Significance
Human health and the microbiota are intricately intertwined. A major interest in manipulating the microbiome has been focused on the use of symbiont microbes to improve human health. However, relatively little has been discovered on the specific molecules from microbes in the microbiota that are immunomodulatory and the mechanisms by which these molecules regulate immunity. Herein we have defined how a symbiont molecule modulates innate immunity. Using polysaccharide A of Bacteroides fragilis as the paradigm for microbiome-induced immune responses, we have discovered important mechanisms by which symbiont molecules induce antiinflammatory responses. We reveal a lipid structure on polysaccharide A that drives host antiinflammatory responses by triggering a complex collaborative integration of Toll-like receptor, C-type lectin-like receptor, and PI3K signaling pathways.
Keywords: symbionts, Bacteroides fragilis, zwitterionic polysaccharides (ZPSs), polysaccharide A, host–microbe interactions
Abstract
The mammalian immune system is tolerized to trillions of microbes residing on bodily surfaces and can discriminate between symbionts and pathogens despite their having related microbial structures. Mechanisms of innate immune activation and the subsequent signaling pathways used by symbionts to communicate with the adaptive immune system are poorly understood. Polysaccharide A (PSA) of Bacteroides fragilis is the model symbiotic immunomodulatory molecule. Here we demonstrate that PSA-dependent immunomodulation requires the Toll-like receptor (TLR) 2/1 heterodimer in cooperation with Dectin-1 to initiate signaling by the downstream phosphoinositide 3-kinase (PI3K) pathway, with consequent CREB-dependent transcription of antiinflammatory genes, including antigen presentation and cosignaling molecules. High-resolution LC-MS/MS analysis of PSA identified a previously unknown small molecular-weight, covalently attached bacterial outer membrane-associated lipid that is required for activation of antigen-presenting cells. This archetypical commensal microbial molecule initiates a complex collaborative integration of Toll-like receptor and C-type lectin-like receptor signaling mechanisms culminating in the activation of the antiinflammatory arm of the PI3K pathway that serves to educate CD4+ Tregs to produce the immunomodulatory cytokine IL-10. Immunomodulation is a key function of the microbiome and is a focal point for developing new therapeutic agents.
From birth to death, animals live with and are exposed to multitudes of microorganisms (1). While pathogenic microorganisms often cause disease, commensal microorganisms colonize their host and usually have beneficial rather than harmful effects (2). A balanced microbiota and the health of the host depend on the ability of the host’s immune system to differentiate between symbionts and pathogens. The cross-talk between resident microorganisms and the host involves signals emitted by the microbes, which are translated by the host into tolerogenic mechanisms that allow peaceful coexistence. Given the substantial impact of symbiotic microbes on the host’s health, the nature of the tolerogenic signals from these microbes and the cellular mechanisms that establish host–microbe interactions need further elucidation.
Bacteroides fragilis is a gram-negative obligate anaerobe inhabiting the lower gastrointestinal tract of humans. B. fragilis confers beneficial immunomodulatory effects on the host immune system through a single molecule, polysaccharide A (PSA), a zwitterionic symbiosis factor that is abundantly expressed in the microbe’s capsular complex (3). Documented beneficial effects of B. fragilis PSA include: 1) stimulation of immune system development and balance (4); 2) protection from pathogenic infections (5); 3) induction of host regulatory T cell (Treg) production of IL-10 during inflammation following T cell activation by plasmacytoid dendritic cells (PDCs) (6); 4) IL-10–dependent protection from intestinal inflammatory diseases (7); and 5) protection from systemic immune-mediated diseases, such as experimental autoimmune encephalomyelitis (EAE, the mouse model of multiple sclerosis in humans) (8, 9). Although PSA-mediated protection against both colitis and EAE requires Toll-like receptor 2 (TLR2) on host antigen-presenting cells (APCs) (6, 10), the innate signaling mechanisms that direct the immune system toward an antiinflammatory rather than a proinflammatory response are not understood.
A recent study reported a genomic screen of gut microbiota components with the genetic potential to encode PSA-like zwitterionic capsular polysaccharides (11). This study showed that a number of bacteria in the orders Bacteroidales and Erysipelotrichales can express PSA-like molecules and that culture lysates of these organisms induce more IL-10 and Tregs in human peripheral-blood mononuclear cells than do lysates of related organisms whose genomes do not encode zwitterionic capsular polysaccharides. An understanding of the molecular mechanisms of PSA-mediated immunomodulation should illuminate the function of other molecules in the gut.
On the basis of our data, we propose a model in which PSA is recognized by the TLR2/TLR1 heterodimer in collaboration with Dectin-1. Downstream, after integration of signals from both these receptors, the phosphoinositide 3-kinase (PI3K) pathway is activated. Upstream signaling through all 3 pathways is required for inactivation of glycogen synthase kinase 3β (GSK3β), which in turn promotes cAMP response element–binding protein (CREB)-dependent antiinflammatory gene transcription. We have discovered that the terminal-reducing end of PSA is composed of a covalently linked small lipid moiety that is critically important for induction of immunoregulatory signaling. We propose that B. fragilis PSA educates the host immune system by initiating multiple signaling mechanisms. These complex signaling mechanisms, working in concert, serve to stimulate a tolerogenic rather than a proinflammatory response in the gut.
Results
PSA Signals through the TLR2/TLR1 Heterodimer.
The hallmark of PSA activation of the immune system is the promotion of antiinflammatory responses (7) via IL-10–producing Tregs (12). PSA signaling through PDCs and the cognate interaction of PDCs with CD4+ T cells are required for PSA’s immunomodulatory effects (6). Sensing of PSA by APCs requires TLR2 for immunoregulation of both local intestinal inflammation and systemic inflammatory diseases (6, 10, 13, 14). This tolerogenic response mediated by PSA is counterintuitive because TLR2 ligation is known to result in NF-κB–mediated proinflammatory responses. However, a number of recent studies show that TLR2 can also regulate antiinflammatory responses (15–17). To compare proinflammatory responses to PSA with those to other TLR ligands, we stimulated cultured bone marrow-derived dendritic cells (BMDCs) for 24 h with PSA, Pam3CSK4 (a TLR2 agonist), or Escherichia coli lipopolysaccharide (LPS; a TLR4 agonist), and we measured the proinflammatory cytokines TNFα and IL-6 in culture supernatants (SI Appendix, Fig. S1). After stimulation by PSA, BMDC culture supernatant levels of both TNFα and IL-6 were significantly lower than those after stimulation of these cultures by E. coli LPS or Pam3CSK4. This result suggested that PSA recognition and signaling might involve other mechanisms in addition to TLR2 ligation. We therefore initiated an in-depth study of the APC receptors and signaling pathways required for PSA to induce T cell production of IL-10.
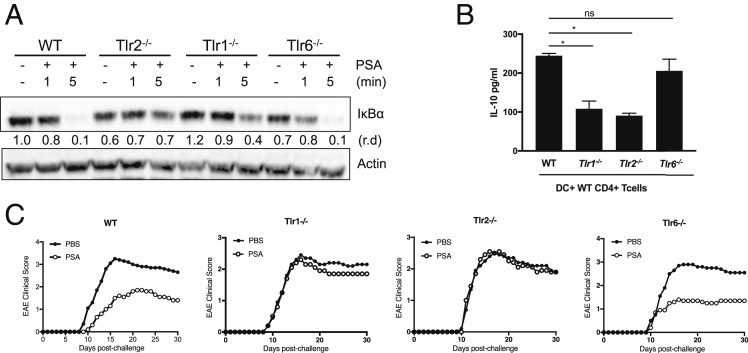
In most microbial product-initiated TLR2-dependent signaling systems, TLR2 heterodimerizes with TLR1 or TLR6. In order to determine whether TLR1 or TLR6 participates in PSA recognition, we used bone marrow-derived macrophages (BMDMs) from WT and TLR-knockout mice to examine degradation of IκBα, the cytoplasmic inhibitor of NF-κB, in response to PSA (Fig. 1A). While degradation of IκBα is complete in WT and TLR6-deficient (Tlr6−/−) macrophages in response to PSA, it is significantly less pronounced in TLR2-deficient (Tlr2−/−) and TLR1-deficient (Tlr1−/−) macrophages. These data suggest that PSA recognition requires the TLR2/TLR1 heterodimer but not the TLR2/TLR6 heterodimer. Furthermore, PSA elicits higher levels of cytokine expression when TLR2 and TLR1 are coexpressed in HEK293 cells than when TLR2 alone is expressed (SI Appendix, Fig. S2). It has been established that the inducible nitric oxide synthase (iNOS) gene is up-regulated in response to PSA in a TLR2-dependent manner (13) and is required for endosomal depolymerization and major histocompatibility class II (MHCII) presentation of PSA (18–20). After 24-h PSA exposure and endosomal processing, iNOS expression is evident and therefore is up-regulated in WT and Tlr6−/− cells but not in Tlr2−/− or Tlr1−/− cells (SI Appendix, Fig. S3).
