Significance
γ-Tubulin is essential for eukaryotic cells to produce new microtubules that are assembled into arrays like mitotic spindles, and its function depends on proteins that form complexes with it. In centrosome-lacking cells, the generation of new microtubules is dependent on formation of the γ-tubulin ring complex (γTuRC). Our experiments reveal γ-tubulin–dependent but γTuRC-independent phenomena of microtubule generation during mitotic cell division in the plants that lack the centrosome. We conclude that the γTuRC plays a critical role in spindle pole organization, but that plant cells still complete cell division in its absence. Therefore, this work in plant cells advances our knowledge of acentrosomal microtubule organization in general.
Keywords: γ-tubulin, microtubule nucleation, mitosis, phragmoplast, spindle
Abstract
γ-Tubulin typically forms a ring-shaped complex with 5 related γ-tubulin complex proteins (GCP2 to GCP6), and this γ-tubulin ring complex (γTuRC) serves as a template for microtubule (MT) nucleation in plants and animals. While the γTuRC takes part in MT nucleation in most eukaryotes, in fungi such events take place robustly with just the γ-tubulin small complex (γTuSC) assembled by γ-tubulin plus GCP2 and GCP3. To explore whether the γTuRC is the sole functional γ-tubulin complex in plants, we generated 2 mutants of the GCP6 gene encoding the largest subunit of the γTuRC in Arabidopsis thaliana. Both mutants showed similar phenotypes of dwarfed vegetative growth and reduced fertility. The gcp6 mutant assembled the γTuSC, while the wild-type cells had GCP6 join other GCPs to produce the γTuRC. Although the gcp6 cells had greatly diminished γ-tubulin localization on spindle MTs, the protein was still detected there. The gcp6 cells formed spindles that lacked MT convergence and discernable poles; however, they managed to cope with the challenge of MT disorganization and were able to complete mitosis and cytokinesis. Our results reveal that the γTuRC is not the only functional form of the γ-tubulin complex for MT nucleation in plant cells, and that γ-tubulin-dependent, but γTuRC-independent, mechanisms meet the basal need of MT nucleation. Moreover, we show that the γTuRC function is more critical for the assembly of spindle MT array than for the phragmoplast. Thus, our findings provide insight into acentrosomal MT nucleation and organization.
Microtubules (MTs) are assembled into physiologically important arrays in eukaryotic cells to accomplish demanding tasks like mitosis and cytokinesis. The production of new MTs is essential for a cell to remodel its MT network, and the MT nucleation event is dependent on γ-tubulin, which forms complexes with γ-tubulin complex proteins (GCPs). In most eukaryotes, the γ-tubulin ring complex (γTuRC) functions as the MT nucleator and is composed of the related GCP2 to GCP6 proteins (1); however, the budding yeast Saccharomyces cerevisiae lacks genes encoding GCP4 to GCP6, so that it only forms the γ-tubulin small complex (γTuSC) composed of γ-tubulin plus GCP2 and GCP3 (2). Other fungi, such as the fission yeast Schizosaccharomyces pombe and the filamentous fungus Aspergillus nidulans, produce GCP4 to GCP6, but the removal of these subunits affects neither γ-tubulin localization to the spindle pole body nor MT nucleation and function, suggesting that γTuSC is sufficient for MT nucleation in these organisms (3, 4).
Compared with fungi, the importance of γTuRC becomes more pronounced in animals. Because the centrosome is the most prominent localization site of the γTuRC, targeting γ-tubulin to this MT-organizing center (MTOC) often represents the functionality of the protein in MT nucleation. Collectively, GCP4 to GCP6 are γTuRC-specific components that contribute to γ-tubulin targeting at the centrosome (5). Consequently, silencing their expression causes defects in centriole duplication, spindle assembly (e.g., formation of monopolar spindles), and ultimately mitotic arrest (5, 6). In the fly Drosophila melanogaster, γTuSC may localize to the centrosome without GCP4 to GCP6 (7); however, the centrosome localization of the γTuSC is insufficient for assembly of the functional spindle apparatus and normal mitosis in flies.
The GCP2 to GCP6 proteins bear the homologous γ-tubulin ring protein (Grip) motifs of Grip1 and Grip2, which interact with other GCP proteins and γ-tubulin, respectively (8–10). While the γTuSC structure is known to have the Y-shaped complex of GCP2 and GCP3 with 2 γ-tubulins associated on top (11), it has been challenging to learn how other homologous GCPs participate in assembly of the γTuRC ring. A serendipitous analysis indicated that GCP4 and GCP5 position at one end of the ring helix, with GCP6 at the other end (10). On the other hand, it is hypothesized that GCP4 and GCP5 rely on GCP6 to join a γ-tubulin complex but not vice versa, as evidenced by the fact that GCP6/GCPF is required for GCP4/GCPD and GCP5/GCPE localization at the spindle pole body in A. nidulans (3). Moreover, in animal cells, GCP6 shows a greater degree of importance for γ-tubulin localization at the centrosome compared with GCP4 and GCP5 (5, 7). The outstanding function of GCP6 may be attributed to the fact that it contains a region of 27-aa repeats between the Grip1 and Grip2 motifs that are absent in other GCPs (1). Nevertheless, the differing degrees of defects on GCP4 to GCP6 depletion has inspired hypotheses of novel γTuSC-like complexes containing 1 or more types of these subunits (1, 9, 12, 13). However, many of these hypotheses regarding the assembly of functional γ-tubulin complexes rely on the criterion of γ-tubulin localization to the centrosome. It is mostly unknown how γ-tubulin–dependent but centrosome-independent MT organization may be altered when the γTuRC assembly is disturbed.
Bipolar spindles can be assembled in the absence of the centrosomes, as is seen in certain reproductive cells of animals and all cells of flowering plants (14, 15). Although silencing GCP4 to GCP6 compromises centrosomal spindle assembly (5, 7), it is unclear whether the defects arise from the abnormality in MT nucleation at the centrosome, noncentrosomal sites, or both. It is now recognized that centrosome-independent MT nucleation events like MT-dependent MT nucleation also make essential contributions to spindle assembly, and these acentrosomal MT nucleation mechanisms have been shown to require the γTuRC (16, 17).
γ-Tubulin decorates all MT arrays during cell division in plant cells (18, 19). Flowering plants produce all proteins in the γTuRC, which is thought to be required for the generation of new MTs in all arrays (20). All the GCPs and their interacting proteins, such as MZT1 and NEDD1, examined to date decorate mitotic MT arrays like γ-tubulin, and GCP2, GCP3, MZT1, and NEDD1 are known to be essential, like γ-tubulin, in the model plant Arabidopsis thaliana (21–25). Interference of GCP4 expression alters MT organization patterns in interphase cells and impairs the assembly of both spindle and phragmoplast arrays in A. thaliana (24). Consequently, the mutant plants exhibit phenotypes of extremely minimal growth when GCP4 is expressed at ∼20% of the wild-type level by artificial microRNA targeted at GCP4 (amiR-GCP4). However, to date, no null homozygous mutant plants have been isolated for the GCP4 to GCP6 genes in plants, and thus their predicted essential functions remain hypothetical.
