Abstract
Lafora progressive myoclonus epilepsy is a fatal rare neurodegenerative disorder characterized by the accumulation of insoluble abnormal glycogen deposits in the brain and peripheral tissues. Mutations in at least two genes are responsible for the disease: EPM2A, encoding the glucan phosphatase laforin, and EPM2B, encoding the RING-type E3-ubiquitin ligase malin. Both laforin and malin form a functional complex in which laforin recruits the substrates to be ubiquitinated by malin. We and others have described that, in cellular and animal models of this disease, there is an autophagy impairment which leads to the accumulation of dysfunctional mitochondria. In addition, we established that the autophagic defect occurred at the initial steps of autophagosome formation. In this work, we present evidence that in cellular models of the disease there is a decrease in the amount of phosphatidylinositol-3P. This is probably due to defective regulation of the autophagic PI3KC3 complex, in the absence of a functional laforin/malin complex. In fact, we demonstrate that the laforin/malin complex interacts physically and co-localizes intracellularly with core components of the PI3KC3 complex (Beclin1, Vps34 and Vps15), and that this interaction is specific and results in the polyubiquitination of these proteins. In addition, the laforin/malin complex is also able to polyubiquitinate ATG14L and UVRAG. Finally, we show that overexpression of the laforin/malin complex increases PI3KC3 activity. All these results suggest a new role of the laforin/malin complex in the activation of autophagy via regulation of the PI3KC3 complex and explain the defect in autophagy described in Lafora disease.
Keywords: Lafora disease, laforin, malin, ubiquitination, autophagy, Beclin1, PI3KC3, protein degradation
1.-. INTRODUCTION
Lafora disease (LD, OMIM254780, ORPHA#501) is a fatal rare form of progressive myoclonus epilepsy characterized by the accumulation of insoluble abnormal glycogen deposits in brain and peripheral tissues, called Lafora bodies (LBs) ([1], [2]). The onset of the disease occurs during childhood or early adolescence and is characterized by generalized tonic-clonic seizures, myoclonus, absences and visual hallucinations. The disease progresses rapidly with amplification of seizures and dementia, leading to death of the patient after a decade from the onset of the first symptoms. Patients are treated initially with general antiepileptic drugs, but soon they become resistant to their effects. Unfortunately, no treatment is known yet that can alleviate the disease ([1], [2]). LD is an autosomal recessive disorder and, so far, mutations in two genes are related to the disease: EPM2A gene, which encodes the glucan phosphatase laforin ([3], [4]), and EPM2B, which encodes the RING-type E3-ubiquitin ligase malin [5]. Both laforin and malin form a functional complex in which laforin has a phosphatase-independent function: it recognizes and recruits the substrates that are then ubiquitinated by malin ([6], [7], [8]). Perhaps, this is the reason why patients carrying mutations in EPM2A or EPM2B show similar pathological phenotypes [9].
Although during the last decade several groups have contributed to the understanding of the pathophysiology of LD, there are still some missing points that need to be addressed. It was described that the laforin/malin complex was a negative regulator of glycogen synthesis: the laforin/malin complex interacted with and regulated the activity of key components of glycogen synthesis such as glycogen synthase, regulatory subunits of type I protein phosphatase, glycogen debranching enzyme, etc. ([6], [7], [8], [10]). In the absence of a functional laforin/malin complex, the synthesis of glycogen is increased and an abnormal glycogen structure that is less branched and more insoluble is accumulated. This would explain the appearance of LBs in different tissues ([6], [7], [8], [10]).
In addition to the role that the laforin/malin complex has in glycogen homeostasis, we and others have described an involvement of the complex in regulating several physiological processes such as proteostasis ([11], [12], [13], [14]), oxidative stress [15] and neuroinflammation [16]. In the case of proteostasis, it has been described that the laforin/malin complex participates in regulating endoplasmic reticulum stress, proteasome function and autophagy. In the latter, it was reported that the laforin/malin complex could regulate the initial steps of autophagosome formation ([14], [17], [18]).
Autophagy is a conserved cellular pathway that mediates the lysosomal degradation of intracellular components ([19], [18], [20]). Three main autophagic pathways have been described: macroautophagy, microautophagy and chaperone-mediated autophagy ([19], [18], [20]). Macroautophagy (hereafter referred to as autophagy) is the most studied of these pathways and consists of the degradation of large cargoes like different types of intracellular organelles (e.g. damaged mitochondria), protein aggregates, etc. It starts with the formation of a double-membrane structure (named phagophore), originated from different cellular membrane sources (e.g. mitochondria, endoplasmic reticulum, Golgi complex, endosomes, plasma membrane…) that surrounds the cell components to be degraded to form the double-membrane vesicle named autophagosome [21]. In the case of selective autophagy (a form of macroautophagy), autophagosomes are loaded with specific cargoes by means of autophagy receptors (e.g., p62/SQSTM1), that recognize the autophagic substrates and certain components of the autophagosome membrane. Autophagosomes are then fused with lysosomes to form autolysosomes, which finally degrade the substrates [21]. Thus, the autophagy flux can be divided into three main steps, initiation, maturation and fusion with lysosomes ([22], [23]). The initiation process starts in response mostly to nutrient deprivation and/or low cellular energy stress and it is mediated by the activation of different signaling pathways (e.g., mTOR and AMPK pathways) that converge on the ULK1/2 complex, which is composed by the ULK1/2 protein kinases, and the proteins ATG13, ATG101 and FIP200 ([19], [18], [20]). The mTOR and the AMPK pathways inhibit or activate, respectively, the ULK1/2 complex by phosphorylation at different Ser residues (Ser757 in the case of mTOR and Ser555 in the case of AMPK). Once activated, the ULK1/2 complex activates the PI3KC3 complex which eventually synthesizes phosphatidylinositol-3P (PtdIns3P). The PI3KC3 complex consists of a core sub-complex composed of Vps34, a class III phosphatidylinositol-3 kinase, a myristoylated Vps15 protein (a Ser/Thr kinase that acts as an anchor of the sub-complex to the membrane), and Beclin1 (a Bcl2-interacting protein). This core sub-complex binds in a mutually exclusive manner either to ATG14L (a Beclin1-associated autophagy-related key regulator) or to UVRAG (UV irradiation resistance-associated gene; Vps38) ([19], [18], [20]). The activity of the PI3KC3 complex is regulated by phosphorylation of specific residues from different components of the core (e.g. ULK1/2 phosphorylates Beclin1 at Ser15; AMPK phosphorylates Beclin1 at Ser93 and Thr388; etc.). The activity of the PI3KC3 complex can also be regulated by ubiquitination. In this sense, it has been reported that Beclin1 is ubiquitinated by different E3-ligases that promote either its degradation by attaching K48-linked ubiquitin chains (e.g., Nedd4, RNF216, KLHL20-Cullin3-ROC1 complex) or its stability and activation by attaching K63-linked ubiquitin chains (e.g., TRAF6, TRIM50, DDB1-Cullin4-AMBRA1 complex) ([24], [25]).
The synthesis of PtdIns3P by the PI3KC3 complex in some parts of the phagophore allows the recruitment of additional components of the autophagy machinery which contribute to its expansion [e.g., DFCP1 (double FYVE domain-containing protein 1), WIPIs (WD-repeat protein interacting with phosphoinositides), ATG9 (a transmembrane protein), ATG12-ATG5-ATG16 complex, LC3 and LC3-phosphatidylethanolamine] and finally, to the closure of the double membrane of the autophagosome ([19], [18], [20]).
In this work, we describe the role of the laforin/malin complex in the regulation of the initiation step of autophagy. This complex interacts with and polyubiquitinates Beclin1 with K63-linked ubiquitin chains, as well as other components of the PI3KC3 complex, resulting in its activation and the initiation of the autophagic flux.
2.-. MATERIAL AND METHODS
2.1.-. Cell culture and preparation of crude extracts
Human embryonic kidney (HEK293) cells (HPA Culture Collection #85120602) and human osteosarcoma U2OS cells (Public Health England #92022711) were grown in DMEM (Lonza, Barcelona, Spain) supplemented with 100 units/mL penicillin, 100 μg/mL streptomycin, 2 mM glutamine and 10% (v/v) inactivated fetal bovine serum (GIBCO, Madrid, Spain), in a humidified atmosphere at 37°C with 5% (v/v) CO2. Cells were transfected with the corresponding plasmids using X-treme GENE HP transfection reagent (Roche Diagnostics, Barcelona, Spain), according to the manufacturer’s instructions. Twenty-four hours after transfection, cells were scraped on ice in lysis buffer [10 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 2.5% (v/v) glycerol, 0.5% (v/v) Nonidet P-40, complete protease inhibitor cocktail (Roche Diagnostics, Barcelona, Spain), 1 mM PMSF, 2.5 mM NaF, 0.5 mM NaVO4 and 2.5 mM Na4P2O7]. Cells were lysed by repeated passage through a 25-gauge needle and clarified by centrifugation at 13,000×g for 10 min at 4°C.
Primary LD fibroblasts were obtained from a compound heterozygous EPM2A Y112X/N163D (LD3) patient (obtained from Dr. Salas, Neurology Dept. Hospital Vall Hebron, Barcelona, Spain) and from a homozygous EPM2A R241X/R241X (LD1) patient (obtained from Dr. José Serratosa, Fundación Jimenez Diaz, Madrid, Spain). The corresponding informed consent for experimentation was obtained from patients or their families. Corresponding control fibroblasts (C1 and C2) (from Coriell Institute for Medical Research, Camden, New Jersey, USA) were matched by sex and age. All experiments were carried out in accordance with the Code of Ethics of the World Medical Association (Declaration of Helsinki). Experiments were performed in human fibroblasts at passage number 7–14 to avoid culture aging effects. Fibroblasts were grown at 37 °C in a humidified atmosphere at 5% (v/v) CO2/air in MEM with Earlés salts, 2 mM L-glutamine, 1x MEM amino acids, 1x MEM non-essential amino acids, 1x MEM vitamins, 100 U/ml penicillin, 100 μg/ml streptomycin and 15% (v/v) fetal bovine serum.
