Adult-onset Still's disease (AOSD) is a rare autoinflammatory condition with unclear aetiology and highly heterogeneous manifestations, such as spiking fever, skin rash and arthralgia (or arthritis).1 2 AOSD is usually the result of both genetic and environmental factors. However, only a few genetic loci have been associated with AOSD, and none of them have reached the threshold for genome-wide significance (GWS) of p<5×10− 8.2 3 For example, HLA-DRB1 was strongly associated with AOSD in previous studies (the p value for the most significant association was 8.60×10− 6).3 HLA-DRB1 also influences the risk of systemic juvenile idiopathic arthritis (JIA),4 which presents similarly to AOSD but differs in age of onset. In addition, functional variations in periodic fever syndrome genes have been identified in some AOSD patients (Bonferroni corrected p values ranged from 2.34×10− 3 to 2.40×10− 4).5
In this study, for the first time, we conducted a genome-wide association study (GWAS) to systematically screen genetic factors influencing susceptibility to AOSD (online supplementary text). Principal component analysis was adopted to evaluate population stratification between cases and controls and to detect outliers for removal (online supplementary figure S1). After quality control and imputation, a total of 3 547 931 variants in 264 AOSD cases and 2420 controls (discovery: 247 cases vs 2163 controls; replication: 17 cases vs 257 controls; online supplementary text and table S1) from China were analysed. The genomic inflation, λGC, was 1.018, indicating the presence of population stratification to a minimal degree (online supplementary figure S2). A total of 1281 variants, from the HLA class I and II regions, exceeded GWS in this analysis (online supplementary figure S3).
annrheumdis-2019-215239supp001.pdf (506.1KB, pdf)
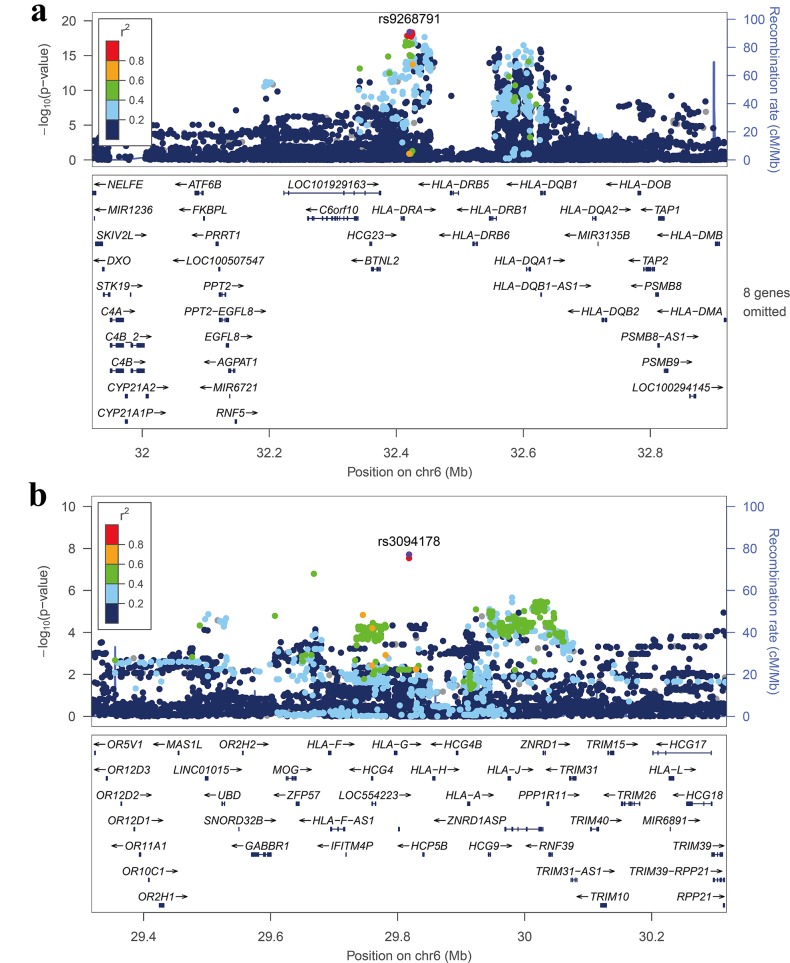
The most significant association was with rs9268791 (OR=2.363, p=4.10×10−19; table 1 and figure 1A), located in the intergenic region between HLA-DRA and HLA-DRB5 (the HLA class II region). For the HLA class I region, the top GWS signal was observed for rs3094178 (OR=2.139, p=1.97×10−8; table 1 and figure 1B), which is located 19 kb downstream of HLA-G. Outside the HLA region, a suggestive association (p<10−5) was observed at VEGFC (rs514410, OR=2.211, p=1.81×10−6; online supplementary table S2, figure S4 and text).
