Significance
In mammals, intrathymic T cell development strictly depends on the activity of the IL-7 cytokine signaling pathway, leading to catastrophic immunodeficiency when even 1 of the components in the pathway fails. Here we report the unexpected observation that the development of T cells in the thymus of the teleost Danio rerio (zebrafish) is instead supported by the collective actions of several cytokine signaling pathways, with IL-7 signaling playing only a minor role. This finding suggests that during the course of evolution, a degenerate network was converted into a state of precarious brittleness, possibly because some of the evolutionarily ancient constitutive cytokine pathways had to be repurposed to meet increased demands for sophisticated regulation during the mammalian immune response.
Keywords: thymus, cytokine signaling, evolution, zebrafish, network structure
Abstract
In mammals, T cell development critically depends on the IL-7 cytokine signaling pathway. Here we describe the identification of the zebrafish ortholog of mammalian IL-7 based on chromosomal localization, deduced protein sequence, and expression patterns. To examine the biological role of il7 in teleosts, we generated an il7 allele lacking most of its coding exons using CRISPR/Cas9-based mutagenesis. il7-deficient animals are viable and exhibit no obvious signs of immune disorder. With respect to intrathymic T cell development, il7 deficiency is associated with only a mild reduction of thymocyte numbers, contrasting with a more pronounced impairment of T cell development in il7r-deficient fish. Genetic interaction studies between il7 and il7r mutants, and il7 and crlf2(tslpr) mutants suggest the contribution of additional, as-yet unidentified cytokines to intrathymic T cell development. Such activities were also ascertained for other cytokines, such as il2 and il15, collectively indicating that in contrast to the situation in mammals, T cell development in the thymus of teleosts is driven by a degenerate multicomponent network of γc cytokines; this explains why deficiencies of single components have little detrimental effect. In contrast, the dependence on a single cytokine in the mammalian thymus has catastrophic consequences in cases of congenital deficiencies in genes affecting the IL-7 signaling pathway. We speculate that the transition from a degenerate to a nonredundant cytokine network supporting intrathymic T cell development emerged as a consequence of repurposing evolutionarily ancient constitutive cytokine pathways for regulatory functions in the mammalian peripheral immune system.
In mice, intrathymic T cell development is critically dependent on the presence of the IL-7 cytokine (1); in Il7-deficient mice, thymic cellularity is reduced by more than 20-fold (2). IL-7 belongs to the so-called γc cytokines, and its receptor is a heterodimer composed of the IL-7 receptor α chain and the common γ chain (γc), also known as IL-2RG (3). In mice mutant for the IL-7 receptor α chain, total thymic cellularity varied between 0.01% and 10% of wild-type levels (4); likewise, in mice mutant for γc, thymic cellularity is reduced by more than 30-fold (5). These findings identify IL-7 as the key cytokine stimulating the proliferation of thymocytes and suggest that it signals through the heterodimeric IL-7R/IL-2RG receptor (3). Likewise, in humans, lack of IL-7 is associated with severe T cell lymphopenia (6), and the same phenotype is observed in IL-7R (7) and IL-2RG (8) deficiencies. Collectively, these observations indicate that IL-7 is part of a nonredundant cytokine signaling network required for the development and maintenance of T cells in mammals. Of note, in contrast to the situation with T cell development, the requirement for IL-7 signaling in B cell development varies and is species-dependent; for instance, whereas B cells fail to develop properly in mice lacking IL-7, B cell development is not affected in humans with IL-7 deficiency (2, 7). These observations suggest a certain degree of functional flexibility in lymphoid-specific cytokine networks.
The results of large-scale forward genetic screens in zebrafish (9, 10) suggest that IL-7 signaling is also important for T cell development in lower vertebrates. In this screen, we identified lines deficient for il7r, jak1, and jak3 genes, all exhibiting reductions in thymocyte numbers, resulting in peripheral lymphopenia; moreover, genetic interaction analyses suggested that il7r, jak1, and jak3 act in the same pathway (10). Others have shown that a deficiency of 1 of 2 zebrafish il2rg genes causes a severe block in T cell development (11). Collectively, these results are compatible with the notion of a strong evolutionary conservation of the IL-7R–IL2-RG–JAK3 signaling pathway in vertebrate T cell development.
When viewed from the perspective of network design, the IL-7–(IL-7R–IL-2RG)–JAK3 signaling axis underlying mammalian T cell development appears to be remarkably brittle, despite the fact that T cells are of central importance for the immune defense of vertebrates. The apparent nonredundancy of the IL-7–(IL-7R–IL-2RG)–JAK3 signaling axis in T cells is mirrored in the nonredundant function of the FOXN1 transcription factor, which is essential for differentiation of the mammalian thymic microenvironment (12, 13), including provision of the IL-7 cytokine (14). Lack of Foxn1 also causes catastrophic immunodeficiency resulting from agenesis of the stromal microenvironment essential for mammalian T cell development.
While these findings suggest that the genetic network of T cell development is essentially nonredundant in mammals, there are indications that this might not be true for T cell development in teleosts. For instance, in contrast to the situation in mammals, the foxn1 transcription factor gene of teleosts is not essential for thymopoiesis; in fact, coexpression in thymic epithelial cells of its paralog, foxn4, directs a substantial amount of lymphoid development in foxn1-deficient fish (15). In addition, il7-deficient fish were conspicuously absent in the collection of mutant lines of our large-scale forward genetic screen (9). Although there are trivial reasons for this outcome, we entertained the possibility that il7 deficiency might have only a mild or no phenotype and thus would not have been detected under the stringent phenotypic selection criteria of the genetic screen. Therefore, we set out to determine the phenotype of il7-deficient zebrafish to examine the possibility that the cytokine signaling network in the thymus of teleosts might exhibit features of robustness not apparent from previous studies.
Here we report that, in contrast to mammals, T cell development in zebrafish is regulated by the collective activity of several γc cytokines, and discuss the adaptive value of this degenerate network design.
Results
Identification of the Zebrafish il7 Gene.
We identified ENSDARG00000092045 on chromosome 24 as a likely candidate for the zebrafish il7 gene, initially based on the evolutionarily conserved synteny between IL7 and ZC2HC1A genes that appears to hold for all jawed vertebrates, from sharks (16) to humans (http://useast.ensembl.org/index.html) (SI Appendix, Fig. S1A). Indeed, conceptual translation of the associated transcripts (SI Appendix, Fig. S1 B and C) indicates a close similarity to previously identified IL-7 homologs in lower vertebrates and to mammalian IL-7 (SI Appendix, Fig. S1 D and E). No other IL-7–related sequence was found in publicly available sequence collections and our own datasets, suggesting that only a single il7-like gene is present in zebrafish.
