Abstract
The developing fetus is exposed to chemicals, which are metabolized to electrophiles that form adducts with nucleophilic Cys34 of human serum albumin (HSA). By measuring these adducts in neonatal blood spots (NBS), we obtain information regarding fetal exposures during the last month of gestation. To discover potential risk factors for childhood leukemia resulting from in utero exposures, we used untargeted adductomics to measure HSA-Cys34 adducts in 782 archived NBS, collected from incident cases of childhood acute lymphoblastic leukemia (ALL) or acute myeloid leukemia (AML) and matched population-based controls. Among a total of 28 Cys34 modifications that were measured, we found no differences in adduct abundances between childhood leukemia cases and controls overall. However, cases of T-cell ALL had higher abundances of adducts of reactive carbonyl species and a Cys34 disulfide of homocysteine was present at lower levels in AML cases. These results suggest that oxidative stress and lipid peroxidation may be etiologic factors of T-cell ALL, and alterations in one-carbon metabolism and epigenetic changes may be predictors of AML. Future replication of the results with larger sample sizes is necessary.
Keywords: Adductomics, Human serum albumin, Newborn blood spots, Childhood leukemia, In utero exposures
1. Introduction
Acute leukemia is the most common cancer among children under the age of 15. Acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML) represent 80% and 15% of childhood leukemia cases, respectively.[1] In the United States, the incidence of childhood leukemia has been increasing by about 1% per year to the current rate of 4.6 cases per 100,000 children per year.[2] Childhood leukemia is a biologically heterogeneous disease with subtypes defined by cell lineage and cytogenetic characteristics, such as chromosome translocations and aneuploidy.[3]
By measuring leukemic translocations in archived newborn blood spots (NBS) or stored cord blood, Greaves and colleagues inferred that many childhood leukemias originated in utero with about 1% of live births harboring preleukemic clones.[4,5] However, because only about 1% of children with these clones eventually developed leukemia, the etiology of this disease appears to involve at least two events, an initiating event in utero followed by postnatal genetic or epigenetic changes that lead to overt leukemia.[4,6] Although the factors contributing to these causal prenatal and postnatal events are not well understood, the increasing incidence of childhood leukemia over time, particularly in affluent countries, suggests contributions from environmental factors.[1,4,7]
Both the fetus and developing child receive myriad chemical exposures from endogenous (e.g., human and microbial metabolism) and exogenous sources (e.g., infections, the diet, xenobiotics, and parental smoking). Although many of these chemicals are stable molecules that can be characterized via metabolomics[8] and proteomics, others are reactive intermediates of metabolism and oxidative stress that can alter physiological processes in early life but cannot be measured in vivo. Since the sulfhydryl group at Cys34 of human serum albumin (HSA) is a powerful scavenger of electrophiles, including reactive oxygen species, in the interstitial space,[9] we adapted an untargeted adductomics method based on nanoflow liquid chromatography-high resolution mass spectrometry (nLC-HRMS)[10] to characterize Cys34 modifications (adducts) in archived NBS.[11] Because HSA has a residence time of 28 days,[12] Cys34 adducts in NBS capture systemic exposures occurring during the last month of gestation.
Our adductomics methodology was validated with 49 archived NBS collected from newborns whose mothers either actively smoked during pregnancy or were nonsmokers.[11] Of the 26 Cys34 adducts that were detected, the Cys34 adduct of cyanide was found to consistently discriminate between newborns of smoking and nonsmoking mothers. Since hydrogen cyanide is a component of cigarette smoke, these results indicate the suitability of NBS-based adductomics for investigating in utero exposures to reactive electrophiles that may influence disease risks later in life.
Here, we extended the investigation of Cys34 adducts in archived NBS to discover potential risk factors for childhood leukemia. Participants included 338 ALL cases and 45 AML cases and matched controls from the California Childhood Leukemia Study (CCLS), a population-based case-control study.[13] Of the 28 adducts measured, some were found to differ in abundance between children who developed specific types of leukemia and healthy controls.
2. Materials and Methods
2.1. Chemicals and Reagents
Acetonitrile (Ultra Chromasolv, LCMS grade), triethylammonium bicarbonate (TEAB) buffer (1 M), ethylenediamine-tetraacetic acid (EDTA, anhydrous), and porcine trypsin were from Sigma-Aldrich (St. Louis, MO). Methanol (Optima, LCMS grade), formic acid (Optima, LCMS grade), and iodoacetamide (IAA) were from Fisher Scientific (Pittsburgh, PA). Purified human hemoglobin was from MP Biomedicals, LLC (Santa Ana, CA). Isotopically labeled T3 (iT3) with sequence AL-[15N,13C-Val]-LIAFAQYLQQCPFEDH-[15N,13C-Val]-K was custom-made (>95%, BioMer Technology, Pleasanton, CA), and the carbamidomethylated iT3 (IAA-iT3)[14] was used as an internal standard to monitor retention time and mass drifts, as well as drift in instrument performance. Water was prepared with a PureLab purification system (18.2 mΩ cm resistivity at 25 °C; Elga LabWater, Woodridge, IL).
2.2. Study Subjects and Specimens
Parents participating in the CCLS[13] provided informed consent and (when appropriate) participating children provided assent according to human-subjects protocols approved by internal review boards of the University of California and the State of California. Incident cases of newly diagnosed ALL and AML among children under age 15 were ascertained from major clinical centers in northern and central California. Controls were randomly selected from birth certificates obtained through the California Office of Vital Records. Cases and controls were individually matched on child’s month and year of birth, sex, Hispanic ethnicity based on birth certificates, and maternal race.[13] Parents were interviewed to collect information on potentially etiologically-relevant exposures.
