Abstract
Skeletal muscle is a heterogeneous tissue. Individual myofibers that make up muscle tissue exhibit variation in their metabolic and contractile properties. Although biochemical and histological assays are available to study myofiber heterogeneity, efficient methods to analyze the whole transcriptome of individual myofibers are lacking. Here, we report on a single-myofiber RNA-sequencing (smfRNA-Seq) approach to analyze the whole transcriptome of individual myofibers by combining single-fiber isolation with Switching Mechanism at 5′ end of RNA Template (SMART) technology. Using smfRNA-Seq, we first determined the genes that are expressed in the whole muscle, including in nonmyogenic cells. We also analyzed the differences in the transcriptome of myofibers from young and old mice to validate the effectiveness of this new method. Our results suggest that aging leads to significant changes in the expression of metabolic genes, such as Nos1, and structural genes, such as Myl1, in myofibers. We conclude that smfRNA-Seq is a powerful tool to study developmental, disease-related, and age-related changes in the gene expression profile of skeletal muscle.
Keywords: muscle, RNA, gene expression, molecular cell biology, molecular biology, SMART-Seq
Introduction
Skeletal muscle is composed of a variety of different cell types, including endothelial cells, fibro/adipogenic cells (FAPs),3 adipocytes, mesenchymal cells, and fibroblasts, among others (1–4). Further heterogeneity of skeletal muscle is also manifested by the diversity in the composition of myofiber types that constitute muscles (5). Skeletal muscle fiber types are often categorized based on their contractile properties, giving two broad categories: fast-twitch muscles and slow-twitch muscles (5, 6). These fiber types can be further subcategorized based on metabolic properties and myosin heavy chain isoforms (5, 7).
Methods of investigating changes in fiber type in response to different stimuli rely on staining and biochemical analyses of individual fibers, biochemical analyses of the whole muscle, or sequencing of entire muscles (8). Standard bulk RNA-Seq is not suited for the analysis of myofibers at high resolution, because it captures the entirety of the muscle tissue, resulting in the pooling of different fiber types in addition to nonmyogenic cells (8). Consequently, in such studies, the myofiber-specific gene signature cannot be inferred based on RNA-Seq of whole muscle tissue. The emergence of single-cell technology provides ample opportunity for further investigation into the heterogeneity of single muscle fibers at the transcriptome level. Here, we combine single-myofiber isolation of the extensor digitorum longus (EDL) in Mus musculus with Switching Mechanism at 5′ end of RNA Template (SMART) technology to analyze the whole transcriptome of individual myofibers (Fig. 1) (9), in a method called single-myofiber RNA sequencing (smfRNA-Seq).
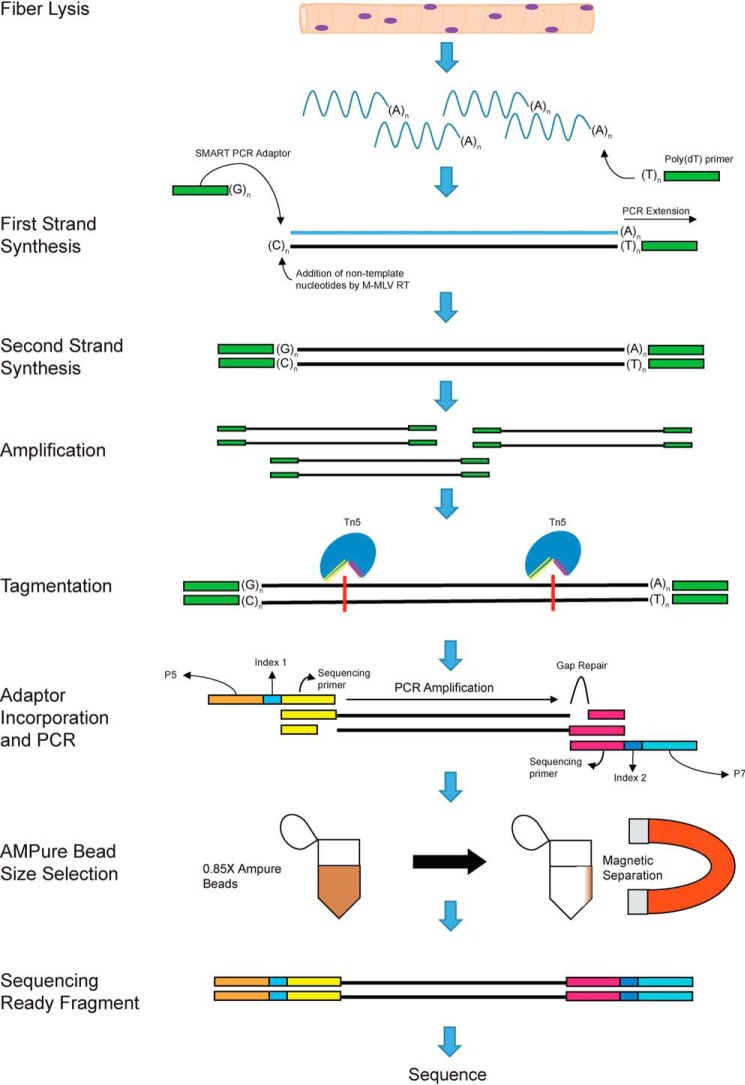
Figure 1.