In this work, we explored whether the γTuRC is the sole functional form of the γ-tubulin complex for acentrosomal MT assembly during cell division using A. thaliana as a reference organism, because MTs are nucleated and organized in the absence of the centrosome in flowering plants. We chose to attack GCP6 because of its outstanding role in targeting the γTuRC proteins to MTOCs according to animal and fungal studies. Our efforts led to the successful isolation of 2 independent loss-of-function gcp6 homozygous mutants that produced offspring. While the mutants suffer from severe defects in γ-tubulin localization in mitotic MT arrays and are challenged in cell division, they nevertheless strive to undergo vegetative and reproductive growth, albeit with significant disadvantages compared with the wild-type control. Our results, summarized below, reveal that γTuRC-independent MT nucleation mechanisms contribute to MT assembly and organization in acentrosomal arrays in plants and perhaps other eukaryotes as well.
Results
The gcp6 Mutants Show Pleiotropic Defects in Growth and Reproduction.
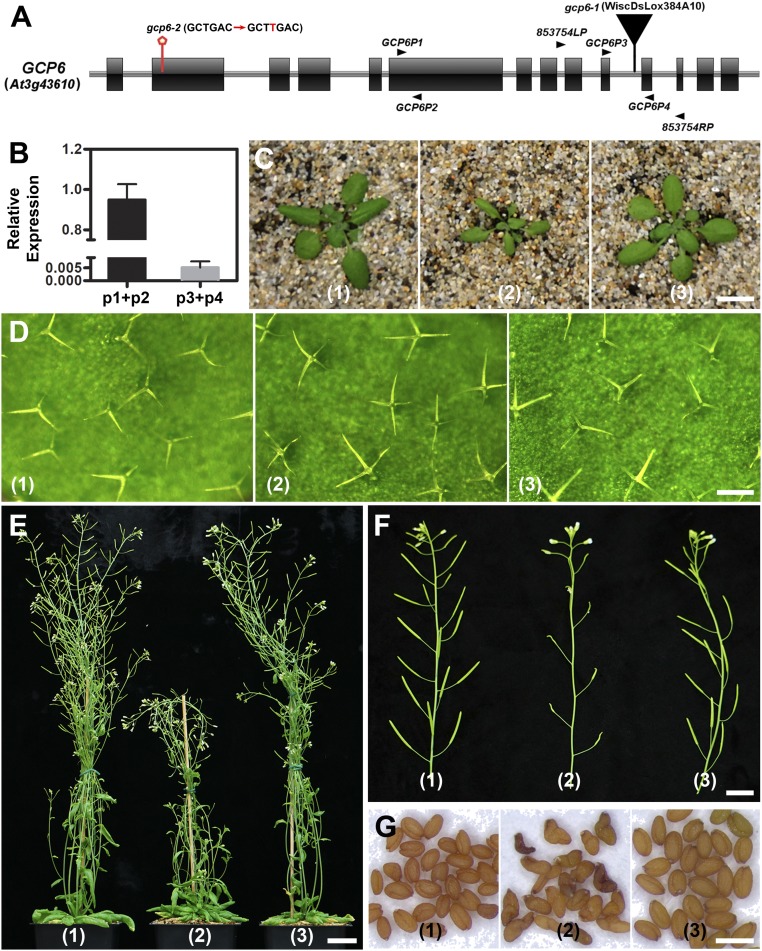
To detect mutations at the GCP6 locus identified as At3g43610 by The Arabidopsis Information Resource, we first examined annotated T-DNA insertional candidates (http://signal.salk.edu/cgi-bin/tdnaexpress). A mutant carrying an insertion at the 10th intron between exons 10 and 11 (Fig. 1A) was recovered from the CS853754 seed pool that gave rise to a homozygous gcp6-1 offspring. To determine how expression of the GCP6 gene was affected in the mutant, we performed quantitative RT-PCR experiments, which showed no expression of full-length GCP6 in the mutant (Fig. 1B). To determine whether the loss of GCP function would result in a similar phenotype, we generated the second allele, gcp6-2, which carries a single base insertion of T detected in the sense strand (SI Appendix, Fig. S1 A–C), recovered using the CRISPR/Cas9-mediated genome editing technique. This insertion led to the loss of a PvuII restriction enzyme site and the introduction of a premature stop codon after the codon encoding amino acid 85 (SI Appendix, Fig. S1 A–D). Therefore, gcp6-2 is most likely a null mutation.
Fig. 1.
GCP6 plays a critical role in vegetative growth and reproduction in A. thaliana. (A) Diagrammatic illustration of the GCP6 (At3g43610) gene with the exons represented by boxes and introns by lines. The diagram also shows the positions of the gcp6-1 and gcp6-2 mutations, as well as the primers used for RT-PCR and detection of the gcp6-1 mutation. (B) Quantitative RT-PCR results using primer pairs of GCP6P1 plus GCP6P2 (p1+p2) and GCP6P3 plus GCP6P4 (p3+p4) indicate that compared with the control plant, only a truncated version of GCP6 is expressed in the gcp6-1 mutant. (C–G) Phenotypic comparison of the control plant (1), the gcp6-1 mutant (2), and the gcp6-1 plant expressing the GCP6-GFP fusion protein (3) during vegetative growth and reproduction. (C) Compared with the control and GCP6-GFP–transformed plants, the gcp6-1 mutant plant exhibits much-reduced growth at 4 wk after germination. (D) The gcp6-1 mutant leaves produce trichomes that are dominated by 4 branches, while those of the control and complemented plants have 3 branches. (E) The mature 7-wk-old gcp6-1 plant shows much-retarded growth compared with the control and complemented plants. (F) The gcp6-1 mutant produces poorly developed siliques compared with those of the control and complemented siliques that are filled with seeds. (G) Compared with the oval-shaped seeds produced by the control and complemented plants, the gcp6-1 seeds show bent or twisted morphologies. (Scale bars: 1 cm in C, 200 µm in D, 2 cm in E, 1 cm in F, and 1 mm in G.)
The 2 homozygous mutants exhibited nearly identical macroscopic defects in both vegetative and reproductive growth compared with the wild-type control (SI Appendix, Fig. S1E). Moreover, the 2 mutants exhibited indistinguishable microscopic defects in γ-tubulin localization and in MT organization during mitosis (see Figs. 3 and 4 and SI Appendix, Figs. S3 and S4). Therefore, we concluded that the gcp6-1 mutation led to the loss of GCP6 function like gcp6-2, and thus carried out most experiments in the gcp6-1 mutation background. To determine whether the growth phenotypes were linked to the mutation, we expressed a GCP6-GFP fusion protein under the control of its native promoter in the gcp6-1 mutant. The mutation was suppressed, as indicated by restored growth indistinguishable from that of the wild-type control (Fig. 1 C–E), further confirming that inactivation of the GCP6 gene caused the gcp6-1 phenotype.
Fig. 3.
