Abstract
In our genomes there are thousands of copies of human endogenous retroviruses (HERVs) originated from the integration of exogenous retroviruses that infected germ line cells millions of years ago, and currently an altered expression of this elements has been associated to the onset, progression and acquisition of aggressiveness features of many cancers. The transcriptional reactivation of HERVs is mainly an effect of their responsiveness to some factors in cell microenvironment, such as nutrients, hormones and cytokines. We have already demonstrated that, under pressure of microenvironmental changes, HERV-K (HML-2) activation is required to maintain human melanoma cell plasticity and CD133+ cancer stem cells survival. In the present study, the transcriptional activity of HERV-K (HML-2), HERV-H, CD133 and the embryonic transcription factors OCT4, NANOG and SOX2 was evaluated during the in vitro treatment with antiretroviral drugs in cells from melanoma, liver and lung cancers exposed to microenvironmental changes. The exposure to stem cell medium induced a phenotype switching with the generation of sphere-like aggregates, characterized by the concomitant increase of HERV-K (HML-2) and HERV-H, CD133 and embryonic genes transcriptional activity. Although with heterogenic response among the different cell lines, the in vitro treatment with antiretroviral drugs affected HERVs transcriptional activity in parallel with the reduction of CD133 and embryonic genes expression, clonogenic activity and cell growth, accompanied by the induction of apoptosis. The responsiveness to antiretroviral drugs treatment of cancer cells with stemness features and expressing HERVs suggests the use of these drugs as innovative approach to treat aggressive tumours in combination with chemotherapeutic/radiotherapy regimens.
Electronic supplementary material
The online version of this article (10.1007/s12307-019-00231-3) contains supplementary material, which is available to authorized users.
Keywords: Endogenous retroviruses, Cancer microenvironment, Phenotype switching, Embryonic transcription factors, Antiretroviral drugs, Cancer hallmarks
Introduction
In human genome there are thousands of copies of HERVs, originated by the integration of infectious exogenous retroviruses in the germ line cells, occurred millions of years ago [1]. The inserted retroviral DNA has been subsequently inherited in a mendelian fashion, residing in every nucleated cell of the organism, where HERVs actually constitute up to 8% of the genome [2]. During evolution, the host co-opted HERVs sequences for own biological functions. Indeed, the long terminal repeats (LTRs) of HERVs contain enhancers, promoters and polyadenylation signals [3] that shape the cellular transcriptome. Under the influence of the surrounding microenvironment, HERVs are differentially expressed during developmental stages and in differentiated tissues [4, 5]. Several HERVs encode active retroviral proteins, which are involved for physiological functions in the host, such as the development of the placenta and pregnancy maintenance [6]. Most of HERVs have been silenced by genetic mechanisms, such as mutations and deletions, as well as epigenetic, including increased methylation, resulting in their inactivation. Although their transcriptional activity and the encoded proteins may exert important physiological functions, HERVs deregulation has been associated with many complex diseases, such as autoimmunity, neurological and psychiatric disorders, infectious diseases and cancer [7–11]. In cancer, HERVs drive tumorigenesis through different mechanisms, such as insertional mutagenesis, activation of downstream oncogenes, expression of HERV-K oncogenic proteins, alteration of cellular checkpoints, fusogenic and immunosuppressive activity [12–14]. Indeed, HERVs expression has been found in tumor tissues as mRNA and proteins and antibodies against HERVs have been detected in sera from patients [11, 12, 15–17]. Besides, HERVs can exert a positive role in cancer by stimulating the innate immune response mainly acting via IFN network stimulation [18].
One of the main features of HERVs is their responsiveness to factors present in the cell microenvironment, such as nutrients, hormones and cytokines, leading to their transcriptional reactivation [11]. In this context, HERVs could represent the link among microenvironment and cancer cells as genetic elements reactive to external stimuli [5]. Several findings demonstrate the crucial role of tumor microenvironment (TME) in tumorigenesis and cancer progression [19, 20]. TME is a complex milieu, including stromal cells, blood vessels, activated cancer associated fibroblasts (CAFs), tumor-associated macrophages (TAMs) and extracellular matrix (ECM). By the release of growth factors, CAFs stimulate the survival of tumor cells and the recruitment of several cell types such as proangiogenic macrophages, neutrophils and endothelial precursor cells [21]. Besides, anti-inflammatory cytokines produced by infiltrating regulatory T cells and TAMs, promote tumor progression inducing an immunosuppressive environment [22]. The complexity of the TME is also due to presence of several macromolecules or metabolites such as collagen, hyaluronan, fibronectin, albumin, lipids and others, that differently contribute to the cancer development especially in stressful conditions [23, 24]. Furthermore, due to the high rate of proliferation and the chaotic and inefficient neo-vasculature, regions of most solid tumors are subjected to hypoxic environment [25]. Taken together, all these components exert a microenvironmental pressure, resulting in tumor cells reprogramming and generation/expansion of cancer stem cells (CSCs), able to self-renew and initiate and maintain the tumor growth [26]. CSCs are also involved in chemoresistance, cancer relapse and poor overall survival, given their ability to self-renew and to differentiate into the heterogeneous lineages of cancer cells also in response to chemotherapeutic agents [27, 28]. Thus, CSCs and TME influence each other, contributing to the malignancy evolution in a dynamic process into the entire life of the tumor. Indeed, differentiated tumor cells under TME pressure can generate CSCs, demonstrating that cell plasticity is maintained throughout tumor progression, a phenomenon called phenotype switching [29, 30].
Different biomarkers have been investigated to distinguish CSCs from the bulk of the tumor cells. Among these markers, the CD133 (also known as prominin-1), a membrane-bound glycoprotein, is one of the most recognized CSCs marker [31] and its expression is associated with negative prognosis in different type of tumors including melanoma [32], liver [33] and lung cancer [34]. CSCs are also characterized by the expression of stem cell-specific embryonic transcription factors, including octamer-binding transcription factor 4 (OCT4), NANOG and SRY- related HMG-box 2 (SOX2) [35]. These transcription factors often work in combinatorial complexes regulating the expression of gene loci involved in tumor transformation and metastasis [36]; indeed, they correlate to poorly differentiated cancer cells [37].
In a previous study, we demonstrated that in human melanoma TVM-A12 cells, with CSCs features and expressing OCT4 and NANOG, HERV-K reactivation under pressure of microenvironmental changes is strictly required to maintain cell plasticity and to sustain the expansion of the CD133+ subpopulation [38]. Starting from our previous experience on melanoma cells, in the present study we widen the assessment of the response to microenvironmental changes and HERVs expression also in cell lines from liver and lung cancer. Moreover, with the aim to identify novel strategy to target aggressive cancer cells expressing HERVs, we analysed the response to antiretroviral drugs treatment in vitro.
Materials and Methods
Cell Lines and Culture Conditions
In this study the human melanoma cell line TVM-A12, stabilized in our laboratory, the hepatocellular carcinoma HepG2 and the lung adenocarcinoma A549 cell lines (all from ATCC, VA, USA) were used. The TVM-A12 cell line was cultured in RPMI-1640 medium, while HepG2 and A549 cell lines were cultured in DMEM medium. Both media were supplemented with 10% (v/v) heat inactivated fetal bovine serum (FBS), L-glutamine (2 mM), Penicillin-Streptomycin (100 mg/ml) (all from Sigma, MO, USA) and hereinafter called standard media (SM). All cell lines were maintained at 37 °C in a humidified 5% CO2 atmosphere and passaged twice weekly after detachment with trypsin (0.05%) and EDTA solution (0.02%) in PBS (Sigma). To assess the response to microenvironmental modifications, all cells were cultured in the serum free stem cells medium X-VIVO™ 15 (Lonza, Verviers, Belgium), named X-VIVO in text and figures, supplemented with Penicillin-Streptomycin (100 mg/ml). Specifically, cells were cultured for 18 h in SM to ensure cell attachment; afterward, cell were washed with PBS and cultivated in SM or X-VIVO for 72 h. Cells were than detached and back cultured with the same fresh media for further 24 h.
