Abstract
The Paenibacillus larvae infecting phage API480 (vB_PlaP_API480) is the first reported podovirus for this bacterial species, with an 58 nm icosahedral capsid and a 12 × 8 nm short, non-contractile tail. API480 encodes 77 coding sequences (CDSs) on its 45,026 bp dsDNA genome, of which 47 were confirmed using mass spectrometry. This phage has got very limited genomic and proteomic similarity to any other known ones registered in public databases, including P. larvae phages. Comparative genomics indicates API480 is a new species as it’s a singleton with 28 unique proteins. Interestingly, the lysis module is highly conserved among P. larvae phages, containing a predicted endolysin and two putative holins. The well kept overall genomic organisation (from the structural and morphogenetic modules to the host lysis, DNA replication and metabolism related proteins) confirms a common evolutionary ancestor among P. larvae infecting phages. API480 is able to infect 69% of the 61 field strains with an ERIC I genotype, as well as ERIC II strains. Furthermore, this phage is very stable when exposed to high glucose concentrations and to larval gastrointestinal conditions. This highly-specific phage, with its broad lytic activity and stability in hive conditions, might potentially be used in the biocontrol of American Foulbrood (AFB).
Subject terms: Environmental biotechnology, Pharmacology
Introduction
American foulbrood disease (AFB) is one of the most devastating bacterial diseases affecting honeybees and it is caused by Paenibacillus larvae, a Gram-positive worldwide-distributed spore forming bacterium. This infection begins when adult bees provide spore-contaminated food to larvae in the initial stages of development (first 36 hours after egg hatching) causing larvae death1,2.
The burning of hives and contaminated material is the compulsory action recommended by authorities to control the proliferation of AFB which causes devastating economic loss to the beekeeping industry and the environment. The use of antibiotics is not advised as they are not active against spores, and they cause further wide-spreading bacterial resistances3,4. They are also forbidden in Europe (Regulation (EEC) 2377/90 and further amendments).
Bacteriophages (phages) are now valuable solutions to this infection control. Phages are bacterial viruses that specifically infect their hosts, relying on the cell biosynthetic machinery to produce new viral particles. Phages are considered self-dosing and self-limiting antibacterial agents. After the host lysis, new phages are able to trigger new infection cycles to the surrounding hosts, resulting in exponential phage growth until no host is available5. An important feature of phages is their inactivity in the extracellular environment, thus being innocuous to animals or plants6. The potential of P. larvae phages as a tool for treating AFB has been explored by evaluating its efficacy both in infected laboratory-raised larvae7–9 and in infected experimental hives10. Up to date, 48 P. larvae phage genome sequences have been described. They all belong to the Siphoviridae family and they mostly encode known integration genes. Their genomes have been grouped into four clusters (with Fern, Harrison, Vegas and Halcyone as representative phages) and one singleton (phage Lily), based on genomic diversity11. All of these 48 phages seem to have a common evolutionary ancestor, showing an overall common structure.
The isolation and genomic characterization of the first podovirus infecting P. larvae is reported here, together with the evaluation of its viability in experimental conditions envisaging the possibility of using this phage in AFB control.
Results
Phage isolation and host range
The isolation of new P. larvae strains was carried out in order to broaden the geographic and genetic diversity of the collection. A field sample collection carried out throughout 2018 allowed the isolation of 45 strains: 29 from hives with visible signs of infection and 16 from apparently healthy brood. All isolated strains exhibited an identical fingerprint pattern after rep-PCR matching those produced by ERIC I reference strains (data not shown).
The phage vB_PlaP_API480 (API480) was isolated from a hive soil sample collected in Guadalajara (Spain).
A panel of 68 P. larvae strains (including reference strains) were used to evaluate the lytic activity of API480 (Table 1). API480 revealed a broad lytic spectrum, infecting 69% of the 61 field strains, of which 57% exhibited EOP scores greater than 10%. All remaining strains (31%) were lysed from without. API480 was also able to infect the ERIC II strain CCUG 48972 (EOP < 10%) and lysed without replication one of ERIC II, one of ERIC III and two of ERIC IV strains. Only the P. larvae strain LMG 16252 (ERIC III) was not lysed by this phage. Additionally, lysis tests in non-P. larvae strains revealed that API480 was able to infect B. circulans and B. coagulans, although those strains do not propagate the phage. All the others were not sensitive to the phage, including the 1st instar larvae commensal strains, L. kunkeei and P. apium alpha 2.2.
Table 1.
API480 lytic spectra and EOP against different strains (P. larvae strains were obtained from honey (01), dead larvae (02) and wax (03). The EOP was scored as 0 (negative), 1 (<10%), 2 (10–100%), 3 (>100%) and LFW (lysis from without). N/A (Non-applicable).
| Specie | Strain | Genotype | Score |
|---|---|---|---|
| Paenibacillus larvae | Pl01–03; Pl02-(23, 30b, 31, 33, 37, 46, 49, 56, 64, 66, 71, 72, 73, 74, 75, 76, 81, 84) | ERIC I | LFW |
| Paenibacillus larvae | CCUG 48973 | ERIC II | LFW |
| Paenibacillus larvae | LMG 15974 | ERIC III | LFW |
| Paenibacillus larvae | LMG 16247, LMG 16250 | ERIC IV | LFW |
| Paenibacillus larvae | LMG 16252 | ERIC III | 0 |
| Paenibacillus larvae | Pl02-(35, 52, 69, 77, 79, 80, 85) | ERIC I | 1 |
| Paenibacillus larvae | CCUG 48972 | ERIC II | 1 |
| Paenibacillus larvae | Pl02-(07, 13, 18, 27, 51, 89) | ERIC I | 2 |
| Paenibacillus larvae | LMG 9820 | ERIC I | 2 |
| Paenibacillus larvae | Pl02-(01, 07b2, 14, 21, 34, 36, 45, 47, 48, 50, 53, 54, 55, 57, 58, 59, 60, 61, 62, 63, 65, 67, 68, 70, 78, 83, 86, 87) | ERIC I | 3 |
| Paenibacillus larvae | Pl03–28 | ERIC I | 3 |
| Lactobacillus pentosus | DSM 20314 | N/A | 0 |
| Lactobacillus rhamnosus | CECT 288 | N/A | 0 |
| Lactobacillus paracasei | CECT 277 | N/A | 0 |
| Lactobacillus casei | CECT 5275 | N/A | 0 |
| Lactobacillus acidophilus | ATCC 4356 | N/A | 0 |
| Lactobacillus kunkeei | LMG 18925 | N/A | 0 |
| Bacillus subtilis | DSMZ 10 | N/A | 0 |
| Bacillus cereus | CEB collection | N/A | 0 |
| Bacillus circulans | CEB collection | N/A | LFW |
| Bacillus coagulans | CECT 12 | N/A | LFW |
| Parasaccharibacter apium | Alpha 2.2 | N/A | 0 |
| Paenibacillus polymyxa | LMG 13294 | N/A | 0 |
| Paenibacillus alvei | LMG 13253 | N/A | 0 |
Phage morphology
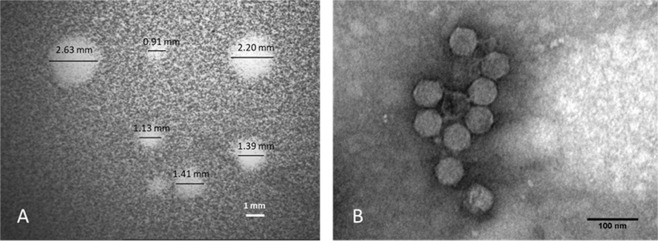
API480 forms clear plaques with diameters ranging from 0.9 to 2.6 mm, in 0.4% (w/v) agar plates (Fig. 1A). TEM images revealed the presence of phage particles with 58 nm diameter icosahedral capsids and 12 × 8 nm short non-contractile tails, belonging to the Podoviridae family (Fig. 1B).
