Significance
The mitochondrial enzyme glutaminase (GLS) is frequently up-regulated in cancer cells, and a GLS-selective inhibitor is being evaluated in clinical trials. Previous screens identified succinylated lysine residues on GLS, but the functional consequences of these posttranslational modifications have remained unclear. Here, we report that the mitochondrial desuccinylase SIRT5 stabilizes GLS. Both GLS and SIRT5 are upregulated during cellular transformation, and high expression of SIRT5 in human breast tumors correlates with poor patient prognosis. Mechanistically, SIRT5-mediated desuccinylation of residue K164 protects GLS from ubiquitination at K158 and from subsequent degradation. These findings reveal an important role for SIRT5 in mammary tumorigenesis and establish a posttranslational mechanism regulating GLS levels. Collectively, they support further investigation of SIRT5 as a potential therapeutic target.
Keywords: sirtuin, glutaminase, cancer, metabolism, SIRT5
Abstract
The mitochondrial enzyme glutaminase (GLS) is frequently up-regulated during tumorigenesis and is being evaluated as a target for cancer therapy. GLS catalyzes the hydrolysis of glutamine to glutamate, which then supplies diverse metabolic pathways with carbon and/or nitrogen. Here, we report that SIRT5, a mitochondrial NAD+-dependent lysine deacylase, plays a key role in stabilizing GLS. In transformed cells, SIRT5 regulates glutamine metabolism by desuccinylating GLS and thereby protecting it from ubiquitin-mediated degradation. Moreover, we show that SIRT5 is up-regulated during cellular transformation and supports proliferation and tumorigenesis. Elevated SIRT5 expression in human breast tumors correlates with poor patient prognosis. These findings reveal a mechanism for increasing GLS expression in cancer cells and establish a role for SIRT5 in metabolic reprogramming and mammary tumorigenesis.
Metabolic reprogramming during tumorigenesis permits sustained biomass accumulation and redox homeostasis, even when nutrient supplies in the tumor microenvironment are fluctuating or limited (1). Transformed cells often exhibit high rates of glucose consumption coupled to lactate secretion regardless of O2 availability (the Warburg effect), as well as increased use of glutamine as a carbon and nitrogen source (1). Deamidation of glutamine to glutamate in the cytosol is catalyzed by amidotransferase enzymes, which require an acceptor metabolite for the amide nitrogen. However, mitochondrial glutaminases (GLS and GLS2) rapidly convert glutamine to glutamate and ammonium, with no need for a nitrogen acceptor substrate. The glutamate product of the glutaminase reaction can be incorporated into glutathione and nonessential amino acid biosynthesis pathways or deaminated to yield the tricarboxylic acid (TCA) cycle intermediate α-ketoglutarate (α-KG) (2, 3). In some contexts, glutaminase activity is essential for the proliferation or survival of cancer cells, and the GLS inhibitor CB-839 is in clinical trials for treatment of various malignancies (4).
Several oncogenic signaling pathways drive up-regulation of GLS at the transcriptional or translational level (5–8), and GLS activity is also influenced by phosphorylation (9, 10). High-throughput mass spectrometry-based screens have identified numerous additional posttranslational modifications (PTMs) on GLS including lysine acylations, the functions of which have not yet been elucidated (11, 12). Acylation modifications of lysine residues, which include acetylation, malonylation, succinylation, glutarylation, myristoylation, and palmitoylation, are regulated by the sirtuins, a family of NAD+-dependent deacylases (13). Collectively, these enzymes modulate diverse cellular processes including gene transcription, DNA repair, stress resistance, and metabolism (13). Because their activity is controlled by the local NAD+/NADH ratio, sirtuins couple the nutritional status of a cell to the protein PTM landscape.
Seven sirtuins, SIRT1–7, are encoded in the human genome, 3 of which (SIRT3–5) are localized primarily in mitochondria. The sirtuins play complex roles in healthy and diseased cells and can act to suppress or support tumorigenesis. For example, loss of SIRT2, SIRT3, SIRT4, or SIRT6 increases tumor incidence in mice (14–17), consistent with tumor suppressor activity, but each of these enzymes is also reported to have onco-supportive activity in specific contexts (18–21). Numerous studies have found tumor-suppressor functions for SIRT1 (22, 23), whereas SIRT7 is widely associated with oncogenic activity (24). The roles of SIRT5 in cancer are less well established, but emerging evidence indicates that SIRT5 can support metabolic reprogramming, favoring the Warburg effect and promoting catabolism of amino acids and fatty acids (25).
SIRT5 has robust lysine desuccinylase activity and is the primary regulator of the mitochondrial succinylome (26, 27). Proteomic screens of mouse liver tissue, mouse embryonic fibroblasts (MEFs), and human cervical carcinoma cells (HeLa) have identified 1,000 to 3,000 lysine succinylation sites across 250 to 800 proteins (11, 12, 26). Specific succinylations on GLS were identified in both MEFs and HeLa cells (11, 12), implicating GLS as a SIRT5 substrate. Here, we report that cellular transformation is accompanied by increased expression of SIRT5, which supports proliferation, anchorage-independent growth, and tumorigenesis. Mechanistically, SIRT5 stabilizes GLS by desuccinylating lysine residue K164, which in turn protects GLS from ubiquitination at residue K158 and subsequent degradation. Thus, SIRT5 supports oncogenesis in part by stabilizing a mitochondrial enzyme with critical roles in metabolic reprogramming.
Results
SIRT5 Regulates GLS Levels in Transformed Cells.