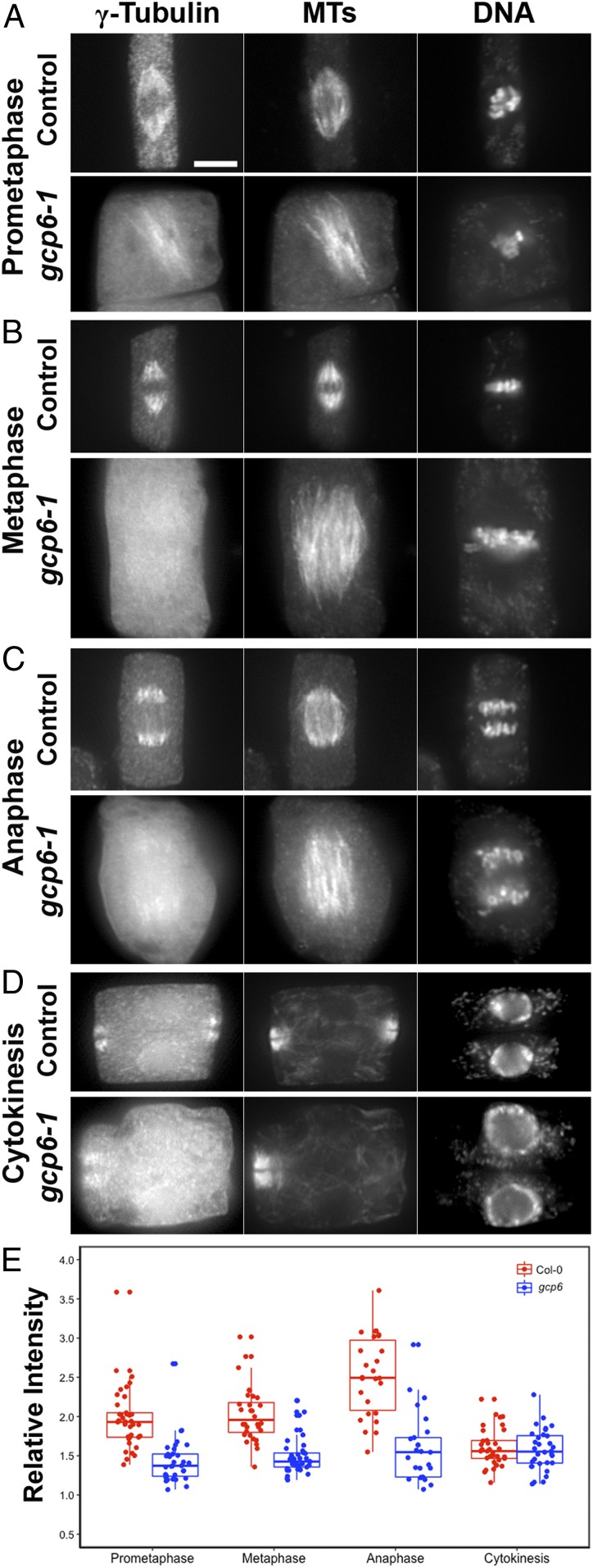
GCP6 is required for conspicuous localization of γ-tubulin on spindle MTs. γ-Tubulin, MTs, and DNA are detected in control and gcp6-1 mutant cells. (A) At prometaphase, γ-tubulin localizes to spindle MTs that reach chromosomes after the nuclear envelope breakdown and stands out from the cytoplasmic signal in the control cell but is detected only weakly on MTs in the gcp6-1 mutant. (B) The prominent localization of γ-tubulin on spindle MTs in the control cell is replaced by the mostly cytosolic diffuse signal in the gcp6-1 mutant at metaphase. (C) At anaphase, γ-tubulin strongly associates with the shortening kinetochore fiber MTs in the control cell, while the gcp6-1 mutant cell has the signal weakly detected on MTs. (D) During cytokinesis, however, γ-tubulin shows a conspicuous association with phragmoplast MTs in both the control and gcp6-1 mutant cells. (E) Quantitative assessment of MT-localized γ-tubulin signal vs. the cytosolic diffuse signal in the wild-type control and the gcp6-1 mutant cells; the number of cells measured is 37, 33, 32, 39, 26, 25, 33, and 32, respectively. Clear decreases in cell numbers are seen during prometaphase, metaphase, and anaphase but not during cytokinesis. (Scale bar: 5 µm.)
Fig. 4.
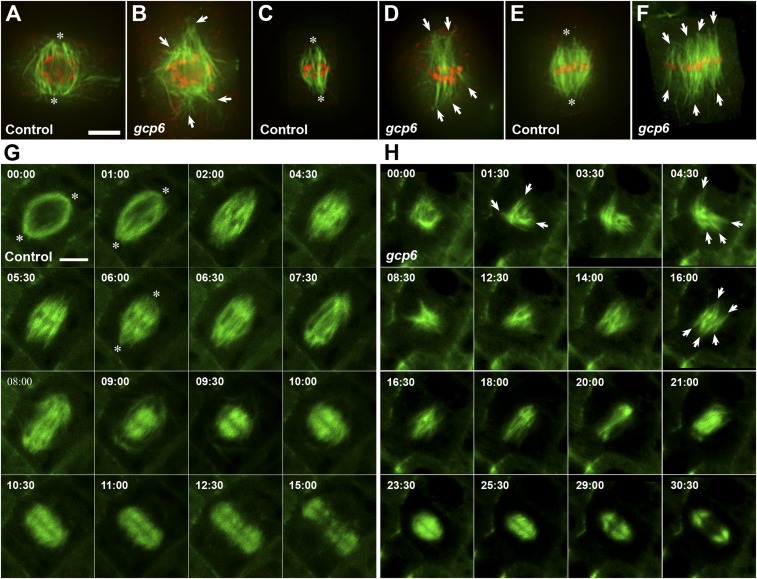
GCP6 plays a critical role in spindle morphogenesis. (A–F) Comparative views of MT organization in wild-type control cells (A, C, and E) and gcp6 mutant cells (B, D, and F) at late prophase (A and B), prometaphase (C and D), and metaphase (E and F). Merged micrographs have MTs in green and DNA in red. (A) MTs formed on the nuclear envelope are organized into a prospindle centered at two poles (asterisks) in a control cell when the preprophase band MTs are barely detected. (B) In the gcp6-2 mutant cell at a similar stage, MT bundles are disorganized and aligned toward discrete foci (arrows). (C) After the nuclear envelope breakdown, MT bundles start to encounter condensed chromosomes and assume the spindle array with convergent poles (asterisks) in the control cell. (D) The gcp6-1 mutant cell also forms MT bundles that have contacts with chromosomes but point at different directions (arrows). (E) When chromosomes are aligned at the metaphase plate, a typical acentrosomal spindle MT array has convergent poles (asterisks). (F) The gcp6-1 mutant cell forms a spindle array with nearly parallel MT bundles (arrows) connected to chromosomes aligned at the metaphase plate. (G and H) Comparative views of cell cycle-dependent MT reorganization in control (G, from Movie S2) and a gcp6 mutant cell (H, from Movie S3) that express the VisGreen-TUB6 fusion protein. The time stamps are in min:s. (G) In the control cell, spindle poles can be discerned (asterisks). At prophase, the prospindle exhibits a clear bipolar configuration on the nuclear envelope (00:00), followed by rigorous MT formation and bundling after the nuclear envelope breakdown (01:00 to 04:30). At 05:30, a clear metaphase spindle is established, followed by shortening of kinetochore fibers at 06:00 to 08:00. In the meantime, rich MTs are formed in the spindle midzone (06:00 to 09:00). These MTs eventually assume a mirrored pattern leaving a dark midline, marking the birth of the phragmoplast MT array (09:00). This phragmoplast array expands toward the cell periphery while becoming shortened (09:30 to 15:00). Eventually, MTs begin to be disassembled from the center (15:00). (H) In the gcp6-1 mutant cell, MTs are formed on the nuclear envelope but not in an obvious prospindle configuration (00:00). Rigorous MT polymerization takes place, resulting in the formation of randomly oriented MT bundles (arrows) following nuclear envelope breakdown (01:00 to 12:30). Eventually, a bipolar array can be seen but without MTs converging toward the poles (14:00). A metaphase spindle is established with more or less parallel kinetochore fibers (arrows; 16:00). At anaphase, the shortening of kinetochore fibers is concomitant with the formation of MTs in the spindle midzone (18:00 to 21:00). These spindle midzone MTs are replaced by a mirrored phragmoplast array with a dark line in the middle (23:30). This cytokinetic array expands while having its MTs shortened and depolymerized from the center, as seen in the control cell (25:30 to 30:30). (Scale bar: 5 µm.)
