SUMMARY
The delivery of mechanical power, a crucial component of animal motion, is constrained by the universal compromise between the force and the velocity of its constituent molecular systems. While the mechanisms of force generation have been studied at the single molecular motor level, there is little understanding of the magnitude of power that can be generated by folding proteins. Here, we use single-molecule force spectroscopy techniques to measure the force-velocity relation of folding titin domains that contain single internal disulfide bonds, a common feature throughout the titin I-band. We find that formation of the disulfide regulates the peak power output of protein folding in an all-or-none manner, providing at 6.0 pN, for example, a boost from 0 to 6,000 zW upon oxidation. This mechanism of power generation from protein folding is of great importance for muscle, where titin domains may unfold and refold with each extension and contraction of the sarcomere.
In Brief
Eckels et al. use single-molecule magnetic tweezers to simultaneously probe the folding dynamics of titin Ig domains and monitor the redox status of single disulfides within the Ig fold. Oxidation of the disulfide bond greatly increases both the folding force and the magnitude of power delivered by protein folding.
Graphical abstract
INTRODUCTION
Mechanical systems are defined by their force-velocity relation, which relates the magnitude of the work performed and the rate at which it is delivered. The inherent trade-off between force and velocity holds true on scales ranging from man-made machines down to single-molecule actuators and determines the peak power that can be delivered by a mechanical system (Ilton et al., 2018; Mahadevan and Matsudaira, 2000). In biological organisms, the magnitude of the mechanical power delivered determines the timescales on which it can respond to its environment—for example, to evade a predator or to capture a prey. For these reasons, evolution has driven biological systems to optimize their power delivery. We know of many molecular scale actuators such as motors (Finer et al., 1994; Gelles et al., 1988; Howard, 1997), filament torsion (Sun et al., 1997; Way et al., 1995), filament assembly (Forscher et al., 1992; Theriot et al., 1992) and filament disassembly (Merz et al., 2000) that can deliver surprisingly large amounts of mechanical power on the scale of ~1,000 zW per molecular subunit. Mechanical power scales in networks of these molecules, reaching levels that are compatible with the environment of the organism (Mahadevan and Matsudaira, 2000). Thus, the delivery of mechanical power is a central issue in understanding biological design, but very little is known about what dictates the mechanics at a molecular level in these power delivery systems.
One important addition was the discovery that protein folding does a large amount of mechanical work, on par with that measured in other molecular systems (Fernandez and Li, 2004; Rivas-Pardo et al., 2016). However, the force-velocity relation for protein folding has never been measured, and the power capabilities of a folding-driven molecular actuator remain unknown. Titin, the giant elastic protein of muscle tissues, is a likely candidate for being an important source of mechanical power derived from folding (Eckels et al., 2018; Rivas-Pardo et al., 2016). While it is generally accepted that folding of a single protein domain can generate more work than the power stroke of a molecular motor, it remains unknown whether protein folding can reach levels of power output that are similar to those of the motors in a muscle sarcomere (Bianco et al., 2016). For titin folding to contribute some energy during a muscle contraction, it is required that (1) titin operate under a tension of several piconewtons during normal muscle function, (2) the magnitude of the work done by titin be of the same magnitude as the myosin motors, (3) titin folding should proceed fast enough so that the myosin motors do not cause titin to go slack, and (4) the rate of work delivery be great enough to account for a substantial fraction of power output of a contracting sarcomere. Although it was originally estimated that the force per titin is in the sub-piconewton range, based on immunoelectron microscopy and passive tension measurements (Trombitás et al., 2003), new in vivo endoscopic imaging experiments have shown that many human muscles operate with sarcomere lengths >3.0 μm during normal extension and flexion (Chen and Delp, 2016; Chen et al., 2016; Lichtwark et al., 2018; Llewellyn et al., 2008; Sanchez et al., 2015), contradicting earlier immunoelectron microscopy measurements. These longer sarcomere lengths equate to muscle passive tensions >10 mN/mm2 and single titin forces >4 pN for many muscle types (Prado et al., 2005; Trombitás et al., 2003). Through quantum dot tracking experiments on titin in rabbit psoas myofibrils, we previously confirmed this force range to be true, thus satisfying requirement 1 for titin folding-unfolding reactions to play an important role in muscle mechanics (Rivas-Pardo et al., 2016).
While we have also measured the work done by titin folding (requirement 2) in our previous work (Rivas-Pardo et al., 2016), the final two questions (requirements 3 and 4) can be answered only through the determination of the force-velocity relation for titin folding. Here, we use ultra-fast single-molecule magnetic tweezers to measure, for the first time, the power output from titin protein folding. We demonstrate that the oxidation of a single disulfide bond in the core of the protein amplifies the power output in an all-or-none manner, thus acting as a power switch. The disulfide bond makes this possible by raising the folding forces of the protein by >8 pN. This is likely an important mechanism in muscle for controlling the folding energetics of titin, which contains an abundance of cysteine residues shown to form disulfide bonds (Giganti et al., 2018; Manteca et al., 2017; Mayans et al., 2001). However, because titin is thought to operate under a constitutive resting force of several piconewtons in vivo, enough to unfold and extend many immunoglobulin (Ig) domains (Linke, 2018; Rivas-Pardo et al., 2016), it is unclear whether there exists a mechanism for the oxidation of disulfide bonds in polypeptides under force. We provide evidence that a typical oxidoreductase enzyme, protein disulfide isomerase (PDI), is able to reversibly induce disulfide formation at forces as high as 5 pN and possesses additional chaperone activity to assist folding. These single-molecule experiments provide the first measurements of the speed and power output of titin protein folding, which are key determinants of whether titin provides an energetic contribution during muscle contraction. While much weight has been given to equivalent optical tweezers force spectroscopy experiments on the muscle myosin II motors in understanding the scaling of force generation to tissue levels, re-evaluation of the relative power contributions of titin and the myosin motors should be considered given the data presented here.
RESULTS
Real-Time Control of Single Ig Domain Oxidation Status
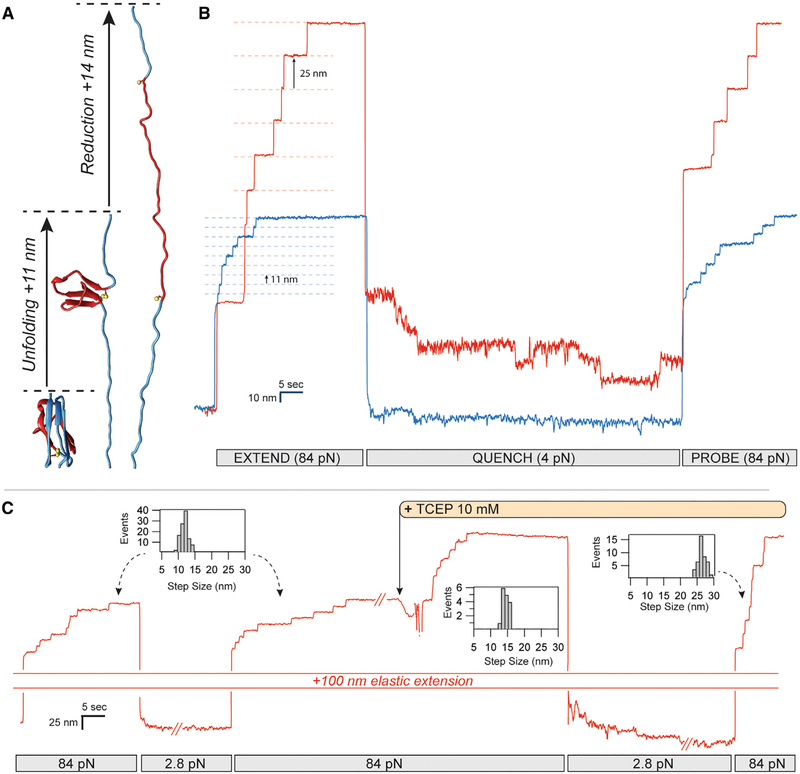
The magnetic tweezers instrument provides a natural way to probe the force-velocity relation of a nanomechanical system because it supplies feedback-free force clamp along with high-resolution tracking of folding trajectories. Here, we measure the force-velocity relation of a tandem modular protein construct containing eight repeats of cardiac titin Ig domain I27 with two cysteines that can form a single disulfide bond (Wiita et al., 2006). The eight identical repeats of the I27 domain in the expressed construct generate an unambiguous finger-print upon unfolding. In the oxidized form (I27OXD), the formed disulfide limits the extension of the amino acid backbone upon unfolding (Figure 1A), resulting in 11-nm upward steps as each Ig domain unfolds up to its disulfide bond (Figure 1B, blue trace) during the extend pulse. In contrast, the same Ig domains with the reduced disulfide (I27RED) demonstrate 25-nm upward steps, identical to those observed in the wild-type I27 domain (Figure 1B, red trace). After unfolding, the force is reduced to allow the polypeptide backbone to collapse during the refold pulse. There is a marked difference in the folding trajectories of the reduced and oxidized Ig domains—the reduced Ig domains demonstrate a gradual downward stepwise trajectory, a combination of six downward events and two upward events, indicating a net total of four folding events. By pulling again at high force in the probe pulse, the four 25-nm unfolding steps confirm the refolding count. By contrast, the oxidized Ig domains show immediate collapse during the refolding pulse and eight 11-nm steps during the probe pulse, demonstrating complete refolding.
Figure 1. Real-Time Control of Cryptic Disulfide Bonds in Single Polyproteins under Force.
(A) Under force, an Ig domain extends up to its buried disulfide bond, exposing it. Upon reduction, a further extension occurs.