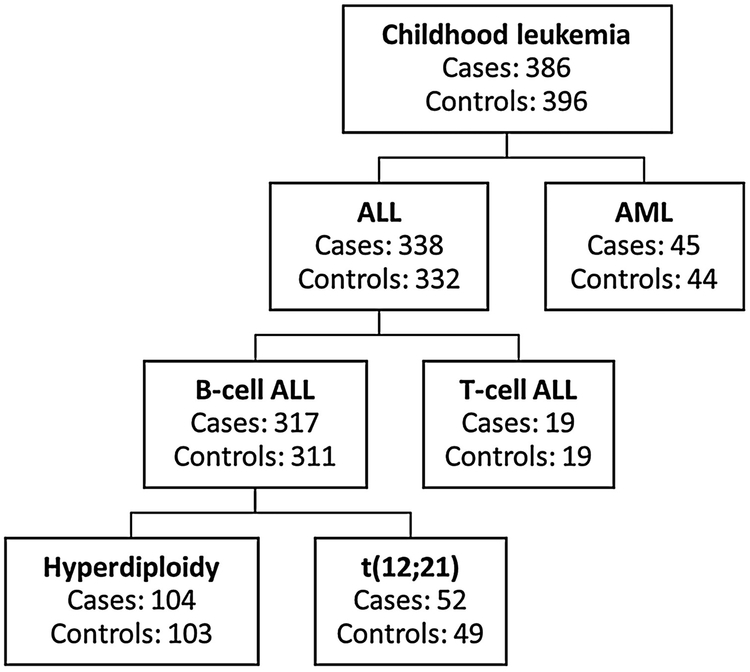
Archived newborn NBS were retrospectively obtained from the California Department of Public Health, Genetic Diseases Screening Branch.[15] The NBS were collected at birth and had been archived at −20 °C for 12 to 33 years prior to the present investigation. We used single, 4.7-mm NBS punches (equivalent to 5 – 8 μL of whole blood) from a total of 782 subjects; including 386 cases (of which 338 were ALL cases and 45 were AML cases) and 396 controls. Figure 1 shows the number of cases and controls for each childhood leukemia subtype.
Figure 1.
Number of cases and controls in each subtype of childhood leukemia of the 782 subjects included in the analysis.
Summary statistics for the 782 subjects included in this analysis are presented in Table 1, with and without stratification by leukemia subtype. As expected from the matched design, cases and controls were similar with respect to sex, ethnicity, birth year, and NBS age, both overall and when considering ALL and AML separately. The mean age at diagnosis was ~6 years for both ALL and AML cases.
Table 1.
Summary characteristics of all subjects overall, and stratified by ALL and AML.
| Overall | ALL | AML | ||||
|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Cases | Controls | |
| Number of samples | 386 | 396 | 338 | 332 | 45 | 44 |
| Child's sex | ||||||
| Male | 222 | 227 | 191 | 188 | 28 | 27 |
| Female | 164 | 169 | 147 | 144 | 17 | 17 |
| Child's ethnicity | ||||||
| Hispanic | 180 | 185 | 158 | 156 | 22 | 22 |
| Non-Hispanic | 206 | 211 | 180 | 176 | 23 | 22 |
| Child's birth year | ||||||
| Mean (SD) | 1996 (4) | 1996 (4) | 1996 (4) | 1996 (4) | 1997 (5) | 1997 (5) |
| Min | 1985 | 1985 | 1985 | 1985 | 1986 | 1986 |
| Max | 2006 | 2006 | 2005 | 2005 | 2006 | 2006 |
| Child's age at diagnosis (y) | ||||||
| Mean (SD) | 5.69 (3.56) | 5.70 (3.45) | 5.64 (4.37) | |||
| Min | 0.00 | 0.00 | 0.00 | |||
| Max | 14.76 | 14.76 | 14.06 | |||
| NBS age (y) | ||||||
| Mean (SD) | 22 (4) | 22 (4) | 22 (4) | 22 (4) | 21 (5) | 21 (5) |
| Min | 12 | 12 | 13 | 13 | 12 | 12 |
| Max | 33 | 33 | 33 | 33 | 32 | 32 |
| Hb (mg/mL) | ||||||
| Mean (SD) | 1.83 (0.51) | 1.86 (0.46) | 1.85 (0.50) | 1.86 (0.46) | 1.68 (0.52) | 1.96 (0.45) |
| Min | 0.48 | 0.41 | 0.48 | 0.41 | 0.74 | 0.71 |
| Max | 3.56 | 3.17 | 3.56 | 3.17 | 3.51 | 3.14 |
2.3. Sample Preparation and nLC-HRMS Analysis
For each subject, one 4.7-mm NBS punch was analyzed to detect and quantify HSA-Cys34 adducts as described previously.[11] Briefly, proteins were extracted from NBS and concentrations of hemoglobin (Hb)[11] were measured to normalize for blood volume using UV-Visible absorption spectroscopy. Hb concentrations were similar between cases and controls (Table 1), indicating that the NBS punches had comparable blood volumes. Methanol was added to enrich HSA by precipitating Hb and other interfering proteins and the supernatant was digested with trypsin and pressure cycling. The digests were analyzed with an Orbitrap Elite HRMS coupled to a Dionex Ultimate 3000 nanoflow LC system with a nano-electrospray ionization source (Thermo Fisher Scientific, Waltham, MA). The MS was operated in data-dependent mode, and tandem MS (MS2) fragmentation spectra were acquired in the linear ion trap. Samples were analyzed in four batches of ~200 samples, and duplicate injections were made for each sample.
2.4. Adduct Identification, Quantification, and Annotation
Cys34 adducts were identified, quantified, and annotated as described previously in detail.[10] Briefly, Cys34 adducts were pinpointed from the MS2 spectra as modifications to the third largest tryptic peptide (T3) with the sequence 21ALVLIAFAQYLQQC34PFEDHVK41 (m/z = 811.7594). Triply charged precursor ions with m/z = 811.7594 ± Δm/z, where Δm/z represents the mass added to the T3-thiolate ion (Cys34-S−), were classified as putative Cys34 adducts. The tryptic peptide adjacent to T3 with sequence 42LVNEVTEFAK51 (doubly charged precursor ion at m/z = 575.3111) was used as a ‘housekeeping peptide’ to adjust for the amount of digested HSA in each sample. An isotopically labeled and carbamidomethylated T3 peptide (IAA-iT3)[14] was used as an internal standard to monitor instrument performance. Adduct abundances were obtained by peak picking and integration using the Xcalibur Processing Method (version 3.0, Thermo Fisher Scientific, Waltham, MA) based on the average monoisotopic masses (MIMs) (5 ppm mass accuracy) and RTs of the putative adducts, as described previously.[11] Putative adducts were annotated with database searches based on the proposed empirical formula.[11] Mass accuracies were estimated by differences between theoretical and observed MIMs.