The gcp6 mutants exhibited pleiotropic growth phenotypes from initial vegetative growth to gametogenesis. Because leaf trichomes are sensitive to disturbances of defects in MT activities, as demonstrated by many mutants, including amiR-GCP4, phenotypes often are expressed in trichome branching (24, 26). We found that leaf trichomes mostly bore 4 branches, in contrast to the overwhelming preponderance of 3-pronged trichomes in the wild-type control and rescued plants expressing GCP6-GFP (Fig. 1D and Table 1). The mutants also exhibited significantly reduced fertility, as indicated by severely shortened siliques (Fig. 1F). Because defects in MTs often cause abortions in male gametophyte development (27), we analyzed whether such low fertility was due in part to male gametogenesis. Phenotypes were detected in pollen collected from open flowers. Among pollen grains produced by the homozygous gcp6-1 mutant, ∼30% were aborted, while the wild-type control hardly ever produced shrunken pollen. Therefore, the gcp6 mutation led to serious abortion of male gametogenesis.
Table 1.
The gcp6-1 mutant shows increased trichome branching
| Strain | Number of trichome branches | ||||
| 2 | 3 | 4 | 5 | n | |
| Control (GCP6), % | 0.21 | 80.84 | 18.74 | 0.21 | 475 |
| gcp6-1, % | 0.30 | 2.74 | 87.20 | 9.76 | 328 |
| GCP6-GFP in gcp6-1, % | 0.20 | 86.15 | 12.83 | 0.81 | 491 |
The sixth and seventh rosette leaves of 21-d-old plants were used for the analysis of trichome phenotypes.
Most of the few seeds produced by the gcp6-1 mutant were misshaped, with a bent or twisted appearance (Fig. 1G). Again, the introduction of the GCP6-GFP fusion into gcp6-1 resulted in the production of oval-shaped seeds. Therefore, we conclude that the function of GCP6 is critical for leaf growth, trichome branching, stem elongation, gametogenesis, and seed development (Fig. 1).
GCP6 Is Associated with Other GCP Proteins in the γTuRC.
Because the GCP6-GFP fusion protein is functional, as confirmed by the genetic suppression of gcp6-1, it functions as the native form. We first tested whether GCP6-GFP interacts with other GCP proteins in A. thaliana. Immunoaffinity purification results indicate that GCP6-GFP was recovered and detected by mass spectrometry (MS)-assisted peptide identification that is comparable to the results of GCP2-GFP purification (Table 2 and SI Appendix, Tables S1 and S2). Together with GCP6-GFP, γ-tubulin and GCP2 to GCP5 were also detected (Table 2 and SI Appendix Tables S1 and S2). These results indicate that GCP6 associates with GCP proteins to form the γTuRC in vivo in A. thaliana.
Table 2.
γTuRC proteins identified after immunopurification
| Protein detected | Bait | |||||||||||
| GCP2 | GCP2 (in gcp6-1) | GCP6 | GCP61-1043 | |||||||||
| Peptides, n | Spectra, n | Coverage, % | Peptides, n | Spectra, n | Coverage, % | Peptides, n | Spectra, n | Coverage, % | Peptides, n | Spectra, n | Coverage, % | |
| γ-Tubulin 1 | 43 | 743 | 90.72 | 16 | 22 | 48.73 | 58 | 161 | 87.97 | 4 | 4 | 10.97 |
| γ-Tubulin 2 | 41 | 661 | 90.72 | 15 | 21 | 42.62 | 58 | 160 | 88.19 | 4 | 4 | 10.97 |
| GCP2 | 36 | 243 | 59.20 | 20 | 25 | 39.32 | 38 | 97 | 55.82 | 2 | 2 | 3.39 |
| GCP3 | 58 | 473 | 74.82 | 17 | 18 | 24.46 | 64 | 160 | 66.95 | 2 | 2 | 2.74 |
| GCP4 | 44 | 238 | 55.97 | — | — | — | 56 | 134 | 61.21 | — | — | — |
| GCP5 | 35 | 165 | 40.10 | — | — | — | 41 | 94 | 41.91 | — | — | — |
| GCP6 | 50 | 181 | 50.29 | — | — | — | 65 | 146 | 47.31 | 43 | 109 | 46.73 |
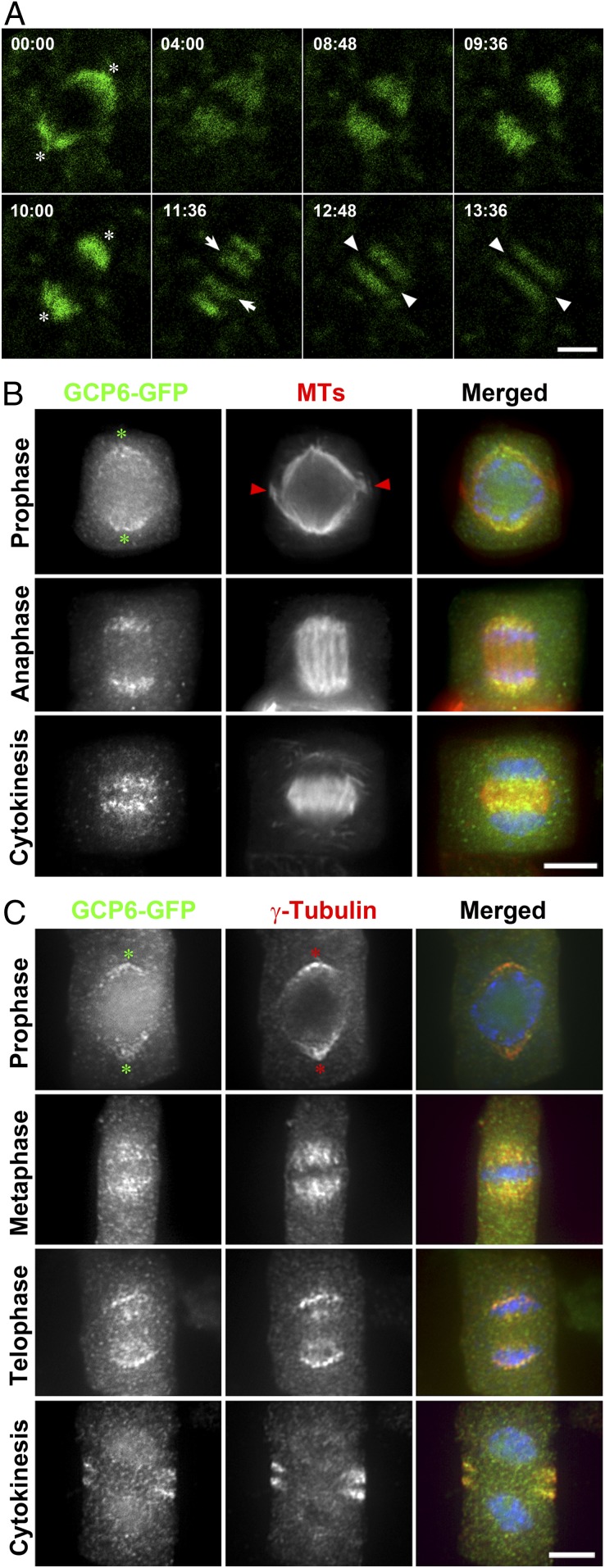
We then observed GCP6-GFP in live cells under a confocal microscope. Although it was difficult to discern its signal from the background autofluorescence in interphase cells, GCP6-GFP was detected in mitotic cells decorating both the spindle and phragmoplast arrays (Movie S1). The signal became eminent at late prophase on the nuclear envelope (00:00; Fig. 2A). The signal became concentrated on bipolar spindles and was particularly prominent toward the spindle poles during later stages of mitosis (04:00 to 10:00; Fig. 2A). Later, decreases in the signal at the spindle poles were accompanied by its appearance in the middle region by leaving the middle zone lacking fluorescent signal (11:36; Fig. 2A). Its localization in a phragmoplast-like pattern was observed afterward (12:48 to 13:36; Fig. 2A). However, the GCP6-GFP signal left a wide gap in the midzone, representing the feature of biased association with MTs toward their minus ends in the phragmoplast.
