Significance
Numerous human cancers exhibit silencing or inactivation of p16INK4 and ARF, while their increased expression is often observed in diseases of aging. The mechanism underlying the regulation of these genes is complex and not fully understood. By utilizing a human p16INK4A reporter cell line and CRISPR libraries targeting noncoding regulatory elements and transcription factors, we discovered a cis-element adjacent to the ARF promoter that interacts with p16INK4A via chromatin looping to mediate p16INK4A expression. Targeting the cis-element or its associated transcription factors disrupts repression and induces p16INK4A transcription. Discovery of such natural noncoding elements responsible for p16INK4A regulation provides additional insight into the mechanism governing the dynamic regulation of this gene.
Keywords: chromatin conformation capture, genome editing, transcriptional regulation, knock-in, CRISPR screen
Abstract
Loss of function of CDKN2A/B, also known as INK4/ARF [encoding p16INK4A, p15INK4B, and p14ARF (mouse p19Arf)], confers susceptibility to cancers, whereas its up-regulation during organismal aging provokes cellular senescence and tissue degenerative disorders. To better understand the transcriptional regulation of p16INK4A, a CRISPR screen targeting open, noncoding chromatin regions adjacent to p16INK4A was performed in a human p16INK4A-P2A-mCherry reporter cell line. We identified a repressive element located in the 3′ region adjacent to the ARF promoter that controls p16INK4A expression via long-distance chromatin interactions. Coinfection of lentiviral dCas9-KRAB with selected single-guide RNAs against the repressive element abrogated the ARF/p16INK4A chromatin contacts, thus reactivating p16INK4A expression. Genetic CRISPR screening identified candidate transcription factors inhibiting p16INK4A regulation, including ZNF217, which was confirmed to bind the ARF/p16INK4A interaction loop. In summary, direct physical interactions between p16INK4A and ARF genes provide mechanistic insights into their cross-regulation.
The 50-kb CDKN2A/B (INK4/ARF) locus on human chromosome 9p21 (mouse chromosome 4) is sequestered within the larger topologically associated domain (TAD) defined by the neighborhood boundary genes MTAP and DMRTA1. The CDKN2A/B gene cluster specifies 3 tumor suppressor proteins: p16INK4A, ARF, and p15INK4B. Although p16INK4A and ARF messenger RNAs (mRNAs) are encoded by the common 3′ exons 2 and 3, their transcription is independently controlled by distinct promoters located 5′ to unique exon1α (p16INK4A) and exon1β (ARF), which reside ∼13 kb apart. The p16INK4A and p15INK5B proteins are canonical cell-cycle inhibitors that bind and inactivate the cyclin-dependent kinases CDK4 and CDK6 to induce G1-phase cell-cycle arrest and contribute to cellular senescence (1–3). In contrast, ARF mainly inhibits the ubiquitin E3 ligase MDM2 to activate p53-dependent transcriptional targets. Inactivation of ARF and p16INK4A in mice induces tumors with complete penetrance (4, 5), and epigenetic silencing or mutational inactivation of these genes is associated with numerous human cancers (6). Moreover, several lines of evidence suggest that naturally increased transcription of p16INK4A and ARF during aging induces senescence of various cell types (7–10). Hence, understanding the regulation of these genes has major implications for cancer and age-associated degenerative disorders.
Efforts to pharmacologically restore p16INK4A expression to suppress cancer progression have been explored through the identification of candidate small molecules and natural compounds enabling p16INK4A reactivation (11–13), and Food and Drug Administration-approved drugs that mimic p16INK4A in inhibiting CDK4 and CDK6 are now in widespread use in human cancer treatment (3). Conversely, generalized age-dependent induction of p16INK4A may cause deleterious effects by inducing senescence of normal tissues. Indeed, p16INK4A-positive senescent cells accumulate in many tissues as animals age, and their elimination in mice tempers age-associated degenerative diseases and extends life span (14, 15).
Genome-wide association studies focusing on cancers and degenerative diseases have identified numerous single-nucleotide polymorphism (SNPs) located upstream of the INK4/ARF locus that fall within a superenhancer cluster of an ∼500-kb region possessing H3K27ac activity (16, 17). To investigate the function of those human aging- and cancer-associated SNPs and noncoding segments, genome editing-based screening, including clustered regularly interspaced short palindromic repeats (CRISPR) screening, provides a powerful approach (18–23). A successful CRISPR screen designed to identify functional regulatory elements of human p16INK4A would be enhanced by a p16INK4A-fluorescent reporter endogenously transcribed by the native p16INK4A promoter in its proper chromosomal context.
There have been several efforts by other groups to derive p16INK4A reporter cell lines. However, minimal INK4A promoter regulatory sequences driving a reporter did not fully mirror endogenous transcriptional regulation (24). Others engineered a large human genomic segment including the entire INK4/ARF gene cluster containing a firefly luciferase gene inserted into the C terminus of the p16INK4A-coding region that was then used to generate transgenic mice (25). Similarly, Demaria et al. (26) engineered a bacterial artificial chromosome containing ∼50 kb of the murine p16Ink4a locus, such that the p16Ink4a promoter drove a trimodal reporter (3MR) to selectively kill senescent cells. Moreover, Baker et al. (14) generated a transgenic mouse strain by using an ∼2.6-kb fragment containing the p16Ink4a promoter to drive the expression of a FKBP-Casp8-IRES-GFP cassette, which could conditionally eliminate senescent cells in vivo. Although all of these transgenic strains could report real-time expression of p16Ink4a under various physical conditions or stresses, these models lack the in vivo chromatin niche that may affect more precise control of transcription when compared with that of the endogenous allele. Most recently, Liu and colleagues described a p16tdTomato reporter allele, enabling the in vivo characterization and purification of cells featuring activation of the p16Ink4a promoter. However, the tdTomato expression in cells depleted of a neomycin selection cassette was weakly detected and correlated less well with endogenous p16Ink4a mRNA (27). Burd et al. (28) targeted the translational start site (TSS) of the endogenous p16Ink4a locus by inserting the firefly luciferase complementary DNA followed by a SV40 polyadenylation signal. The resulting knock-in allele was expected to be null for p16Ink4a expression and yet retain intronic structures surrounding exon 1β (28, 29).
Based on these considerations, we developed a p16INK4A reporter cell line recapitulating endogenous transcriptional activity. We performed a CRISPR screen with a pooled single guide RNA (sgRNA) array targeting Assay for Transposase-Accessible Chromatin Sequencing (ATAC-seq) and H3K27ac marked regions spanning the entire TAD containing p16INK4A, as well as a loss-of-function genetic CRISPR screen targeting 1,639 human transcription factors. We have revealed a mechanism underlying the transcriptional regulation of p16INK4A via a cis-regulatory element adjacent to ARF promoter.
Results
Generation and Characterization of the p16INK4A-P2A-mCherry Reporter Allele.
