Abstract
RAS is the most frequently mutated oncogene in cancer and a critical driver of oncogenesis. Therapeutic targeting of RAS has been a goal of cancer research for more than 30 years due to its essential role in tumor formation and maintenance. Yet the quest to inhibit this challenging foe has been elusive. Although once considered “undruggable”, the struggle to directly inhibit RAS has seen recent success with the development of pharmacological agents that specifically target the KRAS(G12C) mutant protein, which include the first direct RAS inhibitor to gain entry to clinical trials. However, the limited applicability of these inhibitors to G12C-mutant tumors demands further efforts to identify more broadly efficacious RAS inhibitors. Understanding allosteric influences on RAS may open new avenues to inhibit RAS. Here, we provide a brief overview of RAS biology and biochemistry, discuss the allosteric regulation of RAS, and summarize the various approaches to develop RAS inhibitors.
Keywords: RAS inhibitor, cancer, signal transduction, drug discovery, GTPase, Monobody
1. Introduction
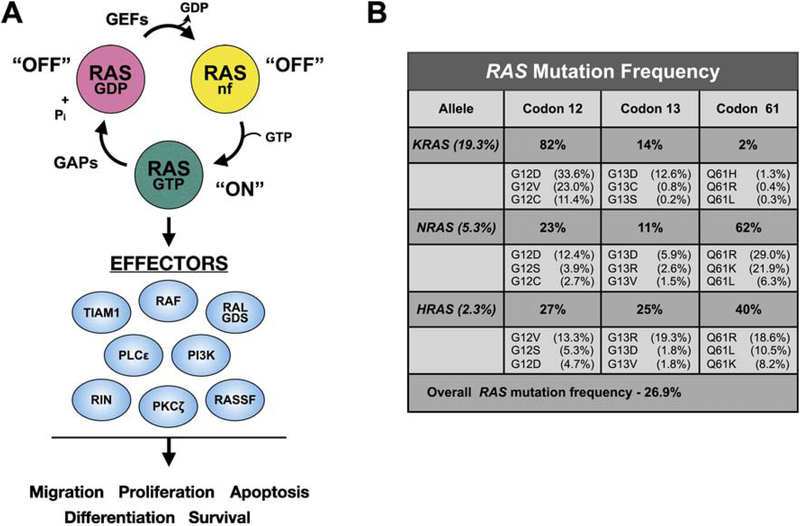
The RAS family of GTPases attracts attention even after more than three decades of discovery due to their central role in cell signaling and tumorigenesis. RAS is a GTP-regulated molecular switch relaying upstream signals from membrane receptors to a variety of downstream effector molecules (Fig. 1A) [1, 2]. Normally, RAS cycles between an inactive GDP-bound state, a transient nucleotide-free state, and the active GTP-bound state that engages effectors to initiate various signaling cascades that generally promote cell growth and survival [3]. The balance between these states is regulated by guanine nucleotide exchange factors (GEFs) that promote the release of GDP, and GTPase activating/accelerating proteins (GAPs) that enhance the relatively poor intrinsic GTPase activity of RAS. Mutational activation of RAS, as well as a number of its effectors, is observed in numerous human cancers. Indeed, mutations in RAS are the most common oncogenic driver mutations in human cancer, present in approximately 30% of all cancers (Fig. 1B)[4]. These oncogenic mutations occur at several “hot spots” (codons 12, 13, and 61; Figs. 1B and 2) which impair GTPase activity and interaction with GAPs, thereby shifting the RAS equilibrium to favor the GTP-bound state, resulting in constitutive engagement and activation of downstream effector pathways [5].
Figure 1. RAS signaling and mutational activation in cancer.
A. GTPase cycle. RAS proteins normally reside in the inactive, GDP-bound state. Mitogenic stimulation results in recruitment of GEFs to the plasma membrane and binding of RAS. This results in destabilization of nucleotide binding leading to release of GDP and creation of a transient nucleotide free state. Due to the high concentration of GTP in cells relative to GDP, RAS proteins load with GTP resulting in the switch to the active state. RAS-GTP recruits and activates a number of downstream targets, including RAF and PI3K. Termination of RAS signaling occurs through hydrolysis of GTP to GDP which is facilitated by GTPase accelerating/activating proteins that enhance the relatively poor intrinsic GTPase activity of RAS by nearly 100-fold, thereby returning RAS to the inactive, GDP-bound state. B. RAS mutation frequency in human tumors. Data were compiled from the Catalogue of Somatic Mutations (COSMIC), v86 [174]. Frequency of mutations in each RAS gene is shown in the first column. The percentage of mutations in each codon hotspot is indicated to the right of each gene. The frequency of the top three most prevalent amino acid substitutions at the indicated codon is indicated below the mutation frequency for that codon hotspot. These mutational hot spots all reside in the effector lobe of RAS.
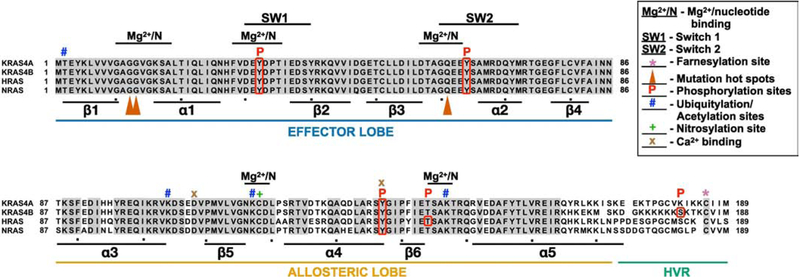
Figure 2. RAS family members.
RAS proteins were aligned with Clustal multiple alignment. KRAS4A and KRAS4B are derived from alternative splicing of the same gene resulting in different C-termini. Grey shading highlights residues that are identical in all four RAS proteins. RAS proteins can be divided into three functional regions: the effector lobe, allosteric lobe, and hypervariable region (HVR). SW1, switch 1 region (aa 30–40); SW2, switch 2 region (aa 60–76); Mg2+/N, magnesium and nucleotide binding regions, *, farnesylation site; , mutation hotspots; P, phosphorylation site; #, ubiquitylation or acetylation sites; +, nitrosylation site; x, Ca2+ binding sites. Alpha helices (α) and beta sheets (β) are indicated below lineup.
Despite decades of effort, the ongoing quest to develop therapeutic inhibitors of oncogenic RAS has met with many challenges. Two primary reasons have been proposed: First, RAS has a picomolar affinity for guanine nucleotide, while the cellular concentration of guanine nucleotides is in the millimolar range making it unfavorable for the binding of nucleotide analogs [6]. Second, outside of the nucleotide binding pocket RAS appears to lack deep pockets amenable to the binding of small molecules [7]. Nonetheless, continued research has led to a number of innovative strategies for targeting allosteric sites on RAS. Below, we describe RAS allostery and the potential therapeutics that have been developed to inhibit RAS through novel mechanisms.
2. RAS Biochemistry at a Glance
Humans have three RAS genes: HRAS, NRAS, and KRAS. Due to alternative splicing in the KRAS gene, the three genes encode four distinct yet highly homologous ~21 kDa proteins: HRAS, NRAS, KRAS4A and KRAS4B, with KRAS4B representing the major KRAS isoform [8]. Although all three RAS oncogenes are potently transforming in model systems, KRAS accounts for 83% of RAS mutations in human cancers, with NRAS mutated in roughly 13% and HRAS 4% of tumors (Fig. 1B). This imbalance is also reflected by a difference in the spectrum of RAS mutations in specific tumor types. For example, KRAS is mutated in nearly 100% of pancreatic ductal adenocarcinoma (PDAC), with frequent mutational activation in lung (30%) and colorectal cancers (CRC)(45%) [9]. In contrast, mutations in HRAS and NRAS are rarely observed in these malignancies. Both KRAS and NRAS mutations are observed at roughly equivalent frequencies (23% and 20%, respectively) in multiple myeloma, whereas NRAS mutations predominate in melanoma (28% vs 0.8% KRAS and 1% HRAS) [9]. In addition, each RAS isoform exhibits a distinct bias in codon mutations: codon 12 mutations predominate in KRAS (82%), whereas NRAS is most frequently mutated at codon 61 (62%) [5]. Interestingly, the distribution of activating mutations at codons 12, 13, and 61 in HRAS are relatively similar (27%, 25%, and 40%, respectively) [5].
Structurally, RAS is comprised of 2 domains: the catalytic or G-domain (residues 1–172) and the highly divergent hyper variable region (HVR; residues 173–188/9)(Fig. 2) [5]. The four isoforms of human RAS proteins differ primarily in the sequence of the HVR. The G domain of RAS is highly conserved, with residues 1–86 invariant among the four RAS proteins (Fig. 2). This region, termed the effector lobe, includes resides critical for RAS function including switch 1 (SW1; residues 30–40) and switch 2 (SW2; residues 60–76) which engage effectors [10]. Residues 87–172 constitute the allosteric lobe, which shares approximately 86% identity across RAS isoforms (Fig. 2; [11]). This region, along with the HVR, interacts with membranes and contains all the isoform specific differences.
Upon GTP loading, SW1 and SW2 undergo marked structural changes, reordering to form an interaction surface that engages specific RAS effector proteins. Switch 1 exists in either an open or closed conformation (called state 1 and state 2, respectively), which differ in their biochemical activities [12]. In state 1, effector binding is diminished, while nucleotide exchange is favored. Conversely, in state 2, effector binding and GTP hydrolysis are promoted. Further, in state 2, SW1 is stabilized by effector binding and mobilizes SW2 to convert between two states corresponding to a catalytically incompetent (T state) and catalytically active (R state) conformation [13]. Stabilizing state 1 conformation should interrupt RAS-effector interaction. Indeed, compounds like Zn-cyclen and Cu-cyclen selectively target the state 1 conformation thereby perturbing RAS-RAF interactions [14, 15]. However, these transition metal cyclenes are not viable therapeutic options for RAS inhibition in vivo due to their non-drug nature.
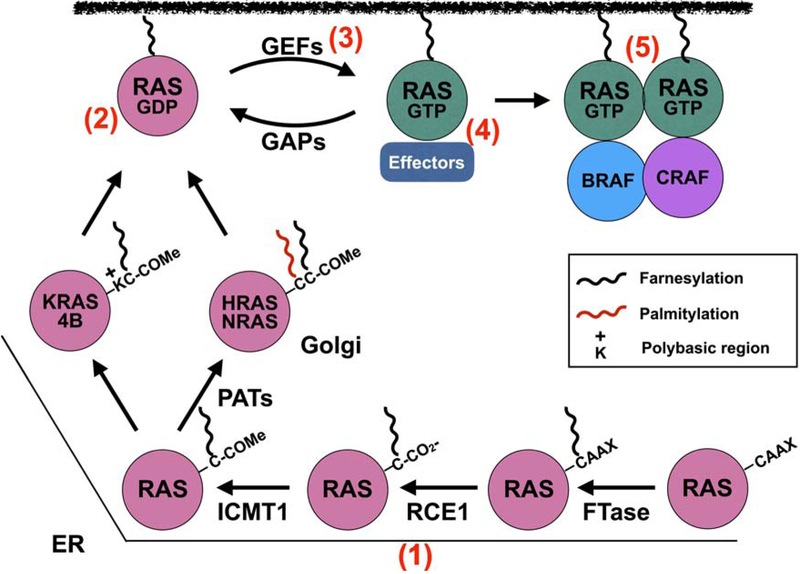
The biological activity of RAS is dependent on localization of the protein to discrete membrane microdomains, dictated in part by the HVR. The COOH-terminus of RAS consists of the conserved CaaX motif (Cys, aliphatic, aliphatic, any residue) that is post-translationally modified by the covalent attachment of a farnesyl group to the Cys of the CaaX by the fanesyl transferase (FTase; Fig. 2) [16]. Farnesyl modified RAS accumulates on the cytoplasmic face of the endoplasmic reticulum (ER) where it encounters RAS-converting enzyme 1 (RCE1), an endoprotease that removes the -aaX amino acids [17]. RAS is then modified by another ER-resident enzyme, isoprenylcysteine carboxylmethyltransferase (ICMT), that catalyzes the methyl esterification of the α- carboxyl group of the farnesylcysteine [17]. The next steps in RAS lipidation are isoform specific and involve elements within the HVR that are upstream of the CaaX motif to palmitoylat HRAS, NRAS, and KRAS4A in contrast to KRAS4B, which possesses a polybasic stretch of amino acids that interact with the negatively charged phospholipids of the plasma membrane [18–20].
RAS regulates a diverse array of cellular processes including gene expression, proliferation, cell survival, and differentiation utilizing a variety of effectors to mediate these diverse biological actions (Fig. 1A). The RAF and PI3 kinase families of RAS effectors are key mediators of oncogenic RAS activity [21, 22]. RAS activation of RAF initiates a phosphorylation cascade that results in phosphorylation and activation of the extracellular signal-regulated kinase (ERK), which in turn phosphorylates multiple cytoplasmic proteins, cytoskeletal proteins, and transcription factors that initiate genetic programs associated with cell growth and survival [23]. Similarly, the p110 catalytic subunits (p110α, β, δ, and γ) of class I phosphoinositide 3-kinases (PI3Ks) generate phosphoinositol triphosphate (PIP3), which activates AKT and downstream transcription factors including NF-κB and forkhead transcription factors (FOXOs) that are involved in cell survival and DNA damage and repair, respectively [24, 25]. In addition to RAF and PI3K, RAS activates a number of additional effectors including several GEFs that regulate additional RAS-family GTPases including the Ral and the RHO families [26]. While RHO family GTPases contribute to many of the morphological changes engendered by RAS activation, RAL has emerged as a critical RAS-stimulated pathway in human tumor cells as part of the oncogenic process [27].
An emerging theme in RAS biology is the context-dependent activation of signaling pathways. For example, RAF, but not PI3K or RalGEF, activation was sufficient for RAS-mediated transformation of NIH/3T3 mouse fibroblasts [28]. However, RalGEF, but not RAF or PI3K, activation was sufficient and necessary for RAS-mediated transformation of HEK human embryonic kidney cells and a variety of other human cells including fibroblasts [28]. RAS isoforms differentially activate downstream signaling cascades. For example, all oncogenic RAS isoforms potently activate ERK-MAPK signaling, however, KRAS activates the lipid kinase PI3Kα/AKT signaling more effectively than H/NRAS [29].
The context dependence of RAS signaling has also been observed under different growth conditions. Deletion of KRAS in a panel of isogenic endometrial lines did not impact MAPK or PI3K signaling in 2D culture conditions yet was sufficient to inhibit anchorage-independent growth of these cells [30]. Indeed, the signaling pattern observed in isogenic cell lines harboring RAS mutations did not match the pattern observed upon ectopic overexpression of RAS. These differences may stem in part from the high levels of RAS overexpression in the transfected lines compared to levels of endogenous RAS in isogenic tumor lines [31]. In addition, different mutations in the same RAS isoform elicit distinct signaling and biological outputs. Pharmacological inhibition of K-RAS(G12C) mutant cells with the G12C-specific inhibitor ARS-1620 suppressed the growth of cells when cultured as spheroids but had varied effects on cells grown under 2D-adherent conditions [32]. We have also observed different patterns of signaling upon RAS inhibition in 2D vs 3D growth conditions [33]. These findings further highlight the context-dependence of RAS signaling and suggest that 2D tissue culture conditions may not accurately reflect the signaling pathways engaged during RAS dependent tumorigenesis.
