Significance
The mammalian gut is a complex set of tissues formed during development by orchestrating the timing of expression of many genes. Here we uncover the identity of these genes, their pathways, and how they change during gut organogenesis. We used RNA-seq profiling in the wild-type mouse gut in both sexes during development (E10.5–E14.5), as well as in a Ret null mouse, a model of Hirschsprung disease (HSCR). These studies have allowed us to expand the universe of genes and developmental processes that contribute to enteric neuronal innervation and to its dysregulation in disease.
Keywords: gut development, transcript profiling, Hirschsprung disease
Abstract
The development of the gut from endodermal tissue to an organ with multiple distinct structures and functions occurs over a prolonged time during embryonic days E10.5–E14.5 in the mouse. During this process, one major event is innervation of the gut by enteric neural crest cells (ENCCs) to establish the enteric nervous system (ENS). To understand the molecular processes underpinning gut and ENS development, we generated RNA-sequencing profiles from wild-type mouse guts at E10.5, E12.5, and E14.5 from both sexes. We also generated these profiles from homozygous Ret null embryos, a model for Hirschsprung disease (HSCR), in which the ENS is absent. These data reveal 4 major features: 1) between E10.5 and E14.5 the developmental genetic programs change from expression of major transcription factors and its modifiers to genes controlling tissue (epithelium, muscle, endothelium) specialization; 2) the major effect of Ret is not only on ENCC differentiation to enteric neurons but also on the enteric mesenchyme and epithelium; 3) a muscle genetic program exerts significant effects on ENS development; and 4) sex differences in gut development profiles are minor. The genetic programs identified, and their changes across development, suggest that both cell autonomous and nonautonomous factors, and interactions between the different developing gut tissues, are important for normal ENS development and its disorders.
Vertebrate organogenesis involves highly regulated and evolutionarily conserved processes leading to the programmed differentiation of diverse cell types and their integration into a tissue or organ. The pathways involved are genetically programmed with genes expressed in precise spatial and temporal patterns and regulated by feedforward and feedback mechanisms, called gene regulatory networks (GRNs) (1, 2). GRNs are modular, comprised of a small number of subcircuit classes, conserved across species, and provide systems-level views of organogenesis (1, 2). They can also be the genetic basis for developmental disorders (3), as for Hirschsprung disease (HSCR, congenital aganglionosis), a multifactorial disorder of gastrointestinal development in which the enteric nervous system (ENS) fails to develop (4).
The mammalian gastrointestinal tract develops from a tube to an organ comprising at least 6 well-characterized cell and tissue types (epithelial, smooth muscle, vascular, neuron, glia, and extracellular matrix) that provide its major barrier function and its integrated physiology involving absorption, secretion, and motility (5). The ontogeny of the gut involves cells that arise stepwise from multiple regimens of differentiation, being dependent on region-specific interactions between the endoderm-derived epithelium and the mesoderm-derived mesenchyme. Signaling between these 2 layers is critical for the differentiation and apoptosis of both epithelial and mesenchymal cells, and their subsequent homeostasis. The major developmental event in completion of early gut development is the migration of neural crest cells (NCCs) into the gut, and their differentiation into enteric neuroblasts (enteric neural crest cells [ENCCs]) and, subsequently, enteric neurons and glia. These neuroblasts colonize the gut mesenchyme to form 2 neuronal networks, the myenteric (Auerbach’s) plexus, between the longitudinal and circular muscles, and the submucosal (Meissner’s) plexus, between the circular muscle and the submucosal layer. The myenteric and submucoal plexuses provide motor innervation to both muscular layers of the gut, and secretomotor innervation of the mucosa nearest the lumen of the gut, respectively (6).
The many stages of gut development require numerous initiating signaling events activating transcription factors (TFs) targeting diverse genes and pathways varying across development (7, 8). To improve our understanding of this process, we conducted gene expression profiling across gut development in wild-type mice. We also examined the homozygote null mouse for Ret (9, 10), which is a major gene for the disorder (4), and a model for HSCR. Ret encodes a receptor tyrosine kinase (RTK) controlling ENCC differentiation and their migration through the gut mesenchyme. RET signaling is mediated by binding of a group of soluble proteins of the glial cell line-derived neurotrophic factor (GDNF) family ligands. RET does not directly bind to its ligand, but requires an additional coreceptor, 1 of 4 GDNF family receptor-α (GFRα) members (11). Mutations leading to Hirschsprung disease has been reported in many of these genes, highlighting the importance of this GRN in disease (12). Ret null mice exhibit complete aganglionosis with transcriptional changes in many of these genes (4), making it an ideal model to study how development is compromised in HSCR (10, 13).
Today, comprehensive transcriptomewide studies allow the creation of a gene atlas from small amounts of developing embryonic tissue and identification of weakly expressing transcripts (14–16). Prior microarray studies of wild-type and Ret-deficient mouse guts late in gut development (embryonic day 15.5 [E15.5]) has shown the dysregulation of many genes consequent to loss of Ret expression, including signaling molecules and TFs (17). However, we wanted to study the early effects of Ret loss uncompromised by secondary and downstream consequences. Here, we characterize the gut developmental “parts list” as a function of developmental time (E10.5, E12.5, and E14.5), sex (male and female), and Ret genotype (wild type and homozygous null) to characterize its normal and HSCR genetic programs in development. We focus on the time period when the gut is differentiating into a mature organ and NCCs are undergoing migration, proliferation, and maturation to ENCCs and enteric neurons (18). We identify bursts of activity of the activation of specific TFs prior to the differentiation and establishment of each major cell type and specialized tissue, and therefore its major regulators. We also demonstrate the dramatic effect of loss of Ret activity, on all major gut cell types. The effect of the gut mesenchyme on ENS development is well known; in distinction, we now show that loss of Ret identifies reverse crosstalk on the gut mesenchyme through multiple muscle-specific markers including Myogenin. Thus, both cell autonomous and nonautonomous genetic effects impact the ENS and its disorders. Finally, HSCR has a profound male sex bias, observed in appropriate mouse models (19), of unknown molecular etiology. This study shows that the gut transcriptional program is similar in males and females, with a few genes with specialized functions like cholesterol efflux and inflammatory response showing higher expression in females. Thus, additional mouse models need to be studied to understand the sex bias in HSCR.
Results
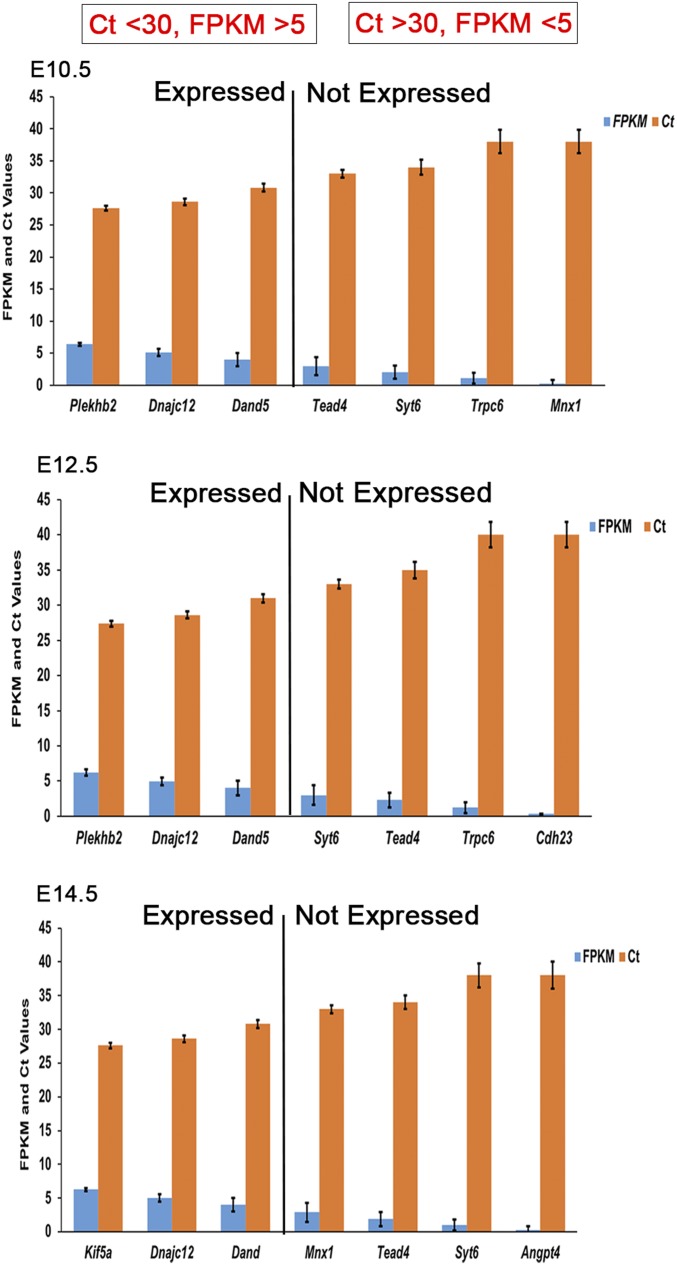
We performed RNA-sequencing (RNA-seq) on dissected whole gut tissue at 3 embryonic stages during mouse development and assessed gene expression patterns using fragments per kilobase of transcript per million mapped reads (FPKM) values (20). Since there is no formal FPKM value that defines “expression,” we defined this quantity by qPCR analysis of selected genes. We randomly selected 7 genes with FPKM values between 0 and 6, at each developmental stage in the wild-type male embryonic gut, on the same RNA samples used for RNA-seq. These results showed that in independent experiments using 40 qPCR cycles, genes with FPKM of <5 consistently had threshold cycle (Ct) values >30 (Fig. 1); thus, FPKM ≥5 was used as the expression threshold. We identified 9,436 and 9,432 genes expressed in the E10.5 wild-type male and female, respectively, or ∼41% of the total transcriptome of 23,235 genes assayed. At subsequent times, only a small ∼3% increase in the numbers of transcribed genes was noted, 10,143 (male) and 10,180 (female) genes at E12.5, and 10,216 (male) and 10,293 (female) genes at E14.5. Thus, increased developmental complexity is not associated with large increases in gene numbers per se.
