Significance
Endoplasmic reticulum aminopeptidase 1 (ERAP1), an enzyme important for regulating adaptive immune responses, is an emerging target for immunotherapy applications. One of the hallmarks of ERAP1 is that it must be able to select substrates out of hundreds of thousands of possible ones. An incomplete understanding of how it performs this selection has been a major barrier to predicting and manipulating its effects on the immune response. We solved high-resolution crystal structures of ERAP1 with substrate analogs, which, along with biochemical and computational analysis, shed light on the mechanism that ERAP1 uses to select its substrates. Our structures constitute an important framework for better predicting epitope selection and immunodominance.
Keywords: enzyme, mechanism, peptide, X-ray crystallography, immune system
Abstract
Endoplasmic reticulum aminopeptidase 1 (ERAP1) is an intracellular enzyme that optimizes the peptide cargo of major histocompatibility class I (MHC-I) molecules and regulates adaptive immunity. It has unusual substrate selectivity for length and sequence, resulting in poorly understood effects on the cellular immunopeptidome. To understand substrate selection by ERAP1, we solved 2 crystal structures of the enzyme with bound transition-state pseudopeptide analogs at 1.68 Å and 1.72 Å. Both peptides have their N terminus bound at the active site and extend away along a large internal cavity, interacting with shallow pockets that can influence selectivity. The longer peptide is disordered through the central region of the cavity and has its C terminus bound in an allosteric pocket of domain IV that features a carboxypeptidase-like structural motif. These structures, along with enzymatic and computational analyses, explain how ERAP1 can select peptides based on length while retaining the broad sequence-specificity necessary for its biological function.
The correct functioning of the human adaptive immune system relies on the presentation on the cell surface of small peptides bound onto major histocompatibility class I (MHC-I) molecules (1). Changes in the nature of these peptides indicate changes in the proteome of the cell and can be interpreted by T lymphocytes as signs of infection or transformation leading to killing of the cell by induced apoptosis (2). Aberrant antigenic peptide generation can allow pathogens to escape detection or allow normal cells to be recognized erroneously as diseased, contributing to such processes as viral or cancer immune evasion and autoimmunity. Antigenic peptides are generated inside the cell by proteolysis of intracellular proteins (3). Endoplasmic reticulum (ER)-resident aminopeptidases, such as ER aminopeptidase 1 (ERAP1), play key roles in the generation of antigenic peptides (4). ERAP1 can help generate antigenic peptides by trimming N-terminally extended precursors to the correct length for MHC-I binding, but it can also destroy them by overtrimming down to lengths that preclude MHC-I binding (5). Through these functions, ERAP1 can affect the cellular immunopeptidome and regulate immune cytotoxic responses (6–9). Additional functions of ERAP1 have also been investigated, including the regulation of blood pressure and innate immune responses, both of which are related to ERAP1’s ability to be secreted from the cell under certain conditions (10, 11).
ERAP1 is polymorphic, and several coding missense single nucleotide polymorphisms (SNPs) in the ERAP1 gene have been associated with predisposition to HLA-associated inflammatory autoimmunity, as well as with the outcome of viral infections and cancer (12–14). ERAP1 polymorphisms have been shown to affect both its enzymatic activity and selectivity and to influence antigen presentation (15, 16). The importance of ERAP1 in regulating immune responses has attracted interest in drug discovery aimed at modulating its activity for applications in cancer immunotherapy or the control of autoimmunity (17). First-generation active site inhibitors for ERAP1 have been shown to have activity in regulating immune responses in model systems, opening opportunities for clinical applications (18, 19).
ERAP1 belongs to the M1 family of aminopeptidases, and its structure consists of 4 domains that form a concave-like structure with a large internal cavity that includes the catalytic site and can accommodate large peptides (20). The crystal structure of ERAP1 with bound active site inhibitors has provided useful insight into the function of the enzyme, but only limited information on how the enzyme recognizes physiologically relevant substrates (21–23). ERAP1 has been crystallized in 2 distinct conformations, one more globular in which domain IV abuts domain II, excluding outside solvent access from a large internal cavity, and the other more open in which domain IV moves away from domain II, exposing the internal cavity. It has been hypothesized that the closed conformation is catalytically active and that the open conformation serves to facilitate large peptide capture, requiring cycling between these 2 conformations during catalytic turnover (21, 24). Based on biochemical and structural data, a putative regulatory site has been proposed to promote long peptide trimming, although its precise location and exact role in the catalytic process have been controversial (21, 25, 26).
Here we report 2 high-resolution crystal structures of ERAP1 at 1.68 Å and 1.72 Å with bound noncleavable peptide analogs that mimic transition state intermediates formed during catalysis. These 2 structures represent snapshots of how ERAP1 recognizes peptide substrates, provide key insight into the substrate specificity of this enzyme, and highlight features of the interactions that underlie the generation of the immunopeptidome of human cells.
Results
Substrate Analogs Provide a Snapshot of the Catalytic Cycle.
To stabilize the peptide–enzyme interaction, we designed 2 noncleavable peptide analogs based on the phosphinic pseudopeptide scaffold described previously (27, 28). The 2 amino-terminal pseudo amino acids were designed to carry homophenylalanine and leucine side chains, as according to previous biochemical and structural studies, these are well accommodated in the ERAP1 active site (18, 29). The remaining peptide sequences were based on epitope precursors known to be processed by ERAP1 (15, 30, 31). In particular, the 15-mer peptide was based on the epitope with sequence GPGRAFVTI from the envelope glycoprotein gp160 of the HIV 1, carrying the natural precursor sequence and shown previously to be processed efficiently by ERAP1 generating the mature epitope (30). The 10-mer peptide was based on the SRHHAFSFR epitope from the aggrecan protein normally presented by HLA-B*27:05, which also has been shown to be efficiently generated by ERAP1 (15, 31). The chemical structures of the peptide analogs are shown in SI Appendix, Fig. S1 A and B.
Due to their transition-state nature, both peptides are potent inhibitors of ERAP1 (SI Appendix, Fig. S1C). Furthermore, the inhibitory potency of both peptide analogs is in the nM range (72 nM for the 15-mer and 128 nM for the 10-mer), more than 2 orders of magnitude better than a phosphinic pseudotripeptide carrying homophenylalanine, leucine, and arginine (identical to the N terminus of the 15-mer peptide and very similar to the 10-mer peptide. which bears lysine at position 3), suggesting that the remaining peptide sequence participates in productive interactions with the enzyme (SI Appendix, Fig. S1C) (32).
ERAP1 Is in a Closed Conformation with Peptides Trapped in an Internal Cavity.
