Significance
We describe a biobank of patient-derived pancreatic cancer organoids, characterized by whole-genome DNA sequencing, RNA sequencing, and histology. The organoid biobank will be made publicly available and thus can serve as a resource for others. Pancreatic cancer organoids have been described previously; however, here we expose organoids to extensive drug screens to reveal unique drug sensitivity profiles for individual organoid lines. These findings underscore the importance of personalized approaches when using targeted agents to treat cancer in the clinic.
Keywords: pancreatic cancer, organoids, personalized medicine, cancer, biobank
Abstract
We report the derivation of 30 patient-derived organoid lines (PDOs) from tumors arising in the pancreas and distal bile duct. PDOs recapitulate tumor histology and contain genetic alterations typical of pancreatic cancer. In vitro testing of a panel of 76 therapeutic agents revealed sensitivities currently not exploited in the clinic, and underscores the importance of personalized approaches for effective cancer treatment. The PRMT5 inhibitor EZP015556, shown to target MTAP (a gene commonly lost in pancreatic cancer)-negative tumors, was validated as such, but also appeared to constitute an effective therapy for a subset of MTAP-positive tumors. Taken together, the work presented here provides a platform to identify novel therapeutics to target pancreatic tumor cells using PDOs.
Pancreatic ductal adenocarcinoma (PDAC) accounts for >7% of all cancer deaths. With an overall survival rate of only 8.5%, PDAC is one of the deadliest forms of cancer, for which treatment options are limited (1). In addition, distal cholangiocarcinomas (dCCs), that originate from the distal bile duct at the site where it passes through the pancreas, were recently described to be molecularly more similar to pancreatic tumors than to those of the liver (2).
Currently, PDAC is treated with either gemcitabine/nab-paclitaxel or FOLFIRINOX (5-fluorouracil, leucovorin, irinotecan, and oxaliplatin), combined with surgery when possible (3, 4). Despite these interventions, response rates remain poor, with overall survival of only 6 to 11 mo in patients ineligible for surgery (3, 4). As alterations in therapeutically targetable molecular pathways are known to contribute to disease pathogenesis (5–8), agents targeting these pathways hold promise to improve the treatment of these tumors. However, variable responses to these therapeutics are expected, and biomarkers to predict response to therapy are lacking. Thus, tools to identify the most effective chemotherapeutic regimens for individual patients, as well as models to develop additional drug treatment strategies, are urgently needed.
Organoid technology allows for the establishment of patient-derived cultures with much higher efficiency than classical 2D cell lines (9). This has resulted in the generation of a range of tumor-derived organoid biobanks that recapitulate tumor characteristics and can be used to address basic and translational research questions (5, 6, 10–15). We and others have previously reported the establishment of patient-derived organoids (PDOs) derived from PDAC (5, 6, 16). Using slightly different protocols, these studies show that PDOs can be established from PDAC with a success rate of 70 to 73%. Here we report an additional pancreas tumor biobank and show the feasibility of this model for personalized drug screening. Organoids derived from several non-PDAC tumor types, such as pancreatic acinar cell carcinoma and distal cholangiocarcinoma, are also included. Organoids were established from tumor-adjacent normal epithelium when available, allowing for the direct comparison of normal and tumor cells from the same patient.
Patient-Derived Organoids Can Be Established from Different Pancreas Tumor Types and Recapitulate the Tissue of the Original Tumor
Tissues from biopsies or surgical resections were obtained and digested as described previously (16). Tumor cells were subsequently grown in 2 types of media, designated tumor medium 1 (TM1) and tumor medium 2 (TM2). TM1 contains all components of complete pancreatic medium (CM) except EGF and PGE2, whereas TM2 lacks PGE2, WNT, and the TGF-β inhibitor A83-01. After initial tumor digestion and plating in extracellular matrix, samples were cultured in both media. If organoids grew out on both media, both cultures were maintained in parallel and compared later. In total, over the course of 3 y, 83 tumor samples were received. Fifty-two grew out in at least 1 of the 2 tumor media (outgrowth efficiency of 62%). Of these 52 PDOs, 31 were analyzed by whole-genome sequencing (WGS) and are described in this study (Fig. 1A and SI Appendix, Table S1). Available organoids are cataloged by Hubrecht Organoid Technology (www.hub4organoids.eu) and can be requested using their assigned HUB codes (SI Appendix, Table S1). With the exception of PDO 30 and 31, which were established from biopsies, all other PDOs were established from surgical resections. The outgrowth efficiency of organoids from biopsies was 31%. For 12 of the 31 established PDOs, tumor samples grew out on both TM1 and TM2 (SI Appendix, Table S1 indicates the samples established on each TM). Sato et al. (5) reported that the dependence on WNT signaling of PDOs correlates with a basal tumor subtype that is characterized by GATA6-dependent gene expression. In line with these findings, we observed an inverse correlation between GATA6 expression and WNT dependency in PDOs (SI Appendix, Fig. S1A). This suggests that medium composition may have a selective impact on PDO outgrowth. In cases where tumor material grew out on both media, no difference in GATA6 expression profile was observed between the 2 lines, indicating that the medium itself does not directly affect the expression of GATA6. This is illustrated by the gene expression heatmap, in which most of the organoid pairs cluster together, indicating similarities in gene expression levels (SI Appendix, Fig. S1B).
Fig. 1.
Patient-derived organoids can be established from different pancreatic tumor types and recapitulate the tissue of the original tumor. (A) Pie chart depicting the characteristics of the tumor biobank described in this work. ACC, acinar cell carcinoma; CC, cholangiocarcinoma; IPMN, intraductal papillary mucinous neoplasm. (B) Brightfield images of 3 PDO cultures, shown in 2 magnifications. (C) Brightfield images of H&E staining of tumor tissue and corresponding organoids showing organoid morphology in culture. (D) IHC staining for TP53 in tumor tissue and corresponding organoids of patient 11. The TP53 staining is consistent with TP53 mutation status of the tumor and organoids and is corresponding in tumor tissue and organoids. (E) IHC for SMAD4 in tumor tissue and brightfield images of corresponding organoid lines, grown in either complete medium or medium lacking A83-01 and Noggin. SMAD4 mutant cells can be functionally selected in organoid cultures by removing TGF-β inhibitors A83-01 and Noggin. (F) qPCR for BMP target genes ID1 and ID3. Induction of BMP signaling by the removal of Noggin and A83-01 resulted in up-regulation of ID1 and ID3 in SMAD4 wild-type PDO 8 and PDO 10 but not in SMAD mutant PDO 23. Expression is shown relative to organoids grown in complete medium. This experiment was performed in technical triplicate.
When available, corresponding tumor-adjacent normal tissue was processed to establish wild-type organoids. We successfully established these matched normal control organoids for 5 patients, corresponding to 7 of the established tumor organoid lines (as in some cases, tumor organoids could be established on both TM1 and TM2). Morphology of tumor organoids differed from cystic (with either a clear or filled lumen) to dense structures (Fig. 1B). Comparison of hematoxylin and eosin (H&E) staining of PDOs and corresponding tumor tissue showed morphological similarities between PDOs and tumor tissue with cellular atypia, including aberrant location of the nuclei, according to pathological assessment (Fig. 1C).