Fig. 1.
Gene expression in the embryonic mouse gut. Comparisons of FPKM from RNA-seq and Ct values after 40 cycles of qPCR of wild-type embryonic male mouse gut cDNA isolated at the developmental stages of E10.5, E12.5, and E14.5. We set FPKM = 5 as the value considered for a gene to be expressed; error bars are the SEMs for 3 biological replicates each.
Relative Effects of Time, Genotype, and Sex on Gut Development.
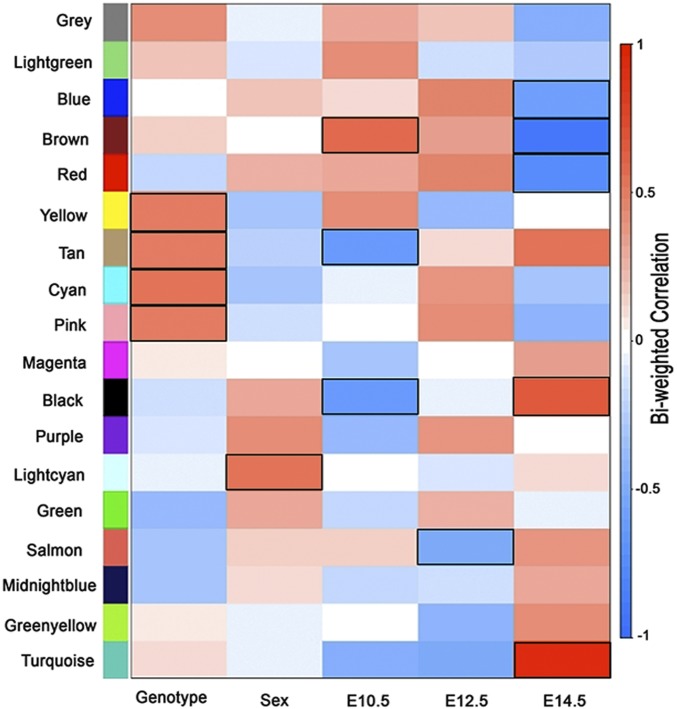
In the first analysis, we attempted to identify groups of coexpressed genes depending on developmental stage, sex, and Ret genotype and their interactions. We used signed network analysis to construct these modules, using weighted gene coexpression network analysis (WGCNA) (21) on 9,327 genes with a mean FPKM of ≥5 across all 36 samples, i.e., 3 time points, 2 sexes, 2 genotypes (Ret wild type and null), and 3 replicates. Expression values were transformed to log2(FPKM+1). Our analysis grouped 7,793 genes into 18 modules (labeled by colors) (SI Appendix, Table S1). Next, we tested associations between the first principal component (the eigengene) of each module’s expression profile with each of 5 binary variables using linear regression (genotype, sex, and 3 pairwise developmental time comparisons), under a Bonferroni correction adjusting for 18 modules (P < 0.00277) (Fig. 2).
Fig. 2.
WGCNA. A correlation matrix using biweight midcorrelation in WGCNA between 18 modules constructed from all expressed genes from 36 different conditions (3 time points, 2 sexes, 2 genotypes, 3 replicates). At a Bonferroni correction threshold adjusting for 18 modules (P < 0.00277), the lightcyan module was found to be associated with sex (P = 0.001), and the tan (P = 0.0015), cyan (P = 0.0007), yellow (P = 0.0026), and pink (P = 0.0027) modules were found to be associated with genotype. The tan (P = 6.18 × 10−5), black (P = 9.7 × 10−5), and brown (P = 0.0002) modules were associated with gene expression at E10.5 as compared to other time points. At E14.5, turquoise (P = 5.2 × 10−16), brown (P = 4.6 × 10−15), red (P = 8.8 × 10−8), black (P = 1.4 × 10−5), and blue (P = 2.7 × 10−4) modules were associated in comparison to other time points; salmon (P = 0.0022, 62 genes) was associated with E12.5 only. All of the associated module for each condition has been highlighted.
First, we identified a small lightcyan module containing 43 genes (0.55% of all genes in modules) associated with sex (P = 0.001) and 4 modules comprising 1,092 genes (14% of all genes in modules) associated with genotype: yellow (P = 0.0026, 694 genes), pink (P = 0.0027, 199 genes), tan (P = 0.0015, 139 genes), and cyan (P = 0.0007, 60 genes). Three modules comprising 1,644 genes (21% of all genes in modules) were associated with early gene expression (E10.5 versus other time points): brown (P = 0.0002, 1,210 genes), black (P = 9.7 × 10−5, 295 genes), and tan (P = 6.18 × 10−5, 139 genes). Only one module, salmon (P = 0.0022, 62 genes, 0.80% of all genes in modules), was associated with E12.5 (versus others); the later (E14.5 versus others) gene expression program was characterized by 5 modules and 5,512 genes (71% of all genes in modules): turquoise (P = 5.2 × 10−16, 2,208 genes), blue (P = 2.7 × 10−4, 1,443 genes), brown (P = 4.6 × 10−15, 1,210 genes), red (P = 8.8 × 10−8, 356 genes), and black (P = 1.4 × 10−5, 295 genes). Note that by virtue of their reciprocal definition, the black and brown modules are associated with both early and late development: the brown/black module contains genes expressed significantly higher/lower early/late in development.
Thus, in quantitative terms, the greatest gene expression changes in gut development were across developmental time (21% and 71% of genes in early versus late programs), followed by Ret genotype (14%) together with a miniscule effect of sex (0.55%). In other words, the developmental parts list is skewed toward later use in development with differentiation into specialized structures and organ growth; early development is characterized by a differentiation program necessary to make the relevant cell types for later use. Thus, to understand the biological roles of these coexpressed genes in the various modules, we performed gene ontology (GO) annotation using DAVID (https://david.ncifcrf.gov/) (22) and assessed the statistical significance of the grouped annotated functions at a false discovery rate (FDR) of 1%.
Genes and Pathways Associated with Developmental Time, Ret, and Sex.
