Abstract
Ligninolytic peroxidases are microbial enzymes involved in depolymerisation of lignin, a plant cell wall polymer found in land plants. Among fungi, only Dikarya were found to degrade lignin. The increase of available fungal genomes allows performing an expert annotation of lignin-degrading peroxidase encoding sequences with a particular focus on Class II peroxidases (CII Prx). In addition to the previously described LiP, MnP and VP classes, based on sequence similarity, six new sub-classes have been defined: three found in plant pathogen ascomycetes and three in basidiomycetes. The presence of CII Prxs could be related to fungal life style. Typically, necrotrophic or hemibiotrophic fungi, either ascomycetes or basidiomycetes, possess CII Prxs while symbiotic, endophytic or biotrophic fungi do not. CII Prxs from ascomycetes are rarely subjected to duplications unlike those from basidiomycetes, which can form large recent duplicated families. Even if these CII Prxs classes form two well distinct clusters with divergent gene structures (intron numbers and positions), they share the same key catalytic residues suggesting that they evolved independently from similar ancestral sequences with few or no introns. The lack of CII Prxs encoding sequences in early diverging fungi, together with the absence of duplicated class I peroxidase (CcP) in fungi containing CII Prxs, suggests the potential emergence of an ancestral CII Prx sequence from the duplicated CcP after the separation between ascomycetes and basidiomycetes. As some ascomycetes and basidiomycetes did not possess CII Prx, late gene loss could have occurred.
Subject terms: Evolution, Bioinformatics
Introduction
Plant cell walls represent the major components of plant biomass. As both a physical barrier and a dynamic compartment, they play important roles in the development of plants and their interactions with the environment, but also in ecology as they are major components of the carbon cycle. Th primary cell wall mainly consists of celluloses, hemicelluloses, pectins and a minority of structural and enzymatic proteins which confer growth abilities to the plant. The secondary cell wall mostly contains cellulose and lignin, a complex polyphenolic polymer responsible for land plant rigidity and associated with the vessels appearance, with notable difference in composition between softwood (coniferous) and hardwood (deciduous). Altogether, they form intricate macromolecular networks that are synthesized by complex processes. Lignocellulosic cell walls are carbon sources which can therefore be degraded by micro-organisms such as some bacteria and more efficiently by certain fungi1,2. Plant cell walls are decayed primarily by wood and litter decomposers that easily digest cellulosic compounds due to enzymatic activities. However, the accessibility of these compounds is restricted by the presence of the more resistant lignin polymers. Therefore, efficient lignin depolymerization capacities are necessary and are developed by wood-decaying basidiomycetes3. In a recent review2, ligninolytic enzymes were inventoried into three categories: phenol oxidases (or laccases), class II peroxidases (CII Prxs thereafter, also known as POD) and Dye-decolorizing peroxidases. These last ones (called DyP-type peroxidases) are more reported in bacteria than in fungi4, and little is known about their physiological role. Historically, wood-decaying fungi have been characterized as white-rot (WR) or brown-rot (BR) fungi due to the color of remaining woody fragments. WR fungi can either degrade both lignin and polysaccharides (simultaneous decay) or preferentially remove lignin, leaving most of the cell wall polysaccharides unaffected (selective decay). As an example, Phanerochaete chrysosporium is a very efficient wood decomposer; that can simultaneously degrade lignin and cellulose5–7. A closely related species, Ceriporiopsis subvermispora, preferentially depolymerizes lignin with reduced cellulose degradation8. Genomes of several WR fungi have been screened for the presence of ligninolytic peroxidases such as Agaricus bisporus or Pleurotus ostreatus, which possess limited copies of CII Prx9,10. BR fungi, like Postia placenta and Serpula lacrymans, are mainly capable of pectin, hemicellulose and cellulose break-down but have only few or no lignin-degrading enzymes such as the CII Prx11–13. BR fungi preferentially attack softwoods i.e. gymnosperm species. The lack of CII Prx is also observed in the ectomycorrhizal (ECM) fungus Laccaria bicolor14 as well as others ECM15 suggesting that the reduced number or absence of CII Prx isoforms could be correlated with the mode of carbon acquisition, and then the type of interaction with the host plants. The hypothesis is that the loss of CII Prx in ECM was initiated in their saprotroph ancestor and therefore preceded the evolution of the ECM habit15. It is also important to note that a no ligninolytic fungi belongs to rhizosphere microbial communities containing ligninolytic fungi which could act synergistically. Phylogenetic analysis of CII Prx from Agaromycetes and from Hymenochaetales suggested that lignin degradation by WR fungi is strongly correlated with the presence of ligninolytic peroxidases and could not only be performed by laccases, the other ligninolytic enzyme group16,17. It was even proposed to limit the “white rot” term to fungi that possess CII Prx18, as many fungi are intermediate between WR and BR in wood decay mechanism18–20.
CII Prxs belong to the non-animal haem peroxidases superfamily together with the intracellular class I peroxidases (CI Prxs) and secretory plant class III peroxidases (CIII Prxs)21. The three classes CI, CII and CIII Prx are all characterized by a prosthetic group and 10 conserved α-helices22. In these classes, various substrates can be oxidized with hydrogen peroxide (H2O2) as the electron acceptor and reduced to water. The amino acids necessary for haem binding and to the H2O2 access channel are conserved between the various peroxidase classes. The CII Prxs, only detected in fungi, are extracellular peroxidases that were initially subdivided into 3 groups based on their catalytic properties: lignin (LiP), manganese (MnP) and versatile (VP) peroxidases23. Additional genomic data allowed the identification of a fourth group found in basidiomycetes, called “generic peroxidases” (GP)24. Three residues (Glu35, Glu39 and Asp179 in the mature PcMnP01) have been identified as implicated in the manganese-dependent activity25. These residues are well conserved within the MnP and VP proteins. VP were originated thanks to the appearance of an exposed tryptophan, implied in oxidation of nonphenolic lignin. Later, the loss of the Mn2+-binding site led to the generation of LiP proteins, which possess higher catalytic efficiency26.
Many investigations were done these last years regarding the evolution and distribution of CII Prxs among fungal genomes15,24,27,28. The resulting hypothesis is that CII Prx exists in the ancestor of Auriculariales and Agaricomycetes among basidiomycetes, with subsequent gene expansion of this family in saprotrophic fungal species involved in wood decay, degrading lignin to obtain their organic carbon sources. These works also indicated that ectomycorrhizal fungal species that later arose from independent lineages similarly present a loss of CII Prx gene as a convergent genome adaptation for symbiosis, thus limiting host immunity responses. Finally, CII Prx could also be a means for pathogenic species to degrade host cell wall for penetration. In ascomycetes, putative CII Prxs have been neither described, nor included in the four previous CII Prx families. Ascomycetes have limited ability to degrade lignin and are mostly responsible for soft rot decay in wet environment29.
Wood decay, and particularly lignin degradation by fungal laccases and peroxidases has a major ecological incidence on the carbon cycle30 and presents industrial interests. However automatic genome annotation has been demonstrated to produce increased mis-predictions in the case of multigenic families and genes containing numerous exons and very short intron/exon sizes31. We mined publicly available basidiomycete and ascomycete genomes to perform an expert annotation of the different Prxs. We set up a protocol coupling automatic and manual annotation for an exhaustive and quality annotation in order to analyze the evolution process of this complex family.