Organoid Culture Media Composition Functionally Selects for Oncogenic Mutations in PDOs
The TP53 status of a subset of PDOs and corresponding tumor tissue was determined by immunohistochemistry (IHC). Accumulation of mutant TP53 protein can occur due to conformational changes that result in increased protein half-life (17); therefore, TP53 IHC staining is used as a clinical parameter to determine TP53 mutation status (18). TP53 IHC staining in organoids correlated with the corresponding tumor tissue results (example shown in Fig. 1D, where tissue is indicated by “TUMOR” and patient-derived organoid is indicated by “PDO”). Loss of SMAD4 is commonly observed in pancreatic tumors (19). Using IHC, the SMAD4 status of a subset of tumors was assessed. Tumors 8 and 10 stained positive for this marker, indicative of wild-type protein, whereas tumor 23 showed loss of SMAD4 expression (Fig. 1E). In organoids, positive SMAD4 IHC was never observed. This may be explained by the presence of Noggin and A83-01 in the culture medium, resulting in inhibition of BMP/TGF-β signaling in the organoids. Although not by IHC, we found that loss of SMAD4 function could be detected in culture. Withdrawal of A83-01 and Noggin from the medium resulted in up-regulation of the TGF-β target genes ID1 and ID3 in SMAD4 wild-type PDO 8 and 10 cells, and resulted in growth cessation after 2 wk in culture. In contrast, SMAD4-mutant PDO 23 was unaffected by Noggin/A83-01 withdrawal and did not show up-regulation of TGF-β target genes (Fig. 1 E and F). Thus, despite the absence of positive SMAD4 IHC in organoids, we conclude that the molecular differences between SMAD4 wild-type and mutant organoid lines are retained in culture. We confirmed molecularly that the SMAD4 mutation status of the PDOs indeed fits with their response to Noggin/A83-01 withdrawal. This finding highlights how culture conditions can alter cell behavior and underscores that these conditions should be carefully considered depending on the application.
We conclude that PDOs retain morphological characteristics of the tissue of origin. Organoids can be manipulated in culture by pharmacologic inhibition or removal/addition of certain growth factors to select for cells with tumor-specific genetic alterations. This holds potential to avoid overgrowth of contaminating wild-type cells in tumor samples, a notorious problem when establishing tumor-derived organoids.
Anecdotal Expansion of Precancerous Cells Found in “Healthy” Pancreas
The selection pressure created by the addition or removal of growth factors allows for enrichment of rare tumor-like cells, which can be present in normal pancreas. For example, organoids established from normal pancreatic tissue (of a patient with pancreatic metastasis of ovarian cancer) could be cultured for at least 15 passages on TM1, lacking EGF. This suggested that these cells were independent of EGF ligand for downstream EGFR signaling. Indeed, these organoids were found to carry an activating G12R KRAS mutation, suggesting a (pre)neoplastic pancreatic lesion (SI Appendix, Fig. S2A). As this mutation was not present in both the ovarian carcinoma tumor tissue and organoids derived thereof, the organoid line was most likely established from a pancreas-derived neoplastic clone, such as pancreatic intraepithelial neoplasia (PanIN), a precursor of PDAC. The established organoid line responded to Nutlin-3 treatment, indicative of TP53 wild-type status (SI Appendix, Fig. S2B). Indeed, this precancerous lesion was detected upon pathological assessment of the biopsy specimen (SI Appendix, Fig. S2C), highlighting the strength of organoid cultures and the artificial selection pressures created by the culture medium to both detect and expand rare (pro)tumorigenic cells.
PDOs Contain Genetic Alterations Typical for Tumor Types from Which They Are Derived.
Thirty-one PDOs were analyzed by WGS. Matching germ line DNA was available for 26 of these PDOs, allowing accurate detection of somatic events. PDO 27 contained a low number of mutations and displayed a stable karyotype. In addition, no oncogenic mutations were detected in this PDO. Therefore, we assumed that PDO 27 was derived from wild-type cells present in the primary resection specimen and excluded this sample from further analysis. Oncogenic events (protein altering point mutations, indels, amplifications, and deep deletions) were characterized in 202 genes previously found to be associated with pancreatic cancer (5–8) (Fig. 2A, Dataset S1, and SI Appendix, Fig. S3). The 5 PDOs for which no germ line DNA was available were functionally annotated with COSMIC. The most commonly altered gene, KRAS, was mutated in 22 of the 25 lines (88%), of which 30% were p.G12D, 23% were p.G12V, 13% were p.G12R, 13% were p.Q61H, and 6% were p.G13D. In the 3 PDOs without a KRAS mutation, we detected a p.V600E mutation in BRAF in PDO 11 and 25 and a p.G12D NRAS mutation in PDO 22. TP53 was mutated or lost in 84% (21 of 25) of the PDOs. Loss of CDKN2A was detected in 80% of the lines (20 of 25). Other commonly found genetic alterations included loss of SMAD4, EEF2A mutations, MYC amplifications, activating mutations in PIK3CA, and ARID1A inactivation.
Fig. 2.
Genomic landscape of pancreatic PDOs recapitulates genetic alterations commonly found in this tumor type. (A) Overview of severe somatic events detected in PDOs in genes commonly altered in PDAC. Here a panel of 220 oncogenic driver genes (marked with an asterisk) and tumor suppressor genes was analyzed for genetic alterations. Different mutation types are depicted with different colors. Five samples lacked a reference DNA, marked with “*no blood ref.,” and pathogenic mutations for these samples were called based on the COSMIC database. The mutation frequency per gene is depicted on the left and is calculated without the inclusion of tumor-only samples. In some cases, multiple organoid lines were established from tissues obtained from 1 patient. Color coding at the bottom of the figure shows whether organoid lines are derived from the same patient. (B) Volcano plots showing the common losses and gains of chromosome arms (annotated by the chromosome number, followed by p or q). The gains and losses were normalized against the mean genome ploidy level.
Organoids derived from non-PDAC tumors included PDOs 1, 2, 22, and 26. PDO 1, derived from a squamous adenocarcinoma, was the sole PDO carrying mutations in only 1 gene (TP53) in the panel of genes analyzed. PDO 22, derived from an acinar cell carcinoma, was found to be mutated in CDKN2A and SMAD4. Finally, cholangiocarcinoma-derived PDO 2 and PDO 16 carried mutations in PDAC driver genes, including TP53, CDKN2A, EEF2, SMAD4, GNAS, and KRAS.
Next, we compared the genetic landscape of PDOs derived from the same patient but established in different culture media (SI Appendix, Fig. S4). All 8 PDO paired sets of organoids showed 3 to 5 shared nonsynonymous mutations in cancer genes, which likely reflect the clonal driver events. However, every PDO also harbored unique genomic events in cancer genes, including substantial nonsynonymous mutations in PDOs 3, 4, and 5; variations on a structural level, such as ERBB2 amplification in PDO 6 but not in PDO 7; and MYC amplification in PDO 9 but not in PDO 10. Likewise, PDO 13 underwent a whole genome duplication, whereas PDO 12 did not, explaining the amplification events of GNAS and ERBB2 observed in PDO 13. Overall, none of the matched PDOs was genetically identical to its counterpart. This indicates that the different tumor media can be used to capture intratumor heterogeneity, which may be of interest in light of drug resistance mechanisms. The finding that none of the matched PDOs was genomically identical to its counterpart underscores the relevance of (intentional or unintentional) in vitro selection that can result in enrichment of specific tumor clones. This is in line with the described genetic heterogeneity of PDAC.