We next examined the expression of the putative zebrafish il7 gene in the thymus and the kidney, the 2 key lymphopoietic sites in adult zebrafish. As expected, strong expression was found in the thymus (Fig. 1A) and in scattered regions of the kidney (Fig. 1B). Collectively, these results strongly suggest that ENSDARG00000092045 represents the il7 homolog of zebrafish.
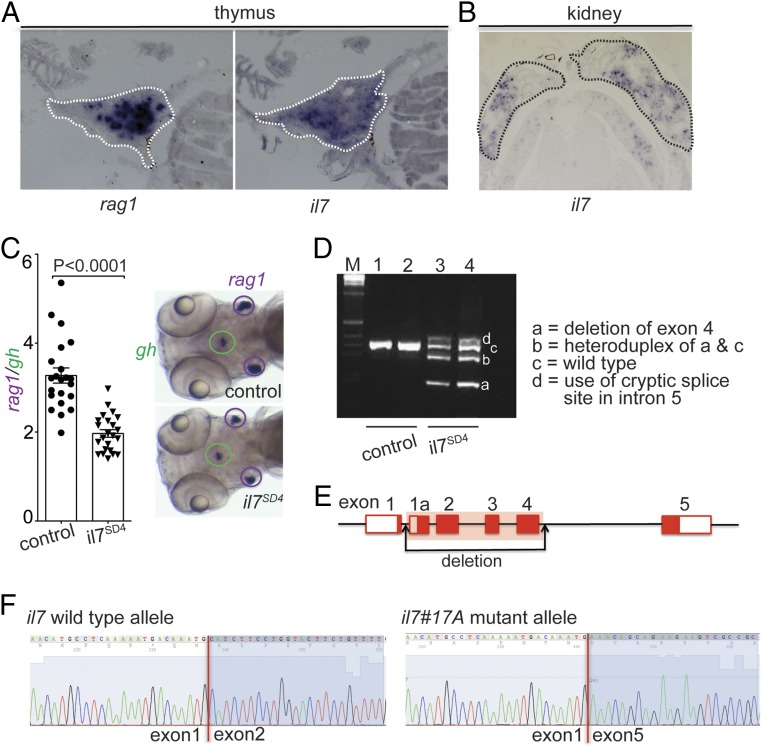
Fig. 1.
Characterization of the zebrafish il7 gene (ENSEMBL ID: ENSDARG00000092045). (A) RNA in situ hybridization on tissue sections of adult zebrafish thymus with probes specific for rag1 and il7. (B) RNA in situ hybridization on tissue sections of adult zebrafish kidney with a probe specific for il7. (C) Reduced thymopoietic activity in il7 morphants as determined by the thymopoietic index (Left), derived from quantitative analysis of whole-mount RNA in situ hybridization experiments (Right). Each data point represents 1 fish; data are mean ± SEM. (D) RT-PCR analysis of il7 cDNA structures resulting from interference with splicing of the pre-mRNA transcripts by an antisense oligonucleotide targeting the splice donor site of exon 4 (SD4). The deletion in variant a does not cause a frame shift, since all exons of the il7 gene are in phase 0; the use of cryptic splice site in the intron between exons 4 and 5 causes a frame shift mutation. (E) Schematic of the il7 gene structure, indicating the deletion introduced by CRISPR/Cas9 mutagenesis. (F) Sequence analysis of cDNAs demonstrating the loss of sequences derived from exons 2 to 4 in transcripts emanating from the il7#17A mutant allele.
Phenotype of il7-Deficient Zebrafish.
In an initial effort to explore the functional role of il7 during T cell development, we examined the phenotype of il7 morphants at 5 d postfertilization (dpf). The RNA in situ signal for rag1-expressing cells in the thymus was normalized to that of growth hormone gene (gh)-expressing cells in the hypophysis, and the extent of thympopoietic activity was expressed as the rag1/gh ratio. The results, shown in Fig. 1C, indicate a surprisingly mild reduction of rag1-expressing cells of ∼30%. However, despite the presence of an antisense morpholino targeting the splice donor site of exon 4, a small fraction of pre-mRNAs was properly spliced (Fig. 1D); thus, we considered the possibility that the phenotype of il7 morphants might be the result of a hypomorphic rather than an amorphic constellation. To circumvent this problem, we generated il7-deficient zebrafish using the CRISPR/Cas9 method of mutagenesis. The il7#17A allele exhibits a large deletion in the ENSDARG00000092045 locus, removing exons 2 to 4 (Fig. 1E and SI Appendix, Figs. S1 B and C and S2A), as demonstrated by cDNA cloning (Fig. 1F) and RNA sequencing (SI Appendix, Fig. S2 B and C).
The il7 mutants were viable and appeared grossly normal (Fig. 2A); this phenotype is in line with the only minor changes seen in the transcriptomes of mutant fish when analyzed at 5 dpf (for whole larvae) and 3 mo (for whole kidney marrow) (Fig. 2D and SI Appendix, Table S1). At 63 h postfertilization (hpf)—that is, approximately 10 h after colonization of the thymic rudiment has begun (17)—the lack of il7 was associated with small reductions of the numbers of ikaros-expressing T cell precursors in the thymic rudiment (Fig. 2B). This phenotype persisted, such that at 5 dpf, the number of rag1-expressing cells in the thymus was still reduced in the mutants (Fig. 2C). This effect is very similar in magnitude to the phenotype of il7 morphants (Fig. 1C).