SMART technology and incorporation of Illumina adaptors to the fiber mRNA. Shown is a schematic displaying the steps and biochemical reactions involved in the generation of sequence ready cDNA fragments. M-MLV, Moloney murine leukemia virus; RT, reverse transcription.
We describe a robust method to extract RNA from a single myofiber followed by generation of sequencing ready libraries and whole transcriptome analysis. Using this technique, we first determined the genes that are found in the whole muscle that are not produced by the myofiber and are instead produced by nonmyogenic cell types. To demonstrate the effectiveness of this technique, we next went on to analyze the differences in the whole transcriptome between myofibers isolated from young and old mice. smfRNA-Seq proved to be a useful tool in determining gene expression changes that occur in myofibers between different conditions.
Results
Isolation of high quality mRNA from single myofibers
One of the intentions of our novel method is to give researchers the ability to sequence RNA from a single myofiber without the confounding presence of other cell types. A potential source of unwanted signal in the sequencing of myofiber RNA is the presence of muscle stem cells, also known as satellite cells, that are physically associated with the fibers. Using our method, we have found that satellite cells can be almost completely removed from the fibers with the addition of trypsin to the collagenase digestion buffer at a final concentration of 0.25%. This process does not damage the fiber itself, with a proper EDL isolation yielding over 200 myofibers per mouse (Fig. S1A). However, this buffer effectively strips the myofibers of their satellite cells as shown by the reduction in the number of PAX7+ cells per fiber (Fig. S1, B–D).
Because muscle fibers are very tough and do not readily break down under normal lysing conditions, extracting the RNA from a single myofiber can prove challenging. With whole muscle, a method to overcome this is by freezing the muscle in liquid nitrogen and grinding it into a powder with mortar and pestle (10). However, this method cannot realistically be done to a single fiber while still collecting all of the RNA. Therefore, we lysed the fiber with lysis buffer in RNase-free water, utilizing osmotic pressure and gentle pipetting to break down the fiber and retrieve the intact RNA. This method proved effective because more than an adequate amount of RNA was recovered, even from a single myofiber, for use with SMART-Seq technology (Table 1).
Table 1.
Total cDNA after SMART cDNA synthesis and amplification and after incorporation of Illumina adaptors and size selection
| Samples | Total cDNA after SMART reaction and amplification | Total cDNA after adding Illumina adapters and size selection | Average fragment size after size selection |
|---|---|---|---|
| ng | ng | bp | |
| Single fiber 1 | 36.38 | 98.25 | 313 |
| Single fiber 2 | 130.56 | 66.9 | 355 |
| Single fiber 3 | 89.76 | 64.5 | 329 |
| Five fibers 1 | 22.78 | 61.2 | 351 |
| Five fibers 2 | 48.62 | 161.85 | 382 |
| Five fibers 3 | 40.97 | 54.15 | 382 |
| Twenty fibers 1 | 24.65 | 46.8 | 486 |
| Twenty fibers 2 | 36.89 | 66.75 | 483 |
| Twenty fibers 3 | 29.07 | 86.55 | 484 |
| Whole muscle 1 | 3.74 | 34.65 | 617 |
| Whole muscle 2 | 4.76 | 63.45 | 528 |
| Whole muscle 3 | 5.1 | 53.25 | 456 |
From the extracted RNA, we successfully generated sequencing-ready cDNA libraries using the DNA SMART-Seq HT kit (Takara Biosciences) in combination with the Nextera XT DNA library preparation kit (Illumina). Final single-myofiber sequencing-ready libraries were of adequate quantity and of ideal size for sequencing, with fragments being of comparable average size as those generated from traditional whole muscle RNA-Seq libraries (Fig. S1, E and F). This implies that our fiber RNA extraction procedure generated high-quality starting material that was compatible with low-input library preparation technologies.
