Abstract
Background/Aim: Circulating mRNA can be a useful source of cancer biomarkers. We took advantage of direct transcriptomic analysis in plasma RNA to identify novel mRNA markers for non-small cell lung cancer (NSCLC). Patients and Methods: Plasma RNA from NSCLC patients and healthy individuals was profiled with cDNA-mediated annealing, selection, extension and ligation (DASL) microarrays. The microarray results were further validated in plasma RNA. Results: Through RNA profiling and online database mining, four gene transcripts were filtered as candidate markers of NSCLC. After validation, the PCTAIRE-1 transcript was identified as a circulating mRNA marker. The diagnostic potential of PCTAIRE-1 was evaluated by receiver operating characteristic curve analysis, which gave a sensitivity and specificity of 60% and 85%, respectively. In addition, high plasma PCTK1 levels were also correlated with poor progression-free survival (p=0.008). Conclusion: Circulating mRNA can be profiled with the DASL assay. From the profile, PCTAIRE-1 RNA in the plasma we discovered as a novel diagnostic/prognostic biomarker and an indicator of poor survival in NSCLC patients.
Keywords: PCTK1/CDK16, biomarker, non-small cell lung cancer, circulating mRNA
The detection of biomarkers in liquid biopsy (i.e., in body fluids) is valuable for the diagnosis and prognosis of malignant diseases, as its sampling is less invasive and can be carried out repeatedly. Some well-known circulating cancer markers, including CEA, PSA, CA125, and CA153, are proteins. These markers do not have sufficient sensitivity and specificity for detecting certain cancers [e.g., non-small cell lung cancer (NSCLC)]. The identification of novel biomarkers in these cancers is thus in great demand. In addition to protein markers, circulating nucleic acids are promising sources for new biomarkers, as circulating nucleic acids provide information regarding the genome or gene expression. In addition, some well-developed technologies, such as polymerase chain reaction (PCR) and high-throughput sequencing, can detect traces of nucleic acids in the circulation, allowing the usage of nucleic acid markers.
Different species of circulating nucleic acid markers, including cell-free DNA (cfDNA), microRNA (miRNA) (1-4), long noncoding RNA (lncRNA) (4-8), and mRNA (9), have been linked with malignant diseases. Among them, mRNA is one of the least investigated species (9). Until now, only a few circulating mRNA markers have been reported. For example, tyrosinase mRNA can be detected in the blood of some melanoma patients (10); telomerase mRNAs can be detected in breast cancer patients (11), CK19, mammaglobin, 5T4, and circulating thyroid-stimulating hormone receptor (TSHR) mRNAs can be detected in thyroid cancer patients (12); 5T4 mRNA can be detected in lung cancer patients (13); and CEA and CK19 mRNAs can be detected in colorectal cancer patients (14). These studies have revealed that circulating mRNAs can be potential biomarkers.
A difficulty in identifying circulating mRNA markers is that the RNA profile in plasma is different from that in tumor tissues. As blood is rich in background RNA from the whole body, most RNA markers found in tumor tissues may be masked by the background and cannot be used. A high-throughput analysis in blood samples could be directly used to discover mRNA markers that are not masked. Another difficulty is that mRNA in plasma is usually fragmented and found at a low abundance. A special amplification technology is required to enrich its amount before conducting subsequent analyses. In our previous study, we have shown that circulating mRNA could be amplified and profiled using the cDNA-mediated annealing, selection, extension, and ligation (DASL) assay (15). In the current study, we applied the DASL assay to identify novel circulating mRNA biomarkers in NSCLC patients.
PCTAIRE-1 (PCTK1), also known as cyclin-dependent kinase 16 (CDK16), belongs to the PCTAIRE family, which comprises a conserved central kinase domain with an amino acid sequence highly similar to that of CDK family members (16). PCTK1 is highly expressed in the brain and testis but not in other tissues (17-20). PCTK1 is involved in a wide range of biological functions, including neurite outgrowth (21), spermatogenesis (22), secretory transport (23,24) and insulin secretion (25). Recently, few studies have revealed the involvement of PCTK1 in cell proliferation or anti-apoptosis in several types of malignancies (26,27).
In this study, we found that the levels of PCTK1 RNA are high in the plasma of NSCLC patients. We also demonstrated that high levels of PCTK1 RNA are correlated with poor survival. NSCLC is the most common type of lung cancer cases and an important cause of death worldwide (28-31). The development of diagnostic and prognostic biomarkers will be helpful in cancer management. The identification of circulating PCTK1 as a novel mRNA biomarker for NSCLC is valuable for the future development of diagnostic or monitoring tools.
Patients and Methods
Patient samples and plasma collection. Forty-three NSCLC patients were recruited from the Chang Gang Memorial Hospital. Another forty-three healthy controls were recruited from the Health Check Center of the same hospital. Written informed consent from each patient was obtained before sample collection. This study was approved by the Chang Gung Memorial Hospital Institution Review Board (with approval numbers 100-2065A3, 102-1749C, 102-1584C1, 103-7926C1, and 104-7490D). The clinical stage of each patient was classified according to the AJCC staging system. All participants were divided into two groups. Five patients (1 male and 4 females, ranging from 49 to 90 years of age, mean age: 69.2±14.9) with primary stage IV NSCLC and six healthy individuals (2 males and 4 females, ranging from 56 to 61 years of age, mean age: 58.1±1.7) were enlisted in the discovery phase. The NSCLC patients provided blood samples before (Nontreatment group) and two weeks after the first treatment with chemotherapy or targeted therapy (Treatment group). The control individuals provided blood samples once (Healthy group). Thirty-eight treatment-naive patients with non-small cell lung cancer regardless of stage (21 males and 17 females, ranging from 38 to 81 years of age, mean age: 61.7±12.2 years) and thirty-seven healthy individuals with a similar distribution of age (28 males and 5 females, ranging from 53 to 79 years of age, mean age: 62.9±7.7 years) were enrolled in the validation phase. Peripheral blood from the participants was collected into an EDTA tube and centrifuged at 1600 × g for 10 min at room temperature. The supernatants (plasma) were separated and stored at –80˚C until further processing.
Isolation of circulating mRNAs. Total RNA was extracted from plasma using TRIzol LS reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. In the discovery phase, RNA from each sample was extracted from 5 ml plasma. In the validation phase, RNA was extracted from 250 μl plasma. Briefly, a 250-μl plasma aliquot was mixed with one milliliter TRIzol LS reagent. After 5 min of incubation, 266 μl of chloroform was added to the mixture. Then, the mixture was centrifuged at 14000 × g for 15 min at 4˚C. The aqueous layer was transferred to a new tube. An equal volume of isopropanol and 1 μl GlycoBlue (Thermo Fisher Scientific) was added to the solution to precipitate RNA. The solution was further centrifuged at 14000 × g for 15 min at 4˚C. The RNA pellet was washed with cold 70% ethanol, air dried for 10 min, and resuspended in 30 μl DEPC-treated water at 56˚C for 10 min. RNA concentration was estimated with a Nanodrop ND-1000 (Thermo Fisher Scientific) and the RiboGreen RNA assay on a Qubit fluorescence reader (Thermo Fisher Scientific).
