Abstract
Here we demonstrate that regulation of the Complement (C’) components of the immune system is an ancient and conserved feature of mammalian pregnancy. Transcript levels were reduced for complement components C3 and C4 throughout pregnancy in a marsupial, Monodelphis domestica. Downstream C’ component transcripts were significantly less abundant relative to non-pregnant controls at the start of pregnancy but increased during late pregnancy, in some cases peaking close to parturition. These results are consistent with observations in human pregnancy that deposition of C5 through C9 on fetal membranes is associated with labor and parturition. Complement regulators CD46 and CD59 are present at the fetomaternal interface during M. domestica pregnancy as well, implying regulation of C’ effector mechanisms is necessary for maintenance of normal marsupial pregnancy. Collectively these results support regulating the complement system may have contributed to the transition from oviparity to viviparity in mammals over 165 million years ago.
1. Introduction
Mechanisms to protect the fetus and fetal membranes from immune injury are critical to successful pregnancy in viviparous animals. This protection is mediated by regulating or inhibiting a variety of immune mechanisms (reviewed in Erlebacher, 2013). One of these mechanisms is the C’ system. C’ is a collection of blood proteins that act in a cascade to mediate a variety of immune responses. C’ is ancient in deuterostomes and highly conserved across vertebrates (Sunyer and Lambris, 1998). The effects of C’ are broad including forming holes in cellular membranes and stimulating apoptosis of both microbial and animal cells, stimulating inflammation, and facilitating phagocytosis (Sarma and Ward, 2011).
C’ has been associated with both normal pregnancy and pregnancy complications. Some C’ components, for example, are associated with normal human trophoblast (Faulk et al., 1980). C’ at the fetomaternal interface, however, also contributes to pathology such as recurrent spontaneous abortions (RSA) and antiphospholipid antibody syndrome (APS) (Girardi et al., 2006; Shamonki et al., 2007). Mutations in C’ genes are also strongly correlated with women who suffer from pregnancy-associated atypical hemolytic uremic syndrome (Fakhouri et al., 2010). Beyond humans, a deleterious mutation of a murine-specific C’ regulator, Crry, results in lethal inflammation in mouse embryos (Xu et al., 2000; Mao et al., 2003).
Regulation of the C’ system during pregnancy is well established in eutherian mammals. This may be a requirement of normal pregnancy since there is evidence that the maternal immune system is immunologically “aware” of the fetal antigens in eutherians. Indeed, anti-paternal antibodies are common in eutherian pregnancy, and have even been shown to be protective to the fetus in some species (Antczak et al., 1984; Chaouat et al., 1985; Orgad et al., 1999). How “aware” the maternal immune system is of fetal tissues during pregnancy in distantly related mammals, such as marsupials, remains a subject of debate. A study in tammar wallabies, Macropus eugenii, found no evidence of anti-paternal antibodies generated by maternal immune systems (Van Oorschot and Cooper, 1988). Therefore it is possible that marsupials may not need to regulate C’ or have other differences at the fetomaternal interface.
Previously we have shown that a large number of genes involved in the immune response are differentially regulated during pregnancy in the opossum, Monodelphis domestica (Hansen et al., 2016, 2017). Analysis of the uterine post-attachment transcriptome, when the yolk sac placental membranes have invaded the maternal endometrium, revealed that pro-inflammatory cytokine transcripts are elevated less than 24 hours before parturition (Hansen et al., 2017). Conversely, transcripts for complement component C3 were significantly reduced than in non-pregnant controls (Freyer et al., 2007; Hansen et al., 2016). Here we follow up on these results focusing on a broader range of C’ components and their regulators throughout pregnancy in the opossum.
2. Materials and Methods
2.1. Animal use and tissue collection
This study was approved under protocol numbers 13-100920-MCC and 15-200334-B-MC from the University of New Mexico Institutional Animal Care and Use Committee. All uterine tissues collected as previously described (Hansen et al., 2017). Briefly, pregnant and post-natal M. domestica uterine horns were excised, dissected, treated with RNALater (Ambion, Carlsbad, California), and stored at −80°C until use. Uterine samples were collected on embryonic days 3 (E3), 9 (E9), 10 (E10), 11 (E11), 12, (E12), 13 (E13), 14 (E14), post-natal day 1 (P1), and from non-pregnant past breeders (NP) as a control.
2.2. cDNA and quantitative PCR
RNA was extracted and cDNA was synthesized as described previously (Hansen et al., 2017). Briefly, RNA was phenol-extracted and isolated. cDNA libraries were generated from 500ng RNA using the SuperScript III First Strand Synthesis Kit (Invitrogen, Carlsbad, California) according to manufacturer’s instructions. All cDNA synthesis reactions were performed in triplicate to minimize bias in cDNA library generation.
qPCR methods performed were as described previously (Hansen et al., 2017). All reactions were performed on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, California). Novel M. domestica-specific primers used in this study were generated according to manufacturer’s recommendations for SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, Hercules, California). Primer pairs were developed for C1q, C3, C4, C5, C6. C7, C9, CD46, CD59 and DAF (Table 1). We were unable to develop primers for C1r, C1s, C2 and C8 that functioned reliably in quantitative PCR. Relative normalized expression levels were calculated in CFX Manager software (Bio-Rad, Hercules, California) by the Vandesompele method using tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta polypeptide (YWHAZ) and TATA box-binding protein (TBP) as reference genes (Vandesompele et al., 2002). These reference genes were chosen due to their relatively uniform transcription levels in both pregnant and non-pregnant uterine transcriptome data (Hansen et al., 2016). Statistical tests and graphs were generated using Prism 7 (GraphPad) and edited for visual clarity in Illustrator (Adobe).
