Abstract
Mucor circinelloides is one of the causal agents of mucormycosis, an emerging and high mortality rate fungal infection produced by asexual spores (sporangiospores) of fungi that belong to the order Mucorales. M. circinelloides has served as a model genetic system to understand the virulence mechanism of this infection. Although the G-protein signaling cascade plays crucial roles in virulence in many pathogenic fungi, its roles in Mucorales are yet to be elucidated. Previous study found that sporangiospore size and calcineurin are related to the virulence in Mucor, in which larger spores are more virulent in an animal mucormycosis model and loss of a calcineurin A catalytic subunit CnaA results in larger spore production and virulent phenotype. The M. circinelloides genome is known to harbor twelve gpa (gpa1 to gpa12) encoding G-protein alpha subunits and the transcripts of the gpa11 and gpa12 comprise nearly 72% of all twelve gpa genes transcript in spores. In this study we demonstrated that loss of function of Gpa11 and Gpa12 led to larger spore size associated with reduced activation of the calcineurin pathway. Interestingly, we found lower levels of the cnaA mRNAs in sporangiospores from the Δgpa12 and double Δgpa11/Δgpa12 mutant strains compared to wild-type and the ΔcnaA mutant had significantly lower gpa11 and gpa12 mRNA levels compared to wild-type. However, in contrast to the high virulence showed by the large spores of ΔcnaA, the spores from Δgpa11/Δgpa12 were avirulent and produced lower tissue invasion and cellular damage, suggesting that the gpa11 and gpa12 define a signal pathway with two branches. One of the branches controls spore size through regulation of calcineurin pathway, whereas virulences is controlled by an independent pathway. This virulence-related regulatory pathway could control the expression of genes involved in cellular responses important for virulence, since sporangiospores of Δgpa11/Δgpa12 were less resistant to oxidative stress and phagocytosis by macrophages than the ΔcnaA and wild-type strains. The characterization of this pathway could contribute to decipher the signals and mechanism used by Mucorales to produce mucormycosis.
Introduction
Mucor circinelloides is a basal fungus that belongs to the Phylum Zygomycota [1]. This organism produces three types of spores, zygospores are produced by mating of hyphae of opposite sex: (+) and (-). Arthrospores are generated by hyphae in submerged cultures after the end of the exponential growth or under unfavorable growth conditions. Sporangiospores are produced by asexual reproduction on solid substrate in large number [2, 3]. Several growth conditions determine whether M. circinelloides sporangiospores (spores) will produce mycelium or yeast; the most well-studied being the presence of carbon source and atmospheric conditions during spore germination [4–6]. M. circinelloides is one of the etiological agents of the mucormycosis, a fungal infection that affects mainly immunocompromised patients [7–8]. This infection has called medical attention as its incidence has risen worldwide recently [9–11], and relatively high mortality rates have been reported [12–13]. In this sense, the fate of vegetative cells in M. circinelloides has implications for infectiveness, as the mycelial stage usually shows virulent morphology [14–15], while the yeast morphology is less virulent [15]. Furthermore, the ability to germinate of the Mucorales sporangiospores in the immunocompromised patient is a critical step for stablishing the infection [12, 16].
Larger spores lead to a faster germination rate that, in turn, correlates with a more virulent phenotype in M. circinelloides [17]. Spore size in M. circinelloides is controlled in part by CnaA, which is the catalytic subunit A of the phosphatase Ca+2-dependent calcineurin [15]. We have previously described that ADP-ribosylation factor-1 (Arf1) dysfunction also increases spore size and virulence and generates defects during the aerobic growth in M. circinelloides [18]. Moreover, additional molecular regulators of dimorphism in M. circinelloides have been reported. Thus, deletion of the calcineurin regulatory subunit CnbB-encoding gene promotes the yeast rather than mycelial form, even in the presence of oxygen, and this mutant is less virulent compared to its wild-type form [15]. Additionally, deletion of arf2 leads to difficulty in the generation of the yeast morphology [18]. In fungi, the protein kinase A (PKA) pathway is a key signaling pathway that controls spore production, spore germination, resistance to different stress conditions, and virulence [19]; deletion of pkaR1, which encodes a regulatory subunit 1 of PKA, leads to a decrease in spore production and shorter hyphae length after yeast-mycelium transition in M. circinelloides [20].
Heterotrimeric G proteins are regulators of the PKA pathway, and their canonical functions are involved in the control of the activity of several effectors; for example, adenylyl cyclase, lipid modifying enzymes like phospholipase C, and ion channels [21–22]. In the ascomycetes Aspergillus nidulans and A. parasiticus, the Gα subunit FadA stimulates the cAMP levels leading to the activation of the PkaC subunit, which ultimately controls the gene transcription involved in sporulation [23–24].
Our group previously reported that the largest number of members of genes encoding heterotrimeric G proteins in the Fungi kingdom is present in M. circinelloides [25]. The twelve Gα-encoding genes (gpa1-12) were grouped into three (I-III) of the four groups of the Gα fungal phylogenetic classification [25–26]. In M. circinelloides, Gpa11 and Gpa12, which belong to group I (Gαs) and group III (Gαi), respectively, showed increased transcript levels during the spore stage (~69% of the total gpa transcript levels), suggesting that these genes have an important role in spore production or germination in this Mucoral [25]. However, the molecular role of these subunits in spore function is still unknown.
The aim of this work was to functionally characterize the role of the heterotrimeric Gα subunits Gpa11 and Gpa12 in sporulation and vegetative growth and determine their role in the virulence of M. circinelloides.
Results
Deletion of gpa11 and gpa12 in M. circinelloides
M. circinelloides exhibits twelve Gα subunits (Gpa1-Gpa12). The mRNA of two of these subunits, Gpa11 and Gpa12, have been reported to accumulate mainly in the spore stage and decrease during vegetative growth [25]. To determine whether these genes are relevant to spore physiology, we created gpa11 and gpa12 deletion mutants. Replacement fragments of gpa11 and gpa12 that contained 5′ upstream and 3′ downstream regions from gpa11 or gpa12 flanking the selective marker pyrG were used to transform protoplasts of the M. circinelloides wild-type strain MU402 (leuA-, pyrG-), which is auxotrophic for leucine and uracil [27], to generate gpa11 and gpa12 deletion strains (Fig 1). Transformants grown in selective medium (MMC) were recovered and subsequent vegetative cycles in selective medium were performed to obtain homokaryotic Δgpa11 and Δgpa12 mutants. After 5 cycles of vegetative growth/sporulation, we obtained two independent transformants for gpa11 and four for gpa12 that showed 100% Ura+ spores, confirming the homokaryosis.
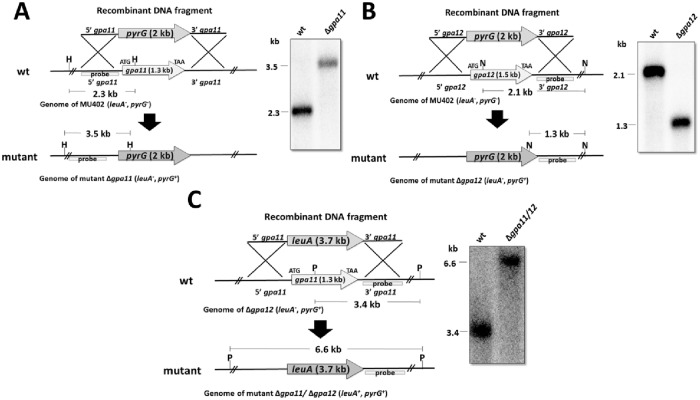
Fig 1. Deletion of gpa11 and gpa12 in M. circinelloides confirmed by Southern-blot.
A) gpa11; B) gpa12; and C) gpa11/gpa12 deletion. The 5′ and 3′ regions (1.1 kb for each) upstream and downstream from the start and stop codons, respectively, of gpa11 or gpa12 were designed to flank the pyrG selection marker for gene deletion (A and B). For the deletion of gpa11 in Δgpa12, the 5′ and 3′ regions (1.1 kb for each) upstream and downstream from the start and stop codons, respectively, of gpa11 were designed to flank leuA (C). For each diagram, the recombinant fragments that were used to transform protoplasts of the strain MU402 (pyrG−, leuA−) are shown. Molecular confirmation by Southern blotting was performed using specific probes for each gpa gene. DNA samples from transformed and parental (MU402) strains were digested with the indicated restriction enzymes (H, HincII; N, NcoI; P, PvuII).
The presence of 2.3-kb and 3.5-kb hybridization fragments in wild-type and Δgpa11, respectively; confirmed the presence of a unique integration event in the desired locus, and homokaryosis of the gpa11 deletant (Fig 1A). Meanwhile, the presence of 2.1-kb and 1.3-kb hybridization fragments in wild-type and gpa12, respectively revealed the mutation in gpa12 (Fig 1B).
A mutant lacking both genes was generated by transformation of the Δgpa12 strain with a replacement fragment containing the selective marker leuA flanked by 5′ upstream and 3′ downstream regions of gpa11. Transformants grown in selective media (without uracil and leucine) were characterized by Southern blot hybridization. A 6.6-kb hybridization band in the selected transformant confirmed the presence of a unique integration event in the desired locus and the homokaryosis of the double Δgpa11/Δgpa12 knockout mutant (Fig 1C).
Expression analysis of gpa11 and gpa12 in spores generated from single and double knockout mutants confirmed the null expression of those genes (Fig 2).
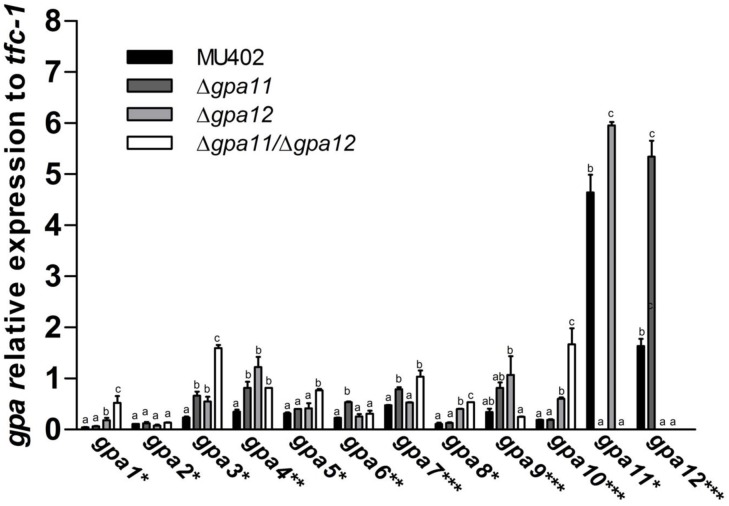
Fig 2. mRNA quantitation of gpa genes in sporangiospores in gpa11 and gpa12 deletion mutants of M. circinelloides.
Total RNA was isolated from spores of the wild-type, Δgpa11, Δgpa12, and Δgpa11/Δgpa12. The mRNA from each gpa gene was determined by qRT-PCR. Asterisks denote the phylogenetic group of the Gpa proteins in M. circinelloides (* I; **-II, and ***-III) [25]. Four independent experiments were performed under the same conditions. Different letters denote statistically significant differences (ANOVA, Fisher, p<0.05).
For the further analysis, we used two independent strains for each gene mutation revealing similar results for these independent strains in all experiment tested. Furthermore, single Δgpa11 and Δgpa12 mutant strains were complemented with their respective wild-type gene by expression in the pEUKA4 vector [28] generating the Δgpa11+gpa11wt and Δgpa12+gpa12wt strains.
