Abstract
In the last years increasing attention has been given to the connection between genotype/phenotype and cardiovascular events in subjects with familial hypercholesterolemia (FH). MicroRNAs (miRs) bound to high-density lipoprotein (HDL) may contribute to better discriminate the cardiovascular risk of FH subjects. Our aim was to evaluate the HDL-miR panel in heterozygous FH (HeFH) patients with an LDLR null or defective mutation and its association with pulse wave velocity (PWV). We evaluated lipid panel, HDL-miR panel and PWV in 32 LDLR null mutation (LDLR-null group) and 35 LDLR defective variant (LDLR-defective group) HeFH patients. HDL-miR-486 and HDL-miR-92a levels were more expressed in the LDLR-null group than the LDLR-defective group. When we further stratified the study population into three groups according to both the LDLR genotype and history of ASCVD (LDLR-null/not-ASCVD, LDLR-defective/not-ASCVD and LDLR/ASCVD groups), both the LDLR/ASCVD and the LDLR-null/not-ASCVD groups had a higher expression of HDL-miR-486 and HDL-miR-92a than the LDLR-defective/not-ASCVD group. Finally, HDL-miR-486 and HDL-miR-92a were independently associated with PWV. In conclusion, the LDLR-null group exhibited HDL-miR-486 and HDL-miR-92a levels more expressed than the LDLR-defective group. Further studies are needed to evaluate these HDL-miRs as predictive biomarkers of cardiovascular events in FH.
Subject terms: Predictive markers, Dyslipidaemias
Introduction
Atherosclerotic process is a progressive inflammatory disease caused by several external and hereditary factors1. Of these, the alterations of lipid and glucose metabolism play a key role in the pathogenesis and progression of atherosclerotic cardiovascular disease (ASCVD)2,3. In particular, an increase of the low-density-lipoprotein (LDL) cholesterol plasma level is causatively associated with ASCVD4. Despite changes in lifestyle and lipid lowering therapies, ASCVD is considered the principal condition of reduced quality-adjusted life years and death5. Thus, other mechanisms are involved in atherosclerosis in addition to LDL cholesterol.
Of note, in the last few years several studies have shown an important role of microRNAs (miRs) in the pathophysiology of atherosclerosis6. MiRs are a class of small noncoding RNAs that inhibit gene expression through the alteration of messenger RNA after transcriptional process into the cell7. Not all mature miRs are developed in the cell; thus, extracellular miRs such as plasmatic miRs may serve as intercellular messenger8. Many biological functions have been attributed to miRs; as concerns the metabolic pathways, they seemed to act as modulators of lipid metabolism, and transport and sustained inflammatory diseases9,10. In subjects characterized by high levels of LDL cholesterol such as familial hypercholesterolemia (FH), several studies demonstrated that a number of circulating miRs were upregulated from childhood, confirming their role in cholesterol homeostasis also in lipid genetic disorders11,12. To avoid plasmatic ribonucleases, miRs are transported by several carriers; in this context, the most crucial and stable transporters of miRs are plasmatic lipoproteins, especially the high-density lipoprotein (HDL) particles13.
Interestingly, Vickers et al. found that the HDL-miR panel was significantly different between homozygous familial hypercholesterolemia and normal subjects14.
In the last few years increasing attention has been given to the connection between genotype/phenotype and ASCVD in familial hypercholesterolemia15–17. In particular, several studies have shown that heterozygous FH (HeFH) subjects with an LDL receptor (LDLR) null mutation had an increased atherosclerotic burden with respect to HeFH subjects with an LDLR defective variant18,19. In this context, it may be helpful to evaluate the HDL-miR panel in HeFH subjects with a different LDLR genotype.
To better describe the link between HDL-miRs and HeFH genotype, in this study we aimed to investigate the plasma levels of several HDL-miRs correlated with lipid homeostasis and atherosclerotic pathway (miR-486, miR-92a, miR-24, miR-223, miR-625*, miR-122)20,21 in HeFH subjects with an LDLR null or defective mutation. Moreover, we evaluated the association of HDL-miRs with pulse wave velocity (PWV), an instrumental parameter of early atherosclerosis largely utilized in clinical practice for cardiovascular risk assessment22.
Results
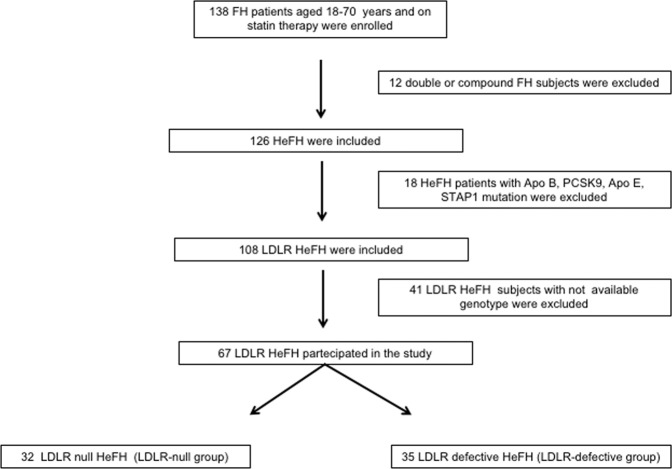
In total, 138 FH patients were assessed and 67 LDLR HeFH patients satisfied the inclusion criteria and were involved in the study (Fig. 1). Partecipants were divided in two groups according to the LDLR genotype: 32 HeFH patients with an LDLR null mutation (LDLR-null group) and 35 HeFH patients with an LDLR defective mutation (LDLR-defective group).
Figure 1.
Enrollment of the Study Population. FH = familial hypercholesterolemia, HeFH = heterozygous familial hypercholesterolemia, LDLR = low-density lipoprotein receptor, ApoB = apolipoprotein B, PCSK9 = proprotein convertase subtilisin/kexin type 9, ApoE = apolipoprotein E, STAP1 = signal transducing adaptor family member 1.
The pretreatment lipid parameters of partecipants are displayed in Table 1. Of corse, the LDLR-null group had higher levels of total, LDL and non-HDL cholesterol than the LDLR-defective group.
Table 1.
Pretreatment lipid values of the Study Population.
| LDLR-null group (n = 32) |
LDLR-defective group (n = 35) |
p Value between two groups |
|
|---|---|---|---|
| Pretreated Lipid Values | |||
| Total cholesterol, mg/dL | 387.62 ± 28.98 | 336.13 ± 29.44 | <0.01 |
| HDL cholesterol, mg/dL | 51.58 ± 10.49 | 52.3 ± 11.23 | 0.85 |
| Triglycerides, mg/dL | 95.5 (71.5–150) | 97.5 (71.75–150) | 0.81 |
| LDL cholesterol, mg/dL | 282.32 ± 23.87 | 233.28 ± 22.43 | <0.01 |
| Non-HDL cholesterol, mg/dL | 316.19 ± 37.92 | 265.83 ± 38.38 | <0.01 |
Data are presented as mean ± standard deviation or median (interquartile range).