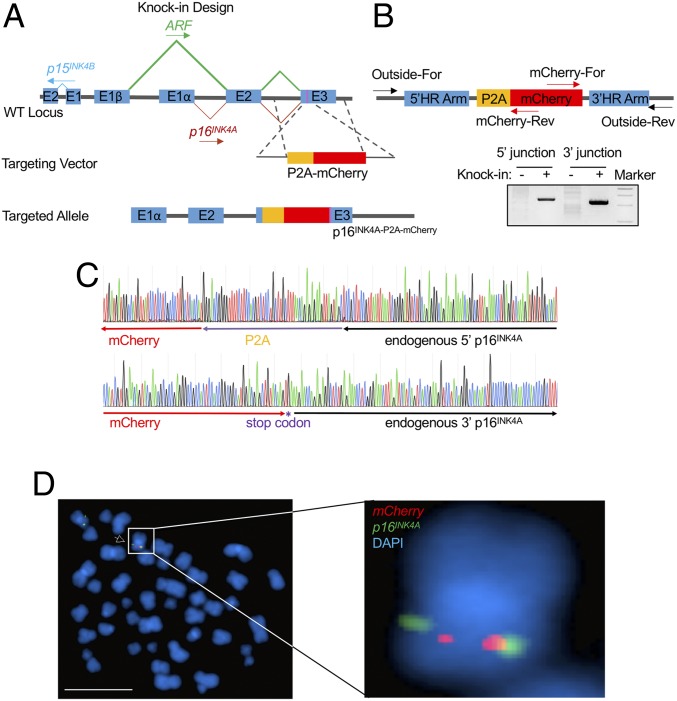
We utilized CRISPR/Cas9-mediated homologous recombination to deliver the P2A-mCherry cassette upstream of the p16INK4A stop codon in a patient-derived human B-ALL cell line, SEM, which maintains an intact INK4/ARF locus (Fig. 1A). The p16INK4A-P2A-mCherry reporter allele was translated in the same reading frame as p16INK4A, but not ARF. Because ribosomes skip the synthesis of the glycyl-prolyl peptide bond at the C terminus of the P2A peptide, translation leads to dissociation of P2A and its immediate downstream p16INK4A protein (30). Therefore, the knock-in allele produced p16INK4A under control of the endogenous promoter and intrinsic cis-regulatory elements, while delivering the dissociated mCherry reporter separately.
Fig. 1.
Generation and characterization of the p16INK4A-P2A-mCherry reporter allele. (A) Schematic diagram of the knock-in design for the p16INK4A-P2A-mCherry reporter allele. The magenta bar in exon 3 indicates the stop codon of p16INK4A. (B) Schematic diagram of the primer design for the genotyping PCR. “Outside” primers bind to endogenous loci outside the 800-bp homology arms and were used to specifically measure successful knock-in events (+) at the 5′ and 3′ boundary regions in combination with primers residing within mCherry. Wild-type SEM cells were used as negative controls (−). (C) Genotyping PCR products from the 5′ and 3′ knock-in boundaries were sequenced to verify the seamless knock-in of the mCherry reporter gene to the endogenous locus. (D) FISH of the P2A-mCherry knock-in cassette in p16mCherry/+ reporter cells. The P2A-mCherry DNA is labeled with red, and an INK4/ARF fosmid clone is labeled with green. Metaphase cells were scored for the correct knock-in events by pairing of red and green signals and for random integration signals. (Scale bar, 10 µm.) DAPI staining indicates the nuclei.
To enable knock-in efficiency in human SEM cells, we used a double-cut nuclease cleavage strategy to release exogenous DNA fragments from a donor vector in vivo (31, 32), followed by serial sorting to enrich knock-in events (33). In brief, we constructed the knock-in vector containing a P2A-mCherry cassette flanked with 5′ and 3′ p16INK4A homology arms (HAs) of ∼800 bp. The HA/knock-in cassette was bordered with a sgRNA and a protospacer adjacent motif (PAM) sequence targeting a DNA sequence 5′ of the stop codon of p16INK4A (Dataset S1). In the presence of Cas9 and the p16INK4A sgRNA, the HA/knock-in cassette was released from the donor vector with 2 nuclease cleavages and delivered to the target genomic region. Individual knock-in clones were derived from a targeted bulk population and characterized by genotyping PCR and Sanger sequencing (Fig. 1 B and C). A heterozygous clone carrying the P2A-mCherry knock-in cassette, hereafter called p16mCherry/+, was selected for use as a reporter cell line for this study. To rule out the possibility of random integration of the reporter in the p16mCherry/+ cell line, fluorescence in situ hybridization (FISH) was performed with a P2A-mCherry DNA probe and a fosmid DNA probe targeting the INK4/ARF locus. Each of 18 metaphase cells harvested from the p16mCherry/+ cell line was identified as heterozygous for the P2A-mCherry knock-in cassette, yielding 2 red signals (P2A-mCherry probe) located adjacent to 2 of the 4 green signals (p16INK4A probe) in each nucleus. No random integration events were observed in other chromosomes (Fig. 1D).
The p16INK4A-P2A-Cherry Reporter Allele Recapitulates Endogenous Transcription of p16INK4A.
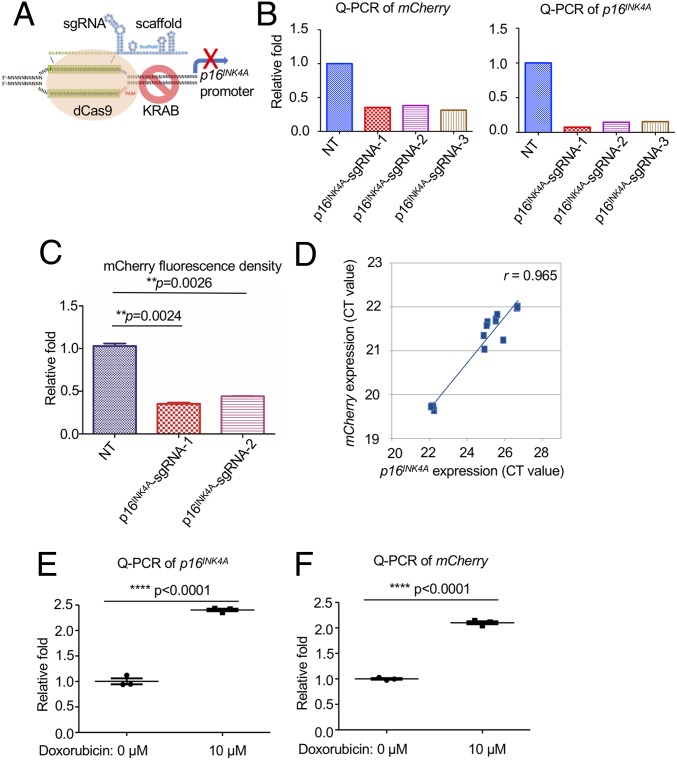
Next, we tested whether the p16INK4A-P2A-mCherry reporter allele would respond to the transcriptional regulation of the cellular p16INK4A promoter. The p16mCherry/+ cells were first infected with lentivirus encoding nuclease-deficient dCas9 fused to the transcriptional repressor domain of KRAB (34) to generate a stable cell line, p16mCherry/+;dCas9-KRAB (Fig. 2A). Three sgRNAs targeting the p16INK4A promoter were cloned and delivered individually into p16mCherry/+;dCas9-KRAB cells. Quantitative RT-PCR (qRT-PCR) and flow cytometry each revealed that mCherry and p16INK4A expression were suppressed by all 3 sgRNAs compared with parental p16mCherry/+;dCas9-KRAB cells (Fig. 2 B and C). As expected, mRNA expression of mCherry strongly correlated with that of p16INK4A when measured in cells targeted with the 3 sgRNAs and dCas9-KRAB (Pearson’s r = 0.965) (Fig. 2D). In p16INK4A-sgRNA-1–targeted cells, p16INK4A expression and translation were significantly reduced, resulting in elevated RB1 phosphorylation and yielding a proliferation advantage when compared with controls (SI Appendix, Fig. S1 A–D). The chemotherapy drug doxorubicin (Dox), which has been shown to activate p16INK4A (35), induced p16INK4A expression and reporter activity following 10 μM Dox treatment for 24 h (Fig. 2 E and F). Taken together, these data confirmed that the p16INK4A-P2A-mCherry reporter allele was expressed under the control of the endogenous promoter.