Interestingly, the single mutation of each gpa gene provoked a significant upregulation of the other. Thus, deletion of gpa11 led to an ~30% increase in gpa12 mRNA levels, while deletion of gpa12 resulted in an ~226% increase of gpa11 mRNA levels, suggesting a compensatory mechanism (Fig 2, S1 Table). Moreover, single deletion of gpa11 and gpa12 led to a significant increase in the mRNA levels of several gpa genes. The expression of some gpa genes was increased more than two folds in Δgpa11 (gpa3, gpa4, gpa6, gpa9, and gpa12), and in Δgpa12 (gpa1, gpa3, gpa4, gpa8, gpa9, and gpa10) (Fig 2, S1 Table). In addition, deletion of both genes led to a further rise in mRNA levels of some gpa genes, including gpa1 (~11.73 folds), gpa3 (~6.8 folds), and gpa8 (~4.78 folds), which encode proteins that belong to group I of the Gα phylogenetic fungal classification (Gαs). Also, the mRNA levels of a gene, gpa10, encoding a protein belonging to group III (GαI) of the phylogenetic fungal classification was highly increased (~8.77 folds) in the double-gpa mutant strain (Fig 2, S1 Table).
Gpa11 and Gpa12 are involved in sporangiospore size and germination rate in M. circinelloides
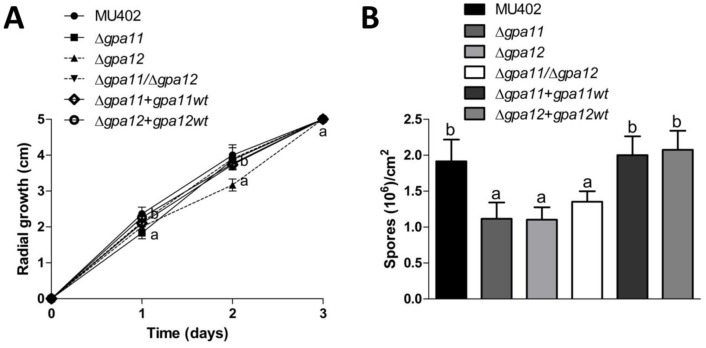
In order to investigate the physiological role of gpa11 and gpa12, radial growth and spore production of the different strains were assessed. The radial growth of all the mutant strains was similar to the wild-type strain at the end of the experiment (Fig 3A). After 5 days of incubation, there was around 42–46% decrease in spore production in all single and double mutant strains compared to the wild-type strain (Fig 3B), indicating that both genes participate in spore production. Additionally, the Δgpa12 strain produced slightly less spores compared to other mutant strain. The Δgpa11+gpa11wt and Δgpa12+gpa12wt complemented strains did not show significant difference in spore production compared to the wild-type strain.
Fig 3. Sporangiospore production in Δgpa11 and Δgpa12 mutants of M. circinelloides.
50 spores were grown in YPG agar plates and A) radial growth was recorded each day during the experiment. B) Spores grown after 5 days were counted. Four independent experiments were performed for each condition. Statistically significant differences are indicated by different letters (ANOVA and Fisher’s tests; p ≤ 0.05).
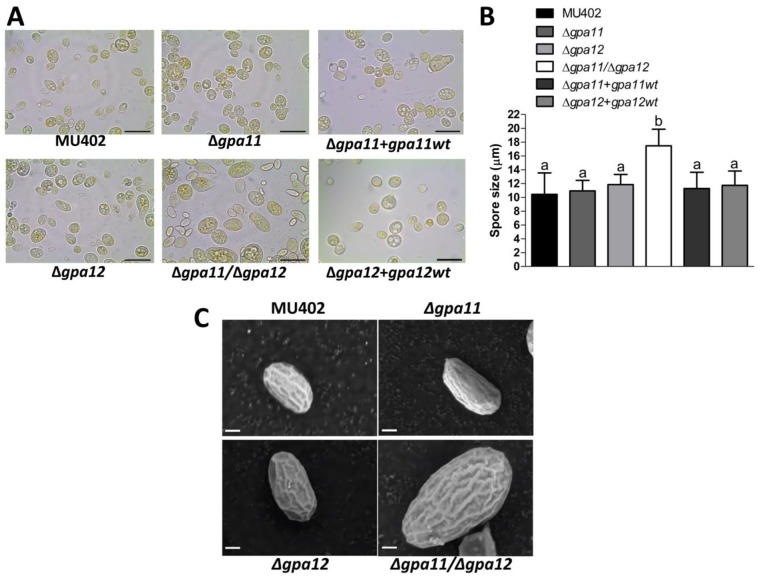
The spores produced on YPG media were observed by light microscopy. The spores from the Δgpa11 mutant strain were morphologically similar (in size) to those produced from the wild-type strain (on average 10.3 and 10.2 μm, respectively) (Fig 4). While the Δgpa12 spores were slightly larger than those of the wild-type strain, at 11.4 μm diameter on average. Interestingly, the Δgpa11/Δgpa12 mutant strain generated the largest spore size, compared to the rest of the strains, at 16.3 μm diameter on average (Fig 4), but some of them were up to 20 μm of size. Interestingly, the Δgpa11+gpa11wt and Δgpa12+gpa12wt complemented strains showed similar size as the wild-type strain (Fig 4A and 4B). The spore size was also evaluated by scanning electron microscopy, confirming that the spores from the Δgpa11/Δgpa12 mutant were larger than those of the other strains and all had a similar surface shape (Fig 4C). Similar results in terms of spore size were observed with independent gpa mutant strains (S1 Fig).
Fig 4. Influence of gpa11 and gpa12 on sporangiospore size in M. circinelloides.
The spores from the different M. circinelloides knockout strains produced in YPG were observed under A) light microscope (100 X), scale bar equal to 20 μm. B) The spore size of each strain was quantified using the Leica Application Suite. C) The spores obtained after five days of incubation on solid YPG media were observed under scanning electron microscope. Representative photographs from the corresponding strains of M. circinelloides under 10,000 magnifications. Scale bar is equal to 1 μm. Statistically significant differences are indicated by different letters (ANOVA and Fisher’s tests; p ≤ 0.05).
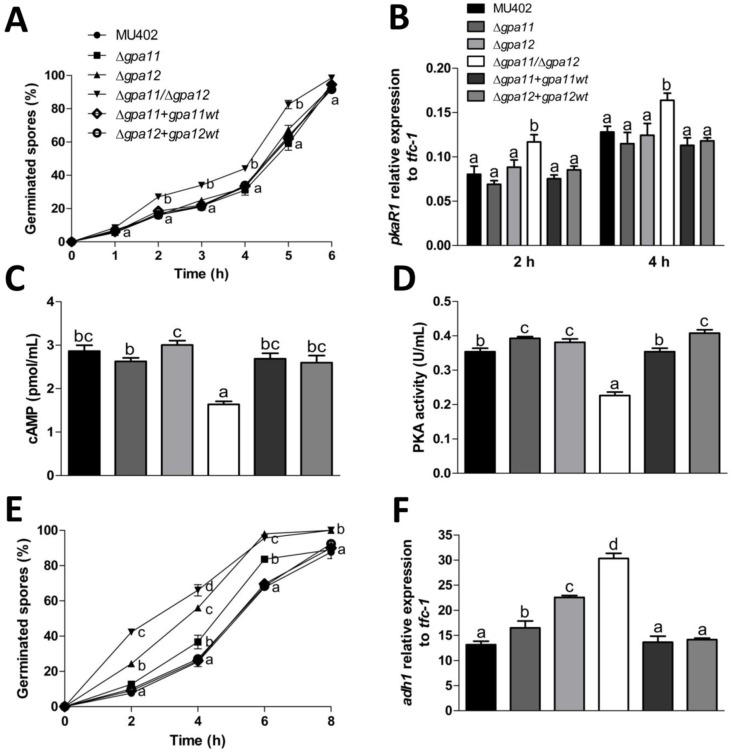
It has been described in M. circinelloides that an increase in spore size correlates with a faster aerobic germination rate [17]. To confirm this assumption, we measured the germination rate of the different strains grown in YPG. The double gpa-mutant strain had a 50% higher germination rate after 2 h of growth compared to the wild-type strain (Fig 5A). However, both single gpa-mutant and complemented strains germinated at very similar rates to the wild-type strain (Fig 5A).
Fig 5. Sporangiospore germination of M. circinelloides gpa11 and gpa12 deletion mutants.
A) Percentage of germinated spores in aerobic conditions from each strain in YPG. Aerobic germinated cells were considered independent of hyphae length or number of mother cells. B) Relative expression of pkaR1, quantified by qRT-PCR using tfc-1 as reference gene from hyphae obtained from each strain grown at 2 and 4 h. ΔCT analysis was performed to compare the mRNA levels between strains. Spores from the different strains from M. circinelloides were inoculated into YPG media and incubated with constant shaking (150 rpm) for 3 h, and C) intracellular cAMP levels, and D) PKA activity were determined. E) Percentage of spores germinated under anaerobic conditions in YPG media. F) Relative expression of adh1 determined from yeast cells at 4 h of growth, ΔCT analysis was performed to compare mRNA levels between strains. Four independent experiments were performed for each condition. Statistically significant differences are indicated by different letters (ANOVA and Fisher’s tests; p ≤ 0.05).
Aerobic germination was corroborated using the gene marker pkaR1, which is a gene expressed preferentially during hyphae growth compared to the yeast or spore stage [29]. Only the Δgpa11/Δgpa12 mutant strain accumulated significantly more pkaR1 mRNA observed in the germinating spores after 2 (40%) and 4 h (28%) in YPG with respect to other strains (Fig 5B). The higher mRNA levels of pkaR1 in the Δgpa11/Δgpa12 mutant strain suggest a repression in the PKA pathway. To probe this hypothesis, we conducted the quantification of cAMP levels and PKA activity in the different strains after 3 h of aerobic germination. In correlation with the mRNA levels, the only strain that showed lower cAMP levels and PKA activity was the Δgpa11/Δgpa12 mutant strain, meanwhile the rest of the strains showed similar levels as the wild-type strain (Fig 5C and 5D).
Germination rate under anaerobic conditions revealed that Δgpa12 and Δgpa11/Δgpa12 mutants germinated faster than the Δgpa12, complemented and wild-type strains. The double gpa-mutant strain demonstrated a higher rate of germination (5.52 folds) after 2 h compared to the wild-type strain (Fig 5E). The mRNA levels of adh1, which is preferentially expressed during anaerobic growth [29–30], were significantly higher in Δgpa11 (21%), Δgpa12 (71%) and Δgpa11/Δgpa12 (130%) strains after 4 h of growth compared to the wild-type strain (Fig 5F).
These results indicated that the products of gpa11 and gpa12 genes are involved in the control of the regulation of sporangiospore size as well as the aerobic and anaerobic germination rates in M. circinelloides.
