Abstract
Enabled by new approaches for rapid identification and selection of human monoclonal antibodies, atomic-level structural information for viral surface proteins, and capacity for precision engineering of protein immunogens and self-assembling nanoparticles, a new era of antigen design and display options has evolved. While HIV-1 vaccine development has been a driving force behind these technologies and concepts, clinical proof-of-concept for structure-based vaccine design may first be achieved for respiratory syncytial virus (RSV), where conformation-dependent access to neutralization-sensitive epitopes on the fusion glycoprotein determines the capacity to induce potent neutralizing activity. Success with RSV has motivated structure-based stabilization of other class I viral fusion proteins for use as immunogens and demonstrated the importance of structural information for developing vaccines against other viral pathogens particularly difficult targets that have resisted prior vaccine development efforts. Solving viral surface structures also supports rapid vaccine antigen design and application of platform manufacturing approaches for emerging pathogens.
Keywords: respiratory syncytial virus, coronavirus, influenza, nanoparticle display, vaccine development, platform technology, immunization, X-ray crystallography, electron microscopy
INTRODUCTION
The goal of vaccination is to create immunity in the host that will prevent disease if the infectious pathogen is encountered. Since vaccination is typically intended for healthy, uninfected subjects, the immunization process must avoid off-target effects and be well tolerated and safe. Vaccines for viral pathogens are usually designed to induce antibody responses that target viral proteins involved in cellular entry to achieve neutralizing activity in serum and mucosal secretions. If sufficient neutralizing activity is present at the time of infection, virus will be blocked from infecting cells or the extent of infection will be limited so that innate and adaptive immune mechanisms have an opportunity to clear virus-infected cells before viral spread and antigen load are sufficient to cause clinical symptoms. Therefore, in its simplest form, vaccine development for viral diseases involves the delivery of antigens that will induce virus-specific neutralizing antibodies and avoid the induction of any off-target antibodies. In addition, antibodies without classical neutralizing activity can sometimes provide effective immunity through Fc-mediated effector functions (1, 2), but would still need to bind viral proteins with high specificity. For many viruses, additional immunological responses may be needed for effective vaccine-induced immunity, including CD4 and CD8 T cells with particular properties and localization (3, 4). For the purposes of this review on the use of atomic-level structural information to guide vaccine antigen design, we will focus on the induction of antibodies by protein antigens. Although generating an antibody response after vaccination is a complex biological process, as technology improves, much of it can be explained using chemistry, geometry, and topology. Thereby, the process of designing proteins to be vaccine immunogens is becoming more of a rational engineering exercise and less fraught with the uncertainties inherent in biology.
CONCEPTS OF ANTIBODY NEUTRALIZATION AND B CELL TARGETING
Antibodies are proteins designed to interact with other biological substances. The basis of that recognition is determined largely by the topological complementarity of the surface contours between elements of the antibody variable domain and the surfaces present on the molecule being recognized (i.e. the antigen). The strength of this interaction is influenced by chemistry and affected by the physical constraints that determine the surface area of interaction. Antibodies are heterodimeric oligomers composed of a heavy chain and a light chain and produced by B lymphocytes and plasma cells. The variable domains, particularly the complementarity-determining regions (CDRs) that form loops at the tips of the antigen-binding fragments (Fab), form the critical surfaces relevant to antigen recognition. The Fab is formed from both heavy and light chains, each having a variable domain with three CDRs. The classes and subclasses of antibodies are determined by the constant regions of the heavy chain (Fc), which also confer functional properties, determine whether the antibody is membrane-bound or secreted, and affect location and half-life (Figure 1).
Figure 1. Relative size and components of an antibody molecule.
(A) Full-length human IgG1 (PDB ID: 1IGT) is a dimer of heterodimers composed of two heavy chains (orange) and two light chains (light orange). One heavy and one light chain are shown in molecular surfaces and the other heavy and light chains are shown in ribbons. The variable (V) and constant (C) domains of both the light (L) and heavy (H) chains are labeled and the loops of the complementarity determining region (CDR) are highlighted in red and pink for the heavy and light chains, respectively. The antigen binding fragment (Fab) is distinguished from the crystallizable fragment (Fc). (B) The prefusion conformation of RSV F (pre-F) is shown with two protomers in grey and white molecular surfaces and a third protomer in ribbons, colored as a rainbow from blue to red, N-to C-terminus, respectively. Full-length IgG from (A) was aligned with the motavizumab Fab and is shown in molecular surfaces binding to pre-F, which is ~11 nm tall. The relative size of the IgG1 compared to the viral surface protein is demonstrated to illustrate how IgG binding to one epitope may partially interfere with binding to other nearby epitopes on the surface.
Structure-based vaccine design seeks to create surfaces on immunogens that will elicit protective antibody responses against the target pathogen. Defining the atomic-level details of key surfaces on antigens accessible on pathogens is a primary requirement for structure-based vaccine design. Knowing which proteins to attack, and which specific sites on those proteins to target with antibodies, is fundamental to initiating a structure-based vaccine project. Therefore, structure-based design often begins by identifying pathogen-specific antibodies with specific properties such as potent neutralization. Neutralizing activity implies that an antibody will reduce the frequency of virus-infected cells. To neutralize, the antibody must bind to a vulnerable site on a surface-exposed viral protein. The strength or potency of neutralization is determined by many factors, but is often linked to specificity or site of protein binding, strength of binding (affinity and avidity), accessibility of the binding site on the virus, and extent of occupancy on the available sites on the virus. The basis for protection can be blocking of viral attachment, prevention of viral entry post-attachment, prevention of virus release from infected cells, fusion inhibition and reduction of cell-to-cell transmission, aggregation of virus to each other or to mucus to effectively reduce inoculum size, diversion to dead-end entry pathway, or phagocytosis of virus. Regardless of the mechanism, antibody binding to the viral protein is essential.