Reverse Transcriptase Inhibitor Treatments
As described above, cells were cultured in SM to ensure cells attachment before starting treatments. Then, cells were washed and cultured in SM or in X-VIVO for 72 h and in presence or not of the nucleoside reverse-transcriptase inhibitor (NRTI) azidothymidine (AZT) (Sigma) at the concentrations 8 and 32 μM or the non-nucleoside reverse transcriptase inhibitor (NNRTI) efavirenz (EFV) (kindly provided by Corrado Spadafora) at the concentration 15 μM. Cells were than detached and cultured with the same fresh media for further 24 h in presence of AZT or EFV at half the concentration previously used.
Morphological Analysis and Cell Growth
At the end of the treatment (96 h) with the antiretroviral drugs, morphological analysis was carried out by phase contrast microscopy, using the Olympus IX51 inverted microscope equipped with a digital camera and 10× lens magnification (Olympus, Tokyo, Japan), and cells were examined through trypan blue dye exclusion test in duplicate.
Flow Cytometry Analysis
For cytofluorimetric analysis cells were detached as previously described, washed twice in PBS, incubated with the phycoerythrin-conjugated antibody CD133/2 (clone 293C3) or phycoerythrin-conjugated isotype antibody used as control (all from Miltenyi Biotec, Bergisch Gladbach, Germany), for 30 min at 4 °C. Cells were then fixed with 1% formaldehyde solution for 5 min at 4 °C, centrifuged for 5 min at 800 g and washed once with PBS.
For apoptosis analysis cells were detached as previously described, washed in PBS, fixed with 70% ethanol (Sigma) for 45 min at 4 °C, washed in PBS and stained with propidium iodide (25 μg/ml diluted in PBS) (Sigma) and treated with RNase A (150 μg/ml) (Sigma). Before the analysis, cells were stored for at least 3 h at 4 °C. Flow cytometer analysis was performed by BD FACS Calibur (Becton Dickinson, NJ, USA) using CellQuest Pro software on a minimum of 5000 events for each sample.
RNA Extraction and RT-Real Time PCR
The expression levels of env gene of HERV-K and HERV-H and of CD133, OCT4, NANOG and SOX2 were evaluated by RT-Real Time PCR, using specific primer pairs (Table 1) [38–40]. In particular, the primers used specifically recognized sequences of the HERV-K (HML2) subgroup (hereinafter indicated as HERV-K). The list of the sequences potential identified by primer pairs is reported in Table 2 for HERV-K [41, 42] and in Table 3 for HERV-H [43–46].
Table 1.
Primers list
| Family | Gene | Forward | Reverse | GeneBank accession number |
|---|---|---|---|---|
| HERV-K [38] | env | 5’-GCCATCCACCAAGAAAGCA-3’ | 5’-AACTGCGTCAGCTCTTTAGTTGT-3’ | AF164614 |
| HERV-H [39] | env | 5’-TTCACTCCATCCTTGGCTAT-3’ | 5’-CGTCGAGTATCTACGAGCAAT-3’ | AJ289711 |
| OCT4 [40] | 5’-TATGCAAAGCAGAAACCCTCGTGC-3’ | 5’-TTCGGGCACTGCAGGAACAAATTC-3’ | NM_002701 | |
| NANOG [40] | 5’-TCCAGCAGATGCAAGAACTCTCCA-3’ | 5’-CACACCATTGCTATTCTTCGGCCA-3’ | NM_024865 | |
| SOX2 [40] | 5’-GCCGAGTGGAAACTTTTGTCG-3’ | 5’-GGCAGCGTGTACTTATCCTTCT-3’ | NM_003106 | |
| CD133 [38] | 5’-TTTCAAGGACTTGCGAACTCTCTT-3’ | 5’-GAACAGGGATGATGTTGGGTCTCA-3’ | NM_001145848.1 | |
| GUSB [39] | 5’-CAGTTCCCTCCAGCTTCAATG-3’ | 5’-ACCCAGCCGACAAAATGC-3’ | NM_000181 |
Table 2.
HERV-K HML-2 provirus sequences potentially recognized by the used primers
| Chromosome localization | Accession number | |
|---|---|---|
| K(I)/ERVK-4 | 3 | AB047209.1 |
| K108 | 7 | AF164614 |
| K(C7) | 7 | Y17832 |
| HML-2.HOM | 7 | AF074086 |
| K115 | 8 | AY037929 |
| K36 | 11 | DQ112101.1 |
| K(C19) | 19 | Y17833 |
| K113 | 19 | NC_022518 |
Table 3.
HERV-H sequences potentially recognized by the used primers
| Chromosome localization | Accession number | |
|---|---|---|
| HERV-H/env59 | 2 | AJ289711.1 |
| HERV-H/env62 | 2 | AJ289709.1 |
| HERV-H 19 | 2 | AC009495.5 |
| HERV-H/env60 | 3 | AJ289710.2 |
| HERV-H 18 env pseudogene | 2 | AF108842.1 |
| Homo sapiens RGH1 retrovirus-like element | 19 | D10083.1 |
| Homo sapiens RGH2 retrovirus-like element | 12 | D11078.1 |
| LTR46 | 17 | AC124319.13 |
Briefly, total RNA was extracted using TRIzol™ reagent (Invitrogen, MA, USA) according to the manufacturer’s instructions, retrotranscribed into cDNA (ImProm-II™, Promega, WI, USA) and amplified by Real Time PCR (CFX96, Bio-Rad Laboratories, CA, USA) using SYBR Green chemistry (Promega). The threshold cycle (Ct) comparative method was used to analyze the relative changes in gene expression of each sample compared with the reference sample, calculating as follows:
where ΔCt (sample) = [Ct (target gene) – Ct (GUSB)] and ΔCt (calibrator) was the mean of ΔCt of TVM-A12, HepG2 or A549 cells maintained in SM. Samples were analyzed in triplicate and for each experiment no template control was included to verify any contamination.
Colony Formation Assay
Colony formation assay was performed by seeding cells at different density in SM in 6-well plates (Corning Incorporated, NY, USA), according to the growth rate of the different cell lines: 200 cells/well for TVM-A12, 300 cells/well for HepG2, 100 cells/well for A549. After overnight incubation to ensure cell attachment, culture medium was replaced with fresh medium, and cells were treated with AZT (8 and 32 μM) or EFV (15 μM), for 10 days. At the end of the treatments, colonies were washed with PBS, fixed in ethanol for 30 min and stained with 0.5% methylene blue solution (Sigma), for 15 min at room temperature. A cluster containing 25 or more cells was scored as colony. The colony-forming efficiency was calculated as plating efficiency (PE) as follow
where PE indicates the percentage of cells seeded into a well that finally grow to form a colony [47]. Digital images of the colonies were obtained using the ChemiDoc XRS+ and Image Lab software (Bio-Rad, CA, USA). Size of colonies were calculated using the OpenCFU software [48].
Statistical Analysis
Data analysis was performed using the SPSS statistical software system (version 20.0, SPSS Inc., Chicago, IL, USA). The comparison of the means was carried out using Bonferroni post-hoc multiple comparison Anova test. To determine any correlation between HERVs transcription levels and embryonic genes expression, Spearman’s rho correlation coefficient was calculated. Statistical probabilities were expressed as p ≤ 0.050 (*) or p < 0.001 (**).
