Significance
During pathogenic invasion, neutralizing antibodies (nAbs) are involved in regulating immune clearance and evoking the host-protective response. We previously reported a highly potent nAb 8C11 against HEV, an RNA virus with an icosahedral capsid and associated with abundant acute hepatitis. Structural analysis demonstrates that the binding of 8C11 to HEV VLPs would result in tremendous spatial clashing with the capsid. Cryo-EM analysis showed that 8C11 binding leads to complete disorder of the outer rim of the VLP at earlier stages (∼15 min) and causes the dissociation of HEV VLPs into homodimer species within 2 h. Similar 8C11-mediated dissociation was observed for the native HEV virion. Our results categorize a viral neutralization mechanism and suggest a strategy to generate 8C11-like antibodies.
Keywords: viral neutralization, antibody, cryo-EM structure, virus dissociation
Abstract
In adaptive immunity, organisms produce neutralizing antibodies (nAbs) to eliminate invading pathogens. Here, we explored whether viral neutralization could be attained through the physical disruption of a virus upon nAb binding. We report the neutralization mechanism of a potent nAb 8C11 against the hepatitis E virus (HEV), a nonenveloped positive-sense single-stranded RNA virus associated with abundant acute hepatitis. The 8C11 binding flanks the protrusion spike of the HEV viruslike particles (VLPs) and leads to tremendous physical collision between the antibody and the capsid, dissociating the VLPs into homodimer species within 2 h. Cryo-electron microscopy reconstruction of the dissociation intermediates at an earlier (15-min) stage revealed smeared protrusion spikes and a loss of icosahedral symmetry with the capsid core remaining unchanged. This structural disruption leads to the presence of only a few native HEV virions in the ultracentrifugation pellet and exposes the viral genome. Conceptually, we propose a strategy to raise collision-inducing nAbs against single spike moieties that feature in the context of the entire pathogen at positions where the neighboring space cannot afford to accommodate an antibody. This rationale may facilitate unique vaccine development and antimicrobial antibody design.
Neutralizing antibodies (nAbs) play key roles in immune clearance and the subsequent evocation of the host-protective response during pathogenic invasion. NAbs mediate viral neutralization by blocking virus attachment and, in turn, prohibiting any conformational changes required for viral membrane fusion or the release of progeny virions (1–5). These complicated procedures initiate an immune response, clearing the virus through the activation of various integrated phagocytotic and degradative host-immune pathways (1–3). Indeed, numerous signaling pathways and effectors are associated with antibody-mediated immune clearance and protection: antibody-mediated endocytosis, antibody-dependent cell-mediated cytotoxicity, and complement-dependent cytotoxicity, among others (4, 6). These processes are energetically demanding and exhaust the host cell’s supplies, rendering the cell susceptible to secondary inflammatory events (7).
Physical collisions at the molecular level often induce dramatic structural and conformational changes and rearrangements in multisubunit protein ensembles (8, 9). Studies have shown that virus dissociation can be influenced in vitro by physical disturbances, such as high pressure or extreme pH (10–12). Indeed, work on the picornaviruses has shown that binding of nAbs destabilizes the virion and releases the viral genome, even though the structural integrity of the resultant capsid is maintained (13–17). This raises the question as to whether a physical collision created while the antibody is engaged on the surface of the pathogen could eliminate the pathogen or render it harmless in situ. We surmised that such a mode of pathogen elimination would be more efficient than the complex pathways activated through conventional immune processes of viral clearance.
The hepatitis E virus (HEV), associated with abundant acute hepatitis, is a positive-sense single-stranded RNA virus, consisting of a nonenveloped icosahedral capsid (27–34 nm) and 7.2 kb of genomic RNA (18). The HEV genome includes 3 ORFs (ORF1–3) of which ORF2 encodes 2 ∼660-amino acid (aa) translated proteins via 2 alternative start codons: the capsid protein (pORF2) (18) and a secreted form of ORF2 (ORF2S) (19). Crystal and cryo-electron microscopy (cryo-EM) structures of the HEV capsid proteins have been extensively elucidated, including the HEV protruding domain (E2s, amino acids 455–602) and the T = 1 and T = 3 VLPs (10, 20–25). Monomeric pORF2 is composed of a shell domain (S domain and aa 118–317), a middle domain (M domain and aa 318–451), and a protrusion domain (P domain and aa 452–606). The E2s domain folds into a typical β-barrel, 2 of which tightly form a homodimeric spike that protrudes from each of the icosahedral 2-fold axes of the capsid. A flexible groove region in E2s is crucial in the binding of nAbs and is proposed to be the cellular receptor-binding region (26).
We previously reported a crystal structure of the HEV E2s domain in the complex with nAb 8C11. This nAb conferred potent neutralizing ability in both our HEV cell model and an animal model through binding to a conformational neutralization epitope flanking the protrusion structure (23). Intriguingly, the light chain of 8C11 Fab in the crystal structure physically collides with the neighboring M domain of pORF2 when the complex structure is superimposed on the whole HEV T = 1 VLP crystal structure (23). We hypothesized that such a collision could render the virus ineffective through physically disrupting its structure. To further understand the consequence of this structural clash on the virus, here, we investigate the dynamic structural variation of HEV VLP during binding with 8C11 Fab using size-based characterization and cryo-EM 3D reconstruction. We found a tremendous structural dissociation of the 8C11-bound VLP that was substantially affected by time and antibody dosage. The binding of 8C11 initiated a complete disordering of the VLP rim structure in the early stages (∼15 min) of the 8C11-VLP interaction with no structure resolvable thereafter. Similar capsid deconstruction was also observed for the native HEV virion as determined using a fluorescence-staining RNA release assay. We, thus, propose an immune-favorable rationale for raising collision-inducing nAbs for virus neutralization and offer a strategy for such antibody generation.
Results
The 8C11 Binding Induces a Physical Collision on HEV VLPs and Dissociates the Particles.