2.2.-. Plasmids
Plasmids pFLAG-laforin, pCMV-HA-laforin, pCMV-Myc-laforin, pECFP-laforin, pEGFP-malin, pFLAG-malin and pcDNA3-HA-malin, have already been described [17]; plasmid pFLAG-malin P69A is described in [26]; plasmids pcDNA3-Myc-Beclin1 and pEGFP-2xFYVE were a generous gift of Dr. Stenmark (Dept. Biochem. Inst. Cancer Research, Norwegian Radium Hospital, Oslo, Norway); pFLAG-TRAF6-wt (#21624) and pVitro2-hygro-N-Myc-hVps34/hVps15-C-V5-his (#24055) were from Addgene (Teddington, UK); plasmid pcDNA3-FLAG-ATG14L was from Dr. Yue (Dep. Neurol. Neurosci. Friedman Brain Institute, Icahn School of Medicine at Mount Sinai, NY, USA); plasmid pFLAG-hUVRAG was from Dr. Liang (Dept. Mol. Microbiol and Immunol. Univ. Southern California, Los Angeles, CA, USA); plasmid pcDNA3-FLAG-Bcl2 was from Dr. Borner (Inst. Biochem, Univ. Freiburg, Germany); plasmid pCLPCX-FLAG-AMBRA1 was from Dr. Fimia (Dept. Epidemiology and Preclin. Res. National Inst. Infectious Dis. IRCCS Lazzaro Spallanzani, Rome, Italy); plasmid pcDNA3.1-HA-ULK1 was from Dr. Minoo Razi (London Research Institute, UK); plasmid pCMV-6xHisUbiq was kindly provided by Dr. Manuel Rodríguez (Proteomics Unit, CIC-BioGUNE, Vizcaya, Spain) and plasmids pCMV-6xHis-Ubiq-K48R and pCMV-6xHis-Ubiq-K63R were a generous gift of Dr. Ch. Blattner (Institute of Toxicology and Genetics, Karlsruhe Institute of Technology, Karlsruhe, Germany). Plasmids pcDNA3-Myc-Beclin1 K117A and K437A were obtained by site-directed mutagenesis using plasmid pcDNA3-Myc-Beclin1 as template, the Quick Change kit (Stratagene) and the corresponding mutagenic oligonucleotides: K117A-F: 5’-GAGAACCTCAGCCGAAGACTGGCGGTCACTGGGGACCTTTTTG-3’, K117A-R: 5’-CAAAAAGGTCCCCAGTGACCGCCAGTCTTCGGCTGAGGTTCTC-3’; K437A-F: 5’-GTTCATGCTGACGAATCTTGCGTGGGGTCTTGCTTGGGTG-3’, K437A-R: 5’-CACCCAAGCAAGACCCCACGCAAGATTCGTCAGCATGAAC-3’. All mutants were sequenced to ensure that additional mutations were not introduced during the mutagenesis procedure.
2.3.-. GFP-trap analysis
HEK293 cells were transfected with specific constructs of laforin, malin, and the protein of interest. Cells were scraped on ice in phosphate-buffered saline (PBS), washed twice in PBS and then lysed using a 25-gauge needle in lysis buffer [10 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% (v/v) Nonidet P-40, complete protease inhibitor cocktail (Roche Diagnostics, Barcelona, Spain), 1 mM PMSF, 2.5 mM NaF, 0.5 mM NaVO4 and 2.5 mM Na4P2O7]. Cell lysates were then centrifuged at 13,000×g for 10 min at 4°C. Supernatants (1.5 mg of total protein) were incubated with Chromotek GFP-trap beads (Chromotek, Planegg-Martinsried, Germany) for ten minutes in a rocking platform at 4°C and the GFP- and CFP-fused proteins were immunoprecipitated and visualized by immunoblotting using specific antibodies (see below). As a negative control, a construct expressing CFP or GFP alone (plasmid pECFP-N1 and pEGFP-N1, respectively) was used to confirm the specificity of the interaction.
2.4.-. Western blot analysis
Western blot was carried out essentially as described [27]. 50 μg of total protein from the soluble fraction of cell lysates were analyzed by SDS-PAGE and proteins were transferred to PVDF membranes (Millipore, Madrid, Spain). Membranes were blocked with 5% (w/v) non-fat milk in Tris-buffered saline Tween20 buffer [TBS-T: 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.1% (v/v) Tween20] for 1 h at room temperature and incubated overnight at 4°C with the corresponding primary antibodies. Then, membranes were probed with suitable secondary antibodies for 1 h at room temperature. Signals were visualized using Lumi-Light Western Blotting Substrate (Roche Applied Science, Barcelona, Spain) or ECL Prime Western Blotting Detection Reagent (GE Healthcare, Barcelona, Spain) and analyzed by chemiluminescence using an image reader LAS-4000 (GE Healthcare, Barcelona, Spain). The following antibodies were used: anti-FLAG tag (F3165), anti-HA tag (H9658), anti-Myc tag (M5546) and anti-tubulin (T6199) from Sigma Chemical Co. (Madrid, Spain); anti-laforin (MABN606, Millipore, Madrid, Spain); anti-GFP (210-PS-1GFP; Immunokontact, Abbingdon, UK); anti-Beclin1 (#3495), anti-Vps34 (#4263), anti-ULK1 (#8054) and anti-Vps34 (#4263) from Cell Signaling Technology (Danvers, MA, USA); anti-V5 (R960–25; Invitrogen, Madrid, Spain); anti-AMBRA1 (26190002; Novus Biologicals, Cenntenial, CO, USA) and anti-ATG14L (#PD026) from Medical and Biological Laboratories (Nagano, Japan).
2.5.-. Immunoprecipitation analysis
Proteins in cell lysates obtained as above (1.5 mg) were immunoprecipitated using either preimmune serum or anti-Beclin1 (#3495, Cell Signaling Technologies) antibody and protein A/G agarose beads (sc-2003, Santa Cruz Biotechnologies, Dallas, TX, USA). Immunoprecipitated proteins were analyzed by western blotting as above using the appropriate antibodies.
2.6.-. Analysis of ubiquitination
To study ubiquitination, we followed the method described in [28]. Briefly, HEK293 cells were transfected with plasmid pCMV-6xHis-Ubiq (encoding a modified form of ubiquitin tagged with 6xHis residues), and plasmids encoding laforin, malin and the protein of interest. After 24 hours of transfection, cells were lysed using a 25-gauge needle in buffer A (6 M guanidinium-HCl, 0.1 M sodium phosphate, 0.1 M Tris-HCl, pH 8.0) to inhibit the action of deubiquitinases. 1.5 mg of protein of a clarified extract (12,000×g, 15 min) were incubated with 150 μl of a TALON cobalt resin (Clontech, Barcelona, Spain) equilibrated in buffer A containing 10 mM imidazole (buffer B), for 3 hours at room temperature on a rocking platform, to purify His-tagged proteins. The resin was then successively washed with 1 mL of buffer B and four times with buffer C (buffer B, but with 8 M urea instead of 6 M guanidinium-HCl). Bound proteins were boiled in 40 μl of 2xLaemmli’s sample buffer and analyzed by Western blotting using the appropriate antibodies. When indicated, plasmids pCMV-6xHis-Ubiq-K48R and pCMV-6xHis-Ubiq-K63R were used in the assay instead of pCMV-6xHis-Ubiq wild type, to determine the topology of the ubiquitin chains.
2.7.-. Immunofluorescence and confocal microscopy
Human osteosarcoma U2OS cells were grown as indicated in the case of HEK293 cells. They were transfected with the indicated plasmids and grown on 12-well plates containing coverslips. Cells were fixed with 4% (w/v) paraformaldehyde in phosphate-buffered saline (PBS) for 10 min at room temperature, washed three times with PBS and permeabilized with 0.2% (v/v) Triton X-100 in PBS for 30 min. Cells were then blocked for one hour with 10% (v/v) fetal bovine serum, 0.5% (w/v) BSA and 0.1% (v/v) Triton X-100 in PBS and then incubated overnight at 4 °C with 1/500 dilution of the indicated antibody [anti-FLAG tag (F3165) from Sigma Chemical Co. (Madrid, Spain), anti-V5 (R960–25) from Invitrogen (Madrid, Spain), anti-Beclin1 (#3495) or anti-Vps34 (#4263) from Cell Signaling Technology (Danvers, USA)] in blocking solution. Samples were washed three times with PBS and incubated for 1 h at room temperature with a 1/1000 dilution of anti-mouse Alexa-Fluor 568 or anti-rabbit Alexa-Fluor 633 (Invitrogen, Madrid, Spain), respectively. Finally, samples were washed three times with PBS and mounted on slides using Aqua-Poly/Mount coverslipping medium (Polysciences, Inc. Eppelheim, Germany). The co-localization assays were performed with a Leica TCS SP8 confocal microscope (Leica, Wetzlar, Germany) using an HCX PL APO 63× 1.4 NA oil objective. Images were treated with the ImageJ 1.49 software (Wayne Rasband, National Institutes of Health, Bethesda, MD, USA).
Primary fibroblasts from control and LD patients grown on coverslips were transiently transfected with pEGFP-2xFYVE plasmid using Lipofectamine 2000 Transfection Reagent (ThermoFisher, Madrid, Spain). 48 hours after transfection, cells were fixed with 4% (w/v) paraformaldehyde in PBS for 10 min at room temperature. Coverslips were mounted using Aqua-Poly/Mount coverslipping medium (Polysciences, Inc. Eppelheim, Germany). Images were acquired as above and the total fluorescence of Z-stack merges of 15 confocal images with a Z-step size of 0.30 μm was determined using the ImageJ software described above. The size and number of the puncta present in the cells was also quantified using Image J.
2.8.-. Statistical analysis
Results are shown as means +/− standard error of the mean (SEM). Differences between paired samples were analyzed by two-tailed Student’s t-tests using Graph Pad Prism version 5.0 statistical software (La Jolla, CA, USA). P values have been considered as ***P< 0.001.
3.-. RESULTS
3.1.-. The laforin/malin complex interacts physically with Beclin1 and promotes its polyubiquitination.
