Significance
Here we demonstrate that IL-1α neutralization reduces the severity of disease in a mouse model of Crohn’s disease. Unexpectedly, IL-1α neutralization exerts its antiinflammatory effects by modulating the gut microbial ecosystem, and corrected the mucosal dysbiosis. Although studies report that the gut microbiome affects cytokine production, few have shown that specific neutralization of a pivotal cytokine such as IL-1α alters the composition and function of the gut microbiome. The study demonstrates a predictive relationship between IL-1α neutralization and abundance of specific bacterial species. The findings have translational significance, since IL-1α treatment prevented DSS-induced mucosal dysbiosis and preserved immunological homeostasis. These preclinical findings support the therapeutic potential of IL-1α neutralization in patients with IBD.
Keywords: IL-1α, Crohn’s disease, IBD, SAMP1/YitFc
Abstract
Crohn’s disease and ulcerative colitis are chronic and progressive inflammatory bowel diseases (IBDs) that are attributed to dysregulated interactions between the gut microbiome and the intestinal mucosa-associated immune system. There are limited studies investigating the role of either IL-1α or IL-1β in mouse models of colitis, and no clinical trials blocking either IL-1 have yet to be performed. In the present study, we show that neutralization of IL-1α by a specific monoclonal antibody against murine IL-1α was highly effective in reducing inflammation and damage in SAMP mice, mice that spontaneously develop a Crohn’s-like ileitis. Anti-mouse IL-1α significantly ameliorated the established, chronic ileitis and also protected mice from developing acute DSS-induced colitis. Both were associated with taxonomic divergence of the fecal gut microbiome, which was treatment-specific and not dependent on inflammation. Anti–IL-1α administration led to a decreased ratio of Proteobacteria to Bacteroidetes, decreased presence of Helicobacter species, and elevated representation of Mucispirillum schaedleri and Lactobacillus salivarius. Such modification in flora was functionally linked to the antiinflammatory effects of IL-1α neutralization, as blockade of IL-1α was not effective in germfree SAMP mice. Furthermore, preemptive dexamethasone treatment of DSS-challenged SAMP mice led to changes in flora composition without preventing the development of colitis. Thus, neutralization of IL-1α changes specific bacterial species of the intestinal microbiome, which is linked to its antiinflammatory effects. These functional findings may be of significant value for patients with IBD, who may benefit from targeted IL-1α–based therapies.
Inflammatory bowel disease (IBD), primarily encompassing ulcerative colitis (UC) and Crohn’s disease (CD), is an idiopathic disorder of the gastrointestinal (GI) tract that results from a complex interplay of environmental factors, an abnormal gut microbiome, and dysregulated immune responses in genetically susceptible individuals (1, 2). Recent advances in next-generation sequencing techniques have started to unravel the contributions of the gut microbiota to the pathogenesis of IBD. Few studies have explored how the microbiome exerts its effects in IBD in terms of the interaction of different microbes with the gut mucosa, and there is no identification of a consistent and distinct microbiome signature in IBD (3, 4). Nevertheless, increasing evidence implicates dysbiosis of the gut microbiome in the pathogenesis of chronic intestinal inflammation (5).
Among the many factors investigated that have a significant impact on the development of IBD, the role of cytokines represents a very active and ongoing area of research, particularly in light of the impressive efficacy of cytokine-targeted biologics, e.g., anti-TNF and anti-interleukin (IL)-23, to treat patients with IBD (6). In fact, the imbalance between pro- and antiinflammatory cytokines limits the resolution of inflammation, leading to the perpetuation of disease and tissue damage (7). These findings require further investigation into cytokine-driven gut inflammatory processes in order to identify the major cytokine targets for the treatment of IBD (8). Among these, IL-1 family members play a pivotal role in the pathogenesis of acute and chronic intestinal inflammatory processes (9) and orchestrate a plethora of immune and physiological responses that regulate both innate and adaptive immunity (10). IL-1 exists in 2 isoforms, IL-1α and IL-1β, which are encoded by distinct genes but bind to the same receptor (IL-1R1) and display similar biological properties. However, IL-1α and IL-1β also have distinct differences. For example, IL-1α is constitutively present as precursor IL-1α in healthy epithelial lining of the entire GI tract, and, when intestinal epithelial cells (IECs) undergo necrosis, e.g., during DSS-induced colitis, the IL-1α precursor is released and triggers the IL-1R in myeloid cells, functioning as an “alarmin” (11). IL-1 β is produced by myeloid cells within the lamina propria (not by IECs) (11, 12). In this context, Bersudsky et al. have shown a differential role for IL-1α versus IL-1β in colonic inflammation and repair, demonstrating that IL-1α released from damaged IECs initiates and propagates colon inflammation, and IL-1α–deficient mice exhibit attenuated disease symptoms with improved recovery. On the contrary, although IL-1β may contribute to inflammation, this group found that the dominant effect of IL-1β in colitis appears to be facilitation of the repair process (13).
Although there are no controlled trials of blocking either IL-1α or IL-1β in IBD, a broad spectrum of inflammatory diseases, both rare hereditary and common, are treated with biologics that reduce IL-1α and IL-1β. The CANTOS trial, a landmark, worldwide trial neutralizing IL-1β in over 10,000 patients, revealed that IL-1β is pathologic in atherosclerosis, gout, osteoarthritis, type 2 diabetes, heart failure, cancer incidence, and cancer death (14). Moreover, a randomized, placebo-controlled trial of anti–IL-1α in 360 patients with advanced metastatic colorectal cancer met its primary and secondary endpoints (15, 16). Although anti–IL-1α has also been used to treat patients with severe hidradenitis suppurativa (17), it has not yet been evaluated in IBD, which may represent an important opportunity as a therapeutic option.
We have previously shown that inbred SAMP1/YitFc (SAMP) mice develop a progressive CD-like ileitis that worsens over time without chemical, genetic, or immunologic manipulation. The resulting ileitis has remarkable phenotypic similarities to human CD, such as disease location, histologic features, and response to conventional CD therapies (18). Additionally, SAMP mice have shown a significant predictive value in regard to response to novel, experimental therapies (18). Therefore, the SAMP model presents an ideal experimental system to investigate the possible therapeutic effects of specific IL-1α neutralization in IBD, and the potential mechanism(s) by which this may occur.
In this study, we demonstrate elevated, inflammatory lesion-specific expression of IL-1α in SAMP mice. Additionally, we show that the anti–IL-1α neutralizing mAb FLO1 induces alterations in the mucosal immunological milieu, leading to significant amelioration of chronic ileitis and preventing the development of acute, DSS-induced colitis in this strain. More importantly, we show that IL-1α neutralization is associated with taxonomic divergence of the intestinal microbiota that is essential for its antiinflammatory function, as the latter is completely abrogated in germfree (GF) SAMP mice. Finally, we demonstrate a predictive relationship between IL-1α neutralization and the presence of specific bacterial species, which is clearly linked to the antiinflammatory effects.
Results
SAMP Mice Express High Levels of Intestinal IL-1α and IL-1β.
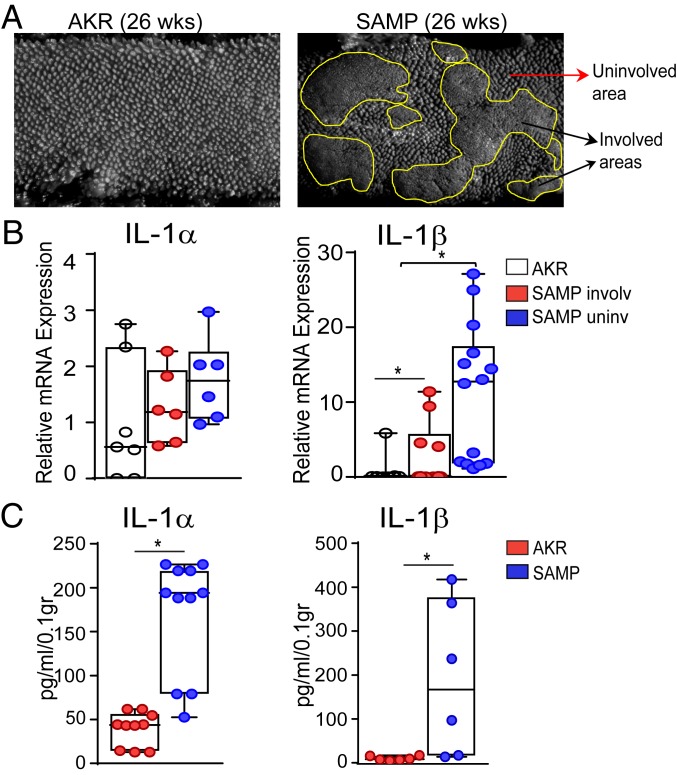
Increased production of IL-1 has been reported in local gut tissues of animal models of IBD (19, 20), as well as in the intestinal mucosa of IBD patients (21). To confirm these findings in our SAMP model, we evaluated IL-1α and IL-1β expression in these mice and compared it to their uninflamed parental controls (i.e., AKR strain). Involved and uninvolved areas from ilea (Fig. 1A) were assayed for IL-1α and IL-β mRNA levels, which revealed that SAMP mice display a trend for increased IL-1α compared to AKR controls (Fig. 1B), and that IL-1β mRNA transcripts in both involved and noninvolved areas from ilea of SAMP mice were markedly elevated compared to AKR mice (Fig. 1B). Furthermore, when mucosal samples from ilea of experimental mice were harvested and cultured for 24 h, protein levels of IL-1α and IL-1β were strikingly increased in SAMP mice compared to AKR controls (Fig. 1C). Taken together, these results show that ileitis-prone SAMP mice express high levels of intestinal IL-1α and IL-1β, suggesting that IL-1 likely plays an important role in this mouse model.
