Significance
As a catabolic program that maintains homeostasis during adversity, autophagy is an immune defense restricting pathogenesis of viruses, including HSV-1. While HSV-1 ICP34.5 interferes with host Beclin1 to suppress autophagy and support virus replication in neurons, whether autophagy is a cell type-specific antiviral response or broadly limits HSV-1 replication in nonneuronal cells is unknown. We show that the HSV-1 Us3 Ser/Thr kinase antagonizes autophagy in nonneuronal cells. Phosphorylation of autophagy regulators ULK1 and Beclin1 was Us3 dependent and Beclin1 was identified as a direct Us3 substrate. Finally, replication of Us3-deficient HSV-1 was enhanced by depleting ULK1 and Beclin1. This establishes that autophagy limits HSV-1 replication in nonneuronal cells and reveals a new function for the Us3 kinase encoded by α-herpesviruses.
Keywords: autophagy, HSV-1 replication, Us3, Beclin1, Akt signaling
Abstract
Autophagy is a powerful host defense that restricts herpes simplex virus-1 (HSV-1) pathogenesis in neurons. As a countermeasure, the viral ICP34.5 polypeptide, which is exclusively encoded by HSV, antagonizes autophagy in part through binding Beclin1. However, whether autophagy is a cell-type–specific antiviral defense or broadly restricts HSV-1 reproduction in nonneuronal cells is unknown. Here, we establish that autophagy limits HSV-1 productive growth in nonneuronal cells and is repressed by the Us3 gene product. Phosphorylation of the autophagy regulators ULK1 and Beclin1 in virus-infected cells was dependent upon the HSV-1 Us3 Ser/Thr kinase. Furthermore, Beclin1 was unexpectedly identified as a direct Us3 kinase substrate. Although disabling autophagy did not impact replication of an ICP34.5-deficient virus in primary human fibroblasts, depleting Beclin1 and ULK1 partially rescued Us3-deficient HSV-1 replication. This shows that autophagy restricts HSV-1 reproduction in a cell-intrinsic manner in nonneuronal cells and is suppressed by multiple, independent viral functions targeting Beclin1 and ULK1. Moreover, it defines a surprising role regulating autophagy for the Us3 kinase, which unlike ICP34.5 is widely encoded by alpha-herpesvirus subfamily members.
Macroautophagy or autophagy is a catabolic survival program that enables eukaryotic cells to sustain homeostasis under adverse conditions by recycling cytoplasmic components (1–3). It is regulated in part by the mechanistic target of rapamycin complex 1 (mTORC1), a multisubunit Ser/Thr kinase that coordinates anabolic or catabolic outputs (4). Whereas mTORC1 activation stimulates protein synthesis and represses autophagy (Fig. 1A), inhibition of mTORC1 by energy, nutrient, or growth factor insufficiency, suppresses protein synthesis and promotes autophagy (4). Inhibiting mTORC1 activates the autophagy regulator ULK1, the Ser/Thr kinase component of the multisubunit ULK complex (5, 6). Subsequent phosphoinositide 3-kinase catalytic subunit type III (PI3KC3) complex activation by phosphorylation of the regulatory subunit Beclin1 initiates autophagosome accumulation (2). Autophagy is also provoked by stress-induced Ser/Thr kinases that arrest protein synthesis by phosphorylating the α-subunit of the critical translation initiation factor eIF2 (7). In contrast, site-specific Beclin1 phosphorylation by Akt limits autophagy (8). Not only does autophagy fuel survival and adaptation during unfavorable circumstances, it also impacts the pathophysiology of aging, cancer, neurodegenerative diseases, inflammation, and infection biology (9–12).
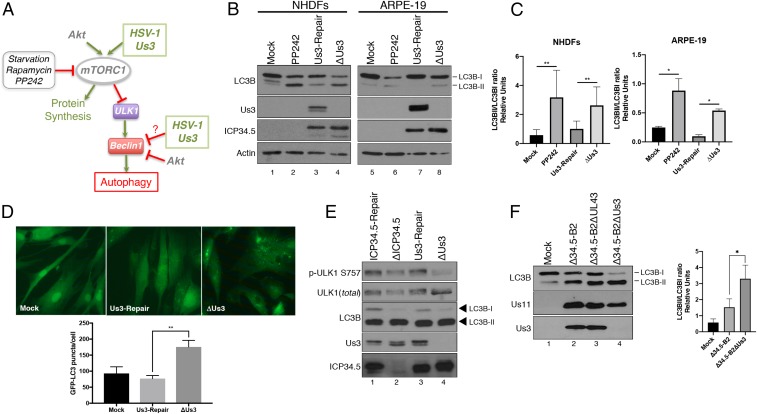
Fig. 1.
Suppression of autophagy in HSV-1–infected cells requires the virus-encoded Us3 Ser/Thr kinase. (A) Illustration depicting the control of autophagy by mTORC1 and the action of cellular and HSV-1–encoded effectors. Activation of proteins or cellular processes is shown by green arrows, while inhibitory actions are shown by perpendicular red lines. (B) NHDFs or ARPE-19 cells were mock infected (+/−PP242) or infected (multiplicity of infection [MOI] = 5) with a Us3-deficient virus (ΔUs3), or a virus where the Us3 deficiency was repaired (Us3-Repair). At 12 hpi, total protein was fractionated by SDS/PAGE and analyzed by immunoblotting with the indicated antibodies. Migration of unmodified LC3B-I and lipidated LC3B-II in this representative image are indicated on the Right of the panel. (C) The abundance of LC3B-I and LC3B-II in independent replicate experiments (n = 4 for NHDFs; n = 3 for ARPE-19) shown in B were quantified using Licor Image Studio software and expressed graphically as the LC3BII/LC3BI ratio. Error bars represent mean ± SEM. **P < 0.01; *P < 0.05 by paired t test. (D) Quantification of GFP-LC3B puncta (autophagosomes) in NHDFs stably expressing GFP-LC3B following infection with the indicated virus for 12 h. Error bars represent the mean ± SEM of at least 3 experiments where >50 cells were analyzed per experiment. **P < 0.01 by Student’s t test. Representative images (63× magnification) are shown above the graph. (E) NHDFs were infected (MOI = 5) with an ICP34.5-deficient virus (ΔICP34.5), a virus where the ICP34.5 deficiency was repaired (ICP34.5-Repair), ΔUs3, or Us3-Repair virus. At 12 hpi, total protein was isolated, fractionated by SDS/PAGE, and analyzed by immunoblotting using the indicated antibodies. (F) NHDFs were mock infected or infected (MOI = 1) with the ICP34.5-deficient parent virus (Δ34.5-B2), an ICP34.5 and UL43 doubly deficient virus (Δ34.5-B2ΔUL43), or an ICP34.5 and Us3 doubly deficient virus (Δ34.5-B2ΔUs3). Total protein harvested at 16 hpi was analyzed as in A. A representative image is shown (Left). The LC3BII/LC3BI ratio was quantified as in C from 3 (n = 3) separate experiments (Right). Bars represent mean ± SEM. *P < 0.05 by Student’s t test.
Although autophagy and autophagic signaling promote replication of some viruses (13), autophagy is also a powerful cell-intrinsic host defense capable of restricting virus pathogenesis (12, 14–18). Unlike proviral examples where autophagy supports virus replication, a broad antiviral capacity of autophagy has been difficult to demonstrate in vitro using cultured cells, suggesting that its impact might be restricted in a cell-type–specific manner (14, 19–26). Autophagy plays a notable role limiting virus pathogenesis in long-lived cell types like neurons (24, 27). Herpes simplex virus (HSV) is significant in this regard as the virus executes its lifecycle within 2 very different cell types. After entering mucosal epithelia, HSV infects peripheral neurons and establishes lifelong latency where virus reproduction and viral genes needed for productive growth are suppressed (28, 29). Physiological stress triggers episodic reactivation, whereby virus gene expression is activated in neurons, productive virus replication ensues, and infectious virus is released back into the epithelial entry site (28). While autophagy limits HSV-1 replication in peripheral neurons (30), how autophagy might impact virus reproduction in a cell-autonomous manner in nonneuronal cells is not understood. This is critical because replication in nonneuronal cells is paramount for HSV-1 spread to new hosts.
The ICP34.5 and Us11 proteins encoded by HSV-1 limit autophagy by preventing eIF2 inactivation (7, 31). In addition, Us11 limits autophagy by interfering with Tank Binding Kinase 1 (TBK-1), whereas ICP34.5 also antagonizes Beclin1 (27, 32, 33). HSV-1 replicated better in ATG5-deficient mouse sensory neurons unable to undergo autophagy, and an HSV-1 encoding an ICP34.5 mutant unable to interact with and inhibit Beclin1 exhibited reduced pathogenesis in adult mice (27, 30, 34). Enhanced destruction of viral proteins and/or virions likely contributed to this in vivo phenotype and is consistent with autophagy acting as a neuron-specific antiviral defense (35, 36). However, replication of an HSV-1 ICP34.5 mutant virus unable to inhibit Beclin1 was paradoxically unaffected in nonneuronal cells even when autophagy was disabled (19). This raised the possibility that other unidentified HSV-1 functions antagonize autophagy in nonneuronal cells.