Table 1.
qPCR primer sequences.
| Gene | Primer Direction |
Primer Sequence |
|---|---|---|
| YWHAZ* | Forward | AAAGACGGAAGGTGCTGAG |
| Reverse | CCTCAGCCAAGTAACGGTAG | |
| TBP* | Forward | GTGCCCGAAATGCTGAATAC |
| Reverse | TTTCCTGGCTGCTAATCTGG | |
| C3 | Forward | TTTGTCTGTGCCCTCAACGG |
| Reverse | GACGCCCAGCCTTCAACATC | |
| C4 | Forward | GCTTCGTCTCTATGTGGCAG |
| Reverse | GCCCTGTAGCAAGAAAGGG | |
| C5 | Forward | GAACATTGAACGGACCCTG |
| Reverse | TGGGAGATGGCTTAGAGTAG | |
| C6 | Forward | AAGTGGAGAACAGGTGTACC |
| Reverse | CAGCCACTCAGTAAACACAG | |
| C7 | Forward | GATGGTCTAGCAGTGGTTG |
| Reverse | CCTTACTTTCCTCCCTTCTG | |
| C9 | Forward | TTCTCCCTGCCATACTGATG |
| Reverse | GCTTGCGGTAATAGGTTCC | |
| CD46 | Forward | TCCAGGTGTCAGGATTCGG |
| Reverse | TGGTCTACTTGTAGCAGGTGG | |
| CD59 | Forward | CTGCATTCTCCTTGGGTTC |
| Reverse | GTACCAGTCATAACCCGAAG | |
| DAF | Forward | CTTCAGAATGTGGTGTGGTC |
| Reverse | GTCTTCTGCTCCCAATAACC |
Reference gene for normalization
3. Results
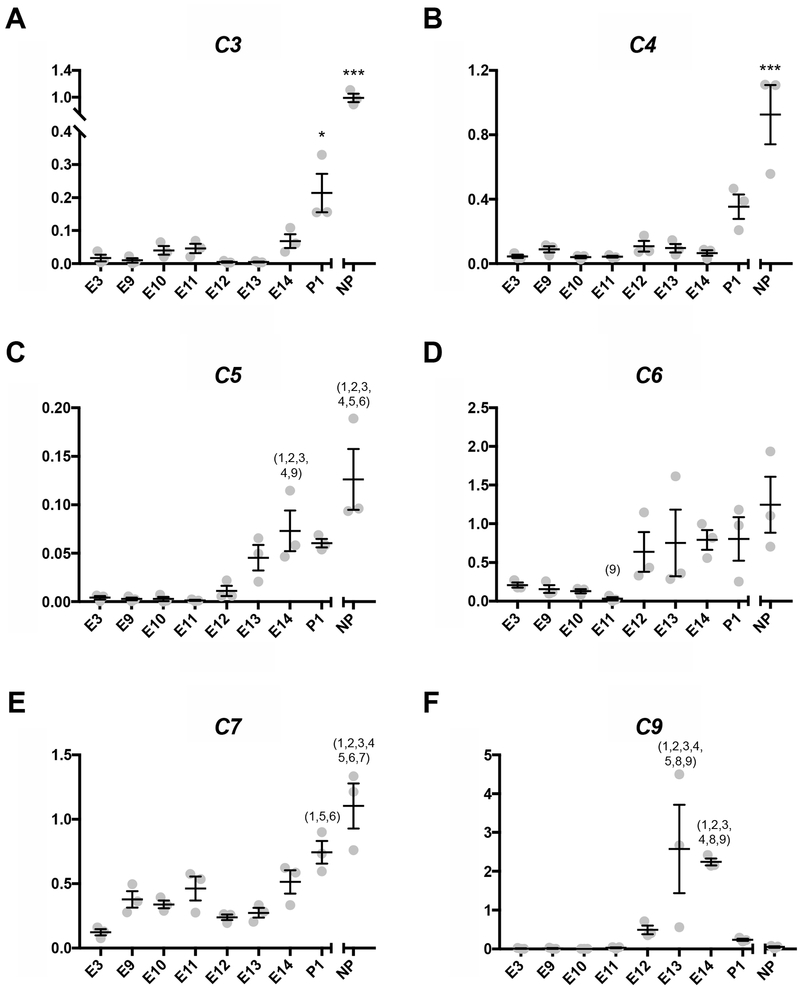
The gene encoding the C’ component C3 was one with significantly reduced uterine transcript levels at terminal pregnancy in the opossum, relative to that of non-pregnant controls (Hansen et al, 2016). C3 along with C4 are among the most ancient and conserved of the C’ components and are central to all three pathways of C’ activation (Al-Sharif et al., 1998; Sarma and Ward, 2011; Smith et al., 1996). Here we addressed whether C3 transcript levels were only reduced in opossum near parturition, or earlier in gestation. As evident from the results presented in Figure 1A and 1B, both C3 and C4 transcript levels were essentially undetectable and significantly lower than non-pregnant uterine control tissues throughout opossum pregnancy. However beginning on postnatal day 1 (P1), transcript levels were increasingly detectable, perhaps returning to non-pregnant (NP) levels (Figure 1A, B). The transcription of C3 and C4 was significantly lower in all pregnant and P1 relative to NP samples.