Figure 1.
Characteristics of API480. (A) Plaque morphology (black lines indicate the diameter of API480 plaques obtained through a SZ40 Zoom Stereo Microscope (Olympus). Scale bar: 1 mm; (B) Transmission electron micrographs showing the virion particle morphology (stained with 2% uranyl acetate). Scale bar: 100 nm.
Phage genomic and proteomic properties
General overview
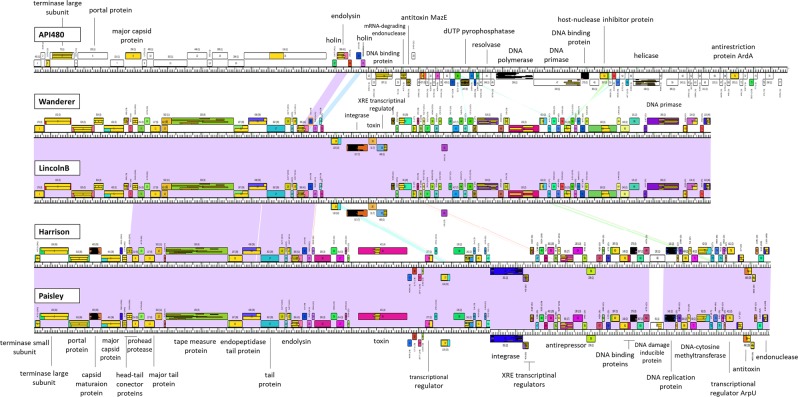
Phage API480 genome, deposited in the GenBank with the accession number MK533143, is a linear dsDNA molecule of 45,026 bp with 39.24% GC content. API480 encodes 77 coding sequences (CDSs), of which 60 have hypothetical function (being 28 unique to this phage) and only 17 with an assigned function (Supplementary Table S2). Genes are tightly packed achieving 1.71 genes per 1,000 bp, with the genome being 91.9% coded. Furthermore, API480’s genome has a translation of 65 proteins that start on ATG codon (84.4%), six on GTG codon (7.8%) and six on TTG codon (7.8%). Although no tRNA or antibiotic resistance genes were identified, ten promoters and eight factor-independent terminators were found, as well as components of the MazEF toxin-antitoxin module, mRNA-degrading endonuclease (gp26) toxin MazF and its antitoxin the MazE (gp27). The API480 genome is composed by a left-to-right followed by a right-to-left transcription module (Fig. 2). The DNA packaging and phage morphogenesis genes are located at the beginning of the left arm, similar to the P. larvae siphoviruses. Only three proteins with assigned function were identified in this region: terminase large subunit (gp4), portal protein (gp6) and the major capsid protein (gp8). The host lysis proteins are located in the middle of the genome. The endolysin (gp18) is predicted to function as a N-acetylmuramoyl-L-alanine amidase. There are two predicted holins downstream and upstream the endolysin, both with transmembrane domains. Homologs of these putative holin_bhlA (gp17) and the phage_holin_5 (gp21) have been found in most of the P. larvae siphoviruses proteins11. The DNA replication, transcription, and metabolism are located in the right arm of the API480 genome: predicted DNA binding protein (gp24), mRNA-degrading endonuclease (gp26), antitoxin MazE (gp27), dUTP pyrophosphatase (gp39), resolvase (gp44), DNA polymerase (gp46), DNA primase (gp47), single-stranded DNA-binding protein (gp48), host nuclease inhibitor protein (gp50), helicase (gp57) and antirestriction protein ArdA (gp70). A similar organization is also observed in the other P. larvae phages, for instance, in Wanderer, LincolnB, Harrison and Paisley (Fig. 2), with the exception of the lysogeny module, which was not identified in the API480 genome.
Figure 2.
Pairwise genome maps. API480 whole-genome was compared with the closest relatives, Wanderer, LincolnB, Harrison and Paisley. Maps were created with Phamerator. Pairwise sequence similarity (minimal BLASTN cut-off E-value is 10−5) is indicated according to colour spectrum where purple and red lines denote regions of highest and lowest nucleotide similarity, respectively. Gene products are labelled with predicted function (phams i.e. proteins members have the same colour, orphams i.e. unique proteins are shown in white). Their positioning either above or below the bar correspond to rightwards or leftwards transcription, respectively.
Comparative analysis
An initial API480 genome alignment by BLASTN revealed that API480 genome is distinct from other available genomes in the GenBank. The highest genome coverage obtained to an E-value of 0.0 was only 5% by LincolnB and Wanderer with an identity of 85.6% and 89.3%, respectively. As mentioned previously, the levels of amino acid identity of API480-encoding proteins are very limited and when existent they are mostly found against P. larvae phage proteins (<55% average amino acid identity), indicating the lack of a close relationship between API480 and any phage currently known.
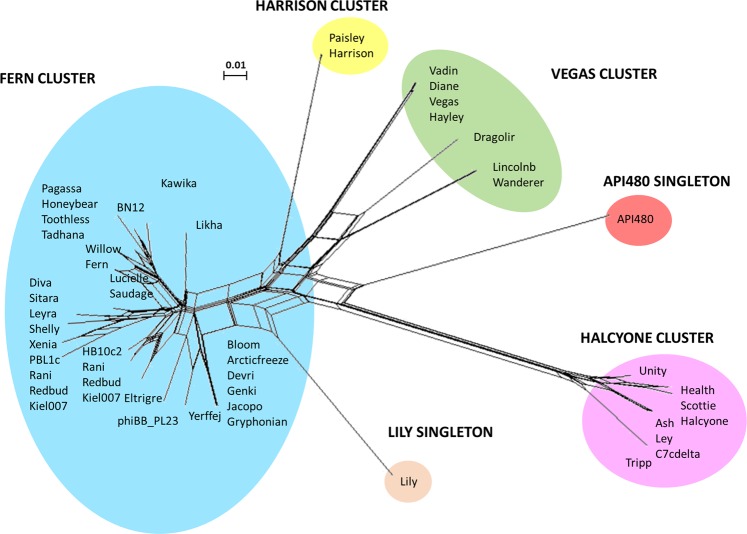
To improve the placement of the API480 genome in the context of the P. larvae phage population, we compared all P. larvae phages through shared gene content. Comparative analysis of all 49 P. larvae phage genomes resulted in the establishment of a new singleton enclosing API480 among four clusters (Fern, Harrison, Vegas and Halcyone) and one singleton (Lily), previously described by Stamereilers et al.11. The four clusters and the two singletons are shown on Fig. 3 and their relationship was represented by the branch lengths. This clustering joins phages that share more than 40% of their proteins with all members of the same cluster (Supplementary Table S3). While Lily still shares a maximum of 39% of its proteins with some phages (Arcticfreeze, Devri, Bloom, Jacopo, Genki and Gryphonian), API480 is an even more distant phage as its only shares 14% or fewer proteins. Wanderer (13.6%), LincolnB (13.6%), Harrison (11.2%) and Paisley (11.2%) can be considered as the closest relatives (Supplementary Table S3).
Figure 3.
Diversity of P. larvae phages genomes. A total of 49 P. larvae phages (48 siphoviruses and 1 podovirus – API480) were compared with Phamerator in 3D and the relationship of shared gene content was visualized into 2D space with Splitstree. Clusters assigned based on sharing >40% gene products are highlighted with colours.
Through the Phamerator analysis it was also possible to see that only 19 out of the 77 predicted API480 proteins are shared with P. larvae siphoviruses (Supplementary Table S4) and that Harrison and Vegas are the most closely related clusters to the API480. While no universal proteins were found, it is clear that the host lysis proteins (holin_bhlA, endolysin and the holin) are mostly conserved. This is evidenced by the presence of these proteins in all phages from different clusters and singletons, except for Halcyone cluster (Supplementary Table S4). It is also visible in the Phamerator genome map that the only area highlighted in purple shading between genomes (nucleotide sequence most similar), refers to the endolysin site (Fig. 2).
