Significance
Complete hydatidiform moles (CHMs) develop from androgenetic conceptuses and are characterized by enhanced proliferation of trophoblast cells and a significantly higher risk of trophoblast tumors. Loss of the maternal genome and duplication of the paternal genome are considered to be responsible for the phenotype, but the detailed mechanism remains unclear. Here, we report the derivation of trophoblast stem (TS) cells from CHMs. These cells have reduced sensitivity to contact inhibition of cell proliferation and exhibit aberrant expression of imprinted genes, which are expressed from only 1 parental allele. We also reveal that the maternally expressed imprinted gene p57KIP2 would be responsible for the enhanced proliferation of CHM-derived TS cells. Our findings provide an insight into the pathogenesis of CHMs.
Keywords: complete hydatidiform mole, genomic imprinting, trophoblast stem cells, p57KIP2, choriocarcinoma
Abstract
A complete hydatidiform mole (CHM) is androgenetic in origin and characterized by enhanced trophoblastic proliferation and the absence of fetal tissue. In 15 to 20% of cases, CHMs are followed by malignant gestational trophoblastic neoplasms including choriocarcinoma. Aberrant genomic imprinting may be responsible for trophoblast hypertrophy in CHMs, but the detailed mechanisms are still elusive, partly due to the lack of suitable animal or in vitro models. We recently developed a culture system of human trophoblast stem (TS) cells. In this study, we apply this system to CHMs for a better understanding of their molecular pathology. CHM-derived TS cells, designated as TSmole cells, are morphologically similar to biparental TS (TSbip) cells and express TS-specific markers such as GATA3, KRT7, and TFAP2C. Interestingly, TSmole cells have a growth advantage over TSbip cells only after they reach confluence. We found that p57KIP2, a maternally expressed gene encoding a cyclin-dependent kinase inhibitor, is strongly induced by increased cell density in TSbip cells, but not in TSmole cells. Knockout and overexpression studies suggest that loss of p57KIP2 expression would be the major cause of the reduced sensitivity to contact inhibition in CHMs. Our findings shed light on the molecular mechanism underlying the pathogenesis of CHMs and could have broad implications in tumorigenesis beyond CHMs because silencing of p57KIP2 is frequently observed in a variety of human tumors.
A complete hydatidiform mole (CHM) is a gestational trophoblastic disease characterized by enhanced trophoblast proliferation, swollen villi, and the absence of embryonic components (1). Whereas the core mesenchyme of normal villi is surrounded by a single layer of cytotrophoblast (CT) cells, CHM villi contain multiple layers (2). Although CHMs are benign in most cases, 15 to 20% of them are followed by malignant gestational trophoblastic neoplasms, including invasive mole and choriocarcinoma (3). CHMs develop from androgenetic conceptuses (4, 5) and can be placed into 2 classes: monospermic and dispermic. The majority of CHMs are monospermic and arise from fertilization of an anucleate egg with a haploid sperm, followed by endoredupliction. Dispermic CHMs account for 4 to 15% of CHMs and develop through fertilization of an anucleate egg with 2 sperm (6).
Whereas most autosomal genes are expressed from both parental alleles, a small subset of genes, known as imprinted genes, are exclusively expressed from 1 parental allele (7). To date, more than 100 imprinted genes have been identified in humans (8), and many of them are expressed in the placenta (9). Aberrant genomic imprinting may be responsible for the pathogenesis of CHMs. However, it remains uncertain which imprinted gene(s) are involved in the overgrowth of trophoblast cells in CHMs, partly due to the lack of suitable animal or in vitro models. For example, androgenetic mouse embryos exhibit severe growth retardation and early lethality. The extraembryonic tissues develop relatively well in these embryos but do not give rise to malignant trophoblastic neoplasms (10, 11). Moreover, some immortalized cell lines have been established from CHMs by transducing oncogenes such as human telomerase reverse transcriptase (12, 13), but forced expression of oncogenes affects cell proliferation and can mask the phenotype of CHMs.
Recently, we have succeeded in establishing human trophoblast stem (TS) cells from cytotrophoblast (CT) cells isolated from first-trimester placentas (14). Here, we apply this culture system to CHMs and reveal that CHM-derived TS cells exhibit resistance to contact inhibition, which may be accounted for by loss of p57KIP2 (CDKN1C) expression. These findings are fundamental to understanding the pathogenesis of CHMs and normal placental development.
Results
Establishment and Molecular Characterization of TS Cells from CHMs.
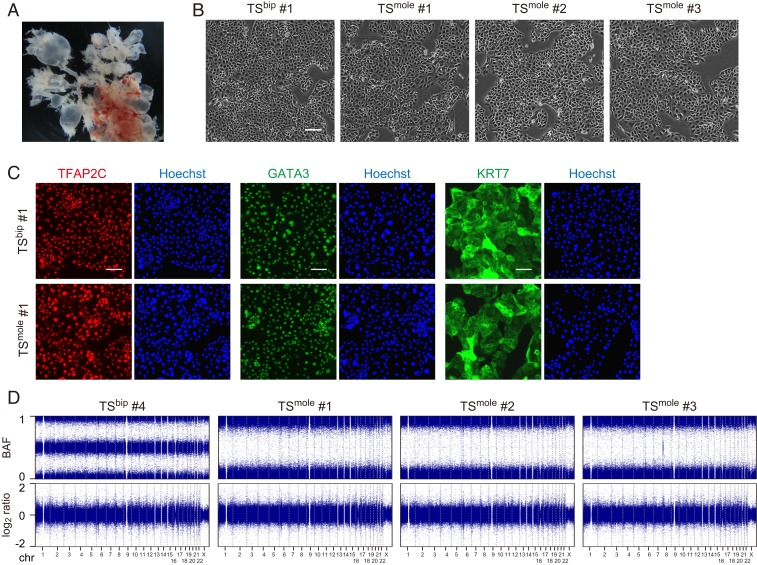
We isolated CT cells from 5 CHM samples and established TS cell lines (Fig. 1A and SI Appendix, Table S1). These cells, designated as TSmole cells, were morphologically similar to biparental TS (TSbip) cells (Fig. 1B) and expressed trophoblast markers such as TFAP2C, GATA3, and KRT7 (Fig. 1C). We performed copy number variation (CNV) analysis of 3 TSmole cell lines with the Japonica array, which is a SNP array optimized for the Japanese population, and revealed genome-wide loss of heterogeneity (Fig. 1D). Thus, these 3 lines may be derived from monospermic fertilization.