Fig. 1.
PSA signals through the TLR2/TLR1 heterodimer. (A) Western blot analysis of protein (50 μg) extracted from BMDMs at different time points after PSA stimulation. Degradation of IκBα was detected with anti-IκBα. Antiactin was used as a loading control. Relative density (r.d.) was measured with ImageJ software. (B) IL-10 liberation from WT CD4+ T cells cocultured with WT, Tlr1−/−, Tlr2−/−, or Tlr6−/− PDCs and incubated for 5 d in the presence of anti-CD3. Cocultures were either treated or not treated with PSA (50 μg/mL). IL-10 levels in supernatants were measured by ELISA normalized by subtracting the medium control. Error bars indicate SD values. Scores were assessed for statistical significance by t test. *P < 0.05; ns, not significant. These data are representative of 2 independent experiments. (C) Daily EAE clinical disease scores of female C57BL/6J mice after induction with the MOG 35−55 peptide. Mice were treated with PBS or 50 μg of purified PSA. Depicted are the combined results of 2 independent experiments where average disease scores were plotted.
Previous studies have shown that PSA induces Tregs to secrete the antiinflammatory cytokine IL-10 (7, 8, 12) and that processing and presentation of PSA by PDCs are essential in vivo (6). In an in vitro PDC–CD4+ T cell coculture system, we tested whether PSA signals through the TLR2/TLR1 heterodimer to induce IL-10–liberating T cells. CD4+ T cells cocultured with WT or Tlr6−/− PDCs liberate significantly higher levels of IL-10 than T cells cocultured with Tlr2−/− or Tlr1−/− PDCs (Fig. 1B). PSA has been shown to be protective in the mouse model for multiple sclerosis (i.e., EAE) (8). PSA protection from EAE requires the presence of TLR2 on the APC (10). However, whether TLR2 coreceptors are required for EAE protection is not known. Since our in vitro data showed that PSA signaling in PDCs requires TLR2 and TLR1 but not TLR6, we assessed the requirement for these receptors in the mediation of PSA protection from EAE in vivo. We treated groups of TLR-deficient C57BL/6J mice (Tlr2−/−, Tlr1−/−, and Tlr6−/−) with orally gavaged PSA 3 times a week throughout the experiment, beginning 2 wk before EAE induction. PSA treatment reduces disease severity (Fig. 1C) and cumulative disease scores (SI Appendix, Fig. S4) in WT and Tlr6−/− mice but not in Tlr2−/− or Tlr1−/− mice. Taken together, these findings reveal that PSA signals and activates APCs through TLR2/TLR1—but not through TLR2/TLR6—to mediate its antiinflammatory effects both in vitro and in vivo. While TLR2 heterodimerization with other TLRs broadens the spectrum of TLR2 ligands, both TLR2/TLR1 and TLR2/TLR6 heterodimers use the same intracellular signaling molecules resulting in NF-κB–dependent proinflammatory responses. Therefore, we reasoned that perhaps other innate signals that modify the canonical TLR2/TLR1/NF-κB–mediated proinflammatory response and channel signaling into an immunomodulatory pathway were being stimulated.
PSA Signaling Requires Collaboration of Dectin-1 with TLR2.
In considering other signaling pathways in addition to TLR2/TLR1 that might be initiated by a carbohydrate and might be important in PSA-mediated immunomodulation, we investigated the C-type lectin-like receptor (CLR) family, which includes specialized receptors that respond to carbohydrate structures, often in collaboration with TLRs (21). Dectin-1, a CLR that has been shown to react with β-glucans, cooperates with TLR2 in response to zymosan (22, 23). Recognition of microbial ligands by Dectin-1 leads to intracellular signaling via spleen tyrosine kinase (Syk) (24). Upon ligation, Dectin-1 directly recruits Syk, which then phosphorylates itself and downstream signaling molecules. Phosphorylation of Syk upon stimulation of macrophages by PSA (SI Appendix, Fig. S5) supports the likelihood that the Dectin-1 pathway is involved in PSA signaling.
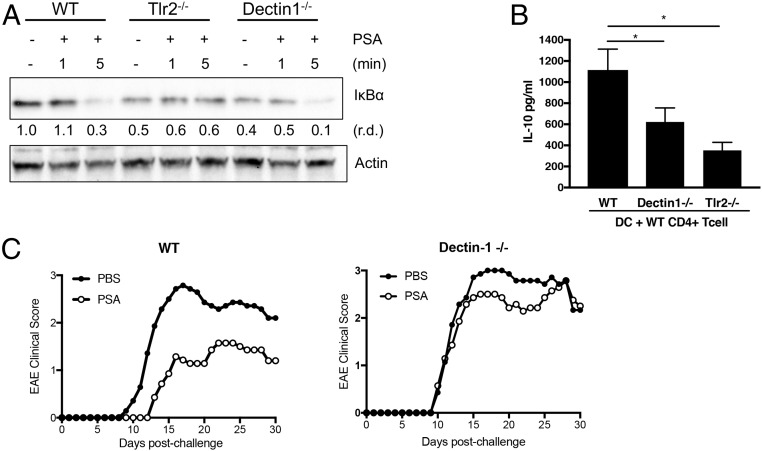
To investigate whether the Dectin-1 pathway is activated in parallel with or upstream of TLR2, we compared degradation of IκBα in WT, Tlr2−/−, and Dectin-1–deficient macrophages (Fig. 2A). While IκBα is not degraded in Tlr2−/− cells, degradation occurs with similar kinetics in WT and Dectin-1–deficient cells. These results imply that the TLR2 and Dectin-1 pathways are activated in parallel with each other and that, if these 2 pathways were integrated, the integration would likely occur downstream at later stages of signaling. We analyzed whether the downstream gene product iNOS is generated in Dectin-1–deficient cells. Western blot analysis of macrophages treated with PSA for 24 h showed that, in addition to TLR2, Dectin-1 is required for production of iNOS (SI Appendix, Fig. S6). To test the requirement for dendritic cell (DC)-specific Dectin-1 in PSA-induced antiinflammatory responses, we used the splenic DC–T cell coculture system described in Fig. 1B. WT and Dectin-1–deficient splenic DCs were cocultured with WT CD4+ T cells during treatment with PSA. IL-10 levels in the culture supernatant were measured by ELISA and normalized by subtracting the medium control. Compared to CD4+ T cells incubated with WT DCs, CD4+ T cells incubated with Dectin-1–deficient DCs produce significantly less IL-10 in response to PSA (Fig. 2B).
Fig. 2.
PSA signaling requires Dectin-1. (A) Western blot analysis of IκB degradation in macrophages at different time points after PSA stimulation. Relative density (r.d.) was measured with ImageJ software. (B) IL-10 liberation from CD4+ T cells cocultured with splenic DCs for 5 d in the presence of anti-CD3. Cocultures were either treated or not treated with PSA (50 μg/mL). IL-10 levels were measured by ELISA of culture supernatants and normalized by subtracting the medium control. Data represent average of 2 experiments. Error bars indicate SD values. Scores were assessed for statistical significance by t test. *P < 0.05. (C) EAE clinical disease scores of female C57BL/6J mice were measured daily for 30 d after induction by the MOG 3I5−55 peptide. Mice were treated with PBS or 50 μg of purified PSA. Depicted are the combined results of 2 independent experiments in which average disease scores were plotted.
Another CLR, DC-SIGN, reportedly responds to PSA in human cells (25). We investigated the role of SIGNR3 (the mouse homolog of DC-SIGN) in PSA-induced signaling. SIGNR3 (CD209d)-deficient DCs exhibit no defect in activating IL-10–producing T cells (SI Appendix, Fig. S7). Therefore, in mouse cells, PSA signaling requires Dectin-1 but not SIGNR3. This finding was validated in vivo when PSA treatment failed to protect Dectin-1–deficient mice against EAE (Fig. 2C and SI Appendix, Fig. S8). Of interest, yeast zymosan is reportedly recognized by both Dectin-1 and TLR2. However, in general, Dectin-1 signaling alone or in collaboration with TLRs regulates proinflammatory responses to fungal infections. Dectin-1 involvement in PSA-mediated immunoregulation suggested that PSA-induced collaboration of Dectin-1 and TLR-2 involves additional signaling pathways that lead to antiinflammatory responses.
