Significance
Astins are natural products that bind to a crucial human regulatory protein, the stimulator of interferon genes (STING), which is a promising new therapeutic target for cancer and immune disorders. Astins have long been regarded as phytochemicals of a Chinese medicinal plant. Here, we show that they are produced by the newly identified fungal endophyte Cyanodermella asteris via a nonribosomal biosynthetic pathway. Moreover, we provide evidence that key astin variants are produced during symbiosis with the aster plant. The production of specific phytochemical variants during symbiotic interactions is poorly studied and might be more widespread than previously expected. These findings pave the way for a cost-effective biotechnological astin production.
Keywords: astin, natural product, medicinal plant, cross-species biosynthesis, NRPS
Abstract
Medicinal plants are a prolific source of natural products with remarkable chemical and biological properties, many of which have considerable remedial benefits. Numerous medicinal plants are suffering from wildcrafting, and thus biotechnological production processes of their natural products are urgently needed. The plant Aster tataricus is widely used in traditional Chinese medicine and contains unique active ingredients named astins. These are macrocyclic peptides showing promising antitumor activities and usually containing the highly unusual moiety 3,4-dichloroproline. The biosynthetic origins of astins are unknown despite being studied for decades. Here we show that astins are produced by the recently discovered fungal endophyte Cyanodermella asteris. We were able to produce astins in reasonable and reproducible amounts using axenic cultures of the endophyte. We identified the biosynthetic gene cluster responsible for astin biosynthesis in the genome of C. asteris and propose a production pathway that is based on a nonribosomal peptide synthetase. Striking differences in the production profiles of endophyte and host plant imply a symbiotic cross-species biosynthesis pathway for astin C derivatives, in which plant enzymes or plant signals are required to trigger the synthesis of plant-exclusive variants such as astin A. Our findings lay the foundation for the sustainable biotechnological production of astins independent from aster plants.
Medicinal plants provide health-promoting or curative properties in versatile ways and have been used since prehistoric times across the world. It is estimated that more than 70% of the world’s population rely on medicinal plants (1). Some medicinal plants are facing extinction by wildcrafting and overcollection as a result (2), making alternative approaches to exploit their beneficial properties desirable.
Medicinal plants contain numerous chemically highly diverse natural products (NPs) with versatile biological functions. Traditional medicine employs predominantly multicomponent mixtures, including brews, decoctions, and tinctures, and the principle active agents are typically inadequately characterized (3). NPs derived from plants are a rich source for successful single-molecule drugs in modern medicine. More than half of the small-molecule pharmaceuticals approved in the last 3 decades were derived from or inspired by nature (4), and a substantial number of these compounds were discovered from plants (5). Prominent examples of plant-derived pharmaceuticals are paclitaxel, vincristine and vinblastine, camptothecin, and artemisinin (5–7).
Plant NPs are generally characterized by low production yields and complex chemical structures, making it challenging to meet market demands by means of plant cultivation or chemical synthesis (8). Instead, successful resupply of plant-derived drugs on the pharmaceutical market relies on biotechnological production that requires knowledge of the usually complex biosynthetic pathways (9). Endophytic microorganisms residing in the interior of plants (10, 11) are increasingly reported to produce NPs originally known from their host plants (12–16). Nevertheless, none of these endophytes has proved suitable for industrial resupply of the desired compound, mainly due to low and unstable production titers (13, 17).
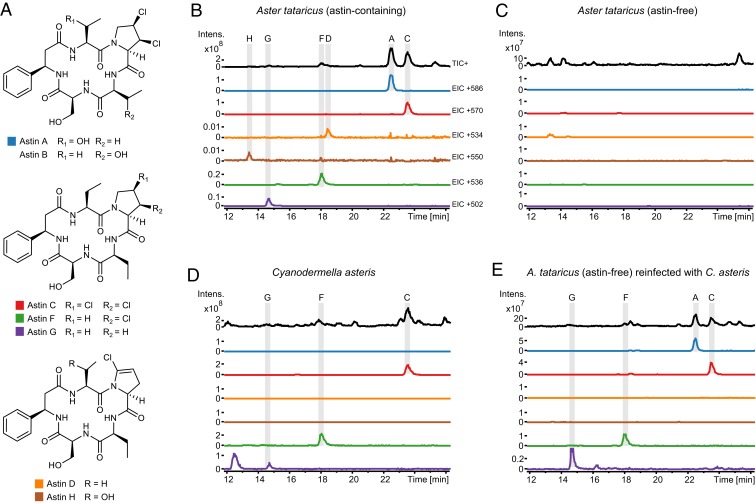
The flowering plant Aster tataricus l. f. (Compositae) has long found use in traditional Chinese medicine for the treatment of cough and bronchial diseases (18). Several studies identified chemically diverse NPs from the roots of A. tataricus (19–21) including astins, macrocyclic peptides that possess anticancer and immunosuppressive activities (22–25). Astins are now known to bind to STING, a crucial cytosolic DNA sensor protein involved in human innate immunity (26). A total of 24 astin chemical variants (20, 27–32) are known, including the structurally related branched tataricins (33) as well as linear asterinins (34, 35). Quantitatively, the astin variants A to C dominate in A. tataricus preparations (18, 31, 36). They consist of 5 amino acids including l-2-aminobutyric acid (2Abu), β-(R)-phenylalanine (βPhe), and the unique 3,4-dichlorinated l-proline (ProCl2) (Fig. 1A). Low natural availability (27, 31, 37) and unsuccessful chemical synthesis approaches (38, 39) of astins prevent a thorough exploration of their biological activity.
Fig. 1.
The production of astins by A. tataricus and C. asteris. (A) Chemical structures of the main astin variants known from A. tataricus. (B–E) HPLC-MS chromatograms of extracts prepared from fresh plant roots and fungal cultures (each one is a representative chromatogram out of at least 3 biological replicates). The chromatograms show astin peaks at characteristic retention times (indicated in capital letters). Extracted ion chromatograms (EIC) depict the respective [M+H]+ ions of astins. TIC+ denotes total ion chromatogram, positive mode. Chromatograms are shown for a representative A. tataricus plant either (B) containing astins or (C) being devoid of astins, (D) for extracts prepared from a C. asteris culture and (E) for extracts prepared from an A. tataricus plant (initially being devoid of astins like in C), 3 mo after reinfection with the endophyte C. asteris. Intens., ion intensity in arbitrary units.
The biosynthetic origins of astins remained unclear, but it is widely assumed that they are made by the plant A. tataricus (40). It is hypothesized that astins are the product of a nonribosomal peptide synthetase (NRPS) biosynthetic pathway (33). NRPSs are large multidomain enzymes predominantly found in bacteria and fungi that catalyze a stepwise peptide synthesis by a thiotemplate mechanism (41). NRPSs are able to utilize a large number of nonproteinogenic amino acid substrates to generate nonribosomal peptides (NRPs) (41). The mycotoxin cyclochlorotine from the fungus Talaromyces islandicus (42) is an NRP showing remarkable structural similarities to astins, suggesting that astins likewise might be produced by a fungus.