(B) The oxidation status and effect of cryptic disulfide bonds are evident in these two traces of polyproteins containing eight repeats of a titin Ig domain. Oxidized polyproteins extending at a high force of 84 pN increase their length in steps of 11 nm (blue trace), whereas reduced polyproteins extend in steps of 25 nm (red trace). While the rate of folding of the reduced polyprotein at 4 pN is slow, oxidation greatly accelerates the folding rate, driving it to completion at the same force. The increased folding probability at 4 pN is measured from the number of steps recovered with a probe pulse at 84 pN.
(C) The status of a single polyprotein can be easily changed from oxidized to reduced by introducing a reducing agent into the solution (TCEP) while the polyprotein is fully unfolded, exposing its thiols to the solution. Step-size histograms record the changes observed from extending a fully oxidized polyprotein ± 2.1 nm), immediately after the addition of TCEP (14.6 ± 1.6 nm) and after refolding the resulting fully reduced polyprotein (25.6 ± 2.2 nm).
The oxidation status of the disulfide bond can be controlled in real time, as depicted in the chart-recorder style single-molecule trace in Figure 1C. In this experiment, the molecule is first stretched at high force (84 pN) to unfold eight oxidized Ig domains, each with a step size of 11 nm (Figure 1C, inset histo-gram). The force is quenched to 2.8 pN to allow for refolding while a buffer containing 10 mM tris(2-carboxyethyl) phosphine (TCEP) is prepared for exchange into the chamber. Unfolding the molecule again at high force demonstrates eight upward steps of 11 nm, after which the reducing buffer is introduced through three successive washes of 100 μL, a volume that is 10-fold higher than the volume contained inside the flow cell. Reduction by TCEP leads to eight successive 14-nm steps caused by cleavage of the disulfide bond and release of the cryptic length sequestered behind the bond (Figure 1A). With the reducing agent still present in the solution, the force on the molecule is again quenched to 2.8 pN, to allow for refolding. A final probe pulse at 84 pN demonstrates eight 25-nm steps, indicating complete refolding of the protein in its reduced state.
A Single Disulfide Bond Shifts Titin Folding to Higher Forces
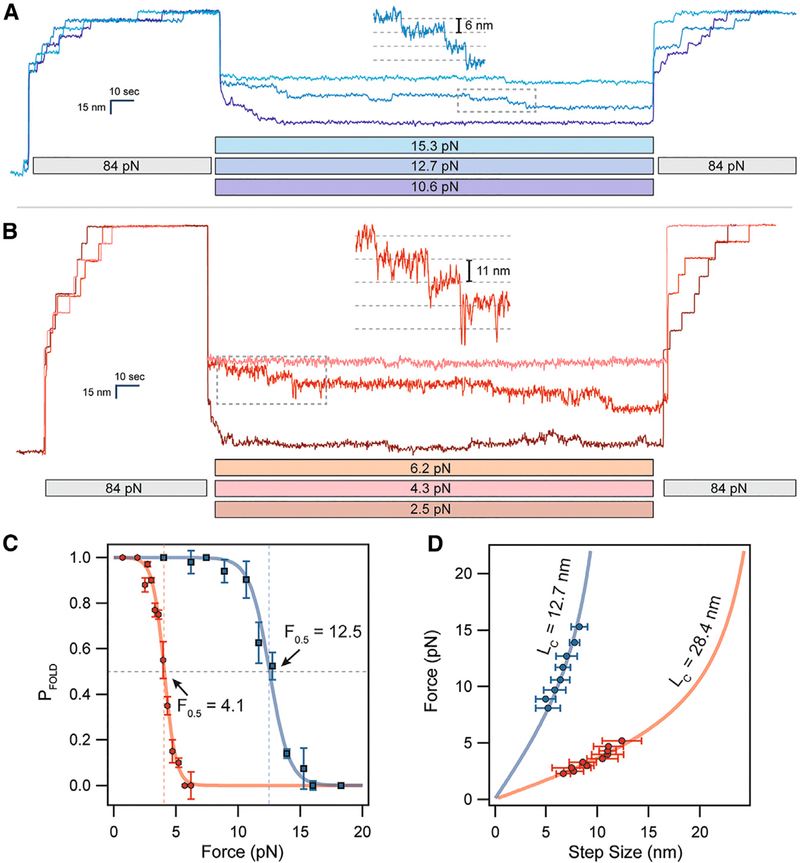
This assay was used to determine the folding probability of the oxidized versus reduced Ig domain over a wide range of forces, calculated as described previously (Rivas-Pardo et al., 2016). After unfolding the I27OXD construct using an extend pulse, the refolding force was clamped at a force ranging from 5 to 18 pN for 100 s, after which the probe pulse revealed how many domains had folded (Figure 2A). The I27OXD domains experienced very little refolding at 15.3 pN, occasionally one domain per octamer, but exhibit seven folding events at a refolding force of 10.6 pN. The step sizes observed during the refolding pulse scale according to the freely jointed chain (FJC) polymer model (Flory, 1953) and match the previously reported value for contour length (Ainavarapu et al., 2007), 12.7 nm, with a slightly larger Kuhn length of 0.65 nm, which is a better fit in the low force regime explored by the magnetic tweezers (Figure 2C). After obtaining the data from the oxidized domain, the protein was reduced with 10 mM TCEP, which was kept in solution to prevent spontaneous re-oxidation or unwanted side reactions of the two reduced cysteine residues while measuring the folding probability. The I27RED construct showed dramatically different folding trajectories, requiring forces as low as 5 pN to begin refolding, and forces down to 2.5 pN for the complete refolding of all eight domains (Figure 2B). The size of refolding events also demonstrated scaling with the FJC model, with a Kuhn length of 1.15 nm and a contour length of 28.4 nm (Figure 1C, red curve). The folding probability as a function of force demonstrates a large, 8.4 pN shift in the midpoint of the folding probability according to the fit with a sigmoid curve (see Method Details). This shift is clearly demonstrated by the raw data, which shows rapid refolding of the I27OXD domains at a force of 10.6 pN but rather slow refolding from I27RED at a force of 4.3 pN (Figure 2A versus 2B). The cause of this increase in the folding kinetics and lowering of the refolding barrier may arise from either the reduced entropy of the backbone imposed by the presence of the disulfide bond or perhaps from some residual structure in the peptide loop delimited by the disulfide bond that serves to nucleate folding.
Figure 2. Cryptic Disulfide Bonds Alter the Folding Dynamics of Titin Ig Domains.
(A) (After full extension, oxidized polyproteins are allowed to fold by quenching the force to 10–16 pN for 100 s. While only a single folding event is observed at 16 pN, complete refolding occurs at 10.6 pN.
(B) Reduced polyproteins fold at a much lower range of forces in the 2–5 pN range.
(C) The folding probability is calculated as the number of refolded domains (probe pulse) divided by eight. Oxidation shifts the midpoint folding probability from 4.1 to 12.5 pN. The data were collected from several different molecules, with n = 11 for I27OXD and n = 9 for I27RED.
(D) The folding steps observed during the quench pulse (insets in A and B) scale in size with the applied force. Oxidized folding steps (blue circles) are fit by the FJC model (solid blue line, see also equations in STAR Methods) using a contour length of 12.7 nm. Reduced folding steps (red circles) are fit using a much larger contour length of 28.4 nm (solid red line). These data came from the same molecules measured in (C).
PDI Mediates Disulfide Reformation under Force
In vivo, disulfide bonds are most commonly introduced via a specialized set of electron shuttling enzymes called oxidoreductases. The most common of these enzymes in mammals is PDI, which consists of four domains with thioredoxin-type folds. Two of the domains, A- and B-, contain a catalytic CXXC motif that can cleave and reform disulfide bonds in substrate proteins, as well as two other domains (A’ and B’) lacking the catalytic motif, thought to possess chaperone activity (Gruber et al., 2006). It is not clear whether the catalytic domains also possess chaperone activity. The magnetic tweezers were recently used to demonstrate that trigger factor, a prokaryotic chaperone, is capable of accelerating folding in substrates under force, which may be a common scenario with polypeptides emerging from the ribosome, the translocon pore, and other molecular tunnels (Haldar et al., 2017). Titin is another case of a protein that is naturally stretched in vivo and experiences a constitutive resting force due to the geometry of the sarcomere (Eckels et al., 2018; Linke, 2018). It is unclear whether the oxidoreductase machinery of the cell is capable of functioning to correctly oxidize disulfide bonds in substrates that experience such forces, but there is evidence that PDI plays an important protective role in cardiac muscle tissue (Severino et al., 2007). Here, we examine the effects of the single catalytic PDI A-domain (referred to as PDI) on the re-oxidation and refolding of the I27 substrate.