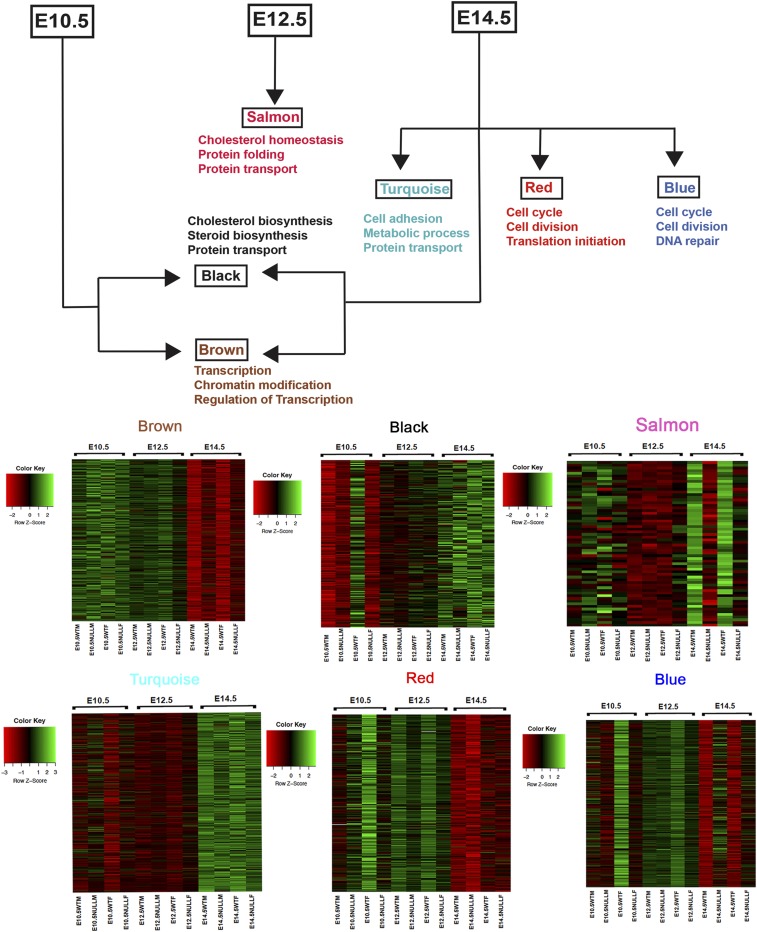
Three modules were associated with developmental time. The brown module, with significantly higher expression in E10.5 versus other times, is enriched for genes controlling transcriptional modulation (Med24, Med22, Nono, etc.), RNA processing (Crnkl1, Supt6, U2af2, etc.), and transcription factors (Foxo4, Gli3, Sox11, etc.) (Fig. 3). In contrast, the black module comprises genes expressed at significantly lower levels in E10.5 versus other times and whose activities control macromolecule and protein transport (see below). The tan module also contains genes with weak expression at E10.5 versus other times; although no functional annotations were significantly enriched, it includes genes of the canonical Wnt signaling pathway (Sfrp5, Fzd5, Tax1bp3, etc.), transcriptional repressors (Snai2, Limd1, etc.), and negative regulators of cell proliferation (Ptprj, Raf1, Asph, etc.). Thus, transcription and cell proliferation are the primary processes underlying gut organogenesis at E10.5. At E12.5, only the salmon module is associated with gene expression and it contains genes for protein translation (Dnajb11, Pdia6, Pdia4, etc.), protein transport (Tmed4, Tomm6, Mcfd2, etc.), and genes involved in steroid metabolism and homeostasis (Npc2, Lamtor1, Ehd1, etc.). Interestingly these processes are still negatively correlated at this stage of gut development, similar to E10.5 (the black module), highlighting the continuity of function or lack thereof of specific genes during gut development (Fig. 3).
Fig. 3.
Biological processes affected by genes changing over developmental time. Various modules are associated with different developmental time points and show a continuity of biological processes over time. The brown module is associated with E10.5 (early gut expressed genes) and the GO biological processes these genes are involved in include mostly transcription factors and chromatin modifiers. The salmon module associated with E12.5 contains genes involved in protein folding and transport which are also seen in the black and turquoise modules, which are associated with E14.5. The red and blue modules associated with E14.5 contain genes involved in the cell cycle, cell division, and DNA repair and are weakly expressed at this stage when compared to early developmental stages. The corresponding heat maps for all modules are also shown.
Two modules (black and turquoise) were associated with higher and 3 (red, blue, and brown) with lower gene expression at E14.5, as compared to earlier stages. Genes in the black module are involved in transport of macromolecules, small molecules, ions/cellular components (Uqcrfs1, Slc35a3, Gpr89, Hook1, etc.), and steroid/cholesterol biosynthesis (Cyp51, ACbd3, Sc5d, etc.), while turquoise module genes include those regulating metabolic processes in cell growth (Ide, Ogdh, Fdft1, etc.), as well as transport of macromolecules, small molecules, ions/cellular components (Slc9a8, Atp1b1, Sec31a, etc.), and cell adhesion (Lima1, Ldha, Tln1, etc.). Thus, these later functions are involved with producing and transporting basic macromolecules necessary for survival, maturation, and growth of the organ (Fig. 3). Genes suppressed during this stage include genes for cell cycle progression (Cks1b, Cetn3, Bex2, etc.), cell division (Spc24, Ccne1, Kif2c, etc.), RNA processing (Raly, Ncbp2, Lsm6, etc.), and translational initiation (Eif3d, Eif4e, Eif3g, etc.) from the red module, and genes controlling cell cycle progression (Dbf4, Kntc1, Cdc16, etc.), cell division (Ccnt2, Cdk19, Tsg101, etc.), and DNA repair (Xrcc5, Xrcc2, Setx, etc.) from the blue module (Fig. 3). Consequently, at E14.5, cell division is not as rapid as earlier in development and the tissue is reaching a morphological developmental equilibrium.
Beyond these qualitative changes, we also identified temporal shifts in the transcription of genes controlling specific gut processes such as cholesterol homeostasis and protein transport. These features are negatively correlated at E10.5 (black) and E12.5 (salmon) but positively correlated at E14.5 (black, turquoise). Understanding the roles of this temporal dynamic behavior in gut disorders may be important.
Gut motility is controlled by the ENS which monitors the state of the lumen and the gut wall by activating intrinsic reflexes that generate peristaltic movements and change blood flow, water, and electrolyte secretions. The ENS arises from vagal NCCs that migrate into the foregut and then migrate caudally (18, 23) and undergo fate transition first to ENCCs and then to enteric neurons. The receptor tyrosine kinase gene Ret is the earliest major and key factor controlling ENCC differentiation, proliferation, and migration. Therefore, it is no surprise that RET is the major gene for HSCR in humans with both rare coding and polymorphic enhancer variants (24–26). Further, Ret gene deletions in the mouse recapitulate most of the HSCR phenotype (6, 10, 13).
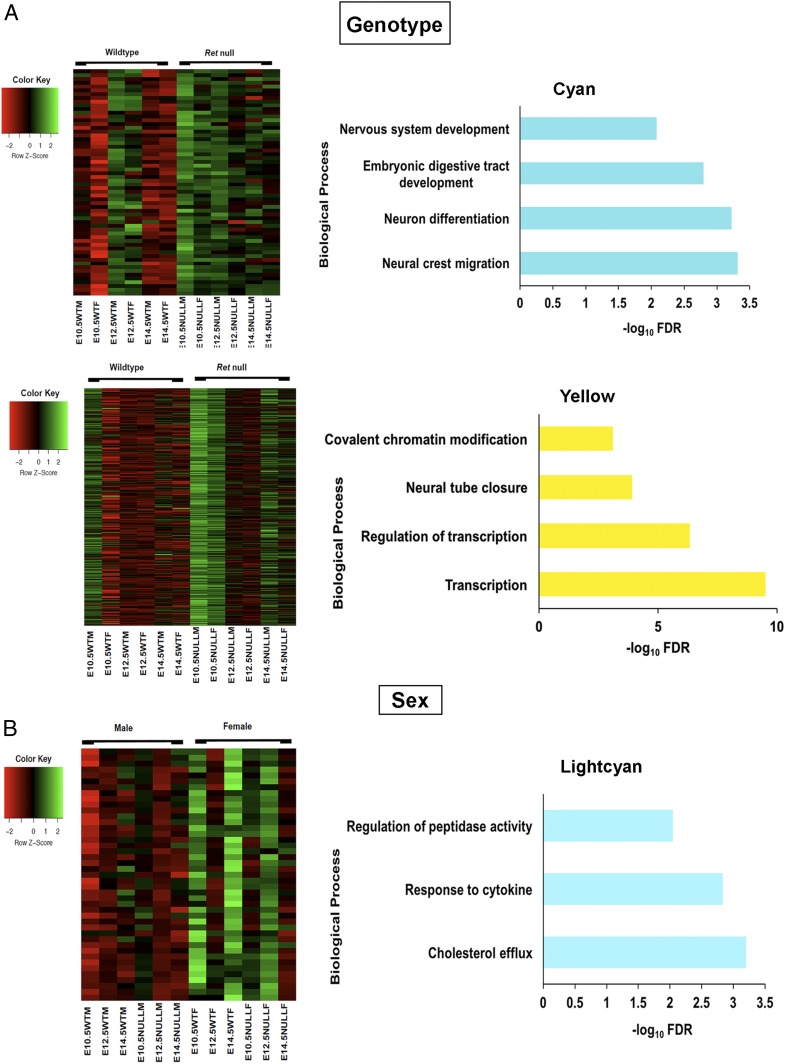
We identified gene modules that are Ret-genotype (wild-type and null homozygotes) dependent. Genes associated with the cyan module, with higher gene expression in Ret null versus wild-type homozygotes, are involved in neural crest cell differentiation (Edn3, Isl1, Gdnf, etc.), enteric neuron differentiation (Hdac5, Gfra1, Isl1, etc.), and transcription factors and activators like Tcf21, Ntf3, Foxf1, etc. (Fig. 4A). These results are consistent with our earlier observations of Gdnf and Gfra1 (4). In contrast, genes in the yellow module, with higher gene expression in Ret wild-type versus null homozygotes, comprise those involved in transcription and its modulation (Zfp335, Med25, Zfp580, etc.) and chromatin modifiers (Ing4, Brd2, L3mbtl2, etc.) (Fig. 4A). Thus, Ret appears to have a significant effect on many early TFs and may explain why Ret mutations have such severe consequences. The pink module contains genes weakly expressed in wild type, as compared to the Ret null homozygote, and includes genes for cell differentiation (Wnt5a, Srpk2, Rbm45, etc.) and nervous system development (Sema5a, Itga8, Efna5, etc.). These results show marginal statistical significance but suggest that certain differentiation and ENS genes may also be suppressed by Ret highlighting its balancing role in ENS development. Similarly, the genes in the tan module are also weakly expressed in wild type and grouped primarily into broad categories like digestive tract development (Fgfr2, Kit, Lgr4, etc.), negative regulation of cell proliferation (Sfrp5, Ptprj, Raf1, etc.) and cytoslelekton reorganization (Pak1, Flna, Parva, etc.). But like the pink module, all functional groups show nominal significance.
