Abstract
CircularRNAs (circRNAs) are non-coding RNAs which compete for microRNA (miRNA) binding, influencing the abundance and stability of other RNA species. Herein we have investigated the effect of circRNAs on the mir200-ZEB1 feedback loop in relationship with the aggressiveness of human melanoma cells. We first compared the level of expression of key factors in the mir200-ZEB1 feedback loop in primary human melanoma cells compared with their matching metastatic one and found a correlation between the aggressiveness of the cells and the level of expression of ZEB1 and SNAI1. We also analyzed factors in the mir200-ZEB1 feedback loop, including circZEB1, during the phenotypic switching of human melanoma cells. Our results showed a correlation between the level of ZEB1 and SNAI1 and the fraction of cancer stem cells in the population. The level of circZEB1 was, however, consistently high during the entire phenotypic transformation. To understand this result we propose a mathematical model of the regulatory circuit. According to the model, the experimental observations can be explained by the presence of a back-splicing factor limiting circRNA production.
Electronic supplementary material
The online version of this article (10.1007/s12307-019-00230-4) contains supplementary material, which is available to authorized users.
Keywords: circRNA, ZEB1, Melanoma, Phenotypic switching
Introduction
Cancer plasticity is an emerging property of tumor cells that is leading us to reconsider the classical strategies for therapeutic intervention [1–5]. Recently, our group showed that human melanoma cells dynamically change their phenotype by expressing epithelial to mesenchymal transition (EMT) markers, in a way that is regulated by complex network of microRNAs (miRNA) [6]. This mechanism allows the tumor to tightly control the number of cancer stem cells (CSC) present in the population, allowing CSCs to either grow or stop growing in order to maintain a specific proportion of EMT-marker expressing cells in the bulk [6]. In particular, we have shown that a cell population completely depleted of CSCs was able to massively switch after 10 days into a CSC-rich population, returning to a steady-state with a low number of CSC within 20 days [6]. Notice that EMT is one of the key processes that cells undergo in order to gain a migratory phenotype and it is thus relevant for metastasis.
Noncoding RNAs, such as miRNAs and circularRNAs (circRNAs) are all recognized to play a key regulatory role in physiological and pathological cellular processes [7]. In this respect, circRNAs, single-strand endogenous noncoding RNAs closed in a loop [8], are widely expressed in mammalian cells and differentially expressed in various tissues and pathological conditions [7–11]. Thanks to their circular form, circRNAs are more stable than linear RNAs and since they have been detected in exosomes and in the blood, they appear to be ideal candidates to act as biomarkers [12–16]. It has been recently demonstrated that circRNAs participate to the complex post-transcriptional regulatory network of the cell, competing with mRNAs for miRNA binding and affecting the abundance and stability of other RNA species [7, 9, 17–19]. The existence of different miRNA targets sharing the same binding sites leads to an indirect, miRNA-mediated, cross-talk between competitive endogenous RNAs (ceRNA) [20–22].
In the present study, we investigated the possible regulative role of circRNAs in a ceRNA circuit involved in the EMT by the combination of experiments, computational models and data analysis. The regulatory core of the EMT process is the double negative feed-back loop that includes ZEB1 and members of mir200 family. Moreover, SNAI1 acts as external transcriptional regulator of the same circuit [23–25]. The human gene ZEB1 can produce multiple functional RNAs including circRNAs [26–29]. One of them, circ-ZEB1.33 (circZEB1) [29], is the product of backsplicing of exons 2 to 4 of ZEB1 transcript variant 1 (NM 001128128) [28, 29]. Furthermore, circZEB1 contains a binding site for hsa-mir200a-3p and hsa-mir141-3p, both belonging to mir200 family [28] which is a well-known post-transcriptional regulator of ZEB1 [23, 30–33]. In the present paper, we compared the level of expression of key factors in the ZEB1 circuit (i.e. ZEB1, SNAI1 and circZEB1) in human primary and metastatic melanoma cells derived from the same patient (WM115/WM266 and IgR39/IgR37 cells, primary and metastatic, respectively). To confirm our experimental results, we checked the level of expression of ZEB1 and SNAI1 in samples of primary and metastatic patients stored on public repositories. To evaluate if our results were specific to melanoma only, we analyzed the level of expression of ZEB1 and SNAI1 in primary and metastatic breast cancer. Finally, we investigated the possible changes for factors in the same ZEB1 circuit in human melanoma IgR39 cells during phenotypic switching. In particular, we considered the time at which cells massively switch into CSCs (T10) and the time at which the population reaches again the steady state (T20), as discussed in [6].
Our experiments also showed that during phenotypic switching the level of circZEB1 was always high and constant, in contrast to what we would have naively expected. To understand the possible mechanism underlying this behavior, we developed a mathematical model of the ZEB1 circuit. A detailed analysis of the model showed that high and constant level of circZEB1 is consistent with the presence of a back-splicing factor limiting circRNA production. This result opens interesting perspectives for further investigations of the dynamics of circZEB1 and its impact on phenotypic switching of cancer cells.
Materials and Methods
Cell Culture
IgR39 and IgR37 cells (primary and metastatic human melanoma cells, respectively) were obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH [6] and WM115 and WM266 (primary and metastatic human melanoma cells, respectively) from ATCC (CRL 1675 and CRL 1676, respectively) [34]. All cell lines were cultured in DMEM, 15% FBS supplemented with 1% MEM vitamin, 1% MEM aminoacid, 1% antibiotics (Penicillin/Streptomycin), 1% L-glutamine (complete medium) at 37∘C in a 5% CO2 humidified environment.
Flow Cytometry
Cells were sorted for phycoerythrin (PE) anti-human CXCR6 (code:FAB699P-025, R& D System, USA) according to [6]. For each flow cytometry evaluation, a minimum of 4 ⋅ 107 cells were stained and at least 5 ⋅ 105 events were collected and analyzed. Flow cytometry sorting and analysis was performed using a FACSAria flow cytometer (Becton, Dickinson and Company, BD, Mountain View, CA). Data were analyzed using FlowJo software (Tree Star, Inc., San Carlos, CA).