DNA copy number analysis revealed aneuploidy in all PDOs with the exception of PDO 25 (SI Appendix, Fig. S5). Of these, 7 PDOs displayed an overall genome ploidy level of 3 or higher, which suggests a whole genome duplication event during tumor development. Next to the chromosomal arm losses as found by Seino et al. (5) (i.e., 6p, 9p, 17p, and 18q), we also detected loss of 3p, 6p, 8p, 19p, 21q and 22q and gain of 20q (Fig. 2B). Overall, copy number profiles showed a high degree of concordance for PDOs derived from the same patient. This consistency was also observed for other large-scale structural alterations, including translocations and inversions (e.g., PDOs 19, 20, and 21) (SI Appendix, Fig. S6).
To investigate the mutational processes operating in pancreatic cancer, we applied an unbiased mutational signature analysis on all detected point mutations (SI Appendix, Fig. S7A) (18). We identified seven signatures that showed high consistency with well-described signatures in human cancer (19). Of these, signatures SBS1, SBS5, and SBS40 were detected in each sample and represent most of the detected mutations (SI Appendix, Fig. S7B). These 3 signatures are related to so-called “clock” signatures that result from mutational aging processes that are also seen in healthy cells (SI Appendix, Fig. S8 and Table S2) (20). Unsupervised clustering of PDOs with 71 previously described primary human pancreatic cancer metastasis samples revealed 5 different clusters, each of which consisted of both PDOs and human pancreatic samples, indicating that the mutational processes from PDOs recapitulate those found in vivo (20).
Cluster 1 (green) is characterized by high mutation burden and strong activity of the mismatch repair deficiency signature in combination with a high number of deletions in repeat context (SI Appendix, Fig. S7 C and D). PDO 25, present in this cluster, also displays a near-diploid karyotype with characteristic features of microsatellite instable tumors (21). Retrospectively, the included patient was diagnosed with a microsatellite instable duodenal adenocarcinoma located in close proximity to the pancreas. Cluster 2 (blue) represents 2 human pancreatic samples that show a large contribution of SBS5 (a clock signature that currently lacks any etiology). Cluster 3 (pink) consists of 2 samples that show strong contributions of SBS17. This signature has recently been linked to 5-FU treatment (22). Cluster 4 (purple) is dominated by mutation patterns (SBS3 and deletions with microhomology) highly characteristic for homologous recombination deficiency (23). The final cluster (orange) is predominantly characterized by the aging signatures (SBS1, SBS5, and SBS40), although some samples also show APOBEC activity (20, 24, 25).
High-Throughput Drug Screening in PDOs Reveals Sensitivities to a Range of Therapeutic Agents
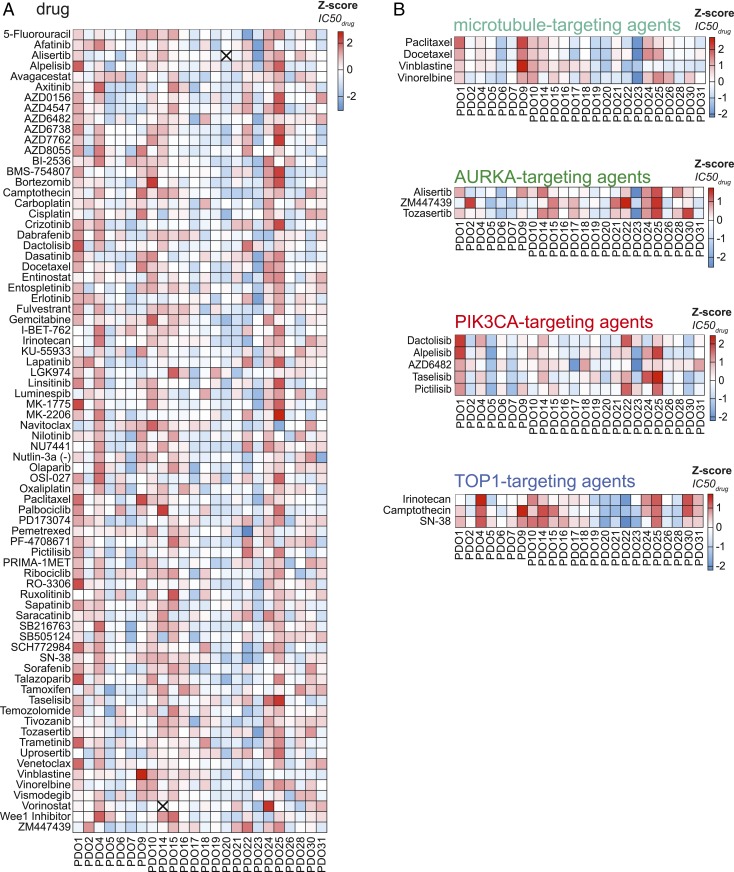
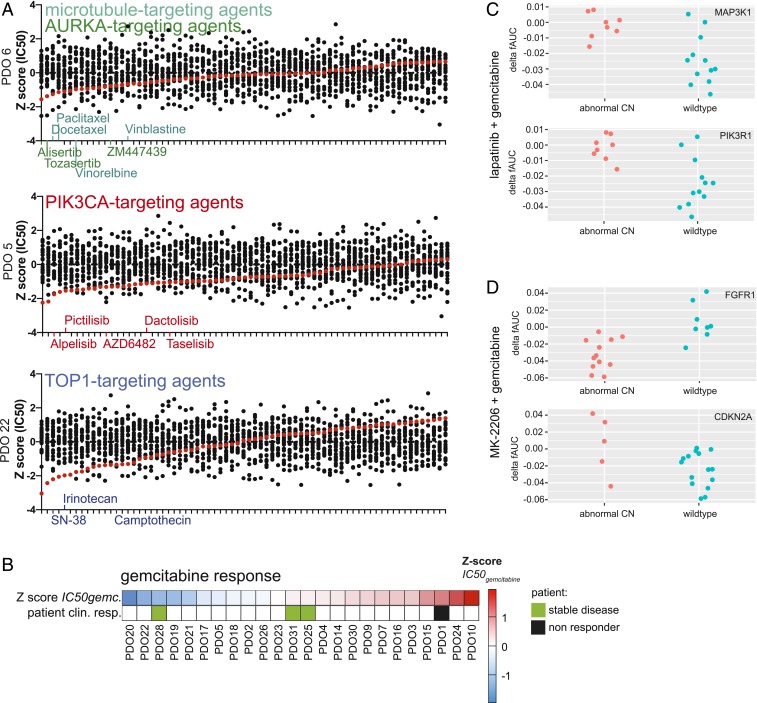
To assess the sensitivity of PDOs to a wide range of chemotherapeutic agents, 24 of the established PDOs were exposed to a panel of 76 therapeutics, including chemotherapeutics currently used in the treatment of PDAC. Different responses were observed for organoids derived from different patients (Fig. 3A). To validate the results obtained using this screening procedure, we compared the responses of organoids when exposed to therapies with similar molecular targets (Fig. 3B and SI Appendix, Fig. S9). As examples, the responses to multiple agents targeting microtubules, aurora kinase A (AURKA), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), and topoisomerase-1 (TOP1), are shown (Fig. 3B). In general, a similar response was observed for agents targeting the same biological process or molecular pathway. As such, these results indicate that observed in vitro responses are indicative for true biological vulnerabilities and are not the consequence of technical variability. For many of the PDOs tested, drugs could be identified for which the individual PDO was more sensitive than all other PDOs tested (Fig. 4A and SI Appendix, Fig. S10). Again, multiple drugs targeting the same molecular pathway were often found among the most effective drugs. Taken together, these findings support the hypothesis that specific targeted therapies will be effective in only a subset of patients, and, as such, a personalized approach will be required to select the right drug for each individual patient.