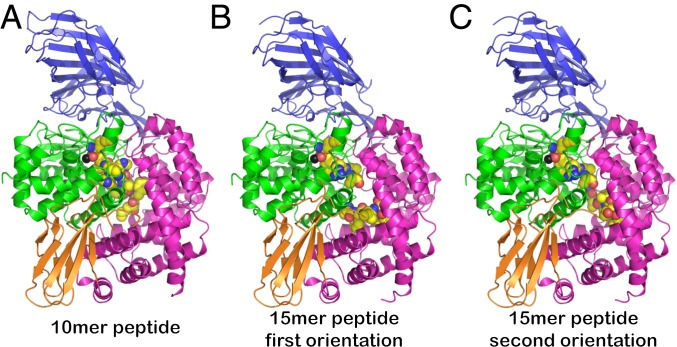
Crystals of ERAP1 in complex with the peptide analogs were obtained after cocrystallization of the purified enzyme with the ligand, as described in Experimental Procedures. The crystals diffracted synchrotron X-ray radiation up to 1.68 Å for the 10-mer peptide and 1.72 Å for the 15-mer peptide. (Data collection and refinement statistics are provided in SI Appendix, Tables S1 and S2.) The structures were solved by molecular replacement, using the closed conformation of ERAP1 (PDB ID code 2YD0) as a search model. Both structures correspond to the closed conformation of ERAP1 with domain IV abutting domain II of the enzyme (Fig. 1). Additional electron density within the internal cavity of ERAP1, starting from the catalytic site that features the Zn(II) atom, was interpreted to correspond to the ligands (SI Appendix, Fig. S2). Although the C-terminal tail of the 15-mer peptide was built in 2 distinct conformations, the overall conformation of the enzyme was unaltered in the 2 complexes (Fig. 1 B and C).
Fig. 1.
Cartoon representations of crystal structures of ERAP1 in complex with peptide analogs. The 4 domains of ERAP1 are shown in blue (domain I), green (domain II), orange (domain III), and magenta (domain IV). The catalytic Zn(II) atom is shown as a black sphere. The ligand is shown in spheres color-coded by atom type (yellow, carbon; blue, nitrogen; red, oxygen; orange, phosphorus). In all cases, the peptide is found trapped inside the internal cavity of the enzyme, with no apparent access to the outside solvent. (A) Crystal structure with the 10-mer peptide. (B and C) Crystal structures with the 15-mer peptide. The C-terminal moiety of the 15-mer was built in 2 distinct conformations. The middle part of the 15-mer peptide did not have a readily traceable electron density and is not shown here.
Configuration of Peptides Inside Cavity.
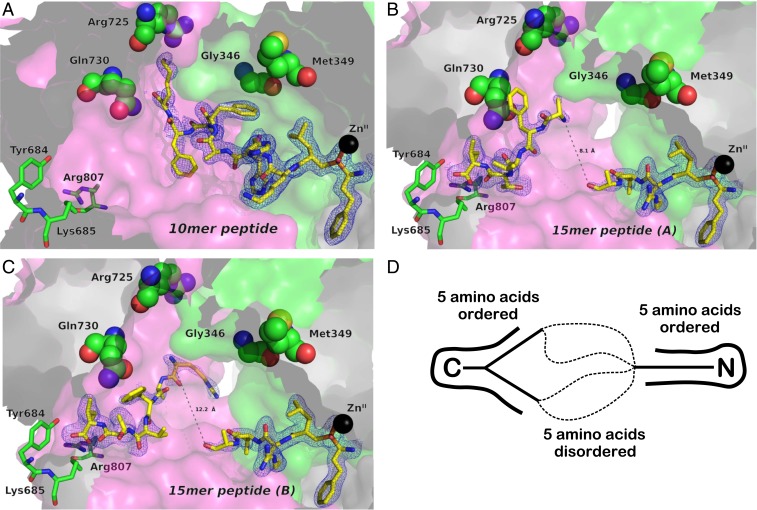
The structure of the 10-mer peptide within the internal cavity of ERAP1 is depicted in more detail in Fig. 2A. The N terminus of the peptide that bears the phosphinic moiety is bound on the catalytic Zn(II) atom in a motif observed previously in crystal structures of homologous aminopeptidases with bound phosphinic groups. The 15-mer peptide had good density near the catalytic site, allowing us to reliably build residues up to the fifth amino acid, suggesting that the peptide becomes progressively disordered as it extends away from the catalytic site through the internal cavity (Fig. 2 B and C). Surprisingly, however, good electron density was also observed for the C-terminal moiety of the peptide that was found anchored at a position distal from the catalytic center and in the base of domain IV. Moving from the C terminus toward the N terminus of the peptide, electron density again became progressively weaker and split into 2 possible directions. To account for this observation, we built 2 distinct conformations of the C-terminal moiety of the peptide. Overall, the 15-mer was clearly visible at its N-terminal end (5 amino acids) and C-terminal end (5 amino acids) but became progressively disordered toward the middle section that traversed the central part of the cavity. In both cases, the distance between the ends of the 2 parts of the built peptide were between 8 and 12 Å, leaving sufficient space to easily accommodate the remaining 5 residues that were disordered and not built in the model (Fig. 2D). Both conformations of the 15-mer peptide are stabilized by electrostatic interactions between its N and C termini and negatively and positively charged residues in the ERAP1 cavity, respectively (SI Appendix, Fig. S3).
Fig. 2.
Schematic representations of the peptide analogs bound in the internal cavity of ERAP1, shown in surface representation. (A) The 10-mer peptide DG056. (B and C) The 2 conformations of the 15-mer peptide DG055. The peptides are shown in sticks color-coded by atom type (yellow, carbon; blue, nitrogen; red, oxygen; orange, phosphorus). Key amino acids from ERAP1 are depicted in sticks or spheres color-coded by atom type (green, carbon; blue, nitrogen; red, oxygen). The catalytic Zn(II) atom is shown as a black sphere. The refined 2Fo-Fc electron density map, contoured at 1σ, is shown as a cyan mesh. Domain II is in green, and domain IV is in pink. (D) Schematic model of how the 15-mer peptide is accommodated in the cavity.
Interactions with Cavity Amino Acids and Potential Role of Disease-Predisposing SNPs.