We previously reported the crystal structure (PDB no. 3RKD) of HEV nAb 8C11 in complex with E2s (P domain) at a binding ratio of 2 Fab to 1 E2s dimer (Fig. 1A) (23). When we fitted the cocrystal structure into the known density maps of HEV T = 1 (generated from the crystal structure of PDB no. 2ZZQ) and T = 3 VLPs (EMDB no. 5173), respectively (24), the 8C11 Fab binding showed enormous clashes with the neighboring pORF2 monomers in both models (Fig. 1 A and B). The Fabs overlaid with the T = 3 capsid shell (Fig. 1B) more conspicuously than with the T = 1 capsid (Fig. 1A) because of a higher volume of T = 3 particle involvement in the structural overlapping and a flatter surface curvature. T = 3 VLPs are believed to be virion-sized particles assembled with the involvement of RNA molecules (24). In the superimposed models, about 4,456 Å3 of each Fab 8C11 (total ∼18,660-Å3 volume) in the E2:8C11 crystal structure overlaps with the T = 1 VLP model, whereas, on the T = 3 map due to the existence of 3 different spatial relationships between Fab and P (and M) domain(s) of the adjacent E2s (24), the volume of overlapping density between the Fab 8C11 and the neighboring pORF2 monomers was 7,721, 4,779, and 2,311 Å3, respectively. These results indicate that the binding modality of 8C11 in the 8C11:E2s cocrystal structure may cause tremendous spatial clashing with its neighboring viral capsid protein, given that it directly binds to the T = 1 or T = 3 icosahedral shell.
Fig. 1.
Physical collision created by 8C11 binding causes HEV VLPs to dissociate. (A and B) Fitting of the crystal structure of the E2s:8C11 complex (PDB no. 3RKD) to density maps of HEV T = 1 (generated from the crystal structure of PDB no. 2ZZQ) (A) and T = 3 (EMDB no. EMD-5173) (B) VLP structures, respectively. The E2s:8C11 complex model, rendered in cartoon mode, was fitted to VLPs according to the E2s part (green), and the resultant overlapping moieties between VLPs and 8C11 are shown in semitransparent surface mode in teal (heavy chain) and marine (light chain). The cartoons above the Right depict structural overlapping domains of p495 with Fab 8C11, T = 1 has a single interaction scenario where the M domain and neighboring P domain are overlapping to Fab with a volume of 4,456 Å3, whereas T = 3 with 2 dimer forms A-B, C-C in its icosahedral lattice has 3 scenarios (24), (1) M domain, neighboring M and P domains creating an overlapping volume of 7,721 Å3, (2) M domain and neighboring P domain, 4,779 Å3, (3) neighboring P domain, 2,311 Å3. (C) HPSEC analysis of HEV VLPs mixed with 8C11 or 3B6 Fab. An extra peak (indicated by the black arrow, named the smaller complex) demonstrates the presence of a species that is larger than 8C11 Fab but far smaller than VLPs in the VLP:8C11 sample. VLPs were, thus, almost depleted as compared with the control. (D) Determination of the molecular weight of the smaller complex in solutions through the sedimentation velocity test. The smaller complex in the VLP:8C11 HPSEC profile was harvested. The c(S) and c(M) profiles suggested the smaller complex fraction is predominantly the p495 dimer in the complex with 2 8C11 Fabs (3.4S, 160.3 kDa) in addition to the surplus Fabs (7.5S, 47.9 kDa). (E) SDS/PAGE analysis of HEV VLPs mixed with 8C11 Fabs. The smaller complex fraction was resolved in lane 4, suggesting a complex comprising both p495 and 8C11 Fab. (F–H) Representative negative-staining micrographs of HEV VLPs (F), VLP: 8C11 (G) and VLP: 3B6 (H), which manifested no detectable particle for the VLP:8C11 sample.
To observe the subsequent events following 8C11 binding to HEV particles, HEV ORF2 aa112-608 (p495) from a genotype 1 strain was expressed and purified from insect cells. The VLPs were homogenous, with an ∼20- to 30-nm diameter in neutral solution (SI Appendix, Fig. S1) as described in our previous studies (27). The obtained HEV VLPs were then used to prepare an immune complex with the antigen-binding (Fab) fragments of nAb 8C11 (VLP:8C11). Another nAb 3B6, which recognizes the top region of the P domain, was recruited to prepare a control immune complex (VLP:3B6) (28). Both monoclonal antibodies (mAbs) 8C11 and 3B6 showed high binding and neutralizing efficiency against HEV VLPs and the native virus before immune complex preparation (SI Appendix, Fig. S2). It should be mentioned that we ever contrived to generate T = 3 VLPs (ORF2 aa14-608) in insect cells according to Li’s protocol (29), however, the purified sample was not abundant and poorly homogeneous for similar biochemical and structural analyses as performed at T = 1 VLPs.
Immune complexes of VLP:8C11 and VLP:3B6 were prepared in a 1:2 molar ratio (p495:Fab) to have a surplus of Fab and incubated at 37 °C for 2 h. The control sample (VLP:3B6) resolved as 2 major peaks in the high-performance size-exclusion chromatography (HPSEC) profile with retention volumes of 6.8 and 8.8 mL corresponding to the immune complex (VLP:3B6) and the excess 3B6 Fab, respectively; this was compared with 7.0 and 9.2 mL for VLP and Fab alone, respectively (Fig. 1C). Strikingly, most of the VLP and 8C11 Fab mixture exhibited a median component (8.3 mL) between the supposed particle complex (6.7 mL, similar with VLP:3B6) and the excess 8C11 Fab (9.1 mL) (Fig. 1C). The amount of the median species was ∼8.7 times that of the supposed particles in terms of integrated peak areas under the HPLC curve. We speculated that the smaller species was the disrupted product generated by the physical collision between 8C11 Fab and VLPs. Moreover, both peaks were confirmed to be immune complexes containing p495 and Fab in the SDS/PAGE (Fig. 1E).
We next examined the morphology of these immune complexes in negative-staining transmission electron microscopy (TEM). The control VLP:3B6 had a larger particle size than the HEV VLPs (Fig. 1 F and H), whereas, only a few VLP-sized particles could be observed in the VLP:8C11 sample (Fig. 1G), consistent with the HPSEC results. The predominantly smaller p495:8C11 complex was further characterized as about 7.5 S (about 160 kDa in c[M] calculation, supposed to be 2 p495 binding with 2 8C11 Fabs) through sedimentation velocity (SV) tests in analytical ultracentrifugation (Fig. 1D and SI Appendix, Fig. S3). Taken together, the binding of 8C11 with HEV VLPs led to the dissociation of VLPs into p495 homodimer, presumably due to the physical collision between the 8C11 and the capsid as hypothesized in the structural analysis.