These context-dependent effects are also observed in vivo. Using CAS9 gene editing approaches, Winslow and colleagues observed distinct profiles of oncogenic codon 12 and 13 mutations in different tissues [34]. The KRAS mutational profile in the pancreas mirrored the mutational frequencies observed in human PDAC patients. However, the mutational spectrum in mouse lung tumors did not correlate with that observed in human lung cancers. Surprisingly, KRAS(G12R) and KRAS(G13R) were more frequent than KRAS(G12C) in mouse lung tumors. These differences in KRAS variants between mouse and human lung cancer are likely a product of both biochemical differences of the various mutants as well as the mutational processes that gave rise to the mutations in vivo. For example, KRAS(G12C) is the most common mutation in current/former smokers whereas KRAS(G12D) is the predominant mutation in non-smokers [34]. Haigis and colleagues demonstrated that KRAS(G12D) and KRAS(A146T) exhibit distinct biologic and signaling outputs in the pancreas and colon suggesting that such tissue specific phenotypes are due to allele-specific signaling properties and engagement of different signaling pathways by various RAS mutants. Thus, all RAS mutants are not created equal [35].
3. Allosteric modulation of RAS function
It is well-established that GAPs and GEFs affect RAS function by accelerating GTP hydrolysis and GDP/GTP exchange, respectively. However, RAS function is modulated through additional, less well appreciated allosteric mechanisms. This section briefly summarizes the current state of knowledge on allosteric regulation of RAS by post-translational modifications (PTMs), ionic interactions, and protein:membrane interactions.
3.1. Post-Translational Modifications
In addition to lipidation and carboxymethylation of the HVR, RAS proteins undergo a number of additional post-translational modifications that may modulate the activity or lifetime of RAS proteins.
3.1.1. Phosphorylation
Phosphorylation is a common mechanism of protein regulation [36]. Although viral RAS undergoes an autophosphorylation reaction in which GTP serves to transfer a phosphate to Thr59, cellular RAS proteins lack such modification due to the presence of Ala at this position [37–39]. Nevertheless, RAS proteins are regulated by phosphorylation. Perhaps the best known example of this is phosphorylation of KRAS by PKC on Ser181 (Figs. 2, 3A). Ser181-phosphorylated KRAS redistributes from the plasma membrane to intracellular membranes, including the endoplasmic reticulum (ER) and mitochondrial outer membrane, where pSer181-KRAS interacts with Bcl-XL to promote apoptosis [40]. The cytotoxicity of pSer181-KRAS was mediated by Bcl-XL-dependent direct interaction of pSer181-KRAS with inositol triphosphate receptors (IP3Rs) on the ER which inhibited Bcl-XL regulated Ca2+ flux between the ER and mitochondria and thereby inhibited mitochondrial respiration [41].
Figure 3. Targeting RAS inhibition.
RAS proteins are initially farnesylated at the ER by farnesyl transferase (FTase) and then HRAS, NRAS, and KRAS4A are translocated to the Golgi where they undergo a second lipidation event mediated by palmitoylacetyl transferases (PATs). KRAS4B possesses a polybasic stretch of Lys residues (K) which function in conjunction with the farnesylated C-term to anchor KRAS4B to the membrane. Many approaches have been utilized to inhibit RAS function from blocking lipidation and membrane association of RAS proteins (e.g., tipifarnib, deltasonamide) (1), interfering with RAS activation by targeting RAS-GDP (e.g., MRTX849, ARS-1620), (2) interaction with GEFs (e.g., DCAI, HBS) (3), interfering with RAS:effector interactions (e.g., ADB7, 3144)(4), and allosteric inhibition of RAS self-association/nanoclustering (e.g. NS1 and K13)(5).
All three RAS isoforms are also phosphorylated on Tyr137 by Abelson tyrosine kinase (ABL) (pY137; Figs. 2, 3A) resulting in a conformational change that enhances binding to RAF [42]. Thus, pY137 may potentiate RAS signaling through long-range allosteric changes communicated between the allosteric lobe and the effector binding region of SW2. RAS activation of its effector, RAS- and RAB-interacting protein 1 (RIN1), activated its own downstream effectors, including ABL, which may serve as a positive feed-back mechanism to enhance RAS signaling [42, 43].
Phosphorylation also negatively regulates RAS signaling. Glycogen synthase kinase 3β (GSK3β) phosphorylated HRAS on Thr144/148 (Fig. 2, 3A), resulting in polyubiquitylation via recruitment of β-TrCP–E3 ligase and subsequent RAS degradation [44]. In another example, Src specifically phosphorylated active GTP-bound H/N-RAS on Tyr32, which reduced binding to RAF and enhanced binding to GAPs, thus accelerating GTP hydrolysis and promoting RAS inactivation [45]. Importantly, this phosphorylation event was reversed by the phosphatase SHP2, which dephosphorylated H/N-RAS and rescued RAS activation (Figs. 2, 3A) [46]. In a recent study, Src was shown to directly phosphorylate KRAS on Tyr32 and Tyr64, altering the conformations of SW1 and SW2, respectively [47]. Interestingly, while this PTM accelerated the intrinsic exchange rate, it also inhibited SOS-mediated nucleotide exchange as well as both intrinsic and GAP-mediated GTP hydrolysis. As a result, phosphorylated KRAS was decoupled from upstream signaling. In addition, di-phosphorylated, GTP-loaded WT and mutant KRAS displayed markedly reduced binding to RAF, suggesting that Src phosphorylation of KRAS inhibited downstream signaling [47]. Conversely, as previously described for H/N-RAS, SHP2-mediated dephosphorylation of KRAS promoted KRAS downstream signaling. These findings firmly support the role of SHP2 as a direct RAS activator. Indeed, SHP2 inhibition reduced the growth of a number of RAS-mutant tumors including glioblastoma, lung cancer, and pancreatic ductal adenocarcinoma, as well as tumors driven by amplification of wild type KRAS such as gastroesophageal cancer (Table 1) [46, 48–51]. Taken together, these data suggest that enhancing phosphorylation of RAS may be a valid strategy to inhibit RAS-dependent tumors. Indeed, several clinical trials have been initiated with SHP2 inhibitors (ClinicalTrials.gov Identifiers: ; ; -see Table 1).
Table 1.
Indirect Inhibitors of RAS.
| Name | Potency | Type | Binding site | Mechanism of action | Model tested | Clinical trials | References |
|---|---|---|---|---|---|---|---|
| II-B08 | IC50 = 5.5 μM | SHP2 inhibitor | Catalytic domain | ↑pY32/↓effector binding | Cell culture; mouse glioblastoma model | [46] | |
| 11a-1 | IC50 = 200 nM | SHP2 inhibitor | Catalytic domain | ↑pY32/↓effector binding | 3D cell culture | [47] | |
| PHPS1 | IC50 = 2.1 μM; Ki = 0.73 μM |
SHP2 inhibitor | Catalytic domain | ↑pY32/↓effector binding | Ex vivo 3D-transdifferentiation assay | [49] | |
| GS493 | IC50 = 71 nM | SHP2 inhibitor | Catalytic domain | ↑pY32/↓effector binding | Cell culture; mouse pancreatic and lung cancer models | [49] | |
| SHP099 | IC50 = 71 nM | SHP2 inhibitor | N-terminal/C-terminal/phosphatase domain interface | ↑pY32/↓effector binding | Cell culture; mouse pancreatic, lung, ovarian, and breast cancer models | [47–51,175] | |
| cmpd #57 | SHP2 inhibitor | SHP2 | ↑pY32/↓effector binding | Cell culture | [48] | ||
| TNO155 | SHP2 inhibitor | SHP2 | ↑pY32/↓effector binding | Phase I clinical trial dose finding study for patients with advanced solid tumors | |||
| JAB-3068 | SHP2 inhibitor | SHP2 | ↑pY32/↓effector binding | Phase I clinical trial dose finding study for patients with advanced solid tumors | |||
| RMC-4630 | SHP2 inhibitor | SHP2 | ↑pY32/↓effector binding | Phase I clinical trial dose finding study for patients with advanced relapsed or refractory solid tumors | |||
| Tipifarnib (R115777) | IC50 = 7.9 nM | FTase inhibitor | FTase | ↓ RAS farnesylation and membrane attachment | Cell culture; mouse models; clinical trials; currently in phase II clinical trials for head and neck tumors | Used in over 80 trials for various cancers; currently: , | [78,81] |
| Lonafarnib (SCH-66336) | IC50 = 1.9 nM | FTase inhibitor | FTase | ↓ RAS farnesylation and membrane attachment | Cell culture; mouse models; cancer clinical trials; progeria clinical trials | Used in nearly 40 clinical trials for various cancers, progeria, and Hep D. progeria: | [79,81,88,89] |
| Deltarasin | Kd = 41 nM | PDEδ inhibitor | Prenyl-binding pocket | ↓ KRAS/PDEδ interaction and RAS trafficking to the plasma membrane | Cell culture; mouse pancreatic cancer model | [99] | |
| Deltazinone 1 | Kd = 8 nM | PDEδ inhibitor | Prenyl-binding pocket | ↓ KRAS/PDEδ interaction and RAS trafficking to the plasma membrane | Cell culture | [100] | |
| Deltasonamide 1 and 2 | Kd = 203 (1) Kd = 385 (2) |
PDEδ inhibitor | Prenyl-binding pocket | ↓ KRAS/PDEδ interaction and RAS trafficking to the plasma membrane | Cell culture | [101] | |
| Bisphosphates/zoledronic acid | FPPS inhibitor | FPPS | ↓ Farnesyl and geranylgeranyl lipid synthesis, RAS lipidation, and membrane localization | Cell culture; mouse models and human trials of pancreatic, prostate, lung, and breast cancer; clinical trials | Used in 100 s of clinical trials, primarily for bone-related maladies | [93–95,176–179] |
3.1.2. Acetylation
Acetylation is another common PTM. The acetylation reaction involves transfer of an acetyl group from the metabolite acetyl-coenzyme A to either the α-amino group on a protein’s N-terminus or the ε-amino group of lysine residues [52]. KRAS was initially reported to be acetylated at lysine 104 although substitution of Ala at Lys104 (non-acetylatable) had no effect on cell proliferation in comparison to cells expressing unsubstituted KRAS(G12V) (Fig. 2)[53]. However, KRAS(G12V) with a K104Q mutation (mimicking acetylation) displayed reduced GEF-induced nucleotide exchange and reduced transforming activity in NIH3T3 cells [53]. Additional studies have reported acetylation sites at Lys 104 and Lys 147 [54]. However, incorporation of acetyl-lysine into KRAS using a genetic-code expansion approach demonstrated that modification of this residue did not alter SOS-mediated nucleotide exchange of the modified protein [54]. These results along with additional studies of Lys 104 mutations suggested that glutamine is a poor mimetic for acetylation at this site [55]. However, the possibility that acetylation regulates RAS functions remains a possibility given the importance of Lys 104 in maintaining the structural integrity of helices 2 and 3 [55]. A recent study reported that KRAS is acetylated on the N-terminal Thr following removal of the initiating Met [56]. This modification interacts with a central beta-sheet to stabilize the N-terminus and switch regions. KRAS lacking lacking this modification and the initiating Met adopts an open, inactive, nucleotide-free conformation. Thus, N-acetylation appears to play an important role in the structural stability and nucleotide binding of RAS proteins.
3.1.3. Ubiquitylation
As mentioned above, RAS is regulated by ubiquitin-mediated degradation. A number of E3 ubiquitin ligases have been implicated in negatively regulating RAS including Rabex-5 [57, 58], leucine zipper-like transcription regulator 1 (LZTR1) [59, 60], and β-TrCP [44]. In human cells, ubiquitylation of RAS by Rabex-5 targeted RAS to the endosome and reduced ERK-MAPK activation. Similarly, LZTR1 recruited the CUL3 E3 ligase to target RAS ubiquitylation resulting in loss of RAS association with the membrane and decreased signaling [59, 60]. Thus, both Rabex5 and LZTR1 were proposed as inhibitors of RAS function.
Conversely, monoubiquitylation may serve to re-localize RAS to specific subcellular compartments in an isoform-specific manner [61], or even potentiate RAS function [62]. KRAS is monoubiquitinated on K147 resulting in enhanced GTP loading and engagement of effectors such as PI3K and RAF (Fig. 2) [63]. Building on this work, Campbell and colleagues showed that increased effector binding to K147-monoubiquitinated KRAS was due to impaired GTP hydrolysis [64]. Interestingly, monoubiquitylation of HRAS on K117 also activated HRAS, but through a distinct mechanism that augmented GTP loading [64].
3.1.4. Nitrosylation
S-nitrosylation is a type of reversible post-translational protein modification that involves nitric oxide (NO) reacting with a cysteine thiol to form nitrosothiol [65]. Unlike other types of post-translational modification, S-nitrosylation does not depend on an enzyme to catalyze the reaction. Rather, it depends on the concentration of NO - as regulated by compartmentalization of the target protein to the nitric oxide synthase (NOS) source of NO - and the reactivity of a given cysteine residue [65]. In the brain, NO was known to upregulate neurogenesis through stimulation of the ERK/MAPK pathway, but the signaling mechanism was unknown [66]. This was resolved in recent work in cultured neuronal stem cells demonstrating that RAS was S-nitrosylated on C118 in the presence of NO, leading to enhanced ERK/MAPK activation and proliferation (Fig. 2)[67]. Conversely, NO-mediated ERK/MAPK activation and cell proliferation was abrogated in cells harboring mutant RAS(C118S) that could not be S-nitrosylated on residue 118. These findings may be particularly relevant in the context of tumor biology, where oxidative stress and formation of reactive oxygen species such as NO are common features of the tumor microenvironment [68]. Thus, even in tumors lacking activating RAS mutations, WT RAS may be activated due to nitrosylation.
3.2. Ionic Allosteric Regulation of RAS
The SW2 states (T & R) discussed earlier are modulated by an allosteric mechanism that depends on Ca2+ binding to a site in the allosteric lobe. Mattos and colleagues showed that Ca2+ was bound by residues D108 and Y137 in Loop 7 and Helix 4, respectively, which created two networks of interactions involving Helix 4, Loop 7, Helix 3, and SW2 that resulted in ordering of SW2 and placement of Q61 in the catalytic domain [69]. This observation suggested that coordinated binding of Ca2+ and RAF helps to order SW1 and SW2 to facilitate GTP hydrolysis. Additionally, crystallographic studies using oncogenic RAS(Q61L) suggested that long-range interactions between the Ca2+ binding region in the allosteric lobe and the RAF interaction surface in the effector lobe were disrupted in such a way that could affect RAS function [70]. These results indicate a requisite role for Ca2+ binding in regulating RAS activity. Another divalent cation, Mg2+, plays a more direct role in in RAS regulation by facilitating nucleotide binding. Mg2+ binds in the active site of RAS and is coordinated by residues in the P-loop [71, 72]. Structural studies with GEFs complexed with RAS indicate that Mg2+ functions to maintain the stability of nucleotides within the binding pocket [73].
3.3. Protein-Membrane Interaction
As described earlier, the HVR of RAS is lipidated to localize RAS to the membrane, and there are differences in the secondary lipid modifications between RAS isoforms. These isoform-specific “second signals” determine the distinct membrane microenvironment of each RAS isoform [74]. These membrane microdomains harbor unique lipid profiles, and therefore isoform differences in the allosteric lobe may promote stabilization of conformational states within the individual microdomains [74]. For example, molecular dynamics simulations of HRAS in a lipid bilayer indicated that GTP-loaded HRAS was stabilized by membrane interactions with the α4 helix, while residues in the HVR stabilized the GDP-bound state [75]. This result suggested that the membrane itself may help GTP-loaded RAS achieve the proper orientation for effector engagement [76, 77].
