Abstract
Cys2/His2‐type (C2H2) zinc finger proteins, such as ZCT1, are an important class of transcription factors involved in growth, development, and stress responses in plants. In the medicinal plant Catharanthus roseus, the zinc finger Catharanthus transcription factor (ZCT) family represses monoterpenoid indole alkaloid (MIA) biosynthetic gene expression. Here, we report the analysis of the ZCT1 promoter, which contains several hormone‐responsive elements. ZCT1 is responsive to not only jasmonate, as was previously known, but is also induced by the synthetic auxin, 1‐naphthalene acetic acid (1‐NAA). Through promoter deletion analysis, we show that an activation sequence‐1‐like (as‐1‐like)‐motif and other motifs contribute significantly to ZCT1 expression in seedlings. We also show that the activator ORCA3 does not transactivate the expression of ZCT1 in seedlings, but ZCT1 represses its own promoter, suggesting a feedback mechanism by which the expression of ZCT1 can be limited.
Keywords: activation sequence‐1‐like, Agrobacterium‐mediated transformation, Catharanthus roseus, Cys2/His2‐type zinc finger transcription factors, ORCA, plant stress response
1. INTRODUCTION
Due to their sessile existence, plants have developed rapid and effective responses to environmental stresses, herbivory, and pathogens. When stressed, plants can shift primary metabolic resources to produce specialized metabolites that act as defense compounds. For instance, the Madagascar periwinkle, Catharanthus roseus, produces numerous monoterpenoid indole alkaloids (MIA) in defense against pathogens and herbivory (Luijendijk, van der Meijden, & Verpoorte, 1996; Dugé de Bernonville et al., 2017). Many of these MIAs are pharmaceutically valuable, including the antihypertensive ajmalicine, the sedative serpentine, and most notably the anticancer compounds vincristine and vinblastine (Van der Heijden, Jacobs, Snoeijer, Hallard, & Verpoorte, 2004; Noble, 1990).
A crucial regulatory mechanism in specialized metabolism is the transcriptional regulation of biosynthetic genes. Several transcription factors (TFs) involved in MIA regulation have been identified in Catharanthus, including the octadecanoid‐responsive Catharanthus AP2‐domain (ORCA) TF family and the zinc finger Catharanthus (ZCT) TF family described below (Menke et al., 1999; Van der Fits & Memelink, 2000; Van der Fits, Zhang, Menke, Deneka, & Memelink, 2000; Sibéril et al., 2001; Chatel et al., 2003; Pauw et al., 2004; Vom Endt, Soares e Silva, Kijne, Pasquali, & Memelink, 2007; Zhang et al., 2011; Suttipanta et al., 2011; Van Moerkercke et al., 2015, 2016; Paul et al., 2016; Liu et al., 2017; Liu, Patra, Pattanaik, Wang, & Yuan, 2019; Patra, Pattanaik, Schluttenhofer, & Yuan, 2017; Pan et al., 2019). Three characterized members (ZCT1, ZCT2, and ZCT3) of the Cys2/His2‐type (C2H2) zinc finger family are expressed by the stress‐induced phytohormone jasmonate (Goklany, Rizvi, Loring, Cram, & Lee‐Parsons, 2013; Pauw et al., 2004). ZCTs repress the expression of at least two of the key MIA biosynthetic genes, strictosidine synthase (STR) and tryptophan decarboxylase (TDC), in transient expression assays (Mortensen et al., 2019; Pauw et al., 2004).
ZCTs potentially limit the extent of MIA biosynthesis induced by jasmonate. For instance, optimum dosages of jasmonate (up to 250 μM; Goklany et al., 2013; Lee‐Parsons, Ertürk, & Tengtrakool, 2004) enhance MIA biosynthesis and are correlated with a high ratio of transcriptional activators (ORCAs) to repressors (ZCTs) levels (Goklany et al., 2013). But higher dosages of jasmonate (>500 μM; Goklany et al., 2013; Lee‐Parsons et al., 2004) inhibit MIA biosynthesis and are correlated with a high ratio of transcriptional repressors (ZCTs) to activator (ORCAs) levels (Goklany et al., 2013). The inhibition of MIA biosynthesis with high jasmonate dosages is potentially mediated through repressors like ZCTs. The repressor activity of the ZCTs is conferred by an ERF‐associated amphiphilic repression (EAR)‐motif (Ohta, Matsui, Hiratsu, Shinshi, & Ohme‐Takagi, 2001). A detailed description of the C. roseus ZCTs can be found in Pauw et al., 2004, and a more general characterization of zinc finger proteins is given in Kiełbowicz‐Matuk, 2012.
Catharanthus roseus ZCTs belong to the C2H2‐type zinc finger family. Analysis of the Arabidopsis thaliana genome revealed that C2H2‐type zinc fingers represent a large family of TFs. Of the 176 C2H2‐type zinc fingers identified in A. thaliana, only 33 are conserved among other eukaryotes, and the rest are plant‐specific; these plant‐specific C2H2‐type zinc fingers result from extensive duplication events (Englbrecht, Schoof, & Böhm, 2004). Conserved C2H2‐type zinc fingers are believed to be involved in ancient biological processes such as RNA metabolism and chromatin remodeling, whereas the duplicated zinc fingers may be involved in species‐specific transcriptional regulation (Englbrecht et al., 2004). In plants, C2H2‐type zinc fingers are involved in stress responses, for example, cold, drought, salt, and oxidative stress responses (Ciftci‐Yilmaz & Mittler, 2008; Kiełbowicz‐Matuk, 2012). The C. roseus zinc finger proteins ZCT1, ZCT2, and ZCT3 have the highest similarity, based on a blastp search, to ZAT8 (AT3G46080), ZAT17 (AT2G28710), and ZAT10/STZ (AT1G27730), respectively, in the A. thaliana genome. All three A. thaliana zinc finger proteins are C1 family and C1‐2i subclass members (Englbrecht et al., 2004). ZAT10 is one of the better studied TFs of the C1‐2i subclass members and is responsive to a number of abiotic stressors such as abscisic acid (ABA), cold temperatures, high light intensities, oxidative stress, salt stress, water deprivation, and wounding (Mittler et al., 2006; Sakamoto, Araki, Meshi, & Iwabuchi, 2000; Sakamoto et al., 2004).
Similarly, C. roseus ZCT1‐3 expression has been shown to be responsive to stressors and is induced by yeast extract, methyl jasmonate (MJ) (Goklany et al., 2013; Pauw et al., 2004), and sodium nitroprusside (SNP), a source of the signaling molecule nitric oxide (Zhou, Zhu, Shao, Wu, & Tang, 2010). Overexpression of ORCA3, one of the key MIA pathway activators, correlates with increased ZCT expression in C. roseus hairy roots, suggesting a potential regulation of ZCTs by ORCA3 (Peebles, Hughes, Shanks, & San, 2009). Otherwise, very little is known about the regulation of ZCTs.
Here, we have chosen to investigate the 1000‐bp upstream region of the ZCT1 coding sequence as the ZCT1 promoter contains many putative hormone‐responsive elements, and ZCT1 expression is highly induced upon MJ treatment in hairy roots (Goklany et al., 2013). Based on these putative regulatory elements and the role of C2H2‐type zinc fingers, we explored whether endogenous ZCT1 expression responds to hormones associated with growth, biotic, and abiotic stress such as auxin, gibberellin (GA), methyl jasmonate (MJ), salicylic acid (SA), and abscisic acid (ABA). We report that auxin (1‐NAA) and MJ induce ZCT1 expression in hairy roots. Through promoter analysis experiments in transiently transformed seedlings, we determined that an activation sequence‐1‐like motif (as‐1‐like motif) contributes significantly to the promoter activity of ZCT1. Further elements within the promoter, including a GA‐responsive complex (GARC), also contribute. Overexpression of ORCA3 did not transactivate the expression of ZCT1 or activate ZCT1 promoter–reporter constructs in seedlings. ZCT1 is capable of repressing its own promoter, suggesting a possible feedback mechanism limiting the expression of ZCT1.
2. MATERIALS AND METHODS
2.1. Cloning, sequencing, and in silico analysis of the ZCT1 promoter
The ZCT1 promoter sequence was obtained with the Universal Genome Walker Kit (Clontech). Approximately 800 bp of the ZCT1 promoter was amplified from C. roseus (Little Bright Eye, NEseeds) genomic DNA using the GSP1 and GSP2 gene‐specific primers (Table S1) matching the ZCT1 coding sequence (GenBank accession AJ632082). This sequence matches the sequence 5′ of the ZCT1 coding region of the published genomes (Franke et al., 2019; Kellner et al., 2015). The promoters of C. roseus ZCT2 and ZCT3 were obtained from the published genomes (Figure S1 and Supplemental Materials).
To identify the transcriptional start site, the SMARTer RACE cDNA Amplification Kit (Clontech) was used to amplify the 5′ cDNA ends using the same GSP1 and GSP2 gene‐specific primers. Promoter sequences were analyzed with the Plant Cis‐Acting Regulatory Elements (PlantCARE; Lescot et al., 2002) and PlantPAN 3.0 databases (Chow et al., 2019). Using the A. thaliana PlantPAN 3.0 database, only exactly matching motifs (similar score of 1) were further considered. Motifs with low information content (e.g., GAT, TF_motif_seq_0237) and frequent occurrences in all tested promoters were excluded. The pyrimidine box matches sequences from the literature (Skriver, Olsen, Rogers, & Mundy, 1991; Rogers, Lanahan, & Rogers, 1994; Gubler et al., 1999).