Fig. 4.
Biological processes affected by genes changing due to genotype and sex. (A) Heat map of GO biological processes for the 2 modules associated with genotype shows genes involved in neural crest migration and neuron differentiation (Cyan), while transcription and chromatin modifiers (yellow) are affected with loss of Ret transcription. (B) The lightcyan module consists of 43 genes for peptidase activity and cholesterol transport and is associated with sex.
We identified only one module (lightcyan) of genes significantly associated with sex, irrespective of genotype and developmental time. This set contains 43 genes broadly classified into those regulating cholesterol efflux (Apoa2, Apoa1, Apoc1, etc.) and regulation of peptidase activity (Ambp, Serpina1b, Serpinf2, etc.). Surprisingly, all show higher expression in females than males (Fig. 4B). Of these, only Cited1 is sex linked showing that sex-linked dosage effects are not the major reason for these differences, suggesting hormonal or sex-biased epigenetic effects instead.
The miniscule sex difference we identified, despite its prominence in developmental disorders of the gut, such as in HSCR, prompted us to look for such differences more carefully within each developmental stage. At E10.5, 270 genes show significant expression differences between wild-type males and females (SI Appendix, Fig. S1A), of which 157 and 113 show higher expression in males and females (SI Appendix, Table S2), respectively, a skew decidedly increased toward males (P = 0.007). Annotation analysis using DAVID showed that male-enriched factors encoded genes for cell morphogenesis and cell motility and neurogenesis (SI Appendix, Fig. S1B). In contrast, the 113 female-enriched factors encoded genes for hemostasis, vasculature development, and blood coagulation (SI Appendix, Fig. S1C). Later at E12.5, 81 and 115 genes show higher expression in males and females, respectively, but now, a skew decidedly favoring females (SI Appendix, Fig. S1D and Table S3), a skew also observed at E14.5 where 133 and 169 show higher expression in males and females, respectively (SI Appendix, Fig. S1G and Table S4). Interestingly, genes expressed significantly at a higher level in males are involved in specific morphological functions like neuronal, musculature, and endothelial development (SI Appendix, Fig. S1 B, E, and H), while female-enriched genes control specific physiological processes like response to wound healing, inflammatory response, and response to hormonal stimulus (SI Appendix, Fig. S1 C, F, and I). Thus, sex differences do occur and occur differentially by functional categories. These results prompted us to search for genetic programs in gut development by more focused pairwise analyses of time and Ret genotype.
Gene Expression Changes during Normal Gut Development over Developmental Time.
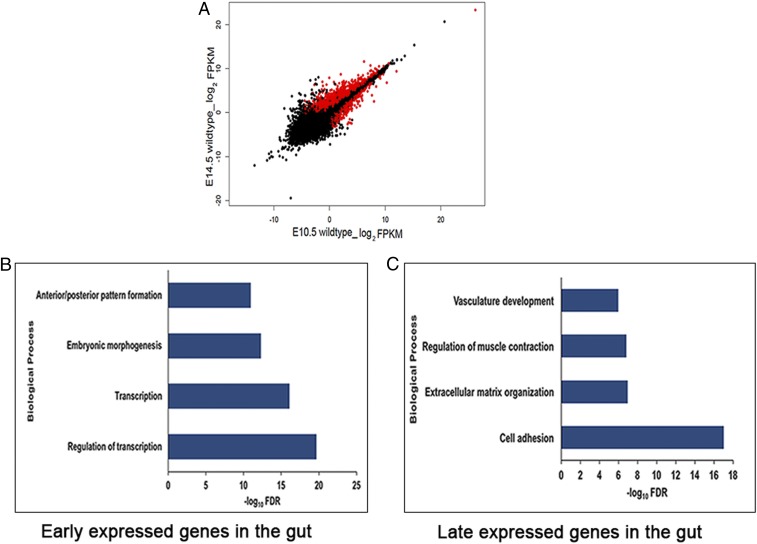
To understand stage-specific gut development, we next compared gene expression changes between E10.5 (early) and E14.5 (late) stages in the wild-type gut to discover 2,697 differentially expressed genes (Fig. 5A and SI Appendix, Table S5), of which 1,227 showed significantly higher expression in E10.5. We performed GO annotation on only the subset of 316 (25.8%) of these genes with 2-fold or greater expression and identified 89 transcription factors (Bach1, Gata2, Barx1, etc.), 73 genes for transcriptional control including various transcription factors like Gli3, Hoxc6, Hoxc9, etc., and 29 for morphogenesis (Crabp2, Prrx1, Col2a1, etc.) (Fig. 5B). Conversely, of 1,470 genes showing significantly higher expression at E14.5, 1,003 genes (68.2%) with 2-fold and greater expression were enriched for biological processes like cell adhesion (76 genes which include Cldn8, Cldn7, Aebp1, etc.), extracellular matrix organization (20 genes including Olfml2b, Eln, Col3a, etc.), regulation of muscle contraction (44 genes including Myl6, Tbx20, Myh7, etc.), and vasculature development (61 genes including Cspg4, Anpep, Cav1, etc.) (Fig. 5C). These results validate our WGCNA analysis that early gut morphogenesis is dictated by transcription factors, whereas later development focuses on specifying specialized tissues. This analysis also identified the specific transcription factors which are candidate genes for genetic disorders of early gut development (see below).
Fig. 5.
Temporal gene expression patterns in the embryonic mouse gut. (A) Scatter plot of log2 FPKM values of genes expressed in wild-type male embryos during early (E10.5) versus late (E14.5) gut development. (B) GO annotations of the transcriptome shows that transcription factors are highly expressed at E10.5. (C) Genes controlling more specialized structures (e.g., vasculature) or processes (e.g., cell adhesion and cell signaling) are prominent at E14.5. Genes marked in red have statistically significant (q value <0.01) expression differences between the two states indicated.
Ret-Dependent Gene Expression in the Gut.
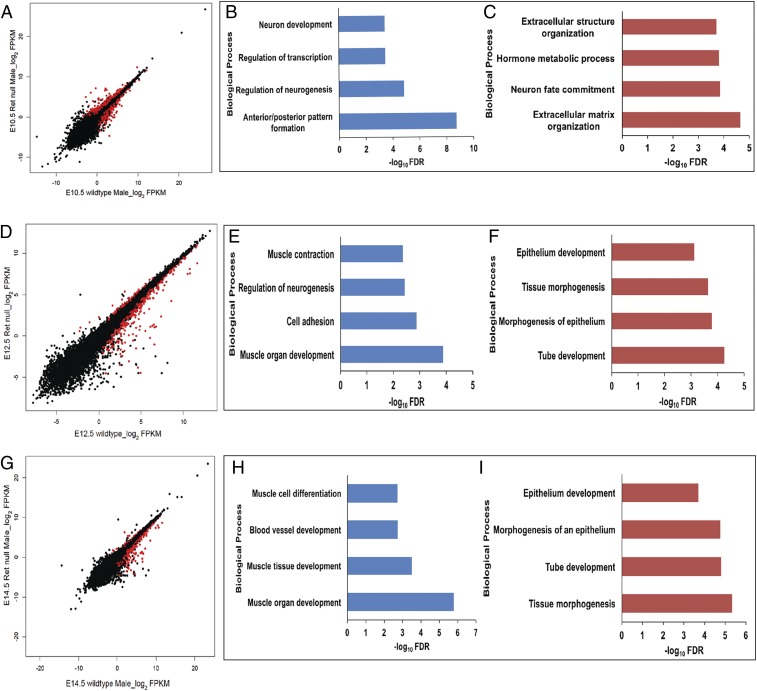
Loss of Ret gene expression in the mouse gut has transcriptional consequences on many genes in the Ret GRN (4), as observed in our WGCNA analysis, but we sought to identify additional temporal patterns of gene expression changes triggered by Ret loss of function by detailed pairwise analysis. At E10.5, when Ret is normally expressed and is the earliest stage where reduction in the numbers of ENCCs and their failure to migrate can be demonstrated in Ret null homozygote mice (27), we identified 763 genes with significant expression differences (Fig. 6A and SI Appendix, Table S6), of which 521 were down-regulated (thus, Ret activated) and 242 were up-regulated (thus, Ret suppressed) in male Ret null embryonic guts as compared to wild type. The Ret-activated genes broadly comprise 3 biological processes: neuronal development (Ret’s primary function), transcriptional regulation, and pattern specification, primarily TFs (Fig. 6B). In contrast, the Ret-suppressed genes are involved in extracellular matrix organization and metabolic processes (Fig. 6C). At E12.5, there are 353 down-regulated and 90 up-regulated genes (Fig. 6D and SI Appendix, Table S7), and, at E14.5, similarly, there are 325 down-regulated and 111 up-regulated genes (Fig. 6G and SI Appendix, Table S8) in Ret null versus wild-type male embryos. The Ret-activated genes at E12.5 are involved in neurogenesis but, surprisingly, also genes for muscle specification (Fig. 6E). This pattern of Ret’s control of gene expression in nonneuronal tissue in the gut is even more pronounced at E14.5 where along with the musculature, genes controlling the vasculature are affected (Fig. 6H), while neurogenesis is largely spared. Thus, this pairwise analysis reveals that there is a gradual shift in the functional classes of genes affected by Ret loss of function, from its known primary role in neurogenesis to its nonautonomous effect on genes in the surrounding tissue in the developing gut, a feature missed by WGCNA analysis. The Ret-suppressed genes, in contrast at E12.5 and E14.5 were those primarily involved in epithelial development and tissue morphogenesis (Fig. 6 F and I), hinting at Ret’s balancing role in controlling nonneuronal tissue development in the developing gut.
