Abstract
Background/Aim: Gremlin1 (GREM1) plays an important role in certain malignancies by antagonising bone morphogenetic proteins and regulating angiogenesis directly/indirectly. The present study aimed to investigate the role of Gremlin1 in the development and progression of gastric cancer (GC). Materials and Methods: Expression of GREM1 in GCs was examined using quantitative real time PCR and The Cancer Genomic Atlas (TCGA) data. Influence on cellular functions was determined in both Gremlin1 knockdown and overexpression cell line models. Results: GREM1 expression was up-regulated in GCs, which was correlated with poorer survival. Increased GREM1 expression was significantly correlated with tumour growth/invasion and lymphatic metastasis. Gremlin1 promoted proliferation and tumourigenic capacity of GC cells in vitro. GREM1 expression was associated with epithelial mesenchymal transition (EMT), angiogenesis and lymphangiogenesis in GC. Conclusion: Increased GREM1 expression in GCs is associated with disease progression and poor prognosis in which EMT, angiogenesis and lymphangiogenesis are likely involved.
Keywords: Gremlin1, bone morphogenetic protein, gastric cancer
Gastric cancer (GC) is one of the leading cancers in China accounting for 42.6% of the new cases diagnosed and 45.0% of the deaths due to the disease worldwide each year (1). Bone morphogenetic proteins (BMPs) play an important part in regulating the homeostasis of the gastric epithelium and tumorigenesis (2,3). BMP signaling regulates the homeostasis of the gastric epithelium through its ability to control the biological functions of the parietal cells (2). Inhibition of BMP signaling in the gastric mucosa leads to severe abnormalities in the proliferation, maturation, and differentiation of several lineages of gastric epithelial cells, and further, form metaplasia, atypical hyperplasia and tumours (2,4). For example, BMP2 inhibits proliferation of GC cells. Methylation-related down-regulation of BMP2 has been observed in GCs (5,6). Other studies have also detected a decrease in the expression of molecules in the BMP signaling pathway in gastric cancer (7), which reflects the inhibitory effect of BMPs on gastric cancer proliferation. BMPs are also related to metastasis of GC. BMP4 expression has been shown to be inversely correlated with the prevalence of metastasis to lymph nodes and also with local invasion (8). However, controversy remains for the exact role played by BMPs in GC. For instance, BMP2 plays a positive role in pathological differentiation and lymph node metastasis through activation of the PI3K/AKT and MAPK pathways (9). It is suggested that GC cells may benefit from either the loss of BMPs for their proliferation or from the BMP-induced invasive traits. Further investigation on the role played by BMPs in GC warrants a better and more comprehensive understanding of the disease.
Gremlin1 acts as an antagonist by preventing binding of bone morphogenetic proteins to their receptors. It is one of the most important direct antagonists of BMP2,4,7, which can block the BMP signalling pathway. It has been reported that Gremlin1 plays an important part in regulating organogenesis, body patterning and tissue differentiation (10,11). Emerging evidence has shown that Gremlin1 is expressed in a number of malignancies including skin cancer, GC, lung cancer, kidney cancer, testicular cancer and so on (12). Immunochemical staining showed that Gremlin1 is positively expressed in GC tumours (12-14). Yamasaki et al. have shown that the expression of Gremlin1 was correlated with shallower tumour depth, smaller tumour size, less nodal involvement, vessel invasion and also a better 5-year survival rate (13). However, the molecular and cellular machinery underlying the involvement of Gremlin1 in the disease progression of GC remains unknown. In this study, in addition to the quantitative analysis of Gremlin1 in two different cohorts of GC tumours, GC cell lines were employed to investigate the influence of knockdown or overexpression of Gemlin1 on cellular functions.
Materials and Methods
GC tumour samples. GC tumours (n=321) and paired adjacent control healthy tissues (n=183) were collected immediately after surgery at the Peking University Cancer Hospital (PUCH) with written consent from all the patients. The specimens were stored at –80˚C until use. The collection and usage of the GC specimens were carried out in accordance with protocols and procedures approved by the Peking University Cancer Hospital Research Ethics Committee.
Analysis of GREM1 expression in human GC tissues using gene expression array data. We evaluated the expression of GREM1 in gastric cancer tissues (n=274) compared to normal gastric tissues (n=33) in the gastric adenocarcinoma cohort of The Cancer Gene Atlas (TCGA) database. The correlations between the GREM1 expression and the clinicopathological parameters and survival were analysed.
Kaplan–Meier survival analysis was also performed to evaluate the relationship of GREM1 with the prognosis of patients with an auto-selected cut-off value.
TCGA database was also employed for the evaluation of the correlation between GREM1 and key genes relevant to the hallmarks of cancer including epithelial mesenchymal transition (EMT), angiogenesis and lymphangiogenesis.
RNA isolation and reverse transcription-polymerase chain reaction (RT-PCR). Total RNA was extracted using TRI Reagent kit (Sigma-Aldrich, Inc., Dorset, UK). Reverse transcription was performed to convert 2 μg of total RNA to complementary DNA using the GoScript™ Reverse Transcription System (Promega, Southampton, UK). Polymerase chain reactions were subsequently carried out for amplification of GREM1 (forward:5’-CTGCTGAAGGGAAAA AGAA; reverse:5’-GATGGATATGCAACGACACT), SNAI1 (forward:5’-CGCTCTTTCCTCGTCAG; reverse:5’-GTTGCAG TATTTGCAGTTGA), SNAI2 (forward:5’-CTCTCCTCTTTCCGG ATACT; reverse:5’-AGCAGTTTTTGCACTGGTAT), TWIST1 (forward:5’-AGCAACAGCGAGGAAGAG; reverse: 5’-GAGGA CCTGGTAGAGGAAGT), ID1 (forward:5’-TCAACGGCGA GATCAG; reverse:5’-GATCGTCCGCAGGAA), ID2 (forward:5’-GAACACGGATATCAGCATC; reverse:5’-ACAGTGCTTTGCT GTCATTT), ID3 (forward:5’-GCGTCATCGACTACATTCTC; reverse:5’-GTCGTTGGAGATGACAAGTT), VEGFA (forward:5’-GAGCCGGAGAGGGAG; reverse:5’-CTGGGACCACTTGGCAT), VEGFC (forward:5’-AGTCGCGACAAACACCTTCT; reverse:5’-CATCCAGCTCCTTGTTTGGT), VEGFD (forward:5’-TGGA ACGATCTGAACAGCAG; reverse:5’-TTCTTCAGGGATCTGGA TGG) and KDR (forward:5’-AACTGAAGACAGGCTACTTG; reverse:5’-GTCGTTCACAATGTTCATCC). After an initial 5-min denaturation step, the target gene was amplified for 25-30 cycles (95˚C for 30 sec, 55˚C for 30 sec and 72˚C for 30 sec), followed by a final extension at 72˚C for 5 min. GAPDH (forward: 5’-GGCTGCTTTTAACTCTGGTA; reverse: 5’-GACTGTGGTCA TGAGTCCTT) was employed as a house-keeping gene.