4. Pharmacological inhibition of RAS
Given the central role of RAS in driving tumor development, there has been significant interest in pharmacologically inhibiting oncogenic RAS. However, despite great efforts aimed at multiple steps in RAS function (Fig. 3), there remains a lack of FDA approved RAS inhibitors. The recent development of small molecule covalent inhibitors of RAS(G12C) has renewed hopes of direct pharmacological inhibition of RAS. In the following sections, we will discuss past and current approaches to inhibit RAS. Figure Tables 1 and 2 summarize indirect and direct inhibitors of RAS, respectively.
Table 2.
Direct RAS inhibitors.
| Name | Potency | Type | Binding site | Mechanism of action | Model tested | Clinical trials | References |
|---|---|---|---|---|---|---|---|
| Sulindac | Small molecule | Unknown | ↓ GEF nucleotide exchange, GAP GTPase, RAF interaction | Cell culture; rat breast cancer model; clinical trials | Nearly 40 clinical trials in various cancers | [102,103] | |
| Cyclorasin 9A5* | IC50 = 0.12 μM | Small molecule | RAS-GTP SW1 loop | ↓ RAF interaction | Cell culture | [104,180] | |
| MCP1 and derivatives | IC50 = 17.9 μM | Small molecule | Unknown | ↓ RAF interaction | Cell culture; mouse tumor models | [106–109] | |
| DCAI | EC50 = 15.8 μM | Small molecule | P1 pocket | ↓ SOS interaction | Cell culture | [110] | |
| Cmpd 11 | IC50 = 5 μM | Small molecule | P1 pocket | ↓ RAF interaction | Cell culture | [113] | |
| Kobe0065 and Kobe2602 | IC50 = 10–20 μM | Small molecule | P1 pocket - state 1 of SW1 | ↓ SOS-mediated nucleotide exchange and RAF interaction | Cell culture; mouse colon carcinoma model | [114] | |
| HBS3 | Kd = 28–158 μM | Small molecule | SOS1 interaction site | ↓ SOS interaction and nucleotide exchange | Cell culture | [116] | |
| SAH-SOSIA* | IC50 = 5–15 μM | Small molecule | SOS1 interaction site | ↓ SOS interaction and nucleotide exchange | Cell culture | [117] | |
| Cmpd2 | EC50 = 2.7 μM | Small molecule | Membrane/P1 pocket | ↓ Effector engagement | Cell culture | [118,119] | |
| 3144 | IC5o = 3.8 μM | Small molecule | SW1/SW2 | ↓ Effector interaction | Cell culture; mouse breast and pancreatic cancer models | [120] | |
| Cmpd 12 | EC50 = 0.32 μM | Small molecule | SII-P/C12 of RAS(G12CJ | Disrupts SW1/2 conformation; traps KRAS in a GDP-bound state; ↓ interaction with effectors and activators | Cell culture | [121] | |
| ARS-1620 | IC50 = 120 nM | Small molecule | SII-P/C12 of RAS(G12C) | Disrupts SW1/2 conformation; traps KRAS in a GDP-bound state; ↓ interaction with effectors and activators | Cell culture; mouse pancreatic and lung cancer models | [32] | |
| MRTX849 | IC50 ≅ 10 nM | Small molecule | SII-P/C12 of RAS(G12C) | Disrupts SW1/2 conformation; traps KRAS in a GDP-bound state; ↓ interaction with effectors and activators | Cell culture; mouse pancreatic cancer model; phase I/II clinical trial for patients with advanced solid tumors with KRAS G12C mutation currently recruiting | [122,123] | |
| AMG 510 | Small molecule | SII-P/C12 of RAS(G12C) | Disrupts SW1/2 conformation; traps KRAS in a GDP-bound state; ↓ interaction with effectors and activators | Phase I/II clinical trial for patients with advanced solid tumors with KRAS G12C mutation currently recruiting | |||
| 2C07 | βME50= 1.10–2.53 mM | Small molecule | SII-G | Stabilizes GDP state; ↓ RAS/SOS interaction and nucleotide exchange and PI3K interaction | None | [181] | |
| BI-2852 | EC50 = 5.8–6.7 μM | Small molecule | Pocket between SW1/SW2 | ↓ GEF, GAP, and effector interactions | Cell culture | [115] | |
| Y13–259 | Monoclonal antibody | HRAS SW2 | Sequestration of RAS in intracellular aggregates | Cell culture | [124–127] | ||
| Anti-p21ser | Monoclonal antibody | Viral KRAS(G12S) residues 5–16 | ↓ GTP loading | Cell culture | [128,129] | ||
| iDab#6 | Kd = 26–180 nM | Intrabody | SW1/SW2 of RAS-GTP | ↓ Effector interaction | Cell culture; mouse fibrosarcoma, colorectal cancer, and lung cancer models | [139–142] | |
| ABD7 | IC50 = 8–10 μM | Small molecule | PI pocket | ↓ PI3K, RAF, and RALGDS interaction | Cell culture | [142,143] | |
| RT11(-i) | Kd = 4–17 nM | Chimeric cell-penetrating antibody | RAS-GTP | ↓ PI3K, RAF, and RALGDS interaction | Cell culture; mouse fibrosarcoma and colorectal cancer models | [144] | |
| R11.1.6 | Kd = 4–40 nM | High affinity scaffold based on sso7d | RAS-GTP SW2 | ↓ GTP hydrolysis and RAF association | Cell culture | [145] | |
| K27 and K55 | Designed ankyrin repeat proteins (DARPins) | K27: RAS-GDP SW1 K55: RAS-GTP SW1/SW2 and prevented RAF interaction | K27: ↓ SOS interaction K55: ↓ RAF interaction | Cell culture | [135] | ||
| NS1 | Kd= 15 nM (HRAS) Kd = 65 nM (KRAS) |
Monobody | α4-α5 dimerization interface | ↓ H/KRAS dimerization and signaling | Cell culture; mouse pancreatic, endometrial, and lung cancer models | [33,136,137] | |
| K13 and K19 | K13: Kd=30 nM K19: Kd=10 nM |
DARPins | KRAS(G12 V) α3-α4 dimerization interface | ↓ RAS dimerization, SOS nucleotide exchange, and RAF, PI3K, and RALGDS interaction | Cell culture | [161] |
4.1. Inhibition of RAS membrane association-indirect RAS inhibition
Given the initial view that RAS may be refractory to inhibition due to the apparent absence of deep pockets amenable to small molecule binding and drug development, attention turned toward inhibiting RAS association with the membrane, which is critical to its biological activity. Inhibition of FTase resulted in encouraging preclinical results, although inhibition was not dependent on RAS mutations [78, 79]. In addition, FTase inhibitor (FTI) treatment did not result in altered KRAS or NRAS membrane localization [80]. Unfortunately, clinical trials with these inhibitors has met with disappointing results due to two important points [81]. First, the preponderance of mutations in human solid tumors occur in KRAS vs HRAS or NRAS (Fig. 1B). Second, KRAS undergoes alternative lipidation by geryanylgeranyl transferases (GGTases) upon inhibition of FTase [80, 82, 83], a fact unknown until the development of FTIs. Thus, KRAS (and NRAS) is refractory to the effects of FTIs. However, all hope for these inhibitors has not been lost. Newer trials with tipifarnib have been initiated for HRAS mutant tumors, such as head and neck tumors (ClinicalTrials.gov Identifier: ) and primary completion is expected by March 2020. In addition, FTIs have been used in other clinical settings such as treating chronic hepatitis D [84] and Hutchinson-Gilford Progeria Syndrome (HGPS), a rare genetic condition arising from persistent farnesylation of lamin A which renders the protein inactive [85–87]. Treatment of HGPS patients with the FTI, lonafarnib, resulted in demonstrable clinical benefit although many symptoms remained [88, 89]. Thus, FTIs may gain eventual approval in treating HGPS.
Due to the alternative lipidation of KRAS (and NRAS) upon FTI treatment, efforts shifted toward dual inhibition of both FTase and GGTase. Although co-treatment with FTI and GGTase-I inhibitors (GGTIs) was effective in preventing prenylation of both K-and N-RAS in 2D culture and 3D xenograft models, dose limiting toxicity of GGTIs was a major concern potentially due to other substrates for GGTase-I as well as the FTase (reviewed in [81]). Despite these concerns, one highly selective GGTI (GGTI-2418) also known as PTX100 was approved for clinical trial and phase 1 results showed the compound to be well tolerated with minimal toxicity. However, its efficacy proved to be limited and the clinical trials were terminated (https://drugs.ncats.io/drug/M67G28K74K).
In addition to the combination of FTIs and GGTIs as described above, considerable efforts have focused on development of dual prenyltransferase inhibitors (DPIs). The most promising of these DPIs, L-778,123, suppressed prenylation of H, K and N-RAS in HL-60 leukemic cells [90]. However, two phase I clinical studies reported that L-778,123 was ineffective to prevent K-RAS prenylation even at toxic doses [91, 92]. An additional approach to block RAS processing has been through targeting the mevalonate pathway with bisphosphonates which inhibit synthesis of farnesyl pyrophosphate (FPP) and geranylgeranylpyrophosphate (GGPP). Aminobisphosphonates, such as zoledronic acid (ZOL), disrupt RAS membrane localization and inhibit RAS-mediated signaling [93]. Although ZOL demonstrated limited efficacy in vitro new nanoparticle formulations exhibit anti-tumor properties in vivo [94, 95]. However, the question remains whether the anti-tumor effects are due to specific inhibition of RAS or numerous other lipidated substrates [96].
Another approach to inhibit RAS membrane association centered on interfering with the recycling of RAS from the plasma membrane (PM) to various intracellular compartments. The phosphodiesterase 6 delta subunit (PDE6δ) binds the farnesylated tails of KRAS, as well as HRAS and NRAS, sequestering the protein from the PM and promoting its recycling [97, 98]. Thus, disrupting PDEδ interaction with KRAS can suppress oncogenic RAS signaling by altering its localization to endomembranes. Indeed, Zimmermann et. al. identified a small molecule inhibitor (Deltarasin) that bound in cells to the prenyl binding pocket of PDEδ with nanomolar affinity (Kd= 41 nM) [99]. Deltarasin inhibited RAS mediated oncogenic signaling and tumor potential of PDACs both in vitro and in vivo. However, non-specific cytotoxicity issues were observed at effective doses (>5 μM), thereby limiting the effectiveness of this compound. These issues were overcome by a second generation pyrazolopyridazinone inhibitor of PDEδ called Deltazinone 1 which was highly selective in inhibiting PDACs [100]. Modification in the chemistry of Deltazinone 1 lead to generation of Deltasonamide 1 and 2, which had improved binding with farnesyl binding pocket of PDEδ [101]. These compounds, though efficacious, had low membrane permeability which reduced their presence in the cytosol. Thus, further modifications will be needed to enhance the cellular uptake of these compounds for therapeutic use.
While the above approaches may lead to inhibition of RAS, their clinical utility may be limited by significant off-target effects due to the inhibition of additional targets that utilize these same pathways. Whether such off-target effects can be successfully minimized through dosing or specific delivery of compounds to tumors remains an unanswered question. The following sections will describe approaches to directly inhibit RAS (Table 2).
4.2. Targeting RAS interaction with activators and effectors
Many groups have focused on inhibiting RAS interaction with activators and effectors as a way of blocking RAS activity. These efforts have led to the isolation of many lead RAS inhibitory compounds. The non-steroidal anti-inflammatory drug (NSAID) sulindac and its derivatives represented the first examples of such compounds [102]. These drugs inhibited RAS activation of RAF and reduced RAS-driven transformation [103]. However, these compounds did not exhibit sufficient potency to move forward in the clinic. A cell penetrating cyclic peptide termed cyclorasin 9A5 blocked RAS-RAF interaction, reduced proliferation, and induced apoptosis of RAS mutant lung tumor cells [104]. However, cyclorasin 9A5 did not exhibit selectivity toward a particular RAS isoform, or WT vs mutant RAS, thus raising the issue of toxicity when used in vivo. Indeed, a recent study suggested that this compound does not actually engage RAS in cells [105]. MCP1 and its derivatives also blocked RAS-RAF interaction but lacked sufficient potency to serve as effective inhibitors [106–109].
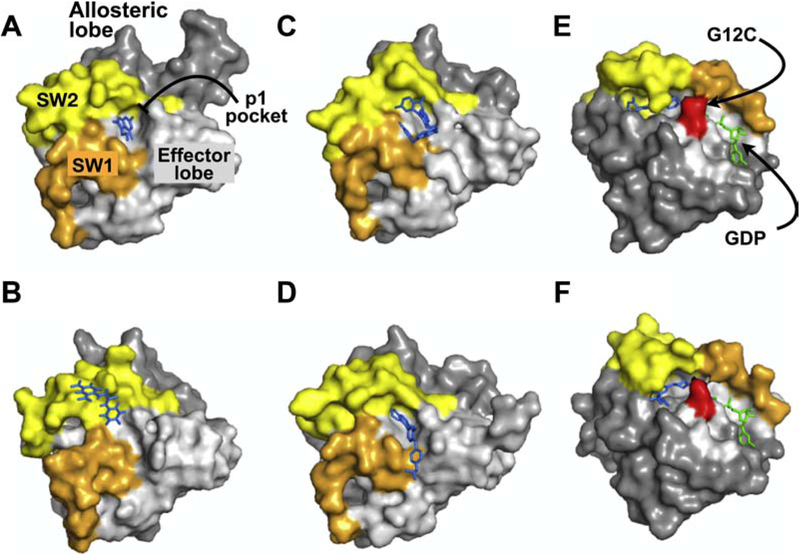
Fragment based screening led to the identification of DCAI which targeted a pocket between SW1 and SW2 (Fig. 4A) [110]. DCAI blocked both nucleotide release and nucleotide exchange with IC50s of 155 uM and 342 uM, respectively, although treatment of cells with DCAI reduced recruitment of the RAF RBD to the plasma membrane with an EC50=15.8 uM. However, it was unclear whether this compound had sufficient potency or selectivity to inhibit RAS-mutant tumor cells in vivo. Interestingly, the same region of RAS targeted by DCAI has been the target of a number of lead RAS inhibitory compounds identified in independent screens as described below.
Figure 4. RAS Structures. X-ray crystral structures of RAS in complex with various inhibitors.
Panels A-D illustrate structures of different compounds that all target the p1 pocket in RAS whereas panels E & F represent covalent RAS inhibitors targeting KRAS(G12C). The effector lobe is shown in light gray, allosteric lobe in dark gray, SW1, orange, and SW2, light yellow. Inhibitors bound to RAS are shown in blue and nucleotide in green. A. KRAS(G12D):DCAI (PDB: 4DST). B. HRAS(T35S):Kobe2601 (PDB: 2LWI). C. KRAS(G12D):BI-2852 (PDB: 6GJ8). D. KRAS(Q61H):ABD7 (PDB: 6FA4). D. KRAS(G12C):ARS-1620 (PDB:5V9U). E. KRAS(G12D):compound 12 (PDB: 6N2K). Structures A-D are shown in the same orientation to highlight the p1 pocket. Structures E and F are shown in a different orientation to highlight the SII-P pocket and the Cys residue (shown in red) targeted by the indicated compounds.
