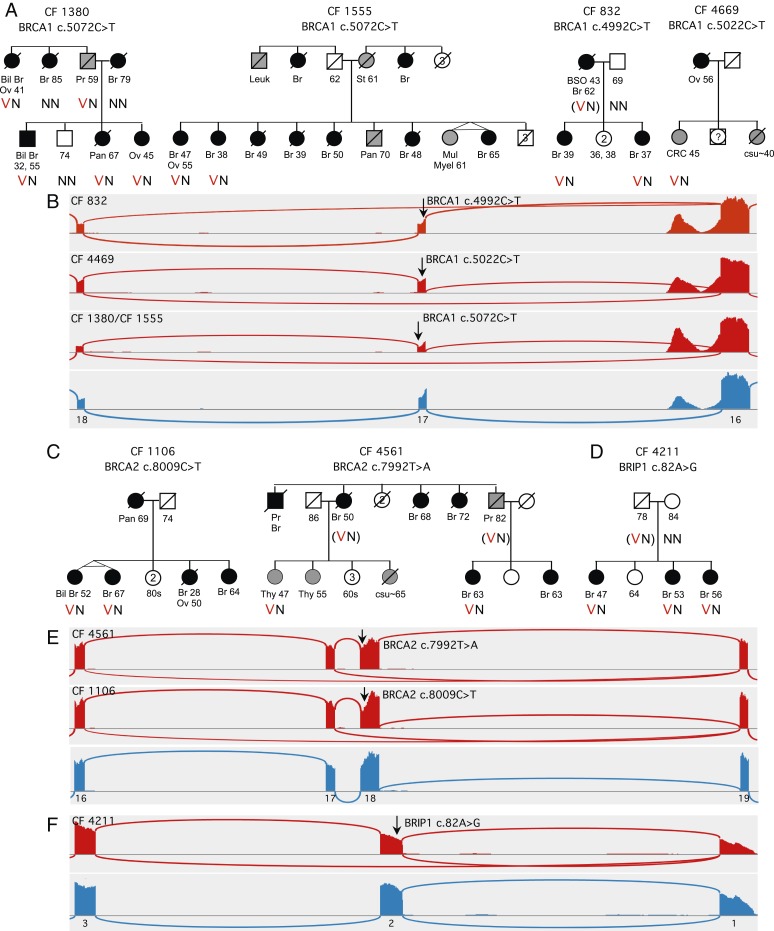
Exonic mutations that alter splicing by destroying exonic splicing enhancers (ESEs) or creating exonic splicing silencers (ESSs) have long been recognized as predisposing to human disease, and their underlying mechanisms have been elegantly explored (22, 23). (APC c.532-1000delGT, described above, is a corresponding intronic mutation that leads to exonification due to loss of an exonic splicing silencer deeply embedded in an intron.) ESE and ESS motifs are common, particularly in small exons, including in BRCA1 and BRCA2. cBROCA can contribute to understanding the roles of exonic mutations by revealing their abnormal splice products and by providing quantitative measures of effects of different mutations in the same exon (Fig. 4).
Fig. 4.
cBROCA analysis of exonic mutations in splicing enhancers and silencers. (A) Abnormal splicing of BRCA1 exon 17 in families CF1380, CF1555, CF832, and CF4469. (B) In family CF832, BRCA1 c.4992C>T deletes a splice enhancer in exon 17, leading to exon skipping in 26% of transcripts from the mutant allele. In family CF4469, BRCA1 c.5022C>T deletes a more 3′ splice enhancer of exon 17, leading to exon skipping in 32% of transcripts from the mutant allele. In families CF1380 and CF1555, BRCA1 c.5072C>T deletes a still more 3′ splice enhancer in exon 17, leading to exon skipping in 61% of transcripts from the mutant allele. In all families, proportions of mutant transcripts are significantly above background (P < 0.0000003 for each comparison). (C) Abnormal splicing of BRCA2 exon 18 in families CF1106 and CF4561. (D) Abnormal splicing of BRIP1 exon 2 in family CF4211. (E) BRCA2 c.7992T>A in family CF4561 and BRCA2 c.8009C>T in family CF1106 both lead to skipping exon 18 and a stop at codon 2702 in 43% and 51% of transcripts, and to skipping exons 17 and 18 and a stop at codon 2645 in 6% and 8% of transcripts, respectively, from the mutant alleles. (F) In family CF4211, BRIP1 c.82A>G leads to skipping exon 2, including the ATG translation start, in 43% of transcripts from the mutant allele. The next in-frame ATG in the mutant transcript is at codon 101 in exon 4, so the mutant protein is predicted to lack residues 1 to 100, including most of the DNA binding domain. Abbreviations are cancers of the breast (Br, including bilateral breast cancer [Bil Br]), ovary (Ov), pancreas (Pan), prostate (Pr), stomach (St), thyroid (Thy), unknown primary site (csu), and colorectal cancer (CRC); multiple myeloma (Mult Mye); and bilateral salpingooophorectomy (BSO). Heterozygosity for a variant allele is indicated by VN and homozygosity for the nonmutant allele by NN.
