Significance
Interactions between proteins normally depend on a range of noncovalent contacts. Under challenging conditions, such as with mechanical force or over long time periods, noncovalent interactions break. Unbreakable protein–protein interactions, linked by covalent bonding, provide many opportunities for robust connection of molecular building blocks, including for biomaterials, enzymes, and vaccines. When evaluating unbreakable interactions, it is important to consider whether reaction happens quickly even at low concentrations. Here we establish a genetically encoded peptide that reacts with its genetically encoded protein partner with a speed close to the limit set by diffusion. We apply a range of biophysical methods to understand the dynamics required for this interaction, demonstrating applicability to rapid and specific detection in a range of species.
Keywords: protein engineering, synthetic biology, mechanobiology, cytoskeleton, nanobiotechnology
Abstract
Much of life’s complexity depends upon contacts between proteins with precise affinity and specificity. The successful application of engineered proteins often depends on high-stability binding to their target. In recent years, various approaches have enabled proteins to form irreversible covalent interactions with protein targets. However, the rate of such reactions is a major limitation to their use. Infinite affinity refers to the ideal where such covalent interaction occurs at the diffusion limit. Prototypes of infinite affinity pairs have been achieved using nonnatural reactive groups. After library-based evolution and rational design, here we establish a peptide–protein pair composed of the regular 20 amino acids that link together through an amide bond at a rate approaching the diffusion limit. Reaction occurs in a few minutes with both partners at low nanomolar concentration. Stopped flow fluorimetry illuminated the conformational dynamics involved in docking and reaction. Hydrogen–deuterium exchange mass spectrometry gave insight into the conformational flexibility of this split protein and the process of enhancing its reaction rate. We applied this reactive pair for specific labeling of a plasma membrane target in 1 min on live mammalian cells. Sensitive and specific detection was also confirmed by Western blot in a range of model organisms. The peptide–protein pair allowed reconstitution of a critical mechanotransmitter in the cytosol of mammalian cells, restoring cell adhesion and migration. This simple genetic encoding for rapid irreversible reaction should provide diverse opportunities to enhance protein function by rapid detection, stable anchoring, and multiplexing of protein functionality.
Transient protein–protein interactions are the norm in living systems. However, stronger interactions between proteins create exciting opportunities to surpass natural assemblies, such as for therapeutics, biomaterials, diagnostics, and vaccines (1). Protein–protein interactions are compared using the equilibrium dissociation constant, Kd. Biologically important protein–protein interactions typically lie in the nanomolar to millimolar range (2, 3). Various approaches have been used to create protein–protein interactions that have no dissociation, with varying features with regard to generality and specificity (4–6). However, an interacting pair is much less useful if it takes days to form such an irreversible complex, when the partners bind and dissociate many times before reaction occurs. Rapid reaction is important for time resolution in biological systems, sensitivity in detection, and also to outcompete other cellular processes such as ligand trafficking or degradation (7). Therefore, Claude Meares introduced the valuable concept of infinite affinity, where an ideal interaction would be irreversible but would also react at the diffusion limit (8). This concept was exemplified by an antibody bearing a nucleophilic Cys binding to an electrophilic small-molecule ligand, where nucleophile/electrophile proximity drove covalent bond formation between the antibody and its target (8). We subsequently established such proximity-driven ligation for a protein–protein interaction (9). Alternative studies have taken this route for covalent ligation via posttranslational chemical modification or unnatural amino acid incorporation (10–12). Nonetheless, such activated complexes require substantial manipulation. Wide application depends upon moving toward infinite affinity for protein–protein interactions where both partners only contain the natural 20 amino acids. This requires a challenging balance between reactivity and specificity.
Here, building on unusual chemistry from Gram-positive bacteria (13), evolution, and computational design, we have established a genetically encoded interaction between a protein and a peptide tag that forms a spontaneous amide bond with close to diffusion-limited kinetics. We carefully analyze the kinetics of docking and reaction. Advanced mass spectrometry approaches enable us to explore the protein dynamics facilitating this rapid reaction. We further establish the broad applicability of this pair for rapid and sensitive detection by flow cytometry and Western blotting in a range of cellular systems. Finally, we apply this technology in concert with the integrin adapter protein talin within the cytoplasm, restoring force transmission between integrin and the cytoskeleton and evaluating the role of interaction stability for cell adhesion and migration.
Results
Engineering Toward Diffusion-Controlled Protein Coupling.
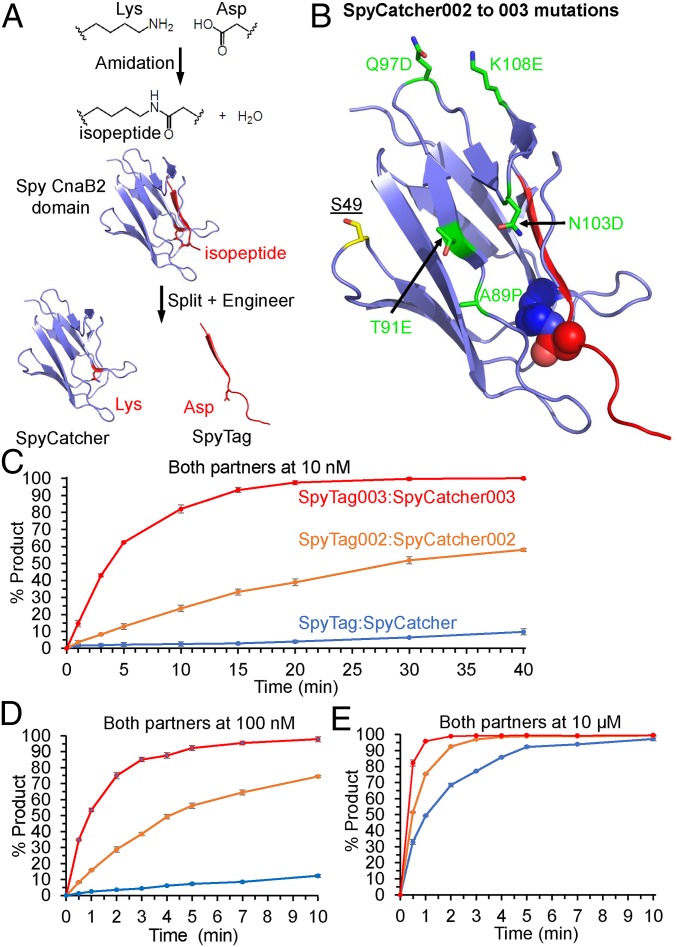
The fibronectin binding protein, FbaB, from Streptococcus pyogenes contains a CnaB2 adhesin domain. CnaB2 is stabilized by a spontaneous reaction of Lys and Asp side chains to form an isopeptide bond (Fig. 1A) (14, 15). We previously achieved genetically encoded and covalent peptide–protein interaction by splitting CnaB2 into the 13-residue SpyTag peptide and the 116-residue SpyCatcher protein (Fig. 1A) (16). SpyTag/SpyCatcher has provided a simple, specific, and genetically encodable method to create a diverse range of biomaterials including hydrogels, vaccines, and thermally stabilized enzymes (1, 17, 18). However, its rate of reaction (1.4 × 103 M−1 s−1) is far below the 105 to 106 M−1 s−1 taken to be the onset of diffusion-controlled protein–protein interactions (19). The rate of reaction was improved >12-fold by phage display selection, yielding SpyTag002 and SpyCatcher002 (see SI Appendix, sequences in SI Appendix, Fig. S1A) (20). With the goal of a further step change in reactivity toward the diffusion limit, we performed further engineering of both the Tag and the Catcher partner.
Fig. 1.
Covalent peptide–protein reaction accelerated by 003 pair. (A) Spontaneous isopeptide bond formation in Spy CnaB2 domain and engineering to give SpyTag and SpyCatcher. The reactive Lys31 and Asp117 are shown in stick format based on PDB 2X5P and 4MLI. (B) Location of SpyCatcher003 mutations. Mutations in SpyCatcher002 to give SpyCatcher003 are marked in green in stick format. The reactive Lys and Asp are shown in spacefill. SpyTag is marked in red. S49 was mutated to Cys in certain constructs for dye attachment. Based on PDB 2X5P and 4MLI. (C) Reaction rate at 10 nM. The 10 nM SpyTag-sfGFP variants were incubated for the indicated time at 25 °C with 10 nM SpyCatcher variants. (D) Reaction rate with each partner at 100 nM, performed as in C. (E) Reaction rate with each partner at 10 µM, performed as in C. Data are mean ± 1 SD, n = 3; some error bars are too small to be visible.
Our design of SpyTag003 started with manual inspection of the SpyTag:SpyCatcher crystal structure (21), combined with leads from phage display screening of SpyTag variants (20). We number Tag and Catcher residues according to the CnaB2 structure, PDB 2X5P (14). In SpyTag, H112 makes water-mediated hydrogen bonding interactions with the side chains of SpyCatcher residues K28 and S30 (21). In SpyTag002, the equivalent residue (T112) would be unable to make the same interactions, and so we made the T112H mutation, which enhanced reaction rate (SI Appendix, Fig. S2A). Going from SpyTag to SpyTag002, addition of positive charge at the Tag’s C terminus enhanced reaction rate (20). In the same vein, we explored introducing additional positive charge at the Tag’s N terminus, which could favor interaction with a patch of negative residues on the Catcher: E20, E21 (unstructured in PDB 4MLI), D22, and E96. Previous hits using our phage display of Tag variants (20) found the R108 mutation, while G109 is the parental sequence from the CnaB2 domain. Appending the RG dipeptide at the N terminus of the Tag also caused a substantial improvement in reaction rate, giving us SpyTag003 (SI Appendix, Fig. S2A).
The 2 key approaches in the design of SpyCatcher003 were loop stabilization and increasing surface polarity. Y84 and E85 within SpyCatcher’s long A79 to A89 loop make interactions with Y119 and K120 of SpyTag (21). Supporting the importance of these contacts, during Tag evolution the 2 wild-type (WT) residues at 119 and 120 were preferred (20). We made the A89P mutation to reduce backbone flexibility in the A79 to A89 loop (22, 23), as previously employed in SpyDock for SpyTag purification (24), and this mutation enhanced reaction speed (SI Appendix, Fig. S2B). To increase the surface polarity and complement the increase in positively charged residues on the Tag, we made 4 mutations (T91E, Q97D, N103D, and K108E) from SpyCatcher002. Q97D and K108E were targeted to improve electrostatic interactions with the N-terminal R of SpyTag003. These charge mutations increased reaction step by step to give the final SpyCatcher003 (SI Appendix, Fig. S2B). Sequences are compared in SI Appendix, Fig. S1A. We mapped the position of SpyCatcher003 mutations on to the structure of the original SpyCatcher (Fig. 1B).