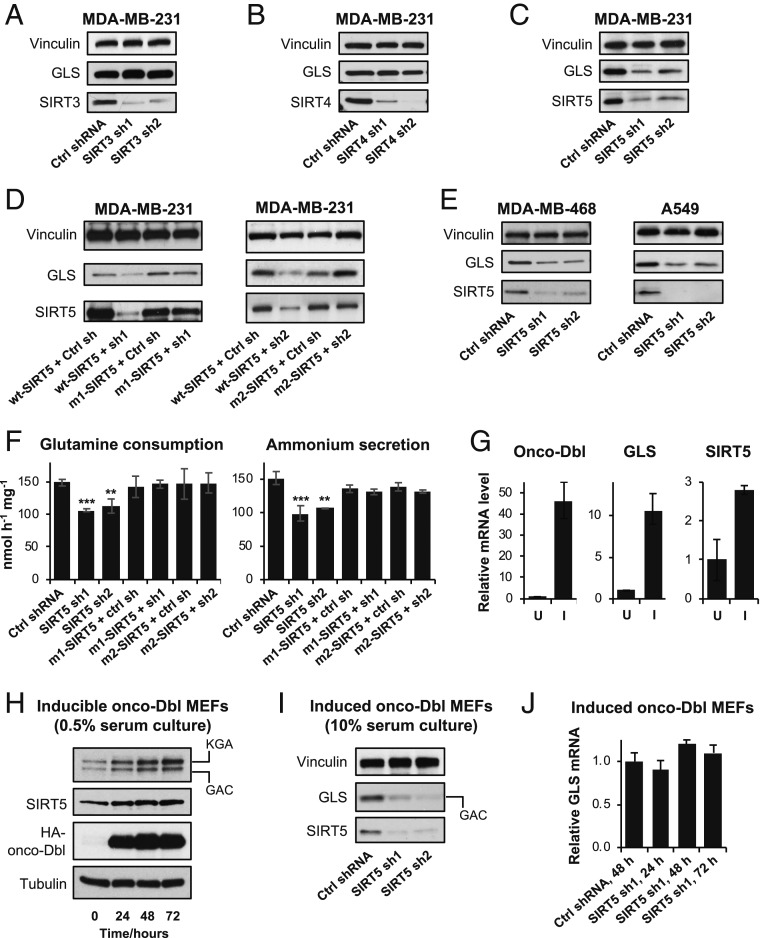
Cellular transformation downstream of diverse oncogenic signals is accompanied by, and dependent on, increased expression of mitochondrial GLS (8, 28, 29). Previous high-throughput screens revealed that specific lysine residues on GLS are subjected to acylations, a class of PTMs regulated by sirtuin-family enzymes (11, 12). To test whether sirtuins influence GLS function in transformed cells, we first used short hairpin RNAs (shRNAs) to knock down each of the mitochondrial family members (SIRT3–5) in MDA-MB-231 breast cancer cells, which express high levels of GLS and are sensitive to GLS-selective inhibitors (30). Western blot analysis showed that depletion of SIRT3 or SIRT4 for 48 h had negligible impact on GLS levels (Fig. 1 A and B). In contrast, knockdown of SIRT5 caused a pronounced decrease in GLS abundance (Fig. 1C). To confirm that this was an on-target, SIRT5-associated effect, we designed RNA interference (RNAi)-resistant SIRT5 constructs (SI Appendix, Fig. S1 A and B). Using these plasmids to restore SIRT5 expression in the presence of the shRNAs was sufficient to rescue GLS levels, indicating that the observed regulation of GLS was specific and SIRT5-mediated (Fig. 1D). In additional cancer cell lines, including KEAP1-mutant A549 lung adenocarinoma cells which, like MDA-MB-231 cells, are highly dependent on GLS activity (7, 31), knockdown of SIRT5 consistently resulted in decreased GLS protein levels (Fig. 1E).
Fig. 1.
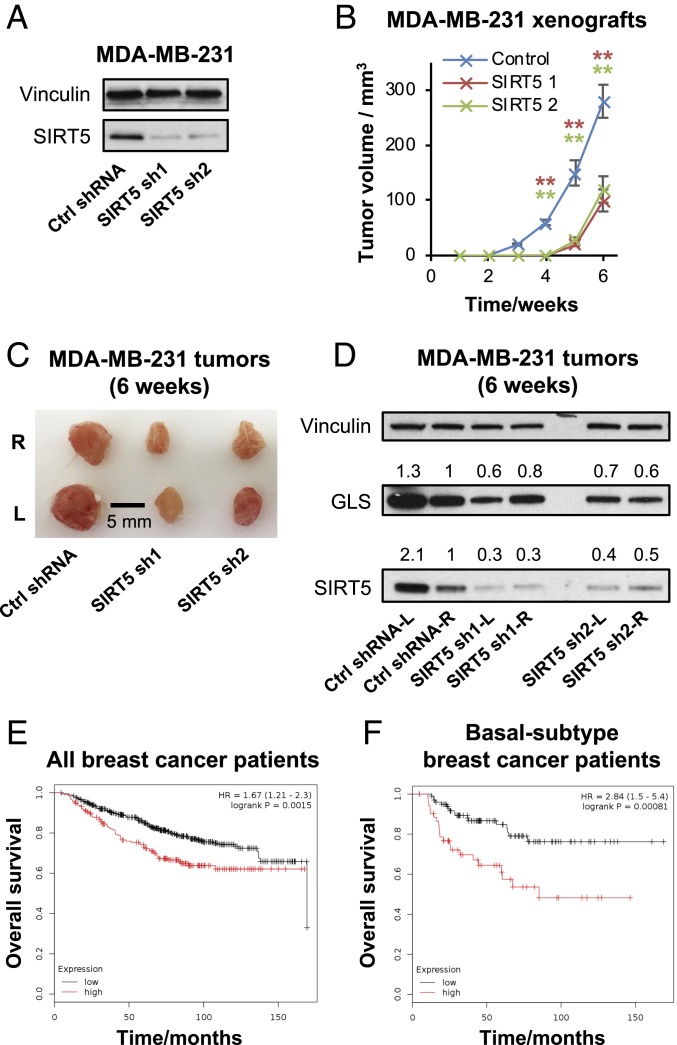
SIRT5 regulates GLS levels in transformed cells. (A–C) Western blots showing GLS levels in MDA-MB-231 breast cancer cells expressing control shRNA or shRNAs selectively targeting each of the mitochondrial sirtuins, SIRT3, SIRT4, and SIRT5. (D) Western blot analysis showing that ectopic expression of RNAi-resistant SIRT5 constructs is sufficient to restore GLS levels in MDA-MB-231 cells in the presence of the SIRT5-targeted shRNAs. (E) Western blot analysis showing the effects of SIRT5 knockdown on GLS levels in additional breast cancer (MDA-MB-468) and lung cancer (A549) cell lines. (F) Plots showing the effect of SIRT5 knockdown on the rate of glutamine consumption (Left) or ammonium secretion (Right) by MDA-MB-231 cells. RNAi-resistant SIRT5 constructs were used to reintroduce SIRT5 in the presence of the shRNAs. Nutrient consumption and secretion rates are shown with units of nanomoles (nmol) per hour per milligram of total cellular protein (nmol⋅h−1⋅mg−1). Mean ± SD from biological triplicates. (G) Quantitative real-time PCR analysis showing transcript levels of oncogenic-Dbl, GLS, and SIRT5 in inducible oncogenic-Dbl MEFs, either uninduced (U) or induced (I) for 24 h. Cells were cultured in low-serum (0.5% FBS) medium. Relative quantification (RQ) is value plotted, with error bars marking RQmax and RQmin. (H) Western blots showing protein levels of GLS (the KGA and GAC splice variants are indicated), SIRT5, and HA-tagged onco-Dbl in uninduced or induced onco-Dbl MEFs. Cells were cultured in low-serum (0.5% FBS) medium. (I) Western blot analysis showing the effects of SIRT5 knockdown on GLS levels in induced (24 h) onco-Dbl MEFs. Cells were cultured in high-serum (10% FBS) medium to minimize cell death resulting from shRNA expression. The high-serum conditions increase the GAC:KGA ratio relative to the blots shown in H. (J) Quantitative real-time PCR analysis showing stable GLS transcript levels following expression of SIRT5-targeted shRNAs in induced onco-Dbl MEFs. RQ value is plotted, with error bars marking RQmax and RQmin. ***P < 0.01. **P < 0.02.