Geometry of antibody binding to an antigen can be an important factor in vaccine design. There are some epitopes or antigenic sites on proteins that can be bound by antibodies from multiple lineages using multiple angles of approach or degrees of rotation (so-called “super sites”(5)). However, there are other critical targets that require an antibody to approach at a particular angle (6) or rotation (7, 8). Viruses also tend to protect sites of vulnerability by frequently mutating surrounding surfaces (e.g. rim of influenza RBD) and adding or removing glycans. These immune evasion mechanisms can therefore require long CDR loops to reach sites of vulnerability (9, 10) or short CDR loops to avoid clashes and allow closer proximity (6). Structure-based design must account for these immune-evasion mechanisms.
HISTORICAL CONTEXT
Induction of selected B cells that can recognize unique surface contours on a viral protein is a 3-dimensional process. Although 2-dimensional protein sequence data and linear epitopes have been sufficient for some prior viral vaccine efforts, many future vaccines will require atomic-level information to assure antibodies that recognize complex epitopes are induced. Most licensed vaccines were developed empirically prior to a basic understanding of immunology and were guided by the induction of antibody responses with a focus on neutralizing activity. These vaccine approaches have been largely based on whole virus, either as a live-attenuated or inactivated virus. More recently, virus-like particles (VLPs) have been successful platforms for hepatitis B and human papilloma virus. The use of whole viruses and VLPs reduces the need for a detailed structural understanding of antibody binding. During the course of virus replication (live-attenuated) or in a fixed virion particle (inactivated virus), the native conformations of viral proteins are likely available for recognition by B cells resulting in functional antibody responses able to neutralize or rapidly clear virus. Recently, a subunit protein vaccine based on glycoprotein E (gE) was licensed for use against herpes varicella zoster (HVZ) and is highly effective against shingles or reactivation (11). There is no structural data published for gE, and although it does induce robust antibody responses, it is thought that much of the efficacy is derived from the CD4 T-cell response. Therefore, it is possible to develop anti-viral vaccines without structural data and detailed understanding of the mechanisms of antibody neutralization, but for the viral targets remaining, the use of traditional vaccine technology will be challenging, in some cases because it has already failed. Additionally, the traditional methodology may be precluded for emerging viruses because the process development time is too long for immediate outbreak response.
One relevant example of vaccine failure involves a formalin-inactivated alum-formulated vaccine for respiratory syncytial virus (FI-RSV), which was tested in the 1960s and resulted in a vaccine-enhanced illness syndrome upon natural RSV infection the following season (12–15). Retrospective studies have shown that compared to natural infection, FI-RSV induced a relatively poor neutralizing antibody response relative to the amount of binding antibody elicited (16, 17). After the structure of the RSV fusion (F) glycoprotein in the prefusion conformation (pre-F) was determined, reagents became available to assess the pre-F conformation and to detect the rearrangement of F to the postfusion 6-helix bundle conformation (post-F). Using antibodies that selectively recognize highly neutralization-sensitive epitopes on pre-F and others that recognize moderately neutralization-sensitive epitopes on the shared surfaces between pre-F and post-F, it was demonstrated that all the F proteins on the FI-RSV preparation had flipped into the post-F conformation (18) rendering it much less immunogenic for inducing functional antibodies. Going forward, structural characterization of vaccine antigens by X-ray crystallography, cryo-electron microscopy, cryo-electron tomography, or antibody-binding assays should eliminate or greatly reduce vaccine failures due to improper antigen conformation.
The current focus on structure-based or structure-guided antigen design is part of a larger technological revolution involving advances in isolation of human monoclonal antibodies, next-generation sequencing, nanoparticle biology, and genetically-engineered animal models (19). This was preceded by conceptual advances involving the term “reverse vaccinology”. This term was first used by Rino Rappuoli in 2000 to describe a process by which complete sequencing of pathogen genomes could be used to identify and down-select surface-expressed or secreted proteins to arrive at new candidate vaccine antigens (20). In 2002, motivated by advances in antibody discovery using phage-display, Dennis Burton used “reverse vaccinology” to describe a process by which identifying antibodies with desirable properties could be used to select or design antigens that elicited the target antibodies (21). The role of structure in the new paradigm for vaccinology has been anticipated for several years (22–24), but the realization is just now happening with successes in RSV and other viral diseases discussed below. The concept of using effective antibodies and structures to guide vaccine antigen design has been repackaged as Reverse Vaccinology 2.0 (25). Structure-based vaccine antigen design enabled by rapid discovery of monoclonal antibodies, high-throughput X-ray crystallography, and high-resolution cryo-EM promises to be a new approach for obtaining immunogens with atomic-level accuracy and epitope-specific targeting (Table 1).
EXAMPLES OF STRUCTURE-BASED VACCINE CONCEPTS
Stabilizing class I fusion proteins to preserve neutralization-sensitive epitopes
Respiratory syncytial virus (RSV).