HDL = high-density lipoprotein, LDL = low-density lipoprotein, LDLR = low-density lipoprotein receptor.
The general features of partecipants are showed in Table 2. No discrepancy of metabolic parameters were showed in the two groups. Moreover, the values of systolic and diastolic BP and the percentage of smokers were the same in the LDLR-null and LDLR-defective groups. Furthermore, hs-CRP values were similar between the two groups. About medication, the LDLR-null group had a more protracted period of statin therapy than the LDLR-defective group (9.5 [2.5–17.5] vs 8 [1.5–12] years, p < 0.05). Furthermore, the majority of patients on ezetimibe were in the LDLR-null group than the LDLR-defective group (65.6% vs 40.0%, p < 0.05). Finally, similar proportions of antihypertensive medication were found between the two groups. In consideration of intensity of statin therapy, no subjects were on low-intensity statins. While a greater number of subjects on moderate-intensity statin were in the LDLR-defective group, the percentage of patients on high-intensity statin was more present in the LDLR-null group than the LDLR-defective group (75.0% vs 28.6%, p < 0.05). Moreover, the LDLR-null group had a greater PWV compared with the LDLR-defective group (9.58 ± 0.92 vs 7.41 ± 0.83 m/s, p < 0.05).
Table 2.
Characteristics of the Study Population.
| LDLR-null group (n = 32) |
LDLR-defective group (n = 35) |
p Value between two groups |
|
|---|---|---|---|
| Demographic Characteristics | |||
| N | 32 | 35 | |
| Age, years | 48.69 ± 14.36 | 51.97 ± 15.47 | 0.37 |
| Men, n (%) | 18 (56.3) | 19 (54.3) | 0.61 |
| Body mass index, kg/m2 | 25.21 ± 3.34 | 25.38 ± 3.79 | 0.84 |
| ASCVD, n (%) | 12 (37.5) | 9 (25.7) | 0.33 |
| Glucose Values | |||
| FPG, mg/dL | 85.7 ± 6.84 | 88.11 ± 6.32 | 0.14 |
| HbA1c, % | 5.42 ± 0.29 | 5.51 ± 0.28 | 0.25 |
| Lipid Values | |||
| Total cholesterol, mg/dL | 188.65 ± 25.26 | 186.5 ± 24.31 | 0.86 |
| HDL cholesterol, mg/dL | 49.16 ± 10.62 | 52.81 ± 8.84 | 0.07 |
| Triglycerides, mg/dL | 87 (80–117) | 88.5 (81–119) | 0.76 |
| LDL cholesterol, mg/dL | 121.54 ± 21.46 | 116.33 ± 20.48 | 0.31 |
| Non-HDL cholesterol, mg/dL | 140.48 ± 22.04 | 135.69 ± 22.07 | 0.16 |
| ApoB, mg/dL | 116.79 ± 25.02 | 110.53 ± 24.71 | 0.11 |
| ApoAI, m g/dL | 133.71 ± 23.34 | 137.62 ± 23.31 | 0.29 |
| ApoB to ApoAI ratio | 0.88 ± 0.34 | 0.81 ± 0.31 | 0.06 |
| Lp(a), nmol/L | 20.8 (10.05–62.95) | 19.2 (9.69–41.05) | 0.45 |
| Risk Factors | |||
| Systolic BP, mmHg | 118.55 ± 9.33 | 119.17 ± 13.37 | 0.83 |
| Diastolic BP, mmHg | 70.48 ± 7.78 | 71.36 ± 9.17 | 0.28 |
| Smoking, n (%) | 10 (31.3) | 13 (37.1) | 0.61 |
| hs-CRP, mg/dL | 0.10 (0.05–0.19) | 0.09 (0.04–0.15) | 0.39 |
| Treatment | |||
| Duration of Statin therapy, years | 9.5 (2.5–17.5) | 8 (1.5–12) | <0.05 |
| Ezetimibe, n (%) | 21 (65.6) | 14 (40.0) | <0.05 |
| Antihypertensive therapy, n(%) | 11 (34.4) | 13 (37.1) | 0.81 |
| Intensity of Statin Therapy | |||
| Low, n (%) | — | — | — |
| Moderate, n (%) | 8 (25.0) | 25 (71.4) | <0.05 |
| High, n (%) | 24 (75.0) | 10 (28.6) | <0.05 |
| Early atherosclerotic biomarker | |||
| PWV, m/s | 9.58 ± 0.92 | 7.41 ± 0.83 | <0.05 |
Data are presented as mean ± standard deviation, percentages, or median (interquartile range).
ASCVD = atherosclerotic cardiovascular disease, FPG = fasting plasma glucose, HbA1c = glycated hemoglobin, HDL = high-density lipoprotein, LDL = low-density lipoprotein, LDLR = low-density lipoprotein receptor, ApoB = apolipoprotein B, ApoAI = apolipoprotein AI, Lp(a) = lipoprotein (a), BP = blood pressure, hs-CRP = high sensitivity C-reactive protein, PWV = pulse wave velocity.