The rate of isopeptide bond formation was analyzed by genetically fusing SpyCatcher003 to the N terminus of superfolder GFP (sfGFP) for reaction with SpyTag003-maltose binding protein (MBP). Fusion to sfGFP enabled the reaction to be monitored by fluorescence scanning of SDS/PAGE at protein concentrations (10 nM) too low for Coomassie staining since sfGFP can remain folded and fluorescent in the presence of SDS (25). Reaction of SpyTag003/SpyCatcher003 with each partner at 10 nM (Fig. 1C), 100 nM (Fig. 1D), and 10 µM (Fig. 1E) occurred substantially faster than for the previous generations, SpyTag/SpyCatcher (16) and SpyTag002/SpyCatcher002 (20). This improved reactivity is most striking at the lowest concentration tested (10 nM), where SpyTag003/SpyCatcher003 reaction went to >90% completion in 15 min, during which time only minimal SpyTag/SpyCatcher reaction occurred. The second-order rate constant for SpyTag003/SpyCatcher003 (SI Appendix, Fig. S1B) was 5.5 ± 0.6 × 105 M−1 s−1 (mean ± 1 SD, n = 3), ∼400-fold faster than the 1.4 × 103 M−1 s−1 previously shown for SpyTag/SpyCatcher (16), and 28-fold faster than the 2.0 × 104 M−1 s−1 for SpyTag002/SpyCatcher002 (20). The overall large improvement depended on a series of marginal gains at each mutational step (SI Appendix, Fig. S2). A sample gel for SpyTag003 reaction is shown in SI Appendix, Fig. S3A. Considering the size of the reacting species (45 kDa for SpyTag003-MBP and 31 kDa for SpyCatcher003-sfGFP), the rate of isopeptide formation for this 003 pair is now within the 105 to 106 M−1 s−1 range described as the onset for diffusion-controlled protein–protein interactions (19).
We further characterized that SpyTag003 and SpyCatcher003 were back-compatible with previous SpyTag/SpyCatcher generations (SI Appendix, Fig. S1 C and D). The enhanced reaction rate depended upon improvements in both Tag and Catcher reactivity (SI Appendix, Fig. S1 C and D).
Analysis of the SpyTag003/SpyCatcher003 System.
Electrospray ionization mass spectrometry (MS) confirmed the expected mass of SpyCatcher003 and of the SpyTag003:SpyCatcher003 adduct, showing loss of H2O upon isopeptide bond formation (SI Appendix, Fig. S4). To test for the maximum achievable conversion, we incubated SpyTag003-MBP and SpyCatcher003, when mixed with a small excess of the other component, and looked for unreacted product or side product using SDS/PAGE with Coomassie staining. This analysis showed >99% completion for SpyTag003 with SpyCatcher003 in excess. Similarly, we found >99% completion for SpyCatcher003 with SpyTag003 in excess (SI Appendix, Fig. S5). We were not able to detect side reactions (SI Appendix, Fig. S5) such as self-reactivity, which occurred with the original SpyCatcher but which was engineered out when SpyCatcher002 was developed (16, 20). Size exclusion chromatography-multiple angle light scattering (SEC-MALS), following Ni-NTA, showed that nearly all SpyCatcher003 eluted in a peak with a solution mass of 15.8 kDa, closely matching the predicted value of 15.6 kDa based on the protein’s sequence (SI Appendix, Fig. S6).
Differential scanning calorimetry (DSC) showed that SpyCatcher003 had a thermal stability (Tm) of 48.3 °C, close to that for SpyCatcher (48.8 °C) and SpyCatcher002 (48.4 °C) (SI Appendix, Fig. S7). The full width at half maximum (FWHM) for SpyCatcher003 (11.0 °C) was smaller than for either SpyCatcher (16.1 °C) or SpyCatcher002 (13.1 °C) (SI Appendix, Fig. S7), indicative of more cooperative protein unfolding. Thus, the improved reactivity of SpyCatcher003 has been achieved with minimal impact on thermal stability. A dramatic increase in Tm to 95.2 °C was seen upon reconstitution of the SpyTag003:SpyCatcher003 complex (SI Appendix, Fig. S7).
The initial noncovalent complex between SpyTag/SpyCatcher was previously estimated to be relatively weak, based upon isothermal titration calorimetry (ITC) on SpyCatcher interaction with a nonreactive SpyTag with the reactive Asp117 mutated to Ala (Kd = 200 nM) (16). We tested SpyTag003 DA-MBP binding to SpyCatcher003 by ITC. ITC showed that the initial binding was indeed improved, but the affinity was too tight to measure by ITC since the isotherm had a very high c value (SI Appendix, Fig. S8 A and B).
We took advantage of the strong noncovalent interaction between SpyTag003 and a nonreactive SpyCatcher variant (SpyDock) for affinity purification using the Spy&Go system (24). This enabled purification of SpyTag003 fusions from cell lysate with 92% purity (SI Appendix, Fig. S8C).
Dissecting Steps in Rapid Covalent Reaction.
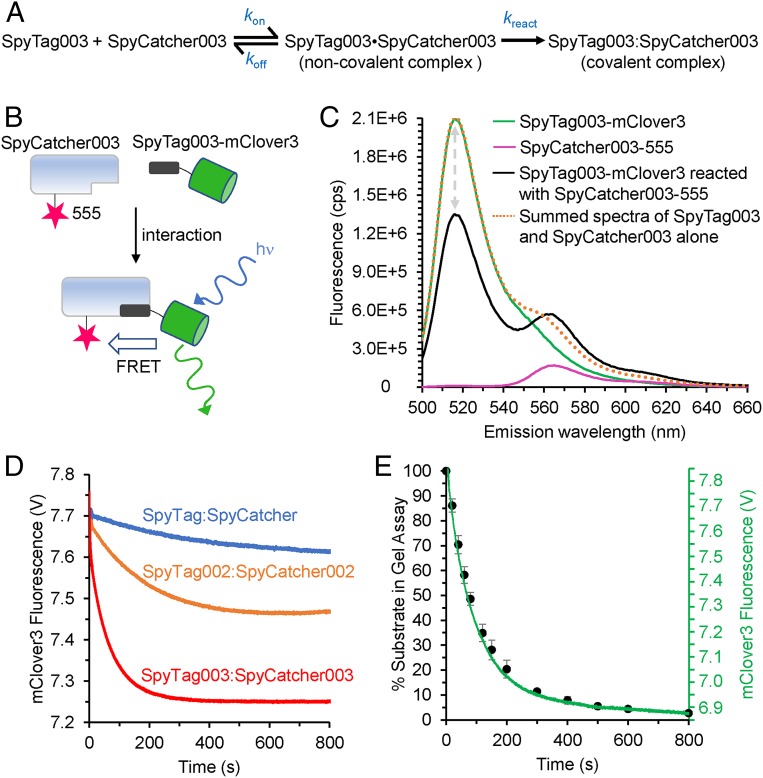
To investigate the process of peptide binding and reaction with its protein partner, we used stopped-flow Förster Resonance Energy Transfer (FRET) analysis. Fig. 2A shows the simplest model where SpyTag003 and SpyCatcher003 form an initial reversible noncovalent interaction (dot indicates a noncovalent complex) and then react to form an irreversible covalent complex (colon indicates a covalent complex). The donor for FRET studies was SpyTag003 linked to the bright green fluorescent protein mClover3 (Fig. 2B). We introduced a unique Cys into SpyCatcher003 through the S49C mutation (Fig. 1B) for labeling using Alexa Fluor 555-maleimide as the fluorescence acceptor. FRET upon interaction of these partners was best observed by the ∼35% decrease in the donor (mClover3) peak fluorescence intensity (Fig. 2C). Donor quenching was used to monitor the kinetics of SpyTag003-mClover3 binding and reacting with SpyCatcher003 under pseudofirst-order conditions (with SpyCatcher003 in excess). Comparison of quenching speed clearly showed that the original and 002 pair interacted much slower than the 003 pair (Fig. 2D). Further dissecting the time course of interaction of the 003 pair, the data were best fit by a triphasic quench (Fig. 2D and SI Appendix, Fig. S9A). This quenching consisted of a rapid first phase (kobs1 0.54 ± 0.03 s−1, mean ± 1 SD, n = 4 for 500 nM SpyCatcher003-555), increasing with protein concentration that accounted for ∼24% of the quench. In addition, there were 2 slower phases: kobs2 0.016 ± 0.001 s−1, mean ± 1 SD, n = 4 for 500 nM SpyCatcher003-555, 64% of the quench and kobs3 0.004 ± 0.0008 s−1, mean ± 1 SD, n = 4 for 500 nM SpyCatcher003-555, 12% of the quench. kobs2 was the spectroscopically dominant process and increased from 0.013 to 0.028 s−1 across the concentration range tested. With increasing pseudofirst-order concentrations of SpyCatcher003-555, the first phase showed linear concentration dependence and represented the bimolecular association of SpyTag003/SpyCatcher003 with an association rate constant (kon) of 6.2 ± 0.5 ×105 M−1s−1 (mean ± 1 SD, n = 4) (SI Appendix, Fig. S9B). This fluorescence change did not depend upon isopeptide bond formation, based upon repeating the experiment using nonreactive SpyTag003 DA-mClover3 with SpyCatcher003-555 (SI Appendix, Fig. S10 A and B). Here a rapid phase, this time spectroscopically dominant, with a similar rate constant (kon = 6.8 ± 0.5 ×105 M−1 s−1, mean ± 1 SD, n = 4) was observed. The subsequent steps (kobs2 ∼ 0.08 s−1 and kobs3 ∼ 0.004 s−1) did not have a detectable concentration dependence. The second-order rate constant for SpyTag003/SpyCatcher003 isopeptide bond formation approaches the value for kon, supporting that this reaction is close to diffusion-controlled. These experiments also revealed that the dissociation rate constant (koff) for the noncovalent SpyTag003•SpyCatcher003 complex was 0.26 ± 0.05 s−1 (mean ± 1 SD, n = 4), assuming that dissociation from SpyCatcher003 is faster than subsequent conformational changes (SI Appendix, Fig. S10B) (26). However, the koff for the SpyTag003 DA•SpyCatcher003 complex is at least 10-fold slower (koff = 0.03 ± 0.01 s−1) (SI Appendix, Fig. S10B).