RSV is an orthopneumovirus in the family Pneumoviridae. It was discovered in 1955 as Chimpanzee coryza agent (26) and was associated with bronchiolitis in young infants in 1956 (27, 28). RSV is the leading cause of hospitalization in children under 5 years of age (29, 30) and causes significant mortality in the frail elderly on a scale similar to seasonal influenza (31, 32). RSV infects all children by the 2nd or 3rd year of life and continues to infect people repeatedly throughout life (33), despite relatively little antigenic variation in the primary target for neutralizing antibody: the F glycoprotein. RSV has multiple mechanisms for immune evasion including 1) infection and virion release at the apical surface of polarized airway epithelium is relatively isolated from systemic immune responses, 2) initial infection in young children occurs during a time of immunological immaturity resulting in nonoptimal priming, 3) nonstructural proteins 1 and 2 inhibit both induction and effector functions of type I interferon (34, 35), 4) the secreted G glycoprotein is immunomodulatory and can alter signaling pathways in dendritic cells (36–38), and 5) the functional, non-triggered, neutralization-susceptible conformation of the F glycoprotein trimer is unstable and rearranges spontaneously into a post-F conformation, minimizing exposure to neutralization-sensitive epitopes only present on the pre-F conformation (Figure 2) (39). The latter mechanism was identified when the crystal structure of the F protein in its prefusion conformation was solved (39) and highly neutralization-sensitive epitopes at the apex of that structure were revealed. Antibodies to pre-F-specific antigenic sites 0, III, and V are particularly potent (Figure 2), often with 10–100 times greater neutralizing activity than the licensed product palivizumab (Synagis®), which is a site II specific mAb (40). Although antibodies directed to antigenic sites II and IV, which are present on the shared surface of both the pre-F and post-F conformations (Figure 2), can achieve high potency, most of them have modest neutralizing activity.
Figure 2. Epitopes present on two major conformations of the RSV F glycoprotein.
The prefusion conformation of RSV F (pre-F) is shown with two protomers in grey and white molecular surfaces and the third protomer in ribbons, colored as a rainbow from blue to red, N- to C- terminus, respectively. Antibodies against sites Ø (red/pink), III (green), and V (orange) bind only to pre-F, whereas antibodies that recognize sites II (yellow) and IV (purple) bind to regions that exist on the shared surfaces of pre-F and post-F. Antibodies to site I (blue) predominantly bind post-F. Antibodies that exclusively bind pre-F are the most potent, those that bind the shared surface tend to have moderate neutralizing activity, and those that bind only to post-F are typically non-neutralizing. Neutralization potency is illustrated by the width of the rainbow-colored triangle.
Protein engineering efforts resulted in authentic stabilized pre-F trimers (41–44) that preserved the key epitopes associated with most neutralizing activity in human sera (45). The RSV pre-F vaccines have been shown to have much greater immunogenicity for induction of neutralizing activity than vaccines based on post-F proteins or historical RSV vaccines (41), and are now being evaluated in clinical trials. It has also been shown that stabilization of RSV F in the prefusion conformation improves the immunogenicity of live chimeric viruses (46, 47), live-attenuated virus (48), virus-like particles (49) and gene-based vectors (50). The concept of stabilizing the prefusion form of F is now being successfully applied to closely related viruses in the Paramyxoviridae family including parainfluenza types 1–4 and Nipah virus. While structure-based antigen design may provide general solutions for pathogens within a family or genus of viruses, as has always been true in biology, there are exceptions and unique ways in which each pathogen evades immune responses that require experimentation to solve. For example, the pre-F structure was recently determined for human metapneumovirus, a Pneumovirus closely related to RSV, but a prefusion-stabilized variant was found to have similar immunogenicity to the post-F form. The current hypothesis for this result is that the antigenic site analogous to site Ø on RSV F is protected by two glycans on hMPV F that block antibody access to the site (51). Therefore, most of the neutralization-sensitive epitopes are present on the shared surface of pre-F and post-F of the hMPV fusion glycoprotein. Nevertheless, grouping viral pathogens by class of fusion proteins is a reasonable way to begin looking for antigen design approaches that are effective across virus families (Figure 3).
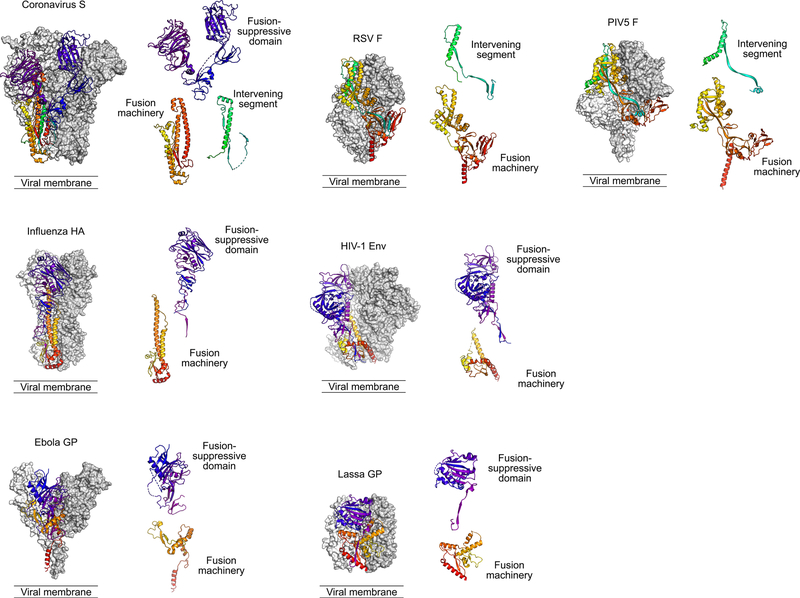
Figure 3. Class I viral fusion proteins.