Fig. 1.
SAMP mice display increased expression of intestinal IL-1α and IL-1β. (A) Fixed ileal specimens analyzed by SM show cobblestoned abnormal mucosa in SAMP (Right) versus AKR (Left) mice. (B) IL-1α and IL-1β mRNA levels in healthy ileal mucosa of AKR mice and involved and uninvolved areas of SAMP mice. Data are presented as relative fold difference compared with AKR (arbitrarily set as 1). (C) IL-1α and IL-1β protein levels in tissue explants from ilea of SAMP versus AKR mice. Data are presented as median ± interquartile range. Statistical significance was determined by 1-way ANOVA followed by Dunn’s post hoc test, with n = 6–13, **P = 0.001 and *P = 0.016 (B); and by 2-tailed unpaired t test, with n = 6–10, *P < 0.01 (C).
Neutralization of IL-1α Ameliorates the Severity of CD-Like Ileitis in Adult SAMP Mice.
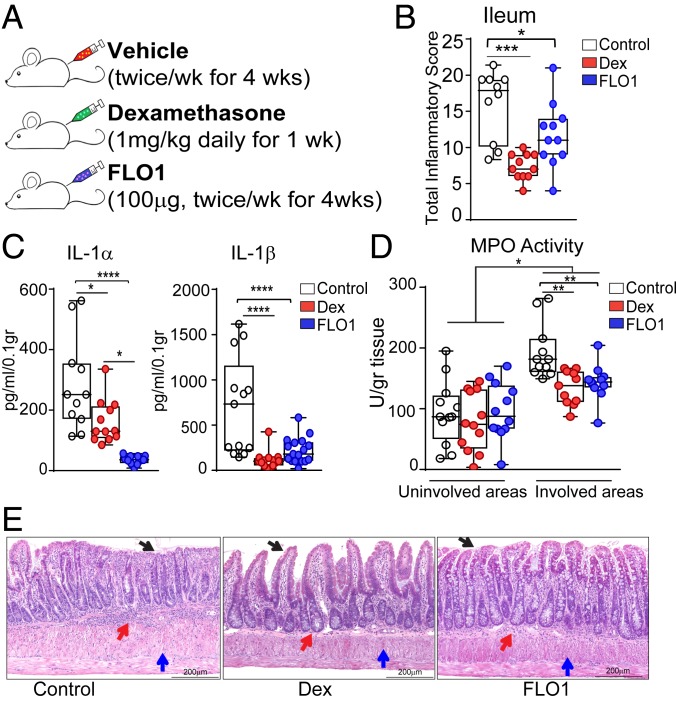
With IL-1 overexpression in SAMP mice indicative of a pathogenic role, we sought to investigate whether specific neutralization of IL-1α, using FLO1, might reduce intestinal inflammation in this model. As shown in Fig. 2A, SAMP mice with established disease (26 wk of age) were treated with FLO1 and compared with vehicle-treated SAMP mice as well as SAMP mice treated with dexamethasone (Dex) as a positive control (22, 23). Interestingly, FLO1-treated SAMP mice demonstrated a marked reduction in total inflammatory scores versus vehicle-treated SAMP mice, and with similar efficacy to Dex-treated SAMP mice (Fig. 2B). SAMP mice treated with a recombinant mouse isotype control antibody (IgG2ak isotype control; XBiotech) show no differences in total inflammatory scores compared to vehicle-treated SAMP mice (SI Appendix, Fig. S2). Along with amelioration of disease, the levels of proinflammatory IL-1α and IL-1β, measured in supernatants of ileal tissue explants cultured for 24 h, were markedly decreased in both FLO1- and Dex-treated mice compared to vehicle-treated controls (Fig. 2C). Additionally, myeloperoxidase (MPO) activity was significantly decreased in both FLO1- and Dex-treated mice compared to vehicle-treated controls (Fig. 2D). Representative histological sections from the 3 experimental groups are shown in Fig. 2E. Overall, these results demonstrate that IL-1α plays a pathologic role in ileitis-prone SAMP mice, and that its specific blockade results in amelioration of disease severity.
Fig. 2.
FLO1 treatment in SAMP mice with established disease results in amelioration of ileitis. (A) Timeline indicating experimental design. (B) Histologic analysis (reported as total inflammatory scores), as well as (C) IL-1α and IL-1β protein levels and (D) MPO activity in involved and uninvolved areas of ileal mucosa from both FLO1- and Dex-treated SAMP mice compared to (vehicle-treated) controls. (E) Representative images of full-thickness H&E-stained ileal tissues from the 3 experimental groups show decreased cellular infiltration (red arrows), preservation of epithelial cell integrity (black arrows), and diminished thickening of the muscularis mucosa (blue arrows) in both FLO1- and Dex-treated mice vs. controls. (Scale bar: 200 µm.) Data are representative of 2 independent experiments and presented as median ± interquartile range. In B, statistical analysis was determined by Kruskal–Wallis followed by Dunn’s post hoc tests, n = 10–11; **P = 0.001 and *P = 0.016. In C and D, statistical analysis was determined by 1-way ANOVA followed by Tukey’s post hoc test, n = 11–17; ****P < 0.0001, **P ≤ 0.005, and *P ≤ 0.049.
FLO1 Treatment Results in Distinct Reproducible Effects on the Fecal Microbiome in Specific Pathogen-Free (SPF) SAMP Mice.
Several studies have linked IBD pathogenesis with characteristic shifts in the composition of the microbiome, reinforcing the concept that IBD results from altered interactions between the gut microbiome and the host mucosal immune system. Prior studies have also shown that specific bacterial pathogens have the ability to bind host-derived proinflammatory cytokines and respond by increasing their growth and/or altering their virulence (24–26). In 1991, Porat et al. showed that recombinant human IL-1β enhanced bacterial growth rates of virulent Escherichia coli in vitro, and that IL-1Ra blocked both the binding and growth-promoting properties of IL-1. These studies suggested the existence of a bacterial binding structure that is recognized by both IL-1β and IL-1Ra (27).
Thus, we hypothesize that alterations in the gut microbiome after IL-1α blockade contribute to its antiinflammatory effects during intestinal inflammation. To test this hypothesis, we performed conventional microbiome 16S rRNA analyses of fecal samples collected from FLO1- as well as Dex-treated SAMP mice with established disease. We also analyzed the microbiome of vehicle-treated SAMP mice. Of note, this analysis was conducted on samples obtained from 2 independent experiments (6 mo apart) in order to determine whether the antiinflammatory effects of FLO1 are reproducible irrespective of the composition of the gut microbiome in the SPF colony, which may be subject to seasonal variability (28). Bacterial 16S rRNA microbiome analyses demonstrated that the fecal microbiome composition was altered due to IL-1α blockade and Dex treatment when compared to vehicle-treated SAMP mice. The results are shown by Venn diagram (SI Appendix, Fig. S3A) and histogram plot (SI Appendix, Fig. S3B). The data display the number of unique and common (core) OTU taxonomic species across the groups in the 2 separate experiments (exp. A, n = 14; and exp. B, n = 13). Most remarkably, principal component analyses (PCAs) showed that microbiome segregation among the 3 groups was consistent and reproducible in the 2 independent experiments (n = 27), highlighting the reproducibility of the treatment effect in regard to microbiota alteration (SI Appendix, Fig. S3C). Although 16S rRNA analyses were conducted to identify variability in potential unique microbial taxa linked to anti–IL-1α treatment, no common microbial species were identified in the 2 separate experiments to which disease amelioration could be significantly linked to an IL-1– or Dex-dependent effect. Altogether, these results clearly demonstrate that FLO1 treatment induces global alterations of the gut microbiome. However, whether anti–IL-1α treatment directly modulates the gut microbiota or these effects are the result of decreased intestinal inflammation remained to be determined.