Mass spectrometry
To confirm API480 gene predictions, its proteome was analysed by mass spectrometry. An SDS-PAGE gel loaded with unpurified phage particles showed several bands (Supplementary Fig. S1). Thirteen gel fragments were extracted and further sequenced. A total of 47 proteins could be identified (Table 2). From the 28 unique proteins from API480, 11 revealed an amino acidic sequence coverage ranging from 24.3% to 69.5%. A total of 29 proteins presented homology to bacterial proteins, from which 20 had a cover range between 7.9% and 93.7%. Finally, from the 20 proteins with homology to other phage, 16 displayed a cover range between 12.5% and 89.2%.
Table 2.
Bacteriophage API480 proteins identified by ESI-MS/MS, after denaturation and phage particle fractionation on SDS-PAGE gel. Identified phage proteins are listed below. SDS-PAGE gel band in which the proteins were identified have been indicated as well as the protein mass, the number of identified unique peptides and the protein sequence that is covered by the peptide (in %).
| Protein | Identified Function | Band no. (most abundant) | Protein MW (kDa) | No. of unique peptides | Sequence coverage (%) |
|---|---|---|---|---|---|
| gp1 | 11,12 (11) | 16.55 | 6 | 50.40 | |
| gp2 | 11,12 (12) | 9.96 | 4 | 58.30 | |
| gp3 | 5,12 (12) | 11.95 | 6 | 55.90 | |
| gp4 | Terminase large subunit | 1,2,3,4,5,6,8,9,10,11 (1) | 55.47 | 14 | 36.66 |
| gp5 | 1,2,5,10,11,12 (12) | 7.05 | 3 | 38.10 | |
| gp6 | Portal protein | 1,2,3,4,5,6,7,8,9,10,11,12 (3) | 68.14 | 35 | 62.68 |
| gp7 | 1,2,3,4,5,6,7,8,9,10,11,12 (6) | 31.45 | 31 | 93.70 | |
| gp8 | Major capsid protein | 1,2,3,4,5,6,7,8,9,10,11,12 (7) | 34.38 | 34 | 89.16 |
| gp9 | 5,9,12,13 (12) | 9.88 | 7 | 67.80 | |
| gp10 | 11,12,13 (12) | 14.40 | 5 | 67.50 | |
| gp11 | 1,2,3,4,12(3) | 75.23 | 12 | 24.26 | |
| gp12 | 1,2,3,4,5,6,8,9,11 (5) | 43.56 | 17 | 50.75 | |
| gp13 | 12 | 11.64 | 7 | 68.00 | |
| gp14 | 2,6,7,8,9,10,11 (6) | 35.47 | 18 | 55.71 | |
| gp15 | 6,7,8,9,10,11,12 (6) | 37.05 | 19 | 63.00 | |
| gp16 | 1,2,3,4,5,6,7,8,9,10,11,12 (2) | 128.32 | 71 | 60.75 | |
| gp17 | Holin | 12 | 9.95 | 4 | 35.00 |
| gp20 | 12 | 12.52 | 3 | 42.90 | |
| gp21 | Holin | 12 | 8.27 | 1 | 26.60 |
| gp24 | DNA binding protein | 1,2,6,9,12 (1) | 36.60 | 5 | 15.58 |
| gp26 | mRNA-degrading endonuclease | 12 | 12.52 | 3 | 28.40 |
| gp30 | 11 | 14.13 | 1 | 13.70 | |
| gp34 | 12 | 9.76 | 1 | 16.30 | |
| gp36 | 10 | 14.43 | 1 | 7.94 | |
| gp37 | 12 | 10.62 | 3 | 59.40 | |
| gp38 | 12 | 12.33 | 2 | 37.40 | |
| gp39 | Deoxyuridine 5′-triphosphate nucleotidohydrolase | 1,2,3,4,5,6,8,9,10,12 (10) | 18.21 | 7 | 49.69 |
| gp40 | 10,11,12 (12) | 9.82 | 6 | 66.30 | |
| gp43 | 12 | 7.15 | 5 | 87.90 | |
| gp44 | Resolvase | 1,2,3,4,5,6,7,8,9,10,11,12 (11) | 19.12 | 8 | 56.90 |
| gp46 | DNA polymerase I | 1,2,3,4 (3) | 87.14 | 28 | 42.96 |
| gp47 | DNA primase | 1,2,3 (1) | 105.39 | 9 | 12.50 |
| gp48 | Dingle-stranded DNA-binding protein | 3,5,8,9,10,11,12 (10) | 17.60 | 11 | 62.10 |
| gp49 | 1,8,9,10,11,12 (9) | 20.18 | 13 | 69.48 | |
| gp50 | Host-nuclease inhibitor | 8,9,10 (8) | 21,75 | 10 | 60.99 |
| gp53 | 11,12 (12) | 8.58 | 1 | 18.40 | |
| gp54 | 12 | 11.83 | 3 | 41.20 | |
| gp55 | 12 | 6.91 | 2 | 53.30 | |
| gp57 | DEAD/DEAH box helicase | 1,2,4,8 (1) | 60.63 | 7 | 20.10 |
| gp59 | 8 | 25,95 | 7 | 44.70 | |
| gp60 | 11,12 (11) | 19.76 | 3 | 18.00 | |
| gp63 | 12 | 9.65 | 2 | 31.30 | |
| gp64 | 12 | 7.95 | 2 | 37.30 | |
| gp65 | 1,11,12 (11) | 15.46 | 6 | 65.39 | |
| gp70 | Antirestriction protein ArdA | 9 | 18,92 | 3 | 19.90 |
| gp76 | 1,11,12 (11) | 17.36 | 7 | 59.60 | |
| gp77 | 12,13 (12) | 8.14 | 4 | 63.40 |
Phage potential for biocontrol application in hives
The potential of the therapeutic use of API480 in apiaries, as far as AFB control is concerned, was investigated through the assessment of the phage growth dynamic, life cycle and stability on field conditions.
Phage integration assays
Although no integrase was identified in API480 genome by the in silico analysis, the ability of the phage to lysogenise its host was investigated to obtain more consistent information about the phage cycle. Assuming that a lysogenized host might become resistant to the recently integrated phage by acquiring phage genetic material, the infectivity of API480 was assessed after lysogeny induction together with the presence of phage genes in host resistant colonies.
Contrary to the original Pl02–27 strain, results revealed that the phage lost the ability to infect R-Pl27 strains, and that the DNA of the same strains allowed the amplification of a 227 bp band (specific for API480) by PCR (Supplementary Fig. S3).
To assess if the use of a phage cocktail could be relevant in supporting API480 in AFB infections, the activity of the other five P. larvae phages from our collection against R-Pl27 colonies was investigated and all of them were infective.
Phage infection parameters
The assessment of the phage generation time and the phage population growth level was accomplished through the determination of phage infection parameters. Adsorption assays revealed that during the first minutes of phage contact with its host, the number of free phages rapidly decreased. After 35 min approximately 85% of the total API480 phage particles were adsorbed to its host (Fig. 4A).
Figure 4.
Phage-host interaction parameters. (A) Percentage of free API480 phages after infection of P. larvae (MOI = 0.1). (B) One-step growth curve of phage API480 in P. larvae Pl02–27. Shown are the PFU per infected cell. Each point represents the average of three independent assays and error bars indicate the standard deviation. Statistical significance, p < 0.05.
As far as phage growth cycle parameters are concerned, the calculated latent period was approximately 30 min and the phage burst size was approximately 3 PFU per infected cell (Fig. 4B).
Phage stability in simulated field conditions
The API480 phage stability in a 50% (w/v) sucrose solution was assessed in vitro for 24 hours. The results revealed that, at least in this time period, the phage viability was not impacted (Supplementary Fig. S3).