Fig. 2.
Mechanistic analysis of enhanced binding and reaction. (A) Simplest kinetic scheme for the association and reaction of SpyTag003 with SpyCatcher003. (B) Scheme of FRET constructs to monitor binding and reaction. After excitation, mClover3 may emit fluorescence or transfer by FRET to Alexa Fluor 555. (C) Steady-state fluorescence spectra of FRET constructs. Emission spectrum upon excitation at 480 nm; SpyTag003-mClover3 and SpyCatcher003-555 were mixed for >1 h, to equilibrate before analysis. The gray arrows indicate the quenching of mClover3 in the presence of the acceptor. cps, counts per second. (D) Rapid-mixing fluorescence analysis for each Tag/Catcher pair. Quenching of mClover3 fluorescence was monitored upon mixing of 100 nM Tag-mClover3 with 400 nM Catcher-555. V, voltage. (E) Comparison for SpyTag003/SpyCatcher003 of stopped-flow fluorescence (green trace) with gel-based reaction (% unreacted SpyTag003) at various times after mixing (black circles, mean ± 1 SD, n = 3). Some error bars are too small to be visible.
To accompany the fluorescence analysis, we analyzed the kinetics of isopeptide bond formation between SpyCatcher003 and SpyTag003-sfGFP (Fig. 2E) under the same buffer, temperature, and protein concentrations as those used in the stopped-flow experiment in Fig. 2D (mClover3 had brighter fluorescence but was less resilient during in-gel analysis than sfGFP). This comparison showed a time course very similar to that of the 2 slow phases in the stopped-flow experiment, with a faster phase leading to the majority (∼80%) of the isopeptide bond formation, followed by a slower phase leading to the remainder of the bond formation. The presence of a minor and slowly reacting form likely explains why, even at high protein concentrations (i.e., 10 μM), SpyTag003/SpyCatcher003 constructs take up to 2 min to react to completion (Fig. 1E), far longer than expected from the fast isopeptide bond formation rate observed here (i.e., ∼12 s from extrapolation of kobs2). Since FRET depends on angle and distance, the fluorescence time courses indicate that the structure of the noncovalent SpyTag003-mClover3•SpyCatcher003-555 complex is likely to change upon isopeptide bond formation. Since isopeptide bond formation is a terminal process with only 1 bond per molecule and no further reaction possible, our data suggest that there are at least 2 conformations of the noncovalent SpyTag003-mClover3•SpyCatcher003-555 complex.
We performed stopped-flow fluorescence with SpyTag003 DA-mClover3 to gain further insight on the heterogeneity of the noncovalent complex with SpyCatcher003-555. Not only are the association kinetics described by more than 1 exponential (SI Appendix, Fig. S10 A and B), but so are the dissociation kinetics (SI Appendix, Fig. S10 C and D). Dissociation kinetics were measured by chasing a 10-fold excess of SpyTag003-MBP in to a preformed complex of SpyTag003 DA-mClover3•SpyCatcher003-555 and monitoring the loss of FRET as a result of SpyTag003 DA-mClover3 dissociation (SI Appendix, Fig. S10 C and D). The biphasic dissociation has rate constants of 0.009 ± 0.001 s−1 (90% of signal change) and 0.002 ± 0.0005 s−1 (10% of signal change) (mean ± 1 SD, n = 4). The presence of more than 1 step on dissociation is a consequence of more than 1 complex conformationally interchanging; thus, the complex has more than 1 dissociation route (27, 28). Hence, this analysis provides further support that the SpyTag003•SpyCatcher003 complex can adopt more than 1 conformation. The presence of such dynamics of the bound state of a complex has previously made calculation of kinetically derived Kd values challenging (28). However, with the 2 dissociation rate constants and the bimolecular value of kon (6.8 ± 0.5 ×105 M−1s−1), this analysis yields estimated Kd values of 3 and 13 nM for each version of the SpyTag003 DA-MBP•SpyCatcher003 complex.
DSC revealed a large increase (46 °C) in Tm of SpyCatcher003 upon reaction with SpyTag003 (SI Appendix, Fig. S7), consistent with a substantial structural rearrangement coupled to binding/reaction. The significant change in FRET coupled to the conversion of the initial bimolecular SpyTag003•SpyCatcher003 complex as it reacts to form the covalently linked SpyTag003:SpyCatcher003 complex is consistent with this structural change. Since the conformational changes occur with the same kinetics as the those of isopeptide bond formation (Fig. 2D), they are likely to be on-pathway induced fit-like conformational changes. An alternative mechanism of conformational selection is possible, involving off-pathway structural changes. Here SpyCatcher003, for example, slowly interconverts between a reactive and an unreactive conformer. The observed 2 slow phases (SI Appendix, Fig. S9A) make it unlikely that these phases can solely be explained by conformational selection. Conformational selection also cannot explain the dissociation of the SpyTag003 DA•SpyCatcher003 complex being biphasic (SI Appendix, Fig. S10 C and D), which is readily compatible with induced fit. Induced fit and conformational selection can be further distinguished by the number of phases when that partner is in excess (29). With induced fit, the same number of phases are observed with either partner in excess (since the bimolecular complex rearranges). However, the phases reporting on conformational selection disappear when the partner undergoing rearrangement is in excess since there is sufficient reactive conformer present. When we repeated the stopped flow in SI Appendix, Fig. S9 (SpyCatcher003-555 in excess), with excess SpyTag003-mClover, the presence of 3 phases is retained (SI Appendix, Fig. S11), suggesting that induced fit is occurring.
Comparison of Rapid Reaction Kinetics of the Different Peptide/Protein Generations.
To provide further insight into the molecular basis of the rate improvements in Fig. 1, we compared the rapid reaction kinetics of SpyTag003/SpyCatcher003 with those of SpyTag002/SpyCatcher002 and SpyTag/SpyCatcher (Fig. 2D). Only for SpyTag002/SpyCatcher002 was the initial rapid phase also of sufficient amplitude to accurately measure the association rate constant (for formation of the noncovalent complex) under the conditions tested. SpyTag002/SpyCatcher002 kon was 4.2 ± 0.5 × 105 M−1 s−1 (mean ± 1 SD, n = 4), only slightly slower than SpyTag003/SpyCatcher003 (6.2 ± 0.5 ×105 M−1 s−1). Instead, the most striking difference between the 3 generations was in the slow phases, where the structural changes are coupled to isopeptide bond formation. The relative pattern of the curves measured here with 100 nM SpyTag-mClover3 variant and 500 nM SpyCatcher-555 variant closely resembles that seen when isopeptide bond formation was followed in Fig. 1D (100 nM of each partner). Thus, the faster 003 reaction mainly arises from improvements in the speed of conformational changes coupled to isopeptide bond formation, rather than simply improving how the Tag and Catcher dock in the first place.
Hydrogen–Deuterium Exchange Analysis of Rapid Reaction of a Split Protein.
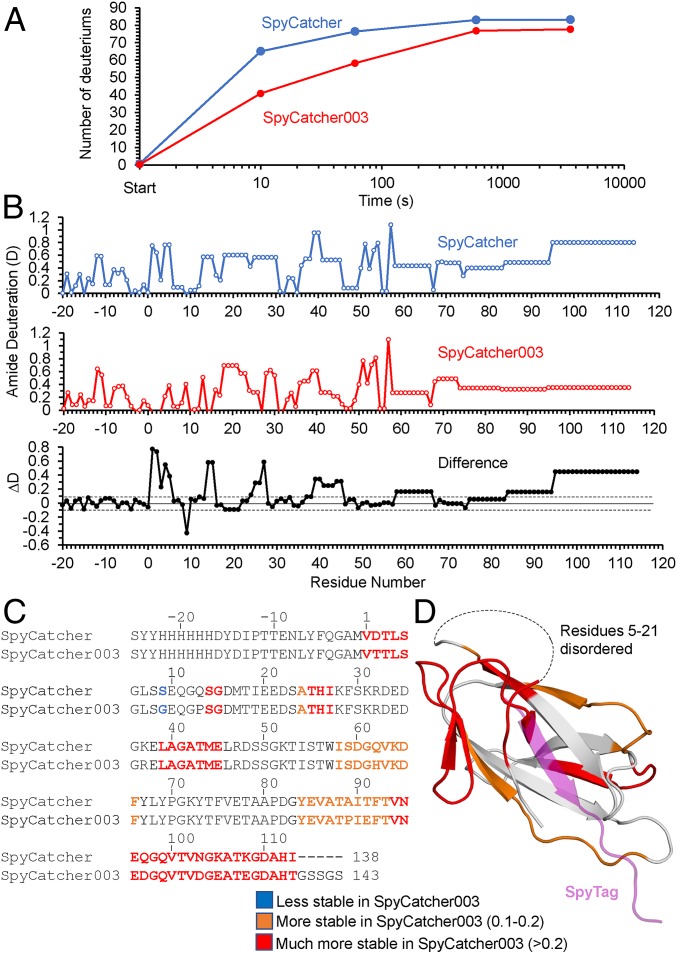
To investigate how the mutations affected the structural dynamics of the split proteins, Hydrogen–Deuterium exchange (HDX) experiments were carried out on SpyCatcher and SpyCatcher003. Here proteins are incubated for varying times in D2O at neutral pH, and the more exposed main-chain NH groups exchange their hydrogen with the solvent faster. Samples are then shifted to acidic pH and cooled, slowing exchange and facilitating subsequent analysis by MS. We focused on SpyCatcher and SpyCatcher003 since they were the start and end points for the development of improved reactivity. The time course for HDX was measured at 4 time points (10, 60, 600, and 3,600 s) for SpyCatcher and SpyCatcher003. The overall results are shown in Fig. 3A, with mass spectra shown in SI Appendix, Fig. S12. HDX makes clear that the structure of SpyCatcher003 is stabilized compared to SpyCatcher, with the biggest structural difference (17 deuterons) at 10 s. Hence, this time point was selected for high-resolution analysis. Online electron transfer dissociation (ETD) fragmentation of intact SpyCatcher variants (top-down approach) was used to characterize the exchange behavior at the amino acid level at the 10-s time point. The mass spectra of representative ETD fragments, along with the spectra of the intact proteins, are shown in SI Appendix, Fig. S13. Although the intact SpyCatcher003 shows a much smaller shift compared to intact SpyCatcher, ETD fragments from the first 22 N-terminal residues of the expressed proteins (c9, c17, and c22 in SI Appendix, Fig. S13) show very little difference. Since these are not part of the SpyCatcher proteins themselves but are N-terminal extensions included for purification purposes and have the same sequence for the 2 proteins, they were expected to have no structural differences between the proteins. In contrast, fragment c54 (SI Appendix, Fig. S13), which includes part of the SpyCatcher domain itself, shows a clear difference in the change of deuteration between SpyCatcher and SpyCatcher003. Thus, this smaller shift for SpyCatcher003 in the fragments as well as the intact proteins shows that the difference in HDX is specific to structural differences in the SpyCatcher domain.