We and others have described in cellular models of LD an impairment of autophagy ([12], [13], [14]) that leads to the accumulation of dysfunctional mitochondria ([29], [30]). We also recently described that in fibroblasts from LD patients carrying mutations in either EPM2A or EPM2B genes there was a severe decrease in the number of endogenous LC3 puncta, especially when autophagy was stimulated [30]. Although our results suggested impairment at the initial steps of autophagosome formation, the molecular details were not fully understood. For this reason, we decided to study in deep the possible involvement of laforin and malin in the initial steps of autophagosome formation.
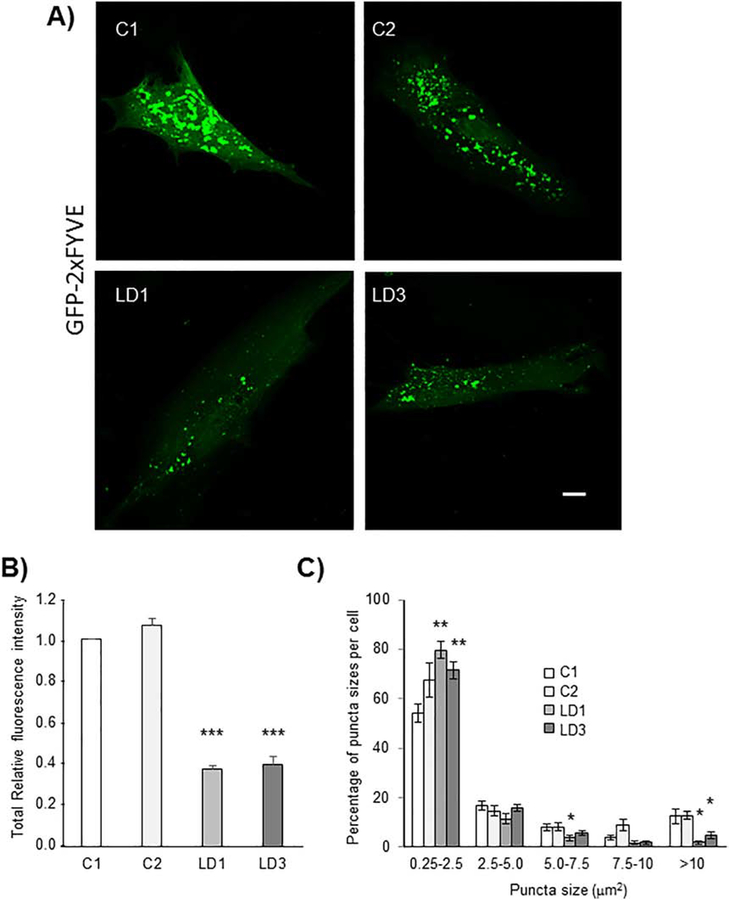
As described at the Introduction, two main complexes play an essential role in autophagy initiation and autophagosome formation: the regulatory ULK1/2 complex (comprising ULK1/2, FIP200, ATG13 and ATG101 proteins) and the PI3KC3 complex [comprising Beclin1, vacuolar protein sorting 34 (Vps34), Vps15 and ATG14L or UVRAG proteins], which produces phosphatidylinositol-3 phosphate (PtdIns3P) on the surface of the initiation membrane ([20],[31], [32]). We started our analyses determining the activity of the PI3KC3 complex in fibroblasts from two LD patients (LD1 and LD3) carrying different mutations in the EPM2A gene and that displayed autophagy impairment [30]. The activity of the PI3KC3 complex was assessed by analyzing the levels of PtdIns3P by fluorescence microscopy analysis based on a PtdIns3P-binding FYVE probe [pEGFP-2xFYVE ([33], [34])]. In Fig. 1A we present images corresponding to Z-stack merges of 15 confocal images with a Z-step size of 0.30 μm (individual selected stacks of the confocal analysis of the cells are shown in Supplementary Fig. S1). When we analyzed the total fluorescence of the cells we observed a severe decrease in the intensity of the probe in both LD fibroblasts in comparison to controls (Fig. 1B). This was due to clear decrease in the proportion of large-size, high-intensity puncta and an increase in the proportion of small-size, low-intensity puncta, which correlates with a decrease in the total fluorescence of the samples (Fig. 1C) [no big differences in the total amount of puncta per cell between controls (from 160 to 90 puncta) and LD fibroblasts (from 140 to 100 puncta)] were observed]. These results indicated that in the absence of a functional laforin/malin complex there was an impairment in the activity of the PI3KC3 complex, which could explain the reduced autophagosome biogenesis.
Fig. 1.-. In the absence of laforin the levels of phosphatidylinositol-3phosphate (PtdIns3P) are reduced.
Primary fibroblasts from two independent control subjects (C1 and C2) and from two LD patients with mutations in EPM2A gene (LD1 and LD3) were transfected with plasmid pEGFP-2xFYVE, which binds to PtdIns3P, and analyzed by fluorescence microscopy using a confocal microscope. A) Representative images corresponding to Z-stack merges of 15 confocal images with a Z-step size of 0.30 μm; Bar: 10 μm. B) The total relative fluorescence intensity per cell was measured using Image J, in at least 50 cells in each sample. C) In the same samples, the number and size of the puncta per cell were measured using Image J. No big differences in the total amount of puncta per cell were observed between controls (from 160 to 90 puncta) and LD fibroblasts (from 140 to 100 puncta). The percentage of puncta sizes per cell is presented. Results are expressed as means +/− SEM (two-tailed Student’s t-tests, *p<0.05, **p<0.01 and ***p<0.001).
Next, we analyzed whether this impairment in PtdIns3P formation was due to altered levels of some components of the PI3KC3 or ULK1/2 complexes. We measured the endogenous protein levels of ULK1, Beclin1, Vps34 and ATG14L in primary fibroblasts from control and an EPM2A patient (LD3) growing in complete medium and treated or not with 10 μM CCCP for 1 hour to induce mitochondria dysfunction and autophagy [30]. However, we did not find apparent differences in the levels of any of these proteins compared to control fibroblasts (Supplementary Fig. S2). In addition, we analyzed the phosphorylation status of different components involved in the initiation of autophagy, such as P-ULK1(Ser757) (a substrate of the mTOR pathway) or P-ULK1(Ser555) and P-Beclin1(Ser93) (substrates of the AMPK pathway), but although we found differences related to the CCCP treatment, no obvious genotype-dependent differences were observed [Supplementary Fig. S3; the activity of the mTOR pathway was assessed measuring the levels of P-p70S6K(Thr389) and the activity of the AMPK pathway was assessed measuring the levels of P-Raptor(Ser792)]. Also, no genotype-dependent changes in the levels of P-Beclin1(Ser15), a substrate of ULK1, were observed (data not shown).
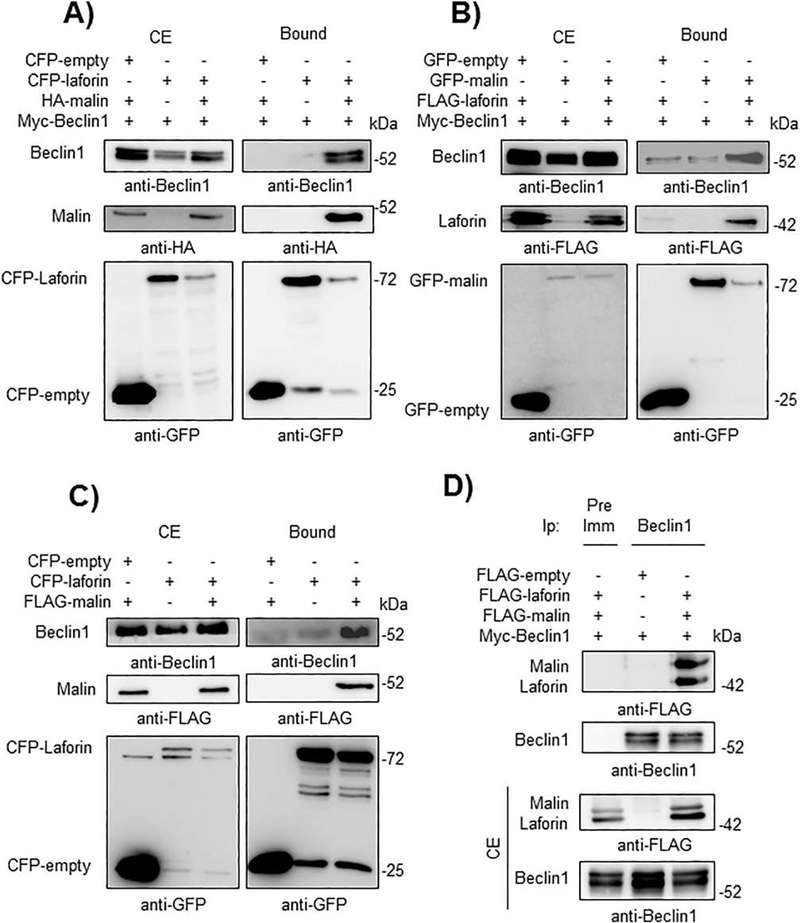
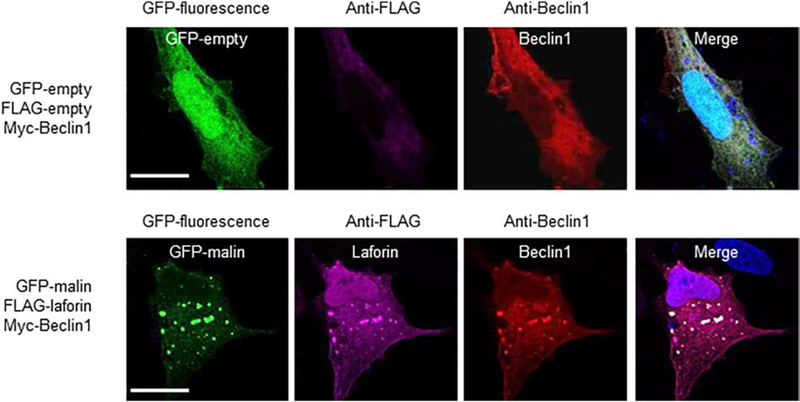
Then, we determined whether laforin or malin could physically interact with members of the PI3KC3 complex by GFP-trap analysis. First, we started with Beclin1. As shown in Fig. 2A and 2B, when laforin and malin were both co-expressed in HEK293 cells, they were able to interact with Beclin1. In the absence of one of the members of the laforin/malin complex a minor interaction with Beclin1 was observed. The laforin/malin complex was also able to pull-down endogenous Beclin1 (Fig. 2C), and when Beclin1 was immunoprecipitated using anti-Beclin1 antibodies, both laforin and malin were present in the immunoprecipitates (Fig. 2D). The interaction between laforin/malin and Beclin1 was confirmed by immunofluorescent studies. As shown in Fig. 3, laforin, malin and Beclin1 co-localized in similar subcellular structures. The change in the expression pattern of Beclin1 when laforin and malin were overexpressed could be due to an enhancement of autophagy, mediated by the laforin/malin complex ([35], [36]). Therefore, all these results indicate a physical interaction between Beclin1 and the laforin/malin complex.