Fig. 1.
Establishment of TSmole cells. (A) Representative CHM image. Swollen villi are visible. (B) Phase-contrast images of TSbip and TSmole cells. (C) Immunostaining of TFAP2C, GATA3, and KRT7. Nuclei were counterstained with Hoechst 33258. (D) CNV analysis of TSbip and TSmole cells. B allele frequency (BAF) and log2 copy number ratio (log2 ratio) are shown. Genome-wide loss of heterogeneity was observed in TSmole cell lines without copy number changes. (Scale bars: B, 200 µm; C, 100 µm.)
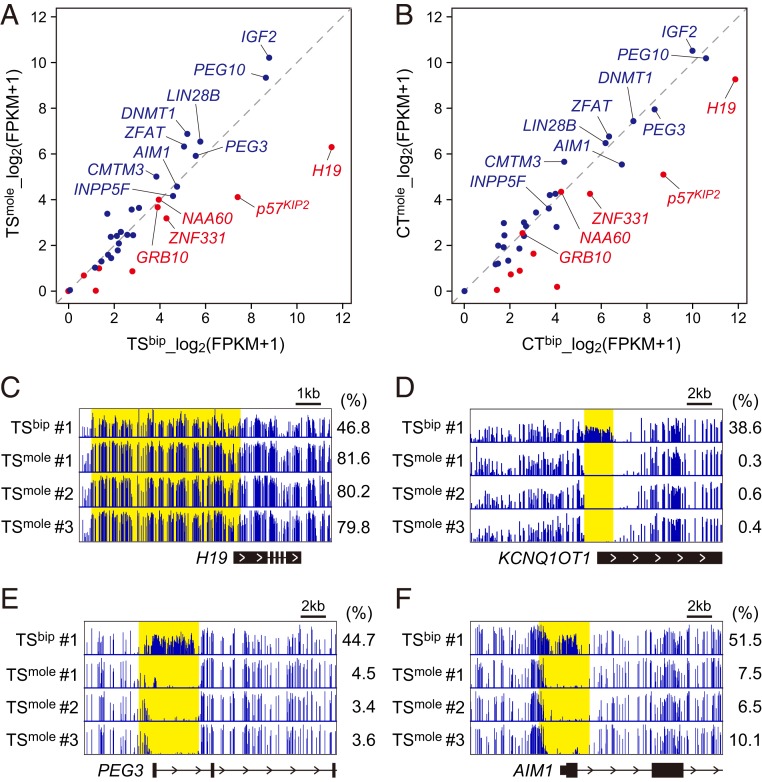
To further characterize TSmole cells, we performed RNA sequencing (RNA-seq) and whole genome bisulfite sequencing (WGBS) on 3 TSmole cell lines. RNA-seq and WGBS data for TSbip cells were obtained from our previous study and used for comparison (14). TSmole and TSbip cells had very similar transcriptome and methylome profiles, and the expression levels of differentiation markers were low in these cells (SI Appendix, Fig. S1 and Dataset S1). However, expression and DNA methylation of imprinted genes were disturbed in TSmole cells (Fig. 2A and Dataset S2). We focused on 10 maternally expressed and 15 paternally expressed genes that maintain allele-specific expression in primary CT cells (15). Among the maternally expressed genes, H19 and p57KIP2 were expressed at the lowest level in TSmole cells compared to TSbip cells. Consistently, the paternally methylated H19 DMR was hypermethylated and the maternally methylated KvDMR1 was unmethylated in TSmole cells (Fig. 2 C and D). KvDMR1 overlaps with the promoter of the paternally expressed noncoding RNA KCNQ1OT1, which may be involved in the repression of neighboring genes such as KCNQ1 and p57KIP2 (16). However, several maternally expressed genes, such as NAA60 and GRB10, showed comparable expression levels in TSmole and TSbip cells (Fig. 2A). This was unlikely to be due to an artifact of in vitro culture because the expression levels of NAA60 and GRB10 were also similar between primary CT cells isolated from CHMs and normal placentas (Fig. 2B).
Fig. 2.
RNA-seq and WGBS of TSmole cells. (A) Expression levels of imprinted genes in TSbip and TSmole cells. Three lines were analyzed for each cell type, and mean expression levels are shown. Maternally and paternally expressed genes are represented in red and blue, respectively. Genes with >10 FPKM in TSbip cells are labeled with their gene names. (B) Expression levels of imprinted genes in CT cells isolated from biparental placentas (CTbip) and CHMs (CTmole). Three CTbip and two CTmole samples were analyzed, and mean expression levels are shown. Genes are labeled as in A. (C) DNA methylation patterns at the H19 DMR. The H19 DMR is shown in yellow, and its methylation levels are indicated on the right. (D) DNA methylation patterns at KvDMR1 (yellow). (E) DNA methylation patterns at the PEG3 DMR (yellow). (F) DNA methylation patterns at the AIM1 DMR (yellow). See also Datasets S1 and S2.