The α-herpesvirus specific Ser/Thr kinase encoded by the Us3 gene is required for HSV-1 neuropathogenesis in mice and stimulates directly or indirectly phosphorylation of numerous viral and cellular substrates (37–40). Despite lacking primary sequence homology to the host kinase Akt, Us3 also behaves as a consitutively activated Akt mimic phosphorylating several Akt substrates including the mTORC1 regulator TSC2 (41). Indeed, Us3 is critical for wild-type (WT) virus replication levels and promotes virus reproduction under stress that restricts mTORC1 activation (42, 43). Here, we show that phosphorylation of the autophagy regulators ULK1 and Beclin1 in virus-infected cells is dependent upon the HSV-1 Us3 Ser/Thr kinase and identify Beclin1 as a direct Us3 kinase substrate. Ectopic Us3 expression suppressed autophagy in uninfected cells, and autophagy was evident in human epithelial cells and fibroblasts infected with Us3-deficient HSV-1. While ICP34.5-deficient virus replication was not influenced by suppressing autophagy, replication of Us3-deficient and Us3-ICP34.5 doubly deficient HSV-1 was partially rescued. This establishes that autophagy broadly restricts HSV-1 reproduction in a cell intrinsic manner in nonneuronal cells. Moreover, it highlights that autophagy is antagonized by multiple, independent HSV-1 functions that target Beclin1 and ULK1 through discrete mechanisms. Finally, it reveals how Beclin1 phosphorylation is subverted in infection biology and an unexpected role for the α-herpesvirus Us3 kinase in regulating autophagy.
Results
Multiple, Independent HSV-1–Encoded Functions Synergize to Coordinately Control Autophagy in Infected Cells.
To determine if Us3 contributes to regulating autophagy in nonneuronal cells infected with HSV-1, ARPE-19 epithelial cells (ARPEs) and normal human dermal fibroblasts (NHDFs) were mock infected or infected with a Us3-deficient virus (ΔUs3) or a virus in which the Us3 mutation was repaired (Us3-Repair). After 12 h, total protein was isolated and levels of unmodified (LC3B-I) vs. lipidated microtubule-associated protein LC3B (LC3B-II), which is required for autophagosome formation and allows direct quantification of autophagic membranes, were evaluated by immunoblotting. As a positive control, mock-infected cultures were treated with the active-site mTOR inhibitor PP242 to induce autophagy. Indeed, reduced LC3B-I and greater LC3B-II abundance was detected in PP242-treated vs. untreated ARPEs and NHDFs (Fig. 1 B and C). Compared to cultures mock infected or infected with a Us3-expressing virus (Us3-Repair), reduced LC3B-I and greater LC3B-II levels were observed in ARPEs and NHDFs infected with a Us3-deficient virus (ΔUs3) (Fig. 1B, compare lane 4 to lanes 1 and 3 and compare lane 8 to lanes 5 and 7; Fig. 1C). Greater numbers of autophagosomes indicated by fluorescent LC3B puncta also selectively accumulated in the cytoplasm of NHDFs stably expressing a GFP-LC3B fusion protein following infection with ΔUs3 compared to cultures mock infected or infected with a Us3-expressing virus (Fig. 1D). This established that the overall reduction of LC3B-I together with the increase of LC3B-II and LC3B-containing autophagosome accumulation in HSV-1–infected epithelial cells and fibroblasts was suppressed in a Us3-dependent manner. It further intimated that Us3 represented a previously undocumented autophagy regulator encoded by HSV-1.
Since Us3 constiutively stimulates mTORC1, which in turn phosphorylates ULK1 on S757 in uninfected cells to repress autophagy, the dependence of ULK1 S757 phosphorylation upon Us3 in HSV-1–infected cells was evaluated. Compared to NHDFs infected with a Us3-expressing virus, impaired accumulation of S757 phosphorylated ULK1 was most apparent in ΔUs3-infected cultures (Fig. 1E, lane 4). Under these conditions, enrichment of LC3B-II compared to LC3B-I in cells infected with ΔUs3 was detected in the presence of the HSV-1 Beclin1 antagonist ICP34.5 (Fig. 1 B and C and Fig. 1E, lane 4). Thus, increased LC3B modification was unlikely to result from reduced ICP34.5 accumulation in cells infected with ΔUs3. Likewise, LC3B-II enrichment relative to LC3B-I in cells infected with ΔICP34.5 was detected in the presence of Us3 (Fig. 1E, lane 2). This suggested that autophagy, like many critical host processes, may be regulated by multiple HSV-1–encoded functions.
To investigate the contribution Us3 and ICP34.5 gene products make to regulate autophagy in HSV-1–infected cells, a Us3 deletion was introduced into an ICP34.5-deficient genome (43). To avoid well-known pleiotropic effects of ICP34.5 on eIF2α phosphorylation that might also impact autophagy (7), the parent ICP34.5-deficient virus genome (Δ34.5-B2) expressed Us11 as an immediate-early gene to prevent PKR activation and limit eIF2α phosphorylation as described previously (44, 45). More LC3B-II was apparent in NHDFs infected with the parental Δ34.5-B2 virus compared to mock-infected cultures (Fig. 1F, compare lanes 1 and 2). A further reduction in LC3B-I and concomitant increase in LC3B-II levels was observed in cells infected with a Δ34.5-B2 virus that contained an introduced Us3 deletion (Fig. 1F, compare lanes 2 and 4). In contrast, LC3B-I and -II levels in cells infected with Δ34.5-B2 containing an introduced deletion into the UL43 gene, which encodes a glycoprotein not essential for growth in culture or pathogenesis in mice (46), remained similar to those detected in NHDFs infected with the parental Δ34.5-B2 virus (Fig. 1F, compare lanes 2 and 3). Thus, deleting Us3 substantially enhanced LC3B-II accumulation in NHDFs infected with ICP34.5-deficient HSV-1. This suggests that both Us3 and ICP34.5 control LC3B lipidation in HSV-1–infected cells and demonstrates that Us3 function is not dependent upon ICP34.5. It further implies that ICP34.5 and Us3 might target independent host functions to synergistically regulate autophagy in infected cells.
Inhibition of Autophagy in HSV-1–Infected Epithelial Cells and Fibroblasts Is Us3 Dependent.
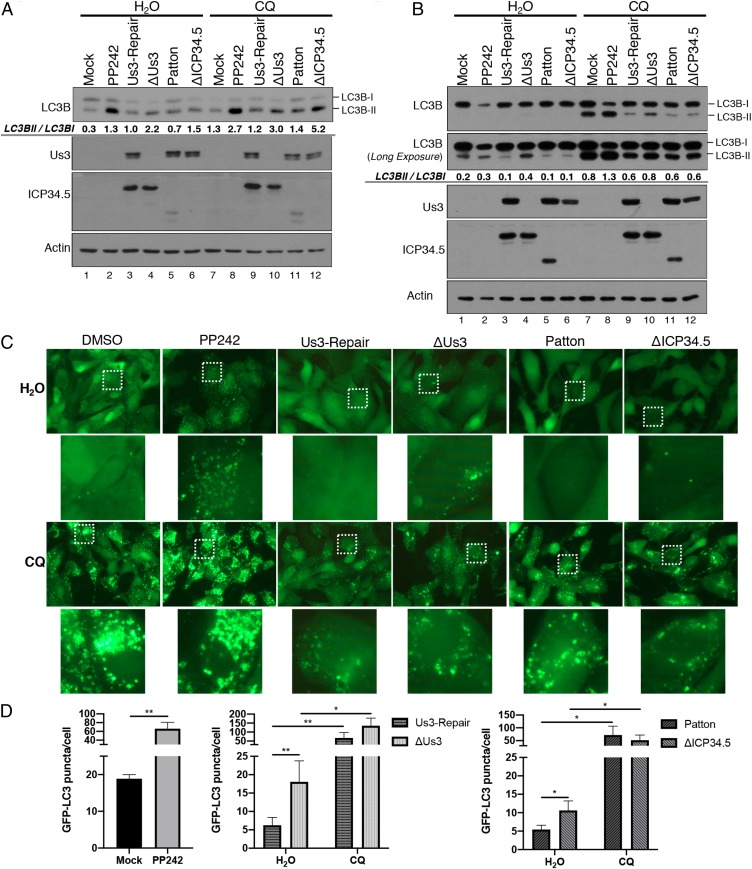
Increased LC3B-II abundance in cells infected with ΔUs3 could either reflect alterations in its production by modification of LC3-I or simply greater steady-state levels resulting from reduced LC3B-II protein turnover. The latter can result from a blockade in flux through the autophagy pathway (47). To distinguish between these possibilities, cells were treated with chloroquine (CQ) to prevent fusion of autophagosomes with lysosomes and inhibit LC3B-II degradation within lysosomes (48). Fig. 2 demonstrates that chloroquine treatment effectively suppressed autophagic flux and increased LC3B-II levels in uninfected (+/−PP242) and HSV-1–infected NHDFs and ARPEs. In particular, LC3B-II levels increased in NHDFs (Fig. 2A, compare lanes 4 through 10) and ARPEs (Fig. 2B, compare lanes 4 through 10) infected with ΔUs3 in response to chloroquine. A similar increase in LCB-II levels upon chloroquine treatment was observed in ΔICP34.5-infected cells (Fig. 2 A and B; compare lanes 6 through 12). To more directly evaluate the impact of Us3 upon autophagosome accumulation in infected cells, differences in LC3B puncta accumulation were quantified in epithelial cells expressing GFP-LC3B. Significantly greater numbers of GFP-LC3B puncta accumulated in cells infected with ΔUs3 compared to a Us3-expressing virus and puncta abundance increased in response to chloroquine (Fig. 2 C and D). Similar findings were observed in cultures infected with ΔICP34.5 as a control (Fig. 2 C and D). This demonstrates that accumulation of LC3B-containing autophagosomes in HSV-1–infected epithelial cells was suppressed in a manner dependent upon the Us3 gene. Moreover, the increased LC3B-II and LC3B puncta accumulation in chloroquine-treated cells infected with Us3-deficient HSV-1 is consistent with greater autophagy induction (production of LC3B-II from LC3B-I) as opposed to inhibition of autophagic flux.