2.2. Creation of a sequence logo for as‐1‐like sequences and C. roseus ZCT1 as‐1‐like sequence
As‐1‐like elements from A. thaliana GST6 (Chen, Chao, & Singh, 1996), A. thaliana PR‐1 (Zhang, Fan, Kinkema, Li, & Dong, 1999), Agrobacterium tumefaciens MAS (Feltkamp, Masterson, Starke, & Rosahl, 1994), A. tumefaciens NOS (Bouchez, Tokuhisa, Llewellyn, Dennis, & Ellis, 1989), A. tumefaciens OCS (Bouchez et al., 1989), Cauliflower mosaic virus 35S (Liu & Lam, 1994), Glycine max GH2/4 (Ulmasov, Hagen, & Guilfoyle, 1994), Nicotiana tabacum GNT1 (van der Zaal et al., 1991), N. tabacum GNT35 (van der Zaal et al., 1991), N. tabacum 103 (Droog, Hooykaas, Libbenga, & Zaal, 1993), N. tabacum PR‐1a (Strompen, Grüner, & Pfitzner, 1998), N. tabacum ParA (Takahashi, Kusaba, Hiraoka, & Nagata, 1991), Silene cucubalis GST (Prändl & Kutchan, 1992), and Triticum aestivum GST‐A1 (Dudler et al., 1991 ; reviewed in Ulmasov et al., 1994 and Krawczyk et al., 2002) were used to create a WebLogo (Crooks et al., 2004). The spacer region was excluded from the WebLogo.
2.3. Cloning ZCT1 promoter constructs for transient evaluation and stable infections
For vector construction, the Golden Gate‐based modular cloning system (MoClo, Weber, Engler, Gruetzner, Werner, & Marillonnet, 2011) was used, with parts from Engler et al. (2014) and Mortensen et al. (2019).
A 1000‐bp (−914 bp to +86 bp, with TSS set to +1) fragment of the ZCT1 promoter was amplified from C. roseus (Little Bright Eye) genomic DNA and cloned into pICH41295 (Weber et al., 2011). The five promoter deletions were generated using standard cloning techniques. The mutant as‐1‐like element is identical to the pZCT1_744 promoter construct, except for the reverse complementary as‐1‐like sites (first and last 5 bp were replaced with reverse complementary sequence) and was purchased from Genewiz in pUC57. The 35S minimal promoter (−46‐ to +6‐bp region) was amplified from pICH51288 (Engler et al., 2014) and cloned into pICH41246 (5U/5U + NT1) and pICH41295 (Pro + 5U; Weber et al., 2011). This allows the fusion of different regulatory elements to the 35S minimal promoter (in pICH41246) for studying its promoter activity isolated from the rest of the ZCT1 promoter. The 35S minimal promoter alone (in pICH41295) serves as a reference. The GA‐responsive complex (GARC; 142 bp) and the GARC with the second INDETERMINATE DOMAIN (IDD) binding site (173 bp) were cloned into pICH41233 (Weber et al., 2011). Promoter elements and deletions were then assembled to a firefly luciferase reporter gene in level 1 vectors as described in Weber et al., (2011). The firefly luciferase (FLUC) reporters were cloned together with a constitutively expressed Renilla luciferase (RLUC) for normalization (Mortensen et al., 2019). The final constructs were assembled in the pSB90 (Mortensen et al., 2019) vector backbone. The pSB90 vector contains VirGN54D in the vector backbone for increased Agrobacterium virulence (as demonstrated in Mortensen et al., (2019)). All newly cloned parts were confirmed by sequencing, and correct assembly was confirmed by restriction enzyme digest, PCR, and/or sequencing. Vector cartoons can be found in Figure S4, and promoter sequences are provided as Supplemental Materials.
2.4. Preparation of C. roseus seedlings for developing transgenic hairy root lines
Catharanthus roseus seeds (Little Bright Eye) were surface‐sterilized in 70% (v/v) ethanol for one minute, followed by 10% (v/v) bleach containing 0.1% (v/v) Triton X‐100 for ten minutes. The seeds were rinsed three times in sterile water and soaked in 1% Plant Preservative Mixture (Caisson Laboratories) in sterile water for 24 hr. The seeds were planted in sterile Magenta GA‐7 boxes on the surface of 1/2 strength Murashige and Skoog media (2.2 g/L Murashige and Skoog basal salts with vitamins, 3% sucrose, 4 g/L Phytoagar, pH 5.7). Seedlings were grown in the dark at 25°C for one week and then transferred to a 16 hr of light (Erligpowht 45W LED Red Blue Lights) photoperiod for approximately 6 weeks before infection with Agrobacterium rhizogenes.
2.5. Development of stable transgenic hairy root lines
Transgenic C. roseus hairy root cultures were generated as previously described (Rizvi et al., 2015). Briefly, six‐week‐old C. roseus seedlings (grown as described above) were transformed by pricking the seedling stem above the cotyledons with modified tweezers dipped into the Agrobacterium rhizogenes R1000 liquid cultures containing pSB119 (Figure S4). This plasmid encodes the expression of TurboGFP (tGFP; Pontellina plumata) driven by the −914‐bp to +86‐bp ZCT1 promoter fragment (pZCT1::tGFP‐I). Hairy roots emerged from the site of infection approximately ~3 weeks after infection. When hairy roots were longer than 5 mm (~6 weeks after infection), hairy root clusters were excised and cultured first on elimination media to remove Agrobacterium, and then on selection media to select for hygromycin‐resistant transgenics. Hairy root cultures were subcultured in liquid media approximately every 28 days and maintained as previously reported (Goklany et al., 2013).
2.6. Hormone treatment of stable pZCT1::tGFP‐I hairy roots
The pZCT1::tGFP‐I hairy roots were cut into pieces approximately 2–3 cm long. Approximately three root pieces were added to each well of 6‐well plates containing 5 ml of liquid Gamborg's media per well and were grown for 7 days. Root pieces were treated with MJ (0.25 or 1 mM; (Goklany et al., 2013)), ABA (0.1, 0.5, or 1 mM; (Wang, Liu, Gao, & Zhang, 2010)), gibberellic acid (GA3; 0.01, 0.03, or 0.1 mM; (Suttipanta et al., 2011)), SA (0.01, 0.1, or 0.25 mM (Kang et al., 2004)), or 1‐NAA (0.01, 0.1, or 1 mM (Goddijn, Kam, Zanetti, Schilperoort, & Hoge, 1992)). As some hormones are dissolved in ethanol, a final concentration of 0.1% ethanol was added to all hormone treatments and the mock treatment. The 6‐well plates were incubated in the dark, at 28°C, and shaking for 24 hr. After 24 hr, samples were collected for GFP quantification. Three independent experiments with at least three biological replicates were performed.
2.7. Image acquisition
Fluorescence microscopy images were acquired with a Nikon Eclipse 80i microscope and 10x/0.30 Nikon objective (Nikon Corporation), equipped with a SPOT RT3 CCD camera (Diagnostic Instruments), controlled by SPOT Advanced imaging software (v. 5.0) with Peripheral Devices and Quantitative Imaging modules. A Nikon Intensilight C‐ HGFI 130‐W mercury lamp, shuttered with a Lambda 10‐B SmartShutter (Sutter Instruments), was used for GFP excitation, and a GFP filter set (470/40 × 495lpxr 525/50 m; Chroma Technologies) was used for detection.
2.8. GFP quantification
Hairy roots expressing TurboGFP were quantified in a fluorescence plate reader after a modified protocol from Marillonnet et al., (2004).
Roots were transferred to filter paper to remove excess culture media. Approximately 100 mg of tissue was transferred into 2‐ml screwcap tubes together with ten 3‐mm glass beads (Fisher), and the fresh weight of the roots was recorded. Samples were frozen in liquid nitrogen and stored at −80°C until analysis. Frozen samples were crushed in a Mini‐BeadBeater‐16 (BioSpec Products Inc.) for 20 s and transferred to ice, and 600 μl of extraction buffer was added (50 mM Na3PO4‐buffer at pH7.2, 1.0 mM EDTA) with fresh 10 mM β‐mercaptoethanol and 0.1% Triton X‐100. After vortexing and centrifugation (21,000 g for 2 min), carefully avoiding cell debris, 200 μl of the supernatant was transferred to a black 96‐well plate and fluorescence was detected with a plate reader (SynergyTM HT, BioTek; excitation 470 nm and emission 509 nm).
2.9. Transient C. roseus transformation
Catharanthus roseus seedlings were transformed with the efficient Agrobacterium‐mediated seedling infiltration (EASI) method (Mortensen et al., 2019). Briefly, 10‐day‐old C. roseus seedlings were transiently transformed with engineered Agrobacterium tumefaciens GV3101 (pMP90) strains by vacuum infiltration, and tissue was collected 3 days after infection. For promoter activity studies, A. tumefaciens GV3101 (pMP90) strains, containing the different promoter driving reporter constructs, were adjusted to OD600 = 0.2 for infiltration into C. roseus seedlings (Figure 4 and 5). Transactivation assays were performed with the reporter strain at OD600 = 0.06 and the effector strain at OD600 = 0.34 (Figure 6, ratio of reporter to effector strain was optimized in Mortensen et al., 2019) and OD600 = 0.4 for transactivation without reporter strain (Figure 7).
2.10. Dual‐luciferase assay
FLUC and RLUC activity were analyzed using the Luc‐PairTM Duo‐Luciferase HT Assay Kit (Genecopoeia) as described in Mortensen et al., (2019).