Material and Methods
Data mining and annotation with Scipio
An exhaustive data mining procedure was performed on the fungal genomes available from BROAD institute, JGI and NCBI to extract all the available ligninase sequences. All sequences used in this study have been annotated by an expert process to discard prediction errors subsequent to automatic annotations. First, a search by keyword and profile number has been done on different databases. These searches have been completed by homology searches against predicted proteomes (BLASTP), transcriptomes (TBLASTN) or whole genomes using well known ligninase sequences from Phanerochaetes. This data mining allowed the creation of initial batches corresponding to the different protein families. Then, a manual and individual curation of each predicted sequence was performed: gene structure (intron number and position), length and the presence of key residues were mandatory controls. When available mainly from NCBI, EST sequences have also been used to confirm annotations. These protein batches were then used to precisely determine the corresponding chromosome positions, gene structures and coding DNA sequences (CDS) with a protocol based on Scipio32. New paralogs, not initially annotated, were found using this procedure. We ended up with 267 sequences from Basidiomycota and 99 from Ascomycota.
Phylogenetic analysis
All protein sequences used for the in silico analyses are available from the RedoxiBase database (http://peroxibase.toulouse.inra.fr)33,34. First, protein sequences were aligned using PRANK35 with default parameters. Then phylogenies were estimated by maximum likelihood using RaxML (version 8.1.5)36, under the PROTGAMMAWAG model, as the substitution model determined by protTest37 was WAG38 and a gamma distribution (4 discrete categories of sites and an estimated alpha parameter). Finally, the trees were edited and analyzed using iTOL (https://itol.embl.de/).
Gene structure analysis
The intron/exon coordinates together with the corresponding genomic sequences of all identified genes were determined with Scipio32, with maximal intron size set to 1000 nt, and minimum percent identity set to 30%. The intron/exon conservation within the different families was verified with CIWOG39 and GECA40. They both analyzed the evolution or conservation of introns between paralogs as well as between species.
Intron size changes were visualized through the graphical representation provided by GECA.
Conserved common introns analysis
Gene structure and common introns (or cintrons) were analyzed from all fungi sequences. First, the protein alignment generated with MAFFT41 was completed with the identification of common introns in the corresponding genes with CIWOG. Cintrons were extracted from the CIWOG database and only those present in one or more sub-classes with a conservation rate higher than 50% were considered as conserved. Finally, the sequences were placed in order of appearance in the phylogenetic tree and the conserved cintrons were highlighted for each sequence.
Duplication analysis
In order to test whether the presence of transposable elements can explain a high duplication rate, RepeatMasker42 version 4.0.3 (with fungi specified as “species”) was run on all analyzed Basidiomycete genomes. No correlation can be made between the number of paralogs in an organism and the number of repeated sequences. Deeper analysis of repeated sequences positions was conducted for Tramete versicolor and Galerina marginata genomes (which possess the highest number of CII Prxs): neither transposable elements nor other repeated sequences were systematically detected nearby to a gene copy.
New PROSITE profiles design and WebLogo
Using a global phylogenetic analysis, different protein clusters have been defined to update the existing PROSITE profiles33 and to design new specific profiles using the silenced residues. These profiles were built from full length alignments of each protein cluster. First, all the sequences from the different protein clusters were aligned with MAFFT. The sequence alignment was split into several sub-alignments according to the cluster definitions. Each cluster alignment contains an annotation line where residues conserved in the whole family are tagged. This annotation line is used to downweight family-conserved columns during the profile construction; therefore only cluster specific residues are taken into consideration. The reliability of each cluster is supported by both the analysis of the gene structures and the presence/absence of the key residues specific to the well described LiP, MnP and VP families. Furthermore, graphical sequence logos were created for each group with Weblogo343 and aligned manually with the others in order to identify the amino acids conserved between the sub-classes.
Results and Discussion
Definition of new sub-classes of ligninases
A high quality of annotation is mandatory to perform a global analysis of multigene families evolution such as those of the CII Prxs44. A set of 150 genomes from ascomycetes, basidiomycetes and early diverging fungi (Table 1) has been carefully annotated for CII Prx encoding sequences and used for phylogeny, clustering analysis and profile design. No ligninase-like sequence has been detected in any early diverging fungi analyzed. The CII Prx numbers and gene structures are highly variable. Between 1 and 15 isoforms can be detected per species and may contain up to 15 introns in a single sequence, with short exons and introns (e.g. 6 nt for the last exon). Characteristic residues necessary for haem binding and electron transport were looked for in all CII Prxs analyzed (Fig. 1). Well conserved clusters have been identified by phylogenetic analysis and the corresponding sequences were used to update the existing profiles and to construct new profiles and sub-profiles. When available, the gene structure (introns/exons) was also used to support the tree topology. This procedure, combining phylogenetic analysis, presence of key residues, gene structure analysis and construction of HMM profiles, allowed identifying mis-classifications (false positives and false negatives) and reassigning them to their appropriate sub-classes.
Table 1.
number of sequences for Ascomycete and Basidiomycete species.
| phylum | order | Trophism | CcP | MnP | LiP | VP | CIIBA | CIIBB | CIIBC | CIIAA | CIIAB | CIIAC | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gonapodya prolifera | Monoblepharid-omycota | Monoblepharidales | S | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mortierella elongata | Mortierellomycotina | Mortierellales | END/S | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mortierella verticillata | Mortierellomycotina | Mortierellales | END/S | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Umbelopsis ramanniana | Mucoromycotina | Umbelopsidales | END/S | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Phycomyces blakesleeanus | Mucoromycotina | Mucorales | S | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mucor circinelloides | Mucoromycotina | Mucorales | S/OP | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Rhizopus delemar | Mucoromycotina | Mucorales | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Piromyces sp | Neocallimastigo-mycota | Neocallimastigales | Sy | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Aspergillus fumigatus | Pezizomycotina | Eurotiales | AP/OP/S | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Aureobasidium pullulans | Pezizomycotina | Dothideales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 3 | |
| Metarhizium acridum | Pezizomycotina | Hypocreales | APE | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| Ascosphaera apis | Pezizomycotina | Onygenales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Penicillium marneffei | Pezizomycotina | Eurotiales | APM | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Geomyces destructans | Pezizomycotina | Leotiomycetes Incertae | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Ajellomyces capsulatus | Pezizomycotina | Onygenales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Arthroderma benhamiae ana. | Pezizomycotina | Onygenales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Coccidioides immitis | Pezizomycotina | Onygenales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Coccidioides posadasii | Pezizomycotina | Onygenales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Paracoccidioides brasiliensis | Pezizomycotina | Onygenales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Trichophyton equinum | Pezizomycotina | Onygenales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Trichophyton rubrum | Pezizomycotina | Onygenales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Trichophyton tonsurans | Pezizomycotina | Onygenales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Trichophyton verrucosum | Pezizomycotina | Onygenales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Blumeria graminis | Pezizomycotina | Erysiphales | BPP/END | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Erysiphe pisi | Pezizomycotina | Erysiphales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Epichloe festucae | Pezizomycotina | Hypocreales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Cercospora zeae-maydis | Pezizomycotina | Capnodiales | HPP | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| Mycosphaerella graminicola | Pezizomycotina | Capnodiales | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 2 | |