Cell lines and cell culture. HGC27 and AGS cell lines were purchased from Sigma. 293T cells were obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA). MKN7, NUGC4, MKN45 cell lines were kindly provided by Peking University Cancer Hospital (PUCH). All cell lines were routinely cultured at 37˚C with 5% CO2 and 95% humidity in Dulbecco’s modified Eagle medium (DMEM; Sigma-Aldrich) supplemented with 5% foetal bovine serum (FBS; HyClone, Fisher Scientific UK Ltd., Loughborough, UK).
Western blot. Cells were lysed when they reached 70%-90% confluence. Protein concentration was measured with a DC protein assay kit (Bio-Rad Laboratories, Watford, UK). Cell lysates of 30 μg total protein for each sample were separated on 10% SDS-PAGE and then electroblotted onto PVDF membranes. The blotted membranes were incubated for overnight at 4˚C with primary antibodies GAPDH (1:2000, sc-47724, Santa Cruz, Princeton, New Jersey, United States) and Gremlin1 (1:500, sc-515877, Santa Cruz), and then with the corresponding anti-mouse secondary antibody (A5278, 1:1000, Sigma-Aldrich) for 1 hour. Protein bands were visualised with a chemiluminescence detection kit (EZ-ECL, Biological Industries, Cromwell, CT, USA) and documented photographed using Syngene imager (Syngene International Ltd., Cambridge, UK).
Gremlin1 knockdown and overexpression. We obtained the lentiviral GREM1 shRNA (CTGAAGCGAGACTGGTGCAAA) and scramble control (CCTAAGGTTAAGTCGCCCTCG) from Cyagen Biosciences (Santa Clara, CA, USA). Both GREM1 shRNA and scramble shRNA plasmid vectors carried two selective markers in mammalian cells, EGFP and neomycin. Lentiviral particles were packaged using 293T cells together with pMD2G and pSPAX2 plasmid vectors. The full-length human Gremlin1 coding sequence was amplified from a cDNA library derived from normal prostate tissue which was subsequently cloned into pEF6/V5-His TOPO TA plasmid vector (Invitrogen Ltd., Inchinnan, Renfrewshire, UK). The recombinant Gremlin1 plasmid vector and empty vector were separately transfected into the AGS cells using electroporation. The empty vectors were employed as controls. G418 (500 μg/ml) and blasticidin (5 μg/ml) were used for the selection. PCR and western blot were used to verify the expression of Gremlin1 in the transfected cells following transduction and transfection.
In vitro cell growth assay. The cells were seeded into 96-well plates at 3000 cells/well in 200 μl medium and cultured for up to 5 days. The cells were then fixed using 4% formaldehyde and stained with 0.1% crystal violet. Excessive crystal violet was rinsed with tap water. The stained crystal violet was dissolved with 10% acetic acid for determination of the absorbance at a wavelength of 540 nm with a spectrophotometer (Bio-Tek Instruments Inc, Winooski, VT, USA).
In vitro clonogenic assay. Three hundred cells were seeded into each well of a 6-well plate. After a 10-days in culture, the cells were fixed with 4% formaldehyde followed by staining with 0.1% crystal violet. The number of colonies were then counted under an inverted microscope.
In vitro cell adhesion assay. A 96-well plate was pre-coated with Matrigel (5 μg/well) (BD Biosciences, Oxford, UK). Cells were seeded into each well at a density of 30,000 cells/200 μl medium. Non-adherent cells were washed off gently with PBS buffer after incubation for 40 min. Adhered cells were then fixed and stained with crystal violet. The absorbance was measured at 540 nm after the stained crystal violet was dissolved with 10% acetic acid.
In vitro cell invasion assay. Transwell inserts (8 μm pore, Greiner Bio-One Ltd., Stonehouse, UK) were coated with a thin layer of Matrigel (50 μg/insert) and air dried. After rehydration with medium, 30,000 cells were seeded per insert followed by incubation for 3 days. The Matrigel coating and non-invaded cells were removed using a cotton swap. Invaded cells were fixed and stained 0.1% crystal violet which was resolved in 10% acetic acid. The absorbance was measured using the spectrophotometer.
In vitro scratch wounding assays. The cells were seeded into a 6-well plate (two million cells per well). After an overnight culture, the cells were then scratched using a 200 μl pipette tip to create a wound. The wound closure was monitored for 5 h using inverted microscopy equipped with a digital camera. The migration was measured using ImageJ.
Statistical analysis. Following a normality check, t-test was employed for normally distributed data whilst non-normally distributed data were analysed using Mann-Whitney test or One-way ANOVA test. Differences were regarded as statistically significant when the p-value was less than 0.05. Kaplan–Meier survival analyses were performed using both an online platform (kmplot.com) (15) and SPSS statistical software (v25, IBM SPSS, Portsmouth, UK).
Results
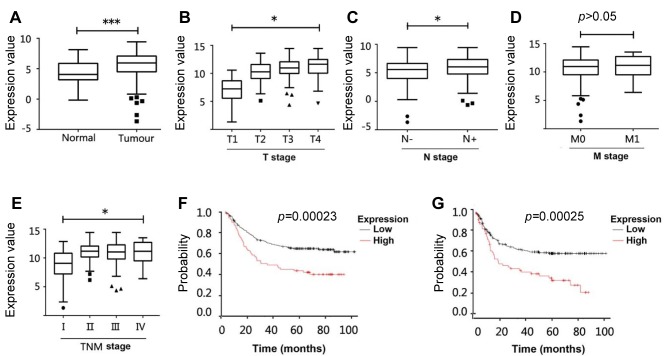
Elevated expression of GREM1 in GCs is associated with poor prognosis. Expression of GREM1 in gastric cancer tissues was quantitatively analysed in comparison with adjacent normal gastric tissues in two GC cohorts from the TCGA and PUCH. As shown in Figure 1A, in the TCGA cohort, the expression of GREM1 in gastric cancer tissues (n=274) was significantly higher than its expression in normal gastric tissues (n=33). Quantitative real time PCR also revealed a higher expression of GREM1 in gastric cancer tissues (n=317) in comparison with adjacent normal gastric tissues (n=181) in the PUCH cohort (Table I).