Fig. 6.
Ret-dependent gene expression in the developing gut. Scatterplots of log2 FPKM values between wild-type and Ret homozygous null embryonic gut at (A) E10.5, (D) E12.5, and (G) E14.5. Genes marked in red in the scatterplots have statistically significant (q value <0.01) expression differences between the indicated states. Genes activated by Ret at E10.5 (B) include neuronal genes and genes involved in transcriptional control, while those suppressed by Ret at E10.5 (C) include genes involved in extracellular matrix formation and in hormonal response. Similar annotation analyses at E12.5 show that along with neuronal genes, Ret also activates genes expressed in the musculature (E) and suppresses genes involved in epithelial development (F). At E14.5, Ret activates muscle-specific and vasculature development genes (H) while suppressing genes involved in epithelial morphogenesis (I).
These results show that Ret has a surprisingly broad cell autonomous role within enteric neurons (27) but even broader nonautonomous roles on the mesenchyme, through which the ENCCs migrate, and the vasculature, which nourishes the tissues and the epithelium in the developing gut. These nonneuronal functions are first observed at E12.5 and persist through E14.5. Further, Ret appears to primarily be a developmental activator since loss of Ret expression largely leads to transcriptional down-regulation. Thus, the 2 major transcriptional programs to emerge from these analyses are: 1) The predominance of multiple classes of transcription factors in early gut development and their control by Ret; and 2) Ret’s cell nonautonomous role in controlling the development of the gut musculature, vasculature, and epithelium. To get a better understanding of these discoveries, we studied them in greater detail.
TFs in Ret-Driven Gangliogenesis.
One of the cardinal features of gut development is the preponderance of TFs and chromatin modifiers at E10.5: 129 (85 down- and 44 up-regulated) TFs show significant expression differences with Ret loss of function (SI Appendix, Table S9) and are 20% of all TFs active at E10.5 (650 genes) involving many TF families. Of these, homeobox (41 members) and zinc finger (29 members) families dominate, of which Gata2 and Sox10 are already known direct activators of Ret (4), suggesting that others in this class may have similar function. With passing developmental time, a dramatically smaller and less-biased TF repertoire is affected: 34 TFs (18 down- and 16 up-regulated) at E12.5, and 28 TFs (12 down- and 16 up-regulated) at E14.5 (SI Appendix, Table S9), primarily belonging to the zinc finger- and T-box–containing families. Interestingly, TF use also varies across gut development with various patterns of regulation. Thus, Sox10, Snai1, Tbx2, Tbx3, and Ascl1 are down-regulated, while Foxp1, Foxp2, and Barx1 are up-regulated in Ret null embryos through development. The effect of Ret loss of function leads to down-regulation of Nfat5, Bnc2, Baz2b, Zfhx4, and Plag1 at E10.5 but they are up-regulated at E12.5 and E14.5 in the Ret null. Finally, Peg3 is down-regulated in Ret null embryonic guts at E10.5 and E12.5 but up-regulated at E14.5 (SI Appendix, Fig. S2). The direct functional consequences of each TF is thus nonconstant through gut development rather than their presumed canonical use as activators or repressors of transcription.
Among the TFs identified, the Hox family members deserve careful scrutiny. Hox factors play critical roles in vertebrate body pattering along the anterior–posterior axis and disruption of these genes is known to have effects on neural tissues, the neural crest, and endodermal and mesodermal derivatives (28–31). Multiple Hox genes are involved in patterning and morphogenesis of the mouse gut (32), especially genes of the Hoxd cluster, since their deletion (from Hoxd4 to Hoxd13) leads to loss of the ileocaecal sphincter and abnormalities in the pyloric and anal sphincter regions (33). Further, deletion of Hoxd1 to Hoxd10 (34) in the mouse leads to agenesis of the cecum. Almost 50% (20 out of 41) of the homeobox genes affected by loss of Ret at E10.5 are Hox genes (SI Appendix, Table S9), including the down-regulation of Hoxd cluster genes (Hoxd4, Hoxd8, Hoxd9, Hoxd10, Hoxd11, and Hoxd13). Thus, Ret loss of function, with consequent induced loss of Hoxd gene expression, may be the causal route to aganglionosis and other gut phenotypes. The literature shows that higher Hoxa4 expression, driven by ectopic expression in a much broader domain, leads to megacolon in the mouse from abnormal mesodermal development (35). Interestingly, Hoxa4 has significantly higher expression in Ret null embryos at E10.5 and is the likely candidate for megacolon, a major unexplained phenotype of HSCR. These results point to an expanded role of Ret in the morphogenesis of the gut than previously thought, its importance in non-ENS phenotypes in HSCR, and in gastrointestinal disorders generally.
Nonautonomous Effects of the ENS and the Mesenchyme.
When enteric neurons in the developing gut migrate through the mesenchyme, their development and activity is dependent on many trophic factors secreted by the mesenchyme, such as Gdnf, Nrtn, and Edn3, well known for their direct effects on ENS development through their cognate receptors (18). Other factors from the surrounding epithelium like Shh and Ihh also have effects on the ENS since their ablation severely reduces ENCC numbers (36, 37). Further, in rat models of HSCR, some regions of the distal colon are constricted with thick muscle and deep epithelial folds, while other regions have thin external muscle with a stretched mucosa (38). This observation is assumed to be an effect of reduced enteric neuron density on the surrounding tissues, but could arise from the effects of mesenchymal defects on the ENS.
Clearly ENS defects affect the mesenchyme. Consider that although at E10.5 there is no significant enrichment of a muscle-expressing genetic program, 11 muscle-specific genes are significantly down-regulated in Ret null embryos at this early stage (Tnnc2, Myh3, Myl1, Myog, Acta1, Tnnc1, Mylpf, Myl4, Actc1, Actn2, and Mybpc1). By E12.5, 18 muscle-expressed genes (Myl6, Actc1, Cryab, Tbx2, Tbx5, Tnc, Eln, Myh6, Tagln2, Tagln3, Tnni1, Tgfb2, Tnnt2, Gpx1, Des, Meox2, Pln, and Unc45b) and by E14.5, 21 muscle-expressed genes (Myl6, Actc1, Acta1, Cryab, Tbx2, Tnc, Tbx5, Eln, Myh6, Tagln2, Ttn, Tagln3, Tnni1, Tgfb2, Tnnt2, Gpx1, Myo18b, Des, Meox2, Pln, and Unc45b) are significantly down-regulated in Ret null embryos and are detectably enriched as a class. The genes affected at E12.5 and E14.5 are near identical, implying a persistent, long-term mesenchymal effect from Ret loss of function. We suspected that this muscle-expressing genetic program has feedback on the ENS.