Fig. 2.-. The laforin/malin complex interacts with Beclin1.
A) CFP-laforin pull-downs overexpressed Beclin1 in the presence of malin. HEK293 cells were co-transfected with the indicated combination of plasmids expressing Myc-Beclin1, HA-malin, CFP (empty) and CFP-laforin. Cells were lysed and 1.5 mg of proteins were incubated with GFP-trap beads, which bind the various GFP forms, including CFP, with high affinity. After washing, beads were boiled in loading buffer and the purified proteins were analyzed by SDS-PAGE and Western blot using anti-GFP, anti-HA or anti-Beclin1 antibodies, as indicated below the different panels. Bound: proteins retained in the beads; CE: crude extracts (50 μg of protein). B) GFP-malin pull-downs overexpressed Beclin1 in the presence of laforin. HEK293 cells were co-transfected with the indicated combination of plasmids expressing Myc-Beclin1, FLAG-laforin, GFP (empty) and GFP-malin. Extracts were analyzed as in A). C) CFP-laforin pull-downs endogenous Beclin1 in the presence of malin. HEK293 cells were co-transfected with the indicated combination of plasmids expressing FLAG-malin, CFP (empty) and CFP-laforin. Extracts were analyzed as in A). D) Beclin1 immunoprecipitates laforin and malin. HEK293 cells were co-transfected with the indicated combination of plasmids expressing Myc-Beclin1, FLAG-malin and FLAG-laforin. Cells were lysed and 1.5 mg of proteins were immunoprecipitated (Ip) with either 1 μl of preimmune serum (Pre-Imm) or 1 μl of anti-Beclin1 antibody. After washing, protein A/G agarose beads were boiled in loading buffer and the purified proteins analyzed by SDS-PAGE and Western blot using anti-FLAG or anti-Beclin1 antibodies. CE: crude extracts (50 μg of protein). In A), B), C) and D) a representative blot of three independent experiments is shown. The positions of different proteins are indicated on the left of the panels, while molecular weight markers are indicated in kDa on the right.
Fig. 3.-. Laforin and malin co-localize with Beclin1.
Representative images of U2OS cells transfected with the plasmids indicated on their left. The subcellular localization of the corresponding proteins was analyzed either by direct fluorescence (GFP and GFP-malin) or by immunofluorescence using anti-FLAG (coupled to an Alexa-Fluor 633 secondary antibody) and anti-Beclin1 (coupled to an Alexa-Fluor 568 secondary antibody) antibodies, as described in Materials and Methods. Fluorescence associated with the corresponding proteins was analyzed by confocal microscopy. DAPI staining was in blue. A merged image of the fluorescent dyes is also shown. Bar: 20 μm.
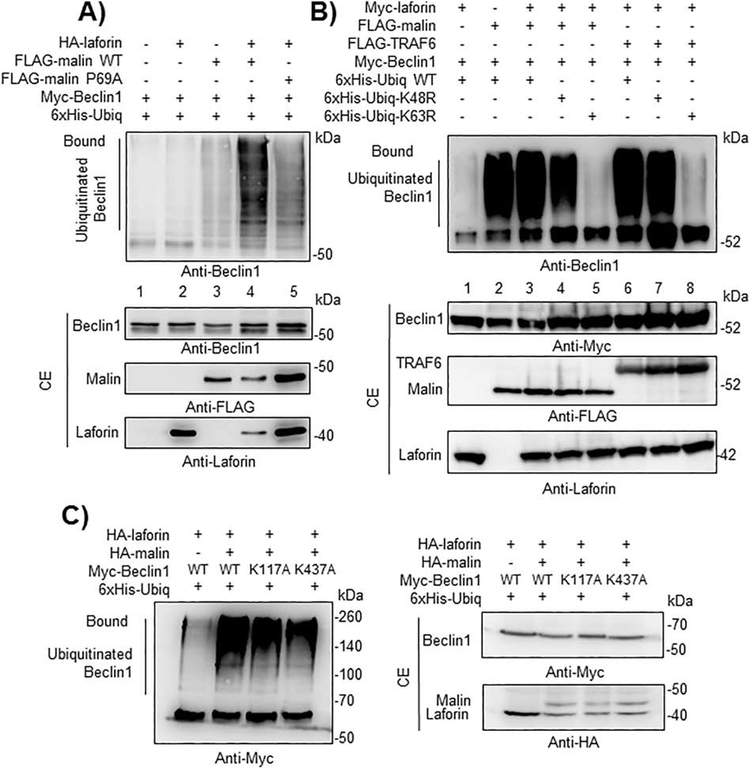
Since the laforin/malin complex recognizes and ubiquitinates different substrates ([6], [7], [8]), next we analyzed whether, as a result of its interaction with Beclin1, it could promote the ubiquitination of this protein. As shown in Fig. 4A, a clear polyubiquitination of Beclin1 was observed when both laforin and malin were present (lane 4), and this modification was dependent on the presence of the E3-ubiquitin ligase malin (lanes 1 and 2) and on the presence of laforin (lanes 1 and 3). In the presence of an inactive form of malin (P69A; being this mutation the most frequent modification found in EPM2B patients) a severe reduction in the ubiquitination of Beclin1 was observed (lane 5). The residual polyubiquitination of Beclin1 observed when no laforin was overexpressed (lane 3) could be due to endogenous levels of laforin present in HEK293 cells, which could form a complex with the overexpressed malin. These results indicate that a functional complex is required for the proper laforin/malin-dependent polyubiquitination of Beclin1. Then, we analyzed the topology of the polyubiquitinated chains present in Beclin1. To perform this analysis we used modified forms of ubiquitin that carried K48R or K63R mutations, which prevent the formation of K48- or K63-linked chains respectively. As indicated in Fig. 4B, the laforin/malin complex promoted the attachment of K63-linked ubiquitin chains, as ubiquitination was severely impaired in cells expressing K63R-ubiquitins (lane 5). On the contrary, in the presence of K48R-ubiquitins, we observed a similar degree of ubiquitination as when wild type ubiquitin was used in the assay (lanes 3 and 4). Similar results were obtained when we used in the assay TRAF6 (Fig. 4B), an E3-ubiquitin ligase that also promotes the attachment of K63-linked polyubiquitin chains to Beclin1 ([24], [37]). As it has been described that residues Lys117 and Lys437 of Beclin1 are involved in the attachment of K63-linked ubiquitin chains to this protein by the action of TRAF6 [37] and DDB1-Cullin4-AMBRA1 complex [38], respectively, we constructed Beclin1 mutants in these residues and observed that the ubiquitination mediated by the laforin/malin complex still took place in them, thus indicating that the modification occurred in alternative residues (Fig. 4C).
Fig. 4.-. A functional laforin/malin complex promotes the ubiquitination of Beclin1, attaching K63-linked polyubiquitin chains.
A) A functional Laforin/malin complex ubiquitinates Beclin1. HEK293 cells were transfected with the indicated plasmids and ubiquitination analysis of Beclin1 was carried out as described in Materials and Methods. The analyses were performed with active (wild type) and inactive (P69A) forms of malin. Proteins present in the bound fraction (Bound: proteins retained in the metal affinity resin) or in the crude extract (CE, 50 μg) were analyzed by Western blotting using the indicated antibodies. B) Topology of the ubiquitination reaction. Ubiquitination reactions were performed as in A) using modified forms of ubiquitin that carried K48R or K63R mutations, which prevent the formation of K48- or K63-linked chains, respectively. As a control, the same reactions were carried out in the presence of the E3-ubiquitin ligase TRAF6 instead of the laforin/malin complex. The position of the ubiquitinated forms of Beclin1 is indicated. C) Laforin/malin complex does not ubiquitinate Beclin1 at Lys117 or Lys437. Ubiquitination of Beclin1 wild type (WT), and their K117A and K437A mutants was performed as in section A) using the indicated plasmids. In A) to C), a representative blot of three independent experiments is shown. Molecular size markers are indicated in kDa on the right of the blots.
In conclusion, the laforin/malin complex interacts physically with Beclin1 and promotes the attachment of K63-linked polyubiquitin chains. This topology is in agreement with the type of modification that the laforin/malin complex introduces in alternative substrates such as R5/PTG [8], AMPK subunits [39], PKM1/2 [40] and p62 [17], and with the functional interaction of the laforin/malin complex with UBE2N [17], an E2-conjugating enzyme that promotes the formation of K63-linked polyubiquitin conjugates.
3.2.-. The laforin/malin complex interacts with and polyubiquitinates additional components of the PI3KC3 complex.