2.11. Monitoring ORCA3 and ZCT1 in transiently transformed seedlings
Expression levels of ORCA3 and ZCT1 were monitored in seedlings transiently overexpressing a β‐glucuronidase (GUS) or ORCA3, or seedlings lacking A. tumefaciens during infiltration, using quantitative real‐time PCR (qRT‐PCR). mRNA was extracted from liquid nitrogen flash‐frozen seedlings (cotyledons from 15 seedlings pooled per biological replicate), stored at −80°C until needed. While still frozen, the cotyledons were crushed by shaking in the Mini‐BeadBeater‐16 (BioSpec) for 15 s with ten 3‐mm glass beads (Fisher). Afterward, RNA was extracted with RNAzol‐RT (Molecular Research Center) and the Direct‐zol RNA Miniprep Plus Kit (Zymo Research) with on‐column DNAse treatment to remove genomic DNA. RNA integrity was assessed using agarose gel electrophoresis, and concentration and purity were quantified with a NanoDrop (ND‐1000 Spectrophotometer; Thermo Scientific). cDNA was synthesized using the SuperScript II First‐Strand Synthesis System (Invitrogen) and oligo‐dT primers with up to 2.5 µg of RNA in a 10 µl reaction, according to the manufacturer's instructions.
cDNA was diluted 1:4, and 1 µl was used in a 10 µl reaction with SYBR Green ROX qPCR Master Mix (Qiagen) on the MX3000P qPCR instrument (Agilent) using the thermocycler protocol previously described with an extension time of 30 s (Goklany et al., 2013). Ct values for each biological replicate were calculated as the average of two technical replicates. Transcript levels were normalized to the housekeeping gene, SAND (Pollier, Vanden Bossche, Rischer, & Goossens, 2014), and fold changes were calculated according to the 2−∆∆Ct method (Livak & Schmittgen, 2001). The qPCR primers (Table S1) were designed previously (Goklany et al., 2013; Pollier et al., 2014), but the amplification efficiency for each gene was confirmed again for this study using Ct values over a range of cDNA dilutions and was 100% ± 10% for each gene monitored. No‐reverse‐transcriptase controls were included for each sample, and SAND Ct values were confirmed to be at least 5 Ct values above the respective experimental sample (Millipore Sigma technical notes).
3. RESULTS
3.1. Sequencing and in silico analysis of the ZCT1, ZCT2, and ZCT3 promoters
To begin to characterize the transcriptional regulation of ZCT1, the promoter for ZCT1 was isolated using a genome‐walking approach and the transcription start site (TSS) was determined using 5′ rapid amplification of cDNA ends (RACE). Six clones were sequenced to determine that the TSS is located 86 bp 5′ of the translational start site (ATG). The cDNA ends were the same for 5 of the 6 clones (this is referred to as the TSS in this study), and the 6th included one additional 5′ bp. Our sequence matches the sequence upstream of the ZCT1 coding region in the sequenced C. roseus genomes (Franke et al., 2019; Kellner et al., 2015). ZCT2 and ZCT3 are induced with jasmonate like ZCT1 and have similar functions as ZCT1 (Chebbi et al., 2014; Goklany et al., 2013; Pauw et al., 2004). Therefore, the promoter sequences of ZCT2 and ZCT3 were obtained from the C. roseus genome and putative regulatory elements in the promoters of ZCT1, ZCT2, and ZCT3 were identified using the Plant Cis‐Acting Regulatory Elements database (PlantCARE) and PlantPAN 3.0. A detailed PlantCARE characterization of the ZCT1, ZCT2, and ZCT3 promoters can be found in Figure S1, and motifs identified with PlantPAN 3.0 are included in the supplementary promoter sequences (.gbk files).
The ZCT1, ZCT2, and ZCT3 promoters share several common hormone‐ and stress‐responsive elements. All three promoters contain at least one wounding‐responsive element within the first 200 bp upstream from the translational start site. Interestingly, the promoters of ZCT1 and ZCT2 both contain two W‐boxes (WRKY binding sites), two abscisic acid‐responsive elements (ABRE), and two ethylene‐responsive elements (ERE). The plant‐specific WRKY TFs (named for the conserved amino acids in WRKY domain) were identified in C. roseus, and WRKY1 is involved in regulation of MIA biosynthesis (Schluttenhofer, Pattanaik, Patra, & Yuan, 2014; Suttipanta et al., 2011).
The ZCT1 promoter contains multiple elements not found in the other promoters, including motifs involved in gibberellin responsiveness (TATC‐box, pyrimidine box, and GA box, which make up the GA‐responsive complex, GARC; Gubler & Jacobsen, 1992; Lanahan, Ho, Rogers, & Rogers, 1992; Rogers & Rogers, 1992) and two IDD motifs, which are recognized by INDETERMINATE DOMAIN (IDD) proteins and co‐activate with the GA‐responsive protein, DELLA (Yoshida et al., 2014). Additionally, there are two CGTCA or TGACG‐motifs in close proximity, which make up an activation sequence‐1‐like element (Krawczyk et al., 2002). We compared the ZCT1 as‐1‐like element to well‐characterized as‐1‐like elements (Figure 1). The binding sites in the ZCT1 promoter are consistent with the as‐1‐like element, except for the nucleotides at positions 6–8, which are complementary to the expected sequence. Unlike the 4 bp spacers in many plants or Agrobacterium genes, or the 6 bp and 9 bp spacers in the as‐1‐like elements of the N. tabacum PR‐1a and A. thaliana PR‐1 genes, the ZCT1 as‐1‐like element has an uncommonly long 14 bp spacer between the two TGACG‐motifs.
Figure 1.
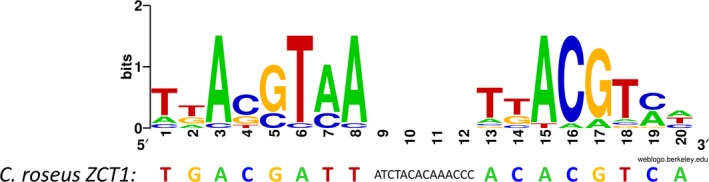
Sequence logo of as‐1‐like sequences and Catharanthus roseus ZCT1 as‐1‐like sequence. As‐1‐like elements were used to create a WebLogo (Crooks et al., 2004). The spacer region (base 9–12 in the WebLogo) was excluded and is usually 4 bp long. The spacer in the ZCT1 promoter is 14 bp
3.2. The ZCT1 promoter responds to several plant hormones
Because we observed several putative hormone‐responsive sites in the ZCT1 promoter (Figure S1), we explored whether the −914‐bp to +86‐bp fragment of ZCT1 responds to auxins (1‐NAA), MJ, SA, ABA, and GA3. As‐1‐like elements are known to be activated by MJ, SA, and auxins (Ulmasov et al., 1994; Xiang, Miao, & Lam, 1996).
To investigate the transcriptional regulation of ZCT1, the −914‐bp to +86‐bp fragment of the ZCT1 promoter was used to create a reporter with GFP (pZCT1::tGFP‐I). This vector was used to create stable transgenic hairy root lines via Agrobacterium‐mediated transformation as previously described (Rizvi et al., 2015). From eight root lines that passed the hygromycin B selection, two independent lines were chosen for a preliminary experiment to confirm the known responsiveness of ZCT1 expression with MJ (Goklany et al., 2013; Rizvi, Weaver, Cram, & Lee‐Parsons, 2016). Both lines showed a strong increase in GFP fluorescence throughout the whole root after treatment with MJ (Figure 2). Line #8 was chosen for treatment with various hormones and concentrations.
Figure 2.
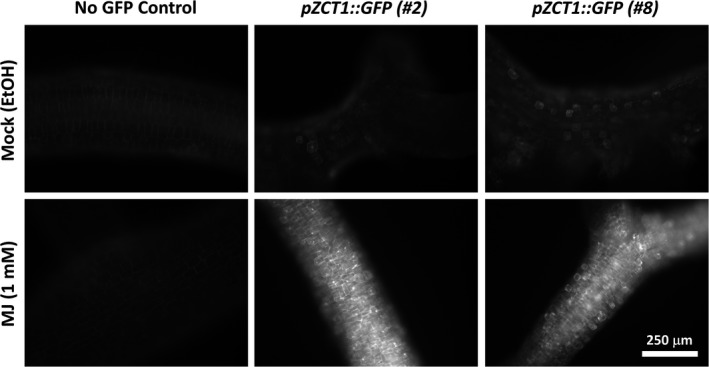
Hairy roots expressing GFP under the control of the ZCT1 promoter show weak basal expression without hormone treatment and increased fluorescence throughout the whole root after treatment with 1 mM MJ. Images show GFP fluorescence of hairy root lines. Hairy roots were treated with ethanol (Mock) or with MJ (1 mM). GFP fluorescence was detected 3 days after the treatment using fluorescence microscopy. The No‐GFP control line lacks a GFP gene and shows weak autofluorescence. pZCT1::GFP (#2 and #8) are two independent hairy root lines with GFP driven by the ZCT1 promoter
Root pieces were treated as described in Materials and Methods. After treatment for 24 hr, GFP expression was quantified using a plate reader. 1‐NAA and MJ induced GFP expression by approximately twofold to sixfold in a dose‐responsive manner, whereas all other hormone treatments (ABA, GA3, and SA) resulted in no significant GFP fluorescence change (Figure 3).
Figure 3.