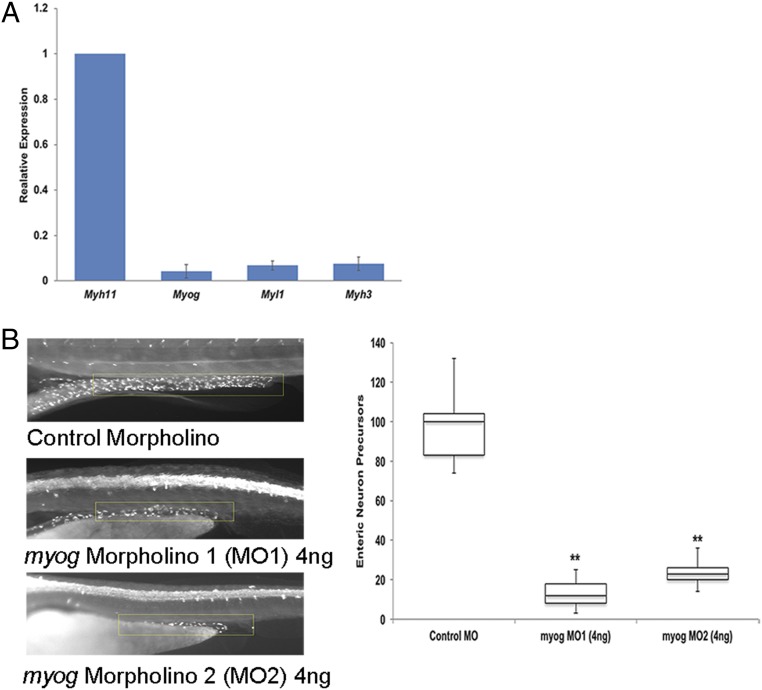
Consider that Myog, a transcription factor so far implicated only in the maturation of skeletal muscle (39), shows transitory expression at E10.5 in the wild-type gut but is significantly down-regulated with loss of Ret at E10.5. We postulated that this expression was specific to smooth muscle cells of the gut. For confirmation, we sorted EGFP-marked smooth muscle cells using florescent activated cell sorting (FACS) from the gut of a Myh11-EGFP transgenic mouse at E10.5 with EGFP expression driven by a 16-kb promoter sequence of the smooth muscle marker Myh11 (40). Taqman qPCR analysis of RNA extracted from these sorted cells show that they express Myh11, as expected, but also Myog, albeit weakly, along with other smooth muscle markers, such as Myl1 and Myh3, confirming the presence of Myog in early smooth muscle cells of the gut (Fig. 7A). We next asked whether loss of Myog would lead to any ENS defects using the zebrafish, a model system known to produce aganglionosis under knockdown of Ret and other HSCR genes (41). Since Myog is expressed at a reduced level and not completely absent in Ret null guts we simulated this effect in zebrafish using 2 splice-blocking morpholinos (MOs) against the intron 2–exon 3 junction of Myog, and marked postmitotic neurons in the developing gut with antibodies against neuronal HuC/HuD antigens to quantify the numbers of neurons (42). At 4 days postfertilization (dpf), with morpholino 1 (MO1), myog morphants showed curved bodies along the anterior–posterior axis, as expected, a hallmark of skeletal muscle defects. Additionally, they also demonstrated a severe loss of migratory enteric neurons as compared to embryos injected with control morpholinos (P = 6.5 × 10−9; Fig. 7B). The anterior–posterior axis curvature was much less pronounced in morpholino 2 (MO2)-injected embryos but loss of enteric neuronal precursors was equally severe (P = 7.8 × 10−9; Fig. 7B). Thus, broader defects within the mesenchyme can also affect the migration of enteric neurons, emphasizing a 2-way crosstalk between the ENS and mesenchyme necessary for normal ENS gangliogenesis.
Fig. 7.
Crosstalk between enteric neurons and their supporting mesenchymal cells. (A) Gene expression qPCR of specific muscle markers was performed using FACS-sorted Mhy11-positive cells from Myh11-EGFP transgenic mouse guts at E10.5. Low-level expression of Myog, Myl1, and Myh3 is observed in the gut smooth muscle cells at this stage of development. (B) Morpholino knockdown of myog in zebrafish leads to a skeletal muscle phenotype (significant body curvature along the anterior–posterior axis). Morpholino knockdown of myog in zebrafish leads to the skeletal muscle phenotype of significant body curvature along the anterior–posterior axis along with significant loss of enteric neuron precursors at 4 dpf. The neuronal numbers, counted in a region of interest starting at the eighth somite and measuring until the end of the gut tube (yellow box), upon sampling 50 embryos each from wild-type and morphant embryos are significantly different (**P < 0.001).
Discussion
The gastrointestinal tract is evolutionarily one of the most primitive organs in the animal kingdom and a rudimentary form is seen in Coelenterates. In the mouse and the human, the gut tube is formed from the primitive endodermal sheet, after a series of morphogenetic movements, followed by the formation of the muscularis layers and the submucosal connective tissue from the mesoderm. Finally, neural crest-derived ENCCs migrate into the forming foregut to innervate the submucosal and myenteric plexus as they form throughout the gut tube, moving in a craniocaudal manner. In the mouse, this innervation starts at E10.5 and is complete by E14.5, which is why we chose this period for developmental transcription profiling. After completion of neurogenesis at E14.5, the gut is still not fully physiologically functional and further development occurs, including after birth and consequent to feeding. However, these other time periods are less relevant to understanding human congenital motility disorders. The studies described here show that the primary feature of the E10.5 to E14.5 genetic programs is the shift from an early burst of transcription factor gene expression, indicative of the generation of cellular diversity by differentiation, to protein translation, indicating the emergence of functions in these diverse cells, to a wider muscle, epithelium, and endothelial developmental program. These trajectories are important for identifying candidate genes for various gut disorders and understanding why certain mutations are more detrimental than others (e.g., occurring earlier than later).
This study also shows that Ret signaling plays a critical and broad role in gut development in 3 major processes: engaging the extensive early regulatory machinery (TFs), initiating ENS development, and inducing neuroblast–mesenchymal interactions. A particularly surprising finding is the feedback effect of Ret on all 3 of these processes, some of which are direct and cell autonomous (e.g., on its own regulator Sox10) while others are indirect and nonautonomous (e.g., on its ligand Gdnf or the mesenchymal TF Myog). Thus, the phenotypic consequences of a Ret deficiency are much broader than its effects on neurogenesis. These findings have 3 corollaries. First, HSCR patients should routinely be examined by a clinical geneticist who is likely to recognize the many additional phenotypic manifestations in HSCR beyond aganglionosis; indeed, ∼32% of HSCR patients have developmental anomalies beyond aganglionosis (12). Second, the HSCR gene universe is considerably larger than currently known or assumed, including genes affecting intestinal smooth muscle and epithelium development. Third, we should not view HSCR as a cell autonomous disorder of the ENS but a multifactorial disorder of the gut involving different cell types.
These studies also provide 2 important lessons for the structure and functions of “systems” guiding organ development, and, therefore, human disease. As first propounded by Eric Davidson (1, 2), development is organized around modular GRNs that provide robustness to the system by buffering developmental noise, epigenetic noise, and the effects of deleterious mutations. Thus, we need to convert our gene lists not only by functional annotations but into GRNs that integrate the genetic and developmental information. These GRNs are the basic “units” of disease, as we have shown for the RET GRN and HSCR (4). Second, GRNs can display instability as well when deleterious mutations occur at certain rate-limiting genes within the GRN and lead to disease by propagating the local biochemical disruption throughout the GRN, thereby amplifying a mutational effect. We hypothesize that this feature partially explains why RET is the major gene for HSCR.
One remaining mystery is that despite the major sex effect in HSCR, gut development is overall not sex biased. However, a small group of 43 genes show sex differences in gene expression, the majority up-regulated in females and involved in diverse physiological processes. Thus, disruptions in the functions of these genes may have a larger genetic effect in males than females by virtue of the females’ higher expression, particularly if expression thresholds are important in development. These genes should be investigated for mutations in HSCR. Nevertheless, we likely have an incomplete picture of the nature and magnitude of sex differences in gut development. The mouse model we used harbors a null allele with no sex difference when aganglionosis is the measured phenotype (10). In contrast, other mouse models segregating a Ret null allele with a hypomorphic Ednrb allele do show strong sex differences in aganglionosis and segment length (19). Thus, studying other phenotypic manifestations in mouse models may be important, for example, by quantifying the numbers and distributions of enteric neurons in the ENS. Finally, the effect of sex on transcription might not be on the whole gut or the entire ENS but on a specific group of cells, an aspect which would not be captured in our bulk transcriptomic assays but require single cell transcription studies.
Materials and Methods
All of the mouse strains described in this study are available from The Jackson Laboratory (Bar Harbor, ME) or by request to A.C. All raw and processed RNA-seq data have been deposited into National Center for Biotechnology Information (NCBI)’s Gene Expression Omnibus and are accessible at GEO (accession no. GSE103070).
Mouse Strains.