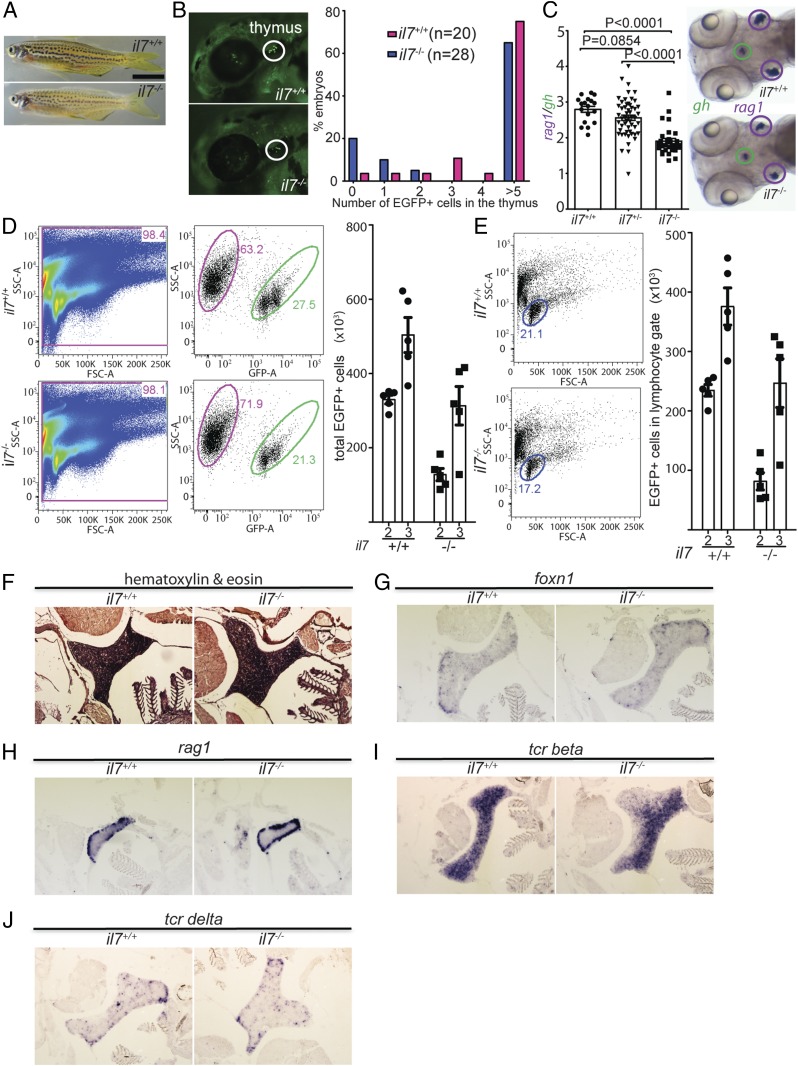
Fig. 2.
Characterization of il7 mutant zebrafish. (A) Normal gross appearance of adult wild-type (il7+/+) and mutant (il7−/−) fish. (B) Reduced number of ikaros-expressing lymphocytes in the thymus of il7-mutant fish also transgenic for an ikaros:eGFP reporter allele. (Left) Representative animals at 72 hpf. (Right) Quantitative assessment of cell numbers in the thymic rudiment. (C) Reduced thymopoietic activity in il7 mutants as determined by the thymopoietic index (Left) derived from quantitative analysis of whole- mount RNA in situ hybridization experiments (Right). Each data point represents 1 fish; data are mean ± SEM. (D, Left) Flow cytometry analysis of GFP-positive cells present in the thymus of wild-type and il7 mutant fish at age 2 mo. (D, Right) Enumeration of the total number of GFP-positive cells in the thymus at age 2 and 3 mo. Each data point represents 1 fish; data are mean ± SEM. (E) Enumeration of the total number of GFP-positive cells with light-scatter characteristics of lymphocytes (encircled; age 2 mo) (Left) in the thymus at age 2 and 3 mo (Right). Each data point represents 1 fish; data are mean ± SEM. A comparison of the data in D and E indicates that the majority of GFP-positive cells in the thymus are lymphocytes. (F) Normal histology of il7 mutant thymus in adult fish. (G–J) RNA in situ hybridization on tissue sections of adult wild-type and il7 mutant zebrafish thymus with probes specific for foxn1 (G), expressed by thymic epithelial cells; rag1 (H), indicative of immature thymocytes; tcr beta (I), indicative of αβ T cells; and tcr delta (J), indicative of γδ T cells.
When examined at successively later stages of development, hematopoietic lineages in mutant fish appeared to be essentially normal, with the exception of slightly reduced numbers of ikaros-expressing lymphoid precursors in the thymus (Fig. 2 D and E). However, in contrast to the situation in mice, where the γδ T cell lineage is absolutely dependent on IL-7 (18), both αβ and γδ T cells appeared to be present in the mutant thymus, as determined by RNA in situ hybridization on tissue sections of adult fish (Fig. 2 F–J). Notably, B cell development in the kidney marrow also appeared to be intact, as demonstrated by the normal numbers of ikaros-expressing cells (Fig. 3 A and B); likewise, no differences were found between wild-type fish and il7 mutants in the numbers of cells expressing rag1 and igm (as markers of the B cell lineage) and tcrb and tcrd (as markers of the 2 principal lineages of T cells) in the kidney, as determined by RNA in situ hybridization of tissue sections (Fig. 3 C–F). Finally, the conclusion of unaltered lymphoid lineage specification in the absence of il7 is supported by the presence of expressed complete assemblies of tcrd, tcrb, igz, and igm antigen receptor genes (Fig. 3G).
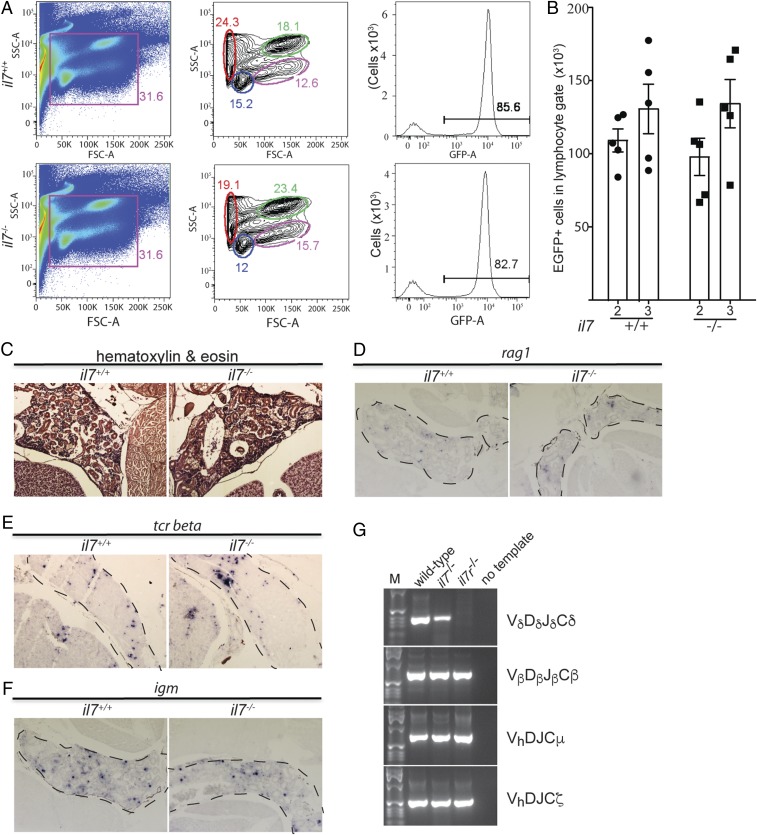
Fig. 3.