Fig. 2.
The p16INK4A-P2A-mCherry reporter allele recapitulates endogenous transcription of p16INK4A. (A) Schematic diagram of the transcriptional repression of p16INK4A by the CRISPR interference system. The dCas9-KRAB fusion protein was guided to the p16INK4A promoter with 3 individual sgRNAs. (B) qRT-PCR was performed on the p16mCherry/+;dCas9-KRAB cells with 3 different p16INK4A-sgRNAs using primers targeting coding sequences of mCherry and p16INK4A. (C) Flow cytometry analysis was performed on the p16mCherry/+;dCas9-KRAB cells with 2 p16INK4A-sgRNAs. The mean value of the mCherry density in each group was calculated and obtained by 3 replicates. The P value was calculated by a 2-tailed t test. (D) The correlation of transcription reduction in mCherry and p16INK4A in response to dCas9-KRAB–mediated transcription repression (from B) was calculated by Pearson’s correlation test (n = 6). (E) qRT-PCR was performed on the p16mCherry/+ cells with or without the treatment of doxorubicin for 24 h by using specific primers targeting the mRNA sequence of p16INK4A. Three biological replicates were performed. The P value was calculated by a 2-tailed t test. (F) qRT-PCR was performed on the p16mCherry/+ cells with or without the treatment of doxorubicin for 24 h by using specific primers targeting the mRNA sequence of mCherry. Three biological replicates were performed. The P value was calculated by a 2-tailed t test.
Noncoding Pooled CRISPR Screening Identified a Distal Repressive Element of p16INK4A Residing within the ARF Locus.
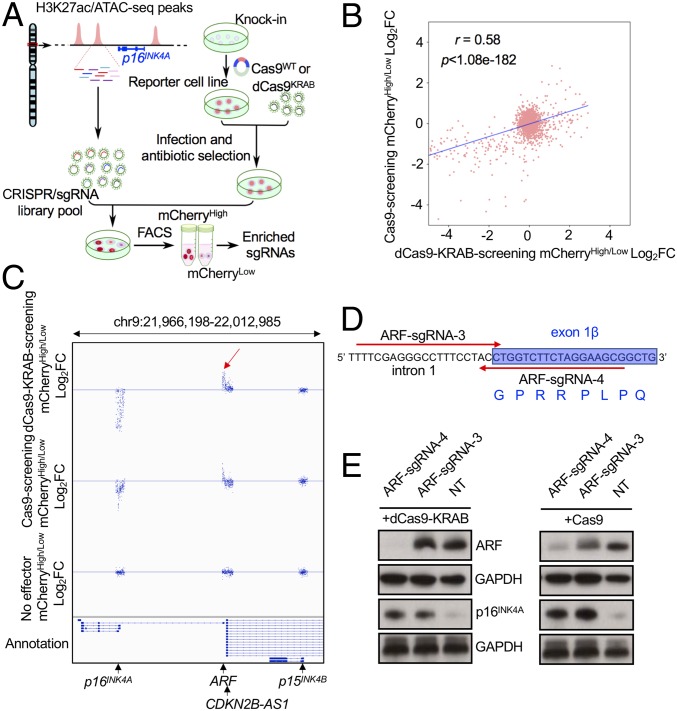
Derivation of a reporter cell line enabled us to comprehensively conduct large-scale screenings to identify regulatory DNA elements and proteins controlling p16INK4A gene expression. Since ATAC-seq measures genome-wide chromatin accessibility corresponding to genomic loci where proteins are bound to DNA, and H3K27ac marks active promoters and enhancers (36, 37), CRISPR screens against these regions could provide detailed information about cis-regulatory functions. Therefore, we synthesized a 2,029-sgRNA array targeting the H3K27ac and ATAC-seq positive peaks defined in human IMR90, HCT116, and SEM cells spanning the entire TAD containing the INK4/ARF gene cluster. Human ARF promoter (+1 to −4,690 bp from TSS) and p16INK4A promoter (+1 to −1,729 from TSS) were defined by molecular cloning and reporter assays (38). An additional 20 nontargeting (NT) sgRNAs were also included as negative controls (Fig. 3A). Quality control of the sgRNA library integrity by deep sequencing revealed 100% coverage of the 2,049 sgRNAs in plasmid libraries as well as HCT116 and IMR90 cells 48 h after lentiviral infection (SI Appendix, Fig. S2 A–C). Subsequently, the same lentiviral library was transduced into p16mCherry/+;dCas9-KRAB cells at a low multiplicity of infection (< 0.3) and then fractionated by flow cytometric sort for mCherry expression (SI Appendix, Fig. S3A). The greatest enrichment of positive control sgRNAs targeting the p16INK4A promoter was observed in the bottom 10% vs. top 10% mCherry sorting gate. Thereafter, the mCherryHigh and mCherryLow populations in replicate screens were selected from the top or bottom 10% sorting gates (SI Appendix, Fig. S3 B and C) and collected for deep sequencing to identify differentially represented sgRNAs, which indicated the corresponding targeted regions associated with transcriptional repression or activation of p16INK4A, respectively. To measure the specificity of the CRISPR/dCas9-KRAB screen, we also sorted the top 10% and bottom 10% of p16mCherry/+ parental cells transduced with the sgRNA library alone (no effector screening).
Fig. 3.
Noncoding pooled CRISPR screening identified a distal repressive element of p16INK4A residing within the ARF locus. (A) Schematic diagram of a working model of dCas9-KRAB– and Cas9-mediated noncoding screening. (B) The global correlation of sgRNA distribution in dCas9-KRAB and Cas9 screens in the p16mCherry/+ reporter cell line. (C) The global distribution of all sgRNAs in a selected region of the INK4/ARF locus from 3 screens in the p16mCherry/+ reporter cell line using dCas9-KRAB, Cas9, and no effector control. The red arrow indicates the most enriched sgRNAs in a 42-bp region of the ARF exon1β and intron 1. (D) Two candidate sgRNAs binding to the plus and minus strands of the most enriched 42-bp core DNA sequence near the ARF promoter were designed for validation (ARF-sgRNA-3 and -4). The red arrows indicate the orientation of the sgRNA binding sequences. (E) Western blotting analysis of ARF and p16INK4A in dCas9-KRAB– and Cas9-expressing SEM cells infected with ARF-sgRNA-3, -4, and the nontargeting sgRNA control.