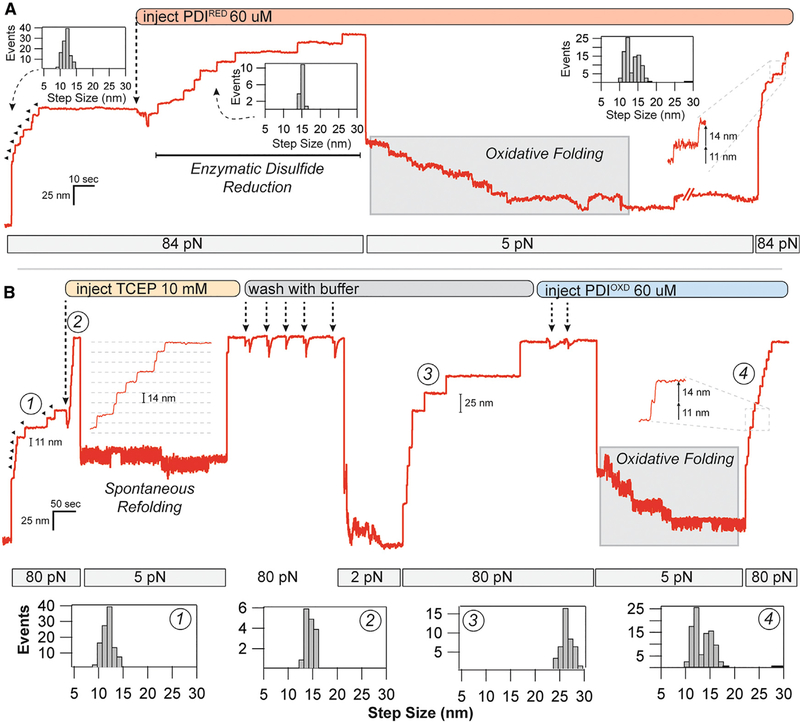
After unfolding all eight I27OXD domains, 60 mM PDIRED was flowed into the chamber to react with the exposed substrate disulfide bonds (Figure 3A). The cleavage of substrate disulfide was marked by eight 14.1-nm upward steps, identical to those seen in the presence of TCEP, with the exception that >200-fold less concentration was used, indicating the highly nucleophilic nature of the catalytic PDI thiols. Upon complete cleavage of the disulfide bonds, the force was reduced to 5 pN to allow the protein to fold. Unlike the situation with TCEP where little to no folding occurs at 5 pN, there is a downward staircase of seven steps, indicating refolding. A subsequent probe pulse of 84 pN reveals both 11- and 14-nm steps, indicating that both disulfide reformation and refolding have occurred. In the cellular environment, PDI is maintained in its oxidized state with the help of several other electron shuttling enzymes. To re-create such an environment, the same assay was repeated with oxidized PDI (see Method Details) and reduced I27 (Figure 3B). I27OXD was first unfolded (Figure 3B, part 1) and reduced through the addition of 10 mM TCEP (Figure 3B, part 2), after which the force was reduced to 5 pN. This confirms that there is little folding activity at 5 pN, as indicated by the single reduced (25 nm) step observed on the following probe pulse. Next, the TCEP was thoroughly removed from the chamber with five washes of 100 mL buffer. Reducing the force to 2 pN and pulling again shows the presence of seven I27RED domains (Figure 3B, part 3). Finally, 60 μM PDIOXD was added to the flow chamber through two washes to create an oxidizing environment. Reduction of the force to 5 pN again shows seven downward steps, in stark contrast to the folding trajectory in the presence of TCEP. The last probe pulse demonstrates seven 11-nm and seven 14-nm steps, indicating re-oxidation of the disulfide bond in addition to refolding (Figure 3B, part 4).
Figure 3. Oxidoreductase Mediated Disulfide Cleavage and Reformation under Force.
(A) After full unfolding of eight I27ox domains (11.4-nm steps), PDIRED was added to the flow cell, causing eight distinct disulfide cleavage events (14.6-nm steps). After quenching the force to 5 pN, a downward folding staircase is observed. Subsequent unfolding at 84 pN reveals a mixture of 11- and 14-nm steps, indicating reformation of the disulfide bond.
(B) Disulfide reformation could also be achieved with oxidized PDI. After full unfolding of eight I27OXD domains, TCEP (10 mM) was added to the flow cell, resulting in eight 14.6-nm steps due to the reduction of the disulfide bond. Quenching the force to 5 pN resulted in only a single refolding event. The protein was stretched again at high force to remove the TCEP with 53 washes using buffer. Reducing the force to 2 pN caused the refolding of 73 × I27RED domains, as indicated by the 25-nm steps. After full unfolding, PDIOXD was added to the flow cell, and the force was reduced to 5 pN again. In the presence of the oxidoreductase, a folding staircase is again observed. Subsequent pulling at 84 pN demonstrates 73 × 11- and 14-nm steps, indicating nearly complete reformation of the disulfide bond at 5 pN. Histograms in (A) and (B) are pooled from all of the oxidative folding experiments performed with the PDI oxidoreductase.
The Catalytic Domain of PDI Possesses Chaperone Activity
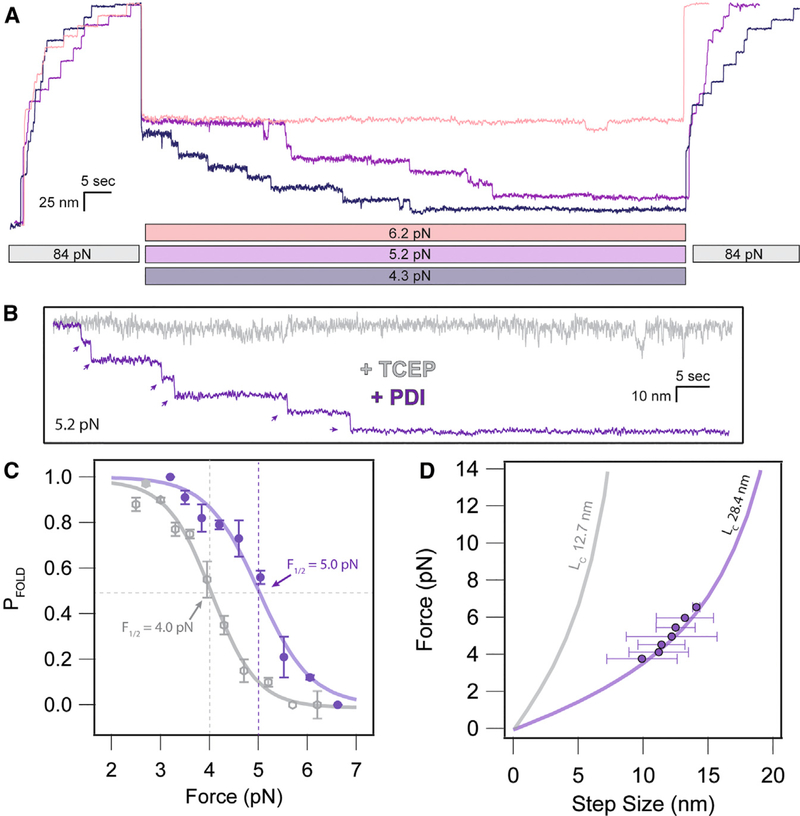
The assay, using I27OXD and PDIRED, allowed for many cycles of enzymatic reduction and oxidative folding to be conducted at different refolding forces (Figure 4A). Scanning the refolding forces from 3.5 to 6.5 pN permitted direct observation of the folding trajectories in the presence of PDI and the ability to control the amount of refolding. The folding probability, calculated as the number of 11-and 25-nm steps in the probe pulse, divided by the number of 11-and 25-nm steps on the extend pulse, shows a clear shift of the sigmoidal folding probability to higher forces (5.0 pN midpoint) in the presence of PDI (Figure 4B). We attribute this to the mixed disulfide formed between PDI and I27 after cleavage of the I27OXD internal disulfide bond, which increases the propensity of the substrate to fold under force. By contrast, folding in the presence of TCEP, which does not form stable adducts with protein thiols, has a midpoint folding force of 4.0 pN. The step sizes measured at each force were collected and averaged and found to fall directly on the FJC curve of folding step sizes when the I27 was maintained in the reduced state by TCEP. For each folding step observed during the quench pulse, there is a corresponding re-oxidized (11 nm) unfolding event. Failure to reform the disulfide bond after folding was exceedingly rare. These data suggest that PDI re-introduces the disulfide bond after the first step of protein folding, collapse of the polypeptide backbone to a high entropy molten globule state. This confirms the conclusions that protein folding drives disulfide formation, inferred from previous single-molecule data that lacked resolution to resolve the individual folding steps (Kahn et al., 2015; Kosuri et al., 2012). Furthermore, the shift in folding probability to higher forces suggests a bona fide chaperone activity of the PDI domain with extended polypeptides, indicating a pathway for disulfide bond introduction into titin even under the resting forces it experiences in muscle.
Figure 4. PDI Introduces Disulfide Bonds after Accelerated Polypeptide Collapse.
(A) Folding trajectories of I27 in the presence of 60 mM PDIRED at 6.2, 5.2, and 4.3 pN. The extend and probe pulses contain both 11- and 14-nm steps due to the initial cleavage of the disulfide bond and subsequent oxidoreductase-mediated reformation of the disulfide bond during the quench.
(B) Comparison of individual folding trajectories of I27RED in the presence of 10 mM TCEP versus I27OX after cleavage by 60 μM PDIRED. The greatly favored folded state in the presence of PDI suggests chaperone activity from the oxidoreductase.
(C) The folding probability of I27 is shifted in favor of the folded state in the presence of PDI. The sigmoidal fits demonstrate a 1.0-pN shift of the midpoint folding probability from 4.0 pN in the presence of TCEP to 5.0 pN in the presence of PDIRED.
(D) The folding step sizes observed in the presence of PDI closely followed the FJC model for the I27RED domain, even if there was disulfide refor-mation. This suggests that the oxidoreductase reintroduces the disulfide bond only after there is collapse of the polypeptide backbone.