Several SNPs in ERAP1 have been associated with disease predisposition and shown to affect enzymatic activity. Recent analysis has assigned 9 ERAP1 SNPs into 10 common human haplotypes (14). In an effort to understand the mechanism by which these amino acid variations may affect substrate recognition by ERAP1, we mapped them onto the 2 structures. Of the 9 known SNPs, 4—Gly346, Met349, Arg725, and Gln730—are located inside or adjacent to the cavity and could participate in interactions with the peptide substrates (Fig. 2, indicated in spheres). Met349 does not appear to make any direct interactions with any part of the 2 peptides in the structure. Gly346 makes van der Waals contact with Phe6 of the 10-mer peptide (Fig. 2A). Since the amino acid change induced by this SNP is Gly/Asp, a nonconservative change, variation at that location may be expected to affect peptide recognition. No direct interaction with the 15-mer peptide can be seen, but its overall configuration could allow interactions with that polymorphic position; however, this cannot be confirmed, due to the lack of density in the middle region of the peptide. Both Arg725 and Gln730, 2 SNPs repeatedly shown to associate with predisposition to autoimmunity and to affect generation of the immunopeptidome in cells (12, 33), are located within interaction distance from residues of both peptides, albeit without any obvious strong favorable interaction. Interestingly, however, both SNPs code for nonconservative amino acid variations (Arg725Gln and Gln730Glu) that modify their charge, affording the opportunity for productive interactions with appropriately located charged or polar side chains in the peptide ligand.
Overall, our mapping suggests that at least 3 of the known ERAP1 SNPs could have functional repercussions by changing direct atomic interactions with the peptide inside the internal cavity of the enzyme, thereby affecting selectivity. Indeed, previous proteomic analyses of the effect of those SNPs on the immunopeptidome of cells are consistent with this observation, demonstrating that the SNPs at position 725 and 730 have effects on the sequence of MHC-presented peptides in cultured cells (34).
Recognition of the C Terminus of the Peptide.
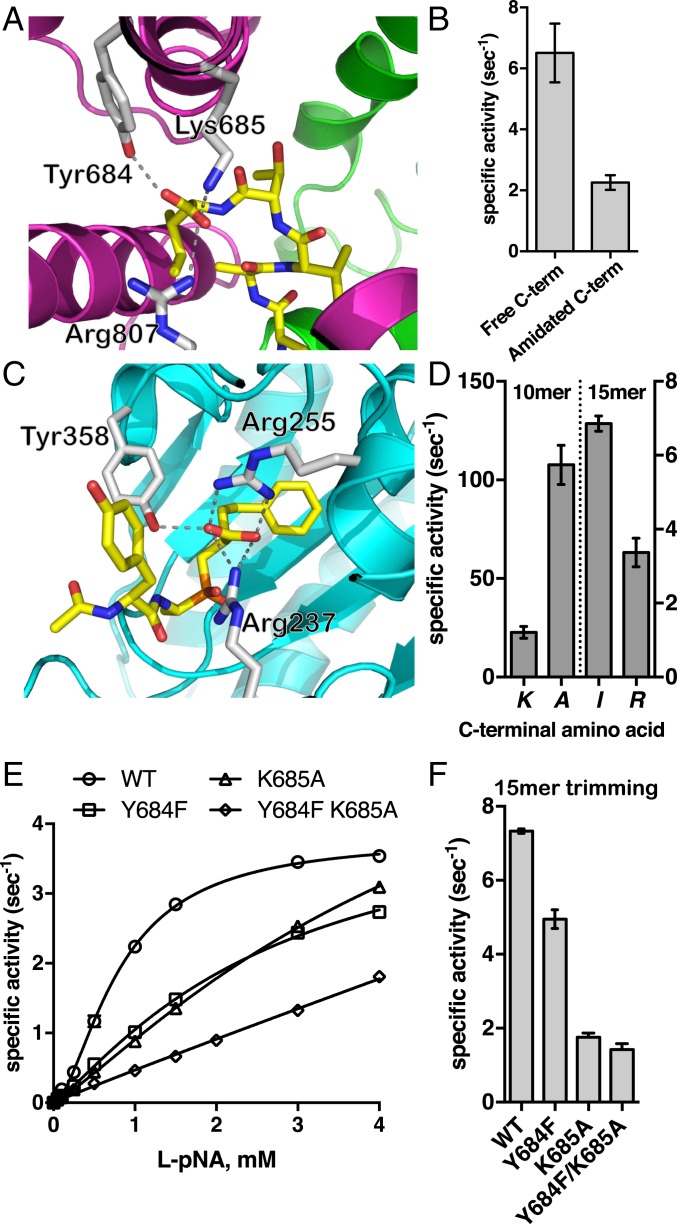
Previous biochemical work has indicated that ERAP1 may be able to recognize the C terminus of peptide substrates, leading to self-activation for longer peptides (21, 35). The exact location and mechanism of this phenomenon has remained elusive, however. Two crystal structures of the isolated C-terminal domain of ERAP1 have suggested a potential site, but in those structures the peptide–protein interaction was found only in the context of an extensive dimerization interface that is unlikely to be physiologically relevant, raising questions about the validity of the proposed site for full-length ERAP1 (25, 26). In our 15-mer peptide-bound structure, both conformations have the C-terminal residue in an identical configuration making specific interactions with 3 residues of domain IV: Tyr684, Lys685, and Arg807. These 3 residues form a carboxyl-recognizing triad that makes salt bridge and hydrogen-bonding interactions with the C terminus of the peptide (Fig. 3A). This ionic interaction is important for substrate recognition, since amidation of the C terminus reduces the rate of trimming of a 15-mer model peptide substrate with an equivalent sequence (Fig. 3B). This interaction appears to be sufficiently potent to conformationally stabilize the peptide, similar to how the transition state analog moiety is stabilized at the active site. Thus, the 15-mer peptide is stably tethered by both ends and is flexible in the middle region, as evidenced by the lack of good density there (Fig. 2D). Interestingly, the recognition of the C terminus of a peptidic ligand in carboxypeptidase A1 (PDB ID code 4UEF) is achieved by a similar triad of residues, 2 positively charged and a tyrosine, that obtain a spatial arrangement reminiscent to that seen in ERAP1 (Fig. 3C). This recognition element is the same as that suggested by ERAP1 domain IV dimer structures, which show interactions between that site and the C terminus tail of an adjacent molecule, an interaction that is probably key to domain IV dimerization and formation of crystals (25, 26). This finding suggests that for sufficiently long peptides, ERAP1 can specifically recognize both the C terminus and the N terminus of the peptide by utilizing structural motifs used by carboxypeptidases and aminopeptidases, respectively. Shorter peptides cannot occupy both locales concurrently, which may limit their trimming. These observations are consistent with the proposed “molecular ruler” mechanism proposed in 2005 by Goldberg and colleagues (35, 36) and can explain the unique ability of ERAP1 to efficiently process peptides >10-aa long.
Fig. 3.
