Gene-edited stem cells help treat skin disorder in mice
Gene editing and stem cell technology treat skin disorder in mice. Image courtesy of iStock/Meletios Verras.
The integrity of human skin is partly ensured by type VII collagen, a protein found in anchoring fibrils in the skin’s basement membrane zone. Mutations in the type VII collagen gene, COL7A1, can lead to recessive dystrophic epidermolysis bullosa (RDEB), a rare disorder that defies treatment and is marked by loss of functional COL7A1, extensive blistering, and open wounds. Joanna Jacków et al. (pp. 26846–26852) used CRISPR-Cas9 to correct mutations in exons 19 and 32 of COL7A1 in induced pluripotent stem cells (iPSCs) derived from skin cells of 2 patients with RDEB. The iPSCs were maintained under chemically defined culture conditions free of animal products, a measure intended to facilitate the cells’ future clinical application. Using a high-fidelity Cas9 attached to a guide RNA delivered as a ribonucleoprotein complex, the authors corrected the mutations in the iPSCs—at up to 58% efficiency—with no detectable off-target effects. Next, the authors generated 2 main cellular components of skin—fibroblasts and keratinocytes—from the gene-edited iPSCs, and used the skin cells to build 3D human skin equivalents, which were grafted onto mice. Two months later, the grafts exhibited significant collagen VII deposition, anchoring fibril formation, and skin integration, similar to wild-type skin. Additionally, mice that received gene-edited skin grafts showed no evidence of tumors 9 months after grafting. Because stem cell reprogramming and gene editing can result in unintended genetic changes, the authors emphasize the need for whole-genome sequencing or other screening methods in future studies to detect any lurking effects. According to the authors, the preclinical findings provide a basis for the clinical development and testing of autologous therapies for RDEB. — P.N.
Effects of splice-altering mutations
Certain genetic mutations can cause disease by altering splicing of RNA transcripts, but such mutations can be difficult to identify and characterize. Silvia Casadei, Suleyman Gulsuner, et al. (pp. 26798–26807) developed a method, called cBROCA, to assess the effects of mutations on RNA splicing. In cBROCA, RNA is isolated from cultured patient lymphoblast cells, reverse-transcribed into complementary DNA, hybridized to a panel of known tumor suppressor genes, and sequenced. Comparison of test and control samples allows identification of altered splicing patterns. The authors applied cBROCA to cancer patients and their relatives from more than 150 families, each having multiple relatives with breast, ovarian, endometrial, or colon cancer, but with negative results from conventional genetic testing. cBROCA analysis identified 123 potential splice-altering mutations in 16 tumor suppressor genes. Mutations were associated with various forms abnormal splicing, including failure to transcribe part or all of an exon and creating new exons. In some cases, a single mutation led to multiple altered transcripts. In addition to altering canonical splice sites, mutations could affect splicing through loss or gain of splicing enhancers or silencers. The authors suggest that cBROCA could enhance understanding of the clinical consequences of rare splice-altering mutations. — B.D.
Methane emissions from natural gas well blowout

Emissions from the gas well blowout in Ohio, March 5, 2018. Image courtesy of Ted Auch (FracTracker Alliance), with Aerial support provided by LightHawk.
Accidental leakages in the oil and natural gas sector can rapidly release large amounts of methane, a major contributor to global warming. Satellite-based instruments that regularly scan the entire globe provide a promising means to detect and quantify accidental methane emissions, which are normally challenging to measure. Sudhanshu Pandey et al. (pp. 26376–26381) used atmospheric methane measurements from the satellite-based Tropospheric Monitoring Instrument to detect and quantify methane emissions from a gas well blowout in Ohio in February 2018. By comparing spatially resolved methane concentrations with atmospheric transport simulations, the authors estimated an emission rate of 120 metric tons per hour on the day of the measurements. This measure is twice the peak emission rate from the Aliso Canyon blowout in California in 2015, which was reported to be the second largest accidental methane release in the United States. Assuming that the detected emissions represent the average rate during the 20 days of methane release, the total estimated emissions would be approximately 60 kt, or a quarter of the reported annual oil and gas sector methane emissions for the entire state of Ohio. According to the authors, the results demonstrate the feasibility of using satellite remote sensing to monitor greenhouse gas emissions from unpredictable events. — B.D.
How bats may avoid collisions during group flight
Bats localize objects by emitting sounds and listening to returning echoes. This process, known as echolocation, allows bats to effectively detect obstacles when flying alone, but group flight can be challenging due to the loud calls produced by neighboring bats, which can mask returning echoes and make neighbor detection difficult. Thejasvi Beleyur and Holger Goerlitz (pp. 26662–26668) used simulations to examine how bats may deal with this challenge. The authors incorporated known properties of echolocation into the model to analyze the effects of group size and structure, as well as call parameters, on neighbor detection. The simulations revealed that a bat flying in the middle of the group can detect most or all of its neighbors with each call when the group size is 10 or less. However, at a group size of 100, it takes the central bat approximately 3 calls to detect 1 neighbor. With increasing group size, the central bat tends to detect only its closest neighbors, especially those immediately in front. Neighbor detection in a large group can be improved by increasing the intercall interval, decreasing the call duration, increasing group density, and increasing the variability of heading direction across individuals. According to the authors, the findings suggest how bats may avoid collisions during flight. — J.W.