Fig. 3.
HDX analysis of SpyCatcher and SpyCatcher003. (A) Time course of deuterium uptake by SpyCatcher (blue) and SpyCatcher003 (orange) after mixing with D2O at 25 °C, determined by mass spectrometry. (B) Combined top-down and middle-down MS analysis of deuterium uptake at the single-residue level for SpyCatcher (Top) and SpyCatcher003 (Middle). Bottom shows the difference in deuteration (ΔD) from SpyCatcher minus SpyCatcher003 values. The dotted lines represent ±0.1 errors in deuteration. (C) Difference in deuterium exchange from B compared with amino acid sequence for SpyCatcher and SpyCatcher003. Blue shows the 1 residue more stable in SpyCatcher than SpyCatcher003. Orange shows residues with 0.1 to 0.2 extra stability in SpyCatcher, and red shows those with >0.2 difference. (D) Difference in deuterium exchange mapped on the domain structure (based on PBD 2X5P), color-coded as in C. SpyTag residues are shown in purple.
We used a combined top-down and middle-down approach, involving digestion of the proteins under conditions that quenched any further HDX (30) to get the fullest coverage of the SpyCatcher variants. This approach was required because of poor ETD cleavage of peptides lacking basic residues. The combined results, representing the averages of 3 experiments, compare the amide deuteration levels for the individual residues in SpyCatcher and SpyCatcher003 (Fig. 3B). Since the uncertainties on the deuterium incorporation of each amide were within ±0.1 (with these boundaries shown in Fig. 3 B, Bottom), we classed only residues with a difference in amide deuteration level (ΔD) larger than 0.1 to be substantially different. In general, where there are differences in the amide deuteration levels, they are lower in SpyCatcher003, implying that there are localized structural differences in SpyCatcher003, possibly through improved local stability. The localized nature of the structural stabilization may explain the observation by DSC that although the Tm values are similar for SpyCatcher and SpyCatcher003, the FWHM is much smaller for SpyCatcher003 (SI Appendix, Fig. S7). Together these results suggest that SpyCatcher003 is a more stably folded protein than the original SpyCatcher.
To simplify interpretation of the current results, we then grouped the residues into 3 classes: 1) residues more stable in SpyCatcher than SpyCatcher003 (blue in Fig. 3 C and D), which comprised only 1 residue (at position 9 where Ser was mutated to Gly); 2) residues with a positive ΔD between 0.1 and 0.2 (a little more stable in SpyCatcher003), which were colored orange (residues 23, 57 to 65, and 83 to 92); and 3) residues with a positive ΔD greater than 0.2 (red in Fig. 3 C and D). The crystal structure of SpyTag:SpyCatcher is disordered for residues 1 to 21 and 103 to 113 (21). Therefore, for structural mapping of the residues, we used the crystal structure of the parental CnaB2 domain truncated at the end of the SpyCatcher domain (residue 113) with the position of the SpyTag overlaid (colored purple) (Fig. 3D). This mapping showed that the structural stabilization is clustered in 2 areas. The more weakly stabilized area is clustered around the C terminus of SpyTag and involves loops of the SpyCatcher that make key interactions with SpyTag (Fig. 3D). Specifically, these are residues immediately adjacent to K37 (mutated to R37 in SpyCatcher003) and include L39 and the loop involving Y84 (the end of this loop has the A89P mutation in SpyCatcher003), which together with Y119 of SpyTag make a network of nonpolar and polar interactions (SI Appendix, Fig. S14A). The more strongly stabilized area involves residues at SpyCatcher’s N terminus (residues 1 to 4, 13 to 14, and 24 to 27) and C terminus (residues 90 to 113), which become colocalized in the folded protein (Fig. 3D) and are the regions disordered in the SpyCatcher crystal structure. I90 and F92 make key nonpolar interactions with I113 and M115 of SpyTag. I90 and F92 are also adjacent to A89 and T91 that are mutated in SpyCatcher003 (SI Appendix, Fig. S14A). This also includes the region containing mutations (Q97D, N103D, K105E, and K108E) involved in the rate enhancement of SpyCatcher to SpyCatcher003 (Fig. 1B) as well as regions (24–27) close to where the N-terminal mutations in SpyTag003 (especially the N-terminal R108) are likely to interact. We compared the HDX differences with the CnaB2 domain B factors, showing that the regions most stabilized by the SpyCatcher003 mutations show some colocalization with the most flexible regions of SpyCatcher (SI Appendix, Fig. S14 B and C). Thus, we suggest that partial prerigidification of SpyCatcher003 reduces the entropic penalty to Tag binding, while favoring productive conformational changes during reaction with SpyTag.
Rapid Reaction of Mammalian Cells Displaying SpyCatcher003.
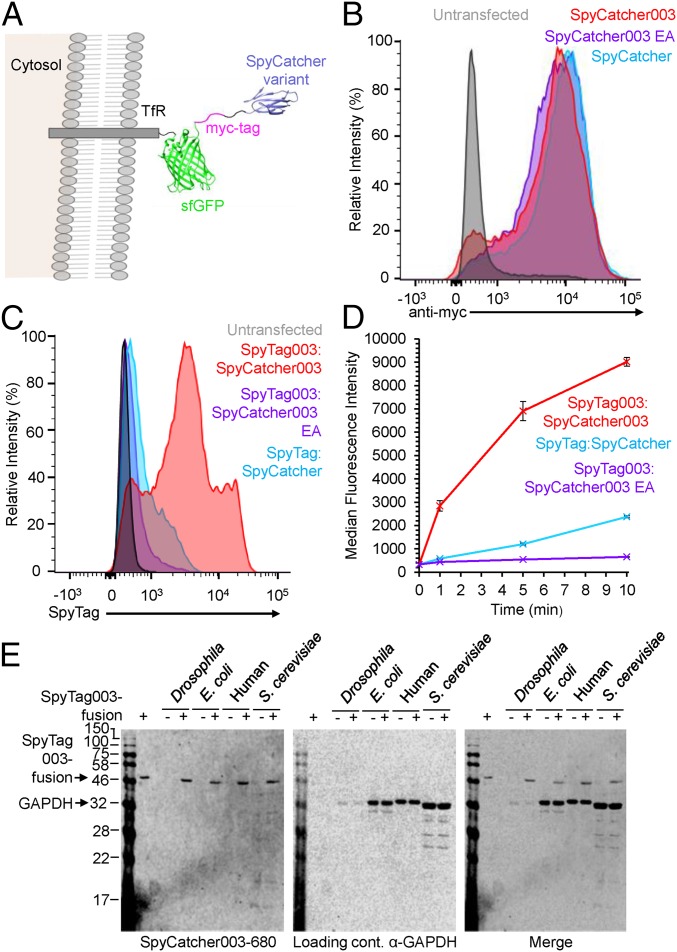
Having established rapid reaction of the 003 pair as purified proteins, we tested their performance in a cellular context. We displayed SpyCatcher003 on the plasma membrane of human Expi293 cells using a transferrin receptor-sfGFP-myc tag fusion (Fig. 4A). Plasma membrane display of the construct was tested by staining for the myc tag by flow cytometry. As a control for the specificity of the SpyTag003/SpyCatcher003 reaction, we analyzed the SpyCatcher003 E77A mutant, preventing reaction with SpyTag constructs (24). Sample dot-plots are shown in SI Appendix, Fig. S15. We found similar strong cell surface expression of SpyCatcher, SpyCatcher003, and SpyCatcher003 EA (Fig. 4B). The interaction of cell surface constructs with SpyTag variants fused to the red fluorescent protein mKate2 was analyzed by flow cytometry (Fig. 4C). SpyTag/SpyCatcher gave a signal only just above background and substantially less than with SpyTag003/SpyCatcher003 (Fig. 4C). We analyzed the speed of this staining by quenching the reaction at different time points with excess nonfluorescent SpyCatcher003, that acts to compete off noncovalently bound SpyTag variants from the cell. Clear staining of SpyCatcher003 was found at the plasma membrane after only 1 min and occurred far more rapidly than for SpyTag/SpyCatcher (Fig. 4D and SI Appendix, Fig. S15). Although SpyCatcher003 EA expressed at the cell surface at similar levels to SpyCatcher003 (Fig. 4B), minimal increase in SpyTag003-mKate2 was observed over time (Fig. 4 C and D) supporting that specific interaction of SpyTag003 and SpyCatcher003 occurred in the cell surface context.
Fig. 4.
Rapid cell surface labeling with 003 pair. (A) Construct for cell surface display of SpyCatcher variants, with Transferrin Receptor (TfR) cytosolic region and transmembrane-helix, sfGFP (PDB 2B3P), a myc-tag, and the SpyCatcher variant (PDB 4MLI). (B) Similar surface expression of different SpyCatcher variants. Expi293 cells were untransfected or transfected with the construct in A bearing SpyCatcher, SpyCatcher003, or SpyCatcher003 EA. Cells were stained for surface expression with an antibody to the myc tag for flow cytometry. (C) Enhanced reactivity of surface SpyCatcher003. SpyCatcher-expressing cells were reacted for 1 min with SpyTag-mKate2, while SpyCatcher003 or SpyCatcher003 EA cells were reacted with SpyTag003-mKate2 for 1 min before flow cytometry. (D) Quantification of surface staining by flow cytometry, after incubation for various times with SpyTag variants as in C (mean ± 1 SD, n = 3). Some error bars are too small to be visible. (E) Testing SpyCatcher003 specificity by Western blotting. Cell lysate from Drosophila melanogaster, Escherichia coli, human Expi293 cells, or Saccharomyces cerevisiae was blotted simultaneously with SpyCatcher003-680 (Left) and anti-GAPDH as a loading control (detected with a secondary antibody at a distinct fluorescence wavelength; Center). The merge of these 2 signals is shown in Right. In + lanes, 3 pmol SpyTag003-MBPa was doped into the lysate as a positive control.