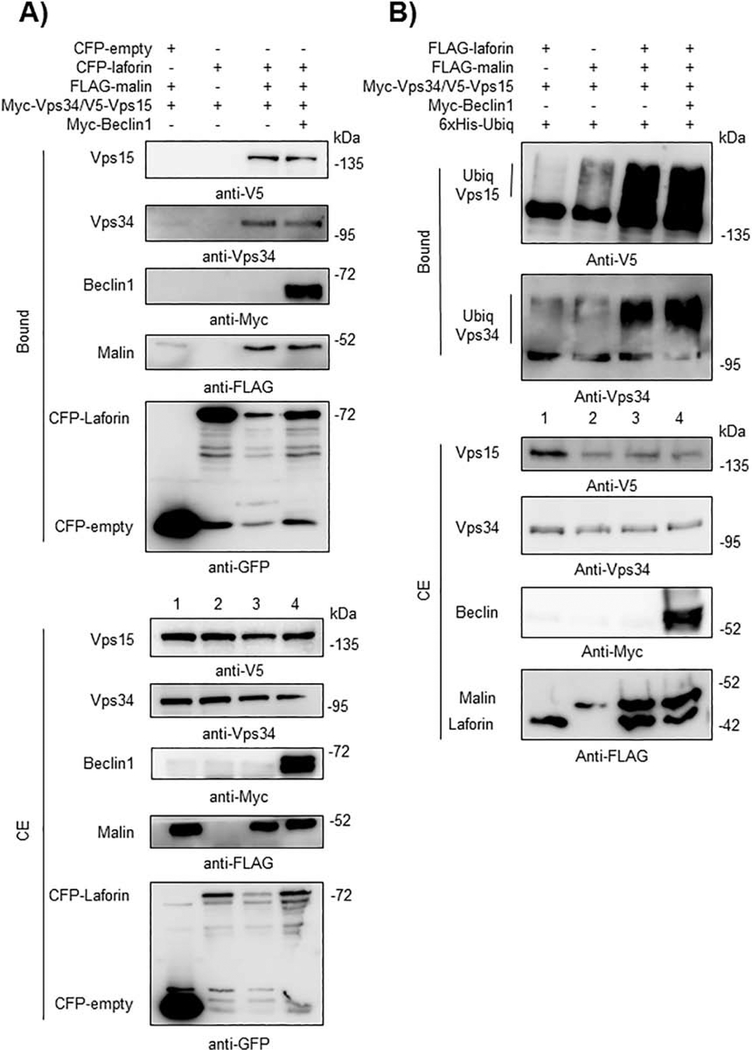
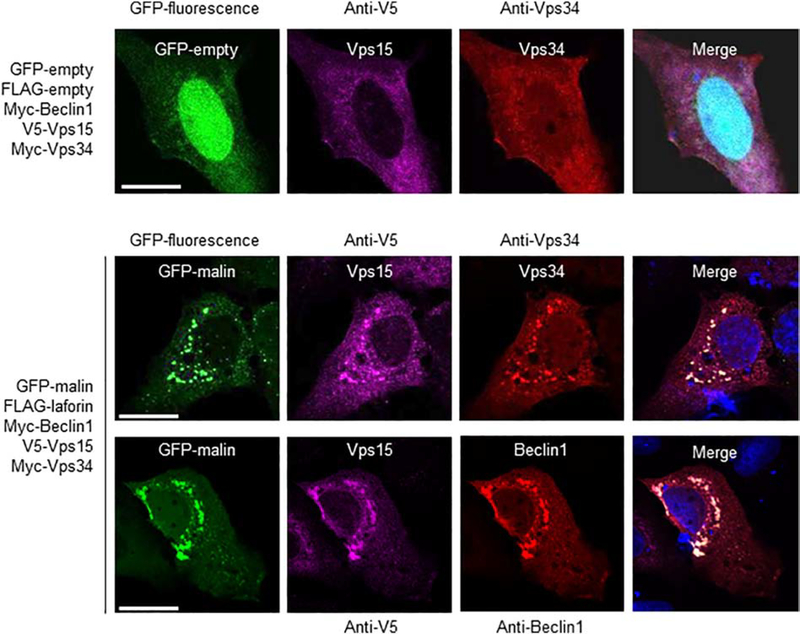
Since Beclin1 is a component of the PI3KC3 complex, we analyzed the possible interaction of laforin and malin with other components of it, such as Vps34 and Vps15. In Fig. 5A we show that only when laforin and malin were present there was a physical interaction with both Vps34 and Vps15 (lane 3). Expression of laforin alone was not enough to promote this interaction (lane 2) and expression of exogenous Beclin1 did not improve the interaction (lane 4), perhaps because the levels of endogenous Beclin1 were high enough to support the interaction of the laforin/malin complex with Vps34 and Vps15. These interactions were confirmed by the co-localization of malin, Vps34 and Vps15 in similar subcellular structures, which also contained Beclin1 (Fig. 6). The change in the expression pattern of Beclin1, Vps34 and Vps15 when laforin and malin were overexpressed could be due to an enhancement of autophagy, mediated by the laforin/malin complex ([35], [36]).
Fig. 5.-. The laforin/malin complex interacts with Vps34 and Vps15 and promotes their ubiquitination.
A) CFP-laforin pull-downs Vps34 and Vps15 in the presence of malin. HEK293 cells were co-transfected with the indicated combination of plasmids expressing Myc-Beclin1, Myc-Vps34/V5-Vps15, FLAG-malin, CFP (empty) and CFP-laforin. Cells were lysed and 1.5 mg of proteins were analyzed as in the legend of Fig. 2A using the appropriated antibodies. B) Laforin/malin complex ubiquitinates Vps34 and Vps15. HEK293 cells were transfected with the indicated plasmids and ubiquitination of Vps34 and Vps15 was analyzed as in the legend of Fig. 4A. In A) and B), a representative blot of three independent experiments is shown. Molecular size markers are indicated in kDa on the right of the blots.
Fig. 6.-. Laforin and malin co-localize with Vps34 and Vps15.
Representative images of U2OS cells transfected with the plasmids indicated on their left. The subcellular localization of the corresponding proteins was analyzed either by direct fluorescence (GFP and GFP-malin) or by immunofluorescence using anti-V5 (coupled to an Alexa-Fluor 633 secondary antibody), and anti-Vps34 or anti-Beclin1 (coupled both to an Alexa-Fluor 568 secondary antibody) antibodies, as described in Materials and Methods. Fluorescence associated with the corresponding proteins was analyzed by confocal microscopy. DAPI staining was in blue. A merged image of the fluorescent dyes is also shown. Bar: 20 μm.
Also, in this case, the laforin/malin complex was able to polyubiquitinate Vps34 and Vps15 (Fig. 5B), even in the absence of overexpressed Myc-Beclin1 (lanes 3 and 4). Therefore, all these results suggest a physical interaction between the laforin/malin complex and different members of the PI3KC3 complex, which results in the attachment to them of polyubiquitin chains.
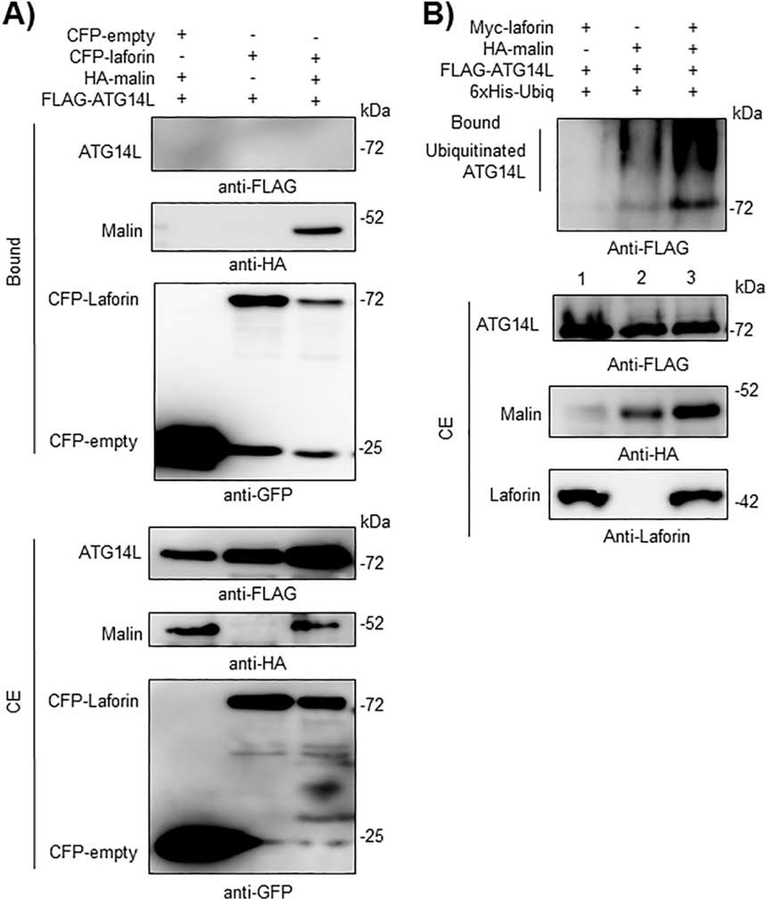
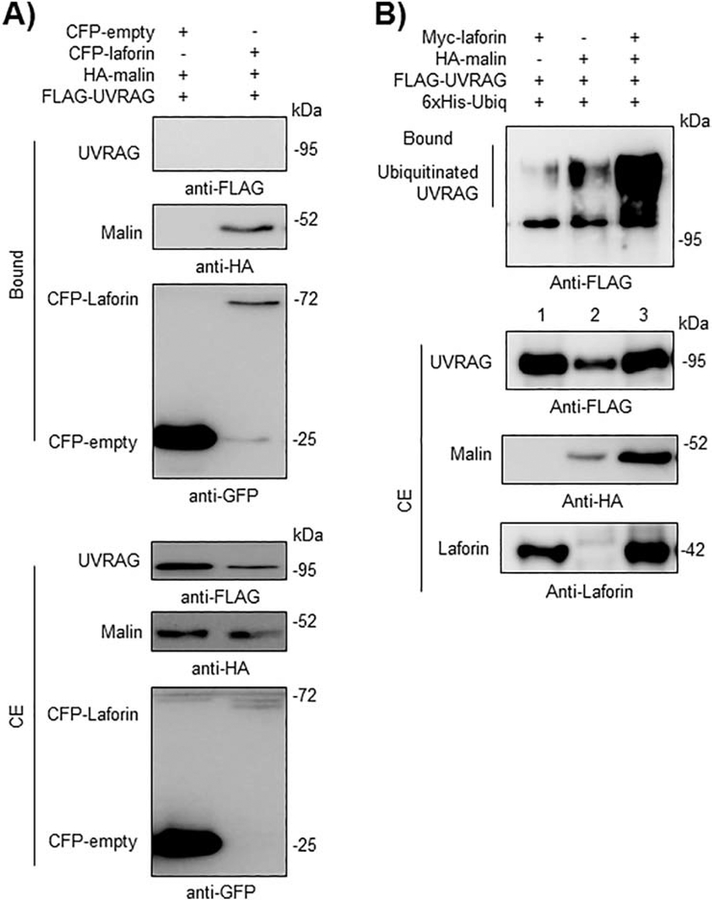
It is known that in a functional PI3KC3 complex, the core Beclin1-Vps34-Vps15 sub-complex interacts in a mutually exclusive manner with either ATG14L or UVRAG proteins. Although we were not able to detect any physical interaction between the laforin/malin complex and either ATG14L (Fig. 7A) or UVRAG (Fig. 8A) in GFP-trap pull-down assays, the laforin/malin complex promoted the polyubiquitination of ATG14L (Fig. 7B, lane 3) and UVRAG (Fig. 8B, lane 3), perhaps through its interaction with the core Beclin1-Vps34-Vps15 sub-complex.