Microarray analysis for circulating mRNA profiling. The DASL assay (Illumina, San Diego, CA, USA) was used to preamplify and label the mRNA transcripts from plasma samples. The pre-amplified cDNA products were then hybridized to a Human HT-12 v4 Expression BeadChip (Illumina), which contains probes for 23,811 coding transcripts. The DASL assay and expression profiling experiments were performed at Genetech Biotech (Taipei, Taiwan). Raw data were processed using Partek Genomic Suite (PGS) software (St. Louis, MO, USA) for bioinformatics analysis (32). The similarities and differences in circulating mRNA profiles between NSCLC patients and healthy controls were assessed by using principal component analysis (PCA) and hierarchical clustering, which group samples according to their expression features. Hierarchical clustering was performed with the Euclidean and Pearson’s dissimilarity algorithms. The differentially expressed genes were selected for further analysis if they met the following criteria: genes that were not low in abundance or with an abnormal pattern in all samples; genes whose average expression levels between patients and controls were significantly different (with p<0.05) in one-way ANOVA; and genes whose mean expression levels in one group was at least two-fold greater than that in the other group.
Online database mining. To confirm the differentially expressed genes that had also been identified in lung tissue mRNA profiles, the selected genes were then filtered in the Oncomine database (http://www.oncomine.org) (33,34). In the Oncomine search for gene expression (cancer vs. normal), a fold change above 1.5 and a p-value below 0.05 were set as the thresholds.
Reverse transcription quantitative PCR (RT-qPCR). To quantify the target mRNA level, the extracted RNA was reverse transcribed into cDNA with MMLV reverse transcriptase (Promega, Madison, WI, USA). Then, 2 μl of the cDNA was used as a template for qPCR, which included Platinum Taq polymerase and its reaction buffer (Thermo Fisher Scientific), 4 dNTPs, primers, and SYBR Green I. qPCR was performed in triplicate with the following thermal cycling conditions: incubation at 95˚C for 3 min, followed by 45 thermal cycles (95˚C for 15 sec, 60˚C for 30 sec and 72˚C for 30 sec), and a final incubation at 72˚C for 3 min. The fluorescent signals were detected at each cycle at 60˚C, which generated an amplification curve of each reaction. The relative level of a target gene to a housekeeping gene, β2-microglobulin (B2M), was calculated from their threshold cycle (Ct) with the equation 2^(CtB2M-Ctgene). The primer sequences for PCTK1 were 5’-GTCAGCCTATCTGAGATTGG-3’ (forward) and 5’-TTCATATTCCAGTCTGATCTC-3’ (reverse). Those for B2M were 5’-CATTCAGACTTGTCTTTCAG-3’ (forward) and 5’-TTCAAACCTCCATGATGC-3’ (reverse).
Immunohistochemical staining. The formalin-fixed, paraffin-embedded (FFPE) tissue sections from NSCLC patients were obtained from the Tissue Bank in Chang Gung Memorial Hospital. One of the serial sections was stained with hematoxylin and eosin to identify the most representative regions of the tumor. Immunohistochemistry was performed according to a protocol from Abcam (https://www.abcam.com/ps/pdf/protocols/ ihc_p.pdf), with slight modifications. Briefly, the tissue section was first treated with heated 0.01 M citrate buffer (pH 6.0) to retrieve antigens, then soaked in 3% H2O2 to inactivate endogenous peroxidase, and in Antibody Diluent solution (Dako, Glostrup, Denmark) to block nonspecific signals. Then, samples were incubated with antibodies against the PCTK1 protein (1:200 dilution; HPA 001366, Sigma-Aldrich, St. Louis, MO, USA) overnight at 4˚C and then with anti-rabbit antibodies conjugated with horseradish peroxidase (1:250 dilution; Calbiochem, Darmstadt, Germany) for 60 min at room temperature. Color was revealed by treatment with DAB chromogen (DAKO) and hematoxylin. The slides were scored for PCTK1 staining under the microscope by a senior pathologist without knowledge of the patients’ histories. The PCTK1 score was calculated according to a simplified H score system (35) based on the percentage of stained cells (3, >90%; 2, 50-89%; 1, 10-49%; and 0, 0-9%) and the intensity of staining (3, strong; 2, moderate; 1, weak; or 0, none). The two scores were multiplied by each other and divided by 3 to obtain the final score. Positive staining was defined as a final score ≥1.
Statistical analysis. Statistical analysis was conducted with SPSS software for Windows version 17.0 (SPSS, Inc., Chicago, IL, USA). A nonparametric Mann-Whitney test was used to compare gene expression levels or IHC scores between different groups. Receiver operating characteristic (ROC) curve and area under curve analyses were used to estimate the discriminative power and determine the cut-off value of the PCTK1 RNA level in specimens. A chi-square test was used to determine the association between circulating PCTK1 levels and clinical-pathological characteristics. The survival curves were plotted with the Kaplan–Meier method and analyzed using the log-rank test. A p-value≤0.05 was regarded as statistically significant.
Results
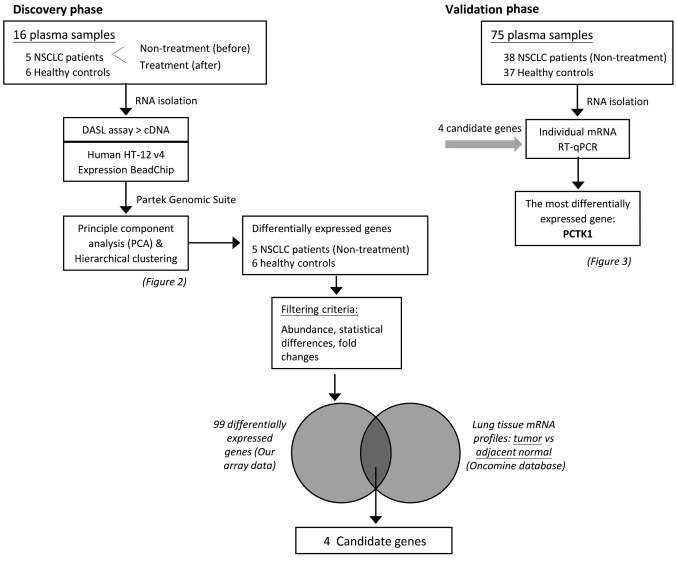
Experimental design. The aim of this study was to identify circulating mRNA markers of NSCLC. The study design included a discovery phase and a validation phase. The whole experimental strategy is simplified in a flow chart in Figure 1.