Figure 1. Gremlin1 expression in GC and its clinical relevance. (A) Expression of GREM1 transcripts in GCs (n=274) and normal tissues (n=33) in the TCGA GC cohort. (B) Correlation between GREM1expression and T staging, including T1 (n=13), T2 (n=70) T3 (n=107) T4 (n=75). (C) GREM1 expression in primary tumours without positive lymph node (n=91) compared with those with lymphatic metastasis (n=171). (D) Expression of GREM1 in tumours presented distant metastases (M1, n=18) in comparison with those had no distant metastasis (M0, n=243). (E) GREM1 and overall TNM staging of the GCs including TNM-I (n=35), TNM-II (n=51), TNM-III (n=155) and TNM-IV (n=18). (F) Kaplan–Meier survival analyses showed correlations between GREM1expression and overall survival of GC patients using the online platform (www.KMplot.com). The cut-off value used in the analysis was 4560. (G) Correlation between GREM1expression and progression free survival (PFS) of GC was analysed (www.KMplot.com), and the cut off value used in the analysis was 4307. *Represents p<0.05, ***represents p<0.001. HR: Hazard ratio.
Table I. The expression of Grem1 transcripts in gastric cancer.
Number in each subgroup are the number of samples which have both gene quantity and clinical information.
Further analyses showed a correlation between GREM1 expression and clinical pathological characteristics of the disease in the TCGA cohort. GREM1 expression was significantly increased in tumours with a more invasive phenotype according to the T stages of the tumours (Figure 1B). The increased expression of GREM1 was also observed in the tumours with lymphatic metastases (Figure 1C). The expression of GREM1 was also associated with the overall TNM staging of the disease, though no marked difference was seen in the tumours that developed distant metastases (Figure 1D and E). However, there was no correlation between the expression of GREM1 and the TNM staging in the PUCH cohort (Table I).
Kaplan–Meier survival analyses showed that overall survival (OS) of patients with high expression of GREM1 (median=15.03 months) was significantly shorter than that in those with lower GREM1 expressing tumours (median=25.87 months) (Figure 1F). Similarly, the elevated expression of GREM1 was also significantly associated with a shorter progression-free survival (PFS, median=9.4 months), compared with those that had lower expression levels of GREM1 (median=13.03 months, p<0.001) (Figure 1G). In addition, Kaplan–Meier survival analysis of GREM1 in the TCGA cohort also showed that patients with higher expression of GREM1 had poorer overall survival (data not shown).
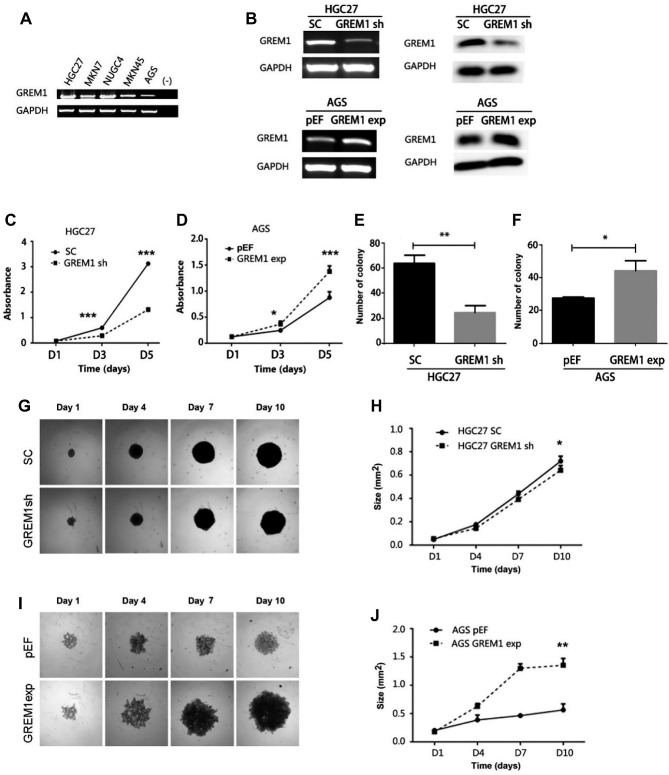
Gremlin1 promotes proliferation of GC cells and tumourigenesis in vitro. In order to explore the biological functions of Gremlin1 in GC cells, we determined the expression of GREM1 in GC cell lines, i.e. HGC27, MKN7, NUGC4, MKN45 and AGS using PCR (Figure 2A). GREM1 is expressed at a relatively lower level in AGS cells in comparison with the other four GC cell lines. Knockdown and overexpression of Gremlin1 were subsequently established in HGC27GREM1sh and AGSGREM1exp using the lentiviral GREM1 shRNA and Gremlin1 overexpression plasmid vectors, respectively (Figure 2B), in comparison with the corresponding controls, i.e. HGC27SC with scrambled shRNA and AGSpEF with the empty plasmid vector.
Figure 2. Gremlin1 regulates proliferation and tumourigenic capacity of GC cells. (A) Gremlin1 was examined for its mRNA expression in GC cell lines using PCR. (B) Gremlin1 knockdown and overexpression in HGC27 and AGS were confirmed using PCR (left) and western blot (right), respectively. Cell proliferation assay was performed for HGC27GREM1sh cells (C) and AGSGREM1exp cells (D) in comparison with the corresponding controls. Colony formation assay was performed on HGC27GREM1sh cells (E) and AGSGREM1exp cells (F). Growth of the HGC27 GREM1sh (G) and AGSGREM1exp (I) cells was examined using a 3D spheroid model. Size of 3-D acini of HGC27 GREM1sh cells (H) and AGSGREM1exp cells (J) was determined using ImageJ compared with their controls. Minimal three independent experiments were performed for all experiments. Shown are representative experimental data. *Represents p<0.05, **represents p<0.01; ***represents p<0.001.
Growth assays showed that Gremlin1 knockdown resulted in reduced proliferation of HGC27GREM1sh cells (Figure 2C), while increased proliferation was observed in AGSGREM1exp cells following the overexpression of Gremlin1 (Figure 2D). Similar impact was seen on the colony formation (Figure 2E and F). The number of colonies was reduced in the HGC27GREM1sh cells, while it was increased in AGSGREM1exp cells compared with the corresponding controls.
The effect of Gremlin1 on cell proliferation was further validated in an in vitro 3D spheroid model (Figure 2G and I). The growth of spheroids formed by the HGC27GREM1sh cells was marginally reduced compared with the HGC27SC control cells (Figure 2G and H). A marked increase was seen in the size of the AGSGREM1exp cell formed spheroids compared with the controls (Figure 2I and J).