The 8C11-Bound VLPs Are Disrupted in a Concentration- and Time-Dependent Manner.
We then investigated the VLP dissociation potential with different starting concentrations of 8C11 Fab. We surmised that the amount of 8C11 Fab input could influence the dissociation efficacy of the VLP:8C11 complex. We found that less 8C11 (p495:8C11 molar ratio = 1:1) resulted in only partial (∼57%) complex dissociation, whereas increased Fab ratios (1:2 and 1:4) led to complete dissociation (Fig. 2A). For the 1:2 Fab ratio, when the incubation time was reduced to 15 min, a predominant VLP-sized peak was retained with the dissociated components accounting for only 1-quarter of the VLP-sized peak in the HPSEC profile (Fig. 2B). Longer reaction times (30 min to 2 h) as expected led to higher rates of dissociation (Fig. 2B). Additionally, the treatment of intact 8C11 antibody induced similar dissociation consequence upon HEV VLPs as its Fab moiety (SI Appendix, Fig. S4).
Fig. 2.
Dissociation extent of HEV VLPs depends on 8C11 concentration and incubation time. (A) The 8C11 concentration-dependent dissociation of HEV VLPs. HEV VLPs were mixed with 8C11 at various p495:8C11 molar ratios (1:1, 1:2, and 1:4) for 2 h and analyzed by HPSEC analysis. Conspicuously, the p495:8C11 ratio of 1:4 completely dissociated HEV VLPs into the smaller species, which reflects law of mass action of 8C11 upon VLP dissociation. (B) Incubation time-dependent dissociation of HEV VLPs. HEV VLPs were mixed with 8C11 at an equal molar ratio (1:1) and incubated for 15 min and 2 h. The dissociated species (retention volume of ∼8.5 mL) became a predominant peak in the HPSEC profiles as the incubation time extended from 15 min to 2 h. (C and D) Cryo-EM analysis of HEV VLPs alone and mixed with mAb 3B6 for 2 h served as a control. (E–H) Cryo-EM analysis of HEV VLPs mixed with 8C11 for 15 min to 2 h. For each cryo-EM analysis, 1 representative raw cryo-EM image (Left) and the Top 2 classes of 2D classification (Right 2 panels) are shown. The number of visible particles in the cryo-EM images decreases with incubation time as does the density volumes of the rim of particle classes.
We next sought to track the dissociation of VLPs at different time points by cryo-EM analysis. HEV VLPs were mixed with Fab 8C11 (p495:8C11, 1:2) for 15, 30, 60 min, and 2 h and then plunge frozen for the cryo-EM image collection (Fig. 2 C–H, Left). HEV VLP in the complex with Fab 3B6 served as a control, and the cryo-EM images and 2D classification results showed identifiable VLPs and complex features (Fig. 2 C and D). Consistent with the HPSEC results shown in Fig. 2B, VLPs in the presence of 8C11 for 15 min showed particle sizes that were overall similar to that of VLPs (Fig. 2C) and substantially smaller than the size of VLP:3B6 (Fig. 2D); albeit there was a higher density of particles at the rim in response to 8C11 binding (Fig. 2E). This binding is consistent with the 2 modes of binding noted for 8C11 (binds to the flank side of protrusion domain, Fig. 1A) and 3B6 [proposed to bind the top of protrusion spike of the particles in terms of its epitope mapping (28)]. With a longer incubation (>15 min), the density of the outside rim of the particle blurred with fewer intact particles screened out from the 2D classification (Fig. 2 E–H and SI Appendix, Table S1). By 2 h, few particles could be detected in both the HPSEC (Fig. 2A) and the cryo-EM images (Fig. 2 H, Left), and the averaged 2D classes from the available particles exhibited poor morphology (Fig. 2H). These results suggest that physical collision imposed by 8C11 binding leads to VLP dissociation within 2 h.
Cryo-EM Reconstruction of the Disassembling Intermediates of the 8C11-Bound VLPs.
The cryo-EM structures of HEV VLP, VLP:8C11 (15-min incubation), and VLP:3B6 were further determined at 3.4-, 3.6-, and 7.2-Å resolutions, respectively, with Relion (30) (SI Appendix, Fig. S5 and Table S2). The structure of VLP:3B6 at a medium resolution showed the proposed binding to the top of the protrusion spike as indicated by epitope mapping in our previous study (28). We then built the atomic model for HEV VLP, which exhibits T = 1 icosahedral symmetry with 60 copies of the p495 subunit arranged into 30 dimeric protruding spikes located at each icosahedral 2-fold axis (Fig. 3A). The densities attributable to the residue backbones and side chains were clearly recognizable for all 3 of the S, M, and P domains (Fig. 3 B and C); these are structurally identical to those previously reported in the crystal structures of the HEV capsid protein (21, 22, 24, 25) (SI Appendix, Fig. S6) and used in our structural analysis of the E2s:8C11 immune complex (Fig. 1A).
Fig. 3.
Cryo-EM structures of HEV VLP, VLP:8C11, and VLP:3B6. (A–C) A 3.4-Å cryo-EM structure of HEV VLPs. (A) The density map of the particle is colored radially, and the particle diameter is indicated. (B) An asymmetric unit (ASU) and corresponding atomic model (P domain, green; M domain, orange; S domain, magenta). (C) Typical cryo-EM map densities of an α-helix and β-sheet at 3.4-Å resolution. (D–F) A 3.6-Å cryo-EM structure of the VLP:8C11 complex incubated for 15 min. (D) The density map of the complex shows a disordered spike region, and the diameter of the core region is indicated. (E) p495 model and crystal structure of 8C11 Fab (blue) were fitted into the density of an ASU of the icosahedron. Few visible density regions can be matched to both the 8C11 Fab and the P domain (highlighted in the dashed square), whereas the M and S domains of the p495 model fit into the VLP:8C11 density map as well as VLP only. (F) Excellent model-fitted densities for the same parts of the α-helix and the β-sheet as (C). (G and H) A 7.2-Å cryo-EM map of the HEV:3B6 complex (G) and an ASU fitted with the model of p495 (green) and 3B6 Fab (red) (H). The diameter of VLP:3B6 is indicated.