For example, cBROCA analysis revealed abnormal splicing of the small (88 bp) exon 17 of BRCA1 in 4 families. Genomic mutations in the 4 families appeared at 3 different mutant sites (SI Appendix, Table S6). All 3 exon 17 mutations yielded transcripts skipping exon 17 with a stop at codon 1672, but with quite different proportions of mutant transcripts. BRCA1 c.5072C>T was evaluated by cBROCA in 4 carriers in 2 families (CF1380, CF1555), with almost identical results; for each participant, the mutant allele yielded 61% ± 1% mutant transcripts [2 × (0.96)(1,409)/4,438] (from SI Appendix, Table S6). The ESE disrupted by this mutation is GAAA(C>T)AG, an enhancer at the 3′ end of exon 17. Cancers in these families included young onset female breast cancer, male breast cancer, and ovarian cancer.
In contrast, BRCA1 c.5022C>T (CF4469) and BRCA1 c.4992C>T (CF832) each produce a lower proportion of exon 17-skipping transcripts from their mutant alleles: 32% and 26%, respectively. These proportions of abnormal transcripts are significantly above background: 6.7 and 5.0 SDs above proportions of exon 17-skipping transcripts in controls. The genomic mutations disrupt ESE motifs CACAT(C>T)ACTTT and ATGCT(C>T)GTG near the 5′ end of exon 17. On ClinVar, interpretations of BRCA1 c.5022C>T and BRCA1 c.4992C>T range from benign to uncertain, based on the absence of any possible protein effect (both are silent mutations) or any in silico-predicted splice effects.
Family histories can be informative in interpreting intermediate proportions of mutant transcripts. Relatives in family CF832 developed breast cancer at young ages, suggesting a damaging effect for BRCA1 c.4992C>T. On the other hand, for family CF4469, very little family history was available so the effect of BRCA1 c.5022C>T is less clear. This mutation is reported on an online database to cooccur in a patient with a BRCA1 frameshift (24), which, if correct ,and in trans with the frameshift, in a person with no signs of Fanconi anemia, would be strong evidence for the mutation being benign. However, the database does not report any details of the genotype or the phenotype. This conundrum reveals the importance of fully reporting particularly informative individuals on online databases.
At BRCA2 exon 18, analysis by cBROCA revealed abnormal splicing in 2 families, based on 2 different genomic mutations: BRCA2 c.7992T>A (CF4561) and BRCA2 c.8009C>T (CF1106) (SI Appendix, Table S6). From both mutant alleles, most abnormal transcripts (43% and 51%, respectively) skipped exon 18, with a stop at codon 2702, and a smaller proportion of abnormal transcripts (6% and 8%, respectively) skipped both exon 17 and exon 18, with a stop at codon 2645. The cancers in the families with these mutations are consistent with their cBROCA results. BRCA2 c.8009C>T is considered pathogenic, in part because of possible effects on protein function of the amino acid substitution S2670L (25). cBROCA analysis suggests that this mutation is damaging to transcription as well. BRCA2 c.7992T>A is silent at the translational level but is similar to BRCA2 c.8009C>T in its effect on transcription. We interpret both mutations as intermediate or hypomorphic alleles.