Mice heterozygous for the conditional Ret allele (Retfl/+) (10, 43), were obtained from David Ginty, Harvard University, Boston, MA. Ret CFP knockin mice (RetCFP/+) were generated by crossing Retfl/+ mice to β-actin Cre mice to remove the RET9 cDNA (SI Appendix, Methods). This mouse was backcrossed >20 generation to C57BL/6 to maintain the line and all experiments were conducted in this isogenic strain. The smMHC/Cre/EGFP mice derived on a C57BL/6 background were obtained from The Jackson Laboratory (40, 44) and mantained in this background by repeated backcossing for >20 generations at the time of the experiments (SI Appendix, Methods). All animal research was approved by the Institutional Animal Care and Use Committee of the Johns Hopkins University School of Medicine.
Mouse Breeding, Dissection, and Genotyping.
Homozygous Ret null mice are neonatal lethals, so Ret heterozygous mice were intercrossed to generate all possible genotypes. Routine mouse genotyping was performed by PCR with the following primer pairs and amplicons: for the wild-type Ret locus, RetF 5′-CAGCGCAGGTCTCTCATCAGTACCGCA-3′ and RetR 5′-CAGCTAGCCGCAGCGACCCGGTTC-3′ resulting in a 449-bp PCR product (slightly modified from ref. 45); for detecting the CFP knockin, we used the primer pairs RetCFP F 5′-ATGGTGAGCAAGGGCGAGGAGCTGTT-3′ and RetCFP R 5′-CTGGGTGCTCAGGTAGTGGTTGTC-3′ to amplify a 615-bp PCR product from RetCFP/+ and RetCFP/CFP embryos. For the Myh11-Cre-EGFP mice, we used the primer pairs CreF 5′-ATGTCCAATTTACTGACCG-3′ and CreR 5′-CGCGCCTGAAGATATAGAAG-3′ to amplify a 241-bp amplicon. The sex of the embryos was determined by PCR with the following primers: Kdm5 forward 5′-CTGAAGCTTTTGGCTTTGAG-3′ and Kdm5 reverse 5′-CCGCTGCCAAATTCTTTGG-3′, mapping to exons 9 and 10 of the Kdm5c/d genes and resulting in 2 331-bp X chromosome-specific bands in females and an additional 301-bp Y chromosome-specific band in males (46).
We dissected the complete gut tube at E10.5 and the developing gut from the stomach to intestine at E12.5 and E14.5 in ice-cold Leibovitz’s L-15 medium (Thermo Fisher Scientific) from wild-type and RetCFP/CFP embryos under a fluorescence stereo zoom microscope (Zeiss AxioZoom.V16) with a suitable filter to detect CFP. The tissues were immediately flash frozen in liquid nitrogen for RNA extraction. The smMHC/Cre/EGFP guts were dissected under a GFP filter before dissociation and cell sorting.
Cell Sorting.
Gut tubes from Myh11-Cre-EFFP embryos at E10.5 were dissociated into single cell using Accumax (Sigma) by repeated pipetting. The cells were filtered serially through a 100-μM and 40-μM cell strainer, and centrifuged at 2,000 rpm for 5 min. The cell pellet was resuspended in 5% FBS, 4 mM EDTA in Leibovitz L-15 medium for cell sorting using MoFlo XDP (BD Biosciences) and a 488-nm laser for EFGP.
mRNA Sequencing Using RNA-Seq.
Total RNA was extracted from each of 3 male and 3 female mouse guts at E10.5, E12.5, and E14.5 from both wild-type and RetCFP/CFP embryos, using TRIzol (Life Technologies) and cleaned on RNeasy columns (Qiagen). At E10.5, owing to the small size of the embryos, 2 each were pooled to create 1 biological replicate and 3 replicates were used. Sample integrity (>9 RNA integrity number [RIN]) was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies) and cDNA were prepared using oligo dT beads as part of the Illumina Tru Seq RNA Sample Preparation protocol followed by standard Illumina library preparation. Libraries were run on a HiSEq. 2000 instrument to a depth of 15 million reads per samples (75 base pairs, paired end), using the manufacturer’s protocols. The resultant reads were mapped to the mouse reference genome (mm10/GRCm38) using Tophat2 (47) with its setting for paired end, nonstrand-specific libraries. Successfully mapped reads were used to assemble transcripts and estimate their abundance using Cufflinks 2.2.1 (48). The resultant data assigned FPKM values for each transcript and gene. The transcript file for each replicate was merged using Cuffmerge and analyzed by Cuffdiff (48) to detect differentially expressed genes between various comparisons. For the purpose of this study we considered all genes with FPKM ≥5 as expressed. All RNA-seq data have been deposited into NCBI’s Gene Expression Omnibus and are accessible at GEO Series accession no. GSE103070.
WGCNA.
We identified modules of coexpressed genes using the WGCNA R package (21). A total of 36 samples were analyzed comprising 3 time points (E10.5, E12.5, and E14.5), 2 sexes (female and male), 2 Ret genotypes (Ret+/+ and RetCFP/CFP), and 3 replicates each. Gene FPKM values were used for analyses after removing those with low quality in any sample (LOWDATA, HIDATA, or FAIL). There were 9,327 genes with a mean FPKM ≥5 across all samples retained for clustering, with expression values transformed to log2(FPKM+1).
We used WGCNA to conduct signed network analysis to construct modules of coexpressed genes, selecting a soft-thresholding power β of 24, the first power with adjusted R2 >0.8, to satisfy the assumption of a scale-free topology (21). Detailed parameters of WGCNA are given in SI Appendix, Methods.
Taqman Gene Expression Assays.
Total RNA was extracted from pooled EGFP FACS-sorted cells from the guts of 3 smMHC/Cre/EGFP embryos at E10.5 using TRIzol (Life Technologies) and cleaned on RNeasy columns (Qiagen). Total RNA (100 ng) was converted to cDNA using SuperScriptIII reverse transcriptase (Life Technologies) and Oligo-dT primers. The total cDNA was subjected to Taqman gene expression analyses (Life Technologies) using transcript-specific probes and primers. For the whole gut experiments to titrate the correlation between FPKM and Ct, 500 ng of RNA from each biological replicate was used to make cDNA. Mouse β-actin (Actb) was used as an internal loading control for normalization. Each assay was performed in triplicate (n = 9 observations per state); the data are presented as means with their SEs. Relative fold changes were calculated based on the 2ΔΔCt method for measuring gene expression, the value for the gene with the highest expression (lowest Ct value) being set to unity. The mouse Taqman probes were obtained from Applied Biosystems and are listed in SI Appendix, Methods.
Zebrafish Maintenance, Embryo Collection, and Morpholino Injections.
Zebrafish (AB strain) were raised and maintained under standard conditions, with embryos collected and staged as previously described (49). All animal research was approved by the Institutional Animal Care and Use Committee of the Johns Hopkins University School of Medicine.
For zebrafish gene knockdown experiments, 2 splice-blocking MOs were designed at the intron 2–exon 3 junction of myog: MO1, 5′-ATCTGAGAAAAGTAGACCACAATGT-3′ and MO2, 5′-GACGACACCTGTCACACACACAC-3′ (Gene Tools, LLC), along with a standard negative control morpholino (5′-CCTCTTACCTCAGTTACAATTTATA-3′) targeting a human beta-globin intron mutation for beta-thalassemia. We also injected a p53 morpholino (5′-GCGCCATTGCTTTGCAAGAATTG-3′) at 2-ng concentration along with our transcript-specific morpholinos because this morpholino has been reported to suppress the apoptotic effects induced by some morpholinos (50). Injections were performed on ∼30 1- to 2-cell zebrafish embryos independently on at least 3 different days (n = 100). Different concentrations (from 1 to 6 ng) of MO were injected to determine the optimal concentration at which a phenotype could be consistently detected. The data shown are for 4 ng where there was a measurable phenotype for enteric neurons at 4 dpf without severe malformation or death of embryos.
Immunostaining, Visualization, and Cell Counting.
Injected zebrafish embryos were fixed at 4 dpf with 4% paraformaldehyde (PFA) for 2 h at room temperature. A monoclonal anti-HuC antibody (Invitrogen A-21271), followed by Alexa Fluor 568 F (ab′)2 fragment of goat anti-mouse IgG secondary antibody (Invitrogen A-11019), was used for fluorescent labeling of enteric neurons, as previously described (41, 51), with mild modifications. The embryos were bleached after fixing in 4% PFA by incubating them in 3% H2O2/0.5% KOH medium until there was complete loss of epidermal pigmentation (∼20 min), followed by a 5-min wash with PBS to stop the bleaching reaction. A total of 50 stained embryos (for both wild type and morphants) were visualized using a Zeiss AxioZoom.V16 fluorescent microscope using a DS red filter to assess the colonization of enteric neurons in the gut of each embryo. Stained neurons were counted using the image-based tool for counting nuclei (ITCN) plugin in ImageJ (42), with the following parameters: width 9 pixels, minimum distance 4.5 pixels, threshold of 1, and using a selected region of interest (ROI). Since enteric neurons in Hirschsprung disease models in zebrafish are mostly lost caudally in the gut, we chose our ROI as 8 somites starting at the caudal end of the gut and moving rostrally. At least 20 embryos were used for cell counting for each concentration of morpholino used. P values were calculated from a pairwise 2-tailed t test.
