Abstract
Oxidation of H3 at lysine 4 (H3K4ox) by lysyl oxidase-like 2 (LOXL2) generates an H3 modification with an unknown physiological function. We find that LOXL2 and H3K4ox are higher in triple-negative breast cancer (TNBC) cell lines and patient-derived xenografts (PDXs) than those from other breast cancer subtypes. ChIP-seq revealed that H3K4ox is located primarily in heterochromatin, where it is involved in chromatin compaction. Knocking down LOXL2 reduces H3K4ox levels and causes chromatin decompaction, resulting in a sustained activation of the DNA damage response (DDR) and increased susceptibility to anticancer agents. This critical role that LOXL2 and oxidized H3 play in chromatin compaction and DDR suggests that functionally targeting LOXL2 could be a way to sensitize TNBC cells to conventional therapy.
Subject terms: Chromosomes, Prognostic markers
Introduction
Histone modifications contribute to gene regulation both by directly affecting chromatin structure and by recruiting effector proteins [1]. Deregulation of this enzymatic system can contribute to diseases, including cancer. The lysyl oxidase family of proteins are copper- and quinone-dependent amine oxidases that oxidize the amino group located in the epsilon-position in lysines, thereby generating an aldehyde group [2]. One of the members of the LOX family, lysyl oxidase-like 2 (LOXL2), deaminates unmethylated and trimethylated lysine 4 in histone H3 (H3K4me3) through an amino-oxidase reaction that uses the Cu(II) ion and the internal cofactor lysine-tyrosylquinone, releasing the amino group and converting K4 into an allysine (H3K4ox) [3, 4]. Generation of this peptidyl aldehyde likely alters the local macromolecular structure of chromatin and the nature of any protein–protein or protein–nucleic acid interactions. This is particularly relevant for gene regulation, as changes in the macromolecular status of histones can affect chromatin conformation [4–6].
LOXL2 is overexpressed in many tumors, and especially in breast cancers [7–9]. In this light, it is intriguing that some breast cancers are intrinsically resistant to chemotherapy; for these subtypes, chemotherapy induces a mesenchymal phenotype through the epithelial-to-mesenchymal transition (EMT) [10]. EMT is likely to be a critical switch for tumor cell invasiveness and cell death resistance [11–13] and to involve chromatin reorganization, as it requires dramatic changes in cellular characteristics and gene expression [6, 14]. Notably, the key transcription factor Snail1 interacts with LOXL2 [15], and LOXL2 H3K4 oxidase activity generates an H3K4ox that regulates the repression of the E-cadherin gene (CDH1) and heterochromatin transcription, which play roles in two essential steps of EMT [6, 16].
Double-strand breaks (DSBs) are a major form of DNA damage and cause a specific signaling response, the DNA damage response (DDR), which can activate cell cycle checkpoint arrest and cell fate decisions, such as apoptosis or senescence. One of the first steps of DDR is the accumulation of DNA repair proteins at the damaged site [17]. Importantly, DDR activation can occur in the absence of DNA damage, by stable association of the repair factors with chromatin [18, 19]. Moreover, increasing evidence suggests that higher-order chromatin structures affect DSB repair and signaling [20]. For example, DDR actively regulates decondensation of chromatin after DSBs [21], and it is amplified when chromatin is in an “open” state [20]. Similarly, DDR signaling is affected by chromatin compaction in a DNA damage-independent manner [18, 19, 22–24].
We addressed the physiological functions of H3K4ox using an in-house generated antibody specific for this modification to analyze the H3K4ox levels in distinct breast cancer subtypes. Intriguingly, mesenchymal triple-negative breast cancer (TNBC) cell lines as well as breast cancer patient-derived xenografts (PDXs) had high H3K4ox levels that correlated with high LOXL2 expression, as compared with other subtypes. Using ChIP-seq to map its genome-wide localization, we found that H3K4ox was enriched in heterochromatin in TNBC cells, which are highly metastatic and resistant to chemotherapy. Decreasing LOXL2 levels reduced the amount of H3K4ox in chromatin, resulting in chromatin decondensation and a sustained activation of DDR. Further, both LOXL2 depletion and treatment of TNBC with chromatin-modifying drugs sensitized cancer cells to conventional treatments. Thus, targeting H3K4ox levels may open a new therapeutic window for this subtype of breast cancer.
Results
Generating an H3K4ox-specific antibody
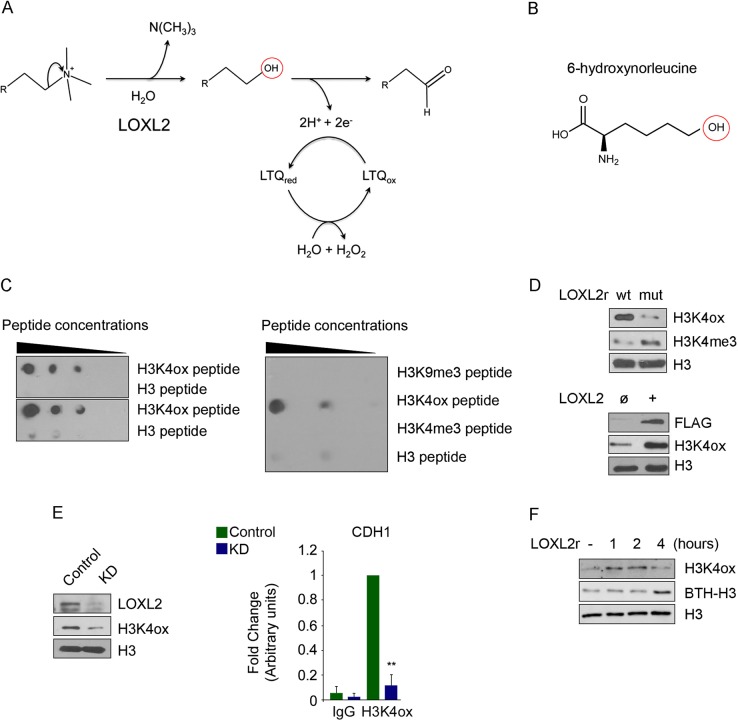
We initially generated a specific antibody for the recently discovered histone modification of H3K4ox, as a prerequisite for studying its physiological function. As the aldehyde group generated by LOXL2 reaction on trimethylated lysine 4 is highly reactive, and hence unfit for immunochemical studies, we hypothesized that a primary alcohol might provide a similar oxygen-bearing functionality that is less reactive, in order to generate a modification-specific antibody that provides a readout of H3K4ox (Fig. 1a). We therefore synthesized a H3 peptide containing a 6-hydroxynorleucine residue as allysine at position 4 and used this for rabbit immunization (Fig. 1b). The resulting H3K4ox antibody was highly specific for the H3K4ox peptide, with very low cross-reactivity for unmodified H3 and no detected cross-reactivity for H3K9me3 or H3K4me3, in a wide range of experimental conditions (dot blots, western blots, and chromatin immunoprecipitation (ChIP) experiments) (Fig. 1c–f). Analysis of purified nucleosomes from 293T cells showed that H3K4ox levels increased, and H3K4me3 levels decreased, when nucleosomes were incubated with wild-type (wt) recombinant LOXL2 but not with a catalytically inactive LOXL2 (LOXL2mut) [3] (Fig. 1d, upper panel). Moreover, the levels of H3K4ox also increased in MCF-7 cells transfected with LOXL2 as compared with cells transfected with the empty vector (Fig. 1d, lower panel). Finally, MDA-MB-231 cells infected with an shRNA targeting the human LOXL2 (LOXL2 knockdown (KD)) showed a specific reduction in H3K4ox levels as compared with cells infected with an irrelevant shRNA (control), in both western blots and ChIP-PCR experiments using the E-cadherin gene promoter (CDH1), which is a well-known LOXL2-mediated H3K4 oxidation target promoter (Fig. 1e) [3, 4]. Kinetics of the reaction using recombinant LOXL2 and nucleosomes revealed that levels of intermediate alcohol were maintained for 2 h, after which they were reduced (Fig. 1f). Biotinylated hydrazide was used to detect the generated aldehyde group, which appears after 4 h of reaction (Fig. 1f). Thus, as the intermediate alcohol is relatively stable, the antibody we generated can be used as a readout of the oxidized histone H3K4, although we cannot distinguish whether it recognizes the intermediate alcohol, the aldehyde group, or both.
Fig. 1.
Quality control of the anti-H3K4ox antibody. a Schematic representation of the LOXL2 reaction. The red circle indicates the intermediate residue that is targeted by the in-house generated anti-H3K4ox antibody. b The artificial amino acid 6-hydroxynorleucine was used in the peptide to generate the anti-H3K4ox antibody. c The anti-H3K4ox antibody was found to be specific in western blot in two replicates of dot blots of dilution series of oxidized histone H3 peptide (H3K4ox) or unmodified H3 peptide (left panel), as well as in a representative dot blot of a dilution series of H3K9me3, H3K4ox, H3K4me3, or H3 peptides (right panel). d Nucleosomes were incubated with recombinant wild-type (wt) LOXL2 or a catalytically inactive LOXL2 (mut) purified from baculovirus to detect H3K4ox/H3K4me3 levels (upper panel). Lysates of MCF-7 cells transfected with an empty vector (ø) or with LOXL2 were analyzed by western blotting, using the indicated antibodies (lower panel). e Western blot for LOXL2, H3K4ox, and total H3 from MDA-MB-231 cells infected with short hairpin RNA (shRNA) as a control, or a knockdown (KD) using a shRNA specific for LOXL2 (LOXL2 KD) (left panel). Anti-H3K4ox ChIP-PCR was used to analyze the E-cadherin gene (CDH1) promoter in MDA-MB-231 cells infected with shRNA for either control (green bar) or LOXL2 KD (blue bar). Data of qPCR amplifications were normalized to the input and to total H3 for each condition. Error bars indicate the SD from at least three independent experiments. **P < 0.01. f Western blot of H3K4ox and biotin incorporation (BTH-H3) in nucleosomes incubated with recombinant LOXL2 purified from baculovirus, after different incubation times
H3K4ox maps to heterochromatin and controls chromatin accessibility in TNBC cells
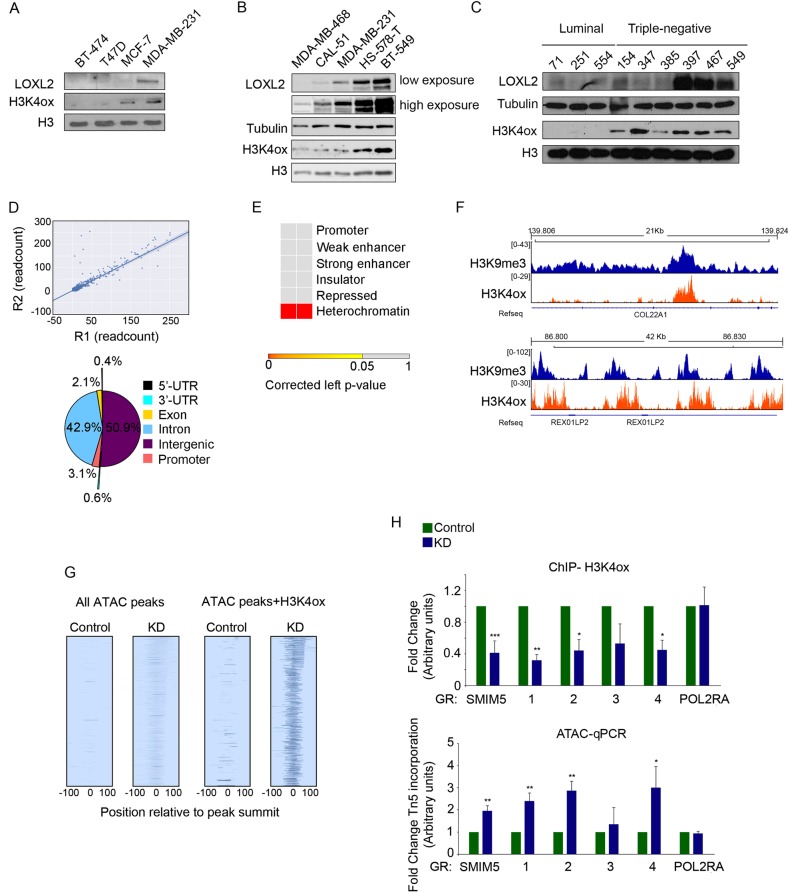
As aberrant expression and activity of LOXL2 have been reported in various cancer types [7–9], we checked the levels of LOXL2 and H3K4ox in several breast cancer cell lines representing different subtypes: luminal A, in the T-47D and MCF-7 cell lines (ER+/HER2–/PR+/–); luminal B, in the BT-474 cell line (ER+/HER2+/PR+/–); and basal TNBC, in the human MDA-MB-231 (ER–/HER2–/PR–) cell line [25]. As compared with the other cell lines, MDA-MB-231 (TNBC) showed high levels of LOXL2 and a corresponding enrichment of H3K4ox (Fig. 2a). Levels of H3K4ox also paralleled increases in LOXL2 expression levels in other TNBC cell lines (e.g., MDA-MB-468, CAL-51, HS-578-T, and BT-549), although with variable LOXL2 expression levels (Fig. 2b). Finally, comparing PDXs from luminal (3 PDXs) or TNBC (6 PDXs) subtypes of breast cancer, we found that in all TNBC PDXs the levels of H3K4ox were higher compared with luminal PDXs. In addition, in four out of six TNBC PDXs H3K4ox levels also correlated with high LOXL2 expression. (Fig. 2c).
Fig. 2.
H3K4ox maps to heterochromatin and controls chromatin accessibility in TNBC cells. Western blot for the indicated antibodies in a panel of breast cancer cell lines (a), TNBC cell lines (b), or PDXs (c). d Pearson correlation between two H3K4ox sequencing replicates. Distribution of all H3K4ox ChIP-seq peaks in MDA-MB-231 cells are given, with the indicated percentages. e Contingency table of the Fisher's exact test showed the statistical overrepresentation of the H3K4ox peaks through different chromatin states. f Genome browser view of H3K4ox and H3K9me3-binding profiles at two representative heterochromatin regions. g Heatmaps show the ATAC signal in all peaks as well as in peaks that overlap with H3K4ox in LOXL2 KD or control cells. h H3K4ox ChIP-PCR validation of the selected genomic regions from the ChIP-seq from control or LOXL2 KD MDA-MB-231 cells. Data of qPCR amplification were normalized to the input and to total H3 (upper panel). ATAC-qPCR validation of the incorporation of the transposase Tn5 at the selected genomic regions from the ChIP-seq from control or LOXL2 KD MDA-MB-231 cells. Data of qPCR amplification were normalized to an unchanging genomic region (the HPRT promoter) and expressed as the fold-change relative to data obtained from control cells, which were set to 1 (lower panel). In both experiments, the RNA polymerase II (POL2RA) promoter was used as a negative control. Error bars indicate the SD from at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001
To elucidate the function of H3K4ox in breast cancer cells, we first performed a ChIP-seq experiment with the anti-H3K4ox antibody to determine the genomic distribution of H3K4ox in the MDA-MB-231 cells. Peaks called using model-based analysis for ChIP-seq (MACS) [26] showed low differences in H3K4ox between two sequencing replicates, with a genome-wide Pearson correlation coefficient of the read count of the two replicates of 0.997 (Fig. 2d, upper panel). We observed that H3K4ox peaks were distributed throughout different genomic elements (Fig. 2d, lower panel). Using the ChromHMM tool [27], we assessed the statistical overrepresentation of the H3K4ox peaks through different chromatin states (promoter, weak or strong enhancer, insulator, repressed, or heterochromatin [nonrepetitive sequences]) and found that H3K4ox peaks were significantly overrepresented in heterochromatin (Fig. 2e, f). As generating an aldehyde in H3 removes a positive charge and creates a very reactive group, we hypothesized that this reaction affects chromatin structure. To test this, we used the assay for transposase-accessible chromatin [28, 29] followed by deep sequencing (ATAC-seq), which exploits the ability of the prokaryotic transposase Tn5 to integrate preferentially into accessible (open) chromatin. ATAC-seq showed an increased ATAC signal in LOXL2 KD cells, but not with control cells, at H3K4ox-positive sites (Fig. 2g). These results were validated in selected regions by ChIP-qPCR and ATAC-qPCR in control and LOXL2 KD cells: H3K4ox enrichment decreased in LOXL2 KD cells, with a correlating increase of ATAC signal, in these regions (Fig. 2h). No changes were observed in a control promoter, POL2RA (Fig. 2h). These data demonstrated that, in the absence of LOXL2, H3K4ox levels decrease and chromatin adopts a more open conformation (Fig. 2g). Thus, our results showed that H3K4ox is enriched in heterochromatin and is directly linked with chromatin accessibility in those regions.
Chromatin structure alterations activate DDR in a LOXL2-dependent manner
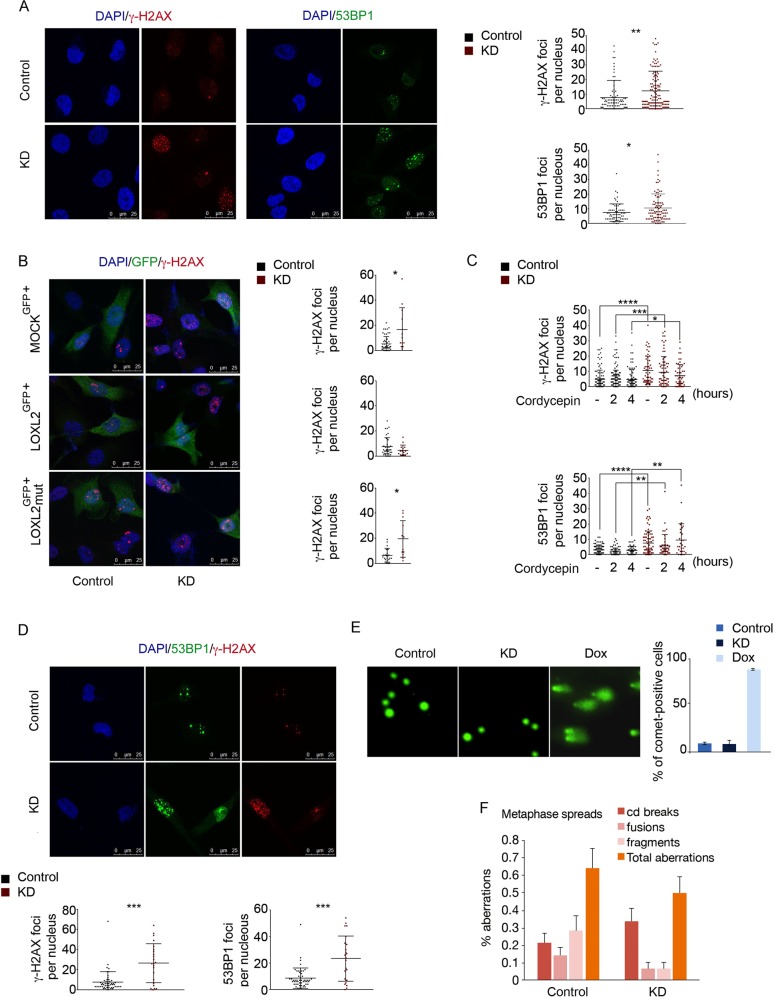
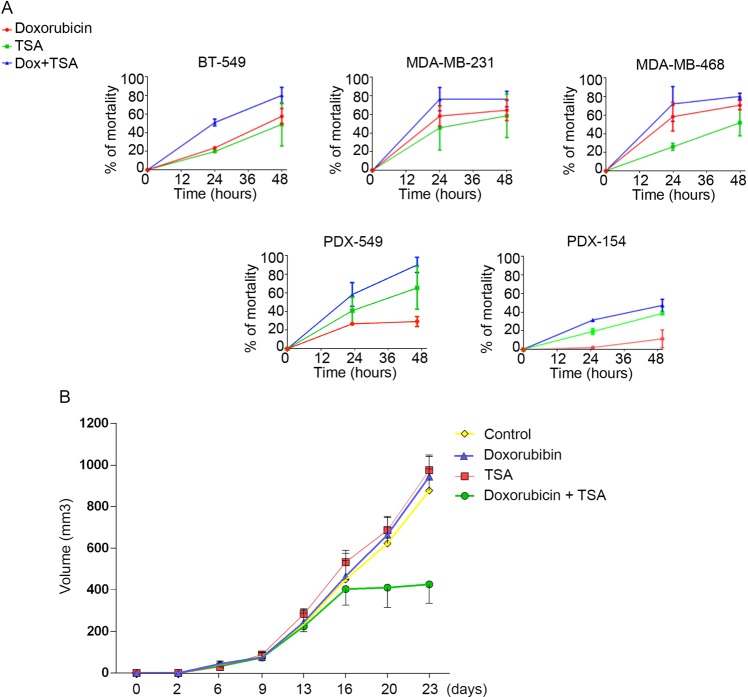
As the chromatin state can influence many aspects of DDR [30], we hypothesized that disruption of LOXL2 expression and impairment of H3K4ox generation might influence DDR by affecting chromatin accessibility. To test this, we analyzed by immunofluorescence MDA-MB-231 cells that had been infected with either LOXL2 KD or control lentiviruses, using two well-established markers of DDR: phosphorylated H2AX (γ-H2AX) and TP53-binding protein 1 (53BP1). Depletion of LOXL2 (using LOXL2 KD) led to more foci of both γ-H2AX and 53BP1 than in control cells, suggesting that LOXL2 KD cells may accumulate DNA breaks and/or activate DDR (Fig. 3a). To determine if the LOXL2 catalytic activity was involved in the observed phenotype, LOXL2 KD cells were complemented by reinfection with ectopic vector expressing either the wt LOXL2-IRES-GFP or a catalytically inactive LOXL2 (LOXL2mut-IRES-GFP), both of which were expressed at similar levels (Fig. S1). Fewer γ-H2AX and 53BP1 foci were observed in LOXL2 KD cells after reintroduction of the wt (but not of the catalytically inactive) LOXL2 (Fig. 3b), establishing that suppressing DDR activation requires both the activity of LOXL2 and H3K4ox generation.
Fig. 3.
Chromatin opening activates DDR via a mechanism that requires catalytically functional LOXL2 but can be independent of DNA damage. a γ-H2AX and 53BP1 staining and foci quantification are shown by immunofluorescence with a specific antibody for γ-H2AX (left image) or for 53BP1 (right image). Dot graphs indicate the number of foci for γ-H2AX (upper graph) and 53BP1 (lower graph) per cell in control and LOXL2 KD conditions. b γ-H2AX staining and foci quantification are shown by immunofluorescence with the indicated antibody after LOXL2 reinfection. MDA-MB-231 cells were first infected with control or LOXL2 KD lentivirus and then, after puromycin selection, again with GFP (MOCKGFP+), wild-type LOXL2-IRES-GFP (LOXL2GFP+), or LOXL2mut-IRES-GFP (LOXL2mutGFP+). Cells were fixed after 24 h. Dot graphs indicate the number of γ-H2AX foci per GFP-positive cells containing MOCKGFP+ (upper graph), LOXL2GFP+ (middle graph), or LOXL2mutGFP+ (lower graph). c Dot graphs indicate the number of γ-H2AX (upper graph) and 53BP1 (lower graph) foci per cell in control and LOXL2 KD cells after treatment with 200 μM cordycepin for the indicated timepoints. d γ-H2AX and 53BP1 staining and foci quantification are shown after immunofluorescence with the indicated antibodies in non-replicative conditions. Dot graphs indicate the number of the γ-H2AX (left graph) and 53BP1 (right graph) foci in control and LOXL2 KD cells. e Representative image showing DNA damage in control or LOXL2 KD MDA-MB-231 cells, visualized by the alkaline comet assay. Cells were treated with 0.3 μM doxorubicin for 24 h. The graph shows the percentage of MDA-MB-231 positive cells. f Chromosome alterations in control and LOXL2 KD MDA-MB-231 cells. Error bars indicate the SD from at least three independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001, ****P < 0.0001
LOXL2 KD activates DDR independently of DNA damage
Increased DDR activation in LOXL2-depleted cells could be a consequence of more DSBs due to reduced H3K4ox levels and/or chromatin decondensation. Notably, aberrant silencing and conflicts between replication forks and transcription, as well as the presence of R-loops (a transcriptional intermediate), can result in DNA damage and are influenced by chromatin state, for example in cells lacking the linker histone H1 [31]. However, our analysis of RNA-seq data revealed that LOXL2 KD cells did not have altered expression of repetitive elements (Table 1), and DDR activation in LOXL2 KD cells was not affected by cordycepin, an inhibitor of RNA synthesis that abolishes R-loop formation [32, 33] (Fig. 3c). As R-loops can generate DNA damage during replication due to fork stalling and collapse [32, 34], we next analyzed γ-H2AX and 53BP1 foci in noncycling LOXL2 KD cells and found DDR activation to also be increased (Fig. 3d). Overall, these data suggested that overexpression of repetitive elements, R-loop formation, and replication fork stalling were not responsible for activating DDR following LOXL2 depletion, when heterochromatin adopts a more open state. As chromatin structure alterations can trigger the DDR, even in the absence of DNA damage [22, 35, 36], we checked for the presence of DNA damage following LOXL2 depletion more directly, using the comet assay, in LOXL2 KD or control cells under alkaline conditions. No increases in DNA damage (due to either single-strand or double-strand DNA breaks) were observed in LOXL2 KD cells (Fig. 3e). Moreover, we did not observe any significant differences in chromosomal lesions between LOXL2 KD or control cells in metaphase spreads (Fig. 3f). Finally, analyzing for mitotic aberrations (anaphase bridges and micronuclei), which can be indicative of replication stress or DNA repair defects, we observed only a mild increase in anaphase bridges in the absence of LOXL2 (Fig. S2). Taken together, our data suggested that the combination of loss of LOXL2 and reduced H3K4ox levels in TNBC cells was sufficient to activate the DDR in the absence of detectable DNA lesions.
Table 1.