Representative trimeric fusion proteins from selected virus families (Coronaviridae, Orthomyxoviridae, Filoviridae, Pneumoviridae, Retroviridae, Arenaviridae, and Paramyxoviridae) are shown with two protomers in grey and white molecular surfaces and a third protomer in ribbons, colored as described below. The panel adjacent to each trimer shows components related to the membrane fusion process, separated by function. These are type I membrane proteins anchored by a C-terminal transmembrane domain (TM). Each is shown in the pre-triggered state, with a free hydrophobic fusion peptide (FP) at the N-terminus of the C-terminal fragment, and two heptad-repeat regions adjacent to the FP and the TM that bring the viral and host-cell membrane together during the rearrangement process. These proteins vary in many ways, including whether there is a fusion-suppressive domain (colored from purple to blue) that must be displaced for the fusion machinery (colored from yellow to red) to function, and the presence or absence of an intervening segment that remains associated with the protein after fusion occurs (colored in cyan to green). Nevertheless, the overall functional similarities may lead to antigen design solutions that are analogous across viral families. Structures shown are derived from PDB IDs HCJ0, 2FK0, 5JQ3, 4MMU, 4ZMP, 5VK2, and 4GIP.
Coronaviruses (CoV).
CoVs are enveloped non-segmented positive-strand RNA viruses that belong to the order Nidovirales. The spike (S) glycoprotein is the largest known class I viral fusion glycoprotein, and it contains both a receptor-binding subunit (S1) and a membrane-fusing subunit (S2) (Figure 3). The S glycoprotein is the major target of neutralizing antibodies, but it is highly glycosylated and antigenically variable (52). Until recently, structural studies, vaccine development, and antibody-isolation efforts have focused on fragments of the S protein that were biochemically tractable, with an emphasis on the receptor-binding domains. Since 2015, advances in cryo-EM have led to the determination of numerous structures of trimeric S glycoprotein ectodomains in the prefusion conformation (53–55). For some of these constructs, the yields were low and the population of proteins was not conformationally homogeneous, with evidence of subunit dissociation and conversion of the S2 subunit to the postfusion conformation (56, 57). By harnessing the structural information, however, several residues were identified in the hinge region of the S2 subunit that when substituted with proline residues led to substantial improvements in yield and conformational homogeneity (57). In addition, the prefusion-stabilized S glycoproteins elicited an improved immune response in mice as compared to the wild-type S glycoproteins. The use of prefusion ectodomains as immunogens should result in a polyclonal response directed against many neutralization-sensitive epitopes located across the large surface of the S glycoprotein, which is preferred to an RBD-exclusive immune response that could be evaded by the antigenic drift of only a few residues.
Human immunodeficiency virus (HIV-1).
HIV-1 is an enveloped single-stranded positive-sense RNA retrovirus. Its virion displays only a few copies of the Envelope fusion protein, which like the CoV S protein is heavily glycosylated. The fusion-suppressive subunit, gp120, binds to host receptors CD4 and CCR5 or CXCR4, resulting in structural changes that allow the gp41 subunit to refold and fuse the viral and host-cell membranes. As the sole target of neutralizing antibodies, HIV-1 Envelope is the focus of intense vaccine development efforts (58), and often serves as the test-bed for new structure-based vaccine-design concepts. The stabilization of the Envelope ectodomain in the prefusion conformation was a tremendous technical achievement that required introduction of a disulfide bond between gp120 and gp41 (59), an isoleucine to proline substitution in gp41 (60), truncation after residue 664 to remove hydrophobic portions of the membrane-proximal external region (MPER) (61), and selection of a specific Envelope sequence from strain BG505 (62). These BG505 SOSIP.664 proteins led to the first near-atomic structures of trimeric prefusion Envelope (63, 64), and the resulting structures have provided the foundation for many additional iterations of structure-based vaccine design. However, unlike RSV F where the major neutralizing escape mechanism is conformational evasion, HIV-1 Envelope also has genetic plasticity and antigenic variability, extensive glycosylation, and immunodominance problems that have only been partially solved. Therefore, structure-based vaccine design is but one part of a multi-dimensional approach to HIV-1 vaccine development.
Scaffolded or chimeric proteins
The use of peptides as immunogens attempts to remove epitopes from viral proteins and present them to the immune system to elicit a focused antibody response. Such an approach is limited to epitopes that are completely, or mostly, contained within a linear stretch of amino acids. Although there are instances of such epitopes, they often exist in the native antigen in a single conformation or an ensemble of related but constrained conformations. When used as immunogens, the isolated peptides are generally flexible and adopt many conformations, only one or a few of which resemble the conformation of the epitope as it exists in the antigen. Consequently, the resulting antibody response from peptide-based immunizations tend to elicit high titers of peptide-directed antibodies, but low titers of antibodies that recognize the native antigen. Early studies attempted to induce neutralizing antibodies against RSV by immunizing with peptides corresponding to antigenic site II on RSV F (65), which is the epitope targeted by palivizumab. Site II was predicted to exist in a helix-loop-helix conformation spanning approximately 25 residues, and peptides comprising this region could bind palivizumab and other neutralizing antibodies. When such peptides were used as immunogens, however, little RSV-neutralizing activity was elicited, suggesting that the conformation of the epitope was not preserved in the peptides.
One structure-based-design approach to address the flexibility of peptide immunogens is referred to as epitope scaffolding or epitope transplantation. Here the goal is to “transplant” an epitope onto a heterologous protein “scaffold” that preserves the conformation of the epitope as it exists in the native antigen. The application of this approach to RSV was inspired by earlier efforts focused on eliciting antibody responses against the MPER of the HIV-1 Envelope gp41 subunit (66). For the RSV epitope scaffolds, a high-resolution crystal structure was initially determined for a peptide comprising antigenic site II of RSV F bound to motavizumab, a more potent derivative of palivizumab (67). With the atomic coordinates of the helix-loop-helix conformation of site II in hand, a computational procedure was performed to identify small proteins whose structures had been previously determined and contained two α-helices that were structurally similar to those in the helix-loop-helix of site II (68). Three proteins were selected, and 13 residues in each were substituted with the corresponding site II residues that contacted motavizumab. Two of those epitope scaffolds could be expressed, and one bound motavizumab with ~100 nM affinity. However, when used to immunize mice, the best epitope scaffold failed to elicit significant RSV-neutralizing activity despite eliciting antibodies that could bind the RSV F protein. To improve upon these first-generation immunogens, a computational approach called Fold From Loops was developed and used to design new epitope scaffolds that contained all of antigenic site II and presented it in a conformation that was nearly identical to that observed in crystal structures of RSV F (69). Interestingly, although these second-generation immunogens also failed to elicit RSV-neutralizing activity in mice, they were able to elicit substantial neutralizing activity in macaques. These results provided a proof-of-principle for epitope-focused vaccine design and have inspired similar efforts for RSV and other pathogens (70, 71). A potential drawback of this approach, however, is that escape from the monoclonal-like antibody response could arise due to the antigenic drift of only a few residues. This could be circumvented by immunization with multiple different epitope scaffolds, although for some viruses this approach would have little benefit over immunizing with the full, properly stabilized viral protein.