Figure 1. Diagram of the study design, clinical sample flow, and process of candidate gene evaluation.
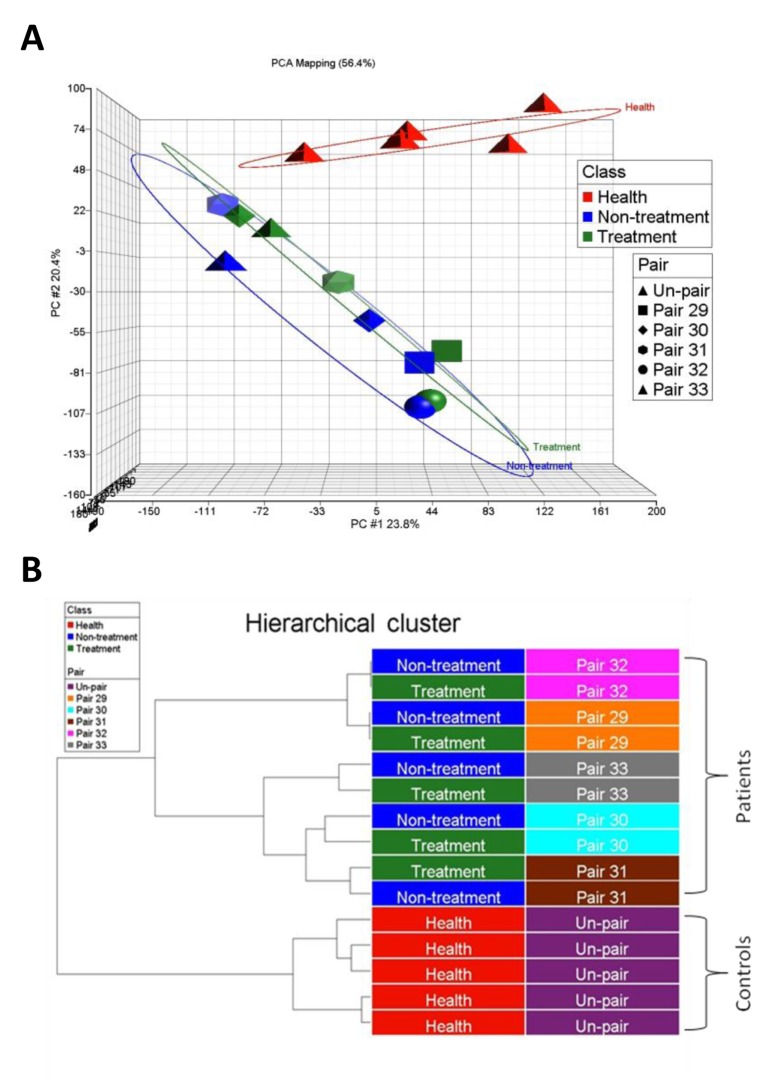
Circulating mRNA profiling in the discovery phase. Five NSCLC patients and six healthy controls were included in the discovery phase. The NSCLC patients provided blood samples before (Nontreatment group) and two weeks after the first treatment with chemotherapy or targeted therapy (Treatment group). The control individuals provided blood samples once (Healthy group). RNA was extracted from the samples and profiled with the DASL assay and an HT-12 bead-based microarray, which were designed to amplify fragmented RNA. The microarray detected approximately 15,000 genes in the plasma RNA of each sample. There was no significant difference in the number of detected genes or the average intensity between samples or between groups. PCA plots demonstrated that the NSCLC patients were grouped together and distinguished from healthy controls (Figure 2A). Hierarchical clustering also confirmed that the NSCLC patients were clustered apart from the healthy controls (Figure 2B). However, in the same patient, the plasma RNA profiles before and after treatment were not significantly different.
Figure 2. Profiles of circulating mRNAs distinguishing NSCLC patients from healthy controls. Transcriptomic profiles of circulating mRNAs in the plasma of NSCLC patients (n=5, paired samples, including nontreatment in blue and treatment in green) and healthy controls (n=6, in red) were analyzed and are presented as PCA plots (A) and hierarchical clustering (B). The results of PCA and hierarchical clustering demonstrate divergent patterns between NSCLC patients and healthy controls.
Selection of candidate genes. We then selected genes whose levels were elevated in the NSCLC patients by comparing the profiles of the Nontreatment group and the Healthy group (the criteria are detailed in the Materials and Methods section). Ninety-nine genes were selected at this stage (not shown). We then determined the levels of these genes in lung cancer tissues in the Oncomine database. Only four genes revealed pronounced elevation in most analyses in the dataset: MAP1B, MMP1, PCTK1 and PTPRF. We then determined the circulating mRNA levels of the four genes in the validation cohort, including 75 plasma samples (38 from NSCLC patients and 37 from healthy controls). Among the four candidate genes, only PCTK1 demonstrated significant elevation in patients’ plasma (Figure 3A).
Figure 3. Identification of PCTK1 as an elevated circulating mRNA in NSCLC patients. (A) The PCTK1 mRNA levels in the plasma were determined by reverse transcription quantitative PCR and normalized to that of the β2-microglobulin (B2M) transcript. (B) Receiver operating characteristic curve analysis of the diagnostic efficacy of plasma PCTK1 RNA levels.