2.5. Data Preprocessing and Normalization
All statistical analyses were performed using the R statistical programming environment as described previously.[11] Briefly, adduct abundances were log-transformed and the means of duplicate measurements were calculated. After averaging the duplicate measurements, missing values were imputed using k-nearest adduct neighbor imputation with k = 3 (see Supplemental Methods in Supporting Information for details). Using the Bioconductor R package ‘scone’[16,17], the optimal scheme for removing unwanted variation used DESeq scaling and adjusted for batch effects, instrument performance, digested HSA, blood volume, and NBS age. Here, ‘batch effects’ refers to the four batches of samples; ‘instrument performance’ was indicated by the abundance of the internal standard (IAA-iT3) in each sample; ‘digested HSA’ was quantified by the abundance of the housekeeping peptide in each sample; ‘blood volume’ was indicated by measurements of Hb in each sample; and ‘NBS age’ (y) was used to account for storage-related differences in the samples.
2.6. Identification of Discriminating Adducts
This exploratory study was designed to discover adducts that are associated with childhood leukemia and can motivate hypotheses and follow-up of potentially causal exposures or pathways. To find robust associations between adduct abundances and case-control status, a combination of regression and classification models - previously described in detail [11] - was used to find adducts that discriminated childhood leukemia cases from matched controls overall and separately for ALL and AML. For ALL, we performed additional analyses stratified by the major subgroups, which included B-cell ALL, B-cell ALL with high-hyperdiploidy (51-67 chromosomes), B-cell ALL with t(12;21) chromosome translocation, and T-cell ALL (Figure 1).
First, the following multivariate linear regression model was fitted:
| (1) |
where Yi represents DESeq scaled and log transformed abundances for the ith adduct; Xcaco is the binary case/control variable, XSex and XEthnicity are binary, matching variables; XBatch is a categorical variable for the four batches; XHK and XIS represent abundances for the housekeeping peptide and the internal standard, respectively; XHb represents Hb concentrations; and XNBS Age represents NBS ages. Adducts were ranked by the nominal p-values of the coefficient β1, and the case-control fold-changes in adduct abundances were calculated as exp(β1). Second, a least absolute shrinkage and selection operator (lasso) logistic regression model was fit with case-control status regressed on the scaled, logged, and normalized adduct abundances of all 28 Cys34 adducts along with the matching variables (i.e., sex, ethnicity). The lasso regression was performed on 500 bootstrapped datasets and adducts were ranked by the proportion of times each adduct was selected into the model out of 500 bootstrap iterations. To ensure robust results, this process was repeated with a range of lasso penalty parameter values. Lastly, adducts were ranked using random forest variable importance measures. A random forest with 500 trees was constructed to predict the case-control status based on all 28 Cys34 adducts and the matching variables. The mean decrease in Gini index was used to rank adducts by their importance in the random forest classifier. This ensemble of variable selection methods was repeated separately for subjects stratified by sex with no apparent differences in results from the combined analyses. Adducts that had a nominal p-value ≤ 0.05 for case control status in Model (1) and were also within the top-10 ranking adducts by lasso and random forest were selected for further investigation.
3. Results
3.1. Data Preprocessing and Normalization
The reproducibility of measurements from duplicate injections was visually assessed using side-by-side boxplots of the differences in duplicate measurements for each subject (Figure S1A) and side-by-side boxplots of relative log abundances (RLA) of duplicate measurements for each subject [18] (Figure S1B). The RLA for a given adduct is defined as the log-ratio of the adduct’s abundance in each sample to the median abundance across samples. Since there were no obvious outliers from the boxplots of the differences in duplicate measurements (Figure S1A) and RLA boxplots (Figure S1B), all samples were kept for downstream analyses. Four adducts that were missing in over half of the samples were removed (Figure S2) after confirming that missing values did not differ significantly between cases and controls (Fisher’s exact test). The remaining missing values were imputed using k-nearest neighbors, with k=3 (Figure S3). This left a total of 782 subjects and 28 adducts of Cys34 and other T3-related peptides for statistical analyses.
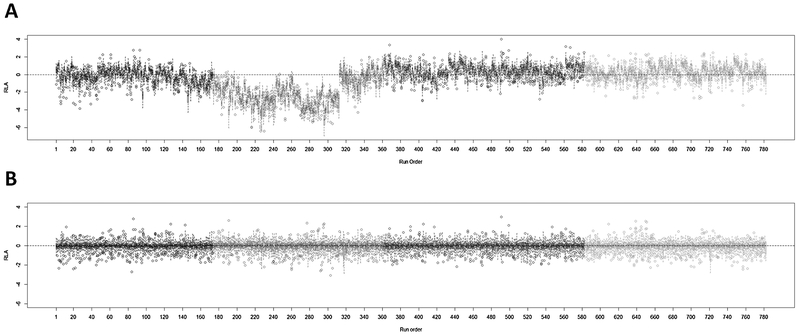
Figure 2 shows RLA boxplots before (Figure 2A) and after normalization (Figure 2B). There were noticeable batch effects that were effectively removed by normalization (Figure 2B). When each of the variables used for normalization (i.e., instrument performance, digested HSA, NBS age and blood volume) was investigated separately by the sample run order (Figure S4), most of the batch-related variation observed in Figure 2A was traced to variation in instrument performance and digested HSA.
Figure 2.
Relative log abundance (RLA) plots (A) before and (B) after normalization.
3.2. Adducts Detected in Archived NBS
Table 2 shows the 28 Cys34 adducts and other T3-related peptides that were detected in the NBS samples. In addition to the unmodified T3 peptide, a variety of adducts was observed, including Cys34 sulfoxidation products, Cys34 disulfides with low-molecular-weight thiols, and Cys34 adducts with reactive carbonyl species and other reactive molecules. Eighteen of the 28 T3-derived analytes are recurring products observed in previous analyses of plasma/serum samples.[10,19-21] Of the remaining 10 adducts, three were new to the present study (unknown adducts 824.41, 862.11 and 862.77) for which representative MS2 spectra are shown in Figure S5, and seven were observed in a previous pilot analysis of archived NBS collected from newborns whose mothers either actively smoked during pregnancy or were nonsmokers.[11] The mean value for scaled, logged, and normalized adduct abundances was 13.8 (range: 11.9 to 16.4) (Figure S6).
Table 2.