Application of SpyCatcher003 to Western Blotting.
To test further the specificity of SpyTag003:SpyCatcher003 interaction, we used Western blotting against lysates from a range of common model organisms. SpyCatcher003 S49C was site-specifically labeled using a maleimide linked to the near-infrared fluorophore DyLight680. We blotted against cell lysate from Escherichia coli, human cells (the Expi293 cell-line), Saccharomyces cerevisiae, and Drosophila melanogaster. As a positive control, each of these cell lysates was doped with a low amount (3 pmol) of a SpyTag003-fusion (31). For each species we saw efficient recognition of the SpyTag003-fusion and minimal cross-reactivity with endogenous cellular proteins (Fig. 4E). The anti-glyceraldehyde-3-phosphate dehydrogenase (α-GAPDH) loading control showed some differential binding to the GADPH between species but equivalent staining with or without the SpyTag003-fusion (Fig. 4E).
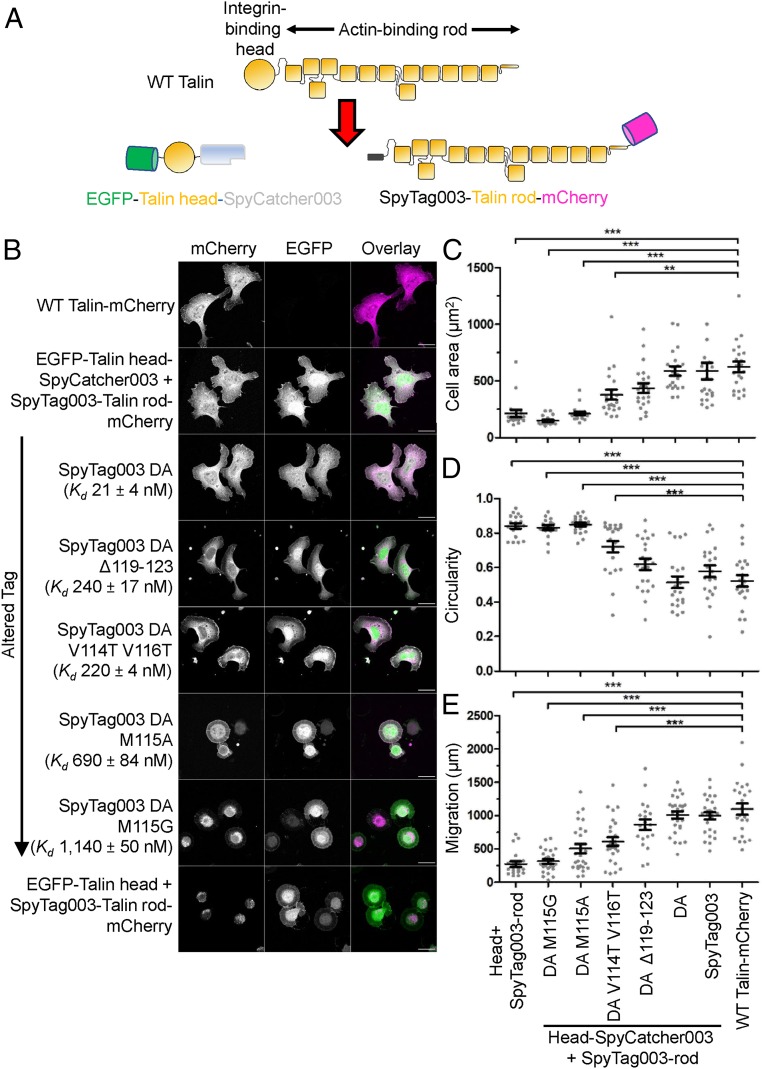
Intracellular Coupling of Split Talin Fragments Reconstitutes Cell Spreading and Mobility.
To test the application of SpyTag003/SpyCatcher003 in the mammalian cytoplasm, we explored whether they can be used to reform a split protein. Rather than using a split-protein biosensor such as split-dihydrofolate reductase or split-GFP, we chose to split a protein where there was an important functional cellular phenotype and mechanical force transduced through the protein. Talin is a scaffold protein in integrin-mediated cell adhesion, bridging the β-integrin tail domains with the actin cytoskeleton, being required for the mechanical connection between a cell and its surroundings (32–35). Talin also interacts with at least 15 adhesion proteins and coordinates their recruitment and release from the adhesion complex (32, 36). Many of these interactions are regulated by the mechanical forces transmitted through the talin protein, making talin a finely regulated cell adhesion mechanosensor (37). Because of the critical role of talin in the regulation of adhesion complex function, alterations in talin’s structural properties can dramatically affect adhesion complex formation, cell attachment, and cell migration (32, 33). To test the recoupling ability, talin was split into its β-integrin–binding head region and actin-binding rod region, with each part genetically fused with either SpyCatcher003 (EGFP-Talin head-SpyCatcher003) or SpyTag003 (SpyTag003-Talin rod-mCherry) (Fig. 5A). We used a mouse embryonic fibroblast (MEF) talin-knock-out strain (deleting both of the talins, 1 and 2) (32, 38) with confocal fluorescence microscopy, to investigate the phenotypes induced by various expression constructs. Talin knock-out cells are rounded and immobile, unable to achieve functional adhesion to the extracellular matrix (32, 38). As a negative control, we cotransfected EGFP-Talin head and SpyTag003-Talin rod-mCherry, which still led to rounded cells (Fig. 5B). However, cells cotransfected with EGFP-Talin head-SpyCatcher003 and SpyTag003-Talin rod-mCherry were restored in their adhesion, producing well-formed lamellipodia similar to cells transfected with WT Talin-mCherry (Fig. 5B). We quantified cell adhesion in terms of cell area and observed statistically significant increase in cell area from the split talin allowing SpyTag003/SpyCatcher003 interaction (P < 0.001, ANOVA with Bonferroni correction, from 17 to 21 cells per group) (Fig. 5C). To understand further the cell morphology, we also quantified cell circularity, we also quantified cell circularity, which showed that SpyTag003/SpyCatcher003 increased cell polarization (Fig. 5D). The knock-out MEFs have no coordinated cell migration, but efficient motility was restored in cells coexpressing the SpyTag003 and SpyCatcher003 split-talin constructs (Fig. 5E).
Fig. 5.
SpyTag003/SpyCatcher003 reconstitution of cell adhesion. (A) Schematic of talin constructs. (B) SpyTag003/SpyCatcher003 reconstituted cell adhesion. Confocal microscopy (maximum intensity projection of z stack) of talin knock-out mouse embryonic fibroblasts transfected with different talin constructs. mCherry is shown in grayscale in Left and magenta in the overlay. EGFP is shown in grayscale in Center and green in the overlay. Magenta/green overlay is displayed as white in Right. (Scale bar, 20 µm.) EGFP-Talin head-SpyCatcher003 was cotransfected with SpyTag003 DA variant-Talin rod-mCherry for rows marked with an arrow; Kd for DA variants binding SpyCatcher003 is shown as mean ± 1 SD, n = 3. (C) Spreading of cells. Quantification of area by microscopy, with each analyzed cell as a dot (mean ± 1 SE, n = 17 to 21 cells from 2 independent experiments). (D) Quantification of cell circularity by microscopy with each cell as a dot (mean ± 1 SE, n = 17 to 21 cells, from 2 independent experiments). (E) Quantification of cell migration in 7 h by microscopy with each cell as a dot (mean ± 1 SE, n = 20 to 28 cells from 2 independent experiments). ***P < 0.001, **P < 0.01.
To test whether Tag/Catcher covalent bond formation was necessary for restored talin function, we tested the nonreactive SpyTag003 DA mutant. Indeed SpyTag003 DA was capable of restoring adhesion and motility similar to WT Talin (Fig. 5 B–E). SpyTag003 DA:SpyCatcher003 still represents a high-affinity interaction: its affinity was too strong to measure by ITC (SI Appendix, Fig. S8B), so we used surface plasmon resonance (SPR) to obtain a Kd = 21 ± 4 nM (SI Appendix, Fig. S16). To further dissect the importance of this interface in talin reconstitution, we then established a series of SpyTag003 DA variants, varying 50-fold in affinity for SpyCatcher003 (SI Appendix, Fig. S16). Peptide mutants with ∼200 nM affinity gave a partial rescue of cell adhesion and motility, whereas mutants with 690 or 1,140 nM affinity gave minimal restoration (Fig. 5 B–E). These findings demonstrate the application of 003 components for assembly of proteins in living cells, generating a complex able to support integrin/talin-mediated mechanotransmission. The experiments also provide a panel of peptides for binding a given protein (SpyCatcher003) with either covalent or broadly tunable noncovalent interaction.
Discussion
We have established a peptide–protein interaction approaching infinite affinity through optimization of both docking and reaction of a split protein. Amide bond formation between an amine and a carboxylic acid here occurs without any activating groups, and half-time for reaction is less than 30 s with 10 µM of each partner. However, even with each partner at a thousand-fold lower concentration, the reaction half-time is less than 5 min. Therefore, this covalent reactivity can be harnessed for the study of proteins over a range of cellular concentrations. There is limited information on the rate of intramolecular isopeptide bond formation in parent Gram-positive adhesins because of the difficulty in separating folding from reaction. Thus, this rate provides insight into the potential speed of natural spontaneous isopeptide formation (13). Other important features of our reactive pair include the absence of Cys in either partner, lack of side reactions, and specificity in a range of cellular systems. Previous iterations have shown broad tolerance of reaction conditions and versatility in the sites of fusion (16, 20, 39).
Interestingly, the 003 mutations were able to improve both the noncovalent association rate and the rate of reaction following this noncovalent docking. Stopped-flow fluorescence illuminated the complexity of the split protein reconstitution, with multiple states and different pathways to reaction. Mutations were distributed around the structure, and all were well separated from the site of reaction. HDX showed that SpyCatcher003 was less dynamic than SpyCatcher at more than 50 of its residues, indicating a widespread reduction in flexibility as split protein reconstitution was accelerated. These areas of flexibility were not well correlated to high B factors in the parent domain structure, emphasizing the importance of structural analysis by HDX of the dynamic Catcher portion. Nevertheless, the cooperative unfolding transition by DSC was similar for SpyCatcher and SpyCatcher003. This suggests that the increased dynamics in SpyCatcher are distinct from the structural transitions which would lead to complete unfolding of the domain, indicating how Tm alone gives a limited picture of conformational flexibility.