Autonomous bioluminescence in single mammalian cells
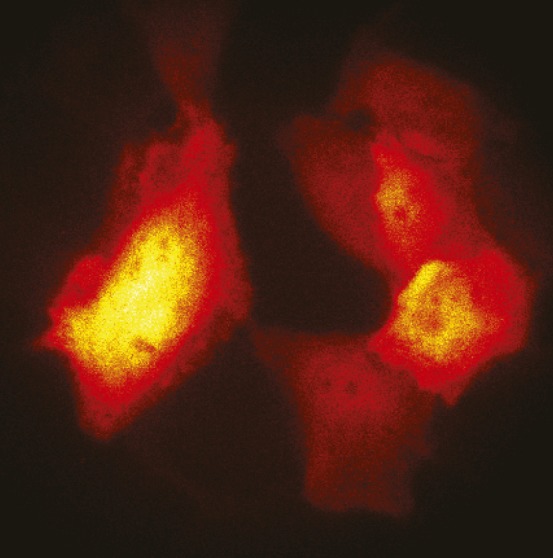
Bioluminescent HeLa cells viewed through a microscope.
Most bioluminescence-based imaging strategies use extraneous compounds, including those inspired by natural bioluminescence systems such as firefly luminescence, to induce light-emitting oxidation reactions in host organisms. Although the genetic instructions for autonomous bioluminescence in bacteria can be encoded in nonbacterial cell types, this approach currently yields insufficient light levels in single mammalian cells. Carola Gregor et al. (pp. 26491–26496) demonstrate that codon optimization applied to 6 genes from the bacterial bioluminescence system can yield autonomous bioluminescence in single mammalian cells at levels rivaling current imaging systems based on external bioluminescence reactants. The study shows that the optimized genes—versions of luxCDABE and frp bacterial genes in ratios optimized for maximum brightness when expressed in mammalian cells—produce bioluminescence levels that outshine wild-type lux genes by at least 3 orders of magnitude. Furthermore, the authors examined the transfected cells for toxicity and found no major effects associated with the expression of the optimized lux genes. The findings offer proof that with further refinement the system could be used for a range of single-cell imaging applications. — T.J.
Sex differences in immune responses
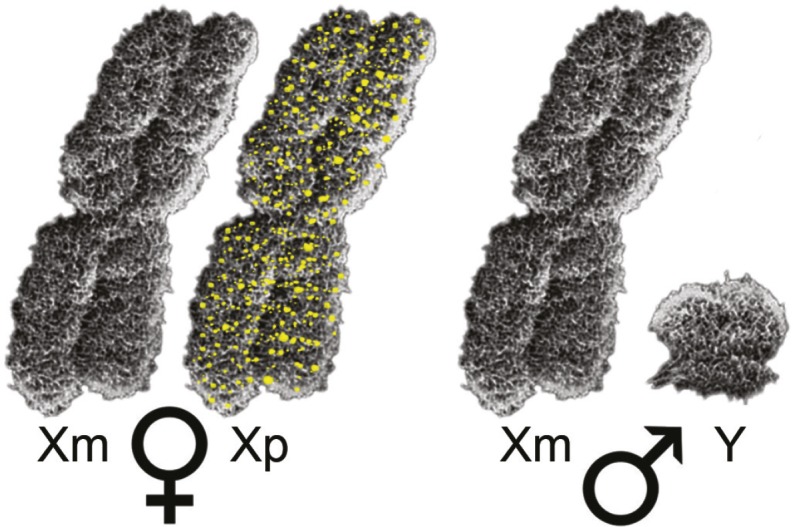
Increased DNA methylation (yellow) when the X chromosome is of paternal origin compared with when it is of maternal origin.
Females in both mice and humans have a higher prevalence of autoimmune diseases and stronger immune responses compared with males. To elucidate the mechanisms underlying the sex differences, Lisa Golden, Yuichiro Itoh, et al. (pp. 26779–26787) analyzed gene expression in a model of C57BL/6 mice that allowed them to study sex chromosome genes without the confounding effects of differences in sex hormones. Genome-wide transcriptome analyses revealed that transcriptomes of auto-antigen-stimulated CD4+ lymphocytes showed a cluster of 5 X genes with higher expression in XY compared with XX mice, a result that was confirmed by qRT-PCR. The authors also found a global increase in DNA methylation when the X chromosome was of paternal origin compared with when it was of maternal origin, a finding consistent with higher expression of X genes in XY cells. Examining the CD4+ T lymphocyte methylome in X-monosomic mice revealed a chromosome-wide increase in methylation at CpG islands in the paternal X chromosome compared with the maternal X, indicating specific silencing of several genes on the paternal X. Parent-of-origin differences in DNA methylation and corresponding gene expression were also found to vary according to the mouse strain used. The authors suggest that parent-of-origin differences in DNA methylation of X genes could play a role in sex differences in immune responses. — S.R.