The evaluation of phage viability in RJ (pH 4.0) revealed a total loss of phage infectivity after 6 hours (Fig. 5A).
Figure 5.
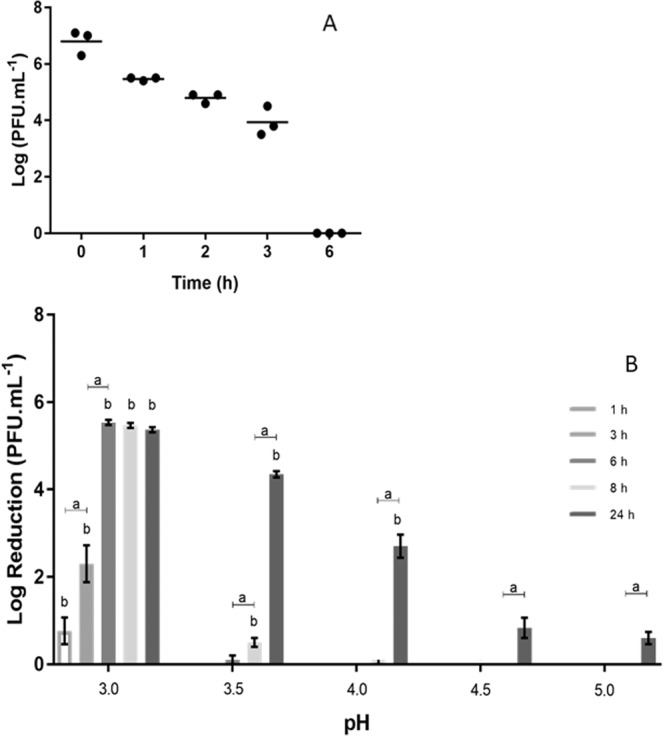
(A) Effect of commercial RJ on the stability of API480 (PFU.mL−1). Data show each of three independent assays. Limit of detection (LOD) = 3 Log; Statistical significance, p < 0.05. (B) Effect of pH (from 3.0 to 5.0) in API480 phage concentration (PFU.mL−1). Each column represents the average of three independent assays and error bars indicate the standard deviation. LOD = 1 Log; Statistical significance, p < 0.05; “a” indicates differences for the same pH; for each timepoint, “b” indicates differences between data from a given pH and the subsequent pH value.
The impact of low pH solutions (3.0, 3.5, 4.0, 4.5 and 5.0) in API480 stability was also assessed. Results, presented in Fig. 5B, revealed that after 6 hours the phage was completely inactivated at pH 3.0 (5.5 Log reduction in average). Interestingly, at pH 3.5, at the same time point, it only caused a slight decrease of 0.1 Log PFU.mL−1, being accentuated (p < 0.05) after 24 hours, with an average reduction of 4.3 Log PFU.mL−1. No effect was observed with the other pH values when assessed for periods lower than 24 hours. Nevertheless, at this time point, the average phage lost was of 2.7, 0.8 and 0.6 Log PFU.mL−1 at pH 4.0, 4.5 and 5.0, respectively.
The assessment of the phage stability in larvae homogenised revealed that a slight and not statistically meaningful phage reduction of 0.4 Log PFU.mL−1 (p > 0.05) was observed, and only after 24 h.
Discussion
The isolation of the first genomic sequence of a P. larvae podovirus, vB_PlaP_API480 (API480) is described here. So far, all P. larvae phages (n = 48) have been reported to be siphoviruses, with a temperate lifestyle and the majority (n = 40) with a wide genetic similarity. The only exception is the Halcyone cluster (n = 8) that is quite diverse from other P. larvae phages11. We showed that API480 has very limited genomic and proteomic relatedness to any phage deposited in public databases, including P. larvae phages. From the 77 CDS predicted in the phage genome, 47 were confirmed by MS/MS. Genomic comparison analysis demonstrated that API480 is a singleton with 28 unique proteins, sharing only 19 of its proteins with other P. larvae siphoviruses, mostly with Harrison and Vegas clusters. This can be interpreted as a recent evolutionary history between API480 and these phages. It was surprising to see how the API480 host lysis proteins seem to be composed of one endolysin and two putative holins and how these proteins are the most conserved among P. larvae phages. Although these putative holins need to be experimentally confirmed, they apparently provide a new way for phages to lyse its host cells at the end of its lytic cycle. As the majority of phages known today, API480 and the other P. larvae phages also have their genomes organised in mosaics, starting with the structural and morphogenetic modules that are followed by the host lysis, DNA replication and metabolism related proteins. They all seem to have an overall common structure despite the limited shared gene content between them. This reinforces the existence of a common evolutionary ancestor among these phages. The low frequency of singletons found in P. larvae phages (from n = 49, 4% are singletons) which differs from other phage populations infecting hosts of similar taxonomic level, such as Bacillus (from n = 83, 18.1% are singletons)12 and Gordonia phages (from n = 79, 17.7% are singletons)13, indicates that the full genetic diversity is still untapped. This can be the result of similar isolation techniques used but not the outcome of low environmental sampling diversity, as P. larvae phages have been isolated from different isolation sources and geographical regions11. Overall, the genomic and proteomic analysis show that API480 is a completely distinct phage, suggesting the creation of a new genus within the Podoviridae family.
The analysis of the firstly reported genetic sequence of API480, widely distinct from the others (maximum genome coverage of 5%), encouraged its detailed characterization considering its use in AFB control.
The evaluation of API480 life cycle was accomplished based on the in silico analysis together with the assessment of the phage ability to integrate the bacterial host genome. The ability to lysogenise bacteria by a temperate phage is based on a gene cluster responsible for integration (integrase) and maintenance (repressors of the lytic cycle) of the prophage. Unlike all the other P. larvae phages no lysogeny module and no integrase gene were found in API480 genome.
The temperate nature of the phage was confirmed by the detection of a phage gene in the host genome and by the conversion of Pl02–27 into a phage-resistant strain. Although the use of temperate phages for therapeutic purposes is accepted, it raises several concerns14. Some strategies are known to safeguard its use for this purpose based on the difficulty of isolating strictly lytic phages, which was already reported for other bacterial species. Recently, Nale et al.15 revealed that all published Clostridium difficile phages are so far temperate and encode an integrase gene. Meader and colleagues (2010)16 anticipated that this is possibly due to the high incidence of prophage genes, revealing resistance to further infections. They also infer that the spore form may favour phage integration into the genome.
The use of optimized phage combinations with distinct and often complementary features (such as host range), as a strategy to lighten the effect of lysogeny and consequent phage resistance, has already been demonstrated14. The susceptibility of R-Pl27 strains to other P. larvae phages from our collection confirmed this idea.
Recent reports on the use of temperate phages for therapy were recently reviewed17. For example, phage ØCD27 was used to control C. difficile infections reducing their presence in the colon and decreasing the toxin expression18. Nale et al.14 reported the reduction of C. difficile colonisation by a cocktail of temperate phages extending the life expectancy of mice. A Pseudomonas aeruginosa systemic infection was treated in flies and in a murine animal model using two temperate phages (MP22 and D3112)19.
As far as AFB control is concerned, spore infected lab-reared larvae were also successfully treated with cocktails of P. larvae phages with a temperate lifestyle8,9.
API480 revealed to have broad lytic spectra, being active against 69% of the isolated field strains. All these strains belong to ERIC I, the far more diverse worldwide genotype20 and the one that causes higher morbidity in hives21. API480 also seems to be able to infect and propagate in ERIC II strains. Nevertheless, no evidence was found that the phage can be used to control infections caused by ERIC III and IV strains, as in this case the lysis occur without phage replication.
According to Yost et al.9 phages show host preference for the ERIC group from which they were isolated, and the preference for ERIC I was also observed in other P. larvae phages.