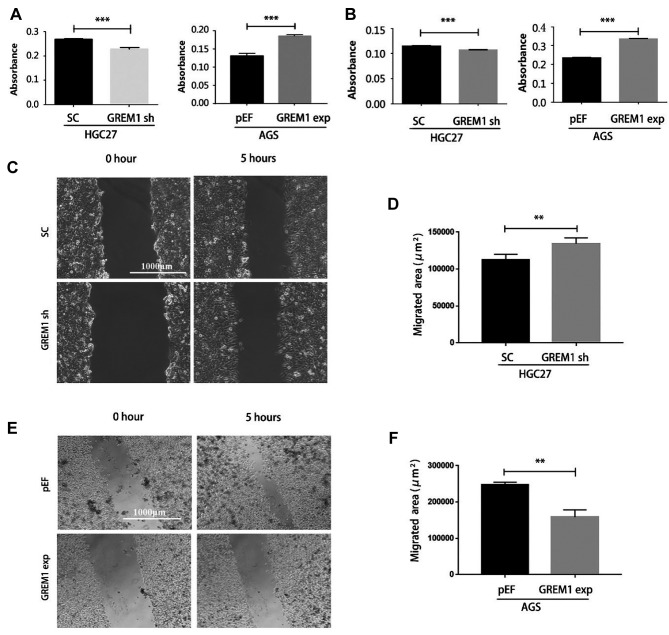
Gremlin1 regulates adhesion, invasion and migration of GC cells. Gemlin1 knockdown resulted in a significant decrease in the adhesion of HGC27GREM1sh cells, while increased adherence was observed following Gremlin1 overexpression in AGS cells (Figure 3A).
Figure 3. Gremlin1 regulates the adhesion, invasion and migration of GC cells. Cell adhesion assay (A) and transwell invasion assay (B) was performed to evaluate the impact of Gremlin1 on the adhesion and invasiveness of GC cell lines. Wounding assays were performed comparing HGC27SC and HGC27GREM1sh with semi-quantification of migration area using Image J (D) and representative images are shown for each cell lines (C). The wounding assay was employed to determine the migration of AGSpEF and AGSGREM1exp followed by semi-quantification of migration area using Image J (E and F). Three independent experiments were performed. **Represent p<0.01, ***represent p<0.001.
As shown in Figure 3B, the AGSGREM1exp cells exhibited enhanced invasiveness, while a marginal decrease was seen in the HGC27 cells following the knockdown of Gremlin1.
Wound assays showed that HGC27GREM1sh cells migrated faster than the control cells (Figure 3C and D), while an obvious reduction was evident in the migration of AGSGREM1exp cells compared with AGSpEF cells (Figure 3E and F).
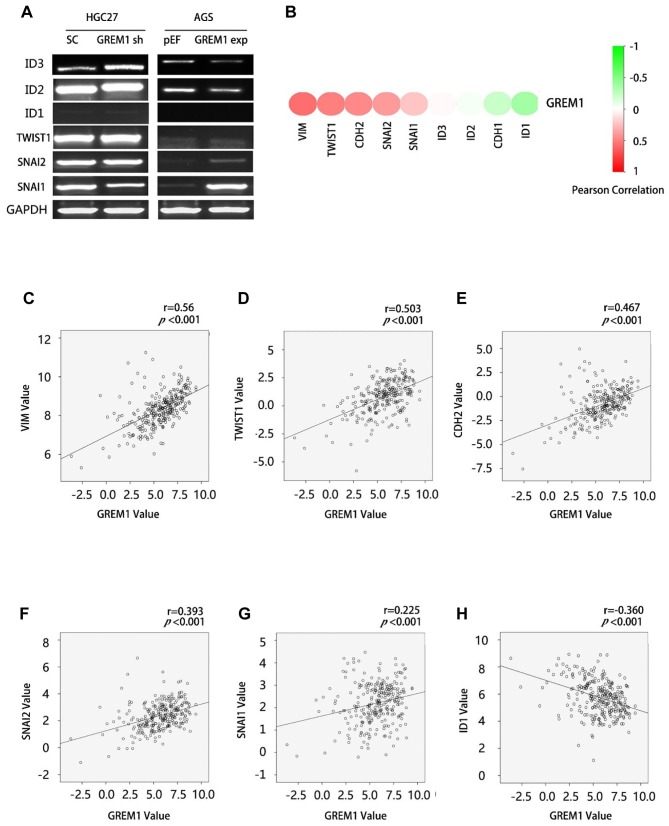
GREM1 and epithelial mesenchymal transition (EMT) in GC. BMP-induced EMT plays an important role in disease progression of solid tumours. Expression of EMT-related genes, particularly of BMP responsive genes were determined in both HGC27GREM1sh and AGSGREM1exp cells compared with their corresponding control cells. An elevated expression of ID3 was seen in the HGC27GREM1sh cells, while a subtle reduction was seen in its expression by the AGSGREM1exp cells (Figure 4A). Similarly, a subtle decrease was seen in the expression of ID2 by the AGSGREM1exp cells. An increased expression was seen for both Snail Family Transcriptional Repressor 2 (SNAI2) and Snail Family Transcriptional Repressor 1 (SNAI1) in the AGSGREM1exp cells.
Figure 4. GREM1 and EMT in GC. (A) PCR were performed to test the expression of EMT markers in the GREM1knockdown HGC27 cells and Gremlin1 overexpression AGS cells. Correlation between GREM1and EMT markers in the TCGA-GC cohort was analysed using Spearman tests, results shown as a heatmap (B) and scatter plots of significantly related EMT markers (C-H).
Furthermore, we analysed the correlation between GREM1 and these EMT-related genes in the TCGA GC cohort. It was found that the expression levels of GREM1 were negatively correlated with the expression of ID1 and ID2, but positively correlated with SNAI1, SNAI2 and the Twist family BHLH transcription factor 1 (TWIST1) (Figure 4).