BRIP1 c.82A>G, p.M28V (CF4211) presents as a missense in the first coding exon of the gene. However, cBROCA analysis indicates that the critical effect is likely to be on transcription: 43% of transcripts from the mutant allele skip exon 2, which includes the ATG translation start. The next in-frame ATG in the mutant transcript is at codon 101 in exon 4, so the mutant protein is predicted to lack residues 1 to 100, which are highly conserved and include a major part of the DNA binding domain. ESE-finder predicts that the mutation disrupts an ESE motif at BRIP1 c.78-87, replacing the ESE with an ESS. Given that BRIP1 confers far lower risks of ovarian or breast cancer than BRCA1 or BRCA2, a BRIP1 hypomorphic allele might be of little consequence. However, 3 sisters with breast cancer in family CF4211 carry this mutation, consistent with a damaging effect.
Evaluation of Genomic Copy Number Variants by cBROCA.
Evaluation of the consequences of genomic copy number amplifications can be challenging because the effect of the amplification depends on whether and how the amplified genomic sequence is spliced into the message. cBROCA can be usefully applied to this problem in 2 ways. Differences in read depth of each exon from RNA of the mutation carrier versus controls indicate which exons are included in mutant transcripts and at what multiplicity. In addition, newly formed splice junctions are supported by cBROCA reads found only in the mutation carrier’s RNA.
Copy number analysis of DNA sequence revealed 4 copies of BRCA2 exons 14 to 24 in 4 families in our series (CF1815, CF4541, CF4748, and CF4755) (Fig. 5A). BROCA sequencing of genomic DNA indicated a triplication of 30 kb with genomic breakpoints at chr13:32,927,735–32,958,445 of one allele, and wild-type copy number of the other allele in all 4 families. Analysis of genomic DNA did not reveal if the triplication was in tandem, or involved additional sequence, or its effect on splicing. So we turned to transcript analysis. cBROCA analysis of RNA revealed inclusion of exons 14 to 24 at significantly greater depth than for controls, with increased depth consistent with 1 normal transcript and 1 transcript with 3 copies of each of exons 14 to 24. The best estimate of the proportion of transcripts from the mutant allele with 3 copies of exons 14 to 24 was 94%. The in-tandem arrangement of the triplicated segment was indicated by the presence of 986 reads spanning BRCA2 c.9256 (last base pair of exon 24) and BRCA2 c.7008 (first base pair of exon 14) in RNA from the mutation carriers, but not from controls. These reads were not aligned to the reference sequence but were detectable in the pool of cBROCA reads. Insertion of 2 additional copies of exons 14 to 24 corresponds to an insertion of 4,498 bp in the BRCA2 message, with a stop at mutant codon 3116. A second minor transcript from the mutant allele is the same as the major mutant transcript but also includes an exonified fragment of intron 24 at chr13:32,958,030–32,958,169 (SI Appendix, Table S7), revealed by 155 reads (not aligned to human reference sequence) spanning the last base pair of the exonified fragment and c.7008 (first base pair of exon 14). The exonified fragment is present in ∼4% of transcripts from CF4541 and ∼1% of control transcripts. It corresponds to an insertion of 139 bp and leads to a stop in both the mutant and normal transcripts.
Fig. 5.
cBROCA analysis of effects of genomic copy number variants on transcription. (A) Genomic triplication of BRCA2 exons 14 to 24 in 4 families leads to greater read depth at splice junctions in cBROCA sequence of mutation carriers compared to unrelated controls (n = 87). Read depth >1.5-fold specifically at exons 14 to 24 suggests the triplication is in tandem. (B) De novo genomic deletion of BRCA2 noncoding exon 1 in family CF4727 leads to lower read depths across all BRCA2 splice junctions for the mutation carrier compared to controls (n = 17). The ratio of BRCA2 reads for CF4727 versus controls was 0.53, suggesting no BRCA2 transcription from the mutant allele. Bil, bilateral; Br, breast cancer; Pan (Panc), pancreatic cancer.