(A) Interactions between the C terminus of bound 15-mer peptide analog and ERAP1. The enzyme is shown in cartoon representation color-coded by domain as in Fig. 1. The peptide is shown as yellow sticks, oxygen atoms are in red, and nitrogen atoms are in blue. Key ERAP1 residues that participate in the interactions are shown as gray sticks. (B) Effect of amidation of the C terminus on the trimming rate of 15-mer peptide LLRIQRGPGRAFVTI. (C) Schematic representation of a phosphinic transition state analog in the catalytic site of carboxypeptidase A1 (PDB ID code 4UEF). Interactions between amino acids in ERAP1 or carboxypeptidase A1 and the C-terminal group of the ligand are indicated by dotted lines. (D) Effect of substituting the C-terminal residue of 10-mer and 15-mer peptide with Ala and Arg, respectively, on trimming rates by ERAP1. (E) MM analysis of ERAP1 variants carrying mutations of key residues of the C terminus recognition site on the hydrolysis of substrate L-pNA. (F) Trimming rate of 15-mer peptide LLRIQRGPGRAFVTI by wild-type ERAP1 and ERAP1 variants Y684F, K685A, and Y684F/K685A.
Surprisingly, the structure of the shorter 10-mer peptide reveals no interaction with this C-terminal docking site. This may be due to the shorter length of the peptide compared to the 15-mer. Indeed, the distance between the N terminus and the C terminus recognition sites is ∼30 Å in the structure of the bound 15-mer peptide (closed conformation) and 35.6 Å in the previously published open conformation of ERAP1 (PDB ID code 3MDJ) (SI Appendix, Fig. S4 A and B). In contrast, the average distance between the N and C termini of HLA-A*02–bound peptides is 24.5 Å, which does not allow them to concurrently occupy both the N terminus and C terminus sites in ERAP1 in similar configurations (SI Appendix, Fig. S4 C and D). Although it is theoretically possible for the 10-mer peptide to extend to configurations spanning 30 Å, it would have to sacrifice other interactions with the central cavity. Interestingly, the 10-mer peptide adopts a pseudohelical configuration parallel to helix H3 and extends toward the interface of domains II and IV, forming a number of hydrogen bonds with residues of both domains. His4 of the 10-mer peptide interacts strongly with Asp435 and Asp429, the main chain carbonyl oxygen of Phe6 of the peptide with Ser342 and its Ser7 with Ser795 and Ser799 of domain IV. The C-terminal lysine of the 10-mer peptide interacts strongly through its side chain amine group with ERAP1 Asp766, which in turn is stabilized by a salt bridge with Arg725. An alternative explanation for the 10-mer not occupying this C-terminus recognition cavity may lie in the nature of the C-terminal residue of the 10-mer, which is a Lys instead of an Ile for the 15-mer peptide. Indeed, it has been previously proposed that ERAP1 prefers to trim antigenic peptide precursors that carry hydrophobic C termini versus positively charged C termini (35). Therefore, it is possible that antigenic peptide precursors that carry positively charged C-terminal residues are processed at a slower rate due to their inability to be recognized productively by the C-terminus docking site, which carries a positive electrostatic potential (SI Appendix, Fig. S3). Indeed, comparing the trimming rates of peptides LLKHHAFSFK and LLKHHAFSFA, the latter is trimmed almost 5-fold faster, suggesting that it is recognized in a different way by the enzyme, possibly involving the C-terminus recognition site (Fig. 3D). Conversely, changing the C-terminal residue of the 15-mer LLRIQRGPGRAFVTI from an Ile to an Arg reduced N-terminal trimming by 2-fold (Fig. 3D). Taken together, these findings suggest that the nature of the C-terminal residue can affect peptide recognition by ERAP1, likely through binding to the C-terminus site.
To confirm the role of C terminus in recognizing residues on the enzymatic activity, we generated ERAP1 variants carrying mutations on residues Tyr684 and Lys685, which are conserved between ERAP1 orthologs in other species (SI Appendix, Table S3). Michaelis–Menten (MM) analysis of variants Y684F, K685A, and double mutant Y684F/K685A using the small chromogenic substrate L-pNA showed reduced activity for both variants carrying a single mutation and an additive effect for the double mutant (Fig. 3E and Table 1). Data were fit to an allosteric MM model as described previously (21). The effect was due primarily to an increase in the Khalf value, which indicates that although these residues lie ∼30 Å from the catalytic site where the small substrate is hydrolyzed, they can influence binding affinity for the substrate. This phenomenon extends to large peptides, since trimming of the 15-mer peptide LLRIQRGPGRAFVTI by the variants was up to 5-fold slower (Fig. 3F). Interestingly, the effect on trimming the 15-mer was dominated by the K685A mutation, with the Y684F mutation having a much smaller effect.
Table 1.
Kinetic parameters calculated from the MM analysis shown in Fig. 3
| Parameter | Wild-type | Y684F | K685A | Y684F/K685A |
| Vmax, s−1 | 3.8 ± 0.1 | 4,6 ± 0.3 | 7.4 ± 0.5 | ND |
| Hill slope | 1.69 ± 0.06 | 1.21 ± 0.05 | 1.19 ± 0.03 | 0.96 ± 0.1 |
| Khalf, mM | 0.80 ± 0.02 | 2.66 ± 0.28 | 5.28 ± 0.50 | ND |
| Kprime, mM | 0.69 ± 0.04 | 3.25 ± 0.27 | 7.2 ± 0.52 | ND |
ND, not determined.
Occupation of the C-Terminal Binding Site Regulates ERAP1 Activity.
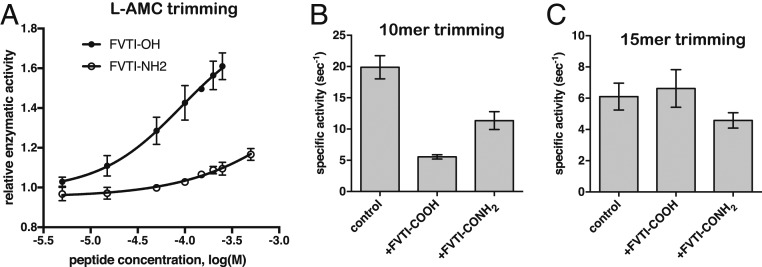
The ability of ERAP1 to hydrolyze small fluorogenic substrates can be enhanced by the presence of some peptides <8-aa long (21). This property has been suggested to arise through a regulatory region in ERAP1, presumably recognizing the C-terminal moiety of the peptide. To test whether the C-terminus recognition site identified here is indeed a regulatory site, we used a small tetrapeptide with the sequence FVTI, taken from the C-terminal sequence of the 15-mer, to test whether it can activate ERAP1-mediated hydrolysis of the fluorogenic substrate L-AMC. The tetrapeptide activated the rate of hydrolysis of L-AMC in a dose-dependent manner, with an ED50 of 92 μΜ (Fig. 4A). To confirm that this effect is due to binding to the same binding site as seen in the crystal structure, we also tested the same tetrapeptide with an amidated C terminus, since amidation abrogates the negative charge and should reduce interactions with Lys685 and Arg807. The amidated tetrapeptide was much less effective in inducing ERAP1 activation, suggesting that the presence of the free C terminus is important for this function (Fig. 4A).