Results
Cancer Cells Undergo Phenotype Switching Accompanied by Transcriptional Activation of HERVs and Cancer Stem Cell-Associated Genes as Result of the Microenvironment Changes
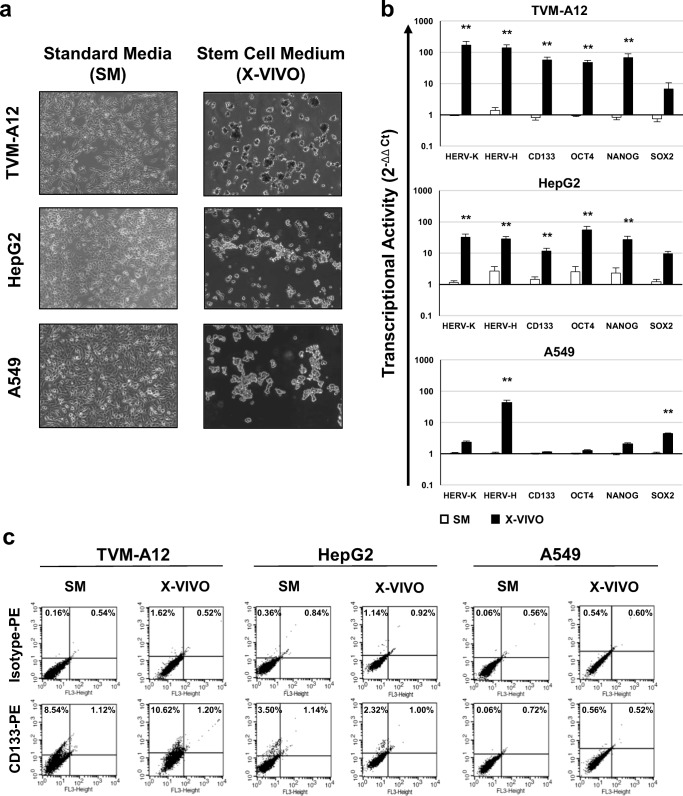
With the purpose to investigate the response of different types of cancer cells to the modifications of the microenvironment, TVM-A12, HepG2 and A549 cells were cultured in SM or in the stem cell medium X-VIVO for 72 h and analysed for morphological changes and for HERVs, embryonic genes and the CD133 stem cell marker expression. As shown in Fig. 1a, all cell lines cultured in SM grew in adhesion and exhibited their typical polygonal morphology. The passage in X-VIVO medium induced the detachment of cells and the formation of cellular sphere-like agglomerates, although in a heterogeneous manner depending on the different cell lines. Indeed, as we have already demonstrated [38], in X-VIVO medium the TVM-A12 cells are able to form dark aggregates, due to the massive melanin production in response to the stress-induced condition of serum deficiency. Interestingly, also in HepG2 and A549 cells the exposure to the same stem cell medium induced sphere-like cell aggregates formation (Fig. 1a).
Fig. 1.
Analysis of morphological changes, transcriptional activity and phenotype of cancer cells cultured in stem cell medium. a Phase contrast microscopy images; on the left panels, TVM-A12 cells cultured in RPMI-1640 medium, HepG2 and A549 cells cultured in DMEM medium, both media named standard media (SM); on the right panels, all the cells lines cultured in the serum-free stem cell medium (X-VIVO). b Relative expression of HERV-K, HERV-H, CD133 and embryonic transcription factors (OCT4, NANOG, SOX2) analyzed by RT-Real time PCR; data are shown as mean ± SE of at least three experiments performed. (*) p ≤ 0.050 or (**) p < 0.001 indicates significant difference compared to SM for each analysed cell lines. c Flow cytometry analysis of the CD133 marker expression after phycoerythrin (PE) conjugated antibodies staining; results of a representative experiment of at least three performed
In previous reports, we already demonstrated the involvement of HERV-K into the acquisition of aggressiveness and CSCs features in human melanoma cells undergoing microenvironmental changes [38, 49]. Indeed, as shown in Fig. 1b, in TVM-A12 cultured in X-VIVO medium, the transcriptional activity of HERV-K, CD133, OCT4 and NANOG was significantly increased when compared to SM (p < 0.001). Herein, as a novelty, we highlight also the increase of HERV-H expression (p < 0.001) and the slight increase of SOX2, although not statistically significant. Likewise, in response to X-VIVO exposure, also HepG2 cells increased the transcriptional activity of both HERV-K and HERV-H, together with the induction of CD133 and the embryonic genes OCT4 and NANOG expression (p < 0.001), and a slight not statistically significant increase of SOX2. Conversely, in A549 cells cultured in X-VIVO, the generation of sphere-like aggregates was accompanied to a poor induction of gene expression with a statistically significant increase only of HERV-H and SOX2 expression (p < 0.001).
According to the data obtained with the RT-Real time PCR, the phenotypic analysis performed by flow cytometry shows that in SM both TVM-A12 and HepG2 cell lines, but not A549, contain a subpopulation of cells expressing CD133 (sum of left and right upper quadrants respect to isotype: TVMA-12 8.96%; HepG2 3.44% and A549 0.16%), that was maintained in X-VIVO medium, particularly in TVM-A12 (9.68%) and, although with less efficiency, in HepG2 (1.26%) cell lines (Fig. 1c). Interestingly, the Spearman correlation analysis of HERVs, CD133 and embryonic transcription factors gene expression (Table 4) demonstrated that, in response to different culture conditions, HERV-K and HERV-H transcriptional activity were high significantly positive correlated in all cell lines (p < 0.001). Moreover, both HERV-K and HERV-H were also significantly positive correlated with the expression of CD133 and of the embryonic genes in all the cell lines. Only SOX2 did not correlate with HERVs expression in TVM-A12 cell line. Moreover, in HepG2 and A549 cells the expression of CD133 correlated positively with OCT4, NANOG and SOX2, while in TVM-A12 CD133 expression correlated positively with OCT4 and NANOG, but not with SOX2 (Supplementary Table 1).
Table 4.
Correlation analysis of HERVs and embryonic genes expression
| TVM-A12 | HepG2 | A549 | |||||
|---|---|---|---|---|---|---|---|
| HERV-K | HERV-H | HERV-K | HERV-H | HERV-K | HERV-H | ||
| HERV-K | Rho | 0.911** | 0.809** | 0.952** | |||
| p | <0.001 | <0.001 | <0.001 | ||||
| CD133 | Rho | 0.877** | 0.741* | 0.835** | 0.741** | 0.976** | 0.929* |
| p | <0.001 | 0.001 | <0.001 | <0.001 | <0.001 | 0.001 | |
| OCT4 | Rho | 0.790** | 0.729* | 0.764** | 0.475* | 0.929* | 0.881* |
| p | <0.001 | 0.001 | <0.001 | 0.034 | 0.001 | 0.004 | |
| NANOG | Rho | 0.784** | 0.676* | 0.856** | 0.530* | 0.930* | 0.952** |
| p | <0.001 | 0.004 | <0.001 | 0.016 | 0.001 | <0.001 | |
| SOX2 | Rho | 0.258 | 0.076 | 0.800** | 0.619* | 0.992** | 0.946** |
| p | 0.366 | 0.778 | <0.001 | 0.004 | <0.001 | <0.001 | |
Rho and p values are shown in bold when significant (p values <0.050)
Antiretroviral Drugs Alter Transcriptional Activity of HERVs and Cancer Stem Cell-Associated Genes during Microenvironmental Changes
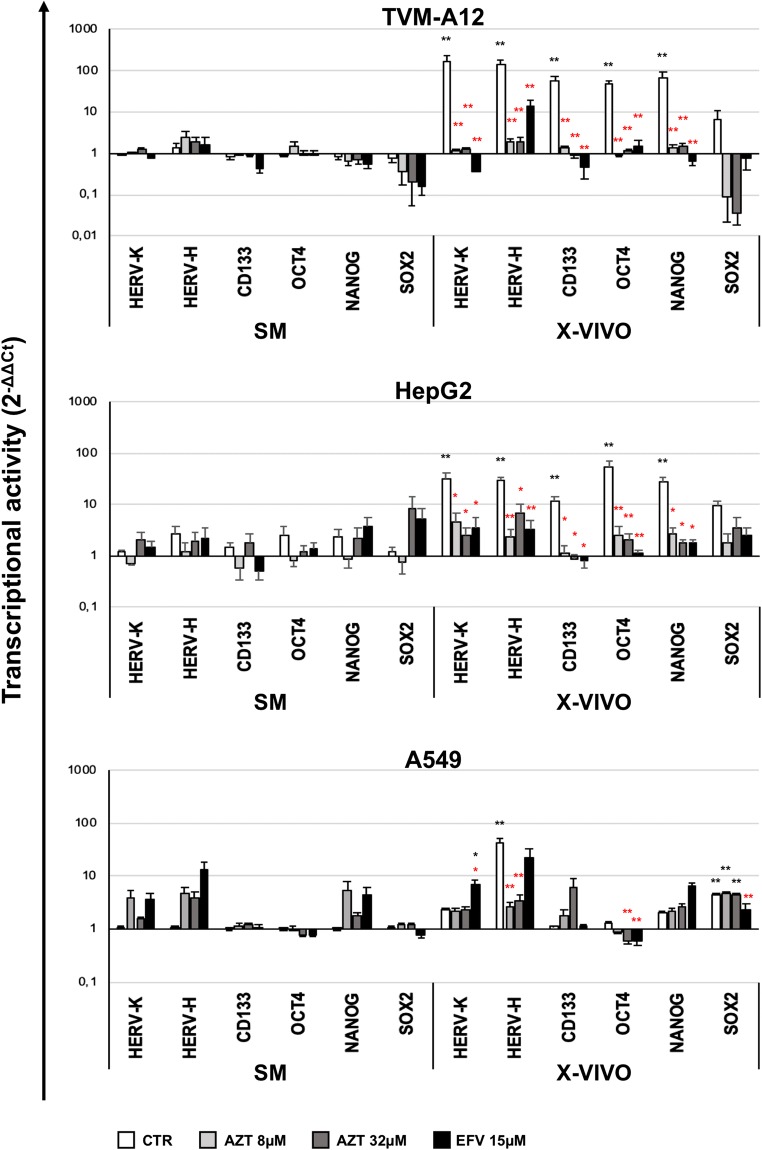
Previously we demonstrated that antiretroviral drugs were able to halt the expansion and maintenance of CD133+ melanoma cells restraining the activation of HERV-K during microenvironmental modification [38]. Thus, we investigated on the effect of the reverse transcriptase inhibitors AZT and EFV on the modulation of gene expression in TVM-A12, HepG2 and A549 cancer cells exposed to microenvironmental changes. By RT-Real time PCR analysis, we assessed the transcriptional activity of HERV-K, HERV-H, CD133 and embryonic factors (OCT4, NANOG, SOX2) on the three selected cell lines, cultured in SM and X-VIVO and treated with AZT (8 and 32 μM) or EFV (15 μM) (Fig. 2). As described above, the untreated TVM-A12 and HepG2 cells grown in X-VIVO, exhibited a high increase of expression of HERV-K, HERV-H, CD133, OCT4 and NANOG genes compared to SM (black asterisks) (all p < 0.001) (Fig. 2). However, in TVM-A12 cultured in X-VIVO, all these genes showed significant reduction of their transcriptional activity after treatment with AZT 8-32 μM or EFV 15 μM when compared to untreated control cells (CTR) (red asterisks) (all p < 0.001) (Fig. 2). Similarly, in HepG2 cultured in X-VIVO, both AZT and EFV were able to significantly decrease HERV-K, CD133 and NANOG (p < 0.050), and to highly significant decrease HERV-H and OCT-4 expression when compared to untreated cells (red asterisks). In A549 cells cultured in X-VIVO, AZT treatment significantly decreased the expression of HERV-H and OCT4, and EFV treatment significantly decreased the expression of OCT4 and SOX2 compared to untreated cells. Conversely, HERV-K was found slightly increased by EFV treatment compared to untreated condition in both SM and X-VIVO medium.