Fig. 7.-. The laforin/malin complex promotes the ubiquitination of ATG14L.
A) CFP-laforin does not pull-down ATG14L in the presence of malin. HEK293 cells were co-transfected with the indicated combination of plasmids expressing FLAG-ATG14L, HA-malin, CFP (empty) and CFP-laforin. Cells were lysed and 1.5 mg of proteins were analyzed as in the legend of Fig. 2A using the appropriated antibodies. B) Laforin/malin complex ubiquitinates ATG14L. HEK293 cells were transfected with the indicated plasmids and ubiquitination of ATG14L was analyzed as in the legend of Fig. 4A. In A) and B), a representative blot of three independent experiments is shown. Molecular size markers are indicated in kDa on the right of the blots.
Fig. 8.-. The laforin/malin complex promotes the ubiquitination of UVRAG.
A) CFP-laforin does not pull-down UVRAG in the presence of malin. HEK293 cells were co-transfected with the indicated combination of plasmids expressing FLAG-UVRAG, HA-malin, CFP (empty) and CFP-laforin. Cells were lysed and 1.5 mg of proteins were analyzed as in the legend of Fig. 2A using the appropriated antibodies. B) Laforin/malin complex ubiquitinates UVRAG. HEK293 cells were transfected with the indicated plasmids and ubiquitination of UVRAG was analyzed as in the legend of Fig. 4A. In A) and B), a representative blot of three independent experiments is shown. Molecular size markers are indicated in kDa on the right of the blots.
3.3.-. The laforin/malin complex does not affect other components involved in autophagosome biogenesis such as Bcl2, AMBRA1 or ULK1.
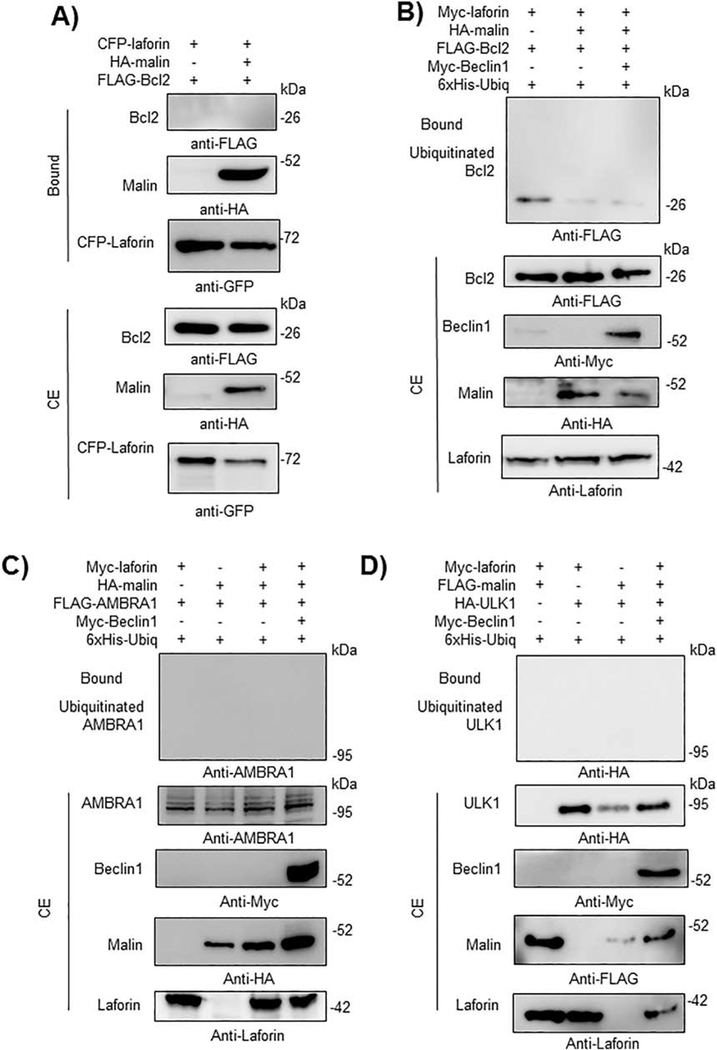
We extended these interaction studies to other proteins which play a role in the regulation of initial steps of autophagosome biogenesis. We analyze the physical interaction of laforin and malin with Bcl2, an inhibitor of the PI3KC3 complex, AMBRA1 and ULK1, but we found no interaction by GFP-trap pull-down assays in any case (Fig. 9A for Bcl2; data not shown for AMBRA1 and ULK1). We also analyzed the ability of the laforin/malin complex to ubiquitinate these proteins but we found no modification for either Bcl2 (Fig. 9B), AMBRA1 (Fig. 9C) or ULK1 (Fig. 9D) even in the presence of overexpressed Myc-Beclin1. These results indicate a specificity of the laforin/malin complex in ubiquitinating some components of the machinery of autophagosome formation but not others.
Fig. 9.-. The laforin/malin complex does not affect Bcl2, AMBRA1 or ULK1.
A) CFP-laforin does not pull-down Bcl2 in the presence of malin. HEK293 cells were co-transfected with the indicated combination of plasmids expressing FLAG-Bcl2, HA-malin, and CFP-laforin. Cells were lysed and 1.5 mg of proteins were analyzed as in the legend of Fig. 2A using the appropriated antibodies. B) Laforin/malin complex does not ubiquitinate Bcl2. HEK293 cells were transfected with the indicated plasmids and ubiquitination of Bcl2 was analyzed as in the legend of Fig. 4A. C) and D) The laforin/malin complex does not ubiquitinate AMBRA1 or ULK1. HEK293 cells were transfected with the indicated plasmids and ubiquitination of AMBRA1 (C) and ULK1 (D) was analyzed as in the legend of Fig. 4A. In A) to D), a representative blot of three independent experiments is shown. Molecular size markers are indicated in kDa on the right of the blots.
Taken together, all these results indicate that the laforin/malin complex regulates some of the initial steps of autophagosome biogenesis by specifically interacting with components of the PI3KC3 complex to promote their polyubiquitination.
3.4.-. The activity of the PI3KC3 complex is enhanced in the presence of a functional laforin/malin complex.
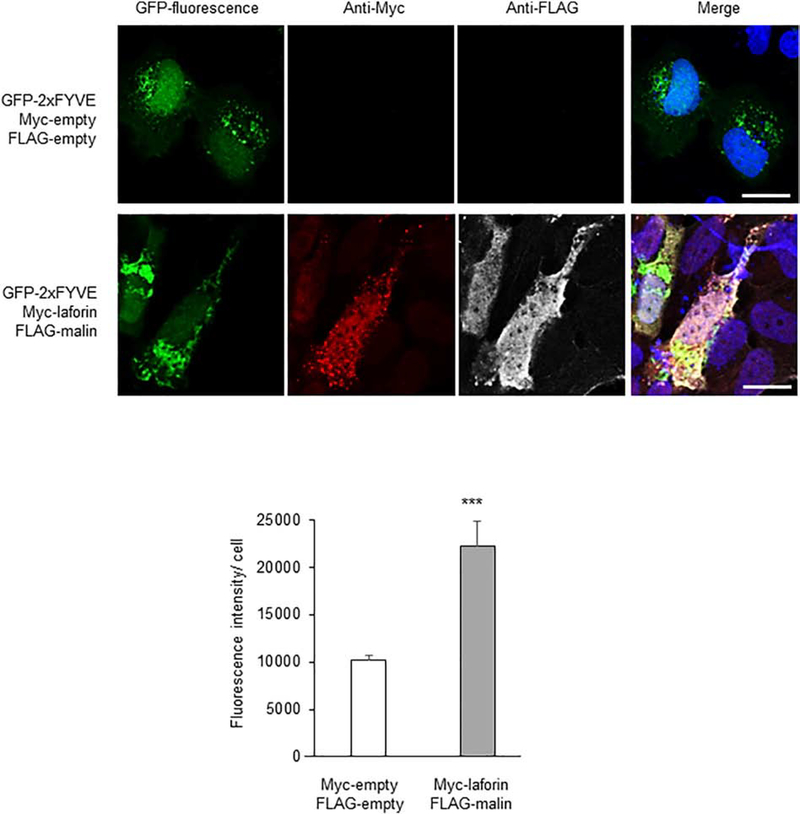
To investigate the functional consequences of the observed interactions and polyubiquitinations of components of the PI3KC3 complex, we overexpressed laforin and malin in U2OS cells and measured the levels of PtdIns3P by fluorescence microscopy analysis based on the PtdIns3P-binding FYVE probe (pEGFP-2xFYVE), as above. In cells expressing at the same time GFP-2xFYVE, laforin and malin, we observed a more than two-fold increase in the levels of PtdIns3P (Fig. 10), indicating a positive effect of the laforin/malin complex on the activity of the PI3KC3 complex.
Fig. 10.-. Overexpression of laforin and malin increases the levels of PtdIns3P.