To complement the dCas9-KRAB and sgRNA library screen, we performed a parallel screen in p16mCherry/+;Cas9 stable SEM cells (Fig. 3A). Comparisons between dCas9-KRAB and wild-type Cas9 screens demonstrated correlation of global sgRNA distribution (r = 0.58, P = 1.08e−182) (Fig. 3B and SI Appendix, Fig. S4 A and B). Data collected from 3 biological replicates were analyzed by the DESeq2 algorithm (39) for each screen. At a stringent cutoff (an adjusted P value of ≤0.01 and a fold-change of log2High/Low ≥1 or ≤−1), we identified 199 differentially represented sgRNAs in the dCas9-KRAB screen (Fig. 3C and SI Appendix, Fig. S4 B and D). Among the 212 positive-control sgRNAs designed to target the p16INK4A promoter, 144 sgRNAs were significantly enriched in the mCherryLow fraction. None of the 190 sgRNAs targeting the p15INK4B promoter were enriched. Notably, we identified 51 sgRNAs that were enriched in the mCherryLow fraction, all of which targeted the ARF promoter and adjacent 3′ regions including exon 1β and a partial sequence of intron 1. As expected, none of the 2,049 sgRNAs were differentially represented in the no-effector screen (SI Appendix, Figs. S4 C and D and S5). Although the ARF bidirectional promoter also controls expression of the antisense long noncoding RNA ANRIL (also named CDKN2B-AS1), none of the 51 enriched sgRNAs resided on exon 1 of ANRIL, which is transcribed in the opposite direction. In addition, the most strongly enriched sgRNAs in the Cas9 screen recognized a minimal 42-bp DNA sequence at the junction of ARF exon 1β and intron 1 (Fig. 3 C and D and SI Appendix, Fig. S4D). Notably, this 42-bp segment includes the ARF exon 1β splice donor site. Therefore, combining the p16INK4A reporter cell line with CRISPR screening identified a previously undiscovered distal repressive regulatory region of p16INK4A in the INK4/ARF locus.
Distal Repressive Element of p16INK4A Composed of DNA Sequences Located 3′ and Adjacent to the ARF Promoter.
To validate the screening results, 2 sgRNAs that resided on the 42-bp sequence between exon 1β and intron 1 (ARF-sgRNA-3 and -4) identified from the CRISPR/Cas9 screen were delivered into SEM cells stably expressing either dCas9-KRAB or Cas9 (Fig. 3 D and E). Consistent with the CRISPR/Cas9 screen in p16mCherry/+ cells, both sgRNAs enhanced p16INK4A expression through synergy with dCas9-KRAB or Cas9. In direct contrast, ARF-sgRNA-4 significantly reduced ARF expression. However, although up-regulating p16INK4A, ARF-sgRNA-3 did not affect ARF expression in SEMdCas9-KRAB cells and only modestly suppressed ARF expression in SEMCas9 cells, demonstrating that down-regulation of ARF protein expression was dispensable for p16INK4A up-regulation (Fig. 3E). Two additional sgRNAs (ARF-sgRNA-1 and -2) identified in the CRISPR screen that bound to exon 1β of ARF and were upstream of the 42-bp core sequence also trigged some increase in p16INK4A expression and reporter activity (SI Appendix, Fig. S6 A and B). Collectively, these data suggest that the DNA sequence 3′ and adjacent to the ARF promoter rather than the encoded protein is a repressive element required for transcriptional regulation of p16INK4A.
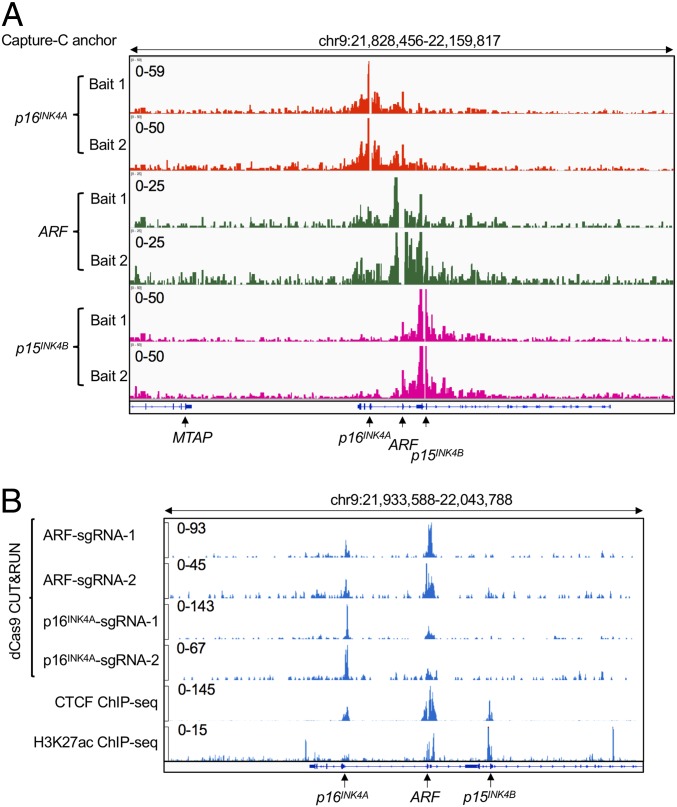
Physical Interactions between ARF, p16INK4A, and p15INK4B Were Detected by Chromatin Conformation Capture.
Given the fact that the ARF promoter and p16INK4A promoters are located ∼13 kb apart in the human genome, the transcriptional repression of p16INK4A by the distal repressive element might require long-distance chromatin interactions. To test this hypothesis, we deployed a multiplex assay of high-resolution chromatin conformation called Capture-C (40, 41), a derivative of the chromatin conformation capture (3C) technique coupled with oligonucleotide enrichment and high-throughput sequencing. This method enables the discovery of distant interacting elements from multiple “bait” sites (or anchor regions) with high resolution. Here, Capture-C was performed using a 3C library prepared from SEM cells. Two biotinylated bait oligonucleotides were designed to hybridize to each of the H3K27ac peaks overlapping the defined p16INK4A, ARF, and p15INK4B promoters, respectively. Strong enrichment of sequences at each bait site confirmed the efficiency of hybridization. Using baits against the p16INK4A promoter, a broad peak encompassing part of the ARF promoter including the identified distal repressive element affecting p16INK4A was identified as a strong interacting region. Similarly, when bait oligonucleotides were hybridized with the p15INK4B promoter, a strong interaction between p15INK4B and ARF was observed. When Capture-C was carried out using the ARF promoter oligonucleotides as bait, both p16INK4A and p15INK4B were captured. These data suggest that in SEM cells the ARF locus physically interacts with both p16INK4A and p15INK4B through long-distance chromatin looping (Fig. 4A). However, since direct contact between p16INK4A and p15INK4B was not observed, we reasoned that ARF/p16INK4A and ARF/p15INK4B associate in separate looping complexes.
Fig. 4.
Physical interactions between ARF, p16INK4A, and p15INK4B were detected by 3C. (A) Next-generation Capture-C was performed on parental human SEM cells for 2 replicates. Two specific anchor probes (Bait 1 and Bait 2) were designed to hybridize to the H3K27ac peaks that overlap each of the promoters of p16INK4A, ARF, and p15INK4B to capture chromatin interactions with the respective promoters within the INK4/ARF locus. (B) CUT&RUN was performed on p16mCherry/+;dCas9-KRAB cells infected individually with 2 sgRNAs targeting the p16INK4A repressive element adjacent to the ARF promoter and 2 sgRNAs targeting the p16INK4A promoter. A Cas9 antibody that recognizes dCas9 was used for a pull-down assay. ChIP-seq tracks of CTCF and H3K27ac were included to indicate the open chromatin status of the locus.