Characterization of hematopoiesis in 3-mo-old zebrafish kidneys. (A) Flow cytometry analysis of cell populations in whole kidney marrow of adult wild-type and il7-mutant fish (Left and Middle). The different cell populations as identified by the characteristic light-scatter characteristics are indicated (red, erythrocytes; blue, lymphocytes; green, myeloid cells; pink, precursor cells) (34). The majority of cells in the lymphocyte gate (blue) are GFP-positive (Right). (B) Enumeration of GFP-positive cells in the lymphocyte gate of wild-type and il7-mutant fish at age 2 and 3 mo. Each data point represents 1 fish; data are mean ± SEM. (C) Normal histology of kidney marrow in il7 mutant adult fish. (D–F) RNA in situ hybridization on tissue sections of adult wild-type and il7 mutant kidney marrow (outlined with dashed lines) with probes specific for rag1 (D), indicative of B cell precursors; tcr beta (E), indicative of αβ T cells; and igm (F), indicative of Β cells. (G) RT-PCR assay for expressed completely assembled antigen receptor genes. Vd-Dd-Jd-Cd assemblies associated with the γδ T cell lineage are present in the whole kidney marrow of il7 mutants but not in that of il7r mutants; assemblies of other lineage-specific antigen receptors are present in both mutant fish lines (Vb-Db-Jb-Cb for αβ T cells, Vh-D-J-Cm for Igμ-expressing B cells, and Vh-D-J-Cz for Igζ-expressing B cells). Representative panels are shown for wild-type fish (n = 5), il7 mutants (n = 4), and il7r mutants (n = 5).
Genetic Network Analysis of il7.
To further substantiate the functional assignment of the il7 gene in the context of cytokine signaling, a genetic interaction analysis was carried out. Genetic interaction is defined as an unexpected phenotype created by combining the effects of 2 genetic variants, where an experimentally observed double-mutant phenotype is less severe (positive or alleviating interaction) or more severe (negative or synthetic interaction) than an expected value predicted from both single mutant phenotypes (19). A positive interaction often indicates that 2 genes operate in the same pathway, as we have shown previously for il7r and jak3 using this method (10).
Again, the thymopoietic activities in il7r;il7 and jak3;il7 double mutants were much greater than expected from the independent activities of the 2 mutations on the outcome of T cell development (Fig. 4 A–D). The observed positive genetic interactions thus indicate that il7, il7r, and jak3 genes jointly act in the same pathway, strongly supporting the notion that the ENSDARG00000092045 gene encodes the zebrafish il7 homolog. However, the genetic interaction analysis described above provided no explanation for the discrepant phenotypes of il7 mutants on the one hand and il7r mutants on the other hand. A possible explanation for the more severe phenotype of the il7r mutant could be that the il7r α chain—in zebrafish as in the mouse—is also part of a second receptor, in complex with a CRLF2-like receptor chain, known as the TSLP receptor in mammals (20).
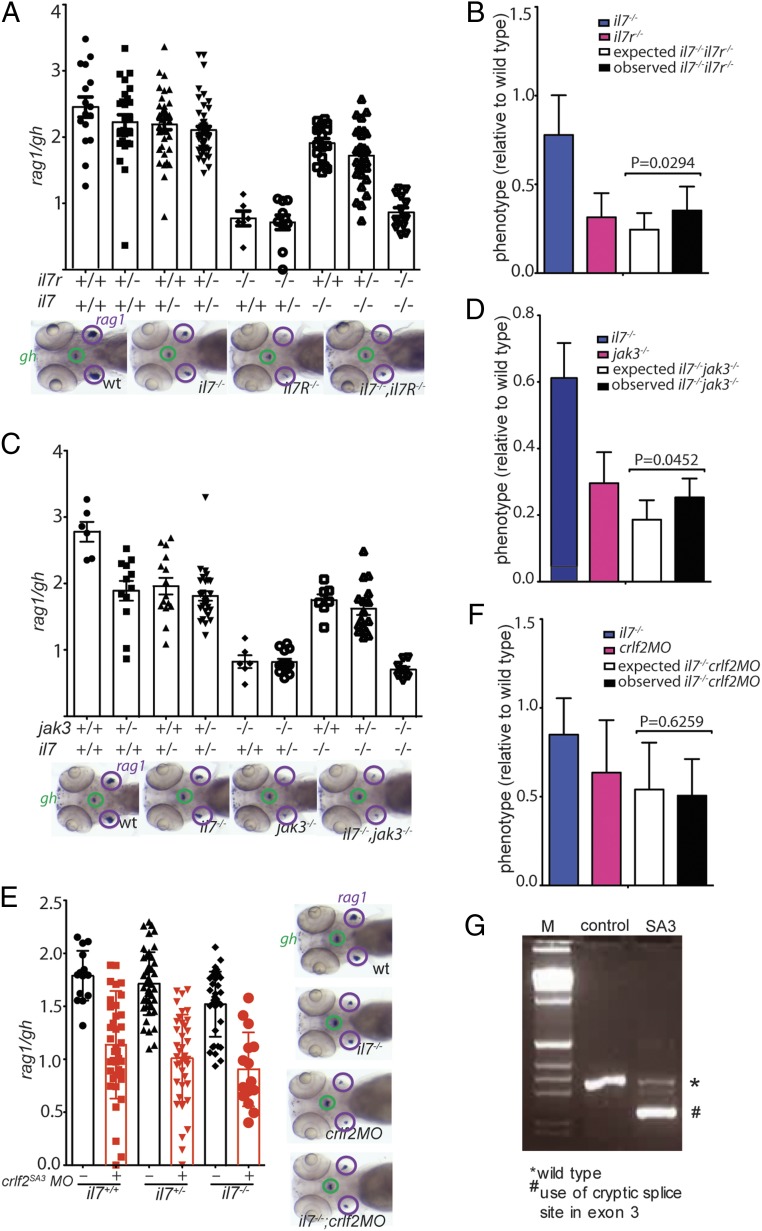
Fig. 4.