Figure 1.
Transcription levels of C’ component genes at the fetomaternal interface. The x-axis represents time points and the y-axis represents relative transcription normalized to reference genes. (*p < 0.05 for difference from all other time points, ***p < 0.0005 for difference from all other time points, 1p < 0.05 different from E3, 2p < 0.05 different from E9, 3p < 0.05 different from E10, 4p < 0.05 different from E11, 5p < 0.05 different from E12, 6p < 0.05 different from E13, 7p < 0.05 different from E14, 8p < 0.05 different from P1, 9p < 0.05 different from NP) (A) Transcription levels of C3. (B) Transcription levels of C4. (C) Transcription levels of C5. (D) Transcription levels of C6. (E) Transcription levels of C7. (F) Transcription levels of C9.
C5 through C9 are the terminal components of C’ cascade and form the membrane attack complex (MAC) that inserts into a cell membrane (Sarma & Ward, 2011; Serna et al., 2016). In the case of C5 there was low to undetectable transcript levels at time points E3 through E11 (Figure 1C). Transcript levels increased slightly on E12, E13, and E14 though not enough to be significant.
C5 transcript numbers remained level after birth on P1. NP levels of C5 transcripts were significantly different from all pregnancy time points but not P1. C5 transcript levels on E14 were significantly different from early the pregnancy time points (E3 through E11).
C6 transcript levels were low E3 through E11 and increased beginning on E12, though not significantly, and remaining relatively unchanged for the rest of pregnancy and parturition (Figure 1D). The only time points when C6 transcript levels were significantly different from each other were E11, when transcription was the lowest, and NP, when transcripts levels were the highest.
C7 transcripts levels generally trended towards increasing between early pregnancy (E3) and parturition (E14) although never reaching non-pregnant levels (Figure 1E). This trend continued into the first postnatal day, P1. NP samples had significantly higher transcript abundance for C7 than all pregnancy time points. P1 was significantly different from only those time points with the lowest C7 transcription: E3, E12, and E13.
C9 transcript levels were low or undetectable for early pregnancy time points E3 through E11 (Figure 1F). On E12 the transcript abundance increased slightly increased and then peaked on E13 and E14. This peak appears to be pregnancy specific since C9 transcript levels fell on P1 and are low in NP tissues.
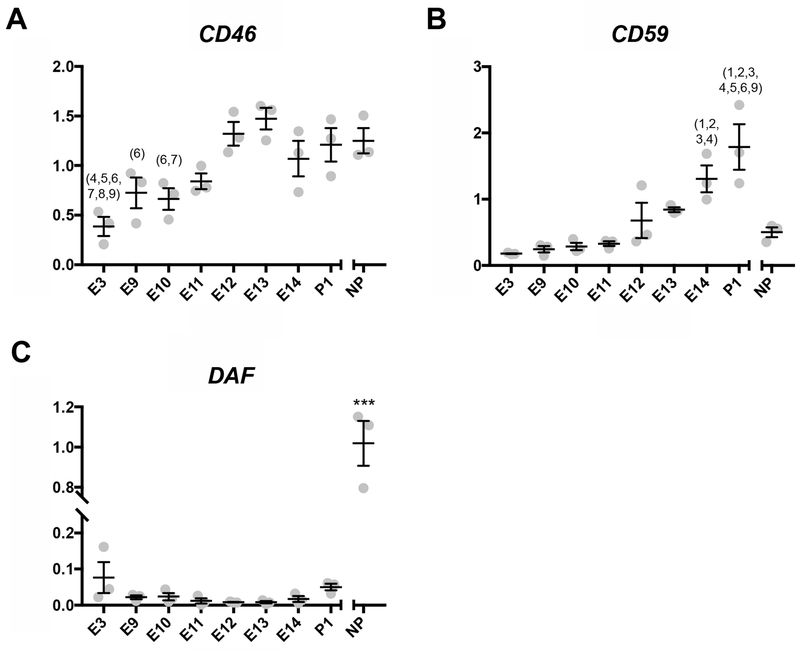
Previous analyses of the opossum uterine transcriptome at terminal pregnancy revealed that transcripts for Decay Accelerating Factor (DAF), an inhibitor of C3 convertase, were relatively low compared to non-pregnant (Hansen et al., 2016). Since CD46 and CD59 are additional C’ regulators that protect human pregnancy we examined transcript levels for these genes as well (Holmes et al., 1992). Between early pregnancy and post-attachment pregnancy the transcription of CD46 generally increased with the peak being at E13 (Figure 2A). CD46 transcription on E3 was the lowest observed and was significantly lower than pregnancy time points E11 through E14 as well as P1 and NP samples. CD59 transcription also increased throughout opossum pregnancy and peaked after parturition on P1 (Figure 2B). NP levels of transcription were most similar to those from early pregnancy though NP was significantly different from only P1. DAF had very low levels of transcription throughout opossum pregnancy and even after parturition (Figure 2C). NP samples had the highest level of DAF transcription and were significantly different from all other observed time points.
