Abstract
Background
Malignant melanoma can most successfully be cured when diagnosed at an early stage in the natural history. However, there is controversy over screening programs and many advocate screening only for high-risk individuals.
Objectives
This study aimed to evaluate the accuracy of an artificial intelligence neural network (Deep Ensemble for Recognition of Melanoma [DERM]) to identify malignant melanoma from dermoscopic images of pigmented skin lesions and to show how this compared to doctors’ performance assessed by meta-analysis.
Methods
DERM was trained and tested using 7,102 dermoscopic images of both histologically confirmed melanoma (24%) and benign pigmented lesions (76%). A meta-analysis was conducted of studies examining the accuracy of naked-eye examination, with or without dermoscopy, by specialist and general physicians whose clinical diagnosis was compared to histopathology. The meta-analysis was based on evaluation of 32,226 pigmented lesions including 3,277 histopathology-confirmed malignant melanoma cases. The receiver operating characteristic (ROC) curve was used to examine and compare the diagnostic accuracy.
Results
DERM achieved a ROC area under the curve (AUC) of 0.93 (95% confidence interval: 0.92–0.94), and sensitivity and specificity of 85.0% and 85.3%, respectively. Avoidance of false-negative results is essential, so different decision thresholds were examined. At 95% sensitivity DERM achieved a specificity of 64.1% and at 95% specificity the sensitivity was 67%. The meta-analysis showed primary care physicians (10 studies) achieve an AUC of 0.83 (95% confidence interval: 0.79–0.86), with sensitivity and specificity of 79.9% and 70.9%; and dermatologists (92 studies) 0.91 (0.88–0.93), 87.5%, and 81.4%, respectively.
Conclusions
DERM has the potential to be used as a decision support tool in primary care, by providing dermatologist-grade recommendation on the likelihood of malignant melanoma.
Keywords: melanoma, artificial intelligence, primary care, detection, identification
Introduction
Malignant melanoma (MM) is less common than basal and squamous cell skin cancer; however, the incidence of MM is increasing faster than that of other forms of cancer and it is responsible for the majority of skin cancer deaths [1]. Early diagnosis of MM (stage 1) has more than 95% five-year relative survival rate compared with 8% to 25% for MM diagnosed at later stages [2].
Current practice guidelines in the United Kingdom recommend appropriately trained health care professionals assess all suspect pigmented lesions using dermoscopy [1,3]. Diagnosis is confirmed with biopsy, histological examination, and specialist pathological interpretation. Pressure to diagnose MM early leads to a high proportion of benign pigmented lesions being referred from primary care to specialist care, and a large proportion of biopsied lesions are found to be benign [4,5]. This creates increased demands on overburdened secondary care and pathology service resources [6]. Improved accuracy of pigmented lesion review in primary care would help reduce this pressure. Techniques such as dermoscopy with classification algorithms, reflectance confocal microscopy, and teledermatology have been reported to improve diagnostic accuracy of MM [7–15]. However, the diagnostic accuracy is still dependent on the degree of experience of the examiners and the equipment required is costly [16].
A large number of smartphone applications for MM detection have been released recently. However, there is little evidence of clinical validation. Kassianos et al reviewed 39 apps that addressed skin cancer issues; 19 involved smartphone photography and 4 provided an estimate of the probability of malignancy. None of these apps had been assessed for diagnostic accuracy [17]. Understandably there is concern about the possible harm to patients that poorly designed, inaccurate, and/or misleading consumer apps may cause [18–20]. However, with appropriate development and suitable evaluation there is no reason why modern electronic technology could not improve diagnostic accuracy. Recently, an artificial intelligence (AI) algorithm categorizing photographs of pigmented lesions has been shown to be capable of classifying MM with a level of competence comparable to that of dermatologists [21]. As Obermeyer and Emanuel state in a recent review, “Machine learning has become ubiquitous and indispensable for solving complex problems in most sciences. The same methods will open up vast new possibilities in medicine” [22]. However, there are ethical issues associated with the clinical applications of AI in medicine that do not apply to current business applications, astronomy, or chemistry, and these cannot be ignored [23].
The primary aim of this study was to evaluate the diagnostic accuracy of an AI algorithm (Deep Ensemble for Recognition of Melanoma [DERM]) developed by Skin Analytics Limited. The secondary aim was to improve the methodology for evaluating an AI diagnostic tool by comparing DERM’s performance with clinical examination by physicians and stratification based on level of expertise and use of dermoscopy using a meta-analysis of diagnostic studies. But it should be noted that this was not designed to be a systematic review such as the recent Cochrane reviews of skin cancer.
Methods
DERM was designed and developed using deep learning techniques that identify and assess features of pigmented lesions that are associated with MM [23–28]. Deep learning differs from earlier machine learning methods by learning features that are associated with MM directly from the data, rather than using features predetermined by a researcher. The algorithm was trained and validated against a dataset of archived dermoscopic images of skin lesions, using 10-fold cross-validation. This approach allows every image to be tested once, while ensuring the same image does not appear in the training and test datasets. Cross-validation is performed by splitting the dataset into several (10) “folds” (datasets). The algorithm is tested against each fold, with the remainder used for training. The results for each fold are then averaged so that the overall performance can be assessed.
The image dataset was collated from several different sources including the PH2 dataset [29], Interactive Atlas of Dermoscopy [30], and ISIC archive [31]. An additional 672 dermoscopic lesion images were collected from a variety of other sources. The ISIC archive contains a large number of images obtained from children, which are easy to classify as benign. Their inclusion in the dataset was found to optimistically bias results so they were excluded from the development work. The ISIC archive also contains a large number of identical and near-identical images which were removed from the dataset. The final dataset consists of a total of 7,102 unique pigmented lesion images, 24% being confirmed as MM by histopathology, though subtype information was not available, the rest being made up of benign and nonbenign lesions.
DERM generates a continuous response to an image with limits of 0 and 1, which reflects its “confidence” that the lesion is MM: a value close to 1 indicates MM and near 0 indicates a benign lesion. A nonparametric receiver operating characteristic (ROC) curve analysis was used to examine the overall diagnostic accuracy of the result using Pepe’s nonparametric methods with bootstrapped estimation [32]. The gold standard for MM was histopathology. We examined different cut-points used by DERM to categorize lesions as positive or negative, ie, illustrating alternative diagnostic rules from the diagnostic model [33]. The methods of Youden [34] and Liu [35] were used, as well as the values that maximized the ROC area, resulted in a sensitivity and a specificity of 95%, and generated less than 1% false negative. The area under the curve (AUC) of the ROC curve, specificity/sensitivity, and diagnostic odds ratios were calculated for each of these cut-points.
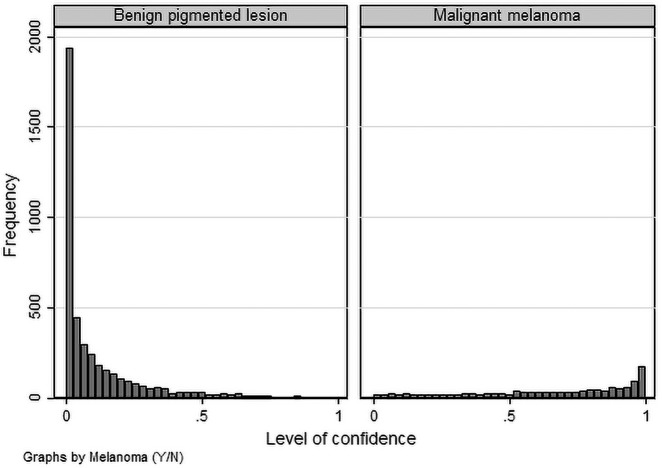
The ROC AUC is not a perfect assessment measure for diagnostic methods when the standard error of the estimator is quite different for the diagnostic alternatives (benign pigmented lesions vs MM), as is the case for DERM (see Figure 1) [36]. This issue was addressed by constructing the Lorenz curve (a mirror image of the ROC curve) with the associated Gini index [37].
Figure 1.
Level of confidence of Deep Ensemble for Recognition of Melanoma (DERM) algorithm by lesion type.