Fig. 2.
Analysis of the effect of antiretroviral drugs treatment on HERVs and cancer stem cell-associated genes expression depending on microenvironmental changes. Relative expression of HERV-K, HERV-H, CD133 and embryonic transcription factors (OCT4, NANOG, SOX2) analyzed by Real-time PCR, in TVM-A12, HepG2 and A549 cells treated with antiretroviral drugs in SM or X-VIVO. Data are shown as mean ± SE of at least three experiments performed. (*) p ≤ 0.050 or (**) p < 0.001. Black asterisks represent comparisons to the untreated control in SM. Red asterisks represent comparisons to the untreated control in X-VIVO
Antiretroviral Drugs Affect Clonogenic Ability, Cell Growth and Apoptosis in TVM-A2, HepG2 and A549 Cell Lines
On the basis of the ability of the reverse transcriptase inhibitors AZT and EFV to modulate the transcriptional activity of HERVs and cancer stem cell associated genes under microenvironmental changes (Fig. 2), we then assessed their effect on the clonogenic ability, cell growth and survival in the same experimental conditions.
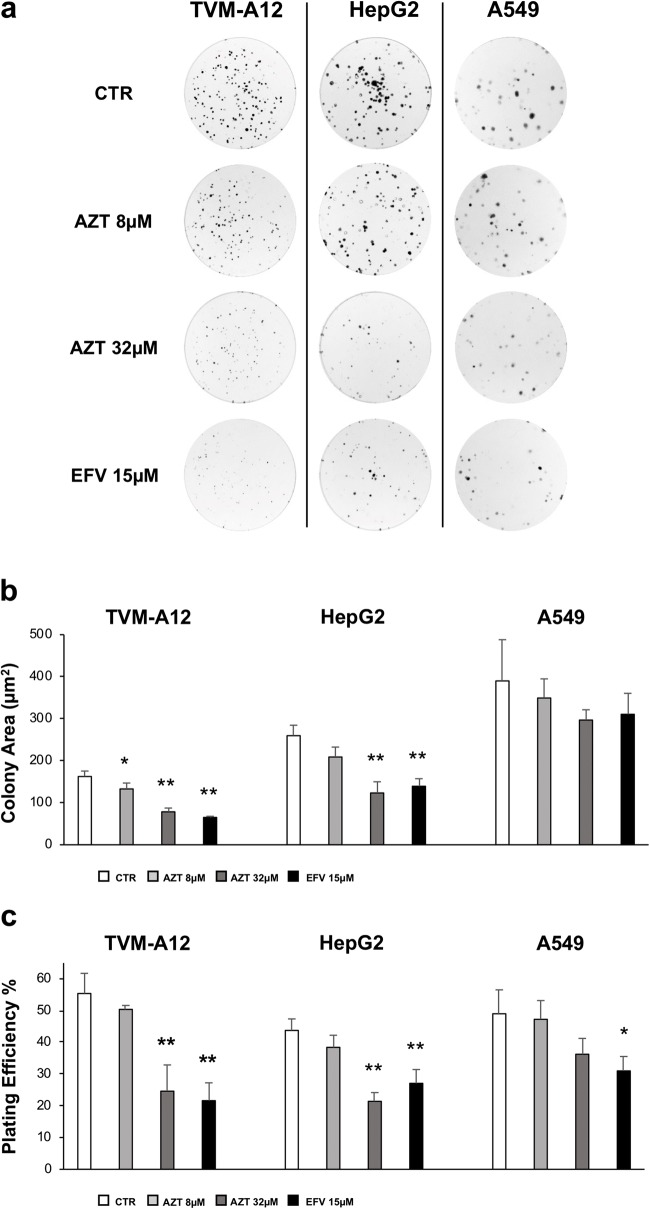
In Fig. 3 is shown the effect of AZT and EFV on the clonogenic activity of cancer cells cultured in SM. In untreated culture condition (CTR), the three cells lines formed colonies of different size (Fig. 3a), as reported by the colony area analysis (Fig. 3b), showing the presence of larger colonies in A549 cells than in TVM-A12 and HepG2 cells (mean values: 389.91, 162.46 and 259.66 μm2, respectively). Antiretroviral drugs significantly reduced the colonies’ size only in TVM-A12 (mean values: AZT 8 μM: 132.87μm2; 32 μM: 78.39μm2; EFV 15 μM: 65.02μm2) and in HepG2 (mean values: AZT 32 μM; 123.3μm2; EFV 15 μM: 139.22μm2) (p < 0.001). The basal plate efficiency (PE) for all the cell lines was in agreement with most of the mammalian cancer cell lines PE, namely between 40 and 80% [47, 50], actually 55.3%, 43.7% and 49% (Fig. 3c) for TVM-A12, HepG2 and A549 respectively. In TVM-A12 cells, the treatment with AZT 32 μM and EFV 15 μM induced a highly significant decrease in PE value (24.62% and 21.62% respectively) (p < 0.001). Similarly, HepG2 cells treated with elevated concentrations of antiretroviral drugs, showed a highly significant reduction of the PE (AZT 32 μM 21.42%, EFV 15 μM 27.1%) (p < 0.001). Interestingly, in A549 cells reverse transcriptase inhibitors induced a significant diminution of PE only at EFV 15 μM 31% (p < 0.050) (Fig. 3c).
Fig. 3.