Analytes derived from the T3 peptide that were detected in archived NBS from childhood leukemia cases and controls.
| Peptide | Median RT (min) |
MIM observed (m/z, 3+) |
MIM theoretical (m/z, 3+) |
ΔMass (ppm) |
Added mass (Da) |
Elemental composition of added mass |
Annotation |
|---|---|---|---|---|---|---|---|
| 796.43a,c,d,e | 25.67 | 796.4297 | 796.4301 | −0.50 | −45.9876 | −CH2S | Cys34→Gly |
| 800.43c,d,e | 26.75 | 800.4296 | 800.4301 | −0.62 | −33.9879 | −SH2 | Cys34→Dehydroalanine |
| 805.76a,c,d,e | 25.46 | 805.7615 | 805.7618 | −0.37 | −17.9922 | −SH2, +O | Cys34→Oxoalanine or formylglycine |
| 810.43a | 25.73 | 810.4336 | −3.9759 | Unknown | |||
| 811.09a | 25.24 | 811.0872 | 811.0875 | −0.37 | −2.0151 | −H2 | Cys34 Sulfenamide |
| 811.76a,b,c,d,e | 26.36 | 811.7589 | 811.7594 | −0.62 | 1.0078 | +H | Unmodified T3f |
| 815.76a | 25.61 | 815.7591 | 815.7594 | −0.37 | 12.0006 | +CH2O, −H2O | CH2 crosslink |
| 816.42a,b,c,d,e | 23.97 | 816.4187 | 816.4191 | −0.49 | 13.9794 | −H2, +O | Cys34 Sulfinamidef |
| 816.43a,b,c,d,e | 26.63 | 816.4310 | 816.4312 | −0.24 | 15.0241 | +CH3 | Methylation (not at Cys34) |
| 819.09a | 26.70 | 819.0870 | 819.0867 | 0.37 | 22.9921 | +Na | S-Sodiation |
| 820.09a,b,c,e | 26.21 | 820.0911 | 820.0911 | 0.00 | 26.0044 | +CN | S-Cyanylation |
| 821.75a,b | 26.13 | 821.7504 | 821.7507 | −0.37 | 30.9823 | −H2, +O2 | Cys34 Sulfonamide |
| 822.42a,b,c,d,e | 25.66 | 822.4222 | 822.4226 | −0.49 | 32.9977 | +HO2 | Cys34 Sulfinic acidf |
| 824.41 | 25.81 | 824.4082 | 38.9557 | Unknown | |||
| 827.09a,b,c | 26.05 | 827.0940 | 827.0945 | −0.60 | 47.0131 | +CH3O2 | S-(O)-O-CH3 |
| 827.75a,b,c,d,e | 25.97 | 827.7536 | 827.7543 | −0.85 | 48.9919 | +HO3 | Cys34 Sulfonic acidf |
| 830.43a,c | 25.90 | 830.4343 | 830.4348 | −0.60 | 57.0340 | +C3H5O | Putative S-addition of acrolein |
| 832.43a | 26.32 | 832.4259 | 832.4262 | −0.36 | 63.0088 | +CH3O3 | Unknown |
| 835.11a,c,e | 26.44 | 835.1070 | 835.1066 | 0.48 | 71.0521 | +C4H7O | S-Addition of crotonaldehydef |
| 845.11a | 26.05 | 845.1092 | 845.1102 | −1.18 | 101.0587 | +C5H9O2 | Unknown |
| 850.10a,c,d,e | 26.59 | 850.0957 | 850.0958 | −0.12 | 116.0182 | +C4H6NOS | S-Addition of hCys (−H2O) |
| 851.43a,b,c,d,e | 24.66 | 851.4269 | 851.4274 | −0.59 | 120.0118 | +C3H6NO2S | S-Addition of Cysf |
| 857.44a | 26.39 | 857.4417 | 857.4419 | −0.23 | 138.0562 | +C7H8NO2 | Unknown |
| 862.11 | 26.89 | 862.1136 | 152.0719 | Unknown | |||
| 862.77 | 25.97 | 862.7735 | 154.0516 | Unknown | |||
| 870.43a,b,c,d,e | 24.14 | 870.4347 | 870.4345 | 0.23 | 177.0352 | +C5H9N2O3S | S-Addition of CysGlyf |
| 913.45a,b,c,d,e | 24.77 | 913.4485 | 913.4487 | −0.22 | 306.0766 | +C10H16N3O6S | S-Addition of GSHf |
| 918.12a,b | 25.14 | 918.1204 | 320.0923 | Unknown |
Cys, cysteine; hCys, homocysteine; CysGly, cysteinylglycine; GSH, glutathione; MIM, monoisotopic mass; RT, retention time.
Also detected by Yano et al. 2018.[11]
Also detected by Liu et al. 2018.[19]
Also detected by Grigoryan et al. 2018.[20]
Also detected by Lu et al. 2017.[21]
Also detected by Grigoryan et al. 2016.[10]
Annotation confirmed with a synthetic standard.
3.3. Adducts Associated with Childhood Leukemia
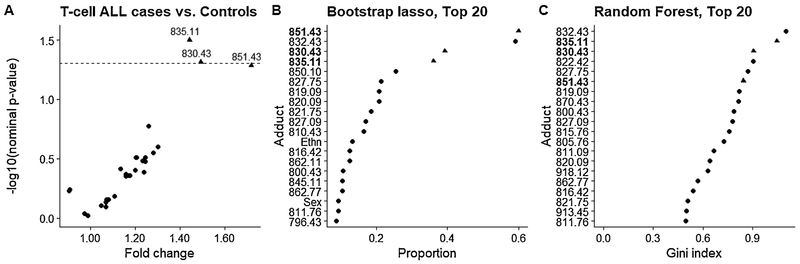
Using a nominal p-value = 0.05 as a cutoff, none of the adducts discriminated between cases and controls for childhood leukemia overall (Figure S7, Table S1), for total ALL (Figure S8, Table S2), for total B-cell ALL (Figure S9, Table S3), or for B-cell ALL subgroups with either high-hyperdiploidy (Figure S10, Table S4) or the t(12;21) translocation (Figure S11, Table S5). In fact, only the small T-cell ALL subgroup (N=19 cases) showed statistical evidence that adduct levels differed between cases and controls (Figure 3, Table S6). For T-cell ALL, adducts 835.11 (S-addition of crotonaldehyde; fold-change = 1.44, nominal p-value = 0.03), 830.43 (putative S-addition of acrolein; fold-change = 1.49, nominal p-value = 0.05), and 851.43 (S-addition of cysteine; fold-change = 1.72, nominal p-value = 0.05) were the top-three adducts ranked by Model (1) and were within the top-6 ranking adducts by lasso and random forest.