Fig. 3.
High-throughput drug screening in PDOs reveals sensitivities to a range of therapeutic agents. (A) A total of 76 compounds were tested in 24 PDOs. The z-scores of obtained IC50 values are depicted in the heatmap. High values (indicating resistance) are depicted in red, and low values (indicating sensitivity) are in blue. An “X” indicates that the data generated for this compound/PDO combination are not present. Compounds are ordered alphabetically. (B) Response of PDOs to compounds targeting the same biological process or pathway, highlighting similar responses observed among the different compounds. High values (indicating resistance) are depicted in red, and low values (indicating sensitivity) are in blue.
Fig. 4.
Individual PDO drug responses indicate clinically relevant therapeutic vulnerabilities and reveal potential biomarkers for therapy response. (A) For PDO 5, PDO 6, and PDO 22, therapeutic compounds are arranged from most effective to least effective. Enrichment of compounds that target the same biological process or pathway is observed. Inhibitors targeting the same target are shown in identical colors, with color-coding as in Fig. 3B. (B) Correlation between gemcitabine response of PDOs and corresponding patients. For patient clinical response, green indicates a response to gemcitabine treatment, and a black box indicates resistance to treatment. Sensitivity to chemotherapy is indicated by the z-score of IC50 values. (C) Response of PDOs treated with lapatinib and gemcitabine, depicted in delta fAUC, plotted for samples either with (red) or without (blue) copy number alteration of MAP3K1 and PIK3R1. (D) Response of PDOs treated with MK-2206 and gemcitabine, depicted in delta fAUC, plotted for samples either with (red) or without (blue) copy number alteration of FGFR1 or CDKN2A.
In Vitro Response of PDOs Indicates Correlation with Clinical Patient Responses in a Limited Number of Patients
Clinical data on given treatment and therapy response was sufficient to assess therapy response in only 4 of the patients. Most patients did not receive treatment at all, or were given treatment in the adjuvant setting (where assessment of therapy response is difficult, since there is no detectable disease at the start of treatment). All 4 patients were treated with gemcitabine. Patient 1 developed distant metastasis during treatment with this chemotherapeutic agent. Indeed, PDO 1 was highly resistant to treatment with gemcitabine in vitro. In contrast, PDO 28 was among the most sensitive PDOs on treatment with gemcitabine. Patient 28 was found to have stable disease with a decrease in distant liver metastasis after gemcitabine treatment, although later, the patient developed distant metastasis and eventually succumbed to the disease. In addition, patients 25 and 31 also presented with stable disease after undergoing gemcitabine treatment. PDO 25 and PDO 31 were among the intermediate responders in our in vitro assays. Although the numbers are small, an overall correlation could be observed between in vitro PDO response and clinical patient response (Fig. 4B). An overview of relevant patient information in given in SI Appendix, Table S3.
DNA Status as a Predictor for In Vitro Drug Response
In addition to single-agent therapy screens, a number of targeted agents were combined with chemotherapeutics that are currently given to pancreatic cancer patients. Since both DNA status and drug response data of PDOs were available, we set out to correlate mutation status with drug response in an unbiased fashion, using both an elastic net model and TANDEM 2-step model. Among the strongest associations detected was the response to the HER2/EGFR inhibitor lapatinib combined with gemcitabine. The response to this combination of agents was measured as the difference between the predicted and observed area under the fitted dose–response curve (delta fAUC). This predicted AUC is based on the additive effect of both single agents. As such, a higher delta fAUC values indicate increased sensitivity. Depending on the mutation status of MAP3K1 and PIK3R1, response to these agents differed significantly between different PDOs (Fig. 4C). PDOs carrying aberrant copy numbers of these kinases downstream of the EGFR receptor (which is the target of lapatinib) showed increased sensitivity to these agents. Other relevant associations were those detected between the response to the AKT-inhibitor MK-2206 combined with gemcitabine and the mutation status of FGFR1 and CDKN2A (Fig. 4D). Since AKT is located downstream of the PI3K family kinases, it is not surprising that here the amplification of MAP3K1 and PIK3R1 did not pop up as a potential biomarker. Moreover, these results indicate the mutation status of these genes might serve as a potential biomarker to predict the response to these therapies. A full report of the analysis performed and the potential biomarkers identified is provided in Dataset S2.
Loss of MTAP Results in Sensitivity to PRMT5 Inhibition
We next focused on a specific therapeutic agent: the selective protein arginine methyltransferase 5 (PRMT5) inhibitor EZP01556 (26). The chromosome 9p21 locus is homozygously deleted in ∼15% of all human cancers, including pancreatic cancer (27). This locus carries the gene cyclin-dependent kinase inhibitor 2A (CDKN2A), encoding the tumor suppressor alternative reading frame (p19-ARF) and inhibitor of CDK4 (p16-INK4) (28, 29). On CDKN2A deletion, nearby genes, such as methylthioadenosine phosphorylase (MTAP), are often codeleted. Indeed, MTAP is deleted in 80 to 90% of the CDKN2A− tumors (30). MTAP plays a crucial role in the methionine salvage pathway by recycling its substrate 5′-methylthioadenosine (MTA), ultimately resulting in regeneration of methionine and adenosine (31). A recent search for therapeutic vulnerabilities in MTAP-deficient cells resulted in the identification of PRMT5 as a synthetic lethal gene in MTAP− cells (32–34). The activity of PRMT5, responsible for methylation of a large number of substrates, including histones (35), is inhibited by high levels of MTA (32). As MTA accumulates in MTAP− cells, exploring the potential of PRMT5 inhibition in MTAP− tumors seems promising (36–38).
CDKN2A and MTAP status of the PDOs were determined by DNA and RNA sequencing (Fig. 5 A and B). DNA and RNA data were concordant for all lines except PDO 8 and 11, where the chromosomal breakpoint was found within the MTAP gene body. RNA status was in concordance with the result of MTAP IHC performed on a subset of the PDACs (SI Appendix, Fig. S11). Importantly, tumor 11 showed a positive MTAP immunostaining, whereas based on the detected DNA alteration, this detected MTAP protein is predicted to be nonfunctional. To quantify the response to EZP015556, the area under the curve (AUC) of all exposed PDOs was calculated. Indeed, MTAP− organoid lines showed increased sensitivity to PRMT5 inhibition (Fig. 5C). The average IC50 was 19.16 μM for MTAP+ lines and 0.68 μM for MTAP− organoid lines. Interestingly, a subset of MTAP+ PDOs also showed sensitivity to PRMT5 inhibition. MTAP+ PDOs 6, 9, 19, 20, 24, and 25 showed AUC values comparable to those of MTAP− organoid lines. These results indicate that PRMT5 inhibition may prove effective in MTAP− and in a subset of MTAP+ tumors. As such, these observations underscore the need for functional testing to identify potentially relevant therapies.