Analysis of the effect of the antiretroviral treatment on colony-forming ability. a Colony-forming ability was assessed in TVM-A12, HepG2 and A549 cells, after 10 days of treatment with AZT (8 and 32 μM) and EFV (15 μM); images of a representative experiment of at least three performed. b Plating efficiency (PE) calculated for each condition as described in materials and methods. c Size of colonies were calculated using OpenCFU software. (*) p ≤ 0.050 or (**) p < 0.001 indicates significant difference compared to untreated cells (CTR). Data are shown as mean ± SD of at least three experiment performed
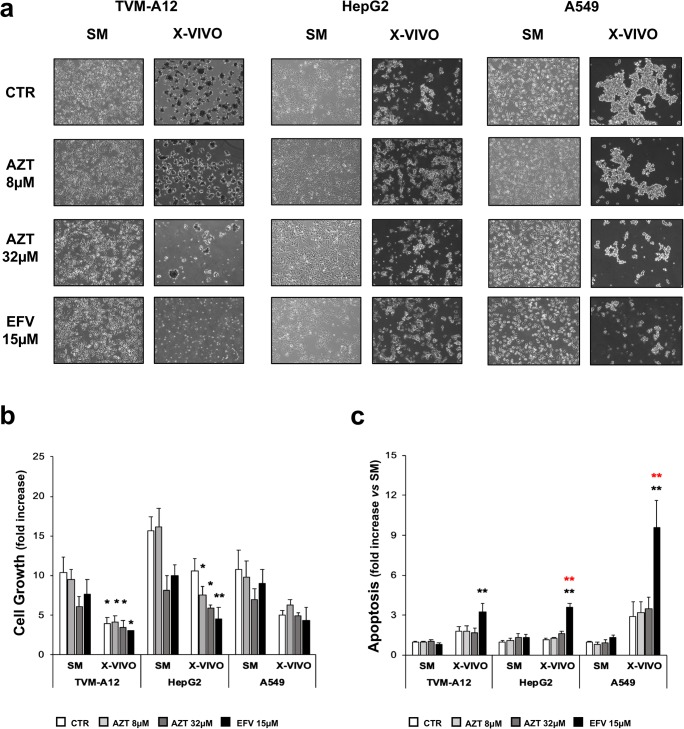
Cell aggregates formation generated in the passage from SM to X-VIVO was reduced both in number and in dimension with AZT 32 μM and EFV 15 μM treatment in all analysed cell lines (Fig. 4a). The exposure to X-VIVO determined a reduction of cell growth in all the cell lines (Fig. 4b). Compared to cell growth in SM, only in HepG2 the treatment with AZT 8 μM, AZT 32 μM and EFV 15 μM caused a significant reduction of cell growth (AZT p < 0.050; EFV p < 0.001).
Fig. 4.
Analysis of the effect of antiretroviral drugs treatment on the sphere-like aggregates formation, cell growth and apoptosis. a Phase contrast microscopy images of adherent cells and grape-like cellular aggregates of TVM-A12, HepG2 and A549 treated with antiretroviral drugs. b Cell growth analyzed by trypan blue dye exclusion test expressed as fold increase respect to seeded cell number. c Apoptosis evaluated by flow cytometry analysis of DNA fragmentation and expressed as fold increase respect to untreated cells in SM. Black asterisks represent comparisons to the untreated control in SM. Red asterisks represent comparisons to the untreated control in X-VIVO. (*) p ≤ 0.050 or (**) p < 0.001. Data are shown as mean ± SE of at least three experiments performed
In TVM-A12 the culture in X-VIVO medium reduced cell growth in both treated and untreated cells compared to SM (p < 0.050). Surprisingly, only the treatment with EFV 15 μM was able to induce a significant increase of apoptosis compared to untreated cells in SM (black asterisks) (p < 0.001), although with different susceptibility among the cell lines. (Fig. 4c). In HepG2 and A549, only EFV 15 μM was able to induce a significant increase of apoptosis compared to untreated cells in X-VIVO (red asterisks) (p < 0.001) (Fig. 4c).
Discussion
In agreement with our previous findings demonstrating the increased aggressiveness of melanoma cells under stress condition depending to HERV-K activation [38, 49], here we show that, also in cells from liver and lung cancers, the response to the exposure to a serum free stem cell medium is associated to phenotype switching towards sphere-like cellular aggregates generation concomitant to HERVs activation. Novelty, present data demonstrate that, in addition to HERV-K, also HERV-H is significantly increased depending on microenvironmental changes in all the analysed cell lines. Noteworthy, tumorigenesis is determined, other than by genetic predisposition and alterations, also by external soluble and cellular factors present in the cancer microenvironment contributing to transformation [20]. In this context, HERVs may exert a role in the crosstalk between cancers microenvironment and cell plasticity [5]. Indeed, depending on microenvironment characteristics, HERVs expression can be regulated by epigenetic mechanisms including the modifications of both DNA and the histones in somatic and embryonic cells [4]. Herein we demonstrated that HERVs expression is different among the cell lines and runs in parallel with the embryonic genes expression. In TVM-A12 and in HepG2 cells, HERV-K and HERV-H reactivation is accompanied by the increased expression of the embryonic transcription factors OCT4 and NANOG, whereas, the phenotype switching in A549 cells is accompanied by slight increase of HERV-H together with SOX2, but not of OCT4 and NANOG. Interestingly, the HERVs expression parallels the expression of embryonic genes. Indeed, the Spearman analysis demonstrated a strong positive correlation among HERV-K and HERV-H transcriptional activity with the embryonic genes expression. It is worth to mention that LTR7/HERV-H, LTR5_HS/HERV-K and L1HS, harbor 99.8% of the candidate regulatory loci with putative transcription factor-binding sites in human embryonic stem cells (hESCs) genome [51], and enhancers and promoters in retroviral LTRs can influence the transcription of neighbouring genes. In hESCs, HERV-H interacts with OCT4 to promote the enhancer activities of LTR7 and nearby regions, modulating genes essential to cell identity [52–54]. Furthermore, transactivation of hypomethylated HERV-K LTRs by OCT4 induces HERV-K expression during normal human embryogenesis [55]. HERV-K transcripts and proteins were found also in pluripotent stem cell lines and induced pluripotent stem cells (iPSC), and their expression were silenced upon differentiation [56]. Thus, from embryos to tumors, the network among embryonic genes and HERVs underlines the role of HERVs activity in the cancer cell stemness and plasticity maintenance. Actually, embryonic factors are involved in CSCs maintenance and are associated with a high presence of CD133+ cells in certain tumors [57, 58]. We already demonstrated that in TVM-A12 cells, HERV-K is strictly required to determine phenotype switching and the expansion of a CD133+ cell subpopulation expressing OCT4 and NANOG [38]. In agreement, here we demonstrate that also in HepG2 cells the culture in X-VIVO induces HERVs and CD133 expression, together with the embryonic transcription factor OCT4 and NANOG. On the other hand, in A549 cells, phenotype switching in X-VIVO is accompanied by HERV-H together with SOX2 activation, but not CD133 that is poorly represented in this cell line, as demonstrated by flow cytometry. Intriguingly, HERV-K upregulation has been associated to cancer progression and poor outcome in HCC patients [59]. Moreover, it has been reported that HBx protein of HBV could increase promoter activity of HERV-W env gene in HepG2 cells through NF-kB [60], suggesting HERVs role as cofactors in HCC etiopathogenesis.
The use of approved antiretroviral drugs to control endogenous retroelements has been already suggested in cancers as well in other diseases [61–63]. Although with heterogeneous response among the different cell lines, herein we demonstrate that antiretroviral drugs are able to inhibit the expression of HERV-K and HERV-H in cancer cells under unfavourable culture condition. Remarkably, HERVs transcriptional activity inhibition by AZT and EFV was accompanied also by the CD133 and embryonic genes expression reduction. The results also demonstrate that antiretroviral drugs could affect the clonogenic ability of all the analysed cancer cell lines. Of note, the colony formation assay highlights the proliferative potential and stem cell-like property of a specific cancer cell line [64]. Moreover, the effects of antiretroviral treatments were evident under pressure of microenvironmental changes due to the exposure to stem cell medium, resulting in sphere-like aggregates reduction both in number and dimension. Intriguingly, the apoptotic response to EFV in X-VIVO is inversely associated to the expression of HERV-K, CD133 and the embryonic genes OCT4 and NANOG, suggesting their interplay in cancer aggressiveness. Actually, embryonic factors are involved in the modulation of signaling pathways to inhibit apoptosis in cancer cells [65, 66]. The cytotoxic effects of AZT have been associated to mitochondrial toxicity and modulation of NF-kB responder genes [67, 68]. Of note, the concentrations of AZT used in the present study have been found to be no cytotoxic in peripheral blood lymphomonocytes in vitro [69], suggesting that the effect in cancer cells could be specific targeting the endogenous RT. For instance, the inhibition of RT activity by NNRTIs such EFV induces a differentiated phenotype in cancer cells [70].