PSA Signaling Leads to Activation of the PI3K Pathway.
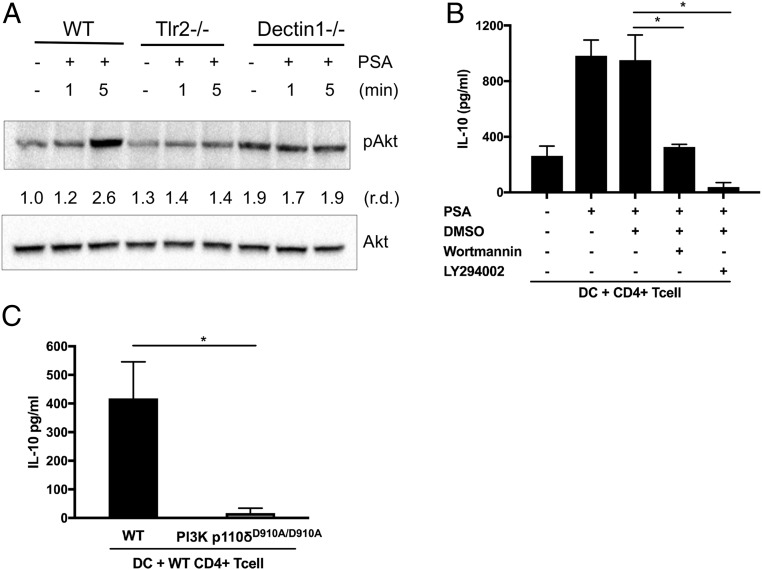
Since PSA triggers the TLR2 and Dectin-1 pathways in parallel and requires both pathways for the transcription of target genes, we considered signaling mechanisms in which these pathways collaborate. One such mechanism known for fine-tuning immune signaling initiated by pattern recognition receptors is the PI3K pathway. Depending on the receptor, the coreceptors, and the strength of the signal, the PI3K pathway can promote either antiinflammatory or proinflammatory responses (26). The existence of cross-talk between TLR2 signaling and the PI3K pathway has been reported (27–29). A number of studies favor a critical role for the PI3K pathway as a negative regulator of TLR signaling and proinflammatory immune responses. DCs from p85α-deficient mice produce more IL-12 than WT DCs upon TLR stimulation in vitro (30), and mice with a kinase-dead mutation in the p110δ catalytic domain (p110δD910A/D910A) develop chronic colitis in vivo (31). It has been shown that TLR signaling results in activation of the PI3K pathway and that TLR2 physically interacts with p85, the regulatory subunit of PI3K (27). To investigate whether the PI3K pathway plays a role in PSA-induced signaling, we first examined phosphorylation of Akt, a key kinase in the PI3K pathway. In macrophages, Akt is phosphorylated in response to PSA (SI Appendix, Fig. S9); thus, the PI3K pathway is also involved in PSA signaling. Phosphorylated and activated Akt is not detectable in either Tlr2−/− or Dectin-1–deficient cells (Fig. 3A); this result shows that, upon PSA stimulation, both TLR2 and Dectin-1 signaling are required for activation of the PI3K pathway. Chemical inhibitors of the PI3K pathway (Wortmannin and LY294002) inhibit PSA-mediated IL-10 production in DC–T cell coculture (Fig. 3B). To avoid nonspecific effects of chemical inhibitors, we tested cells from mice with the kinase-dead mutation (p110δD910A/D910A) (31). DCs from mutant animals do not induce IL-10 production from WT splenic T cells in DC–T cell coculture (Fig. 3C). These results show that the PI3K pathway is activated downstream of Dectin-1 and TLR2 signaling and that the collaboration of these pathways culminates in antiinflammatory responses induced by PSA.
Fig. 3.
PSA signaling leads to activation of the PI3K pathway. (A) Activation of the PI3K pathway was detected by phospho-Akt blotting in macrophages at different time points after PSA stimulation. (B) PSA-induced IL-10 liberation from CD4+ T cells cocultured with splenic DCs. IL-10 levels were measured by ELISA of culture supernatants of splenic DCs cocultured with CD4+ T cells for 5 d in the presence of anti-CD3 and the PI3K inhibitors Wortmannin and LY294002. Cocultures were either treated or not treated with PSA (50 μg/mL). These data are representative of 2 independent experiments. (C) Splenic DCs isolated from either WT or PI3K p110δD910A/D910A animals were cocultured with splenic WT CD4+ T cells and treated with PSA (50 μg/mL). IL-10 levels were measured by ELISA and normalized by subtracting the medium control. Data represent the average of 2 experiments. In B and C, error bars indicate SD values. Scores were assessed for statistical significance by t test. *P < 0.05.
PSA-Induced Immunoregulation Requires CREB-Mediated Transcription.
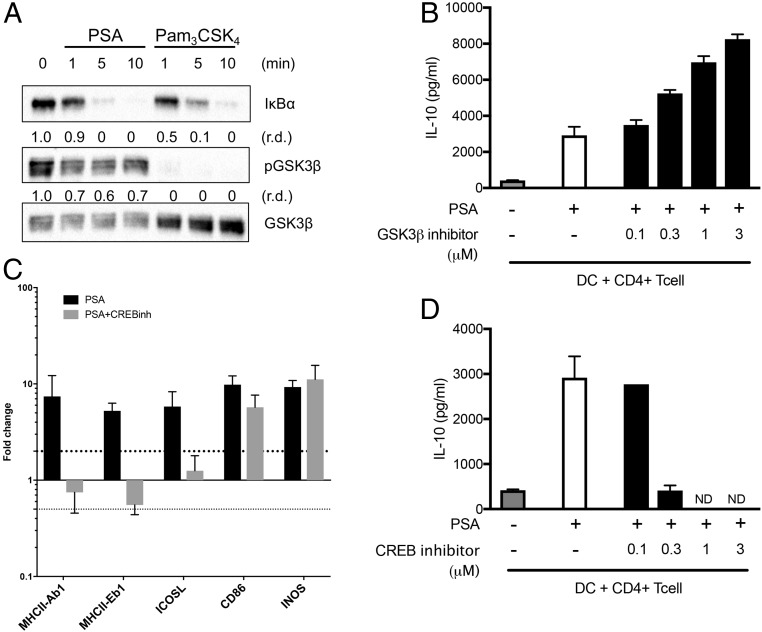
The molecular mechanism of PI3K-dependent antiinflammatory responses depends on inhibition of a key kinase, GSK3β (26). GSK3β is unphosphorylated in its active state and can be inactivated through phosphorylation by other kinases, including Akt (32). Phosphorylation and inactivation of GSK3β result in the inhibition of NF-κB–mediated transcription of proinflammatory cytokines and the promotion of antiinflammatory gene expression mediated by CREB (33). GSK3β favors NF-κB/CREB-binding protein (CBP) association by inhibiting the activation and DNA-binding activity of CREB (34). We compared the activity of GSK3β in macrophages in response to PSA and Pam3CSK4, a typical TLR2/TLR1 ligand for proinflammatory responses. PSA and Pam3CSK4 induce activation of the TLR2 pathway at similar rates, as evidenced by the degradation of the IκB protein (Fig. 4A). However, GSK3β becomes unphosphorylated and active after stimulation with Pam3CSK4, whereas it remains phosphorylated and hence inactive after PSA stimulation (Fig. 4A). Active GSK3β is known to favor binding of NF-κB and the transcription cofactor CBP in promoting induction of proinflammatory genes (35). On the other hand, inactivation of GSK3β results in CBP/CREB-dependent transcription of antiinflammatory genes. Using cocultures of DCs and T cells, we compared IL-10 production in response to PSA in the presence and absence of a GSK3β inhibitor (SB216763). Inhibition of GSK3β increased the level of IL-10 (Fig. 4B) released by CD4+ T cells in response to PSA. To determine whether CREB plays a role in PSA-mediated antiinflammatory responses, we examined the transcription of selected genes required for DC antigen presentation of PSA (iNOS and MHCII) and cognate costimulation of T cells by PSA (ICOSL and CD86). We treated BMDCs with PSA in the presence of a CREB inhibitor (666-15) and used quantitative PCR to analyze the transcription of genes required for PSA function. We found a robust decrease in the transcription of MHCII and ICOSL, whereas the expression of CD86 and iNOS is not notably affected by CREB inhibition (Fig. 4C). It has been shown that the absence of either MHCII or ICOSL is sufficient to essentially eliminate PSA’s ability to induce IL-10 or confer protection in a model of colitis (6). We measured PSA-induced IL-10 levels in DC–T cell cocultures in the presence of the CREB inhibitor 666-15 and found that inhibition of CREB robustly decreases IL-10 production from CD4+ T cells (Fig. 4D). Together, these results show that phosphorylation of GSK3β allows CREB-dependent transcription of essential genes for PSA immunoregulation.