It is striking that the cryo-EM map of the 8C11-bound VLP also demonstrated an overall high resolution of 3.6 Å. However, only the S and M domains were of high resolution with an intact and identical atomic model to that shown in the structure of HEV VLP (SI Appendix, Fig. S6). The moieties corresponding to the P domain and Fab 8C11, on the other hand, were extremely disordered in the density map with some residual, albeit irregular, density around the core shell constituted by the S and M domains (Fig. 3 D–F). By comparison, the cryo-EM structure of VLP:3B6 had intact VLP binding, with 60 separate Fabs and typical resolution of the outward Fab components of the shell (Fig. 3 G and H). These results together indicate that 8C11 binding to VLP bumps against neighboring p495 subunits and leads to a disordered rearrangement of the P domains, destroying their icosahedral symmetry.
To investigate if there was any order to the 8C11-bound P domain, we undertook a structure reconstruction with the same dataset of the VLP:8C11 complex without any icosahedral symmetry (C1). The resultant density map, albeit at a lower resolution (∼25 Å), showed that the 8C11-bound P domains were completely disordered, whereas the core shell comprising the M and S domains remained intact as an icosahedral structure (SI Appendix, Fig. S7). We propose that the status of the 8C11-bound VLP at 15 min may represent VLP disassembling intermediates—8C11 is still bound with the P domain and the P domain still linked to the M domain—and surmise that these intermediates probably further dissociate into smaller species with longer incubation time (Fig. 2A).
The 8C11 Binding Disintegrates Native HEV and Exposes the Genomic RNA.
Next, we sought to examine whether such disassembly induced by 8C11 binding could be replicated with native HEV. To date, we still cannot acquire an adequate sample of native HEV for structure determination due to the low efficiency of the HEV cell culture system (31–33). Nonetheless, we were able to prepare native HEV samples from experimental monkeys challenged with native HEV as described in our previous studies (33). First, we checked the integrity of the native HEV after incubation with 8C11 for 2 h by an ultracentrifugation/precipitation-coupling genome-based qPCR assay; this was based on the presumption that 8C11 binding would disassemble the native HEV caused by physical disruption (Fig. 4A) and prevent the disintegrated viral constituents (with lower sedimentation coefficient than the intact native virion) from pelleting by ultracentrifugation. As expected, more than 90% of the native HEV (1.39 × 107 copies of RNA genome per milliliter as compared with the initial virus input of 1.50 × 107 copies of RNA genome per milliliter) in the control sample (mAb 3B6 and buffer only) was harvested via ultracentrifugation, whereas only 52% of the native HEV (7.93 × 106 copies of RNA genome per milliliter) pelleted in the 8C11-treated HEV sample, significantly less than the control sample (Fig. 4B). These findings indicated that 8C11 may also impose capsid disassembly on native HEV as on VLPs.
Fig. 4.
Property alteration of native HEV while binding with 8C11. (A) Assumed binding modality of 8C11 and 3B6 to native HEV (proposed T = 3) illustrated according to the cocrystal structure of E2s:8C11 and the cryo-EM structures of VLP:3B6 and T = 3 HEV VLP. One E2s monomer is colored green, and the models of binding 8C11 and 3B6 Fabs are colored blue and red, respectively. (B) Virus pelleting assay of native HEV pretreated with 8C11 and 3B6. Native HEV pretreated with 8C11 or 3B6 or alone were pelleted by ultracentrifugation to quantify copy numbers of the RNA genome. Input column in the histogram indicates the RNA genomic copy of native HEV before ultracentrifugation. Results were statistically analyzed by an unpaired Student’s t test. *P ≤ 0.05; ns, not significant (8C11 vs. 3B6, P < 0.0001; 8C11 vs. control, P < 0.001; 3B6 vs. control, P = 0.6752 > 0.05). The decrease in RNA copy number in the 8C11-treated sample as compared with the control suggests that half of the native HEV changes to a smaller material that cannot be pelleted as an intact virus. (C) Real-time PaSTRy assay of native HEV pretreated with 8C11 and 3B6. There is a trend for an increase in fluorescence from the dye bound to the HEV RNA genome exposed by 8C11 binding; this indicates that the HEV capsid is subjected to ongoing perturbation with 8C11 treatment over ∼60 min. The fluorescent signal reaches a plateau early (∼60 min) and remains there until the assay end point (∼150 min). As for the controls, there were no substantial changes in the fluorescence signals of 3B6 or the no-antibody control.
Finally, we monitored the dynamic process of HEV capsid disintegration resulting from 8C11 binding using a particle stability thermal release assay (PaSTRy) (34) in which a dye is used to stain the exposed genomic RNA of the native HEV (Fig. 4C) and is read as changes in the fluorescence signal over incubation time (up to 150 min). We found a gradual time-dependent increase in the exposure of the HEV genome upon 8C11 binding until 90 min that stabilized until the assay end point. The dynamic curve indicated an inflection point at about 90 min that remained stable from then on, almost comparable with the HPSEC analysis of the incubation time required for complete VLP disassembly (2 h; Fig. 2A). In contrast, for both the native virus alone and the 3B6-bound virus, the fluorescence signal was maintained at a lower value below the background level over the 150-min assay. These collective biochemical and structural results support the hypothesis that nAb 8C11 may neutralize HEV through a direct physical collision mechanism.
Discussion
Pathogenic neutralization equips specific hosts with adaptive immunity against infection and/or subsequent disease onset. Immune clearance usually requires immune recognition at vulnerable sites of a virus (e.g., receptor-binding sites, metastable-associated sites, among others); an antibody-mediated immune response; and the presence of adaptive humoral and T-cell immunity against pathogens (5). Despite this capacity, the relatively bigger size of various pathogens (several tens to hundreds of nanometers) generally might exhaust great amounts of the host-immune system resources to clear the pathogen, which could lead to secondary immune events, as represented by various inflammatory responses, such as hepatitis in the case of the hepatitis B virus, hepatisis C virus, and HEV infection. To date, at least 5 viral neutralization mechanisms have been identified, and this includes blocking cell attachment through the use of antibodies that bind to specific viral receptor-binding sites or cellular receptors (2, 3). Recent reports have shown that antibodies can induce viral conformational changes and drive uncoating and release of the genome in enteroviruses (13, 14). For some enveloped viruses, such as HIV-1, antibody-induced viral envelope protein dissociations will interfere with virion attachment to the target cell (12, 35). Unlike these conventional mechanisms of neutralization, here, we show evidence for a vigorous mode of antibody-mediated neutralization induced by the physical disruption of the virus through antibody-imposed structural collisions. Here, we show that the 8C11 nAb physically neutralizes the particulate pathogen into smaller dissociated species, which might possibly be beneficial for the host-immune response clearance as indicated in the immunology literature (7).