Figure 2.
Transcription levels of C’ regulatory protein genes at the fetomaternal interface. The x-axis represents time points and the y-axis represents relative transcription normalized to reference genes. (*p < 0.05 for difference from all other time points, ***p < 0.0005 for difference from all other time points, 1p < 0.05 different from E3, 2p < 0.05 different from E9, 3p < 0.05 different from E10, 4p < 0.05 different from E11, 5p < 0.05 different from E12, 6p < 0.05 different from E13, 7p < 0.05 different from E14, 8p < 0.05 different from P1, 9p < 0.05 different from NP) (A) Transcription levels of CD46. (B) Transcription levels of CD59. (C) Transcription levels of DAF.
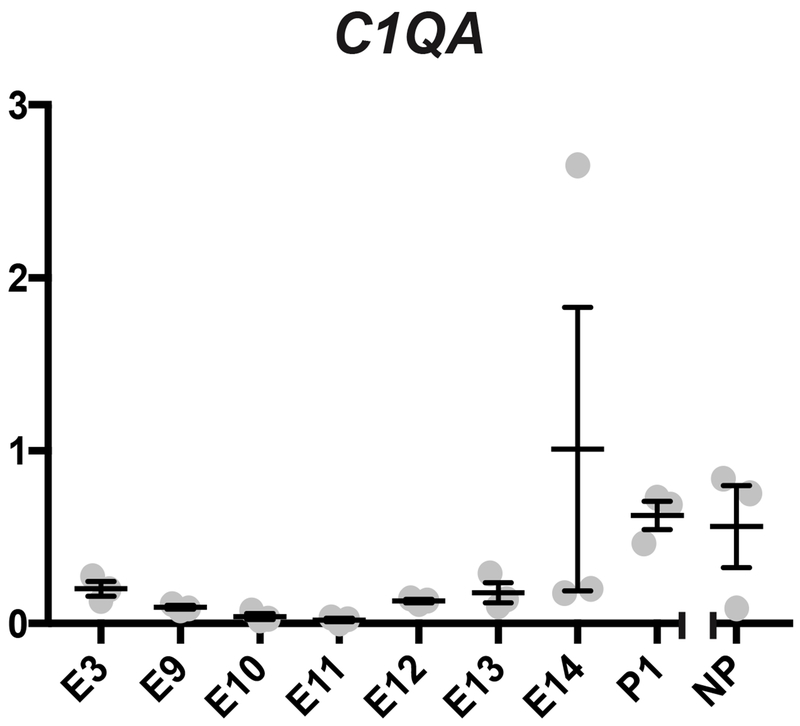
Lastly we also investigated transcripts encoding the C’ component C1q. C1q has been implicated as helpful in establishing a normal placentation in humans (Agostinis et al., 2017). C1q was not among the genes found to be differentially abundant in the analyses of the opossum uterine transcriptome at terminal pregnancy (Hansen et al., 2016). When earlier time points were investigated in the opossum, C1QA transcripts were low to nearly undetectable (Figure 3). There was a detectable rise in transcript levels late in pregnancy, reaching post-pregnancy levels, however there were no significant differences between any time points.
Figure 3.
Transcription of C’ component gene C1QA at the fetomaternal interface. The x-axis represents time points and the y-axis represents relative transcription normalized to reference genes. No significant differences were observed.
4. Discussion
Marsupials provide invaluable model species for exploring the evolution of mammalian reproduction (Laird et al., 2014; Kin et al., 2014; Lynch et al., 2015; Hansen et al., 2016, 2017; Griffiths et al., 2017; Guernsey et al., 2017). They represent a lineage of viviparous mammals that evolved in parallel with eutherian species (such as humans and mice) revealing both novel, lineage-specific innovations as well as ancient, conserved features. Noteworthy is recent research on the role of the immune system in the evolution of implantation, placentation, and parturition (Griffiths et al., 2017, Hansen et al., 2016, 2017). For example, in a recent study of opossum uterine transcriptomes, transcripts from immune related genes were to be found among those that have the greatest variance in abundance between pregnant and non-pregnant tissues (Hansen et al., 2016).
Genes encoding both effector and inhibitory components of the C’ arm of the immune system were among those whose transcripts stood out as being differentially abundant in opossum pregnancy (Hansen et al., 2016). These were considered noteworthy given abnormal C’ action during human pregnancy is associated with complications such as RSA, APS, and preeclampsia (Girardi et al., 2006; Lynch and Salmon, 2010; Tincani et al., 2010). However, systemic increase in C’ activation can be common in normal human pregnancy (Richani et al., 2005). Studying pregnancy related C’ activity in an array of viviparous species could provide insights into the role C’-activation plays in both normal and complicated human pregnancy. Furthermore, C’ genes and proteins are extraordinarily well-conserved across vertebrates, and some components are even present in invertebrate species (Goshima et al., 2016; Nonaka et al., 1999; Ong et al., 2015; Smith et al., 1996; Zhu et al., 2005). Therefore, vertebrate viviparity evolved in the presence of the C’ system and the potential harm it could pose to allogeneic fetal-membranes.