Fig. 2.
Inhibition of autophagy induction, but not autophagic flux, in HSV-1–infected cells is dependent upon Us3. NHDFs (A) or ARPE-19 cells (B) were mock infected or infected (MOI = 5) with HSV-1 ΔUs3 strain F, Us3-Repair strain F, wild-type HSV-1 (strain F or Patton), or ΔICP34.5 strain Patton. At 8 hpi, cells were treated with DMSO, PP242, or chloroquine (CQ) as indicated. At 12 hpi, total protein was isolated, fractionated by SDS/PAGE, and analyzed by immunoblotting with the indicated antibodies. LC3B-I and LC3B-II abundance in each representative images (n = 2) was quantified using Licor Image Studio Software and expressed numerically as the LC3BII/LC3BI ratio below the LC3B immunoblot panel. Failure to detect a 2-fold difference in LC3BII/LC3BI ratio in Fig. 2B lanes 5 and 6 likely reflects the nonlinearity of the chemiluminescent detection and the greater accuracy and linearity of manually counting GFP-LC3 puncta shown in C and D. (C) Representitive images (63× magnification) of GFP-LC3B puncta (autophagosomes) in ARPE-19 cells stably expressing GFP-LC3B that were infected and treated as in B. Higher magnification images (265× magnification) representing white boxed areas are shown below each panel. (D) Quantification of GFP-LC3B puncta in ARPE-19 cells stably expressing GFP-LC3B that were infected and treated as in B. Comparisons were performed between viruses derived from the same parent strain (strain F: ΔUs3 vs. Us3-Repair; strain Patton: WT Patton vs. ΔICP34.5). Error bars represent mean + SEM of triplicate samples with >150 cells analyzed per sample. Statistical analysis was performed by paired t test using the Bonferroni correction for multiple comparisons. *P < 0.05; **P < 0.01.
Suppression of Autophagy in HSV-1–Infected Cells Requires the Virus-Encoded Us3 Ser/Thr Kinase Activity.
To determine whether Us3 Ser/Thr kinase activity was required to control autophagy, NHDFs were infected with WT HSV-1, a Us3-deficient virus (ΔUs3), a virus expressing a kinase-deficient, catalytically inactive Us3 (K220A), or a virus in which the Us3 mutation was repaired (Us3-Repair). After 12 h, total protein was isolated and levels of LC3B-I vs. LC3B-II were evaluated by immunoblotting. Compared to viruses containing a functional Us3 gene (WT, Us3-Repair), cultures infected with ΔUs3 or K220A were enriched for LC3B-II relative to LC3B-I (Fig. 3A, compare lanes 3 and 6 to lanes 4 and 5; Fig. 3B).
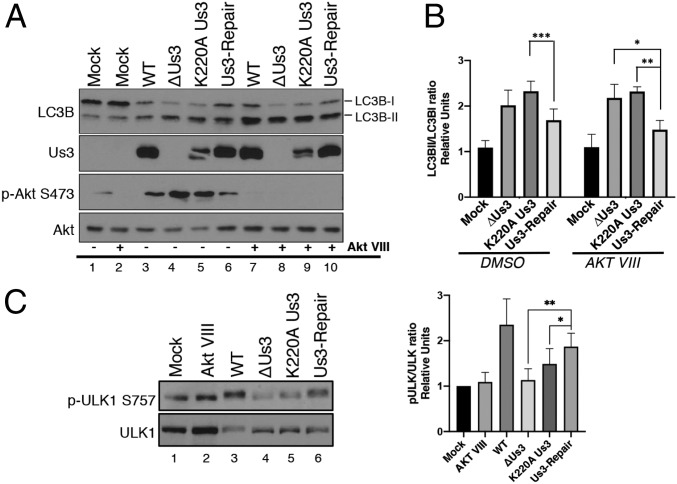
Fig. 3.
Us3 acts downstream of ULK1. (A) NHDFs were mock infected or infected (MOI = 5) with wild-type HSV-1 (WT), a Us3-deficient virus (ΔUs3), a virus expressing a catalytically inactive Us3 protein (K220A Us3), or a virus where the Us3 deficiency was repaired (Us3-Repair). After 1.5 h cells were treated with an Akt inhibitor (Akt VIII) or DMSO. At 12 hpi, total protein was fractionated by SDS/PAGE and analyzed by immunoblotting with the indicated antibodies. Migration of unmodified LC3B-I and lipidated LC3B-II are indicated on the Right of this representative image. (B) The LC3BII/LC3BI ratio from 4 (n = 4) separate experiments (A) was quantified using ImageJ and plotted. Error bars = SEM. *P < 0.05; **P < 0.01; ***P < 0.001 by 2-way ANOVA corrected for multiple comparisons. (C) Phosphorylation of endogenous ULK1 in NHDFs infected as in A (Left shows representative image) and the ratio of phosphorylated p-ULK1 S575 to total ULK1 quantified from 5 separate experiments (n = 5) using Licor Image Studio software (Right). Error bars = SEM. *P < 0.05; **P < 0.005 by 2-way ANOVA corrected for multiple comparisons.
To rule out contributions from the host kinase Akt, a known autophagy regulator activated by HSV-1, cultures were treated with Akt inhibitor VIII (Akt VIII), a highly selective, allosteric Akt activation inhibitor (49). LC3B lipidation was suppressed by Us3-expressing HSV-1 even in the presence of Akt VIII (Fig. 3A, compare lanes 7 and 10 to 8 and 9; Fig. 3B), suggesting that Us3 acts on targets downstream from and independent of Akt, a likely candidate being TSC2 (41). By directly phosphorylating TSC2, Us3 activates mTORC1 in HSV-1–infected cells in an Akt-independent manner (41). As phosphorylation of the major autophagy activator ULK1 on S757 by mTORC1 inhibits ULK1 activity and represses autophagy (5), the impact of Us3 on ULK1 phosphorylation was evaluated by immunoblotting. Phospho-ULK1 was more abundant in cultures infected with viruses expressing functional Us3 compared to ΔUs3 and K220A Us3 (Fig. 3C, compare lanes 3 and 6 to 4 and 5). Taken together, this established that accumulation of LC3B-II in HSV-1–infected NHDFs was suppressed in a manner dependent upon the Us3 gene and its intrinsic Ser/Thr kinase activity. It further demonstrates that the autophagy-promoting form of ULK1 accumulated in cells infected with a Us3-deficient virus and that inhibition of ULK1 in HSV-1–infected cells was dependent upon the Us3 Ser/Thr kinase.
Us3 Targets Both mTOR-Dependent and Independent Autophagy Regulators.