Fig. 2.
GCP6 colocalizes with γ-tubulin in a cell cycle-dependent manner in A. thaliana. (A) Snapshots of GCP6-GFP localization during a mitotic cell division from Movie S1. The time stamps are in min:s. The signal first concentrates at poles (asterisks; 00:00) before appearing in the spindle (04:00 to 08:48). Later, it becomes more concentrated toward spindle poles (09:36 and 10:00). The signal reduction at the poles is concomitant with its appearance in 2 groups in the midzone (arrows; 11:36) that have a wide gap between them and expand toward the cell periphery (arrowheads; 13:36). (B) Dual localization of GCP6-GFP and MTs. In merged images, GCP6-GFP is pseudocolored in green, MTs are in red, and DNA is in blue. GCP6 appears in polar caps (asterisks) marking the poles of prospindle when the remnant of the preprophase band (arrowheads) is still present at late prophase. GCP6 prominently associates with shortening kinetochore fibers at anaphase. GCP6 localizes to phragmoplast MTs with a wide gap in the middle. (C) Colocalization of GCP6-GFP with γ-tubulin. In merged images, GCP6-GFP is pseudocolored in green, γ-tubulin is in red, and DNA is in blue. GCP6 and γ-tubulin show overlapping localization patterns as polar caps (asterisks) in prophase and a metaphase spindle. At telophase, the overlapping signals are most conspicuous at spindle poles with emergence in the middle zone. They both appear in the phragmoplast, leaving a wide gap in the middle. (Scale bars: 5 µm.)
The association of GCP6 with these mitotic MT arrays was further testified by dual localization of GCP6-GFP and MTs, which showed preferential localization toward MT minus ends facing spindle poles and phragmoplast distal ends (Fig. 2B). To compare the GCP6-GFP localization with that of γ-tubulin, we carried out a dual localization experiment using the G9 anti–γ-tubulin monoclonal antibody (28). GCP6-GFP and γ-tubulin colocalized at all stages of mitotic division (Fig. 2C); for example, they marked the prospindle as the “polar caps” at late prophase, decorated half-spindles, and marked the phragmoplast (Fig. 2C).
GCP6 Plays a Critical Role in γ-Tubulin Localization.
To examine how γ-tubulin function might be affected by GCP6 mutation, we first examined its complex formation using GCP2-GFP as the bait for protein purification, because GCP2 and GCP3 are essential for γ-tubulin function in all eukaryotes. Compared with the entire γTuRC purified from the control plants, only γ-tubulin and GCP3 were detected together with GCP2-GFP in the gcp6-1 mutant (Table 2 and SI Appendix, Table S3). This result indicates that γTuSC, but not the γTuRC, was formed in the mutant cells.
Because the gcp6-1 mutation led to incomplete translation of GCP6, we tested whether the hypothetically truncated form of GCP61–1,043 would still form a complex with γ-tubulin or other GCPs. After GCP61–1,043-GFP was expressed under the control of the GCP6 promoter, it was mainly purified by itself with trace amounts of γ-tubulin, GCP2, and GCP3 but not of GCP4 and GCP5 (Table 2 and SI Appendix, Table S4). This result confirms that in the gcp6-1 mutant, the γ-tubulin function did not involve the mutant GCP61–1,043 protein. Furthermore, the GCP61–1,043-GFP mutant protein was no longer detected on MT arrays during cell division (SI Appendix, Fig. S2). These results further support the notion that the gcp6-1 mutation inactivated the corresponding gene.
We then asked how γ-tubulin activities might be affected when GCP6 is inactivated in the gcp6-1 mutant. In the wild-type cells, on nuclear envelope breakdown, γ-tubulin was concentrated along MTs of the spindles before chromosome alignment at the metaphase plate (Fig. 3A). In a gcp6-1 mutant cells at a similar stage, the γ-tubulin was still detected on MTs, but the signal barely stood out against the diffuse signal compared with that in the cytoplasm (Fig. 3A). The difference between the spindle-associated γ-tubulin signal and the cytoplasmic signal was decreased in the gcp6-1 mutant cells at metaphase (Fig. 3B). At anaphase, γ-tubulin became highly concentrated toward the spindle poles in the control cells (Fig. 3C). In contrast, such a pattern was replaced by a faint association of the γ-tubulin signal on MTs of the anaphase spindle in the gcp6-1 mutant cells (Fig. 3C). In the phragmoplast, however, the difference in γ-tubulin localization on MTs was not as obvious between the wild-type control and gcp6-1 mutant cells, as it was enriched along phragmoplast MTs, with a dark gap wider than that of MTs (Fig. 3D). We quantitatively assessed the ratio of the MT-localized γ-tubulin signal and diffuse signal in the cytosol in the control and mutant cells. The difference in the ratio was pronounced during mitosis, especially from prophase to anaphase, while that in cytokinetic cells was not (Fig. 3E).
We also aimed to learn whether the gcp6-2 mutation caused a difference in γ-tubulin localization compared with gcp6-1. At metaphase, the concentrated γ-tubulin signal on spindle MTs seen in the control cells (Fig. 3B) was replaced by diffuse signal and sparse puncta on discrete MT bundles that flanked chromosomes aligned at the metaphase plate (SI Appendix, Fig. S3). This result again indicates that γ-tubulin was still detectable on spindle MTs despite being greatly compromised. In contrast, γ-tubulin was enriched on phragmoplast MTs in the gcp6-2 mutant cells undergoing cytokinesis (SI Appendix, Fig. S3), as was seen in the gcp6-1 cells (Fig. 3D). Therefore, there was no noticeable difference between the 2 gcp6 mutants in terms of compromised γ-tubulin localization.
Defective MT Organization in the Absence of the γTuRC.