Computational methods combined with detailed crystallographic analyses identified potential allosteric sites on RAS [11, 111]. Multiple solvent crystal structures (MSCS) combined with computational solvent mapping (FTMap) identified a total of 13 hot spots, many of which corresponded to effector binding [11]. Gorfe and colleagues identified four potential allosteric pockets in RAS and isolated candidate binders using ensemble docking and virtual screening [111]. This approach was later validated by probe-based molecular dynamics simulations [112]. More recently, they have extended this work and identified compounds predicted to bind the “p1” pocket [111], the same pocket that bound DCAI (Fig. 4A) [113]. Virtual screening yielded 58 potential p1 binding compounds of which 11 were tested in cell-based assays. Compound 11 disrupted RAS-RAF interaction and inhibited RAS mutant cancer cells with a modest IC50 ~1–5 uM [113]. Using in silico screening, Kataoka and colleagues identified Kobe0065 and Kobe2602 which also bound the p1 pocket (Fig. 4B) [114]. The Kobe compounds blocked RAS-RAF interaction by binding state 1 of SW1, disrupted SOS-mediated nucleotide exchange, and reduced signaling and oncogenic transformation by RAS in model systems. However, the potency of these compounds was low (IC50= 10–20 uM). Using a fragment based-NMR screens and structure-based drug discovery, Kessler et al recently reported the isolation of a compound, BI-2852, that binds this p1 pocket blocking interactions with GEFs, GAPs, and effectors thereby inhibiting RAS function (Fig. 4C)[115]. This compound exhibited high nM affinity for KRAS in vitro (450–750 nM) and inhibited KRAS signaling in cells at low uM concentrations (<10uM). Thus, this compound may serve as an additional chemical probe for studying RAS and potentially developing more potent, clinically efficacious inhibitors.
Several groups have attempted to inhibit RAS with synthetic peptides based on the RAS-interacting α-helix of SOS1. Bar-Sagi and colleagues designed one such peptide, HBS3, that incorporated amino acids 929–944 of SOS1 [116]. HBS3 bound preferentially to nucleotide-free RAS vs GDP-bound RAS (Kd=28 uM vs 158 uM, respectively) and reduced both nucleotide exchange in vitro and RAS signaling in cells. However, the potency of this peptide in cells was unclear. In a similar approach, Walensky and colleagues isolated stapled peptides based on the same RAS binding helix of SOS1 [117]. Their lead peptide, SAH-SOS1A bound RAS in vitro with Kd of 100–175 nM, although significantly higher concentrations (5–15 uM) were required to inhibit the viability of RAS-mutant cancer cell lines. However, the specificity of this compound for RAS has also been questioned [105].
Using a novel liposome-based screen to measure prenylated KRAS activation of full length BRAF, Jansen and colleagues identified a small molecule, cmpd2, that inhibited RAS by simultaneously binding phosphatidylserine enriched membrane and the p1 pocket of lipidated KRAS [118]. As a result, cmpd2 promoted an orientation of KRAS that occluded the effector region and blocked engagement of RAS targets [119]. This compound inhibited proliferation and signaling in KRAS mutant cells with an IC50 in the low uM range.
Stockwell and colleagues utilized a computational docking strategy to identify a pan-RAS inhibitor termed 3144 that simultaneously targeted three sites encompassing SW1 and SW2 [120]. Treatment of cells with 3144 reduced interaction of RAS with effectors, inhibited signaling and reduced RAS-driven tumor formation in experimental models. Although the ability of 3144 to selectively inhibit KRAS-driven MEFs vs RASless MEFs driven by an activated BRAF allele was modest (IC50= 3.8 uM vs 11 uM), improvements to this compound may lead to more potent RAS inhibitors in the future.
4.3. A Novel SW2 Binding Pocket
In perhaps the most successful approach thus far in targeting RAS, Shokat and colleagues utilized a novel disulphide tethering approach to specifically target RAS(G12C) given the unique reactivity of the Cys thiol [121]. Based on this approach, they isolated a number of compounds that bound in a previously unrecognized pocket in the SW2 region adjacent to the nucleotide binding pocket, designated as SII-P. Interestingly, these compounds targeted the GDP-bound state of RAS and resulted in significant structural perturbation of the SW1 and SW2 domain that disrupted interactions with both effectors and activators of RAS. Although these compounds targeted the GDP-bound inactive state of RAS(G12C), the compounds nevertheless inhibited RAS(G12C) function in cells, thereby revealing that mutant RAS, at least G12C, continues to cycle in cells.
Building on this work, Janes et al. developed the small molecule ARS-1620 through structure-based design (Fig. 4E)[32]. This compound possessed enhanced reaction kinetics and potency relative to both the initial and second generation SII-P inhibitors. Notably, treatment of KRAS(G12C) mutant cells, but not control cells, with ARS-1620 inhibited RAS/RAF interaction, reduced phosphorylation of relevant downstream target proteins, and inhibited the growth of KRAS(G12C) mutant cell lines [32]. In vivo, ARS-1620 exhibited good pharmacodynamic and pharmacokinetic properties resulting in selective inhibition of KRAS(G12C) mutant tumors but not tumors driven by other KRAS mutants [32]. Based on these successes, Mirati Therapeutics, Inc. has developed additional cysteine-reactive small molecules (Fig. 4F)[122], one of which, MRTX849, demonstrated drug-like cellular potency of ~10 nM, and greater than 1000 fold selectivity for KRAS(G12C) than WT KRAS and broad-spectrum anti-tumor potency in preclinical model [123]. Mirati initiated a Phase 1/2 clinical trial in January of 2019 to test the safety and efficacy of MRTX849, with the aim of treating KRAS(G12C)-positive non-small cell lung cancer and colorectal cancer (ClinicalTrials.gov Identifier: ). Amgen has also developed a compound similar to ARS-1620 that targets the KRAS(G12C) mutant, AMG 510, and has also initiated a Phase 1 clinical trial (ClinicalTrials.gov Identifier: ). These two trials represent the first attempts to directly target mutant RAS in human cancers and their results are eagerly awaited by the RAS community.
4.4. Targeting RAS with biologics
The first direct inhibitor of RAS was the monoclonal antibody Y13–259 which targeted the SW2 region to inhibit oncogenic HRAS-driven proliferation and cell signaling [124–127]. Another RAS monoclonal antibody, anti-p21ser, developed against residues 5–16 of viral KRAS(G12S) also inhibited oncogenic RAS-driven transformation [128]. Interestingly, this antibody targeted the nucleotide-free state of RAS to inhibit GTP loading [129]. Thus, these findings suggest that targeting nucleotide binding may indeed be feasible despite the picomolar affinity of RAS for nucleotide. Indeed, a number of more recent observations further support this possibility. Several RAS mutants exhibit high intrinsic nucleotide exchange rates (e.g., G13D, Q61L, and A146T) suggesting that these RAS variants may transit through a nucleotide-free state more frequently than WT RAS or other RAS mutants [130]. Furthermore, we demonstrated that nucleotide-free RAS negatively regulated a specific PI3K isoform, namely Class 2β, suggesting that nucleotide free RAS is more than just a transient intermediate [131]. In the case of the heterotrimeric G protein Gαq, the cell-permeable compound BIM-46187 inhibited the GTPase by targeting the nucleotide-free state [132]. Thus, targeting nucleotide-free RAS may yet be a viable approach for inhibiting RAS function.
Although the Y13–259 and anti-p21ser monoclonal antibodies were useful tools for defining the importance of RAS in cellular transformation, these reagents were limited by their ability to enter cells and the reducing potential of the intracellular milieu. A number of groups have taken different approaches to target RAS with engineered, synthetic proteins. These include technologies such as Affibodies [133], single chain variable fragments (scFv) [134], DARPins [135], Monobodies [136, 137], and a number of other synthetic proteins [138].
Single domain variable fragments of antibodies (intrabody) were generated that specifically recognized active GTP loaded RAS. One such intrabody, iDab#6, had nanomolar affinity to RAS-GTP [139]. iDab#6 bound oncogenic mutants of all RAS isoforms and abrogated their transforming potential by targeting SW1 and SW2 and impairing effector interaction [140, 141]. Although the use of this intrabody as a therapeutic agent was limited by its size and poor cell penetrance, iDab#6 was employed in a novel competitive screening assay using surface plasmon resonance to identify lead compounds targeting the iDab#6 binding region on RAS [142]. Using a combination of structural biology and medicinal chemistry, a series of compounds (ADB series) were developed that bound KRAS with sub-micromolar affinity, disrupted interaction with effector targets, and inhibited RAS-dependent signaling in human tumor cell lines [142, 143]. Interestingly, ADB7 targeted the same p1 pocket as several other RAS inhibitory compounds (Fig. 4D).
In another approach to improve the cell penetrance of anti-RAS antibodies, the heavy chain fragment (VH) of a RAS specific antibody was used to replace the VH fragment of the cell penetrating antibody, TMab4 [144]. The chimeric IgG1 antibody, RT11, crossed the cell membrane and bound activated WT and mutant versions of all three RAS isoforms with low nM affinity (Kd ~4–17 nM). Though RT11 inhibited RAS mediated growth and signaling in tumor cells by preventing RAS effector association, another layer of specificity was imparted by fusing the RGD10 cyclic peptide to the N-terminus of the light chain of RT11 to generate RT11-i. This modified antibody was targeted to cells expressing tumor associated integrins. RT11-i had better pharmacokinetics and biodistribution in tumors, and inhibited RAS mutant tumors in xenograft models.
R11.1.6, a small, high affinity scaffold based on the DNA binding protein sso7d, had preferential and single digit nanomolar affinity for KRAS(G12D) (Kd= 4 nm) vs WT (Kd= 40nm) in the GTP bound state [145]. Intriguingly, R11.1.6 did not discriminate between mutant KRAS(G12D) and WT in the inactive GDP-bound state. R11.1.6 was not mutation specific as it bound other KRAS mutants (G12C and G12V) as well as HRAS and NRAS with comparable affinities. Structurally, R11.1.6 bound SW2 and reduced intrinsic GTP hydrolysis. Despite locking RAS in the active state, R11.1.6 blocked RAS-driven MAPK activation by sequestering GTP-bound active RAS from effector association.
The K27 and K55 Designed Ankyrin Repeat Proteins (DARPins) also targeted RAS and reduced both ERK and AKT activation as well as anchorage independent growth of KRAS(G13D) mutant HCT116 cells [135]. K27 preferentially bound the inactive GDP form of RAS whereas K55 favored RAS-GTP. Structural analysis of K27 complexed with KRAS(G12V) revealed that K27 bound the SW1 region and occluded interaction with SOS. However, this interaction did not distort the conformation of either SW1 or GDP in contrast to the effects of SOS which destabilizes nucleotide binding. K55 interacted with both SW1 and SW2 GTP-bound KRas(G12V) and prevented interaction with RAF. Thus, these two RAS-inhibitory DARPins functioned by blocking different aspects of RAS function, namely inhibiting nucleotide exchange (K27) and blocking effector interaction (K55).
In the quest to identify novel strategies to inhibit RAS, we employed an unbiased approach using Monobody technology [136]. Monobodies are single-domain synthetic binding proteins of ~95 amino acids that achieve levels of affinity and selectivity similar to antibodies, yet are insensitive to the redox potential of their environment [138]. Thus, Monobodies are ideally suited as genetically encoded tool compounds. We recently isolated a Monobody called NS1 that selectively interacted with HRAS (Kd= ~15 nM) and KRAS (Kd= ~65 nM), but not NRAS (Fig. 5A)[136]. NS1 inhibited oncogenic HRAS and KRAS signaling and transformation both in vitro and in vivo [33, 136, 137]. However, NS1 did not inhibit NRAS or oncogenic proteins downstream of RAS such as BRAF(V600E) or MEK(DD). Furthermore, NS1 did not affect the proliferation of fibroblasts (NIH/3T3 or HEK293) grown in culture (Khan and O’Bryan, unpublished observations). Thus, NS1 specifically inhibited oncogenic RAS with no discernible “off target” effects.
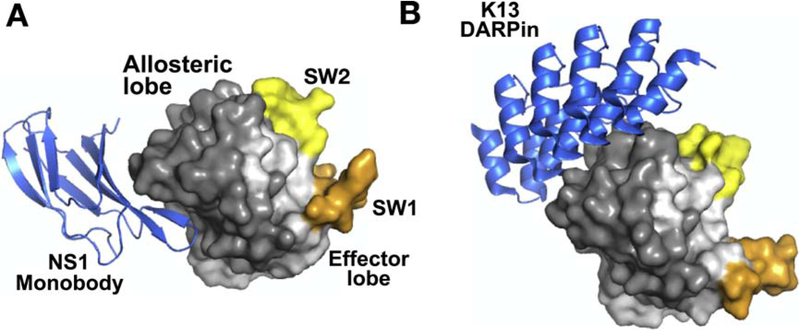
Figure 5. Biologics targeting RAS.
A. Structure of NS1 monobody bound to HRAS WT (PDB:5E95). B. Structure of DARPin K13 bound to KRAS(G12V):GDP (PDB: 6H46). Both structures are shown in the same orientation to illustrate the different regions targeted by NS1 versus K13. Coloring of figure is the same as in Fig. 4. Both NS1 and K13 target the allosteric lobe of RAS.
NS1 bound the α4-α5 region of RAS which has been implicated in RAS dimerization [136, 146, 147]. Indeed, accumulating evidence suggests that RAS dimerization is important for the activation of RAS effectors [148]. RAS oligomerization was first suggested by Santos and colleagues using radiation target analysis [149]. In addition, while RAS bound directly to its effector RAF, this interaction was not sufficient to stimulate RAF kinase activity in vitro (Fig. 6)[150–154]. Interestingly, RAS stimulated RAF dimerization and activation in cells [155, 156], consistent with the obligate role of dimerization in RAF catalytic activation (Fig. 6)[157]. Work from Kaziro and colleagues demonstrated that artificial dimerization of a soluble RAS in vitro or in cells stimulated RAF activation, suggesting that RAS dimerization at the plasma membrane was a necessary step for RAS activation of RAF [158]. Further, RAS dimers were observed in a number of X-ray crystal structures [136, 146, 147], and we identified a specific dimer formed through the α4-α5 interface only in crystal structure of RAS in the active state, as defined by the conformation of SW1 and SW2 [136]. Super-resolution microscopy also revealed the presence of RAS dimers at the plasma membrane, although these structures appeared dependent on the COOH-terminal tails of RAS but independent of the G-domain [159, 160]. NS1, however, has provided significant insight into the role of dimerization in RAS function. When expressed in cells NS1 targeted the α4-α5 region and reduced RAS dimerization and nanoclustering at the plasma membrane leading to decreased proliferation, anchorage-independent growth and tumor formation by KRAS and HRAS, but not NRAS, mutant tumor lines (Fig. 6)[33, 136, 137]. These studies represented the first demonstration of the importance of the α4-α5 interface in RAS function and highlight the potential efficacy of targeting RAS dimerization as a strategy to inhibit oncogenic RAS function (Fig. 6). Recently, Rabbitts and colleagues identified the α3-α4 interface in the allosteric lobe as important for RAS dimerization [161]. They isolated two DARPins, K13 and K19, that selectively bound and inhibited KRAS (Fig. 5B). These DARPins blocked dimerization as well as SOS-mediated nucleotide exchange. Thus, these results suggest that RAS may dimerize through α3-α4 as well as the α4-α5 interface as suggested by previous computer modeling studies and analysis of RAS crystal structures [162–164].
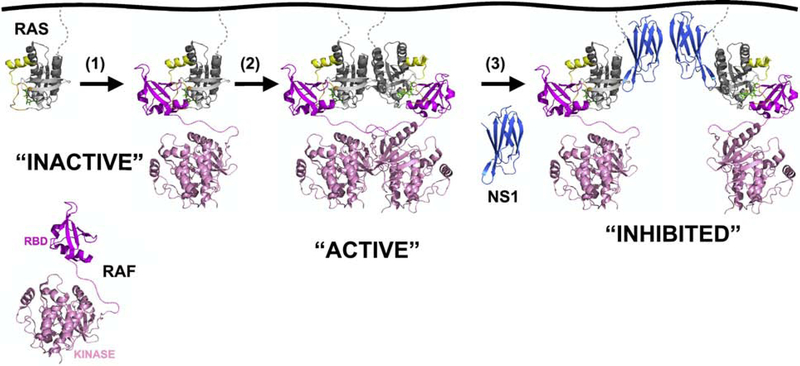
Figure 6. Hypothetical model for RAS activation by dimerization/self-association.