The spike of the HEV capsid, also referred to as the protrusion domain or the E2s domain, is essential for virus-host interactions (19, 21, 22). The spike comprises 2 representative epitopes on the top or flanking side of the protrusion for antibody binding (Fig. 5). mAb 3B6 confers physically preferable binding to the top of the E2s—believed to the viral receptor-binding site—thereby blocking the virus from attaching to the cell surface (21, 36). The 3B6-bound HEV retains its structural integritywith the genome ensconced inside the capsid (Fig. 4 B and C); such a complex is believed to be neutralized by conventional Fc-dependent immune clearance (Fig. 5). The 3B6-like antibodies are usually generated by the immunization of a native virus, a VLP, or an intact/partial capsid protein (28). In comparison, mAb 8C11 binds to the flanking region of the E2s spike with high affinity (5.89 nM) (23). However, this interaction then distorts the icosahedral arrangement of the protrusion (at ∼30 min, Fig. 3D) and squashes the VLP (51.3S) into smaller pieces (7.5S) within 2 h (Fig. 2A and SI Appendix, Fig. S3), exposing the RNA genome (Fig. 4C). Although there is no available structural information for the mode of native virus dissociation upon 8C11 binding, the T = 3 icosahedral structure of the HEV VLPs (24)—believed to be virion-sized particles—showed similar spatial overlapping in the presence of 8C11 binding (Fig. 1B), implicating similar dissociation of the native HEV as that of VLPs. Thus, 8C11 may function via a novel neutralization mechanism, inducing a physical collision that disassembles HEV into smaller pieces for immune clearance (Fig. 5). The containment and elimination of these smaller pieces might require less effort from the host-immune system as compared with the intact virus as noted for 3B6 neutralization.
Fig. 5.
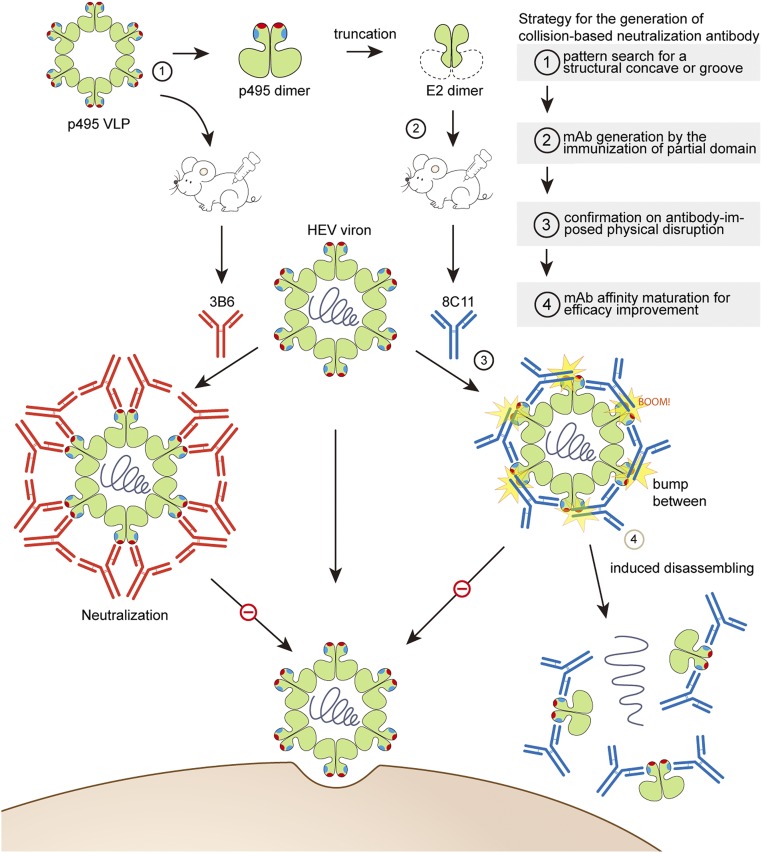
Proposed viral neutralization mechanism of 8C11-mediated physical collision, and a strategy for the generation of 8C11-like mAbs. nAb 3B6, raised from the immunization of HEV VLPs, possesses a conventional neutralization mechanism that directly blocks virus attachment to host cells, which, in some cases, binds to a region that overlaps with receptor binding sites. The nAb-bound virus with no change in the integrity of the capsid then would be cleared by the elaborate host-immune system. The nAb 8C11 was generated by the immunization of the partial capsid protein E2 and found to induce a physical collision when engaged with the whole capsid. Eventually, the virus dissociates into small species and is likely then eliminated by the host-immune system, presumably with less effort. In short, 8C11-like mAbs could be produced by the immunization of a partial moiety of the pathogen, targeting a physically unfavorable binding region (e.g., positions where the neighboring space cannot afford to accommodate an antibody) in the whole virus structure and thereby upsetting pathogenic infection in an immune-favorable way. Proposed strategy for the generation of a collision-based neutralizing antibody is indicated by a step-by-step procedure flowchart in the Upper Right, where every step is directed to the corresponding event that occurs in the antigen immunization and virus neutralization diagram.
The protrusion spike is a common structure on the outermost surfaces of enveloped (e.g., HIV and influenza virus) and nonenveloped (e.g., HEV, rotavirus, and norovirus) viruses. Thus, the use of 8C11-like antibodies for physical collision-mediated neutralization is plausible for a range of viruses. To obtain 8C11-like antibodies that neutralize and eliminate viruses by physical collision, we propose the following structure-guided strategy (Fig. 5) (1): Perform a pattern search on the structures of both the spike moiety and the virus as a whole; i.e., identify a structural concave that is accessible for antibody binding at the flank of the spike but unable to accommodate the entire antibody thereby facilitating the probability of a physical collision; (2) generate mAbs through immunization using only this spike [e.g., E2 in this study (37)], and select mAbs with epitope mapping the flank of the protrusion domain; (3) confirm virus dissociation and neutralization potency through biochemical and virologic analyses; and (4) if possible, enhance the binding affinity—for instance, using an affinity maturation approach—to improve the potency of the antibody-mediated collision for virus dissociation, which point is inferred from the fact that mAb 8C11 confers substantially stronger neutralization against HEV genotype 1 rather than HEV genotype 4 in terms of its binding affinity of genotype 1 having 2 logarithms higher over that of genotype 4 (20, 23). This strategy could be extrapolated for the design of ligands or antibodies that target more varied pathogens not just viruses.