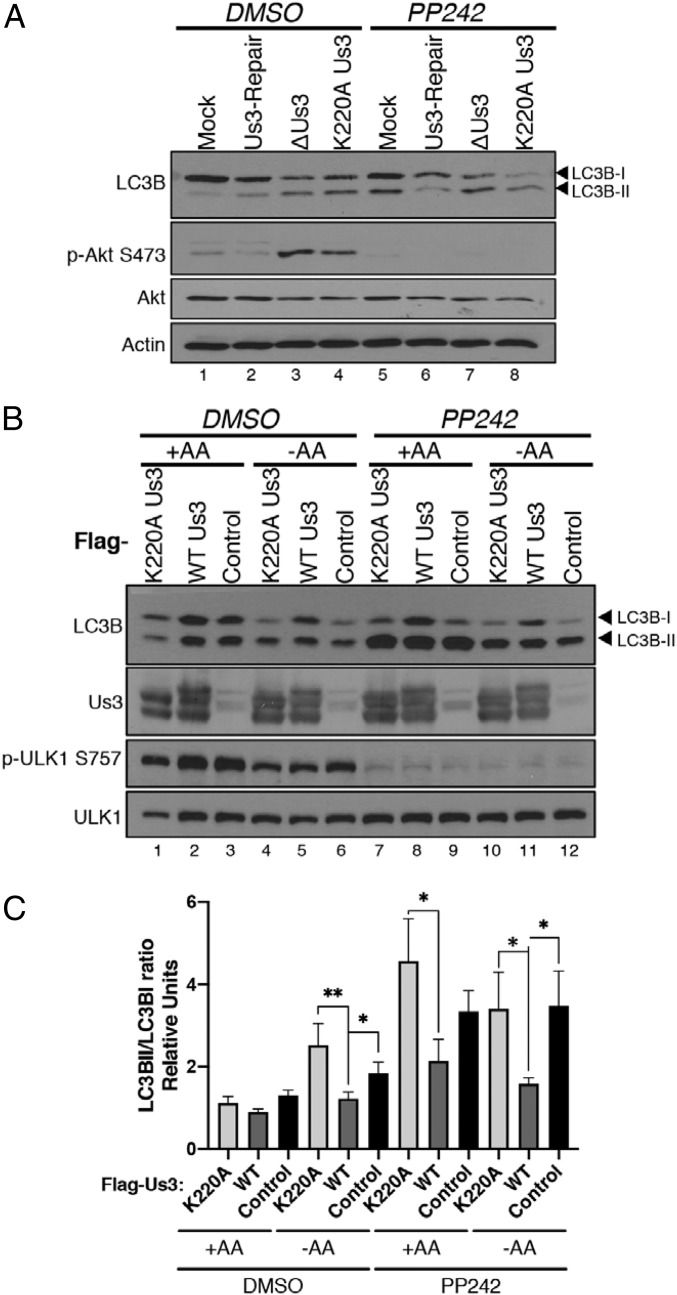
As Us3 activates mTORC1 (41), which inhibits the ULK autophagy-promoting complex (50–52), it was possible that Us3 might only act to suppress autophagy upstream of the ULK complex, or may also act on additional targets downstream of ULK. To investigate this possibility, NHDFs mock infected or infected with either ΔUs3, K220A Us3, or Us3-Repair were treated with DMSO or the mTOR active-site inhibitor PP242. Importantly, lipidated LC3B accumulated in mock-infected, PP242-treated cultures, reflecting activation of ULK1 (Fig. 4A, compare lanes 1 through 5). However, while LC3B-II accumulated in cells infected with Us3-deficient viruses (ΔUs3, K220A), a Us3-expressing virus unexpectedly prevented LC3B-II accumulation in the presence of PP242 (Fig. 4A, compare lane 6 to lanes 7 and 8). This suggested that Us3 might also control autophagy in an mTORC1/2-independent manner. To investigate if this phenotype was dependent upon other viral proteins, uninfected cells transiently transfected with empty vector control or Us3-expressing plasmids were starved for amino acids (AAs) to induce autophagy in the absence or presence of PP242. AA starvation induced LC3B-II accumulation indicative of autophagy that was effectively suppressed by WT, but not K220A Us3 (Fig. 4B, compare lane 5 to lanes 4 and 6; Fig. 4C). In addition, Us3 expression did not detectably stimulate ULK1 S757 phosphorylation in PP242-treated cells compared to untreated cells, implying that ULK1 is not a substrate phosphorylated by Us3 on S757 (Fig. 4B). However, only WT Us3 effectively suppressed PP242-induced autophagy irrespective of the presence of AAs (Fig. 4B, compare lane 11 to lanes 10 and 12; Fig. 4C). This demonstrates that Us3 Ser/Thr kinase activity is necessary and sufficient to suppress starvation-induced autophagy in uninfected cells and acts, both in infected and uninfected cells, in a manner insensitive to an mTOR inhibitor. It further suggests that Us3 can control autophagy downstream of mTORC1.
Fig. 4.
Us3 is sufficient to control autophagy in uninfected cells. (A) NHDFs were mock infected or infected with HSV-1 ΔUs3, K220A Us3, or Us3-Repair virus (MOI = 5). At 8 hpi, the cells were treated with DMSO or PP242. Total protein was harvested at 12 hpi, fractionated by SDS/PAGE, and analyzed by immunoblotting using the indicated antisera. (B) HeLa cells were transfected with plasmids expressing Flag-Us3 (WT), Flag-K220A Us3 or an empty vector (control). After 24 h, AAs were maintained (+) or removed (−) from the media for 4 h in the presence or absence of PP242. Total protein was subsequently harvested and analyzed by immunoblotting with the indicated antibodies. Migration of unmodified LC3B-I and lipidated LC3B-II are indicated on the Right of the panel. A representative image is shown. (C) LC3B-I and LC3B-II abundance was quantified using Licor Image Studio software and the LC3BII/LC3BI ratio from 5 (n = 5) separate experiments shown in B was plotted. Bars represent mean ± SEM. *P < 0.05; **P ≤ 0.01 by paired t test using the Bonferroni correction for multiple comparisons.
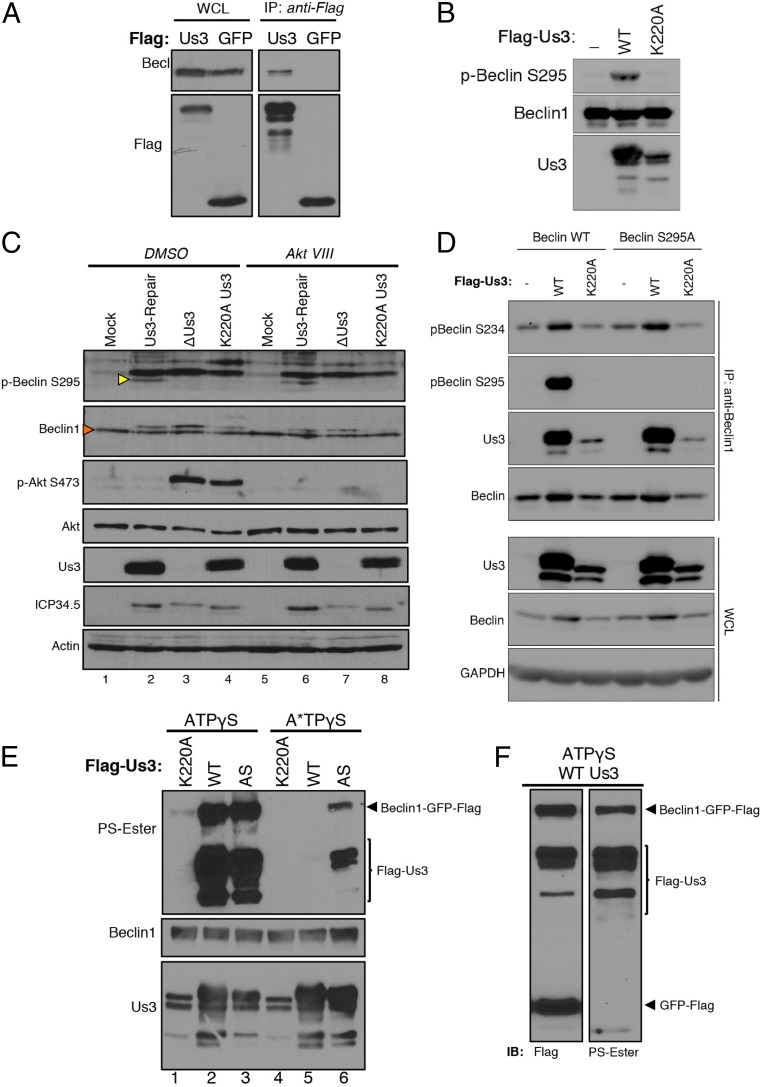
Beclin1 Physically Associates with Us3 and Is a Substrate Directly Phosphorylated by Us3.
Although Us3 does not exhibit significant primary sequence homology to Akt, both kinases share several substrates, including the mTORC1 regulator TSC2 (41). Since Us3, like Akt, can limit autophagy in an mTORC1-independent manner (Fig. 4 A and B), and Akt reportedly suppresses autophagy by phosphorylating Beclin1 in cancer cell lines (8), the capacity of Us3 to target Beclin1 was investigated. Ectopically expressed Us3, but not GFP, physically associated with endogenous Beclin1 in uninfected cells (Fig. 5A). In addition, Beclin1 S295 phosphorylation was stimulated in vitro by immunoprecipitated (IP) WT Us3, but not the kinase-deficient K220A variant (Fig. 5B). This shows that phosphorylation of Beclin1 S295 was dependent upon the Us3 Ser/Thr kinase activity and demonstrates that recognition of phospho-Beclin1 S295 by the antibody requires incubation with kinase-proficient Us3. To determine whether HSV-1 infection influenced Beclin1 phosphorylation and validate these findings in normal primary cells expressing endogenous Beclin1, NHDFs mock infected or infected with viruses expressing wild-type Us3 (Us3-Repair) or unable to express active Us3 (ΔUs3, K220A) were treated with DMSO or Akt VIII and Beclin1 S295 phosphorylation measured by immunoblotting. While total Beclin1 levels remained similar in uninfected and infected cells, phospho-S295 Beclin1 was only detected in infected cultures expressing a functional Us3 kinase (Fig. 5C, compare lane 2 to lanes 3 and 4). Furthermore, phospho-S295 Beclin1 accumulation was insensitive to Akt VIII (Fig. 5C, lane 6). This demonstrates that phospho-S295 Beclin1 accumulation in HSV-1–infected cells can occur under conditions where Akt is inhibited and is dependent upon Us3 and its Ser/Thr kinase activity.