Epistasis analysis of the il7 mutation. (A) Thymopoietic capacities measured in fish of the 9 genotypes arising from an intercross of il7+/−;il7r+/− double-heterozygous parents. Each data point represents 1 fish; data are mean ± SEM. Representative whole-mount RNA in situ hybridization results for the key genotypes are shown at the bottom. (B) Comparison of phenotypes of the 2 single mutants (il7−/− and il7r−/−) and the il7−/−;il7r−/− double mutant; the latter phenotype is compared with the expected phenotype (multiplicative model). The thymopoietic capacity observed in the double mutant is significantly higher than that expected from the combination of the single mutants, calculated under the assumption of no genetic interaction, indicative of alleviating genetic interaction. (C) Thymopoietic capacities measured in fish of the 9 genotypes arising from an intercross of il7+/−;/jak3+/− double-heterozygous parents. Each data point represents 1 fish; data are mean ± SEM. Representative whole-mount RNA in situ hybridization results for the key genotypes are shown at the bottom. (D) Comparison of phenotypes of the 2 single mutants (il7−/− and jak3−/−) and the il7−/−;jak3−/− double mutant with the expected phenotype. The observed thymopoietic capacity observed in the double mutant is significantly higher than that expected from the combination of the single mutants, indicative of alleviating genetic interaction. (E) Thymopoietic capacities measured in crlf2 morphants of 3 different il7 genotypes. Representative whole-mount RNA in situ hybridization results are shown at the right. (F) The degree of reduction of thymopoietic activity in the presence of the crlf2 antisense oligonucleotide is independent of il7 genotype, indicating that the 2 effects are genetically independent (no genetic interaction). (G) RT-PCR analysis of crlf2 cDNA structures resulting from interference with splicing of the pre-mRNA transcripts by an antisense oligonucleotide targeting the splice donor site of exon 3 (SD3).
Since a functional equivalent of mammalian TSLP has not yet been found in zebrafish, we examined the effect of a deficiency of the zebrafish crlf2-like gene (Fig. 4E) on T cell development in the thymus. As shown in Fig. 4 F and G, crlf2-deficiency strongly impairs T cell development in zebrafish larvae, although the magnitude of reduction in T cell development is less than that observed for il7r mutants (10); in contrast, TSLP-signaling contributes only minimally to intrathymic T cell development in mice (21). Since il7 and crlf2 genes do not interact (Fig. 4 F and G), they appear to function independently to regulate T cell development in the zebrafish thymus. Taken together, these observations suggest the presence of an as-yet unidentified cytokine (possibly a functional equivalent of TSLP in zebrafish) that signals through the il7r/crlf2 complex and substantially contributes to intrathymic T cell development. Conceptually, this result suggests an unexpected level of redundancy of cytokine function in zebrafish thymopoiesis instead of the dominant role of IL-7 in mammalian T cell development.
Interestingly, tcrd rearrangements are absent in il7r mutants but not in il7 mutants (Fig. 3D), suggesting that il7 and the putative crlf2 ligand both contribute to the formation of the γδ T cell lineage, and that their simultaneous absence (or the lack of Il7r serving as their common receptor component) is needed to abolish the development of γδ T cells. This contrasts with the situation in mice, where the lack of only IL-7 already results in failure of γδ T cell development (18).
Role of Other γc Cytokines in T Cell Development.
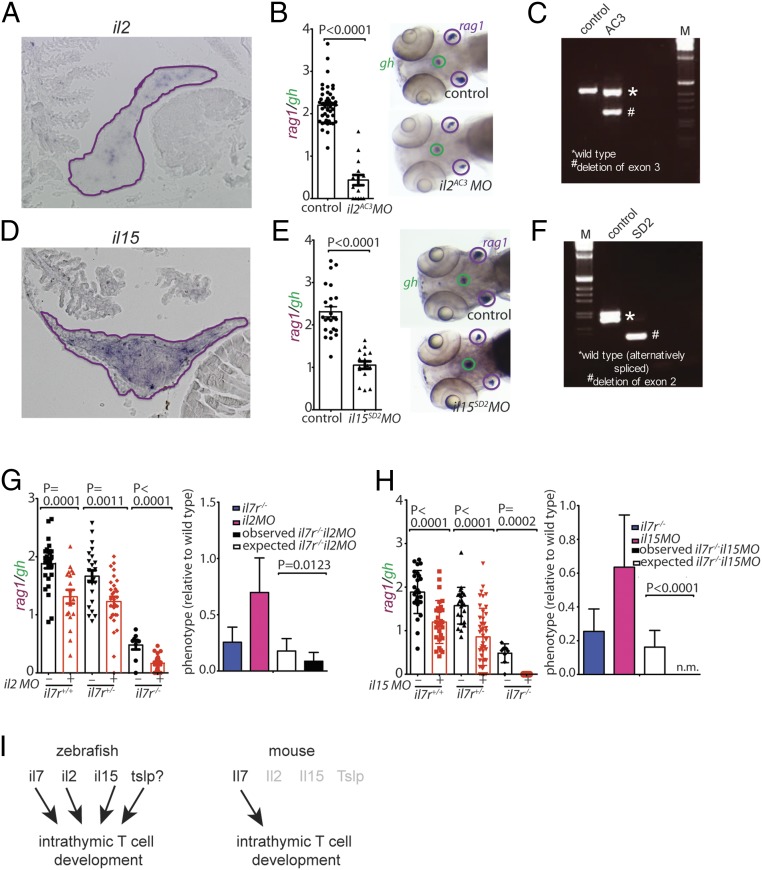
To further explore the landscape of cytokine function in T cell development, we examined whether cytokines other than il7 (and the putative tslp equivalent) have roles in zebrafish T cell development. To this end, we first examined the expression patterns of il2, il15, il15l, and il21 genes in the thymus by RNA in situ hybridization. Whereas expression of il15l could not be detected in the thymus, and only very few il21-positive cells were found, numerous cells expressed il2 and il15 (Fig. 5 A and D). As expected from their expression patterns in the thymus, il15l and il21 morphants exhibited essentially normal T cell development (SI Appendix, Fig. S3 A and B); in contrast, both il2 (Fig. 5 B and C) and il15 (Fig. 5 E and F) morphants showed a strong reduction of the number of rag1-expressing thymocytes, as did il15r morphants (SI Appendix, Fig. S3C). Genetic interaction analyses were carried out between il7r (covering both il7 and the presumptive tslp cytokine receptor complexes), and the 2 thymopoietic cytokine genes il2 and il15. Synthetic interactions were observed for both the il2 and il7r (Fig. 5G) and il15 and il7r (Fig. 5H) combinations. These findings suggest that il7r and the 2 cytokine genes encoding il2 and il15 function in parallel pathways to control T cell development and thus il2 and il15 appear to signal through receptor complexes not involving the il7r α chain.
Fig. 5.