Comparative analysis of whole muscle and single-myofiber RNA sequencing
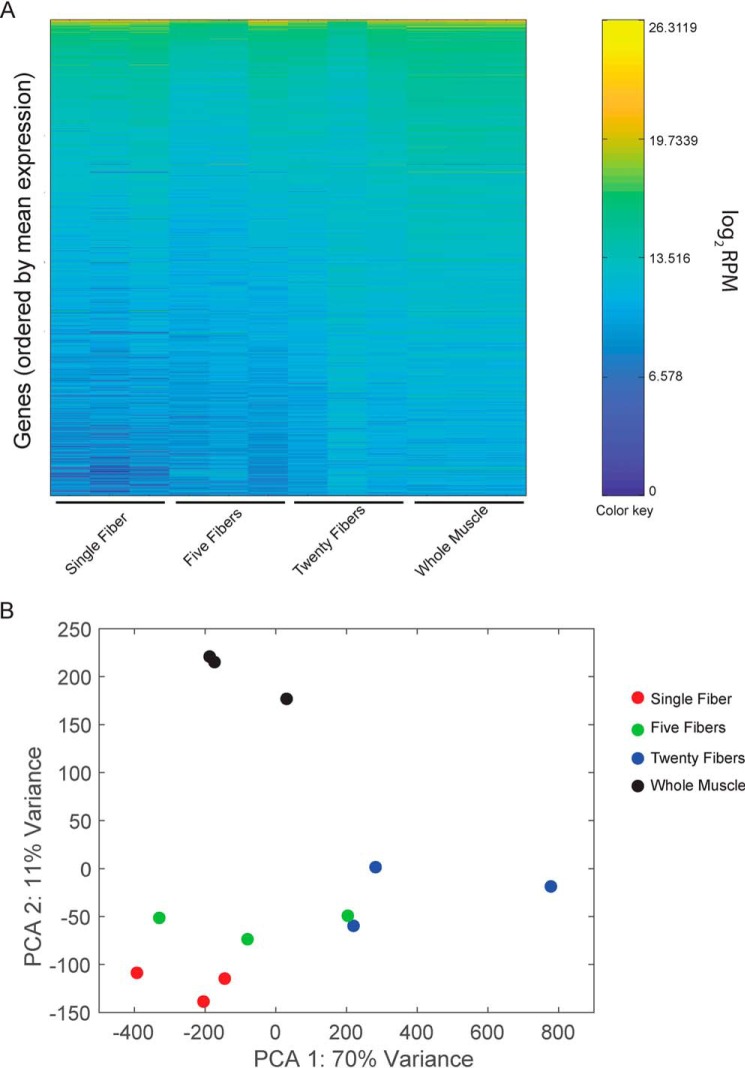
By evaluating the number of unique reads obtained from smfRNA-Seq samples, we found that sequencing depth of single-myofiber libraries was comparable with whole muscle RNA-Seq (Fig. 2A). We obtained an average of 24 million unique reads from single-myofiber samples when multiplexing 12 samples per lane on a NextSeq500 and an average of 35 million unique reads when multiplexing 10 samples per lane. This corresponds to an average overall alignment of 82.42%, with an average unique alignment percentage of 64.47% (Table S3). The overall expression profile of the single myofibers is also very similar to that of the whole muscle, indicating a similar high resolution (Fig. 2A). When performing principal component analysis (PCA), single-fiber samples cluster together on both axes but away from the whole muscle on the PCA2 axis (Fig. 2B). To compare the efficiency of single-cell technologies with increasing input, we also generated RNA-Seq libraries from five and twenty myofibers. When the number of fibers is increased, samples become less similar to both the whole muscle and the single-myofiber samples, with higher variation between technical replicates (Fig. 2, A and B). This is most likely due to the excessive quantity of sample and could be resolved by scaling up the protocol.
Figure 2.
Comparative analysis of whole transcriptome from single myofibers and whole muscle. A, heat map of gene expression in single fibers, groups of five fibers, groups of twenty fibers, and whole muscle, each with three replicates. Colors represent mean gene expression within each sample, from highest expression (yellow) to lowest expression (dark blue). Genes are ordered from top to bottom by their average expression across all samples. B, projection of samples along the first two principal components found by PCA applied to log reads-per-million gene expression.
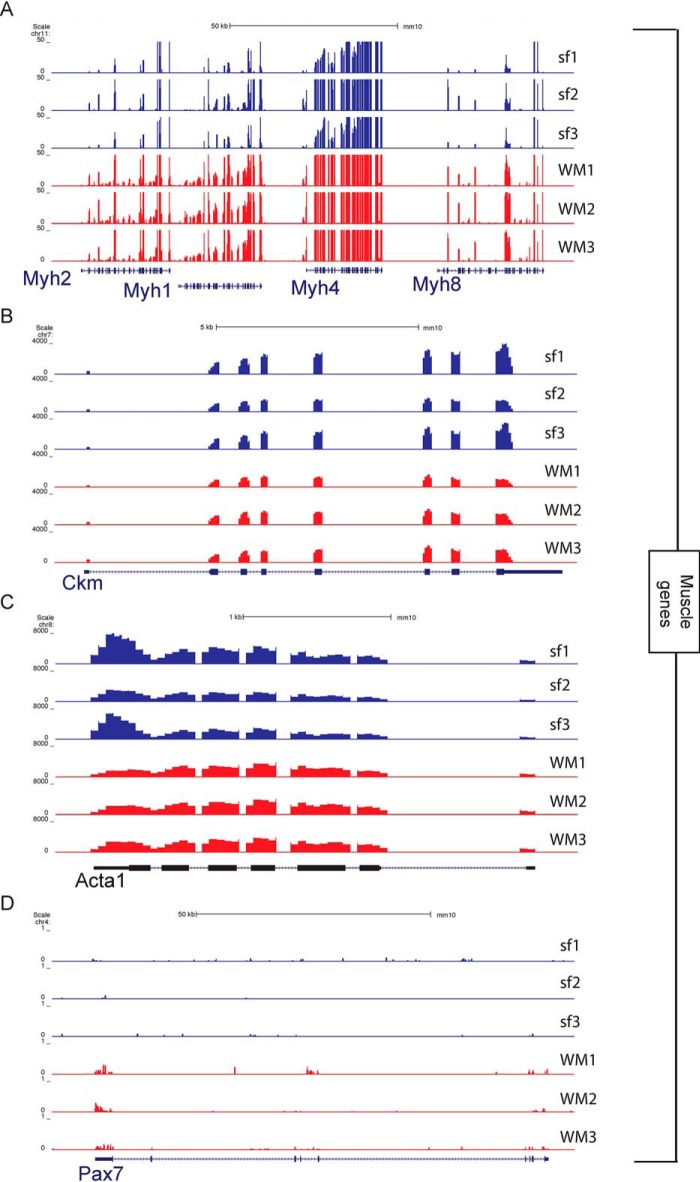
When looking more in depth at individual genes, we see many similarities between the single myofiber and the whole muscle but also crucial differences. Of particular note, we see that muscle-specific genes have similar numbers of reads between the single-fiber and whole-muscle samples (Fig. 3, A–C). The Myh cluster codes for a variety of myosin heavy chain proteins, which are the motor proteins of muscle whose various isoforms are the basis of the different fiber types (7, 11). The similar expression between the single fiber and the whole muscle conclusively shows that the RNA sequenced came from a myofiber alone (Fig. 3A). For further confirmation, we also display Ckm, the muscle-specific creatine kinase (12), and Acta1, which codes for skeletal muscle α-actin (13). These genes have the same pattern as is seen with Myh (Fig. 3, B and C).