| Colletotrichum graminicola | Pezizomycotina | Glomerellales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 1 | 1 | 7 | |
| Magnaporthe grisea | Pezizomycotina | Magnaporthales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 3 | |
| Cochliobolus sativus | Pezizomycotina | Pleosporales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 1 | 5 | |
| Leptosphaeria maculans | Pezizomycotina | Pleosporales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 1 | 4 | |
| Setosphaeria turcica | Pezizomycotina | Pleosporales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 1 | 4 | |
| Melanconium sp. | Mitosporic Ascomycota | mitosporic | NPP | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 1 | 0 | 4 |
| Aplosporella prunicola | Pezizomycotina | Botryosphaeriales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | |
| Botryosphaeria dothidea | Pezizomycotina | Botryosphaeriales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 4 | |
| Cryphonectria parasitica | Pezizomycotina | Diaporthales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | |
| Verticillium dahliae | Pezizomycotina | Glomerellales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 3 | |
| Botrytis cinerea | Pezizomycotina | Helotiales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Sclerotinia sclerotiorum | Pezizomycotina | Helotiales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Fusarium oxysporum | Pezizomycotina | Hypocreales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | |
| Gibberella zeae ana. | Pezizomycotina | Hypocreales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | |
| Nectria haematococca ana. | Pezizomycotina | Hypocreales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | |
| Gaeumannomyces graminis | Pezizomycotina | Magnaporthales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 3 | |
| Alternaria brassicicola | Pezizomycotina | Pleosporales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 1 | 4 | |
| Cochliobolus heterostrophus | Pezizomycotina | Pleosporales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 1 | 5 | |
| Didymella exigua | Pezizomycotina | Pleosporales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 2 | 6 | |
| Parastagonospora nodorum | Pezizomycotina | Pleosporales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 1 | 5 | |
| Pyrenophora tritici-repentis | Pezizomycotina | Pleosporales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 1 | 4 | |
| Cenococcum geophilum | Pezizomycotina | mitosporic | Sy (ECM, ERM, Li) | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| Meliniomyces bicolor | Pezizomycotina | Helotiales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | |
| Meliniomyces variabilis | Pezizomycotina | Helotiales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | |
| Cladonia grayi | Pezizomycotina | Lecanorales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Oidiodendron maius | Pezizomycotina | Leotiomycetes incertae | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Grosmannia clavigera | Pezizomycotina | Ophiostomatales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | |
| Tuber melanosporum | Pezizomycotina | Pezizales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Xanthoria parietina | Pezizomycotina | Teloschistales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Daldinia eschscholzii | Pezizomycotina | Xylariales | END | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| Hypoxylon sp. C14A | Pezizomycotina | Xylariales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 2 | |
| Aspergillus clavatus | Pezizomycotina | Eurotiales | END/S | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Trichoderma asperellum | Pezizomycotina | Hypocreales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Baudoinia compniacensis | Pezizomycotina | Capnodiales | S | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| Zasmidium cellare | Pezizomycotina | Capnodiales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 3 | |
| Aspergillus aculeatus | Pezizomycotina | Eurotiales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Aspergillus wentii | Pezizomycotina | Eurotiales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | |
| Emericella nidulans | Pezizomycotina | Eurotiales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Monascus ruber | Pezizomycotina | Eurotiales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Penicillium chrysogenum | Pezizomycotina | Eurotiales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Talaromyces stipitatus | Pezizomycotina | Eurotiales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Thermoascus aurantiacus | Pezizomycotina | Eurotiales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Acremonium alcalophilum | Pezizomycotina | Glomerellales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | |
| Trichoderma reesei ana | Pezizomycotina | Hypocreales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Hysterium pulicare | Pezizomycotina | Hysteriales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 2 | |
| Amorphotheca resinae | Pezizomycotina | Leotiomycetes incertae | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Uncinocarpus reesii | Pezizomycotina | Onygenales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Ascobolus immersus | Pezizomycotina | Pezizales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Cucurbitaria berberidis | Pezizomycotina | Pleosporales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 2 | |
| Lentithecium fluviatile | Pezizomycotina | Pleosporales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 1 | 3 | 7 | |
| Chaetomium thermophilum | Pezizomycotina | Sordariales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Neurospora crassa | Pezizomycotina | Sordariales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Podospora anserina | Pezizomycotina | Sordariales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | |
| Sordaria macrospora | Pezizomycotina | Sordariales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Sporotrichum thermophile | Pezizomycotina | Sordariales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | |
| Thielavia terrestris | Pezizomycotina | Sordariales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Apiospora montagnei | Pezizomycotina | Xylariales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 2 | |
| Aspergillus flavus | Pezizomycotina | Eurotiales | S/OP | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Aspergillus niger | Pezizomycotina | Eurotiales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Aspergillus oryzae | Pezizomycotina | Eurotiales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Neosartorya fischeri | Pezizomycotina | Eurotiales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Rhytidhysteron rufulum | Pezizomycotina | Hysteriales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 3 | |
| Chaetomium globosum | Pezizomycotina | Sordariales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Trichoderma atroviride | Pezizomycotina | Hypocreales | S/P | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Trichoderma harzianum | Pezizomycotina | Hypocreales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Trichoderma virens | Pezizomycotina | Hypocreales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Cryptococcus gattii | Basidiomycota | Tremellales | APM | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Cryptococcus neoformans | Basidiomycota | Tremellales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Moniliophthora perniciosa | Basidiomycota | Agaricales | HPP | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Amanita muscaria | Basidiomycota | Agaricales | Sy (ECM) | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Hebeloma cylindrosporum | Basidiomycota | Agaricales | 1 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | |
| Laccaria bicolor | Basidiomycota | Agaricales | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | |
| Laccaria amethystina | Basidiomycota | Agaricales | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | |
| Piloderma croceum | Basidiomycota | Atheliales | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | |
| Boletus edulis | Basidiomycota | Boletales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Paxillus involutus | Basidiomycota | Boletales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Paxillus rubicundulus | Basidiomycota | Boletales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Suillus brevipes | Basidiomycota | Boletales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Suillus luteus | Basidiomycota | Boletales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Sebacina vermifera | Basidiomycota | Sebacinales | Sy (OMF/ECM) | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Piriformospora indica | Basidiomycota | Sebacinales | END | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Galerina marginata | Basidiomycota | Agaricales | S (WR) | 1 | 0 | 0 | 0 | 0 | 6 | 16 | 0 | 0 | 0 | 22 |
| Gymnopus luxurians | Basidiomycota | Agaricales | 1 | 3 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 5 | |
| Hypholoma sublateritium | Basidiomycota | Agaricales | 1 | 0 | 0 | 0 | 0 | 10 | 4 | 0 | 0 | 0 | 14 | |
| Pleurotus ostreatus | Basidiomycota | Agaricales | 1 | 0 | 0 | 6 | 3 | 0 | 0 | 0 | 0 | 0 | 9 | |
| Plicaturopsis crispa | Basidiomycota | Amylocorticiales | 1 | 0 | 0 | 1 | 2 | 4 | 0 | 0 | 0 | 0 | 7 | |
| Auricularia delicata | Basidiomycota | Auriculariales | 1 | 0 | 0 | 0 | 17 | 1 | 0 | 0 | 0 | 0 | 18 | |
| Punctularia strigosozonata | Basidiomycota | Corticiales | 1 | 9 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 11 | |
| Sphaerobolus stellatus | Basidiomycota | Geastrales | 1 | 20 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 24 | |
| Bjerkandera adusta | Basidiomycota | Polyporales | 1 | 12 | 0 | 0 | 2 | 5 | 0 | 0 | 0 | 0 | 19 | |
| Ceriporiopsis subvermispora | Basidiomycota | Polyporales | 1 | 12 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 16 | |
| Cerrena unicolor | Basidiomycota | Polyporales | 1 | 0 | 3 | 0 | 4 | 9 | 0 | 0 | 0 | 0 | 16 | |
| Dichomitus squalens | Basidiomycota | Polyporales | 1 | 4 | 0 | 3 | 3 | 2 | 0 | 0 | 0 | 0 | 12 | |
| Ganoderma sp | Basidiomycota | Polyporales | 1 | 0 | 0 | 1 | 4 | 3 | 0 | 0 | 0 | 0 | 8 | |
| Phanerochaete carnosa | Basidiomycota | Polyporales | 1 | 7 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | |
| Phanerochaete chrysosporium | Basidiomycota | Polyporales | 1 | 7 | 10 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 18 | |
| Phlebia brevispora | Basidiomycota | Polyporales | 1 | 6 | 5 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 14 | |
| Phlebiopsis gigantea | Basidiomycota | Polyporales | 1 | 6 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 10 | |
| Polyporus arcularius | Basidiomycota | Polyporales | 1 | 0 | 6 | 2 | 6 | 5 | 0 | 0 | 0 | 0 | 19 | |
| Trametes versicolor | Basidiomycota | Polyporales | 1 | 0 | 11 | 2 | 2 | 9 | 0 | 0 | 0 | 0 | 24 | |
| Heterobasidion annosum | Basidiomycota | Russulales | 1 | 3 | 0 | 0 | 2 | 5 | 0 | 0 | 0 | 0 | 10 | |
| Stereum hirsutum | Basidiomycota | Russulales | 1 | 0 | 0 | 0 | 1 | 0 | 5 | 0 | 0 | 0 | 6 | |
| Tremella mesenterica | Basidiomycota | Tremellales | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Schizophyllum commune | Basidiomycota | Agaricales | S (DND) | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Jaapia argillacea | Basidiomycota | Jaapiales | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | |
| Fistulina hepatica | Basidiomycota | Agaricales | S (BR) | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Coniophora puteana | Basidiomycota | Boletales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Hydnomerulius pinastri | Basidiomycota | Boletales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Serpula lacrymans | Basidiomycota | Boletales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Botryobasidium botryosum | Basidiomycota | Cantharellales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Dacryopinax sp | Basidiomycota | Dacrymycetales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Gloeophyllum trabeum | Basidiomycota | Gloeophyllales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Neolentinus lepideus | Basidiomycota | Gloeophyllales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Daedalea quercina | Basidiomycota | Polyporales | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | |
| Fomitopsis pinicola | Basidiomycota | Polyporales | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | |
| Laetiporus sulphureus | Basidiomycota | Polyporales | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | |
| Postia placenta | Basidiomycota | Polyporales | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | |
| Wolfiporia cocos | Basidiomycota | Polyporales | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | |
| Agaricus bisporus | Basidiomycota | Agaricales | Soil/litter decomposer | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| Amanita thiersii | Basidiomycota | Agaricales | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Coprinopsis cinerea | Basidiomycota | Agaricales | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | |
| Dioszegia cryoxerica | Basidiomycota | Tremellales | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
S: Saprotrophe (Wood Decay: BR = Brown rot; WR = White rot; DND = Decay chemistry Not Defined); PP: Plant Pathogen (NPP: Necrotrophic PP; BPP: Biothophic PP; HPP: Hemibiotrophic PP; OP: Opportunistic Pathogen); AP: Animal Pathogen (APM: Animal Pathogen on Mammals; APE: Animal Pathogen Entomopathogenic); Sy: Symbiotic (ECM: Ectomycorrhizal fungus; ERM: Ericoid Mycorrhizal fungus; OMF: Orchid Mycorrhizal fungus; Li: Lichen); END: endophytic fungus; U:Unicell; Mycel: Mycelium; D: Dimorphic species; T: thallus with rhizoids.
Figure 1.
Weblogo of different CII Prxs from ascomycetes (a) and basidiomycetes (b). basidiomycete and ascomycete CII Prxs were aligned with MAFFT, and then separated into sub-classes. Weblogos were created for each group with Weblogo3 and aligned manually with the others in order to easily identify the conserved AA between the sub-classes highlighted in yellow and those that are specific to one or several sub-classes are highlighted in others colors. AA, AB and AC stand respectively for ascomycete CII Prxs sub-class A, B, and C; BA, BB and BC stand respectively for basidiomycete CII Prxs sub-class A,, B and C,, VP: Versatile peroxidases, MnP: Manganese peroxidases, LiP: Lignin peroxidases. Green highlight: eight cysteines forming four disulfide bridges; blue highlight: two active site histidines; orange highlight: three acidic residues forming the Mn2+ oxidation site; red dot: nine ligands of two structural Ca2+ ions; blue dot: one tryptophan responsible for aromatic substrate oxidation by LiP; dark gray: position specific to one class. *Conserved residues between basidiomycete and ascomycete CII Prx classes.
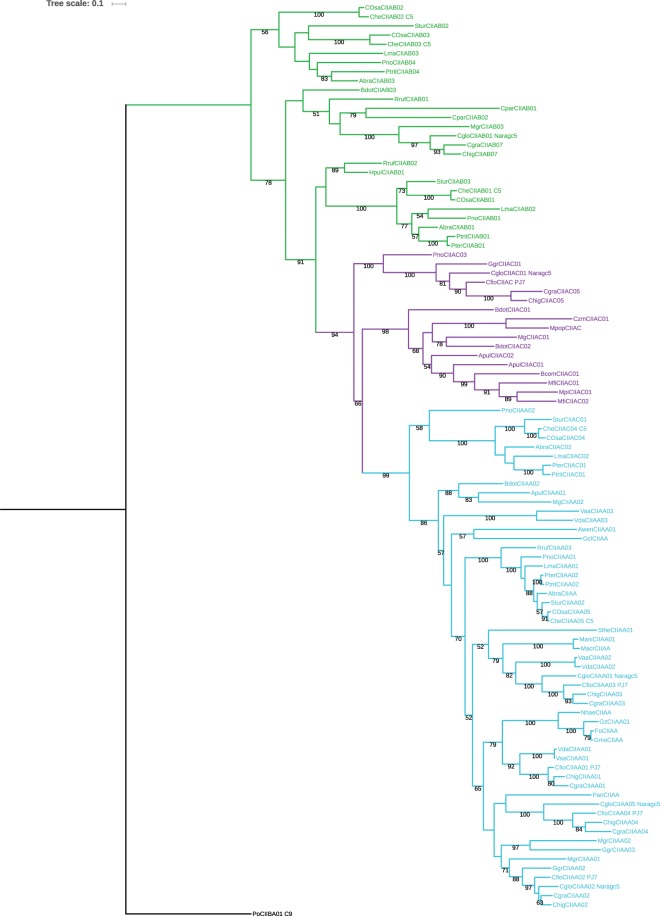
Sequences detected in ascomycetes form a group distinct from the well-described basidiomycete ligninases (Supplementary Fig. S1). The clear distribution of ascomycete ligninases in three clusters (Fig. 2) helps defining and designing three sub-classes of ascomycete ligninases, thereafter referred as ascomycete sub-class A (CIIAA), sub-class B (CIIAB) and sub-class C (CIIAC), and their corresponding profiles (Fig. 1a). By comparing the conserved residues found in basidiomycetes and ascomycetes (Fig. 1), we can see that (i) many residues dispersed throughout the sequences are conserved (~20 aa), between these two very distant phyla; (ii) the 3 sub-classes defined in ascomycetes do not share all of the defined residues with catalytic properties found in basidiomycetes, and miss most of the cysteine residues responsible for protein stability; (iii) the ascomycete CIIAA sub-class has the more divergent sequence profile, which is consistent with its phylogenetic position (Fig. 2). Gene structure analysis doesn’t reveal conserved common introns between the three sub-classes. Extensive changes in the exon-intron structure (intron gain and loss) have already been described for members of the Fusarium clade and appear to be the normality in ascomycetes45.