Mitogenic stimulation results in RAS loading with GTP and recruitment of RAF (1). However, monomeric RAS is not sufficient for activation of RAF. Thus, two RAS protomers either physically interact through specific interfaces, such as the α4-α5 interface (2) or move in sufficiently close proximity to facilitate dimerization of downstream targets such as RAF. Binding of NS1 Monobody (3) in either circumstance provides significant steric hinderance to prevent RAS-mediated dimerization of RAF, thereby inhibiting RAF activation. Given the inability to detect dimerization of purified KRAS on reconstituted membranes [168], additional proteins may be necessary to facilitate RAS dimerization/self-association. Note: the full-length structure of RAF has not been determined. The dimer of the RAF kinase domain was taken from PDB:1UWJ. The RAF RBD structure in complex with HRAS was taken from PDB:3KUD. The hypothetical linkage between these two domains is shown with a dashed line. The HRAS:NS1 structure was taken from PDB:5E95. The HVR of RAS along with the lipidated tail is shown in dashed grey lines.
While these findings clearly underscore the importance of the α4-α5 and α3-α4 interfaces in RAS function, there remains considerable debate regarding the notion of RAS dimerization. Indeed, the isolated G-domain of RAS lacks the intrinsic capacity to dimerize in solution [147, 165–167]. Recently, Groves and colleagues reported that fully processed, native KRAS was incapable of forming oligomers on supported lipid bilayers of varying composition [168]. However, Ambrogio et. al. found that mutations in the α4-α5 interface disrupted RAS dimerization and impaired RAS driven tumorigenesis [169]. Thus, it is possible that RAS itself lacks the intrinsic ability to dimerize and that additional cellular factors are necessary to promote RAS association through the α4-α5 or the α3-α4 regions. Nevertheless, targeting these regions with small biologics, i.e., NS1, is sufficient to impair oncogenic RAS signaling and tumorigenesis [33, 136, 137]. Thus, pharmacological targeting of RAS dimerization could prove useful in blocking RAS-dependent tumors. However, such compounds would need to retain some selectivity in targeting RAS isoforms or discriminating between mutant and WT proteins since loss of all RAS isoforms is not compatible with life [170].
While direct inhibitors of RAS have not yet seen clinical success, the above discussion highlights a number of advances that exemplify progress in the field. Namely, several novel druggable pockets have been identified along with domains that can be targeted for negative allosteric regulation of RAS signaling. Small molecules that directly inhibit KRAS(G12C) are currently the most promising RAS inhibitors with several Phase 1 trials in progress, however, a number of additional leads with broader applicability are not far behind.
5. Future perspectives
Although pharmacological targeting of RAS has been a long and arduous journey filled with many challenges and failures, we are beginning to see light at the end of the tunnel. The recent clinical trials with (G12C) specific inhibitors provide significant hope for the direct inhibition of RAS in human tumors. However, the limited applicability of these inhibitors to only G12C mutant tumors means that the quest to inhibit RAS continues. The need remains for more broadly efficacious inhibitors that target additional RAS mutants such as KRAS(G12D) which predominates in PDAC tumors. Furthermore, the inevitable issue of acquired resistance to such RAS inhibitors will almost certainly need to be addressed. Although the inhibition of pathways downstream of RAS has not proven clinically beneficial in RAS mutant tumors thus far, recent studies provide intriguing insight into potential combination therapies. Groundbreaking results from three separate labs demonstrated that PDAC cell lines with mutations in the RAS signaling pathway up-regulated autophagy in response to inhibition of RAS or its downstream effectors, to promote cell survival [171–173]. Co-inhibition of the RAS pathway and autophagy was synergistically cytotoxic in vitro, and inhibited tumor growth in vivo in PDX mouse models. Perhaps most striking in these reports was a case of compassionate treatment of a PDAC patient with the MEK inhibitor trametinib and the autophagy inhibitor hydroxychloroquine. The patient displayed a marked reduction in tumor marker cancer antigen, overall tumor burden, and pain associated with PDAC. Whether these results will translate into success in a larger clinical setting remains to be seen. However, these studies along with the successful development of RAS-targeted inhibitors, the launch of clinical trials that directly target RAS-mutant cancers, and the abundance of new RAS inhibitors in the developmental pipeline provides great optimism that we will soon be able to conquer the challenge of drugging the “undruggable” RAS.
Supplementary Material
Highlights.
RAS mutations occur in nearly early 30% of human tumors
Different RAS mutations result in context-dependent effects
Indirect inhibitors of RAS target various aspects of RAS processing
Direct inhibitors interfere with RAS activation and effector binding
Newer biologics target RAS activation, effector binding, and self-association
Acknowledgements:
The authors wish to thank Dr Frank Sicheri for helping to generate the hypothetical RAS:RAF heterotetrameric structure in Fig. 6 and Dr. Robert Gemmill for helpful comments on this review. J.P.O. was supported in part by a Merit Review Award (1I01BX002095) from the United States (US) Department of Veterans Affairs Biomedical Laboratory Research and Development Service and NIH awards (CA212608). The contents of this article do not represent the views of the US. Department of Veterans Affairs or the United States Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: The authors have nothing to disclose.
Bibliography
- [1].Rajalingam K, Schreck R, Rapp UR, Albert S, Ras oncogenes and their downstream targets, Biochim Biophys Acta, 1773 (2007) 1177–1195. [DOI] [PubMed] [Google Scholar]
- [2].Karnoub AE, Weinberg RA, Ras oncogenes: split personalities, Nat Rev Mol Cell Biol, 9 (2008) 517–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Downward J, Targeting RAS signalling pathways in cancer therapy, Nat Rev Cancer, 3 (2003) 11–22. [DOI] [PubMed] [Google Scholar]
- [4].Spencer-Smith R, O’Bryan JP, Direct inhibition of RAS: Quest for the Holy Grail?, Semin Cancer Biol, 54 (2019) 138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].O’Bryan JP, Pharmacological targeting of RAS: Recent success with direct inhibitors, Pharmacol Res, 139 (2019) 503–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Waters AM, Der CJ, KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer, Cold Spring Harb Perspect Med, 8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Spiegel J, Cromm PM, Zimmermann G, Grossmann TN, Waldmann H, Small-molecule modulation of Ras signaling, Nat Chem Biol, 10 (2014) 613–622. [DOI] [PubMed] [Google Scholar]
- [8].Cox AD, Der CJ, Ras history: The saga continues, Small GTPases, 1 (2010) 2–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ, Drugging the undruggable RAS: Mission possible?, Nat Rev Drug Discov, 13 (2014) 828–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gorfe AA, Grant BJ, McCammon JA, Mapping the nucleotide and isoform-dependent structural and dynamical features of Ras proteins, Structure, 16 (2008) 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Buhrman G, O’Connor C, Zerbe B, Kearney BM, Napoleon R, Kovrigina EA, Vajda S, Kozakov D, Kovrigin EL, Mattos C, Analysis of binding site hot spots on the surface of Ras GTPase, J Mol Biol, 413 (2011) 773–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Geyer M, Schweins T, Herrmann C, Prisner T, Wittinghofer A, Kalbitzer HR, Conformational transitions in p21ras and in its complexes with the effector protein Raf-RBD and the GTPase activating protein GAP, Biochemistry, 35 (1996) 10308–10320. [DOI] [PubMed] [Google Scholar]
- [13].Johnson CW, Mattos C, The allosteric switch and conformational states in Ras GTPase affected by small molecules, Enzymes, 33 Pt A (2013) 41–67. [DOI] [PubMed] [Google Scholar]
- [14].Spoerner M, Graf T, König B, Kalbitzer HR, A novel mechanism for the modulation of the Ras-effector interaction by small molecules, Biochemical and Biophysical Research Communications, 334 (2005) 709–713. [DOI] [PubMed] [Google Scholar]
- [15].Rosnizeck IC, Graf T, Spoerner M, Tränkle J, Filchtinski D, Herrmann C, Gremer L, Vetter IR, Wittinghofer A, König B, Kalbitzer HR, Stabilizing a Weak Binding State for Effectors in the Human Ras Protein by Cyclen Complexes, Angewandte Chemie International Edition, 49 (2010) 3830–3833. [DOI] [PubMed] [Google Scholar]
- [16].Reiss Y, Goldstein JL, Seabra MC, Casey PJ, Brown MS, Inhibition of purified p21ras farnesyl:protein transferase by Cys-AAX tetrapeptides, Cell, 62 (1990) 81–88. [DOI] [PubMed] [Google Scholar]
- [17].Bergo MO, Ambroziak P, Gregory C, George A, Otto JC, Kim E, Nagase H, Casey PJ, Balmain A, Young SG, Absence of the CAAX Endoprotease Rce1: Effects on Cell Growth and Transformation, Molecular and Cellular Biology, 22 (2002) 171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ahearn IM, Haigis K, Bar-Sagi D, Philips MR, Regulating the regulator: post-translational modification of RAS, Nature Reviews Molecular Cell Biology, 13 (2012) 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Erwin N, Patra S, Dwivedi M, Weise K, Winter R, Influence of isoform-specific Ras lipidation motifs on protein partitioning and dynamics in model membrane systems of various complexity, Biol Chem, 398 (2017) 547–563. [DOI] [PubMed] [Google Scholar]
- [20].Eisenberg S, Henis YI, Interactions of Ras proteins with the plasma membrane and their roles in signaling, Cell Signal, 20 (2008) 31–39. [DOI] [PubMed] [Google Scholar]
- [21].Pacold ME, Suire S, Perisic O, Lara-Gonzalez S, Davis CT, Walker EH, Hawkins PT, Stephens L, Eccleston JF, Williams RL, Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma, Cell, 103 (2000) 931–943. [DOI] [PubMed] [Google Scholar]
- [22].Lu S, Jang H, Gu S, Zhang J, Nussinov R, Drugging Ras GTPase: a comprehensive mechanistic and signaling structural view, Chem Soc Rev, 45 (2016) 4929–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Khosravi-Far R, Der CJ, The Ras signal transduction pathway, Cancer and Metastasis Reviews, 13 (1994) 67–69. [DOI] [PubMed] [Google Scholar]
- [24].Bai D, Ueno L, Vogt PK, Akt-mediated regulation of NFkappaB and the essentialness of NFkappaB for the oncogenicity of PI3K and Akt, Int J Cancer, 125 (2009) 2863–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Farhan M, Wang H, Gaur U, Little PJ, Xu J, Zheng W, FOXO Signaling Pathways as Therapeutic Targets in Cancer, Int J Biol Sci, 13 (2017) 815–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zago G, Biondini M, Camonis J, Parrini MC, A family affair: A Ral-exocyst-centered network links Ras, Rac, Rho signaling to control cell migration, Small GTPases, (2017) 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bodemann BO, White MA, Ral GTPases and cancer: linchpin support of the tumorigenic platform, Nat Rev Cancer, 8 (2008) 133–140. [DOI] [PubMed] [Google Scholar]
- [28].Hamad NM, Elconin JH, Karnoub AE, Bai W, Rich JN, Abraham RT, Der CJ, Counter CM, Distinct requirements for Ras oncogenesis in human versus mouse cells, Genes Dev, 16 (2002) 2045–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Nussinov R, Tsai CJ, Jang H, Oncogenic Ras Isoforms Signaling Specificity at the Membrane, Cancer Res, 78 (2018) 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Vartanian S, Bentley C, Brauer MJ, Li L, Shirasawa S, Sasazuki T, Kim JS, Haverty P, Stawiski E, Modrusan Z, Waldman T, Stokoe D, Identification of mutant K-Ras-dependent phenotypes using a panel of isogenic cell lines, J Biol Chem, 288 (2013) 2403–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hood FE, Klinger B, Newlaczyl AU, Sieber A, Dorel M, Oliver SP, Coulson JM, Bluthgen N, Prior IA, Isoform-specific Ras signaling is growth factor dependent, Mol Biol Cell, 30 (2019) 1108–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Janes MR, Zhang J, Li LS, Hansen R, Peters U, Guo X, Chen Y, Babbar A, Firdaus SJ, Darjania L, Feng J, Chen JH, Li S, Li S, Long YO, Thach C, Liu Y, Zarieh A, Ely T, Kucharski JM, Kessler LV, Wu T, Yu K, Wang Y, Yao Y, Deng X, Zarrinkar PP, Brehmer D, Dhanak D, Lorenzi MV, Hu-Lowe D, Patricelli MP, Ren P, Liu Y, Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor, Cell, 172 (2018) 578–589 e517. [DOI] [PubMed] [Google Scholar]