To compare the accuracy of DERM with that of current diagnostic practices, we decided to conduct a meta-analysis of studies of diagnostic accuracy for MM rather than have a limited panel of dermatologists conduct parallel assessments, as has been done in other studies [21,38]. We chose this approach because biopsy-based histopathology provides the gold standard for MM diagnosis, and a meta-analysis enables comparison to a variety of different clinician experiences and evaluation techniques. This analysis was not intended to be systematic review, but the PRISMA guidelines were followed when appropriate.
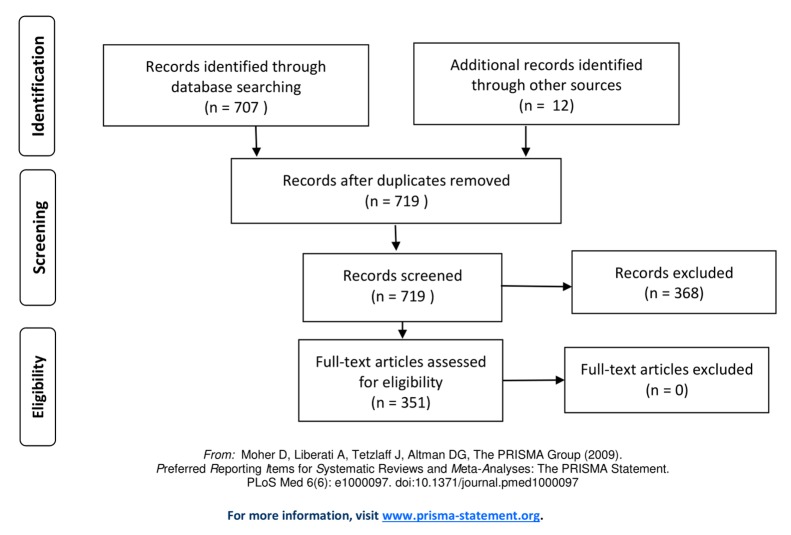
A literature search was conducted for studies reporting diagnostic accuracy data of naked-eye clinical examination, with or without dermoscopy, compared with histologically confirmed diagnosis. MEDLINE (413), Web of Science (707), and EMBASE (322) were searched for the period from January 1, 1990, to September 30, 2017, using terms “accuracy pigmented lesions PLUS melanoma pigmented lesions PLUS detection,” “dermoscopy pigmented lesions PLUS melanoma pigmented lesions PLUS accuracy,” and “melanoma pigmented lesions PLUS diagnosis pigmented lesions PLUS primary care.” Studies included in previous systematic reviews were also included [2,15,39–41]. The PRISMA flow diagram is shown in Figure 2. One author (M.P.) conducted the literature search and extracted counts of true negative; true positive; false negative; false positive; or estimates of sensitivity, specificity, number of lesions examined, and number of MM diagnoses confirmed by histology, from which the counts could be derived. The reports were also examined for information concerning physician experience (general vs specialist physician) and context of use (primary care, secondary care). A meta-analysis from this data was conducted. The Stata user-written packages METANDI [42] and MIDAS [43] were used, and a meta-regression was used to examine associations between diagnostic accuracy and year of study report, level of care, and expertise of the practitioner. Many of the dermoscopy studies reported multiple results for each lesion using different dermoscopic algorithms (eg, ABCD, 7-point checklist, etc. [44]); all of these results were included in the dataset. Since this produces a clustered dataset, violating the statistical assumption of the independence of observations, we conducted a sensitivity analysis. Multiple datasets were generated in which 1 estimate only was randomly included for each study where there were multiple estimates. The results indicated that the initial estimates were not sensitive to the clustering (details of this analysis are not reported here).
Figure 2.
PRISMA flow diagram of publications searched for the meta-analysis.
All analysis was conducted by M.P. using the Stata statistical package (StataCorp. 2015. Stata Statistical Software: Release 15. College Station, TX: StataCorp LP).
Most of the data used to create the algorithm were based on anonymous, publicly available images, and an additional 672 anonymized dermoscopic lesion images were generously made available by clinical dermatologists. The meta-analysis data were derived from published papers that did not include individual patient data. There was no requirement for ethics approval, but the Ethics Committee of Royal Perth Hospital was informed of the study as a courtesy.
Results
Histograms showing the distribution of the DERM value for MM and for benign lesions are shown in Figure 1. The histograms show that the value does not follow a normal distribution and there is a different dispersion of data for the 2 types of lesion. DERM estimated the median level of confidence as 0.059 (interquartile range: 0.016–0.171) when the lesion was a benign pigmented lesion and 0.651 (interquartile range: 0.417–0.849) when the lesion was MM. The equality of the 2 medians was compared by Fisher exact test and found to be significantly different (P < 0.0001).
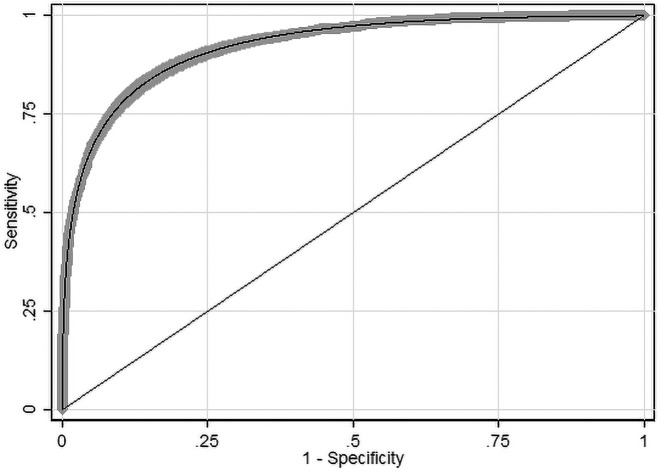
The empirical ROC curve analysis showed that DERM has a high level of accuracy with an AUC of 0.928 (95% confidence interval: 0.922–0.935) and an acceptable goodness-of-fit χ2 = 6,078 (P = 0.98) (Figure 3). The Lorenz curve analysis gave a Gini index of 0.857. The Gini index has an upper limit of 1 and the high value is indicative of high inequality between MM and benign lesions, which supports the ROC analysis.
Figure 3.
The receiver operating characteristic curve of Deep Ensemble for Recognition of Melanoma (DERM) results. Shaded area shows 95% confidence interval.
The Youden, Liu, and maximum AUC methods estimated the same optimum cut-point at a value of 0.272 (95% confidence interval: 0.232–0.313) (Table 1). As the sensitivity increases, the expected loss of specificity occurs, but when the sensitivity is fixed at 95%, specificity is still 64%.
Table 1.
Indices of Diagnostic Accuracy (±95% CI) at Different Cut-Points of the DERM Confidence Value
| Cut-Point | DERM Value | Sensitivity (%) | Specificity (%) | Diagnostic Odds Ratio |
|---|---|---|---|---|
| Optimum (maximum AUC) | 0.272 | 85.0 (83.2–86.7) | 85.3 (84.4–86.3) | 33.0 (28.3–38.4) |
| Confidence ≥0.50 | 0.50 | 67.3 (65.0–69.5) | 95.5 (94.9–96.0) | 43.7 (37.1–51.5) |
| 80% Sensitivity | 0.35 | 80 (fixed) | 90.8 (90.0–91.5) | 37.1 (32.1–42.9) |
| 95% Sensitivity | 0.11 | 95.0 (93.8–96.0) | 64.1 (62.8–65.4) | 33.6 (26.8–42.1) |
| High sensitivity | 0.05 | 98.6 (98.0–99.1) | 46.5 (45.2–47.9) | 62.9 (41.7–95.0) |
| 80% Specificity | 0.21 | 88.2 (86.6–89.7) | 80 (fixed) | 32.7 (27.9–38.4) |
| 95% Specificity | 0.795 | 66.9 (64.3–69.3) | 95% (fixed) | 38.3 (32.1–45.7) |
AUC = area under the curve; CI = confidence interval; DERM = Deep Ensemble for Recognition of Melanoma.