Table 1.
Summary statistics for the genome-wide significant association of single nucleotide polymorphisms with adult-onset Still's disease
| SNP | Nearby gene(s) | Chr | Position (hg19) | Risk/non-risk allele | Stage* | RAF in cases | RAF in controls | OR | SE | P values | P_het |
| rs9268791 | HLA-DRA|HLA-DRB5 | 6 | 32 421 073 | T/C | Meta | – | – | 2.363 | 0.096 | 4.10E−19 | 0.686 |
| Discovery | 0.468 | 0.273 | 2.389 | 0.100 | 2.85E−18 | ||||||
| Replication | 0.382 | 0.224 | 2.054 | 0.360 | 4.54E−02 | ||||||
| rs3094178 | HLA-G | 6 | 29 818 000 | C/G | Meta | – | – | 2.139 | 0.135 | 1.97E−08 | 0.806 |
| Discovery | 0.875 | 0.769 | 2.116 | 0.143 | 1.48E−07 | ||||||
| Replication | 0.767 | 0.568 | 2.366 | 0.432 | 4.62E−02 |
*The discovery data set comprises 247 cases and 2163 controls from northern and central China. The replication data set comprises 17 cases and 257 controls from southern China.
Chr, chromosome;P_het, p value for heterogeneity test across different cohorts using Cochran's Q test; RAF, frequency of risk allele;SNP, single nucleotide polymorphism.
Figure 1.
Regional association plots of the adult-onset Still's disease-associated HLA class I and II regions. (A) HLA class II region and (B) HLA class I region. Purple circles represent the most significantly associated single nucleotide polymorphisms (SNPs) (marker SNPs) in each region in the meta-analysis of discovery and replication. −log10p values (Y-axis) of the SNPs (within the regions spanning 500 kb on either side of the marker SNP) are presented according to the chromosomal positions of the SNPs (X-axis, hg19). SNPs are coloured according to their linkage disequilibrium (LD) with the marker SNP. The LD values were established based on the 1000 Genomes Asian (ASI) Data (March 2012). Estimated recombination rates with samples from the 1000 Genomes Project March 2012 release are shown as blue lines, and the genomic locations of genes within the regions of interest annotated from the University of California, Santa Cruz (UCSC) genome browser are shown as arrows.
We explored regulatory annotations for the GWS SNPs and variants in strong linkage disequilibrium (LD) with them (r2>0.8) using HaploReg6 and RegulomeDB,7 and 94 SNPs had a RegulomeDB score from 1a–1f to 2a–2c, suggesting that they are located in regulatory regions with maximum evidence (online supplementary table S3). Of them, the top SNPs in each region were rs3115628 (p=2.92×10−8; HLA class I) and rs9268832 (p=3.41×10−18; HLA class II). In addition, the expression quantitative trait loci analyses suggested that rs9268832 was significantly associated with the expression of several genes (including HLA-DRB1, HLA-DRB6, HLA-DQA2 and HLA-DQB1; p<5×10−8) and that rs3115628 was significantly correlated with ZFP57 and four HLA pseudogenes (p<5×10−8) in different types of tissues (online supplementary table S4).8 Further studies are required to clarify the mechanisms underlying these associations.
Previous GWAS of JIA on primarily European ancestry samples has also identified three GWS SNPs in the HLA region.9 10 However, none of them overlapped with the AOSD-associated markers identified in this study. Pair-wise LD analysis showed that these SNPs are in low LD (r2<0.1, online supplementary table S5). This finding suggests genetic heterogeneity in these diseases and across different populations.
In summary, we identified the first GWS risk loci and indicated an important role for both HLA class I and II regions in the genetic architecture of AOSD using a genome-wide scan in a Chinese sample. Further replication studies of other populations are necessary to confirm our findings and to investigate the possible ancestral differences.