Figure 3.
Variable selection for T-cell ALL cases and controls using (A) multivariate linear regression, (B) lasso logistic regression on bootstrapped data, and (C) random forest. (A) Volcano plot showing the case/control fold-change for each adduct and the corresponding nominal p-value for each adduct. The dotted line represents a nominal p-value of 0.05. (B) Adducts ranked by the proportion of times each adduct was selected into by bootstrap lasso out of 500 iterations. (C) The top 20 adducts ranked by random forest variable importance (i.e., mean decrease in Gini index).
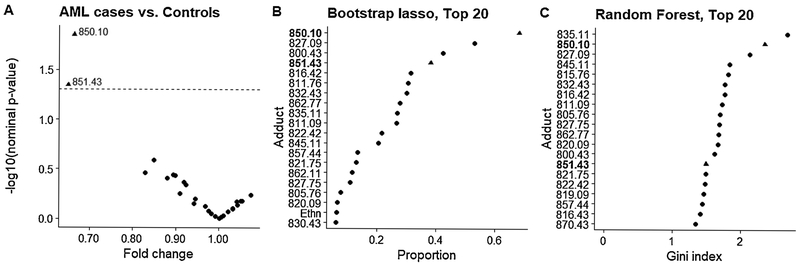
Results of variable selection for AML are summarized in Figure 4 and Table S7. The Cys34 homocysteine adduct with loss of H2O (adduct 850.10) was first-ranked by Model (1) (Figure 4A, fold-change = 0.66, nominal p-value = 0.01) and by lasso (Figure 4B), and second-ranked by random forest (Figure 4C). Although adduct 851.43 (S-addition of cysteine) was second-ranked by Model (1) with a nominal p-value of 0.05 (fold-change = 0.65) and fourth ranked by lasso, this adduct was not highly ranked by random forest.
Figure 4.
Variable selection for AML cases and controls using (A) multivariate linear regression [Model (1)], (B) lasso logistic regression on bootstrapped data, and (C) random forest. (A) Volcano plot showing the case/control fold-change and the corresponding nominal p-value from Model (1) for each adduct. The dotted line represents a nominal p-value of 0.05. (B) Adducts ranked by the proportion of times each adduct was selected by bootstrap lasso out of 500 iterations. (C) The top 20 adducts ranked by random forest variable importance (i.e., mean decrease in Gini index).
4. Discussion
The purpose of this study was to perform untargeted adductomics with archived NBS to characterize in utero exposures to reactive electrophiles that are associated with childhood leukemia. In our previous adductomics study of NBS from newborns of smoking and nonsmoking mothers, we had shown the importance of preprocessing and normalization steps to remove unwanted technical variation prior to downstream statistical analyses for the detection of differential adduct abundance.[11] Results from the current study reinforce this finding. Indeed, over the several months required for analysis of 782 samples in this study, normalization for instrument performance, digested HSA, NBS age and blood volume proved to be essential in removing unwanted variation in adduct levels (Figures 2 and S4). Such technical variation can easily obscure detection of case-control differences.
While no clear associations were detected between adduct abundances and either childhood leukemia or ALL overall, the results from our stratified analyses suggest that distinct etiologies exist across ALL subtypes.[22] In particular, Cys34 modifications by crotonaldehyde (835.11) and putative acrolein (830.43) were more abundant in cases with T-cell ALL than matched controls (Figure 3). However, these results need to be confirmed in future studies since the sample size of the T-cell ALL subgroup was small in this study (19 cases and matched controls). Since crotonaldehyde and acrolein are reactive α, β-unsaturated aldehydes produced by reactions between reactive oxygen species and membrane lipids,[23] elevated levels of these Cys34 adducts of reactive carbonyl species point to involvement of oxidative stress in this ALL subtype. Indeed, increased levels of oxidative damage to proteins, as measured by protein carbonylation, has been observed in ALL cases.[24] Interestingly, the crotonaldehyde adduct (835.11) was also one of the top-ranked features for B-cell ALL with (t12:21) by all three variable-selection methods (Figure S11) even though the nominal p-value = 0.34 for a case-control difference (fold-change = 1.13) was greater than the 0.05 cutoff.