- [33].Khan I, Spencer-Smith R, O’Bryan JP, Targeting the alpha4-alpha5 dimerization interface of K-RAS inhibits tumor formation in vivo, Oncogene, 38 (2019) 2984–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Winters IP, Chiou SH, Paulk NK, McFarland CD, Lalgudi PV, Ma RK, Lisowski L, Connolly AJ, Petrov DA, Kay MA, Winslow MM, Multiplexed in vivo homology-directed repair and tumor barcoding enables parallel quantification of Kras variant oncogenicity, Nat Commun, 8 (2017) 2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Poulin EJ, Bera AK, Lu J, Lin YJ, Strasser SD, Paulo JA, Huang TQ, Morales C, Yan W, Cook J, Nowak JA, Brubaker DK, Joughin BA, Johnson CW, DeStefanis RA, Ghazi PC, Gondi S, Wales TE, Iacob RE, Bogdanova L, Gierut JJ, Li Y, Engen JR, Perez-Mancera PA, Braun BS, Gygi SP, Lauffenburger DA, Westover KD, Haigis KM, Tissue-Specific Oncogenic Activity of KRAS(A146T), Cancer Discov, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Nestler EJ GP, Protein Phosphorylation is of Fundamental Importance in Biological Regulation, in: Siegel GJ AB, Albers RW, et al. (Ed.) Basic Neurochemistry: Molecular, Cellular and Medical Aspects, Lippincott-Raven, Philadelphia, 1999. [Google Scholar]
- [37].Shih TY, Weeks MO, Young HA, Scholnick EM, Identification of a sarcoma virus-coded phosphoprotein in nonproducer cells transformed by Kirsten or Harvey murine sarcoma virus, Virology, 96 (1979) 64–79. [DOI] [PubMed] [Google Scholar]
- [38].Papageorge A, Lowy D, Scolnick EM, Comparative biochemical properties of p21 ras molecules coded for by viral and cellular ras genes, J Virol, 44 (1982) 509–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].John J, Frech M, Wittinghofer A, Biochemical properties of Ha-ras encoded p21 mutants and mechanism of the autophosphorylation reaction, J Biol Chem, 263 (1988) 11792–11799. [PubMed] [Google Scholar]
- [40].Bivona TG, Quatela SE, Bodemann BO, Ahearn IM, Soskis MJ, Mor A, Miura J, Wiener HH, Wright L, Saba SG, Yim D, Fein A, Perez de Castro I, Li C, Thompson CB, Cox AD, Philips MR, PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis, Mol Cell, 21 (2006) 481–493. [DOI] [PubMed] [Google Scholar]
- [41].Sung PJ, Tsai FD, Vais H, Court H, Yang J, Fehrenbacher N, Foskett JK, Philips MR, Phosphorylated K-Ras limits cell survival by blocking Bcl-xL sensitization of inositol trisphosphate receptors, Proc Natl Acad Sci U S A, 110 (2013) 20593–20598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ting PY, Johnson CW, Fang C, Cao X, Graeber TG, Mattos C, Colicelli J, Tyrosine phosphorylation of RAS by ABL allosterically enhances effector binding, FASEB J, 29 (2015) 3750–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Hu H, Bliss JM, Wang Y, Colicelli J, RIN1 is an ABL tyrosine kinase activator and a regulator of epithelial-cell adhesion and migration, Curr Biol, 15 (2005) 815–823. [DOI] [PubMed] [Google Scholar]
- [44].Jeong WJ, Yoon J, Park JC, Lee SH, Lee SH, Kaduwal S, Kim H, Yoon JB, Choi KY, Ras stabilization through aberrant activation of Wnt/beta-catenin signaling promotes intestinal tumorigenesis, Sci Signal, 5 (2012) ra30. [DOI] [PubMed] [Google Scholar]
- [45].Bunda S, Heir P, Srikumar T, Cook JD, Burrell K, Kano Y, Lee JE, Zadeh G, Raught B, Ohh M, Src promotes GTPase activity of Ras via tyrosine 32 phosphorylation, Proc Natl Acad Sci U S A, 111 (2014) E3785–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Bunda S, Burrell K, Heir P, Zeng L, Alamsahebpour A, Kano Y, Raught B, Zhang ZY, Zadeh G, Ohh M, Inhibition of SHP2-mediated dephosphorylation of Ras suppresses oncogenesis, Nat Commun, 6 (2015) 8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kano Y, Gebregiworgis T, Marshall CB, Radulovich N, Poon BPK, St-Germain J, Cook JD, Valencia-Sama I, Grant BMM, Herrera SG, Miao J, Raught B, Irwin MS, Lee JE, Yeh JJ, Zhang Z-Y, Tsao M-S, Ikura M, Ohh M, Tyrosyl phosphorylation of KRAS stalls GTPase cycle via alteration of switch I and II conformation, Nature Communications, 10 (2019) 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Mainardi S, Mulero-Sanchez A, Prahallad A, Germano G, Bosma A, Krimpenfort P, Lieftink C, Steinberg JD, de Wit N, Goncalves-Ribeiro S, Nadal E, Bardelli A, Villanueva A, Bernards R, SHP2 is required for growth of KRAS-mutant non-small-cell lung cancer in vivo, Nat Med, 24 (2018) 961–967. [DOI] [PubMed] [Google Scholar]
- [49].Ruess DA, Heynen GJ, Ciecielski KJ, Ai J, Berninger A, Kabacaoglu D, Gorgulu K, Dantes Z, Wormann SM, Diakopoulos KN, Karpathaki AF, Kowalska M, Kaya-Aksoy E, Song L, van der Laan EAZ, Lopez-Alberca MP, Nazare M, Reichert M, Saur D, Erkan MM, Hopt UT, Sainz B Jr., Birchmeier W, Schmid RM, Lesina M, Algul H, Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase, Nat Med, 24 (2018) 954–960. [DOI] [PubMed] [Google Scholar]
- [50].Fedele C, Ran H, Diskin B, Wei W, Jen J, Geer MJ, Araki K, Ozerdem U, Simeone DM, Miller G, Neel BG, Tang KH, SHP2 Inhibition Prevents Adaptive Resistance to MEK Inhibitors in Multiple Cancer Models, Cancer Discov, 8 (2018) 1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wong GS, Zhou J, Liu JB, Wu Z, Xu X, Li T, Xu D, Schumacher SE, Puschhof J, McFarland J, Zou C, Dulak A, Henderson L, Xu P, O’Day E, Rendak R, Liao WL, Cecchi F, Hembrough T, Schwartz S, Szeto C, Rustgi AK, Wong KK, Diehl JA, Jensen K, Graziano F, Ruzzo A, Fereshetian S, Mertins P, Carr SA, Beroukhim R, Nakamura K, Oki E, Watanabe M, Baba H, Imamura Y, Catenacci D, Bass AJ, Targeting wild-type KRAS-amplified gastroesophageal cancer through combined MEK and SHP2 inhibition, Nat Med, 24 (2018) 968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Drazic A, Myklebust LM, Ree R, Arnesen T, The world of protein acetylation, Biochim Biophys Acta, 1864 (2016) 1372–1401. [DOI] [PubMed] [Google Scholar]
- [53].Yang MH, Nickerson S, Kim ET, Liot C, Laurent G, Spang R, Philips MR, Shan Y, Shaw DE, Bar-Sagi D, Haigis MC, Haigis KM, Regulation of RAS oncogenicity by acetylation, Proc Natl Acad Sci U S A, 109 (2012) 10843–10848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Knyphausen P, Lang F, Baldus L, Extra A, Lammers M, Insights into K-Ras 4B regulation by post-translational lysine acetylation, Biol Chem, 397 (2016) 1071–1085. [DOI] [PubMed] [Google Scholar]
- [55].Yin G, Kistler S, George SD, Kuhlmann N, Garvey L, Huynh M, Bagni RK, Lammers M, Der CJ, Campbell SL, A KRAS GTPase K104Q Mutant Retains Downstream Signaling by Offsetting Defects in Regulation, J Biol Chem, 292 (2017) 4446–4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Dharmaiah S, Tran TH, Messing S, Agamasu C, Gillette WK, Yan W, Waybright T, Alexander P, Esposito D, Nissley DV, McCormick F, Stephen AG, Simanshu DK, Structures of N-terminally processed KRAS provide insight into the role of N-acetylation, Sci Rep, 9 (2019) 10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Xu L, Lubkov V, Taylor LJ, Bar-Sagi D, Feedback regulation of Ras signaling by Rabex-5-mediated ubiquitination, Curr Biol, 20 (2010) 1372–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Yan H, Jahanshahi M, Horvath EA, Liu HY, Pfleger CM, Rabex-5 ubiquitin ligase activity restricts Ras signaling to establish pathway homeostasis in Drosophila, Curr Biol, 20 (2010) 1378–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bigenzahn JW, Collu GM, Kartnig F, Pieraks M, Vladimer GI, Heinz LX, Sedlyarov V, Schischlik F, Fauster A, Rebsamen M, Parapatics K, Blomen VA, Muller AC, Winter GE, Kralovics R, Brummelkamp TR, Mlodzik M, Superti-Furga G, LZTR1 is a regulator of RAS ubiquitination and signaling, Science, 362 (2018) 1171–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Steklov M, Pandolfi S, Baietti MF, Batiuk A, Carai P, Najm P, Zhang M, Jang H, Renzi F, Cai Y, Abbasi Asbagh L, Pastor T, De Troyer M, Simicek M, Radaelli E, Brems H, Legius E, Tavernier J, Gevaert K, Impens F, Messiaen L, Nussinov R, Heymans S, Eyckerman S, Sablina AA, Mutations in LZTR1 drive human disease by dysregulating RAS ubiquitination, Science, 362 (2018) 1177–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Jura N, Scotto-Lavino E, Sobczyk A, Bar-Sagi D, Differential modification of Ras proteins by ubiquitination, Mol Cell, 21 (2006) 679–687. [DOI] [PubMed] [Google Scholar]
- [62].Pfleger CM, Ubiquitin on ras: warden or partner in crime?, Sci Signal, 4 (2011) pe12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Sasaki AT, Carracedo A, Locasale JW, Anastasiou D, Takeuchi K, Kahoud ER, Haviv S, Asara JM, Pandolfi PP, Cantley LC, Ubiquitination of K-Ras enhances activation and facilitates binding to select downstream effectors, Sci Signal, 4 (2011) ra13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Baker R, Wilkerson EM, Sumita K, Isom DG, Sasaki AT, Dohlman HG, Campbell SL, Differences in the regulation of K-Ras and H-Ras isoforms by monoubiquitination, J Biol Chem, 288 (2013) 36856–36862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Martinez-Ruiz A, Cadenas S, Lamas S, Nitric oxide signaling: classical, less classical, and nonclassical mechanisms, Free Radic Biol Med, 51 (2011) 17–29. [DOI] [PubMed] [Google Scholar]
- [66].Contestabile A, Ciani E, Role of nitric oxide in the regulation of neuronal proliferation, survival and differentiation, Neurochem Int, 45 (2004) 903–914. [DOI] [PubMed] [Google Scholar]
- [67].Santos AI, Carreira BP, Izquierdo-Álvarez A, Ramos E, Lourenço AS, Filipa Santos D, Morte MI, Ribeiro LF, Marreiros A, Sánchez-López N, Marina A, Carvalho CM, Martínez-Ruiz A, Araújo IM, S-Nitrosylation of Ras Mediates Nitric Oxide-Dependent Post-Injury Neurogenesis in a Seizure Model, Antioxidants & Redox Signaling, 28 (2018) 15–30. [DOI] [PubMed] [Google Scholar]
- [68].Grek CL, Tew KD, Redox metabolism and malignancy, Current Opinion in Pharmacology, 10 (2010) 362–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Buhrman G, Holzapfel G, Fetics S, Mattos C, Allosteric modulation of Ras positions Q61 for a direct role in catalysis, Proc Natl Acad Sci U S A, 107 (2010) 4931–4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Buhrman G, Wink G, Mattos C, Transformation efficiency of RasQ61 mutants linked to structural features of the switch regions in the presence of Raf, Structure, 15 (2007) 1618–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Pai EF, Kabsch W, Krengel U, Holmes KC, John J, Wittinghofer A, Structure of the guanine-nucleotide-binding domain of the Ha-ras oncogene product p21 in the triphosphate conformation, Nature, 341 (1989) 209–214. [DOI] [PubMed] [Google Scholar]
- [72].Milburn MV, Tong L, deVos AM, Brunger A, Yamaizumi Z, Nishimura S, Kim SH, Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins, Science, 247 (1990) 939–945. [DOI] [PubMed] [Google Scholar]
- [73].Boriack-Sjodin PA, Margarit SM, Bar-Sagi D, Kuriyan J, The structural basis of the activation of Ras by Sos, Nature, 394 (1998) 337–343. [DOI] [PubMed] [Google Scholar]
- [74].Parker JA, Mattos C, The Ras–Membrane Interface: Isoform-Specific Differences in the Catalytic Domain, Molecular Cancer Research, 13 (2015) 595–603. [DOI] [PubMed] [Google Scholar]
- [75].Gorfe AA, Hanzal-Bayer M, Abankwa D, Hancock JF, McCammon JA, Structure and dynamics of the full-length lipid-modified H-Ras protein in a 1,2-dimyristoylglycero-3-phosphocholine bilayer, J Med Chem, 50 (2007) 674–684. [DOI] [PubMed] [Google Scholar]
- [76].Gorfe AA, Mechanisms of allostery and membrane attachment in Ras GTPases: implications for anti-cancer drug discovery, Curr Med Chem, 17 (2010) 1–9. [DOI] [PubMed] [Google Scholar]
- [77].Mazhab-Jafari MT, Marshall CB, Smith MJ, Gasmi-Seabrook GM, Stathopulos PB, Inagaki F, Kay LE, Neel BG, Ikura M, Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site, Proc Natl Acad Sci U S A, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].End DW, Smets G, Todd AV, Applegate TL, Fuery CJ, Angibaud P, Venet M, Sanz G, Poignet H, Skrzat S, Devine A, Wouters W, Bowden C, Characterization of the antitumor effects of the selective farnesyl protein transferase inhibitor R115777 in vivo and in vitro, Cancer Res, 61 (2001) 131–137. [PubMed] [Google Scholar]
- [79].Njoroge FG, Taveras AG, Kelly J, Remiszewski S, Mallams AK, Wolin R, Afonso A, Cooper AB, Rane DF, Liu YT, Wong J, Vibulbhan B, Pinto P, Deskus J, Alvarez CS, del Rosario J, Connolly M, Wang J, Desai J, Rossman RR, Bishop WR, Patton R, Wang L, Kirschmeier P, Ganguly AK, et al. , (+)-4-[2-[4-(8-Chloro-3,10-dibromo-6,11-dihydro-5H-benzo[5, 6]cyclohepta[1,2-b]- pyridin-11(R)-yl)-1-piperidinyl]-2-oxo-ethyl]-1-piperidinecarboxamide (SCH-66336): a very potent farnesyl protein transferase inhibitor as a novel antitumor agent, Journal of medicinal chemistry, 41 (1998) 4890–4902. [DOI] [PubMed] [Google Scholar]
- [80].Whyte DB, Kirschmeier P, Hockenberry TN, Nunez-Oliva I, James L, Catino JJ, Bishop WR, Pai JK, K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors, J Biol Chem, 272 (1997) 14459–14464. [DOI] [PubMed] [Google Scholar]
- [81].Berndt N, Hamilton AD, Sebti SM, Targeting protein prenylation for cancer therapy, Nature Reviews Cancer, 11 (2011) 775–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].James GL, Goldstein JL, Brown MS, Polylysine and CVIM sequences of K-RasB dictate specificity of prenylation and confer resistance to benzodiazepine peptidomimetic in vitro, J Biol Chem, 270 (1995) 6221–6226. [DOI] [PubMed] [Google Scholar]