To independently confirm the physical interaction between ARF and p16INK4A, we deployed a recently developed CUT&RUN strategy (42) in which antibody-targeted controlled cleavage by micrococcal nuclease releases specific protein-DNA complexes into the supernatant for paired-end DNA sequencing. Because CUT&RUN is performed in situ, it enables both quantitative high-resolution chromatin mapping of a local chromatin environment and identification of physical DNA interactions. Here, we performed CUT&RUN in SEM cells stably expressing dCas9-KRAB and sgRNAs targeting the p16INK4A repressive element (ARF-sgRNA-1 and -2). By using a Cas9 antibody that recognizes dCas9 for pull-down, we clearly detected dCas9-binding peaks in the sgRNA-guided sequences in the ARF and p16INK4A loci simultaneously, demonstrating that these regions were physically interacting in situ. Similarly, in SEMdCas9-KRAB cells transduced with sgRNAs targeting the p16INK4A promoter (p16INK4A-sgRNA-1 and -2), we detected a specific peak encompassing the ARF promoter and the identified distal repressive element, as well as the expected peak at the p16INK4A promoter (Fig. 4B). Comparing our Cas9 CUT&RUN data with the CTCF and H3K27ac Chromatin Immunoprecipitation Sequencing (ChIP-seq) data that demarcated compartment boundaries, the interaction between ARF and p16INK4A genes appears to be supported by a convergent looping model mediated by the canonical CTCF/Cohesin complex (43, 44). In summary, these data support the idea that p16INK4A and ARF physically crosstalk by chromatin looping, providing a proximity environment to regulate their respective transcription.
The ARF/p16INK4A Chromatin Interaction and Transcriptional Regulation Is Functionally Detected in Human Neuroblastoma Cells.
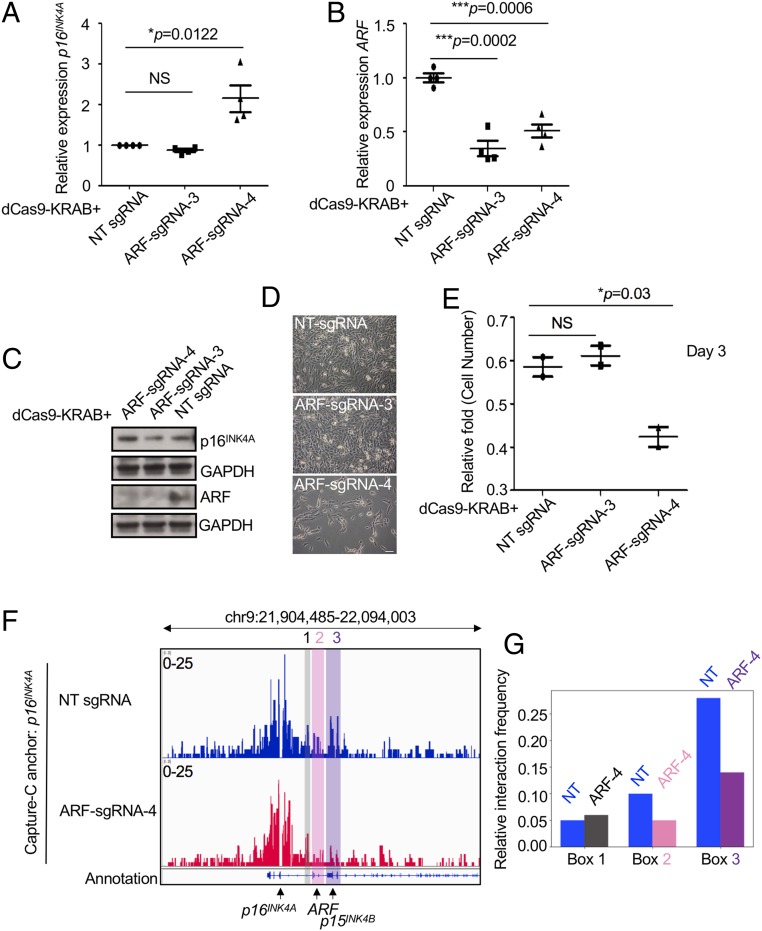
We hypothesized that the same chromatin interaction pattern seen in human B-ALL SEM cells may exist in other cancer cell types that maintain low expression and an intact genomic sequence of p16INK4A. To this end, we transduced lentiviral dCas9-KRAB with ARF-sgRNA-3 and -4 into a human neuroblastoma (NBL) cell line, SK-N-SH. ARF-sgRNA-4, but not sgRNA-3, could significantly up-regulate the expression of p16INK4A consistently at mRNA and protein levels (Fig. 5 A–C). Both sgRNAs conversely decreased ARF expression. During in vitro culture of targeted cells, we observed significant growth retardation using ARF-sgRNA-4, consistent with specific up-regulation of p16INK4A (Fig. 5 D and E). The 3C libraries were prepared from SK-N-SH dCas9-KRAB stable cells targeted with either nontargeting-sgRNA (NT-sgRNA) or ARF-sgRNA-4. When Capture-C was performed using bait oligonucleotides against the p16INK4A promoter on the NT-sgRNA 3C library, the p16INK4A promoter was juxtaposed to both ARF and p15INK4B. However, the ARF/p16INK4A and p16INK4A/p15INK4B looping affinity was significantly reduced in Capture-C using p16INK4A promoter bait oligonucleotides on the ARF-sgRNA-4 3C library (Fig. 5 F and G).
Fig. 5.
The ARF/p16INK4A chromatin interaction and transcriptional regulation is functionally detected in NBL cells. (A) qRT-PCR analysis of p16INK4A in the human NBL cell line SK-N-SH infected with lentiviral dCas9-KRAB and ARF-sgRNAs-3 and -4. NT-sgRNA is a nontargeting sgRNA control. Four biological replicates were performed. The P value was calculated by a 2-tailed t test. (B) qRT-PCR analysis of ARF in the human NBL cell SK-N-SH infected with lentiviral dCas9-KRAB and ARF-sgRNAs-3 and -4. Four biological replicates were performed. The P value was calculated by a 2-tailed t test. (C) Western blotting analysis of ARF and p16INK4A in the human NBL cell line SK-N-SH infected with lentiviral dCas9-KRAB and ARF-sgRNA-3 and -4 compared to NT-infected controls. (D) Representative image of SK-N-SH cells stably expressing dCas9-KRAB and ARF-sgRNAs-3, -4, and NT-sgRNA at day 3. (E) Quantification of cell proliferation by cell number count. Two biological replicates were performed. The P value was calculated by a 2-tailed t test. (F) Next-generation Capture-C was performed on SK-N-SH cells stably expressing dCas9-KRAB and ARF-sgRNAs-3, -4, and NT-sgRNA. Anchor probes that hybridized to the p16INK4A promoter were utilized to capture chromatin interactions. Boxes 2 and 3 highlight ARF and p15INK4B promoters which were interacting with the indicated p16INK4A anchor regions. Box 1 indicates a negative control noncoding region. (G) Quantification of interaction frequency between the anchor regions to Boxes 1, 2, and 3. Signal value from the .bw file of Capture-C was normalized by probe signal for each experiment.
Loss-of-Function CRISPR/Cas9 Screening of Human Transcription Factors in the p16INK4A Reporter Cell Line.