In the smaller set of AML cases and matched controls, a Cys34 adduct of homocysteine (850.10, fold-change = 0.66) was consistently highly-ranked for associations between adducts and case-control status (Figure 4) and offers potential insight into AML etiology. Homocysteine is involved in folate-mediated one-carbon metabolism and is a key intermediate in the methionine recycling pathway.[25] Remethylation of homocysteine produces methionine, which is a precursor for S-adenosylmethionine, a universal methyl group donor that plays an important role in DNA methylation.[26] Since DNA methylation is an important epigenetic mechanism, altered levels of homocysteine could lead to abnormal methylation patterns, which are often seen in cancer cells.[27-29] DNA methylation is regulated by DNA methyltransferases (DNMTs), and DNMT3A is one of the most frequently mutated genes in adult AML.[30] Mutations in epigenetic regulators have been shown to occur as early events in hematopoietic stem cells to produce preleukemic clones that can accrue additional proliferative mutations leading to AML.[31-33] However, Bolouri et al. recently showed that the genomic, transcriptomic, and epigenomic landscapes of adult and childhood AML differ substantially, with mutations in epigenetic regulators, such as DNMT3A, being either absent or less frequent in childhood AML.[30] However, epigenetics of childhood AML is not well characterized compared to adult AML, and epigenetic aberrations involving hypermethylation of CpG island gene promoters and hypomethylation of other regions may contribute to childhood AML.[30,34,35]
Since plasma homocysteine is also an inverse indicator of folate status,[36,37] lower levels of the homocysteine adduct in AML cases may reflect higher folate levels among cases. This is interesting because folate has been shown to protect against cancer initiation while also promoting pre-neoplastic cells and subclinical cancers.[38] Moreover, cancer proliferation is inhibited by limiting the supply of folic acid and its active tetrahydrofolate metabolites and forms the basis for antifolate drugs in cancer therapy.[38] On the other hand, meta-analysis of 12 case-control studies from the Childhood Leukemia International Consortium[39] found a protective effect of maternal folic acid supplementation against both ALL (odds ratio [OR] = 0.80; 95% CI: 0.71, 0.89) and AML (OR = 0.68; 95% CI: 0.48, 0.96).[40] Although no associations were observed for childhood AML and polymorphisms in methylenetetrahydrofolate reductase (MTHFR) that regulates 5-methyl tetrahydrofolate and DNA synthesis, an increased risk of AML was observed for genetic variation in methionine synthase that catalyzes remethylation of homocysteine to methionine, with an OR of 2.74 (95% CI: 1.07, 7.01).[41] Dysfunction in methionine synthase can disrupt the synthesis of purines and thymidine and thereby impact rapidly dividing blood cells in the bone marrow.[25]
In addition to being remethylated to methionine, homocysteine can also be irreversibly degraded via the transsulfuration pathway to cysteine.[25] Homocysteine is metabolized by cystathionine β-synthase (CBS) to cystathionine, which is subsequently converted to cysteine by cystathionine γ-lyase.[25] This relationship between homocysteine and cysteine may explain why the Cys34 cysteinylated adduct (851.43, fold-change = 0.65) was also highly ranked for AML by two of the three variable-selection methods (Figure 4A, B). Moreover, CBS contributes to the endogenous production of hydrogen sulfide, which has been shown to promote cancer progression, possibly by disrupting redox homeostasis or stimulating cell proliferation and survival.[42,43] Thus, while the relationship between CBS activity and leukemogenesis is unknown, it is possible that CBS dysfunction promotes AML through modulation of homocysteine and hydrogen sulfide. Our results, showing lower levels of the homocysteine and cysteine adducts in NBS from AML cases compared to controls, further suggest that development of AML may be mediated by alterations in methionine metabolism prior to birth.
5. Conclusion
We performed untargeted adductomics using 782 NBS from a population-based case-control study to identify HSA-Cys34 adducts associated with childhood leukemia. Although we found no differences in Cys34 adduct abundances overall between cases and controls of either childhood leukemia or ALL, T-cell ALL had higher abundances of adducts of reactive carbonyl species, suggestive of oxidative stress and lipid peroxidation as potentially etiologic factors. Interestingly, there was a 44 percent decrease in abundances of a Cys34 homocysteine adduct in AML cases compared to controls. Since homocysteine is an important intermediate in the folate-methionine metabolism, this may point to alterations in one-carbon metabolism and epigenetic changes as predictors of AML. Future studies should include larger sample sizes of ALL/AML cases and their subtypes to replicate these findings and further explore their implications. We also emphasize that our findings of apparent differences in NBS-adduct abundances between childhood leukemia cases and controls points to potentially causal events occurring prior to birth.
Highlights.
Untargeted adductomics was used to analyze 782 archived newborn dried blood spots
HSA-Cys34 modifications were compared between childhood leukemia cases and controls
T-cell ALL had higher abundances of adducts of reactive carbonyl species
A Cys34 adduct of homocysteine discriminated between AML cases and controls
Acknowledgement
The biospecimens used in this study were obtained from the California Biobank Program, (SIS request number 26), in accordance with Section 6555(b), 17 CCR. The NIEHS, USEPA, and California Department of Public Health are not responsible for the results or conclusions drawn by the authors of this publication.
Funding
This work was supported by the National Institute for Environmental Health Sciences of the U.S. National Institutes of Health (NIEHS grants P01ES018172, P50ES018172, R01ES009137 and P42ES0470518), by the U.S. Environmental Protection Agency (USEPA grants RD83451101 and RD83615901), and by a grant for a pilot project from Children with Cancer, a registered Charity in the U.K.
Abbreviations:
- ALL
acute lymphoblastic leukemia
- AML
acute myeloid leukemia
- CBS
cystathionine β-synthase
- CCLS
California Childhood Leukemia Study
- CI
confidence interval
- cvAUC
cross-validated area under the estimated receiver operating characteristic curve
- Cys34
cysteine at position 34 in human serum albumin
- DNMT
DNA methyltransferase
- Hb
hemoglobin
- HSA
human serum albumin
- MIM
monoisotopic mass
- MS2
tandem mass spectrometry
- m/z
mass-to-charge ratio
- NBS
neonatal blood spot
- nLC-HRMS
nanoflow liquid chromatography-high resolution mass spectrometry
- OR
odds ratio
- RLA
relative log abundance
- RT
retention time
- T3
third largest tryptic peptide of human serum albumin containing Cys34
Footnotes
Ethics approval
The CCLS and Center for Integrative Research on Childhood Leukemia and the Environment were approved by the University of California Committee for the Protection of Human Subjects, the California Health and Human Services Agency Committee for the Protection of Human Subjects, the State of California, and the institutional review boards of all participating hospitals, as appropriate.
Informed consent
Written informed consent was obtained from all adult volunteer subjects and parents of all participating subjects in the CCLS.
Conflict of interest
The authors declare they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Whitehead TP, Metayer C, Wiemels JL, Singer AW, Miller MD, Childhood Leukemia and Primary Prevention, Curr. Probl. Pediatr. Adolesc. Health Care 46 (2016) 317–352. doi: 10.1016/j.cppeds.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Cancer Stat Facts: Childhood Leukemia (Ages 0-19), Natl. Cancer Institute, Surveillance, Epidemiol. End Results Progr (n.d.). https://seer.cancer.gov/statfacts/html/childleuk.html (accessed April 6, 2019).