Corresponding to the depletion of GLS, knockdown of SIRT5 in MDA-MB-231 cells significantly decreased the rate of glutamine consumption from the culture medium, an effect that could be rescued by introducing the RNAi-resistant SIRT5 constructs (Fig. 1 F, Left). The glutaminase reaction releases the amide nitrogen of glutamine as an ammonium ion. Matching previous observations of cultured cancer cells (32–34), ammonium secretion into the culture medium occurred at an approximately 1:1 molar ratio with glutamine consumption (Fig. 1F). Knockdown of SIRT5 decreased the rate of ammonium release, consistent with the down-regulation of GLS, and ectopic expression of RNAi-resistant SIRT5 was again sufficient to rescue this effect (Fig. 1 F, Right). Analysis of intracellular metabolite levels revealed that those downstream of the glutaminase reaction, including glutamate, the TCA cycle intermediates α-KG, malate, and citrate, along with aspartate (a product of glutamate-dependent amination of oxaloacetate), were all significantly decreased by SIRT5 knockdown (SI Appendix, Fig. S1 C and D). In contrast, glycolytic metabolites and essential amino acids were not affected (SI Appendix, Fig. S1E).
SIRT5 Is Up-Regulated during Cellular Transformation.
Oncogenic signaling downstream of hyperactivated small GTPases, including Rho- and Ras-family members, increases expression of GLS (7, 29). Since SIRT5 regulates GLS, we next monitored the level of SIRT5 following expression of oncogenic-Dbl (onco-Dbl), a potent activator of Rho GTPase signaling, in an inducible MEF system. For these signaling experiments we used low-serum [0.5% fetal bovine serum (FBS)] culture conditions in order to maximize the signal-to-background ratio of the oncogenic pathway. Induction for 48 h resulted in a 46-fold increase in onco-Dbl and an 11-fold increase in GLS transcript levels, as reported previously (7), and also a 3-fold increase in the SIRT5 transcript (Fig. 1G). Using Western blot analysis we observed similar changes in expression at the protein level (Fig. 1H), which were consistent across biological replicate clones (SI Appendix, Fig. S2A). Under low-serum culture conditions, both the GAC and KGA splice variants of GLS are detectable in MEFs (Fig. 1H and SI Appendix, Fig. S2A) (7). In a separate MEF system, induction of oncogenic KRAS-G12V also resulted in increased expression of GLS and SIRT5, indicating that these events might be a common feature of cellular transformation (SI Appendix, Fig. S2B).
We then used shRNAs to knock down SIRT5 following induction of onco-Dbl. To minimize cell death, these knockdown experiments were carried out in high-serum culture (10% FBS) conditions, which favor the GAC splice variant of GLS. As in the cancer cell lines, SIRT5 depletion caused a pronounced decrease in the abundance of GLS (Fig. 1I and SI Appendix, Fig. S2C). Notably, levels of the GLS transcript were not affected by SIRT5 depletion, indicating that regulation of GLS by SIRT5 occurs at the translational or posttranslational level (Fig. 1J).
SIRT5 Desuccinylates GLS to Protect It from Ubiquitination.
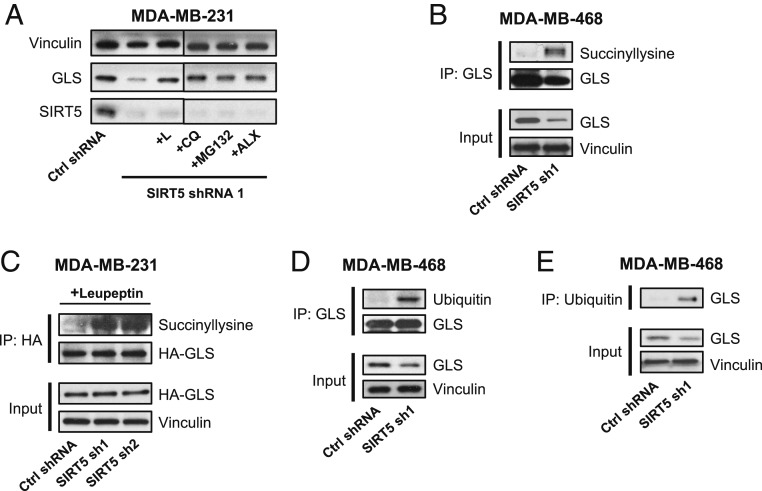
The results above suggest that the mechanism connecting GLS and SIRT5 might involve the regulation of GLS protein stability. To test this possibility, we knocked down SIRT5 for 48 h in MDA-MB-231 cells and simultaneously treated the cells with the proteasome/lysosome inhibitors leupeptin (L), chloroquine (CQ), MG132, or Alx-260–090 (ALX). These treatments abolished the down-regulation of GLS that normally follows SIRT5 knockdown (Fig. 2A).
Fig. 2.
SIRT5 desuccinylates GLS and protects it from ubiquitination. (A) Western blot analysis showing GLS levels in MDA-MB-231 cells expressing a control shRNA or SIRT5-targeted shRNA. Cells were either untreated (left 2 lanes) or treated for 48 h with the proteasome/lysosome inhibitors leupeptin (L), chloroquine (CQ), MG132, or ALX-260–090 to block degradation of GLS. (B) Immunoprecipitation of endogenous GLS from lysates (3 mg total cellular protein per IP) of MDA-MB-468 cells expressing either a control shRNA or SIRT5-targeted shRNA. Western blot analysis using an anti-succinyllysine antibody shows that lysine succinylation on GLS is highly enriched following SIRT5 knockdown. (C) Immunoprecipitation of ectopically expressed HA-tagged GLS from lysates of MDA-MB-231 cells expressing a control shRNA or SIRT5-targeted shRNA and treated with leupeptin to inhibit GLS degradation. Western blot analysis using an anti-succinyllysine antibody shows that succinylated GLS is highly enriched following SIRT5 knockdown. (D) Immunoprecipitation of endogenous GLS from lysates of MDA-MB-468 cells expressing either a control shRNA or a SIRT5-targeted shRNA. Western blot analysis shows that ubiquitinated GLS levels are highly elevated following SIRT5 knockdown, and total GLS levels are depleted. (E) Immunoprecipitation using an anti-ubiquitin antibody from lysates of MDA-MB-468 cells expressing either a control shRNA or a SIRT5-targeted shRNA. Western blot analysis shows that more GLS is immunoprecipitated by the anti-ubiquitin antibody following SIRT5 knockdown despite lower levels of total GLS being present, indicating increased ubiquitination of GLS.
We next wanted to confirm that SIRT5 regulates the lysine succinylation status of GLS. For these experiments, we initially used MDA-MB-468 breast cancer cells, which express high levels of both GLS and SIRT5. After knocking down SIRT5 for 48 h, we immunoprecipitated GLS from whole-cell lysates and probed the pull-downs using an anti-succinyllysine antibody. Although depletion of SIRT5 results in decreased total abundance of GLS, the extent of lysine succinylation on GLS was greatly increased in the SIRT5-knockdown samples (Fig. 2B). We obtained similar results by immunoprecipitating hemagglutinin (HA)-tagged GLS from MDA-MB-231 breast cancer cells, although in this case it was necessary to treat cells with leupeptin to minimize GLS degradation following SIRT5 knockdown (Fig. 2C).