Disulfide Formation Boosts the Work Done by Titin Folding
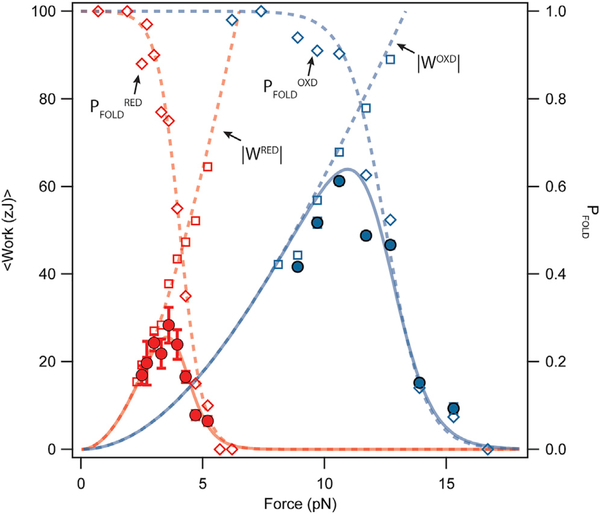
Recent experimental data on muscle fibers suggest that the energetics of titin folding may be important for active muscle contraction, in addition to passive mechanics. From the measurements presented so far, it is not immediately clear whether the presence of an extension-limiting disulfide bond will increase or decrease the mechanical work delivered by a folding Ig domain because higher folding forces and smaller step sizes have opposite effects on work delivery. The maximum work can be calculated simply as the product of the applied force and the mean step size at that force. However, in the force range explored for titin folding, there is an equilibrium between folding and unfolding steps, so the net amount of work delivered by folding a single titin Ig domain is equal to the maximum work (Figure 5, empty squares) times the folding probability (Figure 5, empty diamonds). This quantity is called <W>, the expected value of the work output (Rivas-Pardo et al., 2016) (Figure 5, solid circles). The I27RED folding process generates a peak <W> of only 25.6 zJ at 3.5 pN, whereas I27OXD folding work peaks at a value of 63.9 zJ at 10.9 pN (Figure 5, solid curve). Oxidation of the single disulfide bond in this protein therefore doubles the magnitude of the expected work performed by Ig domain folding. Although these work values are much higher than the average energy delivered by muscle motor proteins (~36 zJ) (Piazzesi et al., 2007), a better assessment of the relevance of protein folding on a physiological scale is the rate at which the energy is delivered.
Figure 5. Oxidation Increases the Work Done by Protein Folding and Shifts Its Peak to Higher Forces.
We calculate the expected value of the work delivered by folding steps as the product of folding work times the folding probability. The expected value of the folding work delivered by a reduced Ig domain peaks at a force of 3.5 pN with a value of 25.6 zJ (solid red line), calculated by multiplying the reduced folding probability (empty red squares, dashed line) times the reduced folding work (empty red diamonds, dashed line). Oxidation causes sizeable effects on the mechanical work delivered by protein folding; the expected value of the folding work delivered by an oxidized Ig domain is now 2.5 times larger (63.9 zJ) and peaks at 11 pN (solid blue line); from the oxidized folding probability (empty blue diamonds, dashed line) times the oxidized folding work (empty blue squares, dashed line). However, in both cases, the delivery of the folding work at their peak is very slow due to measured folding kinetics.
A Model for Power Delivery from Protein Folding
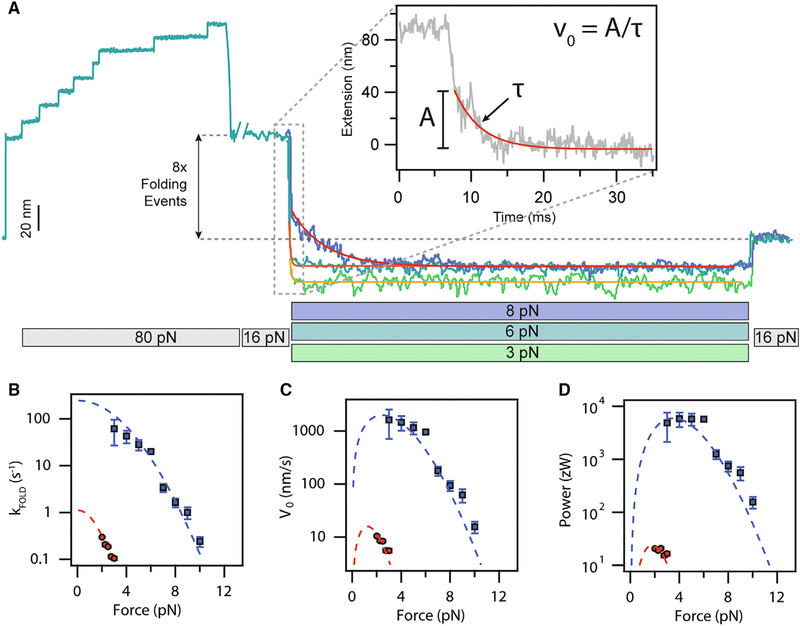
During a muscle contraction, force development occurs within milliseconds after activation of the muscle fiber (Huxley and Simmons, 1971), redistributing some of the load from titin to the crossbridges themselves. Here, we aim to mimic these force changes in titin using ultra-fast force quenches to determine whether there is significant power output from titin on the timescales that are relevant in muscle (Figure 6A). After a high-force extend pulse at 80 pN, the force was ramped down to 16 pN over the course of 0.5 s, where I27OXD does not fold (Figure 2C). After settling at 16 pN for 1 s, the magnets were stepped as quickly as possible (~5 ms) to provide a force in the range of 2–10 pN. Tracking the protein trajectory after these very fast force quenches permits a measurement of the power output due to protein folding. As seen in Figure 6A, the folding trajectories are so rapid that it is difficult to distinguish individual folding steps. The change in length at 16 pN before and after the force quench, as well as the presence of eight steps in the final probe pulse (data not shown), indicate complete refolding of the protein construct. The force ranges over which the folding kinetics could be tracked were 3–10 pN for I27OXD or 2–3 pN for I27RED. The lag introduced by changing the magnet position (<5 ms) and the amplitude of the folding contraction were the limiting factors at lower forces. The effective shortening rate during the refolding pulse was determined by fitting a single exponential to the trajectory (Figure 6A, inset). The amplitude of the exponential was fixed to eight times the step size, as predicted by the freely jointed chain model (Figure 2D), thereby excluding the entropic collapse of the polymer due to the change in force. The observed rate constants plotted for both the oxidized and reduced forms of the protein are shown in Figure 6B, blue and red data points, respectively. The folding rates are fit with a modified exponential model that takes into account the free energy of a polypeptide (Chen et al., 2015; Dudko et al., 2008) modeled as a freely jointed chain polymer extended up to the pulling force:
| (Equation 1) |
Figure 6. Oxidation Acts as a Power Switch Controlling the Delivery of Protein Folding Work.
(A) Force protocol for measuring fast folding contractions at forces at which the folding probability is always one. A fully extended oxidized polyprotein is first quenched to 16 pN, where no folding occurs. A rapid quench follows, resulting in a brief elastic recoil and a very fast folding contraction. A subsequent increase of the force up to 16 pN reveals a much shorter extension, marking the folding of all eight domains. The speed of the folding contraction increases greatly as the force of the quench is reduced. The folding contractions are fit with a single exponential to measure the folding rate.
(B) The force dependency of the folding rate for oxidized domains (blue squares) and reduced domains (red squares). The dashed lines correspond to fits using Equation 1.
(C) Force dependency of the initial velocity of the folding contraction (v0), measured as the amplitude of the contraction divided by its time constant (A, inset). The dashed lines correspond to fits from Equation 2.
(D) The peak power delivered by the folding contraction is calculated as v0 · F. The dashed lines are fits of Equation 3. The figure shows that oxidation of the polyprotein increases the peak output power of its folding contraction by >300-fold, reaching ~6,000 zW. Data were averaged from n = 5 molecules for I27OXD and from n = 3 molecules for I27RED.
Here, ΔLC is the contour length increment and lk is the Kuhn length, as determined from the previous fits to the freely jointed chain (Figure 2D). The curvature of the folding rates due to movement of the folding transition state with force is well accounted for by this model (Figure 6B, dashed fits). These experiments are nearly identical to those performed on muscle fibers to determine the force-velocity relation of muscle, where shortening velocities are recorded after a small step change in the load. The muscle fiber typically exhibits <100-nm shortening per half sarcomere over tens of milliseconds (Caremani et al., 2016; Linari et al., 2015; Piazzesi et al., 2007). Therefore, we consider the initial shortening velocity, v0, of a titin molecule as the physiologically relevant measurable here to mimic the equivalent muscle experiments. This peak velocity, v0, is the slope of the folding trajectory at time zero, and is calculated from the amplitude of the exponential fit multiplied by the force-dependent folding rate, given by
| (Equation 2) |
where N is the number of folding domains and XFJC is the force-dependent step size of folding calculated from the freely jointed chain. Unlike the force-velocity relation of muscle, which does not have a maximum according to the hyperbolic Hill equation, the force-velocity relation shown here is peaked due to the rapidly diminishing folding step size as the force approaches zero. Despite the contour length of the I27OXD being less than half of that of the reduced domain, the incredibly accelerated folding rate causes the folding velocity of I27OXD to be ~300-fold higher at a force of 3 pN. From these data, the peak power output (at the onset of the folding contraction) can be easily calculated by multiplying the applied force by the measured shortening velocity:
| (Equation 3) |
The power output of this disulfide bonded titin construct reaches up to 5,870 zW, measured at a pulling force of 4 pN, with a predicted peak power of 6,000 zW at a force of 3.6 pN, according to the fit by Equation 3. By contrast, the largest power output measured from the reduced domains reaches only 23 zW at a force of 2.5 pN. This is comparable to but slightly under the power output estimated from the wild-type I27 and I10 domains (neither contain disulfide bonds), which reach ~200 zW. It is important to keep in mind that this protein has several different residues from the I27 used in previous studies, which is known to alter folding rates and folding forces. From these measurements, it is clear that the disulfide bond acts as a switch that doubles the amount of work done by folding, and is able to boost the power output of folding by several orders of magnitude.