Fig. 4.
PSA-dependent GSK3β inhibition promotes CREB-mediated transcription. (A) Western blot analysis of phospho-GSK3β in macrophages stimulated with PSA and Pam3CSK4. (B) CD4+ T cells cocultured with splenic DCs for 5 d in the presence of anti-CD3 and GSK3β inhibitor SB216763. Cocultures were either treated or not treated with PSA (50 μg/mL), and IL-10 was measured by ELISA of culture supernatants. These data are representative of 2 independent experiments. (C) Real-time PCR analysis of PSA-stimulated DCs with and without CREB inhibitors for transcript levels of costimulatory molecules. These data areaverage of 2 independent experiments. (D) IL-10 secretion from CD4+ T cells cocultured with splenic DCs in response to PSA in the presence of CREB inhibitor 666-15. These data are representative of 2 independent experiments. In B–D, error bars indicate SD. ND, not detected.
PSA Contains a Bacterial Membrane-Insertion Lipid.
The structure of the lipid moiety (lipid A) of LPS has been extensively studied, and its role in activating the innate immune system has been well defined. In contrast, only a few capsular polysaccharides of gram-negative bacteria have been shown to be anchored in the outer membrane through covalently linked hydrophobic moieties (36, 37). These lipids are conjugated to the reducing end of the polysaccharide. In contrast to immune responses to the lipids of LPS, however, the innate immune responses to lipids on capsular polysaccharides have yet to be revealed.
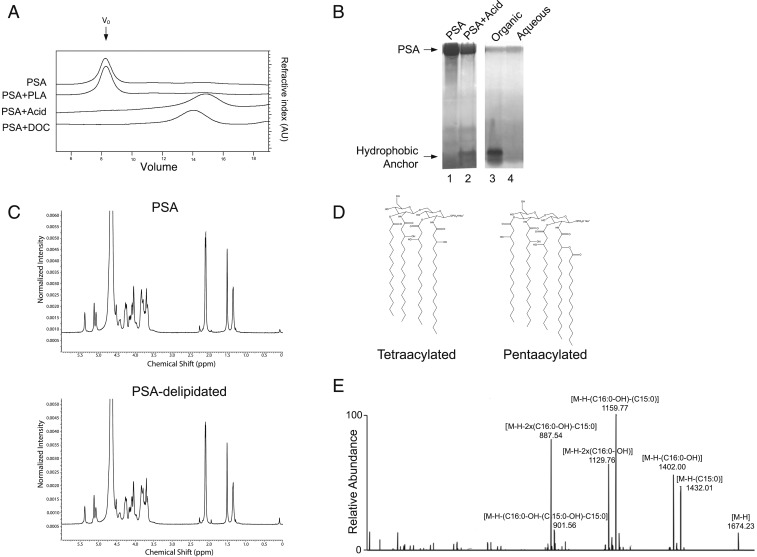
The average molecular size of PSA was determined to be ∼150 kDa by size-exclusion chromatography in 1% sodium deoxycholate buffer (SI Appendix, Fig. S10). However, using a similar column matrix but with a buffer free of detergent, we found that PSA eluted at the column’s void volume (molecular mass > 2,000 kDa) (SI Appendix, Fig. S10). This very large size difference between PSA chromatographed in detergent and that chromatographed in aqueous buffer suggested that hydrophobic lipid moieties that form micelle-like aggregates of the polysaccharide exist and are disrupted in the presence of an amphiphilic detergent, such as 1% deoxycholate.
PSA is a high molecular-weight polysaccharide and does not exhibit the typical lower molecular-mass ladder profile that is characteristic of LPS molecules (SI Appendix, Fig. S11). Extraction of PSA with chloroform/methanol (Bligh-Dyer) shows that no lipid is recovered in the organic phase (SI Appendix, Fig. S12), suggesting that the lipid is covalently linked to the polysaccharide. Many capsule-anchoring lipids of other organisms have been shown to be phospholipids. However, treatment of PSA with phospholipase A2 does not change PSA’s formation of micelle aggregates (Fig. 5A). In contrast, very mild acid hydrolysis (2% acetic acid at 90 °C for 2 h) disrupts the micellar aggregation of PSA. The elution profile of PSA—in the absence of detergent—after mild hydrolysis shifts to the size of the lipidated PSA molecule chromatographed in detergent buffer (∼150 kDa) (Fig. 5A). Analysis of acid-hydrolyzed PSA by zinc gel demonstrated the release of a low molecular-weight band that partitions to the organic phase of chloroform/methanol extraction (Fig. 5B). On the other hand, 1H-NMR analysis of PSA before and after acid hydrolysis showed that the carbohydrate structure of PSA remains unmodified after acid treatment (Fig. 5C). The lipid moiety of PSA before acid hydrolysis was not detected by 1H-NMR analysis of PSA; this finding supports the idea that this moiety is very small compared to the high molecular-weight repeating-unit structure of PSA. After hydrolysis, recovery of lipid in the organic phase of chloroform/methanol extraction showed that it accounts for ∼1% of the dry weight of the original PSA molecule. The identification of this band by liquid chromatography–tandem mass spectrometry (LC-MS/MS) as diacylglucosamine with different degrees of acylation (tri-, tetra-, and penta-) (Fig. 5D and SI Appendix, Fig. S13) further expands the chemical diversity of B. fragilis glycolipids from previously reported lipid A structures (38). MS/MS fragmentation confirmed that the individual acyl chain length of the PSA lipid anchor is mostly C15–C17 (Fig. 5E), which is similar to that of B. fragilis lipid A and longer than that of typical pathogenic lipid A. Further lipid analysis of purified B. fragilis lipooligosaccharide identified similar classes of lipid anchor molecules (SI Appendix, Table S1), implying that these structural variants are shared among lipid-anchored polysaccharides synthesized by B. fragilis.
Fig. 5.
PSA contains a hydrophobic lipid anchor. (A) Gel filtration of 0.2-mg aliquots of PSA on a Superose 6 10/300 GL column running at a 0.5-mL/min flow rate. The elution profiles, from top to bottom, show untreated PSA, PSA treated with phospholipase A2 (PLA), PSA treated with 2% acetic acid, and PSA run in 1% deoxycholate (DOC) buffer. (B) Zinc gel analysis of untreated PSA (lane 1), PSA treated with 2% acetic acid solution at 90 °C for 3 h (lane 2), and the organic (lane 3) and aqueous (lane 4) phases of chloroform/methanol extraction of acid-treated PSA. Release of the acid-labile hydrophobic lipid moiety was detected in the organic phase. (C) 1H-NMR analysis of PSA and delipidated PSA showed no difference in the carbohydrate structure. (D) Representative species of B. fragilis lipid A, such as tetraacylated-monophosphorylated (combined carbon chain length = 64, molecular weight [MW] = 1421.0) and pentaacylated-monophosphorylated (combined carbon chain length = 80, MW = 1675.2), form on the diacylglucosamine backbone. A phosphate group may be attached at C1 or C4′ position, and some of acyl chains may be branched. (E) MS/MS fragmentation assigned the acyl chains as saturated or monohydroxylated C15–C17 fatty acids.
These physical/chemical studies suggest that the lipid linkage to the carbohydrate is acid labile and is likely located at the terminus of PSA because the molecular size of the polysaccharide portion of the molecule does not change significantly after removal of the lipid. To confirm the terminal membrane location of the lipid on the polysaccharide, we suspended live B. fragilis bacteria in 1% acetic acid and incubated this preparation at 65 °C for 2 h, a condition that is sufficient to hydrolyze the bond between the carbohydrate repeating unit and the lipid and to liberate carbohydrate from the lipid terminus (SI Appendix, Fig. S14A). After centrifugation, large molecular-size delipidated PSA was readily detected in the supernatant by Western blot, a result indicating its release from the membrane after very mild acid hydrolysis (SI Appendix, Fig. S14B).