Quantitative Real-Time PCR (qRT-PCR)
SNAI1, ZEB1, circZEB1 and GAPDH specific primers were designed using the software Primer3 [35] and aligned on human genomic transcripts using Blast [36] in order to minimize off-target effects. Briefly, divergent primers encompassing backsplicing site were designed for circZEB1 based on fasta sequence of hsa_circ_0004907 obtained from CircBase [29]. Primers for ZEB1 mRNA were designed on exon 7 of transcript ENST00000446923 (RefSeq NM_001128128) that is shared by 7 of the 9 protein coding transcripts according to Ensembl database (last accessed March 2019 [27]). The following primers were selected to perform qRT-PCR:CircZEB1_F CCAGAAGCCAGTGGTCATGA, CircZEB1_R GTCATCCTCCCAGCAGTTCT,ZEB1_F GAGAAGCCATATGAATGCCCA, ZEB1_R GTATCTGTGGTCGTGTGGGA,SNAI1_F TACAGGACAAAGGCTGACAGA, SNAI1_R CGGGGCATCTCAGACTCTAG,GAPDH_F CACATCGCTCAGACACCATG, GAPDH_R TGACGGTGCCATGGAATTTG.Briefly, total RNA was extracted with the guanidinium thiocyanate-phenol-chloroform extraction with 1 ml of TRIzol Reagent. RNA samples were incubated for 5 minutes at room temperature. After adding 0.2 ml of chloroform in each sample, the tube was vigorously shaked and centrifuged at 12⋅ 103 x g for 15 minutes at 4∘C. The aqueous phase obtained was collected and placed into a new tube and 0.5 mL of 100% isopropanol was added. After 10 minutes at room temperature samples were centrifuged at 12⋅ 103 x g for 10 minutes at 4∘C. The supernatant has been removed from the tube and the pellet washed with 1 ml of 75% ethanol, vortexed briefly and centrifuged. RNA pellet has been left air drying and resuspended in 20 μ l of RNAse-free water. RNA concentration and purity was determined by using Nanodrop (Eppendorf).
Synthesis of cDNA was performed on 1 μ g of total RNA reverse transcribed (RT) using Vilo IV Superscript cDNA synthesis kit (Invitrogen, CA) according to manufacturer’s instructions. Real time q-RT-PCR analysis was performed using ViiA7 Real Time PCR system (Applied Biosystems). Each primer pair was tested at least in six replicates. The PCR-reaction included 25ng of template cDNA, 5μ M of each (forward and reverse) primers, 2μ l of RNAse-free water and 10μ l of LUNA Universal SYBR Green Mastermix (New England Biosystems), in a total volume of 20μ l. Cycling conditions were as follows: 95∘C enzyme activation for 10 min, followed by 50 cycles of amplification: 15” at 95∘C for denaturing, 1 min at 60∘C for annealing/elongation. For each gene, average ΔCt was calculated for each plate using a reference housekeeping the human gene GAPDH. Results were expressed as the average over replicated plates and plotted as 2−ΔCt normalizing over primary melanoma cells using R [37].
GDC Expression Data
Gene expression levels in primary and metastatic tumor samples from patients were obtained from Genomic Data Commons (GDC) data portal [38] RNA-seq data. A total of 468 transcriptome profile from 465 cases classified as melanoma (primary site skin). Samples annotated by GDC as treated with neoadjuvant therapy or not in agreement with the study protocol were excluded. Among these only two samples were classified as primary and metastatic tumors from the same patient (TCGA-ER-A2NF). Regarding breast cancer, 1189 transcriptome profiles were obtained from GDC database for cases that have primary site in breast tissue. Seven of these samples were metastatic and transcriptome of all the corresponding primary tumors were retrieved. ZEB1 expression was estimated using the number of aligned fragments per kilobase of transcript per million reads (FPKM) and was normalized using GAPDH as housekeeping. Note that comparing the relative expression of ZEB1 instead of absolute values allows to eliminate bias due to the normalization procedure used to obtain expression values. Moreover, we considered as additional housekeeping ribosomal protein L19 (RPL19) and PGK2 to verify that the choice of GAPDH does not influence the obtained results (data not shown).
Mathematical Model of the ZEB1 Circuit
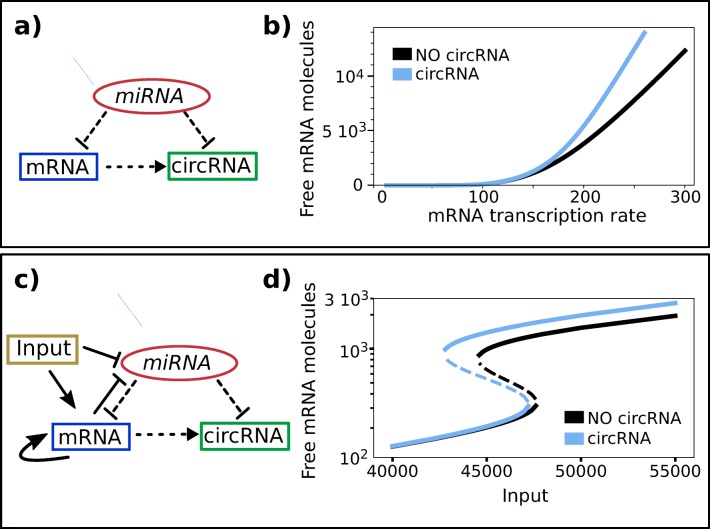
MiRNA-mediated interactions are modeled using a set of differential equations following our previous work [19], details of the model are presented in the Supplementary Materials. Briefly, our model comprises two kinds of post-transcriptional regulation (the binding of mRNA (T) and circRNA (C) by miRNA (μ) and circRNA creation) and two possible transcriptional regulations (promoter silencing and enhancing) (Fig. 5a, c). In general, all these interactions but circRNA creation can involve more than two molecules with multiple binding and partial or reinforced effects (cooperativity of the binding). In accordance with biological evidences, since our aim is to reproduce ZEB1-mir200a circuit, we consider two non competing equivalent biding sites for mir200a on ZEB1 3’UTR and one on circZEB1 [39]. SNAI1 and ZEB1 repression/activation on mir200/ZEB1 promoter was modeled using appropriate Hill functions (see. Suppl.Materials [40, 41]). ZEB1 indirect self-activation [42, 43] was modeled according to [51]. We do not model explicitly SNAI1 production/degradation considering it as a tunable external input (see Fig. 5c).