Here, we show that astins do indeed have a fungal origin. The endophyte Cyanodermella asteris that has been recently isolated from inflorescences of A. tataricus (43) produces astin variants in pure culture, including astin C, one of the 3 main variants isolated from A. tataricus (25, 27, 31, 37). Plants free of C. asteris did not contain any astins, and reinfection with this fungus restored astin production, including plant-exclusive variants such as astin A. We identified the astin biosynthetic gene cluster in the recently sequenced fungal genome (44). C. asteris is characterized by stable and sound production of astins, which is unlike other endophytes that often only poorly and unreliably produce NPs originally known from their host plants (45–48). Our findings pioneer prospective biotechnological production of this valuable NP.
Results
A. tataricus Harbors an Endophytic Community Including the Astin-Producing Fungus C. asteris.
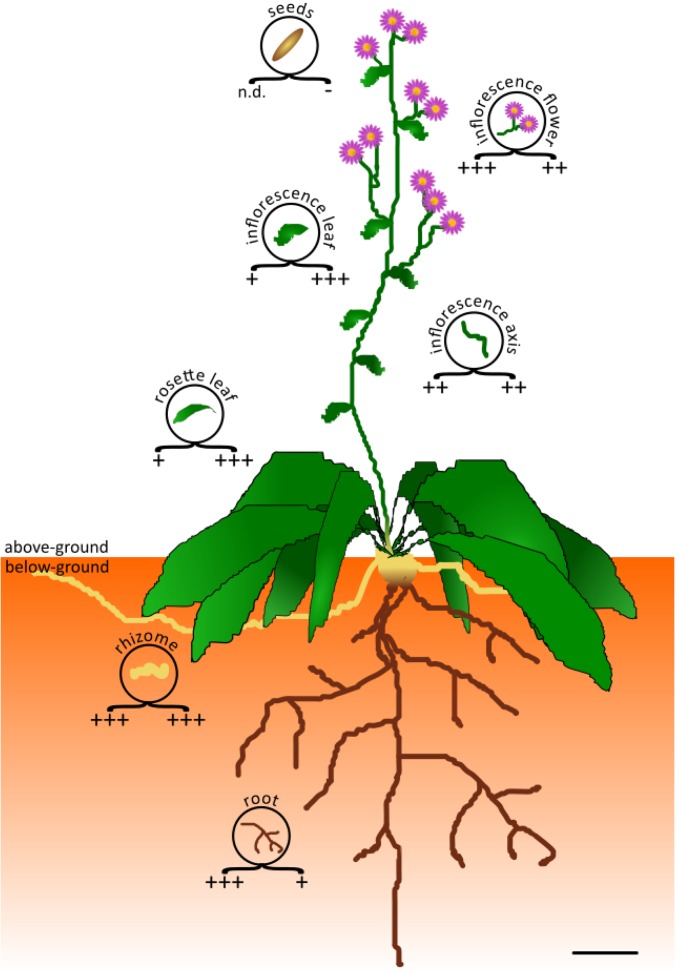
Astins are primarily reported from dried roots and rhizomes of A. tataricus (20, 28, 29, 31). Extracts from fresh plant roots clearly also contained astins, according to high-performance liquid chromatography–mass spectrometry (HPLC-MS) analyses (Fig. 1B). Interestingly, some of the investigated A. tataricus plants did not contain any traces of astins (Fig. 1C), despite cultivation under identical conditions. This information together with structural similarity to the mycotoxin cyclochlorotine of T. islandicus (42) suggested that astins might have a fungal origin. Astins were abundant in inflorescences and leaves in addition to roots and rhizomes (Fig. 2), suggesting that the astin-producing fungus must live in close contact with the plant, for example, as an endophyte, rather than being part of the loosely associated microbial community in the soil environment.
Fig. 2.
Distribution of astins and C. asteris in the plant A. tataricus. The figure serves to illustrate distribution tendencies of astins and the astin-producing endophyte C. asteris. Concentrations in different plant tissues are indicated beneath the circular tissue symbol as plus and minus signs (lower left, astins; lower right, fungus). Astin levels were determined by calculating the peak area of the respective [M+H]+ ions in HPLC-MS extracted ion chromatograms. The signals of astin A and astin C were summed up and assigned to +, ++, or +++ for signal intensities of <5×1011, 5×1011 to 5×1012, or >5×1012, respectively (n.d., not determined). The amount of C. asteris was determined by calculating the ratio of fungal DNA per plant DNA, which was measured by quantitative PCR. Ratios of <0.5%, 0.5 to 5%, and >5% were assigned to +, ++, and +++. Minus (−) indicates no detection of fungal DNA. The measurements were performed for 8 plants, and the most frequent symbol is displayed. (Scale bar, 30 mm.)
To investigate fungal endophytes of A. tataricus, roots were surface-sterilized, dissected, and incubated on agar plates until fungal colonies developed. Several endophytic fungi were isolated and assigned to morphotypes according to macroscopic and microscopic resemblances. A representative isolate of each morphotype was identified by ribosomal internal transcribed spacer (ITS) sequencing (SI Appendix, Tables S1 and S2). Fungal genera isolated and identified included Paraphoma, Colletotrichum, Epiccocum, Cladosporium, and Sacrocladium, all of which have been reported as endophytes or plant-associated fungi from a variety of hosts. Besides these endophytes from roots, we recently reported the isolation of C. asteris (43), a hitherto unknown endophytic fungus, from the inflorescence of A. tataricus. C. asteris is a member of the heterogeneous lichen-forming family Lecanoromycetes (43), and the only member within the small genus Cyanodermella (43, 49) with an endophytic lifestyle.
In total, 22 isolated fungal endophytes of A. tataricus were tested for production of astins. Fermentations in liquid potato dextrose broth (PDB) followed by culture extract screening via HPLC-MS clearly demonstrated the presence of astin C, F, and G in the culture medium of C. asteris (Fig. 1D). Astins were not detected from any of the other endophytes tested.
Growth rate and astin production of C. asteris were highest in malt extract autolysate (MEA) medium (SI Appendix, Fig. S1 and Table S3). Around 63 mg of astin C and 1.5 mg of astin F were isolated to purity from 3 L of culture (SI Appendix, Fig. S2). This allowed confirmation of the chemical structure, including the stereochemistry of both molecules. Their planar structures were established by high-resolution MS analysis and, in addition, either by direct comparison of 1H and 13C nuclear magnetic resonance (NMR) data with literature data or by de novo NMR-based structure elucidation. The absolute configuration was inferred from chiral amino acid analysis, NMR experiments, and comparison of the measured [α]D values with literature data (SI Appendix, Figs. S3–S15 and Tables S4–S9). The dichlorinated astin C, for which antineoplastic and antiinflammatory activity has been reported (22–25, 50), was the main variant of the 3 astins produced by the endophytic fungus C. asteris. Production of astins in axenic culture was stable over time, even after more than a dozen rounds of cultivations.
The Biosynthetic Origins of Astins.