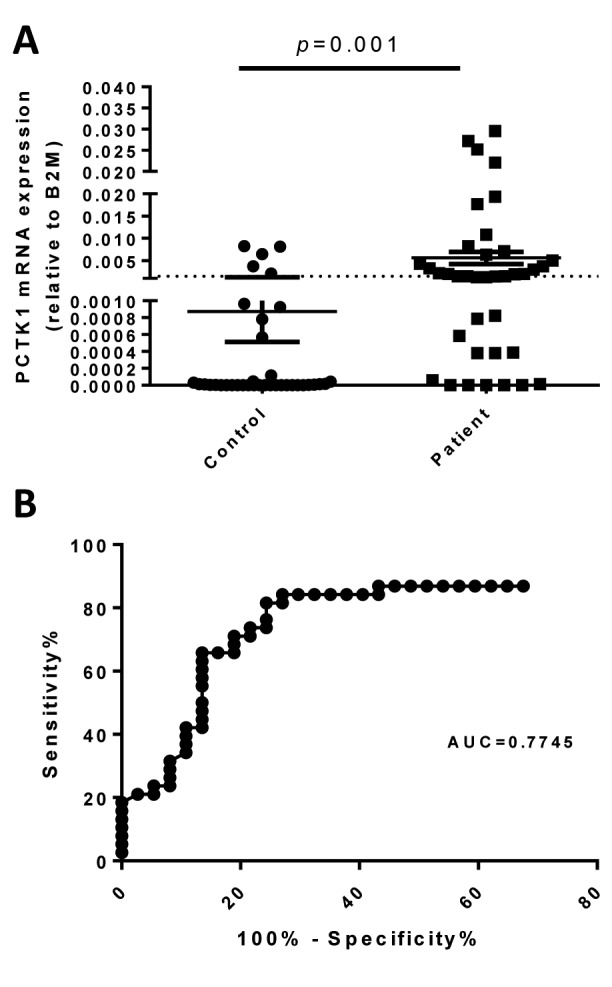
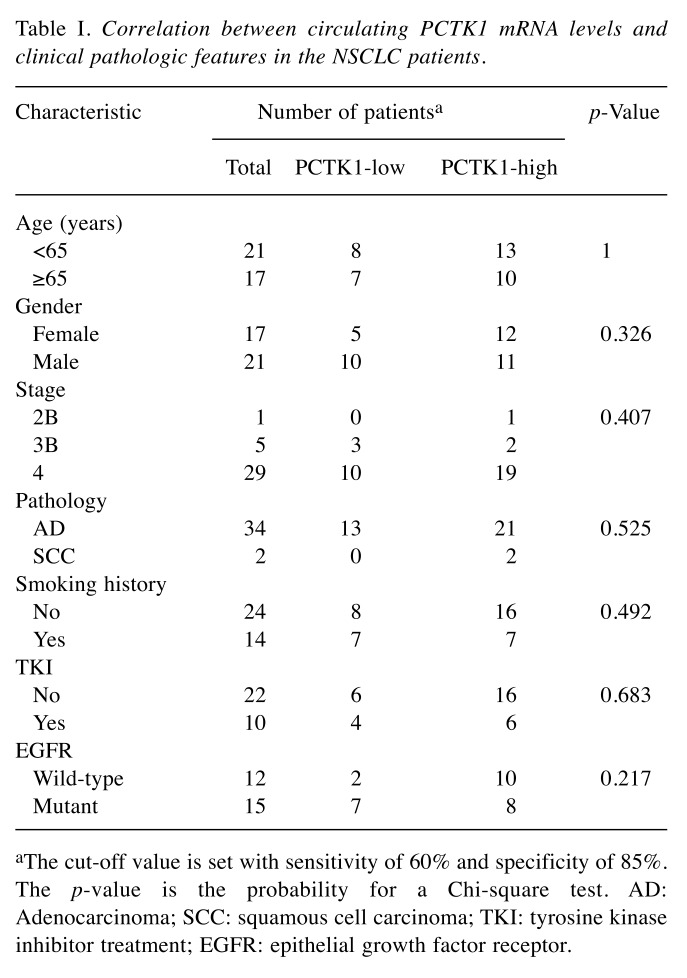
Diagnostic and prognostic value of circulating PCTK1 RNA. A ROC curve was applied to test the ability of PCTK1 RNA in the diagnosis of NSCLC. When the optimal threshold was set, the area under the ROC curve was 0.7745, giving a sensitivity and specificity of 60% and 85%, respectively (Figure 3B). Based on this threshold, the patients were divided into two groups: PCTK1-high and PCTK1-low. In the survival analysis, the PCTK1-high group had significantly (p=0.008) shorter progression-free survival (PFS) (mean=5.5 months) than the PCTK1-low group (mean=14.3 months) in the log-rank test (Figure 4A). The PCTK1-high group also had shorter OS (mean=22.7 months) than the PCTK1-low group (mean=55.5 months), although the difference was not statistically significant (p=0.195) (Figure 4B). In contrast, the PCTK1 levels were not associated with other clinical-pathological factors, such as age, sex, cancer stage, tumor pathology, smoking history, TKI treatment, or EGFR status (Table I).
Figure 4. Association of plasma PCTK1 mRNA levels with patient survival. Kaplan-Meier plots of progression-free survival (PFS) (A) and overall survival (OS) (B) for patients with different plasma PCTK1 mRNA levels (PCTK1-high and PCTK1-low). The median PFS was 5.5 (95%CI=4.5-6.6) months and 14.3 (95%CI=0.7-27.8) months in the PCTK1-high and PCTK1-low groups, respectively; the median OS was 22.7 (95%CI=10.3-35.1) months and 55.5 (95%CI= 28-83) months, respectively.
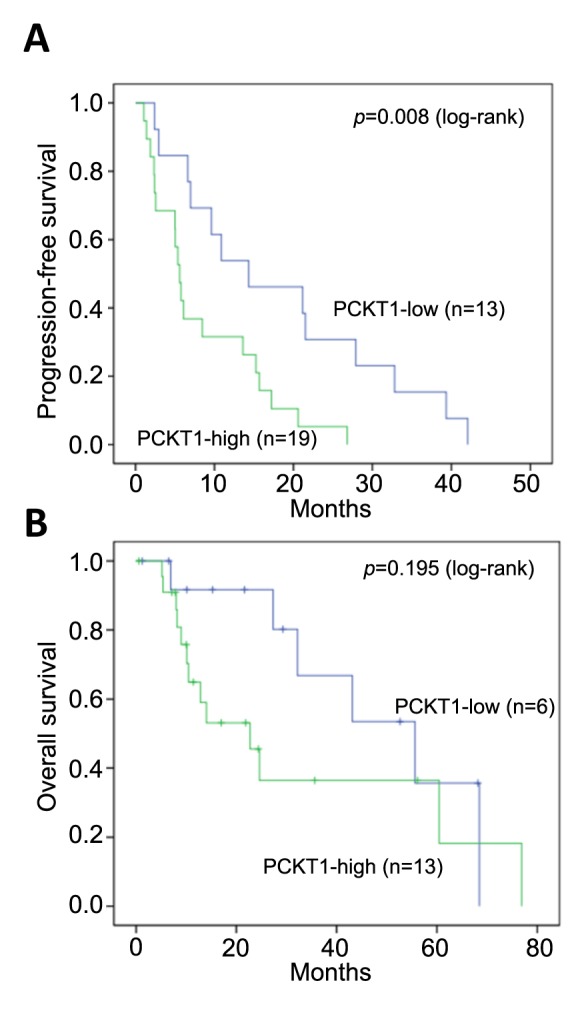
Table I. Correlation between circulating PCTK1 mRNA levels and clinical pathologic features in the NSCLC patients.
aThe cut-off value is set with sensitivity of 60% and specificity of 85%. The p-value is the probability for a Chi-square test. AD: Adenocarcinoma; SCC: squamous cell carcinoma; TKI: tyrosine kinase inhibitor treatment; EGFR: epithelial growth factor receptor.