It is established that genomic instability is a hallmark of cancer cells [71], as well as, a genetic mutation with gain of function in oncogenes or loss of function in tumor suppressor genes [72, 73]. The epigenetic modifications, such as demethylation, can induce activation of embryonic genes promoting the dedifferentiation process and therefore CSCs initiation [74]. On the other hand, in different type of cancers, genomic instability due to genetic and epigenetics factors, is associated to an altered expression HERVs. Recently the activation of HERVs by hypomethylation drugs, such as DNA methyltransferase inhibitors (DNMTi), and histone deacetylase inhibitors (HDACi) has been associated to increased response to immunotherapy [75, 76]. Indeed, the HERVs reactivation induce viral mimicry related to the induction of the IFN response associated to the increase of the expression of the antigens and molecules involved in immune response checkpoint, thus improving the response to the immunotherapy [77]. Single anticancer therapies have always been associated to the selection of residual aggressive and resistant cancer cells, and HERVs reactivation by DNMTi and HDACi could also induce aggressive clones. Thus, the combination of therapy is clearly an indispensable approach to fight aggressive tumors and overcome CSCs mediated therapy resistance in conventional and innovative approaches [78]. In this view, HERVs could represent targets for new combination therapy in cancer.
Conclusions
In response to microenvironmental changes, HERVs could determine cancer cell fate playing a role in genome shaping and instability and driving cancer cells towards the acquisition of stemness features. The responsiveness to antiretroviral drugs treatment of cancer cells with stemness features and expressing HERVs suggests their potential use as innovative approach to treat aggressive tumors in combination with chemotherapeutic/radiotherapy regimens.
Electronic supplementary material
(DOCX 24 kb)
Acknowledgments
This project was supported by the Italian Ministry of University and Research (Research Projects of National Interest), grant no. 2010PHT9NF_001.
Abbreviations
- AZT
Azidothymidine
- EFV
Efavirenz
- CAFs
Cancer associated fibroblasts
- CTR
Control
- CSCs
Cancer stem cells
- DNMTi
DNA methyltransferase inhibitors
- ECM
Extracellular matrix
- Env
Envelope
- FBS
Fetal bovine serum
- GUSB
Beta-glucuronidase
- HBV
Hepatitis B virus
- HCC
Hepatocellular carcinoma
- HDACi
Histone deacetylase inhibitors
- HERVs
Human endogenous retroviruses
- hESC
Human embryonic stem cells
- HML-2
Human-(mouse mammary tumor virus)-like-2
- IFN
Interferon
- iPSC
Induced pluripotent stem cells
- LTRs
Long terminal repeats
- NANOG
DNA binding homeobox transcription factor
- NF-kB
Nuclear factor kappa-light-chain-enhancer of activated B cells
- NNRTI
Non-nucleoside reverse transcriptase inhibitor
- NRTI
Nucleoside reverse-transcriptase inhibitor
- OCT4
Octamer-binding transcription factor 4
- SM
Standard medium
- RT
Reverse-transcriptase
- SOX2
Sex determining region Y-box 2 transcription factor
- TAMs
Tumor-associated macrophages
- TME
Tumor microenvironment
Authors’ Contributions
CM, EB and PSV conceived and designed the study. CM, EB, AAD and AG conceived and designed the experiments. AG, VP, AAD performed the experiments. CM, AG, EB, VP analysed and interpreted the data. CC, MTM supported the experiments and helped to draft the manuscript. SG contributed with conceptualisation the study and critical revision of manuscript. CM, AG, EB and PSV wrote the manuscript. MTM provided the linguistic assistance. PSV and CM provided the financial support and supervised laboratorial processes. All the authors read and approved the final version of the manuscript.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Grandi N, Tramontano E. Type W human endogenous retrovirus (HERV-W) integrations and their mobilization by L1. Machinery: Contribution to the Human Transcriptome and Impact on the Host Physiopathology. Viruses. 2017;9:E162. doi: 10.3390/v9070162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Coffin JM, Hughes SH, Varmus HE. The interactions of retroviruses and their hosts. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. New York: CSHL Press; 1997. pp. 335–341. [Google Scholar]
- 4.Hurst TP, Magiorkinis G. Epigenetic control of human endogenous retrovirus expression: focus on regulation of long-terminal repeats (LTRs) Viruses. 2017;9:E130. doi: 10.3390/v9060130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balestrieri E, Argaw-Denboba A, Gambacurta A, Cipriani C, Bei R, Serafino A, Sinibaldi-Vallebona P, Matteucci C. Human endogenous retrovirus K in the crosstalk between Cancer cells microenvironment and plasticity: a new perspective for combination therapy. Front Microbiol. 2018;9:1448. doi: 10.3389/fmicb.2018.01448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dupressoir A, Lavialle C, Heidmann T. From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation. Placenta. 2012;33:663–671. doi: 10.1016/j.placenta.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Gröger V, Cynis H. Human endogenous retroviruses and their putative role in the development of autoimmune disorders such as multiple sclerosis. Front Microbiol. 2018;9:265. doi: 10.3389/fmicb.2018.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Küry P, Nath A, Créange A, Dolei A, Marche P, Gold J, Giovannoni G, Hartung HP, Perron H. Human endogenous retroviruses in neurological diseases. Trends Mol Med. 2018;24:379–394. doi: 10.1016/j.molmed.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balestrieri E, Arpino C, Matteucci C, Sorrentino R, Pica F, Alessandrelli R, Coniglio A, Curatolo P, Rezza G, Macciardi F, Garaci E, Gaudi S, Sinibaldi-Vallebona P. HERVs expression in autism Spectrum disorders. PLoS One. 2012;7:e48831. doi: 10.1371/journal.pone.0048831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Contreras-Galindo R, Kaplan MH, Contreras-Galindo AC, Gonzalez-Hernandez MJ, Ferlenghi I, Giusti F, Lorenzo E, Gitlin SD, Dosik MH, Yamamura Y, Markovitz DM. Characterization of human endogenous retroviral elements in the blood of HIV-1-infected individuals. J Virol. 2012;86:262–276. doi: 10.1128/JVI.00602-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matteucci C, Balestrieri E, Argaw-Denboba A, Sinibaldi-Vallebona P. Human endogenous retroviruses role in cancer cell stemness. Semin Cancer Biol. 2018;53:17–30. doi: 10.1016/j.semcancer.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Downey RF, Sullivan FJ, Wang-Johanning F, Ambs S, Giles FJ, Glynn SA. Human endogenous retrovirus K and cancer: innocent bystander or tumorigenic accomplice? Int J Cancer. 2015;137:1249–1257. doi: 10.1002/ijc.29003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kassiotis G, Stoye JP. Making a virtue of necessity: the pleiotropic role of human endogenous retroviruses in cancer. Philos Trans R Soc Lond Ser B Biol Sci. 2017;372:20160277. doi: 10.1098/rstb.2016.0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemaître C, Tsang J, Bireau C, Heidmann T, Dewannieux M. A human endogenous retrovirus-derived gene that can contribute to oncogenesis by activating the ERK pathway and inducing migration and invasion. PLoS Pathog. 2017;13:e1006451. doi: 10.1371/journal.ppat.1006451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sauter M, Schommer S, Kremmer E, et al. Human endogenous retrovirus K10: expression of gag protein and detection of antibodies in patients with seminomas. J Virol. 1995;69:414–421. doi: 10.1128/jvi.69.1.414-421.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Contreras-Galindo R, Kaplan MH, Leissner P, Verjat T, Ferlenghi I, Bagnoli F, Giusti F, Dosik MH, Hayes DF, Gitlin SD, Markovitz DM. Human endogenous retrovirus K (HML-2) elements in the plasma of peoplewith lymphoma and breast cancer. J Virol. 2008;82:9329–9336. doi: 10.1128/JVI.00646-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang-Johanning F, Li M, Esteva FJ, Hess KR, Yin B, Rycaj K, Plummer JB, Garza JG, Ambs S, Johanning GL. Human endogenous retrovirus type K antibodies and mRNA as serum biomarkers of early-stage breast cancer. Int J Cancer. 2014;134:587–595. doi: 10.1002/ijc.28389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grandi N, Tramontano E. Human endogenous retroviruses are ancient acquired elements still shaping innate immune responses. Front Immunol. 2018;9:2039. doi: 10.3389/fimmu.2018.02039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Witz IP. The tumor microenvironment: the making of a paradigm. Cancer Microenviron. 2009;2:S9–S17. doi: 10.1007/s12307-009-0025-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maman S, Witz IP. A history of exploring cancer in context. Nat Rev Cancer. 2018;18:359–376. doi: 10.1038/s41568-018-0006-7. [DOI] [PubMed] [Google Scholar]