Here we assessed transcript abundance from C’ components genes C3, C4, C5, C6, C7, and C9 at different stages of opossum pregnancy. We focused primarily on transcription of C3 and downstream C’ components genes because C3 is one of the most central C’ components due to it being involved in the initiation of all three C’ pathways (Sarma and Ward, 2011). Most C’ component genes assessed had low transcription in pre-attachment pregnancy time points, and then started increasing transcription on or around E12 (Figures 1C,D,E,F). C’ components C3 and C4 were an exception with transcription remaining low throughout pregnancy and only significantly increasing after birth (Figures 1A,B). This may indicate that excessive transcription of C3 and C4 could harm opossum pregnancy. One of C3’s cleavage products, C3a, is an anaphylatoxin that can be as dangerous to fetal tissues as the MAC itself (Girardi et al. 2004; 2011). This is a good reason for C’ regulators that inhibit C3 convertases being present at the fetomaternal interface and clearly conserved.
In human trophoblast, C’ regulators DAF, CD46, and CD59 are expressed during pregnancy starting as early as six weeks into gestation (Holmes et al., 1992). CD46 inhibits C3 convertases in all three C’ pathways, including the Alternative Pathway (Liszewski et al., 1996). CD46 is transcribed during later M. domestica pregnancy at similar transcription levels as non-pregnant samples (Figure 2A). Therefore, there may be a need for protection from C3 convertases to an extent at the fetomaternal interface. Significantly lower transcription of CD46 in early pregnancy time points could be attributed to embryos not yet having attached to the maternal endometrium. It is also important to note that what we have called the CD46 gene in M. domestica is technically CD46-like and its nucleotide identity was not similar enough to human CD46 for a genetic analysis to name it as such (Ong et al., 2016). In addition, nucleotide sequence databases such as NCBI and Ensembl have the gene we assessed here as CD46 listed as CD46-like so this gene product may or may not serve the same function in opossums.
CD59 is a C’ regulator that prevents the polymerization of the C9 molecules that form the pore of the MAC (Davies and Lachmann, 1993). Of the three C’ regulators we analyzed here, CD59 showed the most evidence of being important to maintaining an opossum pregnancy. CD59 expression increased between successive opossum pregnancy time points and peaked after parturition on P1 (Figure 2C). CD59 is also expressed by human trophoblast cells as early as week 6 of gestation and in terminal pregnancy trophoblast as well (Holmes et al., 1992; Tedesco et al., 1993; Vanderpuye et al., 1993). Moreover, human trophoblast also expresses CD59 in greater abundance than CD46 and DAF which inhibit C’ action upstream in the C’ pathways (Holmes et al., 1992).
An unexpected result here was that DAF had reduced transcription during pregnancy than non-pregnant controls (Figure 2C). DAF inhibits C3 and C5 convertases in the Classical and Lectin pathways (Liszewski et al., 1996). If there is little to no C3 protein present during M. domestica pregnancy there may not be a need for DAF at the fetomaternal interface. In human trophoblast DAF is primarily expressed on cells that directly contact maternal circulation (Holmes et al., 1992). Given that in M. domestica the trophoblast invades maternal endometrium but does not break into maternal vessels (Freyer et al., 2007), perhaps DAF is not necessary at the fetomaternal interface in opossums, or perhaps other marsupials.
Since there is no current evidence that marsupial mothers produce anti-paternal antibodies capable of activating C’ during pregnancy the Classical Pathway may not be a danger to fetal tissues (van Oorschot & Cooper, 1988). In cross-strain mouse breeding where anti-paternal antibodies are produced in maternal circulation, the antibodies produced did not induce the Classical Pathway of C’ (Bell & Billington, 1980). However, in a mouse model of APS, antiphospholipid (aPL) antibodies were observed activating the Classical Pathway, which lead to pregnancy loss (Girardi et al., 2004). C5, but not C6, was required for aPL-mediated pregnancy loss in mice (Girardi et al., 2003; Redecha et al., 2007). This indicates damage was primarily due to inflammation induced by anaphylatoxin C5a, the cleavage product of C5 that does not become part of the MAC (Girardi et al., 2003; 2011). It is possible that M. domestica and other marsupials generate anti-paternal antibodies during normal pregnancy that simply do not activate the Classical Pathway.
C1q has been implicated as being helpful in establishing a normal placentation in humans and is important to fetal survival in mice (Agostinis et al., 2017; Singh et al. 2011). Indeed, preeclampsia-like symptoms develop in mouse mothers carrying C1q−/− fetuses (Singh et al. 2011). While we did examine the expression of one C1q gene, C1QA, the differences in transcription between any time points were not significant. It is possible that C1q that is not synthesized locally plays a role in placentation or attachment in the opossum. Alternatively, C1q may not be playing a role in early pregnancy in opossums and is not a conserved feature of mammalian viviparity.