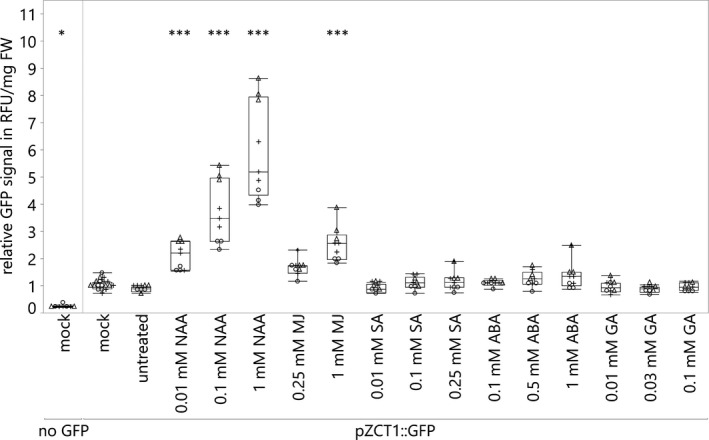
Hairy roots expressing GFP under control of the ZCT1 promoter respond to the auxin 1‐NAA and MJ. Hairy roots expressing GFP under the control of the ZCT1 promoter (pZCT1::GFP) or without GFP transgene (no GFP) were treated with varying concentrations of hormones for 24 hr. The mock treatment contains 0.1% ethanol. No ethanol and no hormones were added to the untreated samples. Each experiment consisted of at least three biological replicates per hormone dosage with a total of three independent assays performed (represented by +, ○, and Δ symbols). For each independent assay, the GFP signal (RFU/mg FW) of each sample is normalized to the average of the pZCT1::GFP mock control. Data were analyzed using a one‐way ANOVA, and significant differences, compared to the pZCT1::GFP mock control, were determined using the Dunnett's method. p‐values < .05 are indicated with one star (*), and p‐values < .001 are indicated with three stars (***). NAA, 1‐naphthalene acetic acid; GA, gibberellin (here GA3); RFU, relative fluorescence units; FW, fresh weight
To determine whether pZCT1::tGFP‐I hairy roots respond to hormones similarly to endogenous ZCT1, wild‐type (WT) hairy root cultures were treated with MJ, ABA, and GA3 for 7 hr, harvested, and analyzed for ZCT1 transcript levels using qRT‐PCR. We have previously shown that ZCT1 expression is responsive to MJ addition by 7 hr (Goklany et al., 2013; Rizvi et al., 2016). In this experiment, ZCT1 expression showed a dose‐responsive increase with MJ, ABA, and a small decrease with GA3 (Figure S2). This response is similar to the response of the pZCT1::tGFP‐I hairy root line to MJ and GA3. However, increased expression with ABA was not observed in the pZCT1::tGFP‐I hairy root line, suggesting that the ABA‐responsive elements might not be captured in the −914‐bp to +86‐bp fragment for the ZCT1 promoter. Overall, these results suggest that the −914‐bp to +86‐bp fragment of the ZCT1 promoter contains many of the elements that confer responsiveness of the endogenous gene to hormone treatments such as 1‐NAA and MJ.
3.3. The as‐1‐like motif contributes to the strong expression from the ZCT1 promoter
ZCT1 is expressed at high levels in cotyledons compared to ORCA3 or the housekeeping gene, SAND. During qRT‐PCR experiments, the basal level of ZCT1 transcripts in cotyledons is high (~20X of SAND), while the basal level of ORCA3 transcripts is very low (<0.1 of SAND; Figure S3). In order to understand how this high level of expression is conferred, we produced a series of mutant ZCT1 promoter constructs driving a FLUC reporter gene and transiently expressed them in C. roseus seedlings using the EASI method (Mortensen et al., 2019), to identify the important elements in the ZCT1 promoter (Figure 4).
Figure 4.
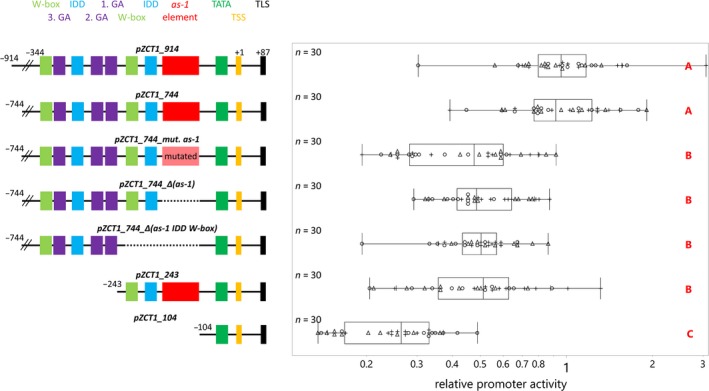
The as‐1‐like element is a major contributor to ZCT1 promoter activity in seedlings. Catharanthus roseus seedlings, at three days after transfer to light, were vacuum‐infiltrated with Agrobacterium tumefaciens (OD600 = 0.2) containing reporter constructs with various ZCT1 promoter deletion‐driven FLUC‐I reporter and the AtuNOS promoter‐driven RLUC‐I normalization reporter. Samples were taken three days postinfection. The relative promoter activity is the ratio of FLUC (firefly luciferase) to RLUC (Renilla luciferase) activity for each sample and then normalized to the ratio of FLUC to RLUC activity of the pZCT1_744 control (set to 1). The experiment was carried out in three independent assays (represented by +, ○, and Δ symbols). Each data point represents the luciferase activity of 2 seedlings. The vertical line of the boxes shows the median, the ends of the boxes show the 1st and 3rd quantile, and whiskers show the lowest and highest data point values within the 1st and 3rd quartile. Detailed information on the promoter sequence and identified cis regulative elements can be found in Figure S1. The data were log‐normal‐transformed to obtain normal distributed data. Data were analyzed using a one‐way ANOVA, and significant differences between groups were determined using the Tukey–Kramer method on log‐normal‐transformed data and were indicated by different letters
Using PlantCARE and PlantPAN 3.0, a high density of motifs was identified within the first 400 bp of the ZCT1 promoter (−400 to 0 bp upstream of the TSS). In particular, we identified a region (−350 to −180 bp upstream of TSS) containing a cluster of GA‐responsive elements making up a GA response complex (GARC; TATC‐box, pyrimidine box, GARE) bound by W‐boxes. The GARC bound by W‐boxes is a regulatory unit associated with the antagonistic regulation of GA and ABA in the amylase promoter in rice (Xie et al., 2006). Even though ZCT1 promoter‐driven GFP expression was not increased with GA3 in transgenic hairy roots, we hypothesized that the GARC bound by W‐boxes might be differently regulated in seedlings where ZCT1 was highly expressed (Figure S3). Also, the structure is likely too complex to occur purely by coincidence. Therefore, we chose to further test this cluster in promoter deletion experiments in transiently transformed seedlings. As‐1‐like elements are associated with jasmonate (JA) and auxin responsiveness and confer high activity of the promoter such as found in the constitutive cauliflower mosaic virus 35S promoter (Bouchez et al., 1989; Liu & Lam, 1994). The as‐1‐like element was chosen for transient promoter deletion experiments, as the pZCT1::GFP expression in transgenic hairy roots was induced by JA and auxin.
The two longest ZCT1 promoter fragments (−914 bp to +86 bp, pZCT1_914, or −744 bp to +86 bp, pZCT1_744) provide similar levels of expression (Figure 4). The pZCT1_914 construct contains several additional stress response elements (STRE), but these do not significantly increase the promoter activity compared to the pZCT1_744 construct under the tested conditions.
To test the importance of the as‐1‐like element, a 58 bp nucleotide deletion (pZCT1_744_ Δ(as‐1)) of the as‐1‐like element and a mutant construct (pZCT1_744_mut. as‐1) were expressed using the EASI method. If the as‐1‐like element is mutated or deleted (pZCT1_744_mut. as‐1 or pZCT1_744_ Δ(as‐1)), promoter activity is reduced by approximately 50%. If one IDD binding site as well as one W‐box is removed in addition to the as‐1‐like element (pZCT1_744_ Δ(as‐1 IDD W‐box)), activity is not reduced compared to the deletion of just the as‐1‐like element (pZCT1_744_ Δ(as‐1)), suggesting that this W‐box is not contributing significantly to the as‐1‐like‐driven promoter activity under the given conditions. A 501 bp 5′ deletion (pZCT1_243) also results in a reduction of promoter activity by approximately 50%. This section contains multiple elements, including the GARC. The pZCT1_104 fragment, which contains the TATA‐box and transcription start site alone, confers ~26% of the promoter activity, suggesting that there are additional regulatory elements within this 104‐bp fragment.
Next, we investigated the GARC without the overshadowing effect of the as‐1‐like element (Figure 5). Fusion of the GARC (including the two IDD binding sites and the W‐boxes) to the 35S minimal promoter (pMinimal35S:GARC + IDD) significantly increases the promoter activity of the 35S minimal promoter. Deletion of one IDD binding site from this fragment (pMinimal35S:GARC) does not significantly affect the activity of the pMinimal35S:GARC + IDD promoter fragment. These results show that the GARC confers promoter activity under the tested conditions, and that the second IDD binding site does not synergistically enhance promoter activity. However, overall, the promoter activity of the 35S minimal promoter fusions is quite low compared to the pZCT1_744 activity.
Figure 5.
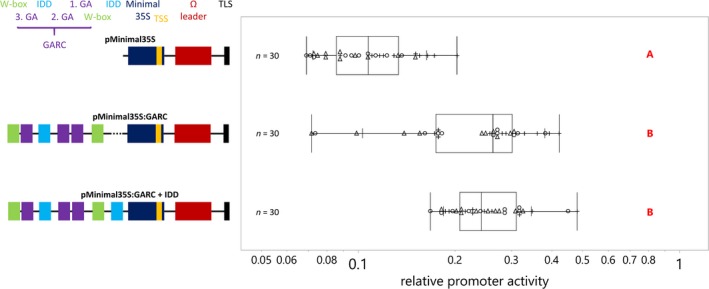
The GA‐responsive complex (GARC) is a contributor to ZCT1 promoter activity under the EASI conditions. Catharanthus roseus seedlings, at three days after transfer to light, were vacuum‐infiltrated with Agrobacterium tumefaciens (OD600 = 0.2) containing reporter constructs with various ZCT1 promoter deletion‐driven FLUC‐I reporter and the AtuNOS promoter‐driven RLUC‐I normalization reporter. Samples were taken three days postinfection. The relative promoter activity is the ratio of FLUC (firefly luciferase) to RLUC (Renilla luciferase) activity for each sample normalized to the ratio of FLUC to RLUC activity of the pZCT1_744 control (set to 1). The experiment was carried out in three independent assays (represented by +, ○, and Δ symbols). Each data point represents the luciferase activity of 2 seedlings. The vertical line of the boxes shows the median, the ends of the boxes show the 1st and 3rd quantile, and whiskers show the lowest and highest data point values within the 1st and 3rd quartile. Detailed information on the promoter sequence and identified cis regulative elements can be found in Figure S1. The data were log‐normal‐transformed to obtain normal distributed data. Data were analyzed using a one‐way ANOVA, and significant differences between groups were determined using the Tukey–Kramer method on log‐normal‐transformed data
Taken together, these results suggest the as‐1‐like element contributes significantly to the expression of ZCT1 in seedlings under the EASI transformation conditions. Other elements in the promoter, including the GARC, also contribute to ZCT1 expression.