Blockade of IL-1α Has No Detectable Antiinflammatory Effect in GF SAMP Mice.
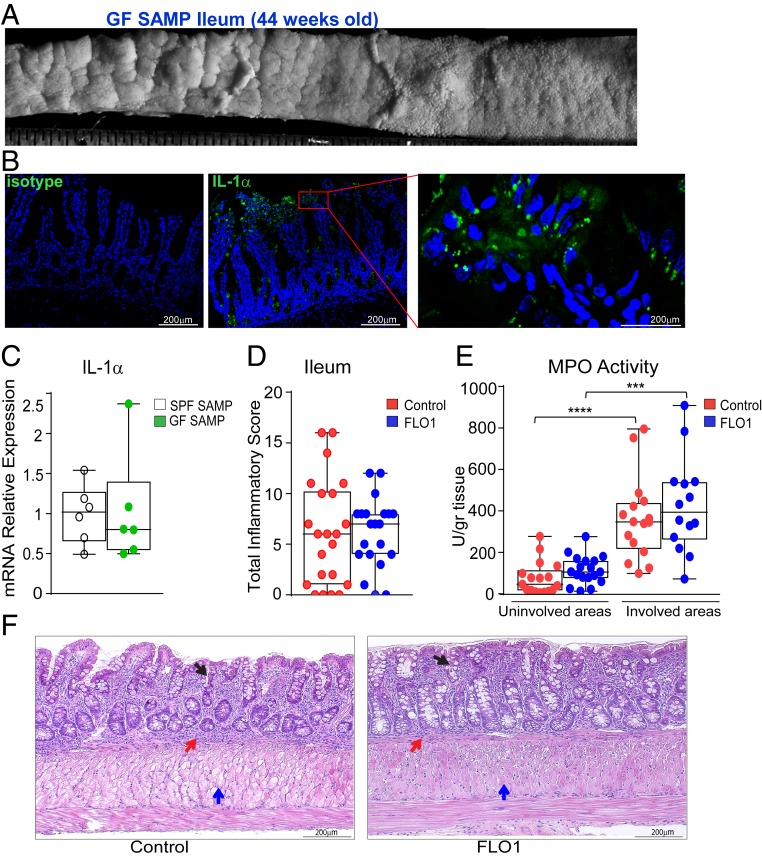
To investigate whether the effects of anti–IL-1α treatment are mediated by functional changes of the gut microbiome, we next performed FLO1 treatment on GF SAMP mice. Previous studies in GF SAMP mice have shown that, although commensal flora are not essential for the generation of ileitis characteristic of this mouse strain (29), they do play an important role in exacerbating gut inflammation. In fact, GF SAMP display an attenuated and delayed ileitis. Nevertheless, they fully develop disease after 30 wk of age under GF conditions, suggesting that bacterial-dependent and -independent mechanisms coexist during the development of ileal inflammation in this model (30). Forty-week-old GF SAMP mice resemble the same ileal 3D-structural inflammatory phenotype as in SPF SAMP mice (Fig. 3A). We first verified the presence of IL-1α in the small intestine of GF SAMP using immunofluorescence confocal microscopy (Fig. 3B), and then we determined that ileal IL-1α mRNA expression was comparable between GF and SPF SAMP mice (Fig. 3C). Next, we treated GF SAMP mice with FLO1 (100 μg; 2 times per week for 4 wk), which were compared to a group of untreated age-/sex-matched GF SAMP mice. IL-1α blockade in GF SAMP mice did not exert the same antiinflammatory changes observed in SPF SAMP mice, supporting our hypothesis that the gut microbiome is essential for the beneficial effects of IL-1α neutralization. Treated and untreated GF SAMP mice did not show much difference in the total inflammatory score (Fig. 3D) and MPO activity in ileal tissues (Fig. 3E). These data demonstrate that not only is IL-1α implicated in the pathogenesis of intestinal inflammation, but IL-1–mediated alterations in composition of the gut microbiome may have important contributions to the antiinflammatory effects of IL-1α blockade.
Fig. 3.
FLO1 treatment in GF SAMP mice has no detectable antiinflammatory effects. (A) Representative 3-D stereomicroscopy of cobblestoned mucosa in ileal tissues from GF SAMP, which, by immunofluorescence staining (B), show abundant nuclear-associated IL-1α protein (green) localized to surface IECs (n = 3). (Scale bars: Middle, 100 μm; Right, 25 μm.) DAPI is shown in blue. (Left) Isotype control staining. (Scale bar: 100 μm.) (C) IL-1α mRNA levels in ileal tissues from SPF and GF SAMP mice. FLO1- vs. vehicle (control)-treated SAMP mice evaluated by (D) total inflammatory score and (E) MPO activity in involved and uninvolved areas. (F) Representative photomicrographs of full-thickness H&E-stained ileal tissue from GF SAMP mice treated with either FLO1 (Right) or vehicle control (Left) show similar cellular infiltration (red arrows), mucosal thickening (blue arrows), and altered epithelial structure (black arrows). (Scale bar: 200 µm.) Data are presented as median ± interquartile range. In C, statistical significance was determined by 2-tailed unpaired t test (n = 6). In D–F, data are representative of 3 independent experiments and analyzed by 2-tailed unpaired t test (D) and Kruskal–Wallis followed by Dunn’s post hoc tests (E and F), n = 14–18; ****P < 0.0001 and ***P = 0.0005.
Young SPF SAMP Mice Pretreated with FLO1 Are Protected from DSS-Induced Colitis.
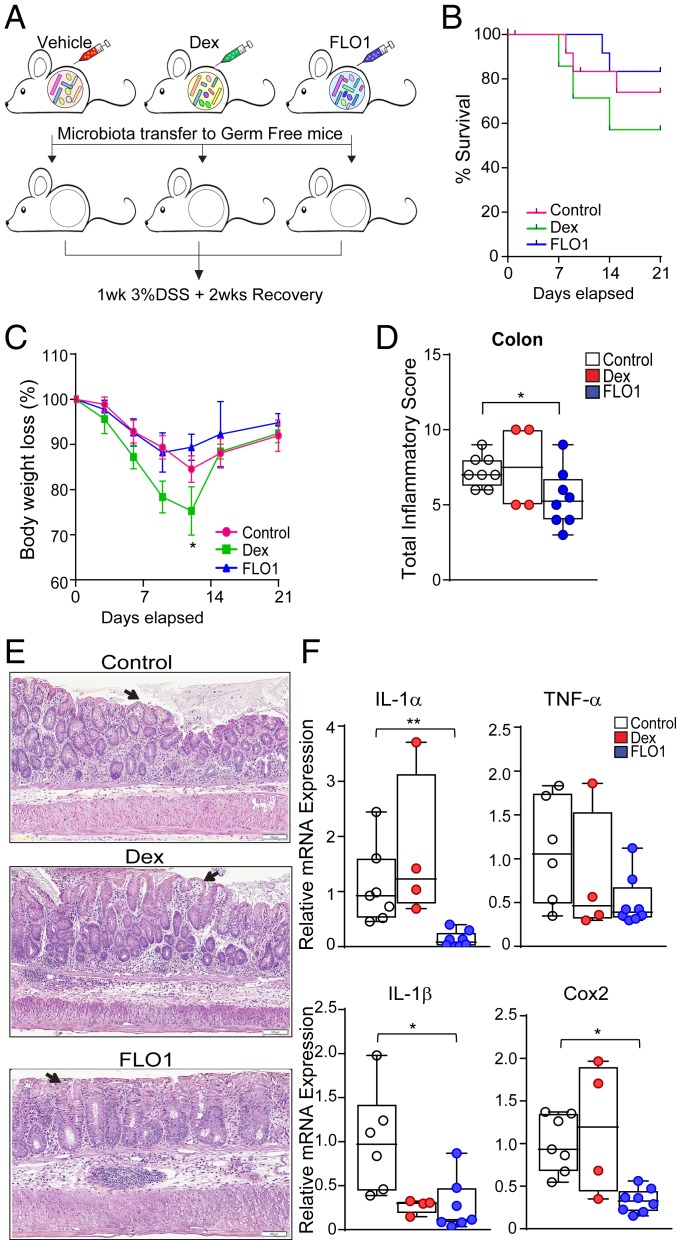
To test whether IL-1α blockade directly modifies the gut microbiome or these effects are a consequence of decreased inflammation and tissue damage, we next investigated the ability of FLO1 to alter the progression of DSS-induced colitis in young SAMP mice before their ileitis onset. By treating 4-wk-old noninflamed SAMP mice, we were able to avoid possible colonic microbiome alterations induced by persistent ileal inflammation. Administration of DSS promotes a breakdown of the gut mucosa and increases colonic permeability, allowing us to determine the functional activity of the colonic microbiome after FLO1 treatment.