Figure 2.
Phylogenetic tree of ascomycetes CII Prx sequences. 99 sequences coming from 26 ascomycetes have been aligned to generate the tree. A basidiomycete sequence was used as outgroup. CIIAA are represented in blue, CIIAB in green and CIIAC in purple. Bootstrap values higher than 50% are indicated.
Members of these 3 new sub-classes are only detected in Pezizomycotina and are absent from the other ascomycete sub-phyla. Moreover, within the Pezizomycotina sub-phylum, ligninase encoding sequences are not detected in all species. They are mainly found in species known to interact with plants: pathogenic ascomycetes, either necrotrophic or hemibiotrophic, possess up to 7 sequences, whereas few or no sequence were detected in saprotrophic species (Table 1).
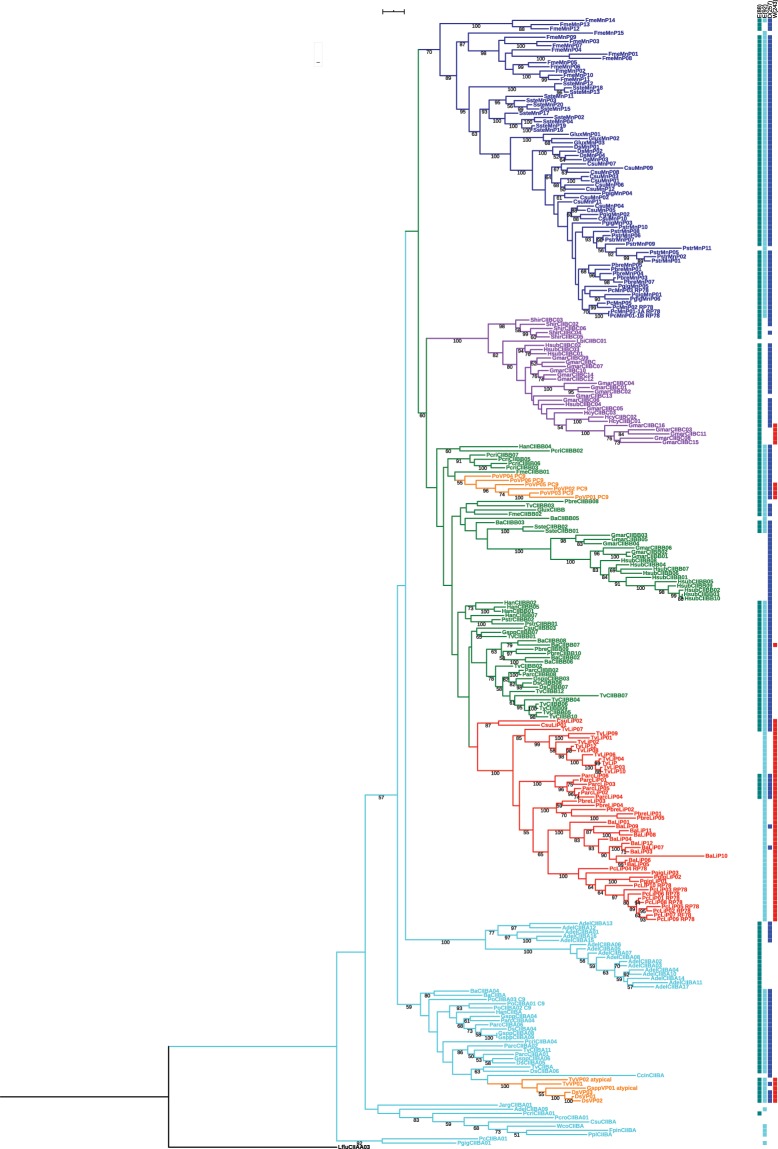
The situation for basidiomycetes is much more complex. The exhaustive mining of 68 basidiomycete genomes has demonstrated the need to redefine the existing profiles and highlighted that some sequences do not belong to the four previously identified groups (LiP, MnP, VP, GP). Three new basidiomycete ligninase sub-classes have been defined, basidiomycete sub-class A (CIIBA), sub-class B (CIIBB) and sub-class C (CIIBC) (Fig. 3). The definition of these 3 new sub-classes also led to re-affect some sequences previously known as MnP or VP. Notably, most sequences of our CII BB class were previously attributed to the MnP-class (mostly short-MnPs), but the phylogenetic analysis suggests a more restricted definition of the MnP class. The analysis of our phylogenetic tree together with the conservation of the catalytic tryptophan (conserved in LiP and VP) and Mn2+ oxidation sites (in MnP and VP) (Fig. 3) clearly show that the sub-classes cannot be resumed to the presence/absence of key residues. It is noteworthy that sequences possessing all the pointed residues and thus susceptible to be VP sequences are scattered in the branches of the CIIBB and CIIBA sub-classes. Interestingly such sequences are also present at the basis of the LiP clade, with two sequences from C. subvermispora, described as phylogenetically and catalytically intermediate between classical LiPs and VPs8. Few specific residue conservations can be detected in each sub-class (Fig. 1b), but the CIIBC sub-class is apparently the more divergent one.
Figure 3.
Phylogenetic tree of basidiomycetes CII Prx sequences. 267 sequences coming from 28 basidiomycetes have been aligned to generate the tree. One Ascomycete sequence was used as outgroup. LiP sequences are represented in red, VP in orange, CIIBB in green, CIIBC in pink, MnP in blue and CIIBA in azure. Bootstrap values higher than 50% are indicated. Presence/absence of 3 conserved residues (E(35), E(39) and D(179)) responsible for Mn2+ oxidation in MnP, as well as the catalytic W typical from LiP are displayed aside, respectively with green, azure, blue and red square.
Phylogenetic and profile analysis were mostly supported by gene structure (intron number and position) conservation in basidiomycetes. Out of 57 common introns (cintrons) detected with CIWOG, 21 were considered conserved since they were present in one or several sub-classes with a conservation rate higher than 50% (Fig. 4). Only seven introns are found in sequences from all families. The family with the more noteworthy pattern of intron composition is the MnP: 3 introns are specific to MnP sequences, and 2 others are only shared with CIIBA sub-class, whereas 3 introns are present in all classes except MnP. The CIIBC sub-class also possesses one intron totally specific and two others very marginal elsewhere. Three introns are mostly common to CIIBB and LiP, which confirms the phylogenetic hypothesis that LiP sequences originate from CIIBB. As to the VP sequences, they are rather intron-rich, with some sequences resembling the CIIBB introns’ pattern, and some others the CIIBA one.
Figure 4.

Conserved common introns from 267 basidiomycete sequences belonging to 28 organisms. Cintrons were extracted from CIWOG’s database and 21 out of 55 were considered conserved since they were present in one or several sub-classes with a conservation rate higher than 50%. Cintrons are highlighted in accordance with the class in which they are in majority. Cintrons common to all classes are highlighted in grey.