U2OS cells were transfected with pEGFP-2xFYVE (which binds to PtdIns3P), pMyc-laforin, and pFLAG-malin, or the corresponding empty plasmids. The levels of PtdIns3P were assessed by measuring the GFP-associated fluorescence by confocal microscopy. The fluorescence intensity per cell was measured in 240 control cells (empty plasmids) and 144 cells expressing at the same time plasmids Myc-laforin and FLAG-malin, which were identified using anti-Myc (coupled to an Alexa-Fluor 568 secondary antibody) and anti-FLAG (coupled to an Alexa-Fluor 633 secondary antibody). DAPI staining was in blue. A representative image of each case and a merged image of the fluorescent dyes are also shown. Results are expressed as arbitrary fluorescence units per cell in the form of means +/− SEM (two-tailed Student’s t-tests, ***p<0.001). Bar: 20 μm.
4.-. DISCUSSION
Although the hallmark of Lafora progressive myoclonus epilepsy is the accumulation of polyglucosan inclusions in brain and peripheral tissues, using animal models of the disease, we and others have demonstrated that there is also an impairment of proteostasis, which included decreased autophagy ([11], [12], [13], [14]). These results were also confirmed in primary cultures of fibroblasts derived from patients harboring mutations in either the EPM2A and EPM2B genes ([12], [14], [30]). These studies also suggested that decreased autophagy in LD was related to an impairment in the initial steps of autophagosome formation, as lower levels of LC3-II were detected in LD samples even after treatment with various lysosomal inhibitors [14]. In agreement with these results, here we provide evidence of a decrease in the levels of PtdIns3P, an essential mediator in the formation of autophagosomes, in primary fibroblasts from LD patients carrying mutations in the EPM2A gene.
Regulation of autophagy is a complex process involving different post-translational modifications such as phosphorylation, ubiquitination, acetylation, etc. Therefore, and in order to understand the molecular basis of the defect present in LD, we first analyzed the phosphorylation status of key components involved in the process of autophagy initiation in LD fibroblasts. However, we could not find any difference with respect to control cells. Then, we explored the possibility of a direct action of the laforin/malin complex on these components. As the activity of the PI3KC3 complex was reduced in LD samples (less PtdIns3P being produced; see above), we started our analysis with Beclin1, one of the core components of the PI3KC3 complex. We observed that Beclin1 interacted physically with the laforin/malin complex and also co-localized with it. More importantly, the laforin/malin complex was able to polyubiquitinate Beclin1 by attaching K63-linked ubiquitin chains, being this process dependent on the presence of an active form of malin. In this way, the laforin/malin complex joins the group of E3-ubiquitin ligases that are able to modify Beclin1 with K63-linked ubiquitin chains, such as TRAF6 [37], the DDB1-Cullin4-AMBRA1 complex [38], and TRIM50 [41]. In these cases, the K63-ubiquitination of Beclin1 has been associated with an activation of the protein and an enhancement of the activity of the PI3KC3 complex ([24], [25]). In agreement with these suggestions, we present evidence that overexpression of the laforin/malin complex enhances the formation of PtdIns3P. Therefore, our results support the hypothesis that the laforin/malin complex regulates Beclin1 by K63-linked polyubiquitination, resulting in an enhancement of the synthesis of PtdIns3P. We also show that the laforin/malin-mediated ubiquitination of Beclin1 occurred mostly at Lys residues different from those already reported in the literature (Lys117 for TRAF6 [37] and Lys437 for DDB1-Cullin4-AMBRA1 complex [38]).
The action of the laforin/malin complex extends to other components of the core of the PI3KC3 complex. Indeed, we present evidence of physical interaction and co-localization of the laforin/malin complex with Vps34 and Vps15, resulting in the ubiquitination of these two proteins. Therefore, the laforin/malin complex joins also the group of other E3-ubiquitin ligases that modify Vps34, such as CHIP which, in combination with the E2-conjugase UBC13/Uev1, attaches K63-linked ubiquitin chains to Vps34, enhancing in this way its stability [42]. In addition, the laforin/malin complex was also able to ubiquitinate ATG14L and UVRAG, but apparently without any physical interaction. Perhaps, the attachment of the laforin/malin complex to the core of the PI3KC3 complex allows it to gain access to these two mutually exclusive members of the whole PI3KC3 complex. All these modifications seem to be specific since other components involved in the process of autophagy initiation such as Bcl2, AMBRA1 or ULK1, were not substrates of the laforin/malin complex.
5.-. CONCLUSIONS
In summary, in this study, we present evidence for the participation of the laforin/malin complex in the initial steps of autophagosome formation. The laforin/malin complex physically interacts with core components of the PI3KC3 system and specifically polyubiquitinates Beclin1, Vps34 and Vps15. In addition, it is also able to polyubiquitinate ATG14L and UVRAG. We suggest that all these polyubiquitinations result in an enhancement of the activity of the PI3KC3 complex, producing more PtdIns3P and therefore increasing the formation of autophagosomes. In Lafora disease, the absence of a functional laforin/malin complex would result in a less activated PI3KC3 complex, leading in this way to the observed impairment of the initial steps of autophagy.
Supplementary Material
HIGHLIGHTS.
Lafora disease cell models produce lower levels of PI3P.
The laforin/malin E3 ligase interacts and ubiquitinates beclin 1.
The laforin/malin complex interacts with additional components of the PI3KC3 complex.
Ubiquitination of the PI3KC3 complex by laforin and malin increase its activity.
6.-. ACKNOWLEDGMENTS
We want to thank Dr. Harald Stenmark (Dept. Biochem. Inst. Cancer Research, Norwegian Radium Hospital, Oslo, Norway) for plasmids pcDNA3-Myc-Beclin1 and pEGFP-2xFYVE, Dr. Chengyu Liang (Dept. Mol. Microbiol and Immunol. Univ. Southern California, Los Angeles, CA, USA) for plasmid FLAG-hUVRAG, Dr. Zhenyu Yue (Dep. Neurol. Neurosci. Friedman Brain Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA) for plasmid pcDNA3-FLAG-ATG14L, Dr. Christoph Borner (Inst. Biochem, Univ. Freiburg, Germany) for plasmid pcDNA3-FLAG-Bcl2, Dr. Gian Maria Fimia (Dept. Epidemiology and Preclin. Res. National Inst. Infectious Dis. IRCCS Lazzaro Spallanzani, Rome, Italy) for plasmid pCLPCX-FLAG-AMBRA1, Dr. Dr. Minoo Razi (London Research Institute, UK) for plasmid pcDNA3.1-HA-ULK1; Dr. Manuel Rodríguez (Proteomics Unit, CIC-BioGUNE, Vizcaya, Spain) for plasmid pCMV-6xHisUbiq; and Dr. Christine Blattner (Institute of Toxicology and Genetics, Karlsruhe Institute of Technology, Karlsruhe, Germany) for plasmids pCMV-6xHis-Ubiq-K48R and pCMV-6xHis-Ubiq-K63R.
This work was supported by grants from the Spanish Ministry of Economy and Competitiveness SAF2014-54604-C3-1-R and SAF2017-83151-R, a grant from Fundación Ramón Areces (CIVP18A3935) and a grant from the National Institutes of Health (NIH-NINDS) P01NS097197, which established the Lafora Epilepsy Cure Initiative (LECI), to PS. We also acknowledge a grant from the Spanish Ministry of Economy and Competitiveness SAF2014-54604-C3-2-R to EK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
7.- DISCLOSURE STATEMENT
None of the authors has any conflict of interest to disclose.