Lysine acylations can regulate protein stability, positively or negatively, by influencing ubiquitination (35, 36). Since SIRT5 catalyzes the desuccinylation of GLS and protects it from degradation, we hypothesized that SIRT5-mediated desuccinylation regulates the ubiquitination status of GLS. To test this, we again immunoprecipitated GLS from control or SIRT5-knockdown MDA-MB-468 cell lysates and then probed the pull-downs using an ubiquitin-targeted antibody. Ubiquitinated GLS was much more abundant in the SIRT5 knockdown samples (Fig. 2D). The reciprocal experiment, i.e., immunoprecipitating total ubiquitinated protein using an anti-ubiquitin antibody and then probing the pull-downs for GLS, also showed that SIRT5 knockdown causes increased ubiquitination of GLS (Fig. 2E). Collectively, these findings suggest that SIRT5 functions to desuccinylate GLS and inhibit its ubiquitination, thus protecting GLS from ubiquitin-mediated degradation.
SIRT5 Desuccinylates GLS Residue K164 to Block Ubiquitination of K158.
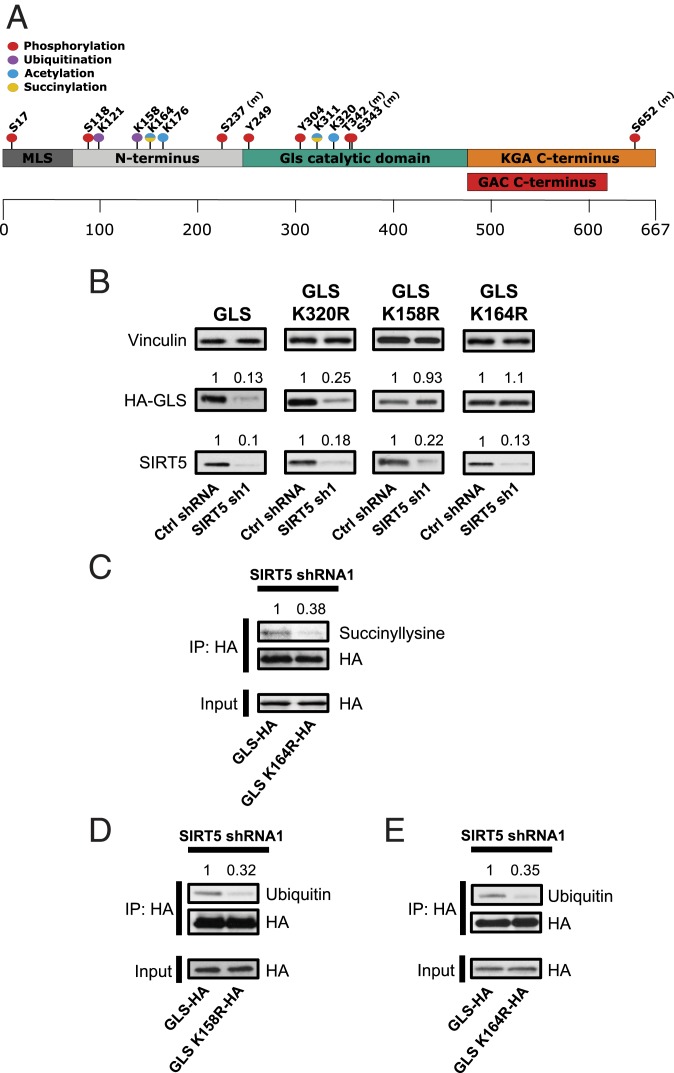
High-throughput mass spectrometry-based screens of MEFs and HeLa cells previously identified several PTMs on GLS (Fig. 3A) (11, 12). To understand the importance of the specific lysine succinylation and ubiquitination sites on GLS, we generated constructs for expressing HA-tagged GLS in which the succinylation site K164 and the neighboring ubiquitination site K158 (SI Appendix, Fig. S3 A and B), as well as the acetylation site K320, were individually changed to arginine. We did not investigate the succinylation site K311, as previous studies have established that this residue is required for oligomerization and activation of GLS (37, 38).
Fig. 3.
Residue K164 of GLS is desuccinylated by SIRT5 to suppress ubiquitination of K158. (A) Diagram showing posttranslational modifications on GLS identified in earlier mass spectrometry-based screens (11, 12). (B) Western blot analysis of lysates prepared from MDA-MB-231 cells ectopically expressing HA-tagged wild-type GLS, K158R-GLS, K164R-GLS, or K320R-GLS, along with a control shRNA or SIRT5-targeted shRNA. The K164R-GLS and K158R-GLS variants are protected from degradation following SIRT5 knockdown. Densitometry analysis of the HA-GLS and SIRT5 bands was performed using ImageJ. (C) Immunoprecipitation of ectopically expressed HA-tagged wild-type GLS or K164R-GLS from lysates of leupeptin-treated MDA-MB-231 cells in which SIRT5 had been knocked down. Western blot analysis using an anti-succinyllysine antibody shows that the K164R-GLS variant has less lysine succinylation than wild-type GLS. (D) Immunoprecipitation of ectopically expressed HA-tagged wild-type GLS or K158R-GLS from lysates of MDA-MB-231 cells in which SIRT5 had been knocked down. Western blot analysis using an anti-ubiquitin antibody shows decreased ubiquitination of the K158R-GLS variant relative to wild-type GLS. (E) Immunoprecipitation of ectopically expressed HA-tagged wild-type GLS or K164R-GLS from lysates of MDA-MB-231 cells in which SIRT5 had been knocked down. Western blot analysis using an anti-ubiquitin antibody shows decreased ubiquitination of the K164R-GLS variant relative to wild-type GLS.
We ectopically expressed the HA-tagged GLS constructs in MDA-MB-231 cells and knocked down SIRT5 for 48 h, at which point whole-cell lysates were prepared and probed by Western blot using an HA-tag–targeted antibody. As expected, knockdown of SIRT5 resulted in reduced levels of wild-type GLS (Fig. 3B). Similarly, the GLS variant K320R (acetylation site) was sharply down-regulated following SIRT5 knockdown. In contrast, the K164R (succinylation site) variant was not affected by SIRT5 knockdown and remained stable when SIRT5 levels were depleted (Fig. 3B). The same was true when we examined the K164R variant in onco-Dbl–expressing MEFs (SI Appendix, Fig. S3C). In addition, the GLS K158R variant, in which the lysine-to-arginine substitution was made at the putative ubiquitination site in close proximity to K164, was similarly unaffected by knockdown of SIRT5 (Fig. 3B). Thus, SIRT5-mediated regulation of GLS stability involves the GLS succinylation site K164, along with the adjacent ubiquitination site K158.
To confirm that K164 is the key SIRT5-regulated succinylation site on GLS, we immunoprecipitated HA-tagged wild-type GLS or K164R-GLS from MDA-MB-231 cells, 48 h after knocking down SIRT5. We then probed the pull-downs by Western blot, using an anti-succinyllysine antibody. This revealed that mutation of K164 results in sharply decreased succinylation of GLS (Fig. 3C), implicating K164 as a primary site for this modification. To confirm that residue K158 is a ubiquitination site, we also immunoprecipitated HA-tagged K158R-GLS and probed the pull-downs using an ubiquitin-targeted antibody. The GLS K158R variant had a much lower ubiquitination level than wild-type GLS (Fig. 3D), supporting a model in which the increased ubiquitination of GLS that follows SIRT5 knockdown is primarily at residue K158.