Nanoparticle display and self-assembling virus-like particles (VLPs)
The immune system is designed to recognize foreign threats to the host. Pathogen-associated molecular patterns (PAMPs) trigger innate mechanisms to initiate host defenses. An ordered array of protein antigens is a PAMP that is particularly effective for activating B-cell responses (72). Prior studies using haptens indicated that antigen spacing of 50–100 Å can effectively activate B cells to produce antibodies. Most viruses are <200 nanometers (nm) in diameter, and vaccines based on particles, whether non-enveloped viral capsids or enveloped VLPs, that are 10–200 nm in size are highly immunogenic. This is best exemplified by the licensed human papillomavirus (HPV) vaccine (73) and demonstrated for other VLP-based experimental vaccines for alphaviruses and polyomaviruses (74). In some cases, the most effective targets for neutralizing antibodies are not a few selected epitopes on viral surface proteins, but quaternary epitopes that determine the particle integrity itself. For example, there are critical cross-reactive neutralizing determinants on dengue viruses for which recognition depends on intact E dimers on mature, fully formed particles (75–78). Rapid vaccine development for Zika virus was reliant on an understanding of flavivirus E dimer and particle structure and antigenicity that was first shown for tick-borne encephalitis (TBE) virus (79) and Dengue virus (80). Similarly, livestock vaccines for the foot-and-mouth disease virus (FMDV), a non-enveloped picornavirus, lose immunogenicity as the icosahedral capsid decays into its pentameric subunits over time. Structural information and molecular dynamics simulations were able to identify mutations to the pentameric interfaces that resulted in increased thermostability and elicitation of higher neutralizing antibody titers after long-term storage of stabilized viruses (81, 82).
Other naturally self-assembling molecules like ferritin (83–85) and lumazine synthase (86) have been exploited to display vaccine antigens and to mimic the ordered symmetrical arrays of antigens on a VLP. Because of advances in cryo-EM, these particles can be structurally defined and authenticated at atomic or near atomic-level resolution, including in complex with antibodies (83). It is now possible to create structurally engineered de novo self-assembling molecules (87, 88), creating the possibility of customized designer nanoparticle vaccines that account for the design of antigens and their display in a coordinated and systematic approach (89).
CONCLUDING REMARKS
The availability of structural information for viral surface proteins has revolutionized vaccine antigen design. This has been fostered by advances in X-ray crystallography and electron microscopy, computational biology, and technologies for isolating human monoclonal antibodies. Using structurally defined probes and reagents likewise improves the discovery of human monoclonal antibodies with given specificities and functions and allows for definition of specific antibody lineages associated with desirable properties to use as molecular targets for vaccine immunogenicity (40, 90–92). Thus, vaccinology has entered a new era with rapidly evolving capabilities for protein engineering and antigen design. The previous era in which one- or two-dimensional protein characteristics and linear epitopes were used to empirically develop candidate vaccines is now based on multidimensional information, conformational epitopes on the infectious forms of proteins and virus particles, and a more methodical engineering and modular approach to vaccine development. Hopefully, this will lead to vaccines for pathogens with a tradition of failed vaccine development efforts and for emerging pathogens that may be susceptible to antigen designs and platform technologies already established for related pathogens.
Although structure-based vaccine design has provided an engineering approach for inducing specific antibody responses, there are still many lessons to learn about antibody elicitation, including achieving the correct angle and rotation of approach for optimal neutralizing activity, and induction of antibodies that interact with or avoid glycans in critical locations. In addition, some epitopes are not easily recognized by B cells, which may be addressed by antigen display approaches or masking of distracting antigenic sites. The problem of antigenic diversity may also find some solutions in antigen display, particularly if multiple related but distinct antigens can be presented simultaneously. Achieving antibodies with the optimal glycosylation, isotype, or other Fc-determined functions may require antigen targeting or special adjuvant formulations to achieve the desired outcome. This may also be true for activating the right B-cell phenotype and maintaining durable antibody levels. The magnitude and localization of antibody may be critical for protecting against some pathogens, so recognizing the optimal structurally defined epitope may have to occur in the right place, making route-of-delivery a key determinant of success. Going forward, successful vaccine development will require structure-guided antigen design, but also advances in antigen display, delivery, and formulation, in addition to improved understanding of lymph node and B-cell biology and more precision in our understanding of viral pathogenesis.
ACKNOWLEDGEMENTS
We thank Kaitlyn Morabito for helpful comments and preparation of the manuscript. We also thank many colleagues and mentors over the years who have contributed thoughts and ideas that have made it into this brief review and apologize for those papers that have not been referenced due to space constraints. This work was supported in part by intramural funding from the National Institutes of Allergy and Infectious Diseases (B.S.G) and grant R01AI127521 (J.S.M).