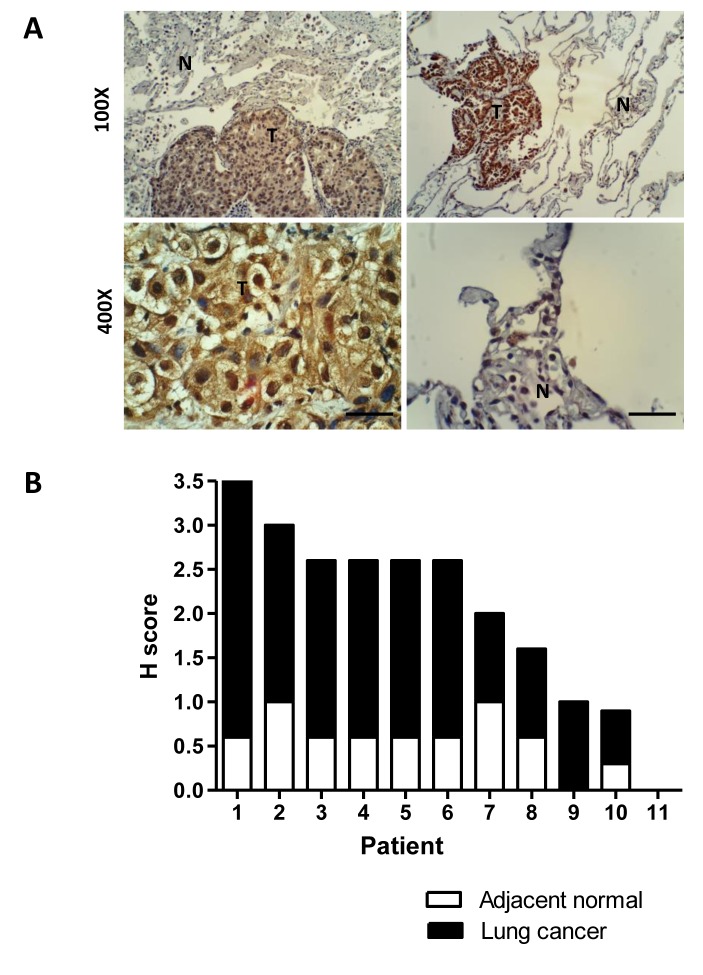
Confirmation of PCTK1 overexpression in NSCLC tissues. Then, the PCTK1 protein levels were determined in 29 FFPE tissues from lung cancer patients. Among them, 18 were from needle biopsies, and 11 were from surgically removed tissues. Only the surgically removed tissues had an obvious normal component in the tissue sections. Immunohistochemical staining showed that cancer tissues had strong positive staining for the PCTK1 protein that was mainly cytoplasmic (Figure 5A). The H score also showed that the tumor tissues had significantly higher staining than the normal tissues (Figure 5B). An H score ≥1 was defined as positive; 93% (27/29) of the tumor tissues were positive, but only 18% (2/11) of the normal tissues were positive.
Figure 5. Overexpression of PCTK1 in lung adenocarcinoma tissues. The expression of PCTK1 (brown color) was investigated by immunohistochemical staining of tissue sections from formalin-fixed, paraffin-embedded NSCLC specimens. (A) Two representative immunohistochemical images (upper two images) of tumors (T) surrounded with normal lung tissues (N) are shown. Typical positive and negative staining was found in the carcinoma (lower left image) and normal lung tissues (lower right images), respectively. Scale bar=50 μm. (B) Bar chart analysis of the histoscores (H scores) of PCTK1 staining in 11 paired tumors and adjacent normal tissues. All the samples show positive staining except for sample number 11, which shows negative staining in both the tumor and normal tissues.
Discussion
In this study, we successfully isolated and profiled circulating mRNAs from NSCLC patients by using the DASL assay. The DASL assay is specialized for the amplification of poor-quality and fragmented RNA extracted from FFPE tissues (15,25,36,37). It can be adapted for the amplification of circulating mRNA in plasma samples. Through the DASL assay and online database mining, we selected certain transcripts as candidate mRNA markers. In the subsequent validation experiments, we confirmed the elevation of PCTK1 in NSCLC patients, both in peripheral blood and in tumor tissues, and that the elevation of circulating PCTK1 RNA is associated with a poor prognosis.
RNA overexpressed in cancer tissues is not necessarily expressed at a high level in the peripheral blood. This is because the tumor mass accounts only for a small portion of the body, and both the tumor tissue and other normal tissues contribute RNAs to the circulation. Most RNAs elevated in tumor tissues are thus masked by background RNAs from normal tissues. Therefore, the RNA candidates overexpressed in tumor tissues are not necessarily good markers in the circulation. Our results confirmed this argument, as only four gene transcripts selected in the plasma matched the overexpressed genes in NSCLC tissues in the Oncomine database. Our study demonstrated that the direct profiling of circulating RNA is a feasible strategy and can be applied to biomarker selection in other diseases.
Recent studies have indicated that cells can release different particles, including exosomes (38,39), microparticles (40) and apoptotic bodies (41). These particles are protected by the plasma membrane and are considered the major sources of circulating RNA. This notion is also confirmed by the observation that nude RNA is very unstable in the blood, which is rich in RNase activity (42). Information about which particle PCTK1 mRNA is derived from will be helpful in the development of better diagnostic methods. For example, different methods have been developed to capture extracellular vesicles (43,44). If PCTK1 mRNA exists in the captured vesicles, these technologies can be applied in the enrichment of circulating PCTK1 RNA in the detection procedure.
In current clinical practice, CEA and CYFRA21-1 are frequently used as biomarkers for the diagnosis or follow-up of NSCLC (45), but their specificity and sensitivity are not satisfactory. Over the past few years, several mRNA biomarkers circulating in peripheral blood mononuclear cells (PBMCs) for NSCLC have been reported. Some of these PBMC RNAs show high specificity and sensitivity in the detection of lung cancer, including angiopoietin-2 (Ang-2) (46), A-kinase anchor protein 4 (AKAP4) (47), cytokeratin 7 (CK7) (48), E74-like factor 3 (ELF3) (48), epidermal growth factor receptor (EGFR) (48), and erythropoietin-producing hepatocellular carcinoma receptor B4 (EphB4) (48). Our current study used plasma RNA instead of PBMC RNA and discovered that PCTK1 is a newly identified plasma biomarker with 85% specificity and 60% sensitivity. In addition, a high level of PCTK1 expression is an indicator of a low survival rate. The combination of these markers and other circulating RNA species, such as miRNA or long lncRNA, may have the potential to make a marker panel for the noninvasive diagnosis or follow-up of NSCLC.