Differential expression analysis between control and LOXL2 knockdown of repetitive elements
| Locus | Ctrl_mean | Loxl2_mean | Prob | log2FC |
|---|---|---|---|---|
| Alu | 3,601,718.96 | 3,714,313.36 | 1.00 | 0.04 |
| RNA | 2,918,334.55 | 3,045,475.41 | 0.99 | 0.06 |
| L1 | 2,837,069.25 | 2,730,921.52 | 0.97 | −0.06 |
| ERV1 | 370,405.10 | 353,869.12 | 0.95 | −0.07 |
| TcMar-Tigger | 239,662.63 | 252,421.97 | 0.87 | 0.07 |
| UCON19 | 0.51 | 1.23 | 0.76 | 1.28 |
| UCON18 | 0.51 | 1.23 | 0.76 | 1.28 |
| hAT-Tip100 | 34,122.72 | 38,990.93 | 0.76 | 0.19 |
| ERVK | 8,958,641.10 | 9,448,789.20 | 0.76 | 0.08 |
| MIR | 673,530.34 | 692,895.61 | 0.76 | 0.04 |
| hAT | 12,908.30 | 16,757.15 | 0.76 | 0.38 |
| TcMar-Mariner | 33,897.47 | 38,475.09 | 0.75 | 0.18 |
| UCON4 | 234.62 | 360.51 | 0.74 | 0.62 |
| hAT-Charlie | 374,265.61 | 380,188.93 | 0.72 | 0.02 |
| UCON16 | 6.58 | 10.85 | 0.72 | 0.72 |
| Penelope | 7.08 | 11.83 | 0.70 | 0.74 |
| ERVL-MaLR | 399,009.16 | 406,080.51 | 0.70 | 0.03 |
| UCON31 | 61.20 | 87.49 | 0.70 | 0.52 |
| MER130 | 15.18 | 21.75 | 0.69 | 0.52 |
| UCON2 | 320.19 | 434.63 | 0.69 | 0.44 |
| UCON10 | 7.09 | 10.93 | 0.67 | 0.63 |
| UCON28c | 10.12 | 14.81 | 0.63 | 0.55 |
| Eulor4 | 1.26 | 1.98 | 0.55 | 0.65 |
| UCON12 | 14.14 | 4.94 | 0.52 | −1.52 |
| TcMar-Tc2 | 12,524.35 | 13,658.11 | 0.51 | 0.13 |
| UCON24 | 1.27 | 0.49 | 0.47 | −1.36 |
| UCON17 | 3.03 | 1.23 | 0.46 | −1.30 |
| ERVL | 174,857.94 | 177,852.71 | 0.44 | 0.02 |
| MamRep605 | 7848.87 | 8610.92 | 0.41 | 0.13 |
| UCON28a | 201.88 | 253.61 | 0.39 | 0.33 |
| Satellite | 28,748.18 | 25,760.18 | 0.37 | −0.16 |
| UCON26 | 532.62 | 618.38 | 0.35 | 0.22 |
| SVA_B | 2696.77 | 3025.33 | 0.31 | 0.17 |
| acro | 14.16 | 10.88 | 0.30 | −0.38 |
| UCON11 | 6.08 | 3.93 | 0.29 | −0.63 |
| UCON15 | 4.29 | 2.25 | 0.29 | −0.93 |
| MuDR | 1610.89 | 1799.90 | 0.28 | 0.16 |
| SVA_F | 3968.24 | 4322.50 | 0.26 | 0.12 |
| Gypsy | 18,034.32 | 16,836.16 | 0.26 | −0.10 |
| DNA | 5850.42 | 6284.72 | 0.25 | 0.10 |
| Dong-R4 | 3696.41 | 3198.15 | 0.25 | −0.21 |
| UCON9 | 18.69 | 12.78 | 0.24 | −0.55 |
| Centre | 11,923.53 | 12,544.21 | 0.22 | 0.07 |
| Merlin | 297.42 | 327.87 | 0.19 | 0.14 |
| SVA_D | 11,475.76 | 11,978.90 | 0.19 | 0.06 |
| UCON28b | 26.33 | 30.80 | 0.18 | 0.23 |
| LTR | 1842.20 | 1987.87 | 0.18 | 0.11 |
| UCON8 | 75.37 | 65.33 | 0.15 | −0.21 |
| TcMar | 8328.59 | 8597.66 | 0.11 | 0.05 |
| SVA_A | 1741.04 | 1827.45 | 0.10 | 0.07 |
| CR1 | 85,288.67 | 84,328.51 | 0.09 | −0.02 |
| Helitron | 5017.41 | 5157.97 | 0.06 | 0.04 |
| UCON6 | 1,277.83 | 1,325.68 | 0.06 | 0.05 |
| RTE | 30,497.19 | 29,901.20 | 0.06 | −0.03 |
| PiggyBac | 4590.52 | 4349.34 | 0.06 | −0.08 |
| UCON22 | 12.14 | 10.47 | 0.06 | −0.21 |
| UCON5 | 11.14 | 9.87 | 0.06 | −0.17 |
| telo | 489.07 | 507.40 | 0.05 | 0.05 |
| L2 | 614,402.54 | 610,776.39 | 0.05 | −0.01 |
| Deu | 2157.42 | 2220.57 | 0.05 | 0.04 |
| hAT-Blackjack | 11,106.77 | 11,287.96 | 0.05 | 0.02 |
| UCON1 | 3.04 | 3.25 | 0.04 | 0.10 |
| UCON27 | 72.82 | 74.65 | 0.03 | 0.04 |
| UCON20 | 12.13 | 10.95 | 0.02 | −0.15 |
| MamRep564 | 756.04 | 712.80 | 0.02 | −0.08 |
| RTE-BovB | 1064.66 | 1011.37 | 0.02 | −0.07 |
| ERV | 440.91 | 422.61 | 0.01 | −0.06 |
| SINE | 4912.94 | 4894.14 | 0.00 | −0.01 |
| UCON25 | 2.27 | 2.20 | 0.00 | −0.04 |
| SVA_E | 1087.33 | 1076.64 | 0.00 | −0.01 |
Alterations in chromatin compaction activate the DDR
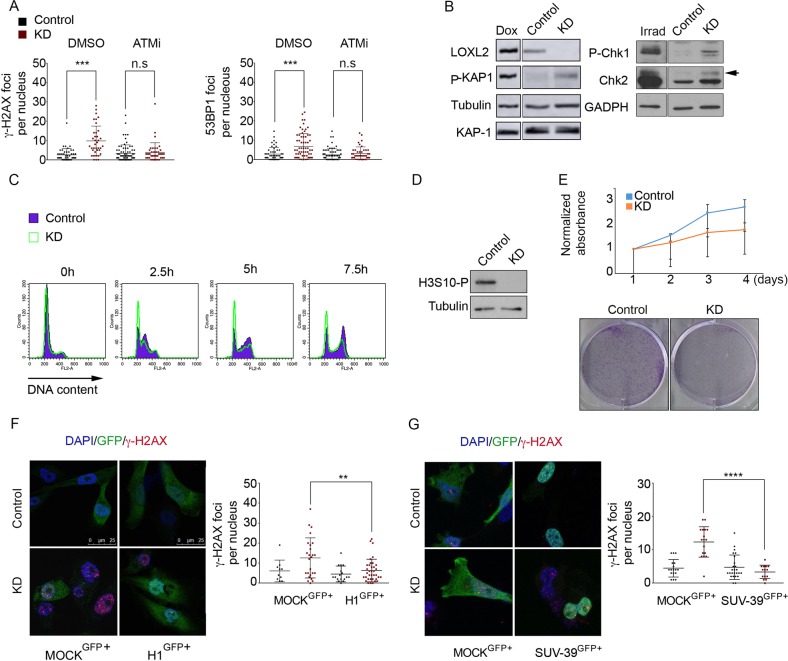
To further address the origin of DDR signaling in LOXL2-depleted cells, we analyzed the behavior of additional DDR signaling components. As both γ-H2AX and 53BP1 foci formation require the ATM kinase in some settings, we treated LOXL2 KD and control cells with the ATM inhibitor KU55933 and analyzed foci formation (Fig. 4a). Decreased foci of both markers were observed upon ATM inhibition, indicating that LOXL2-induced DDR was largely ATM-dependent. Consistent with this, LOXL2 KD cells had increased phosphorylation of several ATM substrates, including KAP-1, CHK1, and CHK2 (Fig. 4b). However, we ruled out that increased DDR signaling was due to apoptosis in LOXL2 KD cells, as no cleaved caspase-3 signal was observed in either LOXL2 KD or control cells (Fig. S3a). As these data suggested that the LOXL2 KD cells activated a checkpoint response, we analyzed cell cycle progression following LOXL2 depletion. After synchronization with a double thymidine block, LOXL2 KD cells were not able to efficiently progress through the cell cycle (Fig. 4c), and western blotting for H3S10-P showed that this histone mark was undetectable in LOXL2 KD cells as compared with control cells. (Fig. 4d). These data strongly suggested that LOXL2 KD cells arrested primarily in G1. Consistent with this possibility, cell proliferation capacity of LOXL2 KD cells was blocked (Fig. 4e, upper panel), and their colony-formation capacity was strongly reduced after only a few passages (Fig. 4e, lower panel). This effect on proliferation was further confirm by gene ontology analysis of the gene expression pattern of LOXL2 KD cells (Fig. S3b; Tables 2 and 3).
Fig. 4.
DDR activation is linked to chromatin decondensation in the absence of LOXL2. a Dot graphs indicate the number of foci with γ-H2AX (left graph) and 53BP1 (right graph) per cell from control and LOXL2 KD cells treated with DMSO or the ATM inhibitor KU55933. b KAP-1 phosphorylation in control or LOXL2 KD cells was analyzed by western blot. Tubulin and total KAP-1 were used as loading controls. As a positive control, MDA-MB-231 cells treated with 0.1 µM doxorubicin for 8 h (to generate DSBs) were used. Intervening lanes were removed as indicated (left panel). Chk1 and Chk2 phosphorylation in control and LOXL2 KD cells were analyzed by western blot. Chk1 phosphorylation was detected using the anti-P(S317) Chk1 antibody. For phosphorylated Chk2, a shift was detected using an anti-total Chk2 antibody. GADPH was used as a loading control. All samples were obtained under the same experimental conditions; in addition, positive control samples (irradiated fibroblasts) were run on the same gel as their corresponding control and LOXL2 KD samples. Intervening lanes in the Chk1/2 and GADPH blots were removed as indicated (right panel). c Cell cycle profile of control and LOXL2 KD cells at 0, 2.5, 5, or 7.5 h after release from a double-thymidine block. Cells were analyzed by fluorescence activated cell sorting (FACS) after propidium iodide staining. d H3S10 phosphorylation levels in control and LOXL2 KD MDA-MB-231 cells were analyzed by western blot. Tubulin was used as a loading control. e Upper panel, MTT assay in control and LOXL2 KD MDA-MB-231 cells; lower panel, colony-survival assay in control and LOXL2 KD MDA-MB-231 cells. γ-H2AX staining and foci quantification are shown by immunofluorescence with the indicated antibody after H1 (f) or SUV-39H1 reinfection (g). MDA-MB-231 cells were infected with control or LOXL2 KD lentivirus, selected with puromycin, and reinfected with GFP (MOCKGFP+), histone 1-GFP (H1GFP+) (f) or SUV-39H1-GFP (SUV-39GFP+) (g). Cells were fixed after 24 h. Dot graphs indicate the number of γ-H2AX foci per GFP-positive cell in each condition. Error bars indicate the SD from at least three independent experiments. **P < 0.01, ***P < 0.001, ****P < 0.0001
Table 2.
Genes upregulated in LOXL2 knockdown
| Locus | logFC | P-Value | FDR | Gene name |
|---|---|---|---|---|
| ENSG00000106541 | 2.557531982 | 0 | AGR2 | |
| ENSG00000185567 | 2.176849204 | 5.15E−299 | 2.25E−295 | AHNAK2 |
| ENSG00000132821 | 2.080483757 | 2.77E−280 | 9.09E−277 | VSTM2L |
| ENSG00000117984 | 1.586454466 | 3.61E−273 | 9.49E−270 | CTSD |
| ENSG00000039139 | 1.913971451 | 1.13E−234 | 1.49E−231 | DNAH5 |
| ENSG00000163083 | 1.827194971 | 3.38E−216 | 4.03E−213 | INHBB |
| ENSG00000184371 | 1.564908962 | 9.87E−215 | 1.08E−211 | CSF1 |
| ENSG00000170373 | 1.445267367 | 7.43E−214 | 7.51E−211 | CST1 |
| ENSG00000183018 | 2.687264202 | 2.37E−207 | 2.22E−204 | SPNS2 |
| ENSG00000148180 | 1.25021061 | 1.35E−204 | 1.18E−201 | GSN |
| ENSG00000187678 | 1.29153331 | 2.72E−191 | 2.23E−188 | SPRY4 |
| ENSG00000109062 | 1.468364079 | 5.45E−183 | 3.97E−180 | SLC9A3R1 |
| ENSG00000124766 | 1.897033792 | 1.36E−178 | 9.39E−176 | SOX4 |
| ENSG00000100994 | 1.081533166 | 2.15E−168 | 1.35E−165 | PYGB |
| ENSG00000181634 | 1.433165343 | 1.64E−154 | 8.26E−152 | TNFSF15 |
| ENSG00000147676 | 1.792602889 | 2.15E−153 | 1.05E−150 | MAL2 |
| ENSG00000107819 | 1.097217673 | 1.35E−152 | 6.33E−150 | SFXN3 |
| ENSG00000167644 | 1.245454774 | 2.18E−150 | 9.88E−148 | C19orf33 |
| ENSG00000172794 | 2.37902321 | 3.41E−147 | 1.49E−144 | RAB37 |
| ENSG00000058085 | 1.09085532 | 1.15E−145 | 4.88E−143 | LAMC2 |
| ENSG00000175130 | 1.074875932 | 2.20E−139 | 9.03E−137 | MARCKSL1 |
| ENSG00000171345 | 1.019257679 | 8.08E−137 | 3.12E−134 | KRT19 |
| ENSG00000167552 | 1.388799485 | 1.60E−136 | 5.82E−134 | TUBA1A |
| ENSG00000132470 | 1.437708599 | 1.07E−135 | 3.79E−133 | ITGB4 |
| ENSG00000008513 | 1.019331762 | 3.84E−133 | 1.29E−130 | ST3GAL1 |
| ENSG00000167642 | 0.963304201 | 1.10E−126 | 3.43E−124 | SPINT2 |
| ENSG00000196154 | 0.94711039 | 2.71E−126 | 8.28E−124 | S100A4 |
| ENSG00000161013 | 1.17382212 | 9.23E−123 | 2.64E−120 | MGAT4B |
| ENSG00000144136 | 0.959244783 | 4.64E−122 | 1.30E−119 | SLC20A1 |
| ENSG00000124145 | 0.935893618 | 1.39E−119 | 3.73E−117 | SDC4 |
| ENSG00000124664 | 1.067904809 | 1.15E−118 | 3.03E−116 | SPDEF |
| ENSG00000108639 | 0.963584447 | 1.29E−117 | 3.32E−115 | SYNGR2 |
| ENSG00000099812 | 2.361113786 | 2.95E−117 | 7.18E−115 | MISP |
| ENSG00000153292 | 1.189984144 | 2.35E−112 | 5.32E−110 | ADGRF1 |
| ENSG00000071054 | 0.884375284 | 1.55E−110 | 3.39E−108 | MAP4K4 |
| ENSG00000163683 | 1.090539229 | 6.92E−109 | 1.49E−106 | SMIM14 |
| ENSG00000069122 | 0.8914899 | 1.23E−107 | 2.48E−105 | ADGRF5 |
| ENSG00000163346 | 1.002881721 | 4.33E−107 | 8.61E−105 | PBXIP1 |
| ENSG00000168032 | 1.546762635 | 6.76E−107 | 1.32E−104 | ENTPD3 |
| ENSG00000010404 | 1.093704162 | 1.94E−106 | 3.74E−104 | IDS |
| ENSG00000026751 | 1.232149717 | 1.10E−100 | 1.95E−98 | SLAMF7 |
| ENSG00000149177 | 0.896255656 | 1.20E−98 | 2.04E−96 | PTPRJ |
| ENSG00000169213 | 2.790765942 | 6.07E−98 | 1.02E−95 | RAB3B |
| ENSG00000148344 | 1.010954162 | 2.71E−97 | 4.51E−95 | PTGES |
| ENSG00000167232 | 1.137723821 | 6.17E−97 | 9.99E−95 | ZNF91 |
| ENSG00000162337 | 0.931691275 | 4.04E−96 | 6.40E−94 | LRP5 |
| ENSG00000013364 | 0.847555049 | 1.19E−94 | 1.87E−92 | MVP |
| ENSG00000143061 | 1.006284582 | 2.26E−94 | 3.49E−92 | IGSF3 |
| ENSG00000119630 | 1.384163447 | 1.38E−93 | 2.11E−91 | PGF |
| ENSG00000138678 | 0.902786259 | 1.84E−90 | 2.65E−88 | GPAT3 |
| ENSG00000125731 | 1.560012238 | 2.27E−90 | 3.24E−88 | SH2D3A |
| ENSG00000101441 | 0.900355704 | 3.67E−90 | 5.18E−88 | CST4 |
| ENSG00000174804 | 1.138994169 | 1.66E−89 | 2.32E−87 | FZD4 |
| ENSG00000135480 | 0.885290783 | 1.96E−89 | 2.71E−87 | KRT7 |
| ENSG00000114554 | 0.782941224 | 6.05E−89 | 8.19E−87 | PLXNA1 |
| ENSG00000131981 | 0.825525944 | 3.76E−88 | 5.04E−86 | LGALS3 |
| ENSG00000205542 | 0.843455136 | 7.86E−87 | 1.02E−84 | TMSB4X |
| ENSG00000118898 | 2.469371901 | 1.48E−86 | 1.91E−84 | PPL |
| ENSG00000114739 | 0.98377701 | 1.49E−86 | 1.91E−84 | ACVR2B |
| ENSG00000172296 | 0.928175781 | 1.83E−86 | 2.31E−84 | SPTLC3 |
| ENSG00000173801 | 0.93691533 | 2.54E−86 | 3.18E−84 | JUP |
| ENSG00000136205 | 0.893027849 | 5.42E−85 | 6.59E−83 | TNS3 |
| ENSG00000078246 | 1.095298427 | 2.01E−84 | 2.40E−82 | TULP3 |
| ENSG00000165757 | 1.886253371 | 7.17E−84 | 8.48E−82 | KIAA1462 |
| ENSG00000158109 | 0.91823575 | 1.48E−82 | 1.69E−80 | TPRG1L |
| ENSG00000115295 | 0.781412289 | 7.32E−82 | 8.29E−80 | CLIP4 |
| ENSG00000151651 | 0.773631649 | 1.07E−81 | 1.20E−79 | ADAM8 |
| ENSG00000183087 | 0.797199229 | 1.18E−80 | 1.32E−78 | GAS6 |
| ENSG00000134531 | 0.763620701 | 1.89E−80 | 2.08E−78 | EMP1 |
| ENSG00000071242 | 1.290957558 | 2.94E−80 | 3.20E−78 | RPS6KA2 |
| ENSG00000074527 | 1.161231478 | 5.89E−80 | 6.29E−78 | NTN4 |
| ENSG00000122359 | 0.732215153 | 8.78E−78 | 9.15E−76 | ANXA11 |
| ENSG00000116478 | 0.727866925 | 1.16E−77 | 1.19E−75 | HDAC1 |
| ENSG00000065613 | 0.732219022 | 1.56E−77 | 1.58E−75 | SLK |
| ENSG00000108846 | 1.870849664 | 1.05E−76 | 1.06E−74 | ABCC3 |
| ENSG00000196975 | 0.868874265 | 5.81E−76 | 5.73E−74 | ANXA4 |
| ENSG00000165476 | 0.732395071 | 1.31E−75 | 1.27E−73 | REEP3 |
| ENSG00000147251 | 0.950209054 | 2.84E−74 | 2.68E−72 | DOCK11 |
| ENSG00000104081 | 1.619563862 | 1.16E−73 | 1.09E−71 | BMF |
| ENSG00000115107 | 0.833533503 | 1.66E−73 | 1.55E−71 | STEAP3 |
| ENSG00000146072 | 0.740689199 | 2.97E−73 | 2.71E−71 | TNFRSF21 |
| ENSG00000118640 | 0.8568247 | 7.92E−73 | 7.18E−71 | VAMP8 |
| ENSG00000198756 | 1.324358789 | 1.14E−72 | 1.02E−70 | COLGALT2 |
| ENSG00000161714 | 0.795194647 | 1.36E−72 | 1.22E−70 | PLCD3 |
| ENSG00000178726 | 0.836487297 | 3.26E−72 | 2.88E−70 | THBD |
| ENSG00000160271 | 0.831966049 | 3.75E−72 | 3.29E−70 | RALGDS |
| ENSG00000102271 | 0.886072448 | 7.99E−72 | 6.95E−70 | KLHL4 |
| ENSG00000112367 | 0.989558706 | 2.33E−71 | 2.01E−69 | FIG4 |
| ENSG00000073849 | 1.156064112 | 7.50E−71 | 6.35E−69 | ST6GAL1 |
| ENSG00000001461 | 0.759211956 | 2.61E−70 | 2.19E−68 | NIPAL3 |
| ENSG00000105778 | 0.716804361 | 7.67E−70 | 6.41E−68 | AVL9 |
| ENSG00000180861 | 2.544226107 | 5.43E−26 | 8.20E−25 | LINC01559 |
| ENSG00000138764 | 0.853001536 | 4.67E−69 | 3.76E−67 | CCNG2 |
| ENSG00000168461 | 0.739918812 | 6.17E−69 | 4.94E−67 | RAB31 |
| ENSG00000165474 | 1.287094496 | 7.37E−69 | 5.87E−67 | GJB2 |
| ENSG00000117472 | 1.402524019 | 3.06E−68 | 2.41E−66 | TSPAN1 |
| ENSG00000124496 | 0.733809105 | 7.91E−68 | 6.04E−66 | TRERF1 |
| ENSG00000171877 | 0.944409394 | 3.72E−67 | 2.73E−65 | FRMD5 |
| ENSG00000126759 | 1.579268489 | 7.91E−67 | 5.74E−65 | CFP |
| ENSG00000124882 | 1.027808793 | 1.06E−66 | 7.64E−65 | EREG |
| ENSG00000102316 | 0.67559959 | 1.29E−66 | 9.23E−65 | MAGED2 |
| ENSG00000075711 | 0.664598245 | 4.29E−66 | 2.99E−64 | DLG1 |
| ENSG00000163435 | 2.261552237 | 1.24E−64 | 8.43E−63 | ELF3 |
| ENSG00000135048 | 0.842294959 | 2.39E−64 | 1.62E−62 | TMEM2 |
| ENSG00000141279 | 0.798544903 | 3.43E−64 | 2.31E−62 | NPEPPS |
| ENSG00000124491 | 1.928383505 | 3.87E−64 | 2.58E−62 | F13A1 |
| ENSG00000136068 | 0.799841098 | 1.04E−63 | 6.88E−62 | FLNB |
| ENSG00000170786 | 1.469743357 | 2.22E−63 | 1.45E−61 | SDR16C5 |
| ENSG00000069702 | 0.968766875 | 2.52E−63 | 1.64E−61 | TGFBR3 |
| ENSG00000069956 | 0.687274901 | 3.83E−63 | 2.48E−61 | MAPK6 |
| ENSG00000183826 | 0.802508399 | 7.36E−63 | 4.72E−61 | BTBD9 |
| ENSG00000137449 | 0.953988726 | 9.13E−63 | 5.82E−61 | CPEB2 |
| ENSG00000160789 | 0.769443927 | 1.58E−62 | 9.90E−61 | LMNA |
| ENSG00000115756 | 0.635371525 | 1.24E−60 | 7.58E−59 | HPCAL1 |
| ENSG00000169439 | 0.774541548 | 2.36E−60 | 1.42E−58 | SDC2 |
| ENSG00000184564 | 2.470980644 | 2.81E−60 | 1.68E−58 | SLITRK6 |
| ENSG00000143816 | 1.634586486 | 5.68E−60 | 3.36E−58 | WNT9A |
| ENSG00000182093 | 0.87376722 | 7.53E−60 | 4.44E−58 | WRB |
| ENSG00000065054 | 0.668246405 | 9.12E−60 | 5.32E−58 | SLC9A3R2 |
| ENSG00000116701 | 1.117244095 | 1.12E−59 | 6.48E−58 | NCF2 |
| ENSG00000100439 | 0.801911634 | 1.37E−59 | 7.90E−58 | ABHD4 |
| ENSG00000101439 | 0.771824808 | 1.52E−59 | 8.73E−58 | CST3 |
| ENSG00000167779 | 0.981583691 | 1.61E−59 | 9.18E−58 | IGFBP6 |
| ENSG00000075618 | 0.742779504 | 1.63E−59 | 9.25E−58 | FSCN1 |
| ENSG00000178585 | 0.715678973 | 1.83E−59 | 1.03E−57 | CTNNBIP1 |
| ENSG00000088836 | 1.218822163 | 4.54E−59 | 2.53E−57 | SLC4A11 |
| ENSG00000006118 | 0.63601116 | 5.56E−59 | 3.08E−57 | TMEM132A |
| ENSG00000140950 | 0.734048999 | 9.06E−59 | 4.98E−57 | TLDC1 |
| ENSG00000114353 | 0.624135935 | 1.44E−58 | 7.86E−57 | GNAI2 |
| ENSG00000189143 | 0.778650604 | 3.07E−58 | 1.65E−56 | CLDN4 |
| ENSG00000166925 | 0.750896396 | 4.93E−58 | 2.63E−56 | TSC22D4 |
| ENSG00000169991 | 0.777045163 | 5.47E−58 | 2.90E−56 | IFFO2 |
| ENSG00000198910 | 0.697815856 | 1.08E−57 | 5.67E−56 | L1CAM |
| ENSG00000142627 | 0.64537199 | 8.67E−57 | 4.47E−55 | EPHA2 |
| ENSG00000119899 | 0.702317857 | 1.04E−56 | 5.35E−55 | SLC17A5 |
| ENSG00000111846 | 0.636366397 | 1.06E−56 | 5.42E−55 | GCNT2 |
| ENSG00000100234 | 0.679728149 | 2.30E−56 | 1.15E−54 | TIMP3 |
| ENSG00000095203 | 0.700479323 | 3.83E−56 | 1.91E−54 | EPB41L4B |
| ENSG00000151276 | 0.736006307 | 6.05E−56 | 3.00E−54 | MAGI1 |
| ENSG00000171310 | 0.607237912 | 1.24E−55 | 6.09E−54 | CHST11 |
| ENSG00000165434 | 0.816777806 | 2.01E−55 | 9.82E−54 | PGM2L1 |
| ENSG00000154930 | 1.233947636 | 5.25E−55 | 2.55E−53 | ACSS1 |
| ENSG00000116285 | 0.720021425 | 7.57E−55 | 3.67E−53 | ERRFI1 |
| ENSG00000106799 | 0.601363577 | 1.04E−54 | 5.00E−53 | TGFBR1 |
| ENSG00000100055 | 1.153795642 | 1.14E−54 | 5.50E−53 | CYTH4 |
| ENSG00000204634 | 0.78091263 | 1.43E−54 | 6.80E−53 | TBC1D8 |
| ENSG00000187764 | 0.934447405 | 3.48E−54 | 1.64E−52 | SEMA4D |
| ENSG00000196611 | 0.697333766 | 6.80E−54 | 3.18E−52 | MMP1 |
| ENSG00000072210 | 0.677662089 | 7.95E−54 | 3.70E−52 | ALDH3A2 |
| ENSG00000166750 | 0.934437232 | 1.24E−53 | 5.75E−52 | SLFN5 |
| ENSG00000160213 | 0.660113084 | 2.58E−53 | 1.19E−51 | CSTB |
| ENSG00000154175 | 1.422923707 | 3.12E−53 | 1.43E−51 | ABI3BP |
| ENSG00000105048 | 0.648949367 | 3.73E−53 | 1.71E−51 | TNNT1 |