Fig. 5.
Computational analysis of ZEB1—circuit Figure shows two schematic representation of miRNA mediated ceRNA circuits (a, c) and numerical prediction of mRNA concentration (b, d) in presence (light blue continuous line) and absence (black line) of the circRNA for an exemplificative set of parameters. a-b) Schematic representation of the miRNA-mediated ceRNA interaction network without mRNA autoactivation and miRNA transcriptional regulation. The model predicts an increase of free mRNA molecules in presence of circRNA as a function of mRNA transcription rate (panel b) and Suppl. Figure S2c, d) Schematic representation of the miRNA-mediated ceRNA interaction network including mRNA autoactivation and miRNA transcriptional regulation and an external regulator (c). This model resembles the ZEB1-mir200-SNAI1 circuit reported in Fig. 1. This kind of circuit can present multistability, as shown in panel (d). For the chosen set of parameters, the circuit presents two stable solutions (continuous lines) and one unstable state (dashed line) at fixed input rate. Total effective transcription rate is reported in the in Suppl. Figure S3. The level of free mRNA increases in presence of co-generate circRNA for increasing transcription rate
Decay rate were set according to experimental estimate as follows. MiRNA half life, experimentally estimated to range from ≈ 8 hours up to days, was set to γμ = 0.001min− 1 while mRNA half life is typically of few hours [44–46] and was set to γT = 0.01min− 1. Since circRNA are much more stable than mRNAs, with typical half life of the order of days, closer to miRNA values [10], we set its decay rate γC = γμ = 0.001min− 1. Protein decay rate was set to 0.01min− 1 corresponding to estimated ZEB1 half life (≈ 2h [40]). For production and interaction rates, we refer to former works and experimentally estimated rates [19, 21, 40, 47].We treated the contributions of the other RNA species as a constant and implicitly including them into the decay rates.
Numerical solutions of equations at steady state and Gillespie simulations were performed using Mathematica v.10 and python Stochpy library. Plots were created using Python [48] and R [37].
Results
Expression of ZEB1 Circuit in Primary and Metastatic Human Melanoma Cells
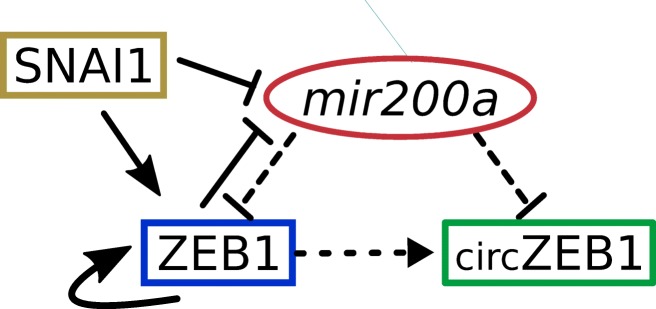
The level of expression of ZEB1, a key regulator of EMT [23–25], is modulated by the external transcription factor SNAI1 and by mir200 family (Fig. 1).
Fig. 1.
ZEB1 circuit. The circuit is composed by a miRNA–TF mutually inhibiting loop involving transcriptional (solid lines) and post-transcriptional (dashed lines) regulation. A solid arrow denotes transcriptional activation, and a solid bar denotes transcriptional inhibition. Dashed arrow connecting ZEB1 to its circRNA indicates co-generation. SNAI1 is considered as an external signal regulating ZEB1 and mir200a at transcriptional level. ZEB1 self-activation is also included
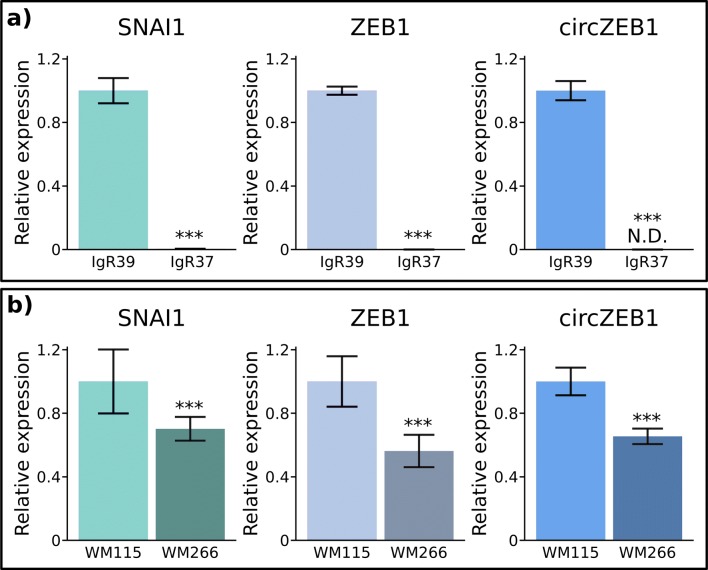
We first checked by qRT-PCR the level of expression of the ZEB1, circZEB1 and SNAI1 RNAs in two different melanoma cell lines obtained from two patients at different stage of aggressiveness, primary or metastatic (Fig. 2). In both cases, we observed a decreased level of expression of SNAI1, ZEB1 and circZEB1 in metastatic cells (Fig. 2). The magnitude of this decrease is, however, dependent on the cell line, possibly representing the specific biological characteristics of each patient.