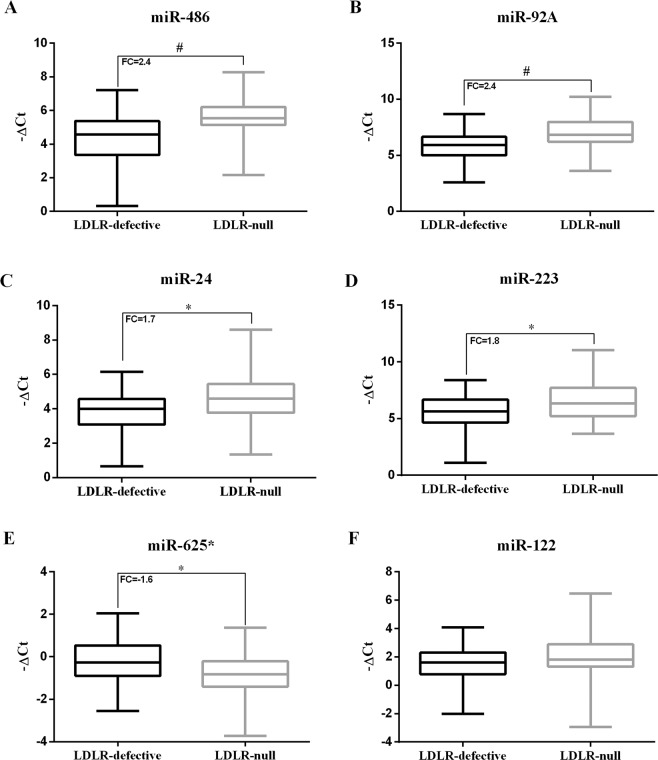
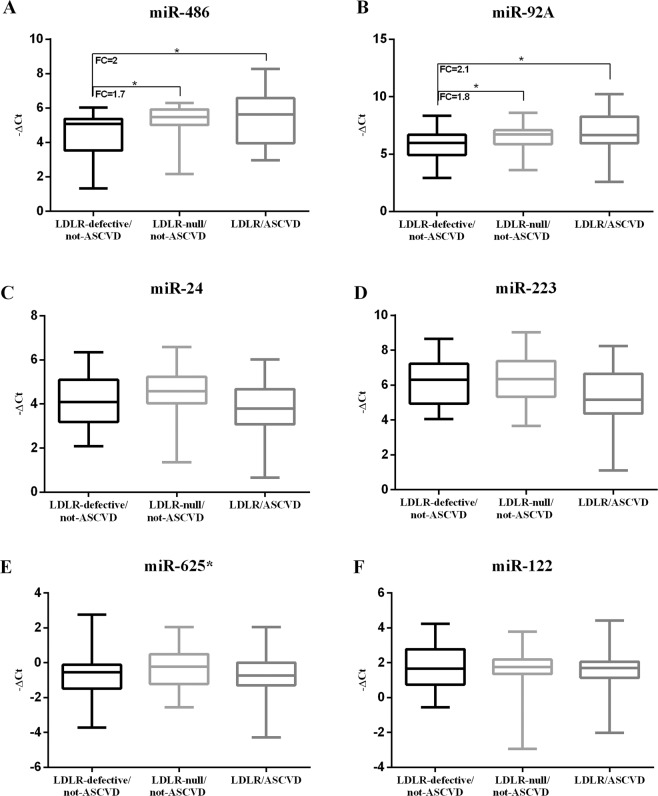
Figure 2 reports the evaluation of HDL-miR panel in the two groups. While the level of HDL-miR-122 was similar between the two groups, a different expression of HDL-miR-486, HDL-miR-92a, HDL-miR-24, HDL-miR-223 and HDL-miR-625* was found between the two groups. In particular, HDL-miR-486 and HDL-miR-92a levels were largely represented in the LDLR-null group than the LDLR-defective group (HDL-miR-486 and HDL-miR-92a fold change +2.4 for both, p < 0.001 for both, Fig. 2A,B). Moreover, to compare the HDL-miR panel in FH patients with or without cardiovascular events, we performed another analysis and stratified the study population into three new groups according to both the LDLR genotype and history of ASCVD: LDLR-null/not-ASCVD group (20 FH patients), LDLR-defective/not-ASCVD group (26 FH patients) and LDLR/ASCVD group (21 FH patients) (Fig. 3).
Figure 2.
HDL-miR panel in the LDLR-null and the LDLR-defective groups. *p value < 0.05 versus LDLR-defective group, #p value < 0.001 versus LDLR-defective group. To test differences of HDL-miR −ΔCt values in the two groups, Student’s t test was used.
Figure 3.
HDL-miR panel in the LDLR-null/not-ASCVD, LDLR-defective/not-ASCVD and the LDLR/ASCVD groups. *p value < 0.05 versus LDLR-defective/not-ASCVD group. To test differences of HDL-miR −ΔCt values in the two groups, Student’s t test was used.
Both the LDLR/ASCVD group and the LDLR-null/not-ASCVD group had a higher expression of HDL-miR-486 and HDL-miR-92a than the LDLR-defective/not-ASCVD group (for LDLR/ASCVD vs LDLR-defective/not-ASCVD group HDL-miR-486 and HDL-miR-92a fold change +2.0 and +2.1 respectively, p < 0.05 for both; for LDLR-null/not-ASCVD vs LDLR-defective/not-ASCVD group HDL-miR-486 and HDL-miR-92a fold change +1.7 and +1.8 respectively, p < 0.05 for both, Fig. 3A,B). Moreover, no difference of HDL-miR panel was found between LDLR/ASCVD and LDLR-null/not-ASCVD groups.
In the simple regression analysis, HDL-miR-92a and HDL-miR-486 were associated with PWV (r = 0.29, p < 0.01 for HDL-miR-92a and r = 0.27, p < 0.05 for HDL-miR-486). Subsequently, in the multiple regression analysis including these two HDL-miRs reaching significance and several cardiovascular risk factors as independent variables, both HDL-miR-92a and HDL-miR-486 remained significantly associated with PWV (p < 0.01 for both) (Table 3).
Table 3.
Multiple regression analysis evaluating PWV as dependent variable.
| Independent Variables | Coefficient β | p Value |
|---|---|---|
| Model* | ||
| HDL-miR-92a, −ΔCt | 0.343 | <0.01 |
| HDL-miR-486, −ΔCt | 0.308 | <0.01 |
*Model was adjusted for age, sex, smoking status, systolic blood pressure, LDL cholesterol and glycated hemoglobin.
Discussion
The improved evaluation of genotype and phenotype of FH have recently focused on the impact of novel cardiovascular risk biomarkers in FH subjects23,24. In this study, we examined the role of HDL-miRs in HeFH subjects with an LDLR null or defective mutation; to our learning, no other studies explored HDL-miR panel in these FH subgroups. We showed that HDL-miR-486 and HDL-miR-92a were more expressed in the LDLR-null group than the LDLR-defective group; moreover, we found that both the LDLR-ASCVD group and the LDLR-null group had significant HDL-miR-486 and HDL-miR-92a levels compared with the LDLR-defective group. Finally, we showed a significant association between HDL-miR-486, HDL-miR-92a and PWV. Our findings are in line with previous studies showing a possible role of these two miRs in lipid homeostasis and the atherosclerotic process; in fact, Liu D. et al. found that miR-486 promoted cholesterol concentration in macrophage-derived foam cells25. Moreover, Zhang et al. showed that miR-486 was higher in patients with acute myocardial infarction than controls26. Liu F. et al. found that also miR-92a was higher in patients with coronary artery disease than controls27; furthermore, several studies showed the possible role of miR-92a in the atherosclerotic process by the promoting endothelial dysfunction and apoptosis in cardiomyocytes28–30. In line with these findings, Niculescu et al. found that miR-486 and miR-92a levels in HDL subfractions may identify subjects at increased risk of recurrent cardiovascular events31; moreover, they showed that the inhibition of miR-486 and miR-92a decreased liver and plasma cholesterol levels by restoring the lipid metabolism liver genes such as ATP binding cassette subfamily G member 4 (ABCG4) and the sterol regulatory element binding transcription factor-1 (SREBF1)32. In this context, miR-486 and miR-92a linked to HDL may be important modulators of HDL functions such as cholesterol efflux, endothelial nitric oxide synthase (eNOS) and nitric oxide (NO) production33.