3.4. ORCA3 does not transactivate the ZCT1 promoter, while ZCT1 represses its own promoter
Previously, elevated ORCA3 expression levels were correlated with increased expression of ZCT1, ZCT2, and ZCT3 in a stable hairy root line, suggesting the regulation of ZCTs by ORCA3 (Peebles et al., 2009). Using the EASI method, we investigated the transactivation of the ZCT1 promoter (pSB135, pZCT1_914::FLUC‐I) by either one of the transcription factors (ORCA3, ZCT1, ZCT2, or ZCT3) or GUS (control) in C. roseus seedlings. The co‐expression of the ORCA3 and the promoter–reporter construct did not result in the activation of the ZCT1 promoter (Figure 6). Overexpression of ORCA3 also did not induce ZCT1 mRNA levels (Figure 7). In comparable experiments, ORCA3 was able to transactivate the STR1 promoter by approximately 10‐fold (Mortensen et al., 2019). These data suggest that the previously observed regulation of ZCT1 by ORCA3 in hairy roots (Peebles et al., 2009) may be indirect or root‐specific. In addition, ZCT1 significantly represses its own promoter (~ 40%), while no significant effect is observed with ZCT2 or ZCT3 (Figure 6). This result suggests a possible mechanism for autoregulation of ZCT1 expression by ZCT1.
Figure 6.
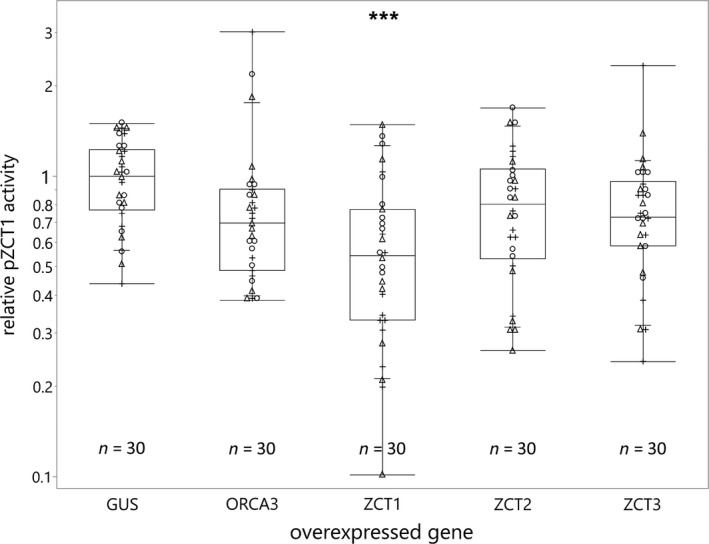
ORCA3 does not activate the ZCT1 promoter, but ZCT1 represses its own promoter. Catharanthus roseus seedlings, at three days after transfer to light, were vacuum‐infiltrated with a combination of two strains of Agrobacterium tumefaciens (total OD600 = 0.4): (I) strain containing the ZCT1 promoter‐driven FLUC‐I reporter and the AtuNOS promoter‐driven RLUC‐I normalization reporter (plasmid pSB135; OD600 = 0.06), and II) strain containing a CaMV2x35s‐driven effector for transactivation (GUS as control—pSB161 (Addgene ID #123197), ORCA3—pSB160 (Addgene ID #123196), or ZCTs—pSB153‐155; OD600 = 0.34). Samples were taken three days postinfection. The ZCT1 promoter activity is the ratio of FLUC (firefly luciferase) to RLUC (Renilla luciferase) activity for each sample normalized to the ratio of FLUC to RLUC activity of the GUS control (set to 1). The experiment was carried out in three independent assays (represented by +, ○, and Δ symbols). Each data point represents luciferase activity of 2 seedlings. The horizontal line of the boxes shows the median, the ends of the boxes show the 1st and 3rd quantile, and whiskers show the lowest and highest data point values within the 1st and 3rd quartile. The data were log‐normal‐transformed to obtain normally distributed data. Data were analyzed using a one‐way ANOVA, and significant differences, compared to the GUS control, were determined using the Dunnett's method on log‐normal‐transformed data. p‐values < .001 are indicated with three stars (***)
Figure 7.
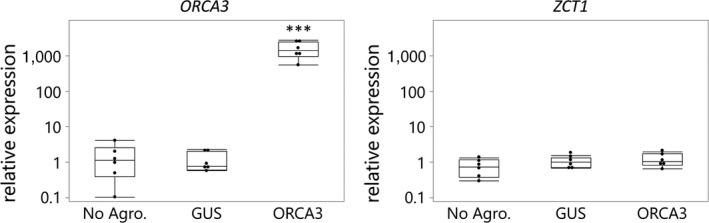
ORCA3 overexpression does not increase ZCT1 expression, and ZCT1 is not significantly induced under the EASI conditions using Agrobacterium tumefaciens strain GV3101. Catharanthus roseus seedlings were transiently transformed, as described in Mortensen et al., (2019), with Agrobacterium tumefaciens containing either a GUS (Addgene ID #123197) or ORCA3 (Addgene ID #123196) overexpression construct (Figure S4). The “No Agro.” condition was treated identical to the other infiltrations but with no Agrobacteria present. Transcript levels were normalized to the housekeeping gene, SAND (Pollier et al., 2014), and fold changes were calculated according to the 2−∆∆Ct method relative to the GUS control (Livak & Schmittgen, 2001). Data were analyzed using a one‐way ANOVA, and significant differences, compared to the GUS control, were determined using the Dunnett's method. p‐values < .001 are indicated with three stars (***)
4. DISCUSSION
In plants, Cys2/His2‐type (C2H2) zinc finger proteins such as ZCT1 are an important class of transcription factors involved in controlling growth, development, and stress‐responsive genes (cold, drought, salt, and oxidative stress; Ciftci‐Yilmaz & Mittler, 2008; Kiełbowicz‐Matuk, 2012). They contain an EAR‐motif involved in active repression (Ciftci‐Yilmaz et al., 2007; Hiratsu, Matsui, Koyama, & Ohme‐Takagi, 2003). Despite their importance, the knowledge is limited on the regulatory networks that control the expression of these factors in response to stresses.
Here, we showed that the promoter of ZCT1 contains several hormone‐responsive elements. Some of these elements are shared in the promoters of other ZCT genes in C. roseus, ZCT2 and ZCT3. However, the C. roseus ZCT1 promoter is particularly interesting due to the high number and arrangement of elements (Figure S1). These in silico observations suggest that ZCT1 may be involved in the crosstalk between phytohormone and defense signaling pathways.
However, little has been shown regarding the hormonal regulation of ZCT1 expression. We previously showed that ZCT1 expression was strongly induced with high dosages of MJ (1 mM) in hairy root cultures (Goklany et al., 2013). Here, we showed that the −914‐bp to + 86‐bp region upstream of ZCT1 also leads to strong induction with auxin (1‐NAA) (Figure 3), as well as with MJ in transgenic hairy roots. ABA also induced ZCT1 expression, but potentially through elements outside of the −914‐bp to + 86‐bp upstream region tested in the transgenic hairy roots (Figure S2).
The responsiveness of ZCT1 expression with high auxin levels suggests their role in decreasing MIA production under growth‐promoting conditions. The synthetic auxin, 2,4‐dichlorophenoxyacetic acid (2,4‐D), repressed the hydroxymethylbutenyl 4‐diphosphate synthase (HDS) gene from the methyl erythritol phosphate (MEP) pathway (Chebbi et al., 2014). The MEP pathway provides isopentenyl diphosphate (IPP) for MIA biosynthesis. ZCT2 was pulled down in a yeast one‐hybrid assay with a region from the HDS promoter and both ZCT1 and ZCT2 repressed the HDS promoter (Chebbi et al., 2014). These results by Chebbi et al. (2014) support our hypothesis that the induction of ZCT1 promoter activity by auxin is leading to a repression of MIA synthesis. Additionally, the responsiveness of the ZCT1 promoter to auxins explains why ZCT1 levels were increased during transient transformation of C. roseus seedlings with A. rhizogenes strain R1000 (Weaver, Goklany, Rizvi, Cram, & Lee‐Parsons, 2014), which transfers genes for auxin biosynthesis into plants (Inzé et al., 1984). ZCT1 levels were not increased during transient transformation of C. roseus seedlings with the A. tumefaciens strain GV3101, which has been disarmed of its endogenous plant hormone biosynthetic genes (Figure 7).