Figure 3.
University of California, Santa Cruz snapshots showing expression of myogenic genes in single myofibers and whole muscle. A, part of the myosin heavy chain (Myh) gene cluster located on chromosome 11. B, muscle creatine kinase (Ckm). C, actin α1 (Acta1) gene. D, paired box 7 (Pax7) gene expressed in the associated satellite cells.
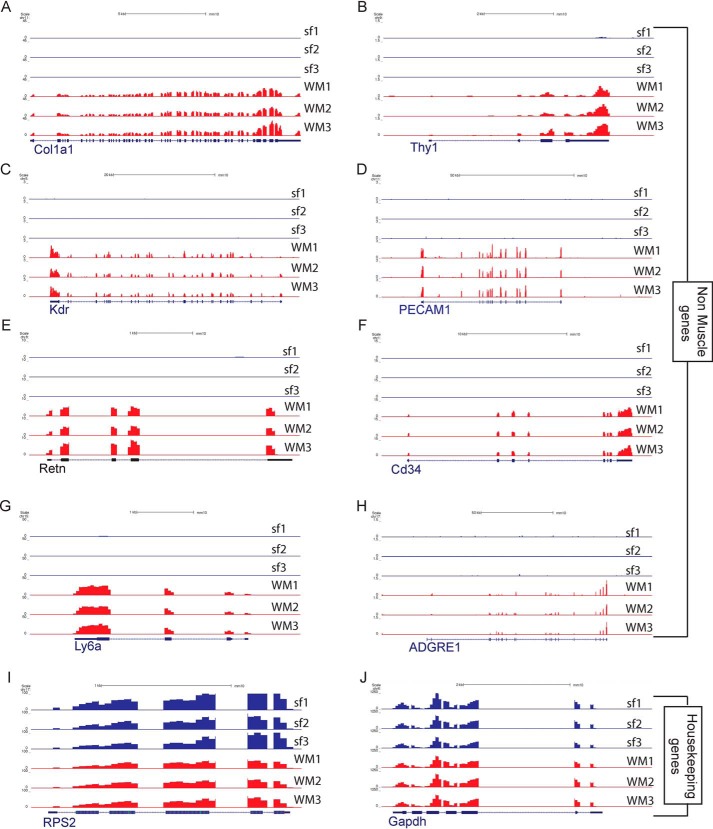
Differences between the single-myofiber and whole-muscle transcriptomes arise from nonmuscle cells. Using smfRNA-Seq, we are capable of completely removing these unwanted cell types to sequence only the myofiber. To demonstrate the removal of cell populations other than the myofiber, we looked at cell-specific genes of a variety of different muscle-resident cell types. We first looked at the expression of the satellite cell marker Pax7 and see that there is no expression of Pax7 in the single-fiber transcriptome despite its presence in whole muscle samples (Fig. 3D). We also analyzed markers for fibroblasts, namely Col1a1 and Thy1 (14, 15). As expected for these genes, they are expressed in the whole muscle but not expressed in the single fiber (Fig. 4, A and B). Endothelial cells are also depleted in single fiber samples, because their markers Kdr and Pecam1 are expressed in whole muscle but not in the single fiber (16, 17) (Fig. 4, C and D). We also analyzed Retn to identify the presence of adipocytes (18), Cd34 for hematopoietic cells (19), Ly6a as a marker for FAPs (2), and Adgre1 for macrophages (20) (Fig. 4, E–J). As expected, in the single-fiber transcriptome there is no expression for any of these genes, all of which were present in the whole muscle. These results clearly demonstrate the purity of smfRNA-Seq in sequencing only the myofiber without confounding cell types.
Figure 4.
University of California, Santa Cruz snapshots showing expression of nonmyogenic genes between single myofibers and whole muscle. A, collagen type 1 α1 chain (Col1a1) gene expressed in fibroblasts. B, CD90 (Thy1) gene expressed in fibroblasts. C, kinase insert domain receptor gene (Kdr) as a marker for endothelial cells. D, CD31 gene (Pecam1) as a marker for endothelial cells. E, resistin (Retn) as a marker for adipocytes. F, Cd34 as a marker for hematopoietic cells. G, Ly6a to detect the presence of FAPs. H, adhesion G protein–coupled receptor E1 (Adgre1) gene for macrophages. I, housekeeping gene Rps2. J, housekeeping gene Gapdh.