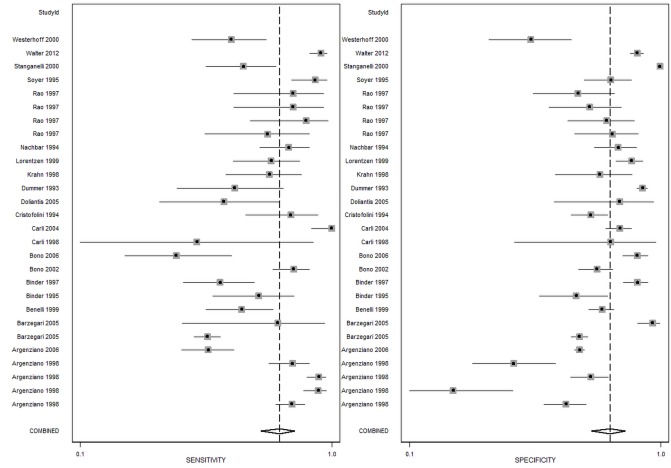
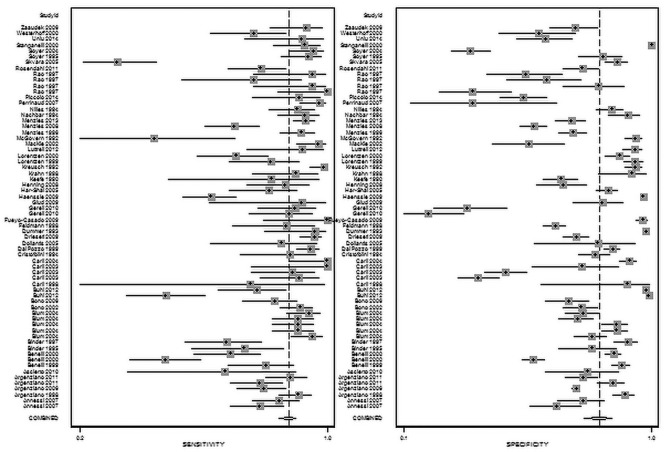
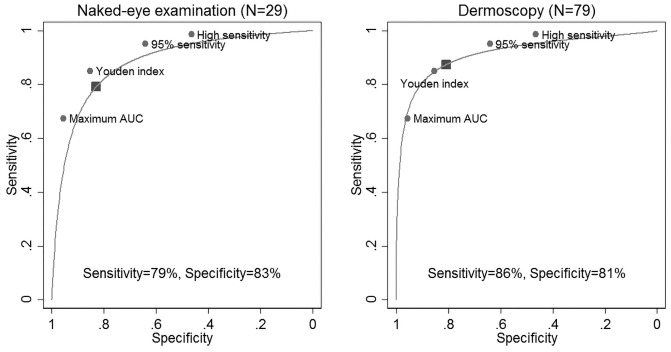
The summary of 82 studies that investigated the diagnostic accuracy of naked-eye examination (n = 29) or dermoscopy (n = 53) for pigmented lesions and MM between 1990 and 2017 is shown in Table 2. A visual guide to the study accuracy is provided in the forest plots in Figures 4 and 5. Table 3 shows the pooled and weighted values of sensitivity, specificity, and diagnostic odds ratio for the studies. The pooled results for all studies are as follows: AUC = 0.90, sensitivity = 85%, and specificity = 82%. The beta value (an indicator of asymmetry of the summary ROC curve) is statistically significant (β = 0.263, P = 0.022), indicating that the diagnostic odds ratio shows variation across the summary ROC curve. For naked-eye examination the pooled results are as follows: AUC = 0.88, sensitivity = 79%, specificity = 83%, β = 0.048, P = 0.81; and for dermoscopy the pooled results are as follows: AUC = 0.91, sensitivity = 86%, specificity = 81%, β = 0.397, P = 0.005.
Table 2.
Studies for Meta-analysis of Diagnostic Accuracy
| Author [Ref] | Date | Total Lesions | No. of Malignant Melanomas (%) | Sensitivity (%) | Specificity (%) | Country of Patients |
|---|---|---|---|---|---|---|
| Annessi [47] | 2007 | 198 | 96 (48.5) | 81.3 | 69.6 | Italy |
| Argenziano [48] | 1998 | 309 | 106 (34.3) | 95.0 | 75.0 | Italy |
| Argenziano [49] | 2006 | 2,528 | 12 (0.475) | 79.2 | 71.8 | Spain, Italy |
| Argenziano [50] | 2011 | 283 | 78 (27.6) | 87.8 | 74.5 | Italy |
| Ascierto [51] | 2010 | 54 | 12 (22.2) | 66.6 | 76.2 | Italy |
| Barzegari [52] | 2005 | 122 | 6 (4.92) | 100 | 90.0 | Iran |
| Benelli [53] | 1999 | 401 | 60 (15.0) | 85.0 | 89.1 | Italy |
| Benelli [54] | 2000 | 600 | 76 (12.7) | 68.8 | 86.0 | Italy |
| Binder [55] | 1995 | 100 | 37 (37.0) | 73.0 | 74.0 | Austria |
| Binder [56] | 1997 | 240 | 58 (24.2) | 63.0 | 91.0 | Austria |
| Blum [57] | 2004 | 269 | 84 (31.2) | 95.2 | 77.8 | Germany |
| Bono [58] | 2002 | 313 | 125 (39.9) | 88.5 | 75.5 | Italy |
| Bono [59] | 2006 | 206 | 76 (36.9) | 63.0 | 80.0 | Italy |
| Carli [60] | 1998 | 15 | 4 (26.7) | 58.5 | 83.5 | Italy |
| Carli [61] | 2003 | 200 | 44 (22.0) | 91.9 | 35.2 | Italy |
| Carli [62] | 2003 | 311 | 28 (9.00) | 100 | 88.5 | Italy |
| Cristofolini [63] | 1994 | 220 | 33 (15.0) | 86.5 | 77.0 | Italy |
| Dal Pozzo [64] | 1999 | 713 | 168 (23.6) | 94.6 | 85.5 | Italy |
| Doliantis [65] | 2005 | 40 | 20 (50.0) | 84.6 | 77.7 | Australia |
| Dreiseitl [66] | 2009 | 458 | 146 (31.9) | 96.0 | 72.0 | Germany |
| Dummer [67] | 1993 | 824 | 25 (3.03) | 80.5 | 95.5 | Germany |
| Feldmann [68] | 1998 | 500 | 30 (6.00) | 88.0 | 64.0 | Austria |
| Fueyo-Casado [69] | 2009 | 303 | 16 (5.28) | 100 | 97.0 | Brazil |
| Gereli [70] | 2010 | 96 | 48 (50.0) | 89.6 | 31.2 | Turkey |
| Glud [71] | 2009 | 83 | 12 (14.5) | 92.0 | 81.0 | Denmark |
| Haenssle [72] | 2010 | 1,219 | 127 (10.4) | 62.0 | 97.0 | Germany |
| Har-Shai [73] | 2005 | 400 | 53 (13.3) | 86.0 | 74.0 | Israel |
| Henning [74] | 2008 | 150 | 50 (33.3) | 92.0 | 38.0 | USA |
| Keefe [75] | 1990 | 222 | 11 (4.95) | 85.7 | 66.5 | Scotland |
| Krähn [76] | 1998 | 80 | 39 (48.8) | 90.0 | 93.0 | Germany |
| Kreusch [77] | 1992 | 317 | 96 (30.3) | 98.9 | 94.1 | Germany |
| Lorentzen [78] | 1999 | 232 | 49 (21.1) | 59.0 | 92.0 | Denmark |
| Lorentzen [79] | 2000 | 258 | 64 (24.8) | 70.7 | 88.0 | Denmark |
| Luttrell [80] | 2012 | 200 | 25 (12.5) | 91.2 | 94.0 | Austria |
| MacKie [81] | 2002 | 126 | 69 (54.8) | 97.0 | 55.0 | Scotland |
| McGovern [82] | 1992 | 237 | 16 (6.75) | 44.0 | 94.0 | USA |
| Menzies [83] | 1996 | 385 | 107 (27.8) | 92.0 | 71.0 | Australia |
| Menzies [84] | 2008 | 497 | 105 (21.1) | 95.0 | 80.0 | Australia |
| Menzies [85] | 2013 | 465 | 217 (46.7) | 93.0 | 70.0 | Australia |
| Nachbar [86] | 1994 | 172 | 69 (40.1) | 92.8 | 91.2 | Germany |
| Nilles [87] | 1994 | 260 | 72 (27.7) | 90.0 | 85.0 | Germany |
| Perrinaud [88} | 2007 | 90 | 78 (86.7) | 98.0 | 37.0 | Switzerland |
| Piccolo [89] | 2014 | 165 | 33 (20.0) | 91.0 | 52.0 | Italy |
| Rao [90] | 1997 | 72 | 51 (70.8) | 91.5 | 59.3 | USA |
| Rosendahl [9] | 2011 | 246 | 79 (32.1) | 82.6 | 80.0 | Australia |
| Skvara [91] | 2005 | 325 | 63 (19.4) | 31.7 | 87.3 | Austria |
| Soyer [92] | 1995 | 159 | 65 (40.9) | 94.0 | 82.0 | Italy |
| Soyer [93] | 2004 | 231 | 68 (29.4) | 96.3 | 32.8 | Italy |
| Stanganelli [94] | 2000 | 3,372 | 55 (1.63) | 80.0 | 99.5 | Italy |
| Unlu [95] | 2014 | 115 | 24 (20.9) | 91.6 | 64.8 | Turkey |
| Walter [96] | 2013 | 1,436 | 36 (2.51) | 91.7 | 33.1 | England |
| Westerhoff [97] | 2000 | 100 | 50 (50.0) | 54.6 | 56.1 | Australia |
| Zalaudek [98] | 2006 | 150 | 44 (29.3) | 94.0 | 71.9 | Multiple |
| Youl [99] | 2007 | 11,116 | 49 (0.441) | 60.0 | 98.0 | Australia |
| All studies | (n = 55) | 32,226 | 3,277 (10.2) |
Figure 4.