Acknowledgments
We appreciated to all the participants, research staffs, and students who took part in this study.
Footnotes
Handling editor: Josef S Smolen
ZL and H-LL contributed equally.
Contributors: ZL, H-LL, YShi and CY designed the study. H-LL, TZ, CL, SL, T-TD, KY, JT, XC, JY, Y-TS, HS, Yue Sun, QH, HC, Zhuochao Zhou, L-JC, JX and LW collected the samples and organized the patient history information. JC, LH, ML, XL, YD, JZ, JJ, CG, XY, DW, KW, C-GL, Yuanchao Sun, YN, Zhaowei Zhou, DP and HN performed or contributed to the experiments. ZL and H-LL conducted data analyses. ZL and H-LL interpretated the results and drafted the manuscript. YShi and CY revised the manuscript. All authors critically reviewed and approved the manuscript.
Funding: This study was supported by National Natural Science Foundation of China (81502016 for H-LL, 81671589 for CY, 81701321 and 81871055 for ZL, 31325014 for YShi, 81421061 for LH, 81501154 for JC, 81772713 for HN), the 973 Program (2015CB559100 for YShi), the National Key R&D Program of China (2016YFC0903402 for YShi and ZL), Taishan Scholar Program of Shandong Province (tsqn201812153 for ZL and tsqn20161077 for HN), Natural Science Foundation of Shandong Province (ZR2019YQ14 for ZL), Program of Shanghai Subject Chief Scientist (15XD1502200 for YShi), Shanghai Municipal Health Commission (ZK2015B01 and 20154011 for YShi), National Program for Support of Top-Notch Young Professionals for YShi.
Competing interests: None declared.
Patient consent for publication: Not required.
Ethics approval: Institutional review board and Institutional Ethical Committee of the AOSD-database partner (Shanghai Jiao Tong University School of Medicine Affiliated Ruijin Hospital, Xinhua Hospital Chongming Branch Affiliated to Shanghai Jiao Tong University School of Medicine, People's hospital of Xinjiang Ugyur Autonomous Region, First Affiliated Hospital of Kunming Medical University, First Affiliated Hospital of Bengbu Medical College).
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Giacomelli R, Ruscitti P, Shoenfeld Y. A comprehensive review on adult onset still's disease. J Autoimmun 2018;93:24–36. 10.1016/j.jaut.2018.07.018 [DOI] [PubMed] [Google Scholar]
- 2. Feist E, Mitrovic S, Fautrel B. Mechanisms, biomarkers and targets for adult-onset Still’s disease. Nat Rev Rheumatol 2018;14:603–18. 10.1038/s41584-018-0081-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Asano T, Furukawa H, Sato S, et al. Effects of HLA-DRB1 alleles on susceptibility and clinical manifestations in Japanese patients with adult onset Still’s disease. Arthritis Res Ther 2017;19 10.1186/s13075-017-1406-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ombrello MJ, Remmers EF, Tachmazidou I, et al. HLA-DRB1*11 and variants of the MHC class II locus are strong risk factors for systemic juvenile idiopathic arthritis. Proc Natl Acad Sci U S A 2015;112:15970–5. 10.1073/pnas.1520779112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sighart R, Rech J, Hueber A, et al. Evidence for genetic overlap between adult onset Still's disease and hereditary periodic fever syndromes. Rheumatol Int 2018;38:111–20. 10.1007/s00296-017-3885-0 [DOI] [PubMed] [Google Scholar]
- 6. Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 2012;40:D930–D934. 10.1093/nar/gkr917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 2012;22:1790–7. 10.1101/gr.137323.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ardlie KG, DeLuca DS, Segre AV, et al. Human genomics. The Genotype-Tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015;348:648–60. 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hinks A, Cobb J, Marion MC, et al. Dense genotyping of immune-related disease regions identifies 14 new susceptibility loci for juvenile idiopathic arthritis. Nat Genet 2013;45:664–9. 10.1038/ng.2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ombrello MJ, Arthur VL, Remmers EF, et al. Genetic architecture distinguishes systemic juvenile idiopathic arthritis from other forms of juvenile idiopathic arthritis: clinical and therapeutic implications. Ann Rheum Dis 2017;76:906–13. 10.1136/annrheumdis-2016-210324 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
annrheumdis-2019-215239supp001.pdf (506.1KB, pdf)