FH patients are characterized by an increased risk of cardiovascular events34; however, ASCVD risk is not the same in FH population. In particular, several studies demonstrated that HeFH patients with an LDLR null mutation had a greater atherosclerotic burden than the HeFH patients with an LDLR defective variant18,19,35; this is probably due to the higher cholesterol burden of LDLR null HeFH compared with LDLR defective HeFH. In our study, all FH patients were on statin therapy and lipid values were similar between the two groups. It is well known that statin treatment has significantly reduced the risk of ASCVD in FH subjects36,37. However, despite statin therapy, the LDLR null HeFH patients have a higher ASCVD risk compared with the LDLR defective HeFH patients on statin therapy38. In line with this consideration, in our study we found that the LDLR null HeFH patients had a higher PWV than the LDLR defective HeFH patients. Moreover, in our population we showed a direct association of HDL-miR-486 and HDL-miR-92a with PWV; thus, these miRs may support the increase of cardiovascular risk and the advance of atherosclerosis in addition to LDL cholesterol burden in FH patients.
Recent clinical trials where increased levels of HDL cholesterol have been obtained, failed to demonstrated a significant reduction of ASCVD in a large population at high cardiovascular risk39,40. These findings strongly support the hypothesis of “HDL quality” and not HDL quantity. Several studies have focused on the alterations of HDL quality in FH subjects, and, in particular, on their decreased ability to support cholesterol efflux from macrophages, and on their diminished anti-inflammatory and anti-oxidant properties41–43. In this context, miRs bound to HDL particles may effectively modulate their function on lipid metabolism and the atherosclerotic process. Recently, several studies have focused attention on miRs as possible targets in FH patients33,44. However, no prospective studies have shown if HDL-miR-486 and HDL-miR-92a could be good predictors for subsequent cardiovascular events in FH subjects; thus, additional studies are required to investigate their role in the progression of the atherosclerotic burden and their possible relationship with ASCVD in a large FH population.
There are several limitations to our study. First, we are unable to establish a causal relationship and temporality between HDL-miR-486 and HDL-miR-92a and possible changes in PWV by the cross-sectional design of this study. The number of studied patients was relatively small; however, we showed a significant difference of HDL-miR panel in the groups and an independent association of HDL-miR-486, HDL-miR-92a and PWV was found. Finally, other cardiovascular parameters such as cholesterol burden were not available and, thus, were not taken into consideration.
In conclusion, HDL-miR-486 and HDL-miR-92a levels were more expressed in the LDLR-null group than the LDLR-defective group; moreover, HDL-miR-486 and HDL-miR-92a were significantly associated with PWV. Our study suggests that these miRs may be helpful to improve cardiovascular risk stratification; additional studies are required to investigate HDL-miRs as predictive biomarkers of cardiovascular events and possible treatment targets in FH patients.
Methods
Study design and population
It was an observational study in subjects with a diagnosis of FH already confirmed by genetic evaluation45,46. The patients were evaluated from the University Hospital of Catania and the Dyslipidemia Center of the Niguarda Hospital in Milan, Italy, two tertiary lipid centers, from April 2017 to December 201816. The age of partecipants were over 18 and under 70 years and assumed statin therapy at the moment of the study. In particular, we only included FH patients with a null or defective LDLR genetic variant; the reason of this restricted criteria was that LDLR mutations were associated with several genotype/phenotype patterns that could contribute to the heterogeneity of FH population. In this context, we aimed to evaluate possible novel cardiovascular biomarkers such as HDL-miRs in these subjects.
The study was accepted by the local ethics committees Catania 2 and Milano Area 3 in accordance with the ethical standards of the institutional and national research committees and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. Informed consent was attained from all partecipants the study.
All subjects obtained a physical examination and clinical biochemistry parameters measured as previously described2. Anthropometric parameters, glycemic status, arterial pressure, medications, ASCVD, advanced renal disease and smoking abits were defined as previously described22,47,48. The severity of liver disease was defined as previously described49. The exclusion criteria were defined as previously described22.
Biochemical analysis
Fasting plasma glucose (FPG), serum total cholesterol, TG, high-density lipoprotein (HDL) cholesterol, hs-CRP, Apolipoprotein B (ApoB), Apolipoprotein A1 (ApoA1) were assessed as previously described50. Levels of lipoprotein (a) [Lp(a)] were measured as previously described22. LDL cholesterol was obtained by the Friedewald formula. Glycated hemoglobin (HbA1c) was measured as previously described51.
HDL purification
HDL particles (d = 1.063–1.21 g/mL) were obtained by sequential ultracentrifugation from plasma collected from all subjects. After separation, HDL particles were dialyzed against sterilized PBS to remove the high-salt KBr solutions and immediately frozen at −80 °C until use52. By using ExoTEST Ready to Use Kit for ELISA Exosome quantification (Hansa BioMed), we assessed that the purified HDL particles were negative for the classic exosomal protein marker CD9.
RNA extraction
HDL-carried miRs were extracted from 400 μl of purified HDL particles as previously described53.
Single taqMan assays
MiR expression levels were evaluated by Single TaqMan MicroRNA Assays as previously described53.
Pulse wave velocity evaluation
PWV was performed as previously described54.
Statistical analysis
The Kolmogorov-Smirnov test was used to assess normality distribution of all variables. Continuous parametric and non parametric data are described as mean ± standard deviation (SD) or median (interquartile range-IQR), respectively; moreover, categorical variables are reported as frequency (percentage) and evaluated by χ2 test. Continuous non-parametric variables (TG, Lpa, duration of statin therapy and hs-CRP) were logarithmically transformed for statistical analysis to diminish dissymetry. MiR expression data are presented as minus Delta Ct values (−ΔCt), calculated according to the following formula: −1*(Threshold Cycle of analysed miRNA − Threshold Cycle of U6 in each sample). MiR expression Fold Changes (FC) were calculated by applying the 2−ΔΔCT method by using small nuclear RNA U6 as reference gene. Student’s t test was performed for clinical and biochemical characteristics. Simple regression analysis was used to evaluate the relation of HDL-miRs with PWV. Subsequently, to analyze a possible independent association with changes of PWV, HDL-miRs reaching significance were included in a multivariate analysis with principal cardiovascular risk factors (age, sex, smoking, systolic BP, HbA1c, and LDL cholesterol). The variance inflation factor (VIF) was performed to assess the problem of multicollinearity in multivariate analysis. All statistical analyses were obtained by IBM SPSS Statistics for Windows version 23. For all tests, p < 0.05 was considered significant.
Ethical approval
This study was approved by the local ethics committee in accordance with the ethical standards of the institutional and national research committees and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors.