Forest plot for naked-eye examination.
Figure 5.
Forest plot for dermoscopy.
Table 3.
Meta-analysis Results
| Subgroup | No. of Estimatesa | No. of Lesions | No. of Malignant Melanoma | Sensitivity (%) (95% CI) | Specificity (%) (95% CI) | sROC Area (95% CI) | |
|---|---|---|---|---|---|---|---|
| All studies | Naked eye | 29 | 23,930 | 2,140 | 79 (72–85) | 83 (76–88) | 0.88 (0.85–0.91) |
| Dermoscopy | 79 | 33,749 | 5,031 | 86 (83–89) | 81 (76–86) | 0.91 (0.88–0.93) | |
| All studies | Nonexperts | 20 | 22,580 | 1,630 | 82 (73–89) | 73 (60–83) | 0.85 (0.82–0.88) |
| Experts | 65 | 29,767 | 3,812 | 84 (79–87) | 85 (80–89) | 0.91 (0.88–0.93) | |
| All studies | Primary care | 10 | 19,152 | 867 | 80 (65–89) | 71 (52–85) | 0.83 (0.79–0.86) |
| Secondary care | 87 | 36,673 | 5,480 | 85 (82–88) | 82 (77–87) | 0.91 (0.88–0.93) | |
| Nonexperts | Naked eye | 9 | 16,304 | 1,045 | 78 (60–90) | 74 (54–88) | 0.83 (0.80–0.86) |
| Dermoscopy | 11 | 6,279 | 585 | 83 (76–89) | 72 (55–84) | 0.86 (0.83–0.89) | |
| Experts | Naked eye | 16 | 7,115 | 922 | 79 (70–86) | 86 (79–91) | 0.90 (0.87–0.92) |
| Dermoscopy | 49 | 22,652 | 2,890 | 85 (79–89) | 85 (77–90) | 0.91 (0.89–0.94) | |
| Primary care | Naked eye | 6 | 14,822 | 595 | 78 (52–92) | 74 (43–91) | 0.83 (0.80–0.86) |
| Dermoscopy | 4 | 4,330 | 272 | 82 (74–87) | 66 (57–74) | 0.83 (0.79–0.86) | |
| Secondary care | Naked eye | 19 | 8,597 | 1,372 | 79 (71–86) | 85 (78–90) | 0.89 (0.86–0.91) |
| Dermoscopy | 68 | 28,076 | 4,108 | 87 (83–90) | 82 (75–87) | 0.91 (0.88–0.93) |
The number of estimates exceeds the number of studies because multiple estimates are made using dermoscopy with alternative diagnostic algorithms.
CI = confidence interval; sROC = summary receiver operating characteristic.
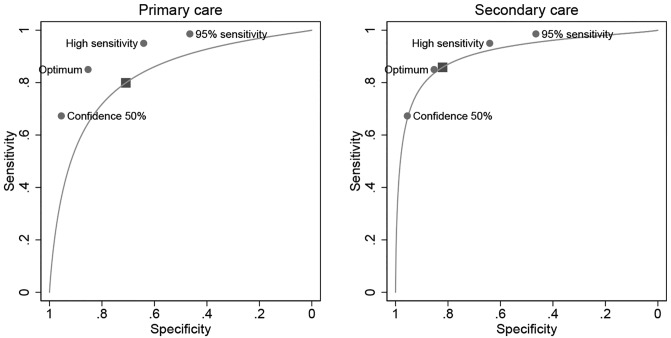
Meta-regression for the year of publication showed no significant association assessed by the combination of sensitivity and specificity for either visual clinical examination (P = 0.25) or dermoscopy (P = 0.18). There was a significant difference between experts and nonexperts both for naked-eye visual clinical examination (P < 0.001) and dermoscopy (P < 0.001), which is reflected in the estimated values shown in Table 3, where experts have both higher sensitivity and specificity than nonexperts, and is most marked for specificity for both methods and for sensitivity only for dermoscopy (Figure 6). The contrast in accuracy is most obvious for primary vs secondary care (P < 0.0001) with the AUC differing by 8% (0.83 vs 0.91) (Figure 7). There was no association between the AUC and year of study publication, suggesting that diagnostic accuracy is not improving over time (P = 0.63).
Figure 6.
Summary receiver operating characteristic curves for naked eye and dermoscopic diagnosis overlaid with the Deep Ensemble for Recognition of Melanoma (DERM) sensitivity and specificity at cut-points from Table 1 (the shaded rectangle shows the summary point from the meta-analysis). AUC = area under the curve.
Figure 7.
Summary receiver operating characteristic curves for primary and secondary care overlaid with the Deep Ensemble for Recognition of Melanoma (DERM) sensitivity and specificity at cut-points from Table 1 (the shaded rectangle shows the summary point from the meta-analysis).
Discussion
Summary
Herewith we present an extensive evaluation of the ability of DERM to identify MM from dermoscopic images of skin lesions. This preliminary analysis demonstrates the ability of an AI-based system to learn features of a skin lesion that are associated with MM, which can then be applied to the identification of MM. We conducted a meta-analysis of MM diagnostic accuracy to generate comparative values from current primary care and specialist dermatologist practices. These results confirm that clinician experience and use of dermoscopy improve accuracy. DERM achieves an AUC of 0.93, sensitivity and specificity of 85% and 85%, respectively, when using the estimated optimum value of 0.28. This is higher than naked-eye visual assessment (0.88, 80% and 71%), and similar to findings for dermatologists with dermoscopy (0.91, 85% and 82%). This is illustrated by plotting a ROC curve of the data from studies in the meta-analysis, and superimposing the DERM data from 4 cut-points (Figures 6 and 7).
A recent comprehensive series of Cochrane reviews concluded that visual inspection alone had a specificity of 42% at a fixed sensitivity of 80% and a sensitivity of 76% at a fixed specificity of 80%, whereas dermoscopy plus visual inspection had a specificity of 92% at a fixed sensitivity of 80% and a sensitivity of 82% at a fixed specificity of 80% [45]. Our meta-analysis showed for visual inspection alone specificity of 83% when sensitivity was 80%; sensitivity of 78% when specificity was 80%; specificity of 86% when sensitivity was 80%; and sensitivity of 87% when specificity was 80%. DERM gave comparable indices of specificity of 89% at sensitivity of 80% and a sensitivity of 90% at specificity of 80%.