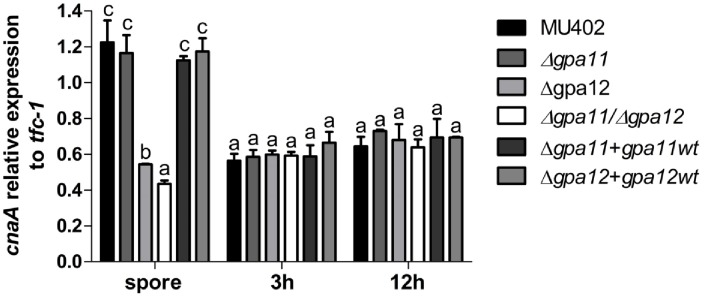
Mutation of gpa12 and the double mutation of gpa11 and gpa12 decreased cnaA mRNA levels in sporangiospores in M. circinelloides
It has been reported that dysfunction of the calcineurin catalytic subunit A (CnaA) leads to an increase in the spore size of M. circinelloides [15]. Based on the larger spore phenotype observed in the Δgpa11/Δgpa12 mutant strain, we performed mRNA quantitation by qRT-PCR of cnaA in spores and during aerobic germination from gpa-mutant strains.
The mRNA levels of cnaA in the wild-type and Δgpa11 mutant spores were similar to each other, while lower mRNA levels were observed in spores from Δgpa12 and Δgpa11/Δgpa12 mutants by around 56 and 64% respectively, compared to Δgpa11, Δgpa11+gpa11wt, Δgpa12+gpa12wt and wild-type strains (Fig 6). Although the mRNA levels of cnaA in mycelium grown for 3 or 12 h in YPG were similar in all strains tested (Fig 6). These results showed that dysfunction of Gpa12 led to down-regulation of cnaA mRNA levels in spores of M. circinelloides. The lower cnaA expression in Δgpa11/Δgpa12 suggested that gpa11 can partially compensate the absence of gpa12.
Fig 6. Quantitation of cnaA mRNA levels in M. circinelloides Δgpa11 and Δgpa12 mutant strains.
Expression of cnaA relative to tfc-1 in total RNA purified from sporangiospores and mycelium grown in aerobic conditions after indicated times in YPG. qRT-PCR was performed to determine the transcript levels of cnaA and a ΔCt analysis was performed to compare the mRNA level between the strains. Four independent experiments were performed for each condition. Statistically significant differences are indicated by different letters (ANOVA and Fisher’s tests; p ≤ 0.05).
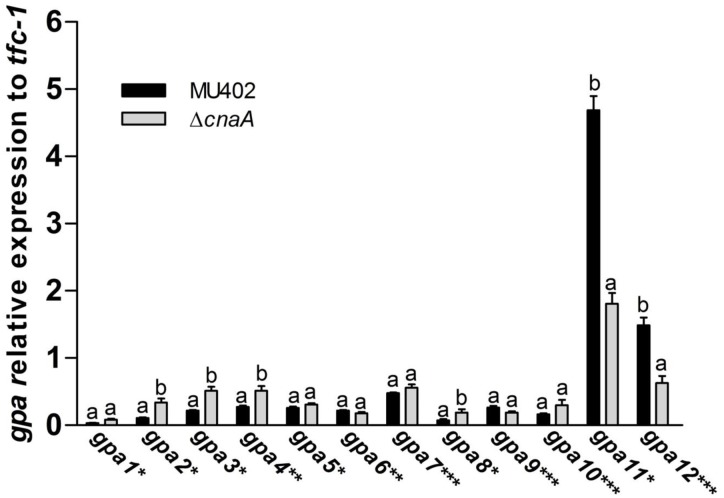
To investigate if cnaA dysfunction affects gpa11 and gpa12 mRNA levels during the spore stage, the transcripts of all gpa were quantified by qRT-PCR in spores of the ΔcnaA mutant form of M. circinelloides. The analysis of transcript levels from all gpa genes revealed that in the spores from the ΔcnaA mutant strain, only gpa11 and gpa12 mRNA levels were significantly decreased (61.4 and 57.9%, respectively) compared to the levels from spores of the wild-type (Fig 7). Furthermore, gpa1, gpa2, gpa3, and gpa8, whose products belong to group I (Gαs), and gpa4 that belongs to group II showed a significant increase in mRNA levels (1.6–2.3 times) compared to the wild-type (Fig 7, S2 Table). Interestingly, the Δgpa11/Δgpa12 mutant strain also showed a significant increase in the mRNA levels of gpa3, and gpa4 (Fig 2). These results suggest a genetic crosstalk between gpa11, gpa12 and cnaA gene products.
Fig 7. Quantitation of gpa mRNA levels in M. circinelloides ΔcnaA.
Changes in levels of mRNA from different gpa genes from total RNA purified from sporangiospores of wild-type (closed bars) and ΔcnaA (open bars). qRT-PCR was carried out to determine the transcript levels of gpa genes, ΔCt analysis was performed to compare the mRNA levels between the strains. Four independent experiments were performed for each condition. Statistically significant differences are indicated by different letters (ANOVA and Fisher’s tests; p ≤ 0.05).
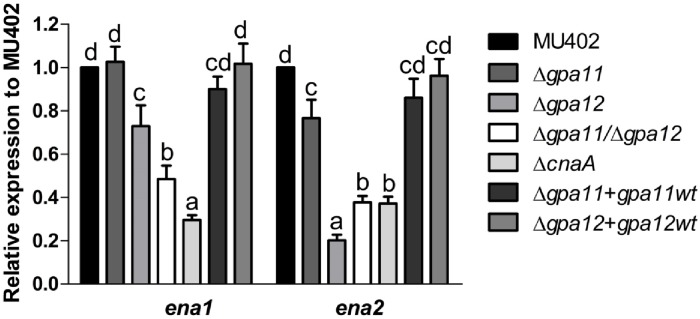
Activation of the calcineurin pathway is downregulated in Δgpa11/Δgpa12 since the dysfunction of Gpa11 and Gpa12 led to the down-regulation of cnaA mRNA levels in spores, we investigated if elements downstream of the calcineurin pathway could also be deregulated in gpa-mutant strains. In the methylotrophic yeast Hansenula polymorpha, the transcription of the gene ena1, which encodes a plasma membrane Na+/K+-ATPase, is induced by the activation of the calcineurin signaling pathway [31]. In order to investigate if a similar scenario could be present in M. circinelloides, we quantified the mRNA levels of ena homologues in spores from all strains. Firstly, we identified in M. circinelloides two Ena homologues, Ena1 and Ena2, with 35.8 and 35.3% identity and 59.2 and 51.6% similarity, respectively, to Ena1 from H. polymorpha and 63.1 and 77.3% identity and similarity between them (S2 Fig). The mRNA levels of ena1 and ena2 from spores of ΔcnaA mutant strain were 70.5 and 62.8% lower, respectively, compared to those levels of the wild-type strain (Fig 8). In Fig 8, marked under expression of ena1 was seen in the double gpa11/gpa12 mutant, while for ena2, loss of gpa12 alone already leads to low expression. For both ena1 and ena2, loss of gpa11 alone does not have a large effect though it is significant already for ena2. In general, this result indicates that ena1 and ena2 mRNA levels are under control of CnaA and Gpa12 in M. circinelloides.
Fig 8. Influence of Gpa11, Gpa12, and CnaA on mRNA levels of ena genes in M. circinelloides.
Levels of A) ena1 and B) ena2 measured by qRT-PCR from total mRNA from sporangiospores. ΔΔCt analysis was performed to compare mRNA levels between strains. Four independent experiments were performed for each condition. Different letters denote statistically significant differences (ANOVA, Fisher, p<0.05).
To determine if the calcineurin signaling pathway could be altered in the Δgpa11/Δgpa12 mutant strain, a calcineurin inhibitor, FK506 was employed. It had been previously reported that FK506 impaired mycelia growth in the ΔcnaA strain [15]. We assessed the capability of spores from the single and double mutant strains in gpa11 and gpa12 to develop mycelia growth in the presence of FK506. Our results showed that only the double mutant Δgpa11/Δgpa12 strain showed a phenotype similar to that of the ΔcnaA strain when grown on a poor nitrogen source (YNB) supplemented with FK506, while gpa-single mutant strains showed a phenotype similar to the wild-type strain independent of the nitrogen source (S3A Fig). It has been shown that the cnaA mutation also leads to increased sensitivity to detergent in a rich YPD media [15]. Our results showed that Δgpa11 and Δgpa12 or double gpa-mutant strains were also more sensitive to the detergent Triton X-100 than the wild type strain in YPG or YNB media (S3B Fig).
The role of gpa11 and gpa12 in the virulence of M. circinelloides
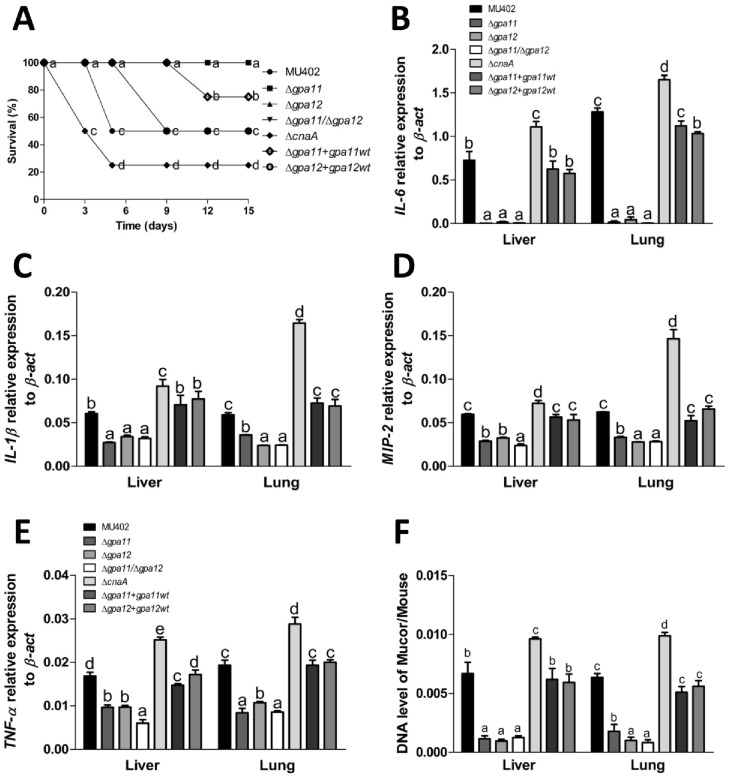
The increase in spore size and faster germination rate are associated with virulence increase in M. circinelloides [17]. Based on the fact that the spores of Δgpa11/Δgpa12 mutant strain are much larger, germinate faster, and cause a decrease in mRNA levels of cnaA compared to the wild-type strain, we performed virulence assays. In the mouse killing assay, diabetic mice were used which simulates one of the risk factors for acquiring mucormycosis [18]. Mice were injected intraperitoneally with 2 × 107 spores/mouse from each strain and mouse survival was monitored each day. Reduced survival was observed only in the Δgpa11+gpa11wt, Δgpa12+gpa12wt, ΔcnaA and wild-type strains. 50 and 75% of the mouse population died after 5 days post-inoculation with the spores from wild-type and ΔcnaA strains, respectively (Fig 9A). We quantified by qRT-PCR the mRNA levels of the inflammation markers IL-6, IL-1β, MIP-2 and TNF-α in the liver and lung of mice infected with the different strains (Fig 9B–9E). The mice infected with the Δgpa11+gpa11wt, Δgpa12+gpa12wt and wild-type strains, showed significantly increased mRNA levels of Il-6 (130 times in liver and 124 times in lung), Il-1β (96% in liver and 118% in lung), MIP-2 (115% in liver and 110% in lung) and TNF-α (109% in liver and 112% in lung) compared to mRNA levels from animals infected with gpa-mutants (Fig 9B–9E). Also, the ΔcnaA mutant strain produced significantly higher mRNA levels of all inflammation markers in liver (21–53%) or lung (28–178%) compared to the wild-type strain (Fig 9B–9E). These results indicate that mice infected with the Δgpa11, Δgpa12, or the Δgpa11/Δgpa12 mutants induce a weaker inflammatory response compared to those infected with the ΔcnaA, Δgpa11+gpa11wt and Δgpa12+gpa12wt or wild-type strains.