The high specificity that this phage revealed for P. larvae is particularly important as far as the first-instar larval commensal bacteria, L. kunkeei and P. apium alpha 2.2 are concerned, as it indicates that this phage would not have a harmful impact on their gut microbiota22,23. While studying the phage growth parameters, API480 revealed a slow adsorption to its host (in 35 min) as reported for example in the C. difficile phage CDHM1 (30 min)24 and a release of as few as three progeny viruses per infected cell. A burst of around eight phages per cell was recently reported in the P. larvae phage HB10c27 and a burst of five and seven phages per cell was obtained in other two temperate phages isolated from C. difficile, φC2 and φC5, respectively25. The latter authors hypothesised that these result could be due to suboptimal growth conditions, explanation also given by Touchon et al.26 while studying life-history traits of temperate phages associated with lysogeny. According to a study carried out by Hadas et al.27 with T4 phage, parameters of phage development and cell lysis are dependent on bacterial growth rate. Correspondingly, slow adsorption rates, small burst size and high latent period were expected in a slow-grower bacterium such as P. larvae28.
The stability of API480 in simulated hive conditions was assessed to learn about its suitability in AFB biocontrol. API480 was very stable when exposed to high glucose concentrations (frequently used for feeding bees) and to pH values higher than 4.0 (often found in the hive).
Moreover, the phage infectivity was almost unaffected by larval fluids (0.4 Log PFU.mL−1 reduction) indicating that, at least for 24 hours, this is a favourable compartment for the host infection. Nevertheless, before reaching the larvae, phages will be mixed with RJ during the crop content regurgitation back to the mouth29 and they will be available in brood combs for larval consumption. According to our results, larvae have no more than 6 hours to ingest phages in a viable state. The decrease in phage viability at some point was an expected result, as Ribeiro et al.30 have recently reported. Some RJ elements may be responsible for this antiviral effect, such as proteinases31 and phenolic compounds32 that probably interact with phage structural proteins, contributing to their inactivation33.
In conclusion, despite being a temperate phage, the API480 broad lytic spectra, the specificity to P. larvae and the behaviour when challenged by simulated hive conditions encouraged its use for therapeutic purposes. Furthermore, the possibility of administering it together with other P. larvae phages which are able to infect API480 resistant strains mitigates the lysogenic nature of the phage.
Materials and Methods
Bacterial strains and cultivation conditions
In this study, 23 previously isolated P. larvae strains were used: 13 field strains (Pl01-(01, 03, 07, 07b2, 13, 14, 18); Pl02-(21, 23, 27, 30b, 31); Pl03–28)34; three strains originally isolated in Spain in 2016 (Guadalajara) (Pl02–86, 87, 89); seven reference strains: LMG 9820, CCUG 48972, CCUG 48973; LMG 15974 and LMG 16252 LMG 16247, LMG 16250.
These strains were cultivated in MYPGP agar (10 g.L−1 Mueller-Hinton Broth (Oxoid); 15 g.L−1 yeast extract (Oxoid); 3 g.L−1 de K2HPO4 (LabKem); 1 g.L−1 de Sodium-pyruvate (Fisher); 2% glucose (Ameresco) and 17 g.L−1 agar (VWR) and incubated at 37 °C under 5% CO2 overnight (O/N).
P. larvae isolation was performed as described by Genersch and Otten (2003)35, from brood samples collected in 132 hives spread over the Portuguese territory (29 with visible signs of infection and 103 apparently non-infected): larvae were emulsified in 500 µL sterile water, heated at 90 °C, 6 min and sewed in MYPGP agar. After incubation for 3 to 6 days at 37 °C, 5% CO2, single colonies were propagated in MYPGP agar and stored at −80 °C with 20% glycerol.
Non-P. larvae strains used to assess phage specificity for P. larvae included: Paenibacillus polymyxa (LMG 13294), Paenibacillus alvei (LMG 13253), Lactobacillus pentosus (DSM 20314), Lactobacillus rhamnosus (CECT 288), Lactobacillus paracasei (CECT 277), Lactobacillus casei (CECT 5275), Lactobacillus acidophilus (ATCC 4356), Bacillus subtilis (DSMZ 10), Bacillus coagulans (CECT 12), Bacillus cereus (CEB collection), Bacillus circulans (CEB collection), Lactobacillus kunkeei (LMG 18925) and Parasaccharibacter apium alpha 2.2 (strain C6)22.
All Lactobacillus spp were cultured in MRS broth (Frilabo) and MRS 15 g.L−1 agar (VWR). Bacillus spp, P. polymyxa and P. alvei were cultured in Nutrient broth ((5 g.L−1 Peptone (Amresco) and 3 g.L−1 Meat extract (Fluka Biochemika)) and Nutrient agar (15 g.L−1 agar). P. apium alpha 2.2 was cultured in Sabouraud dextrose broth (SDB) and sewed in SDB 15 g.L−1 agar (VWR).
The CCUG strains were obtained from the Culture Collection of the Goteborg University, LMG from the BCCM - Belgian Coordinated Collections of Microorganisms, DSMZ from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH and CECT from the Colección Española de Cultivos Tipo.
16S-PCR identification of P. larvae and rep-PCR analysis
The bacterial DNA was purified from bacterial suspensions using the Quick-DNA Fungal/Bacterial Miniprep Kit (Zymo) and amplified using KapaTaq (Kapa Biosystems) according to the manufacturer’s instructions.
The PCR primer sequences and conditions used for P. larvae identification (Supplementary Table S1) were based on the P. larvae 16S rRNA gene36. Positive results revealed a 1,106 bp band in a 1% (w/v) agarose gel under UV light.
The enterobacterial repetitive intergenic consensus (ERIC) genotyping of the previously identified P. larvae was accomplished through genomic fingerprinting as reported in Genersch and Otten (2003)35 (primers and conditions are detailed in Supplementary Table S1). ERIC patterns were visualised in a 2% agarose gel under UV light. LMG 9820, CCUG 48972 LMG 15974 and LMG 16247 were used as standard for ERIC I, II, III and IV profiles, respectively.
Bacteriophage isolation and production
Soil samples from hive surroundings were used for phage isolation. For that, samples were mixed with groups of five different bacterial strains pre-cultured O/N in MYPGP broth (37 °C, 5% CO2). After another O/N incubation, the supernatant was filtered-sterilized through 0.22 µm PES membranes (GE Healthcare) and 10 µL were spotted on the respective bacterial lawn (bellow designated as “spot test”). For lawns preparation 100 µl of the freshly grown strain with 3 mL 0.4% MYPGP agar and poured into agar plates. After O/N incubation at 37 °C, 5% CO2 bacterial inhibition zones were picked and propagated over host bacterial lawns37. After a subsequent incubation, phages were isolated from a single phage plaque. A volume of 2 mL SM buffer (5.8 g.L−1 NaCl (PanReac); 2 g.L−1 MgSO4.7H2O (VWR); 50 mL.L−1 1 M Tris-HCl pH 7.5 (VWR)) was added to the plates. The floating liquid together with the soft-agar were collected, centrifuged (10 min, 9000 × g, 4 °C) and filtered-sterilized through 0.22 µm PES membranes. Phages were stored at 4 °C until use.
For phage propagation, 10 µL of the stored phage suspension were spread evenly on host bacterial lawns38. Plates were incubated O/N at 37 °C with 5% CO2 and treated as described above for phage isolation, until a filter-sterilized high-titre phage suspension around 108 PFU.mL−1 (PFU: phage plaque count) was obtained (stored at 4 °C). The diameter of six individual phage plaques was registered using a SZ40 Zoom Stereo Microscope (Olympus).