We also revealed that paternally expressed genes with high expression levels in TSbip cells tended to show increased expression in TSmole cells (Fig. 2A). However, several genes, including PEG3 and AIM1, did not show increased expression in TSmole cells, which was inconsistent with the methylation patterns of their regulatory elements (Fig. 2 E and F). Intriguingly, increased expression of paternally expressed genes was less apparent in primary CT cells isolated from CHMs (Fig. 2B). Although we analyzed only 2 CT samples from CHMs and more samples are needed to draw a firm conclusion, these data suggest that there may be a compensatory mechanism whereby the expression levels of some imprinted genes are normalized in CHMs. Such a mechanism might also work in TSmole cells but to a lesser extent.
Resistance to Contact Inhibition in TSmole Cells.
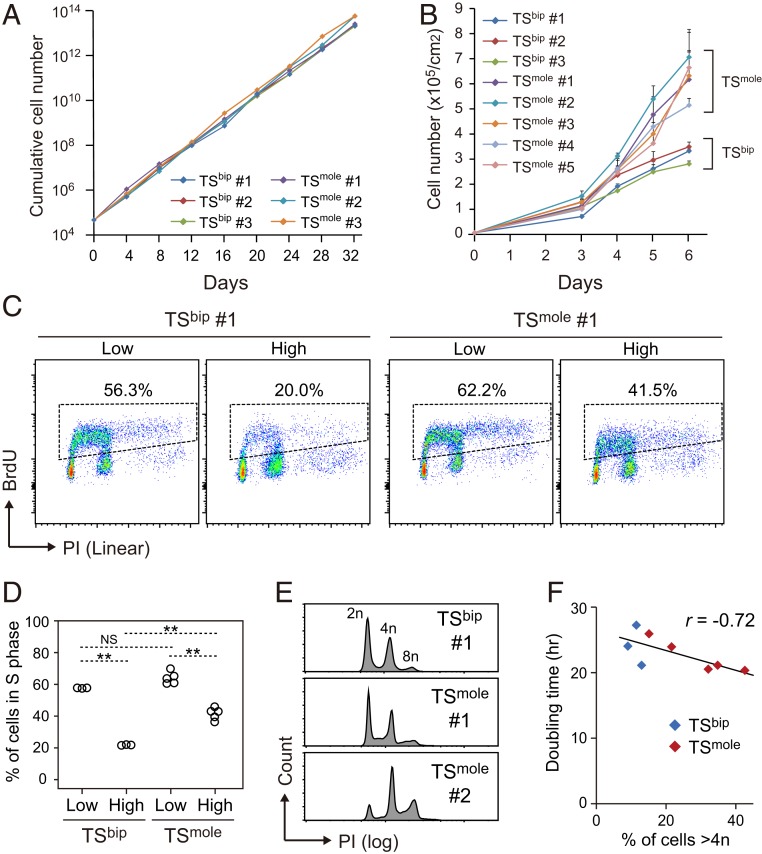
Since CHMs are characterized by trophoblast hypertrophy, we analyzed the proliferation rate of TSmole cells. However, TSmole cells had a proliferation rate comparable to TSbip cells under optimal conditions where the cells were passaged before they reached confluence (Fig. 3A). It has been recognized that although the core mesenchyme of normal villi is surrounded by a single layer of CT cells, CHM villi contain multiple layers (2). This implies that trophoblast cells of CHMs might be resistant to contact inhibition of proliferation. Consistent with this, we found that TSmole cells had a significant growth advantage over TSbip cells after they reached confluence (Fig. 3B).
Fig. 3.
Reduced sensitivity to contact inhibition in TSmole cells. (A) Cumulative growth curves of TSbip and TSmole cells. Three TSbip and three TSmole cell lines were seeded at a density of 5,000 cells/cm2. The cells were passaged when they reached ∼80% confluence and counted every 4 d. (B) Growth curves of TSbip and TSmole cells. Three TSbip and five TSmole cell lines were analyzed. Cells were seeded at a density of 10,000 cells/cm2 and maintained for 6 d without passaging. TSmole cells had significantly higher cell numbers than TSbip at day 6 (P < 0.01; Student’s t test). Data are presented as means + SDs (n = 3). (C) Cell cycle analysis of TSbip and TSmole cells. Cells were seeded at a density of 5,000 (Low) or 100,000 cells/cm2 (High). After 2 d of culture, the cells were labeled with BrdU for an hour and analyzed by flow cytometry. Nuclei were stained with PI. The proportions of cells in S phase are indicated. (D) Summary of the cell cycle analysis in C. Three TSbip and five TSmole cell lines were analyzed. Statistical analysis was performed by the Bonferroni/Dunn test. **P < 0.01; NS, not significant. (E) DNA ploidy analysis of TSbip and TSmole cells. DNA ploidy was measured using cells that were seeded at a high density (100,000 cells/cm2) and cultured for 2 d. (F) Relationship between DNA ploidy and cell doubling time. Three TSbip and five TSmole cell lines were analyzed and are shown in blue and red, respectively. A negative correlation was observed between cell doubling time during the exponential phase and the proportion of cells >4n, which was calculated from the data in E. Pearson’s r was −0.72 (P = 0.02) when all samples were considered and −0.95 (P = 0.05) when only TSmole samples were considered.
Cell cycle analysis using flow cytometry revealed that Bromodeoxyuridine (BrdU)-labeled S phase cells dramatically decreased when TSbip cells were cultured at a high cell density (Fig. 3C). Both G1 and G2 arrest were responsible for the reduction of S phase cells (SI Appendix, Fig. S2 A and B). Cell density-dependent reduction of S phase cells was also observed in TSmole cells, but to a much lesser extent than TSbip cells (Fig. 3 C and D), reinforcing the idea that TSmole cells have reduced sensitivity to contact inhibition. The cell cycle analysis also revealed that some TSmole cell lines contained substantial proportions of cells >4n (Fig. 3E and SI Appendix, Table S1). As cells >8n were negligible, these lines were thought to contain mitotically active tetraploid cells (SI Appendix, Fig. S2C). The proportion of cells >4n was marginally reduced by increased cell density (SI Appendix, Fig. S2D). We found a significant relationship (r = −0.72, P = 0.02) between the proportion of cells >4n and cell doubling time (Fig. 3F), but the sample size was small, and further work is required to establish the negative correlation.