Finally, for patient CF4727, who developed breast cancer at age 53, BROCA analysis of genomic DNA indicated deletion of BRCA2 noncoding exon 1 with break points chr13:32,889,493 and chr13:32,890,429. Genomic sequences of all BRCA2 coding exons (2 to 27) were intact. The deletion was not present in either parent: so de novo in the patient. The effect of the deletion on transcription was evaluated using cBROCA (Fig. 5B). Because the only source of patient RNA was whole blood isolated from a PAXgene RNA tube (Qiagen) after shipment, treatment with puromycin was not possible. Very little RNA could be obtained (2.8 μg total), and quality was marginal (RNA integrity number [RIN] = 6.8). cBROCA analysis was therefore modified so as to test the transcript effects of this mutation. The modified approach was to sequence using cBROCA, then to count the number of reads at every splice junction of BRCA2 in patient RNA and in RNA from 17 individuals with RNA similarly isolated from whole blood from PAXgene RNA tubes (RIN = 6.14 ± 0.91). In order to adjust for differences of RNA quality across samples, we counted the number of reads at every splice junction of ATM, a gene with no mutation in CF4727 or in controls so expected to have similar coverage, then normalized the BRCA2 data for differences in read depths. Normalized read depths for all BRCA2 splice junctions were compared for the proband of CF4727 versus the 17 controls (Fig. 5B). The read depth ratio was 0.53, yielding P = 2.1E−10 by t test for matched pairs (i.e., matched for splice site). The ratio of 0.53 in patient transcripts suggests complete absence of BRCA2 transcription from the mutant allele.
SI Appendix, Table S8 is an index, by gene and genomic position, of all mutations evaluated by cBROCA, with reference to the supplementary table that includes analytic details.
Discussion
Targeted capture and multiplexed sequencing using cBROCA yields qualitative and quantitative assessments of the effects of genomic mutations on transcriptional splicing of tumor suppressor genes. cBROCA has multiple useful features for analysis of candidate splice mutations in patients. Analysis is based directly on patients’ RNA, not on in silico predictions or cellular models. Genomic DNA and RNA can be tested simultaneously by BROCA and cBROCA, respectively. Splicing of multiple genes is tested simultaneously. All mutant transcripts created by each genomic mutation are identified, with very deep coverage providing robust quantitative measures of the proportions of each transcript. cBROCA also serves for mutation discovery because the approach reveals all abnormal transcripts in targeted genes, including mutant transcripts resulting from exonic or intronic genomic variants far from canonical splice sites.
cBROCA analysis of cDNA derived from patient lymphoblasts was used to characterize mutations in tumor suppressor genes. The range of splice alterations detected by cBROCA included complete and partial exon skipping, exonification of intronic sequence, intron retention, alterations of splicing enhancers and silencers in exons or even in introns, and transcript alterations due to genomic amplifications and deletions. Some of these classes of altered splicing can be difficult to identify using PCR-based techniques without prior knowledge of the consequence of the splicing change.
cBROCA incorporates experimental and analytical components of previous methods of targeted RNA sequencing (26–34). Modifications by cBROCA include sequencing all exonic and non-Alu intronic sequences of targeted genes; sequencing with very high coverage to enable detection of low abundance transcripts; and analysis of newly created splice junctions at considerable distances from normal splice sites. A difference from the RNA CaptureSeq protocol (28, 29) is that cBROCA counts only split reads in the first step of analysis. Focusing on split reads reduces background and simplifies calculation of Z scores, which are used to compare numbers of alternate transcripts to expected distributions from controls. The yield is systematic quantification of experimental results.
As mentioned above, a challenge to evaluation of splice-altering mutations is that many transcripts with premature stop codons are subject to nonsense-mediated decay and hence present only at very low abundance in cells. This problem has previously been addressed by high and targeted sequence coverage (35–37), by treating cells with ribosome-binding drugs that inhibit nonsense-mediated decay (38–40), and by labeling and tracking cDNA fragments through library preparation with molecular tags and then sequencing to determine their absolute concentrations (41). cBROCA combines several of these approaches: analyzing cDNA generated from puromycin-treated cells, and very high (>1,000×) coverage made feasible by a small targeted set of critical genes. Even given these treatments, the use of lymphoblast cell lines remains a limitation of cBROCA. Genes are generally expressed in lymphoblast cell lines (42), but, for some genes, it could be necessary to grow cell lines from other tissues.