Fig. 4.
(A) ERAP1 activity in trimming the fluorogenic substrate L-AMC is enhanced by the tetrapeptide FVTI in a dose-dependent manner. The effect is far weaker when the peptide C terminus is amidated (peptide FVTI-NH2). (B) Specific activity against the 10-mer substrate LLKHHAFSFK is reduced in the presence of 200 μM tetrapeptide FVTI and, to a lesser extent, the peptide FVTI-NH2. (C) Specific activity against the 15-mer substrate LLRIQRGPGRAFVTI is not affected by the presence of 200 μM tetrapeptide FVTI.
We also tested the effect of the tetrapeptide FVTI on the trimming rates of 2 peptides that correspond to the sequences of the analogs cocrystallized with ERAP1. Trimming of the N-terminal residue of the 10-mer peptide with the sequence LLKHHAFSFK was significantly reduced in the presence of 200 μM FVTI (Fig. 4B). The C-terminally amidated FVTI peptide (FVTI-NH2) also inhibited trimming of the 10-mer, but to a much lesser extent (Fig. 4B). This apparent inhibition of the 10-mer peptide by the tetrapeptide may arise from steric clashes between the 2 peptides as they attempt to occupy overlapping regions of the ERAP1 cavity (Fig. 2). Interestingly, the FVTI peptide had no discernible effect on trimming of the larger 15-mer with the sequence LLRIQRGPGRAFVTI (Fig. 4C), possibly because the larger peptide already contains this sequence in its C terminus and has a higher affinity, and thus the smaller peptide cannot easily outcompete the larger one at easily achievable concentrations. These observations suggest that the C-terminal binding site can influence the activity of the enzyme in a substrate-dependent manner.
Molecular Dynamics Analysis of Peptide Binding at the C-Terminal Recognition Site.
The effects of the tetrapeptide FVTI on the enzymatic activity of ERAP1 suggest that binding to the C terminus recognition site may influence the active site of the enzyme. Surprisingly, however, comparing the peptide-bound and peptide-free structures indicates only minor structural changes, making it difficult to understand the activation properties of the tetrapeptide (SI Appendix, Fig. S5). We therefore hypothesized that the effect of the substrate’s C terminus on the enzymatic activity could be due to a change in the conformational dynamics of ERAP1. Indeed, comparing the local B-factors to the average B-factor of each of the structures presented here and to a recently published structure of ERAP1 with a transition-state analog in the active site (23) indicates a significantly higher decrease in the thermal parameters of C terminus-interacting residues, in particular for Y684 and K685 (SI Appendix, Fig. S6).
To further investigate this observation, we performed a comparative molecular dynamics (MD) study using the high-resolution structure of the ERAP1 complex in the presence and absence of the C terminus tetrapeptide moiety of the cocrystallized 15-mer peptide (FVTI). Four independent 250-ns MD simulations of ERAP1 were performed in the absence or presence of FVTI with a phosphinic dipeptide transition state analog bound in the active site (SI Appendix, Computational Methods). In agreement with previous MD studies of ERAP1 (36, 37), all our simulations exhibited the stability of the well-folded closed state of the enzyme at this timescale (SI Appendix, Figs. S7 and S8).
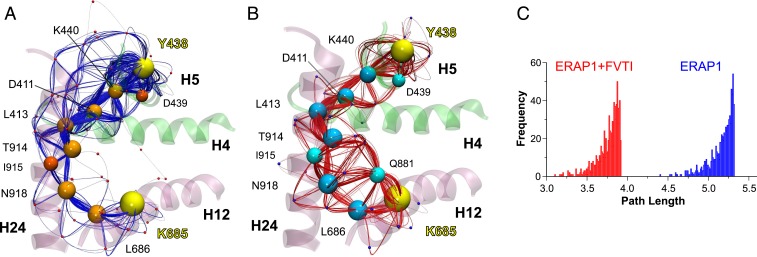
The effect of FVTI in the dynamics of ERAP1 was also investigated through a dynamical network analysis of the MD trajectories using the weighted implementation of suboptimal paths (WISP) method (38). This analysis is based on the dynamic interdependence among nodes that represent the protein substituents (here each residue’s center of mass was defined as a node). The interdependence between 2 nodes is represented by a connecting edge associated with a value representing the strength of their correlation. The functionalized correlation value represents the edge “length” in network space, with larger values for the motions of 2 residues that are highly correlated or anticorrelated. A network of node pairs that are in physical contact (in the Cartesian space) is then calculated between 2 given residues at the distal sites under investigation. Here we calculated a total of 750 paths connecting the catalytic Tyr438 (domain II, sink residue) with Tyr684, Lys685, or Arg807 (domain IV, source residues). These residues were chosen as representative sites of interaction with the N and C termini of the substrate, respectively (Fig. 3A). For Lys685, the correlated motion paths mainly traversed the same residues of helices H12→H24→H4→H5 in either the presence or the absence of FVTI (Fig. 5 A and B), except that binding of FVTI promoted a significant increase in the number of pathways traversing residue Gln881 (node degeneracy of 65%; SI Appendix, Table S4).
Fig. 5.
(A and B) Schematic illustration of calculated paths along networks of correlated residue motions between residues Tyr438 and Lys685 (yellow spheres) that interact with the N and C termini of the 15-mer substrate. The communicating paths are color-coded with blue splines and orange nodes (residues center of mass) for ERAP1 (A) and red splines with cyan nodes for FVTI-bound ERAP1 (B). Residues that belong to the optimal (shortest) pathway are shown using larger spheres, and helices H4 (396 to 411), H5 (435 to 452), H12 (670 to 688), and H24 (906 to 936) are color-coded according to ERAP1 domain. (C) Histograms of the 750 path lengths calculated from the dynamical network analysis of ERAP1 in the absence (blue bars) or presence (red bars) of FVTI. The distribution associated with the MD simulations in the presence of FVTI is shifted toward shorter lengths, indicating a stronger dynamical coupling on FVTI binding. Note that here, “length” is not related to the Cartesian space but rather refers to spans in functionalized correlation space.
Path length analysis revealed that the optimal pathway in the presence of FVTI is shorter, indicating a stronger correlation of residue motions between the 2 sites. In accordance with this finding, comparison of the path length distributions from multiple pathways displayed a notable shift toward shorter values in the simulations of FVTI-bound ERAP1 (Fig. 5C), suggesting that binding of FVTI results in stronger dynamical coupling—more coherent communication—between the 2 sites.