| ENSG00000029534 | 0.948107565 | 3.84E−53 | 1.75E−51 | ANK1 |
| ENSG00000163874 | 0.734961316 | 1.42E−52 | 6.44E−51 | ZC3H12A |
| ENSG00000003249 | 1.44442704 | 3.21E−52 | 1.44E−50 | DBNDD1 |
| ENSG00000059804 | 0.834124252 | 4.53E−52 | 2.00E−50 | SLC2A3 |
| ENSG00000120337 | 1.718937071 | 4.63E−52 | 2.04E−50 | TNFSF18 |
| ENSG00000182022 | 0.664899083 | 5.48E−52 | 2.41E−50 | CHST15 |
| ENSG00000188191 | 0.90765776 | 6.61E−52 | 2.88E−50 | PRKAR1B |
| ENSG00000003147 | 1.021914278 | 1.02E−51 | 4.40E−50 | ICA1 |
| ENSG00000018408 | 0.61449093 | 1.18E−51 | 5.09E−50 | WWTR1 |
| ENSG00000188643 | 0.612623576 | 1.72E−51 | 7.37E−50 | S100A16 |
| ENSG00000106070 | 0.622574635 | 2.67E−51 | 1.14E−49 | GRB10 |
| ENSG00000128342 | 0.665706893 | 3.64E−51 | 1.55E−49 | LIF |
| ENSG00000091409 | 0.586574997 | 1.51E−50 | 6.36E−49 | ITGA6 |
| ENSG00000130477 | 1.980813997 | 2.31E−50 | 9.70E−49 | UNC13A |
| ENSG00000059377 | 1.224196279 | 4.24E−50 | 1.76E−48 | TBXAS1 |
| ENSG00000167106 | 0.797725209 | 5.46E−50 | 2.26E−48 | FAM102A |
| ENSG00000011009 | 0.708291693 | 5.65E−50 | 2.33E−48 | LYPLA2 |
| ENSG00000188042 | 0.587843927 | 1.01E−49 | 4.16E−48 | ARL4C |
| ENSG00000106258 | 3.99646604 | 1.07E−49 | 4.39E−48 | CYP3A5 |
| ENSG00000253719 | 0.641918884 | 1.98E−49 | 7.95E−48 | ATXN7L3B |
| ENSG00000198561 | 0.6435595 | 2.03E−49 | 8.11E−48 | CTNND1 |
| ENSG00000196642 | 0.629620891 | 2.44E−49 | 9.74E−48 | RABL6 |
| ENSG00000162909 | 0.627093674 | 4.17E−49 | 1.66E−47 | CAPN2 |
| ENSG00000166046 | 1.014081966 | 1.05E−48 | 4.14E−47 | TCP11L2 |
| ENSG00000106537 | 1.142647315 | 1.06E−48 | 4.16E−47 | TSPAN13 |
| ENSG00000153179 | 0.654407199 | 1.21E−48 | 4.77E−47 | RASSF3 |
| ENSG00000197442 | 0.704516379 | 1.30E−48 | 5.09E−47 | MAP3K5 |
| ENSG00000138119 | 0.641567321 | 6.81E−48 | 2.62E−46 | MYOF |
| ENSG00000185483 | 0.88479338 | 9.18E−48 | 3.51E−46 | ROR1 |
| ENSG00000184363 | 0.800404785 | 1.32E−47 | 5.02E−46 | PKP3 |
| ENSG00000008083 | 0.690696827 | 1.62E−47 | 6.16E−46 | JARID2 |
| ENSG00000179913 | 1.195221395 | 1.63E−47 | 6.18E−46 | B3GNT3 |
| ENSG00000185127 | 0.709916588 | 1.79E−47 | 6.75E−46 | C6orf120 |
| ENSG00000177106 | 0.686183661 | 4.22E−47 | 1.58E−45 | EPS8L2 |
| ENSG00000188015 | 0.884405539 | 4.58E−47 | 1.71E−45 | S100A3 |
| ENSG00000147400 | 0.694830612 | 1.06E−46 | 3.90E−45 | CETN2 |
| ENSG00000143786 | 0.939975104 | 2.19E−46 | 8.05E−45 | CNIH3 |
| ENSG00000133805 | 0.873822055 | 7.29E−46 | 2.63E−44 | AMPD3 |
| ENSG00000106780 | 0.689300805 | 7.57E−46 | 2.72E−44 | MEGF9 |
| ENSG00000272398 | 1.043194872 | 2.99E−45 | 1.06E−43 | CD24 |
| ENSG00000099998 | 1.065481895 | 6.33E−45 | 2.23E−43 | GGT5 |
| ENSG00000095383 | 0.635665876 | 1.03E−44 | 3.62E−43 | TBC1D2 |
| ENSG00000109436 | 0.653570925 | 1.05E−44 | 3.65E−43 | TBC1D9 |
| ENSG00000198624 | 0.606978398 | 4.62E−44 | 1.58E−42 | CCDC69 |
| ENSG00000153774 | 0.642071127 | 5.74E−44 | 1.96E−42 | CFDP1 |
| ENSG00000130958 | 0.846711873 | 6.23E−44 | 2.11E−42 | SLC35D2 |
| ENSG00000164938 | 0.883485618 | 8.44E−44 | 2.86E−42 | TP53INP1 |
| ENSG00000163947 | 0.753394922 | 9.40E−44 | 3.17E−42 | ARHGEF3 |
| ENSG00000182809 | 1.082621723 | 2.23E−43 | 7.43E−42 | CRIP2 |
| ENSG00000101842 | 2.125670418 | 3.17E−43 | 1.05E−41 | VSIG1 |
| ENSG00000010278 | 0.692915131 | 3.75E−43 | 1.24E−41 | CD9 |
| ENSG00000141753 | 0.652587531 | 4.03E−43 | 1.33E−41 | IGFBP4 |
| ENSG00000071575 | 0.809537681 | 4.69E−43 | 1.53E−41 | TRIB2 |
| ENSG00000041353 | 0.600506668 | 6.85E−43 | 2.23E−41 | RAB27B |
| ENSG00000124942 | 0.878776788 | 8.14E−43 | 2.64E−41 | AHNAK |
| ENSG00000130702 | 0.830408106 | 1.66E−42 | 5.34E−41 | LAMA5 |
| ENSG00000106605 | 0.707072414 | 1.69E−42 | 5.40E−41 | BLVRA |
| ENSG00000160211 | 0.671328183 | 1.71E−42 | 5.47E−41 | G6PD |
| ENSG00000153395 | 0.667145468 | 1.73E−42 | 5.50E−41 | LPCAT1 |
| ENSG00000183778 | 1.193744584 | 3.32E−42 | 1.05E−40 | B3GALT5 |
| ENSG00000131389 | 0.90704002 | 2.13E−41 | 6.66E−40 | SLC6A6 |
| ENSG00000184117 | 0.709961941 | 2.66E−41 | 8.24E−40 | NIPSNAP1 |
| ENSG00000258947 | 1.044442933 | 2.99E−41 | 9.27E−40 | TUBB3 |
| ENSG00000180667 | 0.586841506 | 3.12E−41 | 9.64E−40 | YOD1 |
| ENSG00000157227 | 0.607608231 | 3.40E−41 | 1.05E−39 | MMP14 |
| ENSG00000184557 | 0.61191017 | 1.37E−40 | 4.18E−39 | SOCS3 |
| ENSG00000100106 | 0.606368936 | 2.00E−40 | 6.06E−39 | TRIOBP |
| ENSG00000165233 | 0.647236635 | 2.07E−40 | 6.24E−39 | CARD19 |
| ENSG00000126458 | 0.806490749 | 2.10E−40 | 6.33E−39 | RRAS |
| ENSG00000152217 | 1.432604587 | 3.42E−40 | 1.01E−38 | SETBP1 |
| ENSG00000157796 | 0.64712781 | 4.11E−40 | 1.22E−38 | WDR19 |
| ENSG00000118960 | 0.794746258 | 4.94E−40 | 1.46E−38 | HS1BP3 |
| ENSG00000078269 | 0.598064657 | 1.32E−39 | 3.86E−38 | SYNJ2 |
| ENSG00000133138 | 1.209003045 | 1.35E−39 | 3.94E−38 | TBC1D8B |
| ENSG00000088387 | 0.585401778 | 1.40E−39 | 4.07E−38 | DOCK9 |
| ENSG00000251191 | 2.434425725 | 2.57E−38 | 6.95E−37 | LINC00589 |
| ENSG00000203697 | 4.744840946 | 1.53E−39 | 4.43E−38 | CAPN8 |
| ENSG00000158769 | 0.653902818 | 1.73E−39 | 5.00E−38 | F11R |
| ENSG00000062282 | 0.689520114 | 2.55E−39 | 7.25E−38 | DGAT2 |
| ENSG00000184497 | 0.978103665 | 2.65E−39 | 7.52E−38 | TMEM255B |
| ENSG00000149948 | 0.723373492 | 2.80E−39 | 7.93E−38 | HMGA2 |
| ENSG00000148730 | 0.588607271 | 3.00E−39 | 8.47E−38 | EIF4EBP2 |
| ENSG00000117983 | 2.62243558 | 9.23E−39 | 2.56E−37 | MUC5B |
| ENSG00000105514 | 0.898143653 | 1.15E−38 | 3.16E−37 | RAB3D |
| ENSG00000169432 | 1.168069383 | 1.36E−38 | 3.72E−37 | SCN9A |
| ENSG00000006534 | 0.83694272 | 1.56E−38 | 4.27E−37 | ALDH3B1 |
| ENSG00000074219 | 0.725674533 | 1.92E−38 | 5.24E−37 | TEAD2 |
| ENSG00000255471 | 1.5585602 | 4.89E−14 | 3.52E−13 | AP001528.2 |
| ENSG00000005238 | 0.737875746 | 2.60E−38 | 7.02E−37 | FAM214B |
| ENSG00000205426 | 0.859387072 | 6.01E−38 | 1.59E−36 | KRT81 |
| ENSG00000258088 | 1.553544167 | 6.65E−19 | 6.78E−18 | AC078820.1 |
| ENSG00000138162 | 0.62551312 | 7.91E−38 | 2.08E−36 | TACC2 |
| ENSG00000141524 | 0.709114969 | 8.26E−38 | 2.17E−36 | TMC6 |
| ENSG00000170271 | 2.264074711 | 8.41E−38 | 2.20E−36 | FAXDC2 |
| ENSG00000136720 | 0.700335625 | 8.54E−38 | 2.23E−36 | HS6ST1 |
| ENSG00000162591 | 1.561892509 | 9.75E−38 | 2.53E−36 | MEGF6 |
| ENSG00000173334 | 0.641809108 | 1.16E−37 | 3.02E−36 | TRIB1 |
| ENSG00000110871 | 0.728862751 | 2.35E−37 | 6.03E−36 | COQ5 |
| ENSG00000150471 | 1.085455968 | 2.61E−37 | 6.68E−36 | ADGRL3 |
| ENSG00000133985 | 1.071387544 | 2.67E−37 | 6.82E−36 | TTC9 |
| ENSG00000165806 | 0.615818489 | 3.03E−37 | 7.70E−36 | CASP7 |
| ENSG00000159733 | 0.690251958 | 3.10E−37 | 7.86E−36 | ZFYVE28 |
| ENSG00000125798 | 0.586210217 | 3.58E−37 | 9.07E−36 | FOXA2 |
| ENSG00000146278 | 0.580483004 | 5.25E−37 | 1.32E−35 | PNRC1 |
| ENSG00000131746 | 2.387958603 | 3.96E−36 | 9.78E−35 | TNS4 |
| ENSG00000137767 | 0.667955094 | 5.74E−36 | 1.40E−34 | SQOR |
| ENSG00000054277 | 1.0616567 | 6.32E−36 | 1.54E−34 | OPN3 |
| ENSG00000116016 | 0.598281377 | 1.05E−35 | 2.53E−34 | EPAS1 |
| ENSG00000151690 | 0.658340738 | 1.15E−35 | 2.77E−34 | MFSD6 |
| ENSG00000168487 | 0.664252835 | 1.26E−35 | 3.02E−34 | BMP1 |
| ENSG00000144642 | 0.724358105 | 2.62E−35 | 6.20E−34 | RBMS3 |
| ENSG00000146386 | 0.582324614 | 3.06E−35 | 7.20E−34 | ABRACL |
| ENSG00000109321 | 0.85796308 | 6.09E−35 | 1.42E−33 | AREG |
| ENSG00000185033 | 0.613573094 | 7.11E−35 | 1.65E−33 | SEMA4B |
| ENSG00000124143 | 1.002818698 | 1.31E−34 | 3.02E−33 | ARHGAP40 |
| ENSG00000188766 | 1.007070905 | 1.37E−34 | 3.15E−33 | SPRED3 |
| ENSG00000143198 | 0.665742779 | 1.45E−34 | 3.31E−33 | MGST3 |
| ENSG00000138772 | 0.618561607 | 1.92E−34 | 4.38E−33 | ANXA3 |
| ENSG00000153208 | 0.65916003 | 2.75E−34 | 6.24E−33 | MERTK |
| ENSG00000105379 | 0.650414245 | 3.72E−34 | 8.40E−33 | ETFB |
| ENSG00000174600 | 1.152413998 | 4.99E−34 | 1.12E−32 | CMKLR1 |
| ENSG00000143878 | 0.677873378 | 6.11E−34 | 1.36E−32 | RHOB |
| ENSG00000129636 | 0.586007283 | 7.21E−34 | 1.60E−32 | ITFG1 |
| ENSG00000196358 | 1.157286665 | 7.31E−34 | 1.62E−32 | NTNG2 |
| ENSG00000074855 | 0.727005968 | 1.29E−33 | 2.85E−32 | ANO8 |
| ENSG00000179869 | 1.478231849 | 1.76E−33 | 3.84E−32 | ABCA13 |
| ENSG00000164379 | 0.688194901 | 2.27E−33 | 4.92E−32 | FOXQ1 |
| ENSG00000082458 | 0.603787281 | 4.54E−33 | 9.68E−32 | DLG3 |
| ENSG00000186106 | 0.614744197 | 1.44E−32 | 3.01E−31 | ANKRD46 |
| ENSG00000127863 | 0.807228706 | 1.48E−32 | 3.09E−31 | TNFRSF19 |
| ENSG00000007384 | 0.675083802 | 1.79E−32 | 3.71E−31 | RHBDF1 |
| ENSG00000175938 | 0.991028089 | 1.90E−32 | 3.92E−31 | ORAI3 |
| ENSG00000258245 | 1.54291117 | 1.93E−09 | 9.27E−09 | RPL10P13 |
| ENSG00000032742 | 0.85419356 | 4.05E−32 | 8.32E−31 | IFT88 |
| ENSG00000229512 | 1.425346349 | 8.83E−13 | 5.81E−12 | AC068580.1 |
| ENSG00000187164 | 0.600256967 | 5.52E−32 | 1.12E−30 | SHTN1 |
| ENSG00000154027 | 1.341272323 | 9.88E−32 | 1.99E−30 | AK5 |
| ENSG00000101846 | 0.734435125 | 1.11E−31 | 2.23E−30 | STS |
| ENSG00000206190 | 0.787741284 | 1.70E−31 | 3.36E−30 | ATP10A |
| ENSG00000160255 | 0.651457256 | 2.24E−31 | 4.40E−30 | ITGB2 |
| ENSG00000101213 | 0.990447452 | 4.59E−31 | 8.91E−30 | PTK6 |
| ENSG00000198286 | 0.883579653 | 5.70E−31 | 1.10E−29 | CARD11 |
| ENSG00000106066 | 0.619864757 | 7.17E−31 | 1.38E−29 | CPVL |
| ENSG00000115919 | 0.677292783 | 8.14E−31 | 1.56E−29 | KYNU |
| ENSG00000064787 | 4.909061593 | 9.51E−31 | 1.81E−29 | BCAS1 |
| ENSG00000144218 | 0.616205371 | 1.12E−30 | 2.13E−29 | AFF3 |
| ENSG00000123095 | 0.933963389 | 1.93E−30 | 3.62E−29 | BHLHE41 |
| ENSG00000148671 | 1.02789778 | 2.52E−30 | 4.69E−29 | ADIRF |
| ENSG00000258077 | 1.387892381 | 1.19E−21 | 1.42E−20 | AC078923.1 |
| ENSG00000125534 | 0.590391835 | 7.90E−30 | 1.44E−28 | PPDPF |
| ENSG00000137501 | 0.668063928 | 1.04E−29 | 1.88E−28 | SYTL2 |
| ENSG00000106351 | 0.728533997 | 1.04E−29 | 1.88E−28 | AGFG2 |
| ENSG00000185585 | 0.755238419 | 1.08E−29 | 1.95E−28 | OLFML2A |
| ENSG00000178951 | 0.587643267 | 1.31E−29 | 2.35E−28 | ZBTB7A |
| ENSG00000111110 | 0.952754467 | 1.54E−29 | 2.76E−28 | PPM1H |
| ENSG00000047056 | 0.582933075 | 2.95E−29 | 5.19E−28 | WDR37 |
| ENSG00000227039 | 1.384680791 | 6.64E−17 | 5.90E−16 | ITGB2-AS1 |
| ENSG00000102096 | 0.607774377 | 7.04E−29 | 1.21E−27 | PIM2 |
| ENSG00000158106 | 0.851822142 | 1.01E−28 | 1.74E−27 | RHPN1 |
| ENSG00000133943 | 0.621897984 | 1.44E−28 | 2.45E−27 | C14orf159 |
| ENSG00000132274 | 0.927124554 | 1.44E−28 | 2.46E−27 | TRIM22 |
| ENSG00000167565 | 0.59119894 | 2.89E−28 | 4.87E−27 | SERTAD3 |
| ENSG00000146416 | 0.722563478 | 9.98E−28 | 1.64E−26 | AIG1 |
| ENSG00000102934 | 1.156498397 | 3.17E−27 | 5.08E−26 | PLLP |
| ENSG00000157613 | 0.66005375 | 9.58E−27 | 1.50E−25 | CREB3L1 |
| ENSG00000139636 | 0.759504331 | 1.05E−26 | 1.64E−25 | LMBR1L |
| ENSG00000124313 | 0.956039086 | 1.06E−26 | 1.65E−25 | IQSEC2 |
| ENSG00000167778 | 0.584757438 | 1.37E−26 | 2.12E−25 | SPRYD3 |
| ENSG00000117305 | 0.692063114 | 2.15E−26 | 3.30E−25 | HMGCL |
| ENSG00000177854 | 1.052435249 | 3.25E−26 | 4.95E−25 | TMEM187 |
| ENSG00000175505 | 0.659053782 | 3.46E−26 | 5.27E−25 | CLCF1 |
| ENSG00000135596 | 0.588183448 | 4.27E−26 | 6.47E−25 | MICAL1 |
| ENSG00000259974 | 1.265560114 | 1.32E−08 | 5.80E−08 | LINC00261 |
| ENSG00000039068 | 1.784191229 | 5.61E−26 | 8.46E−25 | CDH1 |
| ENSG00000139044 | 2.17079755 | 6.20E−26 | 9.34E−25 | B4GALNT3 |
| ENSG00000160446 | 0.580686241 | 8.53E−26 | 1.28E−24 | ZDHHC12 |
| ENSG00000112679 | 0.651511201 | 1.02E−25 | 1.53E−24 | DUSP22 |
| ENSG00000158292 | 0.659371473 | 1.14E−25 | 1.69E−24 | GPR153 |
| ENSG00000167470 | 0.746743002 | 1.27E−25 | 1.88E−24 | MIDN |
| ENSG00000100246 | 0.975110479 | 4.43E−25 | 6.45E−24 | DNAL4 |
| ENSG00000116117 | 0.702904005 | 5.86E−25 | 8.50E−24 | PARD3B |
| ENSG00000100290 | 1.23971931 | 7.52E−25 | 1.08E−23 | BIK |
| ENSG00000165949 | 0.802081907 | 1.33E−24 | 1.89E−23 | IFI27 |
| ENSG00000169083 | 0.656064782 | 1.91E−24 | 2.68E−23 | AR |
| ENSG00000109610 | 0.77544685 | 2.24E−24 | 3.13E−23 | SOD3 |
| ENSG00000140931 | 0.704129083 | 2.81E−24 | 3.90E−23 | CMTM3 |
| ENSG00000152128 | 0.697760811 | 3.49E−24 | 4.83E−23 | TMEM163 |
| ENSG00000124406 | 0.610126555 | 4.99E−24 | 6.83E−23 | ATP8A1 |
| ENSG00000106069 | 1.002738235 | 6.13E−24 | 8.36E−23 | CHN2 |
| ENSG00000144455 | 0.66426688 | 6.74E−24 | 9.15E−23 | SUMF1 |
| ENSG00000163898 | 0.637729579 | 7.68E−24 | 1.04E−22 | LIPH |
| ENSG00000120306 | 0.686361467 | 7.96E−24 | 1.07E−22 | CYSTM1 |
| ENSG00000171992 | 0.821352578 | 8.82E−24 | 1.19E−22 | SYNPO |
| ENSG00000275342 | 0.707225979 | 8.83E−24 | 1.19E−22 | PRAG1 |
| ENSG00000140022 | 1.275607654 | 1.75E−23 | 2.33E−22 | STON2 |
| ENSG00000006453 | 0.608949037 | 3.37E−23 | 4.41E−22 | BAIAP2L1 |
| ENSG00000102547 | 0.7411813 | 3.62E−23 | 4.72E−22 | CAB39L |
| ENSG00000162105 | 1.018126629 | 3.82E−23 | 4.98E−22 | SHANK2 |
| ENSG00000150782 | 0.667724476 | 4.97E−23 | 6.41E−22 | IL18 |
| ENSG00000063660 | 0.62014441 | 5.38E−23 | 6.92E−22 | GPC1 |
| ENSG00000162869 | 0.633902575 | 6.97E−23 | 8.91E−22 | PPP1R21 |
| ENSG00000065361 | 0.802225109 | 7.32E−23 | 9.35E−22 | ERBB3 |
| ENSG00000081189 | 0.666188328 | 7.32E−23 | 9.35E−22 | MEF2C |
| ENSG00000122176 | 1.592831251 | 2.05E−22 | 2.55E−21 | FMOD |
| ENSG00000069812 | 1.132970411 | 2.34E−22 | 2.90E−21 | HES2 |
| ENSG00000146530 | 0.946186656 | 2.74E−22 | 3.39E−21 | VWDE |
| ENSG00000249279 | 1.258589847 | 3.01E−07 | 1.13E−06 | LINC02057 |
| ENSG00000151715 | 1.313755414 | 4.48E−22 | 5.44E−21 | TMEM45B |
| ENSG00000100031 | 0.782239579 | 6.38E−22 | 7.72E−21 | GGT1 |
| ENSG00000225339 | 1.250563168 | 1.16E−21 | 1.38E−20 | AL354740.1 |
| ENSG00000230439 | 1.230428653 | 3.86E−30 | 7.17E−29 | AL512488.1 |
| ENSG00000105429 | 0.589326615 | 1.81E−21 | 2.15E−20 | MEGF8 |
| ENSG00000143344 | 0.598376949 | 1.82E−21 | 2.16E−20 | RGL1 |
| ENSG00000114805 | 0.907966339 | 2.84E−21 | 3.32E−20 | PLCH1 |
| ENSG00000105856 | 0.586467521 | 3.20E−21 | 3.71E−20 | HBP1 |
| ENSG00000188153 | 0.668030888 | 3.63E−21 | 4.19E−20 | COL4A5 |
| ENSG00000198513 | 0.787106569 | 3.85E−21 | 4.44E−20 | ATL1 |
| ENSG00000113721 | 1.215644215 | 4.56E−21 | 5.25E−20 | PDGFRB |
| ENSG00000183828 | 0.751113819 | 4.82E−21 | 5.54E−20 | NUDT14 |
| ENSG00000181722 | 0.807166236 | 5.79E−21 | 6.63E−20 | ZBTB20 |
| ENSG00000164976 | 0.825285155 | 6.78E−21 | 7.69E−20 | KIAA1161 |
| ENSG00000148737 | 0.823410224 | 7.52E−21 | 8.51E−20 | TCF7L2 |
| ENSG00000167861 | 0.647865569 | 8.61E−21 | 9.72E−20 | HID1 |
| ENSG00000188747 | 1.012209238 | 1.04E−20 | 1.17E−19 | NOXA1 |
| ENSG00000159314 | 0.882984133 | 1.11E−20 | 1.25E−19 | ARHGAP27 |
| ENSG00000089723 | 0.80542898 | 1.35E−20 | 1.51E−19 | OTUB2 |
| ENSG00000088899 | 0.649788574 | 1.38E−20 | 1.55E−19 | LZTS3 |
| ENSG00000154864 | 0.783916945 | 2.24E−20 | 2.49E−19 | PIEZO2 |
| ENSG00000122515 | 0.582145115 | 2.89E−20 | 3.19E−19 | ZMIZ2 |
| ENSG00000260461 | 1.216719543 | 3.67E−09 | 1.72E−08 | AL133355.1 |
| ENSG00000167105 | 0.61082311 | 3.72E−20 | 4.07E−19 | TMEM92 |
| ENSG00000198429 | 0.697252261 | 3.90E−20 | 4.26E−19 | ZNF69 |
| ENSG00000102452 | 0.765799284 | 4.92E−20 | 5.35E−19 | NALCN |
| ENSG00000108375 | 1.363173418 | 7.55E−20 | 8.14E−19 | RNF43 |
| ENSG00000218014 | 1.189647936 | 1.64E−08 | 7.14E−08 | KRT19P1 |
| ENSG00000133056 | 1.110173291 | 1.42E−19 | 1.51E−18 | PIK3C2B |
| ENSG00000142347 | 1.005039505 | 1.63E−19 | 1.74E−18 | MYO1F |
| ENSG00000267284 | 1.148059389 | 1.95E−09 | 9.39E−09 | AC022031.2 |
| ENSG00000119943 | 0.668952392 | 1.86E−19 | 1.96E−18 | PYROXD2 |
| ENSG00000106003 | 0.591285678 | 1.90E−19 | 2.00E−18 | LFNG |
| ENSG00000240583 | 1.981897469 | 2.32E−19 | 2.43E−18 | AQP1 |
| ENSG00000280303 | 1.116972088 | 5.78E−09 | 2.65E−08 | AC067931.2 |
| ENSG00000125772 | 0.588258566 | 3.68E−19 | 3.83E−18 | GPCPD1 |
| ENSG00000185189 | 0.713266249 | 6.53E−19 | 6.67E−18 | NRBP2 |
| ENSG00000243629 | 1.089891447 | 5.78E−11 | 3.26E−10 | LINC00880 |
| ENSG00000136859 | 0.625902802 | 1.32E−18 | 1.33E−17 | ANGPTL2 |
| ENSG00000170412 | 1.064761214 | 1.47E−18 | 1.48E−17 | GPRC5C |
| ENSG00000115525 | 0.786519457 | 1.70E−18 | 1.70E−17 | ST3GAL5 |
| ENSG00000108551 | 0.634579806 | 2.01E−18 | 1.99E−17 | RASD1 |
| ENSG00000260877 | 1.040510062 | 3.58E−10 | 1.85E−09 | AP005233.2 |
| ENSG00000111452 | 0.797580152 | 3.43E−18 | 3.34E−17 | ADGRD1 |
| ENSG00000164010 | 0.671003676 | 4.05E−18 | 3.92E−17 | ERMAP |
| ENSG00000267013 | 1.031866824 | 1.81E−19 | 1.91E−18 | LINC01929 |
| ENSG00000196517 | 0.601022218 | 7.40E−18 | 7.02E−17 | SLC6A9 |
| ENSG00000184985 | 0.953793323 | 8.27E−18 | 7.82E−17 | SORCS2 |
| ENSG00000073350 | 1.015768255 | 9.33E−18 | 8.78E−17 | LLGL2 |
| ENSG00000160181 | 0.979528456 | 9.78E−18 | 9.19E−17 | TFF2 |
| ENSG00000170214 | 0.985557139 | 1.03E−17 | 9.71E−17 | ADRA1B |
| ENSG00000164050 | 1.092459663 | 1.60E−17 | 1.48E−16 | PLXNB1 |
| ENSG00000232611 | 1.014419347 | 2.24E−07 | 8.55E−07 | AL683813.1 |
| ENSG00000138286 | 1.012503764 | 2.56E−17 | 2.35E−16 | FAM149B1 |
| ENSG00000204852 | 0.586136292 | 2.61E−17 | 2.39E−16 | TCTN1 |
| ENSG00000107968 | 0.672186642 | 2.73E−17 | 2.50E−16 | MAP3K8 |
| ENSG00000103196 | 0.669497228 | 3.21E−17 | 2.93E−16 | CRISPLD2 |
| ENSG00000163694 | 0.664725312 | 3.76E−17 | 3.41E−16 | RBM47 |
| ENSG00000118777 | 0.63684278 | 4.00E−17 | 3.62E−16 | ABCG2 |
| ENSG00000165272 | 0.737450886 | 5.14E−17 | 4.61E−16 | AQP3 |