Characterization of cytokine gene morphants. (A) RNA in situ hybridization on tissue sections of adult zebrafish thymus with probes specific for il2; the positive cells appear to cluster in certain regions of the thymus. (B) Reduced thymopoietic activity in il2 morphants as determined by the thymopoietic index (Left), derived from quantitative analysis of whole-mount RNA in situ hybridization experiments (Right). Each data point represents 1 fish; data are mean ± SEM. (C) RT-PCR analysis of il2 cDNA structures resulting from interference with splicing of the pre-mRNA transcripts by an antisense oligonucleotide targeting the splice acceptor site of exon 3 (AC3). (D) RNA in situ hybridization on tissue sections of adult zebrafish thymus with probes specific for il15. (E) Reduced thymopoietic activity in il15 morphants as determined by the thymopoietic index (Left), derived from quantitative analysis of whole-mount RNA in situ hybridization experiments (Right). Each data point represents 1 fish; data are mean ± SEM. (F) RT-PCR analysis of il15 cDNA structures resulting from interference with splicing of the pre-mRNA transcripts by an antisense oligonucleotide targeting the splice donor site of exon 2 (AC3). (G, Left) Thymopoietic capacities measured in il2 morphants of 3 different il7r genotypes. Each data point represents 1 fish; data are mean ± SEM. (G, Right) The degree of thymopoietic activity in the presence of the il2 antisense oligonucleotide is significantly lower than expected from neutrality, indicating the presence of synthetic genetic interaction between il2 and il7r. (H, Left) Thymopoietic capacities measured in il15 morphants of 3 different il7r genotypes. Each data point represents 1 fish; data are mean ± SEM. (H, Right) The degree of thymopoietic activity in the presence of the il15 antisense oligonucleotide is significantly lower than that expected from neutrality (n.m. denotes the absence of detectable rag1 signal in the double-mutant fish), indicating the presence of a synthetic genetic interaction between il15 and il7r. (I) Schematic representation of the degenerate cytokine network regulating intrathymic T cell development in zebrafish (Left) and the corresponding nonredundant network structure in mouse (Right).
Discussion
The moderate effect of il7 deficiency on T cell development in zebrafish described here has led to the subsequent identification of the degenerate cytokine network underlying the development of T cells in the zebrafish thymus. Our studies show that in addition to il7, intrathymic T cell development in zebrafish requires il2, il15, and possibly other cytokine genes, such as that encoding a cytokine functionally equivalent to mammalian TSLP (Fig. 5I). This degeneracy stands in stark contrast to the brittle network configuration of its mammalian counterpart; here intrathymic T cell development depends almost entirely on the activity of IL-7. Of note, the phenotype of Il2rg/Crlf2 double-deficient mice suggested a minor but recognizable contribution of Tslp to mammalian intrathymic T cell development (21), which we interpret as reflecting an evolutionary trace of the ancient degenerate network configuration.
From a systems perspective, our observations suggest that the degenerate zebrafish network has a higher degree of evolvability than that of mammals (22). Mammalian intrathymic T cell development relies on a nonredundant system of IL-7 signaling (with SCF also making only a minor contribution; ref. 1), since IL-7 deficiency drastically reduces the magnitude of T cell development in the thymus (2, 6). Thus, network functionality in mammals has arrived at an essentially all-or-none configuration. In such a situation, evolutionary adaptations are difficult to achieve, since in the absence of physiologically relevant backup activities, any changes in the ligand–receptor interactions carry the risk of generating catastrophic failure. Therefore, with respect to T cell development in the thymus, the system is essentially locked into the present state. In contrast, the degree of evolvability is much higher in a partially redundant (or degenerate) multicomponent system, in which many elements jointly contribute to the overall outcome of the network. While some components can maintain the original functions, other elements may be modified in response to new physiological circumstances. Such degeneracy appears to be present in the cytokine network regulating intrathymic T cell development in zebrafish, rendering the genetic network underlying the thymopoietic capacity of teleosts much more flexible.
Our present studies raise several questions about the functions of cytokines in the zebrafish immune system. We predict that zebrafish have at least 1 additional cytokine that signals through the il7r/crlf2 receptor pair and speculate that this cytokine is a functional equivalent of the mammalian TSLP cytokine. However, so far we have been unable to identify related sequences in genomic and cDNA sequence collections; possible reasons are a high degree of sequence divergence (which is not uncommon for the rapidly evolving cytokines in general; see SI Appendix, Fig. S1 D and E) and/or low levels of expression. Compared with the situation in the mouse, it appears that the putative zebrafish tslp equivalent has a larger spectrum of activities, since Tslp deficiency in the mouse has little effect on the magnitude of intrathymic T cell development (21) but does affect regulatory functions in the peripheral immune system (20). Likewise, Il2 deficiency in the mouse does not appreciably affect T cell development (23), but instead impairs peripheral immune homeostasis (24). Moreover, although Il15 is important for CD8 T lineage specification in the thymus (25) and memory formation in the periphery (26), the effect of IL-15 and/or IL-15R deficiency on the total number of thymocytes is minimal (27, 28), in contrast to the situation in zebrafish described here. Once the putative zebrafish tslp equivalent is identified, its roles in T and B cell development and the peripheral immune system can be examined and compared with the phenotypes of fish deficient for il2 and il15 genes.
Our results suggest that a degenerate cytokine network of il7, il2, il15, and possibly of the tslp equivalent that collectively supports T cell development in teleosts has evolved into an essentially nonredundant configuration in mammals, in which IL-7 plays a dominant role (Fig. 5I). Whereas other cytokines can contribute to the regulation of T cell development, their impact varies among species; this becomes apparent when comparing the phenotypes of IL-7–deficient (6) and IL-7R–deficient (7) human patients and those of equivalent deficiencies in mice (2, 4).
At present, we can only speculate why the mammalian cytokine network regulating T cell development in the thymus has evolved to rely almost entirely on IL-7 as the lymphopoietic cytokine. It is possible that the higher demands on immune regulation in mammals required the repurposing of cytokines such as IL-2, IL-15, and IL-21 and their corresponding receptors for peripheral functions to achieve a more fine-tuned regulation of the immune response and/or maintenance of tolerance (29). If so, this would suggest that the basic building blocks of immune regulatory cytokines are used in different ways in lower vertebrates and in mammals. In this context, it will be interesting to address the structure of cytokine networks in the immune system of the sister group of jawed vertebrates, lampreys and hagfishes. These jawless vertebrates split off from the lineage giving rise to teleosts and mammals approximately 500 million years ago and exhibit an adaptive immune system based on alternative types of antigen receptors (30). Since the coding capacity of cytokines in this group of vertebrates has not yet been explored, it is difficult to predict whether T cell development in the thymus (31) operates under nonredundant (mammals) and degenerate (teleost) versions of the cytokine network.
Materials and Methods
Animals.