Strengths and Limitations
We trained our algorithm using archived images that have been published to train clinicians. It is likely that biases exist in the datasets (eg, patient demographics, MM subtypes, image capture methods), but it is very difficult to determine whether such biases exist and thus have been introduced into DERM during its development. In addition, it must be emphasized that the algorithm was trained predominantly using images of images rather than images created in a clinical setting. We are currently collecting such images during a clinical trial and plan to report the results in the near future.
By using postbiopsy histology as the gold standard for both DERM and the inclusion criteria for our meta-analysis, images of nonsuspicious lesions have not been included when training or evaluating DERM. We have therefore not shown the ability of DERM (or clinicians) to accurately classify nonsuspicious lesions, which could lead to verification bias as was observed by a study of cancer registry data during a prospective follow-up [46]. However, this bias will apply to both the evaluation of DERM and the meta-analysis results, so it seems unlikely that the comparison of the 2 would be affected, but it remains a possibility.
A strength of our study is that the use of a meta-analysis of naked-eye examination and dermoscopy, the most common current diagnostic methods for MM used in primary care, is based on evaluation of 32,226 pigmented lesions including 3,277 histopathology-confirmed MM.
Comparison With Existing Literature
Recently, 2 other groups who retooled versions of Google’s Inception network for the identification of melanoma showed accuracy equivalent to or better than that of a panel of dermatologists [22,23]. However, this approach is likely to generate issues such as overfitting (because of the small size of the review panel) and a lack of generalization (because of the selected nature of the voluntary reviewers).
A recent addition to the literature was the publication of an extensive systematic review by the Cochrane Collaboration skin group [45]. Four studies were conducted on melanoma diagnosis in adults by visual inspection, dermoscopy with and without visual inspection, reflectance confocal microscopy, and smartphone applications for triaging suspicious lesions. The dates of publication were slightly different from our study dates (up to August 2016 compared with September 2017), they searched more databases, and they did not limit themselves to histology-confirmed pathology as the diagnostic outcome but also included clinical follow-up of benign-appearing lesions, cancer registry follow-up, and “expert opinion with no histology or follow-up.” Despite these differences, the number of studies is very similar. We identified 108 studies (29 visual and 79 dermoscopy) and they identified 104 (24 visual and 86 dermoscopy).
Implications for Research and Practice
Using different cut-points at which DERM defines a lesion as MM, the sensitivity and specificity ranged between 85.0% to 98.6% and 85.3% to 62.9%, respectively. The cut-points calculated by the Youden and Liu methods assume that false-negative and false-positive results have equal importance. This is not the case when dealing with a life-threatening disease, such as MM, where a cut-point that maximizes sensitivity—thus reducing the number of false-negative cases—should be adopted. However, this results in a higher false-positive rate, which has health care and patient costs associated with further investigations. The most appropriate cut-point for use in a clinical setting will need to be determined by consensus agreement taking into account both clinical and economic factors and is likely to be different for different clinical settings and levels of care.
At high levels of sensitivity, DERM offers comparable specificity to dermatologists with dermatoscopes. DERM could therefore provide dermatologist-grade advice on likelihood of MM to general practitioners without the cost and training requirements of dermoscopy. While diagnostic accuracy plays a pivotal role in the clinical evaluation of diagnostic tests, it does not prove that the test improves outcomes in relevant patient populations or that it enhances health care quality, efficiency, and cost-effectiveness. The only way to truly determine a test’s utility in the real-life decision-making setting of clinics is by conducting prospective clinical trials. We are currently conducting clinical validation studies of DERM. To our knowledge, no other AI-based MM diagnostic test is undergoing such extensive clinical utility testing [23,46,47].
Conclusions
Our study demonstrates the ability of an AI-based system to learn features of a skin lesion photograph that are associated with MM. DERM has the potential to be used in primary care to provide dermatologist-grade decision support. It is too early to say deployment of DERM would reduce onward referral, but such clinical validation is ongoing.
Footnotes
Funding: None.
Competing interests: The authors have no conflicts of interest to disclose.
Authorship: All authors have contributed significantly to this publication.
References
- 1.Marsden JR, Newton-Bishop JA, Burrows L, et al. Revised U.K. guidelines for the management of cutaneous melanoma 2010. Br J Dermatol. 2010;163(2):238–256. doi: 10.1111/j.1365-2133.2010.09883.x. [DOI] [PubMed] [Google Scholar]
- 2.Wernli KJ, Henrikson NB, Morrison CC, Nguyen M, Pocobelli G, Whitlock EP. U.S. Preventive Services Task Force Evidence Syntheses, formerly Systematic Evidence Reviews. Rockville, MD: Agency for Healthcare Research and Quality (US); 2016. Screening for Skin Cancer in Adults: An Updated Systematic Evidence Review for the US Preventive Services Task Force [Internet] [PubMed] [Google Scholar]
- 3.National Collaborating Centre for Cancer (UK) National Institute for Health and Care Excellence: Clinical Guidelines. London: National Institute for Health and Care Excellence (UK); 2015. Melanoma: Assessment and Management. [Google Scholar]
- 4.Welch HG, Woloshin S, Schwartz LM. Skin biopsy rates and incidence of melanoma: population based ecological study. BMJ. 2005;331(7515):481. doi: 10.1136/bmj.38516.649537.E0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen SC, Pennie ML, Kolm P, et al. Diagnosing and managing cutaneous pigmented lesions: primary care physicians versus dermatologists. J Gen Intern Med. 2006;21(7):678–682. doi: 10.1111/j.1525-1497.2006.00462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodson AG, Grossman D. Strategies for early melanoma detection: approaches to the patient with nevi. J Am Acad Dermatol. 2009;60(5):719–735. doi: 10.1016/j.jaad.2008.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vestergaard ME, Macaskill P, Holt PE, Menzies SW. Dermoscopy compared with naked eye examination for the diagnosis of primary melanoma: a meta-analysis of studies performed in a clinical setting. Br J Dermatol. 2008;159(3):669–676. doi: 10.1111/j.1365-2133.2008.08713.x. [DOI] [PubMed] [Google Scholar]