Footnotes
DISCLOSURE STATEMENT
B.S.G. is named as an inventor on pending patents for vaccines and/or monoclonal antibodies for RSV, CoV, influenza, Zika, Ebola, and paramyxoviruses. J.S.M. is a named inventor on patents for vaccines and/or monoclonal antibodies for RSV and CoV, has received research funding from MedImmune and Janssen, has been a paid consultant for MedImmune, and is on the scientific advisory board for Calder Biosciences. M.G. is a named inventor on a patent application for single-domain antibodies against RSV F.
LITERATURE CITED
- 1.Schmaljohn AL. 2013. Protective antiviral antibodies that lack neutralizing activity: precedents and evolution of concepts. Curr HIV Res 11: 345–53 [DOI] [PubMed] [Google Scholar]
- 2.Ackerman ME, Alter G. 2013. Opportunities to exploit non-neutralizing HIV-specific antibody activity. Curr HIV Res 11: 365–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sullivan NJ, Hensley L, Asiedu C, et al. 2011. CD8+ cellular immunity mediates rAd5 vaccine protection against Ebola virus infection of nonhuman primates. Nat Med 17: 1128–31 [DOI] [PubMed] [Google Scholar]
- 4.McMichael AJ, Koff WC. 2014. Vaccines that stimulate T cell immunity to HIV-1: the next step. Nat Immunol 15: 319–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kong L, Lee JH, Doores KJ, et al. 2013. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol 20: 796–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Georgiev IS, Gordon Joyce M, Zhou T, Kwong PD. 2013. Elicitation of HIV-1-neutralizing antibodies against the CD4-binding site. Curr Opin HIV AIDS 8: 382–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kallewaard NL, Corti D, Collins PJ, et al. 2016. Structure and Function Analysis of an Antibody Recognizing All Influenza A Subtypes. Cell 166: 596–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corti D, Voss J, Gamblin SJ, et al. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333: 850–6 [DOI] [PubMed] [Google Scholar]
- 9.McLellan JS, Pancera M, Carrico C, et al. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480: 336–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee PS, Arnell AJ, Wilson IA. 2015. Structure of the apo anti-influenza CH65 Fab. Acta Crystallogr F Struct Biol Commun 71: 145–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunningham AL, Lal H, Kovac M, et al. 2016. Efficacy of the Herpes Zoster Subunit Vaccine in Adults 70 Years of Age or Older. N Engl J Med 375: 1019–32 [DOI] [PubMed] [Google Scholar]
- 12.Kim HW, Canchola JG, Brandt CD, et al. 1969. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am J Epidemiol 89: 422–34 [DOI] [PubMed] [Google Scholar]
- 13.Kapikian AZ, Mitchell RH, Chanock RM, et al. 1969. An epidemiologic study of altered clinical reactivity to respiratory syncytial (RS) virus infection in children previously vaccinated with an inactivated RS virus vaccine. Am J Epidemiol 89: 405–21 [DOI] [PubMed] [Google Scholar]
- 14.Fulginiti VA, Eller JJ, Sieber OF, et al. 1969. Respiratory virus immunization. I. A field trial of two inactivated respiratory virus vaccines; an aqueous trivalent parainfluenza virus vaccine and an alum-precipitated respiratory syncytial virus vaccine. Am J Epidemiol 89: 435–48 [DOI] [PubMed] [Google Scholar]
- 15.Chin J, Magoffin RL, Shearer LA, et al. 1969. Field evaluation of a respiratory syncytial virus vaccine and a trivalent parainfluenza virus vaccine in a pediatric population. Am J Epidemiol 89: 449–63 [DOI] [PubMed] [Google Scholar]
- 16.Murphy BR, Alling DW, Snyder MH, et al. 1986. Effect of age and preexisting antibody on serum antibody response of infants and children to the F and G glycoproteins during respiratory syncytial virus infection. J Clin Microbiol 24: 894–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy BR, Walsh EE. 1988. Formalin-inactivated respiratory syncytial virus vaccine induces antibodies to the fusion glycoprotein that are deficient in fusion-inhibiting activity. J Clin Microbiol 26: 1595–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Killikelly AM, Kanekiyo M, Graham BS. 2016. Pre-fusion F is absent on the surface of formalin-inactivated respiratory syncytial virus. Sci Rep 6: 34108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graham BS. 2013. Advances in antiviral vaccine development. Immunol Rev 255: 230–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rappuoli R 2000. Reverse vaccinology. Curr Opin Microbiol 3: 445–50 [DOI] [PubMed] [Google Scholar]
- 21.Burton DR. 2002. Antibodies, viruses and vaccines. Nat Rev Immunol 2: 706–13 [DOI] [PubMed] [Google Scholar]
- 22.Verlinde CL, Merritt EA, Van den Akker F, et al. 1994. Protein crystallography and infectious diseases. Protein Sci 3: 1670–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dormitzer PR, Ulmer JB, Rappuoli R. 2008. Structure-based antigen design: a strategy for next generation vaccines. Trends Biotechnol 26: 659–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dormitzer PR, Grandi G, Rappuoli R. 2012. Structural vaccinology starts to deliver. Nat Rev Microbiol 10: 807–13 [DOI] [PubMed] [Google Scholar]
- 25.Rappuoli R, Bottomley MJ, D’Oro U, et al. 2016. Reverse vaccinology 2.0: Human immunology instructs vaccine antigen design. J Exp Med 213: 469–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blount RE Jr., Morris JA, Savage RE. 1956. Recovery of cytopathogenic agent from chimpanzees with coryza. Proc Soc Exp Biol Med 92: 544–9 [DOI] [PubMed] [Google Scholar]
- 27.Chanock R, Finberg L. 1957. Recovery from infants with respiratory illness of a virus related to chimpanzee coryza agent (CCA). II. Epidemiologic aspects of infection in infants and young children. Am J Hyg 66: 291–300 [DOI] [PubMed] [Google Scholar]
- 28.Chanock R, Roizman B, Myers R. 1957. Recovery from infants with respiratory illness of a virus related to chimpanzee coryza agent (CCA). I. Isolation, properties and characterization. Am J Hyg 66: 281–90 [DOI] [PubMed] [Google Scholar]