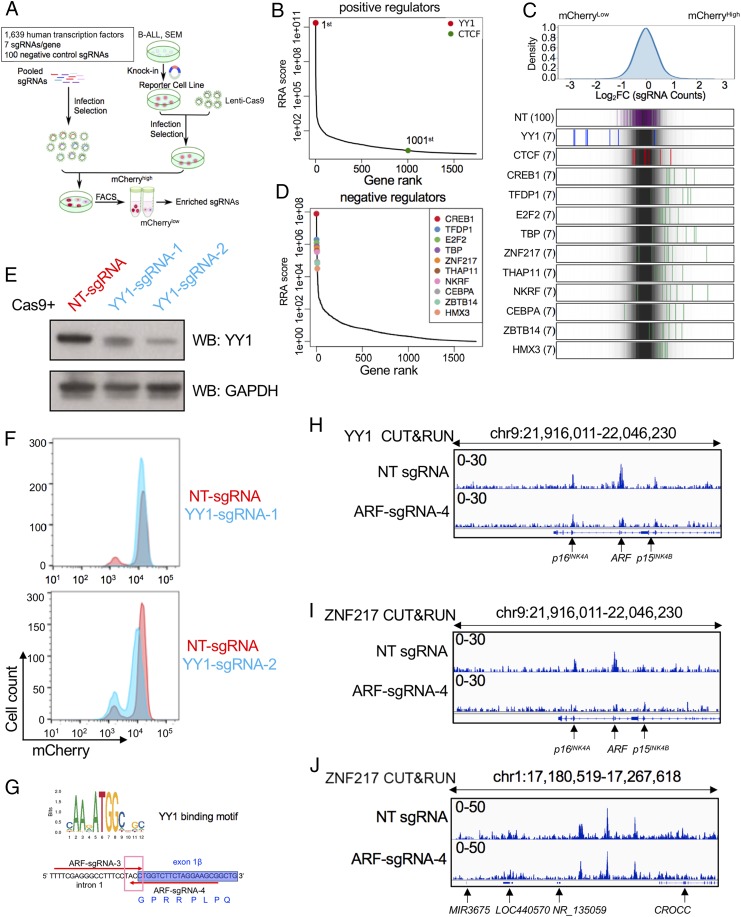
We further undertook a comprehensive loss-of-function CRISPR-Cas9 screen targeting 1,639 human transcription factors in combination with the p16mCherry/+ reporter system to identify putative regulatory effectors. Seven sgRNAs were designed for each transcription factor (TF), and an additional 100 nontargeting sgRNAs were included as negative controls (Fig. 6A). Similar to previous noncoding screening, about 1 million cells from the top 10% (mCherryHigh) and bottom 10% (mCherryLow) sorted populations were collected for deep sequencing to identify differentially represented sgRNAs.
Fig. 6.
Loss-of-function CRISPR screening of human transcriptional factors in the p16INK4A reporter cell line. (A) Schematic diagram of a working model of Cas9-mediated CRISPR screening targeting 1,639 human TFs. (B) Gene ranking of looping factors (YY1, first; CTCF, 1,001st) enriched from screening. The enrichment score of 7 sgRNAs against each TF was combined by MAGeCK algorithm. (C, Top) The overall distribution of all sgRNAs from the screening. (C, Bottom) The Log2[Fold Change (top10/bottom10)] ratio for all sgRNAs targeting CTCF, YY1, and the top 10 negative regulators from D and NT sgRNAs were overlaid on a gray gradient depicting the overall distribution. NT: 100 gRNAs; TF: 7 sgRNAs/each. (D) Gene ranking of top 10 negative candidate regulators enriched from screening analysis by MAGeCK algorithm. (E) Immunoblotting of YY1 in p16mCherry/+;Cas9 cells targeted individually with 2 sgRNAs against the coding sequence of human YY1. GAPDH was probed as a loading control. (F) The p16mCherry/+;Cas9 cells were targeted with Cas9 and 2 sgRNAs identified from TF screening targeting the coding region of human YY1. Flow cytometry analysis of mCherry reporter activity was subsequently conducted in YY1-targeted cells compared with NT control. (G) YY1-binding motif prediction by JASPAR algorithm. (H) CUT&RUN of YY1 was conducted in p16mCherry/+;dCas9-KRAB cells infected with ARF-sgRNA-4 and NT-sgRNA for 2 replicates. Tracks were shown at the viewpoint of the INK4/ARF locus. (I) CUT&RUN of ZNF217 was conducted in p16mCherry/+;dCas9-KRAB cells infected with ARF-sgRNA-4 and NT-sgRNA. Tracks were shown at the viewpoint of the INK4/ARF locus. (J) CUT&RUN of ZNF217 was conducted in p16mCherry/+;dCas9-KRAB cells infected with ARF-sgRNA-4 and NT-sgRNA. Tracks of ZNF217 were shown for viewpoint at a randomly chosen locus.
Surprisingly, the most characterized looping factor, CTCF, was not enriched in the screen (Fig. 6B). Furthermore, we utilized a previously described auxin inducible degron system (45–47) to acutely deplete the CTCF protein in SEM cells (SI Appendix, Fig. S7 A–D) (33). Although the CUT&RUN using CTCF antibody for chromatin pull-down demonstrated loss of CTCF occupancy throughout the INK4/ARF locus (SI Appendix, Fig. S7E), perturbation of CTCF did not alter p16INK4A expression changes at the mRNA or protein levels (SI Appendix, Fig. S6 F and G), suggesting that the ARF/p16INK4A interaction in SEM cells relies primarily on other looping factors.
The top-ranked candidate of positive regulators was Yin Yang 1 (YY1) (Fig. 6 B and C), which belongs to the GLI-Kruppel class of zinc-finger transcription factors and is ubiquitously expressed in most tissue and cell lines. YY1 plays a fundamental role in normal biologic processes such as embryogenesis, differentiation, replication, cellular proliferation, and cancer development (48, 49). Recently, YY1 was also reported to bind to active enhancers and promoter-adjacent elements and to form dimers that facilitate the interactions of these DNA elements (50–52). In the p16mCherry/+;dCas9-KRAB stable cell line, 2 sgRNAs (YY1-sgRNA-1 and -2) targeting the coding sequence of YY1 down-regulated YY1 expression along with a reduction of reporter activity (Fig. 6 E and F). Although shown to act as a positive regulator of p16INK4A from the TF screen, YY1 was identified as a candidate transcription factor for the ARF/p16INK4A interaction due to a conserved YY1-binding motif in the previously identified 42-bp DNA segment within the cis-regulatory element (Fig. 6G). Additionally, YY1 occupancy at both ARF and p16INK4A promoters was confirmed in NT-sgRNA SEMdCas9-KRAB cells by CUT&RUN assay. In ARF-sgRNA-4–targeted cells, YY1-binding affinity to ARF, but not to p16INK4A, was significantly disrupted (Fig. 6H). These paradoxical findings suggest that, while YY1 could facilitate the ARF/p16INK4A chromatin interaction that leads to p16INK4A repression, YY1’s function across the INK4/ARF locus could be more complex due to genome-wide occupancy. Indeed, ChIP-seq data showed that YY1 occupancy extends across the INK4/ARF locus to include the TAD boundaries, the tumor suppressor genes, and the large superenhancer cluster (SI Appendix, Fig. S7A), underscoring that its activity could be widespread across the locus.