From these data, we performed a Gene Ontology (GO) term analysis of genes that are expressed solely in the WM samples, defined as having an RPM value of at least 10 in WM and 0 in single-fiber, indicating that they originate from nonmyogenic cells. Using these criteria, we identified 445 genes that were solely expressed in the WM muscle samples (Table S1). Some of the more up-regulated genes were involved in the formation of the extracellular matrix or immunity (Table S1).
We also attempted to identify genes that were exclusively produced in the myofiber. We defined these genes as being more highly expressed in the single-fiber samples, with a q value < 0.01, being expressed at more than 10 RPM and being expressed at least 10 RPM more than in the WM samples. We identified 622 genes that matched these criteria and performed a GO analysis (Table S2). From this, we see that some of the top pathways associated with these genes are ribosomal proteins and the respiratory chain. However, this is not an exhaustive list, because of the difficulty in identifying genes expressed solely in myofibers when the WM samples are composed primarily of myofibers. For example, Myh4, which is expressed exclusively in muscle fibers, does not pass the criteria laid down and is not part of the list of genes expressed solely in myofibers. On the other hand, dystrophin (Dmd), another myofiber-specific gene, does pass our criteria.
Age effect on the transcriptome of single myofibers
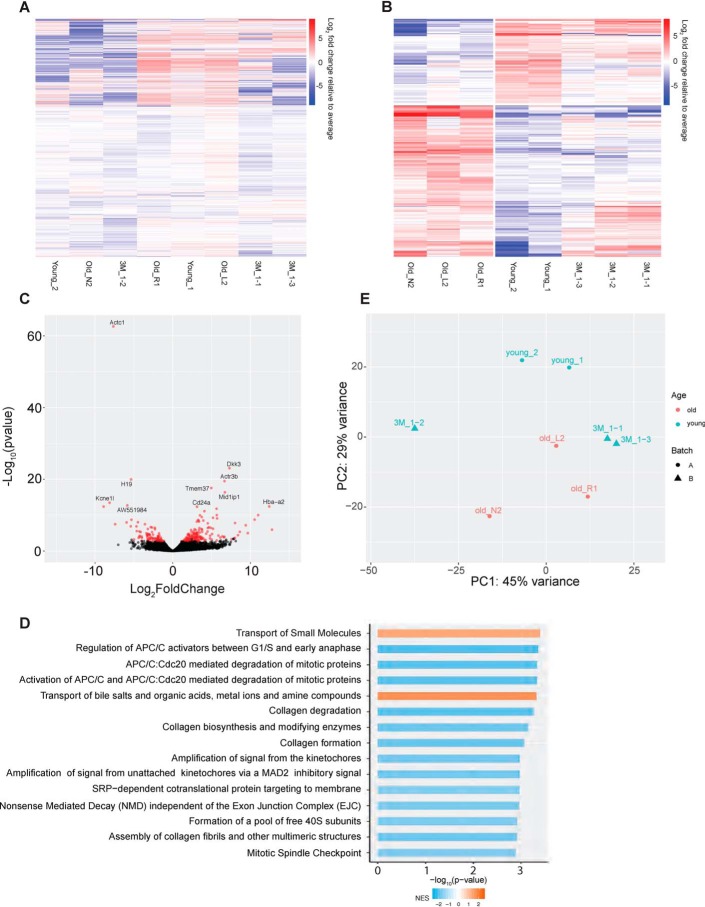
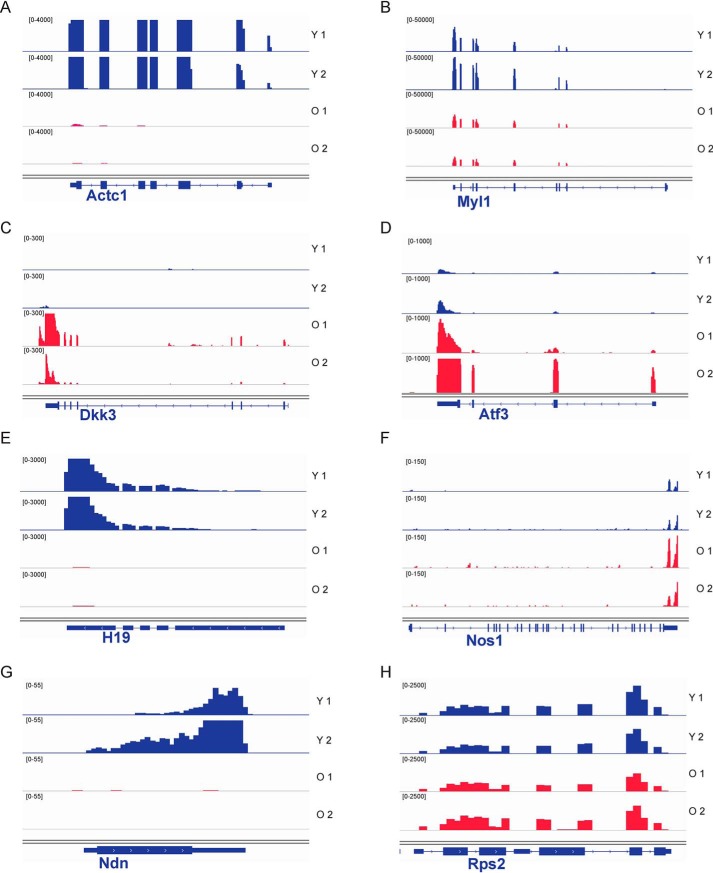
To verify the utility of our new method to analyze variation in the myofiber transcriptome under different conditions, we performed smfRNA-Seq on EDL myofibers isolated from young (1 and 3 months old) and aged (19 months old) mice. Using our technique, we see clear differences in the transcriptome of young and old myofibers. In the old myofibers we see the deregulation of a number of genes, with 181 genes being significantly (padj < 0.05) differentially expressed in old compared with young myofibers (Fig. 5, A and B). PCA demonstrates how young myofibers, isolated from 1-month-old mice, cluster together away from the old myofibers on the PC2 axis (Fig. 5E). Furthermore, the myofibers isolated from 3-month-old mice begin to resemble the 19-month-old myofibers while also maintaining similarities with the young 1-month-old myofibers, demonstrating the gradual change in transcriptome of the myofibers as the mouse ages (Fig. 5E). GO term analysis of the genes with the highest variation between young and old myofibers shows that deregulated pathways in aging include the transport of small molecules such as salts and metal ions and the synthesis of collagens. Additional examples of genes significantly deregulated in aging include Actc1, Myl1, Dkk3, Atf3, H19, Nos1, and Ndn (Fig. 6, A–H). These are genes involved in skeletal muscle structure, metabolism, growth, and maintenance of homeostasis.