Identification of p57KIP2 as a Regulator of Contact Inhibition in TS Cells.
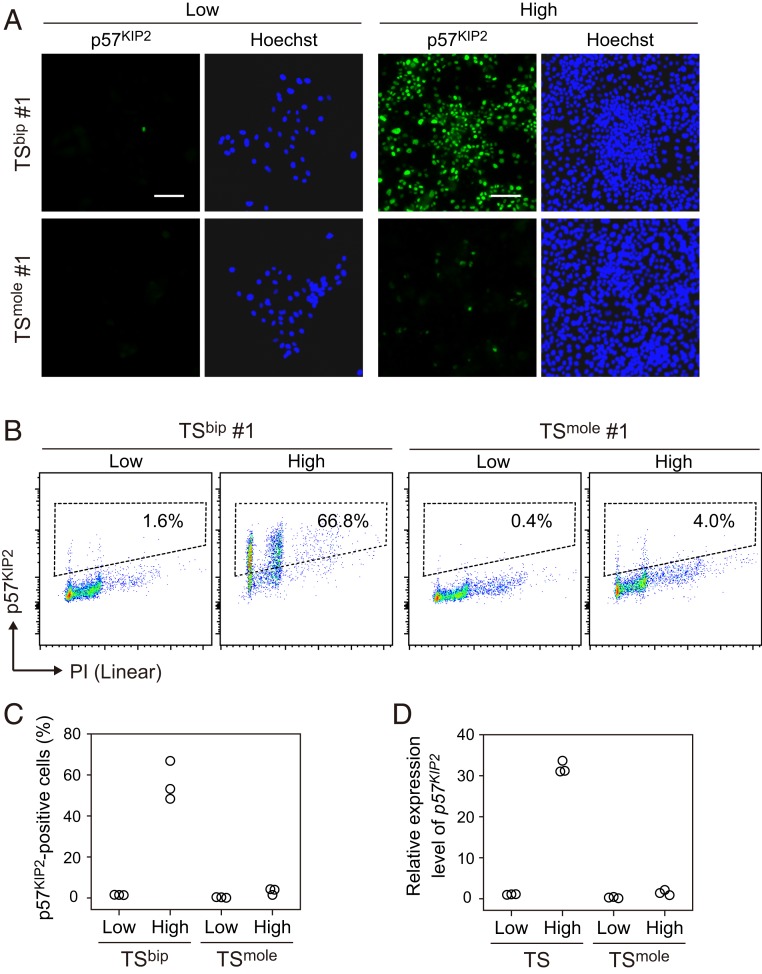
We were next interested in why TSmole cells had reduced sensitivity to contact inhibition. Abnormal expression of imprinted gene(s) is considered to be involved in this phenotype. We focused on p57KIP2 because this gene functions as a cyclin-dependent kinase inhibitor and was expressed at a very low level in TSmole cells (Fig. 2A). Moreover, previous studies have revealed that p57KIP2 expression is absent or very low in CT cells of CHMs and the majority of choriocarcinoma samples associated with CHMs (17, 18). Although few TSbip cells expressed p57KIP2 when they were cultured at a low density, p57KIP2 was strongly induced when cultured at a high density (Fig. 4 A–C). In contrast, p57KIP2-positive cells were almost absent in TSmole cells regardless of cell density. We also observed a ∼30-fold induction of p57KIP2 mRNA in TSbip cells cultured at a high density (Fig. 4D). Among the 14 imprinted genes labeled in Fig. 2A, p57KIP2 was most strongly induced by high cell density (Fig. 4D and SI Appendix, Fig. S3). These data suggest that transcriptional regulation may be an important determinant of the abundance of p57KIP2 in TSbip cells, although we do not exclude the possibility of posttranscriptional regulation.
Fig. 4.
Cell density-dependent induction of p57KIP2 in TSbip cells but not in TSmole cells. (A) Immunostaining images of p57KIP2 in TSbip and TSmole cells. Cells were seeded at a density of 5,000 (Low) or 100,000 cells/cm2 (High). After 2 d of culture, cells were analyzed. Nuclei were stained with Hoechst 33258. (Scale bar: 100 µm.) (B) Flow cytometry analysis of p57KIP2 in TSbip and TSmole cells. Cells cultured as shown in A were analyzed. Nuclei were stained with PI. The proportions of p57KIP2-positive cells are indicated. (C) Summary of flow cytometry analysis of p57KIP2 in B. Three TSbip and three TSmole cell lines were analyzed. (D) Expression levels of p57KIP2 in TSbip and TSmole cells. Three TSbip and three TSmole cell lines were cultured as shown in A and analyzed by quantitative real-time PCR. The data were normalized to GAPDH and B2M.