Lytic spectra determination and efficiency of plating
The lytic activity of the isolated phage vB_PlaP_API480 (API480) was tested against 68 P. larvae strains through spot test, as described above for phage isolation. The presence of bacterial inhibition areas was indicative of host susceptibility to the phage, and these strains were further used to assess efficiency of plating (EOP). A volume of 10 µL of serial phage dilutions was placed in each new bacterial lawn and the drop was allowed to drip along the agar surface to facilitate the phage plaque counting. The relative EOP was calculated by dividing the PFU.mL−1 of each susceptible strain by the titre for the relevant propagating host (Pl02–27)39. The EOP was scored as 0 (negative), 1 (<10%), 2 (10–100%), 3 (>100%) and LFW (Lysis from without) if phage plaques are only visible at the highest dilutions.
Electron microscopy analysis
Phage particles were collected by centrifugation (1 h, 25,000 × g, 4 °C) in a Beckman J2–21 centrifuge with a JA18.1 fixed rotor. The sediment was washed twice in tap water prior to centrifugation as above. Phages were deposited on copper grids with a carbon-coated Formvar film grid, stained with 2% uranyl acetate (pH 4.0) and examined using Jeol JEM 1400 transmission electron microscope (Tokyo, Japan).
DNA isolation, genome sequencing and annotation
P. larvae API480 phage genomic DNA was isolated using the phenol-chloroform-isoamyl alcohol method essentially as described elsewhere39. DNA samples were further used for library construction using the Illumina Nextera XT library preparation kit. The DNA libraries generated were sequenced in the lllumina MiSeq platform, using 250 bp paired-end sequencing reads. An automatic initial treatment was performed on raw sequence data, namely adapters and low quality bases trimming. Demultiplexed reads were de novo assembled into a single contig using Geneious R9 (Biomatters, Newark, NJ, USA).
The assembled genomes were scanned through MyRAST to search for coding regions40 and tRNAscan-SE to search for tRNAs41. To search for function, proteins were analysed through BLASTP42 and HHpred43 using an E-value cutoff of 1 × 10–5 to search for similarities. Identified proteins were also analysed with TMHMM44 and SignalP to predict transmembrane domains and signal peptide cleavage sites45. Putative promoter regions were checked using PromoterHunter from phiSITE46 and were further manually verified. ARNold was used to predict factor-independent terminators47 and the energy was calculated using Mfold48. The total genome or proteome were checked for antibiotic resistance genes through the ResFinder49 and the Resistance Gene Identifier (RGI) of CARD (The Comprehensive Antibiotic Resistance Database). The selected criteria were the display of results with perfect, strict and loose hits. For the search of toxins, the Toxin-Antitoxin Database (TADB) was used with the TAfinder tool50.
Comparative genomic analysis
To determine the relationship of the P. larvae API480 podovirus within all P. larvae phages, all complete genomes sequences deposited at GenBank as of June 2019 (n = 48) were retrieved and analysed as previously described51. Briefly, the shared gene content was analysed with Phamerator52. This program allowed (1) the assignment of all P. larvae phage gene products into phams (proteins with related sequences) or orphams (i.e. unique proteins) using with kclust, an alignment-free algorithm; (2) the generation of pairwise comparison genomic maps; and (3) the identification of conserved domains in all proteins using the NCBI conserved domain database. The resulting protein repertoire relatedness was visualized with SplitsTree53. Phage membership was assigned based on shared gene content a metric recently used to assigned staphylococcal phages51, using a cut-off of 40% of shared genes (phams) to assign phages solely in one cluster.
Protein identification by mass spectrometry (ESI-MS/MS)
Mass spectrometry was performed as described in Oliveira et al.54. Briefly, phage proteins were extracted from a phage stock, prepared as described above (>109 PFU.mL−1) using chloroform:methanol (Acros Organics) (1:1:0.75 in volume). Proteins were subsequently separated by standard SDS-PAGE and stained by Gelcode™ Blue Safe Protein Stain (Thermo Scientific). Bands spanning the entire lane of the gel were cut out and subjected to trypsin digestion. The resulting peptides were then identified by ESI-MS/MS based on a database file containing all predicted phage proteins.
Phage adsorption and One-step growth curve
An O/N grown Pl02–27 culture was harvested by centrifugation (10 min, 8000 × g, 4 °C) and re-suspended in fresh medium to obtain an optical density at 620 nm (OD620) of 0.3 (approximately 3 × 107 CFU.mL−1). The phage was added to the obtained suspension of Pl02–27 with a multiplicity of infection (MOI) of 0.1 and incubated at 37 °C with shaking (120 rpm in a PSU-10i Orbital Shaker, BIOSAN).
To assess the time the phage takes to adsorb to the host, samples of 50 µL were collected every 10 min and for 60 min and immediately chloroform-treated (1:10 (v/v)) and centrifuged (2 min, 8000 × g). The upper phase was serial diluted and the phage was titred (PFU.mL−1) in order to obtain the number of free phages38.
For the one-step growth curve (OSGC) the phage-host adsorption occurred for 35 min (as indicated by the adsorption assay) at 37 °C, 120 rpm. The mixture was then centrifuged (10 min, 8000 × g, 4 °C) and the pellet re-suspended in 10 mL fresh MYPGP broth medium. Samples were taken every 15 min over a period of 60 min and the phage titre was assessed.
The PFU.cell−1 was obtained through the ratio between the PFU.mL−1 in each time point and the initial PFU.mL−1. The burst size was estimated from the resultant sigmoid curve.
Phage life cycle
The evaluation of the phage ability to integrate the bacterial host genome was adapted from a procedure suggested by Kalatzis et al.55. Briefly, an O/N grown culture of the strain Pl02–27 was harvested by centrifugation (10 min, 8,000 × g, 4 °C) and re-suspended in fresh medium to reach an OD620 of 0.3. After a tenfold dilution the host suspension was mixed with the phage to get a MOI of 50 and incubated at 37 °C, 5% CO2 for 24 hours with agitation (120 rpm).
Samples (n = 3) were serial diluted and poured in MYPGP agar (incubation at 37 °C, 5% CO2, O/N). About 10 bacterial colonies were isolated, cultured in solid media and re-cultured again in three serial passages.
The life cycle of API480 was assessed by investigating the presence of the phage into the host genome in each of the isolated colonies. The original Pl02–27 strain was used as control. For that, a specific primer pair for API480 was designed based on the CDS_12: 480_12 Fw 5′-CAGGAACTCAGACCCTACGC -3′ and 480_12 Rev 5′-GCCTGCTGCAAAGTCATACA-3′. A colony PCR reaction was performed in a MJ Mini Personal Thermal Cycler: 10 min at 95 °C followed by 30 cycles of 15 sec at 95 °C; 15 sec at 60 °C and 15 sec at 72 °C and a final extension of 3 min at 72 °C. For the reaction mixture a 1x Xpert Fast Master mix (Grisp) and 0.8 µM of each primer were used. A positive reaction revealed a 227 bp band in a 1% agarose gel.
The API480 activity against the obtained colonies was then investigated trough phage spot test in the respective bacterial lawn. Four Pl02-H27 bacterial clones newly resistant to the phage (designated as R-Pl27) were then tested for phage infection using other P. larvae bacteriophages isolated in previous works: phiIBB_Pl2356; vB_Pl_CEB16, vB_PlCEB_46, vB_PlCEB_51 and vB_PlCEB_55 (unpublished phages from CEB collection).
Phage specificity and stability in simulated field conditions
The phage specificity for P. larvae was evaluated trough spot test on lawns of the above-mentioned non-P. larvae bacterial strains including the first instars larval commensal strains L. kunkeei and P. apium alpha 2.2 (strain C6)22.
API480 stability in simulated hive products/solutions was assessed for 24 hours: phage (final concentration of 107 PFU.mL−1) was incubated in a 50% (w/v) sucrose solution at room temperature (envisaging phage administration in bees’s artificially feed), in royal jelly (RJ) at 37 °C, 5% CO2 (where first–instars larvae lay on) and in solutions with acidic pH values at 37 °C, 5% CO2 (that include those occurring in the hive57,58).