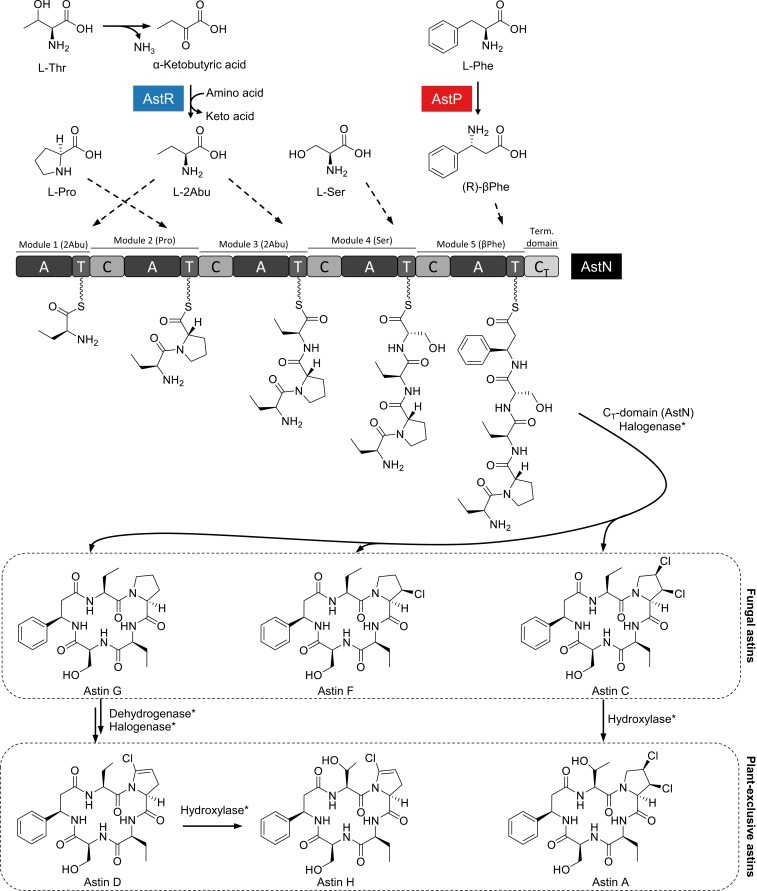
The chemical structures of astins suggest that they are produced on an NRPS enzyme complex. We recently sequenced the genome of C. asteris de novo on an Illumina MiSeq system resulting in an 85× coverage, 37 scaffolds, a genome size of 28.42 Mb, and 10,309 predicted genes (44). The genome encodes 5 putative NPRS enzymes, each being modular type I NRPS systems. Since the fungus is not genetically tractable, we investigated each of these candidate NRPSs in silico in order to identify the one responsible for astin biosynthesis. The large enzyme AstN (614 kDa) is the only NRPS enzyme with the predicted 5-modular composition required for the assembly of the astin pentapeptide. AstN shows remarkably high global sequence similarity (65%, identity 47%) and an analog domain architecture to the NRPS CctN of T. islandicus. CctN is the key enzyme in the cyclochlorotine pathway, as has been experimentally demonstrated by RNA interference-based silencing experiments (51). It can therefore be assumed that AstN is the astin synthetase.
AstN has the deduced domain architecture A−T−(C−A−T)4−CT, where A stands for adenylation domain, T for thiolation domain, C for condensation domain, and CT for terminal condensation-like domain (52). The domain organization implies a stepwise assembly of the 5 amino acid building blocks to a linear precursor pentapeptide. A domains are responsible for selection and activation of the building blocks. Comparison of the active center amino acids of the A domains with the active centers of substrate-assigned A domains of other fungal NPRSs revealed considerable similarities that allow the prediction of the substrate specificity for AstN (SI Appendix, Table S10). Accordingly, 2Abu is incorporated by the first (A1) and the third (A3) A domains. The second building block is proposed to be Pro, due to high similarities of the respective domain (A2) with Pro-activating domains of other fungal NRPS systems (SI Appendix, Table S10). Taking into account the biosynthesis of halogenated NPs in other microorganisms (53–55), chlorination of Pro in astins is assumed to occur during NRPS-mediated peptide assembly. Next, the building block Ser is activated and incorporated by domain A4. The last A domain, A5, is proposed to bind βPhe rather than Phe, because similarities to Phe-activating A domains are only weak (SI Appendix, Table S10). The deduced assembly order of AstN thus is 2Abu−Pro−2Abu−Ser−βPhe (Fig. 3).
Fig. 3.
Biosynthesis model for the main astin variants. In conjunction with in silico analysis of the NRPS adenylation (A) domains and gene function prediction of the ast biosynthetic gene cluster, a biosynthesis model has been deduced. AstR and AstP are proposed here to supply the nonproteinogenic building blocks 2Abu and βPhe, respectively. The NRPS AstN assembles 5 building blocks to a macrocyclic pentapeptide. Dotted line arrows indicate incorporation by the A domains. The products released from AstN are proposed to be the fungal astins G, F, and C, which differ in the degree of chlorination. The plant-exclusive variants A, D, and H likely derive from the fungal astins as a result of a symbiosis between host and endophyte. Asterisk (*) denotes that the enzymes involved in tailoring of the astins are unknown.
The terminal domain in AstN, CT, shows typical features of fungal cyclization domains. The domain has a characteristic variation of the first histidine residue (52) of the highly conserved HHxxxDGxS motif of C domains (motif sequence in AstN: 5302SHSQYDGWS). Moreover, a characteristic signature within the downstream region of fungal CT domains, the so-called WL1−WL2 signature (56), is present in this terminal domain. Thus, the CT domain is expected to cyclize and simultaneously release the macrocyclic pentapeptide from the AstN enzyme.
In comparison with the cyclochlorotine synthetase CctN, the astin synthetase AstN differs primarily in the selection of the first building block during peptide assembly. While the first module of AstN incorporates 2Abu (Table 1), it is Ser in the case of CctN (51), which accounts for the small structural differences between the 2 groups of macrocyclic pentapeptides (SI Appendix, Fig. S16).
Table 1.
Features and putative functions of ast cluster genes
| Locus tag | Gene name | No. of Introns | No. of amino acids | Conserved domain(s)/proposed function | Possible role in astin biosynthesis |
| VA03HOR0624_x10098 | astL | 1 | 353 | Unknown | Unknown |
| VA03HOR0624_x10099 | astM | 5 | 650 | Bromodomain, chromatin-associated protein | Transcriptional regulation |
| VA03HOR0624_x10100 | astN | 2 | 5,584 | NRPS | Astin synthetase |
| VA03HOR0624_x10101 | astO | 0 | 1,530 | ABC-type exporter | Efflux/intercompartmental transport |
| VA03HOR0624_x10102 | astP | 3 | 682 | Aminomutase | Biosynthesis of βPhe* |
| VA03HOR0624_x10103 | astQ | 1 | 79 | Unknown | Unknown |
| VA03HOR0624_x10104 | astR | 1 | 476 | Aminotransferase | Biosynthesis of 2Abu† |
| VA03HOR0624_x10105 | astS | 3 | 320 | DUF3328‡ | Unknown |
| VA03HOR0624_x10106 | astT | 0 | 483 | Transporter (major facilitator superfamily) | Efflux/intercompartmental transport |
| VA03HOR0624_x10107 | astU | 4 | 373 | DUF3328‡ | Unknown |
| VA03HOR0624_x10108 | astV | 1 | 227 | DUF3328‡ | Unknown |
| VA03HOR0624_x10109 | astW | 2 | 1,338 | ABC-type exporter | Efflux/intercompartmental transport |
| VA03HOR0624_x10110 | astX | 1 | 691 | Transcriptional regulator (ICP4 domain); signal transduction (tetratricopeptide repeat) | Transcriptional regulation |
Detailed amino acid sequence analysis of AstP reveals characteristic features of phenylalanine aminomutases according to Heberling et al. (76) (see SI Appendix, Fig. S14 for more information).