Fig. 5.
Beclin1 physically associates with Us3 and is a direct substrate phosphorylated by the HSV-1 kinase. (A) HeLa cells were transfected with a plasmid expressing Flag-Us3 or Flag-GFP. After 48 h, soluble lysates were immunoprecipitated (IP: anti-FLAG) with anti-FLAG antibody. Immune complexes were fractionated by SDS/PAGE and analyzed by immunoblotting with the indicated antibodies. WCL, whole-cell lysates prior to immunoprecipitation. (B) HeLa cells were transiently transfected with plasmids expressing Flag-tagged Beclin1, Flag-Us3 (WT), or Flag-K220A Us3. After 24 h, soluble cell-free fractions from Flag-Us3 (WT or K220A) and Flag-Beclin1–transfected cells were mixed and immunoprecipitated using anti-Flag antibody. After a high-salt wash to reduce nonspecific binding, immune complexes were equilibrated in kinase buffer, incubated with ATP analogs for 30 min at 30 °C, fractionated by SDS/PAGE, and analyzed by immunoblotting with the indicated antibodies. (C) NHDFs mock infected or infected with HSV-1 ΔUs3, K220A Us3, or Us3-Repair virus (MOI = 5) were treated with DMSO or Akt VIII at 1.5 hpi. Total protein was isolated at 12 hpi and analyzed by immunoblotting with the indicated antisera. Additional nonspecific bands representing proteins that migrate slower than Beclin1 were routinely present when endogenous Beclin1 and pBeclin S295 levels were monitored in whole-cell extracts prepared from HSV-1–infected primary fibroblasts. This likely reflects the reduced abundance of endogenous Beclin1 compared to actin, Akt, or virus-encoded proteins in primary NHDFs and/or possible nonspecific cross-reaction of commercially available Beclin1 antibodies with HSV-1 proteins in whole-cell lysates upon immunoblotting. Yellow arrowhead indicates endogenous levels of phosph-S295 Beclin1 (p-Beclin S295) in primary NHDFs. Red arrowhead indicates endogenous total Beclin1 band. The specificity of phospho-specific antibodies used to detect phosphorylated Beclin1 is demonstrated in B and D. (D) HeLa cells were transiently transfected with plasmids expressing Flag-tagged Beclin1 (WT) or Flag-tagged S295A Beclin1. After 24 h, the cells were subsequently transfected with plasmids expressing Flag-Us3 (WT) or Flag-K220A Us3 and treated with Akt inhibitor VIII. Soluble cell-free lysates were prepared after 24 h and immunoprecipitated using anti-Beclin1 antibody (IP: anti-Beclin1). Isolated immune complexes were fractionated by SDS/PAGE and analyzed by immunoblotting with the indicated antibodies. WCL, whole-cell lysates prior to immunoprecipitation. (E) HeLa cells were transiently transfected with plasmids expressing Flag-tagged Beclin-GFP, Flag-Us3 (WT), Flag-K220A Us3, or an analog-sensitive Us3 (AS). After 24 h, soluble cell-free fractions from Flag-Us3 and Beclin1-GFP-Flag–transfected cells were mixed and immunoprecipitated using anti-Flag antibody. After a high-salt wash to reduce nonspecific binding, immune complexes were equilibrated in kinase buffer and incubated with the indicated ATP analogs for 60 min at 30 °C. Following thiophosphate alkylation using p-nitrobenzylmesylate (PNBM), samples were fractionated by SDS/PAGE and analyzed by immunoblotting using the indicated antibodies. (F) As in E, except cells were transiently transfected with plasmids expressing Flag-tagged Beclin1-GFP, Flag-Us3, and Flag-GFP.
Besides S295, Akt also promotes phosphorylation of Beclin1 on S234. To determine if Us3, like Akt, promotes Beclin1 S234 phosphorylation, WT- or S295A-substituted Beclin1 was ectopically expressed by transient transfection of plasmid DNA. Following immunoprecipitation of Beclin1, phospho-Beclin1 S295 and S234 abundance were evaluated by immunoblotting. Fig. 5D shows that WT but not kinase-dead K220A Us3 stimulates accumulation of pBeclin S234 and S295 in cells expressing WT Beclin1. Us3 WT, but not K220A, also effectively stimulated pBeclin S234 phosphorylation in cells expressing Beclin S295A, confirming the selectivity of the pBeclin S295 phospho-specific antibody as Beclin S295A is unable to be phosphorylated on S295. It remains possible that Beclin1 phosphorylation on additional Ser/Thr residues may also be stimulated by Us3. In addition, compared to WT kinase-proficient Us3, less K220A kinase-dead Us3 was detected in anti-Beclin1 immune complexes (Fig. 5D). This demonstrates that Us3 stimulates Beclin1 S234 and S295 phosphorylation and suggests that the Us3 Ser/Thr kinase activity influences binding to Beclin1.
To determine if Beclin1 is a direct Us3 substrate, an engineered Us3 derivative containing a Leu→Gly substitution at residue 259 and capable of accepting a bioorthogonal ATP analog (AS-Us3) unable to be utilized by the endogenous kinome was exploited (41). While nearly all kinases including Us3 can use ATPγS to thiophosphorylate substrates, only analog-sensitive (AS) Us3 can use N6-alkylated ATPγS (A*TPγS) to thiophosphorylate substrates. Thiophosphorylated proteins are subsequently reacted with a thiol-specific alkylating agent that generates a bioorthogonal thiophosphate ester. The resulting labeled proteins are readily detected by immunoblotting, using a thiophosphate ester-specific antibody (53). Following anti-FLAG immunopurification from transfected HeLa cells, both WT and AS-Us3 used ATPγS to thiophosphorylate themselves and a Beclin1-GFP fusion protein (Fig. 5E, lanes 2 and 3). It was necessary to utilize a Beclin1 fusion protein because both Beclin1 and Us3 have similar molecular weights and were not resolvable by SDS/PAGE. Importantly, only the Beclin1-GFP fusion protein, but not free, unfused GFP, was exclusively thiophosphorylated by Us3, indicating that GFP is not a Us3 substrate (Fig. 5F). Both Us3 and Beclin1 phosphorylation were not detected in reactions containing catalytically inactive Us3 K220A (Fig. 5E, lanes 1 and 4), demonstrating that thiophosphorylation required Us3 Ser/Thr kinase activity. Significantly, whereas AS-Us3 accepted ATPγS and A*TPγS to thiophosphorylate both Us3 and Beclin1, wild-type Us3 used ATPγS but not A*TPγS (Fig. 5E, compare lanes 3 and 6 to lanes 2 and 5). This shows that A*TPγS is bioorthogonal, as it was detectably used by only AS-Us3 and not WT Us3. Moreover, it established that Us3 directly phosphorylates the Beclin1-GFP fusion protein, since AS-Us3 is the only kinase in the reaction capable of accepting and using the A*TPγS analog to thiophosphorylate Beclin1-GFP. This identifies the host autophagy regulator Beclin1 as a direct Us3 substrate.
ULK1 and Beclin1 Restrict Replication of Us3-Deficient HSV-1.