Supplementary Material
Acknowledgments
These studies were supported by NIH MERIT (Method to Extend Research in Time) Award HD28088 to A.C.
Footnotes
The authors declare no competing interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE103070).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908756116/-/DCSupplemental.
References
- 1.Davidson E. H., The Regulatory Genome: Gene Regulatory Networks in Development and Evolution (Academic, Burlington, MA, 2006), New Ed p. xi, 289 p.
- 2.Davidson E. H., Emerging properties of animal gene regulatory networks. Nature 468, 911–920 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakravarti A., Turner T. N., Revealing rate-limiting steps in complex disease biology: The crucial importance of studying rare, extreme-phenotype families. BioEssays 38, 578–586 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Chatterjee S., et al. , Enhancer variants synergistically drive dysfunction of a gene regulatory network in Hirschsprung disease. Cell 167, 355–368.e10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furness J. B., Structure of the Enteric Nervous System (Blackwell Publishing, Malden, MA, 2006), pp. 1–28. [Google Scholar]
- 6.Heanue T. A., Pachnis V., Enteric nervous system development and Hirschsprung’s disease: Advances in genetic and stem cell studies. Nat. Rev. Neurosci. 8, 466–479 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Shivdasani R. A., Molecular regulation of vertebrate early endoderm development. Dev. Biol. 249, 191–203 (2002). [DOI] [PubMed] [Google Scholar]
- 8.de Santa Barbara P., van den Brink G. R., Roberts D. J., Development and differentiation of the intestinal epithelium. Cell. Mol. Life Sci. 60, 1322–1332 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakravarti M. A., Lyonnet S., “Hirschsprung disease” in The Metabolic and Molecular Bases of Inherited Disease, Valle B. D. et al. , Eds. (McGraw-Hill, New York, ed. 8, 2001). [Google Scholar]
- 10.Uesaka T., Nagashimada M., Yonemura S., Enomoto H., Diminished Ret expression compromises neuronal survival in the colon and causes intestinal aganglionosis in mice. J. Clin. Invest. 118, 1890–1898 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arighi E., Borrello M. G., Sariola H., RET tyrosine kinase signaling in development and cancer. Cytokine Growth Factor Rev. 16, 441–467 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Tilghman J. M., et al. , Molecular genetic anatomy and risk profile of Hirschsprung’s disease. N. Engl. J. Med. 380, 1421–1432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuchardt A., D’Agati V., Larsson-Blomberg L., Costantini F., Pachnis V., Defects in the kidney and enteric nervous system of mice lacking the tyrosine kinase receptor Ret. Nature 367, 380–383 (1994). [DOI] [PubMed] [Google Scholar]
- 14.Gerrard D. T., et al. , An integrative transcriptomic atlas of organogenesis in human embryos. eLife 5, e15657 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kukurba K. R., Montgomery S. B., RNA sequencing and analysis. Cold Spring Harb. Protoc. 2015, 951–969 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pervouchine D. D., et al. , Enhanced transcriptome maps from multiple mouse tissues reveal evolutionary constraint in gene expression. Nat. Commun. 6, 5903 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heanue T. A., Pachnis V., Expression profiling the developing mammalian enteric nervous system identifies marker and candidate Hirschsprung disease genes. Proc. Natl. Acad. Sci. U.S.A. 103, 6919–6924 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Obermayr F., Hotta R., Enomoto H., Young H. M., Development and developmental disorders of the enteric nervous system. Nat. Rev. Gastroenterol. Hepatol. 10, 43–57 (2013). [DOI] [PubMed] [Google Scholar]
- 19.McCallion A. S., Stames E., Conlon R. A., Chakravarti A., Phenotype variation in two-locus mouse models of Hirschsprung disease: Tissue-specific interaction between Ret and Ednrb. Proc. Natl. Acad. Sci. U.S.A. 100, 1826–1831 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chatterjee S., Chakravarti A., Transcriptome profile of wildtype and Ret homozygote null mouse guts at E10.5, E12.5 and E14.5. NCBI Gene Expression Omnibus (GEO). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE103070. Deposited 24 August 2017.
- 21.Langfelder P., Horvath S., WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang D. W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Le Douarin N. M., Teillet M. A., The migration of neural crest cells to the wall of the digestive tract in avian embryo. J. Embryol. Exp. Morphol. 30, 31–48 (1973). [PubMed] [Google Scholar]
- 24.Emison E. S., et al. , Differential contributions of rare and common, coding and noncoding Ret mutations to multifactorial Hirschsprung disease liability. Am. J. Hum. Genet. 87, 60–74 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emison E. S., et al. , A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature 434, 857–863 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Kapoor A., et al. , Population variation in total genetic risk of Hirschsprung disease from common RET, SEMA3 and NRG1 susceptibility polymorphisms. Hum. Mol. Genet. 24, 2997–3003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Durbec P. L., Larsson-Blomberg L. B., Schuchardt A., Costantini F., Pachnis V., Common origin and developmental dependence on c-ret of subsets of enteric and sympathetic neuroblasts. Development 122, 349–358 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Krumlauf R., Hox genes in vertebrate development. Cell 78, 191–201 (1994). [DOI] [PubMed] [Google Scholar]
- 29.Mallo M., Wellik D. M., Deschamps J., Hox genes and regional patterning of the vertebrate body plan. Dev. Biol. 344, 7–15 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wellik D. M., Hox genes and vertebrate axial pattern. Curr. Top. Dev. Biol. 88, 257–278 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Manley N. R., Capecchi M. R., Hox group 3 paralogs regulate the development and migration of the thymus, thyroid, and parathyroid glands. Dev. Biol. 195, 1–15 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Beck F., Homeobox genes in gut development. Gut 51, 450–454 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zakany J., Duboule D., Hox genes and the making of sphincters. Nature 401, 761–762 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Zacchetti G., Duboule D., Zakany J., Hox gene function in vertebrate gut morphogenesis: The case of the caecum. Development 134, 3967–3973 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Wolgemuth D. J., Behringer R. R., Mostoller M. P., Brinster R. L., Palmiter R. D., Transgenic mice overexpressing the mouse homoeobox-containing gene Hox-1.4 exhibit abnormal gut development. Nature 337, 464–467 (1989). [DOI] [PubMed] [Google Scholar]
- 36.Ramalho-Santos M., Melton D. A., McMahon A. P., Hedgehog signals regulate multiple aspects of gastrointestinal development. Development 127, 2763–2772 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Mao J., Kim B. M., Rajurkar M., Shivdasani R. A., McMahon A. P., Hedgehog signaling controls mesenchymal growth in the developing mammalian digestive tract. Development 137, 1721–1729 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stamp L. A., et al. , Surgical intervention to rescue Hirschsprung disease in a rat model. J. Neurogastroenterol. Motil. 21, 552–559 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sassoon D., et al. , Expression of two myogenic regulatory factors myogenin and MyoD1 during mouse embryogenesis. Nature 341, 303–307 (1989). [DOI] [PubMed] [Google Scholar]
- 40.Xin H. B., Deng K. Y., Rishniw M., Ji G., Kotlikoff M. I., Smooth muscle expression of Cre recombinase and eGFP in transgenic mice. Physiol. Genomics 10, 211–215 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Jiang Q., et al. , Functional loss of semaphorin 3C and/or semaphorin 3D and their epistatic interaction with ret are critical to Hirschsprung disease liability. Am. J. Hum. Genet. 96, 581–596 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gould T. W., Yonemura S., Oppenheim R. W., Ohmori S., Enomoto H., The neurotrophic effects of glial cell line-derived neurotrophic factor on spinal motoneurons are restricted to fusimotor subtypes. J. Neurosci. 28, 2131–2146 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Madsen C. S., et al. , Smooth muscle-specific expression of the smooth muscle myosin heavy chain gene in transgenic mice requires 5′-flanking and first intronic DNA sequence. Circ. Res. 82, 908–917 (1998). [DOI] [PubMed] [Google Scholar]
- 45.Jain S., et al. , Mice expressing a dominant-negative Ret mutation phenocopy human Hirschsprung disease and delineate a direct role of Ret in spermatogenesis. Development 131, 5503–5513 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Clapcote S. J., Roder J. C., Simplex PCR assay for sex determination in mice. Biotechniques 38, 702–706 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Kim D., et al. , TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trapnell C., et al. , Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F., Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 (1995). [DOI] [PubMed] [Google Scholar]
- 50.Robu M. E., et al. , p53 activation by knockdown technologies. PLoS Genet. 3, e78 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuhlman J., Eisen J. S., Genetic screen for mutations affecting development and function of the enteric nervous system. Dev. Dyn. 236, 118–127 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.