The building block 2Abu can be generated from l-threonine via α-ketobutyric acid by the concerted action of a threonine dehydratase and an aminotransferase (77). The cluster-encoded aminotransferase AstR is proposed to catalyze the second step in this reaction.
DUF3328, domain of unknown function 3328. Proteins containing DUF3328 have recently been shown to be involved in fungal NP biosynthesis (78).
The Astin Biosynthetic Gene Cluster Encodes Further Biosynthetic Enzymes, Although There Is No Evident Halogenase.
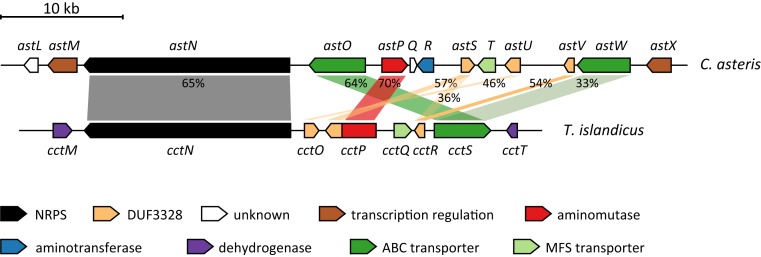
The genetic region around astN contains the astin biosynthetic gene cluster (ast BGC) (Fig. 4). The cluster boundaries were estimated manually based on predicted gene functions and by identifying genetic elements that likely belong to the primary but not to the astin metabolism (SI Appendix, Fig. S17). According to that, the ast BGC consists of 13 distinct genes encoded in an area of about 50 kilo base pairs (kb) in size. Most of the cluster genes show striking sequence similarity to genes of the cyclochlorotine biosynthetic gene cluster (Fig. 4), which strongly supports the conclusion that the proposed genetic region constitutes the ast BGC. For several of the gene products, putative functions in astin biosynthesis are suggested (Table 1). Besides the already described NRPS AstN, these functions are transcriptional regulation (AstM, X), transportation (AstO, T, W), and biosynthesis of astin building blocks (AstR, P).
Fig. 4.
The ast biosynthetic gene cluster in comparison to the cyclochlorotine biosynthetic gene cluster. Percentages illustrate the global amino acid sequence similarity of astin cluster genes to cyclochlorotine cluster genes from T. islandicus (only similarities of >30% are illustrated).
A halogenase is assumed to catalyze the chlorination of the Pro residue in the course of pentapeptide synthesis to yield the dichlorinated astin C variant and the monochlorinated astin F variant. Unexpectedly, no apparent halogenase is encoded in the ast BGC. To our best knowledge, no enzymes are described so far that convert Pro into ProCl2. The regiospecific and stereospecific chlorination of the pyrrolidine ring constitutes a challenging chemical reaction, which is principally only attributed to flavin-dependent and α-ketoglutarate/iron-dependent halogenases (57–59). Remarkably, according to protein function predictions and amino acid sequence comparisons, not a single gene in the whole genome codes for any of these halogenases (SI Appendix, Tables S11 and S12). In view of this, it is likely that a member of a novel type of halogenase, which has yet to be identified, achieves chlorination of the astins.
The Astins in A. tataricus Are Produced by the Endophyte C. asteris and Are Further Metabolized inside of the Host Plant.
A. tataricus plants were investigated for the distribution of the C. asteris endophytes and astins. Plants that contained astins always contained C. asteris. The distribution of astins in the different plant parts of A. tataricus is consistent with the incidence of the endophyte C. asteris (Fig. 2). C. asteris was not detected from plants lacking astins. A. tataricus plants devoid of C. asteris were infected with the endophyte, and HPLC-MS analysis of root extracts prepared 3 mo after infection showed a range of astins comparable to that of other astin-containing plants (Fig. 1E), whereas the noninfected control plant remained totally devoid of astins. These findings clearly demonstrate that the endophyte C. asteris is responsible for astin production in A. tataricus.
Interestingly, a higher number of astin variants could be detected in the host plant than are produced in axenic fungal cultures. Especially, astin A, which is one of the main variants in planta, was not detected in C. asteris culture extracts. It is known that fungal biosynthetic pathways and their produced metabolites are influenced by growth conditions and media compositions (60). Therefore, a large variety of cultivation conditions were tested for C. asteris to induce production of more astin variants (e.g., different cultivation media, temperatures, durations, and supplements; see SI Appendix, Table S13). In addition, the fungus was also cultivated in the presence of phytohormones, plant extracts, and plant pieces (SI Appendix, Table S13) in order to test for plant stimuli. Moreover, since it is possible that astin A arises from incorporation of l-allo-Thr (instead of 2Abu) at module 1 of the NRPS, the fungal culture broth was supplemented with high amounts (500 mg/L) of that building block. However, in all tested approaches, neither astin A nor other hydroxylated variants were produced in the fungal cultures. If, on the contrary, C. asteris was reintroduced into astin-devoid A. tataricus plants, most astins, including the plant-exclusive variant A, reemerged (Fig. 1E). This indicates that some astins are the result of a symbiosis in planta. Apparently, their biosynthesis needs more than one biological partner and requires an inductor or a biosynthetic enzyme from a further party like the host plant itself or endophytes other than C. asteris residing therein.
Discussion
Astins Are Nonribosomal Peptides from a Lecanoromycetes Fungus.
The true origin of plant-derived NPs is often a matter of debate (61). Similar to the recent findings in marine invertebrates, where bacteria and fungi are increasingly made responsible for the production of various NPs (61, 62), there has been a growing number of reports of microbial endophytes able to produce NPs initially known from their host plants (12–16), including the pharmacologically interesting compounds paclitaxel (16), camptothecin (15), and FR900359 (14). Apparently, several plant-derived NPs are not true phytochemicals but are rather made by endophytic microorganisms. This clearly applies to the antitumor pentapeptides astins, which have long been regarded as phytochemicals of the traditional Chinese medicinal plant A. tataricus (40). We could show that they actually originate from C. asteris, a Lecanoromycete fungus living inside the plant as an endophyte. At present, no other microorganisms are reported to produce astins.
The prominent structural analogy between astins and cyclochlorotines from the mold T. islandicus (Eurotiomycetes) is reflected in a related biosynthetic pathway that involves key enzymes with high degrees of similarity. For example, the astN gene is more similar to cctN than to any other gene of a Lecanoromycetes strain from which genome data are available. In light of this and the distant relation between C. asteris and T. islandicus, horizontal gene transfer might be involved in the dissemination of the BGCs encoding the biosynthesis pathways. Notably, however, both BGCs show only a weak gene synteny (Fig. 4), presumably caused by events of gene rearrangements, which is not an unusual observation in fungal BGCs (63).
Astin A and Further Plant-Exclusive Variants Are Only Produced In Planta by a Symbiotic Biosynthesis.
The endophytic fungus C. asteris produces the compounds astin C, F, and G under submerged axenic cultivation conditions. Further variants such as the hydroxylated astin A were only found inside the endophyte-containing plant A. tataricus. Thus, neither the plant nor the fungus alone are able to produce these plant-exclusive astins, but they are a symbiotic product of both organisms.