DISCUSSION
Cysteine residues are widely recognized as molecular switches because of their reversibly reactive nature (Littler et al., 2010; Mϋller et al., 2015), yet the role of disulfide bonds on the dynamics of mechanically active proteins in the physiological force range has never been explored due to instrumental limitations. Magnetic tweezers provide the ability to accurately control the applied forces in the piconewton range, with the additional advantage of being able to exchange buffers throughout the course of an experiment to alter the redox status of protein thiols. Here, we use the magnetic tweezers to demonstrate that disulfide bonding is a switch-like regulator of the power output from Ig domain folding, converting a soft and extensible polypeptide into a potent power-delivering actuator. At a force of 6 pN, the reduced protein domain is completely unable to fold (Figures 2B and 7). Simple oxidation of the disulfide bond allows the protein to fold at the same force of 6 pN with a predicted step size of 4 nm, each step thus delivering 24 zJ of work. With an initial folding velocity of 1,000 nm/s, this equates to a power delivery of ~6,000 zW (Figure 7). The switch-like behavior reported here is easily tuned by the redox status of the muscle, which is determined by antioxidant and oxidoreductase levels, which change with exercise, stress, ischemia, and various homeostatic responses. We further demonstrate that a canonical oxidoreductase enzyme, PDI, is able to introduce disulfide bonds into unfolded titin domains under forces of several piconewtons. These experiments unexpectedly revealed a chaperone-like quality of PDI, shifting the folding to 1.0 pN higher forces.
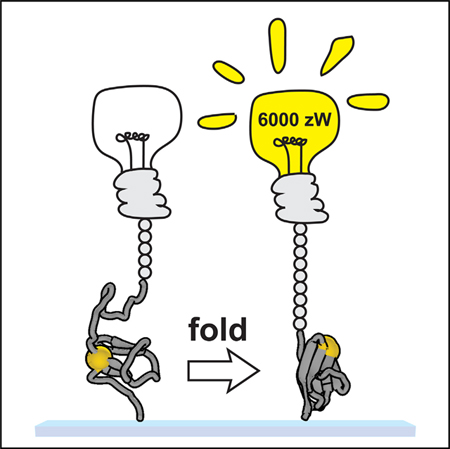
Figure 7. A PDI-Mediated Disulfide Switch of the Power Output from Titin Folding.
Upon initiation of a muscle contraction, the force on titin is lowered through engagement of the crossbridges. Titin domains lacking cysteines or containing reduced disulfide bonds are unable to fold rapidly and therefore contribute very little peak power, reaching a maximum of only 25 zW at a force of 2 pN (left). The oxidation of titin disulfide bonds through the PDI machinery of the cell allows titin domains to fold at higher forces and more rapidly. Despite the collapse step being much larger in the case of the reduced domain, the forces and kinetics from folding the oxidized domain at 4.0 pN are so much higher that the power output is two orders of magnitude greater.
Muscle tissue routinely experiences wide fluctuations in the redox balance of the cytosol, especially during periods of high metabolic demand (e.g., exercise). It is highly likely that the abundant cysteine residues in titin and other sarcomeric proteins (myosin, actin, troponin C, ryanodine receptor, protein kinase C, and myosin binding protein C) regulate muscle mechanics through cysteine chemistry (Steinberg, 2013). The nearly crystalline arrangement of filaments in the sarcomere allows for such dense protein packing that protein thiol concentrations can reach the millimolar range. It is therefore impossible for intracellular antioxidants such as glutathione to completely buffer protein thiols from redox reactions. The role of the abundant cysteines in titin has long remained unclear, and although it had been hypothesized that disulfide bonding (Kosuri et al., 2012; Wiita et al., 2007), disulfide isomerization (Alegre-Cebollada et al., 2011; Giganti et al., 2018), and other post-translational modifications (Alegre-Cebollada et al., 2014) may alter titin elasticity, never before had their role in power output been explored. Additional evidence regarding disulfide bonds comes from myofibril mechanics experiments done in the presence of reducing agents (dithiothreitol [DTT]) or reducing enzymes (human thioredoxin) (Alegre-Cebollada et al., 2014; Grϋtzner et al., 2009). While the marked difference in myofibril passive mechanics under reducing conditions is highly suggestive of the presence of disulfide bonds in titin, it has not yet been possible to image or count the disulfide bonds in muscle tissue, and the actual proportion of formed disulfide bonds in titin remains unknown. Nonetheless, the acquisition of disulfide bonds in the intracellular compartment could be achieved through several different mechanisms, including the formation of intermediates with reactive oxygen species or low-molecular-weight disulfide compounds. While there is evidence that low-molecular-weight thiols can induce disulfide reformation in peptides at zero force (Beedle et al., 2016, 2017, 2018), this is unlikely a viable mechanism at the 4-to 10-pN stretching forces experienced by titin in vivo. A more likely mechanism for the introduction of disulfide bonds is through oxidoreductase enzymes, which have evolved to recognize unfolded substrates containing cysteine residues. PDI is one of the genes that is upregulated in cardiomyocytes in response to myocardial infarction and plays an important role in post-myocardial infarction (MI) remodeling (Severino et al., 2007).
Magnetic tweezers refolding trajectories of I27 in the presence of PDI reaffirm the mechanism of PDI proposed by the previous atomic force microscopy (AFM) studies (Kosuri et al., 2012), namely that re-oxidation of the disulfide bond is preceded by entropic collapse of the I27 polypeptide into a molten globule-like structure, as indicated by the step sizes of folding measured in Figure 4D. What could not be probed in the previous studies was the ability of PDI to bias the folding reaction to the native folded state. While it is intuitive that the presence of a pre-formed disulfide bond allows I27 to fold at higher forces (12.5 pN versus 4.0 pN midpoint), it is not immediately clear why having a mixed disulfide between PDI and I27 also shifts the folding probability of I27 to higher forces (Figures 3B and 4C). One possible explanation is the chaperone activity that has been reported to be a generic property of the thioredoxin fold, independent from its catalytic activity (Quan et al., 1995). The catalytic CXXC motif is surrounded by a small hydrophobic patch that is thought to help thioredoxin-based enzymes recognize and bind to unfolded proteins. It is possible that the hydrophobic patch helps to orga-nize the hydrophobic core of I27 and accelerate folding, as is the case in other typical chaperone proteins (Saio et al., 2014). PDI simply acts as a placeholder so that when the protein collapses to the molten globule state, at 5.0 pN, for example, the free thiol of titin can attack the mixed disulfide with PDI. With the reformed internal disulfide, the natively folded state is then quickly achieved. These studies demonstrate that a substrate such as titin can be successfully re-oxidized by a typical mammalian oxidoreductase enzyme, even though it is under stretching forces of several piconewtons during physiological muscle extensions (Linke, 2018; Rivas-Pardo et al., 2016). It is important to note that the in vivo imaging studies of sarcomere lengths used as the basis for our estimates of the force on titin use a micro-endoscopic needle that slightly perturbs the muscle tissue and must be corrected for in post-imaging analysis (Chen et al., 2016).
These magnetic tweezers experiments further allow for the testing of key hypotheses related to titin protein folding during muscle contraction through direct comparison with equivalent single-molecule assays performed on myosin II motors. Attempts to reconcile full muscle and muscle fiber mechanics experiment data with the trajectories observed from single-molecule optical tweezers experiments on muscle myosin motors demonstrate the complexity of scaling mechanical systems over lengths of many orders of magnitude. Early single-molecule optical tweezers experiments show that single myosin II power strokes generate steps of 12 nm and forces of 3.4 pN on average (Finer et al., 1994). Improvements such as using small ensembles of myosins to enable continuous sliding against a pulling force reported velocities of 500 nm/s at forces of 3 pN per motor (Debold et al., 2005), and the most recent experiments using entire myosin-actin co-filaments extracted from frog muscle observed unloaded velocities of 1,000 nm/s at 1 mM ATP and 20 pN (half the isometric force), providing a power output of 20,000 zW for tens of milliseconds (Kaya et al., 2017). In our case, the power output is not sustained, but instead drops off rapidly as the folding contraction progresses. Because each folding event represents crossing of a single energetic barrier, the rate of the steps and hence the rate of energy delivery fall exponentially. This means that 63% of the work due to folding is delivered within one time constant of the exponential and 95% is delivered within three time constants. When I27OXD folds at 4 pN, the time constant of the folding contraction is ~15 ms, so the average power output over this time period is Pmean ≈ 4,000 zW. This is less than the “burst” power at the onset of the folding contraction, but it is still a substantial power output when compared to the 20,000 zW from single-molecule actomyosin sliding at the half-isometric force (Kaya et al., 2017). It is typical for only the first ~10–20 ms of isotonic velocity transients to be considered during determination of the force-velocity relation in muscle fibers experiments (Caremani et al., 2013, 2016; Reconditi et al., 2004). Over this timescale, titin recovers the majority of its stored energy through folding.
There are several other mechanisms that we speculate could further boost the contribution of titin during active muscle contraction. Calcium binding has been shown to increase the stability of the native I27 fold using high-force AFM measurements (DuVall et al., 2013). It is possible that the presence of calcium could also stabilize some structures in the folding transition state, thereby accelerating the folding rate—an experiment that should be checked using magnetic tweezers. Calcium has also been shown to alter the stiffness of the PEVK fragments of titin and decrease PEVK-actin binding. There are many theories about how this may serve a role during active contraction (DuVall et al., 2017; Herzog et al., 2015; Labeit et al., 2003). It is possible that at the onset of a contraction, high calcium levels induce simultaneous myosin cycling and titin dissociation from actin (Kulke et al., 2001; Linke et al., 2002). Both of these events would work to reduce the force experienced by each titin filament, permitting faster titin refolding, and may also explain some of the observed changes in the spring length and stiffness of titin upon muscle activation. Titin splicing is also a key factor to consider as isoform switching from longer to shorter titin iso-forms, or vice versa, would alter the number of titin Ig domains undergoing unfolding-refolding reactions by altering the passive force on much longer timescales. Titin isoform switching is a documented physiological response in both healthy animal and chronically diseased human hearts (Neagoe et al., 2002; Nelson et al., 2008). Furthermore, different titin Ig domains are likely to have different folding probabilities, depending on the spacing between the cysteine residues in the Ig domain sequence. We anticipate that as the two cysteines move further apart, toward their respective N and C termini, the folding forces should continue to rise due to decreased backbone entropy.