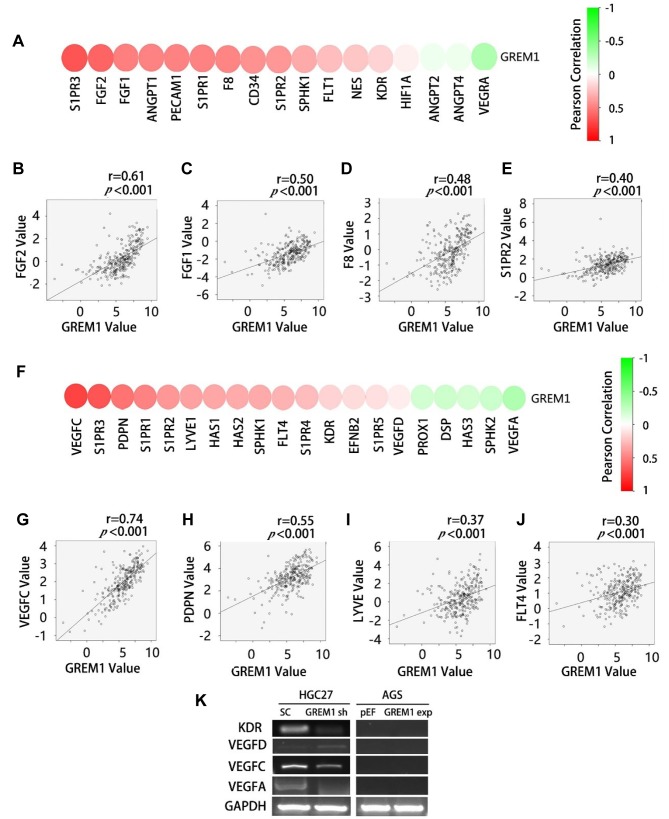
GREM1 and tumour associated angiogenesis and lymphangiogenesis markers. Gremlin1 promotes angiogenesis through an interaction with kinase insert domain receptor (KDR, also known as vascular endothelial growth factor receptor 2, VEGFR2). In the present study, we evaluated the correlation between GREM1 and key genes relevant to angiogenesis and lymphangiogenesis in the TCGA cohort. The expression of GREM1 in GCs was positively correlated with most of the angiogenesis factors/markers, especially fibroblast growth factor-2 (FGF2), fibroblast growth factor-1 (FGF1), factor VIII (F8) and sphingosine 1-phosphate receptor 2 (S1PR2) (Figure 5A-E). Furthermore, a positive correlation was seen between GREM1 and most of the lymphangiogenesis factors, especially vascular endothelial growth factor C (VEGFC), podoplanin (PDPN), lymphatic vessel endothelial hyaluronic acid receptor 1 (LYVE1) and Fms related tyrosine kinase 4 (FLT4, also known as vascular endothelial growth factor receptor 3, VEGFR3) (Figure 5F-J). PCR showed a reduced expression of KDR and VEGFC in HGC27GREM1sh cells after knockdown of Gremlin1 (Figure 5K).
Figure 5. Association between GREM1 and angiogenesis/lymphangiogenesis in GC. Correlation between GREM1and angiogenesis markers were analysed using Spearman tests, results shown as a heatmap (A) and scatter plots (B-E). Correlation between Gremmlin1 and lymphangiogenesis markers were shown as a heatmap (F) and scatter plots (G-J). The expression of angiogenesis/lymphangiogenesis markers in the GREM1 knockdown HGC27 cells and GREM1 overexpression AGS cells was examined using PCR (K).
Discussion
In this study, the expression of Gremlin1 in gastric cancer specimens and normal gastric tissues was determined and analysed. It was found that the expression of Gremlin1 in gastric cancer was significantly elevated compared with normal tissues. The increased expression of Gremlin1 in GC was evident in the TCGA cohort. Further analyses showed that Gremlin1 was positively correlated with tumour growth/invasion and lymph node metastasis. Survival analyses showed that increased expression of Gremlin1 in GCs resulted in poorer OS and PFS. It can thus be suggested that the elevated expression of Gremlin1 in GC is involved in disease progression. Further evaluation using immunohistochemistry will clarify its predictive potential for the prognosis of GC.
To clarify the mechanism of Gemlin1 involvement in GC progression, we determined the influence of Gremlin1 on the properties of GC cell lines. Knockdown of Gremlin1 in HGC27 or its overexpression in AGS cell lines provided contrasting models to examine the biological functions of Gremlin1 in GC cells. Proliferation assay, colony formation and 3D spheroid growth assays revealed that Gremlin1 promoted proliferation, colony formation, and formation and growth of spheroids of GC cells in vitro. The positive impact on the proliferation and tumourigenic capacity of GC cells is in line with its implication in disease progression. This is also supported by a very recently published study in which showed the tumourigenic potential of Gremlin1using an organoid model (16). BMPs have been shown to inhibit proliferation of epithelial cells and epithelium derived cancerous cells through canonical signaling pathways, upon binding to their receptors (17). Such an inhibitory effect on proliferation was also evident for BMP2 in GC (5,6). As a BMP antagonist, the elevated expression of Gremlin1 in GC plays a protective role by antagonizing the inhibition of BMPs on proliferation. However, the most involved BMP is yet to be revealed. Furthermore, the expression profile of different BMP receptors and intracellular downstream molecules should also be considered in the context of tumour microenvironment in order to explain the differential responses to certain BMPs. This could be specific to certain ligands and receptors in cancers (18). Further investigation of other BMPs, BMP receptors, intracellular signaling molecules and regulatory factors in GC will help to build up a comprehensive understanding of the role of BMPs in GC.
Unlike the effect on proliferation, functional assays showed that Gremlin1 knockdown promoted invasion and migration of GC cells by eliminating the antagonistic effect on BMP-induced invasiveness and migration of cancer cells (19). Interestingly, the invasion of AGSGREM1exp cells was also increased in comparison with the AGSpEF cells, rather than decreased. Crosstalk among different growth factors/cytokines and their downstream signal transduction pathways may help to answer this question. For example, BMP and its signalling pathways are intricately linked to many other growth factors and signaling pathways in cancer cells, including epidermal growth factor receptor (EGFR) pathway (RTK/MAPK pathway (20-22), PI3K/Akt pathway (23-25), HGF/MET pathway (26,27) [62, 63]and so on. The change in invasion is the result of the integration of the intricate signal network. Exploration of the interaction between these signaling pathways may help to understand how Gremlin1 regulates the invasion and migration of GC cells.
EMT is an important event during the development and progression of cancer, causing disruption of epithelial homeostasis that may lead to carcinogenesis. It is also a vital part of the transformation of indolent tumour cells into a more aggressive colony, leading to metastasis (28,29). Regulation of EMT by BMPs has been implicated in many studies regarding organ development (30,31) and cancer (32-35). BMPs can induce EMT by both Smad-dependent (36-38) and Smad-independent pathways (39,40). For instance, BMP signalling could directly activate the expression of the EMT transcription factors, Snail, TWIST1 and MSX1/2 (36-38). Regarding the Smad-independent pathway, BMP2 could induce EMT and invasion through regulation of the PI3K/Akt pathway (39,40). In the current study, we found that the expression levels of GREM1were negatively correlated with the expression of ID1, ID2 and ID3, but were positively correlated with Snai1, SNAI2 and TWIST1. It can be seen that GREM1 may also be involved in EMT in gastric cancer, and the specific mechanism needs further exploration.