A thorny problem of cancer genetics is how much reduction of productive transcript of a critical gene is required to cause high risk, or even moderate risk, of the relevant cancer. For cBROCA analysis, statistically significant differences in proportions of altered transcripts were based on differences in read depths at critical sites between test samples and control samples analyzed in the same way. For almost all mutations with statistically significant reductions in normal splicing, PM values were greater than 0.50: that is, reduction of productive transcription from the mutant allele was more than 50%. PM values varied by mutation, gene, and background of naturally occurring alternate transcription. Mutations abrogating canonical splice sites usually, but not always, yielded higher PM values than deeply exonic or intronic mutations.
Clinical consequences of some mutations may be less severe than consequences of complete loss of transcription. Hypomorphic, or intermediate risk, alleles may arise from splice-altering mutations that lead to truncations but retain considerable productive transcript, as with some BRCA1 exon 17 enhancer mutations (Fig. 4A); or from splice altering mutations leading to in-frame loss of nonessential protein domains, as for some PALB2 and ATM mutations (SI Appendix, Table S4); or from mutations with modest effects on protein function, as for BRCA1 p.R1699Q (43). With many patients undergoing testing for mutations in cancer-predisposing genes, it is inevitable that hypomorphic alleles will increasingly frequently appear in clinical practice. We suggest that provision be made in public databases to explicitly document hypomorphic alleles of genes whose complete loss-of-function alleles confer extremely high cancer risks.
Mutation analysis by cBROCA requires generating a lymphoblastoid cell line from patient cells so it is useful to know when this effort and expense is most worthwhile. We found that, for severely affected families with negative (normal) results by conventional genetic testing, cBROCA was most likely to provide valuable information when DNA-based sequencing revealed an extremely rare or private mutation anywhere in a tumor suppressor gene associated with the phenotype of the family. cBROCA was particularly informative when the mutation was not previously reported or remained a variant of unknown significance on ClinVar (25) and similar sites. In our experience and that of others (17, 44), for mutations at canonical splice sites, predictions of normal splicing based on consensus of multiple in silico tools were reliable and led to negative (normal) results using cBROCA (e.g., SI Appendix, Table S3). On the other hand, for mutations at canonical splice sites for which in silico tools predicted altered splicing, cBROCA frequently revealed more complex abnormalities than those predicted (e.g., Fig. 2 and SI Appendix, Table S4). Finally, for deeply exonic or deeply intronic mutations and for many copy number variants, effects on splicing could only be understood by experimental test such as cBROCA (e.g., Figs. 3–5 and SI Appendix, Tables S3–S5).
By now, most changes at or very near canonical splice sites of tumor suppressor genes have been encountered frequently enough to have been tested experimentally, the results reported, and consensus reached. We hope that cBROCA will accelerate this process for newly encountered and still-uncharacterized candidate splice mutations at all genomic sites in critical genes, thereby contributing to accurate mutation interpretation and to precision medicine.
Methods
Study Subjects.
Participants were patients diagnosed with breast, ovarian, endometrial, or colorectal cancer and their informative relatives. Between January 2014 and the present, families with negative (normal) results from conventional genetic testing were enrolled in this project if BROCA analysis of genomic DNA revealed a rare or private variant at any exonic or intronic site in a gene associated with the patient’s or the family’s cancer, or a copy number variant (CNV) with a possible splice effect. The project was approved by the University of Washington Human Subjects Division (Protocol 1583); all participants provided written informed consent.
For each participant, peripheral blood was obtained, lymphocytes isolated, and a lymphoblast cell line created. When the number of cells in suspension culture reached 106 to 107, cell lines were treated with 500 μg/mL puromycin (Sigma-Aldrich) for 5 h prior to harvesting to inhibit nonsense-mediated decay. Total RNA was extracted using TRIzol Reagent (Invitrogen/ThermoFisher) and treated with DNaseI using the RNeasy Mini Kit (QIAGEN) to remove residual genomic DNA. RNA quality was measured on a Tape Station 2200 (Agilent); all samples from cell lines had RNA integrity number (RIN) >8.0. cDNA was generated for 10 μg of RNA (5 μg per library) by a combination of random hexamers and oligo dT priming (Superscript II First Strand cDNA Synthesis System; Invitrogen/ThermoFisher). Double-stranded cDNA was synthesized using the NEBNext mRNA Second Strand Synthesis Module (NEB). For each patient, genomic DNA and double-stranded cDNA adapter-ligated fragments (mean insert size 275 bp) were PCR-amplified for 5 and 10 precapture cycles, respectively; then, a total 550 μg of each genomic and cDNA-derived library were hybridized to the BROCA gene panel (8).