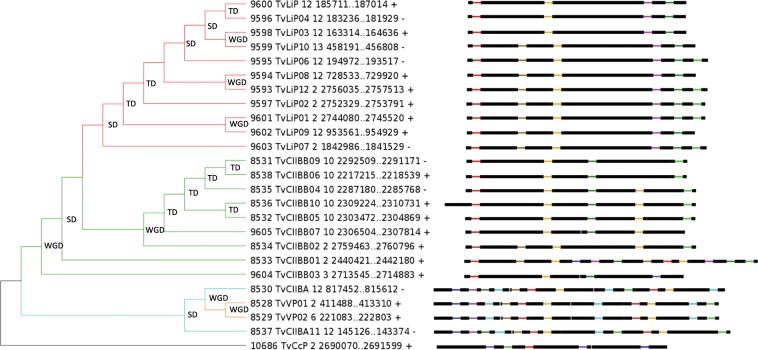
In basidiomycetes, extensive recent gene duplications in LiP, MnP and the three other new sub-classes were identified. The 24 CII Prx sequences of Trametes versicolor distributed among LiP, VP, CIIBA and CIIBB are mainly clustered in 3 genomic regions and are the result of tandem (TD), segmental (SD) and whole genome duplications (WGD) (Fig. 5). These events have been defined as following: TD as successive duplicated genes, SD as blocks of DNA that map to different loci in the same chromosome and WGD as blocks of DNA that map different chromosomes. However, all these duplications are very recent since they form well-supported clusters specific for each species (Fig. 3). It begs the question of duplication events widespread among basidiomycetes but it appears that other peroxidase families such as Cytochrome C peroxidases (CcP) or glutathione peroxidases were less or not subjected to duplication. Besides, no correlation can be made with the distribution of transposable elements. This suggests that these duplications are probably an evolutionary response to selection pressure.
Figure 5.
Phylogenetic analysis, gene localisation, orientation and structure of 24 CII Prxs sequences from Trametes versicolor. Gene structures produced with GECA have been included in front of each branch. LiP sequences are represented in red, CIIBB in green and CIIBA in azure. Tandem duplication (TD), segmental duplication (SD) and whole genome duplication (WGD) events are represented on the base of the branch they occur on. Name in front of each branch contains ID and name attributed in the RedoxiBase, the scaffold number, gene coordinates and orientation.
Working hypothesis of ligninase evolution
Ligninases are detected in ascomycetes and basidiomycetes but are absent from the early diverging fungi analyzed (Choanephora cucurbitarum, Mucor circinelloides, Phycomyces blakesleeanus, Rhizopus oryzae). These two phyla are monophyletic sister groups belonging to Dikarya which emerged from a common ancestral organism. Key residues described in basidiomycete MnP, LiP, and VP are also detected in CII Prx ascomycete sequences (Fig. 1). This suggests that CII Prxs detected in ascomycetes and basidiomycetes could have evolved independently from a similar ancestral sequence following convergent evolution. Notably, CII Prxs belong to the same superfamily as the class I Cytochrome C peroxidases (CcP), and share common key residues23. The taxonomic distribution of the CI Prxs was clarified in order to enable a better understanding of the overall CII Prxs evolution46. CI Prxs are found in plants, fungi, and prokaryotes. Unlike CII and CIII Prxs, they are not glycosylated and do not have signal peptides, calcium ions, or disulfide bridges. They contain five main groups of proteins: (i) Catalase peroxidases (CP) present in prokaryotes and in some eukaryotes following a gene transfer, (ii) Cytochrome c peroxidases (CcP) found in mitochondria containing organisms but not detectable in Viridiplantae, (iii) Ascorbate peroxidases (APx) found only in chloroplastic organisms, and (iv)(v) two hybrid-type peroxidases detected in fungi and different kingdoms. A previous phylogenetic study suggested evolutionary relation between CI Prxs and CII Prxs47.
When comparing ascomycetes and basidiomycetes for the presence of CcP and ligninase encoding sequences (Table 1 and Supplementary Table S1), we can observe that: (i) in most cases, two CcP sequences are detected in fungi which do not possess CII Prx sequences and (ii) only one CcP sequence is detected when the genome contains at least one CII Prx sequence. This suggests that CII Prxs would have emerged from an ancestral sequence that could be a CcP sequence. A similar theory of evolution has already been described for the CIII Prxs46. Indeed, CIII Prxs are only detected in plants, which lack CcP. CIII Prxs and CII Prxs are both subjected to numerous species specific duplications (tandem and segmental duplications), contain highly conserved cysteine residues necessary for disulfite bridges and stability of secreted proteins. As an alternative hypothesis, all fungi would possess at least one CII Prx sequence and loss events occurred more recently. On the principle of maximum parsimony (Supplementary Table S1), this hypothesis seems unlikely since it would require many independent events of gene loss.
In addition, intron positions and numbers are not conserved between the CII Prxs found in ascomycetes and in basidiomycetes. Basidiomycetes contain more introns than ascomycetes (on average 8 and 2 respectively), but intron sizes are higher, on average, in ascomycetes (74 nt) than in basidiomycetes (54 nt). Similar phenomena regarding intron size and number are also observed with other families such as glutathione peroxidases (1 or 2 introns for ascomycetes and 3 to 6 for basidiomycetes). Altogether, these results could suggest independente evolution from two ancestral sequences containing no or few introns. This is in accordance with the hypothesis of a convergent evolution from an existing CI Prx sequence for these two lineages, after the ascomycetes/basidiomycetes separation.
Peroxidase family expansion and recent gene loss processes are likely to be both involved in the history of these genes in fungi. This recent evolutionary history seems particularly driven by fungal life style and leads to numerous adaptive convergence to environment, particularly to host immunity. Typically, necrotrophic or hemibiotrophic fungi, either ascomycetes or basidiomycetes, possess CII Prxs while symbiotic, endophytic or biotrophic fungi mostly do not. WR basidiomycetes and necrotrophic pezizomycetes (ascomycetes) present the highest levels of CII Prxs (Table 1).
Even if numerous key residues are well conserved between the different CII Prx classes, the residues described as necessary for electron transfer are missing in Ascomycetes. Furthermore, the position of the conserved cysteines varies between CII Prxs of ascomycetes and basidiomycetes. These divergences support the independent emergence of CII Prxs in ascomycetes and basidiomycetes and lead to suggest divergent functions (or different catalytic mechanisms). Lignin-degrading fungi possess large batteries of ligninase encoding sequences which enable them to oxidize the lignin polymer and then to use it as a source of carbon. Saprophytic ascomycetes, as well as BR basidiomycetes, described as not being lignivor, present none or low number of ligninase encoding sequences. They probably just use them to depolymerize the lignin by oxidation in order to increase the accessibility to other cell wall components such as cellulose and hemicellulose. Finally, plant pathogenic ascomycetes (either necrotrophic or hemibiotrophic), which contain up to 7 ligninases encoding sequences, use these proteins to depolymerize the lignin in order to access and to infect the host cell. This conclusion is in agreement with the fact that plant pathogen fungi also possess more CAZymes48. A more detailed analysis of the distribution into classes in basidiomycete white rot fungi reveals that most species possess only one enzyme among the 3 mains, i.e. MnP, LiP and VP, and, in our data, 3 species (Auricularia, Hypholoma and Stereum) possess neither of them. Phanerochaete, Phlebia, Phlebiopsis and Ceriporiopsis are the only ones that have MnP and LiP sequences together. All analyzed organisms possess at least one complementary class of ligninase among the sub-classes. This diversity of ligninolytic enzymes is probably a major key for the fungal ability to proliferate on different types of wood2,49.