Angiogenesis is indispensable for the development and progression of both primary and secondary tumours. Our study revealed that GREM1 was positively correlated with angiogenic markers, suggesting that Gremlin1 is likely to participate in the angiogenesis of GC by antagonizing BMP. It has been shown that BMPs can affect directly and indirectly angiogenesis. BMPs and their downstream Smads can regulate proliferation, migration, or tubule formation of vascular endothelial cells (41). On the other hand, BMPs can regulate angiogenesis indirectly through regulation of the expression of VEGF in both cancer cells and osteoblasts (42). For example, BMP2 has been shown to promote tumour-related angiogenesis through up-regulation of VEGF via the p38 pathway in breast cancer (43). BMP9 has been shown to inhibit the proliferation of endothelial cells, as well as to inhibit VEGF mediated angiogenesis, via ACVRL1 and BMPR2 and downstream Smad1/5 signalling (44). In addition, Gremlin1 can directly target VEGF receptors in vascular endothelial cells, promoting angiogenesis as a direct agonist rather than through the interaction with BMP ligands (45). At present, the specific mechanism by which Gremlin1 promotes angiogenesis in gastric cancer is yet to be clarified. However, the expression profile of both BMP/BMP receptors and VEGFs/VEGFRs should be considered.
Lymphangiogenesis plays an important role in cancer metastasis, especially in the dissemination of cancer cells through lymphatic vessels (46). In this study, the expression of GREM1 in GCs with lymph node metastases was significantly higher than that in GC without lymph node metastasis. Furthermore, there was a positive correlation between GREM1 and lymphangiogenesis factors such as VEGFC, PDPN and LYVE. The expression of VEGFC in HGC27GREM1sh cells was indeed lower compared to the control cells. This suggested that Gremlin1 may also be involved in GC by affecting lymphangiogenesis which is yet to explored.
Conclusion
In summary, expression of Gremlin1 is increased in GC, and elevated expression of Gremlin1 is significantly associated with poorer survival of GC patients. Gremlin1 promotes proliferation and tumourigenesis of GC cells in vitro. Furthermore, Gremlin1 may be involved in EMT, angiogenesis and lymphangiogenesis in GC. Further research is required to identify the specific GCs in which Gremlin1 is indispensable for the disease progression, in order to identify novel predictive and/or therapeutic targets.
Conflicts of Interest
The Authors declare that they have no competing interests regarding this study.
Acknowledgements
Dr Zhiwei Sun is a recipient of the Chinese Scholarship from Cardiff University. The authors thank the support for this collaborative research of gastric cancer from both Peking University and Cardiff University.
Copyright
Authors retain copyright. The unrestricted non-commercial use, distribution and reproduction in any medium of CGP articles for academic reasons is allowed, provided that the original work is properly cited.
Authors’ Contributions
LY designed the experiments of the study. ZS and LY wrote the manuscript. ZS, CS and CL performed experiments. ZS, YC, WGJ and LY did the data analyses. ZS, CS, CL, YC, WGJ and LY made contributions to the revision and proof reading.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. PMID: 15761078. DOI: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Todisco A. Regulation of gastric metaplasia, dysplasia, and neoplasia by bone morphogenetic protein signaling. Cell Mol Gastroenterol Hepatol. 2017;3(3):339–347. doi: 10.1016/j.jcmgh.2017.01.014. PMID: 28462376. DOI: 10.1016/j.jcmgh.2017.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willet SG, Mills JC. Stomach organ and cell lineage differentiation: From embryogenesis to adult homeostasis. Cell Mol Gastroenterol Hepatol. 2016;2(5):546–559. doi: 10.1016/j.jcmgh.2016.05.006. PMID: 27642625. DOI: 10.1016/j.jcmgh.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maloum F, Allaire JM, Gagne-Sansfacon J, Roy E, Belleville K, Sarret P, Morisset J, Carrier JC, Mishina Y, Kaestner KH, Perreault N. Epithelial bmp signaling is required for proper specification of epithelial cell lineages and gastric endocrine cells. Am J Physiol Gastrointest Liver Physiol. 2011;300(6):G1065–G1079. doi: 10.1152/ajpgi.00176.2010. PMID: 21415412. DOI:10.1152/ajpgi.00176.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wen XZ, Miyake S, Akiyama Y, Yuasa Y. Bmp-2 modulates the proliferation and differentiation of normal and cancerous gastric cells. Biochem Biophys Res Commun. 2004;316(1):100–106. doi: 10.1016/j.bbrc.2004.02.016. PMID: 15003517. DOI: 10.1016/j.bbrc.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 6.Wen XZ, Akiyama Y, Baylin SB, Yuasa Y. Frequent epigenetic silencing of the bone morphogenetic protein 2 gene through methylation in gastric carcinomas. Oncogene. 2006;25(18):2666–2673. doi: 10.1038/sj.onc.1209297. PMID: 16314833. DOI: 10.1038/sj.onc.1209297. [DOI] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–209. doi: 10.1038/nature13480. PMID: 25079317. DOI: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim SG, Park HR, Min SK, Choi JY, Koh SH, Kim JW, Lee HW. Expression of bone morphogenic protein-4 is inversely related to prevalence of lymph node metastasis in gastric adenocarcinoma. Surg Today. 2011;41(5):688–692. doi: 10.1007/s00595-010-4320-2. PMID: 21533942. DOI: 10.1007/s00595-010-4320-2. [DOI] [PubMed] [Google Scholar]