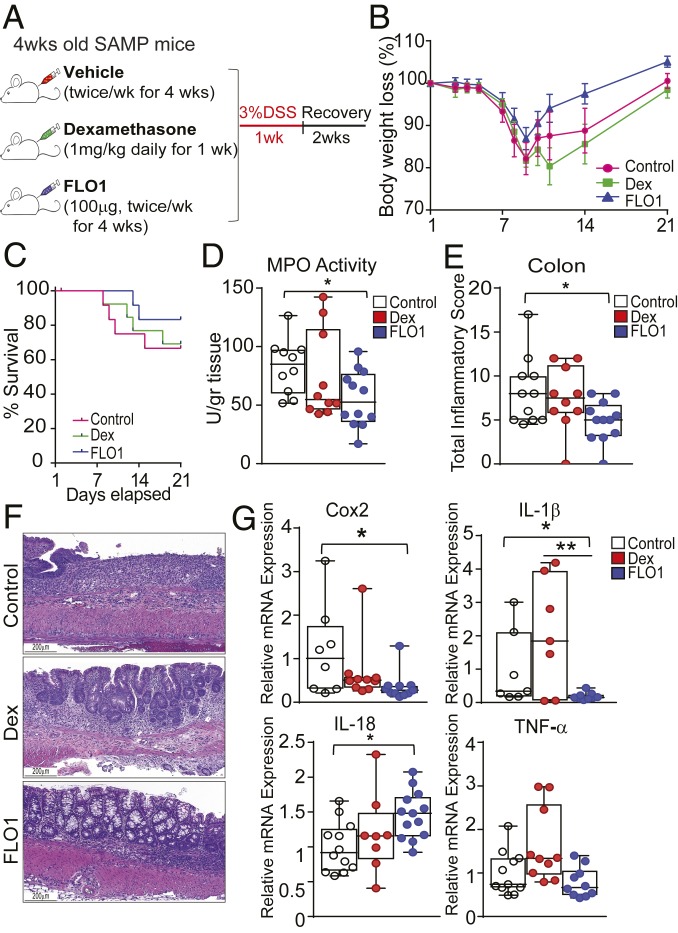
Briefly, noninflamed 4-wk-old SAMP mice were treated with FLO1 as previously described and compared to age-/sex-matched Dex- and vehicle-treated SAMP mice. At the end of treatment, mice were challenged with 7 d of 3% DSS administered in drinking water to induce acute colitis, followed by a 2-wk recovery. The timeline indicating experimental design is depicted in Fig. 4A. The data show that FLO1-treated mice exposed to DSS have a decrease in mortality rate (Fig. 4C) and lost less of their initial body weight when compared to control and Dex-treated mice (Fig. 4B). Consistently, MPO activity measured in freshly isolated colon samples was markedly reduced in FLO1-treated mice compared to control and Dex-treated groups (Fig. 4D). Moreover, histologic analyses provided further support for the efficacy of FLO1 pretreatment in protecting SAMP mice from DSS-induced colitis (Fig. 4 E and F).
Fig. 4.
Pretreatment of young, noninflamed SAMP mice with FLO1 protects them from later challenge with DSS-induced colitis. (A) Schematic representation of study design. (B) Survival and (C) total body weight loss in SAMP mice pretreated with FLO1 compared to Dex-treated and vehicle control mice, which were then subjected to DSS challenge. (D) MPO activity and (E) total inflammatory scores in colonic tissues from FLO1-, Dex-, and control-treated SAMP mice. (F) Representative photomicrographs of full-thickness H&E-stained colonic tissues show preservation of epithelial structure (black arrows) in FLO1- versus Dex- and control-treated SAMP mice. (Scale bar: 200 µm.) (G) mRNA levels of COX2, IL-1β, IL-18, and TNFα in colonic tissues from FLO1-, Dex-, and vehicle-treated SAMP mice subsequently challenged with DSS to induce colitis. Data are presented as relative fold difference compared with control mice (arbitrarily set as 1). All data are representative of 3 independent experiments and presented as median ± interquartile range (n ≥ 9). Statistical significance was determined by Gehan–Breslow–Wilcoxon test (B) and 1-way ANOVA followed by Dunn’s post hoc test (D, E, and G); *P < 0.05 and **P < 0.02.
We next evaluated the mRNA expression of inflammatory markers in colonic tissues from experimental mice (Fig. 4G). COX-2 is recognized as a reliable marker of colonic mucosal inflammation, and its expression has been shown to be markedly higher in IBD patients compared to healthy controls (31). Additionally, multiple studies have demonstrated that the IL-1 signaling pathway is involved in COX-2 up-regulation (32–34). In our studies, colonic COX-2 mRNA expression in FLO1-treated SAMP mice was significantly lower than in Dex-treated and control groups. Finally, we evaluated mRNA levels of proinflammatory cytokines, such as IL-1β and TNFα, which were reduced in FLO1-treated mice compared to the other 2 experimental groups. Recent data obtained from animal models of colitis have suggested that IL-18 can promote intestinal barrier integrity protecting from DSS-induced colitis (35, 36), and, consistent with these findings, we observed that mucosal IL-18 mRNA levels in FLO1-treated mice were markedly increased relative to control and Dex-treated mice. Altogether, these results demonstrate that FLO1 treatment induces bacterial flora modifications that are not inflammation-dependent and are functionally linked to the effects of specifically blocking IL-1α.
IL-1α Neutralization Alters Bacterial Composition in Young SPF SAMP Mice.
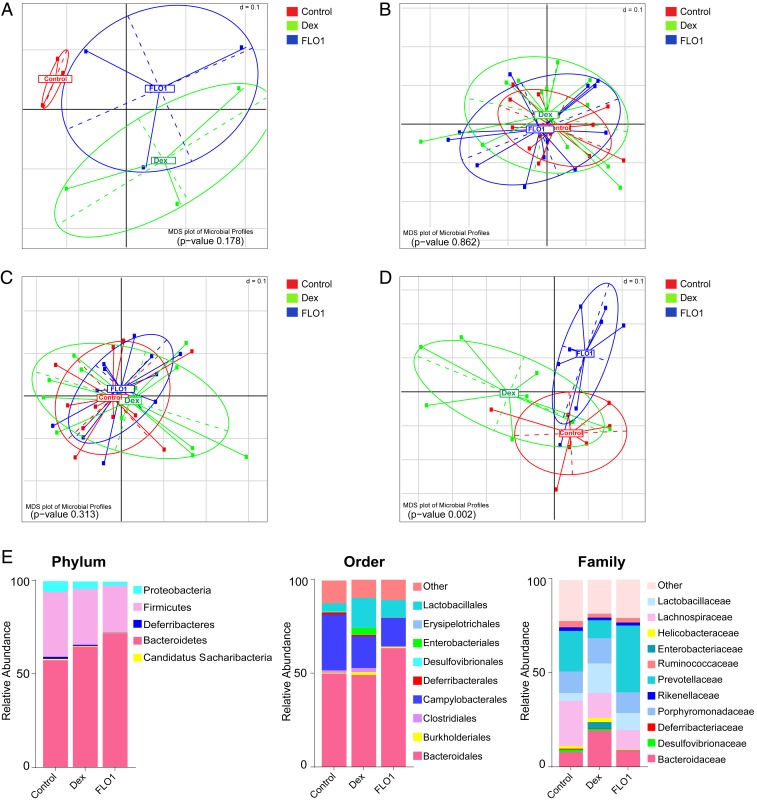
To illustrate the temporal dynamics of the gut microbiome over the course of the DSS treatment, we next profiled stool microbiota of all mice at different time points. Specifically, we collected fecal samples for each experimental group before and after the intersubject preexperimental fecal homogeneization (IsPreFeH) protocol, post-FLO1 and -Dex treatments, and after DSS-induced colitis. A global analysis of β-diversity using multidimensional scaling (MDS) showed that, prior to treatment and after both FLO1 and DEX administration, all experimental groups had a similar microbial community (Fig. 5 B and C), in contrast to the cage–cage variability prior to experimentation (Fig. 5A). DSS-induced colitis produces a profound shift in the composition of intestinal microbiota (37, 38). Consistent with this phenomenon, our studies showed that induction of DSS colitis resulted in clear destabilization of the microbiome composition across the groups (Fig. 5D). Particularly, the segregation of the 3 experimental groups confirmed the same pattern previously observed in the study involving SAMP with established disease in response to FLO1 therapy, with similar MSD plot distribution of experimental groups in Fig. 5D to that of PCA plot group distributions in SI Appendix, Fig. S3C.