PCTK1 is a cyclin-dependent kinase-related kinase that is involved in cell cycle regulation. Initially, PCTK1 has been reported to participate in neurite outgrowth (21) and spermatogenesis (22). However, some studies have discovered its modulatory role in protein exocytosis (24) and the export of secretory proteins from the endoplasmic reticulum to the Golgi apparatus (23). In recent years, several lines of evidence have shown that PCTK1 is involved in various human cancers, such as liver (49), colon, breast and prostate cancers and malignant melanoma (27,50,51). Specifically, Wang et al. have found that PCTK1 is overexpressed in NSCLC and plays a role in cancer cell growth and anti-apoptosis (26). Our finding that PCTK1 is upregulated in lung cancer tissues confirmed their observation. These findings together specify the notable role of PCTK1 in promoting tumorigenesis and make it a potential target for cancer treatment.
Conclusion
Our study demonstrated a strategy for discovering circulating RNA markers and provided a novel biomarker: plasma PCTK1 mRNA. This biomarker can be valuable for the diagnosis/prognosis of NSCLC, and the PCTK1 gene may also be a potential therapeutic target.
Conflicts of Interest
The Authors declare no competing financial interests regarding this study.
Authors’ Contributions
Conceived and designed the experiments: CC Chiou and J Chang. Conducted the experiments: CL Shih, CL Wang, JD Luo, and JJ Hsieh. Analyzed the data: CC Chiou, J Chang, CW Wang, and CJ Yu. Wrote the paper: CC Chiou.
Acknowledgements
This research was supported by the grants from Chang Gung Memorial Hospital (CMRPD1C0321-3, CMRPD1B0091-3, and CMRPG3A0661). The Authors thank Miss Zi-Ming Huang for the assistance of preparing the manuscript.
References
- 1.Imaoka H, Toiyama Y, Fujikawa H, Hiro J, Saigusa S, Tanaka K, Inoue Y, Mohri Y, Mori T, Kato T, Toden S, Goel A, Kusunoki M. Circulating microRNA-1290 as a novel diagnostic and prognostic biomarker in human colorectal cancer. Ann Oncol. 2016;27(10):1879–1886. doi: 10.1093/annonc/mdw279. PMID: 27502702. DOI: 10.1093/annonc/mdw279. [DOI] [PubMed] [Google Scholar]
- 2.Tsukamoto M, Iinuma H, Yagi T, Matsuda K, Hashiguchi Y. Circulating exosomal microRNA-21 as a biomarker in each tumor stage of colorectal cancer. Oncology. 2017;92(6):360–370. doi: 10.1159/000463387. PMID: 28376502. DOI: 10.1159/000463387. [DOI] [PubMed] [Google Scholar]
- 3.Zhu T, Gao W, Chen X, Zhang Y, Wu M, Zhang P, Wang S. A pilot study of circulating microRNA-125b as a diagnostic and prognostic biomarker for epithelial ovarian cancer. Int J Gynecol Cancer. 2017;27(1):3–10. doi: 10.1097/IGC.0000000000000846. PMID: 27636713. DOI: 10.1097/IGC.0000000000000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed N, Bentwich Z, Hod M, Goren Y, Chajut A. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3(9):e3148. doi: 10.1371/journal.pone.0003148. PMID: 18773077. DOI: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan Q, Zuo J, Qiu S, Yu Y, Zhou H, Li N, Wang H, Liang C, Yu M, Tu J. Identification of circulating long non-coding RNA GAS5 as a potential biomarker for non-small cell lung cancer diagnosisnon-small cell lung cancer, long non-coding rna, plasma, GAS5, biomarker. Int J Oncol. 2017;50(5):1729–1738. doi: 10.3892/ijo.2017.3925. PMID: 28339045. DOI: 10.3892/ijo.2017.3925. [DOI] [PubMed] [Google Scholar]
- 6.Li N, Wang Y, Liu X, Luo P, Jing W, Zhu M, Tu J. Identification of circulating long noncoding RNA HOTAIR as a novel biomarker for diagnosis and monitoring of non-small cell lung cancer. Technol Cancer Res Treat. 2017;16(6):1060–1066. doi: 10.1177/1533034617723754. PMID: 28784052. DOI: 10.1177/1533034617723754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang Q, Ni Z, Cheng Z, Xu J, Yu H, Yin P. Three circulating long non-coding RNAs act as biomarkers for predicting nsclc. Cell Physiol Biochem. 2015;37(3):1002–1009. doi: 10.1159/000430226. PMID: 26393913. DOI: 10.1159/000430226. [DOI] [PubMed] [Google Scholar]
- 8.Wen JJ, Ma YD, Yang GS, Wang GM. Analysis of circulating long non-coding rna uca1 as potential biomarkers for diagnosis and prognosis of osteosarcoma. Eur Rev Med Pharmacol Sci. 2017;21(3):498–503. PMID: 28239821. [PubMed] [Google Scholar]
- 9.Otandault A, Anker P, Al Amir Dache Z, Guillaumon V, Meddeb R, Pastor B, Pisareva E, Sanchez C, Tanos R, Tousch G, Schwarzenbach H, Thierry AR. Recent advances in circulating nucleic acids in oncology. Ann Oncol. 2019;30(3):374–384. doi: 10.1093/annonc/mdz031. PMID: 30753271. DOI: 10.1093/annonc/mdz031. [DOI] [PubMed] [Google Scholar]
- 10.Kopreski MS, Benko FA, Kwak LW, Gocke CD. Detection of tumor messenger rna in the serum of patients with malignant melanoma. Clin Cancer Res. 1999;5(8):1961–1965. PMID: 10473072. [PubMed] [Google Scholar]
- 11.Chen XQ, Bonnefoi H, Pelte MF, Lyautey J, Lederrey C, Movarekhi S, Schaeffer P, Mulcahy HE, Meyer P, Stroun M, Anker P. Telomerase rna as a detection marker in the serum of breast cancer patients. Clin Cancer Res. 2000;6(10):3823–3826. PMID: 11051224. [PubMed] [Google Scholar]
- 12.Zhang ZZ, Chen Q, Kong CY, Li ZM, Wang LS. Circulating thyroid stimulating hormone receptor messenger rna and differentiated thyroid cancer: A diagnostic meta-analysis. Oncotarget. 2017;8(4):6623–6629. doi: 10.18632/oncotarget.14251. PMID: 28036261. DOI: 10.18632/oncotarget.14251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kopreski MS, Benko FA, Gocke CD. Circulating rna as a tumor marker: Detection of 5t4 mrna in breast and lung cancer patient serum. Ann N Y Acad Sci. 2001;945:172–178. PMID: 11708475. DOI: 10.1111/j.1749-6632.2001.tb03882.x. [PubMed] [Google Scholar]