Fig 9. Role of gpa11 and gpa12 in the virulence of M. circinelloides.
A) Survival of diabetic mice infected intraperitoneally with 2 × 107 sporangiospores from different M. circinelloides strains recorded each day post-injection. N = 8, two independent experiments. Relative expression of inflammation markers B) IL-6, C) IL-1β, D) MIP-2, E) TNF-α used to monitor tissue damage in the liver and lung of mice infected with spores from the different M. circinelloides strains (N = 4). F) Fungal burden, determined by qPCR, from the liver and lung of infected mice. Three independent experiments were performed for each condition. Statistically significant differences are indicated by different letters (ANOVA and Fisher’s tests; p ≤ 0.05).
Fungal burden was quantified in tissues (liver and lung) of the mice infected with all the strains by qPCR using the tfc-1 as gene marker for M. circinelloides [29]. The quantity of tfc-1 from the Δgpa11, Δgpa12, the Δgpa11/Δgpa12 mutant strains were lower (up to 82% in liver and 87% in lung) than in those infected with the wild-type strain. Moreover, the ΔcnaA strain fungal burden was 26 and 32% more in liver and lung, respectively, compared to the wild-type strain (Fig 9F). These results indicate that mice infected with spores from Δgpa11, Δgpa12, or the Δgpa11/Δgpa12 mutant strains induce a weaker inflammatory response and less mouse tissue invasion compared to those infected with the ΔcnaA, complemented or wild-type strains.
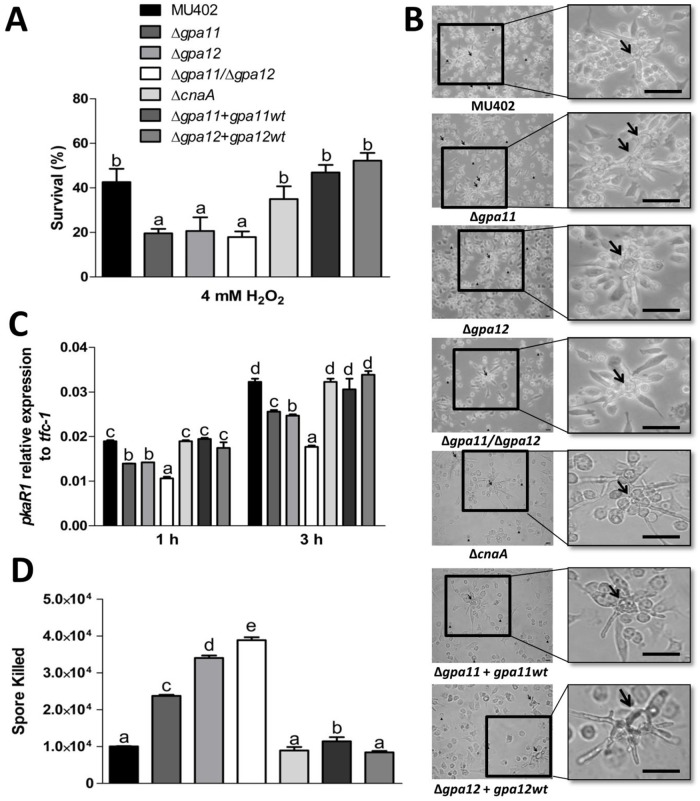
To explain the mechanism of low virulence of the Δgpa11/Δgpa12 spores despite their bigger size, we tested the effect of H2O2 on spore survival as previously reported [32, 33]. The Δgpa11, Δgpa12, and the Δgpa11/Δgpa12 mutant strains showed a significant decrease in spore survival compared to the wild-type strain in YPG (up to 55%) media supplemented with H2O2. The lowest levels of survival were registered for Δgpa11/Δgpa12 spores, compared to the wild-type. While ΔcnaA strain showed no significant difference in survival rate compared to the wild-type strain (Fig 10A), meanwhile the complemented mutant strains showed similar survival rate as the wild-type strain after the challenged with H2O2.
Fig 10. Effect of mutations in gpa11 and gpa12 in the viability of sporangiospores from M. circinelloides in H2O2 and macrophage interaction.
A) Spores from different strains were treated or not with 4 mM H2O2 and incubated at 28 °C on YPG. Survival was obtained from colonies grown for 24 h from the quotient of colonies from treatment versus no treatment. B) Germination of spores during macrophage interaction after 3 h was observed under direct observation by light microscopy (40 X) Scale bar = 20 μm. Arrows indicate germinating hyphae and head of arrows indicate swelling spores. C) The pkaR1 mRNA levels were quantified by qRT-PCR after 1 and 3 h of spore and macrophage interaction. D) Quantitation of killed spores by qPCR using tfc-1 after 3 h of incubation with mouse macrophages. A ΔCt analysis was performed to compare the mRNA and gene levels between the samples. Figures show the average of three independent experiments. Statistically significant differences are indicated by different letters (ANOVA and Fisher’s tests; p ≤ 0.05).
Based on this result, we conducted a confrontation assay with all the M. circinelloides strains against the murine macrophage cell line RAW264.7. After 3 h of interaction of macrophages and spores, we observed spore germination in presence of the macrophages (Fig 10B). The germination rate was quantified indirectly by the mRNA levels of pkaR1 by qRT-PCR, which is used as a molecular marker for hyphae development [28], after 1 or 3 h of macrophage and spore interaction. We observed lower pkaR1 transcript levels after 1 (≈26%) and 3 h (≈22%) in the single gpa-mutant strains compared to the wild-type strain (Fig 10C). The Δgpa11/Δgpa12 mutant strain showed the lowest levels of pkaR1 mRNA after 1 (≈44%) or 3 h (≈45%) compared to the wild-type (Fig 10C), meanwhile the complemented mutant strains showed similar survival rate as the wild-type strain after the challenged with H2O2. Based on a standard curve using different amounts of spores from M. circinelloides, we quantified by qPCR the tfc-1 levels from spores that were digested by macrophages after the interaction as described previously [32, 33]. The spores from the Δgpa11+gpa11wt, Δgpa12+gpa12wt, ΔcnaA and wild-type strains showed fewer spores killed after the interaction with the macrophages (Fig 10D) compared to the Δgpa11, Δgpa12 and Δgpa11/Δgpa12 mutant strains that showed more spores killed in this assay (Fig 10D). These results indicate that the spores from the Δgpa11/Δgpa12 mutant strain are the most susceptible to the macrophages compared to the spores from the ΔcnaA, Δgpa11+gpa11wt, Δgpa12+gpa12wt and wild-type strains. This could be explained, in part, by their higher susceptibility to oxidative damage.
Discussion
M. circinelloides produces several types of spores through its life cycle, and each type of spore has a specific cellular fate [2, 3]. The most well described spore is the sporangiospore, which is an asexual spore and is produced after the mycelium reaches a certain stage on solid surfaces, producing an aerial structure known as the sporangiophore. Several factors have been described that contribute to sporangiospore production in M. circinelloides; for example, light is a positive stimulator of sporulation and carotenogenesis [34]. The detailed molecular mechanism of how sporangiospores are produced in M. circinelloides is still not known. Transcript levels from gpa11 and gpa12 from M. circinelloides, have been shown to mainly accumulate in the sporangiospore, suggesting an important function either in spore formation, production, and/or germination [25].
Our work showed that gpa11 and gpa12 has an important function in the spore since Δgpa12 and specially Δgpa11/Δgpa12 showed increased spore sizes compared to Δgpa11 and wild-type strains. Spore size in M. circinelloides is known to be controlled by the calcineurin pathway. Thus, mutation of cnaA, which encodes the catalytic subunit A of calcineurin, leads to large spore size, faster germination, and increased virulence, among other defects [15]. Interestingly, cnaA mRNA levels in spores of Δgpa12 and Δgpa11/Δgpa12 strains were lower than those of the wild-type strain, suggesting that gpa12 positively controls the levels of cnaA in the spore, a function that can be compensated by gpa11 in absence of gpa12 as indicated the more severe effect observed in Δgpa11/Δgpa12 compared to Δgpa12. Moreover, the mRNA levels of ena1 and ena2, used as a marker for the activated calcineurin pathway [31], were decreased in Δgpa12, Δgpa11/Δgpa12 and ΔcnaA strains compared to the wild-type strain. Additionally, the spore size increase could be result of the low ena1 and ena2 expression because dysfunction of Ena homologues leads to cell swelling in human renal cells, as dysfunction of Na+-K+ ATPases leads to the accumulation of sodium ions in cells, which allows water entry into the cell to compensate the osmotic gradient [35]. Also, Saccharomyces cerevisiae mutants in the regulatory subunit of the calcineurin decreased the expression of ENA1 and accumulated Na+ and Li+ ions [36]. Therefore, it is tempting to speculate that something similar occurs in the Δgpa11/Δgpa12 strain of M. circinelloides, it will be interesting to know if the mutation of ena1 or/and ena2 could altered the spore size in this Mucoral (Fig 11). Overall, these findings suggest that the calcineurin pathway is under the control of Gpa12, and Gpa11 in a lesser extent, in spores from M. circinelloides, and consequently absence of gpa12 and gpa11 or cnaA results similar phenotypes such as spore enlargement (Fig 4), accelerated germination (Fig 5) and sensibility to the calcineurin inhibitor FK506 (S2 Fig). In addition, the double mutation of gpa11 and gpa12 led to lower cAMP levels and PKA activity compared to the the Δgpa11 or Δgpa12 after 3 hours of aerobic growth (Fig 5). Similar levels of cAMP and PKA activity between Δgpa11/Δgpa12 and ΔcnaA were observed (data not shown), indicating a crosstalk in the regulation of PKA pathway between Gpa11/Gpa12 and CnaA (Fig 11). Interestingly, although Δgpa12 decreased the mRNA from cnaA, this mutation did not alter the levels of cAMP and PKA activity, suggesting different pathways in the regulation of activation of PKA by Gpa12 or Gpa11/Gpa12.
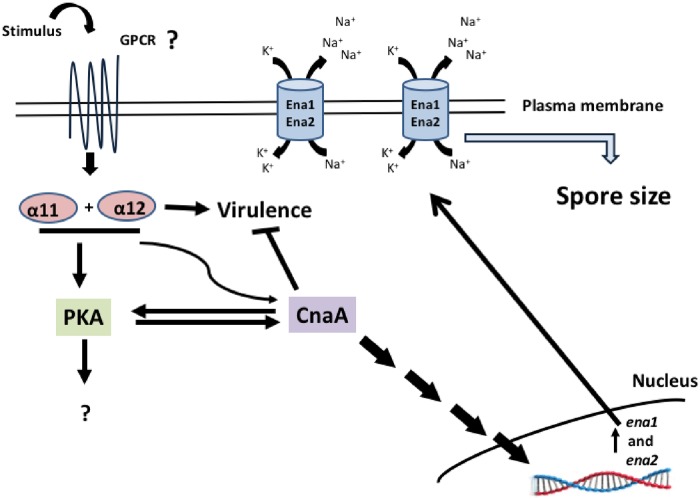
Fig 11. Proposed model of the function of Gpa11 and Gpa12 regulating sporangiospore size and virulence in M. circinelloides.