Fig. 5.
FLO1 treatment modulates bacterial composition in 4-wk-old SAMP mice. Multidimensional scaling (MDS) plot of generalized UniFrac distances showing distribution of fecal microbiota of control-, Dex-, and FLO1-treated mice (A) before IsPreFeH, (B) after IsPreFeH, (C) after FLO1 and Dex treatments, and (D) after DSS. (E) Taxonomic composition of bacterial communities in the 3 experimental groups at phylum, order, and family levels after DSS.
By univariate analysis, Bacteroidetes, at the phylum level, were more abundant in FLO1-treated mice, whereas Firmicutes and Proteobacteria were more abundant in control and Dex-treated mice (Fig. 5E). At the order level, both FLO1- and Dex-treated groups showed more abundance in Lactobacillales and a decrease in Campylobacterales versus the control group. Deferribacterales and Clostridiales were barely detectable in FLO1-treated SAMP in comparison with control and Dex-treated mice. Conversely, Bacteroidales were more abundant in FLO1-treated mice versus the other 2 groups. At the family level, we observed an increase of Lactobacillaceae and Prevotellaceae in FLO1-treated mice versus the control group, whereas Lachnospiraceae and Deferribacteraceae were more abundant in control mice compared to both FLO1 and Dex-treated groups. Interestingly, the Helicobacteraceae family was present in control and Dex-treated mice, but markedly decreased in FLO1-treated mice.
Identification of Bacterial Species That May Predict Response to IL-1α Neutralization.
Because identification of biomarkers for monitoring the IL-1α response would be clinically relevant for humans, we performed multivariable inferential, associative, and predictive statistics to identify the most significant microbial markers associated with baseline SAMP gut commensal status and response to Dex and IL-1α treatments using 104 fecal samples from young mice in study 2. First, using a multivariate reductionist vector analysis, we screened all microbial species that yielded high read counts and homology to the databases at the species level. The most influential species were then selected to conduct linear regression analysis in order to investigate the significant association and interactions between dominant species controlling for collinearity and cofounding variables in full and nested models. The analyses revealed that, in our model, Mucispirillum schaedleri showed a positive association with Ruminococcus gnavus and Helicobacter hepaticus. Additionally, Lactobacillus salivarius showed a significant positive association with Lactococcus garvieae and H. hepaticus. On the contrary, we found a strong negative association between Parabacteroides distasonis and R. gnavus and L. salivarius. SI Appendix, Table S1 illustrates the most important associations and the adjusted significance values indicating the translational potential to identify species that could predict the response to IL-1α therapy.
To evaluate the predictive ability of taxa to differentiate the 3 treatments, we performed a multinomial logistic regression analysis using preselected taxa at the species level that resulted in statistical significance in the vector analysis. Additionally, the model controlled for the 2 experimental replicas and the different phases of treatments. Following iterative analysis, a final model was used to determine the probabilities that selected species could predict the type of treatment received by the mice. The coefficients and significance for all variables and the most important predictive species in the final multinomial logistic model are shown in SI Appendix, Table S2. Briefly, our analysis revealed significant treatment-related changes in FLO1-treated mice compared to the other 2 groups. Particularly, in FLO1-treated mice, M. schaedleri and L. salivarius were significantly increased, with a higher relative risk ratio (RRR) of having these taxa when compared with control mice. Those same taxa had an increased RRR in FLO1 treatment in comparison with Dex treatment, although the RRR for L. salivarius was not significant. Of interest, in this study, H. hepaticus was significantly inhibited by FLO1 treatment compared to both control and Dex groups. Also, R. gnavus was significantly decreased after Dex treatment and also moderately reduced by FLO1 treatment. Ruminococcus flavefaciens showed a higher RRR in FLO1-treated mice versus control and Dex-treated mice. Taken together, these results suggest that IL-1α neutralization in noninflamed SAMP mice directly alters gut microbiome composition, generating an antiinflammatory microbiome. As such, our findings show a unique predictive relationship between FLO1 treatment and specific bacterial species, which may be of translational interest for patients that can benefit from IL-1α therapy.
Transplantation of Gut Microbiome from FLO1-Treated SAMP Mice into GF SAMP Attenuates DSS-Induced Colitis.
To further confirm whether FLO1 treatment leads to distinct changes in the function of the gut microbiome that are essential for its antiinflammatory properties, we also performed FMT experiments, as illustrated in Fig. 6A. Briefly, we transferred fecal homogenates from 4-wk-old SPF SAMP mice treated with either vehicle, Dex, or FLO1 to GF SAMP mice. After FMT, mice were challenged with 7 d of 3% DSS, administered in drinking water, to induce acute colitis, followed by a 2-wk recovery. Strikingly, upon DSS treatment, mice transplanted with the FLO1 microbiota exhibited reduced mortality (Fig. 6B) and decreased body weight loss (Fig. 6C) compared to mice that were colonized with microbiota from vehicle- and Dex-treated SAMP. Consistently, histologic analyses confirmed a reduction in colonic inflammation in recipient mice receiving FLO1 microbiota versus the other 2 experimental groups (Fig. 6 D and E). Finally, mRNA expression of inflammatory markers, such as TNFα, IL-1α, IL-1β, and COX2 in colonic tissues from recipient mice receiving FLO1 microbiota was significantly decreased when compared to the group of mice colonized with the vehicle microbiota (Fig. 6F). Collectively, these results show that transplantation of FLO1 microbiota into GF SAMP mice confers protection from DSS-induced colitis.
Fig. 6.
GF SAMP receiving microbiota from FLO1-treated SAMP donors display attenuated DSS-induced colitis. (A) Study design. (B) Survival, (C) total body weight loss, and (D) total inflammatory scores in colons of GF SAMP recipients transplanted with microbiota from FLO1-treated donors compared to Dex- and vehicle-treated donors after DSS-induced colitis. (E) Representative photomicrographs of full-thickness H&E-stained colonic tissues show preservation of epithelial structure (black arrows) in FLO1 microbiota recipient mice versus the other 2 groups. (Scale bar: 100 µm.) (F) mRNA levels of IL-1α, TNFα, IL-1β, and COX2 in colonic tissues from GF SAMP recipients transplanted with either FLO1, Dex, or vehicle microbiota after DSS-induced colitis. Data are presented as relative fold difference compared with control mice (arbitrarily set as 1). All data are representative of 2 independent experiments and presented as median ± interquartile range, n = 8 for control and FLO1 groups and n = 4 for Dex group due to the high mortality rate. Statistical significance was determined by Gehan–Breslow–Wilcoxon test (C) and 1-way ANOVA followed by Dunn’s post hoc test (D and F); *P < 0.05 and **P < 0.02.
Discussion
Herein, we provide evidence for a pathologic role of IL-1α in the SAMP mouse model of CD-like intestinal inflammation. We demonstrate elevated, inflammatory lesion-specific expression of IL-1α in this strain. Furthermore, we report that specific neutralization of anti–IL-1α with FLO1 induces alterations to the mucosal immunological milieu, leading to significant amelioration of chronic ileitis and preventing the development of acute, DSS-induced colitis in inflammation-prone SAMP mice. More importantly, we show that IL-1α neutralization is associated with taxonomic divergence of the intestinal microbiome, which is downstream and essential for the antiinflammatory properties of FLO1, as the benefit of blocking IL-1α does not occur in GF SAMP mice. Also, we demonstrate a predictive relationship between IL-1α neutralization and the modulation of specific bacterial species, which is clearly linked to the antiinflammatory effects.