- 14.Silva JM, Rodriguez R, Garcia JM, Munoz C, Silva J, Dominguez G, Provencio M, Espana P, Bonilla F. Detection of epithelial tumour rna in the plasma of colon cancer patients is associated with advanced stages and circulating tumour cells. Gut. 2002;50(4):530–534. doi: 10.1136/gut.50.4.530. PMID: 11889075. DOI: 10.1136/gut.50.4.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shih CL, Luo JD, Chang JW, Chen TL, Chien YT, Yu CJ, Chiou CC. Circulating messenger rna profiling with microarray and next-generation sequencing: Cross-platform comparison. Cancer Genomics Proteomics. 2015;12(5):223–230. PMID: 26417025. [PubMed] [Google Scholar]
- 16.Cole AR. Pctk proteins: The forgotten brain kinases. Neurosignals. 2009;17(4):288–297. doi: 10.1159/000231895. PMID: 19816065. DOI: 10.1159/000231895. [DOI] [PubMed] [Google Scholar]
- 17.Charrasse S, Carena I, Hagmann J, Woods-Cook K, Ferrari S. Pctaire-1: Characterization, subcellular distribution, and cell cycle-dependent kinase activity. Cell Growth Differ. 1999;10(9):611–620. PMID: 10511311. [PubMed] [Google Scholar]
- 18.Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, Asplund A, Sjostedt E, Lundberg E, Szigyarto CA, Skogs M, Takanen JO, Berling H, Tegel H, Mulder J, Nilsson P, Schwenk JM, Lindskog C, Danielsson F, Mardinoglu A, Sivertsson A, von Feilitzen K, Forsberg M, Zwahlen M, Olsson I, Navani S, Huss M, Nielsen J, Ponten F, Uhlen M. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13(2):397–406. doi: 10.1074/mcp.M113.035600. PMID: 24309898. DOI: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyerson M, Enders GH, Wu CL, Su LK, Gorka C, Nelson C, Harlow E, Tsai LH. A family of human cdc2-related protein kinases. EMBO J. 1992;11(8):2909–2917. doi: 10.1002/j.1460-2075.1992.tb05360.x. PMID: 1639063. DOI: 10.1002/j.1460-2075.1992.tb05360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Besset V, Rhee K, Wolgemuth DJ. The cellular distribution and kinase activity of the cdk family member pctaire1 in the adult mouse brain and testis suggest functions in differentiation. Cell Growth Differ. 1999;10(3):173–181. PMID: 10099831. [PubMed] [Google Scholar]
- 21.Graeser R, Gannon J, Poon RY, Dubois T, Aitken A, Hunt T. Regulation of the cdk-related protein kinase pctaire-1 and its possible role in neurite outgrowth in neuro-2a cells. J Cell Sci. 2002;115(Pt 17):3479–3490. doi: 10.1242/jcs.115.17.3479. PMID: 12154078. [DOI] [PubMed] [Google Scholar]
- 22.Mikolcevic P, Sigl R, Rauch V, Hess MW, Pfaller K, Barisic M, Pelliniemi LJ, Boesl M, Geley S. Cyclin-dependent kinase 16/pctaire kinase 1 is activated by cyclin y and is essential for spermatogenesis. Mol Cell Biol. 2012;32(4):868–879. doi: 10.1128/MCB.06261-11. PMID: 22184064. DOI: 10.1128/MCB.06261-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmer KJ, Konkel JE, Stephens DJ. Pctaire protein kinases interact directly with the copii complex and modulate secretory cargo transport. J Cell Sci. 2005;118(Pt 17):3839–3847. doi: 10.1242/jcs.02496. PMID: 12091426. DOI: jcs.02496 [pii]10.1242/jcs.02496. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Cheng K, Gong K, Fu AK, Ip NY. Pctaire1 phosphorylates n-ethylmaleimide-sensitive fusion protein: Implications in the regulation of its hexamerization and exocytosis. J Biol Chem. 2006;281(15):9852–9858. doi: 10.1074/jbc.M513496200. PMID: 16461345. DOI: 10.1074/jbc.M513496200. [DOI] [PubMed] [Google Scholar]
- 25.Chen XY, Gu XT, Saiyin H, Wan B, Zhang YJ, Li J, Wang YL, Gao R, Wang YF, Dong WP, Najjar SM, Zhang CY, Ding HF, Liu JO, Yu L. Brain-selective kinase 2 (brsk2) phosphorylation on pctaire1 negatively regulates glucose-stimulated insulin secretion in pancreatic beta-cells. J Biol Chem. 2012;287(36):30368–30375. doi: 10.1074/jbc.M112.375618. PMID: 22798068. DOI: 10.1074/jbc.M112.375618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H, Liu H, Min S, Shen Y, Li W, Chen Y, Wang X. Cdk16 overexpressed in non-small cell lung cancer and regulates cancer cell growth and apoptosis via a p27-dependent mechanism. Biomed Pharmacother. 2018;103:399–405. doi: 10.1016/j.biopha.2018.04.080. PMID: 29674275. DOI: 10.1016/j.biopha.2018.04.080. [DOI] [PubMed] [Google Scholar]
- 27.Yanagi T, Krajewska M, Matsuzawa S, Reed JC. Pctaire1 phosphorylates p27 and regulates mitosis in cancer cells. Cancer Res. 2014;74(20):5795–5807. doi: 10.1158/0008-5472.CAN-14-0872. PMID: 25205104. DOI: 10.1158/0008-5472.CAN-14-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. PMID: 21296855. DOI: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 29.Travis WD. Pathology of lung cancer. Clin Chest Med. 2011;32(4):669–692. doi: 10.1016/j.ccm.2011.08.005. PMID: 22054879. DOI: 10.1016/j.ccm.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 30.Goldstraw P, Chansky K, Crowley J, Rami-Porta R, Asamura H, Eberhardt WE, Nicholson AG, Groome P, Mitchell A, Bolejack V, International Association for the Study of Lung Cancer Staging and Prognostic Factors Committee, Advisory Boards, and Participating Institutions The IASLC lung cancer staging project: Proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM classification for lung cancer. J Thorac Oncol. 2016;11(1):39–51. doi: 10.1016/j.jtho.2015.09.009. PMID: 26762738. DOI: 10.1016/j.jtho.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 31.Rosell R, Karachaliou N. Lung cancer: Maintenance therapy and precision medicine in NSCLC. Nat Rev Clin Oncol. 2013;10(10):549–550. doi: 10.1038/nrclinonc.2013.152. PMID: 23959270. DOI: 10.1038/nrclinonc.2013.152. [DOI] [PubMed] [Google Scholar]
- 32.Downey T. Analysis of a multifactor microarray study using Partek genomics solution. Methods Enzymol. 2006;411:256–270. doi: 10.1016/S0076-6879(06)11013-7. PMID: 16939794. DOI: 10.1016/S0076-6879(06)11013-7. [DOI] [PubMed] [Google Scholar]