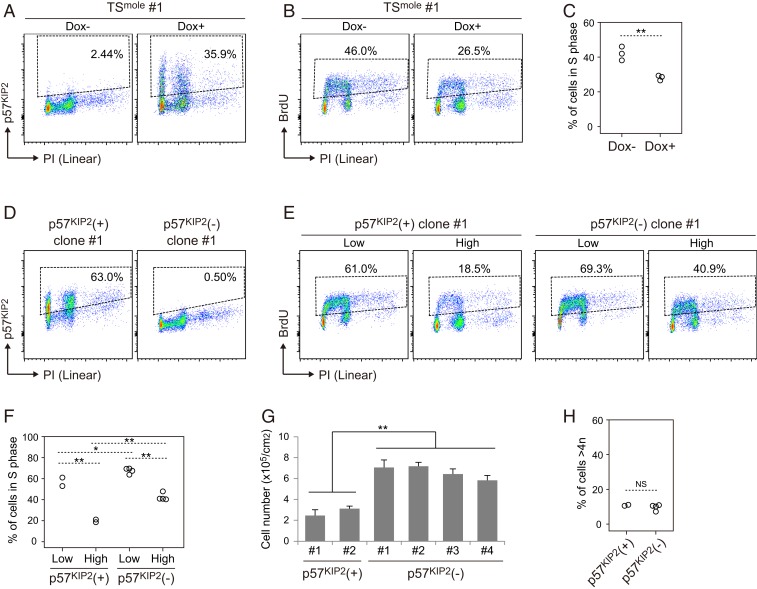
To determine whether loss of p57KIP2 expression is responsible for the reduced susceptibility to contact inhibition in TSmole cells, we utilized the doxycycline (Dox)-inducible Tet-on system (Fig. 5A). Three TSmole cell lines with Dox-inducible p57KIP2 were cultured at a high density, and their cell cycle was analyzed by flow cytometry. The fluorescence intensity of p57KIP2 induced by Dox was comparable to that observed in TSbip cells cultured at a high density (SI Appendix, Fig. S4A). We found that p57KIP2 induction significantly reduced the proportion of cells in S phase (Fig. 5 B and C), which supports the idea that loss of p57KIP2 confers resistance to contact inhibition in TSmole cells. To reinforce this idea, we generated p57KIP2 knockout TS cells using the CRISPR-Cas9 system. We transfected 1 TSbip cells line with lentivirus-expressing Cas9 and gRNA and isolated 14 clones. Two of these were p57KIP2-positive and the others were p57KIP2-negative (Fig. 5D). One of the p57KIP2-positive clones was wild type, and the other was heterozygous, implying that the paternal allele was mutated (SI Appendix, Fig. S4 B and C). These 2 lines were used as controls. We randomly selected 4 of the p57KIP2-negative clones, which were all confirmed to be homozygous knockouts (SI Appendix, Fig. S4C), and analyzed their proliferation. We found that at a high cell density, the proportion of S phase cells was much higher in p57KIP2-negative clones than in p57KIP2-positive clones (Fig. 5 E and F). Consistently, p57KIP2-negative clones had higher proliferative activity than p57KIP2-positive clones after reaching confluence (Fig. 5G). These data reveal that, similar to TSmole cells, p57KIP2 knockout TS cells have reduced sensitivity to contact inhibition. However, the proportion of cells >4n was comparable between p57KIP2-negative and p57KIP2-positive clones, suggesting that loss of p57KIP2 may not be the cause of the genome amplification in TSmole cells (Fig. 5H).
Fig. 5.
Identification of p57KIP2 as a regulator of contact inhibition in TS cells. (A) Overexpression of p57KIP2 in TSmole cells. TSmole cells with Dox-inducible p57KIP2 were seeded at a high density (100,000 cells/cm2) and cultured for 24 h. Then, the cells were treated with 20 ng/mL Dox for 24 h, and p57KIP2 expression was analyzed by flow cytometry. Nuclei were stained with PI. The proportions of p57KIP2-positive cells are indicated. Similar results were obtained with 3 independent TSmole cell lines. (B) p57KIP2-induced reduction of S phase cells. Cells were cultured as shown in A and analyzed by flow cytometry after BrdU treatment. Nuclei were stained with PI. The proportions of cells in S phase are indicated. (C) Summary of the cell cycle analysis in B. Three TSbip and three TSmole cell lines were analyzed. Statistical analysis was performed by Student’s t test. **P < 0.01. (D) Confirmation of loss of p57KIP2 expression in p57KIP2 knockout clones. Cells were seeded at a high density (100,000 cells/cm2). After 2 d of culture, p57KIP2 expression was analyzed by flow cytometry. The proportions of p57KIP2-positive cells are indicated. Representative p57KIP2-positive (+) and p57KIP2-negative (−) clones are shown. (E) Cell cycle analysis of p57KIP2(+) and p57KIP2(−) clones. Cells were seeded at a low (5,000 cells/cm2) or high density (100,000 cells/cm2). After 2 d of culture, the cells were labeled with BrdU and analyzed by flow cytometry. The proportions of cells in S phase are indicated. (F) Summary of the cell cycle analysis in E. Two p57KIP2(+) and four p57KIP2(−) clones were analyzed. Statistical analysis was performed by the Bonferroni/Dunn test. **P < 0.01; *P < 0.05. (G) Reduced sensitivity to contact inhibition in p57KIP2(−) clones. Cells were seeded at a density of 10,000 cells/cm2 as in Fig. 3B and maintained for 5 d without passaging. TSmole cells had significantly higher cell numbers than TSbip cells (P < 0.05; Student’s t test). Data are presented as means + SDs (n = 3). (H) Proportions of cells >4n in p57KIP2(+) and p57KIP2(−) clones. Two p57KIP2(+) and four p57KIP2(−) clones were analyzed. Statistical analysis was performed by Student’s t test. NS, not significant.