Since no incorporation of the hydroxylated building block allo-Thr was observed in C. asteris cultures, we assume that the hydroxylated plant-exclusive variants like astin A and astin H result from a hydroxylation subsequent to the macrocyclic peptide assembly. Similarly, the variants D and H show an alternative chlorination pattern likely originating from dehydrogenation and monochlorination of astin G (Fig. 3). Although it cannot be ruled out that these reactions are catalyzed by C. asteris enzymes that are only expressed in the native environment after receiving a certain trigger from the host plant, all attempts to induce the proposed tailoring reactions by adding plant components to the fungal culture failed. This makes an enzymatic contribution by a species other than C. asteris more likely. It is well known that plants can hydroxylate aliphatic molecules in the course of biotransformation of toxic xenobiotics (64–66). Therefore, the occurrence of modified astin variants is probably best explained by a subsequent modification of the fungal NRPs via plant enzymes. We assume that all of the other variants described from the aster (20, 27–35) arise in a similar way.
Such cross-species metabolic pathways, which require symbiosis between 2 or more biological partners, are quite unexplored. A symbiotic biosynthesis has only been shown for rhizoxin [joint bacterial and fungal biosynthesis (67)]. Furthermore, joint plant and fungal biosynthesis was proposed for one case of camptothecin (68) and for maytansine (69). Taking these examples and our findings on the plant-exclusive astins into consideration, it cannot be ruled out that many further NPs whose biosyntheses are obscure actually are the result of a symbiotic achievement of more than one species.
The ecological benefits of the symbiotic production of plant-exclusive astins are unclear. However, regarding both the relative high concentration of astin A in plants and its increased bioactivity (20, 24, 70) compared to astin C, the modifications would constitute a selective advantage to the host plant by increasing the potency of the fungal peptides. The astins might serve as a defense system against predators or pathogenic opponents.
The Discovery of the Astin Pathway in C. asteris Enables a Biotechnological Production of the Biologically Active Astin C.
While NPs of medicinal plants remain a major source for lead compounds in anticancer therapy and other pharmaceutical applications (13), they are not suited to meet industrial demands, due to slow plant growth, low NP production rates, and time-consuming extraction processes. The increasing findings of endophytes to produce plant-derived NPs (12, 13, 15, 16, 61) raised high hopes regarding the usage of these microorganisms for sustainable NP production in bioreactors (17). However, there have been no major breakthroughs in terms of commercial exploitation for any NP produced by an endophyte (17, 71). Reasons for that are either extremely poor production levels or the substantial decrease in NP production upon repeated subcultivation in axenic conditions (13), which, for example, is commonly observed for endophytic producers of the high-value cancer therapeutics camptothecin and paclitaxel (45–48). Further challenges in achieving biotechnological production using microorganisms arise from missing information on biosynthetic pathways and the encoding genes, which applies even for high-value plant-derived NPs such as paclitaxel, podophyllotoxin, or artemisinin (8, 9, 12).
None of these obstacles is anticipated for microbial astin production. First, C. asteris is characterized by a strong astin C production (20 mg/L). Second, no attenuation of astin production in C. asteris is observed, despite subcultivation for more than a dozen rounds. Third, and perhaps most importantly, the genome sequence of C. asteris is available, and the astin BGC has been elucidated. This opens the door for genetic engineering of astin production in C. asteris as well as heterologous production in more readily accessible microorganisms. Due to the findings reported here, a sustainable biotechnological astin production has now been made possible. Consequently, the prospective pharmaceutical exploitation of astins toward anticancer or immunosuppressive pharmaceuticals has moved a big step forward.
Materials and Methods
General.
Unless otherwise stated, all chemicals were purchased from Sigma-Aldrich, and H2O used was deionized. A. tataricus plants were obtained from the Botanical Garden of the Technische Universität Dresden, and from the company Sarastro Stauden (Ort im Innkreis, Austria). The plants were cultivated in the greenhouse under long-day conditions (16 h light at 23 °C and 8 h dark at 18 °C). A. tataricus dried roots (“Zi Wan”) were purchased from Anguo Mayway Herb Company Ltd. (Hubei, China).
See SI Appendix, Supplementary Methods for information on purification and structural elucidation of astins.
Astin Identification in Plant and Fungal Material by HPLC-ESI-MS and HPLC-MS/MS.
To analyze astins from plant materials, fresh plant tissue was ground with liquid nitrogen and extracted twice with 20 mL of methanol/acetone (1:1) per gram fresh weight by shaking for 30 min followed by one extraction with 20 mL of H2O per gram fresh weight. To prepare astins from commercial dried roots (Zi Wan), 3 g of root material was ground with mortar and pestle and extracted 3 times with 30 mL of ethyl acetate. Astins from fungal material were prepared by extracting 20 mL of a well-grown culture (MEA medium for C. asteris, PDB in case of other fungal endophytes) 3 times with the same volume of ethyl acetate. Alternatively, l-butanol was used as extracting solvent for fungal cultures. If applicable, extracts from the same sample were combined. The extracts were then evaporated in vacuo, resolved in 1 mL of H2O/methanol (1/1) and analyzed by HPLC-MS and/or HPLC-MS/MS using a Zorbax 300SB-C18 column (5 µm, 250 × 2.1 mm, including precolumn: 12.5 × 2.1 mm; Agilent Technologies) coupled to an ESI mass spectrometer (LC/MSD Ultra Trap system XCT 6330; Agilent Technologies). Detection of m/z values was conducted with Agilent DataAnalysis for 6300 series Ion Trap LC/MS 6.1 version 3.4 software (Bruker Daltonics). Solvent A was 0.1% formic acid, and solvent B was 0.06% formic acid in methanol (gradient: 15% B to 50% B in 25 min; flow rate: 0.4 mL/min; column temperature: 40 °C). Electrospray ionization (alternating positive and negative ionization) in Ultra Scan mode with a capillary voltage of 3.5 kV and a drying gas temperature of 350 °C was used for LC-MS analysis. MS/MS experiments were carried out in positive mode.
Astins were initially identified in the Zi Wan sample by means of the masses of their pseudomolecular ions [M+H]+, by the striking isotope pattern arising from the chlorine atoms, and by comparing the MS/MS fragmentation patterns with literature data (37). Astins from further biological samples were identified due to identical retention time values in HPLC and, in particular, on the basis of identical MS/MS fragmentation patterns with the astins from the Zi Wan sample (SI Appendix, Figs. S18–S20).
In the astin distribution experiment (Fig. 2), a Nucleosil 100-C18 column (3 µm, 100 × 2 mm, including precolumn: 10 × 2 mm; Dr. Maisch GmbH) was used with the following parameters: Solvent A was 0.1% formic acid, and solvent B was 0.06% formic acid in acetonitrile (gradient: 10% B to 100% B in 15 min; flow rate: 0.4 mL/min; column temperature: 40 °C). The remaining parameters were as described above.
Isolation and Characterization of Endophytes from Fresh A. tataricus Roots.