The strong induction of ZCT1 promoter activity with auxin and MJ may be due to the as‐1‐like element (Figure 1); as‐1‐like elements are observed to be responsive to auxins, MJ, SA, and further stressors (Ulmasov et al., 1994; Xiang et al., 1996). Initially, it was surprising to us that the pZCT1::GFP line did not respond to treatment with SA (Figure 3), but Van der Does et al. (2013) showed that W‐boxes (which are present in the ZCT1 promoter (Figure 4 and Figure S1)) are enriched in promoter regions of MJ‐inducible genes that are antagonized by SA. This suggests a possible mechanism for repression of MJ‐induced promoter activity by SA‐induced WRKY expression and binding to W‐boxes.
The importance of the as‐1‐like element for ZCT1 promoter activity was shown by the promoter deletions studies in seedlings (Figure 4), as ~50% of the promoter activity was lost if the as‐1‐like element was deleted or mutated. To demonstrate its activity in the absence of the as‐1‐like element, the GARC was fused to the 35S minimal promoter and conferred increased promoter activity in seedlings (Figure 5). However, GA3 did not induce GFP expression in hairy roots (Figure 3); these potential discrepancies could be attributed to a condition‐specific role of the GARC. For instance, in rice aleurone cells, the expression of the amylase gene is regulated by the GARC cluster bounded by W‐boxes, promoting the breakdown of starch in germinating seedlings in the presence of GA3 (Xie et al., 2006).
Peebles et al. (2009) observed increased ZCT1 expression when ORCA3 was overexpressed in hairy roots, suggesting a possible regulation of ZCT1 through ORCA3. We did not observe the induced expression of ZCT1 when ORCA3 was overexpressed in seedlings (Figure 7) nor the transactivation of ZCT1 promoter activity by ORCA3 (Figure 6). This suggests that the overexpression of ORCA3 alone is not sufficient to induce ZCT1. Interestingly, we observed a repression of ZCT1 on its own promoter (Figure 6), suggesting a possible feedback regulation for limiting ZCT1 expression. Key MIA pathway activators, like BIS and ORCA, have been shown to have a positive feedback regulation (Schweizer et al., 2018). The as‐1‐like element had the strongest effect on ZCT1 promoter activity in seedlings (Figure 4). As‐1‐like elements are known to be regulated through the TGA family of basic‐leucine‐zipper (bZIP) transcription factors (Lam & Lam, 1995), providing a target group for further investigation of regulators of the ZCT1 promoter.
In summary, we have identified ZCT1 promoter activity as responsive to auxin (1‐NAA) and MJ. An as‐1‐like element is particularly important for promoter activity and suggests a positive regulation of ZCT1 through TGA transcription factors. In contrast to initial expectations, ORCA3 did not transactivate the ZCT1 promoter, while ZCT1 represses its own promoter. Future investigation into ZCT1 regulation will provide insights into the important biological problem of how phytohormone crosstalk and/or feedback mechanisms are integrated at the level of transcription of regulatory proteins.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
AUTHOR CONTRIBUTIONS
SM, JW, EJC, and CL‐P conceived and designed the research. SM, SS, JW, LC, and NR performed the experiments. SM, SS, JW, LC, NR, EJC, and CL‐P analyzed the data. SM, JW, EJC, and CL‐P wrote the manuscript.
Supporting information
Mortensen S, Weaver JD, Sathitloetsakun S, et al. The regulation of ZCT1, a transcriptional repressor of monoterpenoid indole alkaloid biosynthetic genes in Catharanthus roseus . Plant Direct. 2019;3:1–13. 10.1002/pld3.193
Funding
This work was supported by the National Science Foundation (NSF) MCB Award #1516371 and BBBE Award #1033889 to CL‐P and EJC.
REFERENCES
- Bouchez, D. , Tokuhisa, J. G. , Llewellyn, D. J. , Dennis, E. S. , & Ellis, J. G. (1989). The ocs‐element is a component of the promoters of several T‐DNA and plant viral genes. EMBO Journal, 8(13), 4197–4204. 10.1002/j.1460-2075.1989.tb08605.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel, G. (2003). CrMYC1, a Catharanthus roseus elicitor‐ and jasmonate‐responsive bHLH transcription factor that binds the G‐box element of the strictosidine synthase gene promoter. Journal of Experimental Botany, 54(392), 2587–2588. 10.1093/jxb/erg275 [DOI] [PubMed] [Google Scholar]
- Chebbi, M. , Ginis, O. , Courdavault, V. , Glévarec, G. , Lanoue, A. , Clastre, M. , … Oudin, A. (2014). ZCT1 and ZCT2 transcription factors repress the activity of a gene promoter from the methyl erythritol phosphate pathway in Madagascar periwinkle cells. Journal of Plant Physiology, 171(16), 1510–1513. 10.1016/j.jplph.2014.07.004 [DOI] [PubMed] [Google Scholar]
- Chen, W. , Chao, G. , & Singh, K. B. (1996). The promoter of a H2O2‐inducible, Arabidopsis glutathione S‐transferase gene contains closely linked OBF‐and OBP1‐binding sites. The Plant Journal, 10(6), 955–966. 10.1046/j.1365-313X.1996.10060955.x [DOI] [PubMed] [Google Scholar]
- Chow, C.‐N. , Lee, T.‐Y. , Hung, Y.‐C. , Li, G.‐Z. , Tseng, K.‐C. , Liu, Y.‐H. , … Chang, W.‐C. (2019). PlantPAN3.0: A new and updated resource for reconstructing transcriptional regulatory networks from chip‐seq experiments in plants. Nucleic Acids Research, 47(D1), D1155–D1163. 10.1093/nar/gky1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciftci‐Yilmaz, S. , & Mittler, R. (2008). The zinc finger network of plants. Cellular and Molecular Life Sciences, 65(7–8), 1150–1160. 10.1007/s00018-007-7473-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciftci‐Yilmaz, S. , Morsy, M. R. , Song, L. , Coutu, A. , Krizek, B. A. , Lewis, M. W. , … Mittler, R. (2007). The EAR‐motif of the Cys2/His2‐type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. Journal of Biological Chemistry, 282(12), 9260–9268. 10.1074/jbc.M611093200 [DOI] [PubMed] [Google Scholar]
- Crooks, G. (2004). WebLogo: A sequence logo generator. Genome Research, 14, 1188–1190. 10.1101/gr.849004.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droog, F. N. J. , Hooykaas, P. J. J. , Libbenga, K. R. , & van der Zaal, E. J. (1993). Proteins encoded by an auxin‐regulated gene family of tobacco share limited but significant homology with glutathione S‐transferases and one member indeed shows in vitro GST activity. Plant Molecular Biology, 21(6), 965–972. 10.1007/BF00023595 [DOI] [PubMed] [Google Scholar]
- Dudler, R. (1991). A pathogen‐induced wheat gene encodes a protein homologous to glutathione‐S‐transferases. Molecular Plant‐Microbe Interactions, 4(1), 14–18. 10.1094/MPMI-4-014 [DOI] [PubMed] [Google Scholar]
- Dugé de Bernonville, T. , Carqueijeiro, I. , Lanoue, A. , Lafontaine, F. , Sánchez Bel, P. , Liesecke, F. , … Courdavault, V. (2017). Folivory elicits a strong defense reaction in Catharanthus roseus: Metabolomic and transcriptomic analyses reveal distinct local and systemic responses. Scientific Reports, 7(1), 1–14. 10.1038/srep40453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englbrecht, C. C. , Schoof, H. , & Böhm, S. (2004). Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics, 5, 1–17. 10.1186/1471-2164-5-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler, C. , Youles, M. , Gruetzner, R. , Ehnert, T.‐M. , Werner, S. , Jones, J. D. G. , … Marillonnet, S. (2014). A Golden Gate modular cloning toolbox for plants. ACS Synthetic Biology, 3(11), 839–843. 10.1021/sb4001504 [DOI] [PubMed] [Google Scholar]
- Feltkamp, D. , Masterson, R. , Starke, J. , & Rosahl, S. (1994). Analysis of the involvement of ocs‐like bZip–binding elements in the differential strength of the bidirectional mas1'2′ promoter. Plant Physiology, 105(1), 259–268. 10.1104/pp.105.1.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke, J. , Kim, J. , Hamilton, J. P. , Zhao, D. , Pham, G. M. , Wiegert‐Rininger, K. , … O'Connor, S. E. (2019). Gene discovery in Gelsemium highlights conserved gene clusters in monoterpene indole alkaloid biosynthesis. ChemBioChem, 20(1), 83–87. 10.1002/cbic.201800592 [DOI] [PubMed] [Google Scholar]
- Goddijn, O. J. M. , de Kam, R. J. , Zanetti, A. , Schilperoort, R. A. , & Hoge, J. H. C. (1992). Auxin rapidly down‐regulates transcription of the tryptophan decarboxylase gene from Catharanthus roseus . Plant Molecular Biology, 18(6), 1113–1120. 10.1007/BF00047714 [DOI] [PubMed] [Google Scholar]
- Goklany, S. , Rizvi, N. F. , Loring, R. H. , Cram, E. J. , & Lee‐Parsons, C. W. T. (2013). Jasmonate‐dependent alkaloid biosynthesis in Catharanthus roseus hairy root cultures is correlated with the relative expression of Orca and Zct transcription factors. Biotechnology Progress, 29(6), 1367–1376. 10.1002/btpr.1801 [DOI] [PubMed] [Google Scholar]