- 8.Menzies SW. Evidence-based dermoscopy. Dermatol Clin. 2013;31(4):521–524. vii. doi: 10.1016/j.det.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Rosendahl C, Tschandl P, Cameron A, Kittler H. Diagnostic accuracy of dermatoscopy for melanocytic and nonmelanocytic pigmented lesions. J Am Acad Dermatol. 2011;64(6):1068–1073. doi: 10.1016/j.jaad.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 10.Herschorn A. Dermoscopy for melanoma detection in family practice. Can Fam Physician. 2012;58(7):740–745. e372–e378. [PMC free article] [PubMed] [Google Scholar]
- 11.Creighton-Smith M, Murgia RD, 3rd, Konnikov N, Dornelles A, Garber C, Nguyen BT. Incidence of melanoma and keratinocytic carcinomas in patients evaluated by store-and-forward teledermatology vs. dermatology clinic. Int J Dermatol. 2017;56(10):1026–1031. doi: 10.1111/ijd.13672. [DOI] [PubMed] [Google Scholar]
- 12.Borsari S, Pampena R, Lallas A, et al. Clinical indications for use of reflectance confocal microscopy for skin cancer diagnosis. JAMA Dermatol. 2016;152(10):1093–1098. doi: 10.1001/jamadermatol.2016.1188. [DOI] [PubMed] [Google Scholar]
- 13.Xiong YQ, Ma SJ, Mo Y, Huo ST, Wen YQ, Chen Q. Comparison of dermoscopy and reflectance confocal microscopy for the diagnosis of malignant skin tumours: a meta-analysis. J Cancer Res Clin Oncol. 2017;143(9):1627–1635. doi: 10.1007/s00432-017-2391-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kardynal A, Olszewska M. Modern non-invasive diagnostic techniques in the detection of early cutaneous melanoma. J Dermatol Case Rep. 2014;8(1):1–8. doi: 10.3315/jdcr.2014.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kittler H, Pehamberger H, Wolff K, Binder M. Diagnostic accuracy of dermoscopy. Lancet Oncol. 2002;3(3):159–165. doi: 10.1016/s1470-2045(02)00679-4. [DOI] [PubMed] [Google Scholar]
- 16.Brewer AC, Endly DC, Henley J, et al. Mobile applications in dermatology. JAMA Dermatol. 2013;149(11):1300–1304. doi: 10.1001/jamadermatol.2013.5517. [DOI] [PubMed] [Google Scholar]
- 17.Kassianos AP, Emery JD, Murchie P, Walter FM. Smartphone applications for melanoma detection by community, patient and generalist clinician users: a review. Br J Dermatol. 2015;172(6):1507–1518. doi: 10.1111/bjd.13665. [DOI] [PubMed] [Google Scholar]
- 18.Ferrero NA, Morrell DS, Burkhart CN. Skin scan: a demonstration of the need for FDA regulation of medical apps on iPhone. J Am Acad Dermatol. 2013;68(3):515–516. doi: 10.1016/j.jaad.2012.10.045. [DOI] [PubMed] [Google Scholar]
- 19.Wolfe JA, Ferris LK. Diagnostic inaccuracy of smartphone applications for melanoma detection: reply. JAMA Dermatol. 2013;149(7):885. doi: 10.1001/jamadermatol.2013.4337. [DOI] [PubMed] [Google Scholar]
- 20.Stoecker WV, Rader RK, Halpern A. Diagnostic inaccuracy of smartphone applications for melanoma detection: representative lesion sets and the role for adjunctive technologies. JAMA Dermatol. 2013;149(7):884. doi: 10.1001/jamadermatol.2013.4334. [DOI] [PubMed] [Google Scholar]
- 21.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Obermeyer Z, Emanuel EJ. Predicting the future—big data, machine learning, and clinical medicine. N Engl J Med. 2016;375(13):1216–1219. doi: 10.1056/NEJMp1606181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Char DS, Shah NH. Implementing machine learning in health care—addressing ethical challenges. N Engl J Med. 2018;378(11):981–983. doi: 10.1056/NEJMp1714229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simonyan K, Zisserman A. Very deep convolutional networks. Presented at International Conference on Machine Learning and Applications (IEEE ICMLA’15); Miami, FL. IEEE; 2015. https://arxiv.org/abs/1602.07261. [Google Scholar]
- 25.Szegedy C, Ioffe S, Vanhoucke V, Alemi A. Inception-v4, Inception-ResNet and the impact of residual connections on learning. 2016. https://arxiv.org/abs/1602.07261.
- 26.Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In: Navab N, et al., editors. MICCAI 2015, Part III, LNCS 9351. 2015. pp. 234–241. https://arxiv.org/abs/1505.04597. [DOI] [Google Scholar]
- 27.Codella N, Nguyen Q-B, Pankanti S, et al. Deep learning ensembles for melanoma recognition in dermoscopy images. IBM J Res Dev. 2017;61(4/5):5:1–5:15. https://arxiv.org/abs/1610.04662. [Google Scholar]
- 28.Clevert D, Unterthiner T, Hochreiter S. Fast and accurate deep network learning by exponential linear units (ELUs). Presented at International Conference on Learning Representations; San Juan, Puerto Rico. 2016. https://arxiv.org/abs/1511.07289. [Google Scholar]
- 29.Mendonca T, Ferreira P, Marques J, Marcal A, Rozeira J. PH2—a dermoscopic image database for research and benchmarking. 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society; Osaka, Japan. 2013; https://www.ncbi.nlm.nih.gov/pubmed/24110966. [DOI] [PubMed] [Google Scholar]
- 30.Argenziano G, Soyer P, De Giorgio V, et al. Interactive Atlas of Dermoscopy. Milan, Italy: Edra Medical Publishing and New Media; 2000. p. 208. [Google Scholar]
- 31.ISIC. ADDI Project 2012. PH2 Database. [Accessed 15/5/2017]. Available at: https://www.fc.up.pt/addi/ph2%20database.html.
- 32.Pepe MS. The Statistical Evaluation of Medical Tests for Classification and Prediction. Oxford: Oxford University Press; 2003. [Google Scholar]
- 33.Steyerberg EW, Pencina MJ, Lingsma HF, Kattan MW, Vickers AJ, Van Calster B. Assessing the incremental value of diagnostic and prognostic markers: a review and illustration. Eur J Clin Invest. 2012;42(2):216–228. doi: 10.1111/j.1365-2362.2011.02562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3(1):32–35. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 35.Liu X. Classification accuracy and cut point selection. Stat Med. 2012;31(23):2676–2686. doi: 10.1002/sim.4509. [DOI] [PubMed] [Google Scholar]
- 36.Lee WC. Probabilistic analysis of global performances of diagnostic tests: interpreting the Lorenz curve-based summary measures. Stat Med. 1999;18(4):455–471. doi: 10.1002/(sici)1097-0258(19990228)18:4<455::aid-sim44>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 37.Irwin JR, Hautus MJ. Lognormal Lorenz and normal receiver operating characteristic curves as mirror images. R Soc Open Sci. 2015;2(2):140280. doi: 10.1098/rsos.140280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haenssle HA, Fink C, Schneiderbauer R, et al. Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann Oncol. 2018;29(8):1836–1842. doi: 10.1093/annonc/mdy166. [DOI] [PubMed] [Google Scholar]
- 39.Rajpara SM, Botello AP, Townend J, Ormerod AD. Systematic review of dermoscopy and digital dermoscopy/artificial intelligence for the diagnosis of melanoma. Br J Dermatol. 2009;161(3):591–604. doi: 10.1111/j.1365-2133.2009.09093.x. [DOI] [PubMed] [Google Scholar]
- 40.Harrington E, Clyne B, Wesseling N, et al. Diagnosing malignant melanoma in ambulatory care: a systematic review of clinical prediction rules. BMJ Open. 2017;7(3):e014096. doi: 10.1136/bmjopen-2016-014096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bafounta ML, Beauchet A, Aegerter P, Saiag P. Is dermoscopy (epiluminescence microscopy) useful for the diagnosis of melanoma? Results of a meta-analysis using techniques adapted to the evaluation of diagnostic tests. Arch Dermatol. 2001;137(10):1343–1350. doi: 10.1001/archderm.137.10.1343. [DOI] [PubMed] [Google Scholar]
- 42.Harbord RM, Whiting P. metandi: meta-analysis of diagnostic accuracy using hierarchical logistic regression. Stata Journal. 2009;9(2):211–229. [Google Scholar]