The zebrafish (Danio rerio) wild-type strain TLEK (Tüpfel long fin/Ekkwill) is maintained in the animal facility of the Max Planck Institute of Immunobiology and Epigenetics in Freiburg, Germany. The il7r and jak3 mutant lines have been described previously (10), as has the ikaros:eGFP transgenic reporter (17). All animal experiments were performed in accordance with relevant guidelines and regulations and approved by the Review Committee of the Max Planck Institute of Immunobiology and Epigenetics and the Regierungspräsidium Freiburg, Germany (license Az 35–9185.81/G-14/41).
Morphants.
Morphants were generated by injection of antisense morpholino oligonucleotides (Gene Tools) to block the translation of both maternal and zygotic mRNAs (“ATG morpholinos”) or block splicing of zygotic pre-mRNAs (“splice morpholinos”). Stock solutions were diluted as required; the final concentration in the injection buffer (0.05% vol/vol phenol red and 1× Danieau buffer; http://cshprotocols.cshlp.org/content/2011/7/pdb.rec12467.full) is shown in SI Appendix, Table S2. Approximately 1 to 2 nL of solution was injected into fertilized eggs as described previously (32). The morphants (SI Appendix, Table S2) were analyzed at 5 dpf by RNA in situ hybridization using a combination of rag1- and gh-specific probes, with the results expressed as a thymopoietic index, a dimensionless figure (see below), and by RT-PCR to determine the structure of aberrant splice products, followed by sequence determination of the resulting amplicons (SI Appendix, Table S2).
Thymopoietic Index.
Thymic rag1 gene expression is a marker of ongoing assembly of T cell receptor genes. Thus, the intensity of the RNA in situ signal correlates with the number of differentiating T cells, which we consider to be a measure of T cell development. To provide an internal control (technical, with respect to the hybridization process as such, and biological, with respect to the tissue specificity of the observed genetic effects), we used a probe specific for the growth hormone (gh) gene, which marks a subset of cells in the hypophysis. Determination of rag1/gh ratios was carried out as follows. After RNA in situ hybridization with rag1 and gh probes, ventral images of 4 to 5 dpf zebrafish larvae were obtained with a Leica MZFLIII microscope and a Leica DFC300FX digital camera, essentially generating a 2D projection of the 3D structure. The areas of rag1 and gh signals were measured using ImageJ, and the ratio of average of the rag1-positive area vs. gh-positive area was calculated as a measure of thymopoietic activity. After photographic documentation of the RNA in situ hybridization signal, larvae were processed for genomic DNA extraction for subsequent genotyping.
RNA Extraction and cDNA Synthesis.
Total RNA was extracted using TRI Reagent (Sigma-Aldrich) following the manufacturer’s instructions. After treatment with DNaseI (Promega), RNA extraction using TRI Reagent was repeated. Superscript II Reverse Transcriptase (Invitrogen) and oligo(dT) were used for cDNA synthesis from total RNA.
RNA in Situ Probes.
(a) The probe for il7 was generated by nested RT-PCR using the following primers: first round, il7rt_F1: 5′-TTCCTCAGTCAAATCCTGAATC, il7rt_R1: 5′-CAGTTACGCACTTGACGTTCT; second round, il7rt_F2a: 5′-CTCCGACGATACAATGCGAC, il7rt_R2: 5′-CCTGAATCTTGTGAATGTTGCA. (b) The probe for il2 was generated using the following primers: first round, il2_F1: 5′-CGCACACACTGATGATGATGA, il2_R1: 5′-TTCTGCCTCCATTCGTTCATC; second round, il2_F2: 5′-AGGATGTCTGCTCTACACTG, il2_R2: 5′-ACGTTCTTCAGGAACGTCATG. (c) The probe for il21 was generated using the following primers: first round, il21_F1: 5′-ACGCGCCTCTCGATTACATC, il21_R1: 5′- GAGATTTCACCACACGGTGG; second round, il21_F2: 5′-AGGTGATCGAGCACCTGTGT, il21_R2: 5′-GCTTAGCAGCCCAGTTTCTC. (d) The probe for i15l was generated using the following primers: first round, il15l_F1: 5′-AACAATGAGCGGGTGACGAC, il15l_R1: 5′- CCTCTGGAGAAGCACTTCAG; second round, il15l_F2: 5′-GACGACTCGGCTTTCGATAG, il15l_R2: 5′-CAGAACCGCTCCTTCAGATC. (e) The zebrafish il15 cDNA is equivalent to IMAGE clone 7990051 (GenBank accession no. BC133847.1). (f) The rag1, gh, tcrb, tcrd, and igm probes were as described previously (32).
il7 Mutagenesis.
A deletion in the il7 locus was generated by injection of 2 RNPs containing Cas9 protein and sgRNAs. The injection solution contained (final concentrations) 12.5 to 50 ng/μL sgRNAs, 500 ng/μL Cas9 protein (PNA Bio), and 0.05% (vol/vol) phenol red, in 1× Danieau buffer (http://cshprotocols.cshlp.org/content/2011/7/pdb.rec12467.full). Approximately 1 to 2 nL of the solution was injected into fertilized embryos. The target sequences of the 2 sgRNAs were as follows: sgRNA_1: 5′-AATGACAAATGGTAAGCTGA (position in genome, nt 22,731,430 on chromosome 24) and sgRNA_2: 5′-AAAGGTAACTGAAACAACCC (position in genome, nt 22,733,214 on chromosome 24); coordinates refer to GRCZ11. The sequences around the breakpoint are shown in SI Appendix, Fig. S1; note that a total 16 nontemplated nucleotides were inserted into the genome between the 2 break points.
Analysis of il7 cDNA Isoforms.
Nested RT-PCR was used to examine the structure of the various isoforms of il7 transcripts. For isoforms 1, 2, and 3 (SI Appendix, Fig. S1B), the following primers were used: first round, il7rt_F1: 5′-CAGTTACGCACTTGACGTTCT, il7rt_R2: 5′-CCTGAATCTTGTGAATGTTGCA; second round, il7_F2: 5′-CGAAGCGATATAGCCCCATC, il7rt_R3 5′-TTGTGAATGTTGCAAAGTGGGT. For isoform 4, the following primers were used: first round, il7exon1a_F1: 5′-GCGTACGCTTACCTTGACGA, il7rt_R2: 5′-CCTGAATCTTGTGAATGTTGCA; second round, il7exon1a_F2: 5′-TTTGGAGGAGGACGACAGGA, il7rt_R3: 5′-TTGTGAATGTTGCAAAGTGGGT.
Ag Receptor Gene Assembly.