Healthy looking, fresh roots from astin-containing A. tataricus plants grown in natural soil in sparse mixed forest in Tübingen, Germany (geocoordinates 48.515987, 9.039934) were carefully freed from soil, thoroughly washed 2 to 3 times with H2O, and cut into pieces of ca. 2 to 5 cm length. These root segments were surface-sterilized for 20 min in 5% sodium hypochlorite (NaOCl, in the form of DanKlorix; Colgate-Palmolive), briefly washed in sterile H2O, once again sterilized in 5% NaOCl (10 min), and washed in 20 mL of sterile H2O. Sterilized roots were incubated on tap water agar (15 g of agar per 1 L of tap water) and potato dextrose agar (PD; Carl Roth) for endophyte isolation. Both solid media were supplemented with 0.5 g/L pulverized A. tataricus roots (in order to provide possible essential growth factors) before autoclave and with an antibiotic mixture (ampicillin, streptomycin, tetracycline, each 50 µg/mL) afterward. To control proper surface sterilization of the root material, 1 mL of the second washing water was plated on LB agar plates (Carl Roth). Root surfaces were regarded as sterile if these control plates were free from any growth after 3 d of cultivation at 37 °C. Fungal colonies developing from the surface-sterilized roots were isolated by transferring the endophytes on PD agar plates (including antibiotic mixture; see above), inspected macroscopically and microscopically, and assigned to morphotypes accordingly. For taxonomic characterization, genomic DNA was isolated from liquid fungal cultures (grown for 1 to 2 wk in PDB [Carl Roth] at 150 rpm and 21 °C or 27 °C) of each one representative morphotype using the DNeasy Plant Mini Kit (Qiagen). Ribosomal ITS sequences were amplified using primers ITS1 and ITS4 (72) [as required, primers ITS1F and ITS4B (73) were used alternatively]. PCR products were subcloned into the plasmid pJET1.2/blunt (Thermo Scientific), the ITS sequences were sequenced (Eurofins Genomics), and the sequences were analyzed with BLASTn (against the NCBI nonredundant nucleotide collection) in order to allocate the isolated morphotype to a fungal genus. For analysis of astin production, all isolated fungal endophytes were processed as described above. The morphotype strains can be obtained upon reasonable request.
C. asteris Cultivation.
C. asteris was maintained in a baffled flask with 50 to 100 mL of MEA medium (150 rpm, ca. 22 °C, without lighting). The culture was refreshed every 2 to 4 wk by inoculating fresh medium with 1/100 volume of a gently homogenized preculture (using a homogenizer with glass pistil to force dispersed growth). To test the incorporation of allo-Thr into astins, a C. asteris culture grown in 100 mL of MEA medium was supplemented with 500 mg/L l-allo-Thr or l-Thr at day 7 and cultivated for a further 21 d before extracts were analyzed by HPLC-MS (see above). This experiment was performed twice. To test the influence of host plant extracts on astin production of C. asteris, fungal cultures were supplemented with an aqueous extract of plant tissues. For this, different plant parts (about 5 g each) were ground with liquid nitrogen and extracted with 20 mL of H2O per gram of tissue. The extracts were concentrated 10 times, sterile-filtered, and added to the fungal culture (up to 1% final concentration, experiment performed in triplicates). Fungal extracts were analyzed by HPLC-MS (see above) after 2 to 3 wk of growth. For all other variations performed to elicit astin A production in C. asteris cultures, the different conditions and details are given in SI Appendix, Table S13.
Screening for C. asteris in Plants by qPCR.
A. tataricus total DNA was isolated according to a protocol of Möller et al. (74) with the following modifications: Before DNA precipitation, 150 µL of 5% polyvinylpolypyrrolidone (PVPP) were added to the supernatant. After incubation at room temperature for 30 min with gentle shaking, the sample was centrifuged (16,000 rcf, 4 °C, 10 min) to remove the PVPP. The DNA was precipitated by incubation at −20 °C for up to 2 h.
The extracted plant DNA was then screened for C. asteris DNA by quantitative real-time PCR using the device qTOWER2.2 and the software qPCRsoft 3.2 (both Analytik Jena). Specific primers were used to amplify parts of the gene actin of A. tataricus and ribosomal DNA of C. asteris (SI Appendix, Table S15). The PCR was conducted with only a small amount (5 ng) of DNA in order to minimize a possible inhibition by plant secondary metabolites. PCR settings were 1 cycle with 95 °C for 2 min, then 40 cycles with 95 °C for 5 s, and 64 °C for 25 s, followed by a melting curve analysis for verification of the PCR products. Calibration curves were created for absolute quantification using pure DNA from A. tataricus (noninfected plant tissue) and pure DNA from an axenic C. asteris culture.
Infection of Astin-Free Plants with C. asteris.
Astin-free A. tataricus plants were cultivated in a 2-compartment system (75) in autoclaved soil, where a gauze (pore size 20 µm) separated the plant compartment from the fungal one. The soil of the fungal compartment was infected with 150 mg of freshly homogenized C. asteris tissue. The roots are not able to grow through the gauze, whereas the fungal hyphae can. The 2-compartment system with the plant and infected soil was cultivated in a climate chamber under long-day conditions (16 h light with 23 °C, 8 h dark with 18 °C). Astin content was investigated in plant tissue after 3 mo of growth as described above. Three plants were infected with C. asteris; one noninfected plant served as control.
Data Availability.
All data generated or analyzed during this study are included in this article and SI Appendix.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the financial support by the European Union (European Regional Development Fund, Project Astinprod) and the Free State of Saxony (Grant 100271404 and Grant 100271410). T.S., L.J., L.F., K.-H.v.P., and J.L.-M. were funded by the Sächsische Staatsministerium für Wissenschaft und Kunst and the Sächsische Aufbaubank. T.S. was supported by the Institutional Strategy of the University of Tübingen (Project ZUK63). H.G. was funded by the Bundesministerium für Bildung und Forschung (BMBF) (Grant 031A568A). T.C. was funded by the University of Lille 1. K.S. was funded by the Jane and Aatos Erkko Foundation. W.W. was funded by the BMBF (Grant 0315934). W.J.H.v.B. was supported by the Netherlands Organisation for Scientific Research (Project ACTS 053.80.713). T.W. is supported by the Novo Nordisk Foundation (Grant NNF10CC1016517). We are very grateful to A. Steck, Analytical Services & Applied NMR Development, at Bruker Biospin GmbH, Germany, for the generous support and NMR measurements. We also thank D. Wistuba and her team at the Mass Spectrometry Department, Institute for Organic Chemistry, University of Tübingen, Germany, for HR-MS measurements.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910527116/-/DCSupplemental.