From the TF screen, we additionally identified the top 10 potential negative regulators repressing p16INK4A expression including CREB1, TFDP1, E2F2, TBP, ZNF217, THAP11, NKRF, C/EBPα, ZBTB14, and HMX3 (Fig. 6 C and D). C/EBPα has been recognized as an essential collaborator in Hoxa9/Meis1-mediated mouse leukemogenesis by directly repressing the Cdkn2a/b locus (53). Additionally, the p15INK4B tumor suppressor has also been identified as a direct target of the ZNF217/CoREST transcriptional complex (54). To test if the putative negative regulator ZNF217 interacted with the locus, CUT&RUN using a specific antibody against ZNF217 was performed in SEM cells stably expressing dCas9-KRAB and either NT-sgRNA or ARF-sgRNA-4. ZNF217 occupancy on both the p16INK4A promoter and its distal repressive element in ARF was observed in NT-sgRNA SEM cells (Fig. 6I). However, in ARF-sgRNA-4–targeted cells, ZNF217-binding affinity to ARF and p16INK4A, but not other loci, was significantly disrupted (Fig. 6 I and J), demonstrating that a looped conformation was a prerequisite for ZNF217 binding and suggesting that ZNF217 may play an important role in p16INK4A repression defined by the long-distance chromatin interaction model. Although we specifically addressed ZNF217’s interaction with ARF/p16INK4A, we anticipate that other negative regulators identified in the screen could behave similarly. Collectively, by presenting an unbiased, genome-scale loss-of-function TF screen, we identified plausible transcriptional regulators of p16INK4A.
Discussion
We identified a cis-element located in the 3′ region adjacent to the ARF promoter that represses p16INK4A expression via the formation of long-range chromosomal contacts. The identification of a p16INK4A distal repressive element located 3′ and adjacent to the ARF promoter is in accord with previous observations that p16Ink4a expression was significantly up-regulated in Arf-null mouse embryo fibroblasts, as well as in pre-B lymphocytes and keratinocytes lacking exon 1β (55, 56).
In mammalian cells, each chromosome is hierarchically organized into hundreds of megabase-sized TADs (57). Promoter/enhancer contacts take place within the TAD scaffold, leading to regulated gene expression (58). Intra-TAD chromatin interactions can be facilitated by a pair of CTCF-binding sites engaged in contact with each other when they are in a convergent linear orientation (44, 59). Using a chromosome conformation capture-based PCR assay, Hirosue et al. (11) proposed that CTCF was crucial for higher-order chromatin organization within the INK4/ARF locus and demonstrated that depletion of CTCF disrupted chromatin interactions and reactivated locus transcription in human induced pluripotent stem cells. In our study, by a high-resolution Capture-C assay, we showed that ARF, p16INK4A, and p15INK4B can make physical contacts with each other over long distances. By analyzing publicly available CTCF ChIP-seq data, we found that CTCF showed strong binding affinity to all 3 tumor suppressor promoters at the INK4/ARF locus. However, the loss-of-function results from auxin-inducible degradation of CTCF and the unbiased TF screening did not support the hypothesis that CTCF drives transcriptional regulation within p16INK4A in SEM cells.
Disruption of the ARF/p16INK4A interaction abrogated occupancy of ZNF217 to both ARF and p16INK4A, highlighting that ZNF217 binding is dependent on the ARF/p16INK4A looped chromatin conformation. We propose a 2-step model for chromatin looping regulation of p16INK4A. First, chromatin looping factors (e.g., YY1 or others) bind to the p16INK4A distal regulatory element adjacent to the ARF promoter and p16INK4A simultaneously to facilitate their physical juxtaposition and functional chromatin interaction. Once both genes are in physical contact, repressive effectors such as ZNF217 and/or other transcription factors are recruited to the target sequence to enable p16INK4A transcriptional repression. We further suspect that additional candidate transcription factors identified from the transcription factor CRISPR/Cas9 screen might contribute as well, thereby providing a more comprehensive understanding about how the locus is regulated in human cells.
Materials and Methods
General Methods.
All primers, plasmids, and reagents used in this study are cataloged in Dataset S1. Routine methods regarding cell culture, plasmid construction, transfection, electroporation, flow cytometry, immunoblotting, qRT-PCR, FISH, Capture-C, CUT&RUN, and statistics analysis are described in detail in SI Appendix.
Establishment of a p16INK4A-P2A-mCherry Reporter Cell Line.
SEM were electroporated by using the Nucleofector-2b device (Lonza) with the V-kit and program X-001. For p16INK4A-P2A-mCherry knock-in delivery, 2.5 μg of the donor plasmid and 2.5 μg of the CRISPR/Cas9-p16INK4A-C terminus-sgRNA all-in-one plasmid were used for 5 million SEM cells. Twenty-four hours after transfection, cells were sorted for the CRISPR/Cas9-p16INK4A-C terminus-sgRNA vector GFP fluorescent marker to enrich the transfected cell population. After sorted cells recovered in culture for up to 3 wk, a second sort was performed to select cells for successful knock-in by sorting for cells expressing the mCherry fluorescent marker. Two weeks later, a third sort was repeated, and single-cell–derived colonies were picked up and expanded for knock-in characterization.
CRISPR Library Construction and Screening.
A set of 2,029 sgRNA oligos that target H3K27ac and ATAC-seq positive peaks defined in the TAD containing INK4/ARF in IMR90, HCT116, and SEM cells as well as an additional 20 nontargeting control sgRNAs with no detectable match to the human genome were designed for array-based oligonucleotide synthesis (CustomArray). To construct a sgRNA pooled library targeting human TFs, 7 sgRNAs were designed against each of the 1,639 human TFs. For each library, the synthesized oligo pool was amplified by PCR and cloned into LentiGuide-Puro backbone (#52963) by in-fusion assembly (Clontech). The sgRNA sequences were recovered by genomic PCR and Deep Sequencing using MiSeq for single-end 150-bp read length (Illumina). The primer sequences used for cloning and sequencing are listed in Dataset S1. The sgRNA sequences related to CRISPR screening are described in Datasets S2 and S3.
Capture-C.
Capture-C assay was conducted as described by Davies et al. (41) and Hyle et al. (33). Detailed protocol was provided in SI Appendix.
Supplementary Material
Acknowledgments
This study was initiated with the encouragement of Charles J. Sherr and with salaried support of Judith Hyle and Shaela Wright provided through Sherr’s HHMI laboratory. We thank staffs in the Hartwell Sequencing Core, Cytogenetic Core, and Flow Cytometry and Cell Sorting Shared Resource facility within the Comprehensive Cancer Center of St. Jude Children’s Research Hospital and Drs. Charles J. Sherr, Gerard Zambetti, Keith A. Laycock, Mitchell J. Weiss, and Jun J. Yang for critical comments. This work was funded in part by the Comprehensive Cancer Center development fund Grant NCI-5P30CA021765-37 (to C.L.) and by the American Lebanese Syrian Associated Charities.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. M.S. is a guest editor invited by the Editorial Board.
Data deposition: All plasmids created in this study have been deposited in the Addgene database, https://www.addgene.org/ (accession nos. 136883 and 137026). Raw data collected from CUT&RUN and Capture-C have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus, https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE125842).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909720116/-/DCSupplemental.