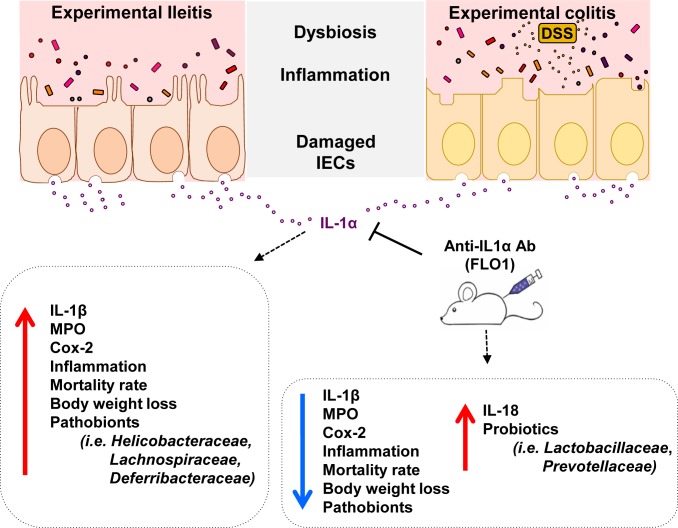
Our findings also support the established role of IL-1α as an alarmin that is released upon IEC injury (39). Particularly in SAMP mice, IL-1α was exclusively up-regulated in areas with mucosal cobblestoning, which correspond to areas of epithelial denudation. Moreover, administration of FLO1 specifically targeted these involved areas, but had no effect on uninvolved ones. Previous studies have shown that IEC necrosis leads to extracellular release of the precursor of IL-1α (40), which can be cleaved to the active form by locally secreted neutrophil serine proteases (41). In turn, extracellular IL-1α induces the recruitment of myeloid cells and release of additional proinflammatory cytokines (9). We provide direct evidence for such mechanism(s) by demonstrating that IL-1α neutralization led to decreased mucosal expression of MPO (i.e., a neutrophil product) and IL-1β primarily secreted by monocytes. Therefore, blockade of IL-1α signaling creates an antiinflammatory mucosal environment and suppresses experimental intestinal inflammation (Fig. 7), as shown both herein and in studies using IL-1α–deficient mice (13).
Fig. 7.
Graphical summary of anti–IL-1α effects in chronic intestinal inflammation. During spontaneous ileitis (Left) and experimental DSS-induced colitis (Right), damaged intestinal epithelial cells (IECs) release large amounts of IL-1α. In turn, IL-1α stimulates the production of IL-1β and other mediators of inflammation (e.g., Cox2, MPO), leading to increased disease, dysbiosis, and epithelial barrier disruption. Following IL-1α blockade with an anti–IL-1α murine-specific monoclonal antibody (FLO1 mAb), the severity of ileitis and colitis is dramatically reduced, along with diminished titer of inflammatory markers (IL-1α, IL-1β, MPO, COX2) and increased levels of IL-18, which is an inducer of intestinal barrier integrity. More importantly, IL-1α blockade is associated with changes in microbiota community structures, including higher abundance of probiotic strains, which may be of translational interest for patients that can potentially benefit from IL-1α therapy.
The present study further expands our knowledge on the role of IL-1α in mucosal immunity by showing that this cytokine displays a unique regulatory role in the function of the intestinal microbiome. This is supported by our findings that administration of FLO1 to SAMP mice was associated with changes in fecal 16S rRNA profiles, which were anti–IL-1α–specific and not a byproduct of concomitant inflammation. First, we obtained highly reproducible results between 2 separate experiments conducted more than 6 mo apart. Second, FLO1 and Dex, although equally antiinflammatory, induced disparate effects on flora composition. Moreover, FLO1 administration to 4-wk-old noninflamed SAMP mice (i.e., prior to the onset of ileitis) has led to distinct changes in the function of the gut microbiome despite the fact that we did not observe significant differences in microbiome composition as detected by 16S rRNA analyses after treatment. These results are consistent with recent studies showing that significant changes in gut microbiome functionality induced by diet or by other interventions are not necessarily associated with significant shifts in gut microbiome composition as analyzed by 16S rRNA sequencing (42). Additionally, our study is in line with recent advances that have shed light on a novel role of cytokines in the pathway(s) of interkingdom signaling occurring between microbial entities and their mammalian host (43). Indeed, prior studies have shown that specific bacterial pathogens have the ability to bind host-derived proinflammatory cytokines and respond by increasing their growth and/or altering their virulence (24–26). Porat et al. showed that human recombinant IL-1β enhanced bacterial growth rates of virulent E. coli and that IL-1Ra was able to block this effect, pointing to the existence of a bacterial binding site that is able to recognize both IL-1β and IL-1Ra (27). Therefore, the possibility exists that anti–IL-1α blockade affects the microbiome indirectly via modification of mucosal IL-1β, consistent with our results showing a marked decrease in intestinal IL-1β following FLO1 treatment. Nevertheless, a direct flora-modifying function of IL-1α may also occur, which can be reverted by its blockade. Our results are also consistent with recent studies by Nunberg et al. showing that IL-1α–deficient mice have an altered microbiome with protective activity against acute DSS-induced colitis (44).
In the present study, we also provide mechanistic evidence for the functional relevance of anti–IL-1α–induced microbiome alterations. Essentially, the antiinflammatory effect of FLO1 was completely abrogated in GF SAMP mice with attenuated ileitis, emphasizing the need for intact bacterial flora for the anti–IL-1α blockade to take effect. Further definitive support is provided by the pretreatment/DSS challenge protocol in noninflamed SAMP mice. Both anti–IL-1α blockade and Dex treatment had profound, although dissimilar, effects on bacterial flora composition, devoid of any confounding effect of active inflammation. On the contrary, only FLO1 administration led to protection of mice from DSS-induced colitis, whereas Dex had no such effect. This observation implies that changes induced by FLO1 administration specifically target flora components that are critical for the induction of intestinal inflammation. In this context, taxonomic profiling revealed increased relative abundance of Bacteroidetes in FLO-treated mice and of Firmicutes and Proteobacteria in control mice. This is in accordance with previous studies reporting that clinical and experimental IBD severity is associated with the increased Proteobacteria:Bacteroidetes ratio (45). Of note, Dex-pretreated mice also had an increased Proteobacteria component. Additionally, FLO1 treatment reduced the abundance of members of the Helicobacteraceae family, including H. hepaticus. This is of particular relevance, since Helicobacter species have been implicated as causative factors in rodent and primate models of colitis, as well as in patients with colorectal cancer (46, 47). Moreover, FLO1-treated mice had a higher RRR for the presence of either M. schaedleri or L. salivarius. M. schaedleri was originally considered a pathobiont that is increased in DSS-induced colitis (48). Nevertheless, recent evidence suggests an antiinflammatory role via the modulation of host immune-related gene expression (49). R. gnavus, which is associated with increased disease activity in IBD patients (3, 50, 51), was quantitatively reduced by FLO1 treatment. By comparison, R. flavefaciens, which was found increased in IBD patients in remission (52), showed a higher RRR in FLO1-treated mice versus control and Dex-treated mice. Overall, these data suggest that IL-1α blockade may modulate the relative abundance of dominant and functionally important microorganisms that are altered in IBD. This demonstrates a predictive relationship between IL-1α neutralization and abundance of specific bacterial species, which also holds translational significance since FLO1 treatment prevented DSS-induced mucosal dysbiosis and preserved immunological homeostasis.
Currently, no clinical trials of which we are aware have tested the efficacy of anti–IL-1 strategies, and, in particular, IL-1α blockade, in patients with IBD. This is quite surprising given the strong conceptual and experimental evidence for such an approach. First, in addition to our present study, others have also reported therapeutic efficacy of anti–IL-1α blockade in experimental colitis. Specifically, IL-1α–deficient mice suffered from a milder form of DSS-induced colitis (13, 44), whereas exogenous administration of the natural inhibitor of IL-1, IL-1Ra, was effective in reducing disease severity in a chemically induced rabbit model of colitis (53). However, the present study reports the antiinflammatory effects of IL-1 blockade in a chronic spontaneous model of CD-like ileitis with high relevance to the human condition. Second, the immunological effects of anti–IL-1α blockade in our study fit well into the common pathogenetic model of IBD. Indeed, FLO1 administration led to mucosal decreases of pivotal proinflammatory factors, including TNFα, IL-1β, and COX2, which all have been linked to the pathogenesis of IBD (54). Of considerably relevance, IL-18 was significantly elevated after FLO1 treatment. This finding aligns well with the dichotomous role of several members of the IL-1 family, which may be protective or proinflammatory depending on temporal associations as well as their cellular sources (9). Of note, the immunological effects of anti–IL-1α blockade were different from those of Dex treatment, which failed to protect noninflamed SAMP mice from DSS-induced colitis, likely due to the deleterious effects of steroids on epithelial cell proliferation and ulcer healing (55, 56). Third, members of the IL-1 family are overexpressed in inflammatory lesions of patients with IBD, indicating a possible contribution to disease pathogenesis (57). Finally, several biologic agents that act by abrogating IL-1 signaling have been developed and approved for the treatment of various autoimmune and autoinflammatory diseases; thus, the feasibility of anti–IL-1 blockade in humans has been tested and validated (58). Importantly, in our study, FLO1 treatment showed comparable therapeutic efficacy to Dex. This is of particular significance since steroids are among the most potent antiinflammatory therapies for IBD, but are associated with serious side effects (59). Nonetheless, target specificity of anti–IL-1α has led to a very acceptable safety profile, which renders this approach a preferable option for patients affected by IBD (15, 58).