The BROCA design includes 2.5 Mb of genomic reference sequence corresponding to 70 complete loci (45), including exons, nonrepetitive intronic sequence, 5′ UTRs and 3′ UTRs, and ∼2 kb upstream and downstream intergenic genomic sequence. Libraries were hybridized in solution to the custom oligonucleotides (Agilent) and then sequenced on an Illumina HiSeq2500 to generate 2 × 101-bp paired-end reads. cDNA and genomic DNA were sequenced using the same protocol. For each sample, 1 genomic DNA library and 2 replicate cDNA libraries were sequenced. For any intermediate or ambiguous cBROCA, cDNA samples from additional affected relatives were sequenced in the same way. Average coverage for targeted regions was 144× for genomic DNA and >1,300× for cDNA.
Bioinformatics Analysis of Genomic DNA Sequence.
Reads were aligned to the human reference genome (hg19) and variants filtered as previously described (8, 46–48). All variants with minor allele frequency <0.01, whether at canonical splice sites or other exonic or intronic sites, including deep intronic regions, were scored using NNSPLICE and MaxEnt to predict disruption or activation of splice sites (49, 50) and by RESCUE-ESE, SPLICEMAN, and HSF to predict effects of the variant on splice enhancer and silencer motifs (51–55). cBROCA revealed both new and known splice mutations, with some of the known mutations yielding more complex consequences than previously reported.
Analysis of cDNA Sequence.
cDNA sequence reads were mapped to the human genome (hg19) using Bowtie, and splicing events were predicted using the TopHat algorithm (4, 5). For each sample, at the donor and acceptor splice sites of each predicted splicing event, 2 ratios were calculated: 1) the number of reads with flanking exonic base pairs adjacent to each other (i.e., a successful splice), divided by the total number of reads including either exonic base pair; and 2) the number of reads including a flanking exonic base pair and the adjacent flanking intronic base pair (i.e., a failed splice), divided by the total number of reads including the exonic base pair. For each sample, the read ratio for each altered transcript was compared to the mean and SD of the analogous ratios for all other samples in the same experiment. The value for the study sample was expressed as the number of SDs from the mean of the control distribution, or Z score. To evaluate multiple altered transcripts from the same genomic mutation, the number of reads was counted for each altered transcript, and the proportion normalized for the proportion of reads representing that transcript in all other samples. Scores were considered significant if Z > 3.0, reflecting test sample read ratios >3 SDs above the mean, but most Z scores were either clearly not significant or >10.0. For each candidate mutation, the parameter “PM” was defined as the proportion of transcripts from the mutant allele that were abnormal. Results were visualized using the Sashimi plot function of the Integrated Genomic Viewer (56). For additional validation, some variants were also tested by mutation-specific experimental methods, including RT-PCR and Sanger sequencing, and/or TOPO TA cloning and Sanger sequencing.
Data Availability Statement.
Results of analyses of all variants have been deposited in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/).
Supplementary Material
Acknowledgments
We thank the families for their participation, their patience, and their loyalty to our project. This work was supported by grants from the NIH (R35CA197458), the Breast Cancer Research Foundation, the Susan G. Komen Foundation, and the American Cancer Society.
Footnotes
Competing interest statement: R.O. is an employee of Color Genomics. Z.T. is an employee of Color Genomics. T.W. consults for Color Genomics.
Data deposition: All variants evaluated in this study have been deposited in the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915608116/-/DCSupplemental.