The results using Tyr684 or Arg807 as the source residues were similar but with less notable shifts of the path length distributions toward shorter values in the presence of FVTI (SI Appendix, Fig. S9 A and B). Specifically, the pathways for Tyr684→Tyr438 were very similar to those calculated for Lys685→Tyr438 (SI Appendix, Fig. S9 C and E), whereas Arg807→Tyr438 displayed the lowest difference in the path length distributions upon binding of FVTI, but also traversed alternative pathways with respect to those calculated when using Tyr684 or Lys685 as the source residue (SI Appendix, Fig. S9 D and F). Interestingly, these results are consistent with the relative effects of mutations in Tyr684 and Lys685 on enzyme kinetics presented above.
To further examine whether the proposed dynamical tightening between the C-terminal recognition sites and the catalytic site is associated with an entropic decrease, we calculated the conformational entropies from the ensemble of configurations by analyzing the dihedral angle fluctuations (SI Appendix, Table S5). On binding of FVTI, the total conformational entropy of ERAP1 decreased by –TΔSconf = 31 ± 8 kcal/mol, with the stabilizing effect of FVTI being more evident in residues lining along helix H12. The stabilizing effect of FVTI was more noticeable in residues lining along helix H12 (SI Appendix, Figs. S10 and S11 and Table S6). Among the residues identified by the dynamical network analysis, Tyr684, Lys685, Leu686, and Gln881 displayed a significant stabilizing effect (–TΔSconf of 0.7, 1.0, 0.5, and 1.0 kcal/mol, respectively), whereas the other residues displayed minor stabilization (–TΔSconf <0.1 kcal/mol). Taken together, our MD results indicate that binding of FVTI at the C-terminus recognition site decreases the conformational entropy of ERAP1 and mediates a stronger dynamical coupling between the N- and C-termini binding sites of the substrate.
Discussion
One hallmark of ERAP1 function in preparing peptide cargo for MHC-I is that it must be able to process a vast variety of peptide sequences. MHC-I also must be capable of binding highly variable peptide sequences, and for this reason, it is highly polymorphic, with several thousand alleles identified in the population. ERAP1 is also polymorphic but to a much lesser extent, and only approximately 10 ERAP1 haplotypes have been identified in the human population (14). Such low polymorphic variation means that it would be impossible to have a tight peptide-binding cleft with strong specificity for amino acid side chains and still be able to process the highly variable peptide sequences that make it into the ER. The crystal structures in conjunction with the biochemical data presented here suggest that the solution to this problem used by ERAP1 is to have a wide peptide-binding site that can accommodate even the largest peptide that may be found in the ER. Specificity still arises by opportunistic interactions with residues in the internal lining of the cavity and shallow pockets, but this specificity is broad enough to allow for the processing of many different peptide sequences and lengths.
ERAP1 has been shown to preferentially select longer peptides (>9- to 10-aa long) as its substrates (35). This property is unusual among the aminopeptidases (36) and is highly desirable for its biological function, since it enhances generation of antigenic peptides of the correct length while limiting destruction from overtrimming. Our present findings clarify the mechanism behind this effect. Longer peptides with hydrophobic C-terminal residues can “reach” into a carboxypeptidase-like C-terminus binding site while also occupying the active site. Occupation of this C-terminus binding site is communicated to the active site through tightening of molecular interactions (i.e., correlated residue motions) that line a path from helix 12 toward helix 5 through helices 24 and 4. Thus, the ERAP1 structure with the 15-mer peptide is perfectly consistent with the “molecular ruler” mechanism put forth more than a decade ago by Goldberg and colleagues. On the other hand, the structure with the 10-mer appears to contradict this “rule”; while in theory, the 10-mer peptide is sufficiently long to occupy concurrently both the active and the C-terminus binding sites, it does not do so, but rather abuts the side of the internal cavity. This observation suggests that for shorter peptides, competition between interaction with the lining of the cavity and the C-terminus site may determine the binding mode.
The sequence of the 10-mer peptide may also be a key factor, since it carries a positively charged amino acid, which makes it incompatible with the mixed hydrophobic positively charged C-terminal binding site with which the 15-mer peptide docks. Indeed, it has been previously observed that ERAP1 is less efficient with substrates that carry positive C-terminal amino acids (35). Most HLA alleles bind peptides with C-terminal anchor residues that are hydrophobic, and thus it appears that ERAP1 has specifically evolved to trim precursors of those antigenic peptides. Accordingly, a lysine-to-alanine mutation in the C terminus of the 10-mer peptide results in an almost 5-fold increase in the trimming rate, suggesting that on substitution of a hydrophobic residue at the specific position, the new peptide is recognized in a different way, possibly through the carboxypeptidase-like C-terminal recognition site, resulting in a higher trimming rate. Nonetheless, other sequences can also be accommodated in the internal cavity, resulting in effective trimming. Thus, it appears that while the “molecular ruler” rule is confirmed by our structures, this is by no means an absolute rule, and it may apply only to peptides with hydrophobic C termini. Peptide substrates with nonhydrophobic C termini and/or shorter length must rely on opportunistic interactions with residues lining the cavity to promote binding and trimming.
The structures presented here can provide a context for another proposed function of ERAP1, namely its ability to be processive, that is, to undergo several rounds of N-terminal excision on the same peptide, leading to final product release. Although the processivity of ERAP1 has not been investigated thoroughly, the Rock group has provided some experimental support (39). Our results provide a structural template in support of a processive mechanism. The peptides are trapped in a closed cavity, and a small channel adjacent to the S1 specificity pocket may be sufficiently large (or enlarged due to local conformational changes) to allow for amino acid escape after cleavage (SI Appendix, Fig. S12) (23). The disordered region in the center of the 15-mer peptide suggests that after a trimming event and dissociation of the generated amino acid, the remaining 14-mer could just “stretch” to reach into the active site without any change in its C terminus. Eventual release of a 9-mer epitope would necessitate a large conformational change toward the open conformation of ERAP1. Slow rates of catalysis for suboptimal substrates would allow them to escape the cavity, whereas fast catalysis for good substrates would proceed via a processive mechanism. This mechanism could explain the observed propensity of ERAP1 to accumulate only specific intermediates of multiple trimming reactions in solution, although substrate selectivity is also a contributing factor (31).