Conclusions
The availability of a large number of fungal genomes (1000 fungal genomes project, https://genome.jgi.doe.gov/programs/fungi/index.jsf)50 allows us to perform exhaustive and expert data mining for the analysis of CII Prxs. They are present in the majority of basidiomycetes, only detected in the Pezizomycotina phylum belonging to ascomycetes whereas absent in early diverging fungi. Sequences found in ascomycetes and basidiomycetes present highly divergent gene structures, and sequence conservation only for key residues scattered throughout the sequences. Altogether, this suggests that actual CII Prxs found in basidiomycetes and ascomycetes probably originate from at least two independent events after the separation between ascomycetes and basidiomycetes. This confirms the conclusion obtained for basidiomycetes with tree reconciliation24. In addition, the sequence similarity with the CI Prx, CcP sequence, and the correlation between lack of the second CcP copy and presence of CII Prx suggest this ancestral sequence could be a CcP.
Four residues already described in MnP, LiP and VP as necessary for electron transfer are sometimes missing in the new closely related ascomycete and basidiomycete sub-classes. The discrepancy between catalytic activity based on few residues and the global sequence conservation is not antagonistic. Indeed, numerous other residues are highly conserved between all CII and within the different sub-classes, suggesting that new CII Prxs could be able to oxidized lignin but with a different electron transfer mechanism. In all cases, the catalytic activity of CIIAA, CIIAB, CIIAC, CIIBA, CIIBB and CIIBC proteins is not yet known and needs to be demonstrated.
In basidiomycetes, the presence of MnP, LiP and VP have been clearly associated with a specific wood material decaying activity thanks to their lignin degradation capacity. CIIBA, CIIBB and CIIBC are mainly detected in wood degrading fungi (white rot), alone or associated with the main CII Prxs (MnP, LiP or VP). But they can also be found alone in brown rot fungi, plant pathogens, litter decomposing fungi and fungi with no defined decaying machinery. On the other hand, CIIAA, CIIAB and CIIAC are found in rather low copy number mainly in plant pathogens ascomycetes. The specific sequence distribution among organisms suggests that these proteins could have two separated purposes: lignin degradation as carbon source and cell penetration.
Early diverging fungi such as Chytridiomycota species are capable to degrade cellulose and pectins which allow them to used wall polymers as a carbon source51. But the lack of CII Prx in all early diverging fungi tested, questions about the accessibility to these carbon source for fungi that do not have the tools to degrade lignin. The absence of ligninases in early divergent fungi has raised the hypothesis of the incidence of microbial on carbon burial at the end of Paleozoic (Floudas et al. 2012), in opposition to a geological hypothesis52,53.
The following questions to address are the respective roles of these different enzymes during wood decay. This would help to better understand the biology of these fungi, and the chemical mechanisms involved in the biological decomposition of wood.
Supplementary Information
Acknowledgements
The authors are thankful to the Paul Sabatier-Toulouse 3 University and to the Centre National de la Recherche Scientifique (CNRS) for granting their work. We are grateful to the Genotoul bioinformatics platform Toulouse Midi-Pyrenees (Bioinfo Genotoul) for providing help and computing and storage resources and to Mark Weber for careful English corrections. The RedoxiBase is hosted by the Toulouse Midi-Pyrénées bioinformatics platform.
Author contributions
C.M. and C.D. designed the study. N.F. and C.D. performed data mining. C.M. and N.F. made bioinformatics and phylogenetic analysis. C.R. and C.D. managed the choice of the species as well as the fungal life style relation. C.M. and C.D. wrote the manuscript. All the authors contributed to manuscript editing.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-56774-4.
References
- 1.Bi F, et al. Carbon regulation of environmental pH by secreted small molecules that modulate pathogenicity in phytopathogenic fungi. Mol. Plant. Pathol. 2016;17:1178–1195. doi: 10.1111/mpp.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janusz G, et al. Lignin degradation: microorganisms, enzymes involved, genomes analysis and evolution. Fems Microbiology Rev. 2017;41:941–962. doi: 10.1093/femsre/fux049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alfaro M, Oguiza JA, Ramírez L, Pisabarro AG. Comparative analysis of secretomes in basidiomycete fungi. J. Proteom. 2014;102:28–43. doi: 10.1016/j.jprot.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Fernández-Fueyo E, et al. Description of the first fungal dye-decolorizing peroxidase oxidizing manganese(II) Appl. Microbiol. Biotechnol. 2015;99:8927–8942. doi: 10.1007/s00253-015-6665-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stajich JE, et al. Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus) Proc. Natl Acad. Sci. USA. 2010;107:11889–11894. doi: 10.1073/pnas.1003391107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandez-Fueyo E, et al. Comparative genomics of Ceriporiopsis subvermispora and Phanerochaete chrysosporium provide insight into selective ligninolysis. Proc. Natl Acad. Sci. USA. 2012;109:5458–5463. doi: 10.1073/pnas.1119912109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hastrup AC, et al. Differences in crystalline cellulose modification due to degradation by brown and white rot fungi. Fungal Biol. 2012;116:1052–1063. doi: 10.1016/j.funbio.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Fernández-Fueyo E, et al. Lignin-degrading peroxidases from genome of selective ligninolytic fungus Ceriporiopsis subvermispora. J. Biol. Chem. 2012;287:16903–16916. doi: 10.1074/jbc.M112.356378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruiz-Dueñas FJ, Fernández E, Martínez MJ, Martínez AT. Pleurotus ostreatus heme peroxidases: an in silico analysis from the genome sequence to the enzyme molecular structure. C. R. Biol. 2011;334:795–805. doi: 10.1016/j.crvi.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Morin E, et al. Genome sequence of the button mushroom Agaricus bisporus reveals mechanisms governing adaptation to a humic-rich ecological niche. Proc. Natl Acad. Sci. USA. 2012;109:17501–17506. doi: 10.1073/pnas.1206847109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martínez AT, Ruiz-Dueñas FJ, Martínez MJ, Del Río JC, Gutiérrez A. Enzymatic delignification of plant cell wall: from nature to mill. Curr. Opin. Biotechnol. 2009;20:348–357. doi: 10.1016/j.copbio.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Eastwood DC, et al. The plant cell wall-decomposing machinery underlies the functional diversity of forest fungi. Science. 2011;333:762–765. doi: 10.1126/science.1205411. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, et al. Localizing gene regulation reveals a staggered wood decay mechanism for the brown rot fungus Postia placenta. Proc. Natl Acad. Sci. USA. 2016;113:10968–10973. doi: 10.1073/pnas.1608454113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin F, Selosse MA. The Laccaria genome: a symbiont blueprint decoded. N. Phytol. 2008;180:296–310. doi: 10.1111/j.1469-8137.2008.02613.x. [DOI] [PubMed] [Google Scholar]
- 15.Kohler A, et al. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat. Genet. 2015;47:410–415. doi: 10.1038/ng.3223. [DOI] [PubMed] [Google Scholar]