References
- 1.Sherr C. J., The INK4a/ARF network in tumour suppression. Nat. Rev. Mol. Cell Biol. 2, 731–737 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Sherr C. J., Ink4-Arf locus in cancer and aging. Wiley Interdiscip. Rev. Dev. Biol. 1, 731–741 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sherr C. J., Beach D., Shapiro G. I., Targeting CDK4 and CDK6: From discovery to therapy. Cancer Discov. 6, 353–367 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serrano M., et al. , Role of the INK4a locus in tumor suppression and cell mortality. Cell 85, 27–37 (1996). [DOI] [PubMed] [Google Scholar]
- 5.Kamijo T., et al. , Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91, 649–659 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Kandoth C., et al. , Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molofsky A. V., et al. , Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature 443, 448–452 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janzen V., et al. , Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature 443, 421–426 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Signer R. A. J., Montecino-Rodriguez E., Witte O. N., Dorshkind K., Aging and cancer resistance in lymphoid progenitors are linked processes conferred by p16Ink4a and Arf. Genes Dev. 22, 3115–3120 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krishnamurthy J., et al. , p16INK4a induces an age-dependent decline in islet regenerative potential. Nature 443, 453–457 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Hirosue A., et al. , Quantitative assessment of higher-order chromatin structure of the INK4/ARF locus in human senescent cells. Aging Cell 11, 553–556 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Rajendran P., et al. , Nrf2 status affects tumor growth, HDAC3 gene promoter associations, and the response to sulforaphane in the colon. Clin. Epigenetics 7, 102 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao R., Choi B. Y., Lee M.-H., Bode A. M., Dong Z., Implications of genetic and epigenetic alterations of CDKN2A (p16INK4a) in cancer. EBioMedicine 8, 30–39 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker D. J., et al. , Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 479, 232–236 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Childs B. G., Durik M., Baker D. J., van Deursen J. M., Cellular senescence in aging and age-related disease: From mechanisms to therapy. Nat. Med. 21, 1424–1435 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasmant E., Sabbagh A., Vidaud M., Bièche I., ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 25, 444–448 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Jeck W. R., Siebold A. P., Sharpless N. E., Review: A meta-analysis of GWAS and age-associated diseases. Aging Cell 11, 727–731 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klann T. S., et al. , CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat. Biotechnol. 35, 561–568 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Callow M. G., et al. , CRISPR whole-genome screening identifies new necroptosis regulators and RIPK1 alternative splicing. Cell Death Dis. 9, 261 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sidik S. M., Huet D., Lourido S., CRISPR-Cas9-based genome-wide screening of Toxoplasma gondii. Nat. Protoc. 13, 307–323 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du J., et al. , A CRISPR/Cas9-based screening for non-homologous end joining inhibitors reveals ouabain and penfluridol as radiosensitizers. Mol. Cancer Ther. 17, 419–431 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Grevet J. D., et al. , Domain-focused CRISPR screen identifies HRI as a fetal hemoglobin regulator in human erythroid cells. Science 361, 285–290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonatopoulos-Pournatzis T., et al. , Genome-wide CRISPR-Cas9 interrogation of splicing networks reveals a mechanism for recognition of autism-misregulated neuronal microexons. Mol. Cell 72, 510–524.e12 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Irelan J. T., et al. , A functional screen for regulators of CKDN2A reveals MEOX2 as a transcriptional activator of INK4a. PLoS One 4, e5067 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamakoshi K., et al. , Real-time in vivo imaging of p16Ink4a reveals cross talk with p53. J. Cell Biol. 186, 393–407 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Demaria M., et al. , An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev. Cell 31, 722–733 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J.-Y., et al. , Cells exhibiting strong p16 INK4a promoter activation in vivo display features of senescence. Proc. Natl. Acad. Sci. U.S.A. 116, 2603–2611 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burd C. E., et al. , Monitoring tumorigenesis and senescence in vivo with a p16(INK4a)-luciferase model. Cell 152, 340–351 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sorrentino J. A., et al. , p16INK4a reporter mice reveal age-promoting effects of environmental toxicants. J. Clin. Invest. 124, 169–173 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim J. H., et al. , High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One 6, e18556 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J. P., et al. , Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 18, 35 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yao X., et al. , Homology-mediated end joining-based targeted integration using CRISPR/Cas9. Cell Res. 27, 801–814 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hyle J., et al. , Acute depletion of CTCF directly affects MYC regulation through loss of enhancer-promoter looping. Nucleic Acids Res. 47, 6699–6713 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilbert L. A., et al. , CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Demaria M., et al. , Cellular senescence promotes adverse effects of chemotherapy and cancer relapse. Cancer Discov. 7, 165–176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Creyghton M. P., et al. , Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 107, 21931–21936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robertson K. D., Jones P. A., The human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and down-regulated by wild-type p53. Mol. Cell. Biol. 18, 6457–6473 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hughes J. R., et al. , Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat. Genet. 46, 205–212 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Davies J. O. J., et al. , Multiplexed analysis of chromosome conformation at vastly improved sensitivity. Nat. Methods 13, 74–80 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skene P. J., Henikoff S., An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Wit E., et al. , CTCF binding polarity determines chromatin looping. Mol. Cell 60, 676–684 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Rao S. S., et al. , A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishimura K., Kanemaki M. T., Rapid depletion of budding yeast proteins via the fusion of an auxin-inducible degron (AID). Curr. Protoc. Cell Biol. 64, 20.9.1–20.9.16 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Natsume T., Kiyomitsu T., Saga Y., Kanemaki M. T., Rapid protein depletion in human cells by auxin-inducible degron tagging with short homology donors. Cell Rep. 15, 210–218 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Nora E. P., et al. , Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169, 930–944.e22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang W., Li D., Sui G., YY1 is an inducer of cancer metastasis. Crit. Rev. Oncog. 22, 1–11 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Chen F., Sun H., Zhao Y., Wang H., YY1 in cell differentiation and tissue development. Crit. Rev. Oncog. 22, 131–141 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Makhlouf M., et al. , A prominent and conserved role for YY1 in Xist transcriptional activation. Nat. Commun. 5, 4878 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beagan J. A., et al. , YY1 and CTCF orchestrate a 3D chromatin looping switch during early neural lineage commitment. Genome Res. 27, 1139–1152 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weintraub A. S., et al. , YY1 is a structural regulator of enhancer-promoter loops. Cell 171, 1573–1588.e28 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Collins C., et al. , C/EBPα is an essential collaborator in Hoxa9/Meis1-mediated leukemogenesis. Proc. Natl. Acad. Sci. U.S.A. 111, 9899–9904 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thillainadesan G., et al. , TGF-β-dependent active demethylation and expression of the p15ink4b tumor suppressor are impaired by the ZNF217/CoREST complex. Mol. Cell 46, 636–649 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Lin A. W., Lowe S. W., Oncogenic ras activates the ARF-p53 pathway to suppress epithelial cell transformation. Proc. Natl. Acad. Sci. U.S.A. 98, 5025–5030 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Randle D. H., Zindy F., Sherr C. J., Roussel M. F., Differential effects of p19(Arf) and p16(Ink4a) loss on senescence of murine bone marrow-derived preB cells and macrophages. Proc. Natl. Acad. Sci. U.S.A. 98, 9654–9659 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dixon J. R., et al. , Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pombo A., Dillon N., Three-dimensional genome architecture: Players and mechanisms. Nat. Rev. Mol. Cell Biol. 16, 245–257 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Vietri Rudan M., et al. , Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 10, 1297–1309 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.