Discussion
Previous studies on mouse androgenetic cell lines have revealed important roles of genomic imprinting in cell proliferation. Similar to TSmole cells, androgenetic mouse embryonic fibroblasts (MEFs) have increased saturation density (19). Interestingly, loss of p57KIP2 expression may not be involved in the enhanced proliferation of androgenetic MEFs, because knockout of p57KIP2 has no effect on the proliferation of biparental MEFs. Alternatively, increased Igf2 and loss of Igf2r, which is a maternally expressed imprinted gene and encodes a decoy receptor for IGF2, seem to stimulate the proliferation of androgenetic MEFs. It is also known that androgenetic mouse TS cells exhibit enhanced proliferation activity. Whereas biparental mouse TS cells require Fgf4 for their proliferation, androgenetic mouse TS cells continue to proliferate for up to 6 d in the absence of Fgf4 (20). Again, p57KIP2 is unlikely to be the major cause of the Fgf4-independent proliferation because p57KIP2 is expressed at similar levels in biparental and androgenetic mouse TS cells. Gab1, which is paternally expressed in the mouse placenta but not imprinted in the human placenta (21), is proposed to be a candidate gene for the Fgf4-independent proliferation (22). These findings suggest that although androgenesis is associated with enhanced cell proliferation, the underlying mechanisms may vary among tissues and species.
p57KIP2 is a putative tumor suppressor gene and most highly expressed in the placenta in both humans and mice (23, 24). Consistent with the function and expression pattern, p57KIP2-deficient mice exhibit placentomegaly (25, 26) and loss of p57KIP2 function is associated with Beckwith–Wiedemann syndrome (BWS) in humans, which is characterized by overgrowth of multiple organs including the placenta (16, 27). However, it has been unclear how the expression and function of p57KIP2 are regulated in the placenta. We showed that p57KIP2 is strongly induced by increased cell density in TSbip cells. This observation is consistent with the expression pattern of p57KIP2 in the placenta. The core mesenchyme of placental villi is surrounded by a single layer of proliferative CT cells that sporadically express p57KIP2 (28). At the tip of villi, CT cells form stratified aggregates known as cell columns, where they exit cell cycle and start to differentiate into extravillous cytotrophoblast (EVT) cells (29). p57KIP2 is strongly expressed in most CT cells that exit cell cycle and also in EVT cells (28). Therefore, increased cell density might be a trigger of p57KIP2 in vivo too. Our data strongly suggest that p57KIP2 is involved in cell cycle arrest in TS cells, but further studies are needed to determine whether p57KIP2 regulates permanent cell cycle exit and EVT differentiation.
Loss of p57KIP2 is observed in both CHMs and most BWS patients (16). Since malignant gestational trophoblastic neoplasms are not clinical characteristics of BWS, loss of p57KIP2 alone cannot account for the high risk of malignancy in CHMs. Polyploidy could be another risk factor. We found that some TSmole cell lines contain high proportions of cells >4n, and the experiments presented suggest that loss of function of p57KIP2 is not responsible for that phenotype. Polyploid cells are also observed in a subset of CHMs in vivo, and such CHMs are suspected to be prone to malignant gestational trophoblastic neoplasms (30, 31). Consistently, most invasive moles and choriocarcinomas exhibit genome amplification (32, 33). However, the frequency of CHMs containing polyploid cells is highly variable among studies, ranging from 2 to 30%, which may be due to inconsistent diagnostic criteria (34, 35). More studies using standardized criteria are needed to determine whether polyploidy is associated with malignant transformation in CHMs. Also, the mechanism underlying the polyploidization remains unknown.
In conclusion, we established and characterized TSmole cells, which led to the identification of p57KIP2 as a regulator of contact inhibition. p57KIP2 is required for normal development of multiple organs, and loss of methylation of KvDMR1, which results in loss of p57KIP2 expression, is common in human adult tumors (36). Therefore, our findings could have broad implications in embryogenesis and tumorigenesis beyond CHMs.
Materials and Methods
Sample Collection.
Placental samples were collected from patients after written informed consent was obtained. Experienced pathologists confirmed the diagnosis of CHMs. This study was approved by the Ethics Committee of Tohoku University Graduate School of Medicine (Research license 2017-1-349).
Culture of TS Cells.
TSmole cells were established as described previously (14). Briefly, CT cells were isolated from CHM tissues and cultured on plates coated with 5–10 µg/mL Col IV (Corning) using TS medium (DMEM/F12 [Wako] supplemented with 0.1 mM 2-mercaptoethanol [Wako], 0.2% FBS [Thermo Fisher Scientific], 0.5% Penicillin-Streptomycin [Thermo Fisher Scientific], 0.3% BSA [Wako], 1% ITS-X supplement [Wako], 1.5 μg/mL l-ascorbic acid [Wako], 50 ng/mL EGF [Wako], 2 μM CHIR99021 [Wako], 0.5 μM A83-01 [Wako], 1 μM SB431542 [Wako], 0.8 mM VPA [Wako], and 5 μM Y27632 [Wako]). TSbip #1, #2, and #3 were established in our previous study (14) and correspond to TSCT #1, #2, and #3, respectively. TSbip #4 was established in this study and used only for CNV analysis. Unless otherwise noted, we used TSmole and TSbip cells passaged 10–20 times for the analysis.
Overexpression of p57KIP2.
Vectors for Dox-inducible p57KIP2 expression were constructed as follows. The Tet-On 3G transactivator of the pTetOne vector (Takara) was PCR-amplified and cloned into the multicloning site of the CS-CA-MCS plasmid (kindly provided by H. Miyoshi, RIKEN BioResource Center, Ibaraki, Japan) using the In-Fusion HD Cloning kit (Takara). The resulting vector was designated as pCS-CA-Tet3G. The CAG promoter of CS-CA-MCS was replaced with the TRE3Gs promoter of pTetOne. The resulting vector was designated as pCS-3G. The coding region of p57KIP2 was PCR-amplified from cDNA prepared from TSbip cells and cloned into pCS-3G to generate pCS-3G-p57KIP2. Sequences of the primers used for vector construction are shown in Dataset S3.