- [3].Greaves M, Childhood leukemia, Bmj. 324 (2002) 283–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Greaves M, A causal mechanism for childhood acute lymphoblastic leukaemia, Nat. Rev. Cancer 18 (2018) 471–484. doi: 10.1038/s41568-018-0015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Greaves M, In utero origins of childhood leukaemia, Early Hum. Dev 81 (2005) 123–129. doi: 10.1016/j.earlhumdev.2004.10.004. [DOI] [PubMed] [Google Scholar]
- [6].Greaves M, Infection, immune responses and the aetiology of childhood leukaemia, Nat. Rev. Cancer 6 (2006) 193–203. doi: 10.1038/nrc1816. [DOI] [PubMed] [Google Scholar]
- [7].Wiemels J, Perspectives on the causes of childhood leukemia, Chem. Biol. Interact 196 (2012) 59–67. doi: 10.1016/j.cbi.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Petrick LM, Schiffman C, Edmands WMB, Yano Y, Perttula K, Whitehead T, Metayer C, Wheelock CE, Arora M, Grigoryan H, Carlsson H, Dudoit S, Rappaport SM, Metabolomics of neonatal blood spots reveal distinct phenotypes of pediatric acute lymphoblastic leukemia and potential effects of early-life nutrition, Cancer Lett. 452 (2019) 71–78. doi: 10.1016/j.canlet.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Aldini G, Vistoli G, Regazzoni L, Gamberoni L, Facino RM, Yamaguchi S, Uchida K, Carini M, Albumin Is the Main Nucleophilic Target of Human Plasma: A Protective Role Against Proatherogenic Electrophilic Reactive Carbonyl Species?, Chem. Res. Toxicol 21 (2008) 824–835. doi: 10.1021/tx700349r. [DOI] [PubMed] [Google Scholar]
- [10].Grigoryan H, Edmands W, Lu SS, Yano Y, Regazzoni L, Iavarone AT, Williams ER, Rappaport SM, Adductomics Pipeline for Untargeted Analysis of Modifications to Cys34 of Human Serum Albumin, Anal. Chem 88 (2016) 10504–10512. doi: 10.1021/acs.analchem.6b02553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yano Y, Grigoryan H, Schiffman C, Edmands W, Petrick L, Hall K, Whitehead T, Metayer C, Dudoit S, Rappaport S, Untargeted adductomics of Cys34 modifications to human serum albumin in newborn dried blood spots, Anal. Bioanal. Chem 411 (2019) 2351–2362. doi: 10.1007/s00216-019-01675-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rappaport SM, Li H, Grigoryan H, Funk WE, Williams ER, Adductomics: Characterizing exposures to reactive electrophiles, Toxicol. Lett 213 (2012) 83–90. doi: 10.1016/j.toxlet.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Metayer C, Zhang L, Wiemels JL, Bartley K, Schiffman J, Ma X, Aldrich MC, Chang JS, Selvin S, Fu CH, Ducore J, Smith MT, Buffler PA, Tobacco Smoke Exposure and the Risk of Childhood Acute Lymphoblastic and Myeloid Leukemias by Cytogenetic Subtype, Cancer Epidemiol. Biomarkers Prev 22 (2013) 1600–1611. doi: 10.1158/1055-9965.EPI-13-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Grigoryan H, Li H, Iavarone AT, Williams ER, Rappaport SM, Cys34 Adducts of Reactive Oxygen Species in Human Serum Albumin, Chem. Res. Toxicol 25 (2012) 1633–1642. doi: 10.1021/tx300096a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].California Department of Public Health, Genetic Disease Screening Program, (n.d.). https://www.cdph.ca.gov/Programs/CFH/DGDS/Pages/default.aspx (accessed October 10, 2018).
- [16].Cole M, Risso D, scone: Single Cell Overview of Normalized Expression data, R Packag. Version 1.2.0 (2018). [Google Scholar]
- [17].Schiffman C, Petrick L, Perttula K, Yano Y, Carlsson H, Whitehead T, Metayer C, Hayes J, Rappaport S, Dudoit S, Filtering procedures for untargeted lc-ms metabolomics data, BMC Bioinformatics. 20 (2019) 1–10. doi: 10.1186/s12859-019-2871-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].De Livera AM, Dias DA, De Souza D, Rupasinghe T, Pyke J, Tull D, Roessner U, McConville M, Speed TP, Normalizing and Integrating Metabolomics Data, Anal. Chem 84 (2012) 10768–10776. doi: 10.1021/ac302748b. [DOI] [PubMed] [Google Scholar]
- [19].Liu S, Grigoryan H, Edmands WMB, Dagnino S, Sinharay R, Cullinan P, Collins P, Chung KF, Barratt B, Kelly FJ, Vineis P, Rappaport SM, Cys34 Adductomes Differ between Patients with Chronic Lung or Heart Disease and Healthy Controls in Central London, Environ. Sci. Technol 52 (2018) 2307–2313. doi: 10.1021/acs.est.7b05554. [DOI] [PubMed] [Google Scholar]
- [20].Grigoryan H, Edmands WMB, Lan Q, Carlsson H, Vermeulen R, Zhang L, Yin S-N, Li G-L, Smith MT, Rothman N, Rappaport SM, Adductomic signatures of benzene exposure provide insights into cancer induction, Carcinogenesis. 39 (2018) 661–668. doi: 10.1093/carcin/bgy042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lu SS, Grigoryan H, Edmands WMB, Hu W, Iavarone AT, Hubbard A, Rothman N, Vermeulen R, Lan Q, Rappaport SM, Profiling the Serum Albumin Cys34 Adductome of Solid Fuel Users in Xuanwei and Fuyuan, China, Environ. Sci. Technol 51 (2017) 46–57. doi: 10.1021/acs.est.6b03955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pui C-H, Carroll WL, Meshinchi S, Arceci RJ, Biology, Risk Stratification, and Therapy of Pediatric Acute Leukemias: An Update, J. Clin. Oncol 29 (2011) 551–565. doi: 10.1200/JCO.2010.30.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lee SE, Park YS, Role of Lipid Peroxidation-Derived α, β -Unsaturated Aldehydes in Vascular Dysfunction, Oxid. Med. Cell. Longev 2013 (2013) 1–7. doi: 10.1155/2013/629028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Battisti V, Maders LDK, Bagatini MD, Santos KF, Spanevello RM, Maldonado P. a., Brulé AO, Araújo MDC, Schetinger MRC, Morsch VM, Measurement of oxidative stress and antioxidant status in acute lymphoblastic leukemia patients, Clin. Biochem 41 (2008) 511–518. doi: 10.1016/j.clinbiochem.2008.01.027. [DOI] [PubMed] [Google Scholar]