Since depletion of SIRT5, a lysine desuccinylase, leads to increased ubiquitination of GLS, we hypothesized that the succinylation of GLS residue K164 following SIRT5 knockdown acts as a marker for subsequent ubiquitination at residue K158. To test this possibility, we used the K164R-GLS variant, which is defective in succinylation (Fig. 3C). As above, we immunoprecipitated HA-tagged wild-type or K164R-GLS from MDA-MB-231 cells in which SIRT5 had been knocked down. We then probed the pull-downs by Western blot using an ubiquitin-targeted antibody. Consistent with our model, the succinylation-defective GLS variant showed substantially less ubiquitination than wild-type GLS (Fig. 3E).
SIRT5 Supports Proliferation and Anchorage-Independent Growth.
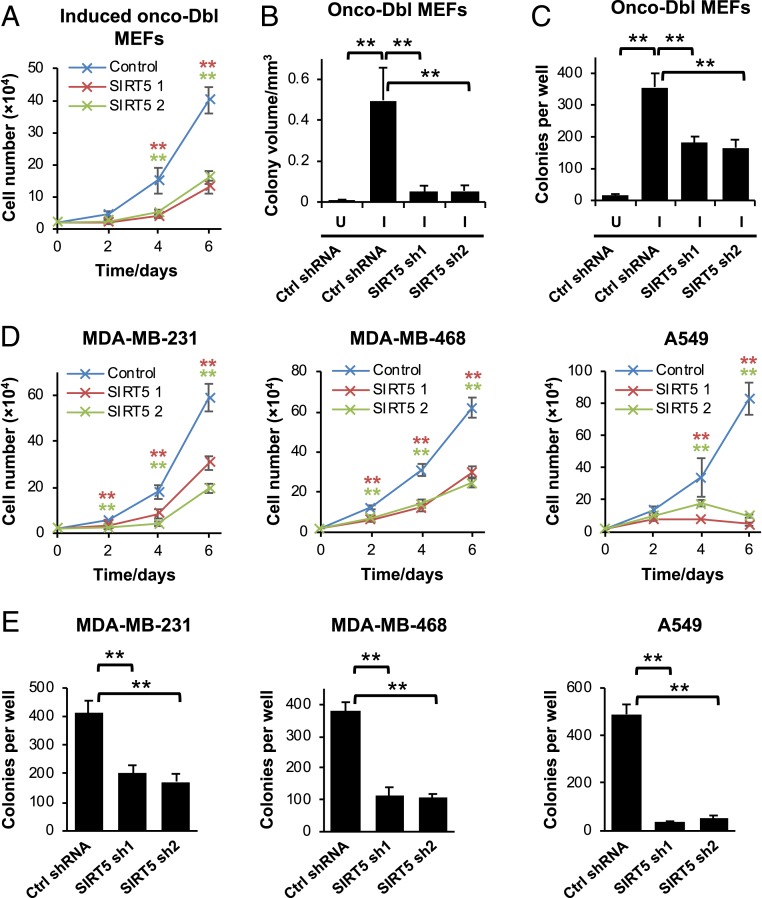
The results above show that SIRT5 is up-regulated during oncogenic transformation and stabilizes GLS, an enzyme with important functions in cancer-cell metabolic reprogramming. We therefore investigated the effects of suppressing SIRT5 expression in transformed cells. Knockdown of SIRT5 in induced onco-Dbl MEFs inhibited cell proliferation (Fig. 4A), with similar results obtained across replicate clones (SI Appendix, Fig. S4A). Knockdown of SIRT5 also decreased the volume and number of colonies formed by the induced onco-Dbl MEFs in anchorage-independent growth assays (Fig. 4 B and C and SI Appendix, Fig. S4 B and C). In breast cancer and lung cancer cell lines, SIRT5 knockdowns again inhibited proliferation and anchorage-independent growth (Fig. 4 D and E). Expression of RNAi-resistant SIRT5 constructs rescued the proliferative defect caused by the SIRT5-targeted shRNAs, indicating that the observed effects were on-target and SIRT5-associated (SI Appendix, Fig. S4D).
Fig. 4.
SIRT5 supports cell proliferation and anchorage-independent growth. (A) Cell proliferation assays showing the inhibitory effect of SIRT5 knockdown in induced onco-Dbl MEFs. Mean ± SD from biological triplicates. (B and C) Anchorage-independent growth assays for uninduced (U) or induced (I) onco-Dbl MEFs expressing either a control shRNA or SIRT5-targeted shRNAs. The colony volume is shown in B, and the total number of colonies formed per well is shown in C. Mean ± SD from biological triplicates. (D) Cell proliferation assays showing the inhibitory effect of SIRT5 knockdown in the breast cancer cell lines MDA-MB-231 and MDA-MB-468 and the lung cancer cell line A549. Mean ± SD from biological triplicates. (E) Anchorage-independent growth assays showing the number of colonies per well formed by the cancer cell lines expressing either a control shRNA or SIRT5-targeted shRNAs. Mean ± SD from biological triplicates. **P < 0.01.
SIRT5 Supports Tumorigenesis In Vivo.
To address whether SIRT5 is important for tumorigenesis in vivo, we transduced MDA-MB-231 breast cancer cells with either a control shRNA or SIRT5-targeted shRNAs and confirmed knockdown of SIRT5 after 48 h (Fig. 5A). We then subcutaneously (s.c.) injected 3 × 106 cells in Matrigel suspension into each flank of female NOD scid gamma (NSG) mice (n = 8 per condition) and measured tumor size every 7 d until 42 d after inoculation. Although tumors formed in all mice, tumor initiation was delayed by ∼2 wk in SIRT5-depleted samples (Fig. 5B). When harvested at 42 d, the SIRT5-shRNA tumors were smaller and weighed less than tumors expressing the control shRNA (Fig. 5C and SI Appendix, Fig. S5A). Western blot analysis of tumor lysates showed that, in the SIRT5-shRNA tumors, knockdown of SIRT5 was still apparent and GLS levels were lower than in tumors expressing control shRNA (Fig. 5D), although GLS levels had partially recovered relative to the depletion seen at early time points in cultured cells.
Fig. 5.
SIRT5 supports mammary tumorigenesis in vivo. (A) Western blots showing SIRT5 depletion in the MDA-MB-231 cells used to inoculate xenograft tumors. (B) Xenograft tumor growth of MDA-MB-231 cells expressing a control shRNA or SIRT5-targeted shRNAs. Cells were transduced with the shRNA constructs in culture. After 48 h, 3 × 106 cells were injected s.c. into each flank of 8-wk-old female NSG mice (n = 4 mice per condition). Tumor size was then measured weekly using calipers over 6 wk. Data are plotted as mean ± SD from the replicate tumors. (C) Image of representative tumors harvested 6 wk after inoculation. (D) Western blot analysis of xenograft tumors formed by MDA-MB-231 cells expressing either a control shRNA or SIRT5-targeted shRNA and harvested 6 wk after inoculation. At this time point, partial knockdown of SIRT5 persists, and partial depletion of GLS is still apparent. (E and F) Kaplan–Meier plots showing overall survival of human breast cancer patients with high (red line) or low (black line) SIRT5 expression (patients split by median expression). E shows data for all patients, and F shows data for patients with basal-subtype breast cancer. Plots were generated using the Kaplan–Meier Plotter (39). **P < 0.01.