Fig. 5.
PRMT5 inhibition is effective in a subset of PDOs. (A) Detection of CDKN2A and MTAP gene body loss in the 25 tumor-derived PDOs for which reference DNA was available. Dark green indicates the presence and light green indicates the absence of gene-coding DNA. For both genes, both alleles are shown. (B) Expression levels of CDKN2A and MTAP as detected by RNA sequencing in PDOs. The heatmap shows log2 values of normalized counts. Red indicates a high value; blue, a low value. (C) Heatmap showing the AUC values of the response to EZP015556 of all tested PDOs and corresponding MTAP DNA status. Low AUC, indicating sensitivity to PRMT5 inhibition, is depicted in blue. High AUC, indicating low sensitivity to PRMT5 inhibition, is depicted in red. MTAP mutation status is indicated in the row below, with black indicating loss of MTAP and white indicating MTAP wild-type status. (D) Induction of MTAP expression in MTAP+ (dark blue), MTAP− (light blue), and MTAP+ EZP01556-sensitive (red) lines. Cells are exposed to EZP015556 either in combination with doxycycline-mediated induction (square symbols, dashed lines) or without (round symbols, solid lines). The experiment was performed in technical triplicate. DOX, doxycycline. (E) MTA levels detected in PDOs, shown in pmol/106 cells. MTA levels were measured in 3 MTAP+ PRMT5 inhibition-resistant lines (blue), 2 MTAP− PRMT5-sensitive lines (green), and 3 MTAP+ PRMT5 inhibition-sensitive lines (red). (F) In MTAP+ PRMT5 inhibition-sensitive PDO 20, MTA levels were measured in a clone infected with the inducible MTAP overexpression construct. MTA levels were measured in both the absence (red bar) and presence (white bar, red outline) of doxycycline, resulting in expression of wild-type MTAP protein. (G) Correlation plot showing correlation (significant, P = 0.0079, Pearson correlation) between MTA levels (x-axis) and sensitivity to EZP015556, depicted by the AUC (y-axis).
Wild-Type MTAP Expression Decreases EZP015556 Sensitivity in MTAP+ Lines
To test the causal role of MTAP dysfunction in PRMT5 inhibitor sensitivity, a lentiviral doxycycline-inducible MTAP expression vector was introduced in both MTAP− and MTAP+ EZP015556-sensitive organoid lines (SI Appendix, Fig. S12A). Functionality of the construct was confirmed both by quantitative PCR (qPCR) for MTAP transcripts and by detection of GFP, whose expression was controlled by the same promotor as MTAP in the construct (SI Appendix, Fig. S12 B and C). Induction of MTAP expression did not change the response of insensitive MTAP+ lines, but reduced EZP015556 sensitivity in MTAP− organoid lines, thereby confirming the causal role of MTAP deletion for EZP015556 sensitivity. Moreover, the induction of MTAP expression decreased EZP015556 sensitivity in MTAP+-sensitive PDOs, suggestive of defective MTAP function in these lines (Fig. 5D).
PDOs Sensitive to PRMT5 Inhibition Are Marked by Elevated MTA Levels
To test whether the mechanism of increased EZP015556 sensitivity was similar in both MTAP− and MTAP+ lines, MTA levels were assessed using proteomics. As reported previously, elevated MTA levels were detected in MTAP− cells when compared to EZP015556-insensitive MTAP+ lines (36–38). In MTAP+ lines sensitive to EZP015556, MTA was detected at levels comparable to that of MTAP− lines (Fig. 5E). MTA levels could be restored by overexpression of wild-type MTAP (Fig. 5F). These results confirm a comparable mechanism of action of EZP015556 in both subtypes of PDOs. Indeed, a correlation was observed between MTA levels and EZP015556 sensitivity (Fig. 5G). These findings suggest that MTA levels could be a better marker than MTAP mutation status to predict response to PRMT5 inhibition.
Discussion
Here a biobank of 30 characterized pancreatic tumor organoids was used to explore the translational potential of organoid technology. This was done in 2 ways. First, established organoids and corresponding primary tissue were histologically assessed and compared for molecular characteristics. Second, after genetic characterization, the established PDOs were exposed to a range of therapeutic agents to identify therapies that effectively killed the pancreatic tumor cells.
Morphology of primary tissue and corresponding organoids revealed similarities. The expression levels of markers currently used in diagnostics were compared between organoids and corresponding primary tissue. It was found that SMAD4 expression was not detectable by IHC in organoids. As mentioned previously, this is likely due to the presence of BMP/TGF-β–inhibiting molecules Noggin and A83-01 in the organoid medium. This result emphasizes the effect that medium composition can have on cell behavior, a fact that is often overlooked. Despite the absence of SMAD4 staining, we showed that SMAD4 wild-type organoids can be distinguished from SMAD4 mutant organoids using functional selection. These findings highlight the importance of using the correct technique to compare organoids and primary tissue to assess translative potential and show that IHC might not always be appropriate.
To assess the potential of this system to identify or validate effective therapies for pancreatic cancer, the generated PDO biobank was exposed to a library of both classical chemotherapies and targeted agents. Although highly dependent on the quality and quantity of the primary material, the average time to establish enough organoids for such an extensive screening procedure is expected to be 2 to 3 mo. However, this time can likely be reduced by decreasing the number of compounds tested (e.g., focusing only on those therapies currently used in first-line treatment).
For most PDOs, we could identify multiple compounds that showed effective tumor killing. Importantly, different drugs killed PDOs derived from different patients. No single therapy could be identified that uniformly resulted in effective tumor killing for all PDOs, indicating that a “personalized” approach is required when using targeted therapies. To identify which therapy is most effective for each individual patient, functional tests such as those described in this study might prove useful. Results indicating that in vitro organoid responses can predict clinical outcome have been published but require further validation before functional testing can be applied in the clinic (6, 13, 39). It will be interesting to see if this correlation holds true in pancreatic cancer and if so, whether organoids can be used to guide therapy decisions in the clinic.
For 4 patients, the available clinical data actually allowed for such a comparison of the response to treatment of both the patient and the matching PDO. PDO 1, derived from tumor material of a patient presenting with progressive disease under gemcitabine treatment, was indeed among the most resistant PDOs in our assays. The 3 other patients were found to have stable disease after receiving gemcitabine treatment. Indeed, matching PDOs showed high or intermediate sensitivity to in vitro gemcitabine exposure. Although these results are encouraging, it should be kept in mind that numbers are small. Moreover, interpretation of clinical data of patients with pancreatic tumors is difficult, as complications are common and progression is quick. In the future, larger correlation studies should be performed to validate the predictive potential of patient-derived pancreatic organoids.