To determine how the loss of the γTuRC due to the absence of GCP6 affects MT behaviors during mitosis, we analyzed how the characteristic MT arrays might be affected. To do so, we first examined MT arrays in cells undergoing mitosis after fixation for antitubulin immunofluorescence in both gcp6-1 and gcp6-2 mutants. Similar results were obtained in both types of cells (Fig. 4 A–F and SI Appendix, Fig. S4). The mutant cells showed great difficulty in establishing spindle poles compared with the control cells, starting from late prophase. Compared with the control cells, in which a bipolar prospindle was formed on the nuclear envelope (Fig. 4A), the loss of GCP6 led to the disorganization of MTs on the nuclear envelope without a bipolar pattern as represented by the gcp6-2 cells (Fig. 4B). At prometaphase, after the nuclear envelope breakdown, MTs appeared mostly in developing kinetochore fibers organized into the fusiform configuration in the control cells (Fig. 4C). In the gcp6-1 mutant cells, however, such an MT convergent pattern was replaced by spindle MTs appearing in discrete bundles acting uncoordinatedly and pointing separately toward 2 ends of the dividing cell (Fig. 4D). At metaphase, while the wild-type cells had kinetochore MT bundles converging toward the poles, the mutant cells had kinetochore fibers in discrete bundles or separately grouped bundles in the bipolar spindles without apparent poles (Fig. 4 E and F). Compared with the significant disturbance of MT organization at prometaphase and disorganized spindle poles, the gcp6-2 mutant cells assembled the preprophase band with dense MT bundles and phragmoplast MT arrays with an obvious bipolar appearance (SI Appendix, Fig. S4).
To further determine how the loss of GCP6 affected mitotic MT reorganization in living cells, a VisGreen (GFP)-TUB6 marker was delivered into the mutant so that MT reorganization during mitotic division could be monitored over time and compared with the control cells expressing the identical marker (Movies S2 and S3). In control cells, mitotic MT reorganization began with the establishment of a prospindle with a clear bipolar configuration on the nuclear envelope (00:00; Fig. 4G). Then rich MTs were assembled and bundled after the nuclear envelope breakdown (01:00 to 04:30; Fig. 4G), which eventually led to the formation of a metaphase spindle (05:30; Fig. 4G). The shortening of kinetochore fibers (06:00 to 08:00; Fig. 4G) was accompanied by rich MT formation in the spindle midzone (06:00 to 09:00; Fig. 4G). These MTs eventually assumed a mirrored pattern leaving a dark midline and developed into the phragmoplast MT array (09:00; Fig. 4G). This phragmoplast array expanded toward the cell periphery (09:30 to 15:00; Fig. 4G). However, the gcp6-1 mutant cells did not have MTs organized into the bipolar configuration on the nuclear envelope (00:00; Fig. 4H), and MTs remained more or less randomly organized (01:00 to 12:30; Fig. 4H). Eventually, a bipolar MT array was established with nearly parallel kinetochore fibers at metaphase (16:00, Fig. 4H). At later stages, the shortening of kinetochore fibers, formation of MTs in the spindle midzone, and organization of the mirrored phragmoplast array took place in the mutant cell to complete cell division. The overall MT pattern was not altered in the gcp6-1 phragmoplasts compared with the control cells (Fig. 4H); therefore, our results indicate that the gcp6 mutant cells strive to carry out both mitosis and cytokinesis, with significant disadvantages in MT organization. In other words, MT reorganization, although seriously disturbed in the absence of GCP6, occurred independently of the γTuRC in these cells, suggesting a γTuRC-independent mechanism that regulates MT nucleation during mitotic cell division.
We also asked whether GCP6 is essential for nucleating MTs at interphase by examining cortical MTs reported by the VisGreen-TUB6 fusion protein in gcp6-1 mutant cells (SI Appendix, Fig. S5). In contrast to the drastic differences in spindle MT arrays established in the mutant and control cells, we found that cortical MTs often assumed parallel patterns in elongated hypocotyl cells with their orientations differing from cell to cell. Therefore, we concluded that the loss of GCP6 does not significantly alter MT nucleation at the cell cortex for assembling cortical MT arrays.
Discussion
Our results reveal the essential function of GCP6 in the assembly of the γTuRC and, consequently, its important contribution to γ-tubulin localization and the organization of spindle MT arrays during mitotic cell division in A. thaliana. However, to our surprise, we found that γ-tubulin–dependent but γTuRC-independent MT nucleation meets the minimal need of generating new MTs for MT reorganization during cell division. Despite the defects in MT organization caused by the gcp6 mutations, the mutants strive to complete mitosis and cytokinesis so that vegetative and reproductive growth can take place, albeit with great disadvantages. Thus, our results suggest that the γTuRC is not the sole functional form of the γ-tubulin complex for acentrosomal MT nucleation during mitosis and cytokinesis in plant cells and perhaps other systems as well.
Assembly of the γ-Tubulin Complex.
Our purification results using GCP6 as the bait recapitulated what we found when GCP2, GCP3, or MZT1 was used, indicating that GCP6 is tightly associated with other GCP proteins in the γTuRC in A. thaliana (25, 29). Here we found that when the truncated GCP61–1,043 protein was used as the bait, it associated with γ-tubulin, GCP2, and GCP3 at only a nearly undetectable level compared with the full-length protein, but not with other GCPs. This truncation removed part of the Grip2 domain toward the C terminus, which likely abolished its interaction with γ-tubulin, which is dependent on Grip2. Furthermore, GCP2 purification from the gcp6 extracts indicates that γTuSC was formed and GCP4 and GCP5 did not associate with GCP2. Therefore, perhaps only the γTuSC functions in the mutant cells. Our results do not rule out the formation of novel γTuSC-like γ-tubulin complexes containing GCP4 and/or GCP5.
In A. thaliana, γ-tubulin and GCP2 are essential proteins, as their losses lead to lethality (30, 31), indicating that γ-tubulin–dependent MT nucleation is essential for plant growth and development. Beyond the subunits of γTuSC, the functions of 2 other γTuRC components have been studied. Down-regulation of GCP4 leads to extremely dwarfism, while no null mutant has been reported, suggesting that GCP4 might be an essential protein as well (24). Moreover, the MZT1 proteins, or GIP1 and GIP2, identified as the interacting partners of GCP3, are also integral components of the γTuRC, decorate spindle and phragmoplast MTs like γ-tubulin itself, and are essential as well (22, 25).
These lines of evidence seem to suggest that the γTuRC is the essential functional form of the MT nucleator; however, our present findings argue against such an assumption, for 2 reasons. First, γTuRC is not the sole form of γ-tubulin–containing MT nucleator. In other words, γ-tubulin likely forms complex(es) without GCP6 that still can nucleate MTs, although perhaps not functioning as robustly as the γTuRC. According to the scenario in fungi, the complex might be γTuSC; however, the findings of the critical functions of GCP4 and MZT1 argue against this idea. Second, we can conclude that the contributions of GCP4 and GCP6 to MT nucleation are different in A. thaliana. The greater importance of GCP6 compared with GCP4 or GCP5 in fungi and animals is measured mostly by formation of the entire γTuRC and/or γ-tubulin targeting to the centrosome or its equivalent MTOCs (3, 5). In fission yeast, the nonessential GCP6/Alp16 and Mzt1 proteins synergistically contribute to the robustness of MT production by targeting the γTuRC to the spindle pole body during spindle assembly and promote the establishment of bipolar spindles (32). This function is not shared by the GCP4/Gfh1 and GCP5/Mod21 in the yeast, again demonstrating the different contributions of the 3 γTuRC-specific subunits. Whether the vital importance of GCP6 is shared for acentrosomal MT nucleation has not been tested previously. Compared with the severe phenotype caused by down-regulation of GCP4 expression (24), the phenotype caused by the gcp6 null mutation suggests that GCP4 plays a more important role than GCP6 in acentrosomal MT nucleation in plants. This conclusion is also supported by the demonstrated importance of GCP4 in mitosis and cytokinesis in the moss Physcomitrella patens by an inducible RNAi experiment (33). A common phenotype of the GCP4 knockdown and gcp6 knockout mutants is that they all led to diminished localization of γ-tubulin along spindle MTs. Such a phenotype contrasts with the recruitment of γ-tubulin to the centrosome when GCP4 was knocked out or GCP5 and GCP6 were knocked down in fly cells (7).