Fig. 2.
Expression of SNAI1, ZEB1 and circZEB1 in primary and metastatic melanoma cell lines. qRT-PCR analysis of SNAI1, ZEB1 and circZEB1 expression was performed in primary (IgR39 and WM115) and corresponding metastatic (WM266 and IgR37) melanoma cell lines according to Materials and Methods section. T*** p < 0.01 versus primary tumor IgR39 or WM115 cells. The results are expressed as 2−ΔΔCt using GAPDH as housekeeping gene and normalized with respect to primary cell line expression
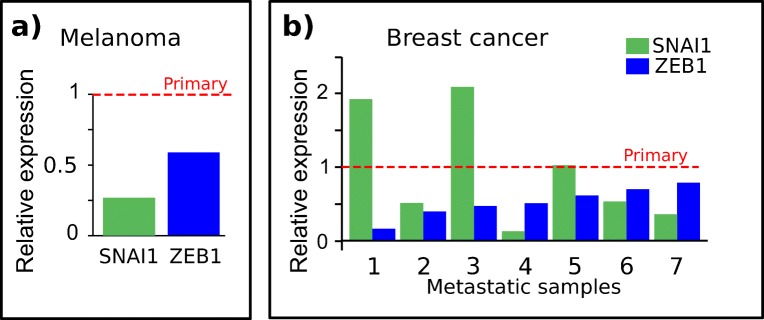
To confirm that our results were not depending on in vitro cell conditions, we analyzed the level of expression of ZEB1 and SNAI1 for data stored in public repositories obtained from tumors classified as primary and metastatic melanoma derived from the same patient [38]. As shown in Fig. 3, we confirmed the decreased level of expression in metastatic samples for ZEB1 and SNAI1. To investigate if this decrease was specific to melanoma, we analyzed the level of expression of ZEB1 and SNAI1 in breast cancer (primary and metastasic) using again public repositories. Figure 3 clearly shows that the level of expression of ZEB1 is lower in the metastatic samples with respect to the matched primary tumors, while for SNAI1 the results are less clear.
Fig. 3.
SNAI1 and ZEB1 mRNA expression in melanoma. a Plot shows the relative expression of SNAI1 and ZEB1 in two paired primary (red dashed line) and metastatic melanoma samples from the same patient (TCGA-ER-A2NF) from GDC database. b Bar plot shows the relative expression of SNAI1 and ZEB1 in seven metastatic breast cancer samples normalized over the corresponding primary tumors from the same patients. Dashed red line indicates the normalized level of expression in primary tumor samples. ZEB1 fold changes are less than one in all the samples, showing that its expression is reduced in metastatic tumors, coherently with what observed in melanoma. Data were downloaded from GDC database as described in materials and methods section. Correspondence between x-axis label and GDC cases: 1-TCGA-BH-A1ES, 2-TCGA-AC-A6IX, 3-TCGA-BH-A1FE, 4-TCGA-E2-A15K, 5-TCGA-E2-A15A, 6-TCGA-E2-A15E, 7-TCGA-BH-A18V
Constant Expression of circZEB1 during Phenotypic Switching
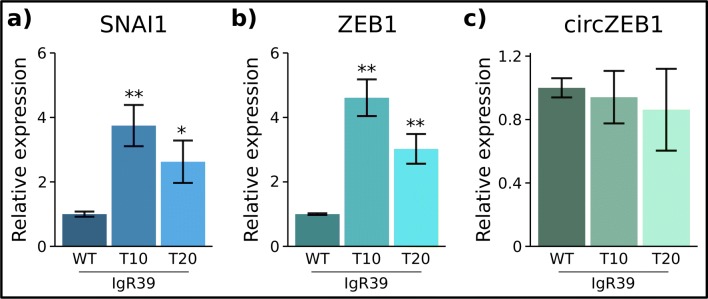
Human melanoma cancer cells negative for CSCs markers can dynamically switch into CSCs in a time dependent manner [6]. Herein, we investigated the expression level of key factors in ZEB1 circuit during the same phenotypic transformation. To this end, we sorted human melanoma cells (IgR39) by flow cytometry and selected a population negative for CSC markers. We then measured the level of expression of ZEB1, SNAI1 and circZEB1 by qRT-PCR at the peak of re-expression of CSC markers (T10, 10 days after the sorting) and when they were back to the steady state (T20, 20 days after the sorting) [6]. As shown in Fig. 4, we observed a significant increase of mRNAs ZEB1 and SNAI1 at the CSC overshoot (T10) and a decrease at T20 towards the steady state (Fig. 4). However, we consistently found high and relatively constant level of circZEB1 both at the overshoot and at the steady state (Fig. 4). This result was unexpected, since in human melanoma and breast cancer cell lines circZEB1 usually follows the changes of ZEB1 mRNA.
Fig. 4.