References
- 1.Farber-Katz S., et al. , Quantitative analysis of BRCA1 and BRCA2 germline splicing variants using a novel RNA-massively parallel sequencing assay. Front. Oncol. 8, 286 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fontes A., Cloning technologies. Methods Mol. Biol. 997, 253–261 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Gaildrat P., et al. , Multiple sequence variants of BRCA2 exon 7 alter splicing regulation. J. Med. Genet. 49, 609–617 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Trapnell C., Pachter L., Salzberg S. L., TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trapnell C., et al. , Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Findlay G. M., et al. , Accurate classification of BRCA1 variants with saturation genome editing. Nature 562, 217–222 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davy G., et al. , Detecting splicing patterns in genes involved in hereditary breast and ovarian cancer. Eur. J. Hum. Genet. 25, 1147–1154 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walsh T., et al. , Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc. Natl. Acad. Sci. U.S.A. 107, 12629–12633 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman L. S., et al. , Novel inherited mutations and variable expressivity of BRCA1 alleles, including the founder mutation 185delAG in Ashkenazi Jewish families. Am. J. Hum. Genet. 57, 1284–1297 (1995). [PMC free article] [PubMed] [Google Scholar]
- 10.Meyer P., Voigtlaender T., Bartram C. R., Klaes R., Twenty-three novel BRCA1 and BRCA2 sequence alterations in breast and/or ovarian cancer families in Southern Germany. Hum. Mutat. 22, 259 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Tesoriero A. A., et al. ; kConFab , Molecular characterization and cancer risk associated with BRCA1 and BRCA2 splice site variants identified in multiple-case breast cancer families. Hum. Mutat. 26, 495 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Serova-Sinilnikova O. M., et al. , BRCA2 mutations in hereditary breast and ovarian cancer in France. Am. J. Hum. Genet. 60, 1236–1239 (1997). [PMC free article] [PubMed] [Google Scholar]
- 13.Hofmann W., Horn D., Hüttner C., Classen E., Scherneck S., The BRCA2 variant 8204G>A is a splicing mutation and results in an in frame deletion of the gene. J. Med. Genet. 40, e23 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shirts B. H., et al. , Improving performance of multigene panels for genomic analysis of cancer predisposition. Genet. Med. 18, 974–981 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Stewart M. D., et al. , BARD1 is necessary for ubiquitylation of nucleosomal histone H2A and for transcriptional regulation of estrogen metabolism genes. Proc. Natl. Acad. Sci. U.S.A. 115, 1316–1321 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wappenschmidt B., et al. , Analysis of 30 putative BRCA1 splicing mutations in hereditary breast and ovarian cancer families identifies exonic splice site mutations that escape in silico prediction. PLoS One 7, e50800 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whiley P. J., et al. ; kConFab Investigators , Splicing and multifactorial analysis of intronic BRCA1 and BRCA2 sequence variants identifies clinically significant splicing aberrations up to 12 nucleotides from the intron/exon boundary. Hum. Mutat. 32, 678–687 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kraus C., et al. , Gene panel sequencing in familial breast/ovarian cancer patients identifies multiple novel mutations also in genes others than BRCA1/2. Int. J. Cancer 140, 95–102 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Meindl A., et al. , Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat. Genet. 42, 410–414 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Golmard L., et al. , Germline mutation in the RAD51B gene confers predisposition to breast cancer. BMC Cancer 13, 484 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corvelo A., Hallegger M., Smith C. W., Eyras E., Genome-wide association between branch point properties and alternative splicing. PLoS Comput. Biol. 6, e1001016 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cartegni L., Chew S. L., Krainer A. R., Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 3, 285–298 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Lee Y., Rio D. C., Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 84, 291–323 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.ClinVar https://www.ncbi.nlm.nih.gov/clinvar/variation/187617/. Accessed 5 December 2019.
- 25.ClinVar https://www.ncbi.nlm.nih.gov/clinvar/variation/52471/. Accessed 8 September 2019.