A range of elegant chemistries have been used for amide bond formation to connect proteins (1). The 2 most widely used methods are sortase or split intein coupling. Sortase coupling has the advantage of small recognition peptides, although application to low-abundance proteins has been limited by the high micromolar Km and the range of peptides able to act as N-terminal substrates (4, 40). The recently engineered sortase CdSrtA3M allows coupling to internal lysines and achieved >95% coupling at 24 h with 100 μM enzyme and 300 μM of each substrate (41). Any approach requiring collision of 3 components for reaction will struggle to reach the same efficiency as one requiring only bimolecular interaction. Along these lines, peptide partners to be ligated by isopeptide bond formation needed to be present at 10 µM for SpyLigase (42), and Km values were ∼10 µM for SnoopLigase (43). Split intein coupling has the advantage of being potentially traceless, but the latest accelerated inteins were analyzed using ∼10 µM of each partner and do not proceed to quantitative yield (5). Therefore, the efficient reaction we see for SpyTag003/SpyCatcher003 in 10 min using 1,000-fold lower concentrations may assist ligation in diverse experimental situations.
SpyTag/SpyCatcher has been referred to as a genetically encodable click chemistry (44), so it is helpful to compare the rate constant for SpyTag003/SpyCatcher003 (5.5 × 105 M−1 s−1) to widely used chemical reactions. The prototypic click reaction, CuI-catalyzed reaction of an azide with an alkyne, has a typical rate constant of 10 to 100 M−1 s−1 (45). Strain-promoted alkyne-azide cycloadditions have a rate constant of ∼10−3 to 1 M−1 s−1 (46). Fast rates have been obtained by inverse electron demand Diels–Alder reactions between tetrazine and transcyclooctene (1 to 106 M−1 s−1), but challenges have been faced in the stability of the fastest reacting variants to storage or side reaction (45), apart from the logistical challenge of incorporating these 2 groups in proteins of interest (47, 48). Native chemical ligation may allow traceless protein coupling (49), but the rate constant is ∼10−1 M−1 s−1 (50). The Tag/Catcher system overcomes the low intrinsic reactivity of the reacting species (amine and carboxylic acid) through a large interaction surface to drive apposition and an environment with shuffling of protons to promote reaction (15). For all protein ligation systems, this study should point to how to evaluate progress on the path to infinite affinity. This work also supports the use of SpyTag003 to enhance the range of applications of a fused protein. Proteins linked to SpyTag and its variants are empowered for Spy&Go affinity purification (24), irreversible anchoring (on magnetic beads or biomaterials) (17, 42, 51), or oligomerization (on coiled-coils or vaccine scaffolds) (24, 48). SpyTag fusion also facilitates multiplexing of function, such as modular linkage of antibodies to fluorophores, toxins, or enzymes, or assembly of polyproteams to stimulate synergistic cell signaling (52, 53). SpyTag variants additionally empower proteins for mechanical analysis by atomic force microscopy (16), optical tweezers (54), or magnetic tweezers (55). Taking Spy interaction toward infinite affinity through this work will further increase these opportunities to extend protein function.
Methods
Plasmids and cloning, protein expression and purification, SEC-MALS, Electrospray Ionization MS, ITC, DSC, SPR, fluorophore conjugation, Western blotting, Spy&Go, and structure visualization are described in SI Appendix, SI Methods.
Isopeptide Bond Formation Assays.
Reactions were carried out at pH 7.0 and 25 °C in succinate–phosphate–glycine buffer (12.5 mM succinic acid, 43.75 mM NaH2PO4, 43.75 mM glycine; pH adjusted to 7.0 using NaOH) (20). Reactions were analyzed by SDS/PAGE on 16% polyacrylamide gels using the XCell SureLock system (Thermo Fisher) at 180 V. The reaction was quenched at 50 °C for 5 min after addition of 6× SDS-loading buffer (0.23 M Tris⋅HCl, pH 6.8, 24% [vol/vol] glycerol, 120 µM bromophenol blue, 0.23 M SDS) in a Bio-Rad C1000 thermal cycler to retain the fluorescence of sfGFP. sfGFP fluorescence in gels was quantified using a Fluorescent Image Analyzer FLA-3000 (FujiFilm) and ImageGauge version 5.21 software.
For measuring the concentration-dependent rates (Fig. 1 and SI Appendix, Fig. S1B), SpyCatcher-sfGFP, SpyCatcher002-sfGFP, and SpyCatcher003-sfGFP were reacted with SpyTag-MBP, SpyTag002-MBP, or SpyTag003-MBP with both partners at 10 nM, 100 nM, or 10 µM. sfGFP fusions enabled reactions to be analyzed at concentrations as low as 10 nM. Percentage isopeptide product formation was calculated by dividing the intensity of the band for the covalent complex by the intensity of all of the bands in the lane and multiplying by 100. In order to correct for differential photobleaching of the sfGFP at different time points, the second-order rate constant for covalent complex formation was determined by monitoring the reduction in the relative intensity of the band for the SpyCatcher-variant-sfGFP to give the change in the concentration of the unreacted SpyCatcher-variant-sfGFP. Time points were analyzed during the linear portion of the reaction curve. 1/[SpyCatcher variant] was plotted against time and analyzed by linear regression using Excel (Microsoft) and Origin 2015 (OriginLab Corporation), including calculation of the SD for the best fit and the square of the correlation coefficient (R2) for line of best fit to the data. Experiments were carried out in triplicate, and the data represent the mean ± 1 SD.
For measuring the reaction of 100 nM SpyCatcher003 with 400 nM SpyTag003-sfGFP in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4), pH 7.4 (Fig. 2E), at 25 °C, experiments were carried out as described above, but the data were plotted as percent SpyTag003-sfGFP remaining. sfGFP was used in place of mClover3 because of sfGFP’s improved resilience, required for SDS/PAGE.
For measuring the completion of SpyCatcher003 and SpyTag003-MBP reaction (SI Appendix, Fig. S5), experiments were performed in succinate–phosphate–glycine buffer at pH 7.0 for 1.5 h at 25 °C, with one partner at 2 µM and the other partner at 2 or 4 µM as indicated. The reaction was quenched at 95 °C for 5 min after addition of 6× SDS-loading buffer in a Bio-Rad C1000 thermal cycler. The gel was stained with InstantBlue (Expedeon) and analyzed with Gel Doc XR imager and Image Lab 5.2 software (Bio-Rad). Percentage completion was determined from the depletion in the band intensity of SpyCatcher003 in the presence of excess SpyTag003-MBP or the depletion in the band intensity of SpyTag003-MBP in the presence of excess SpyCatcher003.
Steady-State Fluorescence.
Spectra were collected at 25 °C in PBS, pH 7.4, using a Horiba-Yvon Fluromax 4 with an excitation wavelength of 480 nm and fluorescence emission collected between 500 and 660 nm using a monochromator. SpyTag003-mClover3 and SpyCatcher003-555 were each individually measured at a concentration of 200 nM. The spectrum of the SpyTag003-mClover3:SpyCatcher003-555 complex was collected after mixing together 200 nM of each protein for at least 1 h at 25 °C.
Stopped-Flow Fluorescence.
Experiments were carried out using an Applied Photophysics SX20 stopped-flow spectrofluorimeter in 1:1 mixing mode at 25 °C in PBS, pH 7.4, using an excitation wavelength of 480 nm, and all fluorescence was monitored above 515 nm through the use of a 515-nm long-pass filter and 2.3-nm slit widths. For association experiments, the final concentration of the SpyTag variant-mClover3 was 100 nM with increasing pseudofirst-order concentrations of SpyCatcher variant-555 from 400 to 1,000 nM.
Data were analyzed using a combination of ProDataSX (Applied Photophysics), Excel (Microsoft), and Origin 2015. Experimental data were fitted to single (Eq. 1), double (Eq. 2), or triple (Eq. 3) exponential equations:
| [1] |
| [2] |
| [3] |
where F is the observed fluorescence; ΔFn is the fluorescence amplitude for the nth step; kobsn is the observed pseudofirst-order rate constant for the nth step, which is equivalent to the inverse relaxation time (1/τ); and Fe is the end-point fluorescence. Fitting of the linear concentration dependence of kobs1 to Eq. 4 yields the bimolecular association rate constant (kon), under the conditions that koff > kforward for subsequent conformational changes:
| [4] |
In the dissociation experiments (SI Appendix, Fig. S10), 400 nM SpyTag003 DA-mClover3 and 500 nM SpyCatcher003-555 in PBS, pH 7.4, were mixed for at least 1 h at 25 °C. Then 6 µM SpyTag003-MBP was added, and fluorescence was monitored at 25 °C. The resulting biphasic fluorescence enhancement was fitted to Eq. 2. R2 was calculated in Excel.
HDX.
HDX samples were prepared by mixing SpyCatcher variants (150 μM in PBS, pH 7.4) with D2O (Cambridge Isotope Laboratories) at a ratio of 2:8 (vol/vol), and the resulting solutions were incubated at 25 °C; 20 µL aliquots were taken out at 10 s, 1 min, 10 min, and 1 h and were quickly quenched by adding 50 mM sodium dihydrogen orthophosphate, pH 2.4 (pH had been adjusted using phosphoric acid). These samples were flash-frozen in liquid nitrogen and stored at −80 °C. For middle-down liquid chromatography-mass spectrometry (LC-MS) experiments, the protein aliquots were quickly thawed and digested at 0 °C with 3 mg/mL pepsin (Sigma-Aldrich) for 1 min, and the sample was injected into the HPLC flow so that the enzyme and peptides/protein were separated. This step also stopped the digestion (56). No pepsin was added for top-down experiments.