For the assay with RJ (supplied by Apiguarda, Portugal), API480 was 1:10 (v/v) diluted in 100 µL of this hive-product and incubated for 0, 1, 3, 6, 8 and 24 hours (3 reaction tubes were prepared for each time point). After the addition of 900 µL SM buffer the mixture was homogenised by vortexing and 200 µL of chloroform were added. Tubes were vortexed and centrifuged (2 min, 14000 × g) and the upper phase was titred (PFU.mL−1).
For the pH assay distinct solutions of universal buffer (UB) (150 mM KCl (PanReac), 10 mM KH2PO4 (PanReac), 10 mM Sodium-Citrate (Thermo Fisher Scientific) and 10 mM H3BO3 (Thermo Fisher Scientific), were adjusted with HCl (Acros Organics) to obtain a pH range between 3.0 to 5.0 with intervals of 0.5 units. API480 was 1:10 (v/v) diluted and incubated. Two controls were used in this experiment: phage in SM buffer (pH 7.4) and phage in UB (pH 7.4). Phage concentration (PFU.mL−1) was assessed at 0, 1, 3, 6, 8 and 24 hours.
The phage infectivity was still performed in 15 healthy larvae gathered from larvae combs, that were weighted, individually homogenised and incubated with API480 for 24 hours: 100 µL of each homogenate with 107 PFU.mL−1.
After, 900 µL of SM buffer and 200 µL of chloroform were added. Phages were collected from the upper phase of the mixture after centrifugation (2 min, 14000 × g) and titred (PFU.mL−1).
Statistical analysis
The statistical analysis of the results was performed using GraphPad Prism 7. In all the assays, means and standard deviations were determined based on 3 independent experiments (n = 3). Results were compared using t-test (phage stability in sucrose 50% (w/v)), one-way ANOVA, with Turkey’s multiple comparison statistical test (phage viability on RJ, phage adsorption and OSGC), and two-way ANOVA, with Turkey’s multiple comparison statistical test (phage stability with acidic pH and in larvae homogenised). All tests were performed with a confidence level of 95%. Differences were considered statistically different if p ≤ 0.05.
Supplementary information
Acknowledgements
The authors would like to aknowledge Dr. Mariano Higes Pascual from Laboratorio de Patología Apícola del Centro de Investigación Apícola y Agroambiental de Marchamalo (CIAPA-IRIAF) for gently providing us P. larvae isolates and the soil sample from which API480 was isolated. This study, carried out under the scope of the project APILYSE, PTDC/CVT-EPI/4008/2014 - POCI-01–0145-FEDER-016598, was supported by FCT - Fundação para a Ciência e a Tecnologia, under the scope of UID/BIO/04469/2019 unit and BioTecNorte operation (NORTE-01-0145-FEDER-000004) funded by the European Regional Development Fund under the scope of Norte2020- Programa Operacional Regional do Norte. HR was supported by FCT through the grant SFRH/BD/128859/2017. MB and RL were supported by KU Leuven through a GOA grant [3E140356].
Author contributions
H.G.R. performed the laboratory work and wrote the manuscript. H.G.R., L.M., J.A. and A.O. analysed the data. R.L., M.B. and J.P.N. performed the by ESI-MS/MS analysis. L.M. did the taxonomic analysis and together with H.O. and H.G.R performed the comparative genome analysis. The work was designed and supervised by A.O. All authors read, reviewed and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-56699-y.
References
- 1.Crailsheim K, Riessberger-Gallé U. Honey bee age-dependent resistance against American foulbrood. Apidologie. 2001;32:91–103. doi: 10.1051/apido:2001114. [DOI] [Google Scholar]
- 2.Genersch E, Ashiralieva A, Fries I. Strain- and Genotype-Specific Differences in Virulence of Paenibacillus larvae subsp. larvae, a Bacterial Pathogen Causing American Foulbrood Disease in Honeybees. Appl. Environ. Microbiol. 2005;71:7551–7555. doi: 10.1128/AEM.71.11.7551-7555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Genersch E. American Foulbrood in honeybees and its causative agent, Paenibacillus larvae. J. Invertebr. Pathol. 2010;103:S10–S19. doi: 10.1016/j.jip.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 4.Miyagi T, et al. Verification of oxytetracycline-resistant American foulbrood pathogen Paenibacillus larvae in the United States. J. Invertebr. Pathol. 2000;75:95–96. doi: 10.1006/jipa.1999.4888. [DOI] [PubMed] [Google Scholar]
- 5.Drulis-Kawa Z, Majkowska-Skrobek G, Maciejewska B. Bacteriophages and phage-derived proteins – Application approaches. Curr. Med. Chem. 2015;22:1757–1773. doi: 10.2174/0929867322666150209152851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atterbury RJ. Bacteriophage biocontrol in animals and meat products. Microb. Biotechnol. 2009;2:601–612. doi: 10.1111/j.1751-7915.2009.00089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beims H, et al. Paenibacillus larvae-Directed bacteriophage HB10c2 and its application in American foulbrood-affected honey bee larvae. Appl. Environ. Microbiol. 2015;81:5411–5419. doi: 10.1128/AEM.00804-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghorbani-Nezami S, LeBlanc L, Yost DG, Amy PS, Jeanne R. Phage therapy is effective in protecting honeybee larvae from American foulbrood disease. J. Insect Sci. 2015;15:1–5. doi: 10.1093/jisesa/iev051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yost DG, Tsourkas P, Amy PS. Experimental bacteriophage treatment of honeybees (Apis mellifera) infected with Paenibacillus larvae, the causative agent of American foulbrood disease. Bacteriophage. 2016;6:e1122698. doi: 10.1080/21597081.2015.1122698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brady TS, et al. Bacteriophages as an alternative to conventional antibiotic use for the prevention or treatment of Paenibacillus larvae in honeybee hives. J. Invertebr. Pathol. 2017;150:94–100. doi: 10.1016/j.jip.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 11.Stamereilers C, et al. Genomic Analysis of 48 Paenibacillus larvae Bacteriophages. Viruses. 2018;10:377. doi: 10.3390/v10070377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grose JH, Jensen GL, Burnett SH, Breakwell DP. Genomic comparison of 93 Bacillus phages reveals 12 clusters, 14 singletons and remarkable diversity. BMC Genomics. 2014;15:855. doi: 10.1186/1471-2164-15-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pope WH, et al. Bacteriophages of Gordonia spp. Display a Spectrum of Diversity and Genetic Relationships. MBio. 2017;8:1–20. doi: 10.1128/mBio.01069-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nale JY, et al. Bacteriophage Combinations Significantly Reduce Clostridium difficile Growth In Vitro and Proliferation In Vivo. Antimicrob. Agents Chemother. 2016;60:968–981. doi: 10.1128/AAC.01774-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nale J, Redgwell T, Millard A, Clokie M. Efficacy of an Optimised Bacteriophage Cocktail to Clear Clostridium difficile in a Batch Fermentation Model. Antibiotics. 2018;7:13. doi: 10.3390/antibiotics7010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meader E, et al. Bacteriophage treatment significantly reduces viable Clostridium difficile and prevents toxin production in an in vitro model system. Anaerobe. 2010;16:549–554. doi: 10.1016/j.anaerobe.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Monteiro R, Pires DP, Costa AR, Azeredo J. Phage Therapy: Going Temperate? Trends Microbiol. 2018;xx:1–11. doi: 10.1016/j.tim.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 18.Meader E, Mayer MJ, Steverding D, Carding SR, Narbad A. Evaluation of bacteriophage therapy to control Clostridium difficile and toxin production in an in vitro human colon model system. Anaerobe. 2013;22:25–30. doi: 10.1016/j.anaerobe.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Chung I, Sim N, Cho Y. Antibacterial Efficacy of Temperate Phage-Mediated Inhibition of Bacterial Group Motilities. Antimicrob. Agents Chemother. 