Level of expression of SNAI1, ZEB1 and circZEB1 during phenotypic switching. qRT-PCR analysis of SNAI1 (a), ZEB1 (b) and circRNA (c) RNA levels was carried out on sorted IgR39 negative for CSC markers 10 days and 20 days after sorting as well as on IgR39 WT as described in Materials and Methods section. *p < 0.1, ** p < 0.05. The results are expressed as 2−ΔΔCt using GAPDH as housekeeping gene and normalized with respect to unsorted cell expression
Mathematical Model for MiRNA-mediated ceRNA Circuit
Steady State Solution of the Model
MiRNA-mediated interactions were modeled using a set of differential equations in agreement with our previous work [19] and as described in details in the materials and methods section and in the Supplementary Materials. We consider two kinds of ceRNA circuits: a more general case with one miRNA (μ) regulating two targets (mRNA, T and circRNA C) without a transcription regulation layer (Fig. 5a) and the specific case of ZEB1-mir200a network comprising transcriptional silencing of the miRNA, self-activation of the target and external transcriptional regulation (input, Fig. 5c). The introduction of self-activation and transcriptional silencing of the miRNA dramatically changes the phenomenology of the model, leading to the possibility of multistable regions (Figs. 5b, d and Fig. S2). Note that multistability of the system, can be achieved also in different models including only one of these interactions, and the number of stable states depends on the chosen parameters [40]. In our model, the mRNA transcription rate can vary in a limited range of values when we consider the circuit in (Fig. 5c), as shown in Fig. S3, while is theoretically unlimited for the more general circuit (Fig. 5a). According to our model, the free mRNA (Teq) and circRNA (Ceq) equilibrium concentrations are given by
| 1 |
where κ and γ are the production and decay rates, μeq is the amount of free miRNA molecules while μT and μC concentrations represent the thresholds determining if C and T are highly influenced by miRNA presence (i.e. when μ ≫ μi, bound state) or are almost free (i.e. when μ ≪ μi). These thresholds are directly proportional to the decay rate γi and inversely proportional to the miRNA-ceRNA affinity. Thus,for a given set of parameters, species with longer half-life and higher miRNA affinity are much more influenced by miRNA, while fast-decaying or low affinity species are less sensitive to variations in miRNA concentration.
The solution reported in Eq. 1 shows that Teq and Ceq are coupled by two factors: the presence of μeq at denominator and the circRNA production rate KC.
Linear Dependence of circRNA on mRNA Concentration
The simplest non-trivial case that can be considered for circRNA production rate is a direct proportionality between KC and the abundance of the associated mRNA (i.e. KC(T) = 𝜖T). This assumption implies that a constant fraction of the total RNA produced is in the form of cirRNA. In this case, it is immediate to verify that circRNA grows faster than linearly with T due to miRNA coupling, until the contribution of miRNA becomes negligible. In this scenario, we obtain that the presence of circRNA-miRNA interaction increases the amount of free mRNA, and, as a consequence, its translation for both the circuits reported in Fig. 5a, c. In fact, circRNA is capable to sequestrate miRNA molecules decreasing the amount of molecules capable to bind mRNA 3’UTR. Note that in the complete circuit in Fig. 5c, the presence of cirRNA increases the range of parameters for which there is multistability. Furthermore, the effect of circRNA on free mRNA increases with mRNA expression, thus increasing the distance between the solutions (Fig. 5d).
Since circRNA is expected to be more stable than its linear counterpart, in our model we can consider μC > μT. Thus, for a given range of transcription rates, mRNA can be in free state μeq < μT while circRNA is bounded (μeq > μC). This effect is even more evident when circRNA affinity for miRNA is higher than its linear counterpart (Fig. S1a). In this case, most of the cirRNA produced will be sequestrated by miRNA and will remain in a bound state (Fig. S1b), while the amount of free circRNA molecules would remain almost undetectable compared to free mRNA increase (Fig. S1a).
Limiting Reagent Model
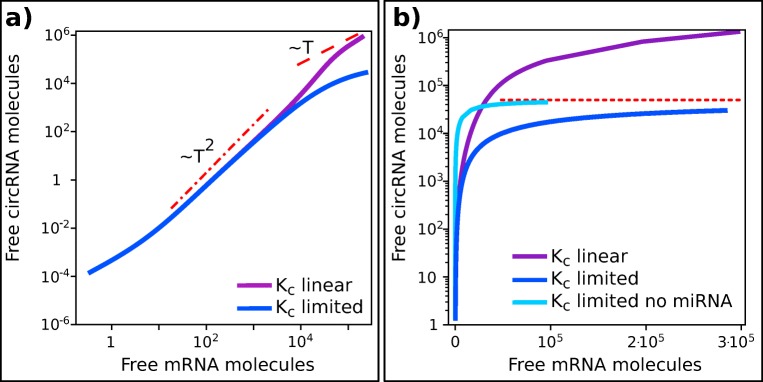
A more complex relationship between circRNA production rate and mRNA level can be obtained under the hypothesis that circRNA production is due to the binding of the linear unspliced transcript with a protein or a complex of proteins (Q) that favor circularization and backsplicing. If the concentration of the protein Q is fixed when the transcription rate increases, the effective circRNA production rate could be expressed in terms of an Hill function: , where 𝜖 depends on the constant concentration of Q and TQ is related to T-Q binding affinity. For small transcription rates, (T ≪ TQ, Q abundant), the dependence of Kc on T is linear KC ≈ 𝜖T/TQ, recovering the linear model presented in the previous section (Fig. 6a). However, increasing mRNA level (T ≫ TQ) the circRNA production rate becomes constant KC ≈ 𝜖. In this limit, the presence of miRNA-mediated crosstalk allows Ceq to increase more than linearly with Teq while it Ceq becomes constant when miRNA contribute becomes negligible (Fig. 6b).
Fig. 6.
CeRNAs levels as a function of mRNA production rate. Figure shows the predicted level of free circRNA as a function of the linear transcript for the model presented in Fig. 5a for two different functional forms of KC(T). a Linear dependency of circRNA production rate on T and Hill-like functional form (limiting reagent) are highly similar in a broad range of values of T (purple and blue lines respectively as in legend). For intermediate values of T both the models give an increase of C that is slightly less than quadratic (dashed-dotted red line) while for large T the two models diverge: the first model increases linearly (red dashed line), while limiting-reagent model shows a sublinear growth of C. b Same data as in panel (a) are plotted in semi-log scale. It is evident that limiting reagent model (blue continuous line) tend to a threshold value (dotted red line), while C increases continuously in linear model (continuous purple line). Note that the behavior of C at intermediate transcription rates is determined by the presence of miRNA. In fact, absence of miRNA implies a faster increase of C towards its limit value (light blue continuous line). All the parameters are maintained constant in the two models, decay rate of T is rescaled in order to make the models comparable. In the limiting model KC has been chosen in order to have 𝜖lim/TQ = 𝜖linear ensuring that C production rate at low T is identical in the two models
Discussion
EMT is a complex physiological process that can help cancer progression and metastasis, involving differential expression of many genes and noncoding RNAs [1, 49–51]. The feedback loop between ZEB1 and the members of mir200 family, involving transcritional and post-transcriptional regulatory is the core of EMT regulatory network [23–25, 41, 51]. High ZEB1 expression in primary tumors and specific subpopulations of cells correlates with the presence of metastasis, drug resistance and poor prognosis [52–56].