| ENSG00000234405 | 1.012989861 | 1.17E−05 | 3.61E−05 | Z69733.1 |
| ENSG00000204282 | 0.999042357 | 2.05E−16 | 1.76E−15 | TNRC6C-AS1 |
| ENSG00000160180 | 1.828562389 | 1.07E−16 | 9.33E−16 | TFF3 |
| ENSG00000091490 | 1.064775 | 1.12E−16 | 9.71E−16 | SEL1L3 |
| ENSG00000174456 | 0.697982291 | 1.56E−16 | 1.35E−15 | C12orf76 |
| ENSG00000224184 | 0.998094957 | 1.45E−12 | 9.33E−12 | MIR3681HG |
| ENSG00000139182 | 0.612905821 | 2.09E−16 | 1.79E−15 | CLSTN3 |
| ENSG00000021762 | 0.664520536 | 2.82E−16 | 2.40E−15 | OSBPL5 |
| ENSG00000024422 | 0.631697665 | 2.90E−16 | 2.47E−15 | EHD2 |
| ENSG00000178209 | 0.59391065 | 4.21E−16 | 3.53E−15 | PLEC |
| ENSG00000213626 | 0.748895824 | 4.77E−16 | 3.99E−15 | LBH |
| ENSG00000203709 | 0.989260567 | 3.38E−19 | 3.53E−18 | C1orf132 |
| ENSG00000120899 | 0.809869697 | 6.04E−16 | 5.03E−15 | PTK2B |
| ENSG00000166171 | 0.634006308 | 6.79E−16 | 5.63E−15 | DPCD |
| ENSG00000119514 | 1.109015536 | 8.36E−16 | 6.87E−15 | GALNT12 |
| ENSG00000166145 | 1.012449601 | 9.25E−16 | 7.55E−15 | SPINT1 |
| ENSG00000139625 | 0.597222522 | 9.56E−16 | 7.80E−15 | MAP3K12 |
| ENSG00000130529 | 1.072374016 | 1.03E−15 | 8.41E−15 | TRPM4 |
| ENSG00000089639 | 0.583026836 | 1.04E−15 | 8.48E−15 | GMIP |
| ENSG00000164362 | 0.96421019 | 1.42E−15 | 1.15E−14 | TERT |
| ENSG00000260265 | 0.980352322 | 6.53E−06 | 2.08E−05 | AC110760.4 |
| ENSG00000260832 | 0.972908743 | 2.37E−05 | 7.03E−05 | AC125793.1 |
| ENSG00000143224 | 0.603143848 | 1.54E−15 | 1.24E−14 | PPOX |
| ENSG00000166387 | 0.768430513 | 1.83E−15 | 1.47E−14 | PPFIBP2 |
| ENSG00000012124 | 0.751559289 | 2.74E−15 | 2.17E−14 | CD22 |
| ENSG00000073282 | 0.941931414 | 2.98E−15 | 2.35E−14 | TP63 |
| ENSG00000234424 | 0.96208621 | 1.12E−05 | 3.44E−05 | AL353743.4 |
| ENSG00000162078 | 1.302273759 | 3.84E−15 | 3.01E−14 | ZG16B |
| ENSG00000170190 | 0.646867941 | 5.64E−15 | 4.38E−14 | SLC16A5 |
| ENSG00000277879 | 0.946436382 | 1.64E−06 | 5.64E−06 | AL391988.1 |
| ENSG00000182578 | 0.871074337 | 7.57E−15 | 5.81E−14 | CSF1R |
| ENSG00000204442 | 0.613852863 | 8.56E−15 | 6.55E−14 | FAM155A |
| ENSG00000084764 | 1.238040706 | 9.29E−15 | 7.09E−14 | MAPRE3 |
| ENSG00000187653 | 0.90344409 | 7.66E−38 | 2.02E−36 | TMSB4XP8 |
| ENSG00000125864 | 0.605676615 | 1.40E−14 | 1.06E−13 | BFSP1 |
| ENSG00000004799 | 0.72648243 | 1.43E−14 | 1.07E−13 | PDK4 |
| ENSG00000164690 | 1.021512855 | 1.43E−14 | 1.08E−13 | SHH |
| ENSG00000158006 | 0.675401351 | 1.52E−14 | 1.14E−13 | PAFAH2 |
| ENSG00000102466 | 1.99530516 | 1.53E−14 | 1.14E−13 | FGF14 |
| ENSG00000107902 | 0.928721406 | 2.36E−14 | 1.74E−13 | LHPP |
| ENSG00000163219 | 1.470672943 | 2.40E−14 | 1.77E−13 | ARHGAP25 |
| ENSG00000105327 | 0.659101322 | 2.47E−14 | 1.82E−13 | BBC3 |
| ENSG00000232931 | 0.879123796 | 1.35E−07 | 5.27E−07 | LINC00342 |
| ENSG00000116885 | 0.944805119 | 2.77E−14 | 2.04E−13 | OSCP1 |
| ENSG00000196542 | 2.000172114 | 3.44E−14 | 2.51E−13 | SPTSSB |
| ENSG00000141469 | 1.257620277 | 4.10E−14 | 2.98E−13 | SLC14A1 |
| ENSG00000207954 | 0.864355996 | 6.57E−05 | 0.0001819 | MIR138-1 |
| ENSG00000269896 | 0.863999249 | 1.98E−06 | 6.72E−06 | AL513477.1 |
| ENSG00000183317 | 1.598192927 | 7.07E−14 | 5.03E−13 | EPHA10 |
| ENSG00000064547 | 1.043247745 | 7.64E−14 | 5.43E−13 | LPAR2 |
| ENSG00000183454 | 1.361311685 | 8.02E−14 | 5.69E−13 | GRIN2A |
| ENSG00000258545 | 0.861885379 | 2.73E−05 | 8.03E−05 | RHOXF1-AS1 |
| ENSG00000171174 | 0.721579865 | 1.15E−13 | 8.07E−13 | RBKS |
| ENSG00000226332 | 0.827459238 | 1.96E−10 | 1.04E−09 | AL354836.1 |
| ENSG00000215769 | 0.819562549 | 6.81E−12 | 4.15E−11 | ARHGAP27P1-BPTFP1-KPNA2P3 |
| ENSG00000069424 | 0.926219371 | 2.49E−13 | 1.70E−12 | KCNAB2 |
| ENSG00000203780 | 1.268825864 | 2.79E−13 | 1.90E−12 | FANK1 |
| ENSG00000147642 | 0.674353182 | 3.40E−13 | 2.30E−12 | SYBU |
| ENSG00000261786 | 0.80298847 | 9.74E−14 | 6.87E−13 | AC006058.1 |
| ENSG00000224086 | 0.750479112 | 5.45E−07 | 1.98E−06 | AC245452.1 |
| ENSG00000251562 | 0.749923658 | 6.90E−14 | 4.92E−13 | MALAT1 |
| ENSG00000108821 | 0.721390767 | 5.89E−13 | 3.92E−12 | COL1A1 |
| ENSG00000163993 | 0.950679444 | 6.17E−13 | 4.10E−12 | S100P |
| ENSG00000119686 | 1.315494173 | 8.81E−13 | 5.80E−12 | FLVCR2 |
| ENSG00000267481 | 0.748507005 | 0.00019344 | 0.000499816 | AC011477.2 |
| ENSG00000161677 | 0.65441487 | 8.86E−13 | 5.82E−12 | JOSD2 |
| ENSG00000168477 | 1.714597499 | 9.18E−13 | 6.02E−12 | TNXB |
| ENSG00000167535 | 0.66107174 | 1.08E−12 | 7.02E−12 | CACNB3 |
| ENSG00000188177 | 1.190091354 | 1.32E−12 | 8.55E−12 | ZC3H6 |
| ENSG00000277778 | 0.730989212 | 3.18E−07 | 1.19E−06 | PGM5P2 |
| ENSG00000099889 | 0.874014164 | 2.18E−12 | 1.39E−11 | ARVCF |
| ENSG00000257315 | 0.726622407 | 2.48E−12 | 1.57E−11 | ZBED6 |
| ENSG00000161395 | 0.681576338 | 2.54E−12 | 1.61E−11 | PGAP3 |
| ENSG00000283646 | 0.715148035 | 5.47E−13 | 3.66E−12 | LINC02009 |
| ENSG00000274307 | 0.705188548 | 0.000339574 | 0.000836261 | AC023449.2 |
| ENSG00000167103 | 1.332705616 | 4.92E−12 | 3.05E−11 | PIP5KL1 |
| ENSG00000159423 | 0.611704098 | 6.38E−12 | 3.90E−11 | ALDH4A1 |
| ENSG00000278897 | 0.70098086 | 0.000273415 | 0.000687648 | AC020951.1 |
| ENSG00000162426 | 1.314896899 | 8.38E−12 | 5.07E−11 | SLC45A1 |
| ENSG00000188064 | 0.633873994 | 1.16E−11 | 6.95E−11 | WNT7B |
| ENSG00000187952 | 0.691227099 | 0.000538949 | 0.001287183 | HS6ST1P1 |
| ENSG00000108641 | 0.676095788 | 1.31E−11 | 7.82E−11 | B9D1 |
| ENSG00000139178 | 0.828600996 | 1.34E−11 | 7.98E−11 | C1RL |
| ENSG00000253669 | 0.690702691 | 6.08E−09 | 2.78E−08 | AP003356.1 |
| ENSG00000281881 | 0.679695644 | 1.99E−05 | 5.96E−05 | SPRY4-IT1 |
| ENSG00000176371 | 0.682561022 | 3.05E−11 | 1.75E−10 | ZSCAN2 |
| ENSG00000066248 | 0.600563626 | 3.22E−11 | 1.85E−10 | NGEF |
| ENSG00000239713 | 0.790898223 | 3.30E−11 | 1.89E−10 | APOBEC3G |
| ENSG00000084710 | 0.814302414 | 4.97E−11 | 2.82E−10 | EFR3B |
| ENSG00000213315 | 0.674927745 | 0.001017341 | 0.002306342 | AL122020.1 |
| ENSG00000233895 | 0.671323329 | 2.93E−05 | 8.55E−05 | AL121761.1 |
| ENSG00000205593 | 1.297364282 | 7.30E−11 | 4.05E−10 | DENND6B |
| ENSG00000173156 | 0.660509744 | 7.44E−11 | 4.13E−10 | RHOD |
| ENSG00000129757 | 1.280060934 | 8.31E−11 | 4.60E−10 | CDKN1C |
| ENSG00000165929 | 1.580465307 | 1.11E−10 | 6.07E−10 | TC2N |
| ENSG00000111319 | 0.916384563 | 1.54E−10 | 8.30E−10 | SCNN1A |
| ENSG00000226137 | 0.662532844 | 2.08E−17 | 1.92E−16 | BAIAP2-AS1 |
| ENSG00000131779 | 0.645897178 | 1.88E−10 | 1.00E−09 | PEX11B |
| ENSG00000149972 | 1.274126912 | 1.95E−10 | 1.04E−09 | CNTN5 |
| ENSG00000250072 | 0.661140207 | 3.69E−05 | 0.000106104 | AC091940.1 |
| ENSG00000158458 | 0.771645473 | 2.17E−10 | 1.15E−09 | NRG2 |
| ENSG00000171443 | 0.620783725 | 2.33E−10 | 1.23E−09 | ZNF524 |
| ENSG00000233452 | 0.657023126 | 0.001884719 | 0.004066423 | STXBP5-AS1 |
| ENSG00000146966 | 0.580752824 | 3.35E−10 | 1.74E−09 | DENND2A |
| ENSG00000245849 | 0.649770435 | 0.001938789 | 0.004172798 | RAD51-AS1 |
| ENSG00000220785 | 0.643682307 | 0.000249588 | 0.000632447 | MTMR9LP |
| ENSG00000160703 | 0.613565082 | 3.93E−10 | 2.02E−09 | NLRX1 |
| ENSG00000006756 | 0.786505348 | 4.19E−10 | 2.15E−09 | ARSD |
| ENSG00000106078 | 1.171724535 | 4.43E−10 | 2.27E−09 | COBL |
| ENSG00000169248 | 0.790213883 | 5.21E−10 | 2.65E−09 | CXCL11 |
| ENSG00000143127 | 0.858887046 | 5.91E−10 | 2.98E−09 | ITGA10 |
| ENSG00000230487 | 0.635330515 | 7.29E−05 | 0.000200601 | PSMG3-AS1 |
| ENSG00000169231 | 0.71986648 | 6.66E−10 | 3.36E−09 | THBS3 |
| ENSG00000264112 | 0.630435915 | 3.88E−13 | 2.61E−12 | AC015813.1 |
| ENSG00000272512 | 0.629649183 | 0.00016484 | 0.000430322 | AL645608.8 |
| ENSG00000248008 | 0.626222703 | 1.44E−15 | 1.16E−14 | NRAV |
| ENSG00000165698 | 1.042720757 | 1.01E−09 | 4.98E−09 | SPACA9 |
| ENSG00000167065 | 0.691874401 | 1.17E−09 | 5.78E−09 | DUSP18 |
| ENSG00000283160 | 0.62478041 | 0.003658265 | 0.007489349 | MIR4521 |
| ENSG00000167306 | 0.684016305 | 1.74E−09 | 8.41E−09 | MYO5B |
| ENSG00000187987 | 1.030707676 | 1.80E−09 | 8.67E−09 | ZSCAN23 |
| ENSG00000185386 | 0.750119972 | 1.89E−09 | 9.10E−09 | MAPK11 |
| ENSG00000281026 | 0.623674242 | 0.00174105 | 0.003777549 | N4BP2L2-IT2 |
| ENSG00000167895 | 1.301863777 | 1.94E−09 | 9.32E−09 | TMC8 |
| ENSG00000270640 | 0.621162265 | 0.003395331 | 0.006999073 | AC104695.3 |
| ENSG00000263155 | 1.349972416 | 2.25E−09 | 1.07E−08 | MYZAP |
| ENSG00000125637 | 0.809041824 | 2.30E−09 | 1.10E−08 | PSD4 |
| ENSG00000178531 | 0.6403658 | 2.47E−09 | 1.18E−08 | CTXN1 |
| ENSG00000006210 | 1.386770722 | 3.03E−09 | 1.43E−08 | CX3CL1 |
| ENSG00000196476 | 0.703193796 | 3.10E−09 | 1.46E−08 | C20orf96 |
| ENSG00000114841 | 0.724964893 | 3.49E−09 | 1.63E−08 | DNAH1 |
| ENSG00000248690 | 0.616225449 | 0.005847093 | 0.011483426 | HAS2-AS1 |
| ENSG00000128849 | 0.993028568 | 3.72E−09 | 1.74E−08 | CGNL1 |
| ENSG00000250903 | 0.616173957 | 7.24E−15 | 5.56E−14 | GMDS-AS1 |
| ENSG00000139192 | 0.677957048 | 4.54E−09 | 2.10E−08 | TAPBPL |
| ENSG00000273888 | 0.615241762 | 0.000253027 | 0.00064005 | FRMD6-AS1 |
| ENSG00000276116 | 0.614111505 | 8.00E−05 | 0.000218844 | FUT8-AS1 |
| ENSG00000078114 | 0.779192836 | 5.73E−09 | 2.63E−08 | NEBL |
| ENSG00000223745 | 0.609457977 | 6.90E−08 | 2.79E−07 | CCDC18-AS1 |
| ENSG00000276476 | 0.595189913 | 3.36E−05 | 9.71E−05 | LINC00540 |
| ENSG00000104332 | 1.024730115 | 6.35E−09 | 2.90E−08 | SFRP1 |
| ENSG00000187688 | 0.74390243 | 6.40E−09 | 2.92E−08 | TRPV2 |
| ENSG00000168961 | 1.249308012 | 7.43E−09 | 3.37E−08 | LGALS9 |
| ENSG00000181444 | 1.289743152 | 7.97E−09 | 3.60E−08 | ZNF467 |
| ENSG00000007516 | 0.7190471 | 9.06E−09 | 4.06E−08 | BAIAP3 |
| ENSG00000074964 | 0.723682951 | 9.90E−09 | 4.42E−08 | ARHGEF10L |
| ENSG00000153233 | 0.860394698 | 1.06E−08 | 4.73E−08 | PTPRR |
| ENSG00000158125 | 0.749227047 | 1.34E−08 | 5.87E−08 | XDH |
| ENSG00000105982 | 0.809043212 | 1.50E−08 | 6.57E−08 | RNF32 |
| ENSG00000130751 | 0.996208061 | 2.12E−08 | 9.07E−08 | NPAS1 |
| ENSG00000187583 | 0.628689467 | 2.15E−08 | 9.21E−08 | PLEKHN1 |
| ENSG00000129295 | 0.804174666 | 3.78E−08 | 1.58E−07 | LRRC6 |
| ENSG00000136167 | 0.72905745 | 4.42E−08 | 1.83E−07 | LCP1 |
| ENSG00000109107 | 0.624763894 | 4.83E−08 | 1.99E−07 | ALDOC |
| ENSG00000102796 | 0.580283788 | 4.92E−08 | 2.02E−07 | DHRS12 |
| ENSG00000169926 | 0.738403193 | 5.42E−08 | 2.22E−07 | KLF13 |
| ENSG00000169583 | 0.867293251 | 6.71E−08 | 2.72E−07 | CLIC3 |
| ENSG00000105204 | 0.693379327 | 6.79E−08 | 2.75E−07 | DYRK1B |
| ENSG00000089127 | 0.795662065 | 8.09E−08 | 3.24E−07 | OAS1 |
| ENSG00000169220 | 0.610978587 | 8.83E−08 | 3.53E−07 | RGS14 |
| ENSG00000160190 | 0.656350516 | 9.89E−08 | 3.94E−07 | SLC37A1 |
| ENSG00000156218 | 0.750817532 | 1.07E−07 | 4.25E−07 | ADAMTSL3 |
| ENSG00000105854 | 0.64320919 | 1.18E−07 | 4.64E−07 | PON2 |
| ENSG00000103034 | 0.978032576 | 1.35E−07 | 5.28E−07 | NDRG4 |
| ENSG00000008517 | 0.607365229 | 1.35E−07 | 5.29E−07 | IL32 |
| ENSG00000163617 | 0.881935404 | 1.36E−07 | 5.33E−07 | CCDC191 |
| ENSG00000184194 | 0.756086964 | 1.44E−07 | 5.61E−07 | GPR173 |
| ENSG00000124116 | 0.704374738 | 1.47E−07 | 5.72E−07 | WFDC3 |
| ENSG00000197279 | 0.61212415 | 1.48E−07 | 5.74E−07 | ZNF165 |
| ENSG00000155265 | 1.06380425 | 1.92E−07 | 7.39E−07 | GOLGA7B |
| ENSG00000196420 | 0.668634419 | 2.03E−07 | 7.76E−07 | S100A5 |
| ENSG00000163995 | 0.914334457 | 2.21E−07 | 8.44E−07 | ABLIM2 |
| ENSG00000173267 | 0.875959067 | 2.32E−07 | 8.85E−07 | SNCG |
| ENSG00000174951 | 0.583579922 | 2.48E−07 | 9.40E−07 | FUT1 |
| ENSG00000166578 | 0.59325713 | 2.97E−07 | 1.12E−06 | IQCD |
| ENSG00000134030 | 0.721745796 | 3.68E−07 | 1.36E−06 | CTIF |
| ENSG00000133640 | 0.867286671 | 3.91E−07 | 1.45E−06 | LRRIQ1 |
| ENSG00000262576 | 0.845341331 | 4.37E−07 | 1.60E−06 | PCDHGA4 |
| ENSG00000160781 | 0.937385076 | 4.56E−07 | 1.67E−06 | PAQR6 |
| ENSG00000050165 | 0.701579384 | 5.28E−07 | 1.92E−06 | DKK3 |
| ENSG00000111863 | 0.721347277 | 6.34E−07 | 2.29E−06 | ADTRP |
| ENSG00000160325 | 0.946677553 | 6.89E−07 | 2.48E−06 | CACFD1 |
| ENSG00000198846 | 0.743926359 | 7.19E−07 | 2.58E−06 | TOX |
| ENSG00000166262 | 1.011056155 | 7.77E−07 | 2.77E−06 | FAM227B |
| ENSG00000239282 | 0.740685304 | 9.03E−07 | 3.20E−06 | CASTOR1 |
| ENSG00000122547 | 0.589219386 | 1.04E−06 | 3.67E−06 | EEPD1 |
| ENSG00000115556 | 0.759404536 | 1.29E−06 | 4.50E−06 | PLCD4 |
| ENSG00000146021 | 0.996087722 | 1.42E−06 | 4.94E−06 | KLHL3 |
| ENSG00000167554 | 0.801922519 | 1.58E−06 | 5.45E−06 | ZNF610 |
| ENSG00000159899 | 0.607690901 | 1.59E−06 | 5.47E−06 | NPR2 |
| ENSG00000085831 | 0.88967755 | 1.86E−06 | 6.33E−06 | TTC39A |
| ENSG00000167600 | 0.949538739 | 1.95E−06 | 6.64E−06 | CYP2S1 |
| ENSG00000135298 | 0.65393669 | 2.08E−06 | 7.03E−06 | ADGRB3 |
| ENSG00000185345 | 1.087332962 | 2.19E−06 | 7.38E−06 | PRKN |
| ENSG00000079974 | 0.704729252 | 2.33E−06 | 7.83E−06 | RABL2B |
| ENSG00000213085 | 0.908492786 | 2.67E−06 | 8.89E−06 | CFAP45 |
| ENSG00000137261 | 1.001309402 | 4.72E−06 | 1.53E−05 | KIAA0319 |
| ENSG00000131620 | 0.893864552 | 5.41E−06 | 1.74E−05 | ANO1 |
| ENSG00000005379 | 0.634150616 | 5.44E−06 | 1.75E−05 | TSPOAP1 |
| ENSG00000170927 | 1.061916014 | 5.77E−06 | 1.85E−05 | PKHD1 |
| ENSG00000183638 | 0.95696744 | 5.87E−06 | 1.88E−05 | RP1L1 |
| ENSG00000185634 | 0.655532658 | 6.41E−06 | 2.04E−05 | SHC4 |
| ENSG00000178026 | 0.704425401 | 7.44E−06 | 2.35E−05 | LRRC75B |
| ENSG00000007237 | 0.94018185 | 7.53E−06 | 2.38E−05 | GAS7 |
| ENSG00000063438 | 0.850087695 | 7.99E−06 | 2.52E−05 | AHRR |
| ENSG00000078081 | 0.943742842 | 1.00E−05 | 3.13E−05 | LAMP3 |
| ENSG00000152779 | 0.893034503 | 1.16E−05 | 3.58E−05 | SLC16A12 |
| ENSG00000105649 | 0.629673856 | 1.17E−05 | 3.59E−05 | RAB3A |
| ENSG00000101670 | 0.641918014 | 1.74E−05 | 5.23E−05 | LIPG |
| ENSG00000153246 | 0.640475 | 2.34E−05 | 6.95E−05 | PLA2R1 |
| ENSG00000197013 | 0.895543676 | 2.42E−05 | 7.15E−05 | ZNF429 |
| ENSG00000100027 | 0.703475726 | 2.55E−05 | 7.54E−05 | YPEL1 |
| ENSG00000222009 | 1.003174533 | 2.78E−05 | 8.16E−05 | BTBD19 |
| ENSG00000077092 | 0.583904079 | 2.82E−05 | 8.25E−05 | RARB |
| ENSG00000138271 | 0.879042367 | 2.88E−05 | 8.41E−05 | GPR87 |
| ENSG00000215788 | 0.630842347 | 4.98E−05 | 0.000140178 | TNFRSF25 |
| ENSG00000111907 | 0.610094964 | 5.19E−05 | 0.000145846 | TPD52L1 |
| ENSG00000148225 | 0.603765317 | 5.24E−05 | 0.000146979 | WDR31 |
| ENSG00000265190 | 0.796463642 | 5.25E−05 | 0.00014743 | ANXA8 |
| ENSG00000166780 | 0.609717829 | 5.44E−05 | 0.00015227 | C16orf45 |
| ENSG00000196917 | 0.687020342 | 5.62E−05 | 0.000157013 | HCAR1 |
| ENSG00000243749 | 0.682870409 | 6.23E−05 | 0.00017307 | TMEM35B |
| ENSG00000100100 | 0.614989243 | 6.46E−05 | 0.000179064 | PIK3IP1 |
| ENSG00000185261 | 0.808099048 | 6.60E−05 | 0.000182782 | KIAA0825 |
| ENSG00000176714 | 0.719708594 | 6.65E−05 | 0.000183929 | CCDC121 |
| ENSG00000189350 | 0.682674219 | 8.27E−05 | 0.00022538 | TOGARAM2 |
| ENSG00000117586 | 0.66232365 | 9.60E−05 | 0.000259115 | TNFSF4 |
| ENSG00000163283 | 0.702973737 | 9.69E−05 | 0.000261336 | ALPP |
| ENSG00000100628 | 0.840396776 | 0.000102783 | 0.000275833 | ASB2 |
| ENSG00000204936 | 0.799723751 | 0.000132176 | 0.000349991 | CD177 |
| ENSG00000249242 | 0.674349922 | 0.00013392 | 0.000354394 | TMEM150C |
| ENSG00000154589 | 0.621650835 | 0.000154203 | 0.000405048 | LY96 |
| ENSG00000137103 | 0.813157861 | 0.000158856 | 0.000416188 | TMEM8B |
| ENSG00000186417 | 0.845874331 | 0.000174472 | 0.000454112 | GLDN |
| ENSG00000168702 | 0.829015848 | 0.000183578 | 0.000476395 | LRP1B |
| ENSG00000177694 | 0.678072111 | 0.000201099 | 0.000518177 | NAALADL2 |
| ENSG00000137460 | 0.621505207 | 0.000215133 | 0.000551848 | FHDC1 |
| ENSG00000233493 | 0.606768387 | 0.000304964 | 0.000759001 | TMEM238 |
| ENSG00000135525 | 0.592393732 | 0.000307346 | 0.00076377 | MAP7 |
| ENSG00000177076 | 0.595537696 | 0.000354308 | 0.000869936 | ACER2 |
| ENSG00000172456 | 0.624018639 | 0.000361318 | 0.000885657 | FGGY |
| ENSG00000112303 | 0.791611125 | 0.000395261 | 0.00096382 | VNN2 |
| ENSG00000174327 | 0.660320887 | 0.000409421 | 0.000994655 | SLC16A13 |
| ENSG00000118997 | 0.801710893 | 0.000462226 | 0.001115511 | DNAH7 |
| ENSG00000255690 | 0.769175757 | 0.000506713 | 0.001215944 | TRIL |
| ENSG00000010295 | 0.685112629 | 0.000525958 | 0.001259364 | IFFO1 |
| ENSG00000145107 | 0.585678306 | 0.000567754 | 0.001349109 | TM4SF19 |
| ENSG00000168026 | 0.683159923 | 0.000690836 | 0.001616374 | TTC21A |
| ENSG00000137285 | 0.737035799 | 0.000813426 | 0.00187585 | TUBB2B |
| ENSG00000156510 | 0.685659195 | 0.000879493 | 0.002015241 | HKDC1 |
| ENSG00000153237 | 0.729462631 | 0.000921302 | 0.002105764 | CCDC148 |
| ENSG00000131849 | 0.669939869 | 0.001076571 | 0.002430196 | ZNF132 |
| ENSG00000169550 | 0.724738148 | 0.0011291 | 0.002537865 | MUC15 |
| ENSG00000253649 | 0.611969408 | 0.001576863 | 0.003452116 | PRSS51 |
| ENSG00000264230 | 0.647657643 | 0.001653031 | 0.003600257 | ANXA8L1 |
| ENSG00000183401 | 0.685190181 | 0.002421385 | 0.005126606 | CCDC159 |
| ENSG00000166816 | 0.611266978 | 0.00242422 | 0.005130953 | LDHD |
| ENSG00000108932 | 0.615640297 | 0.002572756 | 0.00541913 | SLC16A6 |
| ENSG00000106125 | 0.593529032 | 0.003116381 | 0.006461581 | MINDY4 |
| ENSG00000183091 | 0.60347346 | 0.003149988 | 0.006521995 | NEB |
| ENSG00000152582 | 0.655526014 | 0.004260382 | 0.008617234 | SPEF2 |
Table 3.