These data prompted us to examine the prognosis of human breast cancer patients with high versus low tumor SIRT5 mRNA levels (39). Across all patients (n = 626), elevated SIRT5 levels in tumors correlated significantly with decreased overall survival (P = 0.0015, Hazard Ratio = 1.67) (Fig. 5E). This effect was more pronounced in patients with basal subtype breast tumors (n = 153, P = 0.00081, Hazard Ratio = 2.84) (Fig. 5F). In addition, the SIRT5 locus shows a high frequency of copy-number gains or amplifications across a spectrum of human cancer types, including 50 to 60% of ovarian tumors and melanomas and ∼25% of breast tumors (SI Appendix, Fig. S5B). We note that SIRT5 has hundreds of putative substrate proteins (11, 12) and that its biological function is not restricted to the regulation of GLS. Indeed, expression of the succinylation-defective K164R-GLS variant only partially rescued the proliferative defect caused by SIRT5 knockdown (SI Appendix, Fig. S5C).
Taken together, our results show that SIRT5 is up-regulated during cellular transformation and stabilizes GLS by desuccinylating residue K164. This in turn protects GLS from ubiquitination at residue K158 and subsequent degradation. Although SIRT5 likely has numerous functions which collectively promote mitochondrial fitness in proliferating cells, the stabilization of GLS described here is a possible mechanism through which SIRT5 potentiates tumorigenesis by supporting metabolic reprogramming.
Discussion
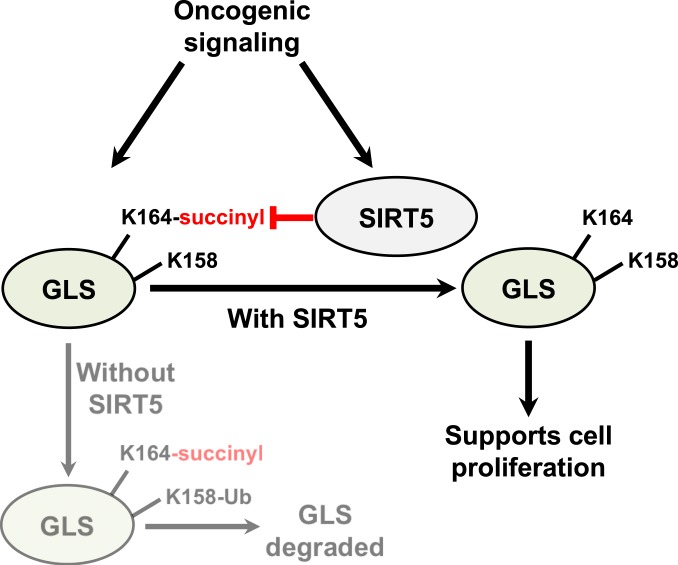
In this study, we report that the mitochondrial lysine deacylase SIRT5 stabilizes GLS via desuccinylation of residue K164 (Fig. 6). Removal of this PTM protects GLS from ubiquitination at residue K158 and subsequent degradation. We further show that SIRT5 is up-regulated during cellular transformation and is important in cancer cells for ex vivo proliferation, anchorage-independent growth, and in vivo tumorigenesis. Consistent with these findings, elevated SIRT5 expression in human breast tumors correlates with decreased patient survival.
Fig. 6.
Schematic of SIRT5-mediated regulation of GLS. Oncogenic signaling drives increased GLS and SIRT5 expression. Succinylation of GLS at residue K164 marks it for ubiquitination at residue K158 and subsequent degradation, and SIRT5-catalyzed desuccinylation of K164 thus stabilizes GLS. Since the glutaminase reaction generates glutamate, which supplies numerous important metabolic processes, stabilization of GLS is one mechanism through which SIRT5 might support cancer cell proliferation.
A by-product of the glutaminase reaction is ammonia, which must be detoxified. It is noteworthy that another key target of SIRT5 is the mitochondrial enzyme carbamoyl phosphate synthetase 1 (CPS1). CPS1 catalyzes the initial reaction of the urea cycle and the production of carbamoyl phosphate from ammonia and carbonate and is activated by SIRT5-mediated deacetylation, deglutarylation, and desuccinylation (40–42). CPS1 is up-regulated and a negative prognosticator in rectal cancers and similarly is up-regulated and essential in LKB1-null non–small-cell lung cancer (NSCLC), where the carbamoyl phosphate product supplies pyrimidine biosynthesis (43). By simultaneously activating GLS and CPS1, SIRT5 couples mitochondrial ammonia production with mitochondrial ammonia detoxification and potentially permits the recycling of some GLS-generated ammonia into nucleotide biosynthesis.
Notably, an earlier study found that SIRT5 negatively regulates autophagy in MDA-MB-231 cells by decreasing the level of ammonia, a metabolite which signals to promote autophagy (44). Western blots in this earlier study indicated a 1.4-fold increase in the abundance of glutaminase following SIRT5 knockdown, leading to the conclusion that SIRT5 is a negative regulator of GLS. One possible explanation for the discrepancy with our results is that the glutaminase antibody used by Polletta et al. (44) is targeted exclusively against the KGA splice variant of GLS, which is expressed at extremely low levels relative to the GAC splice variant in the MDA-MB-231 cell line used. Indeed, the pan-GLS antibody that we used detects both KGA and GAC in MEFs cultured in low-serum conditions (Fig. 1H), but does not yield a significant signal for KGA relative to GAC in any of the cancer cell lines that we examined (Fig. 1). It is possible that decreased ammonia secretion following SIRT5 overexpression, as described by Polletta et al. (44), results from increased activity of the SIRT5 substrate CPS1, which is present and readily detectable in MDA-MB-231 cells (SI Appendix, Fig. S6).
Sirtuin-mediated lysine deacylation is coupled to the hydrolysis of NAD+ to nicotinamide, meaning that sirtuin activities are regulated by the NAD+/NADH ratio. Since numerous metabolic enzymes are regulated by lysine acylation (45), sirtuins play an important homeostatic role by sensing the NAD+/NADH ratio and correspondingly modulating the activity of metabolic pathways. Of the 3 mitochondrial sirtuins, both SIRT3 and SIRT4 exhibit tumor suppressor activity, with SIRT3 knockout mice spontaneously developing mammary tumors and SIRT4 knockout mice developing lung tumors (14, 15). In contrast, the SIRT5 locus shows a high frequency of copy number gain or amplification in many human cancers, including 50 to 60% of ovarian tumors and melanomas and ∼25% of breast tumors (SI Appendix, Fig. S5B). Furthermore, SIRT5 has onco-supportive activity in NSCLC, colorectal cancer, and hepatocellular carcinoma (46–48), as well as in breast cancer as described here.