Relative to non-pregnant uterus, transcription of key components of the C’ system, C3 and C4, appear to be suppressed in the pregnant opossum. This apparent active down-regulation of the C’ genes appears to start early in gestation and is maintained throughout pregnancy. However downstream C’ components C5, C6, and C7 have low transcription in early pregnancy and then increase after embryo attachment on E12. C9, the C’ component that forms the actual pore of the MAC, has a spike of expression near terminal pregnancy on E13 and E14. Transcription of the complement component that most directly inhibits C9, CD59, also has increased expression just before and after parturition. Immediately following parturition transcription of many of the C’ components return to “normal” or non-pregnant state. All this implies that there is potential harm to the embryo and/or fetus by the C’ system in marsupials and regulating C’ was part of the evolutionary process leading to viviparity early in mammals.
Highlights:
Many of the effector components of the complement system, such as C3 and C4, are expressed at very low levels in the opossum uterus during pregnancy, compared to non-pregnant tissue.
Some regulators of Complement activity such as CD59 are upregulated late in opossum pregnancy.
C1q, found to be relevant in human pregnancy, is not upregulated, a feature not conserved across mammals.
Results are consistent with some aspects of the immune system requiring regulation during pregnancy in a marsupial, addressing an old question of whether the maternal immune system requires regulation in a marsupial which lack an invasive placenta and having short pregnancy.
Features of marsupial pregnancy, a lineage of mammals last sharing a common ancestor with eutherians 160 million years ago, are conserved and ancient.
Acknowledgements
We would like to thank the staff of the UNM Biology Animal Research Facility for their assistance with husbandry and care of the M. domestica colony. This research was funded in part by a National Science Foundation award IOS-13531232. Research reported in this publication was also supported in part by the National Institute of General Medical Sciences of the National Institutes of Health under award P30 GM110907. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
The authors declare no conflicts of interests.
References
- Agostinis C, Tedesco F, and Bulla R (2017). Alternative functions of the complement protein C1q at embryo implantation site. J. Reprod. Immunol 119:74–80. doi: 10.1016/j.jri.2016.09.001. [DOI] [PubMed] [Google Scholar]
- Al-Sharif WZ, Sunyer JO, Lambris JD, and Smith LC (1998). Sea urchin coelomocytes specifically express a homologue of the complement component C3. J. Immunol 160, 2983–2997. [PubMed] [Google Scholar]
- Antczak DF, Miller JM, and Remick LH (1984). Lymphocyte alloantigens of the horse II. Antibodies to ELA antigens produced during equine pregnancy. J. Reprod. Immunol 6:283–297. doi: 10.1016/0165-0378(84)90028-7. [DOI] [PubMed] [Google Scholar]
- Baker ML, Wares JP, Harrison GA, and Miller RD (2004). Relationships among the families and orders of marsupials and the major mammalian lineages based on recombination activating gene-1. J. Mamm. Evol 11:1–16. doi: 10.1023/B:JOMM.0000029143.39776.ec. [DOI] [Google Scholar]
- Bell SC, and Billington WD (1980). Major anti-paternal alloantibody induced by murine pregnancy is non-complement-fixing IgG1. Nature. 288:387–388. doi: 10.1038/288387a0. [DOI] [PubMed] [Google Scholar]
- Bulla R, Bossi F, Radillo O, De Seta F and Tedesco F (2003). Placental trophoblast and endothelial cells as target of maternal immune response. Autoimmunity. 36:11–18. doi: 10.1080/0891693031000067331. [DOI] [PubMed] [Google Scholar]
- Chaouat GE, Kolb JP, Kiger NI, Stanislawski MA, and Wegmann TG (1985). Immunologic consequences of vaccination against abortion in mice. J. Immunol 134, 1594–1598. [PubMed] [Google Scholar]
- Croix DA, Samples NK, Vandeberg JL, and Stone WH (1989). Immune response of a marsupial (Monodelphis domestica) to sheep red blood cells. Dev. Comp. Immunol 13:73–78. doi: 10.1016/0145-305X(89)90019-0. [DOI] [PubMed] [Google Scholar]
- Cunningham DS, and Tichenor JR (1995). Decay-accelerating factor protects human trophoblast from complement-mediated attack. Clin. Immunol. Immunopathol 74:156–161. doi: 10.1006/clin.1995.1023. [DOI] [PubMed] [Google Scholar]
- Davies A, and Lachmann PJ (1993). Membrane defence against complement lysis: the structure and biological properties of CD59. Immunol. Res 12: 258–275. doi: 10.1007/BF02918257. [DOI] [PubMed] [Google Scholar]
- Denny KJ, Woodruff TM, Taylor SM, and Callaway LK (2013). Complement in pregnancy: a delicate balance. Am. J. Reprod. Immunol 69:3–11. doi: 10.1111/aji.12000. [DOI] [PubMed] [Google Scholar]
- Dishaw LJ, Smith SL, and Bigger CH (2005). Characterization of a C3-like cDNA in a coral: phylogenetic implications. Immunogenetics. 57:535–548. doi: 10.1007/s00251-005-0005-1. [DOI] [PubMed] [Google Scholar]