Informed consent
Informed consent was obtained from each participant enrolled in the study.
Acknowledgements
F.P. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. All authors approved the final version. The authors wish to thank the Department of Clinical and Experimental Medicine for financial support in the context of the 2016/2018 Department Research Plan of University of Catania (project #A). The authors wish to thank the Lipid TransPort Disorders Italian Genetic Network (LIPIGEN) study for financial support to carry out genetic analysis. The authors wish to thank the Scientific Bureau of the University of Catania for language support. This research did not receive any specific grant from funding agencies in the commercial or not-for-profit sectors.
Author contributions
R.S. contributed to the study design, researched data, contributed to the discussion, and wrote the article. A.D.P. contributed to the study design, researched data, contributed to the discussion, and reviewed and edited the article. C.P., A.O., A.S., A.A., S.D.M., A.S., F.U., A.F., researched data, contributed to the discussion, and reviewed and edited the article. S.P., A.M.R., L.C., contributed to the study design and discussion, and reviewed and edited the article. F.P. designed the study, researched data, contributed to the discussion, and reviewed and edited the article.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ross R. Atherosclerosis–an inflammatory disease. N. Engl. J. Med. 1999;340:115–26. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 2.Scicali R., Di Pino A., Platania R., Purrazzo G., Ferrara V., Giannone A., Urbano F., Filippello A., Rapisarda V., Farruggia E., Piro S., Rabuazzo A.M., Purrello F. Detecting familial hypercholesterolemia by serum lipid profile screening in a hospital setting: Clinical, genetic and atherosclerotic burden profile. Nutrition, Metabolism and Cardiovascular Diseases. 2018;28(1):35–43. doi: 10.1016/j.numecd.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Di Pino A, et al. HbA1c Identifies Subjects With Prediabetes and Subclinical Left Ventricular Diastolic Dysfunction. J. Clin. Endocrinol. Metab. 2017;102:3756–3764. doi: 10.1210/jc.2017-00954. [DOI] [PubMed] [Google Scholar]
- 4.Ference BA, et al. Low-density lipoproteins cause atherosclerotic cardiovascular disease. 1. Evidence from genetic, epidemiologic, and clinical studies. A consensus statement from the European Atherosclerosis Society Consensus Panel. Eur. Heart J. 2017;38:2459–2472. doi: 10.1093/eurheartj/ehx144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Backer Guy, Jankowski Piotr, Kotseva Kornelia, Mirrakhimov Erkin, Reiner Željko, Rydén Lars, Tokgözoğlu Lale, Wood David, De Bacquer Dirk, De Backer G., Jankowski P., Kotseva K., Mirrakhimov E., Reiner Z., Rydén L., Tokgözoğlu L., Wood D., De Bacquer D., Kotseva K., De Backer G., Abreu A., Aguiar C., Badariene J., Bruthans J., Castro Conde A., Cifkova R., Crowley J., Davletov K., Bacquer D. De, De Smedt D., De Sutter J., Deckers J.W., Dilic M., Dolzhenko M., Druais H., Dzerve V., Erglis A., Fras Z., Gaita D., Gotcheva N., Grobbee D.E., Gyberg V., Hasan Ali H., Heuschmann P., Hoes A.W., Jankowski P., Lalic N., Lehto S., Lovic D., Maggioni A.P., Mancas S., Marques-Vidal P., Mellbin L., Miličić D., Mirrakhimov E., Oganov R., Pogosova N., Reiner Ž., Rydén L., Stagmo M., Störk S., Sundvall J., Tokgözoğlu L., Tsioufis K., Vulic D., Wood D., Wood D.A., Kotseva K., Jennings C., Adamska A., Adamska S., Rydén L., Mellbin L., Tuomilehto J., Schnell O., Druais H., Fiorucci E., Glemot M., Larras F., Missiamenou V., Maggioni A., Taylor C., Ferreira T., Lemaitre K., Bacquer D. De, De Backer G., Raman L., Sundvall J., DeSmedt D., De Sutter J., Willems A.M., De Pauw M., Vervaet P., Bollen J., Dekimpe E., Mommen N., Van Genechten G., Dendale P., Bouvier C.A., Chenu P., Huyberechts D., Persu A., Dilic M., Begic A., Durak Nalbantic A., Dzubur A., Hadzibegic N., Iglica A., Kapidjic S., Osmanagic Bico A., Resic N., Sabanovic Bajramovic N., Zvizdic F., Vulic D., Kovacevic-Preradovic T., Popovic-Pejicic S., Djekic D., Gnjatic T., Knezevic T., Kovacevic-Preradovic T., Kos Lj, Popovic-Pejicic S., Stanetic B., Topic G., Gotcheva N., Georgiev Borislav, Terziev A., Vladimirov G., Angelov A., Kanazirev B., Nikolaeva S., Tonkova D., Vetkova M., Milicic D., Reiner Ž., Bosnic A., Dubravcic M., Glavina M., Mance M., Pavasovic S., Samardzic J., Batinic T., Crljenko K., Delic-Brkljacic D., Dula K., Golubic K., Klobucar I., Kordic K., Kos N., Nedic M., Olujic D., Sedinic V., Blazevic T., Pasalic A., Percic M., Sikic J., Bruthans J., Cífková R., Hašplová K., Šulc P., Wohlfahrt P., Mayer O., Cvíčela M., Filipovský J., Gelžinský J., Hronová M., Hasan-Ali H., Bakery S., Mosad E., Hamed H.B., Ibrahim A., Elsharef M.A., Kholef E.F., Shehata A., Youssef M., Elhefny E., Farid H., Moustafa T.M., Sobieh M.S., Kabil H., Abdelmordy A., Lehto S., Kiljander E., Kiljander P., Koukkunen H., Mustonen J., Cremer C., Frantz S., Haupt A., Hofmann U., Ludwig K., Melnyk H., Noutsias M., Karmann W., Prondzinsky R., Herdeg C., Hövelborn T., Daaboul A., Geisler T., Keller T., Sauerbrunn D., Walz-Ayed M., Ertl G., Leyh R., Störk S., Heuschmann P., Ehlert T., Klocke B., Krapp J., Ludwig T., Käs J., Starke C., Ungethüm K., Wagner M., Wiedmann S., Tsioufis K., Tolis P., Vogiatzi G., Sanidas E., Tsakalis K., Kanakakis J., Koutsoukis A., Vasileiadis K., Zarifis J., Karvounis C., Crowley J., Gibson I., Houlihan A., Kelly C., O'Donnell M., Bennati M., Cosmi F., Mariottoni B., Morganti