Genes downregulated in LOXL2 knockdown
| Locus | logFC | P-Value | FDR | Gene name |
|---|---|---|---|---|
| ENSG00000134013 | −3.053434126 | 0 | 0 | LOXL2 |
| ENSG00000122545 | −1.493781116 | 6.6E−272 | 1.45E−268 | SEP-T7 |
| ENSG00000137801 | −1.65563595 | 1.11E−262 | 2.08E−259 | THBS1 |
| ENSG00000112062 | −1.367605967 | 1.43E−252 | 2.34E−249 | MAPK14 |
| ENSG00000146281 | −1.644281336 | 6.67E−244 | 9.73E−241 | PM20D2 |
| ENSG00000105971 | −1.358844865 | 4.29E−189 | 3.31E−186 | CAV2 |
| ENSG00000117500 | −1.227263597 | 2.54E−176 | 1.67E−173 | TMED5 |
| ENSG00000172380 | −1.135404374 | 1.81E−162 | 1.08E−159 | GNG12 |
| ENSG00000152558 | −1.10232016 | 1.28E−160 | 7.33E−158 | TMEM123 |
| ENSG00000100462 | −1.134694357 | 2.31E−160 | 1.26E−157 | PRMT5 |
| ENSG00000213281 | −1.098589526 | 6.65E−160 | 3.49E−157 | NRAS |
| ENSG00000162521 | −1.195180927 | 7.41E−138 | 2.95E−135 | RBBP4 |
| ENSG00000196396 | −1.038088551 | 1.5E−136 | 5.64E−134 | PTPN1 |
| ENSG00000182400 | −1.288279943 | 9.25E−134 | 3.2E−131 | TRAPPC6B |
| ENSG00000115339 | −1.421874178 | 1.01E−132 | 3.31E−130 | GALNT3 |
| ENSG00000105810 | −1.03878264 | 1.67E−132 | 5.35E−130 | CDK6 |
| ENSG00000105849 | −1.177181646 | 7.96E−125 | 2.38E−122 | TWISTNB |
| ENSG00000176853 | −0.991116504 | 4.73E−124 | 1.38E−121 | FAM91A1 |
| ENSG00000113742 | −1.271922708 | 3.08E−120 | 8.43E−118 | CPEB4 |
| ENSG00000156017 | −1.215741594 | 1.39E−117 | 3.52E−115 | CARNMT1 |
| ENSG00000154429 | −1.346899288 | 2.62E−117 | 6.5E−115 | CCSAP |
| ENSG00000101974 | −1.060939839 | 1.68E−116 | 4.02E−114 | ATP11C |
| ENSG00000073712 | −0.991731657 | 6.59E−116 | 1.54E−113 | FERMT2 |
| ENSG00000162104 | −1.444767007 | 1.64E−113 | 3.77E−111 | ADCY9 |
| ENSG00000064042 | −1.111524916 | 1.06E−110 | 2.36E−108 | LIMCH1 |
| ENSG00000087086 | −0.890490359 | 4.05E−108 | 8.57E−106 | FTL |
| ENSG00000106460 | −1.091704144 | 6.11E−108 | 1.27E−105 | TMEM106B |
| ENSG00000095752 | −1.227515654 | 8.77E−108 | 1.8E−105 | IL11 |
| ENSG00000135521 | −1.168276042 | 5.89E−106 | 1.12E−103 | LTV1 |
| ENSG00000187908 | −1.211596715 | 7.57E−106 | 1.42E−103 | DMBT1 |
| ENSG00000176788 | −1.162731668 | 1.53E−103 | 2.82E−101 | BASP1 |
| ENSG00000104375 | −1.261014996 | 2.57E−102 | 4.69E−100 | STK3 |
| ENSG00000146143 | −1.074303191 | 9.78E−102 | 1.76E−99 | PRIM2 |
| ENSG00000145284 | −1.323720519 | 4.77E−100 | 8.34E−98 | SCD5 |
| ENSG00000113645 | −0.916653973 | 2.11E−99 | 3.65E−97 | WWC1 |
| ENSG00000163161 | −1.018873601 | 3.41E−97 | 5.59E−95 | ERCC3 |
| ENSG00000184007 | −0.822555541 | 1.26E−96 | 2.02E−94 | PTP4A2 |
| ENSG00000143977 | −0.987888082 | 2.7E−93 | 4.07E−91 | SNRPG |
| ENSG00000141994 | −1.367808305 | 2.61E−92 | 3.9E−90 | DUS3L |
| ENSG00000172954 | −1.050710392 | 5.52E−92 | 8.14E−90 | LCLAT1 |
| ENSG00000180694 | −1.033774023 | 2.99E−91 | 4.36E−89 | TMEM64 |
| ENSG00000101003 | −1.071829328 | 3.19E−89 | 4.36E−87 | GINS1 |
| ENSG00000123689 | −0.87877304 | 8.1E−88 | 1.07E−85 | G0S2 |
| ENSG00000108561 | −0.906168536 | 7.24E−87 | 9.51E−85 | C1QBP |
| ENSG00000175348 | −1.039373012 | 2.22E−85 | 2.75E−83 | TMEM9B |
| ENSG00000011201 | −1.309694851 | 3.72E−85 | 4.57E−83 | ANOS1 |
| ENSG00000097033 | −0.780495141 | 1.21E−84 | 1.46E−82 | SH3GLB1 |
| ENSG00000103495 | −0.812954992 | 2.8E−83 | 3.28E−81 | MAZ |
| ENSG00000109084 | −1.036628155 | 7.56E−83 | 8.79E−81 | TMEM97 |
| ENSG00000170248 | −0.756074633 | 1.33E−82 | 1.53E−80 | PDCD6IP |
| ENSG00000154734 | −0.993464022 | 2.81E−80 | 3.07E−78 | ADAMTS1 |
| ENSG00000198959 | −0.78388823 | 3.88E−80 | 4.17E−78 | TGM2 |
| ENSG00000109270 | −1.102716006 | 5.57E−79 | 5.9E−77 | LAMTOR3 |
| ENSG00000171033 | −1.511641065 | 3.88E−78 | 4.07E−76 | PKIA |
| ENSG00000262919 | −1.286604883 | 1.12E−77 | 1.16E−75 | FAM58A |
| ENSG00000076003 | −0.895743892 | 1.38E−76 | 1.39E−74 | MCM6 |
| ENSG00000104164 | −0.880674477 | 1.48E−76 | 1.47E−74 | BLOC1S6 |
| ENSG00000112149 | −1.349898585 | 1.17E−75 | 1.15E−73 | CD83 |
| ENSG00000173110 | −1.621696267 | 2.04E−75 | 1.97E−73 | HSPA6 |
| ENSG00000196865 | –1.052977006 | 1.1E−74 | 1.06E−72 | NHLRC2 |
| ENSG00000164209 | –0.79794111 | 1.25E−74 | 1.19E−72 | SLC25A46 |
| ENSG00000092853 | –0.745590308 | 2.04E−73 | 1.89E−71 | CLSPN |
| ENSG00000163513 | –0.719014341 | 2.71E−73 | 2.48E−71 | TGFBR2 |
| ENSG00000127314 | –0.959251127 | 2.17E−72 | 1.93E−70 | RAP1B |
| ENSG00000005020 | –0.909609789 | 2.38E−71 | 2.04E−69 | SKAP2 |
| ENSG00000003989 | –1.062977014 | 3.54E−71 | 3.02E−69 | SLC7A2 |
| ENSG00000076248 | –0.818890935 | 7.73E−70 | 6.43E−68 | UNG |
| ENSG00000110104 | –0.886356935 | 8.21E−70 | 6.74E−68 | CCDC86 |
| ENSG00000106034 | -2.258112177 | 2.58E−69 | 2.1E−67 | CPED1 |
| ENSG00000139793 | –0.822257564 | 3.6E−69 | 2.92E−67 | MBNL2 |
| ENSG00000122870 | –1.265770623 | 1.9E−68 | 1.5E−66 | BICC1 |
| ENSG00000188811 | –1.270791643 | 4.79E−68 | 3.75E−66 | NHLRC3 |
| ENSG00000140262 | –0.845128144 | 4.91E−68 | 3.81E−66 | TCF12 |
| ENSG00000171791 | –1.383509833 | 6.73E−68 | 5.2E−66 | BCL2 |
| ENSG00000085449 | –0.807391186 | 7.7E−68 | 5.92E−66 | WDFY1 |
| ENSG00000163527 | –0.680451793 | 8.38E−68 | 6.36E−66 | STT3B |
| ENSG00000101856 | –0.725495533 | 9.47E−68 | 7.15E−66 | PGRMC1 |
| ENSG00000198689 | –0.855931203 | 1.38E−67 | 1.04E−65 | SLC9A6 |
| ENSG00000139921 | –0.707226796 | 1.81E−67 | 1.35E−65 | TMX1 |
| ENSG00000146047 | –0.927456112 | 2.17E−67 | 1.61E−65 | HIST1H2BA |
| ENSG00000171867 | –0.693918536 | 2.33E−67 | 1.72E−65 | PRNP |
| ENSG00000100625 | –0.866022602 | 5.2E−67 | 3.79E−65 | SIX4 |
| ENSG00000160208 | –0.734399871 | 3.02E−66 | 2.15E−64 | RRP1B |
| ENSG00000153132 | –0.846027423 | 3.16E−66 | 2.24E−64 | CLGN |
| ENSG00000151151 | –1.049984787 | 3.44E−66 | 2.43E−64 | IPMK |
| ENSG00000198948 | –1.324799997 | 4.2E−66 | 2.95E−64 | MFAP3L |
| ENSG00000213160 | –1.153831739 | 4.97E−66 | 3.45E−64 | KLHL23 |
| ENSG00000168615 | –0.756543804 | 7.94E−66 | 5.48E−64 | ADAM9 |
| ENSG00000198964 | –0.81917694 | 1.48E−65 | 1.02E−63 | SGMS1 |
| ENSG00000111371 | –0.655358043 | 6.25E−65 | 4.28E−63 | SLC38A1 |
| ENSG00000138756 | –0.727021119 | 3.81E−64 | 2.55E−62 | BMP2K |
| ENSG00000274997 | –0.669796522 | 2.01E−63 | 1.33E−61 | HIST1H2AH |
| ENSG00000166128 | –0.743923809 | 3.52E−63 | 2.29E−61 | RAB8B |
| ENSG00000153130 | –0.71036132 | 7.22E−63 | 4.65E−61 | SCOC |
| ENSG00000058056 | –0.791211898 | 9.57E−63 | 6.07E−61 | USP13 |
| ENSG00000183598 | –0.937984455 | 1.22E−62 | 7.72E−61 | HIST2H3D |
| ENSG00000157657 | –1.331037104 | 6.49E−62 | 4.06E−60 | ZNF618 |
| ENSG00000109586 | –0.806715459 | 3.03E−61 | 1.89E−59 | GALNT7 |
| ENSG00000144354 | –1.284264102 | 8.96E−61 | 5.55E−59 | CDCA7 |
| ENSG00000164211 | –0.821415196 | 9.8E−61 | 6.04E−59 | STARD4 |
| ENSG00000099901 | –0.797049931 | 1.04E−60 | 6.37E−59 | RANBP1 |
| ENSG00000103342 | –0.665272247 | 1.86E−60 | 1.13E−58 | GSPT1 |
| ENSG00000273703 | –0.759808174 | 1.89E−60 | 1.14E−58 | HIST1H2BM |
| ENSG00000162613 | –0.785667152 | 3.15E−60 | 1.88E−58 | FUBP1 |
| ENSG00000156802 | –0.643909783 | 3.42E−60 | 2.03E−58 | ATAD2 |
| ENSG00000107854 | –0.712659437 | 8.75E−60 | 5.13E−58 | TNKS2 |
| ENSG00000030304 | –1.71173144 | 1.31E−59 | 7.55E−58 | MUSK |
| ENSG00000264364 | –0.726395847 | 2.1E−59 | 1.19E−57 | DYNLL2 |
| ENSG00000131016 | –0.791953337 | 2.29E−59 | 1.29E−57 | AKAP12 |
| ENSG00000115364 | –0.778545058 | 2.65E−59 | 1.48E−57 | MRPL19 |
| ENSG00000111666 | –0.804368606 | 8.4E−59 | 4.64E−57 | CHPT1 |
| ENSG00000106771 | –0.644642299 | 1.24E−58 | 6.77E−57 | TMEM245 |
| ENSG00000154553 | -2.010549714 | 1.69E−58 | 9.16E−57 | PDLIM3 |
| ENSG00000180957 | –0.842836946 | 2.11E−58 | 1.14E−56 | PITPNB |
| ENSG00000164081 | –0.810106876 | 4.81E−58 | 2.58E−56 | TEX264 |
| ENSG00000112118 | –0.639768587 | 5.17E−58 | 2.75E−56 | MCM3 |
| ENSG00000189057 | –0.826299148 | 7.77E−58 | 4.09E−56 | FAM111B |
| ENSG00000116095 | –0.885698839 | 4.64E−57 | 2.43E−55 | PLEKHA3 |
| ENSG00000113083 | –1.085829177 | 4.8E−57 | 2.5E−55 | LOX |
| ENSG00000165156 | –0.739479342 | 5.04E−57 | 2.62E−55 | ZHX1 |
| ENSG00000113448 | –0.76465591 | 5.39E−57 | 2.79E−55 | PDE4D |
| ENSG00000166260 | –0.917921133 | 1.07E−56 | 5.44E−55 | COX11 |
| ENSG00000203668 | –0.623298669 | 1.52E−56 | 7.69E−55 | CHML |
| ENSG00000105698 | –0.737929868 | 1.71E−56 | 8.62E−55 | USF2 |
| ENSG00000176890 | –0.736138571 | 2.01E−56 | 1.01E−54 | TYMS |
| ENSG00000164466 | –0.68314086 | 2.1E−56 | 1.05E−54 | SFXN1 |
| ENSG00000197312 | –0.84172031 | 7.43E−56 | 3.67E−54 | DDI2 |
| ENSG00000132581 | –0.797529275 | 1.08E−55 | 5.31E−54 | SDF2 |
| ENSG00000105281 | –0.76012066 | 1.34E−54 | 6.44E−53 | SLC1A5 |
| ENSG00000101773 | –0.684417581 | 1.43E−54 | 6.8E−53 | RBBP8 |
| ENSG00000125166 | –0.64746293 | 2.47E−54 | 1.17E−52 | GOT2 |
| ENSG00000065615 | –0.766061917 | 3.65E−54 | 1.72E−52 | CYB5R4 |
| ENSG00000106366 | –0.690600823 | 4.53E−54 | 2.12E−52 | SERPINE1 |
| ENSG00000138675 | –1.055137884 | 2.97E−53 | 1.37E−51 | FGF5 |
| ENSG00000166801 | –0.679971393 | 4.81E−53 | 2.18E−51 | FAM111A |
| ENSG00000125430 | –1.119040342 | 1.8E−52 | 8.13E−51 | HS3ST3B1 |
| ENSG00000136986 | –0.64205774 | 3.04E−52 | 1.37E−50 | DERL1 |
| ENSG00000143507 | –0.824435143 | 3.91E−52 | 1.75E−50 | DUSP10 |
| ENSG00000132646 | –0.659226998 | 4.16E−52 | 1.85E−50 | PCNA |
| ENSG00000092470 | –0.776503363 | 4.36E−52 | 1.93E−50 | WDR76 |
| ENSG00000125870 | –0.692582004 | 6.21E−52 | 2.72E−50 | SNRPB2 |
| ENSG00000130830 | –1.001331439 | 7.01E−52 | 3.05E−50 | MPP1 |
| ENSG00000169193 | –1.233765255 | 1.18E−51 | 5.09E−50 | CCDC126 |
| ENSG00000115159 | –0.745203813 | 1.55E−51 | 6.63E−50 | GPD2 |
| ENSG00000089597 | –0.608914143 | 4.59E−51 | 1.95E−49 | GANAB |
| ENSG00000138448 | –0.679300714 | 2.25E−50 | 9.46E−49 | ITGAV |
| ENSG00000261609 | –0.838562302 | 3.25E−50 | 1.36E−48 | GAN |
| ENSG00000118596 | –0.91558522 | 3.25E−50 | 1.36E−48 | SLC16A7 |
| ENSG00000104388 | –0.77698668 | 8.4E−50 | 3.46E−48 | RAB2A |
| ENSG00000111725 | –0.851677774 | 1.59E−49 | 6.47E−48 | PRKAB1 |
| ENSG00000070214 | –0.695894872 | 1.61E−49 | 6.52E−48 | SLC44A1 |
| ENSG00000149289 | –0.717233099 | 1.62E−49 | 6.53E−48 | ZC3H12C |
| ENSG00000196323 | –0.700131838 | 1.75E−49 | 7.04E−48 | ZBTB44 |
| ENSG00000182504 | –0.657051624 | 8.72E−49 | 3.46E−47 | CEP97 |
| ENSG00000082269 | –0.721562816 | 2.1E−48 | 8.21E−47 | FAM135A |
| ENSG00000055208 | –0.688892188 | 2.44E−48 | 9.51E−47 | TAB2 |
| ENSG00000083720 | –0.85708639 | 3.3E−48 | 1.28E−46 | OXCT1 |
| ENSG00000110031 | –0.750834355 | 3.95E−48 | 1.53E−46 | LPXN |
| ENSG00000121966 | –0.877794416 | 4.44E−48 | 1.72E−46 | CXCR4 |
| ENSG00000081923 | –0.788155108 | 8.6E−48 | 3.3E−46 | ATP8B1 |
| ENSG00000133026 | –0.759331662 | 3.21E−47 | 1.21E−45 | MYH10 |
| ENSG00000167645 | –0.945756835 | 3.93E−47 | 1.47E−45 | YIF1B |
| ENSG00000048392 | –0.69447929 | 5.19E−47 | 1.93E−45 | RRM2B |
| ENSG00000157978 | –0.881827291 | 9.73E−47 | 3.61E−45 | LDLRAP1 |
| ENSG00000169429 | –0.814790283 | 1.09E−46 | 4.01E−45 | CXCL8 |
| ENSG00000253304 | –1.283748113 | 2.32E−46 | 8.48E−45 | TMEM200B |
| ENSG00000139278 | –0.735325809 | 3.22E−46 | 1.18E−44 | GLIPR1 |
| ENSG00000164070 | –0.61500254 | 3.29E−46 | 1.2E−44 | HSPA4L |
| ENSG00000116406 | –0.582479865 | 3.37E−46 | 1.22E−44 | EDEM3 |
| ENSG00000205302 | –0.632567484 | 6.01E−46 | 2.18E−44 | SNX2 |
| ENSG00000119917 | –0.598753864 | 1.72E−45 | 6.12E−44 | IFIT3 |
| ENSG00000094804 | –0.61234381 | 2.51E−45 | 8.93E−44 | CDC6 |
| ENSG00000180998 | –1.39647951 | 2.96E−45 | 1.05E−43 | GPR137C |
| ENSG00000166741 | –1.457281718 | 1.07E−44 | 3.74E−43 | NNMT |
| ENSG00000113070 | –0.759564729 | 1.28E−44 | 4.43E−43 | HBEGF |
| ENSG00000135250 | –0.766365183 | 1.37E−44 | 4.74E−43 | SRPK2 |
| ENSG00000106683 | –0.770624188 | 2.57E−44 | 8.9E−43 | LIMK1 |
| ENSG00000214517 | –0.683773756 | 2.59E−44 | 8.93E−43 | PPME1 |
| ENSG00000100479 | –0.857727695 | 2.8E−44 | 9.64E−43 | POLE2 |
| ENSG00000130175 | –0.636724289 | 5.94E−44 | 2.02E−42 | PRKCSH |
| ENSG00000170185 | –0.607338959 | 1.22E−43 | 4.08E−42 | USP38 |
| ENSG00000123908 | –0.609276107 | 1.75E−43 | 5.86E−42 | 37469 |
| ENSG00000064666 | –0.604102394 | 1.93E−43 | 6.43E−42 | CNN2 |
| ENSG00000130024 | –0.624632149 | 4.42E−43 | 1.45E−41 | PHF10 |
| ENSG00000135272 | –0.779876332 | 1.35E−42 | 4.34E−41 | MDFIC |
| ENSG00000138646 | –0.611105347 | 1.87E−42 | 5.96E−41 | HERC5 |
| ENSG00000178904 | –0.821536016 | 4.05E−42 | 1.28E−40 | DPY19L3 |
| ENSG00000116984 | –0.592452679 | 6.84E−42 | 2.15E−40 | MTR |
| ENSG00000151233 | –0.696734208 | 1.24E−41 | 3.89E−40 | GXYLT1 |
| ENSG00000049130 | –1.363755139 | 2.4E−41 | 7.47E−40 | KITLG |
| ENSG00000131943 | –0.819671981 | 4.72E−41 | 1.45E−39 | C19orf12 |
| ENSG00000180730 | –0.972346863 | 6.99E−41 | 2.14E−39 | SHISA2 |
| ENSG00000151239 | –0.646933517 | 1.79E−40 | 5.44E−39 | TWF1 |
| ENSG00000132819 | –0.768136766 | 2.14E−40 | 6.44E−39 | RBM38 |
| ENSG00000163577 | –0.691799144 | 2.24E−40 | 6.7E−39 | EIF5A2 |
| ENSG00000145743 | –0.871378383 | 2.77E−40 | 8.28E−39 | FBXL17 |
| ENSG00000137168 | –0.580135569 | 9.42E−40 | 2.77E−38 | PPIL1 |
| ENSG00000038210 | –0.645756993 | 1.75E−39 | 5.06E−38 | PI4K2B |
| ENSG00000146263 | –0.647666403 | 1.86E−39 | 5.35E−38 | MMS22L |
| ENSG00000116717 | –0.675446031 | 1.93E−39 | 5.56E−38 | GADD45A |
| ENSG00000167842 | –0.621928208 | 1.97E−39 | 5.64E−38 | MIS12 |
| ENSG00000114520 | –0.763326165 | 2.34E−39 | 6.7E−38 | SNX4 |
| ENSG00000154678 | –0.944300821 | 2.5E−39 | 7.13E−38 | PDE1C |
| ENSG00000164402 | –0.637231639 | 4.65E−39 | 1.31E−37 | 39692 |
| ENSG00000162073 | –0.831857412 | 5.7E−39 | 1.6E−37 | PAQR4 |
| ENSG00000144369 | –0.757917112 | 8.73E−39 | 2.43E−37 | FAM171B |
| ENSG00000123572 | –1.320658507 | 8.85E−39 | 2.46E−37 | NRK |
| ENSG00000129691 | –0.662466725 | 1.08E−38 | 2.99E−37 | ASH2L |
| ENSG00000002822 | –0.607342874 | 1.85E−38 | 5.06E−37 | MAD1L1 |
| ENSG00000101412 | –0.797853061 | 2.45E−38 | 6.64E−37 | E2F1 |
| ENSG00000156162 | –0.585388505 | 2.98E−38 | 8.02E−37 | DPY19L4 |
| ENSG00000112144 | –0.601617859 | 3.85E−38 | 1.03E−36 | ICK |
| ENSG00000125734 | –0.622236713 | 3.88E−38 | 1.04E−36 | GPR108 |
| ENSG00000101544 | –0.625762283 | 3.98E−38 | 1.06E−36 | ADNP2 |
| ENSG00000198860 | –0.623117034 | 4.95E−38 | 1.31E−36 | TSEN15 |
| ENSG00000091986 | –0.628100732 | 5.14E−38 | 1.36E−36 | CCDC80 |
| ENSG00000114982 | –0.612077259 | 9.28E−38 | 2.42E−36 | KANSL3 |
| ENSG00000145604 | –0.706553217 | 1.28E−37 | 3.32E−36 | SKP2 |
| ENSG00000187051 | –0.687870702 | 1.56E−37 | 4.04E−36 | RPS19BP1 |
| ENSG00000163626 | –0.701696175 | 2.3E−37 | 5.91E−36 | COX18 |
| ENSG00000125885 | –0.583920903 | 2.3E−37 | 5.91E−36 | MCM8 |
| ENSG00000157557 | –0.662994439 | 4.57E−37 | 1.15E−35 | ETS2 |
| ENSG00000127564 | –0.830827192 | 4.99E−37 | 1.26E−35 | PKMYT1 |
| ENSG00000172260 | –0.882667662 | 5.77E−37 | 1.45E−35 | NEGR1 |
| ENSG00000198478 | –0.913187859 | 7.59E−37 | 1.9E−35 | SH3BGRL2 |
| ENSG00000152455 | –0.651372389 | 9.5E−37 | 2.37E−35 | SUV39H2 |
| ENSG00000181744 | –0.857381192 | 3.74E−36 | 9.25E−35 | C3orf58 |
| ENSG00000105825 | –0.683206742 | 4.19E−36 | 1.03E−34 | TFPI2 |
| ENSG00000185129 | –0.650813837 | 4.66E−36 | 1.14E−34 | PURA |
| ENSG00000131153 | –0.830497048 | 5.14E−36 | 1.26E−34 | GINS2 |
| ENSG00000104361 | –1.128506294 | 9.17E−36 | 2.23E−34 | NIPAL2 |
| ENSG00000142552 | -2.164888397 | 1.32E−35 | 3.17E−34 | RCN3 |
| ENSG00000118971 | –0.828403671 | 1.65E−35 | 3.94E−34 | CCND2 |
| ENSG00000162437 | –0.911533859 | 1.86E−35 | 4.42E−34 | RAVER2 |
| ENSG00000072609 | –0.725569758 | 2.69E−35 | 6.35E−34 | CHFR |
| ENSG00000135750 | –1.097089858 | 2.74E−35 | 6.47E−34 | KCNK1 |
| ENSG00000198108 | –1.330709511 | 3.6E−35 | 8.47E−34 | CHSY3 |
| ENSG00000139438 | –1.36818776 | 5.27E−35 | 1.23E−33 | FAM222A |
| ENSG00000176170 | –1.061371315 | 1.42E−34 | 3.25E−33 | SPHK1 |
| ENSG00000172159 | –0.719708766 | 1.54E−34 | 3.53E−33 | FRMD3 |
| ENSG00000115380 | –0.592899689 | 2.06E−34 | 4.68E−33 | EFEMP1 |
| ENSG00000109511 | –1.823866992 | 2.43E−34 | 5.52E−33 | ANXA10 |
| ENSG00000198780 | –0.894526335 | 2.96E−34 | 6.7E−33 | FAM169A |
| ENSG00000132970 | –0.851435716 | 1.4E−33 | 3.08E−32 | WASF3 |
| ENSG00000136111 | –0.603181977 | 1.96E−33 | 4.27E−32 | TBC1D4 |
| ENSG00000225830 | –0.709999482 | 2.48E−33 | 5.35E−32 | ERCC6 |
| ENSG00000109255 | –1.175751743 | 3.49E−33 | 7.47E−32 | NMU |
| ENSG00000121058 | –0.618925976 | 4.93E−33 | 1.05E−31 | COIL |
| ENSG00000164508 | –1.194336423 | 7.79E−33 | 1.64E−31 | HIST1H2AA |
| ENSG00000123570 | –1.292915486 | 7.81E−33 | 1.65E−31 | RAB9B |
| ENSG00000176834 | –0.652497435 | 8.39E−33 | 1.76E−31 | VSIG10 |
| ENSG00000186416 | –0.589099004 | 8.99E−33 | 1.89E−31 | NKRF |
| ENSG00000106789 | –0.67263713 | 1.3E−32 | 2.72E−31 | CORO2A |
| ENSG00000172379 | –0.788111039 | 1.74E−32 | 3.62E−31 | ARNT2 |
| ENSG00000112592 | –0.651455796 | 1.74E−32 | 3.62E−31 | TBP |
| ENSG00000159259 | –0.683175408 | 3.54E−32 | 7.29E−31 | CHAF1B |
| ENSG00000226742 | –1.146129313 | 6.12E−32 | 1.24E−30 | HSBP1L1 |
| ENSG00000129173 | –0.786345274 | 7.03E−32 | 1.43E−30 | E2F8 |
| ENSG00000170689 | –0.891856708 | 1.2E−31 | 2.41E−30 | HOXB9 |
| ENSG00000112294 | –1.176881508 | 1.51E−31 | 2.99E−30 | ALDH5A1 |
| ENSG00000174021 | –0.597920622 | 2E−31 | 3.95E−30 | GNG5 |
| ENSG00000164199 | –0.796504879 | 2.27E−31 | 4.46E−30 | ADGRV1 |
| ENSG00000153037 | –0.672607947 | 3.19E−31 | 6.24E−30 | SRP19 |
| ENSG00000170734 | –0.586607195 | 3.55E−31 | 6.92E−30 | POLH |
| ENSG00000179454 | –0.750172183 | 3.78E−31 | 7.35E−30 | KLHL28 |
| ENSG00000168564 | –0.588203808 | 4.93E−31 | 9.54E−30 | CDKN2AIP |
| ENSG00000112031 | –0.762313636 | 6.71E−31 | 1.29E−29 | MTRF1L |
| ENSG00000180773 | –0.580924737 | 8.38E−31 | 1.61E−29 | SLC36A4 |
| ENSG00000164161 | –1.167949712 | 8.5E−31 | 1.63E−29 | HHIP |
| ENSG00000170085 | –1.010340626 | 8.52E−31 | 1.63E−29 | SIMC1 |
| ENSG00000213047 | –0.852741909 | 9.45E−31 | 1.8E−29 | DENND1B |
| ENSG00000178695 | –0.608721797 | 1.3E−30 | 2.45E−29 | KCTD12 |
| ENSG00000162694 | –0.581927567 | 1.4E−30 | 2.63E−29 | EXTL2 |
| ENSG00000153993 | –1.881252549 | 2.69E−30 | 5.02E−29 | SEMA3D |
| ENSG00000121236 | –0.974421588 | 3.3E−30 | 6.14E−29 | TRIM6 |
| ENSG00000157869 | –0.655287956 | 4.29E−30 | 7.95E−29 | RAB28 |
| ENSG00000135966 | –0.618380605 | 6.88E−30 | 1.26E−28 | TGFBRAP1 |
| ENSG00000121005 | –1.212412618 | 8.16E−30 | 1.49E−28 | CRISPLD1 |
| ENSG00000067177 | –0.643679511 | 1.08E−29 | 1.95E−28 | PHKA1 |
| ENSG00000104643 | –0.792913093 | 1.57E−29 | 2.8E−28 | MTMR9 |
| ENSG00000163818 | –0.711190839 | 1.7E−29 | 3.03E−28 | LZTFL1 |
| ENSG00000145545 | –0.582387628 | 2.79E−29 | 4.91E−28 | SRD5A1 |
| ENSG00000143376 | –0.70665512 | 3.54E−29 | 6.2E−28 | SNX27 |
| ENSG00000276043 | –0.609555567 | 5.15E−29 | 8.95E−28 | UHRF1 |
| ENSG00000176406 | –1.738298125 | 1.08E−28 | 1.85E−27 | RIMS2 |
| ENSG00000101871 | –0.597281832 | 1.53E−28 | 2.6E−27 | MID1 |
| ENSG00000197223 | –0.808372429 | 4.86E−28 | 8.12E−27 | C1D |
| ENSG00000214357 | –0.84131965 | 5.59E−28 | 9.31E−27 | NEURL1B |
| ENSG00000137462 | –1.051657592 | 7.74E−28 | 1.28E−26 | TLR2 |
| ENSG00000040199 | –0.614077873 | 1.01E−27 | 1.65E−26 | PHLPP2 |
| ENSG00000135185 | –0.890686199 | 1.23E−27 | 2.01E−26 | TMEM243 |
| ENSG00000128591 | –0.771115601 | 1.28E−27 | 2.1E−26 | FLNC |
| ENSG00000113356 | –0.70152701 | 1.33E−27 | 2.17E−26 | POLR3G |
| ENSG00000112218 | –1.31975116 | 1.38E−27 | 2.25E−26 | GPR63 |
| ENSG00000166250 | –0.584854006 | 1.41E−27 | 2.3E−26 | CLMP |
| ENSG00000123892 | –0.709596504 | 2.05E−27 | 3.31E−26 | RAB38 |
| ENSG00000126215 | –0.59285666 | 3.95E−27 | 6.32E−26 | XRCC3 |
| ENSG00000111331 | –0.709429851 | 4.04E−27 | 6.45E−26 | OAS3 |
| ENSG00000179348 | –0.959652747 | 5.77E−27 | 9.16E−26 | GATA2 |
| ENSG00000135045 | –0.580419299 | 7.97E−27 | 1.25E−25 | C9orf40 |
| ENSG00000128923 | –0.640712113 | 9.41E−27 | 1.48E−25 | MINDY2 |
| ENSG00000151150 | -1.00381512 | 1.76E−26 | 2.72E−25 | ANK3 |
| ENSG00000167513 | –0.659977018 | 3.21E−26 | 4.9E−25 | CDT1 |
| ENSG00000084676 | –0.598801314 | 4.32E−26 | 6.55E−25 | NCOA1 |
| ENSG00000174405 | –0.594636053 | 7.09E−26 | 1.06E−24 | LIG4 |
| ENSG00000134056 | –0.815574482 | 1.05E−25 | 1.57E−24 | MRPS36 |
| ENSG00000137965 | –0.582015156 | 1.21E−25 | 1.79E−24 | IFI44 |
| ENSG00000157510 | –0.701292502 | 1.38E−25 | 2.04E−24 | AFAP1L1 |
| ENSG00000173083 | –0.723996154 | 2.26E−25 | 3.32E−24 | HPSE |
| ENSG00000175197 | –0.673342058 | 5.14E−25 | 7.46E−24 | DDIT3 |
| ENSG00000108960 | –0.58542233 | 6.22E−25 | 8.99E−24 | MMD |
| ENSG00000100916 | –0.624568654 | 1.56E−24 | 2.22E−23 | BRMS1L |
| ENSG00000181222 | –0.726152084 | 1.6E−24 | 2.27E−23 | POLR2A |
| ENSG00000187123 | –0.956022878 | 2.07E−24 | 2.9E−23 | LYPD6 |
| ENSG00000124788 | –0.590242443 | 2.42E−24 | 3.37E−23 | ATXN1 |
| ENSG00000129354 | –0.691992358 | 2.83E−24 | 3.93E−23 | AP1M2 |
| ENSG00000078401 | –0.592475007 | 4E−24 | 5.52E−23 | EDN1 |
| ENSG00000121316 | –1.050558857 | 6.3E−24 | 8.57E−23 | PLBD1 |
| ENSG00000121211 | –0.582342226 | 6.37E−24 | 8.66E−23 | MND1 |
| ENSG00000135776 | –0.585970667 | 1.14E−23 | 1.52E−22 | ABCB10 |
| ENSG00000162599 | –0.648028127 | 1.15E−23 | 1.53E−22 | NFIA |
| ENSG00000143195 | –0.652324164 | 1.34E−23 | 1.78E−22 | ILDR2 |
| ENSG00000182010 | –0.810382882 | 1.94E−23 | 2.57E−22 | RTKN2 |
| ENSG00000196083 | –0.719308824 | 1.98E−23 | 2.62E−22 | IL1RAP |