- [83].Rowell CA, Kowalczyk JJ, Lewis MD, Garcia AM, Direct demonstration of geranylgeranylation and farnesylation of Ki-Ras in vivo, J Biol Chem, 272 (1997) 14093–14097. [DOI] [PubMed] [Google Scholar]
- [84].Koh C, Canini L, Dahari H, Zhao X, Uprichard SL, Haynes-Williams V, Winters MA, Subramanya G, Cooper SL, Pinto P, Wolff EF, Bishop R, Ai Thanda Han M, Cotler SJ, Kleiner DE, Keskin O, Idilman R, Yurdaydin C, Glenn JS, Heller T, Oral prenylation inhibition with lonafarnib in chronic hepatitis D infection: a proof-of-concept randomised, double-blind, placebo-controlled phase 2A trial, Lancet Infect Dis, 15 (2015) 1167–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Merideth MA, Gordon LB, Clauss S, Sachdev V, Smith AC, Perry MB, Brewer CC, Zalewski C, Kim HJ, Solomon B, Brooks BP, Gerber LH, Turner ML, Domingo DL, Hart TC, Graf J, Reynolds JC, Gropman A, Yanovski JA, Gerhard-Herman M, Collins FS, Nabel EG, Cannon RO 3rd, Gahl WA, Introne WJ, Phenotype and course of Hutchinson-Gilford progeria syndrome, N Engl J Med, 358 (2008) 592–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].De Sandre-Giovannoli A, Bernard R, Cau P, Navarro C, Amiel J, Boccaccio I, Lyonnet S, Stewart CL, Munnich A, Le Merrer M, Levy N, Lamin a truncation in Hutchinson-Gilford progeria, Science, 300 (2003) 2055. [DOI] [PubMed] [Google Scholar]
- [87].Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, Erdos MR, Robbins CM, Moses TY, Berglund P, Dutra A, Pak E, Durkin S, Csoka AB, Boehnke M, Glover TW, Collins FS, Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome, Nature (London), 423 (2003) 293–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Gordon LB, Shappell H, Massaro J, D’Agostino RB Sr., Brazier J, Campbell SE, Kleinman ME, Kieran MW, Association of Lonafarnib Treatment vs No Treatment With Mortality Rate in Patients With Hutchinson-Gilford Progeria Syndrome, JAMA, 319 (2018) 1687–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Gordon LB, Kleinman ME, Miller DT, Neuberg DS, Giobbie-Hurder A, Gerhard-Herman M, Smoot LB, Gordon CM, Cleveland R, Snyder BD, Fligor B, Bishop WR, Statkevich P, Regen A, Sonis A, Riley S, Ploski C, Correia A, Quinn N, Ullrich NJ, Nazarian A, Liang MG, Huh SY, Schwartzman A, Kieran MW, Clinical trial of a farnesyltransferase inhibitor in children with Hutchinson-Gilford progeria syndrome, Proc Natl Acad Sci U S A, 109 (2012) 16666–16671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Morgan MA, Onono FO, Spielmann HP, Subramanian T, Scherr M, Venturini L, Dallmann I, Ganser A, Reuter CW, Modulation of anthracycline-induced cytotoxicity by targeting the prenylated proteome in myeloid leukemia cells, J Mol Med (Berl), 90 (2012) 149–161. [DOI] [PubMed] [Google Scholar]
- [91].Lobell RB, Liu D, Buser CA, Davide JP, DePuy E, Hamilton K, Koblan KS, Lee Y, Mosser S, Motzel SL, Abbruzzese JL, Fuchs CS, Rowinsky EK, Rubin EH, Sharma S, Deutsch PJ, Mazina KE, Morrison BW, Wildonger L, Yao SL, Kohl NE, Preclinical and clinical pharmacodynamic assessment of L-778,123, a dual inhibitor of farnesyl:protein transferase and geranylgeranyl:protein transferase type-I, Mol Cancer Ther, 1 (2002) 747–758. [PubMed] [Google Scholar]
- [92].Britten CD, Rowinsky EK, Soignet S, Patnaik A, Yao SL, Deutsch P, Lee Y, Lobell RB, Mazina KE, McCreery H, Pezzuli S, Spriggs D, A phase I and pharmacological study of the farnesyl protein transferase inhibitor L-778,123 in patients with solid malignancies, Clin Cancer Res, 7 (2001) 3894–3903. [PubMed] [Google Scholar]
- [93].Santini D, Caraglia M, Vincenzi B, Holen I, Scarpa S, Budillon A, Tonini G, Mechanisms of disease: Preclinical reports of antineoplastic synergistic action of bisphosphonates, Nat Clin Pract Oncol, 3 (2006) 325–338. [DOI] [PubMed] [Google Scholar]
- [94].Porru M, Zappavigna S, Salzano G, Luce A, Stoppacciaro A, Balestrieri ML, Artuso S, Lusa S, De Rosa G, Leonetti C, Caraglia M, Medical treatment of orthotopic glioblastoma with transferrin-conjugated nanoparticles encapsulating zoledronic acid, Oncotarget, 5 (2014) 10446–10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Salzano G, Zappavigna S, Luce A, D’Onofrio N, Balestrieri ML, Grimaldi A, Lusa S, Ingrosso D, Artuso S, Porru M, Leonetti C, Caraglia M, De Rosa G, Transferrin-Targeted Nanoparticles Containing Zoledronic Acid as a Potential Tool to Inhibit Glioblastoma Growth, J Biomed Nanotechnol, 12 (2016) 811–830. [DOI] [PubMed] [Google Scholar]
- [96].Konstantinopoulos PA, Karamouzis MV, Papavassiliou AG, Post-translational modifications and regulation of the RAS superfamily of GTPases as anticancer targets, Nat Rev Drug Discov, 6 (2007) 541–555. [DOI] [PubMed] [Google Scholar]
- [97].Chandra A, Grecco HE, Pisupati V, Perera D, Cassidy L, Skoulidis F, Ismail SA, Hedberg C, Hanzal-Bayer M, Venkitaraman AR, Wittinghofer A, Bastiaens PI, The GDI-like solubilizing factor PDEdelta sustains the spatial organization and signalling of Ras family proteins, Nat Cell Biol, 14 (2011) 148–158. [DOI] [PubMed] [Google Scholar]
- [98].Schmick M, Vartak N, Papke B, Kovacevic M, Truxius DC, Rossmannek L, Bastiaens PIH, KRas Localizes to the Plasma Membrane by Spatial Cycles of Solubilization, Trapping and Vesicular Transport, Cell, 157 (2014) 459–471. [DOI] [PubMed] [Google Scholar]
- [99].Zimmermann G, Papke B, Ismail S, Vartak N, Chandra A, Hoffmann M, Hahn SA, Triola G, Wittinghofer A, Bastiaens PIH, Waldmann H, Small molecule inhibition of the KRAS-PDEδ interaction impairs oncogenic KRAS signalling, Nature (London), 497 (2013) 638–642. [DOI] [PubMed] [Google Scholar]
- [100].Papke B, Murarka S, Vogel HA, Martin-Gago P, Kovacevic M, Truxius DC, Fansa EK, Ismail S, Zimmermann G, Heinelt K, Schultz-Fademrecht C, Al Saabi A, Baumann M, Nussbaumer P, Wittinghofer A, Waldmann H, Bastiaens PIH, Identification of pyrazolopyridazinones as PDEδ inhibitors, Nature Communications, 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Martin-Gago P, Fansa EK, Klein CH, Murarka S, Janning P, Schurmann M, Metz M, Ismail S, Schultz-Fademrecht C, Baumann M, Bastiaens PI, Wittinghofer A, Waldmann H, A PDE6δ-KRas Inhibitor Chemotype with up to Seven H-Bonds and Picomolar Affinity that Prevents Efficient Inhibitor Release by Arl2, Angewandte Chemie, 56 (2017) 2423–2428. [DOI] [PubMed] [Google Scholar]
- [102].Thompson HJ, Jiang C, Lu J, Mehta RG, Piazza GA, Paranka NS, Pamukcu R, Ahnen DJ, Sulfone metabolite of sulindac inhibits mammary carcinogenesis, Cancer Res, 57 (1997) 267–271. [PubMed] [Google Scholar]
- [103].Herrmann C, Block C, Geisen C, Haas K, Weber C, Winde G, Moroy T, Muller O, Sulindac sulfide inhibits Ras signaling, Oncogene, 17 (1998) 1769–1776. [DOI] [PubMed] [Google Scholar]
- [104].Upadhyaya P, Qian Z, Selner NG, Clippinger SR, Wu Z, Briesewitz R, Pei D, Inhibition of Ras signaling by blocking Ras-effector interactions with cyclic peptides, Angewandte Chemie, 54 (2015) 7602–7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Ng S, Juang Y-C, Chandramohan A, Kaan HYK, Sadruddin A, Yuen TY, Ferrer FJ, Lee XEC, Xi L, Johannes CW, Brown CJ, Kannan S, Aronica PG, Berglund N, Verma CS, Liu L, Stoeck A, Sawyer TK, Partridge AW, Lane DP, De-risking drug discovery of intracellular targeting peptides: screening strategies to eliminate false-positive hits, bioRxiv, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Kato-Stankiewicz J, Hakimi I, Zhi G, Zhang J, Serebriiskii I, Guo L, Edamatsu H, Koide H, Menon S, Eckl R, Sakamuri S, Lu Y, Chen QZ, Agarwal S, Baumbach WR, Golemis EA, Tamanoi F, Khazak V, Inhibitors of Ras/Raf-1 interaction identified by two-hybrid screening revert Ras-dependent transformation phenotypes in human cancer cells, Proc Natl Acad Sci U S A, 99 (2002) 14398–14403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Lu Y, Sakamuri S, Chen QZ, Keng YF, Khazak V, Illgen K, Schabbert S, Weber L, Menon SR, Solution phase parallel synthesis and evaluation of MAPK inhibitory activities of close structural analogues of a Ras pathway modulator, Bioorg Med Chem Lett, 14 (2004) 3957–3962. [DOI] [PubMed] [Google Scholar]
- [108].Skobeleva N, Menon S, Weber L, Golemis EA, Khazak V, In vitro and in vivo synergy of MCP compounds with mitogen-activated protein kinase pathway- and microtubule-targeting inhibitors, Mol Cancer Ther, 6 (2007) 898–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Gonzalez-Perez V, Reiner DJ, Alan JK, Mitchell C, Edwards LJ, Khazak V, Der CJ, Cox AD, Genetic and functional characterization of putative Ras/Raf interaction inhibitors in C. elegans and mammalian cells, J Mol Signal, 5 (2010) 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Maurer T, Garrenton LS, Oh A, Pitts K, Anderson DJ, Skelton NJ, Fauber BP, Pan B, Malek S, Stokoe D, Ludlam MJ, Bowman KK, Wu J, Giannetti AM, Starovasnik MA, Mellman I, Jackson PK, Rudolph J, Wang W, Fang G, Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity, Proc Natl Acad Sci U S A, 109 (2012) 5299–5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Grant BJ, Lukman S, Hocker HJ, Sayyah J, Brown JH, McCammon JA, Gorfe AA, Novel allosteric sites on Ras for lead generation, PLoS One, 6 (2011) e25711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Prakash P, Hancock JF, Gorfe AA, Binding hotspots on K-ras: consensus ligand binding sites and other reactive regions from probe-based molecular dynamics analysis, Proteins, 83 (2015) 898–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].McCarthy MJ, Pagba CV, Prakash P, Naji AK, van der Hoeven D, Liang H, Gupta AK, Zhou Y, Cho KJ, Hancock JF, Gorfe AA, Discovery of High-Affinity Noncovalent Allosteric KRAS Inhibitors That Disrupt Effector Binding, ACS Omega, 4 (2019) 2921–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Shima F, Yoshikawa Y, Ye M, Araki M, Matsumoto S, Liao J, Hu L, Sugimoto T, Ijiri Y, Takeda A, Nishiyama Y, Sato C, Muraoka S, Tamura A, Osoda T, Tsuda K, Miyakawa T, Fukunishi H, Shimada J, Kumasaka T, Yamamoto M, Kataoka T, In silico discovery of small-molecule Ras inhibitors that display antitumor activity by blocking the Ras-effector interaction, Proc Natl Acad Sci U S A, 110 (2013) 8182–8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Kessler D, Gmachl M, Mantoulidis A, Martin LJ, Zoephel A, Mayer M, Gollner A, Covini D, Fischer S, Gerstberger T, Gmaschitz T, Goodwin C, Greb P, Haring D, Hela W, Hoffmann J, Karolyi-Oezguer J, Knesl P, Kornigg S, Koegl M, Kousek R, Lamarre L, Moser F, Munico-Martinez S, Peinsipp C, Phan J, Rinnenthal J, Sai J, Salamon C, Scherbantin Y, Schipany K, Schnitzer R, Schrenk A, Sharps B, Siszler G, Sun Q, Waterson A, Wolkerstorfer B, Zeeb M, Pearson M, Fesik SW, McConnell DB, Drugging an undruggable pocket on KRAS, Proc Natl Acad Sci U S A, 116 (2019) 15823–15829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Patgiri A, Yadav KK, Arora PS, Bar-Sagi D, An orthosteric inhibitor of the Ras-Sos interaction, Nat Chem Biol, 7 (2011) 585–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Leshchiner ES, Parkhitko A, Bird GH, Luccarelli J, Bellairs JA, Escudero S, Opoku-Nsiah K, Godes M, Perrimon N, Walensky LD, Direct inhibition of oncogenic KRAS by hydrocarbon-stapled SOS1 helices, Proc Natl Acad Sci U S A, 112 (2015) 1761–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Jansen JM, Wartchow C, Jahnke W, Fong S, Tsang T, Pfister K, Zavorotinskaya T, Bussiere D, Cheng JM, Crawford K, Dai Y, Dove J, Fang E, Feng Y, Florent JM, Fuller J, Gossert AD, Hekmat-Nejad M, Henry C, Klopp J, Lenahan WP, Lingel A, Ma S, Meyer A, Mishina Y, Narberes J, Pardee G, Ramurthy S, Rieffel S, Stuart D, Subramanian S, Tandeske L, Widger S, Widmer A, Winterhalter A, Zaror I, Hardy S, Inhibition of prenylated KRAS in a lipid environment, PLoS One, 12 (2017) e0174706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Fang Z, Marshall CB, Nishikawa T, Gossert AD, Jansen JM, Jahnke W, Ikura M, Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site, Cell Chem Biol, (2018). [DOI] [PubMed] [Google Scholar]
- [120].Welsch ME, Kaplan A, Chambers JM, Stokes ME, Bos PH, Zask A, Zhang Y, Sanchez-Martin M, Badgley MA, Huang CS, Tran TH, Akkiraju H, Brown LM, Nandakumar R, Cremers S, Yang WS, Tong L, Olive KP, Ferrando A, Stockwell BR, Multivalent Small-Molecule Pan-RAS Inhibitors, Cell, 168 (2017) 878–889 e829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM, K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions, Nature, 503 (2013) 548–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Fell JB, Fischer JP, Baer BR, Ballard J, Blake JF, Bouhana K, Brandhuber BJ, Briere DM, Burgess LE, Burkard MR, Chiang H, Chicarelli MJ, Davidson K, Gaudino JJ, Hallin J, Hanson L, Hee K, Hicken EJ, Hinklin RJ, Marx MA, Mejia MJ, Olson P, Savechenkov P, Sudhakar N, Tang TP, Vigers GP, Zecca H, Christensen JG, Discovery of Tetrahydropyridopyrimidines as Irreversible Covalent Inhibitors of KRAS-G12C with In Vivo Activity, ACS Med Chem Lett, 9 (2018) 1230–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].KRAS G12C Inhibitor (MRTX849), https://www.mirati.com/mrtx849/ (2019).