γTuRC-Independent MT Nucleation.
Because the γTuRC is no longer formed in the gcp6 mutant, the γTuSC and perhaps other novel γTuSC-like complexes takes part in MT nucleation in those cells. In fungi like budding yeast in which γTuSC, but not the γTuRC, is present, the spindle pole body protein Spc110p interacts with γTuSC and plays an essential role in assembling it into a higher-order complex competent for MT nucleation (13, 34, 35). Because the nucleation function of γTuSC depends on oligomerization by its receptor/activator-like Spc110p (34), such a factor remains to be discovered if a similar mechanism is used for γTuSC activation in the gcp6 mutant cells in A. thaliana. Both pericentrin and proteins like centrosomin bear a protein domain, namely centrosomin motif 1 (CM1), which is responsible for γTuSC activation (36). Some of the CM1-containing proteins function in MT nucleation at sites other than the centrosome or spindle pole body (2); however, these so-called γTuSC receptors with CM1 do not have obvious homologs in plants (2, 15).
The dispersal of the γ-tubulin signal to the cytosol in the gcp6 mutant cells undergoing cell division indicates that its association with spindle MTs is primarily γTuRC-dependent in plant cells. The reminiscent signal of γ-tubulin along MTs in GCP knockdown mutants often has been attributed to the function of remaining proteins that form fewer γTuRC complexes compared with controls. Because the mutant cells lack a functional GCP6, our results show that weak γ-tubulin association with spindle MTs can occur in a γTuRC-independent manner in acentrosomal plant cells. The question is whether the nucleation is dependent solely on γTuSC or on γTuSC together with γTuSC-like complexes. Because the amiR-GCP4 mutant shows an obviously more severe phenotype than the gcp6 mutants demonstrated here (24), it is likely that γTuSC-like complexes containing GCP4 take part in MT nucleation. Such a prediction is consistent with the finding in fly cells that knockdown of γTuRC-specific subunits together results in additive phenotypes in γ-tubulin localization and MT nucleation (7).
The predominant association of γ-tubulin with spindle MTs is dependent on the augmin complex in both plant and animal cells, as compromised augmin function leads to diminished γ-tubulin signal on spindles (33, 37–41). Augmin recruits the γTuRC to preexisting MTs so that new MT nucleation events occur in either parallel or branched forms (42). In the absence of the γTuRC due to the absence of GCP6, the interaction with augmin no longer occurs, so that the γ-tubulin signal becomes diffuse in the cytosol. Although γ-tubulin is displaced from spindle MTs in both augmin and gcp mutants, their phenotypes in spindle MT organization are different. Unlike the broadening of spindle poles and disengagement of MT bundles seen in the gcp6 cells, augmin mutants produce elongated spindles with nicely converged spindle poles in plants (39, 40). This implies that the γTuRC also takes part in MT nucleation not associated with preexisting MTs during spindle assembly. In the gcp6 mutant cells, MTs nucleated independently of the γTuRC encountered great difficulty in converging toward unified spindle poles implying that the γTuRC also plays a critical role in organizing MTs after they are nucleated.
The γTuRC Contributes to MT Organization Differently in Different Mitotic MT Arrays.
The gcp6 mutant exhibited a striking phenotype in MT organization at prophase. Typically, MTs are organized into a bipolar prospindle array with well-defined and focused poles decorated by γ-tubulin in a pattern described as the polar cap (43). Without the γTuRC in the absence of GCP6, ample MTs were still produced but did not form polar caps. Because they could not be organized into a bipolar prospindle, we concluded that the γTuRC also plays a critical role in MT organization before their encounters with chromosomes. Although it is often tempting to postulate that prospindles could be the precursor of spindles after the nuclear envelope breakdown, artificial disassembly of prospindles by pharmacologic means does not stop plant cells from establishing bipolar spindles with chromosomes aligned at the metaphase plate (44). In moss cells, cytoplasmic MTOCs, designated as gametosomes, are established at prophase in a γ-tubulin–dependent manner and play a more critical role in division plane orientation than in establishment of the bipolar spindle array (44). Organization of spindle poles often is attributed to collective functions of MT minus end-directed motors as demonstrated in both plant and animal cells (45, 46). Here our results indicate that the γTuRC is essential for defining the poles of prospindles, and that MT motors alone are insufficient for organizing prospindles.
In the gcp6 mutant cells, phragmoplast MTs exhibited clear bipolar patterns with γ-tubulin associated in a pattern comparable to that in the control cells, in contrast to the striking defects in spindle MT organization. γ-Tubulin plays a critical role in phragmoplast expansion by generating new MTs toward the expanding edge during cytokinesis (47, 48). Although such a function is often attributed to the action of the entire γTuRC, our results demonstrate that there is a robust γTuRC-independent mechanism for the association of γ-tubulin with phragmoplast MTs to nucleate new MTs. Our results also indicate that effective phragmoplast expansion could take place in a γTuRC-independent manner. Therefore, these findings prompted us to investigate γ-tubulin–dependent but γTuRC-independent mechanisms that regulate acentrosomal MT nucleation and organization in plant cells that are perhaps applicable in other systems as well.
Methods
Plant materials, growth conditions and transformation, recombinant DNA techniques and plasmid construction, RNA extraction and real-time quantitative RT-PCR, protein isolation and analysis by mass spectrometry, and microscopic observation are described in SI Appendix, Materials and Methods.
Data Availability.
The data and associated protocols are included in SI Appendix. Experimental materials will be made available on request to qualified researchers for their own use.
Supplementary Material
Acknowledgments
We thank Dr. Takashi Hashimoto (Nara Institute of Science and Technology), Dr. Tetsuya Horio (Nippon Sport Science University), and Dr. Tsuyoshi Nakagawa (Shimane University) for providing seeds, the G9 antibody, and vectors used in this study. This work was supported by the NSF (Grants MCB-1616076 and MCB-1920358) and a University of California Davis Academic Senate Research Grant. B.L. is supported by the US Department of Agriculture, National Institute of Food and Agriculture under an Agricultural Experiment Station Hatch Project (CA-D-PLB-2536-H).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912240116/-/DCSupplemental.