- Gubler, F. , & Jacobsen, J. V. (1992). Gibberellin‐responsive elements in the promoter of a barley high‐pl alpha‐amylase gene. The Plant Cell, 4, 1435–1441. 10.2307/3869514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler, F. , Raventos, D. , Keys, M. , Watts, R. , Mundy, J. , & Jacobsen, J. V. (1999). Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant Journal, 17(1), 1–9. 10.1046/j.1365-313X.1999.00346.x [DOI] [PubMed] [Google Scholar]
- Hiratsu, K. , Matsui, K. , Koyama, T. , & Ohme‐Takagi, M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain in Arabidopsis . Plant Journal, 34(5), 733–739. 10.1046/j.1365-313X.2003.01759.x [DOI] [PubMed] [Google Scholar]
- Inzé, D. , Follin, A. , Van Lijsebettens, M. , Simoens, C. , Genetello, C. , Van Montagu, M. , & Schell, J. (1984). Genetic analysis of the individual T‐DNA genes of Agrobacterium tumefaciens; further evidence that two genes are involved in indole‐3‐acetic acid synthesis. MGG Molecular and General Genetics, 194(1–2), 265–274. 10.1007/BF00383526 [DOI] [Google Scholar]
- Kang, S.‐M. , Jung, H.‐Y. , Kang, Y.‐M. , Yun, D.‐J. , Bahk, J.‐D. , Yang, J.‐K. , & Choi, M.‐S. (2004). Effects of methyl jasmonate and salicylic acid on the production of tropane alkaloids and the expression of PMT and H6H in adventitious root cultures of Scopolia parviflora . Plant Science, 166(3), 745–751. 10.1016/j.plantsci.2003.11.022 [DOI] [Google Scholar]
- Kellner, F. , Kim, J. , Clavijo, B. J. , Hamilton, J. P. , Childs, K. L. , Vaillancourt, B. , … O'Connor, S. E. (2015). Genome‐guided investigation of plant natural product biosynthesis. Plant Journal, 82(4), 680–692. 10.1111/tpj.12827 [DOI] [PubMed] [Google Scholar]
- Kiełbowicz‐Matuk, A. (2012). Involvement of plant C2H2‐type zinc finger transcription factors in stress responses. Plant Science, 185–186, 78–85. 10.1016/j.plantsci.2011.11.015 [DOI] [PubMed] [Google Scholar]
- Krawczyk, S. (2002). Analysis of the spacing between the two palindromes of activation sequence‐1 with respect to binding to different TGA factors and transcriptional activation potential. Nucleic Acids Research, 30(3), 775–781. 10.1093/nar/30.3.775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, E. , & Lam, Y. K. (1995). Binding site requirements and differential representation of TGA factors in nuclear ASF‐1 activity. Nucleic Acids Research, 23(18), 3778–3785. 10.1093/nar/23.18.3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanahan, M. B. , Ho, T. H. , Rogers, S. W. , & Rogers, J. C. (1992). A Gibberellin response complex in cereal alpha‐amylase gene promoters. The Plant Cell, 4(2), 203–211. 10.2307/3869573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee‐Parsons, C. W. T. , Ertürk, S. , & Tengtrakool, J. (2004). Enhancement of ajmalicine production in Catharanthus roseus cell cultures with methyl jasmonate is dependent on timing and dosage of elicitation. Biotechnology Letters, 26(20), 1595–1599. 10.1023/B:BILE.0000045825.37395.94 [DOI] [PubMed] [Google Scholar]
- Lescot, M. (2002). PlantCARE, a database of plant cis‐acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research, 30(1), 325–327. 10.1093/nar/30.1.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Gao, F. , Ren, J. , Lu, X. , Ren, G. , & Wang, R. (2017). A Novel AP2/ERF transcription factor CR1 regulates the accumulation of vindoline and serpentine in Catharanthus roseus . Frontiers in Plant Science, 8, 1–11. 10.3389/fpls.2017.02082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , & Lam, E. (1994). Two binding sites for the plant transcription factor ASF‐1 can respond to auxin treatments in transgenic tobacco. Journal of Biological Chemistry, 269(1), 668–675. [PubMed] [Google Scholar]
- Liu, Y. , Patra, B. , Pattanaik, S. , Wang, Y. , & Yuan, L. (2019). GATA and phytochrome interacting factor transcription factors regulate light‐induced vindoline biosynthesis in Catharanthus roseus . Plant Physiology, 180(3), 1336–1350. 10.1104/pp.19.00489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K. J. , & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real‐time quantitative PCR and the 2‐ΔΔC T method. Methods, 25(4), 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Luijendijk, T. J. C. , van der Meijden, E. , & Verpoorte, R. (1996). Involvement of strictosidine as a defensive chemical in Catharanthus roseus . Journal of Chemical Ecology, 22(8), 1355 10.1007/BF02027718 [DOI] [PubMed] [Google Scholar]
- Marillonnet, S. , Giritch, A. , Gils, M. , Kandzia, R. , Klimyuk, V. , & Gleba, Y. (2004). In planta engineering of viral RNA replicons: Efficient assembly by recombination of DNA modules delivered by Agrobacterium . PNAS, 101(18), 6852–6857. 10.1073/pnas.0400149101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke, F. L. H. (1999). A novel jasmonate‐ and elicitor‐ responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate‐ and elicitor‐ inducible AP2‐ domain transcription factor, ORCA2. The EMBO Journal, 18(16), 4455–4463. 10.1093/emboj/18.16.4455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler, R. , Kim, Y. , Song, L. , Coutu, J. , Coutu, A. , Ciftci‐Yilmaz, S. , … Zhu, J.‐K. (2006). Gain‐ and loss‐of‐function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Letters, 580(28–29), 6537–6542. 10.1016/j.febslet.2006.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen, S. , Bernal‐Franco, D. , Cole, L. F. , Sathitloetsakun, S. , Cram, E. J. , & Lee‐Parsons, C. W. T. (2019). EASI transformation: An efficient transient expression method for analyzing gene function in Catharanthus roseus seedlings. Frontiers in Plant Science, 10(June), 1–17. 10.3389/fpls.2019.00755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble, R. L. (1990). The discovery of the vinca alkaloids—chemotherapeutic agents against cancer. Biochemistry and Cell Biology, 68(12), 1344–1351. 10.1139/o90-197 [DOI] [PubMed] [Google Scholar]
- Ohta, M. , Matsui, K. , Hiratsu, K. , Shinshi, H. , & Ohme‐Takagi, M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell Online, 13(8), 1959–1968. 10.1105/tpc.13.8.1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Q. , Wang, C. , Xiong, Z. , Wang, H. , Fu, X. , Shen, Q. , … Tang, K. (2019). CrERF5, an AP2/ERF transcription factor, positively regulates the biosynthesis of bisindole alkaloids and their precursors in Catharanthus roseus . Frontiers in Plant Science, 10, 10.3389/fpls.2019.00931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra, B. , Pattanaik, S. , Schluttenhofer, C. , & Yuan, L. (2017). A network of jasmonate‐responsive bHLH factors modulate monoterpenoid indole alkaloid biosynthesis in Catharanthus roseus . New Phytologist, 217(4), 1566–1581. 10.1111/nph.14910 [DOI] [PubMed] [Google Scholar]
- Paul, P. , Singh, S. K. , Patra, B. , Sui, X. , Pattanaik, S. , & Yuan, L. (2016). A differentially regulated AP2/ERF transcription factor gene cluster acts downstream of a MAP kinase cascade to modulate terpenoid indole alkaloid biosynthesis in Catharanthus roseus . New Phytologist, 213(3), 1107–1123. 10.1111/nph.14252 [DOI] [PubMed] [Google Scholar]
- Pauw, B. , Hilliou, F. A. O. , Martin, V. S. , Chatel, G. , de Wolf, C. J. F. , Champion, A. , … Memelink, J. (2004). Zinc finger proteins act as transcriptional repressors of alkaloid biosynthesis genes in Catharanthus roseus . Journal of Biological Chemistry, 279(51), 52940–52948. 10.1074/jbc.M404391200 [DOI] [PubMed] [Google Scholar]
- Peebles, C. A. M. , Hughes, E. H. , Shanks, J. V. , & San, K.‐Y. (2009). Transcriptional response of the terpenoid indole alkaloid pathway to the overexpression of ORCA3 along with jasmonic acid elicitation of Catharanthus roseus hairy roots over time. Metabolic Engineering, 11(2), 76–86. 10.1016/j.ymben.2008.09.002 [DOI] [PubMed] [Google Scholar]
- Pollier, J. , Vanden Bossche, R. , Rischer, H. , & Goossens, A. (2014). Selection and validation of reference genes for transcript normalization in gene expression studies in Catharanthus roseus . Plant Physiology and Biochemistry, 83, 20–25. 10.1016/j.plaphy.2014.07.004 [DOI] [PubMed] [Google Scholar]
- Prändl, R. , & Kutchan, T. M. (1992). Nucleotide sequence of the gene for a glutathione S‐transferase from cell suspension cultures of Silene cucubalus . Plant Physiology, 99(4), 1729–1731. 10.1104/pp.99.4.1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi, N. F. , Cornejo, M. , Stein, K. , Weaver, J. , Cram, E. J. , & Lee‐Parsons, C. W. T. (2015). An efficient transformation method for estrogen‐inducible transgene expression in Catharanthus roseus hairy roots. Plant Cell, Tissue and Organ Culture (PCTOC), 120(2), 475–487. 10.1007/s11240-014-0614-1 [DOI] [Google Scholar]