- 29.Hall CB, Weinberg GA, Iwane MK, et al. 2009. The burden of respiratory syncytial virus infection in young children. N Engl J Med 360: 588–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graham BS, Anderson LJ. 2013. Challenges and opportunities for respiratory syncytial virus vaccines. Curr Top Microbiol Immunol 372: 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walsh EE, Falsey AR. 2012. Respiratory syncytial virus infection in adult populations. Infect Disord Drug Targets 12: 98–102 [DOI] [PubMed] [Google Scholar]
- 32.Falsey AR, McElhaney JE, Beran J, et al. 2014. Respiratory syncytial virus and other respiratory viral infections in older adults with moderate to severe influenza-like illness. J Infect Dis 209: 1873–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glezen WP, Taber LH, Frank AL, Kasel JA. 1986. Risk of primary infection and reinfection with respiratory syncytial virus. Am J Dis Child 140: 543–6 [DOI] [PubMed] [Google Scholar]
- 34.Lo MS, Brazas RM, Holtzman MJ. 2005. Respiratory syncytial virus nonstructural proteins NS1 and NS2 mediate inhibition of Stat2 expression and alpha/beta interferon responsiveness. J Virol 79: 9315–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barik S 2013. Respiratory syncytial virus mechanisms to interfere with type 1 interferons. Curr Top Microbiol Immunol 372: 173–91 [DOI] [PubMed] [Google Scholar]
- 36.Johnson TR, Johnson JE, Roberts SR, et al. 1998. Priming with secreted glycoprotein G of respiratory syncytial virus (RSV) augments interleukin-5 production and tissue eosinophilia after RSV challenge. J Virol 72: 2871–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson TR, McLellan JS, Graham BS. 2012. Respiratory syncytial virus glycoprotein G interacts with DC-SIGN and L-SIGN to activate ERK1 and ERK2. J Virol 86: 1339–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tripp RA, Jones LP, Haynes LM, et al. 2001. CX3C chemokine mimicry by respiratory syncytial virus G glycoprotein. Nat Immunol 2: 732–8 [DOI] [PubMed] [Google Scholar]
- 39.McLellan JS, Chen M, Leung S, et al. 2013. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science 340: 1113–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilman MS, Castellanos CA, Chen M, et al. 2016. Rapid profiling of RSV antibody repertoires from the memory B cells of naturally infected adult donors. Sci Immunol 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McLellan JS, Chen M, Joyce MG, et al. 2013. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 342: 592–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stewart-Jones GB, Thomas PV, Chen M, et al. 2015. A Cysteine Zipper Stabilizes a Pre-Fusion F Glycoprotein Vaccine for Respiratory Syncytial Virus. PLoS One 10: e0128779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joyce MG, Zhang B, Ou L, et al. 2016. Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV. Nat Struct Mol Biol 23: 811–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krarup A, Truan D, Furmanova-Hollenstein P, et al. 2015. A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat Commun 6: 8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ngwuta JO, Chen M, Modjarrad K, et al. 2015. Prefusion F-specific antibodies determine the magnitude of RSV neutralizing activity in human sera. Sci Transl Med 7: 309ra162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang B, Surman S, Amaro-Carambot E, et al. 2015. Enhanced Neutralizing Antibody Response Induced by Respiratory Syncytial Virus Prefusion F Protein Expressed by a Vaccine Candidate. J Virol 89: 9499–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X, Liang B, Ngwuta J, et al. 2017. Attenuated Human Parainfluenza Virus Type 1 Expressing the Respiratory Syncytial Virus (RSV) Fusion (F) Glycoprotein from an Added Gene: Effects of Prefusion Stabilization and Packaging of RSV F. J Virol 91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stobart CC, Rostad CA, Ke Z, et al. 2016. A live RSV vaccine with engineered thermostability is immunogenic in cotton rats despite high attenuation. Nat Commun 7: 13916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cullen LM, Blanco JC, Morrison TG. 2015. Cotton rat immune responses to virus-like particles containing the pre-fusion form of respiratory syncytial virus fusion protein. J Transl Med 13: 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Widjojoatmodjo MN, Bogaert L, Meek B, et al. 2015. Recombinant low-seroprevalent adenoviral vectors Ad26 and Ad35 expressing the respiratory syncytial virus (RSV) fusion protein induce protective immunity against RSV infection in cotton rats. Vaccine 33: 5406–14 [DOI] [PubMed] [Google Scholar]
- 51.Battles MB, Mas V, Olmedillas E, et al. 2017. Structure and immunogenicity of pre-fusion-stabilized human metapneumovirus F glycoprotein. Nat Commun 8: 1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buchholz UJ, Bukreyev A, Yang L, et al. 2004. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc Natl Acad Sci U S A 101: 9804–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kirchdoerfer RN, Cottrell CA, Wang N, et al. 2016. Pre-fusion structure of a human coronavirus spike protein. Nature 531: 118–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walls AC, Tortorici MA, Bosch BJ, et al. 2016. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature 531: 114–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walls AC, Tortorici MA, Frenz B, et al. 2016. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat Struct Mol Biol 23: 899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuan Y, Cao D, Zhang Y, et al. 2017. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun 8: 15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pallesen J, Wang N, Corbett KS, et al. 2017. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc Natl Acad Sci U S A 114: E7348–e57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burton DR, Ahmed R, Barouch DH, et al. 2012. A Blueprint for HIV Vaccine Discovery. Cell Host Microbe 12: 396–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Binley JM, Sanders RW, Clas B, et al. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. Journal of Virology 74: 627–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanders RW, Vesanen M, Schuelke N, et al. 2002. Stabilization of the Soluble, Cleaved, Trimeric Form of the Envelope Glycoprotein Complex of Human Immunodeficiency Virus Type 1. Journal of Virology 76: 8875–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klasse PJ, Depetris RS, Pejchal R, et al. 2013. Influences on trimerization and aggregation of soluble, cleaved HIV-1 SOSIP envelope glycoprotein. J Virol 87: 9873–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sanders RW, Derking R, Cupo A, et al. 2013. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 9: e1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lyumkis D, Julien JP, de Val N, et al. 2013. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342: 1484–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Julien JP, Cupo A, Sok D, et al. 2013. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342: 1477–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lopez JA, Andreu D, Carreno C, et al. 1993. Conformational constraints of conserved neutralizing epitopes from a major antigenic area of human respiratory syncytial virus fusion glycoprotein. J Gen Virol 74 (Pt 12): 2567–77 [DOI] [PubMed] [Google Scholar]
- 66.Ofek G, Guenaga FJ, Schief WR, et al. 2010. Elicitation of structure-specific antibodies by epitope scaffolds. Proc Natl Acad Sci U S A 107: 17880–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McLellan JS, Chen M, Kim A, et al. 2010. Structural basis of respiratory syncytial virus neutralization by motavizumab. Nat Struct Mol Biol 17: 248–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McLellan JS, Correia BE, Chen M, et al. 2011. Design and characterization of epitope-scaffold immunogens that present the motavizumab epitope from respiratory syncytial virus. J Mol Biol 409: 853–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Correia BE, Bates JT, Loomis RJ, et al. 2014. Proof of principle for epitope-focused vaccine design. Nature 507: 201–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hollingshead S, Jongerius I, Exley RM, et al. 2018. Structure-based design of chimeric antigens for multivalent protein vaccines. Nat Commun 9: 1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Herve PL, Deloizy C, Descamps D, et al. 2017. RSV N-nanorings fused to palivizumab-targeted neutralizing epitope as a nanoparticle RSV vaccine. Nanomedicine 13: 411–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chackerian B, Durfee MR, Schiller JT. 2008. Virus-like display of a neo-self antigen reverses B cell anergy in a B cell receptor transgenic mouse model. J Immunol 180: 5816–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schiller JT, Lowy DR. 2012. Understanding and learning from the success of prophylactic human papillomavirus vaccines. Nat Rev Microbiol 10: 681–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chang LJ, Dowd KA, Mendoza FH, et al. 2014. Safety and tolerability of chikungunya virus-like particle vaccine in healthy adults: a phase 1 dose-escalation trial. Lancet 384: 2046–52 [DOI] [PubMed] [Google Scholar]
- 75.Dowd KA, Mukherjee S, Kuhn RJ, Pierson TC. 2014. Combined effects of the structural heterogeneity and dynamics of flaviviruses on antibody recognition. J Virol 88: 11726–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pierson TC, Diamond MS. 2012. Degrees of maturity: the complex structure and biology of flaviviruses. Curr Opin Virol 2: 168–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Swanstrom JA, Plante JA, Plante KS, et al. 2016. Dengue Virus Envelope Dimer Epitope Monoclonal Antibodies Isolated from Dengue Patients Are Protective against Zika Virus. MBio 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.de Alwis R, Smith SA, Olivarez NP, et al. 2012. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc Natl Acad Sci U S A 109: 7439–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rey FA, Heinz FX, Mandl C, et al. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 375: 291–8 [DOI] [PubMed] [Google Scholar]
- 80.Kuhn RJ, Zhang W, Rossmann MG, et al. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108: 717–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kotecha A, Seago J, Scott K, et al. 2015. Structure-based energetics of protein interfaces guides foot-and-mouth disease virus vaccine design. Nat Struct Mol Biol 22: 788–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Scott KA, Kotecha A, Seago J, et al. 2017. SAT2 Foot-and-Mouth Disease Virus Structurally Modified for Increased Thermostability. J Virol 91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yassine HM, Boyington JC, McTamney PM, et al. 2015. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med 21: 1065–70 [DOI] [PubMed] [Google Scholar]
- 84.Kanekiyo M, Wei CJ, Yassine HM, et al. 2013. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 499: 102–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Georgiev IS, Joyce MG, Chen RE, et al. 2018. Two-Component Ferritin Nanoparticles for Multimerization of Diverse Trimeric Antigens. ACS Infect Dis 4: 788–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jardine J, Julien JP, Menis S, et al. 2013. Rational HIV immunogen design to target specific germline B cell receptors. Science 340: 711–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Burkhard P, Lanar DE. 2015. Malaria vaccine based on self-assembling protein nanoparticles. Expert Rev Vaccines 14: 1525–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.King NP, Sheffler W, Sawaya MR, et al. 2012. Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science 336: 1171–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.King NP, Bale JB, Sheffler W, et al. 2014. Accurate design of co-assembling multi-component protein nanomaterials. Nature 510: 103–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu X, Zhou T, Zhu J, et al. 2011. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333: 1593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Joyce MG, Wheatley AK, Thomas PV, et al. 2016. Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses. Cell 166: 609–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Goodwin E, Gilman MSA, Wrapp D, et al. 2018. Infants Infected with Respiratory Syncytial Virus Generate Potent Neutralizing Antibodies that Lack Somatic Hypermutation. Immunity 48: 339–49 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]