- 43.Dwamena B. Statistical Software Components S456880. Department of Economics, Boston College; Stata module for meta-analytical integration of diagnostic test accuracy studies. https://ideas.repec.org/c/boc/bocode/s456880.html. [Google Scholar]
- 44.Lee JB, Hirokawa D. Dermatoscopy: facts and controversies. Clin Dermatol. 2010;28(3):303–310. doi: 10.1016/j.clindermatol.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 45.Dinnes J, Deeks JJ, Chuchu N, et al. Dermoscopy, with and without visual inspection, for diagnosing melanoma in adults. Cochrane Database Syst Rev. 2018;12:CD011901. doi: 10.1002/14651858.CD011902.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Begg C, Greenes R. Assessment of diagnostic tests when disease verification is subject to selection bias. Biometrics. 1983;39(1):207–215. [PubMed] [Google Scholar]
- 47.Annessi G, Bono R, Sampogna F, Faraggiana T, Abeni D. Sensitivity, specificity, and diagnostic accuracy of three dermoscopic algorithmic methods in the diagnosis of doubtful melanocytic lesions: the importance of light brown structureless areas in differentiating atypical melanocytic nevi from thin melanomas. J Am Acad Dermatol. 2007;56(5):759–767. doi: 10.1016/j.jaad.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 48.Argenziano G, Fabbrocini G, Carli P, De Giorgi V, Sammarco E, Delfino M. Epiluminescence microscopy for the diagnosis of doubtful melanocytic skin lesions: comparison of the ABCD rule of dermatoscopy and a new 7-point checklist based on pattern analysis. Arch Dermatol. 1998;134(12):1563–1570. doi: 10.1001/archderm.134.12.1563. [DOI] [PubMed] [Google Scholar]
- 49.Argenziano G, Puig S, Zalaudek I, et al. Dermoscopy improves accuracy of primary care physicians to triage lesions suggestive of skin cancer. J Clin Oncol. 2006;24(12):1877–1882. doi: 10.1200/JCO.2005.05.0864. [DOI] [PubMed] [Google Scholar]
- 50.Argenziano G, Longo C, Cameron A, et al. Blue-black rule: a simple dermoscopic clue to recognize pigmented nodular melanoma. Br J Dermatol. 2011;165(6):1251–1255. doi: 10.1111/j.1365-2133.2011.10621.x. [DOI] [PubMed] [Google Scholar]
- 51.Ascierto PA, Palla M, Ayala F, et al. The role of spectrophotometry in the diagnosis of melanoma. BMC Dermatol. 2010;10:5. doi: 10.1186/1471-5945-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barzegari M, Ghaninezhad H, Mansoori P, Taheri A, Naraghi ZS, Asgari M. Computer-aided dermoscopy for diagnosis of melanoma. BMC Dermatol. 2005;5:8. doi: 10.1186/1471-5945-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benelli C, Roscetti E, Dal Pozzo V, Gasparini G, Cavicchini S. The dermoscopic versus the clinical diagnosis of melanoma. Eur J Dermatol. 1999;9(6):470–476. [PubMed] [Google Scholar]
- 54.Benelli C, Roscetti E, Pozzo VD. The dermoscopic (7FFM) versus the clinical (ABCDE) diagnosis of small diameter melanoma. Eur J Dermatol. 2000;10(4):282–287. [PubMed] [Google Scholar]
- 55.Binder M, Schwarz M, Winkler A, et al. Epiluminescence microscopy: a useful tool for the diagnosis of pigmented skin lesions for formally trained dermatologists. Arch Dermatol. 1995;131(3):286–291. doi: 10.1001/archderm.131.3.286. [DOI] [PubMed] [Google Scholar]
- 56.Binder M, Puespoeck-Schwarz M, Steiner A, et al. Epiluminescence microscopy of small pigmented skin lesions: short-term formal training improves the diagnostic performance of dermatologists. J Am Acad Dermatol. 1997;36:197–202. doi: 10.1016/s0190-9622(97)70280-9. [DOI] [PubMed] [Google Scholar]
- 57.Blum A, Clemens J, Argenziano G. Three-colour test in dermoscopy: a re-evaluation. Br J Dermatol. 2004;150(5):1040. doi: 10.1111/j.1365-2133.2004.05941.x. [DOI] [PubMed] [Google Scholar]
- 58.Bono A, Bartoli C, Cascinelli N, et al. Melanoma detection. Dermatology. 2002;205(4):362–366. doi: 10.1159/000066436. [DOI] [PubMed] [Google Scholar]
- 59.Bono A, Tolomio E, Trincone S, et al. Micro-melanoma detection: a clinical study on 206 consecutive cases of pigmented skin lesions with a diameter ≤3 mm. Br J Dermatol. 2006;155(3):570–573. doi: 10.1111/j.1365-2133.2006.07396.x. [DOI] [PubMed] [Google Scholar]
- 60.Carli P, De Giorgi V, Naldi L, Dosi G. Reliability and inter-observer agreement of dermoscopic diagnosis of melanoma and melanocytic naevi: Dermoscopy Panel. Eur J Cancer Prev. 1998;7(5):397–402. doi: 10.1097/00008469-199810000-00005. [DOI] [PubMed] [Google Scholar]
- 61.Carli P, Quercioli E, Sestini S, et al. Pattern analysis, not simplified algorithms, is the most reliable method for teaching dermoscopy for melanoma diagnosis to residents in dermatology. Br J Dermatol. 2003;148(5):981–984. doi: 10.1046/j.1365-2133.2003.05023.x. [DOI] [PubMed] [Google Scholar]
- 62.Carli P, Mannone F, de Giorgi V, Nardini P, Chiarugi A, Giannotti B. The problem of false-positive diagnosis in melanoma screening: the impact of dermoscopy. Melanoma Res. 2003;13(2):179–182. doi: 10.1097/00008390-200304000-00011. [DOI] [PubMed] [Google Scholar]
- 63.Cristofolini M, Zumiani G, Bauer P, Cristofolini P, Boi S, Micciolo R. Dermatoscopy: usefulness in the differential diagnosis of cutaneous pigmentary lesions. Melanoma Res. 1994;4(6):391–394. doi: 10.1097/00008390-199412000-00008. [DOI] [PubMed] [Google Scholar]
- 64.Dal Pozzo V, Benelli C, Roscetti E. The seven features for melanoma: a new dermoscopic algorithm for the diagnosis of malignant melanoma. Eur J Dermatol. 1999;9(4):303–308. [PubMed] [Google Scholar]
- 65.Dolianitis C, Kelly J, Wolfe R, Simpson P. Comparative performance of 4 dermoscopic algorithms by nonexperts for the diagnosis of melanocytic lesions. Arch Dermatol. 2005;141(8):1008–1014. doi: 10.1001/archderm.141.8.1008. [DOI] [PubMed] [Google Scholar]
- 66.Dreiseitl S, Binder M, Hable K, Kittler H. Computer versus human diagnosis of melanoma: evaluation of the feasibility of an automated diagnostic system in a prospective clinical trial. Melanoma Res. 2009;19(3):180–184. doi: 10.1097/CMR.0b013e32832a1e41. [DOI] [PubMed] [Google Scholar]
- 67.Dummer W, Doehnel KA, Remy W. Videomicroscopy in differential diagnosis of skin tumors and secondary prevention of malignant melanoma [in German] Hautarzt. 1993;44(12):772–776. [PubMed] [Google Scholar]
- 68.Feldmann R, Fellenz C, Gschnait F. The ABCD rule in dermatoscopy: analysis of 500 melanocytic lesions [in German] Hautarzt. 1998;49(6):473–476. doi: 10.1007/s001050050772. [DOI] [PubMed] [Google Scholar]
- 69.Fueyo-Casado A, Vázquez-Lopez F, Sanchez-Martin J, Garcia-Garcia B, Pérez-Oliva N. Evaluation of a program for the automatic dermoscopic diagnosis of melanoma in a general dermatology setting. Dermatol Surg. 2009;35(2):257–262. doi: 10.1111/j.1524-4725.2008.34421.x. [DOI] [PubMed] [Google Scholar]
- 70.Gereli MC, Onsun N, Atilganoglu U, Demirkesen C. Comparison of two dermoscopic techniques in the diagnosis of clinically atypical pigmented skin lesions and melanoma: seven-point and three-point checklists. Int J Dermatol. 2010;49(1):33–38. doi: 10.1111/j.1365-4632.2009.04152.x. [DOI] [PubMed] [Google Scholar]
- 71.Glud M, Gniadecki R, Drzewiecki KT. Spectrophotometric intracutaneous analysis versus dermoscopy for the diagnosis of pigmented skin lesions: prospective, double-blind study in a secondary reference centre. Melanoma Res. 2009;19(3):176–179. doi: 10.1097/CMR.0b013e328322fe5f. [DOI] [PubMed] [Google Scholar]