Through one or more GPCRs, Gpa11 and Gpa12 in the spores of M. circinelloides activate CnaA, which in turn activates the calcineurin pathway. The calcineurin pathway leads to the expression of genes regulated by this pathway, like ena1 and ena2. The dysfunction of Ena1 and Ena2, which are Na+-K+ ATPases, could increase cytosolic levels of sodium ions, allowing the influx of water, leading to an increase in spore size in M. circinelloides as was observed in the Δgpa11/Δgpa12 strain. The CnaA negatively regulates the virulence and stimulates the PKA pathway, meanwhile Gpa11 or Gpa12 positively regulate the virulence and activate the PKA pathway during aerobic germination, but unknown elements are implied in the avirulent phenotype by the mutations in gpa11 and gpa12.
Additionally to the regulation of the calcineurin pathway by gpa12 and gpa11, the calcineurin pathway also regulates the expression of gpa12 and gpa11 in the spore since their mRNA levels was down regulated in the ΔcnaA strain (Fig 7). This observation revealed a regulatory crosstalk between gpa12 and gpa11 and the calcineurin pathway. Alteration of this regulatory system by deletion of some key element seems to affect the expression of other gpa genes. Thus, ΔcnaA strain showed a significant mRNA increase of gpa2-4, which codes for products belonging to Group I (Gαs) and Group II of the Gα phylogeny. The mRNA levels of gpa3 and gpa4 were also increased in the Δgpa12 and Δgpa11/Δgpa12 mutant strains, suggesting a correlation between gpa11 with gpa12 and cnaA in terms of regulation of gpa3 and gpa4 mRNA levels in spores from M. circinelloides, although more experiments are needed to clarify this possible correlation.
Deletion of M. circinelloides in cnaA showed an elevated virulence (Fig 9) that has been attributed in part to the larger spore size that result in increased germination rate, allowing faster destruction of the host [17]. Surprisingly, single and double mutants of gpa11 and gpa12 were completely avirulent in a diabetic mouse model, despite the Δgpa11/Δgpa12 displayed low cnaA mRNA levels and mutant produce large spores that show high germination rate compared to the wild-type strain (Fig 5). Moreover, the inability to infect mice could not be attributed to a poor growth of the gpa11 and gpa12 mutants (Fig 3). The defect in virulence of gpa11 and gpa12 mutants and the higher virulence of the mutant of cnaA were confirmed by the figures of fungal burden and the levels of chemokines (IL-6, IL-1β, TNF-α, and MIP-2) known to be induced under filamentous infections [37–39], including M. circinelloides [40].
The single and double mutants of gpa11 and gpa12 did not show a significant increase in the mRNA levels of these inflammation markers in diabetic mice tissues compared to the levels from the wild-type or ΔcnaA strains, despite larger spore size and higher germination rate of Δgpa11/Δgpa12 mutant compared to the wild-type strain. Despite these similarities, spores of Δgpa11/Δgpa12 and ΔcnaA strains behave in a totally different manner in terms of H2O2 resistance, since ΔcnaA spores are more resistant to H2O2 than those from Δgpa11/Δgpa12, and also single mutants of this genes (Fig 10). Moreover, it has been also reported that the spores from ΔcnaA strain are more resistant to phagocytosis [15], but the single and double gpa-mutant strains were more efficiently killed by macrophages than spores from the wild-type strain. This result could in part explain that gpa11 and gpa12 mutants are unable to infect diabetic mice (Fig 9) The combination of large spores and inability to cause infection shown by mutants of gpa11 and gpa12, especially the double mutant, suggests that both phenotypes are controlled by the same regulatory pathway (Fig 11). This regulatory pathway in some point diverge in two branches, one that controls spore size through the calcineurin pathway and another that regulates the expression of genes important for the virulence. This model could explain the higher virulence of cnaA mutants that keep functioning the virulence-specific regulatory branch and simultaneously produce large spores with accelerated germination, it seems that a total lack of CnaA is needed to display a virulent phenotype. Therefore, gpa11 and gpa12 define a signal transduction pathway that play pivotal role in fungal infection in M. circinelloides. The dissection of this pathway with the identification of receptor to unknown signals, other components of the signal cascade, and targets genes will provide a better knowledge of the understudied process of fungal colonization by Mucorales.
Our work is important to the fungi field revealing new hypothesis to test. Fungal spore size has been found to be an important virulence determinant in fungal pathogens including ascomycetes (Beauveria bassiana, Botrytis cinerea), basiodiomycetes (Cryptococcus neoformans) or mucorales (Mucor circinelloides). In the entomopathogenic fungus B. bassiana, when the vacuolar ATPase subunit H encoding gene was disrupted, conidia diameter was reduced by 16% when compared to the wild-type strain. Moreover, this mutant displayed significantly attenuated virulence against the Galleria mellonella larvae [41]. In both fungal phytopathogens B. cinera and M. fructicola, environmental conditions regulate spore size. It has been reported that in B. cinerea, when grown on culture medium that contains 30% glucose (w/v), spores were about 50% larger than those grown on 2% glucose (w/v). Notably, these larger spores had enhanced aggressiveness against the flowers of Rosa hybrida [42]. Comparative analysis of spore sizes from different C. neoformans strains revealed that spores from serotype A are around 10% larger in diameter when compared to the serotype D [43]. Noteworthy, serotype A is the major responsible of C. neoformans infections (95%) and it has been associated with more severe infections in immunocompromised patients [44]. Interestingly, to the best of our knowledge, the first report for larger fungal spores (>10 μm in diameter) In M. circinelloides that does not imply a more virulent phenotype when compared to the wild-type strain.
Our work provides evidence for a genetic and functional linkage between gpa11 and gpa12 with the calcineurin pathway in M. circinelloides, suggesting that cnaA is under control of gpa11 and gpa12 and these elements are associated with a larger spore phenotype. However, larger spore phenotype by itself is not a prerequisite for virulence, suggesting that gpa11 and gpa12 also regulate a set of genes that are specifically involved in virulence.
Methods
Fungal strains, culture media, and growth conditions
M. circinelloides f. lusitanicus MU402 (leuA-, pyrG-) [27] was used for the gpa11 and gpa12 gene deletion studies. Additionally, the MU636 strain (leuA-, pyrG+) derived from M. circinelloides MU402 was also employed in this work [45], although it is not shown, this strain exhibits similar phenotypes as the wild-type strain MU402. A M. circinelloides ΔcnaA mutant strain [15] derived from MU402 was kindly provided by Dr. Soo Chan Lee (University of Texas at San Antonio, USA). Yeast–peptone–glucose (YPG) medium (pH 4.5); 1 L medium containing 3 g yeast extract (BD Bioxon, USA), 10 g gelatin peptone (BD Bioxon, USA), 20 g glucose (BD Bioxon, USA). Yeast nitrogen base (YNB) (Sigma, USA) or MMC (minimal medium with casamino acids), which contained 10 g/L casamino acids (BD Difco, USA), 20 g/L glucose, and 0.5 g/L yeast nitrogen base, supplemented with uracil (200 mg/L) (Sigma, USA) as required; 15 g/L agar (BD Bioxon, USA) was added when solid media were prepared. For spore production and culture conditions, germination under self-anaerobic and aerobic atmosphere, as well as the quantitation of growth, spore size, and hyphae length protocols described previously were followed with no modifications [18, 46].
100 spores from the different strains were treated or not with 4 mM H2O2 for 1 h at 4 °C and spread on YNB or YPG plates and incubated for 24 h at 28 °C. YNB or YPG solid media were supplemented or not with 0.005% Triton X-100 and 100 spores from different strains were spread on the plates, incubation was carried out for 1 day at 28 °C.
Targeted deletion in M. circinelloides
Targeted deletion of gpa11
Using genomic DNA from MU402 wild-type strain of M. circinelloides 1.1-kb fragments that correspond to the 5′ upstream or 3′ downstream regions from the start or stop translation codon from gpa11, respectively, were amplified by PCR. The pair of oligonucleotides Gpa11-P-Fwd and Gpa11-P-Sac-Rev was used to amplify the 5′ region, with a Sacl site added to the 3′ end of the Gpa11-P-Sac-Rev primer. The oligonucleotides Gpa11-T-Sac-Fwd and Gpa11-T-Xba-Rev were used to amplify the 3′ region, with a SacI site added to the 5′ end of the Gpa11-T-Sac-Fwd primer and an Xbal site to the 3′ end of the Gpa11-T-Xba-Rev primer. A 2-kb fragment that corresponds to the pyrG gene was PCR-amplified using the primers PyrG-Sac-Fwd and PyrG-Sac-Rev and SacI sites were added to its 5′ end and 3′ end (S3 Table).
The 5′ and 3′ fragments from gpa11 and pyrG were independently cloned into pJET1.2/blunt resulting in the pGpa11-5′ (for the gpa11 5´-region), pGpa11-3′ (for the gpa11 3′-region), and pPyrG (for the pyrG gene) vectors. The Sacl sites in the different fragments allowed the subcloning of pyrG into pGpa11-5′, resulting in pGpa115′-pyrG. Finally, 1.1-kb from the 3′ region of gpa11 was cut using the Sacl and Xbal enzymes, and subcloned into pGpa115′-pyrG—the resulting plasmid was named pGpa115′-pyrG-3′. This resulting plasmid was used as template to obtain the recombinant fragment (Gpa115′-pyrG-3′ of 4.2 kb) by PCR with the oligonucleotides Gpa11-P-Fwd and Gpa11-T-Xba-Rev. All recombinant plasmids were confirmed by PCR and restriction enzyme patterns.
Targeted deletion of gpa12
The gpa12 recombinant fragment coupled with the pyrG selection gene was generated through overlapping-PCR. In this process, three PCR fragments containing overlapping sequences between them were PCR-amplified using the following oligonucleotides (S3 Table): 12pUFow-Pstl and 12pURev-pyrG, which amplified 1-kb from the 5′ upstream region of the start translation codon of gpa12; and oligonucleotides 12pDRev-Notl and 12pDFow-pyrG, which amplified 1-kb from the 3′ downstream region of the stop translation codon of gpa12. In oligonucleotide 12pURev-pyrG, an added nucleotide sequence at the 3' end hybridized to the 5′ end of pyrG; and in oligonucleotide 12pDFow-pyrG, an added nucleotide sequence at its 5' end hybridized to the 3' end of pyrG. The gene pyrG was PCR-amplified using the oligonucleotides PyrG-Sac-Fwd and PyrG-Sac-Rev, with pMAT1700 [47] as a template. Three individual PCR reactions were performed to obtain 3 DNA fragments. The gpa12 5′ region, pyrG (2 kb), and the gpa12 3′ region, which were all purified. Overlapping-PCR was performed using 100 ng/μL of each template; 10 μM each of the oligonucleotides 12pUFow-Pstl and 12pDRev-Notl and 1 μL of Herculase II Fusion Enzyme (Agilent Technologies, USA), at a molar ratio of 1:1:1, were used to generate the recombinant fragment of gpa12.