M., Cherubini A., Di Lenarda A., Radini D., Ramani F., Francese M.G., Gulizia M.M., Pericone D., Davletov K., Aigerim K., Zholdin B., Amirov B., Assembekov B., Chernokurova E., Ibragimova F., Kodasbayev A., Markova A., Mirrakhimov E., Asanbaev A., Toktomamatov U., Tursunbaev M., Zakirov U., Abilova S., Arapova R., Bektasheva E., Esenbekova J., Neronova K., Asanbaev A., Baigaziev K., Toktomamatov U., Zakirov U., Baitova G., Zheenbekov T., Erglis A., Andrejeva T., Bajare I., Kucika G., Labuce A., Putane L., Stabulniece M., Dzerve V., Klavins E., Sime I., Badariene J., Gedvilaite L., Pečiuraite D., Sileikienė V., Skiauteryte E., Solovjova S., Sidabraite R., Briedis K., Ceponiene I., Jurenas M., Kersulis J., Martinkute G., Vaitiekiene A., Vasiljevaite K., Veisaite R., Plisienė J., Šiurkaitė V., Vaičiulis Ž., Jankowski P., Czarnecka D., Kozieł P., Podolec P., Nessler J., Gomuła P., Mirek-Bryniarska E., Bogacki P., Wiśniewski A., Pająk A., Wolfshaut-Wolak R., Bućko J., Kamiński K., Łapińska M., Paniczko M., Raczkowski A., Sawicka E., Stachurska Z., Szpakowicz M., Musiał W., Dobrzycki S., Bychowski J., Kosior D.A., Krzykwa A., Setny M., Kosior D.A., Rak A., Gąsior Z., Haberka M., Gąsior Z., Haberka M., Szostak-Janiak K., Finik M., Liszka J., Botelho A., Cachulo M., Sousa J., Pais A., Aguiar C., Durazzo A., Matos D., Gouveia R., Rodrigues G., Strong C., Guerreiro R., Aguiar J., Abreu A., Cruz M., Daniel P., Morais L., Moreira R., Rosa S., Rodrigues I., Selas M., Gaita D., Mancas S., Apostu A., Cosor O., Gaita L., Giurgiu L., Hudrea C., Maximov D., Moldovan B., Mosteoru S., Pleava R., Ionescu M., Parepa I., Pogosova N., Arutyunov A., Ausheva A., Isakova S., Karpova A., Salbieva A., Sokolova O., Vasilevsky A., Pozdnyakov Y., Antropova O., Borisova L., Osipova I., Lovic D., Aleksic M., Crnokrak B., Djokic J., Hinic S., Vukasin T., Zdravkovic M., Lalic N.M., Jotic A., Lalic K., Lukic L., Milicic T., Macesic M., Stanarcic Gajovic J., Stoiljkovic M., Djordjevic D., Kostic S., Tasic I., Vukovic A., Fras Z., Jug B., Juhant A., Krt A., Kugonjič U., Chipayo Gonzales D., Gómez Barrado J.J., Kounka Z., Marcos Gómez G., Mogollón Jiménez M.V., Ortiz Cortés C., Perez Espejo P., Porras Ramos Y., Colman R., Delgado J., Otero E., Pérez A., Fernández-Olmo M.R., Torres-LLergo J., Vasco C., Barreñada E., Botas J., Campuzano R., González Y., Rodrigo M., de Pablo C., Velasco E., Hernández S., Lozano C., González P., Castro A., Dalmau R., Hernández D., Irazusta F.J., Vélez A., Vindel C., Gómez-Doblas J.J., García Ruíz V., Gómez L., Gómez García M, Jiménez-Navarro M., Molina Ramos A., Marzal D., Martínez G., Lavado R., Vidal A., Rydén L., Boström-Nilsson V., Kjellström B., Shahim B., Smetana S., Hansen O., Stensgaard-Nake E., Deckers J.W., Klijn A.J., Mangus T.J.P., Peters R.J.G., Scholte op Reimer W., Snaterse M., Aydoğdu S., Ç Erol, Otürk S., Tulunay Kaya C., Ahmetoğlu Y., Ergene O., Akdeniz B., Çırgamış D., Akkoyun H Kültürsay S., Kayıkçıoğlu M., Çatakoğlu A.B., Çengel A., Koçak A.A., Ağırbaşlı M.A., Açıksarı G., Çekin M.E., Tokgözoğlu L., Kaya E.B., Koçyiğit D., Öngen Z., Özmen E., Sansoy V., Kaya A., Oktay V., Temizhan A., Ünal S., İ Yakut, Kalkan A.K., Bozkurt E., Kasapkara H.A., Dolzhenko M., Faradzh C., Hrubyak L., Konoplianyk L., Kozhuharyova N., Lobach L., Nesukai V., Nudchenko O., Simagina T., Yakovenko L., Azarenko V., Potabashny V., Bazylevych A., Bazylevych M., Kaminska K., Panchenko L., Shershnyova O., Ovrakh T., Serik S., Kolesnik T., Kosova H., Wood D., Adamska A., Adamska S., Jennings C., Kotseva K., Hoye P Atkin A., Fellowes D., Lindsay S., Atkinson C., Kranilla C., Vinod M., Beerachee Y., Bennett C., Broome M., Bwalya A., Caygill Lindsay, Dinning L., Gillespie A., Goodfellow R., Guy J., Idress T., Mills C., Morgan C., Oustance N., Singh N., Yare M., Jagoda J.M., Bowyer H., Christenssen V., Groves A., Jan A., Riaz A., Gill M., Sewell T.A., Gorog D., Baker M., De Sousa P., Mazenenga T., Porter J., Haines F., Peachey T., Taaffe J., Wells K., Ripley D.P., Forward H., McKie H., Pick S.L., Thomas H.E., Batin P.D., Exley D., Rank T., Wright J., Kardos A., Sutherland S.-B., Wren L., Leeson P., Barker D., Moreby B., Sawyer J., Stirrup J., Brunton M., Brodison A., Craig J., Peters S., Kaprielian R., Bucaj A., Mahay K., Oblak M., Gale C., Pye M., McGill Y., Redfearn H., Fearnley M. Management of dyslipidaemia in patients with coronary heart disease: Results from the ESC-EORP EUROASPIRE V survey in 27 countries. Atherosclerosis. 2019;285:135–146. doi: 10.1016/j.atherosclerosis.2019.03.014. [DOI] [PubMed] [Google Scholar]
- 6.Jones Buie Joy N., Goodwin Andrew J., Cook James A., Halushka Perry V., Fan Hongkuan. The role of miRNAs in cardiovascular disease risk factors. Atherosclerosis. 2016;254:271–281. doi: 10.1016/j.atherosclerosis.2016.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–24. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 8.Boon Reinier A., Vickers Kasey C. Intercellular Transport of MicroRNAs. Arteriosclerosis, Thrombosis, and Vascular Biology. 2013;33(2):186–192. doi: 10.1161/ATVBAHA.112.300139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vickers KC, et al. MicroRNA-223 coordinates cholesterol homeostasis. Proc. Natl. Acad. Sci. USA. 2014;111:14518–23. doi: 10.1073/pnas.1215767111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Mauro Stefania, Scamporrino Alessandra, Petta Salvatore, Urbano Francesca, Filippello Agnese, Ragusa Marco, Di Martino Maria T., Scionti Francesca, Grimaudo Stefania, Pipitone Rosaria M., Privitera Graziella, Di Pino Antonino, Scicali Roberto, Valenti Luca, Dongiovanni Paola, Fracanzani Anna, Rabuazzo Agata M., Craxì Antonio, Purrello Michele, Purrello Francesco, Piro Salvatore. Serum coding and non‐coding RNAs as biomarkers of NAFLD and fibrosis severity. Liver International. 2019;39(9):1742–1754. doi: 10.1111/liv.14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martino F, et al. Circulating miR-33a and miR-33b are up-regulated in familial hypercholesterolaemia in paediatric age. Clin. Sci. 2015;129:963–972. doi: 10.1042/CS20150235. [DOI] [PubMed] [Google Scholar]