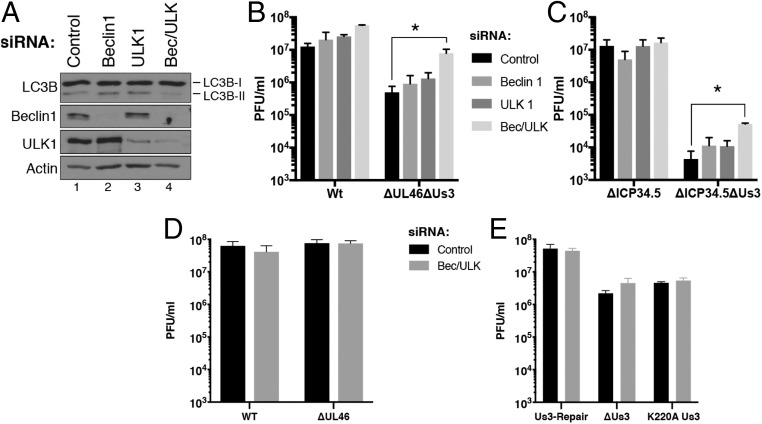
Having shown that autophagy in HSV-1–infected cells was suppressed by Us3 and ICP34.5, how this finding might impact virus reproduction in nonneuronal cells remained unknown. To define how these 2 viral factors acting through both Beclin1 and ULK1 might influence virus replication, NHDFs were transfected with control, nonsilencing RNA, Beclin1 siRNA, ULK1 siRNA, or a mixture of both Beclin1 and ULK1 siRNAs (Fig. 6A) and the growth of WT or specific mutant HSV-1 derivatives, including those lacking ICP34.5 and/or Us3 was compared. Notably, depletion of both Beclin1 and ULK1 was required to significantly limit LC3B-II accumulation in uninfected, primary human fibroblasts (Fig. 6A). While depletion of Beclin1 and or ULK1 did not detectably alter WT KOS37 growth, depleting both Beclin1 and ULK1 together increased replication of the isogenic derivative ΔUs3ΔUL46 doubly deficient virus (Fig. 6B) but had no detectable impact on replication of ΔUs3, Us3K220A, or ΔUL46 single mutant viruses (Fig. 6 D and E). The UL46 mutation was required to genetically prevent Akt activation in HSV-1–infected cells (54). Growth of the Δ34.5-B2 Patton strain was similarly unaffected by ULK1 and or Beclin1 depletion (Fig. 6C). However, growth of the Us3-deficient derivative that is otherwise isogenic to the Δ34.5-B2 parent virus was increased by 10-fold upon codepletion of both ULK1 and Beclin1 (Fig. 6C). This establishes that host ULK1 and Beclin1 autophagy pathways restrict productive HSV-1 growth in normal human cells and are antagonized primarily by Us3.
Fig. 6.
Beclin1- and ULK1-dependent autophagy restricts productive HSV-1 replication and are antagonized primarily by Us3. (A) NHDFs were treated with nonsilencing (control), Beclin1-specific (Beclin1), ULK1-specific (ULK1), or a mixture of Beclin1- and ULK1-specific siRNAs (Bec/ULK). After 72 h, total protein was isolated, fractionated by SDS/PAGE, and analyzed by immunoblotting with the indicated antibodies. (B) NHDFs treated as in A were infected (MOI = 2.5 × 10−3) with WT HSV-1 Kos37, or a Kos37 derivative doubly deficient for Us3 and VP11/12 (ΔUs3ΔUL46). After 3 d, infectious virus produced was quantified by plaque assay in Vero cells. Bars represent mean + SEM of at least 3 experiments. Statistical analysis was performed using the Student’s t test. *P ≤ 0.05. (C) As in B except cells were infected with a Patton strain HSV-1 doubly deleted for ICP34.5 and Us3 (Δ34.5-B2ΔUs3) or the ICP34.5-deficient parental virus (Δ34.5-B2). (D) NHDFs treated with nonsilencing (control) siRNA or a mixture of Beclin1- and ULK1-specific siRNAs (Bec/ULK) were infected (MOI = 2.5 × 10−3) with WT HSV-1 Kos37 or UL46-deficient Kos37 derivative virus (ΔUL46). (E) As in D except NHDFs were infected with the indicated HSV-1 F-strain Us3-deficient or Us3-repaired virus. Infectious virus was quantified as in B. Bars represent mean + SEM of at least 3 experiments and statistical differences were not detected between control and Bec/ULK siRNA-treated samples using a Student’s t test.
Discussion
While autophagy protects against neurotrophic virus infection in model organisms, evidence demonstrating an antiviral role for autophagy at the cellular level is lacking (18, 19). Whereas an HSV-1 encoding a mutant ICP34.5 unable to bind Beclin1 was attenuated in adult mice, replication defects in many cell types were not evident and disabling autophagy did not impact ICP34.5-deficient virus reproduction in nonneuronal cells (30). Here, we establish that the HSV-1 Us3 Ser/Thr kinase regulates autophagy in infected cells and synergizes with ICP34.5 to limit autophagy. Unlike ICP34.5-deficient viruses, replication of a Us3-deficient virus and a ICP34.5-Us3 doubly deficient virus were restored in part when autophagy was disabled in cultured cells. Moreover, we show that Us3 suppresses autophagy by antagonizing both ULK1 and Beclin1. Us3 constitutively activates mTORC1 to enforce inhibition of ULK1 and effectively mimics Akt by directly phosphorylating Beclin1. This establishes a role for hijacking Akt signaling to repress Beclin1-dependent autophagic functions in infection biology and shows this mechanism is not simply confined to restricting autophagy in cancer cells.
Although ICP34.5 is only encoded by HSV type 1 and 2, protein kinases related to Us3 are encoded only by α-herpesvirus subfamily members, many of which are neurotrophic. Us3-like kinases may allow viruses lacking ICP34.5 to limit autophagy. Not all α-herpesviruses,however, limit autophagy, as autophagy is induced in cells infected with varicella-zoster virus (55, 56). The overall extent to which the substrate specificity of other α-herpesvirus Us3 homologs overlaps with HSV-1 Us3, however, remains to be defined and could be a defining determinant that limits autophagy.
The identification of Beclin1 as a direct HSV-1 Us3 substrate defines an unexpected regulatory function for this family of α-herpesvirus-specific Ser/Thr kinases. It strengthens the hypothesis that Us3 mimics activated Akt, despite not sharing significant primary sequence homology. Finally, it illustrates how harnessing a constitutively activated Ser/Thr kinase enables HSV-1 to commandeer control over disparate, critical host regulatory pathways that stimulate anabolic and repress catabolic processes through a single virus-encoded gene product.
The sheer amount of conserved viral genome coding capacity devoted to limiting autophagy reflects the importance of preventing this outcome in HSV-infected cells. Autophagy triggered by HSV-1 influences innate immune responses in part via tripartite motif (TRIM) proteins that control levels of the DNA sensor cGAS and TBK1, both of which control type I IFN production (57–59). At least 3 independent HSV-1–encoded functions can antagonize autophagy. The Us3 Ser/Thr kinase joins ICP34.5 as another direct modulator of host autophagy pathways, together with other independent viral functions that prevent eIF2 phosphorylation (7, 31–33), which can inhibit infected cell protein synthesis and promote autophagy. Besides encoding redundant functions to restrict autophagy, Us3 targets mTORC1-dependent and independent regulatory steps and both Us3 and ICP34.5 antagonize Beclin1 via discrete mechanisms. The function of this redundancy might simply ensure from a stochastic perspective that autophagy is limited in infected cells or enable different functions to predominate in different cell types. Replication in nonneuronal cells, where Beclin1 and ULK1 might restrict virus reproduction, represents a critical step in the virus productive growth cycle that enables HSV-1 to spread to new hosts. Finally, identifying Us3 as 1 of several redundant functions controlling autophagy in HSV-1–infected cells resolves prior paradoxical findings that pathogenesis was dependent upon virus-encoded autophagy antagonists, but not reproduction in cultured cells (7, 18, 19, 27).
Materials and Methods
Cell Culture, Viruses, and Chemicals.
NHDFs (Lonza, Walkersville, MD) and HeLa cells were propagated in 5% CO2 with DMEM (Corning; 10-013-CV) supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin, and 5% (vol/vol) FBS. Vero cells (ATCC) were cultured in DMEM supplemented with 5% (vol/vol) calf serum. ARPE-19 cells (kindly provided by D. Tortorella, Icahn School of Medicine at Mount Sinai, New York, NY) were grown in DMEM: F12 medium (Gibco, 12634-010) supplemented with 10% (vol/vol) FBS. F-strain WT, Us3-deficient (VRR1202), K220A Us3 mutant (VRR1204), and Us3-Repair (VRR1202-Repair) HSV-1 were provided by Rich Roller (University of Iowa, Iowa City, IA) and described previously (60). F-strain ICP34.5-deficient (R3616) and repair viruses (R3616R) were provided by B. Roizman, The University of Chicago, Chicago, IL. Patton strain ICP34.5-deficient virus was described previously. Kos37 strain HSV-1 WT, ΔUL46, and ΔUs3ΔUL46 were provided by Jim Smiley (University of Alberta, Edmonton, Alberta, Canada) (54). Isolation of the Patton strain HSV-1 Δ34.5 genome as a bacterial artificial chromosome (B2 BAC) that expresses immediate-early (IE) Us11, allelic replacement in Escherichia coli to construct recombinant HSV-1 genomes, and infectious virus production following transfection of BAC genomes into permissive Vero cells was described (44). For allelic replacement of Us3 or UL43, a PCR product containing the kanamycin resistance (KanR) gene flanked on each side by 42 nucleotides homologous to sequences in the Us3 or UL43 gene was then generated as described (44) by PCR using the following primers: for Us3[Us3pKD13P1-135392 (5′-CACCACACCACCCAGCGAGGCCGAGCGCCTGTGTCATCTGCAGTGTAGGCTGGAGCTGCTTC-3′)] and [Us3pKD13P4-136550 (5′-GCGGGTCCAGGCCGGTCGGGTGTACGGCGGGCGATTGTTCCCATTCCGGGGATCCGTCGACC)]; for UL43 [UL43pKD13P1-95151 (5′-CGGCCCCGGGCCCGGCCACCCAGTGCCTGGCACTGGGCGCCGGTGTAGGCTGGAGCTGCTTC-3′)] and [UL43PKD1P4-96094 (5′-ACCGGCGGTCTTCGGGACTAATGCCTTTTATTGAAAAATATAATTCCGGGGATCCGTCGACC-3′)]. Proper integration of the KanR cassette was validated by PCR as described (43) using the following primer pairs for Us3 [Us3-UpSt-135321 (5′-GGCCCCTTTTATACCCCAGC-3′); Us3-DnSt-136621 (5′-GGGCGAAGCGCGCCGTCGAA-3′)] and UL43 [UL43-UpSt-95090 (5′-GCCGGTGGAGCGGCCCTTGT-3′); UL43-DnSt-96159 (5′-TCCCCTTCCGGAATTTATAC-3′)]. Akt inhibitor VIII was purchased from Calbiochem (124018); PP242 (P0037) and ATPγS (A1388) were purchased from Sigma; PNBM from Abcam (ab138910); and N6-phenylethyl-ATPγS was from BioLog (P026).