To generate lentivirus expressing Tet-On 3G, pCS-CA-Tet3G was cotransfected with pCMV-VSV-G-RSV-Rev and pCAG-HIVgp (kindly provided by H. Miyoshi, RIKEN BioResource Center) into 293T cells using Lipofectamine LTX (Thermo Fisher Scientific). Ten micomolar Forskolin (Wako) was added after 24 h of transfection. The supernatant was collected after 3 d of transfection, concentrated with Lenti-X Concentrator (Takara), and stored at −80 °C. Lentivirus-expressing p57KIP2 was also generated using pCS-3G-p57KIP2. TSmole cells harboring Dox-inducible p57KIP2 were generated by adding lentivirus-expressing Tet-On 3G and p57KIP2 to the culture medium. p57KIP2 expression was induced by adding 20 ng/mL Dox for 24 h.
Knockout of p57KIP2.
Cas9 was amplified with PCR from the Alt-R S.p. Cas9 Expression Plasmid (IDT) and cloned into pCS-3G to generate pCS-3G-Cas9. The CAG promoter of CS-CA-MCS was replaced with the human U6 (hU6) promoter, and the gRNA scaffold sequence was cloned downstream of the hU6 promoter. The resulting vector was designated as pCS-hU6. A gRNA target sequence for p57KIP2 (5′-GGC GAC AAG ACG CTC CAT CG-3′), which was designed and evaluated using CHOPCHOP (37), was cloned between the hU6 promoter and the gRNA scaffold to generate pCS-hU6-p57KIP2.
Lentivirus-expressing Cas9 and the gRNA was prepared as described above and transduced into TSbip cells. Cas9 was transiently expressed by adding 100 ng/mL Dox for 24 h, and single cells were cloned by limiting dilution. During lentivirus transduction and single-cell cloning, we cultured TS cells on plates coated with 0.5 µg/mL iMatrix-511 (Nippi) using a modified TS medium (DMEM/F12 supplemented with 1% KSR [Thermo Fisher Scientific], 0.5% Penicillin-Streptomycin, 0.15% BSA, 1% ITS-X supplement, 200 µM l-ascorbic acid, 50 ng/mL EGF, 2 μM CHIR99021, 5 μM A83-01, 0.8 mM VPA, and 2.5 μM Y27632). This medium ameliorated the toxicity of lentivirus and improved cloning efficiency. Obtained clones were maintained in the original TS medium and analyzed by flow cytometry and Sanger sequencing. Sequences of the primers used for vector construction and Sanger sequencing are shown in Dataset S3.
External Data.
The RNA-seq and WGBS data for TSbip (Japanese Genotype-phenotype Archive [JGA] accession no. JGAS00000000117) and CTbip, EVTbip, and SynTbip (JGA accession no. JGAS00000000038) were from our previous studies (14, 15).
Statistical Analyses.
Results are presented as means + SDs. The statistical methods used are described in the figure legends. A P value of less than 0.05 was considered statistically significant. P values less than 0.05 and 0.01 were marked by 1 and 2 asterisks, respectively. Statistical analysis was performed using Statcel software (OMS) or R (version 3.3.1).
Other Methods.
The procedures for immunostaining, CNV analysis, RNA-seq, real-time PCR, WGBS, and flow cytometory are provided in SI Appendix.
Data Availability.
All sequencing data reported in this paper are available at JGA (accession no. JGAS00000000207) (38).
Supplementary Material
Acknowledgments
We thank all the individuals and their families who participated in this study; Dr. Hitoshi Hiura, Dr. K. Nakayama, Dr. R. Funayama, Ms. N. Miyauchi, Ms. M. Tsuda, Ms. M. Kikuchi, Ms. M. Nakagawa, and Mr. K. Kuroda for technical assistance; and the Biomedical Research Core of Tohoku University Graduate School of Medicine for technical support. This work was supported by Japan society for the promotion of science Grants-in-Aid for Scientific Research (JSPS KAKENHI) Grants 19H05757 and 18K09216, Japan Agency for Medical Research and Development (AMED) Grant JP18bm0704021, and the Naito Foundation (to H.O.), and the Core Research for Evolutional Science and Technology from AMED Grants JP17gm0510011 and JP19gm1310001, KAKENHI Grant 17H04335, the Uehara Memorial Foundation, and Takeda Science Foundation (to T.A.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: All sequencing data reported in this paper have been deposited in Japanese Genotype-phenotype Archive (JGA) (accession no. JGAS00000000207).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1916019116/-/DCSupplemental.