| ENSG00000135698 | –0.685340754 | 2.6E−23 | 3.41E−22 | MPHOSPH6 |
| ENSG00000122435 | –0.73861782 | 4.38E−23 | 5.68E−22 | TRMT13 |
| ENSG00000099256 | –0.742089516 | 5.07E−23 | 6.54E−22 | PRTFDC1 |
| ENSG00000173926 | –0.725353141 | 5.49E−23 | 7.06E−22 | MARCH3 |
| ENSG00000171016 | –0.635078785 | 6.62E−23 | 8.48E−22 | PYGO1 |
| ENSG00000166908 | –0.610595678 | 1.01E−22 | 1.28E−21 | PIP4K2C |
| ENSG00000254535 | –1.198870344 | 1.2E−22 | 1.52E−21 | PABPC4L |
| ENSG00000117481 | –0.628980367 | 1.35E−22 | 1.69E−21 | NSUN4 |
| ENSG00000049192 | –0.719589951 | 1.56E−22 | 1.96E−21 | ADAMTS6 |
| ENSG00000133739 | –0.688520659 | 1.78E−22 | 2.23E−21 | LRRCC1 |
| ENSG00000083454 | –1.060807972 | 1.96E−22 | 2.44E−21 | P2RX5 |
| ENSG00000154319 | –0.887801464 | 2.23E−22 | 2.77E−21 | FAM167A |
| ENSG00000023892 | –0.675886174 | 2.38E−22 | 2.95E−21 | DEF6 |
| ENSG00000138795 | –0.843440962 | 3.04E−22 | 3.75E−21 | LEF1 |
| ENSG00000079156 | –0.842535226 | 3.07E−22 | 3.78E−21 | OSBPL6 |
| ENSG00000170396 | –0.805881831 | 3.81E−22 | 4.67E−21 | ZNF804A |
| ENSG00000135211 | –0.747849147 | 3.86E−22 | 4.73E−21 | TMEM60 |
| ENSG00000165323 | –0.725561816 | 6.07E−22 | 7.35E−21 | FAT3 |
| ENSG00000152495 | –0.751284296 | 6.71E−22 | 8.1E−21 | CAMK4 |
| ENSG00000124813 | –0.681157995 | 7.41E−22 | 8.93E−21 | RUNX2 |
| ENSG00000109738 | –0.716767424 | 1.08E−21 | 1.3E−20 | GLRB |
| ENSG00000082438 | –1.400231606 | 1.49E−21 | 1.77E−20 | COBLL1 |
| ENSG00000138463 | –0.692070234 | 1.91E−21 | 2.25E−20 | DIRC2 |
| ENSG00000148680 | –0.6672409 | 1.96E−21 | 2.31E−20 | HTR7 |
| ENSG00000169750 | –0.921289251 | 3.24E−21 | 3.75E−20 | RAC3 |
| ENSG00000114698 | –1.130704953 | 3.84E−21 | 4.42E−20 | PLSCR4 |
| ENSG00000112319 | –0.822410931 | 6.03E−21 | 6.89E−20 | EYA4 |
| ENSG00000115392 | –0.58789192 | 6.16E−21 | 7.02E−20 | FANCL |
| ENSG00000185818 | –1.163457685 | 6.72E−21 | 7.63E−20 | NAT8L |
| ENSG00000143942 | –0.728072397 | 6.72E−21 | 7.63E−20 | CHAC2 |
| ENSG00000185862 | –0.776759935 | 1.13E−20 | 1.27E−19 | EVI2B |
| ENSG00000164619 | –1.07660645 | 1.2E−20 | 1.35E−19 | BMPER |
| ENSG00000058091 | –0.700546181 | 2.42E−20 | 2.67E−19 | CDK14 |
| ENSG00000175556 | –0.6405596 | 2.9E−20 | 3.19E−19 | LONRF3 |
| ENSG00000176641 | –0.725354894 | 2.93E−20 | 3.22E−19 | RNF152 |
| ENSG00000135414 | –0.646867873 | 6.7E−20 | 7.23E−19 | GDF11 |
| ENSG00000134569 | –0.789074753 | 6.71E−20 | 7.24E−19 | LRP4 |
| ENSG00000080823 | –0.602706451 | 7.6E−20 | 8.18E−19 | MOK |
| ENSG00000188312 | –0.722540228 | 8.34E−20 | 8.96E−19 | CENPP |
| ENSG00000163568 | –0.962591724 | 1.46E−19 | 1.56E−18 | AIM2 |
| ENSG00000160392 | –0.610014247 | 2.03E−19 | 2.14E−18 | C19orf47 |
| ENSG00000119042 | –0.612708814 | 3.03E−19 | 3.17E−18 | SATB2 |
| ENSG00000166532 | –0.673619231 | 3.77E−19 | 3.91E−18 | RIMKLB |
| ENSG00000100483 | –0.677631261 | 8.66E−19 | 8.8E−18 | VCPKMT |
| ENSG00000080493 | –1.327083425 | 2.48E−18 | 2.44E−17 | SLC4A4 |
| ENSG00000248487 | –0.969369054 | 2.57E−18 | 2.52E−17 | ABHD14A |
| ENSG00000153956 | –1.026853205 | 3.17E−18 | 3.09E−17 | CACNA2D1 |
| ENSG00000204767 | –0.615679238 | 4.62E−18 | 4.46E−17 | FAM196B |
| ENSG00000166582 | –0.670746514 | 5.34E−18 | 5.15E−17 | CENPV |
| ENSG00000121578 | –0.646426771 | 5.55E−18 | 5.33E−17 | B4GALT4 |
| ENSG00000175183 | –0.603690194 | 7.81E−18 | 7.4E−17 | CSRP2 |
| ENSG00000134508 | –0.584140679 | 7.84E−18 | 7.42E−17 | CABLES1 |
| ENSG00000154237 | –0.591563665 | 1.09E−17 | 1.02E−16 | LRRK1 |
| ENSG00000118257 | –0.661860339 | 1.27E−17 | 1.19E−16 | NRP2 |
| ENSG00000136160 | –0.728515383 | 1.31E−17 | 1.22E−16 | EDNRB |
| ENSG00000172889 | –0.78025385 | 2.36E−17 | 2.17E−16 | EGFL7 |
| ENSG00000170498 | –1.543019875 | 2.8E−17 | 2.56E−16 | KISS1 |
| ENSG00000112394 | –1.707112548 | 4.7E−17 | 4.24E−16 | SLC16A10 |
| ENSG00000151229 | –0.746387031 | 6.14E−17 | 5.48E−16 | SLC2A13 |
| ENSG00000091129 | –1.696794072 | 6.83E−17 | 6.05E−16 | NRCAM |
| ENSG00000171388 | –1.043380952 | 8.76E−17 | 7.71E−16 | APLN |
| ENSG00000228300 | –0.706210371 | 9.96E−17 | 8.73E−16 | C19orf24 |
| ENSG00000149970 | –0.855419499 | 1.04E−16 | 9.09E−16 | CNKSR2 |
| ENSG00000162711 | –0.776561197 | 1.06E−16 | 9.24E−16 | NLRP3 |
| ENSG00000156795 | –0.622341733 | 1.18E−16 | 1.03E−15 | WDYHV1 |
| ENSG00000169684 | –0.74358858 | 2.58E−16 | 2.21E−15 | CHRNA5 |
| ENSG00000197147 | –0.671838185 | 2.76E−16 | 2.36E−15 | LRRC8B |
| ENSG00000206538 | –0.84083006 | 3.21E−16 | 2.72E−15 | VGLL3 |
| ENSG00000133101 | –0.714667229 | 3.28E−16 | 2.78E−15 | CCNA1 |
| ENSG00000107518 | –0.601853491 | 3.48E−16 | 2.94E−15 | ATRNL1 |
| ENSG00000155495 | –1.359915587 | 4.29E−16 | 3.61E−15 | MAGEC1 |
| ENSG00000181754 | –0.612040358 | 5.47E−16 | 4.56E−15 | AMIGO1 |
| ENSG00000168685 | –0.629731176 | 6.4E−16 | 5.31E−15 | IL7R |
| ENSG00000154274 | –0.654221576 | 6.52E−16 | 5.41E−15 | C4orf19 |
| ENSG00000112425 | –0.870586941 | 6.91E−16 | 5.72E−15 | EPM2A |
| ENSG00000128610 | –1.168896695 | 6.95E−16 | 5.75E−15 | FEZF1 |
| ENSG00000154914 | –0.915019891 | 7.14E−16 | 5.9E−15 | USP43 |
| ENSG00000082014 | –1.08393885 | 8.58E−16 | 7.05E−15 | SMARCD3 |
| ENSG00000105929 | –0.876658986 | 3.48E−15 | 2.74E−14 | ATP6V0A4 |
| ENSG00000169570 | –0.772019802 | 4.97E−15 | 3.88E−14 | DTWD2 |
| ENSG00000131480 | –1.0578589 | 7.95E−15 | 6.09E−14 | AOC2 |
| ENSG00000127311 | –0.757865784 | 8.99E−15 | 6.87E−14 | HELB |
| ENSG00000169851 | –0.775603189 | 9.22E−15 | 7.04E−14 | PCDH7 |
| ENSG00000026103 | –0.72906724 | 1.21E−14 | 9.14E−14 | FAS |
| ENSG00000235194 | –0.79485048 | 3.52E−14 | 2.56E−13 | PPP1R3E |
| ENSG00000105894 | –0.782561566 | 3.9E−14 | 2.83E−13 | PTN |
| ENSG00000162545 | –1.145665693 | 4.19E−14 | 3.04E−13 | CAMK2N1 |
| ENSG00000150764 | –0.704440404 | 4.83E−14 | 3.49E−13 | DIXDC1 |
| ENSG00000018236 | –1.47880204 | 4.91E−14 | 3.53E−13 | CNTN1 |
| ENSG00000187231 | –0.644113059 | 5.52E−14 | 3.97E−13 | SESTD1 |
| ENSG00000110852 | –0.662960751 | 6.52E−14 | 4.66E−13 | CLEC2B |
| ENSG00000170577 | –0.939095059 | 7.26E−14 | 5.16E−13 | SIX2 |
| ENSG00000175893 | –0.614669715 | 1.1E−13 | 7.72E−13 | ZDHHC21 |
| ENSG00000084734 | –1.185795317 | 1.34E−13 | 9.33E−13 | GCKR |
| ENSG00000166689 | –0.674253903 | 1.62E−13 | 1.13E−12 | PLEKHA7 |
| ENSG00000152669 | –0.704229096 | 1.78E−13 | 1.23E−12 | CCNO |
| ENSG00000069667 | –0.817249087 | 1.88E−13 | 1.3E−12 | RORA |
| ENSG00000167941 | –1.822789729 | 2.12E−13 | 1.46E−12 | SOST |
| ENSG00000152270 | –0.627826609 | 2.26E−13 | 1.55E−12 | PDE3B |
| ENSG00000152527 | –0.65848094 | 3.22E−13 | 2.19E−12 | PLEKHH2 |
| ENSG00000070759 | –0.583708292 | 4.71E−13 | 3.16E−12 | TESK2 |
| ENSG00000068615 | –0.935596963 | 4.71E−13 | 3.16E−12 | REEP1 |
| ENSG00000163328 | –1.081666267 | 5.61E−13 | 3.75E−12 | GPR155 |
| ENSG00000005249 | –0.989574304 | 7.85E−13 | 5.19E−12 | PRKAR2B |
| ENSG00000120279 | –1.086387629 | 8.3E−13 | 5.48E−12 | MYCT1 |
| ENSG00000140450 | –0.633642782 | 8.3E−13 | 5.48E−12 | ARRDC4 |
| ENSG00000197415 | –0.649632825 | 1.08E−12 | 7.02E−12 | VEPH1 |
| ENSG00000170417 | –0.93939747 | 1.48E−12 | 9.5E−12 | TMEM182 |
| ENSG00000126803 | –1.11469054 | 1.54E−12 | 9.92E−12 | HSPA2 |
| ENSG00000170899 | –0.684484123 | 1.7E−12 | 1.09E−11 | GSTA4 |
| ENSG00000168077 | –1.165454147 | 2.15E−12 | 1.37E−11 | SCARA3 |
| ENSG00000181418 | –1.58931145 | 2.26E−12 | 1.44E−11 | DDN |
| ENSG00000154040 | –0.938234977 | 2.4E−12 | 1.52E−11 | CABYR |
| ENSG00000188060 | –1.258637511 | 2.57E−12 | 1.62E−11 | RAB42 |
| ENSG00000108852 | –0.843142689 | 2.7E−12 | 1.71E−11 | MPP2 |
| ENSG00000166823 | –0.986678684 | 4.18E−12 | 2.61E−11 | MESP1 |
| ENSG00000104154 | –0.597478232 | 4.45E−12 | 2.77E−11 | SLC30A4 |
| ENSG00000106025 | –0.593643936 | 6.72E−12 | 4.1E−11 | TSPAN12 |
| ENSG00000163472 | –0.745176085 | 1.25E−11 | 7.49E−11 | TMEM79 |
| ENSG00000178343 | –0.948648227 | 1.45E−11 | 8.61E−11 | SHISA3 |
| ENSG00000140044 | –0.769542054 | 1.64E−11 | 9.7E−11 | JDP2 |
| ENSG00000166813 | –0.590817747 | 1.72E−11 | 1.01E−10 | KIF7 |
| ENSG00000143382 | –1.022866716 | 1.85E−11 | 1.09E−10 | ADAMTSL4 |
| ENSG00000280670 | –0.649067763 | 2.08E−11 | 1.22E−10 | CCDC163 |
| ENSG00000143320 | –0.873020398 | 2.19E−11 | 1.28E−10 | CRABP2 |
| ENSG00000090020 | –0.695130894 | 2.54E−11 | 1.47E−10 | SLC9A1 |
| ENSG00000116141 | –0.722567149 | 2.54E−11 | 1.48E−10 | MARK1 |
| ENSG00000165655 | –0.867909098 | 2.67E−11 | 1.54E−10 | ZNF503 |
| ENSG00000128694 | –0.624482372 | 2.9E−11 | 1.67E−10 | OSGEPL1 |
| ENSG00000135378 | –0.665705024 | 4.18E−11 | 2.39E−10 | PRRG4 |
| ENSG00000135333 | –1.203520059 | 4.22E−11 | 2.42E−10 | EPHA7 |
| ENSG00000008277 | –0.724949048 | 4.27E−11 | 2.44E−10 | ADAM22 |
| ENSG00000134321 | –0.688580763 | 4.43E−11 | 2.53E−10 | RSAD2 |
| ENSG00000196843 | –0.607550863 | 4.54E−11 | 2.59E−10 | ARID5A |
| ENSG00000186205 | –0.637588673 | 4.66E−11 | 2.65E−10 | MARC1 |
| ENSG00000171365 | –0.672055612 | 5.69E−11 | 3.21E−10 | CLCN5 |
| ENSG00000128872 | –0.584078549 | 5.78E−11 | 3.26E−10 | TMOD2 |
| ENSG00000221890 | –0.782195142 | 6.9E−11 | 3.84E−10 | NPTXR |
| ENSG00000234602 | –0.995961314 | 7.14E−11 | 3.97E−10 | MCIDAS |
| ENSG00000157303 | –0.96944952 | 7.23E−11 | 4.02E−10 | SUSD3 |
| ENSG00000164414 | –0.591096299 | 7.69E−11 | 4.27E−10 | SLC35A1 |
| ENSG00000178401 | –0.94668282 | 7.84E−11 | 4.35E−10 | DNAJC22 |
| ENSG00000139973 | –0.875379862 | 8.35E−11 | 4.61E−10 | SYT16 |
| ENSG00000182118 | –0.593515976 | 8.35E−11 | 4.61E−10 | FAM89A |
| ENSG00000184254 | –0.62334185 | 8.41E−11 | 4.64E−10 | ALDH1A3 |
| ENSG00000130244 | –0.638701689 | 9.48E−11 | 5.2E−10 | FAM98C |
| ENSG00000011332 | –0.682979301 | 1.07E−10 | 5.86E−10 | DPF1 |
| ENSG00000151468 | –0.953297034 | 1.3E−10 | 7.06E−10 | CCDC3 |
| ENSG00000162999 | –1.755455161 | 1.6E−10 | 8.58E−10 | DUSP19 |
| ENSG00000116396 | –0.896224276 | 1.67E−10 | 8.97E−10 | KCNC4 |
| ENSG00000105290 | –0.98909701 | 1.99E−10 | 1.06E−09 | APLP1 |
| ENSG00000184838 | –0.709702383 | 2.05E−10 | 1.09E−09 | PRR16 |
| ENSG00000131471 | –1.508965698 | 2.07E−10 | 1.1E−09 | AOC3 |
| ENSG00000141404 | –1.039495113 | 2.15E−10 | 1.14E−09 | GNAL |
| ENSG00000125384 | –1.217017506 | 2.82E−10 | 1.47E−09 | PTGER2 |
| ENSG00000061918 | –0.775981343 | 3.62E−10 | 1.87E−09 | GUCY1B3 |
| ENSG00000156172 | –0.844143968 | 3.84E−10 | 1.98E−09 | C8orf37 |
| ENSG00000100092 | –0.612795457 | 5.7E−10 | 2.89E−09 | SH3BP1 |
| ENSG00000175175 | –0.668290059 | 6.19E−10 | 3.13E−09 | PPM1E |
| ENSG00000150893 | –0.822911314 | 7.32E−10 | 3.67E−09 | FREM2 |
| ENSG00000173890 | –0.725043577 | 9.66E−10 | 4.79E−09 | GPR160 |
| ENSG00000129595 | –0.756168514 | 1.32E−09 | 6.44E−09 | EPB41L4A |
| ENSG00000180739 | –1.212317922 | 1.43E−09 | 0.000000007 | S1PR5 |
| ENSG00000104369 | –0.713893389 | 1.49E−09 | 7.28E−09 | JPH1 |
| ENSG00000163884 | –1.001721809 | 2.22E−09 | 1.06E−08 | KLF15 |
| ENSG00000172543 | –1.443971336 | 2.6E−09 | 1.23E−08 | CTSW |
| ENSG00000034533 | –0.602893943 | 2.98E−09 | 1.41E−08 | ASTE1 |
| ENSG00000016391 | –1.454323229 | 0.000000003 | 1.42E−08 | CHDH |
| ENSG00000163412 | –1.455095924 | 3.22E−09 | 1.51E−08 | EIF4E3 |
| ENSG00000130821 | –0.69419495 | 3.28E−09 | 1.54E−08 | SLC6A8 |
| ENSG00000135929 | –0.792267569 | 3.68E−09 | 1.72E−08 | CYP27A1 |
| ENSG00000174370 | –1.005785086 | 4.1E−09 | 1.91E−08 | C11orf45 |
| ENSG00000025423 | –0.998326574 | 4.63E−09 | 2.14E−08 | HSD17B6 |
| ENSG00000111199 | –1.268393502 | 5.71E−09 | 2.62E−08 | TRPV4 |
| ENSG00000155974 | –0.67183537 | 6.04E−09 | 2.76E−08 | GRIP1 |
| ENSG00000147606 | –1.027590206 | 6.16E−09 | 2.81E−08 | SLC26A7 |
| ENSG00000065675 | –1.501715725 | 6.81E−09 | 3.09E−08 | PRKCQ |
| ENSG00000179841 | –0.749380697 | 7.31E−09 | 3.31E−08 | AKAP5 |
| ENSG00000260822 | –0.582080039 | 0.000000227 | 0.000000864 | AC004656.1 |
| ENSG00000156265 | –0.79220517 | 8.14E−09 | 3.67E−08 | MAP3K7CL |
| ENSG00000079215 | –0.679149885 | 9.51E−09 | 4.25E−08 | SLC1A3 |
| ENSG00000221944 | –0.600202219 | 9.68E−09 | 4.32E−08 | TIGD1 |
| ENSG00000101680 | –0.905321065 | 1.11E−08 | 4.94E−08 | LAMA1 |
| ENSG00000126785 | –0.618262682 | 1.15E−08 | 5.08E−08 | RHOJ |
| ENSG00000205628 | –0.582080325 | 0.000251743 | 0.000637414 | LINC01446 |
| ENSG00000223773 | –0.583523523 | 0.008232673 | 0.015658057 | CD99P1 |
| ENSG00000282826 | –0.583636435 | 0.008093028 | 0.015414788 | FRG1CP |
| ENSG00000241544 | –0.586635592 | 0.001586262 | 0.0034698 | LINC02029 |
| ENSG00000122420 | –0.7597314 | 1.73E−08 | 7.48E−08 | PTGFR |
| ENSG00000111879 | –0.681429858 | 1.79E−08 | 7.72E−08 | FAM184A |
| ENSG00000186162 | –0.589358402 | 4.94E−16 | 4.13E−15 | CIDECP |
| ENSG00000225793 | –0.598603858 | 0.002182824 | 0.004653786 | AL080250.1 |
| ENSG00000164674 | –0.834186124 | 2.35E−08 | 0.0000001 | SYTL3 |
| ENSG00000163710 | –0.74510529 | 2.55E−08 | 0.000000108 | PCOLCE2 |
| ENSG00000138741 | –1.124387768 | 2.62E−08 | 0.000000111 | TRPC3 |
| ENSG00000147003 | –0.945694847 | 2.92E−08 | 0.000000123 | TMEM27 |
| ENSG00000168874 | –1.171454266 | 3.51E−08 | 0.000000147 | ATOH8 |
| ENSG00000100077 | –0.645354414 | 4.29E−08 | 0.000000178 | GRK3 |
| ENSG00000206652 | –0.600319045 | 0.0041797 | 0.008468391 | RNU1-1 |
| ENSG00000056998 | –1.159597278 | 4.96E−08 | 0.000000204 | GYG2 |
| ENSG00000110811 | –1.364088188 | 5.18E−08 | 0.000000212 | P3H3 |
| ENSG00000234155 | –0.603589219 | 0.0000182 | 0.0000545 | AL135903.2 |
| ENSG00000132718 | –0.675745243 | 5.96E−08 | 0.000000243 | SYT11 |
| ENSG00000262202 | –0.611008242 | 0.00012338 | 0.000327359 | AC007952.4 |
| ENSG00000263535 | –0.612042556 | 0.004466662 | 0.009001187 | AC134669.1 |
| ENSG00000260966 | –0.619021953 | 0.000673059 | 0.001578768 | AP001486.2 |
| ENSG00000100321 | –1.052508227 | 0.000000103 | 0.000000411 | SYNGR1 |
| ENSG00000201558 | –0.622105346 | 6.01E−11 | 3.37E−10 | RNVU1-6 |
| ENSG00000139173 | –1.036479375 | 0.000000114 | 0.00000045 | TMEM117 |
| ENSG00000273320 | –0.627836339 | 0.001733515 | 0.003763066 | AC007032.1 |
| ENSG00000175764 | –0.612825531 | 0.000000149 | 0.000000581 | TTLL11 |
| ENSG00000196405 | –0.658711721 | 0.000000158 | 0.000000615 | EVL |
| ENSG00000169085 | –0.59715125 | 0.000000164 | 0.000000633 | C8orf46 |
| ENSG00000255389 | –0.628197788 | 0.004805773 | 0.009610814 | Z97989.1 |
| ENSG00000272821 | –0.628905849 | 0.00000876 | 0.0000274 | U62317.3 |
| ENSG00000222365 | –0.632247461 | 0.00000032 | 0.0000012 | SNORD12B |
| ENSG00000245146 | –0.63578959 | 0.002320634 | 0.004924648 | LINC01024 |
| ENSG00000184271 | –0.985548652 | 0.000000242 | 0.000000921 | POU6F1 |
| ENSG00000027075 | –0.603546465 | 0.000000268 | 0.00000101 | PRKCH |
| ENSG00000249456 | –0.640283418 | 0.002467147 | 0.00521089 | AL731577.2 |
| ENSG00000159263 | –0.785381395 | 0.000000284 | 0.00000107 | SIM2 |
| ENSG00000268001 | –0.641308425 | 0.000000459 | 0.00000168 | CARD8-AS1 |
| ENSG00000272779 | –0.642724602 | 3.69E−15 | 2.9E−14 | AC245060.4 |
| ENSG00000261175 | –0.644864636 | 0.001089512 | 0.00245603 | LINC02188 |
| ENSG00000259959 | –0.649511362 | 0.00000882 | 0.0000276 | AC107068.1 |
| ENSG00000204228 | –0.962404554 | 0.000000335 | 0.00000125 | HSD17B8 |
| ENSG00000114270 | –0.589453951 | 0.00000035 | 0.0000013 | COL7A1 |
| ENSG00000226887 | –1.126238941 | 0.000000395 | 0.00000146 | ERVMER34-1 |
| ENSG00000117425 | –0.660152077 | 0.000000436 | 0.0000016 | PTCH2 |
| ENSG00000250299 | –0.650539806 | 0.000742332 | 0.001725842 | MRPS31P4 |
| ENSG00000055732 | –0.706315288 | 0.000000475 | 0.00000174 | MCOLN3 |
| ENSG00000233382 | –0.654372192 | 0.001896591 | 0.004089349 | NKAPP1 |
| ENSG00000175745 | –0.605230114 | 0.000000521 | 0.0000019 | NR2F1 |
| ENSG00000232093 | –0.658713398 | 0.004308088 | 0.008706828 | DCST1-AS1 |
| ENSG00000165716 | –0.84644461 | 0.000000701 | 0.00000252 | FAM69B |
| ENSG00000131187 | –0.649231899 | 0.00000073 | 0.00000261 | F12 |
| ENSG00000170775 | –0.603129994 | 0.000000804 | 0.00000286 | GPR37 |
| ENSG00000164749 | –0.714196763 | 0.000000822 | 0.00000292 | HNF4G |
| ENSG00000256663 | –0.661542619 | 0.00000329 | 0.0000109 | AC112777.1 |
| ENSG00000228742 | –0.662633295 | 0.000530017 | 0.001268157 | AC002384.1 |
| ENSG00000198515 | –1.13484411 | 0.00000108 | 0.00000381 | CNGA1 |
| ENSG00000127324 | –0.591605084 | 0.00000113 | 0.00000397 | TSPAN8 |
| ENSG00000237870 | –0.666779213 | 5.37E−08 | 0.00000022 | AC073130.1 |
| ENSG00000145040 | –0.999709672 | 0.00000138 | 0.0000048 | UCN2 |
| ENSG00000197959 | –0.728336114 | 0.00000158 | 0.00000543 | DNM3 |
| ENSG00000237187 | –0.673528302 | 1.09E−14 | 8.29E−14 | NR2F1-AS1 |
| ENSG00000158050 | –0.888282466 | 0.00000182 | 0.0000062 | DUSP2 |
| ENSG00000272269 | –0.674471909 | 0.000000168 | 0.000000648 | AL138724.1 |
| ENSG00000162733 | –0.85420253 | 0.00000199 | 0.00000675 | DDR2 |
| ENSG00000170458 | –0.753322548 | 0.00000203 | 0.00000689 | CD14 |
| ENSG00000275131 | –0.675077093 | 8.09E−09 | 3.65E−08 | AC241952.1 |
| ENSG00000246228 | –0.676059031 | 3.1E−20 | 3.4E−19 | CASC8 |
| ENSG00000254887 | –0.676505955 | 0.001307319 | 0.002905128 | AC010247.1 |
| ENSG00000230445 | –0.683831123 | 0.0000288 | 0.0000842 | LRRC37A6P |
| ENSG00000204335 | –0.655622518 | 0.00000381 | 0.0000125 | SP5 |
| ENSG00000121207 | –1.100166986 | 0.00000424 | 0.0000138 | LRAT |
| ENSG00000151687 | –0.657707604 | 0.00000479 | 0.0000156 | ANKAR |
| ENSG00000153162 | –0.713261029 | 0.00000516 | 0.0000166 | BMP6 |
| ENSG00000120693 | –0.923295877 | 0.0000056 | 0.000018 | SMAD9 |
| ENSG00000169330 | –0.583313432 | 0.00000629 | 0.0000201 | KIAA1024 |
| ENSG00000250731 | –0.68687386 | 0.000258139 | 0.000651726 | TPM3P6 |
| ENSG00000130675 | –0.922679936 | 0.00000653 | 0.0000208 | MNX1 |
| ENSG00000117318 | –0.846439456 | 0.00000656 | 0.0000209 | ID3 |
| ENSG00000206052 | –0.91285047 | 0.00000696 | 0.0000221 | DOK6 |
| ENSG00000119699 | –0.79719179 | 0.00000704 | 0.0000223 | TGFB3 |
| ENSG00000100867 | –0.695196402 | 0.00000731 | 0.0000231 | DHRS2 |
| ENSG00000244486 | –0.991783568 | 0.00000738 | 0.0000233 | SCARF2 |
| ENSG00000166963 | –1.102639672 | 0.00000789 | 0.0000249 | MAP1A |
| ENSG00000254615 | –0.690954391 | 9.46E−10 | 4.7E−09 | AC027031.2 |
| ENSG00000163009 | –0.691160202 | 0.000483706 | 0.0011635 | C2orf48 |
| ENSG00000109705 | –0.700668566 | 0.00000911 | 0.0000284 | NKX3-2 |
| ENSG00000178662 | –0.707748281 | 0.00000999 | 0.0000311 | CSRNP3 |
| ENSG00000248121 | –0.691464153 | 0.001506607 | 0.003312113 | SMURF2P1 |
| ENSG00000254893 | –0.826566134 | 0.0000103 | 0.0000321 | AC113404.3 |
| ENSG00000164841 | –0.622993759 | 0.0000104 | 0.0000323 | TMEM74 |
| ENSG00000215210 | –0.702924697 | 0.001207828 | 0.002698653 | RBMXP2 |
| ENSG00000267383 | –0.703518554 | 0.0000101 | 0.0000314 | AC011447.3 |
| ENSG00000277072 | –0.70804624 | 0.000396301 | 0.000965996 | STAG3L2 |
| ENSG00000111012 | –1.073265175 | 0.0000114 | 0.000035 | CYP27B1 |
| ENSG00000147036 | –0.759385987 | 0.0000114 | 0.000035 | LANCL3 |
| ENSG00000186019 | –0.7083644 | 0.000754381 | 0.001750497 | AC021092.1 |
| ENSG00000259673 | –0.709890208 | 6.48E−10 | 3.27E−09 | IQCH-AS1 |
| ENSG00000258429 | –0.624426456 | 0.0000121 | 0.0000371 | |
| ENSG00000141391 | –0.60827287 | 0.0000122 | 0.0000373 | PRELID3A |
| ENSG00000211448 | –0.972541619 | 0.0000124 | 0.0000381 | DIO2 |
| ENSG00000115616 | –0.665234335 | 0.0000133 | 0.0000408 | SLC9A2 |
| ENSG00000112599 | –0.735375549 | 0.0000157 | 0.0000474 | GUCA1B |
| ENSG00000204792 | –0.712824082 | 0.00000285 | 0.00000946 | LINC01291 |
| ENSG00000170629 | –0.718917939 | 7.44E−08 | 0.0000003 | DPY19L2P2 |
| ENSG00000270607 | –0.721426273 | 0.0000854 | 0.000231989 | AC009549.1 |
| ENSG00000166415 | –0.915490775 | 0.0000193 | 0.0000577 | WDR72 |
| ENSG00000188985 | –0.721464876 | 0.000000975 | 0.00000344 | DHFRP1 |
| ENSG00000270696 | –0.72171295 | 5.63E−13 | 3.76E−12 | AC005034.3 |
| ENSG00000261087 | –0.722660516 | 0.000910157 | 0.00208174 | AP003469.4 |
| ENSG00000224167 | –0.72658689 | 1.62E−08 | 7.06E−08 | AL390729.1 |
| ENSG00000115738 | –0.875366803 | 0.0000248 | 0.0000733 | ID2 |
| ENSG00000007968 | –0.997190263 | 0.0000264 | 0.0000779 | E2F2 |
| ENSG00000133687 | –0.885164289 | 0.0000268 | 0.000079 | TMTC1 |
| ENSG00000260442 | –0.727912721 | 0.000664516 | 0.001559564 | ATP2A1-AS1 |
| ENSG00000178184 | –0.791109251 | 0.0000279 | 0.0000819 | PARD6G |
| ENSG00000276672 | –0.733771157 | 3.09E−18 | 3.03E−17 | AL161891.1 |
| ENSG00000177465 | –0.817978872 | 0.0000291 | 0.0000851 | ACOT4 |
| ENSG00000264350 | –0.736318413 | 0.0000175 | 0.0000525 | AC090897.1 |
| ENSG00000238142 | –0.738440741 | 0.00083095 | 0.001913575 | BX284668.5 |
| ENSG00000143028 | –0.730875189 | 0.0000305 | 0.0000888 | SYPL2 |
| ENSG00000173320 | –0.595801591 | 0.000032 | 0.0000929 | STOX2 |
| ENSG00000260000 | –0.738881279 | 0.0000167 | 0.0000505 | AL133338.1 |
| ENSG00000177822 | –0.741978242 | 0.00003 | 0.0000875 | AC098864.1 |
| ENSG00000156140 | –0.669690954 | 0.0000439 | 0.000124705 | ADAMTS3 |
| ENSG00000241360 | –0.647292344 | 0.0000467 | 0.000132129 | PDXP |
| ENSG00000149646 | –0.661096929 | 0.0000498 | 0.000140129 | CNBD2 |
| ENSG00000164920 | –0.58797654 | 0.0000546 | 0.000152834 | OSR2 |
| ENSG00000273033 | –0.764755287 | 1.97E−08 | 8.47E−08 | LINC02035 |
| ENSG00000116819 | –0.594037737 | 0.0000587 | 0.000163798 | TFAP2E |
| ENSG00000274213 | –0.767601803 | 0.000644991 | 0.001516995 | AC015912.3 |