References
- 1.Teixidó-Travesa N., Roig J., Lüders J., The where, when and how of microtubule nucleation: One ring to rule them all. J. Cell Sci. 125, 4445–4456 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Lin T. C., Neuner A., Schiebel E., Targeting of γ-tubulin complexes to microtubule organizing centers: Conservation and divergence. Trends Cell Biol. 25, 296–307 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Xiong Y., Oakley B. R., In vivo analysis of the functions of γ-tubulin-complex proteins. J. Cell Sci. 122, 4218–4227 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anders A., Lourenço P. C. C., Sawin K. E., Noncore components of the fission yeast γ-tubulin complex. Mol. Biol. Cell 17, 5075–5093 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cota R. R., et al. , MZT1 regulates microtubule nucleation by linking γTuRC assembly to adapter-mediated targeting and activation. J. Cell Sci. 130, 406–419 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Bahtz R., et al. , GCP6 is a substrate of Plk4 and required for centriole duplication. J. Cell Sci. 125, 486–496 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Vérollet C., et al. , Drosophila melanogaster γ-TuRC is dispensable for targeting γ-tubulin to the centrosome and microtubule nucleation. J. Cell Biol. 172, 517–528 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiese C., Zheng Y., Microtubule nucleation: γ-tubulin and beyond. J. Cell Sci. 119, 4143–4153 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Tovey C. A., Conduit P. T., Microtubule nucleation by γ-tubulin complexes and beyond. Essays Biochem. 62, 765–780 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farache D., et al. , Functional analysis of γ-tubulin complex proteins indicates specific lateral association via their N-terminal domains. J. Biol. Chem. 291, 23112–23125 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kollman J. M., et al. , The structure of the γ-tubulin small complex: Implications of its architecture and flexibility for microtubule nucleation. Mol. Biol. Cell 19, 207–215 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oakley B. R., Paolillo V., Zheng Y., γ-Tubulin complexes in microtubule nucleation and beyond. Mol. Biol. Cell 26, 2957–2962 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kollman J. M., Merdes A., Mourey L., Agard D. A., Microtubule nucleation by γ-tubulin complexes. Nat. Rev. Mol. Cell Biol. 12, 709–721 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meunier S., Vernos I., Acentrosomal microtubule assembly in mitosis: The where, when, and how. Trends Cell Biol. 26, 80–87 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Yamada M., Goshima G., Mitotic spindle assembly in land plants: Molecules and mechanisms. Biology 6, E6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prosser S. L., Pelletier L., Mitotic spindle assembly in animal cells: A fine balancing act. Nat. Rev. Mol. Cell Biol. 18, 187–201 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Petry S., Mechanisms of mitotic spindle assembly. Annu. Rev. Biochem. 85, 659–683 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu B., Joshi H. C., Palevitz B. A., Experimental manipulation of γ-tubulin distribution in Arabidopsis using anti-microtubule drugs. Cell Motil. Cytoskeleton 31, 113–129 (1995). [DOI] [PubMed] [Google Scholar]
- 19.Liu B., Marc J., Joshi H. C., Palevitz B. A., A γ-tubulin-related protein associated with the microtubule arrays of higher plants in a cell cycle-dependent manner. J. Cell Sci. 104, 1217–1228 (1993). [DOI] [PubMed] [Google Scholar]
- 20.Hashimoto T., A ring for all: γ-tubulin-containing nucleation complexes in acentrosomal plant microtubule arrays. Curr. Opin. Plant Biol. 16, 698–703 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Zeng C. J., Lee Y. R., Liu B., The WD40 repeat protein NEDD1 functions in microtubule organization during cell division in Arabidopsis thaliana. Plant Cell 21, 1129–1140 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janski N., et al. , The GCP3-interacting proteins GIP1 and GIP2 are required for γ-tubulin complex protein localization, spindle integrity, and chromosomal stability. Plant Cell 24, 1171–1187 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seltzer V., et al. , Arabidopsis GCP2 and GCP3 are part of a soluble gamma-tubulin complex and have nuclear envelope targeting domains. Plant J. 52, 322–331 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Kong Z., Hotta T., Lee Y. R., Horio T., Liu B., The γ-tubulin complex protein GCP4 is required for organizing functional microtubule arrays in Arabidopsis thaliana. Plant Cell 22, 191–204 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakamura M., et al. , Arabidopsis GCP3-interacting protein 1/MOZART 1 is an integral component of the γ-tubulin-containing microtubule nucleating complex. Plant J. 71, 216–225 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Buschmann H., Lloyd C. W., Arabidopsis mutants and the network of microtubule-associated functions. Mol. Plant 1, 888–898 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Liu B., Ho C. M., Lee Y. R., Microtubule reorganization during mitosis and cytokinesis: Lessons learned from developing microgametophytes in Arabidopsis thaliana. Front. Plant Sci. 2, 27 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horio T., Basaki A., Takeoka A., Yamato M., Lethal level overexpression of γ-tubulin in fission yeast causes mitotic arrest. Cell Motil. Cytoskeleton 44, 284–295 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Nakamura M., Ehrhardt D. W., Hashimoto T., Microtubule and katanin-dependent dynamics of microtubule nucleation complexes in the acentrosomal Arabidopsis cortical array. Nat. Cell Biol. 12, 1064–1070 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Pastuglia M., et al. , γ-tubulin is essential for microtubule organization and development in Arabidopsis. Plant Cell 18, 1412–1425 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamura M., Hashimoto T., A mutation in the Arabidopsis γ-tubulin-containing complex causes helical growth and abnormal microtubule branching. J. Cell Sci. 122, 2208–2217 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Masuda H., Toda T., Synergistic role of fission yeast Alp16GCP6 and Mzt1MOZART1 in γ-tubulin complex recruitment to mitotic spindle pole bodies and spindle assembly. Mol. Biol. Cell 27, 1753–1763 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakaoka Y., et al. , An inducible RNA interference system in Physcomitrella patens reveals a dominant role of augmin in phragmoplast microtubule generation. Plant Cell 24, 1478–1493 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyon A. S., et al. , Higher-order oligomerization of Spc110p drives γ-tubulin ring complex assembly. Mol. Biol. Cell 27, 2245–2258 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knop M., Schiebel E., Spc98p and Spc97p of the yeast γ-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. EMBO J. 16, 6985–6995 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawin K. E., Lourenco P. C., Snaith H. A., Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Curr. Biol. 14, 763–775 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Goshima G., Mayer M., Zhang N., Stuurman N., Vale R. D., Augmin: A protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol. 181, 421–429 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ho C. M., et al. , Augmin plays a critical role in organizing the spindle and phragmoplast microtubule arrays in Arabidopsis. Plant Cell 23, 2606–2618 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hotta T., et al. , Characterization of the Arabidopsis augmin complex uncovers its critical function in the assembly of the acentrosomal spindle and phragmoplast microtubule arrays. Plant Cell 24, 1494–1509 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee Y. J., et al. , The mitotic function of augmin is dependent on its microtubule-associated protein subunit EDE1 in Arabidopsis thaliana. Curr. Biol. 27, 3891–3897.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Uehara R., et al. , The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc. Natl. Acad. Sci. U.S.A. 106, 6998–7003 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goshima G., Kimura A., New look inside the spindle: Microtubule-dependent microtubule generation within the spindle. Curr. Opin. Cell Biol. 22, 44–49 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Lee Y. J., Liu B., Microtubule nucleation for the assembly of acentrosomal microtubule arrays in plant cells. New Phytol. 222, 1705–1718 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Kosetsu K., et al. , Cytoplasmic MTOCs control spindle orientation for asymmetric cell division in plants. Proc. Natl. Acad. Sci. U.S.A. 114, E8847–E8854 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goshima G., Nédélec F., Vale R. D., Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J. Cell Biol. 171, 229–240 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marcus A. I., Li W., Ma H., Cyr R. J., A kinesin mutant with an atypical bipolar spindle undergoes normal mitosis. Mol. Biol. Cell 14, 1717–1726 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murata T., et al. , Mechanism of microtubule array expansion in the cytokinetic phragmoplast. Nat. Commun. 4, 1967 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee Y. R., Liu B., The rise and fall of the phragmoplast microtubule array. Curr. Opin. Plant Biol. 16, 757–763 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and associated protocols are included in SI Appendix. Experimental materials will be made available on request to qualified researchers for their own use.