Figure 5.
Comparative analysis of whole transcriptome between single myofibers from young and old mice. A, heat map of all genes expressed in old and young myofibers. B, heat map of differentially expressed genes between the old and young myofibers. Colors indicate Log2 fold change relative to average per gene, with red indicating a higher expression and blue as a lower expression between young (1 and 3 months) and old (19 months) myofibers. C, volcano plot of the differentially expressed genes between young and old myofibers. Points in red indicate there is a significant difference between the two groups. D, GO term analysis of the top 15 differentially regulated pathways. E, projection of samples along first two principal components found by PCA applied to log reads-per-million gene expression of young (1 and 3 months) and old (19 months) myofibers.
Figure 6.
IGV snapshots showing the expression of selected differentially expressed genes between young and old myofibers. Young myofiber tracks are labeled in blue, and old are in red. A, actin α cardiac muscle 1 (Actc1). B, myosin light chain 1 (Myl1). C, dickkopf WNT signaling pathway inhibitor 3 (Dkk3). D, activating transcription factor 3 (Atf3). E, H19, imprinted maternally expressed transcript (H19). F, nitric-oxide synthase 1 (Nos1). G, necdin (Ndn). H, housekeeping gene ribosomal protein S2 (Rps2). IGV, Integrative Genomics Viewer.
Discussion
smfRNA-Seq is a powerful new technique that allows high-resolution whole transcriptome sequencing of a single myofiber. Here, we have demonstrated that RNA can be successfully isolated from a single myofiber at a suitable concentration and quality to be used with SMART-Seq technology, generating a high-quality cDNA library for sequencing.
What distinguishes smfRNA-Seq from single cell RNA-Seq is that sequencing depth is comparable with large-scale bulk RNA-Seq (Fig. 2). However, as shown by PCA, single-myofiber samples are very similar to whole muscle samples with regards to PCA1 but dissimilar with regards to PCA2. By analyzing individual genes, we can interpret the similarities as being due to muscle-specific genes, whereas the lack of other cell types allows the single fibers to form a distinct cluster away from the whole muscle. Indeed, comparison of single myofiber to whole-muscle samples identified a list of genes expressed only in the whole muscle, which are likely responsible for this distinction (Table S1). This dissociation of the myofiber transcriptome from the transcriptome of other muscle-resident cell types will be of critical importance to tease out the contribution of the myofiber when a gene of interest is expressed in multiple cell types or when a treatment condition has a whole-muscle effect.
All of the fibers analyzed in this study were of the fast type. This is because the fibers were isolated from the EDL, which is composed of over 90% fast fibers (21). We confirmed that they were fast fibers by analyzing the levels of transcript for Tnnt3 and Tnni2, which are expressed in the fast muscle, as well as Tnnt1, which is only expressed in slow muscle fibers (22, 23). We see that there is no expression of Tnnt1 and a high expression of the Tnnt3 and Tnni2 genes (Fig. S2, A–C). Because we could identify these fibers as fast-twitch, the method would be appropriate to distinguish differences in the transcriptome between fast and slow fiber types.