Targeted deletion of gpa11 in Δgpa12 strain: To generate the Δgpa11/Δgpa12 double-mutant strain, the plasmid pGpa115′-pyrG-3’-previously obtained for the gpa11 mutation was used. The gene leuA, which is required for leucine synthesis [48], was obtained by PCR with the oligonucleotides leuA-Sac-Fwd and leuA-Sac-Rev, and SacI restriction sites were added to both oligonucleotides at their 5′ ends (S3 Table), and then annealed into the plasmid pLeu4 [49]. Both leuA (3.7 kb) and the pGpa113′-pyrG-5′ vector were digested with SacI, and both products were ligated using the enzyme T4 DNA ligase (Promega, USA); the resulting plasmid was named pGpa115′-leuA-3′. The plasmid pGpa115′-leuA-3′ was used as a template and using the oligonucleotides Gpa11-P-Fwd and Gpa11-T-Xba-Rev, PCR amplification was carried out to obtain the recombinant fragment (5.9-kb) to delete gpa11 in the Δgpa12 strain. All recombinant plasmids were identified by PCR and restriction fragment patterns.
Protoplasts obtained from M. circinelloides MU402 were transformed with the recombinant DNA fragments (3 μg/transformation) according to a previously described protocol [50]. Transformants were obtained under selective conditions on MMC agar plates lacking uracil for single deletion mutants or YNB agar plates lacking uracil and leucine for the double mutants. Monosporic cultures were obtained after 5 cycles of sporulation in either selective medium or rich medium, at which point, 100% of the spores could grow on the selective medium, indicating the transformation of all nuclei.
Molecular identification of gpa11 and gpa12 mutant strains from M. circinelloides
PCR confirmation of gpa11 gene deletion
Transformed strains were identified by PCR using the oligonucleotides 11-C-Fwd (forward) and pyrG-R2 (reverse) (S3 Table); 11-C-Fwd hybridized outside the recombination fragment used to delete the gpa11 gene and pyrG-R2 (S3 Table) that hybridize in the selectable marker sequence to determine a homologous integration event, a positive recombination event was determined by PCR amplification of 1.4-kb band. A lack of amplification band indicated the wild-type genotype (S4 Fig).
PCR confirmation of gpa12 gene deletion
Using a similar approach, the transformants were identified by PCR with the pair of oligonucleotides 12-C-Fwd, which hybridizes outside the recombination fragment used to delete gpa12 gene, and pyrG-R2 (S3 Table), which hybridizes in the selectable marker sequence. Those which generated an amplicon of 1.3-kb was the mutant genotype, but non-amplification confirmed a wild-type genotype. (S4 Fig).
PCR confirmation of double mutant Δgpa11/Δgpa12
Using the oligonucleotides 11-C-Fwd (forward) and leuA-C-Rev (reverse) (S3 Table); 11-C-Fwd hybridized outside the recombination fragment used to delete the gpa11 gene and leuA-C-Rev (S3 Table) hybridized in the selectable marker sequence to determine a homologous integration event. A positive recombination event was determined by PCR amplification of 2.2-kb band, while no amplification confirmed the wild-type genotype (S4 Fig).
As a positive control for DNA integrity that was used for PCR reactions, the pair of oligonucleotides, Gpa11-T-Sac-Fwd / Gpa11-T-Xba-Rev, which hybridized with the 3′-end region of gpa11 were used for Δgpa11 and a 1.1-kb amplicon indicated a positive reaction. For Δgpa12 the pair of primers, 12pUFow-Pstl / 12pURev-pyrG, which hybridized with the 5’ region of gpa12 were used and 1.1 kb was considered as positive control. The pair of oligonucleotides 11-C-Fwd / Gpa11-P-Sac-Rev was used in a PCR reaction for positive controls for Δgpa11/Δgpa12 and a 1.3 kb amplicon indicated a positive PCR reaction; the wild-type strain also amplified an identical PCR size band as well as each mutant strain (S4 Fig).
Southern blot to identify the deletions of gpa11 and gpa12
A total of 1 μg of genomic DNA from MU402 and the mutants of M. circinelloides were digested with indicated restriction enzyme. DNA was electrophorized and transferred to nylon membranes (HybondTM-N+, Amersham Biosciences, UK) following the recommended protocol [51]. Southern blot hybridization was performed under stringent conditions. DNA probes were labelled with [α-32P] dCTP using Ready-To-Go Labeling Beads (GE Healthcare Life Science). For Southern blot experiments, a DNA probe (1 kb) was directly amplified from genomic DNA from M. circinelloides using the primer pairs that amplified the 5′ region upstream of the start translation codon from gpa11, for gpa12 identification a fragment (1 kb) corresponding to the 3′ region downstream from the stop translation codon was used as a probe and finally the primer pairs that amplified the 3′ region downstream from the stop translation codon (1 kb) from gpa11 for the double mutation.
Complementation of Δgpa11 and Δgpa12 strains
The gpa11 and gpa12 wild-type ORFs were PCR-amplified using the genomic DNA of M. circinelloides MU402 as the template and the oligonucleotides gpa11-FWR-XhoI/gpa11-REV-NotI and gpa12-FWR-SalI/gpa11-REV-NotI (S3 Table), which introduced restriction sites for XhoI and NotI at the 5' and 3' ends, respectively. Wild-type gpa11 and gpa12 genes were cloned in pEUKA4 vector [28] under the control of gpd1 promoter from M. circinelloides in the XhoI and NotI sites; the resulting plasmids were confirmed based on restriction patterns and DNA sequencing. Both recombinant plasmids were used to transform the Δgpa11 and Δgpa12 strains and selection was performed in YNB media (Difco, Franklin Lakes, NJ, USA) medium.
qRT-PCR confirmation
Total RNA isolated from gpa11 and gpa12 single and double mutant strains was used as a template and qRT-PCR was performed to determine all gpa transcript levels, as described previously [25].
Specimen preparation for scanning electron microscope (SEM) imaging of spores
SEM was performed according to Oshell 1997 [52], 4 h post-fixation with osmium tetroxide (1% diluted in cacodylate buffer 0.01 M, pH 7.4). The samples were dehydrated in ethanol solutions of 60, 70, 80 and 90%, and three times in 100% ethanol for 15 min each.
For hexamethyldisilazane (HMDS) drying the samples were rinsed in ethanol- HMDS in proportions of 2:1, 1:1, 1:2, 1:3, and HMDS 100% for 15 min each. The samples were transferred to a desiccator for 24 h. Samples were mounted on stubs and examined with a Zeiss GeminiSEM scanning electron microscope.
Ethics statement
The ethics agreement was made for the mouse virulence model protocol following the recommendations of the Mexican Federal Regulations for the Use and Care of Laboratory Animals (NOM-062-ZOO-1999) (Especificaciones técnicas para la producción, cuidado y uso de los animales de laboratorio/Technical specifications for production, use and care of laboratory animals) [53]. The Internal Biosecurity and Bioethics Committee of Instituto de Investigaciones Químico Biológicas de la Universidad Michoacana de San Nicolás de Hidalgo firstly reviewed the experimental protocols and authorized them (trade number 06-13/2016).
Mouse virulence assays
To assess the virulence of M. circinelloides strains, we used methods described previously [18, 54] with some modifications. Briefly, each group consisting of eight male BALB/c mice (12–16 weeks old, weighing ~20 g, obtained from CINVESTAV, Zacatenco. México) were treated with streptozotocin (200 mg/kg) (Sigma, USA) to induce a diabetic state (>300 mg/dL glucose in blood), and then independent mice groups were inoculated with 2×107 spores from each strain of M. circinelloides. The spores were suspended in saline solution and injected intraperitoneally into mice, and survival of mice was monitored daily. Three independent assays were conducted for each group.
Macrophages spore killing assay
Mouse monocyte/macrophage RAW 267.4 (TIB-71) was purchased from American Type Culture Collection (ATCC). Cells were maintained in DMEM medium supplemented with 10% FBS (Sigma, USA), 100 U/mL penicillin (Sigma, USA), and 100 μg/mL streptomycin (Sigma, USA) (basal medium) at 5% CO2 and 37 °C for 24 h before treatments.
Macrophages were seeded at 8 × 105 cells/well in DMEM basal medium (GIBCO, Thermo Fisher Scientific, USA) without antibiotics and incubated at 37 °C in 6-well plates. After incubation overnight, the basal medium in each well was replaced with fresh medium, then the macrophages were co-cultured with 2 × 105 spores produced in YPG for 1 or 3 h. Immediately, the supernatants and cells and spores were removed and placed in independent tubes. Then, cells and spores were centrifuged at 1,500 x g for 5 min. Pellet was recovered and frozen until nucleic acid extraction was performed.
Total M. circinelloides RNA and DNA isolation from in vitro cultures, from macrophages or from mouse tissues
Total RNA and genomic DNA from M. circinelloides, mice tissues and macrophages were isolated using RNAeasy mini kit and QIAamp DNA Mini Kit, respectively (Qiagen, Venlo, Netherlands).
M. circinelloides cultures were obtained by filtration in 5 μm membrane filters (Millipore, USA), the mouse tissues were collected following infection with M. circinelloides after 15 days of spore inoculation. Approximately 25–50 mg of tissue sample from each mouse was transferred into a tube with MagNA Lyser Green Beads (Roche, Switzerland) pre-cooled on ice. Denaturing RLT buffer (700 μL) from the RNeasy Mini Kit or buffer ATL (180 μL) of QIAamp DNA Mini Kit were added immediately before homogenization to obtain total RNA or genomic DNA, respectively. For cell disruption, tubes were placed in the MagNA Lyser Instrument (Roche, Switzerland) and processed twice at 5,500 x g for 40 s with cooling on ice for 1 min between each step. Then, samples were centrifuged for 1 min at 20,000 x g (Eppendorf 5417) and the supernatants were used for total RNA or genomic DNA isolation using the corresponding kit, according to the manufacturer’s instructions. To eliminate DNA contamination from RNA samples, samples were treated with DNase I (Promega, USA) according to the manufacturer’s protocol. The RNA was eliminated from DNA samples using RNAse A (Roche, Switzerland). RNA or genomic samples were separated on non-denaturing 2% and 1% agarose gel, respectively stained with ethidium bromide (Sigma, USA), visualized using a Gel Doc XR+ Imager (Bio-Rad, Hercules, CA, USA), and quantified using a SmartSpec Plus spectrophotometer (Bio-Rad).
Determination of abundance of DNA from M. circinelloides by qPCR
Abundance of M. circinelloides from mice tissues or macrophages was compared based in a spore standard curve of M. circinelloides. MU402 spore concentrations between 1 x 102 and 1 x 107 mL-1 by 10-fold serial dilutions were employed. Consequently, DNA was extracted as previously reported, to generate a spore standard curve by qPCR by detection of the validated tfc-1 nuclear gene.
Oligonucleotide design and quantitative reverse transcription polymerase chain reaction (qRT-PCR)
The primers and hydrolysis probe for the calcineurin catalytic subunit (cnaA, NCBI gene bank accession number AGJ95088.1); the Na+-K+ ATPases ena1 (ID number 105213), and ena2 (ID number 113279) DNA sequences were obtained from M. circinelloides genome database [55]. Design of the primers and hydrolysis probes for each gene was performed using Biosearch Technologies software (www.biosearchtech.com) to ensure the specificity of all detections during qRT-PCR assays (S4 Table). To evaluate the relative gene expression of gpa, pkar1, and adh1 from M. circinelloides protocols described previously were followed [25, 29]. Genomic DNA and total RNA isolation were performed as described previously [18].