- 33.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, Varambally S, Ghosh D, Chinnaiyan AM. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–180. doi: 10.1593/neo.07112. PMID: 17356713. DOI: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Oncomine: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. PMID: 15068665. DOI: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ravn V, Rasmussen BB, Hojholt L, Barfoed M, Heiberg I, Thorpe SM. Reproducibility of subjective immunohistochemical estrogen- and progesterone receptor determination in human endometrium. Pathol Res Pract. 1993;189(9):1015–1022. doi: 10.1016/s0344-0338(11)80674-6. PMID: 8302719. DOI: 10.1016/S0344-0338(11)80674-6. [DOI] [PubMed] [Google Scholar]
- 36.Ravo M, Mutarelli M, Ferraro L, Grober OM, Paris O, Tarallo R, Vigilante A, Cimino D, De Bortoli M, Nola E, Cicatiello L, Weisz A. Quantitative expression profiling of highly degraded rna from formalin-fixed, paraffin-embedded breast tumor biopsies by oligonucleotide microarrays. Lab Invest. 2008;88(4):430–440. doi: 10.1038/labinvest.2008.11. PMID: 18305565. DOI: 10.1038/labinvest.2008.11. [DOI] [PubMed] [Google Scholar]
- 37.Setlur SR, Mertz KD, Hoshida Y, Demichelis F, Lupien M, Perner S, Sboner A, Pawitan Y, Andren O, Johnson LA, Tang J, Adami HO, Calza S, Chinnaiyan AM, Rhodes D, Tomlins S, Fall K, Mucci LA, Kantoff PW, Stampfer MJ, Andersson SO, Varenhorst E, Johansson JE, Brown M, Golub TR, Rubin MA. Estrogen-dependent signaling in a molecularly distinct subclass of aggressive prostate cancer. J Natl Cancer Inst. 2008;100(11):815–825. doi: 10.1093/jnci/djn150. PMID: 18505969. DOI: 10.1093/jnci/djn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17(7):879–887. doi: 10.1093/intimm/dxh267. PMID: 15908444. DOI: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 39.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mrnas and micrornas is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. PMID: 17486113. DOI: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 40.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT Jr., Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport rna and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476. doi: 10.1038/ncb1800. PMID: 19011622. DOI: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, Hristov M, Koppel T, Jahantigh MN, Lutgens E, Wang S, Olson EN, Schober A, Weber C. Delivery of microrna-126 by apoptotic bodies induces cxcl12-dependent vascular protection. Sci Signal. 2009;2(100):ra81. doi: 10.1126/scisignal.2000610. PMID: 19996457. DOI: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 42.Tsui NB, Ng EK, Lo YM. Stability of endogenous and added rna in blood specimens, serum, and plasma. Clin Chem. 2002;48(10):1647–1653. PMID: 12324479. [PubMed] [Google Scholar]
- 43.Hong CS, Funk S, Whiteside TL. Isolation of biologically active exosomes from plasma of patients with cancer. Methods Mol Biol. 2017;1633:257–265. doi: 10.1007/978-1-4939-7142-8_16. PMID: 28735492. DOI: 10.1007/978-1-4939-7142-8_16. [DOI] [PubMed] [Google Scholar]
- 44.Shih CL, Chong KY, Hsu SC, Chien HJ, Ma CT, Chang JW, Yu CJ, Chiou CC. Development of a magnetic bead-based method for the collection of circulating extracellular vesicles. N Biotechnol. 2016;33(1):116–122. doi: 10.1016/j.nbt.2015.09.003. PMID: 26409934. DOI: 10.1016/j.nbt.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 45.Okamura K, Takayama K, Izumi M, Harada T, Furuyama K, Nakanishi Y. Diagnostic value of cea and cyfra 21-1 tumor markers in primary lung cancer. Lung Cancer. 2013;80(1):45–49. doi: 10.1016/j.lungcan.2013.01.002. PMID: 23352032. DOI: 10.1016/j.lungcan.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 46.Coelho AL, Araujo A, Gomes M, Catarino R, Marques A, Medeiros R. Circulating ang-2 mrna expression levels: Looking ahead to a new prognostic factor for nsclc [corrected] PLoS One. 2014;9(2):e90009. doi: 10.1371/journal.pone.0090009. PMID: 24587185. DOI: 10.1371/journal.pone.0090009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gumireddy K, Li A, Chang DH, Liu Q, Kossenkov AV, Yan J, Korst RJ, Nam BT, Xu H, Zhang L, Ganepola GA, Showe LC, Huang Q. Akap4 is a circulating biomarker for non-small cell lung cancer. Oncotarget. 2015;6(19):17637–17647. doi: 10.18632/oncotarget.3946. PMID: 26160834. DOI: 10.18632/oncotarget.3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu XM, Wu YC, Liu X, Huang XC, Hou XX, Wang JL, Cheng XL, Mao WM, Ling ZQ. Cell-free RNA content in peripheral blood as potential biomarkers for detecting circulating tumor cells in non-small cell lung carcinoma. Int J Mol Sci. 2011;17(11) doi: 10.3390/ijms17111845. PMID: 27827952. DOI: 10.3390/ijms17111845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, Qin X, Guo T, Liu P, Wu P, Liu Z. Up-regulation of cdk16 by multiple mechanisms in hepatocellular carcinoma promotes tumor progression. J Exp Clin Cancer Res. 2017;36(1):97. doi: 10.1186/s13046-017-0569-2. PMID: 28716136. DOI: 10.1186/s13046-017-0569-2. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Yanagi T, Reed JC, Matsuzawa S. Pctaire1 regulates p27 stability, apoptosis and tumor growth in malignant melanoma. Oncoscience. 2014;1(10):624–633. doi: 10.18632/oncoscience.86. PMID: 25593992. DOI: 10.18632/oncoscience.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yanagi T, Shi R, Aza-Blanc P, Reed JC, Matsuzawa S. Pctaire1-knockdown sensitizes cancer cells to tnf family cytokines. PLoS One. 2015;10(3):e0119404. doi: 10.1371/journal.pone.0119404. PMID: 25790448. DOI: 10.1371/journal.pone.0119404. [DOI] [PMC free article] [PubMed] [Google Scholar]