Muscle is a complex tissue that functions by scaling the motions of nanoscopic actuators into macroscopic movements. With so many constituent proteins contributing to the overall structure, energetics, and material properties of the muscle, it is difficult to tease apart their relative contributions with muscle fiber experiments alone. This comparison between single-molecule data on motors and folding Ig domains is therefore central to the question of their relative contributions to the energy released during muscle contraction. The magnetic tweezers data presented here demonstrate that a truncated chain of only eight di-sulfide-containing Ig domains are capable of folding at speeds of up to 1,900 nm/s and power output of 6,000 zW, which should renew the debate over how to incorporate titin dynamics into models of muscle contraction.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Edward Eckels (ece2117@columbia.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Lines
Proteins were expressed using E. coli strains BL21 Star DE3 (Thermo Fisher) for the I27 construct, and ERL (Sauer Lab) for the HaloTag construct. Transfection was achieved by adding DNA to chemically competent E. coli. All growth was done in LB media at 37 C until IPTG induction, after which the cells were grown at 25 C.
METHOD DETAILS
Magnetic Tweezers Instrumentation
The magnetic tweezers is built on top of a Axiovert 135 TV (Zeiss, Germany) with epi-illumination from a cold white collimated LED lamp (MCWHL5, Thorlabs USA). Imaging is achieved with a 63x oil-immersion objective with no further magnification in the optical path. The image is focused by the microscope tube lens onto a high-speed USB 3.0 CMOS camera (model XiQ, Ximea GmbH Germany) that is para-focal with the microscope eyepieces. A piezo objective positioner (PIFOC P-725, Physik Instrumente Germany) is used to calibrate the focal stack for every magnetic bead, reference bead pair prior to an experiment. A stack of 5x N52 magnets (cylindrical 3/16” × 3/16” from KJ Magnetics, USA) are positioned by a high speed voice coil (Equipment Solutions, USA) that contains an optical encoder with sub-micron resolution. Positioning is achieved via a custom-built PID feedback circuit and amplifier that compares the commanded position with the signal from the optical encoder. The feedback is tuned to provide accurate displacements over a range of 8 mm, which is verified using a Heidenhain optical template (Heidenhain, USA).
A low pass filter in the feedback circuit typically limits the commanded position to 20 Hz. This is done to prevent vibration of the microscope due to the inertia of the voice coil during positioning. However, the force protocol depicted in Figure 6 of the main text requires only small amplitude motions of the magnets. For these experiments, the low pass filter of the feedback circuit was bypassed and a separate analog filter module (SIM965, Stanford Research Systems USA) at 200 Hz was used to achieve ~5 ms response times. All data acquisition (NI-6341, National Instruments USA) and position tracking was implemented in real time using a custom C++ software (Microsoft Visual Studio C++ Compiler 2017). The software was run on PC with an i7 processor, enabling frame rates of up to ~1,500, depending on camera sensor cropping. Forces were previously calibrated via determination of an exponential force-position magnet law for the instrument(Popa et al., 2016; Yu et al., 2014).
Magnetic Tweezers Flow Cell
Flow cells were constructed from a laser-cut piece of parafilm sandwiched between two microscope coverslips. The bottom 24×40 mm coverslips were cleaned and silanized as previously described(Chen et al., 2015; Popa et al., 2016). Briefly, the coverslips were sonicated for 20 minutes each in 1% Hellmanex, Acetone, and Methanol, with copious rinsing with distilled water in between each step. The coverslips were dried at 100 C and then activated with air plasma for ten minutes. After activation the coverslips were immediately submerged in 1% aminopropyltrimethoxy-silane (APTMS, Sigma Aldrich USA) in anhydrous ethanol. After 20 minutes of silanization, the coverslips were rinsed with ethanol and cured at 100 C until dry. The coverslips could then be stored under vacuum in a desiccator for up to two months.
To construct the flow cell, a single parafilm spacer was sandwiched between the amino-silanized coverslip and a 22×22 mm coverslip that had been treated with Repel-Silane (GE Healthcare USA). The flow cell was warmed at 80 C on a hotplate for 10 minutes to create a seal with the parafilm. After cooling, a solution of 1% glutaraldehyde (Sigma Aldrich) in PBS was passed through each cell and allowed to react for 1 hour. Amino functionalized beads (2.89 micron, Spherotech USA) were added to the chambers for fifteen additional minutes. The glutaraldehyde solution and any unattached beads were rinsed from the flow cell with several washes of PBS. HaloTag O4 ligand with an amino terminal group was diluted 500-fold into PBS and was allowed to sit in the chambers overnight. The next day, the chambers were blocked with a solution of 1% BSA in PBS for a minimum of four hours at 4 C, where the chambers were then stored until use.
Molecular Biology
All multimeric constructs were created from multiple rounds of digestion and ligation using BamHI/BglII restriction sites as described previously(Wiita et al., 2006). The sequence of the disulfide containing I27 domain has been reported previously: two cysteine residues were introduced via QuickChange at positions G32 and A75, while the native cysteine residues at positions C47 and C63 were mutated to alanines (Ainavarapu et al., 2007). The N-terminal SpyCatcher and C-terminal Avi expression construct was designed in the pQE-80L (QIAGEN) plasmid for these experiments. The N-terminal HaloTag and C-terminal SpyTag was designed in the pFN18a plasmid (Promega).
Protein Expression and Purification
The I27OXD plasmid was modified to contain an N-terminal SpyCatcher (Zakeri et al., 2012) and a C-terminal AviTag (Avidity). The octameric I27 repeat was expressed in BL21 star (DE3) E. coli strain overnight at room temperature using standard LB media and IPTG induction protocols. Cells were harvested the next day following standard procedures and lysed using a French Pressure Cell. After high speed centrifugation to remove cellular debris, the supernatant was incubated with Ni-NTA agarose and eluted with 250 mM imidazole. Disulfide bond formation was guaranteed by overnight incubation of the eluted protein in 0.03% hydrogen peroxide. The following day, the buffer was exchanged for PBS using cellulose dialysis tubing to ensure efficient biotinylation (Fairhead and Howarth, 2015) of the C-terminal AviTag by the BirA enzyme (Avidity). The biotinylation reaction followed the protocol recommended by the manufacturer, and was allowed to proceed for one hour at 30 C. Immediately after biotinylation, the protein was purified via size exclusion on a Superdex 200 column (GE Healthcare). SDS-PAGE gel was used to confirm the molecular weight of the purified protein.
The HaloTag-SpyTag construct was expressed in a custom ERL strain of bacteria (Dr. Robert Sauer, MIT) overnight at room temperature. After elution from the nickel resin, the protein was immediately purified by size exclusion and stored at −80 C until needed. To reconstitute a full protein for a magnetic tweezers experiment, the HaloTag-SpyTag construct and SpyCatcher-Avi constructs were mixed in a 1:1 molar ratio for 1 hour at 4 C before further dilution to ~10 nM in PBS supplemented with 10 mM ascorbic acid for application to the flow cell.
Single-Molecule Recordings
A solution containing ~10 nM I27OXD was added to the flow cell for 20 minutes and was washed out with an experimental buffer con-sisting of PBS buffer pH 7.4 containing 10 mM ascorbate to prevent nonspecific oxidation of cysteine thiols and other side chains (Valle-Orero et al., 2017). Streptavidin beads were blocked the previous night with 1% BSA in PBS, which was exchanged to the experimental PBS/ascorbate buffer immediately before the experiment. 50 μL of the streptavidin beads were flowed through the chamber and allowed to settle on the bottom coverslip, after which the permanent magnets were brought down to a resting position that applies 4 pN to the beads. Fresh experimental buffer was frequently exchanged into the flow cell, every ~30 minutes, due to evaporative losses from the open ends of the flow cell.
QUANTIFICATION AND STATISTICAL ANALYSIS
Freely Jointed Chain
The experimental data were fit using nonlinear least-squares (Levenberg-Marquart) fitting procedures provided in Igor Pro 7.0 (Wave-metrics Inc), weighted by the standard deviation where appropriate. The number of molecules or individual observations are reported in figure legends. The freely jointed chain model (Flory, 1953) predicts the extension of the polymer, x, as a function of the force, F:
Where ΔLC is the contour length increment upon unfolding, lk is the Kuhn segment length, and kBT is the thermal energy and is approximately equal to 4.11 pN-nm. The fits for the oxidized and reduced forms of the protein are summarized in Table S1.
Sigmoidal Fits
The folding probability as a function of force follows a sigmoidal dependence as described before. The following equation and parameters were fit. See Table S2 for the fit parameters.
Calculation of Expected Work
The folding work was calculated as the step size multiplied by the folding work. The expected value of the work was then equal to the folding work multiplied by the folding probability. The value of the peak work and the force at which in the oxidized and reduced proteins can be found in Table S3.