- 9.Kang MH, Oh SC, Lee HJ, Kang HN, Kim JL, Kim JS, Yoo YA. Metastatic function of bmp-2 in gastric cancer cells: The role of pi3k/akt, mapk, the nf-kappab pathway, and mmp-9 expression. Exp Cell Res. 2011;317(12):1746–1762. doi: 10.1016/j.yexcr.2011.04.006. PMID: 21570392. DOI: 10.1016/j.yexcr.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 10.van Vlodrop IJ, Baldewijns MM, Smits KM, Schouten LJ, van Neste L, van Criekinge W, van Poppel H, Lerut E, Schuebel KE, Ahuja N, Herman JG, de Bruine AP, van Engeland M. Prognostic significance of gremlin1 (grem1) promoter cpg island hypermethylation in clear cell renal cell carcinoma. Am J Pathol. 2010;176(2):575–584. doi: 10.2353/ajpath.2010.090442. PMID: 2808066. DOI: 10.2353/ajpath.2010.090442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khokha MK, Hsu D, Brunet LJ, Dionne MS, Harland RM. Gremlin is the bmp antagonist required for maintenance of shh and fgf signals during limb patterning. Nat Genet. 2003;34(3):303–307. doi: 10.1038/ng1178. PMID: 12808456. DOI: 10.1038/ng1178. [DOI] [PubMed] [Google Scholar]
- 12.Laurila R, Parkkila S, Isola J, Kallioniemi A, Alarmo EL. The expression patterns of gremlin 1 and noggin in normal adult and tumor tissues. Int J Clin Exp Pathol. 2013;6(7):1400–1408. PMID: 23826422. [PMC free article] [PubMed] [Google Scholar]
- 13.Yamasaki Y, Ishigami S, Arigami T, Kita Y, Uchikado Y, Kurahara H, Kijima Y, Maemura K, Natsugoe S. Expression of gremlin1 in gastric cancer and its clinical significance. Med Oncol. 2018;35(3):30. doi: 10.1007/s12032-017-1073-4. PMID: 5797189. DOI: 10.1007/s12032-017-1073-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honma R, Sakamoto N, Ishikawa A, Taniyama D, Fukada K, Hattori T, Sentani K, Oue N, Tanabe K, Ohdan H, Yasui W. Clinicopathological and prognostic significance of epithelial gremlin1 expression in gastric cancer. Anticancer Res. 2018;38(3):1419–1425. doi: 10.21873/anticanres.12366. PMID: 29491067. DOI: 10.21873/anticanres.12366. [DOI] [PubMed] [Google Scholar]
- 15.Szasz AM, Lanczky A, Nagy A, Forster S, Hark K, Green JE, Boussioutas A, Busuttil R, Szabo A, Gyorffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7(31):49322–49333. doi: 10.18632/oncotarget.10337. PMID: 27384994. DOI: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urbischek M, Rannikmae H, Foets T, Ravn K, Hyvonen M, de la Roche M. Organoid culture media formulated with growth factors of defined cellular activity. Sci Rep. 2019;9(1):6193. doi: 10.1038/s41598-019-42604-0. PMID: 30996238. DOI: 10.1038/s41598-019-42604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zabkiewicz C, Resaul J, Hargest R, Jiang WG, Ye L. Bone morphogenetic proteins, breast cancer, and bone metastases: Striking the right balance. Endocr Relat Cancer. 2017;24(10):R349–R366. doi: 10.1530/ERC-17-0139. PMID: 28733469. DOI: 10.1530/ERC-17-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye L, Lewis-Russell JM, Kyanaston HG, Jiang WG. Bone morphogenetic proteins and their receptor signaling in prostate cancer. Histol Histopathol. 2007;22(10):1129–1147. doi: 10.14670/HH-22.1129. PMID: 17616940. DOI: 10.14670/HH-22.1129. [DOI] [PubMed] [Google Scholar]
- 19.Scherberich A, Tucker RP, Degen M, Brown-Luedi M, Andres AC, Chiquet-Ehrismann R. Tenascin-w is found in malignant mammary tumors, promotes alpha8 integrin-dependent motility and requires p38mapk activity for bmp-2 and tnf-alpha induced expression in vitro. Oncogene. 2005;24(9):1525–1532. doi: 10.1038/sj.onc.1208342. PMID: 15592496. DOI: 10.1038/sj.onc.1208342. [DOI] [PubMed] [Google Scholar]
- 20.Lehmann K, Janda E, Pierreux CE, Rytomaa M, Schulze A, McMahon M, Hill CS, Beug H, Downward J. Raf induces tgfbeta production while blocking its apoptotic but not invasive responses: A mechanism leading to increased malignancy in epithelial cells. Genes Dev. 2000;14(20):2610–2622. doi: 10.1101/gad.181700. PMID: 316988. DOI: 10.1128/aac.16.6.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oft M, Peli J, Rudaz C, Schwarz H, Beug H, Reichmann E. Tgf-beta1 and ha-ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev. 1996;10(19):2462–2477. doi: 10.1101/gad.10.19.2462. PMID: 8843198. DOI: 10.1101/gad.10.19.2462. [DOI] [PubMed] [Google Scholar]
- 22.Yue J, Mulder KM. Requirement of ras/mapk pathway activation by transforming growth factor beta for transforming growth factor beta 1 production in a smad-dependent pathway. J Biol Chem. 2000;275(40):30765–30773. doi: 10.1074/jbc.M000039200. PMID: 10843986. DOI: 10.1074/jbc.M000039200. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Liao J, Lu Y, Duan X, Sun W. Activation of the pi3k/akt pathway mediates bone morphogenetic protein 2-induced invasion of pancreatic cancer cells panc-1. Pathol Oncol Res. 2011;17(2):257–261. doi: 10.1007/s12253-010-9307-1. PMID: 20848249. DOI: 10.1007/s12253-010-9307-1. [DOI] [PubMed] [Google Scholar]
- 24.Bakin AV, Tomlinson AK, Bhowmick NA, Moses HL, Arteaga CL. Phosphatidylinositol 3-kinase function is required for transforming growth factor beta-mediated epithelial to mesenchymal transition and cell migration. J Biol Chem. 2000;275(47):36803–36810. doi: 10.1074/jbc.M005912200. PMID: 10969078. DOI: 10.1074/jbc.M005912200. [DOI] [PubMed] [Google Scholar]
- 25.Lamouille S, Derynck R. Cell size and invasion in tgf-beta-induced epithelial to mesenchymal transition is regulated by activation of the mtor pathway. J Cell Biol. 2007;178(3):437–451. doi: 10.1083/jcb.200611146. PMID: 2064840. DOI: 10.1083/jcb.200611146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye L, Lewis-Russell JM, Sanders AJ, Kynaston H, Jiang WG. Hgf/sf up-regulates the expression of bone morphogenetic protein 7 in prostate cancer cells. Urol Oncol. 2008;26(2):190–197. doi: 10.1016/j.urolonc.2007.03.027. PMID: 18312940. DOI: 10.1016/j.urolonc.2007.03.027. [DOI] [PubMed] [Google Scholar]
- 27.Ye L, Lewis-Russell JM, Davies G, Sanders AJ, Kynaston H, Jiang WG. Hepatocyte growth factor up-regulates the expression of the bone morphogenetic protein (bmp) receptors, bmpr-ib and bmpr-ii, in human prostate cancer cells. Int J Oncol. 2007;30(2):521–529. PMID: 17203235. [PubMed] [Google Scholar]