REFERENCES
- [1].Turnbull J, Tiberia E, Striano P, Genton P, Carpenter S, Ackerley CA, Minassian BA, Lafora disease, Epileptic Disord, 18 (2016) 38–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Garcia-Gimeno MA, Knecht E, Sanz P, Lafora Disease: A Ubiquitination-Related Pathology, Cells, 7 (2018) 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Minassian BA, Lee JR, Herbrick JA, Huizenga J, Soder S, Mungall AJ, Dunham I, Gardner R, Fong CY, Carpenter S, Jardim L, Satishchandra P, Andermann E, Snead OC 3rd, Lopes-Cendes I, Tsui LC, Delgado-Escueta AV, Rouleau GA, Scherer SW, Mutations in a gene encoding a novel protein tyrosine phosphatase cause progressive myoclonus epilepsy, Nat Genet, 20 (1998) 171–174. [DOI] [PubMed] [Google Scholar]
- [4].Serratosa JM, Gomez-Garre P, Gallardo ME, Anta B, de Bernabe DB, Lindhout D, Augustijn PB, Tassinari CA, Malafosse RM, Topcu M, Grid D, Dravet C, Berkovic SF, de Cordoba SR, A novel protein tyrosine phosphatase gene is mutated in progressive myoclonus epilepsy of the Lafora type (EPM2), Hum Mol Genet, 8 (1999) 345–352. [DOI] [PubMed] [Google Scholar]
- [5].Chan EM, Young EJ, Ianzano L, Munteanu I, Zhao X, Christopoulos CC, Avanzini G, Elia M, Ackerley CA, Jovic NJ, Bohlega S, Andermann E, Rouleau GA, Delgado-Escueta AV, Minassian BA, Scherer SW, Mutations in NHLRC1 cause progressive myoclonus epilepsy, Nat Genet, 35 (2003) 125–127. [DOI] [PubMed] [Google Scholar]
- [6].Vilchez D, Ros S, Cifuentes D, Pujadas L, Valles J, Garcia-Fojeda B, Criado-Garcia O, Fernandez-Sanchez E, Medrano-Fernandez I, Dominguez J, Garcia-Rocha M, Soriano E, Rodriguez de Cordoba S, Guinovart JJ, Mechanism suppressing glycogen synthesis in neurons and its demise in progressive myoclonus epilepsy, Nature Neurosci, 10 (2007) 1407–1413. [DOI] [PubMed] [Google Scholar]
- [7].Worby CA, Gentry MS, Dixon JE, Malin decreases glycogen accumulation by promoting the degradation of protein targeting to glycogen (PTG), J Biol Chem, 283 (2008) 4069–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Solaz-Fuster MC, Gimeno-Alcaniz JV, Ros S, Fernandez-Sanchez ME, Garcia-Fojeda B, Criado Garcia O, Vilchez D, Dominguez J, Garcia-Rocha M, Sanchez-Piris M, Aguado C, Knecht E, Serratosa J, Guinovart JJ, Sanz P, Rodriguez de Cordoba S, Regulation of glycogen synthesis by the laforin-malin complex is modulated by the AMP-activated protein kinase pathway, Hum Mol Genet, 17 (2008) 667–678. [DOI] [PubMed] [Google Scholar]
- [9].Gómez-Abad C, Gómez-Garre P, Gutiérrez-Delicado E, Saygi S, Michelucci R, Tassinari CA, Rodriguez de Cordoba S, Serratosa JM, Lafora disease due to EPM2B mutations. A clinical and genetic study., Neurology, 64 (2005) 982–986. [DOI] [PubMed] [Google Scholar]
- [10].Cheng A, Zhang M, Gentry MS, Worby CA, Dixon JE, Saltiel AR, A role for AGL ubiquitination in the glycogen storage disorders of Lafora and Cori’s disease, Genes Dev, 21 (2007) 2399–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Vernia S, Rubio T, Heredia M, Rodriguez de Cordoba S, Sanz P, Increased endoplasmic reticulum stress and decreased proteasomal function in lafora disease models lacking the phosphatase laforin, PLoS One, 4 (2009) e5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Aguado C, Sarkar S, Korolchuk VI, Criado O, Vernia S, Boya P, Sanz P, de Cordoba SR, Knecht E, Rubinsztein DC, Laforin, the most common protein mutated in Lafora disease, regulates autophagy, Hum Mol Genet, 19 (2010) 2867–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Puri R, Ganesh S, Laforin in autophagy: a possible link between carbohydrate and protein in Lafora disease?, Autophagy, 6 (2010) 1229–1231. [DOI] [PubMed] [Google Scholar]
- [14].Criado O, Aguado C, Gayarre J, Duran-Trio L, Garcia-Cabrero AM, Vernia S, San Millan B, Heredia M, Roma-Mateo C, Mouron S, Juana-Lopez L, Dominguez M, Navarro C, Serratosa JM, Sanchez M, Sanz P, Bovolenta P, Knecht E, Rodriguez de Cordoba S, Lafora bodies and neurological defects in malin-deficient mice correlate with impaired autophagy, Hum Mol Genet, 21 (2012) 1521–1533. [DOI] [PubMed] [Google Scholar]
- [15].Roma-Mateo C, Aguado C, Garcia-Gimenez JL, Ibanez-Cabellos JS, Seco-Cervera M, Pallardo FV, Knecht E, Sanz P, Increased oxidative stress and impaired antioxidant response in lafora disease, Mol Neurobiol, 51 (2015) 932–946. [DOI] [PubMed] [Google Scholar]
- [16].Lopez-Gonzalez I, Viana R, Sanz P, Ferrer I, Inflammation in Lafora Disease: Evolution with Disease Progression in Laforin and Malin Knock-out Mouse Models, Mol Neurobiol, 54 (2017) 3119–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sanchez-Martin P, Roma-Mateo C, Viana R, Sanz P, Ubiquitin conjugating enzyme E2-N and sequestosome-1 (p62) are components of the ubiquitination process mediated by the malin-laforin E3-ubiquitin ligase complex, Int J Biochem Cell Biol, 69 (2015) 204–214. [DOI] [PubMed] [Google Scholar]
- [18].Menzies FM, Fleming A, Caricasole A, Bento CF, Andrews SP, Ashkenazi A, Fullgrabe J, Jackson A, Jimenez Sanchez M, Karabiyik C, Licitra F, Lopez Ramirez A, Pavel M, Puri C, Renna M, Ricketts T, Schlotawa L, Vicinanza M, Won H, Zhu Y, Skidmore J, Rubinsztein DC, Autophagy and Neurodegeneration: Pathogenic Mechanisms and Therapeutic Opportunities, Neuron, 93 (2017) 1015–1034. [DOI] [PubMed] [Google Scholar]
- [19].Bento CF, Renna M, Ghislat G, Puri C, Ashkenazi A, Vicinanza M, Menzies FM, Rubinsztein DC, Mammalian Autophagy: How Does It Work?, Annu Rev Biochem, 85 (2016) 685–713. [DOI] [PubMed] [Google Scholar]
- [20].Dikic I, Elazar Z, Mechanism and medical implications of mammalian autophagy, Nature Rev Mol Cell Biol, 19 (2018) 349–364. [DOI] [PubMed] [Google Scholar]
- [21].Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F, Choi AM, Chu CT, Codogno P, Colombo MI, Cuervo AM, Debnath J, Deretic V, Dikic I, Eskelinen EL, Fimia GM, Fulda S, Gewirtz DA, Green DR, Hansen M, Harper JW, Jaattela M, Johansen T, Juhasz G, Kimmelman AC, Kraft C, Ktistakis NT, Kumar S, Levine B, Lopez-Otin C, Madeo F, Martens S, Martinez J, Melendez A, Mizushima N, Munz C, Murphy LO, Penninger JM, Piacentini M, Reggiori F, Rubinsztein DC, Ryan KM, Santambrogio L, Scorrano L, Simon AK, Simon HU, Simonsen A, Tavernarakis N, Tooze SA, Yoshimori T, Yuan J, Yue Z, Zhong Q, Kroemer G, Molecular definitions of autophagy and related processes, EMBO J, 36 (2017) 1811–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lumkwana D, du Toit A, Kinnear C, Loos B, Autophagic flux control in neurodegeneration: Progress and precision targeting-Where do we stand?, Prog Neurobiol, 153 (2017) 64–85. [DOI] [PubMed] [Google Scholar]
- [23].Herzig S, Shaw RJ, AMPK: guardian of metabolism and mitochondrial homeostasis, Nature Rev Mol Cell Biol, 19 (2018) 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Boutouja F, Brinkmeier R, Mastalski T, El Magraoui F, Platta HW, Regulation of the Tumor-Suppressor BECLIN 1 by Distinct Ubiquitination Cascades, Int J Mol Sci, 18 (2017) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hill SM, Wrobel L, Rubinsztein DC, Post-translational modifications of Beclin 1 provide multiple strategies for autophagy regulation, Cell Death Diff, 26 (2019) 617–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Couarch P, Vernia S, Gourfinkel-An I, Lesca G, Gataullina S, Fedirko E, Trouillard O, Depienne C, Dulac O, Steschenko D, Leguern E, Sanz P, Baulac S, Lafora progressive myoclonus epilepsy: NHLRC1 mutations affect glycogen metabolism, J Mol Med, 89 (2011) 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Moruno-Manchon JF, Perez-Jimenez E, Knecht E, Glucose induces autophagy under starvation conditions by a p38 MAPK-dependent pathway, Biochem J, 449 (2013) 497–506. [DOI] [PubMed] [Google Scholar]
- [28].Kaiser P, Tagwerker C, Is this protein ubiquitinated?, Methods Enzymol, 399 (2005) 243–248. [DOI] [PubMed] [Google Scholar]
- [29].Upadhyay M, Agarwal S, Bhadauriya P, Ganesh S, Loss of laforin or malin results in increased Drp1 level and concomitant mitochondrial fragmentation in Lafora disease mouse models, Neurobiol Dis, 100 (2017) 39–51. [DOI] [PubMed] [Google Scholar]
- [30].Lahuerta M, Aguado C, Sanchez-Martin P, Sanz P, Knecht E, Degradation of altered mitochondria by autophagy is impaired in Lafora disease, The FEBS journal, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cadwell K, Crosstalk between autophagy and inflammatory signalling pathways: balancing defence and homeostasis, Nat Rev Immunol, 16 (2016) 661–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Grumati P, Dikic I, Ubiquitin signaling and autophagy, J Biol Chem, 293 (2018) 5404–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Platta HW, Abrahamsen H, Thoresen SB, Stenmark H, Nedd4-dependent lysine-11-linked polyubiquitination of the tumour suppressor Beclin 1, Biochem J, 441 (2012) 399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nascimbeni AC, Codogno P, Morel E, Local detection of PtdIns3P at autophagosome biogenesis membrane platforms, Autophagy, 13 (2017) 1602–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Garyali P, Siwach P, Singh PK, Puri R, Mittal S, Sengupta S, Parihar R, Ganesh S, The malin-laforin complex suppresses the cellular toxicity of misfolded proteins by promoting their degradation through the ubiquitin-proteasome system, Hum Mol Genet, 18 (2009) 688–700. [DOI] [PubMed] [Google Scholar]
- [36].Puri R, Suzuki T, Yamakawa K, Ganesh S, Dysfunctions in endosomal-lysosomal and autophagy pathways underlie neuropathology in a mouse model for Lafora disease, Hum Mol Genet, 21 (2012) 175–184. [DOI] [PubMed] [Google Scholar]
- [37].Shi CS, Kehrl JH, TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin1 to control TLR4-induced autophagy, Sci Signal, 3 (2010) ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Xie Y, Kang R, Sun X, Zhong M, Huang J, Klionsky DJ, Tang D, Posttranslational modification of autophagy-related proteins in macroautophagy, Autophagy, 11 (2015) 28–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Moreno D, Towler MC, Hardie DG, Knecht E, Sanz P, The laforin-malin complex, involved in Lafora disease, promotes the incorporation of K63-linked ubiquitin chains into AMP-activated protein kinase beta subunits, Mol Biol Cell, 21 (2010) 2578–2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Viana R, Lujan P, Sanz P, The laforin/malin E3-ubiquitin ligase complex ubiquitinates pyruvate kinase M1/M2, BMC Biochem, 16 (2015) 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Fusco C, Mandriani B, Di Rienzo M, Micale L, Malerba N, Cocciadiferro D, Sjottem E, Augello B, Squeo GM, Pellico MT, Jain A, Johansen T, Fimia GM, Merla G, TRIM50 regulates Beclin 1 proautophagic activity, Biochim Biophys Acta, 1865 (2018) 908–919. [DOI] [PubMed] [Google Scholar]
- [42].Liu J, Li M, Li L, Chen S, Wang X, Ubiquitination of the PI3-kinase VPS-34 promotes VPS-34 stability and phagosome maturation, J Cell Biol, 217 (2018) 347–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.