Nested RT-PCR assays were used to examine the presence of transcribed assembled antigen receptor genes as described previously (32).
RNA-Seq Analyses.
For differential gene expression analysis, RNA was extracted from whole kidney marrow (wild-type and il7-deficient animals; n = 3 each). The libraries were sequenced in paired-end 75-bp mode on an Illumina HiSeq 2500 instrument. The high-throughput RNA sequencing analysis pipeline and the relevant statistical models used are essentially as described previously (9). RNA-seq data have been deposited in the Gene Expression Omnibus database (33).
Statistical Analysis.
Two-tailed t tests were used to determine the significance levels of the differences between the means of 2 independent samples, considering equal or unequal variances as determined by the F test. For multiple tests, Bonferroni correction was applied.
Supplementary Material
Acknowledgments
We thank Christine Strohmeier for help with RNA in situ hybridization experiments and the staff of the Deep Sequencing Unit of the Max Planck Institute of Immunobiology and Epigenetics for next-generation sequencing. This work was supported by the Max Planck Society and the European Research Council (ERC) under the European Union’s Seventh Framework Programme (FP7/2007-2013), ERC Grant Agreement 323126.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE136250).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915223116/-/DCSupplemental.
References
- 1.Di Santo J. P., Rodewald H.-R., In vivo roles of receptor tyrosine kinases and cytokine receptors in early thymocyte development. Curr. Opin. Immunol. 10, 196–207 (1998). [DOI] [PubMed] [Google Scholar]
- 2.von Freeden-Jeffry U., et al. , Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J. Exp. Med. 181, 1519–1526 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin J. X., Leonard W. J., The common cytokine receptor γ chain family of cytokines. Cold Spring Harb. Perspect. Biol. 10, a028449 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peschon J. J., et al. , Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J. Exp. Med. 180, 1955–1960 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DiSanto J. P., Müller W., Guy-Grand D., Fischer A., Rajewsky K., Lymphoid development in mice with a targeted deletion of the interleukin 2 receptor gamma chain. Proc. Natl. Acad. Sci. U.S.A. 92, 377–381 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horev L., et al. , Generalized verrucosis and HPV-3 susceptibility associated with CD4 T-cell lymphopenia caused by inherited human interleukin-7 deficiency. J. Am. Acad. Dermatol. 72, 1082–1084 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Puel A., Ziegler S. F., Buckley R. H., Leonard W. J., Defective IL7R expression in T(-)B(+)NK(+) severe combined immunodeficiency. Nat. Genet. 20, 394–397 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Noguchi M., et al. , Interleukin-2 receptor gamma chain mutation results in X-linked severe combined immunodeficiency in humans. Cell 73, 147–157 (1993). [PubMed] [Google Scholar]
- 9.Iwanami N., et al. , Forward genetic screens in zebrafish identify pre-mRNA-processing pathways regulating early T cell development. Cell Rep. 17, 2259–2270 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwanami N., et al. , Genetic evidence for an evolutionarily conserved role of IL-7 signaling in T cell development of zebrafish. J. Immunol. 186, 7060–7066 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Sertori R., et al. , Conserved IL-2Rγc signaling mediates lymphopoiesis in zebrafish. J. Immunol. 196, 135–143 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Nehls M., Pfeifer D., Schorpp M., Hedrich H., Boehm T., New member of the winged-helix protein family disrupted in mouse and rat nude mutations. Nature 372, 103–107 (1994). [DOI] [PubMed] [Google Scholar]
- 13.Nehls M., et al. , Two genetically separable steps in the differentiation of thymic epithelium. Science 272, 886–889 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Calderón L., Boehm T., Synergistic, context-dependent, and hierarchical functions of epithelial components in thymic microenvironments. Cell 149, 159–172 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Swann J. B., et al. , Conversion of the thymus into a bipotent lymphoid organ by replacement of FOXN1 with its paralog, FOXN4. Cell Rep. 8, 1184–1197 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Venkatesh B., et al. , Elephant shark genome provides unique insights into gnathostome evolution. Nature 505, 174–179 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hess I., Boehm T., Intravital imaging of thymopoiesis reveals dynamic lympho-epithelial interactions. Immunity 36, 298–309 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Moore T. A., von Freeden-Jeffry U., Murray R., Zlotnik A., Inhibition of gamma delta T cell development and early thymocyte maturation in IL-7−/− mice. J. Immunol. 157, 2366–2373 (1996). [PubMed] [Google Scholar]
- 19.Baryshnikova A., Costanzo M., Myers C. L., Andrews B., Boone C., Genetic interaction networks: Toward an understanding of heritability. Annu. Rev. Genomics Hum. Genet. 14, 111–133 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Ziegler S. F., Liu Y.-J., Thymic stromal lymphopoietin in normal and pathogenic T cell development and function. Nat. Immunol. 7, 709–714 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Al-Shami A., et al. , A role for thymic stromal lymphopoietin in CD4(+) T cell development. J. Exp. Med. 200, 159–168 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tononi G., Sporns O., Edelman G. M., Measures of degeneracy and redundancy in biological networks. Proc. Natl. Acad. Sci. U.S.A. 96, 3257–3262 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schorle H., Holtschke T., Hünig T., Schimpl A., Horak I., Development and function of T cells in mice rendered interleukin-2 deficient by gene targeting. Nature 352, 621–624 (1991). [DOI] [PubMed] [Google Scholar]
- 24.Sadlack B., et al. , Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell 75, 253–261 (1993). [DOI] [PubMed] [Google Scholar]
- 25.Etzensperger R., et al. , Identification of lineage-specifying cytokines that signal all CD8+-cytotoxic-lineage-fate “decisions” in the thymus. Nat. Immunol. 18, 1218–1227 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Surh C. D., Sprent J., Regulation of mature T cell homeostasis. Semin. Immunol. 17, 183–191 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Kennedy M. K., et al. , Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J. Exp. Med. 191, 771–780 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lodolce J. P., et al. , IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity 9, 669–676 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Leonard W. J., Lin J. X., O’Shea J. J., The γc family of cytokines: Basic biology to therapeutic ramifications. Immunity 50, 832–850 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Pancer Z., et al. , Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature 430, 174–180 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Bajoghli B., et al. , A thymus candidate in lampreys. Nature 470, 90–94 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Schorpp M., et al. , Conserved functions of Ikaros in vertebrate lymphocyte development: Genetic evidence for distinct larval and adult phases of T cell development and two lineages of B cells in zebrafish. J. Immunol. 177, 2463–2476 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Edgar R., Domrachev M., Lash A. E., Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Traver D., et al. , Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat. Immunol. 4, 1238–1246 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.