Comparing the distinct functions of the mitochondrial sirtuins is informative for understanding the different impacts that these enzymes have on tumorigenesis. The tumor suppressor SIRT3 opposes cancer cell metabolic reprogramming in general, and the Warburg effect in particular, in part by deacetylating and activating the E1α subunit of the pyruvate dehydrogenase complex (PDC), along with components of the mitochondrial electron transport chain (45). These actions promote pyruvate flux into the TCA cycle coupled with oxidative phosphorylation, a metabolic phenotype characteristic of differentiated, nonproliferating cells. SIRT4 can also counter metabolic reprogramming by suppressing the flux of glutamine-derived carbon into the TCA cycle (49).
In several respects, SIRT5 opposes the actions of SIRT3 and SIRT4 and consequently supports metabolic reprogramming. SIRT5 suppresses cellular respiration by inhibiting both the PDC and succinate dehydrogenase (25, 48). These actions block entry of pyruvate into the TCA cycle and result in decreased capacity for oxidative phosphorylation. Moreover, whereas SIRT3 suppresses glycolytic flux by destabilizing HIF1α (50), the demalonylase activity of SIRT5 activates glycolytic enzymes (51). By simultaneously suppressing oxidative phosphorylation and activating glycolysis, SIRT5 might enhance the Warburg effect. We show here that SIRT5 also supports enhanced hydrolysis of glutamine to glutamate, another hallmark of cancer cell metabolism. A recent study found that the deglutarylation activity of SIRT5 contributes to colorectal carcinogenesis by activating glutamate dehydrogenase 1 (GLUD1), a mitochondrial enzyme catalyzing the conversion of glutamate into the TCA cycle intermediate α-KG (52). By positively regulating both GLS and GLUD1, SIRT5 coordinately stimulates a metabolic pathway for glutamine-mediated TCA cycle anaplerosis.
An important question for future studies will be to determine how the succinylation of GLS “primes” it for ubiquitination and degradation. It is possible that succinylation acts as a marker to recruit an E3 ubiquitin ligase, and it will be of interest to determine whether this represents a general mechanism by which SIRT5 stabilizes substrate proteins. The supportive role of SIRT5 in cancer cell metabolic reprogramming, in conjunction with the fact that SIRT5-null mice are viable with relatively subtle metabolic deficiencies (40), supports further investigation of SIRT5 as a potential therapeutic target.
Materials and Methods
Inducible Oncogenic-Dbl and Oncogenic-KRAS MEF System.
The isogenic inducible onco-Dbl MEF line was described previously (7, 53). Briefly, the onco-Dbl gene or the KRAS-G12V gene was subcloned into plasmid pTRE-HA (Clontech) and cotransfected with plasmid pMET-Puro into parental MEFs containing the transcriptional transactivator tTA (Clontech). After puromycin selection, individual colonies were isolated and tested for inducible onco-Dbl or KRAS-G12V expression. Cells were maintained in Dulbecco’s Modified Eagle Media (DMEM) (Gibco) supplemented with 10% FBS (Gibco) and 0.6 μg⋅ml−1 doxycycline, and onco-Dbl or KRAS-G12V expression was induced by replating cells in doxycycline-free medium.
Cell Culture.
Inducible Dbl-MEFs and inducible KRAS-G12V MEFs were cultured in DMEM (Gibco) supplemented with 10% FBS (Gibco) ± 0.6 μg⋅ml−1 doxycycline. For signaling experiments, low-serum (0.5% FBS) DMEM was used. The breast cancer and lung cancer cell lines were obtained from American Type Culture Collection and maintained in RPMI medium 1640 (Gibco) supplemented with 10% FBS (Gibco). All cells were cultured at 37 °C in a 5% CO2 atmosphere.
Antibodies and Inhibitors.
The antibodies used in this study, and the dilutions for Western blot analysis, were the following: anti-GLS (custom-made rabbit polyclonal antibody, antigen peptide CKLDPRREGGDQRHS, 1:1,000); anti-GLS (C-term) (Abgent, AP8809B, 1:8,000); anti-glutaminase (Abcam, ab156876, 1:10,000); anti–HA-tag (Cell Signaling Technology, 3742, 1:5,000); anti-SirT3 (Cell Signaling Technology, 5490, 1:1,000); anti-SIRT4 (LSBio, LS-C403546, 1:1,000); anti-SirT5 (Cell Signaling Technology, 8782, 1:1,000); anti-ubiquitin (Santa Cruz Biotechnology, sc-8017, 1:1,000); anti-succinyllysine (PTM Biolabs, PTM-419, 1:1,000); anti-vinculin (Sigma, V9131, 1:10,000); anti-tubulin α/β (Cell Signaling, 2148, 1:5,000); and anti-CPS1 (Abcam, ab45956, 1:1,000). Inhibitors used were the following: leupeptin (Sigma, L-2884, final concentration: 20 μg/mL); chloroquine (InvivoGen, tlrl-chq, final concentration: 5 μM); MG132 (ApexBio, A2585, final concentration: 20 nM); and ALX-260–090 (Enzo Life Sciences, ALX-260–090, final concentration: 20 nM).
Western Blot Analysis.
Details about Western blot analysis are provided in SI Appendix.
Immunoprecipitation.
Details about immunoprecipitations are provided in SI Appendix.
Cell Proliferation and Anchorage-Independent Growth Assays.
Details about cell proliferation and anchorage-independent growth assays are provided in SI Appendix.
Quantitative Real-Time PCR Analysis.
Details about the quantitative real-time PCR analysis are provided in SI Appendix.
Lentivirus System for SIRT5 Knockdown and Ectopic Expression of GAC.
Details about the lentiviral system for SIRT5 knockdown and ectopic expression of GAC are provided in SI Appendix.
Tumor Xenograft Experiments.
The MDA-MB-231 cells (3 × 106) transduced 48 h previously with either control shRNA vector or SIRT5-targeted shRNA vector were injected s.c. into each flank of female NSG mice, aged 8 wk. Tumor size was measured using calipers every 7 d, and tumor volume was calculated using the formula 0.5 × (length × width2) (54). Tumors from NSG mice were harvested 42 d after xenograft inoculation. All animal experiments were performed in compliance with Cornell University Institutional Animal Care and Use Committee regulations (protocol number 2003–0097).
Glutamine Consumption and Ammonium Secretion Measurements.
Details about glutamine consumption and ammonium secretion measurements are provided in SI Appendix.
Metabolite Extraction and Measurements.
Details about metabolite extraction and measurements are provided in SI Appendix.
Statistical Analysis.
Student’s t test was used for all statistical analyses.
Data Availability.
All data described are presented in the figures, and all reagents and materials are described in this section.
Supplementary Material
Acknowledgments
We thank Cindy Westmiller for her assistance with manuscript preparation. This study was supported by NIH Grants R35 GM122575, R01 CA201402, and U54 CA210184 (to R.A.C.). M.J.L. acknowledges a postdoctoral research award from the Breast Cancer Coalition of Rochester.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. R.J.D. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911954116/-/DCSupplemental.