The Lipid Anchor of PSA Is Required for Its Immunomodulatory Activities.
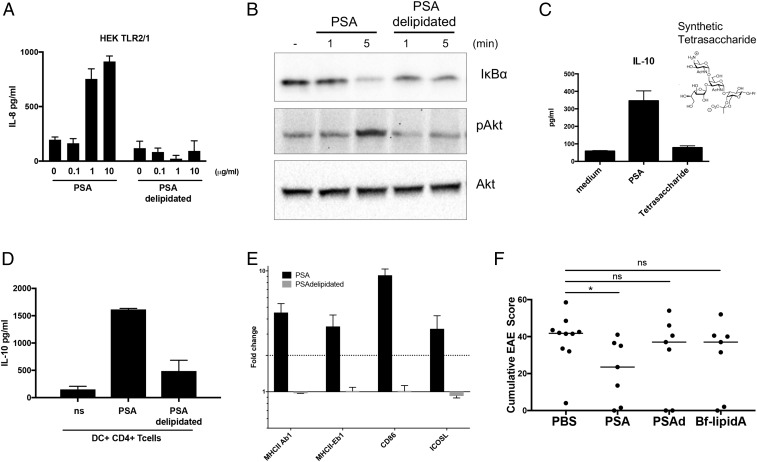
To determine the role of the lipid anchor in PSA function, we compared the induction of TLR2/TLR1-expressing HEK cells by PSA with that by delipidated PSA. IL-8 expression in response to PSA is significantly reduced when PSA is delipidated by acid treatment with subsequent chloroform/methanol extraction to remove the lipid (Fig. 6A). Degradation of IkB and phosphorylation of Akt are not observed when the cells are stimulated with delipidated PSA; thus TLR and PI3K pathway signaling (Fig. 6B) is also abrogated when PSA does not contain its lipid anchor. In addition, expression of the NF-κB–dependent gene product iNOS is substantially diminished when the cells are incubated with delipidated PSA (SI Appendix, Fig. S15). We performed the DC–T cell coculture assay, using a chemically synthesized tetrasaccharide repeating unit of PSA (39). This tetrasaccharide is not sufficient to induce IL-10 production by CD4+ T cells (Fig. 6C). Furthermore, chloroform extraction of acid-treated PSA, but not of native PSA (preacid treatment), abrogates IL-10 production in DC–T cell cocultures (Fig. 6D). We have previously shown that PSA-mediated cognate DC–CD4+ T cell interactions depend on MHCII and costimulatory molecules like ICOSL and CD86. Intact PSA robustly induces transcription of a number of important costimulatory molecules as well as MHCII. When PSA is delipidated by acid hydrolysis, induction of MHCII and costimulatory molecules is robustly down-regulated (Fig. 6E). Furthermore, delipidated PSA eliminates PSA-mediated protection against EAE in vivo (Fig. 6F). These data demonstrate that the very small amount of covalently bound lipid in PSA is critical for the initiation of antiinflammatory immune responses. To determine whether the lipid alone is sufficient for PSA activity, we isolated lipid A from an acapsular mutant of B. fragilis that does not synthesize the polysaccharide portion of PSA. By LC/MS analysis, we found that B. fragilis lipid A from this mutant had a composition similar to that of the covalently attached anchoring lipid A of PSA (SI Appendix, Table S1). In the EAE model, B. fragilis lipid A alone was not sufficient to protect animals from disease (Fig. 6F). Our data show that PSA’s function depends on both the lipid A and the polysaccharide.
Fig. 6.
Immunomodulation by PSA requires its lipid anchor. (A) ELISA analysis of IL-8 production from HEK cells stably expressing the TLR2/TLR1 heterodimer and stimulated with PSA or delipidated PSA for 24 h. Error bars indicate SD. These data are average of 2 independent experiments. (B) Western blot analysis of 50 μg of protein extract from BMDMs stimulated with PSA or delipidated PSA. Degradation of IκB was detected with antibody to IκBα and phosphorylation of Akt with antibody to pAkt. Anti-Akt was used as a loading control. (C and D) IL-10 liberation from CD4+ T cells cocultured with splenic DCs. IL-10 levels were measured by ELISA of culture supernatants of splenic DCs cocultured with CD4+ T cells for 5 d in the presence of anti-CD3. Error bars indicate SD values. These data are representative of 2 independent experiments. (E) Real-time PCR analysis of PSA- and delipidated PSA–stimulated DCs for expression levels of costimulatory molecules. Error bars indicate SEM values. These data are average of 2 independent experiments. (F) Cumulative EAE scores of female C57BL/6J mice treated with PBS, PSA, delipidated PSA, and B. fragilis lipid A. Bars indicate the median. Scores were assessed for statistical significance by t test. *P < 0.05; ns, not significant. These data are average of 2 independent experiments.
Discussion
Until recently, host–microbe interactions have been studied primarily from the perspective of infectious diseases; thus, these studies have focused on the microbial products of pathogens. However, the molecular basis and mechanisms responsible for host–symbiont interactions have not been elucidated. In the last decade, high-throughput metagenomic projects have associated a number of microbial taxa in the gut with a wide range of localized and systemic diseases (40). Recently, the field of microbiome research has turned its attention to identifying specific organisms or molecules that modulate disease or treatment (41–43). In addition to the latter approach, which will help to move the field from association to causation, an understanding of the molecular mechanisms of symbiotic molecules is essential in developing safe therapeutic agents derived from symbiotic microbes.
Collaboration of Pattern Recognition Receptors in Response to a Symbiosis Factor.
A number of host defense mechanisms have been discovered wherein the immune response to pathogenic antigens and secreted virulence factors leads to inflammation. While the immune system robustly reacts to pathogenic microbes in order to eliminate them, it peacefully coexists with trillions of symbiotic organisms inhabiting the body. It was originally thought that the main mechanism of tolerance was microbial avoidance of detection on mucosal surfaces. However, more recent studies have revealed the existence of complex molecular communication between the symbionts and the host immune system (44).
PSA from B. fragilis is the archetypal symbiosis factor from the human gut microbiota. Our previous studies have shown that PSA induces immunologic tolerance via IL-10–producing Tregs and confers protection in animal models of inflammation in both the gut and the central nervous system (6–9). Prior reports from our group and others have demonstrated the TLR2-dependence of PSA-mediated protection in animal models of disease. Because TLR2-mediated responses are canonically proinflammatory, we sought to understand the signaling mechanisms leading to TLR2 dependence with an antiinflammatory/tolerogenic outcome. We show here that PSA requires the TLR2/TLR1 heterodimer—but not TLR2/TLR6—to activate APCs for the downstream induction of Tregs that produce IL-10. However, TLR2/TLR1 heterodimerization alone does not explain why PSA elicits antiinflammatory responses. In general, TLR2/TLR1 heterodimer-induced activation of the NF-κB pathway results in proinflammatory responses. Therefore, we predicted that PSA might be stimulating other receptors and signaling pathways to produce the set of signals required for an antiinflammatory response. One family of receptors recognizing carbohydrate structures is the C-type lectin receptor family. In human cells, DC-SIGN—a member of the CLR family—responds to PSA (25). We found that the murine homolog of human DC-SIGN, SIGNR3 (CD209d), is not required for PSA-mediated immune responses. This discrepancy may be due to differences between the innate receptors on human vs. mouse cells or to the criteria used to determine activation. Instead, our study shows that PSA activates the Dectin-1 pathway as well as TLR2/TLR1, that the 2 pathways are activated independently but in parallel with each other, and that both pathways are required for immunomodulatory activity in vitro and in an in vivo EAE model. Despite the questionable purity of yeast zymosan preparations, they are reported to be recognized by both Dectin-1 and TLR2, with the consequent induction of NF-κB–dependent proinflammatory responses (22). This observation suggests that PSA uses other mechanisms in addition to TLR2 and Dectin-1 collaboration. We found that PSA induces the PI3K pathway and that activation of this pathway requires both TLR2 and Dectin-1. In the presence of PI3K inhibitors or a kinase-dead mutation (p110δD910A/D910A) in APCs, PSA-induced IL-10 production from T cells is abrogated. This finding is consistent with studies showing that the PI3K pathway can act as a negative regulator of TLR signaling and proinflammatory immune responses (30). In addition, a critical role for the PI3K pathway in the gut has been documented in p110δD910A/D910A mice, which develop chronic colitis in vivo (31). Our findings suggest that PSA-like symbiosis factors contribute to PI3K-dependent protection against gut inflammation. Simultaneous activation of different pattern recognition receptors by different ligands, resulting in synergistic collaboration of different pathways, has been shown previously (45). However, PSA is unique as an example of a single bacterial molecule concurrently activating TLR2/TLR1 and Dectin-1 to initiate a complex signaling cascade involving the PI3K pathway to induce antiinflammatory responses.