Finally, we used pancreatic cancer organoids to confirm that PRMT5 inhibition effectively targets MTAP− tumors. Our results indicate that a subset of MTAP+ tumors also might be susceptible to PRMT5 inhibition. Further research is needed to unravel the mechanisms underlying this sensitivity, but our initial results indicate that this is caused by lack of function of the endogenous MTAP protein. Potentially, expression analysis of organoids that are undergoing treatment with PRMT5 inhibitors will result in elucidation of the underlying mechanisms. Regardless, these findings highlight the fact that genetic testing might not always be sufficient to identify therapy responders. Clinical trials with PRMT5 inhibitors are currently ongoing (NCT03573310, NCT02783300, and NCT03614728). It will be of interest to see if the MTAP (or CDKN2A) status of the tumors can be correlated to patient response.
Conclusion
Here we report the establishment of a biobank of PDOs grown from pancreatic cancer and distal cholangiocarcinoma and characterized by histology, RNA sequencing, DNA sequencing, and drug response. Organoids retain histological features of primary tumors and carry genetic alterations commonly found in this tumor type. High-throughput drug screening using a panel of 76 compounds identified a range of targeted therapies with efficacy in PDOs. In line with personalized medicine approaches, therapy responses differed among the PDOs. The established PDO model was used to validate PMRT5 inhibition as a potential therapeutic approach for PDAC. We show that PRMT5-inhibitor EZP015556 can be effective in both MTAP− and a subset of MTAP+ PDOs, both of which are characterized by high MTA levels.
Methods
Human Material for Organoid Cultures.
The collection of patient data and tissue for the generation and distribution of organoids was done in accordance with the guidelines of the European Network of Research Ethics Committees following European, national, and local law. The Biobank Research Ethics Committee of University Medical Center Utrecht (TCBio) approved the biobanking protocol (12–093 HUB-Cancer) under which this research was performed. All donors participating in this study provided signed informed consent. Available organoids are cataloged at www.hub4organoids.eu and can be requested at info@hub4organoids.eu.
Tissue Processing.
Patient material was collected in Advanced DMEM/F12 (Life Technologies; 12634-034), supplemented with 1× GlutaMAX (adDMEM/F12; Life Technologies; 12634-034), penicillin-streptomycin (Life Technologies; 15140-122) and 10 mM Hepes (Life Technologies; 15630-056) (designated +/+/+ hereinafter). For collection of patient material, 100 µg/mL Primocin (InvivoGen; ant-pm1) was added. Material was cut into small fragments. Random pieces of ∼5 mm3 were stored at −20 °C for DNA isolation or fixed in formalin for histopathological analysis. Fragments were incubated at 37 °C in 1 mg/mL collagenase (Sigma-Aldrich; C9407). Digested tissue was sheared using 5-mL pipettes. Cell suspension was diluted with 10 mL of +/+/+ and strained over a 100-µm EasyStrainer filter (Greiner; 542000) and centrifuged at 300 × g. The resulting pellet was resuspended in ice-cold 70% 10 mg·mL−1 cold Cultrex growth factor reduced BME type 2 (Trevigen; 3533-010-02) in organoid medium. Droplets of ∼10 µL were plated in preheated suspension culture plates (Greiner; M9312). Plates were inverted and maintained at 37 °C to let the BME solidify. After 30 min, prewarmed organoid medium was added to the plates. For the first week, 10 µM rho-associated kinase (ROCK) inhibitor (Y-27632; Abmole Bioscience; M1817) was added to the medium.
Organoid Culture.
Organoids were grown in +/+/+ supplemented with different subsets of growth factors, depending on whether wild-type or tumor organoids were established. For organoids derived from wild-type tissue, medium consisted of Wnt3a-conditioned medium (50% vol/vol), plus +/+/+ containing 1× B27 supplement (Life Technologies; 17504-044), 1,25 mM N-acetyl-l-cysteine (Sigma-Aldrich; A9165), 10 mM nicotinamide (Sigma-Aldrich; N0636), 50 ng/mL human EGF (PeproTech; AF-100-15), 500 nM A83-01, 100 ng/mL human FGF10 (PeproTech; 100-26), 1 µM prostaglandin E2 (Tocris Bioscience; 2296), 10 nM gastrin (R&D Systems; 3006), 4% (vol/vol) RSPO, and Noggin (produced via the r-PEX protein expression platform at U-Protein Express BV). This medium was termed complete medium (CM).
Tumor organoids were grown in parallel in 2 types of tumor organoid media, tumor medium 1 (TM1) and tumor medium 2 (TM2). TM1 was identical to CM, the only difference being the absence of EGF and PGE2. The difference between TM2 and CM was the absence of PGE2, A83-01, and Wnt3a-conditioned medium.
For passaging, organoids were collected, washed, and disrupted either by mechanical shearing or digestion with TrypLE Express (Life Technologies; 12605-010). After passaging, organoid fragments were replated in fresh BME, and 10 µM Y-27632 was added to prevent cell death. The organoid passaging procedure is described in detail in SI Appendix.
RNA Isolation and RNA Sequencing.
Total RNA was isolated from organoids that had been passaged 4 to 6 d earlier using the Qiagen RNeasy Mini Kit. The quality and quantity of isolated RNA were checked and measured using the Bioanalyzer 2100 RNA Nano 6000 Kit (Agilent; 5067-1511). Library preparation was performed with 500 ng of total input RNA using the Truseq Stranded Total RNA Kit with Ribo-Zero Human/Mouse/Rat Sets A and B (Illumina; RS-122-2201 and RS-122-2202). Library quality was checked using the Agilent High-Sensitivity DNA Kit (5067-4626) and the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific; Q32854). Libraries were pooled to a final concentration of 2 nM. Library pools (1.0 to 1.4 pM) were loaded and sequenced on an Illumina NextSeq system with 2 × 75-bp high output. Samples were sequenced to an average depth of 22.2 ± 7.2 million fragments. After quality control, mapping and counting analyses were performed using our in-house RNA analysis pipeline v2.1.0 (https://github.com/UMCUGenetics/RNASeq), based on best practices guidelines (https://software.broadinstitute.org/gatk/documentation/article.php?id=3891).
RNA Isolation, cDNA Synthesis, and qPCR.
Organoids were collected from tissue culture plates and washed twice in 10 mL of +/+/+. RNA was extracted using The Qiagen RNeasy Mini Kit according to protocol. RNA was measured with a NanoDrop spectrophotometer (Thermo Fisher Scientific). For cDNA synthesis, GoScript Reverse Transcriptase (Promega; A5003) was used according to protocol. qPCR reactions were performed in 384-well format using IQ SYBR Green (Bio-Rad; 1708880). More details are provided in SI Appendix.
IHC Staining.
Tissue and organoids were fixed in 4% paraformaldehyde, followed by dehydration, paraffin- embedding, sectioning, and standard H&E staining. Staining on tissue was performed at the University Medical Center Utrecht using the following antibodies: anti-SMAD4 antibody (Santa Cruz Biotechnology; mouse monoclonal, clone B-8, catalog no. sc7966, diluted 1:300, 1 h at room temperature), anti-TP53 antibody (Thermo Fisher Scientific; mouse monoclonal, clone D07+BP53-12, catalog no. MS-738, ARS pH 9, diluted 1:2,000, 1 h at room temperature) or anti-MTAP antibody (Abcam ab126770, rabbit monoclonal, clone ERP689, catalog no. ab126770, ARS pH 6, diluted 1:1,000, overnight incubation at 4 °C). More details on staining procedures are given in the SI Appendix.