Calculation of the Power
Collapse trajectories were fit using a single exponential function with a fixed amplitude equal to the number of domains times the step size xFJC at the collapse force:
Where N is equal to eight domains for all recordings considered and kFJC, the rate constant, is the only free fit parameter. The folding rates were fit with a rate equation(Chen et al., 2015; Dudko et al., 2008) determined by escape from a potential with the shape of a freely jointed chain polymer under a pulling force:
Where the Kuhn length lk and the increment in contour length △LC are determined by the experimental fits of the step size. The only free parameter in this equation is k0. The initial velocity of the folding contraction is determined by the slope at time zero of the exponential fit and can be calculated by taking the derivative of the rate equation at time zero:
Finally the power output of the collapsing protein chain can be calculated by multiplying the initial velocity by the applied load
The values of the rate constants of folding, maximum measured folding velocities, and maximum calculated power output from folding are summarized in Table S4
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| ERL strain, E. coli | Robert Sauer Lab (MIT) | N/A |
| BL21 Star (DE3) | ThermoFisher | C6010-03 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| BirA biotin-protein ligase standard reaction kit | Avidity | BirA-500 |
| HaloTag Ligand Amine O4 | Promega | P6741 |
| (3-Aminopropyl) trimethoxysilane | Sigma-Aldrich | 281778 |
| Repel-Silane | GE Healthcare | 17-1332-01 |
| Glutaraldehyde solution (Grade I, 25% in H2O) | Sigma-Aldrich | G5882 |
| Tris(2-carboxyethyl)phosphine hydrochloride | Sigma-Aldrich | C4706 |
| L-Ascorbic Acid | Sigma Aldrich | A7506 |
| Hydrogen Peroxide 30% in H2O | Fisher Scientific | H325 |
| Bovine Serum Albumin, sulfhydryl blocked | Lee Biosolutions | 100-10SB |
| Recombinant DNA | ||
| Halo-SpyTag | This study | N/A |
| SpyCatcher-I278(32C/75C) | This study | N/A |
| pFN18A HaloTag T7 Flexi Vector | Promega | G2751 |
| pQE80L vector | QIAGEN | 32932 |
| Software and Algorithms | ||
| Magnetic Tweezers Software (MT 2.2) | Fernandez Lab | http://fernandezlab.biologycolumbia.edu/downloads |
| Other | ||
| Dynabeads M-270 Streptavidin | ThermoFisher | 65305 |
| Amino-polystyrene particles (2.5–2.9 micron) | Spherotech | AP-25-10 |
| xiQ CMOS camera | Ximea | MQ013MG-ON |
| PIFOC objective nano-positioner | Physik Instrumente | P-725.CDD |
| Collimated LED lamp | Thorlabs | MCWHL5 |
| N52 magnets 3/16’’ diameter | K&J Magnetics | D33-N52 |
| Analog filter module | Stanford Research Instruments | SIM965 |
Highlights.
Real-time observation of disulfide reduction and oxidation using magnetic tweezers
Oxidation of titin Ig domains shifts the midpoint folding force from 4.1 to 12.5 pN
An 8-repeat oxidized titin construct reaches folding speeds of up to 1,900 nm/s
Peak power output of the same oxidized titin construct is 6,000 zW
ACKNOWLEDGMENTS
This work was funded by the generous support of NIH grant R01HL61228 (to J.M.F.) and fellowship F30HL129662 (to E.C.E.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.04.046.
DATA AND SOFTWARE AVAILABILITY
The magnetic tweezers instrumentation software executable can be downloaded from: http://fernandezlab.biology.columbia.edu/sites/default/files/MT_2_N52_Installer.zip
REFERENCES
- Ainavarapu SRK, Brujić J, Huang HH, Wiita AP, Lu H, Li L, Walther KA, Carrion-Vazquez M, Li H, and Fernandez JM (2007). Contour length and refolding rate of a small protein controlled by engineered disulfide bonds. Biophys. J 92, 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alegre-Cebollada J, Kosuri P, Rivas-Pardo JA, and Fernández JM (2011). Direct observation of disulfide isomerization in a single protein. Nat. Chem 3, 882–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alegre-Cebollada J, Kosuri P, Giganti D, Eckels E, Rivas-Pardo JA, Hamdani N, Warren CM, Solaro RJ, Linke WA, and Fernández JM (2014). S-glutathionylation of cryptic cysteines enhances titin elasticity by blocking protein folding. Cell 156, 1235–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beedle AEM, Lynham S, and Garcia-Manyes S. (2016). Protein S-sulfenylation is a fleeting molecular switch that regulates non-enzymatic oxidative folding. Nat. Commun 7, 12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beedle AEM, Mora M, Lynham S, Stirnemann G, and Garcia-Manyes S. (2017). Tailoring protein nanomechanics with chemical reactivity. Nat. Commun 8, 15658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beedle AEM, Mora M, Davis CT, Snijders AP, Stirnemann G, and Garcia-Manyes S. (2018). Forcing the reversibility of a mechanochemical reaction. Nat. Commun 9, 3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco P, Reconditi M, Piazzesi G, and Lombardi V. (2016). Is muscle powered by springs or motors? J. Muscle Res. Cell Motil 37, 165–167. [DOI] [PubMed] [Google Scholar]
- Caremani M, Melli L, Dolfi M, Lombardi V, and Linari M. (2013). The working stroke of the myosin II motor in muscle is not tightly coupled to release of orthophosphate from its active site. J. Physiol 591, 5187–5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caremani M, Pinzauti F, Reconditi M, Piazzesi G, Stienen GJM, Lombardi V, and Linari M. (2016). Size and speed of the working stroke of cardiac myosin in situ. Proc. Natl. Acad. Sci. USA 113, 3675–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, and Delp SL (2016). Human soleus sarcomere lengths measured using in vivo microendoscopy at two ankle flexion angles. J. Biomech 49, 4164–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Yuan G, Winardhi RS, Yao M, Popa I, Fernandez JM, and Yan J. (2015). Dynamics of equilibrium folding and unfolding transitions of titin immunoglobulin domain under constant forces. J. Am. Chem. Soc 137, 3540–3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Sanchez GN, Schnitzer MJ, and Delp SL (2016). Changes in sarcomere lengths of the human vastus lateralis muscle with knee flexion measured using in vivo microendoscopy. J. Biomech 49, 2989–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debold EP, Patlak JB, and Warshaw DM (2005). Slip sliding away: load-dependence of velocity generated by skeletal muscle myosin molecules in the laser trap. Biophys. J 89, L34–L36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudko OK, Hummer G, and Szabo A. (2008). Theory, analysis, and interpretation of single-molecule force spectroscopy experiments. Proc. Natl. Acad. Sci. USA 105, 15755–15760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuVall MM, Gifford JL, Amrein M, and Herzog W. (2013). Altered mechanical properties of titin immunoglobulin domain 27 in the presence of calcium. Eur. Biophys. J 42, 301–307. [DOI] [PubMed] [Google Scholar]
- DuVall MM, Jinha A, Schappacher-Tilp G, Leonard TR, and Herzog W. (2017). Differences in titin segmental elongation between passive and active stretch in skeletal muscle. J. Exp. Biol 220, 4418–4425. [DOI] [PubMed] [Google Scholar]
- Eckels EC, Tapia-Rojo R, Rivas-Pardo JA, and Fernández JM (2018). The Work of Titin Protein Folding as a Major Driver in Muscle Contraction. Annu. Rev. Physiol 80, 327–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairhead M, and Howarth M. (2015). Site-Specific Biotinylation of Purified Proteins Using BirA In Site-Specific Protein Labeling, Gautier A. and Hinner MJ, eds. (Springer; ), pp. 171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez JM, and Li H. (2004). Force-clamp spectroscopy monitors the folding trajectory of a single protein. Science 303, 1674–1678. [DOI] [PubMed] [Google Scholar]
- Finer JT, Simmons RM, and Spudich JA (1994). Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature 368, 113–119. [DOI] [PubMed] [Google Scholar]
- Flory PJ (1953). Principles of Polymer Chemistry (Cornell University Press; ). [Google Scholar]
- Forscher P, Lin CH, and Thompson C. (1992). Novel form of growth cone motility involving site-directed actin filament assembly. Nature 357, 515–518. [DOI] [PubMed] [Google Scholar]
- Gelles J, Schnapp BJ, and Sheetz MP (1988). Tracking kinesin-driven movements with nanometre-scale precision. Nature 331, 450–453. [DOI] [PubMed] [Google Scholar]
- Giganti D, Yan K, Badilla CL, Fernandez JM, and Alegre-Cebollada J. (2018). Disulfide isomerization reactions in titin immunoglobulin domains enable a mode of protein elasticity. Nat. Commun 9, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber CW, Čemažar M, Heras B, Martin JL, and Craik DJ (2006). Protein disulfide isomerase: the structure of oxidative folding. Trends Biochem. Sci 31, 455–464. [DOI] [PubMed] [Google Scholar]
- Grϋtzner A, Garcia-Manyes S, Kötter S, Badilla CL, Fernandez JM, and Linke WA (2009). Modulation of titin-based stiffness by disulfide bonding in the cardiac titin N2-B unique sequence. Biophys. J 97, 825–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar S, Tapia-Rojo R, Eckels EC, Valle-Orero J, and Fernandez JM (2017). Trigger factor chaperone acts as a mechanical foldase. Nat. Commun 8, 668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog W, Powers K, Johnston K, and Duvall M. (2015). A new paradigm for muscle contraction. Front. Physiol 6, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J. (1997). Molecular motors: structural adaptations to cellular functions. Nature 389, 561–567. [DOI] [PubMed] [Google Scholar]