In recent years, an increasing number of circRNAs have been recognized to play a role in the regulation of gene expression. One of the main mechanisms of action of circRNAs is the capability to bind miRNAs competing with their canonical targets [9, 14, 17, 18]. Moreover, circRNAs have been found deregulated in different tumors and their altered expression seems to be related to tumor prognosis and aggressiveness [11, 15–17, 57–60]. Recently, it has been shown that circZEB1 actively interacts with mir200a [61], but his specific role in the regulation of EMT was not investigated.
In this context, we have analyzed the level of expression of ZEB1 in human primary melanoma with respect to metastasis in public repositories (GDC database) and in two cell lines obtained from two distinct paired melanoma patients. Our results show that the level of expression of ZEB1 is consistently reduced in metastatic melanoma compared to their corresponding primary tumors. Interestingly, the same feature is observed in breast cancer, which is much more studied so that many more matched samples from primary and metastatic tumors in the same patient are available in the TCGA database. The result suggests that this expression pattern is a general feature of metastasis and is not restricted to melanoma. The decreased expression level of mesenchymal markers in samples obtained from metastasic tumors indicates that cells switch to an epithelial phenotype when they reach the metastatic site [52, 55].
We then investigated the possible dynamic change of ZEB1 circuit during phenotypic switching of IgR39 melanoma cells. In fact, we have recently showed that melanoma cells negative for CSCs markers are able to dynamically re-express the CSC’s markers through a tight regulatory network involving mainly EMT-related genes [6]. The phenotypic switching does not happen gradually but through an overshoot in which cancer cells switch massively into CSCs. In this way, the tumor is able to regulate the number of CSCs in its bulk [6]. Accordingly, herein, we found that the level of ZEB1 increases at the overshoot in IgR39 human melanoma cells, confirming their plastic expression of EMT markers found in [6]. However, surprisingly, we found a constant and high level of expression for circZEB1. To better understand the biological significance of the this finding, we investigated by a computational model two possible scenarios: 1) a direct proportionality between the total transcription rate and the amount of circRNA produced (linear model); 2) the presence of a third factor that favors back-splicing (limiting reagent model). This second scenario is biologically justified since it has been recently reported that canonical spliceosome proteins appear to play an active role in circRNA biogenesis [62–64].
In the first scenario, the presence of ceRNA interactions between the species would predict a superlinear relationship between total free mRNA amount and circRNA amount [19], even in presence of a transcriptional feedback loop. In the second scenario, at low transcription rates the direct proportionality between mRNA and and circRNA production rate is preserved and this model can not be distinguished from the first scenario. The fold change reduction of circZEB1 observed in metastatic cells (IgR37 and WM266) compared to the primary ones are thus coherent with both scenarios. On the other hand, at high transcription rate, the back-splicing factor can become limiting and the two models diverge, predicting a constant level of circRNA at increasing mRNA expression. This corresponds very closely to our observations in IgR39 cells during phenotyping switching where the basal level of ZEB1 is high and increases even more at the overshoot. Hence, our model suggests that the regulation of circZEB1 depends on a back-splicing factor and that the constant level of circZEB1 during phenotypic switching can be explained by our model under the assumption of a high ZEB1 transcription rate.
All together, our findings show that under specific biological conditions the cells mantain high level of circZEB1 probably involving the expression of a back-splicing factor [62–64]. This evidence opens new interesting avenues for further investigation of this regulatory process during dynamic and temporally dependent processes such as phenotypic switching.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The project was supported by funded from the Center for Complexity and Biosystems of UniMI. The research leading to these results was funded by AIRC under IG2018-ID 21558 project-PI Pusch Michael.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.La Porta CAM, Zapperi S. Semin Cancer Biol. 2018;53:42. doi: 10.1016/j.semcancer.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 2.La Porta CAM, Zapperi S. Semin Cancer Biol. 2017;44:3. doi: 10.1016/j.semcancer.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Kreso A, O’Brien CA, van Galen P, Gan OI, Notta F, Brown AMK, Ng K, Ma J, Wienholds E, Dunant C, Pollett A, Gallinger S, McPherson J, Mullighan CG, Shibata D, Dick JE. Science. 2013;339(6119):543. doi: 10.1126/science.1227670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, Wong KK, Brandstetter K, Wittner B, Ramaswamy S, Classon M, Settleman J. Cell. 2010;141(1):69. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Easwaran H, Tsai HC, Baylin SB. Mol Cell. 2014;54(5):716. doi: 10.1016/j.molcel.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sellerio AL, Ciusani E, Ben-Moshe NB, Coco S, Piccinini A, Myers CR, Sethna JP, Giampietro C, Zapperi S, La Porta CAM. Sci Rep. 2015;5:15464. doi: 10.1038/srep15464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jia Chan J, Tay Y. Int J Mol Sci. 2018;19:1310. doi: 10.3390/ijms19051310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen LL. Nat Rev Mol Cell Biol. 2016;17(4):205. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 9.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 10.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. 2013;19(2):141. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen T, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73(18):5609. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 12.Li Yan, Zheng Qiupeng, Bao Chunyang, Li Shuyi, Guo Weijie, Zhao Jiang, Chen Di, Gu Jianren, He Xianghuo, Huang Shenglin. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Research. 2015;25(8):981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PloS one. 2015;10(10):e0141214. doi: 10.1371/journal.pone.0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang S, Yang B, Chen B, Bliim N, Ueberham U, Arendt T, Janitz M. The emerging role of circular RNAs in transcriptome regulation. Genomics. 2017;109(5-6):401. doi: 10.1016/j.ygeno.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, Lyu D, Zheng B, Xu Y, Long Z, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clinica Chimica Acta. 2015;444:132. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 17.Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y, Yang S, Zeng Z, Liao W, Ding YQ, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7(18):26680. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, Liang L, Gu J, He X, Huang S (2016) Nature Commun 7. 10.1038/ncomms11215 [DOI] [PMC free article] [PubMed]
- 19.Fumagalli MR, Zapperi S, La Porta CA. Impact of the cross-talk between circular and messenger RNAs on cell regulation. J Theor Biol. 2018;454:386. doi: 10.1016/j.jtbi.2018.06.024. [DOI] [PubMed] [Google Scholar]
- 20.Salmena L, Poliseno L, Tay Y, Kats L, Paolo Pandolfi P. Cell. 2011;146:353. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bosia C, Pagnani A, Zecchina R. Modelling competing endogenous RNA networks. PLoS ONE. 2013;8(6):1. doi: 10.1371/journal.pone.0066609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Figliuzzi M, Marinari E, De Martino A. Biophys J. 2013;104:1203. doi: 10.1016/j.bpj.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park SM, Gaur AB, Lengyel E, Peter ME. Genes Dev. 2008;22:894. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. EMBO Rep. 2008;9(6):582. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang G, Guo X, Hong W, Liu Q, Wei T, Lu C, Gao L, Ye D, Zhou Y, Chen J, Wang J, Wu M, Liu H, Kang J. Proc Natl Acad Sci USA. 2013;110(8):2858. doi: 10.1073/pnas.1212769110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Consortium TU. Nucleic Acids Res. 2015;43:D204. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zerbino Daniel R, Achuthan Premanand, Akanni Wasiu, Amode M Ridwan, Barrell Daniel, Bhai Jyothish, Billis Konstantinos, Cummins Carla, Gall Astrid, Girón Carlos García, Gil Laurent, Gordon Leo, Haggerty Leanne, Haskell Erin, Hourlier Thibaut, Izuogu Osagie G, Janacek Sophie H, Juettemann Thomas, To Jimmy Kiang, Laird Matthew R, Lavidas Ilias, Liu Zhicheng, Loveland Jane E, Maurel Thomas, McLaren William, Moore Benjamin, Mudge Jonathan, Murphy Daniel N, Newman Victoria, Nuhn Michael, Ogeh Denye, Ong Chuang Kee, Parker Anne, Patricio Mateus, Riat Harpreet Singh, Schuilenburg Helen, Sheppard Dan, Sparrow Helen, Taylor Kieron, Thormann Anja, Vullo Alessandro, Walts Brandon, Zadissa Amonida, Frankish Adam, Hunt Sarah E, Kostadima Myrto, Langridge Nicholas, Martin Fergal J, Muffato Matthieu, Perry Emily, Ruffier Magali, Staines Dan M, Trevanion Stephen J, Aken Bronwen L, Cunningham Fiona, Yates Andrew, Flicek Paul. Ensembl 2018. Nucleic Acids Research. 2017;46(D1):D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Yu-Chen, Li Jian-Rong, Sun Chuan-Hu, Andrews Erik, Chao Rou-Fang, Lin Feng-Mao, Weng Shun-Long, Hsu Sheng-Da, Huang Chieh-Chen, Cheng Chao, Liu Chun-Chi, Huang Hsien-Da. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Research. 2015;44(D1):D209–D215. doi: 10.1093/nar/gkv940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glažar Petar, Papavasileiou Panagiotis, Rajewsky Nikolaus. circBase: a database for circular RNAs. RNA. 2014;20(11):1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Y, Lu J, Li X, Zhu H, Fan X, Zhu S, Wang Y, Guo Q, Wang L, Huang Y, Zhu M, Wang Z. BMC Cancer. 2014;14(1):85. doi: 10.1186/1471-2407-14-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iliopoulos D, Polytarchou C, Hatziapostolou M, Kottakis F, Maroulakou I, Struhl K, Tsichlis P. Sci Signal. 2009;2:ra62. doi: 10.1126/scisignal.2000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pichler M, Ress A L, Winter E, Stiegelbauer V, Karbiener M, Schwarzenbacher D, Scheideler M, Ivan C, Jahn S W, Kiesslich T, Gerger A, Bauernhofer T, Calin G A, Hoefler G. MiR-200a regulates epithelial to mesenchymal transition-related gene expression and determines prognosis in colorectal cancer patients. British Journal of Cancer. 2014;110(6):1614–1621. doi: 10.1038/bjc.2014.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guan T, Dominguez CX, Amezquita RA, Laidlaw BJ, Cheng J, Henao-Mejia J, Williams A, Flavell RA, Lu J, Kaech SM. J Exp Med. 2018;215(4):1153. doi: 10.1084/jem.20171352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Facchetti F, Previdi S, Ballarini M, Minucci S, Perego P, Porta C. Apoptosis. 2004;9(5):573. doi: 10.1023/B:APPT.0000038036.31271.50. [DOI] [PubMed] [Google Scholar]
- 35.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Camacho Christiam, Coulouris George, Avagyan Vahram, Ma Ning, Papadopoulos Jason, Bealer Kevin, Madden Thomas L. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10(1):421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.R Core Team R (2015) Language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