Materials and Methods
Cells and Viruses.
HepG2 (HB-8065, obtained from the ATCC, MD, USA) and Huh7 S10-3 cells (kindly provided by Dr. Suzanne Emerson, NIH) were grown in Eagle's minimum essential medium (MEM, GIBCO) containing 10% FBS (GIBCO, Australia) and antibiotics (10- U/mL ampicillin and 100-U/mL streptomycin) (Lukang; Shandong, China) at 37 °C with 5% CO2. The HEV infectious clone of the Kernow-C1 p6 (genotype 3) was kindly provided by Suzanne Emerson (NIH). HEV stocks were generated by transfecting Huh7 S10-3 cells with in vitro transcribed anisotropic HEV genomic RNA.
Cloning, Expressing, and Purification.
The ORF2 gene fragment of HEV genotype 1, encoding residues 112–606, was cloned into pFastBac plasmid (Invitrogen). Sf9 cells were transfected by lipofection to produce recombinant viruses. Tn5 cells in ESF 921 were infected with the recombinant baculoviruses for 4 d at 30 °C. The target proteins (p495) were mainly secreted into the culture medium. p495 in the cell culture supernatant was concentrated by incubation with 8% polyethylene glycol 6,000 in the presence of 0.4-M NaCl at 4 °C overnight. The precipitate was resuspended in 20-mM phosphate buffer pH 6.0 (PB6.0) and purified using a diethylaminoethyl (DEAE)-5PW column.
Antibody and Fab Preparation.
The nAbs used in this study were produced in mouse ascites fluid and were affinity purified with a Protein A column. Fab fragments were prepared from purified mouse monoclonal antibodies by papain cleavage. A reducing l-cysteine buffer was used to activate the papain, and mAbs were mixed with papain at an appropriate molar ratio and then incubated overnight at 37 °C. The reaction was stopped by the addition of iodoacetamide, and the product was analyzed by SDS/PAGE. The Fab fragments were purified using a TSK-GEL DEAE-5PW column. The Fc fragments and uncleaved mAbs were trapped in the column, whereas, the Fab fragments were collected in the flow-through component.
ELISA.
The 96-well plates were coated with 100-ng/well p495 diluted in 20-mM PB6.0 and, subsequently, incubated at 4 °C for 12 h. The plate was then washed 5 times with PBS containing 0.05% Tween-20 (PBST buffer). The wells were blocked with 200-μL/well blocking buffer (PBS + 0.25% casein +1% gelatin +0.05% proclin-300) for 4 h at 37 °C. The plate was spin dried and incubated with 100 μL/well of serially diluted antibody for 30 min at 37 °C. The wells were washed 5 times with PBST and then incubated with 100 μL/well of HRP-conjugated goat–anti-mouse IgG (1:5,000) for 30 min at 37 °C. The wells were again washed, incubated with substrate for color development, and the absorbance read at 450 nm in a plate reader.
Neutralization Assay.
Antibodies were incubated with the virus for 30 min at 37 °C before HepG2 cell inoculation. After 6 h, the inoculum was removed, and the cells were washed 3 times with PBS, followed by incubation with DMEM supplemented with 10% FBS and 2% DMSO. After 5 d, the cells were stained with anti-HEV-ORF2 monoclonal antibody. The neutralization efficiency was determined by the number of infected cells in each condition. The data were analyzed by nonlinear regression (GraphPad Prism, Inc.) to determine the neutralization obtained with a given antibody concentration and the IC50.
PaSTRy.
Thermofluor experiments (34) were performed with a MX2005p RT-PCR instrument (Agilent). Environment-sensitive fluorescent dye SYTO9 (Invitrogen, S34854) was used to detect the presence of RNA. Each 50-μL reaction was set up in a thin-walled PCR plate, containing 1.0-μg HEV native virion, 5-μM SYTO9, and 3× SYPROred in PBS. The temperature ramped from 25 to 99 °C with the fluorescence level recorded in triplicate at 0.5-°C intervals. In addition, a similar assay was also performed on virus-mAb complexes with 1.0-μg HEV native virion preincubated with 2.5-μg antibodies. The RNA release (Tr) and melting temperature (Tm) were taken as the minima of the negative first derivative of the RNA exposure and protein denaturation curves, respectively.
Virus Precipitation Assay.
HEV virus from Rhesus monkey stools was prepared in PBS. Viruses of 1.5 × 107 RNA copies were incubated with 10-µg mAb in 1-mL PBS at 37 °C for 1 h. The mixtures were centrifuged on SW60 rotors at 31,000 rpm for 2 h at 4 °C. Pellets were suspended with 200-µL PBS to determine the HEV RNA copy number. HEV RNA was purified from 50 µL of each sample and the HEV RNA copy number was determined by quantitative real-time reverse transcription–PCR (RT-PCR) as previously reported (38).
HPSEC.
Purified HEV VLPs or complexes were subjected to HPLC (Agilent Technologies 1200 series; Santa Clara, CA) through a TSK Gel G5000 pwxl 7.8 × 300-mm column (TOSOH, Tokyo, Japan) equilibrated in 20-mM PB6.0. The column flow rate was maintained at 0.5 mL/min, and proteins in the eluents were detected at 280 nm. To prepare the 8C11:VLP complex in the serial mixing ratio, we maintained the same final amount of VLPs in the various mixtures, whereas for the control, the 3B6 antibody was just added to the VLP sample with an approximate molar ratio of 1:2 (p495 monomer:3B6 Fab).
Analytical Ultracentrifugation.
SV was used to assess the homogeneity of p495 at 20 °C using a Beckman XL-A analytical ultracentrifuge equipped with absorbance optics and an An60-Ti rotor. The samples were diluted in 20-mM PB6.0–0.83 OD280 nm. The speed was set at 30,000 rpm for analysis. The sedimentation coefficient was obtained using the c(s) method with the Sedifit software (kindly provided by Dr. P. Schuck, National Institutes of Health, http://www.analyticalultracentrifugation.com). As for the analysis of the smaller complex fraction, the molecular weights were calculated from the c(M) profile that was converted from the corresponding c(s) curve. Datasets were processed using the program Origin for nonlinear least squares fit.