Quantification of intracellular cAMP levels and protein kinase A activity
Spores from MU402, Δgpa11, Δgpa12, Δgpa11/ Δgpa12, ΔcnaA, Δgpa11+gpa11wt or Δgpa12+gpa12wt strains were inoculated into YPG media and incubated with constant shaking (150 rpm) for 3 h. Biological samples that were obtained under these conditions were used in either cAMP or PKA measurements.
cAMP levels
Cells were separated from media and frozen in liquid nitrogen. Upon nitrogen evaporation, 60 mg of biological samples were weighed, and resuspended in 1 mL of 0.1 M HCl, and 250 μL of glass beads (0.1 mm) (BioSpec Products, Inc., Bartlesville, OK, USA) were added. Samples were homogenized by receiving three pulses at full speed for 30 s, with 1 min incubation on ice intermittently, in a beadbeater (607-BioSpec Products, Inc., Bartlesville, OK, United States). The supernatants were used for cAMP measurements using the Direct cAMP ELISA kit (Enzo Life Science, San Diego, USA). Protocols were followed accordingly to the manufacturer’s instructions. The OD was measured at 405 nm on a microplate reader (Bio-Rad Benchmark; Bio-Rad Laboratories, Hercules, CA, USA).
PKA activity
We obtained crude protein extract (0.5 μg) from each sample, and a PKA-specific kinase activity was quantified by using the PKA Colorimetric Activity Kit (Invitrogen, Carlsbad, CA, United States), as described previously [15].
Statistical analysis
All data were evaluated by analysis of variance (ANOVA, level of statistically significant difference at α<0.05). Fisher post-hoc test was used.
Supporting information
The spores from the independent M. circinelloides knockout strains produced in YPG were observed under A) light microscope (100 X), scale bar is equal to 20 μm. B) Under scanning electron microscope. Representative photographs from the corresponding strains of M. circinelloides (1000 X), scale bar is equal to 10 μm.
(DOCX)
Ena1 from Hansenulla polymorpha was used as bait to find Ena homologues in M. circinelloides. A) Clustal W alignment analysis. Identical and similar amino acid residues are shown in black and grey boxes, respectively. B) Identity analysis of Ena homologues in M. circinelloides is shown as identity/similarity in percentage, Mc: M. circinelloides; Hp: H. polymorpha.
(DOCX)
A) Effect of inhibitor of calcineurin FK506 on the growth of M. circinelloides. Radial growth of spores grown on YPG or YNB agar plates recorded each day during the experiment. Representative photos show radial growth after 3 days. Three independent experiments were performed for each condition. B) 100 spores from each strain were inoculated by spreading on YPG and YNB plates supplemented with 0.005% Triton X-100. Plates were incubated for 1 day. The bars represent the number of colonies formed with treatment versus the colonies formed without treatment. Three independent experiments were performed for each condition. *Statistically significant difference (ANOVA, Fisher, p<0.05).
(DOCX)
A) gpa11; B) gpa12 and C) gpa11/gpa12 mutation strategy. The 5′ and 3′ regions upstream and downstream from the start and stop translation codons, respectively, were used to flank the pyrG (single mutants) or leuA (double mutant) selective marker. The diagrams show the recombinant fragments used to delete the gpa genes in protoplasts of M. circinelloides MU402 wild-type strain. The photographs show the molecular confirmation by PCR with specific primers for each gene showing specific PCR-bands that indicate recombination in the corresponding gpa loci. M: Molecular size markers (kb). C+: PCR control for positive amplification (wt: 1.1 kb amplicon to identify the wt gpa11 or gpa12 genes); Δgpa11: 1.4 kb amplicon to identify the deletion of gpa11 gene; Δgpa12: 1.3 kb amplicon to identify the deletion of gpa12 gene; and Δgpa11/Δgpa12: 2.2 kb amplicon to identify the deletion of gpa11 gene in the mutant Δgpa12.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
V M-C, MI R-D Coordinación de la Investigación Científica, Universidad Michoacana de San Nicolás de Hidalgo, México (2.6, 2.35). https://www.cic.umich.mx V M-C, MI R-D, J C-G Consejo Nacional de Ciencia y Tecnología, México (CONACYT; 181747, 167071, and 256119). https://www.conacyt.gob.mx V G Fundación Séneca-Agencia de Ciencia y Tecnología de la Región de Murcia, Spain (19339/PI/14). https://www.fseneca.es The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wijayawardene NN, Pawłowska J, Letcher PM, Kirk PM, Humber RA, Schüßler A et al. Notes for genera: basal clades of fungi (including Aphelidiomycota, Basidiobolomycota, Blastocladiomycota, Calcarisporiellomycota, Caulochytriomycota, Chytridiomycota, Entomophthoromycota, Glomeromycota, Kickxellomycota, Monoblepharomycota, Mortierellomycota, Mucoromycota, Neocallimastigomycota, Olpidiomycota, Rozellomycota and Zoopagomycota). Fungal Divers. 2018; 92: 43–129. 10.1007/s13225-018-0409-5. [DOI] [Google Scholar]
- 2.Orlowski M. Mucor dimorphism. Microbiol Rev. 1991; 55(2): 234–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morin-Sardin S, Nodet P, Coton E, Jany JL. A Janus-faced fungal genus with human health impact and industrial applications. Fungal Biol Rev. 2017; 31(1):12–32. 10.1016/j.fbr.2016.11.002. [DOI] [Google Scholar]
- 4.McIntyre M, Breum J, Arnau J, Nielsen J. Growth physiology and dimorphism of Mucor circinelloides (syn. racemosus) during submerged batch cultivation. Appl Microbiol Biotechnol. 2002; 58:495–502. 10.1007/s00253-001-0916-1 [DOI] [PubMed] [Google Scholar]
- 5.Lübbehüsen TL, Nielsen J, McIntyre M. Characterization of the Mucor circinelloides life cycle by on-line image analysis. J Appl Microbiol. 2003; 95:1152–60. 10.1046/j.1365-2672.2003.02098.x [DOI] [PubMed] [Google Scholar]
- 6.Lübbehüsen T L, Nielsen J, McIntyre M. Morphology and physiology of the dimorphic fungus Mucor circinelloides (syn. M. racemosus) during anaerobic growth. Mycol Res. 2003; 107:223–30. 10.1017/S0953756203007299 [DOI] [PubMed] [Google Scholar]
- 7.Hassan MIA, Voigt K. Pathogenicity patterns of mucormycosis: epidemiology, interaction with immune cells and virulence factors. Med Mycol. 2019; 1;57(Supplement_2):S245–S256. 10.1093/mmy/myz011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baldin C, Ibrahim AS. Molecular mechanisms of mucormycosis-The bitter and the sweet. PLoS Pathog. 2017; 3;13(8):e1006408 10.1371/journal.ppat.1006408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeong W, Keighley C, Wolfe R, Lee WL, Slavin MA, Kong DCM, et al. The epidemiology and clinical manifestations of mucormycosis: a systematic review and meta-analysis of case reports. Clin Microbiol Infect. 2019; 25(1):26–34. 10.1016/j.cmi.2018.07.011 [DOI] [PubMed] [Google Scholar]
- 10.Chander J, Kaur M, Bhalla M, Punia RS, Singla N, Bhola K et al. Changing Epidemiology of Mucoralean Fungi: Chronic Cutaneous Infection Caused by Mucor irregularis. Mycopathologia 2015; 180(3–4):181–6. 10.1007/s11046-015-9908-z [DOI] [PubMed] [Google Scholar]
- 11.Lewis RE, Kontoyiannis DP. Epidemiology and treatment of mucormycosis. Future Microbiol. 2012; 8(9):1163–1175. 10.2217/fmb.13.78 [DOI] [PubMed] [Google Scholar]
- 12.Skiada A, Lass-Floerl C, Klimko N, Ibrahim A, Roilides E, Petrikkos G. Challenges in the diagnosis and treatment of mucormycosis. Med Mycol. 2018;1;56(suppl_1):93–101. 10.1093/mmy/myx101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenks JD, Reed SL, Seidel D, Koehler P, Cornely OA et al. Rare mould infections caused by Mucorales, Lomentospora prolificans and Fusarium, in San Diego, CA: the role of antifungal combination therapy. Int J Antimicrob Agents. 2018; 52(5):706–12. 10.1016/j.ijantimicag.2018.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ibrahim AS, Spellberg B, Walsh TJ, Kontoyiannis DP. Pathogenesis of mucormycosis. Clin Infect Dis. 2012; 54 Suppl 1:S16–22. 10.1093/cid/cir865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SC, Li A, Calo S, Heitman J. Calcineurin plays key roles in the dimorphic transition and virulence of the human pathogenic zygomycete Mucor circinelloides. PLoS Pathog. 2013; 9: e1003625 10.1371/journal.ppat.1003625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sephton-Clark PCS, Muñoz JF, Ballou ER, Cuomo CA, Voelz K. Pathways of Pathogenicity: Transcriptional Stages of Germination in the Fatal Fungal Pathogen Rhizopus delemar. mSphere. 2018; 3(5). 10.1128/mSphere.00403-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li CH, Cervantes M, Springer DJ, Boekhout T, Ruiz-Vazquez RM, Torres-Martinez SR, et al. Sporangiospore size dimorphism is linked to virulence of Mucor circinelloides. PLoS Pathog. 2011; 7(6): e1002086 10.1371/journal.ppat.1002086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patiño-Medina JA., Maldonado-Herrera G, Pérez-Arques C, Alejandre-Castañeda V, Reyes-Mares NY, Valle-Maldonado MI, et al. Control of morphology and virulence by ADP-ribosylation factors (Arf) in Mucor circinelloides. Curr Genet. 2018; 64:853–69. 10.1007/s00294-017-0798-0 [DOI] [PubMed] [Google Scholar]
- 19.Fuller KK, Rhodes JC. Protein kinase A and fungal virulence: a sinister side to a conserved nutrient sensing pathway. Virulence. 2012; 3(2):109–21. 10.4161/viru.19396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ocampo J, Fernandez Nuñez L, Silva F, Pereyra E, Moreno S, Garre V, et al. A subunit of protein kinase A regulates growth and differentiation in the fungus Mucor circinelloides. Eukaryot Cell. 2009; 8:933–44. 10.1128/EC.00026-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tesmer JJ. The quest to understand heterotrimeric G protein signaling. Nat Struct Mol Biol. 2010; 17: 650–2. 10.1038/nsmb0610-650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Syrovatkina V, Alegre KO, Dey R, Huang XY. Regulation, Signaling, and Physiological Functions of G-Proteins. J Mol Biol. 2016; 428(19):3850–68. 10.1016/j.jmb.2016.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hicks JK, Yu JH, Keller NP, Adams TH. Aspergillus sporulation and mycotoxinproduction both require inactivation of the FadA G alpha protein-dependent signaling pathway. EMBO J. 1997. 16(16):4916–23. 10.1093/emboj/16.16.4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roze LV, Beaudry RM, Keller NP, Linz JE. Regulation of aflatoxin synthesis by FadA/cAMP/protein kinase A signaling in Aspergillus parasiticus. Mycopathologia. 2004; 158: 219–32. 10.1023/B:MYCO.0000041841.71648.6e [DOI] [PubMed] [Google Scholar]