- 38.Grossman R, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, Staudt LM. N Engl J Med. 2016;375:1109. doi: 10.1056/NEJMp1607591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brabletz S, Brabletz T. EMBO Rep. 2010;11:670. doi: 10.1038/embor.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu M, Jolly MK, Levine H, Onuchic JN, Ben-Jacob E. MicroRNA-based regulation of epithelial-hybrid-mesenchymal fate determination. Proc Natl Acad Sci. 2013;110(45):18144. doi: 10.1073/pnas.1318192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu Mingyang, Jolly Mohit Kumar, Gomoto Ryan, Huang Bin, Onuchic José, Ben-Jacob Eshel. Tristability in Cancer-Associated MicroRNA-TF Chimera Toggle Switch. The Journal of Physical Chemistry B. 2013;117(42):13164–13174. doi: 10.1021/jp403156m. [DOI] [PubMed] [Google Scholar]
- 42.Hill Louise, Browne Gareth, Tulchinsky Eugene. ZEB/miR-200 feedback loop: At the crossroads of signal transduction in cancer. International Journal of Cancer. 2012;132(4):745–754. doi: 10.1002/ijc.27708. [DOI] [PubMed] [Google Scholar]
- 43.Preca BT, Bajdak K, Mock K, Sundararajan V, Pfannstiel J, Maurer J, Wellner U, Hopt UT, Brummer T, Brabletz S, et al. A self-enforcing CD44s/ZEB1 feedback loop maintains EMT and stemness properties in cancer cells. Int J Cancer. 2015;137(11):2566. doi: 10.1002/ijc.29642. [DOI] [PubMed] [Google Scholar]
- 44.Bail S, Swerdel M, Liu H, Jiao X, Goff LA, Hart RP, Kiledjian M. Differential regulation of microRNA stability. Rna. 2010;16(5):1032. doi: 10.1261/rna.1851510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rüegger S, Großhans H. MicroRNA turnover: when, how, and why? Rüegger. Trends Biochem Sci. 2012;37(10):436. doi: 10.1016/j.tibs.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Z, Qin YW, Brewer G, Jing Q. MicroRNA degradation and turnover: regulating the regulators. Wiley Interdisciplinary Reviews: RNA. 2012;3(4):593. doi: 10.1002/wrna.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martirosyan A, De Martino A, Pagnani A, Marinari E (2017) Sci Rep 7. 10.1038/srep43673 [DOI] [PMC free article] [PubMed]
- 48.Hunter JD. Comput Sci Eng. 2007;9(3):90. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 49.Kalluri R, Weinberg R. J Clin Invest. 2009;119:1420. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Font-Clos F, Zapperi S, La Porta CA. Topography of epithelial-mesenchymal plasticity. Proc Natl Acad Sci. 2018;115(23):5902. doi: 10.1073/pnas.1722609115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jolly MK, Boareto M, Huang B, Jia D, Lu M, Ben-Jacob E, Onuchic JN, Levine H (2015) Front Oncol. 10.3389/fonc.2015.00155 [DOI] [PMC free article] [PubMed]
- 52.Kurahara H, Takao S, Maemura K, Mataki Y, Kuwahata T, Maeda K, Ding Q, Sakoda M, Iino S, Ishigami S, et al. Epithelial- mesenchymal transition and mesenchymal-epithelial transition via regulation of ZEB-1 and ZEB-2 expression in pancreatic cancer. J Surg Oncol. 2012;105(7):655. doi: 10.1002/jso.23020. [DOI] [PubMed] [Google Scholar]
- 53.Richard G, Dalle S, Monet MA, Ligier M, Boespflug A, Pommier RM, de la Fouchardière A, Perier-Muzet M, Depaepe L, Barnault R, et al. ZEB1-mediated melanoma cell plasticity enhances resistance to MAPK inhibitors. EMBO Mol Med. 2016;8(10):1143. doi: 10.15252/emmm.201505971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sakata J, Utsumi F, Suzuki S, Niimi K, Yamamoto E, Shibata K, Senga T, Kikkawa F, Kajiyama H. Inhibition of ZEB1 leads to inversion of metastatic characteristics and restoration of paclitaxel sensitivity of chronic chemoresistant ovarian carcinoma cells. Oncotarget. 2017;8(59):99482. doi: 10.18632/oncotarget.20107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kahlert C, Lahes S, Radhakrishnan P, Dutta S, Mogler C, Herpel E, Brand K, Steinert G, Schneider M, Mollenhauer M, et al. Overexpression of ZEB2 at the invasion front of colorectal cancer is an independent prognostic marker and regulates tumor invasion in vitro. Clinical Cancer Res. 2011;17(24):7654. doi: 10.1158/1078-0432.CCR-10-2816. [DOI] [PubMed] [Google Scholar]
- 56.Eger A, Aigner K, Sonderegger S, Dampier B, Oehler S, Schreiber M, Berx G, Cano A, Beug H, Foisner R. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24(14):2375. doi: 10.1038/sj.onc.1208429. [DOI] [PubMed] [Google Scholar]
- 57.Kristensen L, Hansen T, Venø M, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shang Xingchen, Li Guanzhen, Liu Hui, Li Tao, Liu Juan, Zhao Qi, Wang Chuanxi. Comprehensive Circular RNA Profiling Reveals That hsa_circ_0005075, a New Circular RNA Biomarker, Is Involved in Hepatocellular Crcinoma Development. Medicine. 2016;95(22):e3811. doi: 10.1097/MD.0000000000003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sand M, Bechara FG, Gambichler T, Sand D, Bromba M, Hahn SA, Stockfleth E, Hessam S. Circular RNA expression in cutaneous squamous cell carcinoma. J Dermatol Sci. 2016;83(3):210. doi: 10.1016/j.jdermsci.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 60.Jiang LH, Sun DW, Hou JC, Ji ZL, et al. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1. doi: 10.1007/s12282-017-0793-9. [DOI] [PubMed] [Google Scholar]
- 61.Gong Y, et al. Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6. Cancer Cell Int. 2018;18(1):116. doi: 10.1186/s12935-018-0602-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A. Exon circularization requires canonical splice signals. Cell Rep. 2015;10(1):103. doi: 10.1016/j.celrep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 63.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 64.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biology. 2015;12(4):381. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.