- 12.D’Agostino M, et al. Circulating miR-200c is up-regulated in paediatric patients with familial hypercholesterolaemia and correlates with miR-33a/b levels: implication of a ZEB1-dependent mechanism. Clin. Sci. (Lond). 2017;131:2397–2408. doi: 10.1042/CS20171121. [DOI] [PubMed] [Google Scholar]
- 13.Vickers KC, Remaley AT. Lipid-based carriers of microRNAs and intercellular communication. Curr. Opin. Lipidol. 2012;23:91–7. doi: 10.1097/MOL.0b013e328350a425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011;13:423–33. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santos Raul D., Maranhao Raul C. What is new in familial hypercholesterolemia? Current Opinion in Lipidology. 2014;25(3):183–188. doi: 10.1097/MOL.0000000000000073. [DOI] [PubMed] [Google Scholar]
- 16.Averna, M. et al. Familial hypercholesterolemia: The Italian Atherosclerosis Society Network (LIPIGEN). Atheroscler. Suppl. 29 (2017). [DOI] [PubMed]
- 17.Pirillo A, et al. Spectrum of mutations in Italian patients with familial hypercholesterolemia: New results from the LIPIGEN study. Atheroscler. Suppl. 2017;29:17–24. doi: 10.1016/j.atherosclerosissup.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 18.Ten Kate G-JR, et al. The effect of LDLR-negative genotype on CT coronary atherosclerosis in asymptomatic statin treated patients with heterozygous familial hypercholesterolemia. Atherosclerosis. 2013;227:334–41. doi: 10.1016/j.atherosclerosis.2012.12.016. [DOI] [PubMed] [Google Scholar]
- 19.Bourbon M, et al. Mutational analysis and genotype-phenotype relation in familial hypercholesterolemia: The SAFEHEART registry. Atherosclerosis. 2017;262:8–13. doi: 10.1016/j.atherosclerosis.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Wagner J, et al. Characterization of levels and cellular transfer of circulating lipoprotein-bound microRNAs. Arterioscler. Thromb. Vasc. Biol. 2013;33:1392–400. doi: 10.1161/ATVBAHA.112.300741. [DOI] [PubMed] [Google Scholar]
- 21.Fichtlscherer S, Zeiher AM, Dimmeler S. Circulating MicroRNAs: biomarkers or mediators of cardiovascular diseases? Arterioscler. Thromb. Vasc. Biol. 2011;31:2383–2390. doi: 10.1161/ATVBAHA.111.226696. [DOI] [PubMed] [Google Scholar]
- 22.Scicali R, et al. Analysis of S100A12 plasma levels in hyperlipidemic subjects with or without familial hypercholesterolemia. Acta Diabetol. 2019;56:899–906. doi: 10.1007/s00592-019-01338-1. [DOI] [PubMed] [Google Scholar]
- 23.Paquette M, Dufour R, Baass A. The Montreal-FH-SCORE: A new score to predict cardiovascular events in familial hypercholesterolemia. J. Clin. Lipidol. 2017;11:80–86. doi: 10.1016/j.jacl.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 24.Farnier M, et al. How to implement clinical guidelines to optimise familial hypercholesterolaemia diagnosis and treatment. Atheroscler. Suppl. 2017;26:25–35. doi: 10.1016/S1567-5688(17)30022-3. [DOI] [PubMed] [Google Scholar]
- 25.Liu D, et al. MiR-486 regulates cholesterol efflux by targeting HAT1. Biochem. Biophys. Res. Commun. 2016;472:418–24. doi: 10.1016/j.bbrc.2015.11.128. [DOI] [PubMed] [Google Scholar]
- 26.Zhang R, et al. Expression of circulating miR-486 and miR-150 in patients with acute myocardial infarction. BMC Cardiovasc. Disord. 2015;15:51. doi: 10.1186/s12872-015-0042-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu F, Li R, Zhang Y, Qiu J, Ling W. Association of plasma MiR-17-92 with dyslipidemia in patients with coronary artery disease. Medicine (Baltimore). 2014;93:e98. doi: 10.1097/MD.0000000000000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Y, et al. Circulating miR-92a expression level in patients with essential hypertension: a potential marker of atherosclerosis. J. Hum. Hypertens. 2017;31:200–205. doi: 10.1038/jhh.2016.66. [DOI] [PubMed] [Google Scholar]
- 29.Shang F, et al. MicroRNA-92a Mediates Endothelial Dysfunction in CKD. J. Am. Soc. Nephrol. 2017;28:3251–3261. doi: 10.1681/ASN.2016111215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song Y-S, et al. Bone marrow mesenchymal stem cell-derived vascular endothelial growth factor attenuates cardiac apoptosis via regulation of cardiac miRNA-23a and miRNA-92a in a rat model of myocardial infarction. PLoS One. 2017;12:e0179972. doi: 10.1371/journal.pone.0179972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niculescu LS, et al. MiR-486 and miR-92a Identified in Circulating HDL Discriminate between Stable and Vulnerable Coronary Artery Disease Patients. PLoS One. 2015;10:e0140958. doi: 10.1371/journal.pone.0140958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niculescu LS, et al. Inhibition of miR-486 and miR-92a decreases liver and plasma cholesterol levels by modulating lipid-related genes in hyperlipidemic hamsters. Mol. Biol. Rep. 2018;45:497–509. doi: 10.1007/s11033-018-4186-8. [DOI] [PubMed] [Google Scholar]