Transcriptional Regulation of Antiinflammatory Responses by PSA-Driven Signaling Pathways.
The kinase Akt is a key player in the PI3K pathway. Phosphorylation and inactivation of the transcription factor GSK3β by Akt result in the inhibition of NF-κB–mediated transcription of proinflammatory cytokines and in an increase in the production of IL-10 (33). GSK3β inhibits the association of CBP and CREB and promotes NF-κB/CBP interaction for proinflammatory gene transcription. We have found that PSA is distinguished from Pam3CSK4 via inactivation of GSK3β. GSK3β remains phosphorylated and inactive in response to PSA, whereas Pam3CSK4 results in GSK3β that is not phosphorylated and is therefore active. The latter situation skews the transcriptional response from NF-κB/CBP-dependent proinflammatory genes to CREB/CBP-dependent antiinflammatory genes. We have previously reported that costimulatory molecules (CD86 and ICOSL) as well as MHCII are essential for PSA immunomodulation (6). Here we show that the transcription of MHCII and ICOSL, both of which have CREB binding sites in their promoter regions, is CREB-dependent. In contrast, transcription of CD86 and iNOS, which do not have CREB binding sites, does not require CREB.
Structural Features of Capsular Polysaccharides of Symbionts.
Capsular polysaccharides are critical to bacterial colonization, virulence, and fitness in the organisms’ physical niche and to bacterial interactions with the host immune system (46–49). PSA is the most abundantly expressed of the 8 polysaccharides that form the capsular complex of B. fragilis (50). Four sugars compose a single repeating unit of PSA (51); each molecule, as produced by the microbe, has 100 to 200 of these repeating units, with an average molecular size of ∼150 kDa. Many gram-negative bacteria have 2 important polysaccharides on their outer surface: An LPS and a capsular polysaccharide. The LPS—a nearly universal structure in gram-negative bacteria—has a lipid terminus (lipid A) that is inserted into the outer membrane and is the canonical TLR4 agonist. Among capsules, a membrane-inserted lipid has been resolved in only a few instances. These membrane-inserted lipids (typically phospholipids) anchor the hydrophilic polysaccharide capsule to the outer membrane by covalently bound lipid moieties located at the reducing end of the polysaccharide (52–55).
It is interesting that PSA is anchored through a modified lipid A structure and yet appears as a large molecular-size capsule on the organism’s surface and does not have the typical LPS ladder pattern on SDS gels. Structurally, the lipid anchor of PSA belongs to the lipid A family, but it has a number of structural features that distinguish it from classic E. coli lipid A. The latter is uniform in structure, hexacylated, and diphosphorylated and has 14-carbon straight acyl chains. The lipid anchor of PSA has a high degree of structural variability, with many analogs existing in culture. These molecules vary in acylation (penta-, tetra-, and tri-), phosphorylation (no phosphate groups or only 1), and acyl chain length (15 to 18 carbons), and the acyl chains can be branched. Such structural variability has also been reported in other gut or oral commensals (56). Significant structural unpredictability might explain the discrepancies observed among TLR2/TLR4-mediated responses to the lipid A of B. fragilis and the lipid A of other symbionts (57).
Recently, it was reported that symbiont-originated glycolipids belonging to the lipid A family are crucial for bacterial fitness in the host gut (58) and have an important role in modulating host immunity (59). The specific structural differences between symbiont lipid A and pathogen lipid A are likely to be key factors in the vast differences in biologic activity. Although lipid A of LPS from pathogenic bacteria has been extensively studied for inflammatory functions, B. fragilis lipid A is known to have little proinflammatory capacity (60). Bacteroides dorei lipid A, which is structurally similar to B. fragilis lipid A, actually antagonizes inflammation induced by E. coli LPS (59). We have now found that B. fragilis lipid A makes a key mechanistic contribution to the capsule’s immunologic function. While many TLR2 ligands contain carbohydrate moieties, the crystal structure of TLR2 would best appear to bind lipids (61, 62), and it is usually the lipid portion of TLR2 ligands that activates downstream signaling. B. fragilis lipid A is required for activation of APCs through TLR2; because TLR2 activation is required for activation of downstream signaling pathways, B. fragilis lipid A is required for activation of the downstream pathways by PSA. In our proposed model, TLR2/TLR1-dependent activation by the B. fragilis lipid A moiety of PSA and Dectin-1–dependent activation by the carbohydrate portion of PSA ultimately result in transcription of a number of genes (including those for iNOS, MHCII, and costimulatory molecules like CD86) that are essential for the processing of PSA and its presentation to CD4+ T cells by APCs. Expression of these proteins is critical for MHCII pathway-mediated processing and presentation of the carbohydrate portion of PSA to Tregs. Tregs are key cells in the production of IL-10, the major antiinflammatory outcome of PSA stimulation of the immune system. In future studies, we will elucidate the structure–function relations of different chemical variations of this lipid moiety.
PSA’s unusual structure (i.e., a zwitterionic carbohydrate repeating unit conjugated to a lipid that belongs to the lipid A family) defines a class of symbiosis factors occurring in different bacterial phyla commonly found in the microbiota. A recent report identifies a number of diverse bacteria that are putative producers of PSA-like molecules, and our findings are most likely applicable to these molecules (11). We have uncovered a lipid anchor of a prominent symbiosis factor belonging to the lipid A family of gram-negative bacterial membrane-inserted lipids that are essential for immunologic signaling. Our study reveals that B. fragilis lipid A collaboratively initiates innate signaling mechanisms leading to a tolerogenic response that diminishes the severity of inflammatory disease. Since similar lipids are found in many Bacteroidetes, this paradigm for microbe–host communication would appear to qualify as a pattern (i.e., a symbiont-associated molecular pattern) that the immune system recognizes in symbionts. Further structural and functional investigation of this lipid moiety will continue to unravel the host-protective mechanisms of some prototypic symbiotic factors. Insights into the mechanisms used by the microbiota to modulate immune responses in health and disease have important implications for the development of new immunotherapies.
Materials and Methods
Animals.
C57BL/6J and TLR2-knockout mice were purchased from the Jackson Laboratory and maintained at Harvard Medical School’s animal facilities. All experiments with animals were approved by the Harvard Medical Area Standing Committee on Animals (animal protocol no. 8604781). TLR1- and TLR6-knockout mice were obtained from Egil Lien, University of Massachusetts Medical School, Worcester, MA. Organs and cells from PI3K-dead (p110δD910A/D910A) animals were obtained from S.E.P., University of North Carolina, Chapel Hill, NC. SIGNR3 (C57BL/6-Cd209dtm1.1Cfg/Mmucd) knockout mice were purchased from the Mutant Mouse Resource Research Center at the Jackson Laboratory. Dectin-1 organs and tissues were obtained from Hidde Ploegh, Harvard Medical School, Boston, MA. Dectin-1 knockout animals and immortalized macrophage cell lines were obtained from Stuart Levitz, University of Massachusetts Medical School, Worcester, MA, with the permission of Gordon Brown, University of Aberdeen, Scotland.
B. fragilis PSA Purification.
PSA was purified from a mutant B. fragilis strain overexpressing PSA (∆mpiM44) (1) by previously described methods (2). The purity of PSA was confirmed by SDS/PAGE, 1H-NMR spectroscopy, and UV wavelength scans.
Cell Culture.