- 28.Larue L, Bellacosa A. Epithelial-mesenchymal transition in development and cancer: Role of phosphatidylinositol 3’ kinase/akt pathways. Oncogene. 2005;24(50):7443–7454. doi: 10.1038/sj.onc.1209091. PMID: 16288291. DOI: 10.1038/sj.onc.1209091. [DOI] [PubMed] [Google Scholar]
- 29.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. doi: 10.1038/nrm3758. PMID: 4240281. DOI: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakajima Y, Yamagishi T, Hokari S, Nakamura H. Mechanisms involved in valvuloseptal endocardial cushion formation in early cardiogenesis: Roles of transforming growth factor (tgf)-beta and bone morphogenetic protein (bmp) Anat Rec. 2000;258(2):119–127. doi: 10.1002/(SICI)1097-0185(20000201)258:2<119::AID-AR1>3.0.CO;2-U. PMID: 10645959. DOI: 10.1002/(SICI)1097-0185(20000201)258:2<119::AID-AR1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 31.Romano LA, Runyan RB. Slug is an essential target of tgfbeta2 signaling in the developing chicken heart. Dev Biol. 2000;223(1):91–102. doi: 10.1006/dbio.2000.9750. PMID: 10864463. DOI: 10.1006/dbio.2000.9750. [DOI] [PubMed] [Google Scholar]
- 32.Yang S, Zhong C, Frenkel B, Reddi AH, Roy-Burman P. Diverse biological effect and smad signaling of bone morphogenetic protein 7 in prostate tumor cells. Cancer Res. 2005;65(13):5769–5777. doi: 10.1158/0008-5472.CAN-05-0289. PMID: 15994952. DOI: 10.1158/0008-5472.CAN-05-0289. [DOI] [PubMed] [Google Scholar]
- 33.Montesano R. Bone morphogenetic protein-4 abrogates lumen formation by mammary epithelial cells and promotes invasive growth. Biochem Biophys Res Commun. 2007;353(3):817–822. doi: 10.1016/j.bbrc.2006.12.109. PMID: 17189614. DOI: 10.1016/j.bbrc.2006.12.109. [DOI] [PubMed] [Google Scholar]
- 34.Piek E, Moustakas A, Kurisaki A, Heldin CH, ten Dijke P. Tgf-(beta) type i receptor/alk-5 and smad proteins mediate epithelial to mesenchymal transdifferentiation in nmumg breast epithelial cells. J Cell Sci. 1999;112(Pt 24):4557–4568. doi: 10.1242/jcs.112.24.4557. PMID: 10574705. [DOI] [PubMed] [Google Scholar]
- 35.Yang S, Du J, Wang Z, Yuan W, Qiao Y, Zhang M, Zhang J, Gao S, Yin J, Sun B, Zhu T. Bmp-6 promotes e-cadherin expression through repressing deltaef1 in breast cancer cells. BMC Cancer. 2007;7:211. doi: 10.1186/1471-2407-7-211. PMID: 2217560. DOI: 10.1186/1471-2407-7-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luna-Zurita L, Prados B, Grego-Bessa J, Luxan G, del Monte G, Benguria A, Adams RH, Perez-Pomares JM, de la Pompa JL. Integration of a notch-dependent mesenchymal gene program and bmp2-driven cell invasiveness regulates murine cardiac valve formation. J Clin Invest. 2010;120(10):3493–3507. doi: 10.1172/JCI42666. PMID: 2947227. DOI: 10.1172/JCI42666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma L, Lu MF, Schwartz RJ, Martin JF. Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development. 2005;132(24):5601–5611. doi: 10.1242/dev.02156. PMID: 16314491. DOI: 10.1242/dev.02156. [DOI] [PubMed] [Google Scholar]
- 38.Dyer L, Lockyer P, Wu Y, Saha A, Cyr C, Moser M, Pi X, Patterson C. Bmper promotes epithelial-mesenchymal transition in the developing cardiac cushions. PLoS One. 2015;10(9):e0139209. doi: 10.1371/journal.pone.0139209. PMID: 4587915. DOI: 10.1371/journal.pone.0139209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang MH, Kang HN, Kim JL, Kim JS, Oh SC, Yoo YA. Inhibition of pi3 kinase/akt pathway is required for bmp2-induced emt and invasion. Oncol Rep. 2009;22(3):525–534. doi: 10.3892/or_00000467. PMID: 19639199. DOI: 10.3892/or_00000467. [DOI] [PubMed] [Google Scholar]
- 40.Kang MH, Kim JS, Seo JE, Oh SC, Yoo YA. Bmp2 accelerates the motility and invasiveness of gastric cancer cells via activation of the phosphatidylinositol 3-kinase (pi3k)/akt pathway. Exp Cell Res. 2010;316(1):24–37. doi: 10.1016/j.yexcr.2009.10.010. PMID: 19835871, DOI: 10.1016/j.yexcr.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 41.Langenfeld EM, Langenfeld J. Bone morphogenetic protein-2 stimulates angiogenesis in developing tumors. Mol Cancer Res. 2004;2(3):141–149. PMID: 15037653. [PubMed] [Google Scholar]
- 42.Ye L, Jiang WG. Bone morphogenetic proteins in tumour associated angiogenesis and implication in cancer therapies. Cancer Lett. 2016;380(2):586–597. doi: 10.1016/j.canlet.2015.10.036. PMID: 26639195. DOI: 10.1016/j.canlet.2015.10.036. [DOI] [PubMed] [Google Scholar]
- 43.Raida M, Clement JH, Leek RD, Ameri K, Bicknell R, Niederwieser D, Harris AL. Bone morphogenetic protein 2 (bmp-2) and induction of tumor angiogenesis. J Cancer Res Clin Oncol. 2005;131(11):741–750. doi: 10.1007/s00432-005-0024-1. PMID: 16136355. DOI: 10.1007/s00432-005-0024-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scharpfenecker M, van Dinther M, Liu Z, van Bezooijen RL, Zhao Q, Pukac L, Lowik CW, ten Dijke P. Bmp-9 signals via alk1 and inhibits bfgf-induced endothelial cell proliferation and vegf-stimulated angiogenesis. J Cell Sci. 2007;120(Pt 6):964–972. doi: 10.1242/jcs.002949. PMID: 17311849. DOI: 10.1242/jcs.002949. [DOI] [PubMed] [Google Scholar]
- 45.Stabile H, Mitola S, Moroni E, Belleri M, Nicoli S, Coltrini D, Peri F, Pessi A, Orsatti L, Talamo F, Castronovo V, Waltregny D, Cotelli F, Ribatti D, Presta M. Bone morphogenic protein antagonist drm/gremlin is a novel proangiogenic factor. Blood. 2007;109(5):1834–1840. doi: 10.1182/blood-2006-06-032276. PMID: 17077323. DOI: 10.1182/blood-2006-06-032276. [DOI] [PubMed] [Google Scholar]
- 46.Al-Rawi MA, Jiang WG. Lymphangiogenesis and cancer metastasis. Front Biosci (Landmark Ed) 2011;16:723–739. doi: 10.2741/3715. PMID: 21196198. [DOI] [PubMed] [Google Scholar]