- Rizvi, N. F. , Weaver, J. D. , Cram, E. J. , & Lee‐Parsons, C. W. T. (2016). Silencing the transcriptional repressor, ZCT1, illustrates the tight regulation of terpenoid indole alkaloid biosynthesis in Catharanthus roseus hairy roots. PLoS ONE, 11(7), 1–17. 10.1371/journal.pone.0159712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, J. C. , Lanahan, M. B. , & Rogers, S. W. (1994). The cis‐acting gibberellin response complex in high‐pl alpha‐amylase gene promoters. Requirement of a coupling element for high‐level transcription. Plant Physiology, 105(1), 151–158. 10.1104/pp.105.1.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, J. C. , & Rogers, S. W. (1992). Definition and functional implications of gibberellin and abscisic acid cis‐acting hormone response complexes. The Plant Cell, 4(11), 1443–1451. 10.2307/3869515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, H. , Araki, T. , Meshi, T. , & Iwabuchi, M. (2000). Expression of a subset of the Arabidopsis Cys2His2‐type zinc‐finger protein gene family under water stress. Gene, 248, 23–32. 10.1016/S0378-1119(00)00133-5 [DOI] [PubMed] [Google Scholar]
- Sakamoto, H. , Maruyama, K. , Sakuma, Y. , Meshi, T. , Iwabuchi, M. , Shinozaki, K. , & Yamaguchi‐Shinozaki, K. (2004). Arabidopsis Cys2/His2‐type zinc‐finger proteins function as transcription repressors under drought. Society, 136, 2734–2746. 10.1104/pp.104.046599.2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluttenhofer, C. , Pattanaik, S. , Patra, B. , & Yuan, L. (2014). Analyses of Catharanthus roseus and Arabidopsis thaliana WRKY transcription factors reveal involvement in jasmonate signaling. BMC Genomics, 15(1), 1–20. 10.1186/1471-2164-15-502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer, F. , Colinas, M. , Pollier, J. , Van Moerkercke, A. , Vanden Bossche, R. , de Clercq, R. , & Goossens, A. (2018). An engineered combinatorial module of transcription factors boosts production of monoterpenoid indole alkaloids in Catharanthus roseus . Metabolic Engineering, 48, 150–162. 10.1016/j.ymben.2018.05.016 [DOI] [PubMed] [Google Scholar]
- Sibéril, Y. , Benhamron, S. , Memelink, J. , Giglioli‐Guivarc'h, N. , Thiersault, M. , Boisson, B. , … Gantet, P. (2001). Catharanthus roseus G‐box binding factors 1 and 2 act as repressors of strictosidine synthase gene expression in cell cultures. Plant Molecular Biology, 45(4), 477–488. 10.1023/a:1010650906695 [DOI] [PubMed] [Google Scholar]
- Skriver, K. , Olsen, F. L. , Rogers, J. C. , & Mundy, J. (1991). cis‐acting DNA elements responsive to gibberellin and its antagonist abscisic acid. Proceedings of the National Academy of Sciences, 88(16), 7266–7270. 10.1073/pnas.88.16.7266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strompen, G. , Grüner, R. , & Pfitzner, U. M. (1998). An as‐1‐like motif controls the level of expression of the gene for the pathogenesis‐related protein 1a from tobacco. Plant Molecular Biology, 37(5), 871–883. 10.1023/A:1006003916284 [DOI] [PubMed] [Google Scholar]
- Suttipanta, N. , Pattanaik, S. , Kulshrestha, M. , Patra, B. , Singh, S. K. , & Yuan, L. (2011). The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus . Plant Physiology, 157(4), 2081–2093. 10.1104/pp.111.181834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, Y. , Kusaba, M. , Hiraoka, Y. , & Nagata, T. (1991). Characterization of the auxin‐regulated par gene from tobacco mesophyll protoplasts. The Plant Journal, 1(3), 327–332. 10.1046/j.1365-313X.1991.t01-2-00999.x [DOI] [PubMed] [Google Scholar]
- Ulmasov, T. , Hagen, G. , & Guilfoyle, T. (1994). The ocs element in the soybean GH2/4 promoter is activated by both active and inactive auxin and salicylic acid analogues. Plant Molecular Biology, 26(4), 1055–1064. 10.1007/BF00040688 [DOI] [PubMed] [Google Scholar]
- Van der Does, D. , Leon‐Reyes, A. , Koornneef, A. , Van Verk, M. C. , Rodenburg, N. , Pauwels, L. , … Pieterse, C. M. J. (2013). Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1‐JAZ by targeting GCC promoter motifs via transcription factor ORA59. The Plant Cell, 25(2), 744–761. 10.1105/tpc.112.108548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Fits, L. , & Memelink, J. (2000). ORCA3, a jasmonate‐responsive transcriptional regulator of plant primary and secondary metabolism. Science, 295, 295–297. 10.1126/science.289.5477.295 [DOI] [PubMed] [Google Scholar]
- Van der Fits, L. , Zhang, H. , Menke, F. L. , Deneka, M. , & Memelink, J. (2000). A Catharanthus roseus BPF‐1 homologue interacts with an elicitor‐responsive region of the secondary metabolite biosynthetic gene Str and is induced by elicitor via a JA‐independent signal transduction pathway. Plant Molecular Biology, 44(5), 675–685. 10.1023/A:1026526522555 [DOI] [PubMed] [Google Scholar]
- Van der Heijden, R. , Jacobs, D. I. , Snoeijer, W. , Hallard, D. , & Verpoorte, R. (2004). The Catharanthus alkaloids: Pharamacognosy and biotechnology. Current Medicinal Chemistry, 11, 607–628. [DOI] [PubMed] [Google Scholar]
- Van der Zaal, E. J. , Droog, F. N. J. , Boot, C. J. M. , Hensgens, L. A. M. , Hoge, J. H. C. , Schilperoort, R. A. , & Libbenga, K. R. (1991). Promoters of auxin‐induced genes from tobacco can lead to auxin‐inducible and root tip‐specific expression. Plant Molecular Biology, 16(6), 983–998. 10.1007/BF00016071 [DOI] [PubMed] [Google Scholar]
- Van Moerkercke, A. , Steensma, P. , Gariboldi, I. , Espoz, J. , Purnama, P. C. , Schweizer, F. , … Goossens, A. (2016). The basic helix‐loop‐helix transcription factor BIS2 is essential for monoterpenoid indole alkaloid production in the medicinal plant Catharanthus roseus . Plant Journal, 88, 3–12. 10.1111/tpj.13230 [DOI] [PubMed] [Google Scholar]
- Van Moerkercke, A. , Steensma, P. , Schweizer, F. , Pollier, J. , Gariboldi, I. , Payne, R. , … Goossens, A. (2015). The bHLH transcription factor BIS1 controls the iridoid branch of the monoterpenoid indole alkaloid pathway in Catharanthus roseus . Proceedings of the National Academy of Sciences of the United States of America, 112(26), 201504951 10.1073/pnas.1504951112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vom Endt, D. , Soares e Silva, M. , Kijne, J. W. , Pasquali, G. , & Memelink, J. (2007). Identification of a bipartite jasmonate‐responsive promoter element in the Catharanthus roseus ORCA3 transcription factor gene that interacts specifically with AT‐Hook DNA‐binding proteins. Plant Physiology, 144(3), 1680–1689. 10.1104/pp.107.096115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C.‐T. , Liu, H. , Gao, X.‐S. , & Zhang, H.‐X. (2010). Overexpression of G10H and ORCA3 in the hairy roots of Catharanthus roseus improves catharanthine production. Plant Cell Reports, 29(8), 887–894. 10.1007/s00299-010-0874-0 [DOI] [PubMed] [Google Scholar]
- Weaver, J. , Goklany, S. , Rizvi, N. , Cram, E. J. , & Lee‐Parsons, C. W. T. (2014). Optimizing the transient fast agro‐mediated seedling transformation (FAST) method in Catharanthus roseus seedlings. Plant Cell Reports, 33(1), 89–97. 10.1007/s00299-013-1514-2 [DOI] [PubMed] [Google Scholar]
- Weber, E. , Engler, C. , Gruetzner, R. , Werner, S. , & Marillonnet, S. (2011). A modular cloning system for standardized assembly of multigene constructs. PLoS ONE, 6(2), 10.1371/journal.pone.0016765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, C. , Miao, Z. , & Lam, E. (1996). Coordinated activation of as‐1‐type elements and a tobacco glutathione S‐transferase gene by auxins, salicylic acid, methyl‐jasmonate and hydrogen peroxide. Plant Molecular Biology, 32, 415–426. [DOI] [PubMed] [Google Scholar]
- Xie, Z. , Zhang, Z.‐L. , Zou, X. , Yang, G. , Komatsu, S. , & Shen, Q. J. (2006). Interactions of two abscisic‐acid induced WRKY genes in repressing gibberellin signaling in aleurone cells. Plant Journal, 46(2), 231–242. 10.1111/j.1365-313X.2006.02694.x [DOI] [PubMed] [Google Scholar]
- Yoshida, H. , Hirano, K. , Sato, T. , Mitsuda, N. , Nomoto, M. , Maeo, K. , … Ueguchi‐Tanaka, M. (2014). DELLA protein functions as a transcriptional activator through the DNA binding of the INDETERMINATE DOMAIN family proteins. Proceedings of the National Academy of Sciences, 111(21), 7861–7866. 10.1073/pnas.1321669111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Hedhili, S. , Montiel, G. , Zhang, Y. , Chatel, G. , Pré, M. , … Memelink, J. (2011). The basic helix‐loop‐helix transcription factor CrMYC2 controls the jasmonate‐responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus . Plant Journal, 67(1), 61–71. 10.1111/j.1365-313X.2011.04575.x [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Fan, W. , Kinkema, M. , Li, X. , & Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR‐1 gene. Proceedings of the National Academy of Sciences of the United States of America, 96(11), 6523–6528. 10.1073/pnas.96.11.6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, M.‐L. , Zhu, X.‐M. , Shao, J.‐R. , Wu, Y.‐M. , & Tang, Y.‐X. (2010). Transcriptional response of the catharanthine biosynthesis pathway to methyl jasmonate/nitric oxide elicitation in Catharanthus roseus hairy root culture. Applied Microbiology and Biotechnology, 88(3), 737–750. 10.1007/s00253-010-2822-x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