- Huxley AF, and Simmons RM (1971). Proposed mechanism of force generation in striated muscle. Nature 233, 533–538. [DOI] [PubMed] [Google Scholar]
- Ilton M, Bhamla MS, Ma X, Cox SM, Fitchett LL, Kim Y, Koh J, Krishnamurthy D, Kuo C-Y, Temel FZ, et al. (2018). The principles of cascading power limits in small, fast biological and engineered systems. Science 360, eaao1082. [DOI] [PubMed] [Google Scholar]
- Kahn TB, Fernández JM, and Perez-Jimenez R. (2015). Monitoring Oxidative Folding of a Single Protein Catalyzed by the Disulfide Oxidoreductase DsbA. J. Biol. Chem 290, 14518–14527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya M, Tani Y, Washio T, Hisada T, and Higuchi H. (2017). Coordinated force generation of skeletal myosins in myofilaments through motor coupling. Nat. Commun 8, 16036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuri P, Alegre-Cebollada J, Feng J, Kaplan A, Inglés-Prieto A, Badilla CL, Stockwell BR, Sanchez-Ruiz JM, Holmgren A, and Fernández JM (2012). Protein folding drives disulfide formation. Cell 151, 794–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulke M, Fujita-Becker S, Rostkova E, Neagoe C, Labeit D, Manstein DJ, Gautel M, and Linke WA (2001). Interaction between PEVK-titin and actin filaments: origin of a viscous force component in cardiac myofibrils. Circ. Res 89, 874–881. [DOI] [PubMed] [Google Scholar]
- Labeit D, Watanabe K, Witt C, Fujita H, Wu Y, Lahmers S, Funck T, Labeit S, and Granzier H. (2003). Calcium-dependent molecular spring elements in the giant protein titin. Proc. Natl. Acad. Sci. USA 100, 13716–13721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtwark GA, Farris DJ, Chen X, Hodges PW, and Delp SL (2018). Microendoscopy reveals positive correlation in multiscale length changes and variable sarcomere lengths across different regions of human muscle. J. Appl. Physiol 125, 1812–1820. [DOI] [PubMed] [Google Scholar]
- Linari M, Brunello E, Reconditi M, Fusi L, Caremani M, Narayanan T, Piazzesi G, Lombardi V, and Irving M. (2015). Force generation by skeletal muscle is controlled by mechanosensing in myosin filaments. Nature 528, 276–279. [DOI] [PubMed] [Google Scholar]
- Linke WA (2018). Titin Gene and Protein Functions in Passive and Active Muscle. Annu. Rev. Physiol 80, 389–411. [DOI] [PubMed] [Google Scholar]
- Linke WA, Kulke M, Li H, Fujita-Becker S, Neagoe C, Manstein DJ, Gautel M, and Fernandez JM (2002). PEVK domain of titin: an entropic spring with actin-binding properties. J. Struct. Biol 137, 194–205. [DOI] [PubMed] [Google Scholar]
- Littler DR, Harrop SJ, Goodchild SC, Phang JM, Mynott AV, Jiang L, Valenzuela SM, Mazzanti M, Brown LJ, Breit SN, and Curmi PM (2010). The enigma of the CLIC proteins: ion channels, redox proteins, enzymes, scaffolding proteins? FEBS Lett. 584, 2093–2101. [DOI] [PubMed] [Google Scholar]
- Llewellyn ME, Barretto RPJ, Delp SL, and Schnitzer MJ (2008). Minimally invasive high-speed imaging of sarcomere contractile dynamics in mice and humans. Nature 454, 784–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan L, and Matsudaira P. (2000). Motility powered by supramolecular springs and ratchets. Science 288, 95–100. [DOI] [PubMed] [Google Scholar]
- Manteca A, Schönfelder J, Alonso-Caballero A, Fertin MJ, Barruetabeña N, Faria BF, Herrero-Galá n E, Alegre-Cebollada J, De Sancho D, and Perez-Jimenez R. (2017). Mechanochemical evolution of the giant muscle protein titin as inferred from resurrected proteins. Nat. Struct. Mol. Biol 24, 652–657. [DOI] [PubMed] [Google Scholar]
- Mayans O, Wuerges J, Canela S, Gautel M, and Wilmanns M. (2001). Structural evidence for a possible role of reversible disulphide bridge formation in the elasticity of the muscle protein titin. Structure 9, 331–340. [DOI] [PubMed] [Google Scholar]
- Merz AJ, So M, and Sheetz MP (2000). Pilus retraction powers bacterial twitching motility. Nature 407, 98–102. [DOI] [PubMed] [Google Scholar]
- Mϋller H-M, Steringer JP, Wegehingel S, Bleicken S, Mϋnster M, Dimou E, Unger S, Weidmann G, Andreas H, García-Sáez AJ, et al. (2015). Formation of Disulfide Bridges Drives Oligomerization, Membrane Pore Formation and Translocation of Fibroblast Growth Factor 2 to Cell Surfaces. J. Biol. Chem 290, 8925–8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neagoe C, Kulke M, del Monte F, Gwathmey JK, de Tombe PP, Hajjar RJ, and Linke WA (2002). Titin isoform switch in ischemic human heart dis-ease. Circulation 106, 1333–1341. [DOI] [PubMed] [Google Scholar]
- Nelson OL, Robbins CT, Wu Y, and Granzier H. (2008). Titin isoform switching is a major cardiac adaptive response in hibernating grizzly bears. Am. J. Physiol. Heart Circ. Physiol 295, H366–H371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazzesi G, Reconditi M, Linari M, Lucii L, Bianco P, Brunello E, De-costre V, Stewart A, Gore DB, Irving TC, et al. (2007). Skeletal muscle performance determined by modulation of number of myosin motors rather than motor force or stroke size. Cell 131, 784–795. [DOI] [PubMed] [Google Scholar]
- Popa I, Rivas-Pardo JA, Eckels EC, Echelman DJ, Badilla CL, Valle-Orero J, and Fernández JM (2016). A HaloTag Anchored Ruler for Week-Long Studies of Protein Dynamics. J. Am. Chem. Soc 138, 10546–10553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado LG, Makarenko I, Andresen C, Krϋger M, Opitz CA, and Linke WA (2005). Isoform diversity of giant proteins in relation to passive and active contractile properties of rabbit skeletal muscles. J. Gen. Physiol 126, 461–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan H, Fan G, and Wang CC (1995). Independence of the chaperone activity of protein disulfide isomerase from its thioredoxin-like active site. J. Biol. Chem 270, 17078–17080. [DOI] [PubMed] [Google Scholar]
- Reconditi M, Linari M, Lucii L, Stewart A, Sun Y-B, Boesecke P, Narayanan T, Fischetti RF, Irving T, Piazzesi G, et al. (2004). The myosin motor in muscle generates a smaller and slower working stroke at higher load. Nature 428, 578–581. [DOI] [PubMed] [Google Scholar]
- Rivas-Pardo JA, Eckels EC, Popa I, Kosuri P, Linke WA, and Ferná n-dez, J.M. (2016). Work Done by Titin Protein Folding Assists Muscle Contraction. Cell Rep 14, 1339–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saio T, Guan X, Rossi P, Economou A, and Kalodimos CG (2014). Structural basis for protein antiaggregation activity of the trigger factor chaperone. Science 344, 1250494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez GN, Sinha S, Liske H, Chen X, Nguyen V, Delp SL, and Schnitzer MJ (2015). In Vivo Imaging of Human Sarcomere Twitch Dynamics in Individual Motor Units. Neuron 88, 1109–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severino A, Campioni M, Straino S, Salloum FN, Schmidt N, Herbrand U, Frede S, Toietta G, Di Rocco G, Bussani R, et al. (2007). Identification of protein disulfide isomerase as a cardiomyocyte survival factor in ischemic cardiomyopathy. J. Am. Coll. Cardiol 50, 1029–1037. [DOI] [PubMed] [Google Scholar]
- Steinberg SF (2013). Oxidative stress and sarcomeric proteins. Circ. Res 112, 393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Footer M, and Matsudaira P. (1997). Modification of Cys-837 identifies an actin-binding site in the beta-propeller protein scruin. Mol. Biol. Cell 8, 421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot JA, Mitchison TJ, Tilney LG, and Portnoy DA (1992). The rate of actin-based motility of intracellular Listeria monocytogenes equals the rate of actin polymerization. Nature 357, 257–260. [DOI] [PubMed] [Google Scholar]
- Trombitás K, Wu Y, McNabb M, Greaser M, Kellermayer MSZ, Labeit S, and Granzier H. (2003). Molecular basis of passive stress relaxation in human soleus fibers: assessment of the role of immunoglobulin-like domain unfolding. Biophys. J 85, 3142–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle-Orero J, Rivas-Pardo JA, Tapia-Rojo R, Popa I, Echelman DJ, Haldar S, and Fernández JM (2017). Mechanical Deformation Accelerates Protein Ageing. Angew. Chem. Int. Ed. Engl 56, 9741–9746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way M, Sanders M, Garcia C, Sakai J, and Matsudaira P. (1995). Sequence and domain organization of scruin, an actin-cross-linking protein in the acrosomal process of Limulus sperm. J. Cell Biol 128, 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiita AP, Ainavarapu SRK, Huang HH, and Fernandez JM (2006). Force-dependent chemical kinetics of disulfide bond reduction observed with single-molecule techniques. Proc. Natl. Acad. Sci. USA 103, 7222–7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiita AP, Perez-Jimenez R, Walther KA, Gräter F, Berne BJ, Holmgren A, Sanchez-Ruiz JM, and Fernandez JM (2007). Probing the chemistry of thioredoxin catalysis with force. Nature 450, 124–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Dulin D, Cnossen J, Köber M, van Oene MM, Ordu O, Berghuis BA, Hensgens T, Lipfert J, and Dekker NH (2014). A force calibration standard for magnetic tweezers. Rev. Sci. Instrum 85, 123114. [DOI] [PubMed] [Google Scholar]
- Zakeri B, Fierer JO, Celik E, Chittock EC, Schwarz-Linek U, Moy VT, and Howarth M. (2012). Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. USA 109, E690–E697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.