One condition that leads to changes in the numbers and in the gene expression profile of various muscle-resident cell types is aging (24, 25). Because of the changes occurring in multiple cell types, including immune cells (26, 27), FAPs (28), and satellite cells (29, 30), whole-muscle RNA-Seq would be inappropriate to distinguish age-induced changes in myofibers alone. Using smfRNA-Seq, we were able to uncover differences in the transcriptome of old and young myofibers, without any added noise from age-related alterations in other cell types. Among the genes deregulated in aging myofibers are genes involved in muscle growth and structure, suggesting that some of these altered genes may be responsible for age-related muscle atrophy, also called sarcopenia. Of note, we see that Actc1 and Myl1 are greatly down-regulated in old compared with young myofibers (Fig. 6, A and B). Despite not reaching significance, there is also a moderate decrease in Acta1 with a log2 fold change of −0.84. Because actins and myosins play a role in myofiber structure, force, and contractile properties, we suspect that the down-regulation of these genes may play a role in the weakening in contractile strength in aged muscles (31, 32). A gene that is significantly up-regulated in aged myofibers is Dkk3, which encodes a secreted inhibitor of WNT signaling (Fig. 6C) (33). This finding is in line with a previous study demonstrating that increased Dkk3 in aged muscle tissue leads to muscle atrophy through autocrine signaling (33) while also confirming that the myofiber is the source of this secreted factor. There is also a down-regulation in Ndn in the old myofibers (Fig. 6G). Ndn produces Necdin, which is important in muscle regeneration, because it promotes the differentiation of satellite cells, and Necdin knockout mice have a significant impairment in muscle regeneration (34). Necdin was also shown to protect skeletal muscle from tumor-induced muscle wasting, also called cachexia, in part by binding to and inhibiting P53 (35). It would be of interest to see whether Necdin decrease in aging myofibers is also a cause of age-induced muscle atrophy.
In addition to genes involved in muscle structure and growth, our data also show that multiple genes involved in key metabolic processes and maintenance of homeostasis are also deregulated in old compared with young myofibers. For example, we detect a loss in the expression of the long noncoding RNA H19 (Fig. 6E). Although the role of H19 during myogenesis has been studied (36), namely in controlling myoblast proliferation, its role in fully differentiated myotubes is still unclear. However, H19 has also been shown to be involved in glucose metabolism and consequently, insulin sensitivity (37, 38). We speculate that H19 reduction in old myofibers may therefore be a contributor to metabolic dysfunction, which is a common feature of aging (38).
In old myofibers, we also see an up-regulation of Atf3, which is a stress factor that is known to be up-regulated in muscle after a disturbance in homeostasis, such as after exercise (39) (Fig. 6D). Interestingly, Atf3 was shown to dampen the expression of pro-inflammatory cytokines after exercise (40). We speculate that its up-regulation in aging myofibers may occur in response to an age-related disturbance in homeostasis, such as the stress induced by chronic low-grade inflammation (41). Another gene that is up-regulated in aging is Nos1, which produces NO and plays a role in skeletal muscle metabolism (Fig. 6F). Notably, Nos1 is known to increase glucose uptake and inhibit mitochondrial respiration (42), which may be a contributor to age-related metabolic dysfunctions. In addition to these top deregulated genes, GO term analysis revealed that genes involved in collagen synthesis, assembly, and degradation are all perturbed in aging myofibers (Fig. 5D). Because collagens are important components of the muscle extracellular matrix, our data suggest that old myofibers may contribute to changes in extracellular matrix stiffness (43, 44).
Altogether, we were able to demonstrate the effectiveness of smfRNA-Seq by sequencing the transcriptome of single myofibers from young and old mice. This method allowed us to conclude that aging does have an effect on myofibers at the level of the transcriptome, without the confounding signal from nonmyogenic cell types. This comparative analysis proves that smfRNA-Seq can thus be used to study other developmental or muscle-wasting disorders, such as muscular dystrophy. Additionally, smfRNA-Seq has the potential to be adapted to a larger scale to perform high-throughput analysis of numerous myofibers, which will be important in the study of myofiber heterogeneity.
Materials and methods
Animal care
All procedures that were performed on animals were approved by the McGill University Animal Care Committee.
Accession numbers
All gene expression data reported in this study are available through GEO accession number GSE138591.
Commercial kits
The following commercial kits were used in this experiment: SMART-Seq HT kit (Takara catalog no. 634437), Nextera XT DNA library preparation kit (Illumina catalog no. FC-131-1024), and Nextera XT index kit (Illumina catalog no. FC-131-1001).
Buffers
Myofiber digestion buffer was prepared using 1000 units/ml of collagenase from Clostridium histolyticum (Sigma catalog no. C0130) in unsupplemented DMEM (Invitrogen catalog no. 11995073). Myofiber immunofluorescence blocking buffer is composed of 5% horse serum (Wisent catalog no. 065-250), 2% bovine serum albumin (Sigma catalog no. A8022), 1% Triton X-100 (Sigma catalog no. T9284), in PBS (Wisent catalog no. 311-425-CL). RNA extraction buffer is made using 19 μl of the 10× lysis buffer plus 1 μl of RNase inhibitor from the SMART-Seq HT kit. 1 μl of the previously composed 10× lysis buffer is added to 9 μl of RNase-free water to make 1× lysis buffer.
Dissection of the EDL
The EDL was dissected from the hindlimb in the following manner: the skin of the hindlimb was removed by cutting around the ankle with a pair of scissors, and an incision was made along the ventral side of the leg. The epimysium around the tibialis anterior (TA) was removed, and the tendon of the TA was cut at the ankle while making sure to only cut the top tendon because the bottom tendon belongs to the EDL. Using a pair of forceps, the TA was gently peeled off the leg up to the knee and was then cut out as close to the knee as possible with a pair of scissors. To expose the EDL tendon at the knee, the biceps femoris was first removed with a pair of forceps. The EDL was removed by cutting from tendon to tendon with a pair of scissors.