- 33.Ganjali S, et al. HDL abnormalities in familial hypercholesterolemia: Focus on biological functions. Prog. Lipid Res. 2017;67:16–26. doi: 10.1016/j.plipres.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 34.Santos RD, et al. Defining severe familial hypercholesterolaemia and the implications for clinical management: a consensus statement from the International Atherosclerosis Society Severe Familial Hypercholesterolemia Panel. lancet. Diabetes Endocrinol. 2016;4:850–61. doi: 10.1016/S2213-8587(16)30041-9. [DOI] [PubMed] [Google Scholar]
- 35.Junyent M, et al. Impact of low-density lipoprotein receptor mutational class on carotid atherosclerosis in patients with familial hypercholesterolemia. Atherosclerosis. 2010;208:437–41. doi: 10.1016/j.atherosclerosis.2009.07.058. [DOI] [PubMed] [Google Scholar]
- 36.Smilde T, et al. Effect of aggressive versus conventional lipid lowering on atherosclerosis progression in familial hypercholesterolemia (ASAP): a prospective, randomised, double-blind trial. Lancet. 2001;357:577–581. doi: 10.1016/S0140-6736(00)04053-8. [DOI] [PubMed] [Google Scholar]
- 37.Scicali Roberto, Di Pino Antonino, Ferrara Viviana, Urbano Francesca, Piro Salvatore, Rabuazzo Agata Maria, Purrello Francesco. New treatment options for lipid-lowering therapy in subjects with type 2 diabetes. Acta Diabetologica. 2017;55(3):209–218. doi: 10.1007/s00592-017-1089-4. [DOI] [PubMed] [Google Scholar]
- 38.Benn M, Watts GF, Tybjaerg-Hansen A, Nordestgaard BG. Familial hypercholesterolemia in the danish general population: prevalence, coronary artery disease, and cholesterol-lowering medication. J. Clin. Endocrinol. Metab. 2012;97:3956–64. doi: 10.1210/jc.2012-1563. [DOI] [PubMed] [Google Scholar]
- 39.Lincoff AM, et al. Evacetrapib and Cardiovascular Outcomes in High-Risk Vascular Disease. N. Engl. J. Med. 2017;376:1933–1942. doi: 10.1056/NEJMoa1609581. [DOI] [PubMed] [Google Scholar]
- 40.HPS3/TIMI55–REVEAL Collaborative Group et al. Effects of Anacetrapib in Patients with Atherosclerotic Vascular Disease. N. Engl. J. Med. 2017;377:1217–1227. doi: 10.1056/NEJMoa1706444. [DOI] [PubMed] [Google Scholar]
- 41.Schaefer JR, et al. In vivo metabolism of apolipoprotein A-I in a patient with homozygous familial hypercholesterolemia. Arterioscler. Thromb. a J. Vasc. Biol. 1992;12:843–8. doi: 10.1161/01.ATV.12.7.843. [DOI] [PubMed] [Google Scholar]
- 42.Ansell BJ, Watson KE, Fogelman AM, Navab M, Fonarow GC. High-Density Lipoprotein Function. J. Am. Coll. Cardiol. 2005;46:1792–1798. doi: 10.1016/j.jacc.2005.06.080. [DOI] [PubMed] [Google Scholar]
- 43.Nenseter MS, et al. Cholesterol efflux mediators in homozygous familial hypercholesterolemia patients on low-density lipoprotein apheresis. J. Clin. Lipidol. 2013;7:109–116. doi: 10.1016/j.jacl.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 44.Creemers EE, Tijsen AJ, Pinto YM. Circulating MicroRNAs. Circ. Res. 2012;110:483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 45.Casula M, et al. Evaluation of the performance of Dutch Lipid Clinic Network score in an Italian FH population: The LIPIGEN study. Atherosclerosis. 2018;277:413–418. doi: 10.1016/j.atherosclerosis.2018.08.013. [DOI] [PubMed] [Google Scholar]
- 46.Stone NJ, et al. 2013 ACC/AHA Guideline on the Treatment of Blood Cholesterol to Reduce Atherosclerotic Cardiovascular Risk in Adults. J. Am. Coll. Cardiol. 2014;63:2889–2934. doi: 10.1016/j.jacc.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 47.Calanna Salvatore, Scicali Roberto, Di Pino Antonino, Knop Filip Krag, Piro Salvatore, Rabuazzo Agata Maria, Purrello Francesco. Alpha- and beta-cell abnormalities in haemoglobin A1c-defined prediabetes and type 2 diabetes. Acta Diabetologica. 2014;51(4):567–575. doi: 10.1007/s00592-014-0555-5. [DOI] [PubMed] [Google Scholar]
- 48.Ryan H, Trosclair A, Gfroerer J. Adult current smoking: differences in definitions and prevalence estimates–NHIS and NSDUH, 2008. J. Environ. Public Health. 2012;2012:918368. doi: 10.1155/2012/918368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spadaro Luisa, Noto Davide, Privitera Graziella, Tomaselli Tania, Fede Giuseppe, Scicali Roberto, Piro Salvatore, Fayer Francesca, Altieri Ida, Averna Maurizio, Purrello Francesco. Apolipoprotein AI and HDL are reduced in stable cirrhotic patients with adrenal insufficiency: a possible role in glucocorticoid deficiency. Scandinavian Journal of Gastroenterology. 2015;50(3):347–354. doi: 10.3109/00365521.2014.985707. [DOI] [PubMed] [Google Scholar]
- 50.Calanna, S. et al. Lipid and liver abnormalities in haemoglobin A1c-defined prediabetes and type 2 diabetes. Nutr. Metab. Cardiovasc. Dis. 24 (2014). [DOI] [PubMed]
- 51.Di Pino, A. et al. High intake of dietary advanced glycation end-products is associated with increased arterial stiffness and inflammation in subjects with type 2 diabetes. Nutr. Metab. Cardiovasc. Dis. 27 (2017). [DOI] [PubMed]
- 52.Ossoli A, et al. Recombinant LCAT (Lecithin:Cholesterol Acyltransferase) Rescues Defective HDL (High-Density Lipoprotein)-Mediated Endothelial Protection in Acute Coronary Syndrome. Arterioscler. Thromb. Vasc. Biol. 2019;39:915–924. doi: 10.1161/ATVBAHA.118.311987. [DOI] [PubMed] [Google Scholar]
- 53.Di Mauro S, et al. Intracellular and extracellular miRNome deregulation in cellular models of NAFLD or NASH: Clinical implications. Nutr. Metab. Cardiovasc. Dis. 2016;26:1129–1139. doi: 10.1016/j.numecd.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 54.Di Pino A, et al. Cardiovascular Risk Profile in Subjects With Prediabetes and New-Onset Type 2 Diabetes Identified by HbA 1c According to American Diabetes Association Criteria. Diabetes Care. 2014;37:1447–1453. doi: 10.2337/dc13-2357. [DOI] [PubMed] [Google Scholar]