In conclusion, this study provides significant data supporting a central role of IL-1α in the pathogenesis of experimental IBD. Moreover, these preclinical findings strongly support the therapeutic potential of IL-1α neutralization in patients with IBD. IL-1α blockade exerts its antiinflammatory effects by modulating the gut microbial ecosystem, thus correcting the mucosal dysbiosis that takes place in IBD. Further research is needed to define the specific bacterial strains that could functionally mediate the antiinflammatory effects of anti–IL-1α therapy in IBD, and the underlying immunological pathways that are affected. Clarification of such bacteria–host interactions will pave the way for novel and targeted therapeutic strategies for patients affected by these devastating conditions.
Materials and Methods
All methods employed in this article are routinely used in our laboratories. Fully detailed procedures are described in SI Appendix, SI Materials and Methods. All materials, data, and associated protocols, including data and methods included in the SI Appendix, will be made available to readers upon request.
Experimental Animals.
SAMP and AKR mice were maintained under SPF conditions, fed with standard laboratory chow, and kept on 12-h light/dark cycles in the animal resource core (ARC) facility of Case Western Reserve University (CWRU). GF SAMP mice have been propagated in our ARC facility at CWRU since 2011 (60, 61). All protocols were approved by the institutional animal care and use committee of CWRU.
Intersubject Preexperimental Fecal Homogeneization (IsPreFeH).
Fecal homogenization was achieved using the IsPreFeH strategy (29, 61).
Anti–IL-1α Treatment.
SPF and GF mice were injected intraperitoneally with FLO1 (XBiotech), an anti–IL-1α monoclonal antibody (100 μg; 2 times per week for 4 wk). A full description of treatment studies and antibody validation (SI Appendix, Fig. S1) is provided in SI Appendix, SI Materials and Methods.
DSS-Induced Colitis Model.
Induction of acute colitis was achieved in SAMP mice with 7 d of 3% dextran sodium sulfate (DSS; batch no. DB001-37; TdB Consultancy) in drinking water.
Fecal Microbiota Transplantation (FMT).
Fresh fecal samples were obtained and pooled at equal amounts from 8-wk-old SAMP mice after 4 wk of treatment with either FLO1, Dex, or vehicle control. Pooled samples were suspended (1:10, wt/vol) in sterile saline containing 20% of glycerol, snap-frozen, and stored at −80 °C until use. A 200-μL inoculum was administered to each group of GF SAMP mice by oral gavage once a week for 3 consecutive weeks before induction of DSS colitis.
Histology.
Ilea and colons were removed from mice and histologically evaluated as previously described (22).
Tissue Culture.
Both involved and uninvolved areas (29) were collected and cultured in complete RPMI medium 1640 for 24 h. Supernatants were subsequently collected and stored at −80 °C for further assays.
IL-1α Staining.
Paraffin sections of mouse ilea were stained with goat polyclonal Ab against murine IL-1α.
MPO Activity Assay.
Colon specimens and both involved/uninvolved areas from ilea were assayed for myeloperoxidase (MPO) activity as previously described (29).
qRT-PCR.
Colon samples and SM-microdissected samples from ilea were placed in RNAlater, left at 4 °C overnight, then stored at −80 °C. Total RNA was isolated using established protocols, and RTqPCR amplification was subsequently performed.
16S rRNA Gene Microbiome Analysis.
Following fecal DNA extraction, microbiome amplification for the 16s rRNA gene V4 regions and high-throughput 16S rRNA gene microbiome sequencing and analysis were conducted using the well-established Illumina and analytical protocols as previously described (62, 63).
Statistical Analysis.
All experiments were performed at least twice. All data are presented as box-and-whisker plot showing the minimum to maximum with all data points and the horizontal line representing the median; a P value smaller than 0.05 was considered significant. Student’s t tests and/or 1-way ANOVA were used to compare continuous data across all available experimental groups, provided the data fulfilled the assumptions for parametric statistics. Also, nonparametric tests were used for data with nonfulfilled distribution assumptions due to some normalized data. Statistical and graphical analyses were performed using GraphPad Prism version 6 (GraphPad), R studio, and STATA.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants DK091222, DK055812, DK042191, and AI015614. We also acknowledge the Mouse Models Core and the Histology/Imaging Core of the Cleveland Digestive Disease Research Core Center (DK097948). We thank also Natalia Aladyshkina and Mathew Conger for assistance with animal husbandry.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915043116/-/DCSupplemental.
References
- 1.Wallace K. L., Zheng L. B., Kanazawa Y., Shih D. Q., Immunopathology of inflammatory bowel disease. World J. Gastroenterol. 20, 6–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Souza H. S., Fiocchi C., Immunopathogenesis of IBD: Current state of the art. Nat. Rev. Gastroenterol. Hepatol. 13, 13–27 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Nishida A., et al. , Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin. J. Gastroenterol. 11, 1–10 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Kostic A. D., Xavier R. J., Gevers D., The microbiome in inflammatory bowel disease: Current status and the future ahead. Gastroenterology 146, 1489–1499 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hold G. L., et al. , Role of the gut microbiota in inflammatory bowel disease pathogenesis: What have we learnt in the past 10 years? World J. Gastroenterol. 20, 1192–1210 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katsanos K. H., et al. , Biological therapies in inflammatory bowel disease: Beyond anti-TNF therapies. Clin. Immunol. 206, 9–14 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Neurath M. F., Cytokines in inflammatory bowel disease. Nat. Rev. Immunol. 14, 329–342 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Waldner M. J., Neurath M. F., Novel cytokine-targeted therapies and intestinal inflammation. Curr. Opin. Pharmacol. 9, 702–707 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Lopetuso L. R., Chowdhry S., Pizarro T. T., Opposing functions of classic and novel IL-1 family members in gut Health and disease. Front. Immunol. 4, 181 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van de Veerdonk F. L., Netea M. G., New insights in the immunobiology of IL-1 family members. Front. Immunol. 4, 167 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scarpa M., et al. , The epithelial danger signal IL-1α is a potent activator of fibroblasts and reactivator of intestinal inflammation. Am. J. Pathol. 185, 1624–1637 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Paolo N. C., Shayakhmetov D. M., Interleukin 1α and the inflammatory process. Nat. Immunol. 17, 906–913 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bersudsky M., et al. , Non-redundant properties of IL-1α and IL-1β during acute colon inflammation in mice. Gut 63, 598–609 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Ridker P. M., et al. ; CANTOS Trial Group , Antiinflammatory therapy with Canakinumab for atherosclerotic disease. N. Engl. J. Med. 377, 1119–1131 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Hickish T., et al. , MABp1 as a novel antibody treatment for advanced colorectal cancer: A randomised, double-blind, placebo-controlled, phase 3 study. Lancet Oncol. 18, 192–201 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Hong D. S., et al. , MABp1, a first-in-class true human antibody targeting interleukin-1α in refractory cancers: An open-label, phase 1 dose-escalation and expansion study. Lancet Oncol. 15, 656–666 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Kanni T., et al. , MABp1 targeting IL-1α for moderate to severe hidradenitis suppurativa not eligible for adalimumab: A randomized study. J. Invest. Dermatol. 138, 795–801 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Pizarro T. T., et al. , SAMP1/YitFc mouse strain: A spontaneous model of Crohn’s disease-like ileitis. Inflamm. Bowel Dis. 17, 2566–2584 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cominelli F., et al. , Interleukin 1 (IL-1) gene expression, synthesis, and effect of specific IL-1 receptor blockade in rabbit immune complex colitis. J. Clin. Invest. 86, 972–980 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCall R. D., et al. , Tissue interleukin 1 and interleukin-1 receptor antagonist expression in enterocolitis in resistant and susceptible rats. Gastroenterology 106, 960–972 (1994). [DOI] [PubMed] [Google Scholar]