References
- 1.David B., Wolfender J. L., Dias D. A., The pharmaceutical industry and natural products: Historical status and new trends. Phytochem. Rev. 14, 299–315 (2015). [Google Scholar]
- 2.Cordell G. A., Sustainable medicines and global health care. Planta Med. 77, 1129–1138 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Ekor M., The growing use of herbal medicines: Issues relating to adverse reactions and challenges in monitoring safety. Front. Pharmacol. 4, 177 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newman D. J., Cragg G. M., Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 75, 311–335 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kinghorn A. D., Pan L., Fletcher J. N., Chai H., The relevance of higher plants in lead compound discovery programs. J. Nat. Prod. 74, 1539–1555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klayman D. L., et al. , Isolation of artemisinin (qinghaosu) from Artemisia annua growing in the United States. J. Nat. Prod. 47, 715–717 (1984). [DOI] [PubMed] [Google Scholar]
- 7.Cragg G. M., Newman D. J., Natural products: A continuing source of novel drug leads. Biochim. Biophys. Acta 1830, 3670–3695 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atanasov A. G., et al. , Discovery and resupply of pharmacologically active plant-derived natural products: A review. Biotechnol. Adv. 33, 1582–1614 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miralpeix B., et al. , Metabolic engineering of plant secondary products: Which way forward? Curr. Pharm. Des. 19, 5622–5639 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Hardoim P. R., et al. , The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 79, 293–320 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kusari S., Hertweck C., Spiteller M., Chemical ecology of endophytic fungi: Origins of secondary metabolites. Chem. Biol. 19, 792–798 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Chandra S., Endophytic fungi: Novel sources of anticancer lead molecules. Appl. Microbiol. Biotechnol. 95, 47–59 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Venugopalan A., Srivastava S., Endophytes as in vitro production platforms of high value plant secondary metabolites. Biotechnol. Adv. 33, 873–887 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Crüsemann M., et al. , Heterologous expression, biosynthetic studies, and ecological function of the selective Gq-signaling inhibitor FR900359. Angew. Chem. Int. Ed. Engl. 57, 836–840 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Puri S. C., Verma V., Amna T., Qazi G. N., Spiteller M., An endophytic fungus from Nothapodytes foetida that produces camptothecin. J. Nat. Prod. 68, 1717–1719 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Stierle A., Strobel G., Stierle D., Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of Pacific yew. Science 260, 214–216 (1993). [DOI] [PubMed] [Google Scholar]
- 17.Kusari S., Singh S., Jayabaskaran C., Biotechnological potential of plant-associated endophytic fungi: Hope versus hype. Trends Biotechnol. 32, 297–303 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Yu P., et al. , Expectorant, antitussive, anti-inflammatory activities and compositional analysis of Aster tataricus. J. Ethnopharmacol. 164, 328–333 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Ng T. B., Liu F., Lu Y., Cheng C. H., Wang Z., Antioxidant activity of compounds from the medicinal herb Aster tataricus. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 136, 109–115 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Morita H., Nagashima S., Takeya K., Itokawa H., Astins A and B, antitumor cyclic pentapeptides from Aster tataricus. Chem. Pharm. Bull. (Tokyo) 41, 992–993 (1993). [DOI] [PubMed] [Google Scholar]
- 21.Cheng D., Shao Y., Terpenoid glycosides from the roots of Aster tataricus. Phytochemistry 35, 173–176 (1994). [PubMed] [Google Scholar]
- 22.Cozzolino R., et al. , Antineoplastic cyclic astin analogues kill tumour cells via caspase-mediated induction of apoptosis. Carcinogenesis 26, 733–739 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Morita H., et al. , Cyclic peptides from higher plants. XXVIII. Antitumor activity and hepatic microsomal biotransformation of cyclic pentapeptides, astins, from Aster tataricus. Chem. Pharm. Bull. (Tokyo) 44, 1026–1032 (1996). [DOI] [PubMed] [Google Scholar]
- 24.Rossi F., et al. , New antitumour cyclic astin analogues: Synthesis, conformation and bioactivity. J. Pept. Sci. 10, 92–102 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Shen Y., et al. , Mitochondria-dependent apoptosis of activated T lymphocytes induced by astin C, a plant cyclopeptide, for preventing murine experimental colitis. Biochem. Pharmacol. 82, 260–268 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Li S., et al. , The cyclopeptide astin c specifically inhibits the innate immune CDN sensor STING. Cell Rep. 25, 3405–3421.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Kosemura S., Ogawa T., Totsuka K., Isolation and structure of asterin, a new halogenated cyclic pentapeptide from Aster tataricus. Tetrahedron Lett. 34, 1291–1294 (1993). [Google Scholar]
- 28.Morita H., Nagashima S., Shirota O., Takeya K., Itokawa H., Two novel monochlorinated cyclic pentapeptides, astin D and astin E from Aster tataricus. Chem. Lett. 11, 1877–1880 (1993). [Google Scholar]
- 29.Morita H., Nagashima S., Takeya K., Itokawa H., Cyclic peptides from higher plants 8: 3 novel cyclic pentapeptides, astin F, astin G and astin H from Aster tataricus. Heterocycles 38, 2247–2252 (1994). [Google Scholar]
- 30.Morita H., Nagashima S., Takeya K., Itokawa H., A novel cyclic pentapeptide with a beta-hydroxy-gamma-chloroproline from Aster tataricus. Chem. Lett. 11, 2009–2010 (1994). [Google Scholar]
- 31.Xu H. M., et al. , Six new chlorinated cyclopentapeptides from Aster tataricus. Tetrahedron 69, 7964–7969 (2013). [Google Scholar]
- 32.Morita H., Nagashima S., Takeya K., Itokawa H., Structure of a new peptide, astin J, from Aster tataricus. Chem. Pharm. Bull. (Tokyo) 43, 271–273 (1995). [DOI] [PubMed] [Google Scholar]
- 33.Xu H. M., et al. , Tataricins A and B, two novel cyclotetrapeptides from Aster tataricus, and their absolute configuration assignment. Tetrahedron Lett. 54, 1380–1383 (2013). [Google Scholar]
- 34.Cheng D., Shao Y., Hartman R., Roder E., Zhao K., Oligopeptides from Aster tataricus. Phytochemistry 36, 945–948 (1994). [DOI] [PubMed] [Google Scholar]
- 35.Cheng D. L., Shao Y., Zhao K., Hartmann R., Roeder E., Pentapeptides from the roots of Aster tataricus. Pharmazie 51, 185–186 (1996). [PubMed] [Google Scholar]
- 36.Zhao D. X., Hu B. Q., Zhang M., Zhang C. F., Xu X. H., Simultaneous separation and determination of phenolic acids, pentapeptides, and triterpenoid saponins in the root of Aster tataricus by high-performance liquid chromatography coupled with electrospray ionization quadrupole time-of-flight mass spectrometry. J. Sep. Sci. 38, 571–575 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Liu X., Cao P., Zhang C., Xu X., Zhang M., Screening and analyzing potential hepatotoxic compounds in the ethanol extract of Asteris Radix by HPLC/DAD/ESI-MS(n) technique. J. Pharm. Biomed. Anal. 67-68, 51–62 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Li F., et al. , Design and synthesis of plant cyclopeptide Astin C analogues and investigation of their immunosuppressive activity. Bioorg. Med. Chem. Lett. 28, 2523–2527 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Schumacher K. K., et al. , First total synthesis of astin G. Tetrahedron Lett. 40, 455–458 (1999). [Google Scholar]