SMAD4 Signaling Detection.
To detect the presence of SMAD4, organoids were cultured for 2 wk in CM lacking A83-01 and Noggin. Suring this 2-wk period, organoids were passaged once. After 14 d, organoids were collected for RNA isolation and cDNA synthesis as described elsewhere.
DNA Isolation and KRAS PCR of PDOs.
DNA was isolated according to protocol, using the ReliaPrep gDNA Tissue Miniprep System (Promega; A2052) according to protocol. qPCR for exon 2 of human KRAS was performed using forward primer 5′-ACACGTCTGCAGTCAACTGG-3′ and reverse primer 5′- TAACTTGAAACCCAAGGTAC-3′, with GoTaq DNA Polymerase (Promega; M3008) according to protocol at an annealing temperature of 58 °C. A 1% Agarose gel was used to purify the qPCR product, and gel extraction was performed using the QIAquick Gel Extraction Kit (Qiagen; 28706). DNA was sequenced (using both forward and reverse sequencing primers) by Macrogen.
DNA Isolation and WGS of Organoid Lines.
Organoids were dissociated and DNA was isolated using the QiaSymphony DSP DNA Mini Kit (Qiagen; 937236). Libraries were prepared using the Illumina TruSeq DNA Nano Library Prep Kit (20015964). Paired-end sequencing of the organoid lines was performed (2 × 150 bp) on the generated libraries with 30× coverage using the Illumina HiSeq X Ten sequencing system at the Hartwig Medical Foundation.
Somatic Mutation Calling.
This procedure is described in detail in SI Appendix.
High-Throughput Drug Screens.
A total of 24 PDOs were screened in the drug screening facility of the Sanger Institute. In short, organoids were dissociated into single cells and plated in 384-well plates 4 d later. Test compounds were added 24 h later. After 72 h, cell viability was determined using a CellTiter-Glo cell viability assay (Promega). Details on organoid drug screening and cell viability calculations are provided in SI Appendix.
Drug Screen Analysis and z-Score Calculation.
IC50 values were normalized to a z-score, using the formula z = (x − μ)/σ, where x is the IC50 value of the PDO of interest, µ is the average IC50 for all PDOs tested, and σ is the standard of the IC50 values for all PDOs tested. All calculations were performed with these values to visualize differences in drug response between PDOs, in contrast to differences in IC50 values between different compounds.
PRMT5 Inhibitor Drug Screens.
For exposure to EZP015556, cells were plated in BME droplets and exposed to a gradient of this compound for 14 d. The medium was refreshed every 3 to 4 d. Readout was performed as previously described, using CellTiter-Glo. Technical details and cell viability calculations are provided in SI Appendix.
Organoid Infection with MTAP Lentivirus.
MTAP lentivirus was produced in HEK293 cells (SI Appendix). For infection, organoids were collected and then disrupted into small clumps/single cells using TrypLE. After washing with 10 mL of +/+/+, the organoid pellet was resuspended in 150 μL of virus suspension, 1 μg/mL polybrene was added, and the mixture was transferred to a 48-well plate. After a 1-h centrifugation (600 × g, 32 °C), cells were left to incubate for 6 h with the virus. Then the organoid pellet was collected, washed twice with 10 mL of +/+/+, and plated as usual. After recovery (3 to 5 d), organoids were cultured in the presence of 1 μg/mL puromycin (InvivoGen; 58-58-2) to select for infected organoids. Infected organoids were expanded as usual. When ready, organoids were exposed to PRMT5 inhibitor as described previously in either the presence or absence of 3 μg/mL doxycycline.
Sample Preparation for Liquid Chromatography-Mass Spectrometry Measurements.
Organoids were passaged and cultured for 7 d. On the day of collection, 2 U/mL dispase was added to the wells and left to incubate for 40 min at 37 °C. Subsequently, organoids were washed 3 times in ice-cold PBS and then resuspended in 1 mL of +/+/+. A 10-μL sample was taken for cell counting and transferred to a tube containing 1 mL of TrypLE. After a 5-min incubation, cells were counted. The remaining intact organoids were collected for metabolomic analysis. For this, organoid samples were quenched in 500 µL of dry-ice cold methanol/water solution (80%/20%; vol/vol; dry-ice cold), and then stored at −80 °C. Details on liquid chromatography-mass spectrometry sample preparation and measurements are provided in SI Appendix.
Identification of Synergistic Drug Combinations and Associated Biomarkers.
A combination drug screen was performed using 9 drugs (MK-2206, trametinib, linsitinib, lapatinib, mk-1775, taselisib, 5-fluorouracil, sorafenib, and SCH772984) in combination with 1 of 2 anchor drugs (gemcitabine and trametinib), for a total of 11 unique combinations. Library compounds were screened using a 7-point dose–response curve with a half-log dilution series covering a 1,000-fold range over a 72-h period, together with an anchor compound at fixed concentration. Drug combination synergy is measured as the difference between the expected and observed areas under the fitted dose–response curve (delta fAUC). The area under the curve is calculated using trapezoidal integration (AUC) and integrating the curve using the estimated parameters (fAUC). Synergy is present when the median delta fAUC across biological replicates is positive.
We implemented a drug response model to integrate multiple molecular data types (SNPs/indels, copy number, and mRNA expression) when predicting drug combination synergy using TANDEM (40). TANDEM differs from conventional models by using a 2-step approach that first fits an elastic net to predict synergy based on copy number and mutation data, then fits a second elastic net model to predict the residuals from the first model. This approach maximizes the interpretability by emphasizing genetic perturbation of upstream pathways. Features selected by TANDEM were ranked according to magnitude of the elastic net coefficient, scaled by the variance in predicted outcomes. The features depicted represent a selection from the top-ranked genetic events (mutations and copy number aberrations). Full details on the complete analysis are provided in SI Appendix.
Data Availability.
The organoid DNA and RNA sequencing data have been deposited at the European Genome-phenome Archive (https://www.ebi.ac.uk/ega/studies/EGAS00001000369; accession no. EGAS00001003369). WGS data of metastatic pancreatic cancer patients were obtained from the Hartwig Medical Foundation and are available under data request number DR-010. This WGS data are freely available for academic use from the Hartwig Medical Foundation through standardized procedures; a request form is available at https://www.hartwigmedicalfoundation.nl.