In the top-down approach, SpyCatcher proteins were analyzed by LC-MS using a C4 analytical column (2.0 mm × 30 mm, Phenomenex). Protein elution was conducted at −20 °C using the subzero temperature technology developed previously, which can reduce the back-exchange to as low as 2% (30). The Ultra-Performance Liquid Chromatography system was coupled to an Orbitrap Fusion mass spectrometer equipped with ETD capability (Thermo Fisher). The instrumental parameters for the Orbitrap were spray voltage 3,500 V (positive), transfer tube temperature 275 °C, vaporizer temperature 275 °C, sheath gas 25, auxiliary gas 10, S-lens radiofrequency 60. The automatic gain control target was set at 2 × 105, while the ETD reagent target value was 3 × 105. Online ETD experiments were done by selecting 1 charge state (16+) of the SpyCatcher variants in 1 LC-MS run, with an isolation window of 5 m/z units in the quadrupole. The column, accessories, injector, and extensively coiled solvent delivery lines were embedded in an ice bucket to minimize H/D back-exchange. The syringe used for injection was chilled on ice as well. The mobile phase was 0.1% (vol/vol) formic acid (A) and 99.9% (vol/vol) acetonitrile/0.1% (vol/vol) formic acid (B). The proteins were eluted by 15 to 50% B in a 10-min binary solvent gradient with a flow rate of 200 µL/min. Detection of the intact proteins in the LC-MS experiments was performed over an m/z range of 300 to 2,000. In the ETD experiments, fluoranthene radical anions (Thermo Fisher) were introduced into the ion trap over 50 ms, and the ETD reaction time was 10 ms. ETD fragment ions were detected in the Orbitrap using a scan range of 150 to 2,000. In the peptide-based bottom-up LC-MS experiments, peptides were separated by 5 to 70% B in a 12-min gradient. The MS survey scan was carried out with a mass resolution of 120,000 FWHM. The Orbitrap detection was calibrated to be <3 parts per million (ppm) error using Pierce FlexMix Calibration Solution (Thermo Fisher) (30).
LC-MS and ETD data were processed using Xcalibur 2.0 software (Thermo Fisher), and the generated ETD peak lists were searched against the sequence of SpyCatcher or SpyCatcher003 using Protein Prospector (http://prospector.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msproduct) with a 10-ppm fragment tolerance. Matched ions were also checked by manual inspection. Peptide LC-MS/MS data were searched against a custom-generated FASTA database which contained the targeted protein sequences. The mass shift of the peptides and intact proteins and the deuteration status of individual amides were determined based on their centroid m/z values before and after H/D exchange, according to the method developed previously (30, 57). All HDX data were normalized to 100% D2O content (80% D exchange-in buffer for all of the time points). Percent deuterium incorporation values were obtained by comparing the number of acquired deuteriums to the total number of amide hydrogens contained in the peptide/protein.
Mammalian Cell Expression of SpyCatcher.
TfR-sfGFP-myc tag-SpyCatcher variants were expressed in suspension Expi293 cells (Thermo Fisher) cultured in Expi293 Expression media (Thermo Fisher) supplemented with 50 U/mL penicillin/streptomycin (Thermo Fisher). Cells were grown in a humidified Multitron Cell incubator (Infors HT) at 37 °C with 7% CO2, rotating at 110 to 125 rpm. Cells at a density of 3.0 × 106 cells/mL were transfected with 2.7 μL ExpiFectamine 293 Reagent per 1 μg of plasmid DNA. ExpiFectamine transfection enhancers (Thermo Fisher) were added 16 to 22 h after transfection. Cells were grown for 48 h and then analyzed.
Flow Cytometry.
Cells were washed twice in FACS buffer (PBS, pH 7.5, 1 mM EDTA, 1% BSA, 0.1% [wt/vol] sodium azide) by centrifugation at 300 g at 4 °C for 3 min. For labeling cells with anti-myc-Alexa Fluor 647 antibody (Thermo Fisher), 0.5 to 1 × 106 cells were incubated with the antibody at 5 μg/mL for 20 min on ice in FACS buffer, followed by washing twice in FACS buffer; 20 nM SpyTag variant-mKate2 was incubated with 0.5 to 1 × 106 cells in 2 mL FACS buffer on ice for 1, 5 or 10 min. Reaction was stopped by adding 10 µM unlabeled SpyCatcher003, followed by washing the cells twice in FACS buffer. SI Appendix, Fig. S15B, represents cells stained for 1 min. Cells were maintained at 4 °C before analysis. Cells were analyzed on a BD Fortessa X20, gating on live cells using forward-scatter, side-scatter, and DAPI staining. Settings were 405 nm laser and 450/50 nm emission filter for DAPI, 488 nm laser and 530/30 nm emission filter for sfGFP, 561 nm laser and 610/20 nm emission filter for mKate2, and 640 nm laser and 670/30 nm emission filter for Alexa Fluor 647. Data were analyzed using FlowJo version 9.0. Experiments were carried out in triplicate. Data represent the mean ± 1 SD.
Immunostaining and Microscopy for Talin Cell Experiments.
The Tln1−/−Tln2−/− MEF cell line has been previously described (38). Cells were maintained in high-glucose Dulbecco’s Modified Eagle Medium supplemented with 10% (vol/vol) FBS and 1% (vol/vol) GlutaMax (Thermo Fisher) in a 37 °C, 5% CO2 incubator. Tln1−/−Tln2−/− MEF cells were transfected with expression constructs using the Neon Transfection System electroporator (Thermo Fisher) according to the manufacturer’s instructions. Cells were plated on Zeiss high-performance 170-µm-thick coverslips coated with 10 µg/mL sterile-filtered fibronectin (purified from human plasma using a gelatin-affinity column) in PBS, pH 7.4, at 37 °C for 1 h and washed with PBS, pH 7.4. After 24 h, nontransfected cells were removed by washing twice with PBS, pH 7.4, and cells were either used for time-lapse imaging or fixed with 4% (wt/vol) paraformaldehyde in PBS, pH 7.4, for 20 min at 25 °C. Fixed cells were washed twice with PBS, pH 7.4; mounted with ProLong Diamond (Thermo Fisher) with DAPI; and stored at 4 °C until imaging. Cells were imaged with 25× or 63× oil immersion objectives and a LSM780 or LSM800 confocal unit (Zeiss); 488- and 568-nm lasers were used for exciting EGFP and mCherry. Emission filters were 499 to 579 nm for EGFP and 585 to 712 nm for mCherry. Zeiss Zen Black software, ImageJ 1.50e, and FIJI were used in image analysis (58). Quantification was only performed on transfected cells, showing EGFP and/or mCherry signal above background.
Time-lapse images were taken with Cell IQ (CM technologies) equipped with 37 °C and 5% CO2 incubator, with 20× objective, for 20 h and with 8-min intervals. The resulting image stacks were analyzed with ImageJ (58) and MTrackJ plugin (59). Cell morphology was analyzed from the bright-field image by cell area (a region of interest was drawn manually on the cell boundaries). Circularity was analyzed by 4π × [Area]/[Perimeter]2, with a value of 1.0 indicating a perfect circle and lower values indicating an increasingly complex shape. Imaging parameters were kept constant for all samples within each experiment. Statistical analysis was performed by ANOVA with Bonferroni correction in GraphPad Prism 5. Area and circularity were quantified from 17 to 21 cells per condition from 2 independent experiments. Migration was determined from 20 to 28 cells per condition from 2 independent experiments.
Data Availability.
Amino acid sequences of each version of SpyTag and SpyCatcher are available in SI Appendix, Fig. S1A. Amino acid sequences of other constructs are available in GenBank as described in SI Appendix, SI Methods, Plasmids and Cloning. Plasmids encoding pDEST14 SpyCatcher003, pJ404-SpyCatcher003-sfGFP, pDEST14-SpyCatcher003 S49C, pET28a-SpyTag003-MBP, pENTR4-TfR-sfGFP-myc tag-SpyCatcher003, pET28-SpyTag003-mKate2, pET28-SpyTag003-sfGFP, pET28-SpyTag003-mClover3, pEGFP-C1 EGFP-Talin head-SpyCatcher003, and pEGFP-C1 SpyTag003-Talin rod-mCherry were deposited in the Addgene repository (https://www.addgene.org/Mark_Howarth/). Further information and request for resources and reagents should be directed to and will be fulfilled by the lead contact, M.H.
Supplementary Material
Acknowledgments
Funding was provided by the European Research Council (ERC-2013-CoG 615945-PeptidePadlock) (A.H.K., S.S., and M.H.). I.N.A.K.A. was funded by Yayasan Khazanah, Oxford Centre for Islamic Studies, and St. John’s College Oxford. We thank Dr. David Staunton of the University of Oxford Department of Biochemistry Biophysical Suite and Dr. Michal Maj of the Sir William Dunn School of Pathology Flow Cytometry Facility for assistance. We acknowledge Dr. Anthony Tumber of the University of Oxford Department of Chemistry for assistance with MS, supported by the Biotechnology and Biological Sciences Research Council (grant BB/R000344/1). We thank Dr. Jingxi Pan (NovoAb Bioanalytics Inc.) for assistance with HDX. We acknowledge Academy of Finland for financial support (grant 290506 for P.T. and V.P.H.) and Biocenter Finland for infrastructure support. R.R. acknowledges the Finnish Cultural Foundation and the Foundations’ Post Doc Pool for grant 00191229.
Footnotes
Competing interest statement: M.H. and A.H.K. are authors on a patent application covering sequences for enhanced isopeptide bond formation (UK Intellectual Property Office 1706430.4). M.H. is an author on a patent for isopeptide bond formation (EP2534484) and a SpyBiotech cofounder, shareholder, and consultant.
This article is a PNAS Direct Submission. J.W.B. is a guest editor invited by the Editorial Board.
Data deposition: Amino acid sequences have been deposited in GenBank (accession nos. MN433887, MN433888, MN433889, MN433890, MN433891, MN527523, and MN527524). Plasmids encoding pDEST14 SpyCatcher003, pJ404-SpyCatcher003-sfGFP, pDEST14-SpyCatcher003 S49C, pET28a-SpyTag003-MBP, pENTR4-TfR-sfGFP-myc tag-SpyCatcher003, pET28-SpyTag003-mKate2, pET28-SpyTag003-sfGFP, pET28-SpyTag003-mClover3, pEGFP-C1 EGFP-Talin head-SpyCatcher003, and pEGFP-C1 SpyTag003-Talin rod-mCherry were deposited in the Addgene repository (https://www.addgene.org/Mark_Howarth/).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909653116/-/DCSupplemental.