DCs were isolated from spleens of untreated mice with CD11c microbeads (Miltenyi). To prepare PDCs, BMDCs were grown in complete medium in the presence of Flt3L (RandD). PDCs were isolated with mPDCA-1 microbeads (Miltenyi). CD4+ T cells were isolated with a Mouse T Cell CD4 Subset Column Kit (R&D Systems). For coculture experiments, 2 × 104 DCs were mixed with 105 T cells in the presence of anti-CD3 (1 μg/mL), and the cells were cultured with PSA (50 to 100 μg/mL) for 5 d at 37 °C in 5% CO2 in 96-well round-bottom plates. To prepare BMDCs, bone marrow was collected from femurs and tibias of 6- to 8-wk-old WT or knockout mice. Cells were grown in complete RPMI medium (Gibco) containing granulocyte-macrophage colony-stimulating factor (GM-CSF, 10 ng/mL; Invitrogen). After 4 d of culture, the old medium was removed and fresh medium containing GM-CSF (10 ng/mL) was added to the cells. On day 8, adherent and nonadherent cells were collected and used in the experiments. To prepare BMDMs, bone marrow was collected as described for BMDCs. The cells were grown in DMEM containing d-glucose (4.5 mg/liter), l-glutamine (4 mM), and sodium pyruvate (110 mg/L; Invitrogen) and supplemented with 10% heat-inactivated FBS and 20% cell line L929-conditioned medium for 5 to 6 d, with addition of fresh medium on day 2. HEK cells were purchased from InvivoGen and maintained according to the company’s instructions.
Western Blot.
Cells were lysed in lysis buffer (20 mM Tris at pH 7.6, 150 mM NaCl, 25 mM glycerophosphate, 2 mM EDTA, 10% glycerol, 1% Triton X-100, 1 mM DTT, 1 mM NaVO4, and protease inhibitors); total protein extracts were separated by SDS/PAGE and transferred to a PVDF membrane. Antibodies used for immunoblotting were as follows: anti-IκBα, anti-actin, and anti-iNOS (all from Santa Cruz Biotechnology); and anti-phosphoSyk, anti-phosphoAkt, anti-Akt, anti-phosphoGSK3β, and anti-GSK3β (all from Cell Signaling Technology). Relative density was measured with ImageJ software.
Real-Time PCR Analysis.
Total RNA from BMDCs was isolated from unfrozen cells with an RNeasy Mini Kit, dissolved in 20 μL of RNase-free water, and stored at −20 °C. RNA was quantified spectrophotometrically with a NanoDrop ND-1000 photometer. Expression of mRNA was assessed by 2-step SYBR Green I relative real-time PCR (iCycler iQ System, Bio-Rad). In brief, total RNA (1 μg) was converted into first-strand cDNA with the SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen) according to the manufacturer’s protocol. Use of random hexamers leads to specific transcriptions of mRNA. The PCR amplification reactions contained 4 μL of first-strand cDNA diluted at 1.3 ng/µL and mixed with 5 μL of iQ SYBR Green Supermix (Bio-Rad; master mixture: 2× mix containing SYBR Green I dye, hot-start iTaq DNA polymerase, optimized buffer, and dNTPs qualified for quantitative PCR) and 1 µL of each of the specific primer pairs at 10 µM in a final reaction volume of 10 μL. All reactions were performed in duplicate. No-template controls were included on each plate for each primer pair. The PCR profile consisted of initial denaturation for 5 min at 95 °C followed by 45 cycles of 10 s at 95 °C (denaturing), 10 s at 60 °C (annealing), and 10 s at 72 °C (extension). Cumulative fluorescence was measured at the end of the extension phase of each cycle. Product-specific amplification was confirmed by melting curve analysis. The expression of all genes was calculated with the β-actin gene ACTB as a reference (2[−Cttarget gene]/2[−CtACTB]) and recorded as fold-changes in comparison to PBS-treated samples.
Zinc Staining.
PSA was run on a 10% or 16.5% Tris/Tricine SDS polyacrylamide gel (BioRad #345-0063). The gel was incubated first in 100 °C water with gentle rocking for a total of 3 × 15 min and then in 200 mL of 10 mM zinc sulfate (Sigma #83265) for 15 min. The zinc solution was discarded, and 100 mL of a 200 mM imidazole (Sigma #I202) solution was added to the gel. The gel was placed on a black surface and photographed with white light.
EAE Induction and PSA Treatment.
Female C57BL/6J mice (Jackson Laboratory) were purchased at 6 to 8 wk of age and maintained in our facility for at least 1 wk before experiments. Mice were treated with PSA, and EAE was induced with MOG (myelin–oligodendrocyte–glycoprotein), as previously reported (8, 9). In brief, mice received 50 to 100 μg of purified PSA by oral gavage 3 times a week throughout the experiment, starting 2 wk before EAE induction. Mice were challenged subcutaneously with 200 μg of MOG (Peptides International) in 250 mL of complete Freund’s adjuvant (Sigma) fortified with Mycobacterium tuberculosis H37 Ra antigen (2.5 mg/mL; BD, Difco). On days 0 and 2, challenged mice received 300 ng of Bordetella pertussis toxin by intraperitoneal. injection (List Biological). Disease progression was scored daily (0, no clinical signs; 0.5, partially limp tail; 1, paralyzed tail; 2, loss of coordination and hind-limb paresis; 2.5, paralysis of 1 hind limb; 3, paralysis of both hind limbs; 3.5, paralysis of both hind limbs and forelimb weakness; 4, forelimb and hind-limb paralysis; and 5, moribund state) (63). Mice with a score of 4 were killed.
Structural Analysis of the Lipid Anchor of PSA with LC-MS/MS.
Purified PSA was hydrolyzed with 2% acetic acid at 90 °C for 2 h. The sample was neutralized and extracted with chloroform. The lower organic phase was collected, dried under nitrogen, and resuspended in 67% in isopropanol solution. Reverse-phase ultra-HPLCy (Thermo Vanquish)/MS/MS (Thermo Q Exactive Orbitrap) equipped with Phenomenex Kintex C8 (75 × 2.1mm × 2.6u) was used for separation and analysis. Water:isopropanol gradients of 65:35 to 85:15, with 10 mM ammonium formate as buffer, were applied for separation and elution of lipid A species in 10 mM formic acid.
Statistical Analysis.
P values were calculated by the 2-tailed nonparametric Mann–Whitney U test; *P < 0.05, **P < 0.01, and ***P < 0.001.
Data Availability.
All data are contained in the paper.
Supplementary Material
Acknowledgments
This work was supported by Grants K01-DK102771 (National Institute of Diabetes and Digestive and Kidney Diseases), U19AI109764 (NIH-Centers of Excellence for Translaational Research), and W81XWH1910626 (Department of Defense), and by funding from the European Union’s Horizon 2020 Research and Innovation Programme under Marie Skłodowska Curie Grant Agreement 661138.
Footnotes
The authors declare no competing interest.
See Profile on page 26144.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915978116/-/DCSupplemental.