SDS/PAGE.
Protein samples mixed with 2× loading buffer (100-mM Tris⋅HCl, PH6.8, 4% SDS, 0.2% bromophenol blue, 200-mM DTT, 4% SDS, and 20% glycerol) were subjected to 12% gradient SDS-polyacrylamide gel electrophoresis (SDS/PAGE). The proteins were stained by Coomassie Brilliant Blue (Nacalai Tesque, Kyoto, Japan), according to standard laboratory protocols.
Negative-Staining Electron Microscopy.
The purified HEV VLP and immune complexes were diluted in PBS (pH 7.4) and then absorbed onto 200 mesh carbon-coated copper grids (Quantifoil Micro Tools) for 1 min. The grids were washed twice with double-distilled water and negatively stained with 2% phosphotungstic acid (pH 6.4) for 30 s. Specimens were evaluated and imaged with the FEI Tecnai T12 TEM at 25,000× magnification.
Cryo-EM Sample Preparation and Data Acquisition.
VLP-Fab complexes were prepared by incubating Fabs with VLPs at 37 °C. Aliquots (3 μL) of HEV VLP or its complexes were deposited onto glow discharged holey carbon Quantifoil Cu grids (R2/2, 200 mesh, Quantifoil Micro Tools) inside the FEI Mark IV Vitrobot at a humidity level of 100%, blotted with filter papers for 6 s. The grids were examined in a Tecnai G2 F30 TEM at 300 keV. The images were recorded by an FEI Falcon II detector in the 17-frame movie mode with an electron dose of ∼25 e-/Å2 at a calibrated magnification of 93,000.
Data Processing.
Movie frames are aligned to correct the drift and beam-induced motion with the program MotionCor2 (39), and the contrast transfer function of each aligned micrograph was estimated with the program Gctf (40). Particles from several representative micrographs were manually boxed using the “e2boxer.py” program from the EMAN 2.1 package (41) as a reference for autopicking particles of the whole dataset with the program Relion 2.0 (30). All extracted particle images were subjected to multiple rounds of 2D classifications, and the classes exhibited identifiable secondary structures, and those that were rated as “good” were used to generate initial 3D models with the random model method in the program AUTO3DEM (42). Particles were further processed for 3D image reconstructions with Relion2.0. The resolutions of the maps were estimated using gold-standard criteria with the Fourier shell correlation cutoff = 0.143 (43). Local resolution variations were estimated using the program ResMap (44).
Model Building and Refinement.
The atomic model of the HEV VLP was built using the initial templates of the p495 crystal structure (PDB no. 2ZZQ). An initial model generated from molecular modeling was docked into an asymmetric unit (ASU) of the final cryo-EM map of HEV VLP using Chimera (45). Manual model building and automatic refinement were alternately and iteratively performed in Coot (46) and the module phenix_real_space_refine in PHENIX (46, 47). The refined protomer was docked into the density of 6 neighboring ASUs, which were then treated as a whole model for further optimization to avoid clashes between ASUs. Model statistics, including bond lengths, bond angles, all atom clashes, rotamer statistics, and Ramachandran plot statistics, etc., were closely inspected with Coot. The final models were validated with Molprobity (48). The atomic coordinates and structure factors of HEV VLP and VLV:8C11 were deposited in the Protein Data Bank (PDB) with accession nos. 6LAT (49) and 6LB0 (50). The cryo-EM maps were deposited in the Electron Microscopy Data Bank (EMDB) with accession nos. EMD-0863 (HEV VLP) (51), EMD-0866 (VLP:8C11) (52), and EMD-0861 (VLP:3B6) (53). Figures were prepared with Chimera (45).
Supplementary Material
Acknowledgments
This research was supported by grants from the Natural Science Foundation of Fujian Province (Grant 2017J07005), the National Science and Technology Major Project of Infectious Diseases (Grant 2018ZX10101001-002), and the National Natural Science Foundation of China (Grants 81871247, 81991490, and 81571996).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. B.V.V.P. is a guest editor invited by the Editorial Board.
Data Deposition: Coordinates and EM maps are available at the Protein Data Bank (PDB) and the Electron Microscopy Data Bank (EMDB), respectively, under the accession nos. 6LAT (HEV VLP), 6LB0 (VLP:8C11), EMD-0863 (HEV VLP), EMD-0866 (VLP:8C11), and EMD-0861 (VLP:3B6). All other data to support the conclusions are in the main paper or supplementary materials.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1916028116/-/DCSupplemental.