References
- 1.Banerjee A., Howarth M., Nanoteamwork: Covalent protein assembly beyond duets towards protein ensembles and orchestras. Curr. Opin. Biotechnol. 51, 16–23 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Houk K. N., Leach A. G., Kim S. P., Zhang X., Binding affinities of host-guest, protein-ligand, and protein-transition-state complexes. Angew. Chem. Int. Ed. Engl. 42, 4872–4897 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Dubacheva G. V., Curk T., Frenkel D., Richter R. P., Multivalent recognition at fluid surfaces: The interplay of receptor clustering and superselectivity. J. Am. Chem. Soc. 141, 2577–2588 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Antos J. M., Truttmann M. C., Ploegh H. L., Recent advances in sortase-catalyzed ligation methodology. Curr. Opin. Struct. Biol. 38, 111–118 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stevens A. J., et al. , Design of a split intein with exceptional protein splicing activity. J. Am. Chem. Soc. 138, 2162–2165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rashidian M., Dozier J. K., Distefano M. D., Enzymatic labeling of proteins: Techniques and approaches. Bioconjug. Chem. 24, 1277–1294 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Squires T. M., Messinger R. J., Manalis S. R., Making it stick: Convection, reaction and diffusion in surface-based biosensors. Nat. Biotechnol. 26, 417–426 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Chmura A. J., Orton M. S., Meares C. F., Antibodies with infinite affinity. Proc. Natl. Acad. Sci. U.S.A. 98, 8480–8484 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holm L., Moody P., Howarth M., Electrophilic affibodies forming covalent bonds to protein targets. J. Biol. Chem. 284, 32906–32913 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsukiji S., Miyagawa M., Takaoka Y., Tamura T., Hamachi I., Ligand-directed tosyl chemistry for protein labeling in vivo. Nat. Chem. Biol. 5, 341–343 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Lu Y., Huang F., Wang J., Xia J., Affinity-guided covalent conjugation reactions based on PDZ-peptide and SH3-peptide interactions. Bioconjug. Chem. 25, 989–999 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Chen X. H., et al. , Genetically encoding an electrophilic amino acid for protein stapling and covalent binding to native receptors. ACS Chem. Biol. 9, 1956–1961 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang H. J., Baker E. N., Intramolecular isopeptide bonds: Protein crosslinks built for stress? Trends Biochem. Sci. 36, 229–237 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Oke M., et al. , The Scottish Structural Proteomics Facility: Targets, methods and outputs. J. Struct. Funct. Genomics 11, 167–180 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagan R. M., et al. , NMR spectroscopic and theoretical analysis of a spontaneously formed Lys-Asp isopeptide bond. Angew. Chem. Int. Ed. Engl. 49, 8421–8425 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zakeri B., et al. , Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. U.S.A. 109, E690–E697 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun F., Zhang W. B., Mahdavi A., Arnold F. H., Tirrell D. A., Synthesis of bioactive protein hydrogels by genetically encoded SpyTag-SpyCatcher chemistry. Proc. Natl. Acad. Sci. U.S.A. 111, 11269–11274 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schoene C., Bennett S. P., Howarth M., SpyRing interrogation: Analyzing how enzyme resilience can be achieved with phytase and distinct cyclization chemistries. Sci. Rep. 6, 21151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schreiber G., Haran G., Zhou H. X., Fundamental aspects of protein-protein association kinetics. Chem. Rev. 109, 839–860 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keeble A. H., et al. , Evolving accelerated amidation by SpyTag/SpyCatcher to analyze membrane dynamics. Angew. Chem. Int. Ed. Engl. 56, 16521–16525 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L., Fierer J. O., Rapoport T. A., Howarth M., Structural analysis and optimization of the covalent association between SpyCatcher and a peptide Tag. J. Mol. Biol. 426, 309–317 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu H., Grimsley G. R., Razvi A., Scholtz J. M., Pace C. N., Increasing protein stability by improving beta-turns. Proteins 77, 491–498 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trevino S. R., Schaefer S., Scholtz J. M., Pace C. N., Increasing protein conformational stability by optimizing beta-turn sequence. J. Mol. Biol. 373, 211–218 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khairil Anuar I. N. A., et al. , Spy&Go purification of SpyTag-proteins using pseudo-SpyCatcher to access an oligomerization toolbox. Nat. Commun. 10, 1734 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pédelacq J. D., Cabantous S., Tran T., Terwilliger T. C., Waldo G. S., Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Bagshaw C. R., Biomolecular Kinetics: A Step-by-Step Guide (CRC Press, London, 2017). [Google Scholar]
- 27.Keeble A. H., Hemmings A. M., James R., Moore G. R., Kleanthous C., Multistep binding of transition metals to the H-N-H endonuclease toxin colicin E9. Biochemistry 41, 10234–10244 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Keeble A. H., Kleanthous C., The kinetic basis for dual recognition in colicin endonuclease-immunity protein complexes. J. Mol. Biol. 352, 656–671 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Gianni S., Dogan J., Jemth P., Distinguishing induced fit from conformational selection. Biophys. Chem. 189, 33–39 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Pan J., Zhang S., Parker C. E., Borchers C. H., Subzero temperature chromatography and top-down mass spectrometry for protein higher-order structure characterization: Method validation and application to therapeutic antibodies. J. Am. Chem. Soc. 136, 13065–13071 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Dovala D., Sawyer W. S., Rath C. M., Metzger L. E. IV, Rapid analysis of protein expression and solubility with the SpyTag–SpyCatcher system. Protein Expr. Purif. 117, 44–51 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Rahikainen R., Öhman T., Turkki P., Varjosalo M., Hytönen V. P., Talin-mediated force transmission and talin rod domain unfolding independently regulate adhesion signaling. J. Cell Sci. 132, jcs226514 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Rahikainen R., et al. , Mechanical stability of talin rod controls cell migration and substrate sensing. Sci. Rep. 7, 3571 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X., et al. , Talin depletion reveals independence of initial cell spreading from integrin activation and traction. Nat. Cell Biol. 10, 1062–1068 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang G., Giannone G., Critchley D. R., Fukumoto E., Sheetz M. P., Two-piconewton slip bond between fibronectin and the cytoskeleton depends on talin. Nature 424, 334–337 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Klapholz B., Brown N. H., Talin—The master of integrin adhesions. J. Cell Sci. 130, 2435–2446 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Goult B. T., Yan J., Schwartz M. A., Talin as a mechanosensitive signaling hub. J. Cell Biol. 217, 3776–3784 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Theodosiou M., et al. , Kindlin-2 cooperates with talin to activate integrins and induces cell spreading by directly binding paxillin. eLife 5, e10130 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keeble A. H., Howarth M., Insider information on successful covalent protein coupling with help from SpyBank. Methods Enzymol. 617, 443–461 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Chen I., Dorr B. M., Liu D. R., A general strategy for the evolution of bond-forming enzymes using yeast display. Proc. Natl. Acad. Sci. U.S.A. 108, 11399–11404 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McConnell S. A., et al. , Protein labeling via a specific lysine-isopeptide bond using the pilin polymerizing sortase from Corynebacterium diphtheriae. J. Am. Chem. Soc. 140, 8420–8423 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fierer J. O., Veggiani G., Howarth M., SpyLigase peptide-peptide ligation polymerizes affibodies to enhance magnetic cancer cell capture. Proc. Natl. Acad. Sci. U.S.A. 111, E1176–E1181 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buldun C. M., Jean J. X., Bedford M. R., Howarth M., SnoopLigase catalyzes peptide-peptide locking and enables solid-phase conjugate isolation. J. Am. Chem. Soc. 140, 3008–3018 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Sun F., Zhang W.-B., Unleashing chemical power from protein sequence space toward genetically encoded click chemistry. Chin. Chem. Lett. 28, 2078–2084 (2017). [Google Scholar]
- 45.Oliveira B. L., Guo Z., Bernardes G. J. L., Inverse electron demand Diels-Alder reactions in chemical biology. Chem. Soc. Rev. 46, 4895–4950 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Gordon C. G., et al. , Reactivity of biarylazacyclooctynones in copper-free click chemistry. J. Am. Chem. Soc. 134, 9199–9208 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chin J. W., Expanding and reprogramming the genetic code of cells and animals. Annu. Rev. Biochem. 83, 379–408 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Brune K. D., Howarth M., New routes and opportunities for modular construction of particulate vaccines: Stick, click, and glue. Front. Immunol. 9, 1432 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kent S. B., Total chemical synthesis of proteins. Chem. Soc. Rev. 38, 338–351 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Saito F., Noda H., Bode J. W., Critical evaluation and rate constants of chemoselective ligation reactions for stoichiometric conjugations in water. ACS Chem. Biol. 10, 1026–1033 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Wieduwild R., Howarth M., Assembling and decorating hyaluronan hydrogels with twin protein superglues to mimic cell-cell interactions. Biomaterials 180, 253–264 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Pardee K., et al. , Portable, on-demand biomolecular manufacturing. Cell 167, 248–259.e12 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Veggiani G., et al. , Programmable polyproteams built using twin peptide superglues. Proc. Natl. Acad. Sci. U.S.A. 113, 1202–1207 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bartsch T. F., et al. , Elasticity of individual protocadherin 15 molecules implicates tip links as the gating springs for hearing. Proc. Natl. Acad. Sci. U.S.A. 116, 11048–11056 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu P., et al. , Accurate computational design of multipass transmembrane proteins. Science 359, 1042–1046 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Konermann L., Pan J., Liu Y. H., Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem. Soc. Rev. 40, 1224–1234 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Pan J., Han J., Borchers C. H., Konermann L., Hydrogen/deuterium exchange mass spectrometry with top-down electron capture dissociation for characterizing structural transitions of a 17 kDa protein. J. Am. Chem. Soc. 131, 12801–12808 (2009). [DOI] [PubMed] [Google Scholar]
- 58.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meijering E., Dzyubachyk O., Smal I., Methods for cell and particle tracking. Methods Enzymol. 504, 183–200 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Amino acid sequences of each version of SpyTag and SpyCatcher are available in SI Appendix, Fig. S1A. Amino acid sequences of other constructs are available in GenBank as described in SI Appendix, SI Methods, Plasmids and Cloning. Plasmids encoding pDEST14 SpyCatcher003, pJ404-SpyCatcher003-sfGFP, pDEST14-SpyCatcher003 S49C, pET28a-SpyTag003-MBP, pENTR4-TfR-sfGFP-myc tag-SpyCatcher003, pET28-SpyTag003-mKate2, pET28-SpyTag003-sfGFP, pET28-SpyTag003-mClover3, pEGFP-C1 EGFP-Talin head-SpyCatcher003, and pEGFP-C1 SpyTag003-Talin rod-mCherry were deposited in the Addgene repository (https://www.addgene.org/Mark_Howarth/). Further information and request for resources and reagents should be directed to and will be fulfilled by the lead contact, M.H.