Immunoblotting and Antibodies.
Total protein was collected by lysis in Laemmli buffer (60 μM Tris⋅HCl, pH 6.8, 2% SDS, 10% glycerol, 710 mM 2-mercaptoethanol) and immunoblotting was performed as previously described (61). Anti-Akt (no. 9272), Akt-pSer473 (no. 9271), ULK1 (no. 8054), ULK1-pSer757 (no. 6888), mouse anti-Beclin1 (no. 4122), rabbit anti-GAPDH (no. 2118), and anti-actin (no. 3700) were purchased from Cell Signaling; anti-thiophosphate ester (PS-Ester) was from Epitomics (2686-1); anti-LC3B (L7543) and anti-FlagM2 were from Sigma; and rabbit anti-Beclin1 (sc-48341) was from Santa Cruz Biotechnology. Anti-Beclin1-pSer295 (p117-295) and Beclin1-pSer234 (p117-234) were purchased from PhosphoSolutions. Anti-Us3 and anti-Us11 were generously provided by B. Roizman. Anti-ICP34.5 was described previously (45). Licor image studio software or ImageJ was used to quantify the abundance of modified and unmodified LC3B (LC3B-I and LC3B-II), which were used to subsequently calculate the LC3BII/LC3BI ratio.
GFP-LC3B Expression and Imaging.
GFP-LC3B was cloned into pLenti-Neo plasmid as previously described (62) and verified by DNA sequencing. Lentivirus particles were produced by transient transfection of 293LTV cells (Cell Biolabs, LTV-100) as described (63). Briefly, 293TLTV cells were transfected with psPAX2 (Addgene, 12260), pMDG.2 (Addgene, 12259), and the transfer vector pLenti-GFP-LC3B using Lipofectamine 2000. NHDFs or ARPE-19 cells transduced with the lentivirus were selected with puromycin (2 μg/mL). Cells stably expressing GFP-LC3B were grown on coverslips and treated and/or infected with HSV-1 strains as indicated. After fixing with 4% paraformaldehyde for 15 min, DNA was stained with 4′,6′-diamidino-2-phenylindole (DAPI) and images were captured using a Leica DM5000 microscope equipped with Leica imaging software.
Multicycle Virus Growth Assay and siRNA Transfections.
NHDFs were seeded at 1 × 105 cells per well in 12-well dishes. The next day, cells were transfected with AllStars negative control siRNA (Qiagen), Beclin1 siRNA (Qiagen, SI05126534), ULK1 siRNA (Dharmacon, D-005049-01-0002), or a combination of Beclin1 and ULK1 siRNAs at 20 nM using RNAimax (Invitrogen, 13778075) as described (64). Three days posttransfection, the cells were washed with PBS and infected with 250 plaque-forming units (PFU) of the indicated HSV-1 strain in 0.3 mL media per well for 1.5 h, after which the virus inoculum was removed and replaced with fresh media. Infected cultures were collected at 72 h postinfection (hpi), freeze –thawed 3 times, and the infectious virus produced was quantified by plaque assay in Vero cells.
Plasmid Transfections.
pcDNA4-Beclin1-Flag (Addgene, 24388) was used to construct pcDNA4-Beclin1-GFP-Flag. pcDNA3-Us3-Flag, K220A, and L256G were kindly provided by R. Roller and described previously (41). Plasmids were transfected in HeLa cells using Lipofectamine 2000 (Invitrogen, 11668019) according to the manufacturer’s protocol. The amount of DNA and Lipofectamine 2000 was adjusted to achieve similar expression levels of Us3 and K220A.
Immunoprecipitation.
HeLa cells were first transfected with pcDNA4-Beclin1-Flag or pcDNA4-S295A-Beclin1-Flag. After 24 h, cells were subsequently transfected with pcDNA3-Us3Flag or pcDNA3-K220A-Us3-Flag and treated with Akt VIII (5 μM). After an additional 24 h, cells were rinsed with ice-cold PBS and lysed in IP lysis buffer for 30 min at 4 °C (20 mM Hepes-KOH, pH 7.9, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 1× phosphatase inhibitor mixture (Roche), 1× protease inhibitor mixture (Roche). Following clarification by centrifugation (14,000 × g, 10 min, 4 °C), anti-Beclin1 antibody was added to the soluble fraction for at least 4 h at 4 °C. Protein A/G Sepharose (30 μL of a 50% vol/vol slurry, Santa Cruz Biotechnology) was subsequently added while the tube was rotated end over end for at least 1 h at 4 °C. Sepharose pellets were collected by brief centrifugation in a microfuge, washed 3× with cold IP lysis buffer, suspended in 30 to 40 μL gel loading buffer, boiled for 3 min, fractionated by SDS/PAGE, and analyzed by immunoblotting with indicated antibodies.
In Vitro Kinase Assay.
HeLa cells (10-cm dishes) were transfected with pcDNA4-Beclin1-Flag or pcDNA4-Beclin1-GFP-Flag as indicated, pcDNA3-Us3Flag, pcDNA3-K220AUs3Flag, pcDNA3-L256GUs3Flag, or pcDNA3-GFP-Flag. Twenty-four hours posttransfection, Beclin1-FLAG– or Beclin1-GFP-Flag–expressing cells were treated with Akt VIII (5 μM for 4 h). Cells were lysed in 50 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholate, and 0.1% SDS plus Complete Mini protease inhibitor (Roche), and extracts were clarified by centrifugation (12,000 × g, for 10 min at 4 °C). Soluble fractions from Beclin1-FLAG or Beclin1-GFP-Flag–transfected cells were mixed 1:1 with an equivalent fraction from Us3Flag, L256GUs3Flag, or K220AUs3-Flag–transfected cells. A total of 25 μL of settled bed volume of Anti-Flag M2 affinity gel (Sigma, A2220) was added and the mixture was rocked (2 h at 4 °C). Immune complexes were collected by brief centrifugation, resuspended in buffer (10 mM Tris⋅HCl pH 8.0, 1 M NaCl, 0.2% Nonidet P-40), rocked gently (15 min at 4 °C), and then washed 3 times with 0.5 mL of Us3 kinase buffer (50 mM Tris⋅HCl pH 9.0, 20 mM MgCl2, 0.1% Nonidet P-40). Kinase reactions were performed in 30 μL of Us3 kinase buffer supplemented with 10 μM ATP, 0.5 mM DTT for 30 min at 30 °C, and stopped by adding 30 mL of 2× SDS gel loading buffer. Where indicated, 250 μM ATPγS or N6-phenylethyl ATPγS, was substituted for ATP and reactions were stopped by adding EDTA to 40 mM. Following alkylation with 2.5 mM p-nitrobenzyl mesylate (PNBM) for 2 h at room temperature, samples were fractionated by SDS/PAGE and analyzed by immunoblotting. Alkylated proteins were detected using anti-thiophosphate ester (PS-ester) antibody.