- [124].Furth ME, Davis LJ, Fleurdelys B, Scolnick EM, Monoclonal antibodies to the p21 products of the transforming gene of Harvey murine sarcoma virus and of the cellular ras gene family, J Virol, 43 (1982) 294–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Mulcahy LS, Smith MR, Stacey DW, Requirement for ras proto-oncogene function during serum-stimulated growth of NIH 3T3 cells, Nature (London), 313 (1985) 241–243. [DOI] [PubMed] [Google Scholar]
- [126].Lacal JC, Aaronson SA, Monoclonal-Antibody Y13–259 Recognizes an Epitope of the P21 Ras Molecule Not Directly Involved in the Gtp-Binding Activity of the Protein, Molecular and Cellular Biology, 6 (1986) 1002–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Cardinale A, Lener M, Messina S, Cattaneo A, Biocca S, The mode of action of Y13–259 scFv fragment intracellularly expressed in mammalian cells, Febs Letters, 439 (1998) 197–202. [DOI] [PubMed] [Google Scholar]
- [128].Clark R, Wong G, Arnheim N, Nitecki D, McCormick F, Antibodies specific for amino acid 12 of the ras oncogene product inhibit GTP binding, Proc Natl Acad Sci U S A, 82 (1985) 5280–5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Feramisco JR, Clark R, Wong G, Arnheim N, Milley R, McCormick F, Transient reversion of ras oncogene-induced cell transformation by antibodies specific for amino acid 12 of ras protein, Nature (London), 314 (1985) 639–642. [DOI] [PubMed] [Google Scholar]
- [130].Smith MJ, Marshall CB, Theillet FX, Binolfi A, Selenko P, Ikura M, Real-time NMR monitoring of biological activities in complex physiological environments, Curr Opin Struct Biol, 32C (2015) 39–47. [DOI] [PubMed] [Google Scholar]
- [131].Wong KA, Russo A, Wang X, Chen Y-J, Lavie A, O’Bryan JP, A new dimension to Ras function: a novel role for nucleotide-free Ras in Class II phosphatidylinositol 3-kinase beta (PI3K-C2b) regulation, PLoS ONE, 7 (2012) e45360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Schmitz AL, Schrage R, Gaffal E, Charpentier TH, Wiest J, Hiltensperger G, Morschel J, Hennen S, Haussler D, Horn V, Wenzel D, Grundmann M, Bullesbach KM, Schroder R, Brewitz HH, Schmidt J, Gomeza J, Gales C, Fleischmann BK, Tuting T, Imhof D, Tietze D, Gutschow M, Holzgrabe U, Sondek J, Harden TK, Mohr K, Kostenis E, A cell-permeable inhibitor to trap Gαq proteins in the empty pocket conformation, Chem Biol, 21 (2014) 890–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Lofblom J, Feldwisch J, Tolmachev V, Carlsson J, Stahl S, Frejd FY, Affibody molecules: engineered proteins for therapeutic, diagnostic and biotechnological applications, FEBS Lett, 584 (2010) 2670–2680. [DOI] [PubMed] [Google Scholar]
- [134].Yang JL, Liu DX, Zhen SJ, Zhou YG, Zhang DJ, Yang LY, Chen HB, Feng Q, A novel anti-p21Ras scFv antibody reacting specifically with human tumour cell lines and primary tumour tissues, BMC Cancer, 16 (2016) 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Guillard S, Kolasinska-Zwierz P, Debreczeni J, Breed J, Zhang J, Bery N, Marwood R, Tart J, Overman R, Stocki P, Mistry B, Phillips C, Rabbitts T, Jackson R, Minter R, Structural and functional characterization of a DARPin which inhibits Ras nucleotide exchange, Nat Commun, 8 (2017) 16111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Spencer-Smith R, Koide A, Zhou Y, Eguchi RR, Sha F, Gajwani P, Santana D, Gupta A, Jacobs M, Herrero-Garcia E, Cobbert J, Lavoie H, Smith M, Rajakulendran T, Dowdell E, Okur MN, Dementieva I, Sicheri F, Therrien M, Hancock JF, Ikura M, Koide S, O’Bryan JP, Inhibition of RAS function through targeting an allosteric regulatory site, Nat Chem Biol, 13 (2017) 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Spencer-Smith R, Li L, Prasad S, Koide A, Koide S, O’Bryan JP, Targeting the alpha4-alpha5 interface of RAS results in multiple levels of inhibition, Small GTPases, (2017) 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Sha F, Salzman G, Gupta A, Koide S, Monobodies and other synthetic binding proteins for expanding protein science, Protein Sci, 26 (2017) 910–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Tanaka T, Lobato MN, Rabbitts TH, Single domain intracellular antibodies: a minimal fragment for direct in vivo selection of antigen-specific intrabodies, J Mol Biol, 331 (2003) 1109–1120. [DOI] [PubMed] [Google Scholar]
- [140].Tanaka T, Rabbitts TH, Interfering with RAS-effector protein interactions prevent RAS-dependent tumour initiation and causes stop-start control of cancer growth, Oncogene, 29 (2010) 6064–6070. [DOI] [PubMed] [Google Scholar]
- [141].Tanaka T, Williams RL, Rabbitts TH, Tumour prevention by a single antibody domain targeting the interaction of signal transduction proteins with RAS, Embo J, 26 (2007) 3250–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Quevedo CE, Cruz-Migoni A, Bery N, Miller A, Tanaka T, Petch D, Bataille CJR, Lee LYW, Fallon PS, Tulmin H, Ehebauer MT, Fernandez-Fuentes N, Russell AJ, Carr SB, Phillips SEV, Rabbitts TH, Small molecule inhibitors of RAS-effector protein interactions derived using an intracellular antibody fragment, Nat Commun, 9 (2018) 3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Bery N, Cruz-Migoni A, Bataille CJ, Quevedo CE, Tulmin H, Miller A, Russell A, Phillips SE, Carr SB, Rabbitts TH, BRET-based RAS biosensors that show a novel small molecule is an inhibitor of RAS-effector protein-protein interactions, Elife, 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Shin SM, Choi DK, Jung K, Bae J, Kim JS, Park SW, Song KH, Kim YS, Antibody targeting intracellular oncogenic Ras mutants exerts anti-tumour effects after systemic administration, Nat Commun, 8 (2017) 15090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Kauke MJ, Traxlmayr MW, Parker JA, Kiefer JD, Knihtila R, McGee J, Verdine G, Mattos C, Wittrup KD, An engineered protein antagonist of K-Ras/B-Raf interaction, Sci Rep, 7 (2017) 5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Guldenhaupt J, Rudack T, Bachler P, Mann D, Triola G, Waldmann H, Kotting C, Gerwert K, N-Ras forms dimers at POPC membranes, Biophys J, 103 (2012) 1585–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Kovrigina EA, Galiakhmetov AR, Kovrigin EL, The Ras G Domain Lacks the Intrinsic Propensity to Form Dimers, Biophys J, 109 (2015) 1000–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Santos E, Dimerization opens new avenues into Ras signaling research, Sci Signal, 7 (2014) pe12. [DOI] [PubMed] [Google Scholar]
- [149].Santos E, Nebreda AR, Bryan T, Kempner ES, Oligomeric structure of p21 ras proteins as determined by radiation inactivation, J Biol Chem, 263 (1988) 9853–9858. [PubMed] [Google Scholar]
- [150].Zhang XF, Settleman J, Kyriakis JM, Takeuchi-Suzuki E, Elledge SJ, Marshall MS, Bruder JT, Rapp UR, Avruch J, Normal and oncogenic p21ras proteins bind to the amino-terminal regulatory domain of c-Raf-1, Nature (London), 364 (1993) 308–313. [DOI] [PubMed] [Google Scholar]
- [151].Vojtek AB, Hollenberg SM, Cooper JA, Mammalian Ras interacts directly with the serine/threonine kinase Raf, Cell, 74 (1993) 205–214. [DOI] [PubMed] [Google Scholar]
- [152].Moodie SA, Willumsen BM, Weber MJ, Wolfman A, Complexes of Ras. GTP with Raf-1 and mitogen-activated protein kinase kinase, Science, 260 (1993) 1658–1661. [DOI] [PubMed] [Google Scholar]
- [153].Van Aelst L, Barr M, Marcus S, Polverino A, Wigler M, Complex formation between RAS and RAF and other protein kinases, Proc Natl Acad Sci U S A, 90 (1993) 6213–6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Warne PH, Viciana PR, Downward J, Direct interaction of Ras and the amino-terminal region of Raf-1 in vitro, Nature (London), 364 (1993) 352–355. [DOI] [PubMed] [Google Scholar]
- [155].Weber CK, Slupsky JR, Kalmes HA, Rapp UR, Active Ras induces heterodimerization of cRaf and BRaf, Cancer Res, 61 (2001) 3595–3598. [PubMed] [Google Scholar]
- [156].Rushworth LK, Hindley AD, O’Neill E, Kolch W, Regulation and role of Raf-1/B-Raf heterodimerization, Mol Cell Biol, 26 (2006) 2262–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Rajakulendran T, Sahmi M, Lefrancois M, Sicheri F, Therrien M, A dimerization-dependent mechanism drives RAF catalytic activation, Nature (London), 461 (2009) 542–545. [DOI] [PubMed] [Google Scholar]
- [158].Inouye K, Mizutani S, Koide H, Kaziro Y, Formation of the Ras dimer is essential for Raf-1 activation, J Biol Chem, 275 (2000) 3737–3740. [DOI] [PubMed] [Google Scholar]
- [159].Nan X, Tamguney TM, Collisson EA, Lin LJ, Pitt C, Galeas J, Lewis S, Gray JW, McCormick F, Chu S, Ras-GTP dimers activate the Mitogen-Activated Protein Kinase (MAPK) pathway, Proc Natl Acad Sci U S A, 112 (2015) 7996–8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Nan X, Collisson EA, Lewis S, Huang J, Tamguney TM, Liphardt JT, McCormick F, Gray JW, Chu S, Single-molecule superresolution imaging allows quantitative analysis of RAF multimer formation and signaling, Proc Natl Acad Sci U S A, 110 (2013) 18519–18524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Bery N, Legg S, Debreczeni J, Breed J, Embrey K, Stubbs C, Kolasinska-Zwierz P, Barrett N, Marwood R, Watson J, Tart J, Overman R, Miller A, Philips C, Minter R, Rabbitts TH, KRAS-specific inhibition using a DARPin binding to a site in the allosteric lobe, Nat Commun, 10 (2019) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Prakash P, Sayyed-Ahmad A, Cho KJ, Dolino DM, Chen W, Li H, Grant BJ, Hancock JF, Gorfe AA, Computational and biochemical characterization of two partially overlapping interfaces and multiple weak-affinity K-Ras dimers, Sci Rep, 7 (2017) 40109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Sarkar-Banerjee S, Sayyed-Ahmad A, Prakash P, Cho KJ, Waxham MN, Hancock JF, Gorfe AA, Spatiotemporal Analysis of K-Ras Plasma Membrane Interactions Reveals Multiple High Order Homo-oligomeric Complexes, J Am Chem Soc, 139 (2017) 13466–13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Muratcioglu S, Chavan TS, Freed BC, Jang H, Khavrutskii L, Freed RN, Dyba MA, Stefanisko K, Tarasov SG, Gursoy A, Keskin O, Tarasova NI, Gaponenko V, Nussinov R, GTP-Dependent K-Ras Dimerization, Structure, 23 (2015) 1325–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].O’Connor C, Kovrigin EL, Global conformational dynamics in ras, Biochemistry, 47 (2008) 10244–10246. [DOI] [PubMed] [Google Scholar]
- [166].Kraulis PJ, Domaille PJ, Campbell-Burk SL, Van Aken T, Laue ED, Solution structure and dynamics of ras p21.GDP determined by heteronuclear three- and four-dimensional NMR spectroscopy, Biochemistry, 33 (1994) 3515–3531. [DOI] [PubMed] [Google Scholar]
- [167].Ito Y, Yamasaki K, Iwahara J, Terada T, Kamiya A, Shirouzu M, Muto Y, Kawai G, Yokoyama S, Laue ED, Walchli M, Shibata T, Nishimura S, Miyazawa T, Regional polysterism in the GTP-bound form of the human c-Ha-Ras protein, Biochemistry, 36 (1997) 9109–9119. [DOI] [PubMed] [Google Scholar]
- [168].Chung JK, Lee YK, Denson JP, Gillette WK, Alvarez S, Stephen AG, Groves JT, K-Ras4B Remains Monomeric on Membranes over a Wide Range of Surface Densities and Lipid Compositions, Biophysical journal, 114 (2018) 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Ambrogio C, Kohler J, Zhou ZW, Wang H, Paranal R, Li J, Capelletti M, Caffarra C, Li S, Lv Q, Gondi S, Hunter JC, Lu J, Chiarle R, Santamaria D, Westover KD, Janne PA, KRAS Dimerization Impacts MEK Inhibitor Sensitivity and Oncogenic Activity of Mutant KRAS, Cell, 172 (2018) 857–868 e815. [DOI] [PubMed] [Google Scholar]
- [170].Simanshu DK, Nissley DV, McCormick F, RAS Proteins and Their Regulators in Human Disease, Cell, 170 (2017) 17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Bryant KL, Stalnecker CA, Zeitouni D, Klomp JE, Peng S, Tikunov AP, Gunda V, Pierobon M, Waters AM, George SD, Tomar G, Papke B, Hobbs GA, Yan L, Hayes TK, Diehl JN, Goode GD, Chaika NV, Wang Y, Zhang GF, Witkiewicz AK, Knudsen ES, Petricoin EF 3rd, Singh PK, Macdonald JM, Tran NL, Lyssiotis CA, Ying H, Kimmelman AC, Cox AD, Der CJ, Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer, Nat Med, 25 (2019) 628–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Kinsey CG, Camolotto SA, Boespflug AM, Guillen KP, Foth M, Truong A, Schuman SS, Shea JE, Seipp MT, Yap JT, Burrell LD, Lum DH, Whisenant JR, Gilcrease GW 3rd, Cavalieri CC, Rehbein KM, Cutler SL, Affolter KE, Welm AL, Welm BE, Scaife CL, Snyder EL, McMahon M, Protective autophagy elicited by RAF-->MEK-->ERK inhibition suggests a treatment strategy for RAS-driven cancers, Nat Med, 25 (2019) 620–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Lee CS, Lee LC, Yuan TL, Chakka S, Fellmann C, Lowe SW, Caplen NJ, McCormick F, Luo J, MAP kinase and autophagy pathways cooperate to maintain RAS mutant cancer cell survival, Proc Natl Acad Sci U S A, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, Stefancsik R, Harsha B, Kok CY, Jia M, Jubb H, Sondka Z, Thompson S, De T, Campbell PJ, COSMIC: somatic cancer genetics at high-resolution, Nucleic Acids Res, 45 (2017) D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Kano Y, Gebregiworgis T, Marshall CB, Radulovich N, Poon BPK, St-Germain J, Cook JD, Valencia-Sama I, Grant BMM, Herrera SG, Miao J, Raught B, Irwin MS, Lee JE, Yeh JJ, Zhang ZY, Tsao MS, Ikura M, Ohh M, Tyrosyl phosphorylation of KRAS stalls GTPase cycle via alteration of switch I and II conformation, Nat Commun, 10 (2019) 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Chen YN, LaMarche MJ, Chan HM, Fekkes P, Garcia-Fortanet J, Acker MG, Antonakos B, Chen CH, Chen Z, Cooke VG, Dobson JR, Deng Z, Fei F, Firestone B, Fodor M, Fridrich C, Gao H, Grunenfelder D, Hao HX, Jacob J, Ho S, Hsiao K, Kang ZB, Karki R, Kato M, Larrow J, La Bonte LR, Lenoir F, Liu G, Liu S, Majumdar D, Meyer MJ, Palermo M, Perez L, Pu M, Price E, Quinn C, Shakya S, Shultz MD, Slisz J, Venkatesan K, Wang P, Warmuth M, Williams S, Yang G, Yuan J, Zhang JH, Zhu P, Ramsey T, Keen NJ, Sellers WR, Stams T, Fortin PD, Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases, Nature (London), 535 (2016) 148–152. [DOI] [PubMed] [Google Scholar]
- [177].Caraglia M, Marra M, Naviglio S, Botti G, Addeo R, Abbruzzese A, Zoledronic acid: an unending tale for an antiresorptive agent, Expert Opin Pharmacother, 11 (2010) 141–154. [DOI] [PubMed] [Google Scholar]
- [178].Salzano G, Marra M, Porru M, Zappavigna S, Abbruzzese A, La Rotonda MI, Leonetti C, Caraglia M, De Rosa G, Self-assembly nanoparticles for the delivery of bisphosphonates into tumors, Int J Pharm, 403 (2011) 292–297. [DOI] [PubMed] [Google Scholar]
- [179].Marra M, Salzano G, Leonetti C, Porru M, Franco R, Zappavigna S, Liguori G, Botti G, Chieffi P, Lamberti M, Vitale G, Abbruzzese A, La Rotonda MI, De Rosa G, Caraglia M, New self-assembly nanoparticles and stealth liposomes for the delivery of zoledronic acid: a comparative study, Biotechnol Adv, 30 (2012) 302–309. [DOI] [PubMed] [Google Scholar]
- [180].Marra M, Salzano G, Leonetti C, Tassone P, Scarsella M, Zappavigna S, Calimeri T, Franco R, Liguori G, Cigliana G, Ascani R, La Rotonda MI, Abbruzzese A, Tagliaferri P, Caraglia M, De Rosa G, Nanotechnologies to use bisphosphonates as potent anticancer agents: the effects of zoledronic acid encapsulated into liposomes, Nanomedicine, 7 (2011) 955–964. [DOI] [PubMed] [Google Scholar]
- [181].Upadhyaya P, Bedewy W, Pei D, Direct Inhibitors of Ras-Effector Protein Interactions, Mini Rev Med Chem, 16 (2016) 376–382. [DOI] [PubMed] [Google Scholar]
- [182].Mirati Therapeutics, KRAS G12C Inhibitor (MRTX849), https://www.mirati.com/mrtx849/ (2019).
- [183].Gentile DR, Rathinaswamy MK, Jenkins ML, Moss SM, Siempelkamp BD, Renslo AR, Burke JE, Shokat KM, Ras Binder Induces a Modified Switch-II Pocket in GTP and GDP States, Cell Chem Biol, 24 (2017) 1455–1466 e1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.