References
- 1.Reading S. A., Dimmock N. J., Neutralization of animal virus infectivity by antibody. Arch. Virol. 152, 1047–1059 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Klasse P. J., Sattentau Q. J., Occupancy and mechanism in antibody-mediated neutralization of animal viruses. J. Gen. Virol. 83, 2091–2108 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Marasco W. A., Sui J., The growth and potential of human antiviral monoclonal antibody therapeutics. Nat. Biotechnol. 25, 1421–1434 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burton D. R., Antibodies, viruses and vaccines. Nat. Rev. Immunol. 2, 706–713 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Lu L. L., Suscovich T. J., Fortune S. M., Alter G., Beyond binding: Antibody effector functions in infectious diseases. Nat. Rev. Immunol. 18, 46–61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hangartner L., Zinkernagel R. M., Hengartner H., Antiviral antibody responses: The two extremes of a wide spectrum. Nat. Rev. Immunol. 6, 231–243 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Murphy K., Weaver C., Janeway’s Immunobiology (Garland Science, ed. 9, 2017). [Google Scholar]
- 8.Lazear E., et al. , Antibody-induced conformational changes in herpes simplex virus glycoprotein gD reveal new targets for virus neutralization. J. Virol. 86, 1563–1576 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng Q., et al. , Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization. Nat. Microbiol. 4, 124–133 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li T. C., et al. , Essential elements of the capsid protein for self-assembly into empty virus-like particles of hepatitis E virus. J. Virol. 79, 12999–13006 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santos J. L., Bispo J. A., Landini G. F., Bonafe C. F., Proton dependence of tobacco mosaic virus dissociation by pressure. Biophys. Chem. 111, 53–61 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Silva J. L., Weber G., Pressure-induced dissociation of brome mosaic virus. J. Mol. Biol. 199, 149–159 (1988). [DOI] [PubMed] [Google Scholar]
- 13.Dong Y., et al. , Antibody-induced uncoating of human rhinovirus B14. Proc. Natl. Acad. Sci. U.S.A. 114, 8017–8022 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plevka P., et al. , Neutralizing antibodies can initiate genome release from human enterovirus 71. Proc. Natl. Acad. Sci. U.S.A. 111, 2134–2139 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brioen P., Rombaut B., Boeyé A., Hit-and-run neutralization of poliovirus. J. Gen. Virol. 66, 2495–2499 (1985). [DOI] [PubMed] [Google Scholar]
- 16.Delaet I., Boeyé A., Monoclonal antibodies that disrupt poliovirus only at fever temperatures. J. Virol. 67, 5299–5302 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCullough K. C., et al. , Conformational alteration in foot-and-mouth disease virus virion capsid structure after complexing with monospecific antibody. Immunology 60, 75–82 (1987). [PMC free article] [PubMed] [Google Scholar]
- 18.Tam A. W., et al. , Hepatitis E virus (HEV): Molecular cloning and sequencing of the full-length viral genome. Virology 185, 120–131 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin X., et al. , Origin, antigenicity, and function of a secreted form of ORF2 in hepatitis E virus infection. Proc. Natl. Acad. Sci. U.S.A. 115, 4773–4778 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu Y., et al. , Structural basis for the neutralization of hepatitis E virus by a cross-genotype antibody. Cell Res. 25, 604–620 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guu T. S., et al. , Structure of the hepatitis E virus-like particle suggests mechanisms for virus assembly and receptor binding. Proc. Natl. Acad. Sci. U.S.A. 106, 12992–12997 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li S., et al. , Dimerization of hepatitis E virus capsid protein E2s domain is essential for virus-host interaction. PLoS Pathog. 5, e1000537 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang X., et al. , Structural basis for the neutralization and genotype specificity of hepatitis E virus. Proc. Natl. Acad. Sci. U.S.A. 108, 10266–10271 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xing L., et al. , Structure of hepatitis E virion-sized particle reveals an RNA-dependent viral assembly pathway. J. Biol. Chem. 285, 33175–33183 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamashita T., et al. , Biological and immunological characteristics of hepatitis E virus-like particles based on the crystal structure. Proc. Natl. Acad. Sci. U.S.A. 106, 12986–12991 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schofield D. J., Glamann J., Emerson S. U., Purcell R. H., Identification by phage display and characterization of two neutralizing chimpanzee monoclonal antibodies to the hepatitis E virus capsid protein. J. Virol. 74, 5548–5555 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xing L., et al. , Recombinant hepatitis E capsid protein self-assembles into a dual-domain T = 1 particle presenting native virus epitopes. Virology 265, 35–45 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Zhao M., et al. , A comprehensive study of neutralizing antigenic sites on the hepatitis E virus (HEV) capsid by constructing, clustering, and characterizing a tool box. J. Biol. Chem. 290, 19910–19922 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li T. C., et al. , Expression and self-assembly of empty virus-like particles of hepatitis E virus. J. Virol. 71, 7207–7213 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scheres S. H., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi M., et al. , Prolonged fecal shedding of hepatitis E virus (HEV) during sporadic acute hepatitis E: Evaluation of infectivity of HEV in fecal specimens in a cell culture system. J. Clin. Microbiol. 45, 3671–3679 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka T., et al. , Development and characterization of a genotype 4 hepatitis E virus cell culture system using a HE-JF5/15F strain recovered from a fulminant hepatitis patient. J. Clin. Microbiol. 47, 1906–1910 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shukla P., et al. , Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus-host recombinant. Proc. Natl. Acad. Sci. U.S.A. 108, 2438–2443 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walter T. S., et al. , A plate-based high-throughput assay for virus stability and vaccine formulation. J. Virol. Methods 185, 166–170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poignard P., Fouts T., Naniche D., Moore J. P., Sattentau Q. J., Neutralizing antibodies to human immunodeficiency virus type-1 gp120 induce envelope glycoprotein subunit dissociation. J. Exp. Med. 183, 473–484 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He S., et al. , Putative receptor-binding sites of hepatitis E virus. J. Gen. Virol. 89, 245–249 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Zhang J., et al. , Analysis of hepatitis E virus neutralization sites using monoclonal antibodies directed against a virus capsid protein. Vaccine 23, 2881–2892 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Halbur P. G., et al. , Comparative pathogenesis of infection of pigs with hepatitis E viruses recovered from a pig and a human. J. Clin. Microbiol. 39, 918–923 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z., et al. , Crystal structures of two immune complexes identify determinants for viral infectivity and type-specific neutralization of human papillomavirus. MBio 8, e00787-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang G., et al. , EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Yan X., Sinkovits R. S., Baker T. S., AUTO3DEM–An automated and high throughput program for image reconstruction of icosahedral particles. J. Struct. Biol. 157, 73–82 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheres S. H., Chen S., Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swint-Kruse L., Brown C. S., Resmap: Automated representation of macromolecular interfaces as two-dimensional networks. Bioinformatics 21, 3327–3328 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Pettersen E. F., et al. , UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen V. B., et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng Q., et al. , The cryo-EM structure of HEV VLP. Protein Data Bank. http://www.rcsb.org/structure/6LAT. Deposited 14 November 2019.
- 50.Zheng Q., et al. , The cryo-EM structure of HEV VLP in complex with Fab 8C11. http://www.rcsb.org/structure/6LB0. Deposited 14 November 2019.
- 51.Zheng Q., et al. , The cryo-EM structure of HEV VLP. The Electron Microscopy Data Bank. http://www.ebi.ac.uk/pdbe/entry/EMD-0863. Deposited 14 November 2019.
- 52.Zheng Q., et al. , The cryo-EM structure of HEV VLP in complex with Fab 8C11. The Electron Microscopy Data Bank. http://www.ebi.ac.uk/pdbe/entry/EMD-0866. Deposited 14 November 2019.
- 53.Zheng Q., et al. , The cryo-EM structure of HEV VLP in complex with Fab 3B6. The Electron Microscopy Data Bank. http://www.ebi.ac.uk/pdbe/entry/EMD-0861. Deposited 14 November 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.