Supplementary Material
Acknowledgments
We thank all the employees of U-PORT UMC Utrecht for patient inclusion and tissue acquisition, Kim Boonekamp for her help with MTAP experiments, Fjodor Yousef Yengej for his help with qPCR experiments, Jens Puschhof and Maarten Geurts for their valuable input on drug screen analysis, and the Genomics of Drug Sensitivity in Cancer screening team. This publication and the underlying study were made possible partly on the basis of the data that the Hartwig Medical Foundation and the Center of Personalized Cancer Treatment made available to the study. This work was funded by the Oncode Institute (partly financed by the Dutch Cancer Society), the gravitation program CancerGenomiCs.nl from the Netherlands Organization for Scientific Research, and a Stand Up to Cancer International Translational Cancer Research Grant, a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research (SU2C-AACR-DT1213) and a ZonMw grant (116.006.103). M.J.G. received funding from the Wellcome Trust (206194) and Cancer Research UK (C44943/A22536). The research of L.A.A.B. is funded by the Dutch Digestive Foundation (MLDS CDG 14-020).
Footnotes
Competing interest statement: H.C. is an inventor listed on several patents related to organoid technology. H.C. is the (unpaid) Chief Scientific Officer of Hubrecht Organoid Technology, a cofounder of Surrozen, and a scientific advisory board member for Kallyope, Merus, and Decibel. H. Clevers is a nonexecutive board member of Roche and Genentech and a scientific advisor for Life Sciences Partners. E.D. is an inventor on a patent related to organoid technology.
Data deposition: The DNA and RNA sequences described in this work have been deposited in the European Genome-phenome Archive (https://www.ebi.ac.uk/ega/studies/EGAS00001000369; accession no. EGAS00001003369).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911273116/-/DCSupplemental.
References
- 1.Howlader N., Noone A., Krapcho M., Miller D., Bishop K., Kosary C. et al. , “SEER Cancer Statistics Review, 1975–2014” (National Cancer Institute, 2016).
- 2.Schmuck R. B., de Carvalho-Fischer C. V., Neumann C., Pratschke J., Bahra M., Distal bile duct carcinomas and pancreatic ductal adenocarcinomas: Postulating a common tumor entity. Cancer Med. 5, 88–99 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conroy T., Desseigne F., Ychou M., Bouche O., Guimbaud R., Becouarn Y. et al. , FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl J Med. 364, 1817–1825 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Von Hoff D. D., Ervin T., Arena F. P., Chiorean E. G., Infante J., Moore M et al. , Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N. Engl J Med. 369, 1691–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seino T., et al. , Human pancreatic tumor organoids reveal loss of stem cell niche factor dependence during disease progression. Cell Stem Cell 22, 454–467.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Tiriac H., et al. , Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov. 8, 1112–1129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailey M. H., et al. , Comprehensive characterization of cancer driver genes and mutations. Cell. 173, 371–385.e18 (2018). Erratum in: Cell174, 1034–1035 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamborero D., et al. , Cancer Genome Interpreter annotates the biological and clinical relevance of tumor alterations. Genome Med. 10, 25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drost J., Clevers H., Organoids in cancer research. Nat. Rev. Cancer. 18, 407–418 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Sachs N., et al. , A living biobank of breast cancer organoids captures disease heterogeneity. Cell 172, 373–386.e10 (2018). [DOI] [PubMed] [Google Scholar]
- 11.van de Wetering M., et al. , Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vlachogiannis G., et al. , Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359, 920–926 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Driehuis E., et al. , Oral mucosal organoids as a potential platform for personalized cancer therapy. Cancer Discov. 9, 852–871 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Lee S. H., et al. , Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell 173, 515–528.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kopper O., et al. , An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat Med. 25, 838–849 2019). [DOI] [PubMed] [Google Scholar]
- 16.Boj S. F., et al. , Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bosari S., et al. , p53 gene mutations, p53 protein accumulation and compartmentalization in colorectal adenocarcinoma. Am. J. Pathol. 147, 790–798 (1995). [PMC free article] [PubMed] [Google Scholar]
- 18.Freed-Pastor W. A., Prives C., Mutant p53: One name, many proteins. Genes Dev. 26, 1268–1286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bailey P., et al. ; Australian Pancreatic Cancer Genome Initiative , Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531, 47–52 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Blokzijl F., et al. , Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538, 260–264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor A. M., et al. ; Cancer Genome Atlas Research Network , Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell 33, 676–689.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christensen S., et al. , 5-Fluorouracil treatment induces characteristic T→G mutations in human cancer. Nat Commun. 10, 4571 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davies H., et al. , HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat. Med. 23, 517–525 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexandrov L. B., et al. , Clock-like mutational processes in human somatic cells. Nat. Genet. 47, 1402–1407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jager M., et al. , Deficiency of nucleotide excision repair explains mutational signature observed in cancer. bioRxiv 221168; 10.1101/221168 (November 15, 2018). [DOI] [PMC free article] [PubMed]
- 26.Chan-Penebre E., et al. , A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat. Chem. Biol. 11, 432–437 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Beroukhim R., et al. , The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamijo T., et al. , Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91, 649–659 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Serrano M., Hannon G. J., Beach D., A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 366, 704–707 (1993). [DOI] [PubMed] [Google Scholar]
- 30.Zhang H., Chen Z. H., Savarese T. M., Codeletion of the genes for p16INK4, methylthioadenosine phosphorylase, interferon-alpha1, interferon-beta1, and other 9p21 markers in human malignant cell lines. Cancer Genet. Cytogenet. 86, 22–28 (1996). [DOI] [PubMed] [Google Scholar]
- 31.Zappia V., Della Ragione F., Pontoni G., Gragnaniello V., Carteni-Farina M., Human 5'-deoxy-5'-methylthioadenosine phosphorylase: Kinetic studies and catalytic mechanism. Adv. Exp. Med. Biol. 250, 165–177 (1988). [DOI] [PubMed] [Google Scholar]
- 32.Kryukov G. V., et al. , MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science 351, 1214–1218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mavrakis K. J., et al. , Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science 351, 1208–1213 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Marjon K., et al. , MTAP deletions in cancer create vulnerability to targeting of the MAT2A/PRMT5/RIOK1 axis. Cell Rep. 15, 574–587 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Karkhanis V., Hu Y. J., Baiocchi R. A., Imbalzano A. N., Sif S., Versatility of PRMT5-induced methylation in growth control and development. Trends Biochem. Sci. 36, 633–641 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevens A. P., et al. , Direct and tumor microenvironment-mediated influences of 5′-deoxy-5′-(methylthio)adenosine on tumor progression of malignant melanoma. J. Cell. Biochem. 106, 210–219 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Limm K., et al. , Deregulation of protein methylation in melanoma. Eur. J. Cancer 49, 1305–1313 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Basu I., et al. , A transition state analogue of 5′-methylthioadenosine phosphorylase induces apoptosis in head and neck cancers. J. Biol. Chem. 282, 21477–21486 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Vlachogiannis G., et al. , Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359, 920–926 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aben N., Vis D. J., Michaut M., Wessels L. F. A., TANDEM: A two-stage approach to maximize interpretability of drug response models based on multiple molecular data types. Bioinformatics 32, i413–i420 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The organoid DNA and RNA sequencing data have been deposited at the European Genome-phenome Archive (https://www.ebi.ac.uk/ega/studies/EGAS00001000369; accession no. EGAS00001003369). WGS data of metastatic pancreatic cancer patients were obtained from the Hartwig Medical Foundation and are available under data request number DR-010. This WGS data are freely available for academic use from the Hartwig Medical Foundation through standardized procedures; a request form is available at https://www.hartwigmedicalfoundation.nl.