Acknowledgments
We are grateful to Paola Teegarden and Dr. Matthew Mulvey for introducing Us3-and UL43-deficient alleles into the HSV-1 Δ34.5-B2 BAC and Dan Depledge for advice regarding statistical analysis. In addition, we thank J. Smiley, R. Roller, D. Tortorella, and B. Roizman for generously providing mutant viruses, plasmid DNA, antisera, and cell lines. Finally, we thank Chris Bianco and Hannah Burgess for critically reading the manuscript and members of the I.M. laboratory and Angus Wilson for many stimulating discussions. This work was supported in part by awards to R.M.R. from Consejo Nacional de Ciencia y Tecnologia (Mexico) and a Vilcek Fellowship, and grants from the NIH to I.M. (AI073898 and GM056927). The funders had no role in study design, data collection, and interpretation, or in the decision to submit the work for publication.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
References
- 1.Yang Z., Klionsky D. J., Eaten alive: A history of macroautophagy. Nat. Cell Biol. 12, 814–822 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamb C. A., Yoshimori T., Tooze S. A., The autophagosome: Origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 14, 759–774 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Galluzzi L., Pietrocola F., Levine B., Kroemer G., Metabolic control of autophagy. Cell 159, 1263–1276 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saxton R. A., Sabatini D. M., mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim J., Kundu M., Viollet B., Guan K. L., AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russell R. C., et al. , ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 15, 741–750 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tallóczy Z., et al. , Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 99, 190–195 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang R. C., et al. , Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science 338, 956–959 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cadwell K., Crosstalk between autophagy and inflammatory signalling pathways: Balancing defence and homeostasis. Nat. Rev. Immunol. 16, 661–675 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi A. M., Ryter S. W., Levine B., Autophagy in human health and disease. N. Engl. J. Med. 368, 651–662 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Virgin H. W., Levine B., Autophagy genes in immunity. Nat. Immunol. 10, 461–470 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi Y., Bowman J. W., Jung J. U., Autophagy during viral infection - a double-edged sword. Nat. Rev. Microbiol. 16, 341–354 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirkegaard K., Subversion of the cellular autophagy pathway by viruses. Curr. Top. Microbiol. Immunol. 335, 323–333 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Dong X., Levine B., Autophagy and viruses: Adversaries or allies? J. Innate Immun. 5, 480–493 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson W. T., Viruses and the autophagy pathway. Virology 479-480, 450–456 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jordan T. X., Randall G., Manipulation or capitulation: Virus interactions with autophagy. Microbes Infect. 14, 126–139 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paul P., Münz C., Autophagy and mammalian viruses: Roles in immune response, viral replication, and beyond. Adv. Virus Res. 95, 149–195 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Yordy B., Iwasaki A., Autophagy in the control and pathogenesis of viral infection. Curr. Opin. Virol. 1, 196–203 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexander D. E., Ward S. L., Mizushima N., Levine B., Leib D. A., Analysis of the role of autophagy in replication of herpes simplex virus in cell culture. J. Virol. 81, 12128–12134 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beatman E., et al. , West Nile virus growth is independent of autophagy activation. Virology 433, 262–272 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delpeut S., Rudd P. A., Labonté P., von Messling V., Membrane fusion-mediated autophagy induction enhances morbillivirus cell-to-cell spread. J. Virol. 86, 8527–8535 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gannagé M., et al. , Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe 6, 367–380 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee H. K., et al. , In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity 32, 227–239 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orvedahl A., et al. , Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe 7, 115–127 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H., et al. , Cellular autophagy machinery is not required for vaccinia virus replication and maturation. Autophagy 2, 91–95 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Z., et al. , Coronavirus replication does not require the autophagy gene ATG5. Autophagy 3, 581–585 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Orvedahl A., et al. , HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 1, 23–35 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Bloom D. C., Alphaherpesvirus latency: A dynamic state of transcription and reactivation. Adv. Virus Res. 94, 53–80 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Roizman B., Zhou G., The 3 facets of regulation of herpes simplex virus gene expression: A critical inquiry. Virology 479-480, 562–567 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yordy B., Iijima N., Huttner A., Leib D., Iwasaki A., A neuron-specific role for autophagy in antiviral defense against herpes simplex virus. Cell Host Microbe 12, 334–345 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lussignol M., et al. , The herpes simplex virus 1 Us11 protein inhibits autophagy through its interaction with the protein kinase PKR. J. Virol. 87, 859–871 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X., Matrenec R., Gack M. U., He B., Disassembly of the TRIM23-TBK1 complex by the Us11 protein of herpes simplex virus 1 impairs autophagy. J. Virol. 93, e00497-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Connell D., Liang C., Autophagy interaction with herpes simplex virus type-1 infection. Autophagy 12, 451–459 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilcox D. R., Wadhwani N. R., Longnecker R., Muller W. J., Differential reliance on autophagy for protection from HSV encephalitis between newborns and adults. PLoS Pathog. 11, e1004580 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexander D. E., Leib D. A., Xenophagy in herpes simplex virus replication and pathogenesis. Autophagy 4, 101–103 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tallóczy Z., Virgin H. W. 4th, Levine B., PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy 2, 24–29 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Meignier B., Longnecker R., Mavromara-Nazos P., Sears A. E., Roizman B., Virulence of and establishment of latency by genetically engineered deletion mutants of herpes simplex virus 1. Virology 162, 251–254 (1988). [DOI] [PubMed] [Google Scholar]
- 38.Morimoto T., et al. , Differences in the regulatory and functional effects of the Us3 protein kinase activities of herpes simplex virus 1 and 2. J. Virol. 83, 11624–11634 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sagou K., Imai T., Sagara H., Uema M., Kawaguchi Y., Regulation of the catalytic activity of herpes simplex virus 1 protein kinase Us3 by autophosphorylation and its role in pathogenesis. J. Virol. 83, 5773–5783 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kato A., Kawaguchi Y., Us3 protein kinase encoded by HSV: The precise function and mechanism on viral life cycle. Adv. Exp. Med. Biol. 1045, 45–62 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Chuluunbaatar U., et al. , Constitutive mTORC1 activation by a herpesvirus Akt surrogate stimulates mRNA translation and viral replication. Genes Dev. 24, 2627–2639 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vink E. I., Smiley J. R., Mohr I., Subversion of host responses to energy insufficiency by Us3 supports herpes simplex virus 1 replication during stress. J. Virol. 91, e00295-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vink E. I., Lee S., Smiley J. R., Mohr I., Remodeling mTORC1 responsiveness to amino acids by the herpes simplex virus UL46 and Us3 gene products supports replication during nutrient insufficiency. J. Virol. 92, e01377-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mulvey M., Arias C., Mohr I., Maintenance of endoplasmic reticulum (ER) homeostasis in herpes simplex virus type 1-infected cells through the association of a viral glycoprotein with PERK, a cellular ER stress sensor. J. Virol. 81, 3377–3390 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mulvey M., Poppers J., Sternberg D., Mohr I., Regulation of eIF2alpha phosphorylation by different functions that act during discrete phases in the herpes simplex virus type 1 life cycle. J. Virol. 77, 10917–10928 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.MacLean C. A., Efstathiou S., Elliott M. L., Jamieson F. E., McGeoch D. J., Investigation of herpes simplex virus type 1 genes encoding multiply inserted membrane proteins. J. Gen. Virol. 72, 897–906 (1991). [DOI] [PubMed] [Google Scholar]
- 47.Klionsky D. J., et al. , Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mauthe M., et al. , Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 14, 1435–1455 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Calleja V., Laguerre M., Parker P. J., Larijani B., Role of a novel PH-kinase domain interface in PKB/Akt regulation: Structural mechanism for allosteric inhibition. PLoS Biol. 7, e17 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ganley I. G., et al. , ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284, 12297–12305 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hosokawa N., et al. , Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 20, 1981–1991 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jung C. H., et al. , ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20, 1992–2003 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Allen J. J., et al. , A semisynthetic epitope for kinase substrates. Nat. Methods 4, 511–516 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eaton H. E., Saffran H. A., Wu F. W., Quach K., Smiley J. R., Herpes simplex virus protein kinases US3 and UL13 modulate VP11/12 phosphorylation, virion packaging, and phosphatidylinositol 3-kinase/Akt signaling activity. J. Virol. 88, 7379–7388 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buckingham E. M., et al. , Autophagic flux without a block differentiates varicella-zoster virus infection from herpes simplex virus infection. Proc. Natl. Acad. Sci. U.S.A. 112, 256–261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buckingham E. M., Girsch J., Jackson W., Cohen J. I., Grose C., Autophagy quantification and STAT3 expression in a human skin organ culture model for innate immunity to herpes zoster. Front. Microbiol. 9, 2935 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen M., et al. , TRIM14 inhibits cGAS degradation mediated by selective autophagy receptor p62 to promote innate immune responses. Mol. Cell 64, 105–119 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Sparrer K. M. J., et al. , TRIM23 mediates virus-induced autophagy via activation of TBK1. Nat. Microbiol. 2, 1543–1557 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ahmad L., et al. , Human TANK-binding kinase 1 is required for early autophagy induction upon herpes simplex virus 1 infection. J. Allergy Clin. Immunol. 143, 765–769.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Ryckman B. J., Roller R. J., Herpes simplex virus type 1 primary envelopment: UL34 protein modification and the US3-UL34 catalytic relationship. J. Virol. 78, 399–412 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walsh D., Mohr I., Phosphorylation of eIF4E by Mnk-1 enhances HSV-1 translation and replication in quiescent cells. Genes Dev. 18, 660–672 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jáuregui P., Logue E. C., Schultz M. L., Fung S., Landau N. R., Degradation of SAMHD1 by Vpx is independent of uncoating. J. Virol. 89, 5701–5713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bianco C., Mohr I., Restriction of human Cytomegalovirus replication by ISG15, a host effector regulated by cGAS-STING double-stranded-DNA sensing. J. Virol. 91, e02483-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burgess H. M., Mohr I., Cellular 5′-3′ mRNA exonuclease Xrn1 controls double-stranded RNA accumulation and anti-viral responses. Cell Host Microbe 17, 332–344 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]