2012;56:5612–5617. doi: 10.1128/AAC.00504-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morrissey BJ, et al. Biogeography of Paenibacillus larvae, the causative agent of American foulbrood, using a new multilocus sequence typing scheme. Environ. Microbiol. 2015;17:1414–1424. doi: 10.1111/1462-2920.12625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rauch S, Ashiralieva A, Hedtke K, Genersch E. Negative Correlation between Individual-Insect-Level Virulence and Colony-Level Virulence of Paenibacillus larvae, the Etiological Agent of American Foulbrood of Honeybees. Appl. Environ. Microbiol. 2009;75:3344–3347. doi: 10.1128/AEM.02839-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corby-Harris V, et al. Origin and effect of Alpha 2.2 Acetobacteraceae in honey bee larvae and description of Parasaccharibacter apium gen. nov., sp. nov. Appl. Environ. Microbiol. 2014;80:7460–7472. doi: 10.1128/AEM.02043-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vojvodic S, Rehan SM, Anderson KE. Microbial Gut Diversity of Africanized and European Honey Bee Larval Instars. PLoS One. 2013;8:1–11. doi: 10.1371/journal.pone.0072106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thanki AM, et al. Unravelling the Links between Phage Adsorption and Successful Infection in Clostridium difficile. Viruses. 2018;10:411. doi: 10.3390/v10080411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goh S, Riley TV, Chang BJ. Isolation and Characterization of Temperate Bacteriophages of Clostridium difficile. Appl. Environ. Microbiol. 2005;71:1079–1083. doi: 10.1128/AEM.71.2.1079-1083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Touchon M, Bernheim A, Rocha EPC. Genetic and life-history traits associated with the distribution of prophages in bacteria. ISME J. 2016;10:2744–2754. doi: 10.1038/ismej.2016.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hadas, H., Einav, M. & Zaritsky, A. Bacteriophage T4 development depends on the physiology of its host Escherichia coli. Microbiology 179–185 (1997). [DOI] [PubMed]
- 28.Garcia-Gonzalez E, et al. Paenibacillus larvae Chitin-Degrading Protein PlCBP49 Is a Key Virulence Factor in American Foulbrood of Honey Bees. PLoS Pathog. 2014;10:e1004284. doi: 10.1371/journal.ppat.1004284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hrncir Michael, Jarau Stefan. Food Exploitation By Social Insects. 2009. Social Insects and the Exploitation of Food Sources; pp. 323–330. [Google Scholar]
- 30.Ribeiro, H. G., Correia, R., Moreira, T., Boas, D. V. & Azeredo, J. Bacteriophage biodistribution and infectivity from honeybee to bee larvae using a T7 phage model. Sci. Rep. 1–8 10.1038/s41598-018-36432-x (2019). [DOI] [PMC free article] [PubMed]
- 31.Chen C. Changes in protein components and storage stability of royal jelly under various conditions. Food Chem. 1995;54:195–200. doi: 10.1016/0308-8146(95)00031-D. [DOI] [Google Scholar]
- 32.Pasupuleti VR, Sammugam L, Ramesh N, Gan SH. Honey, propolis, and royal jelly: A comprehensive review of their biological actions and health benefits. Oxid. Med. Cell. Longev. 2017;2017:1–21. doi: 10.1155/2017/1259510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ali H, Alli I, Ismail A, Kermasha S. Protein-phenolic interactions in food. Eurasian J. Anal. Chem. 2012;7:123–133. [Google Scholar]
- 34.Oliveira A, et al. The First Paenibacillus larvae Bacteriophage Endolysin (PlyPl23) with High Potential to Control American Foulbrood. PLoS One. 2015;10:e0132095. doi: 10.1371/journal.pone.0132095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Genersch E, Otten C. The use of repetitive element PCR fingerprinting (rep-PCR) for genetic subtyping of German field isolates of Paenibacillus larvae subsp. larvae. Apidologie. 2003;34:195–206. doi: 10.1051/apido:2003025. [DOI] [Google Scholar]
- 36.Dobbelaere W, de Graaf DC, Peeters JE. Development of a fast and reliable diagnostic method for American foulbrood disease (Paenibacillus larvae subsp. larvae) using a 16S rRNA gene based PCR. Apidologie. 2001;32:363–370. doi: 10.1051/apido:2001136. [DOI] [Google Scholar]
- 37.Pires D, Sillankorva S, Faustino A, Azeredo J. Use of newly isolated phages for control of Pseudomonas aeruginosa PAO1 and ATCC 10145 biofilms. Res. Microbiol. 2011;162:798–806. doi: 10.1016/j.resmic.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 38.Sillankorva S, Neubauer P, Azeredo J. Isolation and characterization of a T7-like lytic phage for Pseudomonas fluorescens. BMC Biotechnol. 2008;8:80. doi: 10.1186/1472-6750-8-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melo LDR, et al. Isolation and characterization of a new Staphylococcus epidermidis broad-spectrum bacteriophage. J. Gen. Virol. 2014;95:506–515. doi: 10.1099/vir.0.060590-0. [DOI] [PubMed] [Google Scholar]
- 40.Aziz RK, et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 43.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Käll L, Sonnhammer EL. Reliability of transmembrane predictions in whole-genome data. FEBS Lett. 2002;532:415–418. doi: 10.1016/S0014-5793(02)03730-4. [DOI] [PubMed] [Google Scholar]
- 45.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 46.Klucar L, Stano M, Hajduk M. phiSITE: database of gene regulation in bacteriophages. Nucleic Acids Res. 2010;38:D366–D370. doi: 10.1093/nar/gkp911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naville M, Ghuillot-Gaudeffroy A, Marchais A, Gautheret D. ARNold: A web tool for the prediction of Rho-independent transcription terminators. RNA Biol. 2011;8:11–13. doi: 10.4161/rna.8.1.13346. [DOI] [PubMed] [Google Scholar]
- 48.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zankari E, et al. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie Y, et al. TADB 2.0: An updated database of bacterial type II toxin-antitoxin loci. Nucleic Acids Res. 2018;46:D749–D753. doi: 10.1093/nar/gkx1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oliveira H, et al. Staphylococci phages display vast genomic diversity and evolutionary relationships. BMC Genomics. 2019;20:1–14. doi: 10.1186/s12864-019-5647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cresawn SG, et al. Phamerator: A bioinformatic tool for comparative bacteriophage genomics. BMC Bioinformatics. 2011;12:395. doi: 10.1186/1471-2105-12-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huson DH. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 54.Oliveira H, et al. Functional Analysis and Antivirulence Properties of a New Depolymerase from a Myovirus That Infects Acinetobacter baumannii Capsule K45. J. Virol. 2018;93:1–16. doi: 10.1128/JVI.01163-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kalatzis PG, et al. Stumbling across the same phage: Comparative genomics of widespread temperate phages infecting the fish pathogen Vibrio anguillarum. Viruses. 2017;9:1–19. doi: 10.3390/v9050122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oliveira A, Melo LDR, Kropinski AM, Azeredo J. Complete genome sequence of the broad-host-range Paenibacillus larvae phage phiIBB_Pl23. Genome Announc. 2013;1:e00438-13–e00438-13. doi: 10.1128/genomeA.00438-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anderson KE, et al. Microbial ecology of the hive and pollination landscape: Bacterial associates from floral nectar, the alimentary tract and stored food of honey bees (Apis mellifera) PLoS One. 2013;8:e83125. doi: 10.1371/journal.pone.0083125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Colibar O, Popovici D, Eugeniu C, Korodi G. The effect of acidifiant on the development of bee families (Apis mellifica) Med. Vet. 2010;43:296–299. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.