| ENSG00000240929 | –0.770831237 | 1.19E−19 | 1.28E−18 | HIST2H2BB |
| ENSG00000184005 | –0.872567649 | 0.0000797 | 0.000218143 | ST6GALNAC3 |
| ENSG00000247556 | –0.7791386 | 8.2E−70 | 6.74E−68 | OIP5-AS1 |
| ENSG00000016402 | –0.767097162 | 0.0000806 | 0.000220457 | IL20RA |
| ENSG00000140465 | –0.704109542 | 0.000082 | 0.000223701 | CYP1A1 |
| ENSG00000240024 | –0.786762956 | 3.5E−29 | 6.14E−28 | LINC00888 |
| ENSG00000128596 | –0.690766509 | 0.0000891 | 0.000241372 | CCDC136 |
| ENSG00000176125 | –0.855443147 | 0.0000949 | 0.00025624 | UFSP1 |
| ENSG00000165730 | –0.595042604 | 0.0000969 | 0.000261295 | STOX1 |
| ENSG00000119915 | –0.779823964 | 0.000105488 | 0.00028257 | ELOVL3 |
| ENSG00000251350 | –0.79220264 | 0.0000027 | 0.00000897 | LINC02475 |
| ENSG00000177181 | –0.894622588 | 0.000124158 | 0.00032929 | RIMKLA |
| ENSG00000280079 | –0.80571522 | 5.25E−09 | 2.41E−08 | AC011447.7 |
| ENSG00000279059 | –0.827433939 | 0.0000225 | 0.000067 | AC007485.2 |
| ENSG00000184486 | –0.606547058 | 0.000159816 | 0.000418622 | POU3F2 |
| ENSG00000138395 | –0.702091541 | 0.00016143 | 0.000422511 | CDK15 |
| ENSG00000095637 | –0.610468574 | 0.000162344 | 0.000424481 | SORBS1 |
| ENSG00000275185 | –0.828054896 | 0.00000027 | 0.00000102 | AC130324.3 |
| ENSG00000135643 | –0.61786301 | 0.000180308 | 0.000468496 | KCNMB4 |
| ENSG00000205002 | –0.777190629 | 0.000186764 | 0.000483708 | AARD |
| ENSG00000229368 | –0.82873561 | 0.0000576 | 0.000160817 | AC090587.2 |
| ENSG00000123096 | –0.61800128 | 0.00020928 | 0.000537779 | SSPN |
| ENSG00000160219 | –0.713815817 | 0.000226571 | 0.000579039 | GAB3 |
| ENSG00000119737 | –0.654203266 | 0.00024644 | 0.000625436 | GPR75 |
| ENSG00000263272 | –0.832845527 | 0.0000107 | 0.0000332 | AC004148.2 |
| ENSG00000204789 | –0.836003991 | 1.67E−10 | 8.96E−10 | ZNF204P |
| ENSG00000279207 | –0.839738829 | 0.00000227 | 0.00000765 | AC015813.6 |
| ENSG00000278291 | –0.844858432 | 0.000507367 | 0.001217292 | AL161772.1 |
| ENSG00000233117 | –0.857807409 | 2.24E−13 | 1.54E−12 | LINC00702 |
| ENSG00000164778 | –0.858184757 | 0.000294259 | 0.000735145 | EN2 |
| ENSG00000173376 | –0.799990294 | 0.000322836 | 0.000799238 | NDNF |
| ENSG00000186340 | –0.612224671 | 0.000327986 | 0.000810917 | THBS2 |
| ENSG00000226608 | –0.86563425 | 2.31E−08 | 9.86E−08 | FTLP3 |
| ENSG00000150510 | –0.635813794 | 0.000354515 | 0.000870281 | FAM124A |
| ENSG00000129167 | –0.767846935 | 0.000362165 | 0.000887408 | TPH1 |
| ENSG00000255224 | –0.871481211 | 0.000139626 | 0.000368601 | AC109322.1 |
| ENSG00000132932 | –0.844803117 | 0.000408508 | 0.000992987 | ATP8A2 |
| ENSG00000179546 | –0.62615245 | 0.000418009 | 0.001014768 | HTR1D |
| ENSG00000124772 | –0.758950016 | 0.000448513 | 0.001084408 | CPNE5 |
| ENSG00000132016 | –0.595302993 | 0.000460125 | 0.001110729 | C19orf57 |
| ENSG00000227908 | –0.87629061 | 0.0000218 | 0.0000647 | FLJ31104 |
| ENSG00000272462 | –0.889371251 | 0.000000306 | 0.00000115 | U91328.2 |
| ENSG00000232368 | –0.891946046 | 8.72E−10 | 4.34E−09 | FTLP2 |
| ENSG00000274020 | –0.902527943 | 0.000000219 | 0.000000837 | LINC01138 |
| ENSG00000167705 | –0.656826415 | 0.00055993 | 0.001333171 | RILP |
| ENSG00000163596 | –0.716182102 | 0.000585479 | 0.001386711 | ICA1L |
| ENSG00000162601 | –0.652501753 | 0.000599957 | 0.0014187 | MYSM1 |
| ENSG00000250132 | –0.906972213 | 0.000126211 | 0.000334668 | AC004803.1 |
| ENSG00000143494 | –0.629613078 | 0.000659815 | 0.001549361 | VASH2 |
| ENSG00000276107 | –0.914373714 | 3.98E−09 | 1.85E−08 | AC037198.2 |
| ENSG00000238266 | –0.920917248 | 2.39E−11 | 1.39E−10 | LINC00707 |
| ENSG00000128203 | –0.656798742 | 0.000682964 | 0.001599429 | ASPHD2 |
| ENSG00000187391 | –0.775390286 | 0.000695998 | 0.001627632 | MAGI2 |
| ENSG00000198157 | –0.672184208 | 0.000719226 | 0.001676278 | HMGN5 |
| ENSG00000266709 | –0.92347604 | 1.49E−15 | 1.2E−14 | AC005224.4 |
| ENSG00000269927 | –0.941179574 | 0.0000116 | 0.0000356 | AC004817.3 |
| ENSG00000224080 | –0.957073973 | 0.00000138 | 0.00000479 | UBE2FP1 |
| ENSG00000171119 | –0.732024185 | 0.000859632 | 0.001973741 | NRTN |
| ENSG00000182795 | –0.713257 | 0.000893473 | 0.002045361 | C1orf116 |
| ENSG00000235859 | –0.981085776 | 1.34E−11 | 7.99E−11 | AC006978.1 |
| ENSG00000184058 | –0.781722778 | 0.000947799 | 0.002160685 | TBX1 |
| ENSG00000137959 | –0.589383242 | 0.000982869 | 0.00223288 | IFI44L |
| ENSG00000249485 | –0.994236206 | 0.0000105 | 0.0000326 | RBBP4P1 |
| ENSG00000188848 | –0.612794187 | 0.00106899 | 0.00241599 | BEND4 |
| ENSG00000235180 | –1.024177912 | 1.2E−09 | 5.92E−09 | LINC00601 |
| ENSG00000280123 | –1.03782143 | 2.74E−12 | 1.73E−11 | AC023632.6 |
| ENSG00000176912 | –1.038065676 | 3.47E−10 | 1.79E−09 | TYMSOS |
| ENSG00000134317 | –0.746307481 | 0.001354336 | 0.003003002 | GRHL1 |
| ENSG00000130208 | –0.676215853 | 0.001374168 | 0.003039733 | APOC1 |
| ENSG00000250462 | –1.061344837 | 4.22E−32 | 8.64E−31 | LRRC37BP1 |
| ENSG00000155465 | –0.67282726 | 0.001515904 | 0.00333088 | SLC7A7 |
| ENSG00000230316 | –1.096386598 | 3.09E−10 | 1.61E−09 | FEZF1-AS1 |
| ENSG00000115318 | –0.601269693 | 0.001614401 | 0.003523135 | LOXL3 |
| ENSG00000125931 | –0.665048797 | 0.001714986 | 0.003725307 | CITED1 |
| ENSG00000261534 | –1.102227861 | 7.9E−10 | 3.95E−09 | AL596244.1 |
| ENSG00000214719 | –1.106140664 | 1.01E−16 | 8.81E−16 | AC005562.1 |
| ENSG00000234456 | –1.116449494 | 4.03E−22 | 4.92E−21 | MAGI2-AS3 |
| ENSG00000092200 | –0.685034084 | 0.001895898 | 0.004088527 | RPGRIP1 |
| ENSG00000216775 | –1.169568862 | 1.42E−39 | 4.14E−38 | AL109918.1 |
| ENSG00000260686 | –1.172038199 | 0.000000482 | 0.00000176 | AC008669.1 |
| ENSG00000226943 | –1.177930323 | 1.53E−08 | 6.66E−08 | ALG1L5P |
| ENSG00000188185 | –1.185262956 | 0.000001 | 0.00000354 | LINC00265 |
| ENSG00000113389 | –0.597567416 | 0.002456821 | 0.005192422 | NPR3 |
| ENSG00000248890 | –1.205696789 | 6.08E−18 | 5.82E−17 | HHIP-AS1 |
| ENSG00000170270 | –0.68387924 | 0.002558896 | 0.005392531 | GON7 |
| ENSG00000119508 | –0.711455187 | 0.002680663 | 0.005624765 | NR4A3 |
| ENSG00000140905 | –0.636524948 | 0.002992587 | 0.006224563 | GCSH |
| ENSG00000050730 | –0.636431758 | 0.003037792 | 0.006309593 | TNIP3 |
| ENSG00000170989 | –0.64441633 | 0.003150496 | 0.006522018 | S1PR1 |
| ENSG00000256304 | –1.233661403 | 2.46E−13 | 1.68E−12 | CCDC150P1 |
| ENSG00000237424 | –1.291854855 | 1.19E−11 | 7.12E−11 | FOXD2-AS1 |
| ENSG00000227359 | –1.292403358 | 0.000000113 | 0.000000447 | AC017074.1 |
| ENSG00000230615 | –1.294100524 | 2.92E−32 | 6.03E−31 | AL139220.2 |
| ENSG00000244300 | –1.311706541 | 6.67E−08 | 0.000000271 | GATA2-AS1 |
| ENSG00000183762 | –0.599097723 | 0.004686398 | 0.009396411 | KREMEN1 |
| ENSG00000168405 | –1.328267416 | 4.82E−09 | 2.23E−08 | CMAHP |
| ENSG00000144834 | –0.620458505 | 0.004968065 | 0.00989472 | TAGLN3 |
| ENSG00000279519 | –1.3382021 | 3.67E−12 | 2.3E−11 | AC007382.1 |
| ENSG00000105371 | –0.591348772 | 0.00761156 | 0.014575965 | ICAM4 |
| ENSG00000128536 | –0.58264386 | 0.007717241 | 0.014763271 | CDHR3 |
| ENSG00000226806 | –1.402676685 | 0.000000047 | 0.000000194 | AC011893.1 |
| ENSG00000257732 | –1.507011758 | 2.49E−14 | 1.83E−13 | AC089983.1 |
To test whether the effects on chromatin in LOXL2 KD cells directly activated ATM-dependent DDR signaling, we forced chromatin condensation in these cells by expressing the linker histone H1 or the H3K9 methyltransferase SUV-39H1 (as green fluorescent protein (GFP)-labeled proteins) and counted the number of γ-H2AX foci in GFP-positive cells. Notably, overexpression of either H1GFP or SUV-39GFP in LOXL2 KD cells reduced the number of γ-H2AX foci as compared with LOXL2 KD cells that expressed GFP alone (+MOCKGFP) (Fig. 4f, g). This suggested that lack of LOXL2 and reduced H3K4ox levels affected the regulation of chromatin condensation (leading to decondensation) and activated DDR, even in the absence of DNA damage.
LOXL2 reduction enhances chemosensitivity of TNBC cells
We found that reducing H3K4ox levels via LOXL2 depletion led to chromatin decondensation, triggered DDR activation, and induced cell cycle arrest, suggesting that LOXL2 inhibition could be interesting as a breast cancer treatment. As no LOXL2-specific inhibitors are currently available, we therefore tested whether reducing the levels of functional LOXL2 would increase the sensitivity of breast cancer cells to chemotherapy. For this, we treated three different TNBC cell lines with doxorubicin (a topoisomerase inhibitor that generates DSBs) either alone or together with trichostatin A (TSA) (a general HDCA inhibitor [37] that increases chromatin accessibility [38]). Indeed, we observed that in all cases, treatment of doxorubicin together with TSA increased the percentage of cell death—indicative of an increased cell sensitivity to chemotherapy (Fig. 5a). Similar results were obtained with two TNBC_PDXs (PDX-549 and PDX-154) (Fig. 5a). To determine if these results were reproducible in vivo, PDX-549 cells were first grown ex vivo, and then 106 cells were subcutaneously implanted in nude mice. After tumor formation, mice were treated with TSA, doxorubicin, doxorubicin plus TSA, or (as a control) vehicle for 25 days. In agreement with our previous results, we observed that tumor growth was substantially impaired with the combined TSA/doxorubicin treatment (Fig. 5b).
Fig. 5.
LOXL2 suppresses sensitivity in TNBC cells. a Cell viability was measured at different timepoints using an MTT assay of cultured different breast cancer cells and PDXs. The effects of doxorubicin, TSA, and doxorubicin plus TSA were analyzed. b Dissociated cells from PDX-549 were orthotopically implanted into NOD/SCID mice and injected intraperitoneally twice weekly with TSA (0.25 mg/kg), doxorubicin (2 mg/kg mouse weight), or doxorubicin plus TSA. Tumor volumes were measured twice a week and are given as averages. Results are given as averages of six independent tumors ± SEM
Discussion
In this work, we found that several TNBC cell lines and PDXs express high levels of LOXL2 as compared with those of the luminal breast cancer subtype. With the use of a newly generated antibody, we were able to detect the H3K4ox modification produced by LOXL2 [3] and we observed how this histone H3 modification was higher in all TNBC cell lines and PDXs as compared with luminal subtypes. This H3 modification is enriched in heterochromatin and is required to maintain the condensed heterochromatin state. It is possible that during tumor progression, some cancer cells undergo the EMT program and start to express LOXL2. The transcription factor Snail1, together with LOXL2, would participate in downregulating the CDH1 gene and other heterochromatin transcripts, giving rise to transformation of cancer epithelial cells into mesenchymal cells (through EMT) [6, 39–41]. Recently, two different groups have suggested that EMT is dispensable for lung and pancreas metastasis but contributes significantly to the formation of recurrent metastasis after chemotherapy [42, 43]. This finding is in agreement with our results, in which we observed that the most aggressive and resistant subtype of breast cancer (TNBC) has a mesenchymal phenotype and is enriched for H3K4ox in its heterochromatin. We observed that reducing H3K4ox levels perturbed the balance between condensed and decondensed chromatin and activated the DDR. Similar phenotypes have been previously observed after depletion of SUV-39, PRDM2, or HP1, which are required for heterochromatin maintenance and cell survival after DNA damage [23, 24, 44, 45]. H3K4ox levels have been correlated to the repression of CDH1 and major satellites (heterochromatin transcripts) during the EMT program [3, 4, 6]. CDH1 is upregulated in TNBC cells when LOXL2 is knocked down and the levels of H3K4ox are decreased; however, the H3K4ox modification does not seem to be involved in repression of heterochromatin transcripts, as we did not observe increased levels of heterochromatin transcripts in LOXL2 KD conditions. Notably, downregulation of heterochromatin transcripts during EMT occur at a specific timepoint during chromatin reorganization, after which the levels of transcription recovered despite the presence of H3K4ox. Intriguingly, although upstream DDR signaling is sustained when chromatin is maintained in a condensed state [22, 46, 47], we found that forcing chromatin decondensation was sufficient to activate DDR signaling in the absence of any detectable DNA lesions. It seems that an abnormal, unbalanced ratio between condensation and decondensation leads to persistent DDR signaling. As LOXL2 has multiple substrates besides histones [5], we cannot discard an additional putative role for LOXL2 in the direct regulation of the DDR machinery. However, we observed that the DDR induced in the absence of LOXL2 can be inhibited by forcing chromatin to condense using either of two different approaches, suggesting that the main molecular mechanism is the regulation per se of the chromatin compaction state.
While a correlation between chromatin compaction and DDR has been previously reported [22, 46, 47], we demonstrated here that H3 oxidation by LOXL2 is another molecular mechanism that maintains compaction. Notably, LOXL2 has been widely linked to cancer and the acquisition of cellular malignancy, as it is overexpressed in many tumors [15, 48–51] and has an important role in tumor formation [52].
Induction of chromatin compaction has been suggested as a potential therapeutic tool in gliomas, in which chromatin compaction limits DDR [46] and promotes damage resistance [53]. However, this does not appear to apply to TNBC cells. In support of this, we have now shown that several TNBC PDX tumors treated with TSA, a histone deacetylase inhibitor that generates an open chromatin state, enhanced tumor killing with doxorubicin in these cells. These observations suggest a rationale for studying combinations of “chromatin opening drugs,” including inhibitors of histones deacetylases, EZH2, or LSD1, as a strategy for overcoming resistance to treatment in TNBC.
Materials and methods
Cell lines, transfections, and infections
Most of the cell lines used in this work (the TNBC lines MDA-MB-468 (ATCC HTB-132), CAL-51 (CSC-C0382), HS-578T (ATCC HTB-126), and MDA-MB-231 (ATCC HTB-26), the luminal A T-47D (ATCC HTB-133), MCF-7 (ATCC HTB-22D), and luminal B BT-474 (ATCC HTB-20)) as well as cells isolated from PDXs, were maintained in Dulbecco’s modified Eagle’s medium (Biowest; L0106-500); the TNBC line BT-549 (ATCC HTB 122) was maintained in Roswell Park Memorial Institute medium (Biowest; L0501-500). All cell cultures were supplemented with 10% fetal bovine serum (Gibco; 10270106), 2 mM l-glutamine (Biowest; X0550-100), and 1% penicillin-streptomycin (Gibco; 15140122) at 37 °C in 5% CO2. All cell lines were regularly tested for the absence of mycoplasma using standard polymerase chain reaction (PCR). For LOXL2-Flag overexpression assays, MCF-7 cells were seeded for 24 h and transfected with 10 µg pcDNA3-hLOXL2-Flag vector using polyethylenimine polymer (Polysciences Inc; 23966-1). Lentiviral particles were produced in HEK293T cells (ATCC CRL-3216) for lentiviral infections to produce LOXL2 KD cells. Cells were grown to 70% confluency and then transfected by dropwise addition of NaCl (150 mM), a DNA mixture of pLKO-short hairpin control (shCT)/shLOXL2 (7.5 µg), pCMV-VSVG (1.5 µg), pMDLg/pRRE (4.5 µg), and pRSV-Rev (1.5 µg), and polyethylenimine polymer (1 mg/ml) (Polysciences Inc), which had been preincubated for 15 min at room temperature. Transfection medium was replaced with fresh medium after 24 h (day 1). The supernatant from the transfection medium at days 2 and 3 was filtered with 0.45 μm filter unit (Merck Millipore) and stored at 4 °C (both were mixed after day 2). The supernatant mixture was concentrated using Lenti-X Concentrator product (Clontech) and centrifuged at 1500 × g for 45 min at 4 °C. The pellet was resuspended in 1 ml fresh medium, aliquoted (100 μl), and stored at –20 °C. Aliquots were used to infect MDA-MB-231 cells [6]. For retroviral infections, HEK293 gag–pol cells were used to produce retroviral particles. Cells were transfected as for HEK293T cells with a mixture of NaCl (150 mM), DNA (2.5 µg of pCMV-VSV-G and 7.5 µg of pMSCV, pMSCV-LOXL2 wt-FLAG or pMSCV-LOXL2 mutFLAG IRES–GFP vectors) and polyethylenimine polymer (1 mg/ml) (Polysciences Inc; 23966-1) that had been preincubated for 15 min at room temperature; transfection medium was replaced with fresh medium at 24 and 48 h and processed as explained above. Viral particles were concentrated using Retro-X Concentrator product (Clontech; 631456) and then used to infect MDA-MB-231 cells. HEK293T cells were used to produce lentiviral particles for histone H1(H1)-GFP expression. Cells were first grown to 70% confluency and transfected with a mixture of NaCl (150 mM), a DNA mixture of FUGW-H1-empty vector/FUGW-H1-GFP (7.5 µg), pCMV-VSVG (1.5 µg), pMDLg/pRRE (4.5 µg), and pRSV-Rev (1.5 µg), and polyethylenimine polymer (1 mg/ml) (Polysciences Inc; 23966-1), which had been preincubated for 15 min at room temperature and processed as explained above. Viral particles were concentrated using Lenti-X Concentrator product (Clontech; 631232) and used to infect MDA-MB-231 cells. For SUV-39H1-GFP rescue experiments, MDA-MB-231 cells infected with either shCT or shLOXL2 were selected for 48 h with puromycin (Puro) (2.5 μg/ml). After selection, cells were seeded for 24 h and transfected with CGA-pCAGGS-SUV39H1-EGFP-IRES-Puro vector (2.5 µg) using TransIT-X2 Dynamic Delivery System (Mirus Bio LLC; MIR600Q).
Antibodies and other reagents
The following antibodies were used: anti-FLAG (F7425, Sigma), anti-LOXL2 (NP1-32954, Novus), anti-H3K4me3 (07-473, EMD Millipore), anti-H3K9me3 (07–442, Millipore), anti-phospho-histone H2AX (S139) clone JBW301 (05–636, EMD Millipore), anti-GFP (ab6556, Abcam), anti-53BP1 (NB100–904, Novus Biol.), anti-phospho-CHK1 (S317) (A300-163A, Bethyl), anti-CHK2 clone 7 (05–649, EMD Millipore), anti-cleaved caspase 3 (Asp175) (9661, Cell Signaling), anti-KAP-1 (ab10484, Abcam), anti-Phospho-KAP-1 (S824) (A300-767A, Bethyl), anti-tubulin (T9026, Sigma), anti-phospho-H3 (S10) (06–570, Millipore), and anti-histone H3 (ab1791, Abcam). The following chemical reagents were used: doxorubicin hydrochloride (Sigma; D1515), cordycepin (Sigma; C3394), and the ATM inhibitor KU55933 (Sigma; SML1109). An anti-H3K4ox antibody was generated from a synthetic peptide in which fluorenylmethoxycarbonyl protecting group (Fmoc)-6-hydroxynorleucine (BAA1117, Iris Biotech) rather than Fmoc-lysine(Boc) was incorporated at position 4. The peptide was purified (>95%) by reversed-phase high-performance liquid chromatography and its identity was confirmed by mass spectrometry, after which it was coupled to keyhole limpet hemocyanin (KLH) for antibody production. Antibody information can be found in the Supplementary Table 1.
Cell extracts, histone isolation, and PDX extraction
To obtain nuclear fractions of LOXL2-Flag–transfected MCF-7 cells and HEK293T cells, cells were first lysed in soft-lysis buffer (50 mM Tris, 2 mM EDTA, 0.1% NP-40, and 10% glycerol, supplemented with protease and phosphatase inhibitors) for 5 min on ice. Samples were centrifuged at 900 × g for 15 min, and the supernatant was discarded. The nuclear pellet was lysed in high-salt lysis buffer (20 mM HEPES pH 7.4, 350 mM NaCl, 1 mM MgCl2, 0.5% Triton X-100, and 10% glycerol, supplemented with protease and phosphatase inhibitors) for 30 min at 4 °C, and samples were centrifuged at 16,000 × g for 10 min supernatant NaCl concentration was reduced to 300 mM NaCl by adding balance buffer (20 mM HEPES pH 7.4, 1 mM MgCl2, and 10 mM KCl). Cells extracts from the different breast cancer cell lines were obtained with SDS lysis buffer (2% SDS, 50 mM Tris-HCl, and 10% glycerol). Histones were isolated from the different breast cancer cell lines using acid precipitation. For this, cell monolayers were first scraped in 1 ml of lysis buffer (10 mM Tris pH 6.5, 50 mM sodium bisulfite, 10 mM MgCl2, 1% Triton X-100, 8.6% sucrose, and 10 mM sodium butyrate). Working at 4 °C, pellets were then purified by three rounds of centrifugation (16,000 × g for 15 s) and resuspension by vortexing, in lysis buffer for the first two rounds and washing buffer (10 mM Tris pH 7.4, 13 mM EDTA) for the final round; supernatant fractions were discarded. The obtained pellets containing chromatin were resuspended in 0.4 N sulfuric acid, the mixture was incubated at 4 °C for 1 h and then centrifuged at 16,000 × g for 10 min, and the histone-containing supernatants were kept. Samples were incubated with acetone (1:9) to block the acid overnight at –20 °C and then centrifuged at 16,000 × g for 10 min histone pellets were air dried for 5 min and then resuspended in water for analysis. Cell extracts of PDX samples were obtained by disrupting tissue using a pellet pestle (Sigma; Z359947) with lysis buffer (50 mM Tris-HCl pH 8, 10 mM EDTA, 1% SDS, and 1 mM DTT). Proteins were separated by SDS-polyacrylamide gel electrophoresis gel and analyzed with the indicated antibodies.