The structures presented in this study provide a rationale for the effect of known coding SNPs of ERAP1 on its function. Approximately one-half of the disease-associated polymorphisms in ERAP1 either line the internal cavity and have direct interactions with a cocrystallized peptide or are nearby and affect the position of residues that make contact with peptide indirectly. This seems to be especially true for positions 725 and 730, 2 positions repeatedly identified in genome-wide association studies (GWAS) to associate with disease predisposition and to affect peptide trimming in functional studies. Interestingly, the Q730E SNP has been found to reduce the preference of ERAP1 for longer peptides, possibly due to the introduction of a negative charge adjacent to the C-terminus recognition site that hinders the approach of the substrate’s C terminus (36). All 4 SNPs that line the internal cavity of ERAP1 are conserved among other species (SI Appendix, Table S3), suggesting that their polymorphic nature may be new for humans and a result of balancing selection due to host–pathogen interactions, as previously suggested regarding the role of ERAP1 in resistance to HIV infection (40). Of note, the lysine C-terminal residue of the 10-mer peptide docks in an aspartate residue adjacent to position 725, suggesting that, at least for peptides bearing a positively charged C terminus, the R725Q SNP could be of significant importance. In contrast, position 528, which has also been repeatedly identified in GWAS and can affect enzymatic activity, is located away from the internal lining of the cavity and cannot make direct contacts with the peptide. However, it does lie on a major pathway that connects the C-terminal binding site to the active site and could affect activation by C-terminal binding, consistent with its proposed effects on the conformational dynamics of ERAP1 (36, 37). Regarding position 127, despite being on the surface of ERAP1 and particularly at domain I, an important functional role should be taken into consideration. As our crystal structure shows, Arg127 is located at the tip of a flexible loop and extends above the S1 specificity pocket, near the putative exit tunnel of the hydrolyzed amino acid residues. Further experiments are needed to unequivocally prove a processive mechanism for ERAP1, however.
Although ERAP1, like all members of the M1 family of aminopeptidases, can trim peptides in solution, it has also been proposed to trim peptides while bound onto MHC-I molecules (41). While no evidence for a direct ERAP1–MHC-I interaction has been reported to date, this mode of action is controversial, since it has been difficult to reconcile onto-MHC trimming with known ERAP1 structures and catalytic mechanism due to major steric limitations (42). Accordingly, the crystal structures presented here are not consistent with the onto-MHC trimming model, given that the peptide is trapped in a fully closed cavity and has no opportunity to concurrently bind (even partially) onto the MHC-I groove (43). Thus, the structures presented here are both consistent with and provide a structural basis for understanding all the main hallmarks of ERAP1 that underlie its biological function: limited sequence selectivity with lack of a specific sequence motif, length preferences, SNP effects, and C-terminal preferences. In this context, more complex mechanistic models of ERAP1 activity might not be necessary to explain its biological function.
In summary, we describe 2 high-resolution structures of ERAP1, an enzyme important for human adaptive immunity that is an emerging target for cancer immunotherapy, with bound substrate analogs. These structures, along with biochemical and computational analysis, support a multistep mechanism for ERAP1-mediated trimming of antigenic peptide precursors. The nature of the interactions revealed by these structures can explain the complex effects of ERAP1 in the cellular immunopeptidome and may be useful in the design of modulators that regulate ERAP1 activity for therapeutic applications.
Experimental Procedures
Detailed experimental procedures for the synthesis of the peptide analogs, protein construct, crystallization and data collection, structure determination and refinement, enzymatic assays, and computational methods are available in the SI Appendix.
Data Availability Statement.
All data discussed in the paper are available to readers.
Supplementary Material
Acknowledgments
We thank the beamline scientists of EMBL-Hamburg for their assistance during data collection at the P13 beamline of PETRA III (Hamburg, Germany). This research was financed by the Harry J. Lloyd Charitable Trust through a grant to E.S. and by the project “National Centre for Scientific Research Demokritos–Institute of Nuclear & Radiological Sciences and Technology, Energy & Safety Research Activities in the Framework of the National RIS3” (MIS 5002559), implemented under the “Action for the Strategic Development on the Research and Technological Sector” program, funded by the Operational Program “Competitiveness, Entrepreneurship and Innovation” (Grant NSRF 2014-2020) and cofinanced by Greece and the European Union (European Regional Development Fund). A.M. and A.P. acknowledge support by the General Secretariat for Research and Technology and the Hellenic Foundation for Research and Innovation (Postdoctoral Grant no. 303). This work was also supported by the project “INSPIRED–The National Research Infrastructures on Integrated Structural Biology, Drug Screening Efforts and Drug Target Functional Characterization” (Grant MIS 5002550), implemented under the “Reinforcement of the Research and Innovation Infrastructure” program funded by the Operational Program “Competitiveness, Entrepreneurship and Innovation” (Grant NSRF 2014-2020) and cofinanced by Greece and the European Union (European Regional Development Fund). Synchrotron access was supported by iNEXT Project 653706 (Proposal ID 5589), funded by the Horizon 2020 Program of the European Union and by European Molecular Biology Laboratory-Hamburg (Proposal MX-619).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Atomic coordinates and structure factors have been deposited in the Protein Data Bank, http://www.rcsb.org/ (PDB ID codes 6RQX and 6RYF). Enzymatic analysis and molecular dynamics data have been deposited on Mendeley Data, http://dx.doi.org/10.17632/5sg9m3wgkf.1.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912070116/-/DCSupplemental.