- [25].Blom HJ, Smulders Y, Overview of homocysteine and folate metabolism. With special references to cardiovascular disease and neural tube defects, J. Inherit. Metab. Dis 34 (2011) 75–81. doi: 10.1007/s10545-010-9177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Strickland KC, Krupenko NI, Krupenko SA, Molecular mechanisms underlying the potentially adverse effects of folate, Clin. Chem. Lab. Med 51 (2013) 607–616. doi: 10.1515/cclm-2012-0561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Rush LJ, Plass C, Alterations of DNA methylation in hematologic malignancies, Cancer Lett. 185 (2002) 1–12. doi: 10.1016/S0304-3835(02)00288-4. [DOI] [PubMed] [Google Scholar]
- [28].Yamashita Y, Yuan J, Suetake I, Suzuki H, Ishikawa Y, Choi YL, Ueno T, Soda M, Hamada T, Haruta H, Takada S, Miyazaki Y, Kiyoi H, Ito E, Naoe T, Tomonaga M, Toyota M, Tajima S, Iwama A, Mano H, Array-based genomic resequencing of human leukemia, Oncogene. 29 (2010) 3723–3731. doi: 10.1038/onc.2010.117. [DOI] [PubMed] [Google Scholar]
- [29].Ehrlich M, DNA methylation in cancer: too much, but also too little, Oncogene. 21 (2002) 5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- [30].Bolouri H, Farrar JE, Triche T, Ries RE, Lim EL, Alonzo TA, Ma Y, Moore R, Mungall AJ, Marra MA, Zhang J, Ma X, Liu Y, Liu Y, Auvil JMG, Davidsen TM, Gesuwan P, Hermida LC, Salhia B, Capone S, Ramsingh G, Zwaan CM, Noort S, Piccolo SR, Kolb EA, Gamis AS, Smith MA, Gerhard DS, Meshinchi S, The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions, Nat. Med 24 (2018) 103–112. doi: 10.1038/nm.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Corces-Zimmerman MR, Hong W-J, Weissman IL, Medeiros BC, Majeti R, Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission, Proc. Natl. Acad. Sci. 111 (2014) 2548–2553. doi: 10.1073/pnas.1324297111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, Kennedy JA, Schimmer AD, Schuh AC, Yee KW, McLeod JL, Doedens M, Medeiros JJF, Marke R, Kim HJ, Lee K, McPherson JD, Hudson TJ, Pan-Leukemia TH Gene Panel Consortium, Brown AMK, Yousif F, Trinh QM, Stein LD, Minden MD, Wang JCY, Dick JE, Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia, Nature. 506 (2014) 328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Grimwade D, Ivey A, Huntly BJP, Molecular landscape of acute myeloid leukemia in younger adults and its clinical relevance, Blood. 127 (2016) 29–41. doi: 10.1182/blood-2015-07-604496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Newcombe AA, Gibson BES, Keeshan K, Harnessing the potential of epigenetic therapies for childhood acute myeloid leukemia, Exp. Hematol 63 (2018) 1–11. doi: 10.1016/j.exphem.2018.03.008. [DOI] [PubMed] [Google Scholar]
- [35].Liang D, Liu H, Yang C, Jaing T, Hung I, Yeh T, Chen S, Hou J, Huang Y, Shih Y, Huang Y, Lin T, Shih L, Cooperating gene mutations in childhood acute myeloid leukemia with and DNMT3A, Blood. 121 (2013) 2988–2995. doi: 10.1182/blood-2012-06-436782.The. [DOI] [PubMed] [Google Scholar]
- [36].Ueland PM, Hustad S, Homocysteine and Folate Status in an Era of Folic Acid Fortification: Balancing Benefits, Risks, and B-vitamins, Clin. Chem 54 (2008) 779–781. doi: 10.1373/clinchem.2008.103218. [DOI] [PubMed] [Google Scholar]
- [37].Pfeiffer CM, Caudill SP, Gunter EW, Osterloh J, Sampson EJ, Biochemical indicators of B vitamin status in the US population after folic acid fortification: results from the National Health and Nutrition Examination Survey 1999–2000, Am. J. Clin. Nutr. 82 (2005) 442–450. doi: 10.1093/ajcn/82.2.442. [DOI] [PubMed] [Google Scholar]
- [38].Smith AD, Kim Y-I, Refsum H, Is folic acid good for everyone?, Am. J. Clin. Nutr 87 (2008) 517–533. doi: 10.1093/ajcn/87.3.517. [DOI] [PubMed] [Google Scholar]
- [39].Metayer C, Milne E, Clavel J, Infante-Rivard C, Petridou E, Taylor M, Schüz J, Spector LG, Dockerty JD, Magnani C, Pombo-de-Oliveira MS, Sinnett D, Murphy M, Roman E, Monge P, Ezzat S, Mueller BA, Scheurer ME, Armstrong BK, Birch J, Kaatsch P, Koifman S, Lightfoot T, Bhatti P, Bondy ML, Rudant J, O’Neill K, Miligi L, Dessypris N, Kang AY, Buffler PA, The Childhood Leukemia International Consortium, Cancer Epidemiol. 37 (2013) 336–347. doi: 10.1016/j.canep.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Metayer C, Milne E, Dockerty JD, Clavel J, Pombo-De-Oliveira MS, Wesseling C, Spector LG, Schüz J, Petridon E, Ezzat S, Armstrong BK, Rudant J, Koifman S, Kaatsch P, Moschovi M, Rashed WM, Selvin S, McCauley K, Hung RJ, Kang AY, Infante-Rivard C, Maternal supplementation with folic acid and other vitamins and risk of leukemia in offspring: A childhood leukemia internotionol consortium study, Epidemiology. 25 (2014) 811–822. doi: 10.1097/EDE.0000000000000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lightfoot TJ, Johnston WT, Painter D, Simpson J, Roman E, Skibola CF, Smith MT, Allan JM, Taylor GM, Genetic variation in the folate metabolic pathway and risk of childhood leukemia, Blood. 115 (2010) 3923–3929. doi: 10.1182/blood-2009-10-249722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zhu H, Blake S, Chan KT, Pearson RB, Kang J, Cystathionine β -Synthase in Physiology and Cancer, Biomed Res. Int 2018 (2018) 1–11. doi: 10.1155/2018/3205125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wu D, Si W, Wang M, Lv S, Ji A, Li Y, Hydrogen sulfide in cancer: Friend or foe?, Nitric Oxide. 50 (2015) 38–45. doi: 10.1016/j.niox.2015.08.004. [DOI] [PubMed] [Google Scholar]