- Erlebacher A (2013). Immunology of the maternal-fetal interface. Annu. Rev. Immunol 31:387–411. doi: 10.1146/annurev-immunol-032712-100003. [DOI] [PubMed] [Google Scholar]
- Fakhouri F, Roumenina L, Provot F, Sallée M, Caillard S, Couzi L, Essig M, Ribes D, Dragon-Durey MA, Bridoux F and Rondeau E (2010). Pregnancy-associated hemolytic uremic syndrome revisited in the era of complement gene mutations. J. Am. Soc. Nephrol 21:859–867. doi: 10.1681/ASN.2009070706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulk WP, Jarret R, Keane M, Johnson PM, and Boackle RJ (1980). Immunological studies of human placentae: complement components in immature and mature chorionic villi. Clin. Exp. Immunol 40, 299–305. [PMC free article] [PubMed] [Google Scholar]
- Freyer C, Zeller U and Renfree MB (2007). Placental function in two distantly related marsupials. Placenta. 28:249–257. doi: 10.1016/j.placenta.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Girardi G, Berman J, Redecha P, Spruce L, Thurman JM, Kraus D, Hollmann TJ, Casali P, Caroll MC, Wetsel RA and Lambris JD (2003). Complement C5a receptors and neutrophils mediate fetal injury in the antiphospholipid syndrome. J. Clin. Invest 112:1644–1654. doi: 10.1172/JCI200318817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardi G, Bulla R, Salmon JE, and Tedesco F (2006). The complement system in the pathophysiology of pregnancy. Mol. Immunol 43:68–77. doi: 10.1016/j.molimm.2005.06.017. [DOI] [PubMed] [Google Scholar]
- Girardi G, Prohászka Z, Bulla R, Tedesco F, and Scherjon S (2011). Complement activation in animal and human pregnancies as a model for immunological recognition. Mol. Immunol 48:1621–1630. doi: 10.1016/j.molimm.2011.04.011. [DOI] [PubMed] [Google Scholar]
- Girardi G, Redecha P, and Salmon JE (2004). Heparin prevents antiphospholipid antibody-induced fetal loss by inhibiting complement activation. Nat. Med 10:1222–1226. doi: 10.1038/nm1121. [DOI] [PubMed] [Google Scholar]
- Goshima M, Sekiguchi R, Matsushita M, and Nonaka M (2016). The complement system of elasmobranches revealed by liver transcriptome analysis of a hammerhead shark, Sphyrna zygaena. Dev. Comp. Immunol 61:13–24. doi: 10.1016/j.dci.2016.03.009. [DOI] [PubMed] [Google Scholar]
- Hansen VL, Faber LS, Salehpoor AA, and Miller RD (2017). A pronounced uterine pro-inflammatory response at parturition is an ancient feature in mammals. Proc. R. Soc. Lond. B. Biol. Sci. 284: 20171694. doi: 10.1098/rspb.2017.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen VL, Schilkey FD, and Miller RD (2016). Transcriptomic changes associated with pregnancy in a marsupial, the gray short-tailed opossum Monodelphis domestica. PLoS One. 11:e0161608. doi: 10.1371/journal.pone.0161608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes CH, Simpson KL, Okada H, Okada N, Wainwright SD, Purcell DFJ, and Houlihan JM (1992). Complement regulatory proteins at the feto-maternal interface during human placental development: distribution of CD59 by comparison with membrane cofactor protein (CD46) and decay accelerating factor (CD55). Eur. J. Immunol 22:1579–1585. doi: 10.1002/eji.1830220635. [DOI] [PubMed] [Google Scholar]
- Koppenheffer TL, Spong KD, and Falvo HM (1998). The complement system of the marsupial Monodelphis domestica. Dev. Comp. Immunol 22:231–237. doi: 10.1016/S0145-305X(98)00015-9. [DOI] [PubMed] [Google Scholar]
- Liszewski MK, Farries TC, Lublin DM, Rooney IA, and Atkinson JP (1996). Control of the complement system. Adv. Immunol 61, 201–283. [DOI] [PubMed] [Google Scholar]
- Lynch AM, and Salmon JE (2010). Dysregulated complement activation as a common pathway of injury in preeclampsia and other pregnancy complications. Placenta. 31:561–567. doi: 10.1016/j.placenta.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao D, Wu X, Deppong C, Friend LD, Dolecki G, Nelson DM, and Molina H (2003). Negligible role of antibodies and C5 in pregnancy loss associated exclusively with C3-dependent mechanisms through complement alternative pathway. Immunity. 19:813–822. doi: 10.1016/S1074-7613(03)00321-2. [DOI] [PubMed] [Google Scholar]
- Medawar PB (1953). Some immunological and endorinological problems raised by the evolution of vivparity in vertebrates. Symp. Soc. Exp. Biol 7, 320–338. [Google Scholar]
- Meredith RW, Janečka JE, Gatesy J, Ryder OA, Fisher CA, Teeling EC, Goodbla A, Eizirik E, Simão TL, Stadler T and Rabosky DL (2011). Impacts of the cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science. 334:521–524. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
- Nonaka M, Azumi K, Ji X, Namikawa-Yamada C, Sasaki M, Saiga H, Dodds AW, Sekine H, Homma MK, Matsushita M and Endo Y (1999). Opsonic complement component C3 in the solitary ascidian, Halocynthia roretzi. J. Immunol 162, 387–391. [PubMed] [Google Scholar]