References
- 1.Rock K. L., Reits E., Neefjes J., Present yourself! By MHC class I and MHC class II molecules. Trends Immunol. 37, 724–737 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weimershaus M., Evnouchidou I., Saveanu L., van Endert P., Peptidases trimming MHC class I ligands. Curr. Opin. Immunol. 25, 90–96 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Rock K. L., Farfán-Arribas D. J., Colbert J. D., Goldberg A. L., Re-examining class-I presentation and the DRiP hypothesis. Trends Immunol. 35, 144–152 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evnouchidou I., Papakyriakou A., Stratikos E., A new role for Zn(II) aminopeptidases: Antigenic peptide generation and destruction. Curr. Pharm. Des. 15, 3656–3670 (2009). [DOI] [PubMed] [Google Scholar]
- 5.York I. A., et al. , The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8-9 residues. Nat. Immunol. 3, 1177–1184 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Ham-mer G. E., Gonzalez F., James E., Nolla H., Shastri N., In the absence of aminopeptidase ERAAP, MHC class I molecules present many unstable and highly immunogenic peptides. Nat. Immunol. 8, 101–108 (2007). [DOI] [PubMed] [Google Scholar]
- 7.James E., Bailey I., Sugiyarto G., Elliott T., Induction of protective antitumor immunity through attenuation of ERAAP function. J. Immunol. 190, 5839–5846 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Ham-mer G. E., Gonzalez F., Champsaur M., Cado D., Shastri N., The aminopeptidase ERAAP shapes the peptide repertoire displayed by major histocompatibility complex class I molecules. Nat. Immunol. 7, 103–112 (2006). [DOI] [PubMed] [Google Scholar]
- 9.York I. A., Brehm M. A., Zendzian S., Towne C. F., Rock K. L., Endoplasmic reticulum aminopeptidase 1 (ERAP1) trims MHC class I-presented peptides in vivo and plays an important role in immunodominance. Proc. Natl. Acad. Sci. U.S.A. 103, 9202–9207 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hisatsune C., et al. , ERp44 exerts redox-dependent control of blood pressure at the ER. Mol. Cell 58, 1015–1027 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Aldhamen Y. A., et al. , Autoimmune disease-associated variants of extracellular endoplasmic reticulum aminopeptidase 1 induce altered innate immune responses by human immune cells.J. Innate Immun. 7, 275–289 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.López de Castro J. A., et al. , Molecular and pathogenic effects of endoplasmic reticulum aminopeptidases ERAP1 and ERAP2 in MHC-I-associated inflammatory disorders: Towards a unifying view. Mol. Immunol. 77, 193–204 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Stratikos E., Stamogiannos A., Zervoudi E., Fruci D., A role for naturally occurring alleles of endoplasmic reticulum aminopeptidases in tumor immunity and cancer pre-disposition. Front. Oncol. 4, 363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ombrello M. J., Kastner D. L., Rem-mers E. F., Endoplasmic reticulum-associated amino-peptidase 1 and rheumatic disease: Genetics. Curr. Opin. Rheumatol. 27, 349–356 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evnouchidou I., et al. , Cutting edge: Coding single nucleotide polymorphisms of endoplasmic reticulum aminopeptidase 1 can affect antigenic peptide generation in vitro by influencing basic enzymatic properties of the enzyme. J. Immunol. 186, 1909–1913 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reeves E., Edwards C. J., Elliott T., James E., Naturally occurring ERAP1 haplotypes encode functionally distinct alleles with fine substrate specificity. J. Immunol. 191, 35–43 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stratikos E., Modulating antigen processing for cancer immunotherapy. OncoImmunology 3, e27568 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zervoudi E., et al. , Rationally designed inhibitor targeting antigen-trimming aminopeptidases enhances antigen presentation and cytotoxic T-cell responses. Proc. Natl. Acad. Sci. U.S.A. 110, 19890–19895 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Georgiadis D., Mpakali A., Koumantou D., Stratikos E., Inhibitors of ER aminopeptidase 1 and 2: From design to clinical application. Curr. Med. Chem. 26, 2715–2729 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Stratikos E., Stern L. J., Antigenic peptide trimming by ER aminopeptidases—Insights from structural studies. Mol. Immunol. 55, 212–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen T. T., et al. , Structural basis for antigenic peptide precursor processing by the endoplasmic reticulum aminopeptidase ERAP1. Nat. Struct. Mol. Biol. 18, 604–613 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kochan G., et al. , Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming. Proc. Natl. Acad. Sci. U.S.A. 108, 7745–7750 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giastas P., Neu M., Rowland P., Stratikos E., High-resolution crystal structure of endoplasmic reticulum aminopeptidase 1 with bound phosphinic transition-state analogue inhibitor. ACS Med. Chem. Lett. 10, 708–713 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papakyriakou A., Stratikos E., The role of conformational dynamics in antigen trimming by intracellular aminopeptidases. Front. Immunol. 8, 946 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gandhi A., Lakshminarasimhan D., Sun Y., Guo H. C., Structural insights into the molecular ruler mechanism of the endoplasmic reticulum aminopeptidase ERAP1. Sci. Rep. 1, 186 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sui L., Gandhi A., Guo H. C., Crystal structure of a polypeptide’s C-terminus in complex with the regulatory domain of ER aminopeptidase 1. Mol. Immunol. 80, 41–49 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mpakali A., et al. , Crystal structure of insulin-regulated aminopeptidase with bound substrate analogue provides insight on antigenic epitope precursor recognition and processing. J. Immunol. 195, 2842–2851 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Mpakali A., et al. , Structural basis for antigenic peptide recognition and processing by endoplasmic reticulum (ER) aminopeptidase 2. J. Biol. Chem. 290, 26021–26032 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zervoudi E., et al. , Probing the S1 specificity pocket of the aminopeptidases that generate antigenic peptides. Biochem. J. 435, 411–420 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saveanu L., et al. , Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat. Immunol. 6, 689–697 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Georgiadou D., et al. , Placental leucine aminopeptidase efficiently generates mature antigenic peptides in vitro but in patterns distinct from endoplasmic reticulum aminopeptidase 1. J. Immunol. 185, 1584–1592 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kokkala P., et al. , Optimization and structure-activity relationships of phosphinic pseudotripeptide inhibitors of aminopeptidases that generate antigenic peptides. J. Med. Chem. 59, 9107–9123 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Brown M. A., Kenna T., Wordsworth B. P., Genetics of ankylosing spondylitis—Insights into pathogenesis. Nat. Rev. Rheumatol. 12, 81–91 (2016). [DOI] [PubMed] [Google Scholar]
- 34.López de Castro J. A., How ERAP1 and ERAP2 shape the peptidomes of disease-associated MHC-I proteins. Front. Immunol. 9, 2463 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang S. C., Momburg F., Bhutani N., Goldberg A. L., The ER aminopeptidase, ERAP1, trims precursors to lengths of MHC class I peptides by a “molecular ruler” mechanism. Proc. Natl. Acad. Sci. U.S.A. 102, 17107–17112 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stamogiannos A., Koumantou D., Papakyriakou A., Stratikos E., Effects of polymorphic variation on the mechanism of endoplasmic reticulum aminopeptidase 1. Mol. Immunol. 67, 426–435 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Stamogiannos A., et al. , Critical role of interdomain interactions in the conformational change and catalytic mechanism of endoplasmic reticulum aminopeptidase 1. Biochemistry 56, 1546–1558 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Wart A. T., Durrant J., Votapka L., Amaro R. E., Weighted implementation of suboptimal paths (WISP): An optimized algorithm and tool for dynamical network analysis. J. Chem. Theory Comput. 10, 511–517 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hearn A., York I. A., Rock K. L., The specificity of trimming of MHC class I-presented peptides in the endoplasmic reticulum. J. Immunol. 183, 5526–5536 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cagliani R., et al. , Genetic diversity at endoplasmic reticulum aminopeptidases is maintained by balancing selection and is associated with natural resistance to HIV-1 infection. Hum. Mol. Genet. 19, 4705–4714 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Chen H., et al. , ERAP1-ERAP2 dimers trim MHC I-bound precursor peptides; implications for understanding peptide editing. Sci. Rep. 6, 28902 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mpakali A., Maben Z., Stern L. J., Stratikos E., Molecular pathways for antigenic peptide generation by ER aminopeptidase 1. Mol. Immunol. 113, 50–57 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Papakyriakou A., et al. , The partial dissociation of MHC class I-bound peptides exposes their N terminus to trimming by endoplasmic reticulum aminopeptidase 1. J. Biol. Chem. 293, 7538–7548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data discussed in the paper are available to readers.