References
- 1.Pavlova N. N., Thompson C. B., The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lukey M. J., Katt W. P., Cerione R. A., Targeting amino acid metabolism for cancer therapy. Drug Discov. Today 22, 796–804 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cluntun A. A., Lukey M. J., Cerione R. A., Locasale J. W., Glutamine metabolism in cancer: Understanding the heterogeneity. Trends Cancer 3, 169–180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garber K., Cancer anabolic metabolism inhibitors move into clinic. Nat. Biotechnol. 34, 794–795 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Gao P., et al. , c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458, 762–765 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Csibi A., et al. , The mTORC1/S6K1 pathway regulates glutamine metabolism through the eIF4B-dependent control of c-Myc translation. Curr. Biol. 24, 2274–2280 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Lukey M. J., Greene K. S., Erickson J. W., Wilson K. F., Cerione R. A., The oncogenic transcription factor c-Jun regulates glutaminase expression and sensitizes cells to glutaminase-targeted therapy. Nat. Commun. 7, 11321 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hao Y., et al. , Oncogenic PIK3CA mutations reprogram glutamine metabolism in colorectal cancer. Nat. Commun. 7, 11971 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thangavelu K., et al. , Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism. Proc. Natl. Acad. Sci. U.S.A. 109, 7705–7710 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han T., et al. , Phosphorylation of glutaminase by PKCε is essential for its enzymatic activity and critically contributes to tumorigenesis. Cell Res. 28, 655–669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinert B. T., et al. , Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 4, 842–851 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Park J., et al. , SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell 50, 919–930 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bheda P., Jing H., Wolberger C., Lin H., The substrate specificity of sirtuins. Annu. Rev. Biochem. 85, 405–429 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Jeong S. M., et al. , SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell 23, 450–463 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim H. S., et al. , SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 17, 41–52 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park S.-H., et al. , SIRT2 is a tumor suppressor that connects aging, acetylome, cell cycle signaling, and carcinogenesis. Transl. Cancer Res. 1, 15–21 (2012). [PMC free article] [PubMed] [Google Scholar]
- 17.Sebastián C., et al. , The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell 151, 1185–1199 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jing H., et al. , A SIRT2-selective inhibitor promotes c-Myc oncoprotein degradation and exhibits broad anticancer activity. Cancer Cell 29, 297–310 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y., et al. , Sirtuin-3 (SIRT3), a therapeutic target with oncogenic and tumor-suppressive function in cancer. Cell Death Dis. 5, e1047 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee N., et al. , SIRT6 depletion suppresses tumor growth by promoting cellular senescence induced by DNA damage in HCC. PLoS One 11, e0165835 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeong S. M., Hwang S., Seong R. H., SIRT4 regulates cancer cell survival and growth after stress. Biochem. Biophys. Res. Commun. 470, 251–256 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Lin Z., Fang D., The roles of SIRT1 in cancer. Genes Cancer 4, 97–104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Latifkar A., et al. , Loss of Sirtuin 1 alters the secretome of breast cancer cells by impairing lysosomal integrity. Dev. Cell 49, 393–408.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barber M. F., et al. , SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 487, 114–118 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bringman-Rodenbarger L. R., Guo A. H., Lyssiotis C. A., Lombard D. B., Emerging roles for SIRT5 in metabolism and cancer. Antioxid. Redox Signal. 28, 677–690 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rardin M. J., et al. , SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 18, 920–933 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du J., et al. , Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334, 806–809 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J. B., et al. , Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell 18, 207–219 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Son J., et al. , Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 496, 101–105 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukey M. J., et al. , Liver-type glutaminase GLS2 is a druggable metabolic node in luminal-subtype breast cancer. Cell Rep. 29, 76–88.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muir A., et al. , Environmental cystine drives glutamine anaplerosis and sensitizes cancer cells to glutaminase inhibition. eLife 6, e27713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friday E., Oliver R. III, Welbourne T., Turturro F., Glutaminolysis and glycolysis regulation by troglitazone in breast cancer cells: Relationship to mitochondrial membrane potential. J. Cell. Physiol. 226, 511–519 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Acosta M. L., Sánchez A., García F., Contreras A., Molina E., Analysis of kinetic, stoichiometry and regulation of glucose and glutamine metabolism in hybridoma batch cultures using logistic equations. Cytotechnology 54, 189–200 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang C., et al. , Glioblastoma cells require glutamate dehydrogenase to survive impairments of glucose metabolism or Akt signaling. Cancer Res. 69, 7986–7993 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li K., et al. , Acetylation of WRN protein regulates its stability by inhibiting ubiquitination. PLoS One 5, e10341 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang W., et al. , Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol. Cell 43, 33–44 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferreira A. P. S., et al. , Active glutaminase C self-assembles into a supratetrameric oligomer that can be disrupted by an allosteric inhibitor. J. Biol. Chem. 288, 28009–28020 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y., et al. , Mechanistic basis of glutaminase activation: A key enzyme that promotes glutamine metabolism in cancer cells. J. Biol. Chem. 291, 20900–20910 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Györffy B., et al. , An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 123, 725–731 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Yu J., et al. , Metabolic characterization of a Sirt5 deficient mouse model. Sci. Rep. 3, 2806 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan M., et al. , Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 19, 605–617 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakagawa T., Lomb D. J., Haigis M. C., Guarente L., SIRT5 deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell 137, 560–570 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim J., et al. , CPS1 maintains pyrimidine pools and DNA synthesis in KRAS/LKB1-mutant lung cancer cells. Nature 546, 168–172 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Polletta L., et al. , SIRT5 regulation of ammonia-induced autophagy and mitophagy. Autophagy 11, 253–270 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar S., Lombard D. B., Mitochondrial sirtuins and their relationships with metabolic disease and cancer. Antioxid. Redox Signal. 22, 1060–1077 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang L., et al. , SIRT5 promotes cell proliferation and invasion in hepatocellular carcinoma by targeting E2F1. Mol. Med. Rep. 17, 342–349 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Lu W., Zuo Y., Feng Y., Zhang M., SIRT5 facilitates cancer cell growth and drug resistance in non-small cell lung cancer. Tumour Biol. 35, 10699–10705 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Du Z., et al. , Targeting a Sirt5-positive subpopulation overcomes multidrug resistance in wild-type Kras colorectal carcinomas. Cell Rep. 22, 2677–2689 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Csibi A., et al. , The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell 153, 840–854 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finley L. W. S., et al. , SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell 19, 416–428 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nishida Y., et al. , SIRT5 regulates both cytosolic and mitochondrial protein malonylation with glycolysis as a major target. Mol. Cell 59, 321–332 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y. Q., et al. , Sirtuin5 contributes to colorectal carcinogenesis by enhancing glutaminolysis in a deglutarylation-dependent manner. Nat. Commun. 9, 545 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stalnecker C. A., et al. , Mechanism by which a recently discovered allosteric inhibitor blocks glutamine metabolism in transformed cells. Proc. Natl. Acad. Sci. U.S.A. 112, 394–399 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jensen M. M., Jørgensen J. T., Binderup T., Kjaer A., Tumor volume in subcutaneous mouse xenografts measured by microCT is more accurate and reproducible than determined by 18F-FDG-microPET or external caliper. BMC Med. Imaging 8, 16 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data described are presented in the figures, and all reagents and materials are described in this section.