- Ong OT, Young LJ, and Old JM (2015). Detection of an active complement system in red-tailed phascogales (Phascogale calura). Comp. Clin. Path 24:1527–1534. doi: 10.1007/s00580-015-2111-2. [DOI] [Google Scholar]
- Ong OT, Young LJ, and Old JM (2016). Preliminary genomic survey and sequence analysis of the complement system in non-eutherian mammals. Aust. Mammal 38:80–90. doi: 10.1071/AM15036. [DOI] [Google Scholar]
- Orgad S, Loewenthal R, Gazit E, Sadetzki S, Novikov I, and Carp H (1999). The prognostic value of anti-paternal antibodies and leukocyte immunizations on the proportion of live births in couples with consecutive recurrent miscarriages. Hum. Reprod 14:2974–2979. doi: 10.1093/humrep/14.12.2974. [DOI] [PubMed] [Google Scholar]
- Richani K, Soto E, Romero R, Espinoza J, Chaiworapongsa T, Nien JK, Edwin S, Kim YM, Hong JS and Mazor M (2005). Normal pregnancy is characterized by systemic activation of the complement system. J. Matern. Fetal Neonatal Med 17:239–245. doi: 10.1080/14767050500072722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodger JC, Fletcher TP, and Tyndale-Biscoe CH (1985). Active anti-paternal immunization does not affect the success of marsupial pregnancy. J. Reprod. Immunol 8:249–256. doi: 10.1016/0165-0378(85)90044-0. [DOI] [PubMed] [Google Scholar]
- Rollins SA, and Sims PJ (1990). The complement-inhibitory activity of CD59 resides in its capacity to block incorporation of C9 into membrane C5b-9. J. Immunol 144, 3478–3483. [PubMed] [Google Scholar]
- Samstein RM, Josefowicz SZ, Arvey A, Treuting PM, and Rudensky AY (2012). Extrathymic generation of regulatory T cells in placental mammals mitigates maternal-fetal conflict. Cell. 150:29–38. doi: 10.1016/j.cell.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma JV, and Ward PA (2011). The complement system. Cell Tissue Res. 343:227–235. doi: 10.1007/s00441-010-1034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Ahmed A, and Girardi G (2011). Role of complement component C1q in the onset of preeclampsia in mice. Hypertension. 70:111. doi: 10.1161/HYPERTENSIONAHA.111.175919. [DOI] [PubMed] [Google Scholar]
- Segura-Cervantes E, Mancilla-Ramirez J, Zurita L, Paredes Y, Arredondo JL, and Galindo-Sevilla N (2015). Blood SC5b-9 complement levels increase at parturition during term and preterm labor. J. Reprod. Immunol 109:24–30. doi: 10.1016/j.jri.2015.02.008. [DOI] [PubMed] [Google Scholar]
- Serna M, Giles JL, Morgan BP, and Bubeck D (2016). Structural basis of complement membrane attack complex formation. Nat. Commun. 7:10587. doi: 10.1038/ncomms10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LC, Chang L, Britten RJ, and Davidson EH (1996). Sea urchin genes expressed in activated coelomocytes are identified by expressed sequence tags. Complement homologues and other putative immune response genes suggest immune system homology within the deuterostomes. J. Immunol 156, 593–602. [PubMed] [Google Scholar]
- Sunyer JO, and Lambris JD (1998). Evolution and diversity of the complement system of poikilothermic vertebrates. Immunol. Rev 166:39–57. doi: 10.1111/j.1600-065X.1998.tb01251.x. [DOI] [PubMed] [Google Scholar]
- Tedesco F, Narchi G, Radillo O, Meri S, Ferrone S, and Betterle C (1993). Susceptibility of human trophoblast to killing by human complement and the role of the complement regulatory proteins. J. Immunol 151, 1562–1570. [PubMed] [Google Scholar]
- Tincani A, Cavazzana I, Ziglioli T, Lojacono A, De Angelis V, and Meroni P (2010). Complement activation and pregnancy failure. Clin. Rev. Allergy Immunol 39:153–159. doi: 10.1007/s12016-009-8183-5. [DOI] [PubMed] [Google Scholar]
- Van Oorschot RAH, and Cooper DW (1988). Lack of evidence for complement-dependent cytotoxic antibodies to fetal paternally derived antigens in the marsupial Macropus eugenii (tammar wallaby). Am. J. Reprod. Immunol. Microbiol 17:145–148. doi: 10.1111/j.1600-0897.1988.tb00219.x. [DOI] [PubMed] [Google Scholar]
- Vanderpuye OA, Labarre CA, and McIntyre JA (1993). Expression of CD59, a human complement system regulator protein, in extraembryonic membranes. Int. Arch. Allergy Immunol 101:376–384. doi: 10.1159/000236480. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, and Speleman F (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:research0034-1. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz GH, and Westfall SA (1967). Immune complement of the opossum. Immunochemistry. 4:61–63. doi: 10.1016/0019-2791(67)90198-X. [DOI] [PubMed] [Google Scholar]
- Xu C, Mao D, Holers VM, Palanca B, Cheng AM, and Molina H (2000). A critical role for murine complement regulator Crry in fetomaternal tolerance. Science. 287(5452), 498–501. doi: 10.1126/science.287.5452.498. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Thangamani S, Ho B, and Ding JL (2005). The ancient origin of the complement system. EMBO J. 24:382–394. doi: 10.1038/sj.emboj.7600533. [DOI] [PMC free article] [PubMed] [Google Scholar]