- 72.Haenssle HA, Korpas B, Hansen-Hagge C, et al. Seven-point checklist for dermatoscopy: performance during 10 years of prospective surveillance of patients at increased melanoma risk. J Am Acad Dermatol. 2010;62(5):785–793. doi: 10.1016/j.jaad.2009.08.049. [DOI] [PubMed] [Google Scholar]
- 73.Har-Shai Y, Glickman YA, Siller G, et al. Electrical impedance scanning for melanoma diagnosis: a validation study. Plast Reconstr Surg. 2005;116(3):782–790. doi: 10.1097/01.prs.0000176258.52201.22. [DOI] [PubMed] [Google Scholar]
- 74.Henning JS, Stein JA, Yeung J, et al. CASH algorithm for dermoscopy revisited. Arch Dermatol. 2008;144(4):554–555. doi: 10.1001/archderm.144.4.554. [DOI] [PubMed] [Google Scholar]
- 75.Keefe M, Dick DC, Waleel RA. A study of the value of the seven-point checklist in distinguishing benign pigmented lesions from melanoma. Clin Exp Dermatol. 1990;15(3):167–171. doi: 10.1111/j.1365-2230.1990.tb02064.x. [DOI] [PubMed] [Google Scholar]
- 76.Krähn G, Gottlöber P, Sander C, Peter RU. Dermatoscopy and high frequency sonography: two useful non-invasive methods to increase preoperative diagnostic accuracy in pigmented skin lesions. Pigment Cell Res. 1998;11(3):151–154. doi: 10.1111/j.1600-0749.1998.tb00725.x. [DOI] [PubMed] [Google Scholar]
- 77.Kreusch J, Rassner G, Trahn C, Pietsch-Breitfeld B, Henke D, Selbmann HK. Epiluminescent microscopy: a score of morphological features to identify malignant melanoma. Pigment Cell Res. 1992;(Suppl 2):295–298. doi: 10.1111/j.1600-0749.1990.tb00388.x. [DOI] [PubMed] [Google Scholar]
- 78.Lorentzen H, Weismann K, Petersen CS, Larsen FG, Secher L, Sk⊘dt V. Clinical and dermatoscopic diagnosis of malignant melanoma: assessed by expert and non-expert groups. Acta Derm Venereol. 1999;79(4):301–304. doi: 10.1080/000155599750010715. [DOI] [PubMed] [Google Scholar]
- 79.Lorentzen H, Weismann K, Kenet RO, Secher L, Larsen FG. Comparison of dermatoscopic ABCD rule and risk stratification in the diagnosis of malignant melanoma. Acta Derm Venereol. 2000;80(2):122–126. [PubMed] [Google Scholar]
- 80.Luttrell MJ, McClenahan P, Hofmann-Wellenhof R, Fink-Puches R, Soyer HP. Laypersons’ sensitivity for melanoma identification is higher with dermoscopy images than clinical photographs. Br J Dermatol. 2012;167(5):1037–1041. doi: 10.1111/j.1365-2133.2012.11130.x. [DOI] [PubMed] [Google Scholar]
- 81.Mackie RM, Fleming C, McMahon AD, Jarrett P. The use of the dermatoscope to identify early melanoma using the three-colour test. Br J Dermatol. 2002;146(3):481–484. doi: 10.1046/j.1365-2133.2002.04587.x. [DOI] [PubMed] [Google Scholar]
- 82.McGovern TWM, Litaker MSM. Clinical predictors of malignant pigmented lesions: a comparison of the Glasgow seven-point checklist and the American Cancer Society’s ABCDs of pigmented lesions. J Dermatol Surg Oncol. 1992;18(1):22–26. doi: 10.1111/j.1524-4725.1992.tb03296.x. [DOI] [PubMed] [Google Scholar]
- 83.Menzies SW, Ingvar C, Crotty KA, McCarthy WH. Frequency and morphologic characteristics of invasive melanomas lacking specific surface microscopic features. Arch Dermatol. 1996;132(10):1178–1182. [PubMed] [Google Scholar]
- 84.Menzies SW, Kreusch J, Byth K, et al. Dermoscopic evaluation of amelanotic and hypomelanotic melanoma. Arch Dermatol. 2008;144(9):1120–1127. doi: 10.1001/archderm.144.9.1120. [DOI] [PubMed] [Google Scholar]
- 85.Menzies SW, Moloney FJ, Byth K, et al. Dermoscopic evaluation of nodular melanoma. JAMA Dermatol. 2013;149(6):699–709. doi: 10.1001/jamadermatol.2013.2466. [DOI] [PubMed] [Google Scholar]
- 86.Nachbar F, Stolz W, Merkle T, et al. The ABCD rule of dermatoscopy: high prospective value in the diagnosis of doubtful melanocytic skin lesions. J Am Acad Dermatol. 1994;30(4):551–559. doi: 10.1016/s0190-9622(94)70061-3. [DOI] [PubMed] [Google Scholar]
- 87.Nilles M, Boedeker RH, Schill WBS. Surface microscopy of naevi and melanomas—clues to melanoma. Br J Dermatol. 1994;130(3):349–355. doi: 10.1111/j.1365-2133.1994.tb02932.x. [DOI] [PubMed] [Google Scholar]
- 88.Perrinaud A, Gaide O, French LE, Saurat J-H, Marghoob AA, Braun RP. Can automated dermoscopy image analysis instruments provide added benefit for the dermatologist? A study comparing the results of three systems. Br J Dermatol. 2007;157(5):926–933. doi: 10.1111/j.1365-2133.2007.08168.x. [DOI] [PubMed] [Google Scholar]
- 89.Piccolo D, Crisman G, Schoinas S, Altamura D, Peris K. Computer-automated ABCD versus dermatologists with different degrees of experience in dermoscopy. Eur J Dermatol. 2014;24(4):477–481. doi: 10.1684/ejd.2014.2320. [DOI] [PubMed] [Google Scholar]
- 90.Rao BK, Marghoob AA, Stolz W, et al. Can early malignant melanoma be differentiated from atypical melanocytic nevi by in vivo techniques? Skin Res Technol. 1997;3(1):8–14. doi: 10.1111/j.1600-0846.1997.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 91.Skvara H, Teban L, Fiebiger M, Binder M, Kittler H. Limitations of dermoscopy in the recognition of melanoma. Arch Dermatol. 2005;141(2):155–160. doi: 10.1001/archderm.141.2.155. [DOI] [PubMed] [Google Scholar]
- 92.Soyer HP, Smolle J, Leitinger G, Rieger E, Kerl H. Diagnostic reliability of dermoscopic criteria for detecting malignant melanoma. Dermatology. 1995;190(1):25–30. doi: 10.1159/000246629. [DOI] [PubMed] [Google Scholar]
- 93.Soyer HP, Argenziano G, Zalaudek I, et al. Three-point checklist of dermoscopy: a new screening method for early detection of melanoma. Dermatology. 2004;208(1):27–31. doi: 10.1159/000075042. [DOI] [PubMed] [Google Scholar]
- 94.Stanganelli I, Serafini M, Bucch L. A cancer-registry-assisted evaluation of the accuracy of digital epiluminescence microscopy associated with clinical examination of pigmented skin lesions. Dermatology. 2000;200(1):11–16. doi: 10.1159/000018308. [DOI] [PubMed] [Google Scholar]
- 95.Unlu E, Akay BN, Erdem C. Comparison of dermatoscopic diagnostic algorithms based on calculation: the ABCD rule of dermatoscopy, the seven-point checklist, the three-point checklist and the CASH algorithm in dermatoscopic evaluation of melanocytic lesions. J Dermatol. 2014;41(7):598–603. doi: 10.1111/1346-8138.12491. [DOI] [PubMed] [Google Scholar]
- 96.Walter FM, Prevost AT, Vasconcelos J, et al. Using the 7-point checklist as a diagnostic aid for pigmented skin lesions in general practice: a diagnostic validation study. Br J Gen Pract. 2013;63(610):e345–e353. doi: 10.3399/bjgp13X667213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Westerhoff K, McCarthy WH, Menzies SW. Increase in the sensitivity for melanoma diagnosis by primary care physicians using skin surface microscopy. Br J Dermatol. 2000;143(5):1016–1020. doi: 10.1046/j.1365-2133.2000.03836.x. [DOI] [PubMed] [Google Scholar]
- 98.Zalaudek I, Argenziano G, Soyer HP, et al. Three-point checklist of dermoscopy: an open internet study. Br J Dermatol. 2006;154(3):431–437. doi: 10.1111/j.1365-2133.2005.06983.x. [DOI] [PubMed] [Google Scholar]
- 99.Youl PH, Baade PD, Janda M, Del Mar CB, Whiteman DC, Aitken JF. Diagnosing skin cancer in primary care: how do mainstream general practitioners compare with primary care skin cancer clinic doctors? Med J Aust. 2007;187(4):215–220. doi: 10.5694/j.1326-5377.2007.tb01202.x. [DOI] [PubMed] [Google Scholar]