- 40.Tan N. H., Zhou J., Plant cyclopeptides. Chem. Rev. 106, 840–895 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Walsh C. T., O’Brien R. V., Khosla C., Nonproteinogenic amino acid building blocks for nonribosomal peptide and hybrid polyketide scaffolds. Angew. Chem. Int. Ed. Engl. 52, 7098–7124 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizutani K., Hirasawa Y., Sugita-Konishi Y., Mochizuki N., Morita H., Structural and conformational analysis of hydroxycyclochlorotine and cyclochlorotine, chlorinated cyclic peptides from Penicillium islandicum. J. Nat. Prod. 71, 1297–1300 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Jahn L., et al. , Cyanodermella asteris sp. nov. (Ostropales) from the inflorescence axis of Aster tataricus. Mycotaxon 132, 107–123 (2017). [Google Scholar]
- 44.Jahn L., et al. , Linking secondary metabolites to biosynthesis genes in the fungal endophyte Cyanodermella asteris: The anti-cancer bisanthraquinone skyrin. J. Biotechnol. 257, 233–239 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Kusari S., Zühlke S., Spiteller M., An endophytic fungus from Camptotheca acuminata that produces camptothecin and analogues. J. Nat. Prod. 72, 2–7 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Li J. Y., et al. , The induction of taxol production in the endophytic fungus–Periconia sp. from Torreya grandifolia. J. Ind. Microbiol. Biotechnol. 20, 259–264 (1998). [Google Scholar]
- 47.Pu X., et al. , Camptothecin-producing endophytic fungus Trichoderma atroviride LY357: Isolation, identification, and fermentation conditions optimization for camptothecin production. Appl. Microbiol. Biotechnol. 97, 9365–9375 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Staniek A., Woerdenbag H. J., Kayser O., Taxomyces andreanae: A presumed paclitaxel producer demystified? Planta Med. 75, 1561–1566 (2009). [DOI] [PubMed] [Google Scholar]
- 49.van Nieuwenhuijzen E. J., et al. , Wood staining fungi revealed taxonomic novelties in Pezizomycotina: New order Superstratomycetales and new species Cyanodermella oleoligni. Stud. Mycol. 85, 107–124 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Itokawa H., Takeya K., Hitotsuyanagi Y., Morita H., “Antitumor compounds isolated from higher plants” in Studies in Natural Products Chemistry (Part E), Atta-ur-Rahman, Ed. (Elsevier, 2000), vol. 24, pp. 269–350. [Google Scholar]
- 51.Schafhauser T., et al. , The cyclochlorotine mycotoxin is produced by the nonribosomal peptide synthetase CctN in Talaromyces islandicus (‘Penicillium islandicum’). Environ. Microbiol. 18, 3728–3741 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Gao X., et al. , Cyclization of fungal nonribosomal peptides by a terminal condensation-like domain. Nat. Chem. Biol. 8, 823–830 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kittilä T., et al. , Halogenation of glycopeptide antibiotics occurs at the amino acid level during non-ribosomal peptide synthesis. Chem. Sci. (Camb.) 8, 5992–6004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dorrestein P. C., Yeh E., Garneau-Tsodikova S., Kelleher N. L., Walsh C. T., Dichlorination of a pyrrolyl-S-carrier protein by FADH2-dependent halogenase PltA during pyoluteorin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 102, 13843–13848 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin S., Van Lanen S. G., Shen B., Regiospecific chlorination of (S)-beta-tyrosyl-S-carrier protein catalyzed by SgcC3 in the biosynthesis of the enediyne antitumor antibiotic C-1027. J. Am. Chem. Soc. 129, 12432–12438 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caradec T., et al. , Prediction of monomer isomery in florine: A workflow dedicated to nonribosomal peptide discovery. PLoS One 9, e85667 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neumann C. S., Fujimori D. G., Walsh C. T., Halogenation strategies in natural product biosynthesis. Chem. Biol. 15, 99–109 (2008). [DOI] [PubMed] [Google Scholar]
- 58.van Pée K. H., Enzymatic chlorination and bromination. Methods Enzymol. 516, 237–257 (2012). [DOI] [PubMed] [Google Scholar]
- 59.Weichold V., Milbredt D., van Pée K. H., Specific enzymatic halogenation–From the discovery of halogenated enzymes to their applications in vitro and in vivo. Angew. Chem. Int. Ed. Engl. 55, 6374–6389 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Bode H. B., Bethe B., Höfs R., Zeeck A., Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem 3, 619–627 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Newman D. J., Cragg G. M., Endophytic and epiphytic microbes as “sources” of bioactive agents. Front Chem. 3, 34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Piel J., Metabolites from symbiotic bacteria. Nat. Prod. Rep. 26, 338–362 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Martín J. F., Liras P., Evolutionary formation of gene clusters by reorganization: The meleagrin/roquefortine paradigm in different fungi. Appl. Microbiol. Biotechnol. 100, 1579–1587 (2016). [DOI] [PubMed] [Google Scholar]
- 64.Bártíková H., et al. , Xenobiotic-metabolizing enzymes in plants and their role in uptake and biotransformation of veterinary drugs in the environment. Drug Metab. Rev. 47, 374–387 (2015). [DOI] [PubMed] [Google Scholar]
- 65.Berthiller F., et al. , Masked mycotoxins: A review. Mol. Nutr. Food Res. 57, 165–186 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Riechers D. E., Kreuz K., Zhang Q., Detoxification without intoxication: Herbicide safeners activate plant defense gene expression. Plant Physiol. 153, 3–13 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scherlach K., Busch B., Lackner G., Paszkowski U., Hertweck C., Symbiotic cooperation in the biosynthesis of a phytotoxin. Angew. Chem. Int. Ed. Engl. 51, 9615–9618 (2012). [DOI] [PubMed] [Google Scholar]
- 68.Kusari S., Zühlke S., Spiteller M., Effect of artificial reconstitution of the interaction between the plant Camptotheca acuminata and the fungal endophyte Fusarium solani on camptothecin biosynthesis. J. Nat. Prod. 74, 764–775 (2011). [DOI] [PubMed] [Google Scholar]
- 69.Kusari P., et al. , Cross-species biosynthesis of maytansine in Maytenus serrata. RSC Adv. 6, 10011–10016 (2016). [Google Scholar]
- 70.Saviano G., et al. , Influence of conformational flexibility on biological activity in cyclic astin analogues. Biopolymers 76, 477–484 (2004). [DOI] [PubMed] [Google Scholar]
- 71.Kusari S., Singh S., Jayabaskaran C., Rethinking production of Taxol® (paclitaxel) using endophyte biotechnology. Trends Biotechnol. 32, 304–311 (2014). [DOI] [PubMed] [Google Scholar]
- 72.White T. J., Bruns T., Lee S., Taylor J. W., “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics” in PCR Protocols: A Guide to Methods and Applications, Innis M. A., Gelfand D. H., Sninsky J. J., White T. J., Eds. (Academic, New York, 1990), pp. 315–322. [Google Scholar]
- 73.Gardes M., Bruns T. D., ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118 (1993). [DOI] [PubMed] [Google Scholar]
- 74.Möller E. M., Bahnweg G., Sandermann H., Geiger H. H., A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res. 20, 6115–6116 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fitze D., Wiepning A., Kaldorf M., Ludwig-Müller J., Auxins in the development of an arbuscular mycorrhizal symbiosis in maize. J. Plant Physiol. 162, 1210–1219 (2005). [DOI] [PubMed] [Google Scholar]
- 76.Heberling M. M., et al. , Ironing out their differences: Dissecting the structural determinants of a phenylalanine aminomutase and ammonia lyase. ACS Chem. Biol. 10, 989–997 (2015). [DOI] [PubMed] [Google Scholar]
- 77.Fotheringham I. G., Grinter N., Pantaleone D. P., Senkpeil R. F., Taylor P. P., Engineering of a novel biochemical pathway for the biosynthesis of L-2-aminobutyric acid in Escherichia coli K12. Bioorg. Med. Chem. 7, 2209–2213 (1999). [DOI] [PubMed] [Google Scholar]
- 78.Ye Y., et al. , Unveiling the biosynthetic pathway of the ribosomally synthesized and post-translationally modified peptide ustiloxin B in filamentous fungi. Angew. Chem. Int. Ed. Engl. 55, 8072–8075 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this article and SI Appendix.