References
- 1.Erturk-Hasdemir D., Kasper D. L., Resident commensals shaping immunity. Curr. Opin. Immunol. 25, 450–455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chung H., et al. , Gut immune maturation depends on colonization with a host-specific microbiota. Cell 149, 1578–1593 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erturk-Hasdemir D., Kasper D. L., Finding a needle in a haystack: Bacteroides fragilis polysaccharide A as the archetypical symbiosis factor. Ann. N. Y. Acad. Sci. 1417, 116–129 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Mazmanian S. K., Liu C. H., Tzianabos A. O., Kasper D. L., An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Sommese L., et al. , Evidence of Bacteroides fragilis protection from Bartonella henselae-induced damage. PLoS One 7, e49653 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dasgupta S., Erturk-Hasdemir D., Ochoa-Reparaz J., Reinecker H.-C., Kasper D. L., Plasmacytoid dendritic cells mediate anti-inflammatory responses to a gut commensal molecule via both innate and adaptive mechanisms. Cell Host Microbe 15, 413–423 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazmanian S. K., Round J. L., Kasper D. L., A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Ochoa-Repáraz J., et al. , A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal Immunol. 3, 487–495 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Ochoa-Repáraz J., et al. , Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J. Immunol. 185, 4101–4108 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Wang Y., et al. , An intestinal commensal symbiosis factor controls neuroinflammation via TLR2-mediated CD39 signalling. Nat. Commun. 5, 4432 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neff C. P., et al. , Diverse intestinal bacteria contain putative zwitterionic capsular polysaccharides with anti-inflammatory properties. Cell Host Microbe 20, 535–547 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Round J. L., Mazmanian S. K., Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. U.S.A. 107, 12204–12209 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q., et al. , A bacterial carbohydrate links innate and adaptive responses through Toll-like receptor 2. J. Exp. Med. 203, 2853–2863 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen Y., et al. , Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe 12, 509–520 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nair S., et al. , The PPE18 of Mycobacterium tuberculosis interacts with TLR2 and activates IL-10 induction in macrophage. J. Immunol. 183, 6269–6281 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Danne C., et al. , A large polysaccharide produced by Helicobacter hepaticus induces an anti-inflammatory gene signature in macrophages. Cell Host Microbe 22, 733–745.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mishima Y., et al. , Microbiota maintain colonic homeostasis by activating TLR2/MyD88/PI3K signaling in IL-10-producing regulatory B cells. J. Clin. Invest. 130, 3702–3716 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cobb B. A., Wang Q., Tzianabos A. O., Kasper D. L., Polysaccharide processing and presentation by the MHCII pathway. Cell 117, 677–687 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duan J., Avci F. Y., Kasper D. L., Microbial carbohydrate depolymerization by antigen-presenting cells: Deamination prior to presentation by the MHCII pathway. Proc. Natl. Acad. Sci. U.S.A. 105, 5183–5188 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duan J., Kasper D. L., Oxidative depolymerization of polysaccharides by reactive oxygen/nitrogen species. Glycobiology 21, 401–409 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sancho D., Reis e Sousa C., Signaling by myeloid C-type lectin receptors in immunity and homeostasis. Annu. Rev. Immunol. 30, 491–529 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gantner B. N., Simmons R. M., Canavera S. J., Akira S., Underhill D. M., Collaborative induction of inflammatory responses by dectin-1 and Toll-like receptor 2. J. Exp. Med. 197, 1107–1117 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dillon S., et al. , Yeast zymosan, a stimulus for TLR2 and dectin-1, induces regulatory antigen-presenting cells and immunological tolerance. J. Clin. Invest. 116, 916–928 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerrigan A. M., Brown G. D., Syk-coupled C-type lectins in immunity. Trends Immunol. 32, 151–156 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bloem K., et al. , Interaction of the capsular polysaccharide A from Bacteroides fragilis with DC-SIGN on human dendritic cells is necessary for its processing and presentation to T cells. Front. Immunol. 4, 103 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hazeki K., Nigorikawa K., Hazeki O., Role of phosphoinositide 3-kinase in innate immunity. Biol. Pharm. Bull. 30, 1617–1623 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Arbibe L., et al. , Toll-like receptor 2-mediated NF-kappa B activation requires a Rac1-dependent pathway. Nat. Immunol. 1, 533–540 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Santos-Sierra S., et al. , Mal connects TLR2 to PI3Kinase activation and phagocyte polarization. EMBO J. 28, 2018–2027 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin M., et al. , Role of the phosphatidylinositol 3 kinase-Akt pathway in the regulation of IL-10 and IL-12 by Porphyromonas gingivalis lipopolysaccharide. J. Immunol. 171, 717–725 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Fukao T., et al. , PI3K-mediated negative feedback regulation of IL-12 production in DCs. Nat. Immunol. 3, 875–881 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Uno J. K., et al. , Altered macrophage function contributes to colitis in mice defective in the phosphoinositide-3 kinase subunit p110δ. Gastroenterology 139, 1642–1653, 1653.e1–1653.e6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cross D. A. E., Alessi D. R., Cohen P., Andjelkovich M., Hemmings B. A., Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 378, 785–789 (1995). [DOI] [PubMed] [Google Scholar]
- 33.Martin M., Rehani K., Jope R. S., Michalek S. M., Toll-like receptor-mediated cytokine production is differentially regulated by glycogen synthase kinase 3. Nat. Immunol. 6, 777–784 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grimes C. A., Jope R. S., CREB DNA binding activity is inhibited by glycogen synthase kinase-3 beta and facilitated by lithium. J. Neurochem. 78, 1219–1232 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woodgett J. R., Ohashi P. S., GSK3: An in-Toll-erant protein kinase? Nat. Immunol. 6, 751–752 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Corcoran A. T., Annuk H., Moran A. P., The structure of the lipid anchor of Campylobacter jejuni polysaccharide. FEMS Microbiol. Lett. 257, 228–235 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Tzeng Y.-L., et al. , Translocation and surface expression of lipidated serogroup B capsular Polysaccharide in Neisseria meningitidis. Infect. Immun. 73, 1491–1505 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weintraub A., Zähringer U., Wollenweber H. W., Seydel U., Rietschel E. T., Structural characterization of the lipid A component of Bacteroides fragilis strain NCTC 9343 lipopolysaccharide. Eur. J. Biochem. 183, 425–431 (1989). [DOI] [PubMed] [Google Scholar]
- 39.Pragani R., Seeberger P. H., Total synthesis of the Bacteroides fragilis zwitterionic polysaccharide A1 repeating unit. J. Am. Chem. Soc. 133, 102–107 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Belizário J. E., Napolitano M., Human microbiomes and their roles in dysbiosis, common diseases, and novel therapeutic approaches. Front. Microbiol. 6, 1050 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vetizou M., et al. , Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sivan A., et al. , Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Surana N. K., Kasper D. L., Moving beyond microbiome-wide associations to causal microbe identification. Nature 552, 244–247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Round J. L., Mazmanian S. K., The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9, 313–323 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trinchieri G., Sher A., Cooperation of Toll-like receptor signals in innate immune defence. Nat. Rev. Immunol. 7, 179–190 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Coyne M. J., et al. , Polysaccharide biosynthesis locus required for virulence of Bacteroides fragilis. Infect. Immun. 69, 4342–4350 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coyne M. J., Chatzidaki-Livanis M., Paoletti L. C., Comstock L. E., Role of glycan synthesis in colonization of the mammalian gut by the bacterial symbiont Bacteroides fragilis. Proc. Natl. Acad. Sci. U.S.A. 105, 13099–13104 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Comstock L. E., Kasper D. L., Bacterial glycans: Key mediators of diverse host immune responses. Cell 126, 847–850 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Comstock L. E., Importance of glycans to the host-bacteroides mutualism in the mammalian intestine. Cell Host Microbe 5, 522–526 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Krinos C. M., et al. , Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature 414, 555–558 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Baumann H., Tzianabos A. O., Brisson J. R., Kasper D. L., Jennings H. J., Structural elucidation of two capsular polysaccharides from one strain of Bacteroides fragilis using high-resolution NMR spectroscopy. Biochemistry 31, 4081–4089 (1992). [DOI] [PubMed] [Google Scholar]
- 52.Willis L. M., et al. , Conserved glycolipid termini in capsular polysaccharides synthesized by ATP-binding cassette transporter-dependent pathways in Gram-negative pathogens. Proc. Natl. Acad. Sci. U.S.A. 110, 7868–7873 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gotschlich E. C., Fraser B. A., Nishimura O., Robbins J. B., Liu T. Y., Lipid on capsular polysaccharides of gram-negative bacteria. J. Biol. Chem. 256, 8915–8921 (1981). [PubMed] [Google Scholar]
- 54.Kuo J. S., Doelling V. W., Graveline J. F., McCoy D. W., Evidence for covalent attachment of phospholipid to the capsular polysaccharide of Haemophilus influenzae type b. J. Bacteriol. 163, 769–773 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neu T. R., Significance of bacterial surface-active compounds in interaction of bacteria with interfaces. Microbiol. Rev. 60, 151–166 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Slocum C., et al. , Distinct lipid a moieties contribute to pathogen-induced site-specific vascular inflammation. PLoS Pathog. 10, e1004215 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alhawi M., Stewart J., Erridge C., Patrick S., Poxton I. R., Bacteroides fragilis signals through Toll-like receptor (TLR) 2 and not through TLR4. J. Med. Microbiol. 58, 1015–1022 (2009). [DOI] [PubMed] [Google Scholar]
- 58.Cullen T. W., et al. , Gut microbiota. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science 347, 170–175 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vatanen T., et al. ; DIABIMMUNE Study Group , Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 165, 842–853 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kasper D. L., Chemical and biological characterization of the lipopolysaccharide of Bacteroides fragilis subspecies fragilis. J. Infect. Dis. 134, 59–66 (1976). [DOI] [PubMed] [Google Scholar]
- 61.Jin M. S., et al. , Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell 130, 1071–1082 (2007). [DOI] [PubMed] [Google Scholar]
- 62.Kang J. Y., et al. , Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer. Immunity 31, 873–884 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Stromnes I. M., Goverman J. M., Active induction of experimental allergic encephalomyelitis. Nat. Protoc. 1, 1810–1819 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained in the paper.