- 26.Levin J. Z., et al. , Targeted next-generation sequencing of a cancer transcriptome enhances detection of sequence variants and novel fusion transcripts. Genome Biol. 10, R115 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howald C., et al. , Combining RT-PCR-seq and RNA-seq to catalog all genic elements encoded in the human genome. Genome Res. 22, 1698–1710 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mercer T. R., et al. , Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat. Biotechnol. 30, 99–104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mercer T. R., et al. , Targeted sequencing for gene discovery and quantification using RNA CaptureSeq. Nat. Protoc. 9, 989–1009 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Ueno T., et al. , High-throughput resequencing of target-captured cDNA in cancer cells. Cancer Sci. 103, 131–135 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halvardson J., Zaghlool A., Feuk L., Exome RNA sequencing reveals rare and novel alternative transcripts. Nucleic Acids Res. 41, e6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merkin J., Russell C., Chen P., Burge C. B., Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science 338, 1593–1599 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cabanski C. R., et al. , cDNA hybrid capture improves transcriptome analysis on low-input and archived samples. J. Mol. Diagn. 16, 440–451 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cieslik M., et al. , The use of exome capture RNA-seq for highly degraded RNA with application to clinical cancer sequencing. Genome Res. 25, 1372–1381 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagy E., Maquat L. E., A rule for termination-codon position within intron-containing genes: When nonsense affects RNA abundance. Trends Biochem. Sci. 23, 198–199 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Maquat L. E., Tarn W. Y., Isken O., The pioneer round of translation: Features and functions. Cell 142, 368–374 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Popp M. W., Maquat L. E., RNA. A TRICK’n way to see the pioneer round of translation. Science 347, 1316–1317 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Carter M. S., et al. , A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem. 270, 28995–29003 (1995). [DOI] [PubMed] [Google Scholar]
- 39.Noensie E. N., Dietz H. C., A strategy for disease gene identification through nonsense-mediated mRNA decay inhibition. Nat. Biotechnol. 19, 434–439 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Drechsel G., et al. , Nonsense-mediated decay of alternative precursor mRNA splicing variants is a major determinant of the Arabidopsis steady state transcriptome. Plant Cell 25, 3726–3742 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu G. K., et al. , Molecular indexing enables quantitative targeted RNA sequencing and reveals poor efficiencies in standard library preparations. Proc. Natl. Acad. Sci. U.S.A. 111, 1891–1896 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mrozek-Gorska P., et al. , Epstein-Barr virus reprograms human B lymphocytes immediately in the prelatent phase of infection. Proc. Natl. Acad. Sci. U.S.A. 116, 16046–16055 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moghadasi S., et al. , The BRCA1 c. 5096G>A p.Arg1699Gln (R1699Q) intermediate risk variant: Breast and ovarian cancer risk estimation and recommendations for clinical management from the ENIGMA consortium. J. Med. Genet. 55, 15–20 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Vallée M. P., et al. , Adding in silico assessment of potential splice aberration to the integrated evaluation of BRCA gene unclassified variants. Hum. Mutat. 37, 627–639 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.University of Washington BROCA Cancer Risk Panel https://testguide.labmed.uw.edu/public/view/BROCA. Accessed 8 September 2019.
- 46.Walsh T., et al. , Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc. Natl. Acad. Sci. U.S.A. 108, 18032–18037 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Norquist B. M., et al. , Inherited mutations in women with ovarian carcinoma. JAMA Oncol. 2, 482–490 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walsh T., et al. , Genetic predisposition to breast cancer due to mutations other than BRCA1 and BRCA2 founder alleles among Ashkenazi Jewish women. JAMA Oncol. 3, 1647–1653 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reese M. G., Eeckman F. H., Kulp D., Haussler D., Improved splice site detection in Genie. J. Comput. Biol. 4, 311–323 (1997). [DOI] [PubMed] [Google Scholar]
- 50.Yeo G., Burge C. B., Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 11, 377–394 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Cartegni L., Wang J., Zhu Z., Zhang M. Q., Krainer A. R., ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 31, 3568–3571 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith P. J., et al. , An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum. Mol. Genet. 15, 2490–2508 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Lim K. H., Ferraris L., Filloux M. E., Raphael B. J., Fairbrother W. G., Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. Proc. Natl. Acad. Sci. U.S.A. 108, 11093–11098 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lim K. H., Fairbrother W. G., Spliceman–A computational web server that predicts sequence variations in pre-mRNA splicing. Bioinformatics 28, 1031–1032 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Desmet F. O., et al. , Human splicing finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 37, e67 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Katz Y., et al. , Quantitative visualization of alternative exon expression from RNA-seq data. Bioinformatics 31, 2400–2402 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Results of analyses of all variants have been deposited in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/).