- 21.Ligumsky M., Simon P. L., Karmeli F., Rachmilewitz D., Role of interleukin 1 in inflammatory bowel disease–Enhanced production during active disease. Gut 31, 686–689 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burns R. C., et al. , Antibody blockade of ICAM-1 and VCAM-1 ameliorates inflammation in the SAMP-1/Yit adoptive transfer model of Crohn’s disease in mice. Gastroenterology 121, 1428–1436 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Reuter B. K., et al. , Spontaneous, immune-mediated gastric inflammation in SAMP1/YitFc mice, a model of Crohn’s-like gastritis. Gastroenterology 141, 1709–1719 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo G., Niesel D. W., Shaban R. A., Grimm E. A., Klimpel G. R., Tumor necrosis factor alpha binding to bacteria: Evidence for a high-affinity receptor and alteration of bacterial virulence properties. Infect. Immun. 61, 830–835 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLaughlin R. A., Hoogewerf A. J., Interleukin-1beta-induced growth enhancement of Staphylococcus aureus occurs in biofilm but not planktonic cultures. Microb. Pathog. 41, 67–79 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Zav’yalov V. P., et al. , Specific high affinity binding of human interleukin 1 beta by Caf1A usher protein of Yersinia pestis. FEBS Lett. 371, 65–68 (1995). [DOI] [PubMed] [Google Scholar]
- 27.Porat R., Clark B. D., Wolff S. M., Dinarello C. A., Enhancement of growth of virulent strains of Escherichia coli by interleukin-1. Science 254, 430–432 (1991). [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez-Palacios A., et al. , The artificial sweetener splenda promotes gut proteobacteria, dysbiosis, and myeloperoxidase reactivity in Crohn’s disease-like ileitis. Inflamm. Bowel Dis. 24, 1005–1020 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez-Palacios A., et al. , Stereomicroscopic 3D-pattern profiling of murine and human intestinal inflammation reveals unique structural phenotypes. Nat. Commun. 6, 7577 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bamias G., et al. , Commensal bacteria exacerbate intestinal inflammation but are not essential for the development of murine ileitis. J. Immunol. 178, 1809–1818 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Nair P. P., et al. , Markers of inflammation and lineage on exfoliated colonic cells in pediatric inflammatory bowel disease. J. Gastrointest. Dig. Syst. 8, 1–6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu W., et al. , Cyclooxygenase-2 is up-regulated by interleukin-1 beta in human colorectal cancer cells via multiple signaling pathways. Cancer Res. 63, 3632–3636 (2003). [PubMed] [Google Scholar]
- 33.Ogata S., et al. , Signaling pathways regulating IL-1alpha-induced COX-2 expression. J. Dent. Res. 86, 186–191 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Di Mari J. F., Mifflin R. C., Adegboyega P. A., Saada J. I., Powell D. W., IL-1alpha-induced COX-2 expression in human intestinal myofibroblasts is dependent on a PKCzeta-ROS pathway. Gastroenterology 124, 1855–1865 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Salcedo R., et al. , MyD88-mediated signaling prevents development of adenocarcinomas of the colon: Role of interleukin 18. J. Exp. Med. 207, 1625–1636 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaki M. H., et al. , The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity 32, 379–391 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Fazio L., et al. , Longitudinal analysis of inflammation and microbiota dynamics in a model of mild chronic dextran sulfate sodium-induced colitis in mice. World J. Gastroenterol. 20, 2051–2061 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eichele D. D., Kharbanda K. K., Dextran sodium sulfate colitis murine model: An indispensable tool for advancing our understanding of inflammatory bowel diseases pathogenesis. World J. Gastroenterol. 23, 6016–6029 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen G. Y., Nuñez G., Sterile inflammation: Sensing and reacting to damage. Nat. Rev. Immunol. 10, 826–837 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cohen I., et al. , Differential release of chromatin-bound IL-1alpha discriminates between necrotic and apoptotic cell death by the ability to induce sterile inflammation. Proc. Natl. Acad. Sci. U.S.A. 107, 2574–2579 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Afonina I. S., et al. , Granzyme B-dependent proteolysis acts as a switch to enhance the proinflammatory activity of IL-1α. Mol. Cell 44, 265–278 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu G. D., et al. , Comparative metabolomics in vegans and omnivores reveal constraints on diet-dependent gut microbiota metabolite production. Gut 65, 63–72 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hughes D. T., Sperandio V., Inter-kingdom signalling: Communication between bacteria and their hosts. Nat. Rev. Microbiol. 6, 111–120 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nunberg M., et al. , Interleukin 1α-deficient mice have an altered gut microbiota leading to protection from dextran sodium sulfate-induced colitis. mSystems 3, e00213-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Somineni H. K., Kugathasan S., The microbiome in patients with inflammatory diseases. Clin. Gastroenterol. Hepatol. 17, 243–255 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Hansen R., Thomson J. M., Fox J. G., El-Omar E. M., Hold G. L., Could Helicobacter organisms cause inflammatory bowel disease? FEMS Immunol. Med. Microbiol. 61, 1–14 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Fox J. G., Ge Z., Whary M. T., Erdman S. E., Horwitz B. H., Helicobacter hepaticus infection in mice: Models for understanding lower bowel inflammation and cancer. Mucosal Immunol. 4, 22–30 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rooks M. G., et al. , Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J. 8, 1403–1417 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loy A., et al. , Lifestyle and horizontal gene transfer-mediated evolution of Mucispirillum schaedleri, a core member of the murine gut microbiota. mSystems 2, e00171-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hall A. B., et al. , A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 9, 103 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berry D., Reinisch W., Intestinal microbiota: A source of novel biomarkers in inflammatory bowel diseases? Best Pract. Res. Clin. Gastroenterol. 27, 47–58 (2013). [DOI] [PubMed] [Google Scholar]
- 52.De Cruz P., et al. , Association between specific mucosa-associated microbiota in Crohn’s disease at the time of resection and subsequent disease recurrence: A pilot study. J. Gastroenterol. Hepatol. 30, 268–278 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Ferretti M., et al. , Neutralization of endogenous IL-1 receptor antagonist exacerbates and prolongs inflammation in rabbit immune colitis. J. Clin. Invest. 94, 449–453 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bevivino G., Monteleone G., Advances in understanding the role of cytokines in inflammatory bowel disease. Expert Rev. Gastroenterol. Hepatol. 12, 907–915 (2018). [DOI] [PubMed] [Google Scholar]
- 55.van Meeteren M. E., Meijssen M. A., Zijlstra F. J., The effect of dexamethasone treatment on murine colitis. Scand. J. Gastroenterol. 35, 517–521 (2000). [DOI] [PubMed] [Google Scholar]
- 56.Ocón B., et al. , The glucocorticoid budesonide has protective and deleterious effects in experimental colitis in mice. Biochem. Pharmacol. 116, 73–88 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Youngman K. R., et al. , Localization of intestinal interleukin 1 activity and protein and gene expression to lamina propria cells. Gastroenterology 104, 749–758 (1993). [DOI] [PubMed] [Google Scholar]
- 58.Dinarello C. A., Simon A., van der Meer J. W., Treating inflammation by blocking interleukin-1 in a broad spectrum of diseases. Nat. Rev. Drug Discov. 11, 633–652 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salice M., Rizzello F., Calabrese C., Calandrini L., Gionchetti P., A current overview of corticosteroid use in active ulcerative colitis. Expert Rev. Gastroenterol. Hepatol. 13, 557–561 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Rodriguez-Palacios A., et al. , Clinical effects of Gamma-radiation-resistant Aspergillus sydowii on germ-free mice immunologically prone to inflammatory bowel disease. J. Pathogens 2016, 5748745 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodriguez-Palacios A., et al. , ‘Cyclical Bias’ in microbiome research revealed by a portable germ-free housing system using nested isolation. Sci. Rep. 8, 3801 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gillevet P., Sikaroodi M., Keshavarzian A., Mutlu E. A., Quantitative assessment of the human gut microbiome using multitag pyrosequencing. Chem. Biodivers. 7, 1065–1075 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lagkouvardos I., Fischer S., Kumar N., Clavel T., Rhea: A transparent and modular R pipeline for microbial profiling based on 16S rRNA gene amplicons. PeerJ 5, e2836 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.