- 25.Valle-Maldonado MI, Jácome-Galarza IE, Díaz-Pérez AL, Martínez-Cadena G, Campos-García J, Ramírez-Díaz MI, et al. Phylogenetic analysis of fungal heterotrimeric G protein-encoding genes and their expression during dimorphism in Mucor circinelloides. Fungal Biol. 2015; 119(12):1179–93. 10.1016/j.funbio.2015.08.009 [DOI] [PubMed] [Google Scholar]
- 26.Bölker M. Sex and crime: heterotrimeric G proteins in fungal mating and pathogenesis. Fungal Genet Biol. 1998; 25(3):143–56. 10.1006/fgbi.1998.1102 [DOI] [PubMed] [Google Scholar]
- 27.Nicolás FE, de Haro JP, Torres-Martínez S, Ruiz-Vázquez RM. Mutants defective in a Mucor circinelloides dicer-like gene are not compromised in siRNA silencing but display developmental defects. Fungal Genet Biol. 2007; 44(6):504–16. 10.1016/j.fgb.2006.09.003 [DOI] [PubMed] [Google Scholar]
- 28.Wolff A. M., Arnau J. Cloning of Glyceraldehyde-3-phosphate Dehydrogenase-Encoding Genes in Mucor circinelloides (Syn. racemosus) and Use of the gpd1 Promoter for Recombinant Protein Production. Fungal Genet Biol. 2002; 35 (1): 21–29. 10.1006/fgbi.2001.1313 [DOI] [PubMed] [Google Scholar]
- 29.Valle-Maldonado MI, Jácome-Galarza IE, Gutiérrez-Corona F, Ramírez-Díaz MI, Campos-García J, Meza-Carmen V. Selection of reference genes for quantitative real time RT-PCR during dimorphism in the zygomycete Mucor circinelloides. Mol Biol Rep; 2015; 42(3):705–11. 10.1007/s11033-014-3818-x [DOI] [PubMed] [Google Scholar]
- 30.Rangel-Porras RA, Meza-Carmen V, Martinez-Cadena G, Torres-Guzmán JC, González-Hernández GA, Arnau J, et al. Molecular analysis of an NAD-dependent alcohol dehydrogenase from the zygomycete Mucor circinelloides. Mol Genet Genomics. 2005; 274(4):354–63. 10.1007/s00438-005-0025-4 [DOI] [PubMed] [Google Scholar]
- 31.Rodríguez C, Galindo LR, Siverio JM. Nitrogen-dependent calcineurin activation in the yeast Hansenula polymorpha. Fungal Genet Biol. 2013; 53:34–41. 10.1016/j.fgb.2013.01.007 [DOI] [PubMed] [Google Scholar]
- 32.Patiño-Medina J. A., Vargas-Tejedaa D., Valle-Maldonado M. I., Alejandre-Castañed V, Jácome-Galarz I. E., Villegas-Moreno J., et al. Sporulation on blood serum increases the virulence of Mucor circinelloides. Microbial Pathogenesis. 2019; 137: 103737 10.1016/j.micpath.2019.103737 [DOI] [PubMed] [Google Scholar]
- 33.Díaz-Pérez SP, Patiño-Medina JA, Valle-Maldonado MI, López-Torres A, Jacome-Galarza IE, Anaya Martínez V, et al. Alteration of fermentative metabolism enhances Mucor circinelloides virulence. Infect Immun; 2019. (In press) 10.1128/IAI.00434-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navarro E, Lorca-Pascual JM, Quiles-Rosillo MD, Nicolás FE, Garre V, Torres-Martínez S, et al. A negative regulator of light-inducible carotenogenesis in Mucor circinelloides. Mol Genet Genomics. 2001; 266(3):463–70. 10.1007/s004380100558 [DOI] [PubMed] [Google Scholar]
- 35.Yordy MR, Bowen JW. Na,K-ATPase expression and cell volume during hypertonic stress in human renal cells. Kidney Int. 1993; 43(4):940–8. 10.1038/ki.1993.132 [DOI] [PubMed] [Google Scholar]
- 36.Mendoza I, Rubio F, Rodriguez-Navarro A, Pardo JM. The protein phosphatase calcineurin is essential for NaCl tolerance of Saccharomyces cerevisiae. J Biol Chem. 1994; 269(12):8792–6. [PubMed] [Google Scholar]
- 37.Peng XD, Zhao GQ, Lin J, Jiang N, Xu Q, Zhu CC, et al. Fungus induces the release of IL-8 in human corneal epithelial cells, via Dectin-1-mediated protein kinase C pathways. Int J Ophthalmol. 2015; 8(3):441–7. 10.3980/j.issn.2222-3959.2015.03.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chamilos G, Lewis RE, Lamaris G, Walsh TJ, Kontoyiannis DP. Zygomycetes hyphae trigger an early, robust proinflammatory response in human polymorphonuclear neutrophils through toll-like receptor 2 induction but display relative resistance to oxidative damage. Antimicrob Agents Chemother. 2008; 52:722–4. 10.1128/AAC.01136-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wüthrich M, Deepe GS Jr, Klein B. Adaptive immunity to fungi. Annu Rev Immunol. 2012; 30:115–48. 10.1146/annurev-immunol-020711-074958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.López-Muñoz A, Nicolás FE, García-Moreno D, Pérez-Oliva AB, Navarro-Mendoza MI, Hernández-Oñate MA et al. An Adult Zebrafish Model Reveals that Mucormycosis Induces Apoptosis of Infected Macrophages. Sci Rep. 2018; 8(1):12802 10.1038/s41598-018-30754-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu J, Zhu XG, Ying SH, Feng MG. Effect of vacuolar ATPase subunit H (VmaH) on cellular pH, asexual cycle, stress tolerance and virulence in Beauveria bassiana. Fungal Genet Biol. 2017; 98:52–60. 10.1016/j.fgb.2016.12.004 [DOI] [PubMed] [Google Scholar]
- 42.Phillips DJ, Margosan DA, Mackey BE. Size, nuclear number, and aggressiveness of Botrytis cinerea on media of varied glucose concentrations. Phytopathology. 1987; 77: 1606–1608. [Google Scholar]
- 43.Velagapudi R, Hsueh YP, Geunes-Boyer S, Wright JR, Heitman J. Spores as infectious propagules of Cryptococcus neoformans. Infect Immun 2009; 77(10):4345–55. 10.1128/IAI.00542-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Desnos-Ollivier M, Patel S, Raoux-Barbot D, Heitman J, Dromer F. Cryptococcosis serotypes impact outcome and provide evidence of Cryptococcus neoformans speciation. mBio. 2015; 6(3): e00311–15. 10.1128/mBio.00311-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Navarro-Mendoza MI, Pérez-Arques C, Panchal S, Nicolás FE, Mondo SJ, Ganguly P, et al. Early Diverging Fungus Mucor circinelloides Lacks Centromeric Histone CENP-A and Displays a Mosaic of Point and Regional Centromeres. Curr Biol. 2019; 29(22):3791–3802. 10.1016/j.cub.2019.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salcedo-Hernandez R, Ruiz-Herrera J. Isolation and characterization of a mycelial cytochrome aa3-deficient mutant and the role of mitochondria in dimorphism of Mucor rouxii. Exp Mycol. 1993; 17:142–54. 10.1006/emyc.1993.1013 [DOI] [Google Scholar]
- 47.Trieu TA, Navarro-Mendoza MI, Pérez-Arques C, Sanchis M, Capilla J, Navarro-Rodriguez P, et al. RNAi-Based Functional Genomics Identifies New Virulence Determinants in Mucormycosis. PLoS Pathog. 2017; 13: e1006150 10.1371/journal.ppat.1006150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Heeswijck R, Roncero MIG. High frequency transformation of Mucor with recombinant plasmid DNA. Carlsberg Research Communications. 1984; 49:691–702. 10.1007/BF02907500. [DOI] [Google Scholar]
- 49.Arnau J, Strøman P. Gene replacement and ectopic integration in the zygomycete Mucor circinelloides. Curr Genet. 1993; 23(5–6):542–6. 10.1007/BF00312649 [DOI] [PubMed] [Google Scholar]
- 50.Gutiérrez A, López-García S, Garre V. High reliability transformation of the basal fungus Mucor circinelloides by electroporation. J Microbiol Methods. 2011; 84:442–6. 10.1016/j.mimet.2011.01.002 [DOI] [PubMed] [Google Scholar]
- 51.Sambrook JF, Russell DW. Molecular cloning: A laboratory manual, 3 ed Ed. Cold Spring Harbor Press; 2001. [Google Scholar]
- 52.Oshel P. HMDS and specimen drying for SEM. Micros today. 1997; 5(4): 16–7 10.1017/S155192950006140X. [DOI] [Google Scholar]
- 53.Secretaría de Agricultura, Ganadería, Desarrollo Rural, Pesca y Alimentación (SAGARPA) NORMA Oficial Mexicana NOM-062- ZOO-1999. Especificaciones técnicas para la producción, cuidado y uso de los animales de laboratorio. Diario Oficial de la Federación 2. 2001; 107–67 (In Spanish).
- 54.Rodríguez-Andrade E, Hernández-Ramírez KC, Díaz-Peréz SP, Díaz-Magaña A, Chávez-Moctezuma MP, Meza-Carmen V et al. Genes from pUM505 plasmid contribute to Pseudomonas aeruginosa virulence. Antonie Van Leeuwenhoek. 2016; 109: 389–96. 10.1007/s10482-015-0642-9 [DOI] [PubMed] [Google Scholar]
- 55.Corrochano LM, Kuo A, Marcet-Houben M, Polaino S, Salamov A, Villalobos-Escobedo JM, et al. Expansion of Signal Transduction Pathways in Fungi by Extensive Genome Duplication. Curr Biol. 2016; 26(12):1577–84. 10.1016/j.cub.2016.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The spores from the independent M. circinelloides knockout strains produced in YPG were observed under A) light microscope (100 X), scale bar is equal to 20 μm. B) Under scanning electron microscope. Representative photographs from the corresponding strains of M. circinelloides (1000 X), scale bar is equal to 10 μm.
(DOCX)
Ena1 from Hansenulla polymorpha was used as bait to find Ena homologues in M. circinelloides. A) Clustal W alignment analysis. Identical and similar amino acid residues are shown in black and grey boxes, respectively. B) Identity analysis of Ena homologues in M. circinelloides is shown as identity/similarity in percentage, Mc: M. circinelloides; Hp: H. polymorpha.
(DOCX)
A) Effect of inhibitor of calcineurin FK506 on the growth of M. circinelloides. Radial growth of spores grown on YPG or YNB agar plates recorded each day during the experiment. Representative photos show radial growth after 3 days. Three independent experiments were performed for each condition. B) 100 spores from each strain were inoculated by spreading on YPG and YNB plates supplemented with 0.005% Triton X-100. Plates were incubated for 1 day. The bars represent the number of colonies formed with treatment versus the colonies formed without treatment. Three independent experiments were performed for each condition. *Statistically significant difference (ANOVA, Fisher, p<0.05).
(DOCX)
A) gpa11; B) gpa12 and C) gpa11/gpa12 mutation strategy. The 5′ and 3′ regions upstream and downstream from the start and stop translation codons, respectively, were used to flank the pyrG (single mutants) or leuA (double mutant) selective marker. The diagrams show the recombinant fragments used to delete the gpa genes in protoplasts of M. circinelloides MU402 wild-type strain. The photographs show the molecular confirmation by PCR with specific primers for each gene showing specific PCR-bands that indicate recombination in the corresponding gpa loci. M: Molecular size markers (kb). C+: PCR control for positive amplification (wt: 1.1 kb amplicon to identify the wt gpa11 or gpa12 genes); Δgpa11: 1.4 kb amplicon to identify the deletion of gpa11 gene; Δgpa12: 1.3 kb amplicon to identify the deletion of gpa12 gene; and Δgpa11/Δgpa12: 2.2 kb amplicon to identify the deletion of gpa11 gene in the mutant Δgpa12.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.