- 21.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 22.Balkwill FR, Capasso M, Hagemann T. The tumor microenvironment at a glance. J Cell Sci. 2012;125:5591–5596. doi: 10.1242/jcs.116392. [DOI] [PubMed] [Google Scholar]
- 23.Sainio A, Järveläinen H. Extracellular matrix macromolecules: potential tools and targets in cancer gene therapy. Mol Cell Ther. 2014;2:14. doi: 10.1186/2052-8426-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gouirand V, Guillaumond F, Vasseur S. Influence of the tumor microenvironment on Cancer cells metabolic reprogramming. Front Oncol. 2018;8:117. doi: 10.3389/fonc.2018.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrova V, Annicchiarico-Petruzzelli M, Melino G, Amelio I. The hypoxic tumour microenvironment. Oncogenesis. 2018;7:10. doi: 10.1038/s41389-017-0011-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Da Silva-Diz V, Lorenzo-Sanz L, Bernat-Peguera A, Lopez-Cerda M, Muñoz P. Cancer cell plasticity: impact on tumor progression and therapy response. Semin Cancer Biol. 2018;53:48–58. doi: 10.1016/j.semcancer.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 27.Agliano A, Calvo A, Box C. The challenge of targeting cancer stem cells to halt metastasis. Semin Cancer Biol. 2017;44:25–42. doi: 10.1016/j.semcancer.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Ahmed N, Escalona R, Leung D, Chan E, Kannourakis G. Tumour microenvironment and metabolic plasticity in cancer and cancer stem cells: perspectives on metabolic and immune regulatory signatures in chemoresistant ovarian cancer stem cells. Semin Cancer Biol. 2018;53:265–281. doi: 10.1016/j.semcancer.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 29.Poli V, Fagnocchi L, Zippo A. Tumorigenic cell reprogramming and Cancer plasticity: interplay between signaling, microenvironment, and epigenetics. Stem Cells Int. 2018;2018:4598195–4598116. doi: 10.1155/2018/4598195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.La Porta CAM, Zapperi S. Explaining the dynamics of tumor aggressiveness: at the crossroads between biology, artificial intelligence and complex systems. Semin Cancer Biol. 2018;53:42–47. doi: 10.1016/j.semcancer.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 31.Glumac PM, LeBeau AM. The role of CD133 in cancer: a concise review. Clin Transl Med. 2018;7:18. doi: 10.1186/s40169-018-0198-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El-Khattouti A, Selimovic D, Haïkel Y, Megahed M, Gomez CR, Hassan M. Identification and analysis of CD133(+) melanoma stem-like cells conferring resistance to taxol: an insight into the mechanisms of their resistance and response. Cancer Lett. 2009;343:123–133. doi: 10.1016/j.canlet.2013.09.024. [DOI] [PubMed] [Google Scholar]
- 33.Vilchez V, Turcios L, Zaytseva Y, Stewart R, Lee EY, Maynard E, Shah MB, Daily MF, Tzeng CWD, Davenport D, Castellanos AL, Krohmer S, Hosein PJ, Evers BM, Gedaly R. Cancer stem cell marker expression alone and in combination with microvascular invasion predicts poor prognosis in patients undergoing transplantation for hepatocellular carcinoma. Am J Surg. 2016;212:238–245. doi: 10.1016/j.amjsurg.2015.12.019. [DOI] [PubMed] [Google Scholar]
- 34.Bertolini G, Roz L, Perego P, Tortoreto M, Fontanella E, Gatti L, Pratesi G, Fabbri A, Andriani F, Tinelli S, Roz E, Caserini R, Lo Vullo S, Camerini T, Mariani L, Delia D, Calabro E, Pastorino U, Sozzi G. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci U S A. 2009;106:16281–16286. doi: 10.1073/pnas.0905653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hadjimichael C, Chanoumidou K, Papadopoulou N, Arampatzi P, Papamatheakis J, Kretsovali A. Common stemness regulators of embryonic and cancer stem cells. World J Stem Cells. 2009;7:1150–1184. doi: 10.4252/wjsc.v7.i9.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kashyap V, Rezende NC, Scotland KB, Shaffer SM, Persson JL, Gudas LJ, Mongan NP. Regulation of stem cell pluripotency and differentiation involves a mutual regulatory circuit of the NANOG, OCT4, and SOX2 pluripotency transcription factors with polycomb repressive complexes and stem cell microRNAs. Stem Cells Dev. 2009;18:1093–1108. doi: 10.1089/scd.2009.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Argaw-Denboba A, Balestrieri E, Serafino A, Cipriani C, Bucci I, Sorrentino R, Sciamanna I, Gambacurta A, Sinibaldi-Vallebona P, Matteucci C. HERV-K activation is strictly required to sustain CD133+ melanoma cells with stemness features. J Exp Clin Cancer Res. 2017;36:20. doi: 10.1186/s13046-016-0485-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Balestrieri E, Pica F, Matteucci C, Zenobi R, Sorrentino R, Argaw-Denboba A, Cipriani C, Bucci I, Sinibaldi-Vallebona P. Transcriptional activity of human endogenous retroviruses in human peripheral blood mononuclear cells. Biomed Res Int. 2015;2015:164529–164529. doi: 10.1155/2015/164529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao J, Li J, Geng P, Li Y, Chen H, Zhu Y. Knockdown of a HIF-2α promoter upstream long noncoding RNA impairs colorectal cancer stem cell properties in vitro through HIF-2α downregulation. Onco Targets Ther. 2015;25:3467–3474. doi: 10.2147/OTT.S81393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subramanian RP, Wildschutte JH, Russo C, Coffin JM. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology. 2011;8:90. doi: 10.1186/1742-4690-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mayer J, Sauter M, Rácz A, Scherer D, Mueller-Lantzsch N, Meese E. An almost-intact human endogenous retrovirus K on human chromosome 7. Nat Genet. 1999;21:257–258. doi: 10.1038/6766. [DOI] [PubMed] [Google Scholar]
- 43.de Parseval N, Casella J, Gressin L, Heidmann T. Characterization of the three HERV-H proviruses with an open envelope reading frame encompassing the immunosuppressive domain and evolutionary history in primates. Virology. 2001;279:558–569. doi: 10.1006/viro.2000.0737. [DOI] [PubMed] [Google Scholar]
- 44.Vargiu L, Rodriguez-Tomé P, Sperber GO, et al. Classification and characterization of human endogenous retroviruses; mosaic forms are common. Retrovirology. 2016;13:7. doi: 10.1186/s12977-015-0232-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindeskog M, Mager DL, Blomberg J. Isolation of a human endogenous retroviral HERV-H element with an open env Reading frame. Virology. 1999;258:441–450. doi: 10.1006/viro.1999.9750. [DOI] [PubMed] [Google Scholar]
- 46.Hirose Y, Takamatsu M, Harada F. Presence of env genes in members of the RTVL-H family human endogenous retrovirus-like elements. Virology. 1993;192:52–61. doi: 10.1006/viro.1993.1007. [DOI] [PubMed] [Google Scholar]