- 16.Morgenstern I, Klopman S, Hibbett DS. Molecular evolution and diversity of lignin degrading heme peroxidases in the Agaricomycetes. J. Mol. Evol. 2008;66:243–257. doi: 10.1007/s00239-008-9079-3. [DOI] [PubMed] [Google Scholar]
- 17.Morgenstern I, Robertson DL, Hibbett DS. Characterization of three mnp genes of Fomitiporia mediterranea and report of additional class II peroxidases in the order hymenochaetales. Appl. Env. Microbiol. 2010;76:6431–6440. doi: 10.1128/AEM.00547-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riley R, et al. Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc. Natl Acad. Sci. USA. 2014;111:9923–9928. doi: 10.1073/pnas.1400592111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Floudas D, et al. Evolution of novel wood decay mechanisms in Agaricales revealed by the genome sequences of Fistulina hepatica and Cylindrobasidium torrendii. Fungal Genet. Biol. 2015;76:78–92. doi: 10.1016/j.fgb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu, N. et al. Comparative analysis of the secretomes of Schizophyllum commune and other wood-decay basidiomycetes during solid-state fermentation reveals its unique lignocellulose-degrading enzyme system. Biotechnology For Biofuels9, 10.1186/s13068-016-0461-x (2016). [DOI] [PMC free article] [PubMed]
- 21.Passardi F, et al. Prokaryotic origins of the non-animal peroxidase superfamily and organelle-mediated transmission to eukaryotes. Genomics. 2007;89:567–579. doi: 10.1016/j.ygeno.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Welinder KG. Superfamily of plant, fungal and bacterial peroxidases. Curr. Opin. Struct. Biol. 1992;2:388–393. doi: 10.1016/0959-440X(92)90230-5. [DOI] [Google Scholar]
- 23.Welinder KG, Mauro JM, Nørskov-Lauritsen L. Structure of plant and fungal peroxidases. Biochem. Soc. Trans. 1992;20:337–340. doi: 10.1042/bst0200337. [DOI] [PubMed] [Google Scholar]
- 24.Floudas D, et al. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science. 2012;336:1715–1719. doi: 10.1126/science.1221748. [DOI] [PubMed] [Google Scholar]
- 25.Sundaramoorthy M, Youngs H, Gold M, Poulos T. High-resolution crystal structure of manganese peroxidase: Substrate and inhibitor complexes. Biochemistry. 2005;44:6463–6470. doi: 10.1021/bi047318e. [DOI] [PubMed] [Google Scholar]
- 26.Ayuso-Fernandez, I., Martinez, A. & Ruiz-Duenas, F. Experimental recreation of the evolution of lignin-degrading enzymes from the Jurassic to date. Biotechnology For Biofuels10, 10.1186/s13068-017-0744-x (2017). [DOI] [PMC free article] [PubMed]
- 27.Nagy LG, et al. Comparative Genomics of Early-Diverging Mushroom-Forming Fungi Provides Insights into the Origins of Lignocellulose Decay Capabilities. Mol. Biol. Evol. 2016;33:959–970. doi: 10.1093/molbev/msv337. [DOI] [PubMed] [Google Scholar]
- 28.Shah F, et al. Ectomycorrhizal fungi decompose soil organic matter using oxidative mechanisms adapted from saprotrophic ancestors. N. Phytol. 2016;209:1705–1719. doi: 10.1111/nph.13722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mäkelä MR, et al. Aromatic metabolism of filamentous fungi in relation to the presence of aromatic compounds in plant biomass. Adv. Appl. Microbiol. 2015;91:63–137. doi: 10.1016/bs.aambs.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Falkowski P, et al. The global carbon cycle: a test of our knowledge of earth as a system. Science. 2000;290:291–296. doi: 10.1126/science.290.5490.291. [DOI] [PubMed] [Google Scholar]
- 31.Fawal N, Li Q, Mathé C, Dunand C. Automatic multigenic family annotation: risks and solutions. Trends Genet. 2014;30:323–325. doi: 10.1016/j.tig.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 32.Keller, O., Odronitz, F., Stanke, M., Kollmar, M. & Waack, S. Scipio: Using protein sequences to determine the precise exon/intron structures of genes and their orthologs in closely related species. BMC Bioinformatics9, doi: 27810.1186/1471-2105-9-278 (2008). [DOI] [PMC free article] [PubMed]
- 33.Koua D, et al. PeroxiBase: a database with new tools for peroxidase family classification. Nucleic Acids Res. 2009;37:D261–D266. doi: 10.1093/nar/gkn680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fawal N, et al. PeroxiBase: a database for large-scale evolutionary analysis of peroxidases. Nucleic Acids Res. 2013;41:D441–444. doi: 10.1093/nar/gks1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Löytynoja A, Goldman N. An algorithm for progressive multiple alignment of sequences with insertions. Proc. Natl Acad. Sci. USA. 2005;102:10557–10562. doi: 10.1073/pnas.0409137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 38.Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 39.Wilkerson M. D., Ru Y., Brendel V. P. Common introns within orthologous genes: software and application to plants. Briefings in Bioinformatics. 2009;10(6):631–644. doi: 10.1093/bib/bbp051. [DOI] [PubMed] [Google Scholar]
- 40.Fawal, N., Savelli, B., Dunand, C. & Mathé, C. GECA: a fast tool for gene evolution and conservation analysis in eukaryotic protein families. Bioinformatics28, 1398-1399, doi:bts153 [pii] 10.1093/bioinformatics/bts153 (2012). [DOI] [PubMed]
- 41.Katoh K, Standley DM. MAFFT: iterative refinement and additional methods. Methods Mol. Biol. 2014;1079:131–146. doi: 10.1007/978-1-62703-646-7_8. [DOI] [PubMed] [Google Scholar]
- 42.Smit, A., Hubley, R. & Green, P. RepeatMasker Open-4.0, (2013-2015).
- 43.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roure B, Baurain D, Philippe H. Impact of missing data on phylogenies inferred from empirical phylogenomic data sets. Mol. Biol. Evol. 2013;30:197–214. doi: 10.1093/molbev/mss208. [DOI] [PubMed] [Google Scholar]
- 45.Croll D, McDonald BA. Intron gains and losses in the evolution of Fusarium and Cryptococcus fungi. Genome Biol. Evol. 2012;4:1148–1161. doi: 10.1093/gbe/evs091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Passardi F, et al. Phylogenetic distribution of catalase-peroxidase: area there patches of order in chaos? Gene. 2007;97:101–13. doi: 10.1016/j.gene.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 47.Zámocký, M., Furtmüller, P. G. & Obinger, C. Evolution of structure and function of Class I peroxidases. Arch Biochem Biophys500, 45-57, doi:S0003-9861(10)00122-0 [pii] 10.1016/j.abb.2010.03.024 (2010). [DOI] [PubMed]
- 48.Zheng Zhi-Liang, Zhao Yihong. Transcriptome comparison and gene coexpression network analysis provide a systems view of citrus response to ‘Candidatus Liberibacter asiaticus’ infection. BMC Genomics. 2013;14(1):27. doi: 10.1186/1471-2164-14-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Janusz G, et al. Comparative transcriptomic analysis of Cerrena unicolor revealed differential expression of genes engaged in degradation of various kinds of wood. Microbiological Res. 2018;207:256–268. doi: 10.1016/j.micres.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 50.Grigoriev IV, et al. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014;42:D699–704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berbee ML, James TY, Strullu-Derrien C. Early Diverging Fungi: Diversity and Impact at the Dawn of Terrestrial Life. Annu. Rev. Microbiol. 2017;71:41–60. doi: 10.1146/annurev-micro-030117-020324. [DOI] [PubMed] [Google Scholar]
- 52.Nelsen MP, DiMichele WA, Peters SE, Boyce CK. Delayed fungal evolution did not cause the Paleozoic peak in coal production. Proc. Natl Acad. Sci. USA. 2016;113:2442–2447. doi: 10.1073/pnas.1517943113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Godderis Y, et al. Onset and ending of the late Palaeozoic ice age triggered by tectonically paced rock weathering. Nat. Geosci. 2017;10:382–+. doi: 10.1038/NGEO2931. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.