Recombinant LOXL2 purification
LOXL2-encoding baculovirus were amplified, and LOXL2-Flag recombinant proteins (wt and mutant) were produced in Sf9 cells according to standard procedures [3]. Cell lysis was performed as previously described [54]. Cell extracts were incubated with Flag M2 beads for 4 h at 4 °C and then washed 4× times with 20 mM HEPES pH 7.4, 1 mM MgCl2, 300 mM NaCl, 10 mM KCl, 10% glycerol, and 0.2% Triton X-100. Recombinant proteins were eluted with the Flag peptide (1 µg/µl) for 1 h at 4 °C.
Oxidation reaction
Nucleosomes (2 µg) were incubated with recombinant LOXL2 purified protein in oxidation buffer, 20 mM HEPES pH 7.4, 100 mM NaCl, 1 mM MgCl2, and 1 mM DTT. Reactions were carried out for the indicated times at 37 °C and then analyzed by SDS/PAGE and western blotting. To detect the aldehyde group, biotin hydrazide (BTH) (5 mM) was added after the oxidation reaction for 2 h at 25 °C. Finally, biotinylated histones were immunoprecipitated with streptavidin beads, and oxidized H3 was detected by western blot.
Dot blot assay
For dot blot assays, 1 µg of each peptide (in 10 µl of sample) was applied under low vacuum to a prewetted nitrocellulose membrane (Amersham Protran 0.45 nitrocellulose, GE Healthcare) using a dot blot apparatus (HYBRI-DOT Manifold; Life Technologies). After blocking the entire blot in 15 mL of 5% nonfat dry milk and 0.1% Tween-20 Tris-buffered saline for 1 h at room temperature, the blot was probed with the indicated antibodies.
ChIP experiments
For ChIP experiments, cells were first crosslinked in 1% formaldehyde for 10 min at 37 °C. Crosslinking was stopped by adding glycine to a final concentration of 0.125 M for 2 min at room temperature. Cell monolayers were scraped in cold soft-lysis buffer (50 mM Tris pH 8.0, 10 mM EDTA, 0.1% NP-40, and 10% glycerol), and incubated 20 min on ice. Nuclei pellets were lysed with SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris pH 8.0), and extracts were sonicated to generate 200–1500-bp DNA fragments. For immunoprecipitation, supernatants were diluted 1:10 with dilution buffer, and samples were incubated with rotation overnight at 4 °C with primary antibody or irrelevant immunoglobulin G. Samples were then treated with elution buffer (100 mM Na2CO3 and 1% SDS) for 1 h at 37 °C and incubated at 65 °C overnight after addition of NaCl to a final concentration of 200 mM, to reverse formaldehyde crosslinking. After proteinase K treatment for 1 h at 55 °C, DNA was purified with MinElute PCR purification kit (Qiagen; 28006) and eluted in Milli-Q water. Genomic regions (GRs) were detected by quantitative staining with PCR SYBR Green (Quantabio; 95073), and the ChIP results were quantified relative to the input amount and the amount of H3 immunoprecipitated in each condition.
Peaks of H3K4ox were called from sequence reads detected through ChIP-seq using the MACS2 tool [26]. The chromatin state files for HepG2 and HMEC cells were computed by the ENCODE project using the ChromHMM tool [27] from https://genome.ucsc.edu/cgi-bin/hgFileUi?db=hg19&g=wgEncodeBroadHmm. Statistical overrepresentation of H3K4ox peaks detected by ChIP-seq was assessed from the two cell lines across several chromatin states: heterochromatin, repressed, insulator, strong enhancer (sum of states 4 and 5), poised/weak enhancer (sum of states 6 and 7), promoter (sum of states 1 and 2), and poised promoter. The contingency table of the Fisher's test carried out for this contained the number of nucleotides within peaks, chromatin states, intersections thereof, and the remaining portion of the genome (computed as the difference from the effective genome size for ChIP-seq peaks calling). The same procedure was applied to detect H3K4ox peak overrepresentation in lamin-associated domains of chromatin, obtained from [55]. ChIP-PCR used the following GR number and peak positions: GR#1: chr 14 (79003315–79007307); GR#2: chr 5 (151967551–151971984); GR#3: chr 1 (230657019–230658589); GR#4: chr 2 (41741709–41745780); GR#5: chr 17 (53951879–53955135); GR#6: chr 4 (62611008–62615505); GR#7: chr 6 (67550237–67554488); GR#8: chr 5 (66894215–66899021); and GR#9: chr 13 (89334968–89336774). The sequences of the primers used can be found in Supplementary Table 2.
ATAC-seq and ATAC-qPCR experiments
ATAC experiments were performed as described previously [28]. Cells were harvested and treated with transposase Tn5 (Nextera DNA Library Preparation Kit, Illumina; FC-121-1030). DNA was purified using MinElute PCR purification kit (Qiagen), samples were amplified by PCR using NEBNext High-Fidelity 2× PCR Master Mix (New England Biolabs; M5041), and DNA was again purified with the MinElute PCR purification kit. Reads produced by ATAC sequencing of two control (Control) replicates and two LOXL2 KD sequencing replicates (LOXL2 KD) were aligned to the hg19 build of the reference human genome using Bowtie 2 [56] with default parameters for pair-end sequencing. ATAC peaks were then called by combining aligned reads of both replicates of the control and the KD using MACS2. To allow for false discovery rate (FDR) threshold selection further downstream in the analysis, no FDR restrictions were imposed on the ATAC peak calling. For ATAC-qPCR experiments, the final elution product of the ATAC protocol was diluted 1:50. Incorporation of the transposase Tn5 to GRs was detected by quantitative staining with PCR SYBR Green (Quantabio; 95073). ATAC-qPCR results for the selected GRs were normalized to the same unaffected GR (HPRT promoter) for each condition. The sequences of the primers used can be found in Supplementary Table 2.
RNA-seq analysis
Reads produced by RNA-seq of the same replicates as described above of two controls (Control) and two LOXL2 KDs were aligned to the hg19 build of the reference human transcriptome using TopHat2 [57] with default parameters for pair-end sequencing. Aligned reads were then analyzed using a standard Cufflinks pipeline [58] to detect differentially expressed genes between the two conditions (LOXL2 KD and Control).
Differential expression analysis of transposable elements (TEs)
The trimmed RNA-seq reads from the two controls and the two LOXL2 samples were processed with the TEtools program [59] using the hg19 annotation of TEs. The obtained count table was then processed with NOIseq [60] to perform a differential expression analysis of LOXL2 against the control. Expression was normalized using the TMM method, whereby TEs with a probability higher than 0.95 of being differentially expressed were considered as statistically significant.
Replicate correlation
The read count (coverage) at each position of the hg19 human reference genome was computed for each replicate of the H3K4ox ChIP-seq and the control and LOXL2 KD ATAC sequencing, using the BEDtools genomecov capability [61]. Genomic positions with zero-read counts were filtered out. Replicate files of each experiment were merged to produce a single file aligned by genomic position, and the corresponding Pearson’s correlation coefficient of read counts was computed. For the graphical representation of the correlation, 100,000 genomic positions were randomly selected.
Analysis of ATAC peaks that overlap with H3K4ox peaks
All significant (P < 10–5) ATAC peaks (LOXL2 KD versus control) and H3K4ox peaks were first intersected with the BEDtools intersect program [61]. Based on this intersection, ATAC peaks were classified as overlapping (if they intersected an H3K4ox peak) or orphan (if not). Only intersections involving more than 95% of the sequence of ATAC peaks were considered. Control and LOXL2 KD read counts over all genomic positions (see above) were intersected with both overlapping and orphan peaks. Read counts over genomic positions of control and experiment replicates were averaged. To carry out the heatmap representation, peak sequences (overlapping or orphan) were aligned by their summits. For linear representation, the average experimental read counts at each downstream and upstream position were summed for both the experimental and the control counts. Position-wise sums were then divided by the read count sum value obtained for the summit of control read counts, thus making all sums relative to the maximum control value.
Integrated analysis of H3K4ox and ATAC peaks, and differentially expressed genes
The differentially expressed genes detected through the RNA-seq analysis of control and LOXL2 KD cells were selected if they were in close vicinity (either up- or downstream) to overlapping ATAC peaks. Two different distance thresholds (0.5 and 1 Mb) were used to detect close differentially expressed genes.
Inhibition of RNA synthesis, ATM kinase, and cell cycle analysis
MDA-MB-231-infected cells selected for 48 h with puromycin (2.5 μg/ml) were seeded and then maintained under puromycin selection.
For inhibition of RNA synthesis, cells were treated at 48 h with 200 µM cordycepin (Sigma; C3394) and fixed with 100% cold ethanol at designated timepoints.
For inhibition of the ATM kinase, cells were incubated at the 24-h timepoint with KU55933 (Sigma; SML1109) for a further 24 h at two different concentrations (either 5 or 10 µM), or for 2 h with doxorubicin (1 µM) as a positive control.
For cell cycle analysis, cells at 48 h after puromycin selection were first synchronized through the double thymidine block protocol. Specifically, cells were seeded to 50% confluency, incubated for 14 h with complete growth medium (supplemented with 2 mM thymidine), washed 2× with phosphate-buffered saline (PBS), and released by a 9-h incubation with complete medium growth. Cells were washed again 2× with PBS, incubated 14-h with 2-mM thymidine, and released with complete growth medium. Cells were then harvested at the designated timepoints and fixed with 100% cold ethanol. After two days, fixed cells were stained with propidium iodide (PI) and analyzed by flow cytometry using BD FACSCalibur (Becton Dickinson). Results were analyzed using BD CellQuest Pro software.
Non-replicative cell experiment
MDA-MB-231 cells were seeded in coverslips and maintained during all the experiment in Dulbecco’s modified Eagle’s medium (Biowest; L0106-500) with 0.5% fetal bovine serum (Gibco; 10270106) at 37 °C in 5% CO2. After 24 h, cells were infected with lentiviral particles for LOXL2 KD. After 96 h under selection, cells were fixed with paraformaldehyde 4% with PBS.
Comet assay
MDA-MB-231-infected cells under puromycin selection (see above) were seeded at the 48-h timepoint and grown another 48 h more (still with puromycin at 2.5 µg/ml). DNA strand breaks were measured at the single-cell level for MDA-MB-231 control or LOXL2 KD cells using an alkaline comet assay with the CometAssay kit (Trevigen, Gaithersburg, MD) following the manufacturer's instructions. Briefly, 5000 cells in 50 μl PBS were combined with 500 μl molten LMAgarose (at 37 °C), and 50 μl of this was immediately transferred to a CometSlide. After a 10-min incubation at 4 °C, slides were immersed in 4 °C lysis solution and incubated overnight at 4 °C. Slides were then immersed in freshly prepared alkaline solution (200 mM NaOH, 1 mM EDTA pH > 13) for 1 h at 4 °C in the dark. For electrophoresis, slides were placed in an electrophoresis slide tray containing alkaline solution and incubated with voltage (21 V) for 30 min. After that, slides were immersed twice in H2O for 5 min and then once in 70% ethanol for 5 min. Samples were dried at 37 °C and stained with SYBR Green staining solution (SYBR Green I, Invitrogen) using a 1:10000 dilution in TE (10 mM Tris-HCl pH 7.5, 1 mM EDTA) for 30 min Finally, slides were dried at 37 °C, and cells were imaged using an Olympus BX61 microscope.
Rescue experiments
MDA-MB-231-infected cells under puromycin selection (see above) were seeded at the 48-h timepoint, incubated under puromycin selection for an addition 24 h, and then transfected with the SUV-39H1-EGFP vector or reinfected with retroviral particles for LOXL2-FLAG or lentiviral particles for H1 expression. After 24 h, cells were fixed for immunofluorescence.
Immunofluorescence, image acquisition, and analysis
Cells were fixed with 4% paraformaldehid for 15 min at room temperature, blocked for 1 h with 1% PBS-bovine serum albumin, incubated at room temperature for 2 h with primary antibody, washed 3× with PBS, and then incubated for 1 h at room temperature with the secondary antibody. Cells were washed again 3× with PBS, incubated for 5 min with 4′,6-diamidino-2-phenylindole (DAPI) (0.25 mg/ml) for cell nuclei staining, and then mounted with fluoromount. Fluorescence images corresponding to DAPI, GFP, γ-H2AX and 53BP1 were acquired in a Leica TCS SPE microscope using a Leica DFC300 FX camera and the Leica IM50 software.
Metaphase spreads
For metaphase spread preparations, cells were treated with colcemid (0.1 ug/ml) for 4 h. Cells were trypsinized, hypotonically swollen in 0.075 M KCl for 15 min at 37 °C, and then fixed (75% MeOH and 25% acetic acid, ice cold). Metaphase preparations were spread on glass slides, stained with 10% Giemsa stain (Sigma), and mounted in DPX mounting medium (PanReac). Images were taken using a Leica DM6000 microscope (Leica, Wetzlar, Germany) and analyzed using Fiji Software (https://fiji.sc/).
Cellular viability experiment
Breast cancer cell lines and PDX cells were seeded in 96-well plates. Cellular viability was analyzed using Thiazolyl Blue Tetrazolium Bromide (MTT) (Sigma; M5655) at different timepoints. The concentrations used were: 0.1 µM doxorubicin for all samples; 250-nM TSA for the breast cancer cell lines; and 500-nM TSA for the PDXs cells. Absorbance was detected at 565 nm on Infinite® 200 PRO Series Multimode Reader (Tecan Group Ltd.) and analyzed with i-control™ Microplate Reader Software (Tecan Group Ltd.)
Cancer PDXs and treatments
Samples from patients with breast cancer were obtained from the operating room and transferred to the pathology department, where breast cancer samples were collected, transferred to the animal facility, and implanted into mice, to generate PDXs. All samples were implanted within 60 to 90 min after surgical removal. All patients willingly signed an informed consent, and the study was approved by the Ethics Committee of the Vall d'Hebron Hospital.
For in vivo experiments, a tumor from an established PDX was dissociated into single cells by enzymatic digestion (collagenase at 300 U/ml and hyaluronidase at 200 U/ml) during 1 h at 37 °C on a rotating wheel. The solution of digested tumor was treated with 0.025% trypsin and filtered sequentially using 100 and 40 µm strainers. Isolated cells were plated in culture dishes with DMEM-F12 supplemented with FBS, glutamine, and penicillin/streptomycin. Once the culture was established, 106 cells were injected into the number four fat pad of 6-week old NOD.CB17PrkdcSCID/J (NOD/SCID) female mice (Charles River) with Matrigel. For this, mice were anesthetized and shaved, the fourth and fifth sets of nipples were localized, and an inverted Y incision was made from the midline point between the fourth set of nipples, ending between the fourth and fifth sets to expose the fourth and fifth fat pads on one side. After the injection, animals were sutured, and analgesics injected. Animals were kept in a clean cage with drinking water supplemented with 1 µM 17-β-estradiol. Tumor xenografts were measured with callipers every 3 days, and tumor volume was determined using the formula: (length × width2) × (pi/6). At the end of the experiment, animals were anesthetized with a 1.5% isoflorane–air mixture and were killed by cervical dislocation.
Treatments were administered intraperitoneally twice weekly. One week after cell injection, mice were randomized and treated with TSA (0.25 mg/kg), doxorubicin (2 mg/kg mouse weight), or doxorubicin plus TSA. The control group was injected with sterile PBS.
Mice were maintained and treated in accordance with institutional guidelines of Vall d'Hebron University Hospital Care and Use Committee.
Supplementary information
Acknowledgements
We would like to thank R. Peña and J. Valle for technical assistance, V.A. Raker for manuscript editing, G. Gil for manuscript reading and advice, A. Jordan for H1-GFP constructs, T. Jenuwein for SUV-39 constructs, and H. Galvez-Garcia for ATAC protocol implementation. This work was supported by grants from Instituto de Salud Carlos III (ISCIII) FIS/FEDER (PI12/01250; CP08/00223; PI16/00253; and CB16/12/00449), MINECO (SAF2013-48849-C2-1-R) to SP, BFU2015-68354 to THS, Breast Cancer Research Foundation (BCRF-17-008) to JA, AGL2014-52395-C2-2-R to DA, Worldwide Cancer Research, Red Temática de Investigación Cooperativa en Cáncer (RD012/0036/005), Fundación Científica de la Asociación Española contra el Cáncer, and Fundació La Marató TV3. THS was supported by institutional funding (MINECO) through the Centres of Excellence Severo Ochoa award and the CERCA Programme of the Catalan Government, and SS-B, by a Fundació La Caixa fellowship. We thank La Caixa Foundation and Cellex Foundation for provide research facilities and equipment. GV has received funding from the MINECO (a “Juan de la Cierva Incorporation” fellowship; IJCI-2014-20723). SP was a recipient of a Miguel Servet contract (ISCIII/FIS), and AI, JPC-C, LP-G, and GS-B are supported by contracts from Worldwide Cancer Research, Fundació La Marató TV3, Fundació FERO, and a FI Fellowship from the Generalitat de Catalunya, respectively.
Data availability
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41388-019-0969-1) contains supplementary material, which is available to authorized users.
References
- 1.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–95. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williamson PR, Kagan HM. Reaction pathway of bovine aortic lysyl oxidase. J Biol Chem. 1986;261:9477–82. [PubMed] [Google Scholar]
- 3.Herranz N, Dave N, Millanes-Romero A, Pascual-Reguant L, Morey L, Diaz VM, et al. Lysyl oxidasE−like 2 (LOXL2) oxidizes trimethylated lysine 4 in histone H3. FEBS J. 2016;283:4263–73. doi: 10.1111/febs.13922. [DOI] [PubMed] [Google Scholar]
- 4.Iturbide A, Garcia de Herreros A, Peiro S. A new role for LOX and LOXL2 proteins in transcription regulation. FEBS J. 2015;282:1768–73. doi: 10.1111/febs.12961. [DOI] [PubMed] [Google Scholar]
- 5.Iturbide A, Pascual-Reguant L, Fargas L, Cebria JP, Alsina B, Garcia de Herreros A, et al. LOXL2 oxidizes methylated TAF10 and controls TFIID-dependent genes during neural progenitor differentiation. Mol Cell. 2015;58:755–66. doi: 10.1016/j.molcel.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 6.Millanes-Romero A, Herranz N, Perrera V, Iturbide A, Loubat-Casanovas J, Gil J, et al. Regulation of heterochromatin transcription by Snail1/LOXL2 during epithelial-to-mesenchymal transition. Mol Cell. 2013;52:746–57. doi: 10.1016/j.molcel.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 7.Barker HE, Cox TR, Erler JT. The rationale for targeting the LOX family in cancer. Nat Rev Cancer. 2012;12:540–52. doi: 10.1038/nrc3319. [DOI] [PubMed] [Google Scholar]
- 8.Cano A, Santamaria PG, Moreno-Bueno G. LOXL2 in epithelial cell plasticity and tumor progression. Future Oncol. 2012;8:1095–108. doi: 10.2217/fon.12.105. [DOI] [PubMed] [Google Scholar]
- 9.Ahn SG, Dong SM, Oshima A, Kim WH, Lee HM, Lee SA, et al. LOXL2 expression is associated with invasiveness and negatively influences survival in breast cancer patients. Breast Cancer Res Treat. 2013;141:89–99. doi: 10.1007/s10549-013-2662-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Almendro V, Cheng YK, Randles A, Itzkovitz S, Marusyk A, Ametller E, et al. Inference of tumor evolution during chemotherapy by computational modeling and in situ analysis of genetic and phenotypic cellular diversity. Cell Rep. 2014;6:514–27. doi: 10.1016/j.celrep.2013.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 12.Escriva M, Peiro S, Herranz N, Villagrasa P, Dave N, Montserrat-Sentis B, et al. Repression of PTEN phosphatase by Snail1 transcriptional factor during gamma radiation-induced apoptosis. Mol Cell Biol. 2008;28:1528–40. doi: 10.1128/MCB.02061-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vega S, Morales AV, Ocana OH, Valdes F, Fabregat I, Nieto MA. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 2004;18:1131–43. doi: 10.1101/gad.294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDonald OG, Wu H, Timp W, Doi A, Feinberg AP. Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nat Struct Mol Biol. 2011;18:867–74. doi: 10.1038/nsmb.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peinado H, Moreno-Bueno G, Hardisson D, Perez-Gomez E, Santos V, Mendiola M, et al. Lysyl oxidase-like 2 as a new poor prognosis marker of squamous cell carcinomas. Cancer Res. 2008;68:4541–50. doi: 10.1158/0008-5472.CAN-07-6345. [DOI] [PubMed] [Google Scholar]
- 16.Herranz N, Pasini D, Diaz VM, Franci C, Gutierrez A, Dave N, et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol. 2008;28:4772–81. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, et al. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soutoglou E, Misteli T. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 2008;320:1507–10. doi: 10.1126/science.1159051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonilla CY, Melo JA, Toczyski DP. Colocalization of sensors is sufficient to activate the DNA damage checkpoint in the absence of damage. Mol Cell. 2008;30:267–76. doi: 10.1016/j.molcel.2008.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gursoy-Yuzugullu O, House N, Price BD. Patching broken DNA: nucleosome dynamics and the repair of DNA breaks. J Mol Biol. 2016;428:1846–60. doi: 10.1016/j.jmb.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8:870–6. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 22.Burgess RC, Burman B, Kruhlak MJ, Misteli T. Activation of DNA damage response signaling by condensed chromatin. Cell Rep. 2014;9:1703–17. doi: 10.1016/j.celrep.2014.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ayrapetov MK, Gursoy-Yuzugullu O, Xu C, Xu Y, Price BD. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc Natl Acad Sci USA. 2014;111:9169–74. doi: 10.1073/pnas.1403565111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khurana S, Kruhlak MJ, Kim J, Tran AD, Liu J, Nyswaner K, et al. A macrohistone variant links dynamic chromatin compaction to BRCA1-dependent genome maintenance. Cell Rep. 2014;8:1049–62. doi: 10.1016/j.celrep.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holliday DL, Speirs V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011;13:215. doi: 10.1186/bcr2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–6. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–8. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsompana M, Buck MJ. Chromatin accessibility: a window into the genome. Epigenetics Chromatin. 2014;7:33. doi: 10.1186/1756-8935-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burgess RC, Misteli T. Not all DDRs are created equal: non-canonical DNA damage responses. Cell. 2015;162:944–7. doi: 10.1016/j.cell.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bayona-Feliu A, Casas-Lamesa A, Reina O, Bernues J, Azorin F. Linker histone H1 prevents R-loop accumulation and genome instability in heterochromatin. Nat Commun. 2017;8:283. doi: 10.1038/s41467-017-00338-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tuduri S, Crabbe L, Conti C, Tourriere H, HoltgrevE−Grez H, Jauch A, et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol. 2009;11:1315–24. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Rubio ML, Perez-Calero C, Barroso SI, Tumini E, Herrera-Moyano E, Rosado IV, et al. The Fanconi anemia pathway protects genome integrity from R-loops. PLoS Genet. 2015;11:e1005674. doi: 10.1371/journal.pgen.1005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gan W, Guan Z, Liu J, Gui T, Shen K, Manley JL, et al. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 2011;25:2041–56. doi: 10.1101/gad.17010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 36.Kaidi A, Jackson SP. KAT5 tyrosine phosphorylation couples chromatin sensing to ATM signalling. Nature. 2013;498:70–4. doi: 10.1038/nature12201. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–9. [PubMed] [Google Scholar]
- 38.Toth KF, Knoch TA, Wachsmuth M, Frank-Stohr M, Stohr M, Bacher CP, et al. Trichostatin A-induced histone acetylation causes decondensation of interphase chromatin. J Cell Sci. 2004;117:4277–87. doi: 10.1242/jcs.01293. [DOI] [PubMed] [Google Scholar]
- 39.Peinado H, Del Carmen Iglesias-de la Cruz M, Olmeda D, Csiszar K, Fong KS, Vega S, et al. A molecular role for lysyl oxidasE−like 2 enzyme in snail regulation and tumor progression. EMBO J. 2005;24:3446–58. doi: 10.1038/sj.emboj.7600781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schietke R, Warnecke C, Wacker I, Schodel J, Mole DR, Campean V, et al. The lysyl oxidases LOX and LOXL2 are necessary and sufficient to repress E-cadherin in hypoxia: insights into cellular transformation processes mediated by HIF-1. J Biol Chem. 2010;285:6658–69. doi: 10.1074/jbc.M109.042424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voloshenyuk TG, Landesman ES, Khoutorova E, Hart AD, Gardner JD. Induction of cardiac fibroblast lysyl oxidase by TGF-beta1 requires PI3K/Akt, Smad3, and MAPK signaling. CytoAbsorbance was detected atkine. 2011;55:90–7. doi: 10.1016/j.cyto.2011.03.024. [DOI] [PubMed] [Google Scholar]
- 42.Fischer Kari R., Durrans Anna, Lee Sharrell, Sheng Jianting, Li Fuhai, Wong Stephen T. C., Choi Hyejin, El Rayes Tina, Ryu Seongho, Troeger Juliane, Schwabe Robert F., Vahdat Linda T., Altorki Nasser K., Mittal Vivek, Gao Dingcheng. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527(7579):472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng Xiaofeng, Carstens Julienne L., Kim Jiha, Scheible Matthew, Kaye Judith, Sugimoto Hikaru, Wu Chia-Chin, LeBleu Valerie S., Kalluri Raghu. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527(7579):525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baldeyron C, Soria G, Roche D, Cook AJ, Almouzni G. HP1alpha recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J Cell Biol. 2011;193:81–95. doi: 10.1083/jcb.201101030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soria G, Polo SE, Almouzni G. Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol Cell. 2012;46:722–34. doi: 10.1016/j.molcel.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 46.Murga M, Jaco I, Fan Y, Soria R, Martinez-Pastor B, Cuadrado M, et al. Global chromatin compaction limits the strength of the DNA damage response. J Cell Biol. 2007;178:1101–8. doi: 10.1083/jcb.200704140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–77. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 48.Fong SF, Dietzsch E, Fong KS, Hollosi P, Asuncion L, He Q, et al. Lysyl oxidase-like 2 expression is increased in colon and esophageal tumors and associated with less differentiated colon tumors. Genes Chromosomes Cancer. 2007;46:644–55. doi: 10.1002/gcc.20444. [DOI] [PubMed] [Google Scholar]
- 49.Moreno-Bueno G, Salvador F, Martin A, Floristan A, Cuevas EP, Santos V, et al. Lysyl oxidase-like 2 (LOXL2), a new regulator of cell polarity required for metastatic dissemination of basal-like breast carcinomas. EMBO Mol Med. 2011;3:528–44. doi: 10.1002/emmm.201100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Torres S, Garcia-Palmero I, Herrera M, Bartolome RA, Pena C, Fernandez-Acenero MJ, et al. LOXL2 is highly expressed in cancer-associated fibroblasts and associates to poor colon cancer survival. Clin Cancer Res. 2015;21:4892–902. doi: 10.1158/1078-0432.CCR-14-3096. [DOI] [PubMed] [Google Scholar]
- 51.Wong CC, Tse AP, Huang YP, Zhu YT, Chiu DK, Lai RK, et al. Lysyl oxidase-like 2 is critical to tumor microenvironment and metastatic niche formation in hepatocellular carcinoma. Hepatology. 2014;60:1645–58. doi: 10.1002/hep.27320. [DOI] [PubMed] [Google Scholar]
- 52.Martin A, Salvador F, Moreno-Bueno G, Floristan A, Ruiz-Herguido C, Cuevas EP, et al. Lysyl oxidase-like 2 represses Notch1 expression in the skin to promote squamous cell carcinoma progression. EMBO J. 2015;34:1090–109. doi: 10.15252/embj.201489975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 54.Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, et al. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol. 2008;28:7337–44. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–51. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 56.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lerat E, Fablet M, Modolo L, Lopez-Maestre H, Vieira C. TEtools facilitates big data expression analysis of transposable elements and reveals an antagonism between their activity and that of piRNA genes. Nucleic Acids Res. 2017;45:e17. doi: 10.1093/nar/gkw953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tarazona S, Furio-Tari P, Turra D, Pietro AD, Nueda MJ, Ferrer A, et al. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 2015;43:e140. doi: 10.1093/nar/gkv711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.