References
- 1.Candelier J. J., The hydatidiform mole. Cell Adhes. Migr. 10, 226–235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fock V., et al. , Trophoblast subtype-specific EGFR/ERBB4 expression correlates with cell cycle progression and hyperplasia in complete hydatidiform moles. Hum. Reprod. 30, 789–799 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Seckl M. J., Sebire N. J., Berkowitz R. S., Gestational trophoblastic disease. Lancet 376, 717–729 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Kajii T., Ohama K., Androgenetic origin of hydatidiform mole. Nature 268, 633–634 (1977). [DOI] [PubMed] [Google Scholar]
- 5.Ohama K., et al. , Dispermic origin of XY hydatidiform moles. Nature 292, 551–552 (1981). [DOI] [PubMed] [Google Scholar]
- 6.Altieri A., Franceschi S., Ferlay J., Smith J., La Vecchia C., Epidemiology and aetiology of gestational trophoblastic diseases. Lancet Oncol. 4, 670–678 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Ferguson-Smith A. C., Genomic imprinting: The emergence of an epigenetic paradigm. Nat. Rev. Genet. 12, 565–575 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Zink F., et al. , Insights into imprinting from parent-of-origin phased methylomes and transcriptomes. Nat. Genet. 50, 1542–1552 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Frost J. M., Moore G. E., The importance of imprinting in the human placenta. PLoS Genet. 6, e1001015 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGrath J., Solter D., Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 37, 179–183 (1984). [DOI] [PubMed] [Google Scholar]
- 11.Surani M. A., Barton S. C., Norris M. L., Nuclear transplantation in the mouse: Heritable differences between parental genomes after activation of the embryonic genome. Cell 45, 127–136 (1986). [DOI] [PubMed] [Google Scholar]
- 12.Steinberg K. M., et al. , Single haplotype assembly of the human genome from a hydatidiform mole. Genome Res. 24, 2066–2076 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto E., et al. , Establishment and characterization of cell lines derived from complete hydatidiform mole. Int. J. Mol. Med. 40, 614–622 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okae H., et al. , Derivation of human trophoblast stem cells. Cell Stem Cell 22, 50–63.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Hamada H., et al. , Allele-specific methylome and transcriptome analysis reveals widespread imprinting in the human placenta. Am. J. Hum. Genet. 99, 1045–1058 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eggermann T., et al. , CDKN1C mutations: Two sides of the same coin. Trends Mol. Med. 20, 614–622 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Chilosi M., et al. , Differential expression of p57kip2, a maternally imprinted cdk inhibitor, in normal human placenta and gestational trophoblastic disease. Lab. Invest. 78, 269–276 (1998). [PubMed] [Google Scholar]
- 18.Sebire N. J., et al. , p57(KIP2) immunohistochemical staining of gestational trophoblastic tumours does not identify the type of the causative pregnancy. Histopathology 45, 135–141 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Hernandez L., Kozlov S., Piras G., Stewart C. L., Paternal and maternal genomes confer opposite effects on proliferation, cell-cycle length, senescence, and tumor formation. Proc. Natl. Acad. Sci. U.S.A. 100, 13344–13349 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogawa H., et al. , Cell proliferation potency is independent of FGF4 signaling in trophoblast stem cells derived from androgenetic embryos. J. Reprod. Dev. 62, 51–58 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okae H., et al. , Re-investigation and RNA sequencing-based identification of genes with placenta-specific imprinted expression. Hum. Mol. Genet. 21, 548–558 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Suzuki D., Morimoto H., Yoshimura K., Kono T., Ogawa H., The differentiation potency of trophoblast stem cells from mouse androgenetic embryos. Stem Cells Dev. 28, 290–302 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Matsuoka S., et al. , p57KIP2, a structurally distinct member of the p21CIP1 Cdk inhibitor family, is a candidate tumor suppressor gene. Genes Dev. 9, 650–662 (1995). [DOI] [PubMed] [Google Scholar]
- 24.Lee M. H., Reynisdóttir I., Massagué J., Cloning of p57KIP2, a cyclin-dependent kinase inhibitor with unique domain structure and tissue distribution. Genes Dev. 9, 639–649 (1995). [DOI] [PubMed] [Google Scholar]
- 25.Zhang P., Wong C., DePinho R. A., Harper J. W., Elledge S. J., Cooperation between the Cdk inhibitors p27(KIP1) and p57(KIP2) in the control of tissue growth and development. Genes Dev. 12, 3162–3167 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi K., Kobayashi T., Kanayama N., p57(Kip2) regulates the proper development of labyrinthine and spongiotrophoblasts. Mol. Hum. Reprod. 6, 1019–1025 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Tunster S. J., Van de Pette M., John R. M., Fetal overgrowth in the Cdkn1c mouse model of Beckwith-Wiedemann syndrome. Dis. Model. Mech. 4, 814–821 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korgun E. T., et al. , Location of cell cycle regulators cyclin B1, cyclin A, PCNA, Ki67 and cell cycle inhibitors p21, p27 and p57 in human first trimester placenta and deciduas. Histochem. Cell Biol. 125, 615–624 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Bischof P., Irminger-Finger I., The human cytotrophoblastic cell, a mononuclear chameleon. Int. J. Biochem. Cell Biol. 37, 1–16 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Martin D. A., et al. , DNA content as a prognostic index in gestational trophoblastic neoplasia. Gynecol. Oncol. 34, 383–388 (1989). [DOI] [PubMed] [Google Scholar]
- 31.Sugimori H., Kashimura Y., Kashimura M., Taki I., Nuclear DNA content of trophoblastic tumors. Acta Cytol. 22, 542–545 (1978). [PubMed] [Google Scholar]
- 32.Nishiya I., Moriya S., Yamashita K., Kikuchi T., Cytophotometric DNA determination of trophoblastic neoplasia. Gynecol. Oncol. 5, 103–108 (1977). [DOI] [PubMed] [Google Scholar]
- 33.Kashimura Y., A quantitative study of nuclear DNA of trophoblastic cells: Proliferation kinetics. Gynecol. Oncol. 16, 374–382 (1983). [DOI] [PubMed] [Google Scholar]
- 34.Fukunaga M., Flow cytometric and clinicopathologic study of complete hydatidiform moles with special reference to the significance of cytometric aneuploidy. Gynecol. Oncol. 81, 67–70 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Sundvall L., et al. , Tetraploidy in hydatidiform moles. Hum. Reprod. 28, 2010–2020 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Scelfo R. A., et al. , Loss of methylation at chromosome 11p15.5 is common in human adult tumors. Oncogene 21, 2564–2572 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Montague T. G., Cruz J. M., Gagnon J. A., Church G. M., Valen E., CHOPCHOP: A CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 42, W401–W407 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okae H., Kobayashi K., Methylome and transcriptome profiling of CHM-derived TS cells. Japanese Genotype-phenotype Archive. https://ddbj.nig.ac.jp/jga/viewer/view/study/JGAS00000000207. Deposited 24 October 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data reported in this paper are available at JGA (accession no. JGAS00000000207) (38).