- 47.Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1:2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 48.Geissmann Q. OpenCFU, a new free and open-source software to count cell colonies and other circular objects. PLoS One. 2013;8:e54072. doi: 10.1371/journal.pone.0054072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Serafino A, Balestrieri E, Pierimarchi P, Matteucci C, Moroni G, Oricchio E, Rasi G, Mastino A, Spadafora C, Garaci E, Vallebona PS. The activation of human endogenous retrovirus K (HERV-K) is implicated in melanoma cell malignant transformation. Exp Cell Res. 2009;315:849–862. doi: 10.1016/j.yexcr.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 50.Harada K, Nonaka T, Hamada N, Sakurai H, Hasegawa M, Funayama T, Kakizaki T, Kobayashi Y, Nakano T. Heavy-ion-induced bystander killing of human lung cancer cells: role of gap junctional intercellular communication. Cancer Sci. 2009;100:684–688. doi: 10.1111/j.1349-7006.2009.01093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Glinsky GV. Transposable elements and DNA methylation create in embryonic stem cells human-specific regulatory sequences associated with distal enhancers and noncoding RNAs. Genome Biol Evol. 2015;7:1432–1454. doi: 10.1093/gbe/evv081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Santoni FA, Guerra J, Luban J. HERV-H RNA is abundant in human embryonic stem cells and a precise marker for pluripotency. Retrovirology. 2012;9:111. doi: 10.1186/1742-4690-9-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu X, Sachs F, Ramsay L, Jacques PÉ, Göke J, Bourque G, Ng HH. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat Struct Mol Biol. 2014;21:423–425. doi: 10.1038/nsmb.2799. [DOI] [PubMed] [Google Scholar]
- 54.Schlesinger S, Goff SP. Retroviral transcriptional regulation and embryonic stem cells: war and peace. Mol Cell Biol. 2015;35:770–777. doi: 10.1128/MCB.01293-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grow EJ, Flynn RA, Chavez SL, Bayless NL, Wossidlo M, Wesche DJ, Martin L, Ware CB, Blish CA, Chang HY, Reijo Pera RA, Wysocka J. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522:221–225. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fuchs NV, Loewer S, Daley GQ, Izsvák Z, Löwer J, Löwer R. Human endogenous retrovirus K (HML-2) RNA and protein expression is a marker for human embryonic and induced pluripotent stem cells. Retrovirology. 2013;10:115. doi: 10.1186/1742-4690-10-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu A, Yu X, Liu S. Pluripotency transcription factors and cancer stem cells: small genes make a big difference. Chin J Cancer. 2013;32:483–487. doi: 10.5732/cjc.012.10282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen YC, Hsu HS, Chen YW, Tsai TH, How CK, Wang CY, Hung SC, Chang YL, Tsai ML, Lee YY, Ku HH, Chiou SH. Oct-4 expression maintained cancer stem-like properties in lung cancer-derived CD133-positive cells. PLoS One. 2008;3:e2637. doi: 10.1371/journal.pone.0002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma W, Hong Z, Liu H, Chen X, Ding L, Liu Z, Zhou F, Yuan Y. Human endogenous retroviruses-K (HML-2) expression is correlated with prognosis and Progress of hepatocellular carcinoma. Biomed Res Int. 2016;2016:8201642–8201649. doi: 10.1155/2016/8201642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu C, Liu L, Wang X, Liu Y, Wang M, Zhu F. HBV X protein induces overexpression of HERV-W env through NF-κB in HepG2 cells. Virus Genes. 2017;53:797–806. doi: 10.1007/s11262-017-1479-2. [DOI] [PubMed] [Google Scholar]
- 61.Sinibaldi-Vallebona P, Matteucci C, Spadafora C. Retrotransposon-encoded reverse transcriptase in the genesis, progression and cellular plasticity of human cancer. Cancers (Basel) 2011;3:1141–1157. doi: 10.3390/cancers3011141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tyagi R, Li W, Parades D, Bianchet MA, Nath A. Inhibition of human endogenous retrovirus-K by antiretroviral drugs. Retrovirology. 2017;14:21. doi: 10.1186/s12977-017-0347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Contreras-Galindo R, Dube D, Fujinaga K, Kaplan MH, Markovitz DM. Susceptibility of human endogenous retrovirus type K to reverse transcriptase inhibitors. J Virol. 2017;91:e01309–e01317. doi: 10.1128/JVI.01309-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khan GN, Kim EJ, Shin TS, Lee SH. Heterogeneous cell types in single-cell-derived clones of MCF7 and MDA-MB-231 cells. Anticancer Res. 2017;37:2343–2354. doi: 10.21873/anticanres.11572. [DOI] [PubMed] [Google Scholar]
- 65.Hu T, Liu S, Breiter DR, Wang F, Tang Y, Sun S. Octamer 4 small interfering RNA results in cancer stem cell-like cell apoptosis. Cancer Res. 2008;68:6533–6540. doi: 10.1158/0008-5472.CAN-07-6642. [DOI] [PubMed] [Google Scholar]
- 66.Jia X, Li X, Xu Y, Zhang S, Mou W, Liu Y, Liu Y, Lv D, Liu CH, Tan X, Xiang R, Li N. SOX2 promotes tumorigenesis and increases the anti-apoptotic property of human prostate cancer cell. J Mol Cell Biol. 2011;3:230–238. doi: 10.1093/jmcb/mjr002. [DOI] [PubMed] [Google Scholar]
- 67.Matteucci C, Minutolo A, Balestrieri E, Marino-Merlo F, Bramanti P, Garaci E, Macchi B, Mastino A. Inhibition of NF-κB activation sensitizes U937 cells to 3′-azido-3′-deoxythymidine induced apoptosis. Cell Death Dis. 2010;1:e81. doi: 10.1038/cddis.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matteucci C, Minutolo A, Marino-Merlo F, Grelli S, Frezza C, Mastino A, Macchi B. Characterization of the enhanced apoptotic response to azidothymidine by pharmacological inhibition of NF-kB. Life Sci. 2015;127:90–97. doi: 10.1016/j.lfs.2015.01.038. [DOI] [PubMed] [Google Scholar]
- 69.Matteucci C, Minutolo A, Balestrieri E, Ascolani A, Grelli S, Macchi B, Mastino A. Effector caspase activation, in the absence of a conspicuous apoptosis induction, in mononuclear cells treated with azidothymidine. Pharmacol Res. 2009;59:125–133. doi: 10.1016/j.phrs.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 70.Sciamanna I, Landriscina M, Pittoggi C, Quirino M, Mearelli C, Beraldi R, Mattei E, Serafino A, Cassano A, Sinibaldi-Vallebona P, Garaci E, Barone C, Spadafora C. Inhibition of endogenous reverse transcriptase antagonizes human tumor growth. Oncogene. 2005;24:3923–3931. doi: 10.1038/sj.onc.1208562. [DOI] [PubMed] [Google Scholar]
- 71.Hanahan D, Weinberg RA. Hallmarks of Cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 72.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bryant KL, Mancias JD, Kimmelman AC, Der CJ. KRAS: feeding pancreatic cancer proliferation. Trends Biochem Sci. 2014;39:91–100. doi: 10.1016/j.tibs.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zagorac S, Alcala S, Fernandez Bayon G, Bou Kheir T, Schoenhals M, Gonzalez-Neira A, Fernandez Fraga M, Aicher A, Heeschen C, Sainz B. DNMT1 inhibition reprograms pancreatic Cancer stem cells via upregulation of the miR-17-92 cluster. Cancer Res. 2016;76:4546–4558. doi: 10.1158/0008-5472.CAN-15-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brocks D, Schmidt CR, Daskalakis M, Jang HS, Shah NM, Li D, Li J, Zhang B, Hou Y, Laudato S, Lipka DB, Schott J, Bierhoff H, Assenov Y, Helf M, Ressnerova A, Islam MS, Lindroth AM, Haas S, Essers M, Imbusch CD, Brors B, Oehme I, Witt O, Lübbert M, Mallm JP, Rippe K, Will R, Weichenhan D, Stoecklin G, Gerhäuser C, Oakes CC, Wang T, Plass C. DNMT and HDAC inhibitors induce cryptic transcription start sites encoded in long terminal repeats. Nat Genet. 2017;49:1052–1060. doi: 10.1038/ng.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Attermann AS, Bjerregaard AM, Saini SK, Grønbæk K, Hadrup SR. Human endogenous retroviruses and their implication for immunotherapeutics of cancer. Ann Oncol. 2018;18:2183–2191. doi: 10.1093/annonc/mdy413. [DOI] [PubMed] [Google Scholar]
- 77.Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote NS, Cope LM, Snyder A, Makarov V, Buhu S, Slamon DJ, Wolchok JD, Pardoll DM, Beckmann MW, Zahnow CA, Merghoub T, Chan TA, Baylin SB, Strick R. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;162:974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Steinbichler TB, Dudás J, Skvortsov S, Ganswindt U, Riechelmann H, Skvortsova II. Therapy resistance mediated by cancer stem cells. Semin Cancer Biol. 2018;53:156–167. doi: 10.1016/j.semcancer.2018.11.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 24 kb)