Myofiber isolation
The dissected EDL was placed in a 1.5-ml Eppendorf tube with 800 μl of myofiber digestion buffer. Trypsin was added to a final concentration of 0.25% to remove the associated satellite cells. The EDL was incubated at 37 °C and 5% CO2 for at least 1 h.
To disassociate the myofibers, we transferred the EDL to a 6-well plate with 2 ml of unsupplemented DMEM that had previously been coated with 10% horse serum (HS) in DMEM for at least 30 min. The EDL was gently pipetted up and down with a large bore pipette coated in HS until no more myofibers could be retrieved.
Immunofluorescence of myofibers
Briefly, freshly isolated myofibers were fixed at T0 using 4% paraformaldehyde in PBS for 5 min. They were washed three times with 0.1% Triton X-100 in PBS, then permeabilized with 0.1% Triton X-100 and 0.1 m glycine in PBS for 15 min, and again washed three times with 0.1% Triton X-100 in PBS. Myofibers were blocked for 1 h with blocking buffer, followed by incubation with a Pax7 hybridoma primary antibody (DSHB catalog no. AB_528428) at 1:100 dilution in blocking buffer overnight at 4 °C. The next day, the fibers were washed three times with 0.1% Triton X-100 in PBS, before incubating the Alexa Fluor 488 goat anti-mouse IgG1 secondary antibody (Invitrogen catalog no. A21121) at a 1:400 dilution in blocking buffer for 1 h. Fibers were washed three times with 0.1% Triton X-100 in PBS and mounted on a microscope slide with Prolong Gold antifade reagent with 4′,6′-diamino-2-phenylindole (Invitrogen catalog no. P3695).
Single-myofiber RNA extraction
Using a small bore glass pipette coated with HS, myofibers were transferred to a 6-well plate with 2 ml of PBS to wash the fibers. A single myofiber was transferred by using a coated small bore pipette to a 0.2-ml PCR tube, and the excess PBS was removed using a pipette. Next, we added 10 μl of lysis buffer and gently pipetted the myofiber up and down for 3 min and then incubated the fiber on ice for 5 min while periodically vortexing and spinning down the sample. The residual fiber pieces were removed by spinning down the sample and transferring the supernatant to a fresh PCR tube.
Whole-muscle RNA extraction
The whole muscle from the hindlimb of a mouse was dissected, frozen in liquid nitrogen, and ground into a powder using a mortar and pestle. RNA from whole muscle was extracted using TRIzol reagents (Ambion catalog no. 15596018).
cDNA library preparation
cDNA was constructed using the DNA SMART-Seq HT kit and following the manufacturer's recommendations. For a single fiber, we used 12 cycles of PCR amplification on the cDNA. The cDNA was then purified using AMPure XP beads (Beckman Coulter catalog no. A63880) at a 1:1 ratio and quantified using the Quant-iT PicoGreen dsDNA assay kit (ThermoFisher catalog no. P11496).
For the addition of Illumina sequencing adapters, we used 250 pg of cDNA in 1.25 μl of water as a starting material and followed the directions provided with the Nextera XT DNA library preparation kit but reduced all quantities by 4× volume. The libraries were size-selected using AMPure XP beads at 0.85× of the sample volume, to remove all fragments below 200 bp.
Sequencing and gene expression analysis from a single fiber and whole muscle
The Illumina NextSeq 500 high-output flow cell was used for sequencing. The sequenced reads were then mapped to the mouse mm10 genome by using HISAT2 (45), using an index downloaded from the HISAT2 website that jointly indexes the mm10 genome and the ENSEMBL transcriptome definition (46).
Differential expression analysis between old and young single myofiber from mouse
RNA-Seq raw reads from young and old single myofiber were included in the analysis. The reads were mapped to the mouse genome assembly (mm10) using HISAT2 (45). The number of aligned reads per gene was obtained with HTSeq (47), using gene annotations from GENCODE M23 (48). Genes with an average read counts smaller than 10 were filtered out. Differentially expressed genes between young and old samples were identified using the R package DESeq2 (49). Log fold changes were calculated, and their associated p values were corrected by independent hypothesis weighting (50).
Gene set enrichment analysis
To obtain the pathways affected between old and young mouse fiber, we used the implementation of Gene Set Enrichment Analysis of the fgsea R package (48). We used the Log fold changes calculated by DESeq2 to create a preranked genelist. The Reactome database was used as a reference for pathways (51).
Author contributions
D. M. B., F. L., A. H. C., T. J. P., H. S. N., and V. D. S. formal analysis; D. M. B. and F. L. validation; D. M. B., F. L., and V. D. S. investigation; D. M. B. and F. L. visualization; D. M. B. and F. L. methodology; D. M. B. and F. L. writing-original draft; V. D. S. conceptualization; V. D. S. supervision; V. D. S. funding acquisition.
Supplementary Material
This work was supported by Canadian Institute of Health Research Grant PJT-15087 and a Natural Sciences and Engineering Research Council grant (to V. D. S.). We would like to acknowledge Compute Canada allocations (to H. S. N. and T. J. P.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1–S3 and Figs. S1 and S2.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE138591.
- FAP
- fibro/adipogenic cell
- smf
- single-myofiber
- RNA-Seq
- RNA sequencing
- EDL
- extensor digitorum longus
- SMART
- switching mechanism at the 5′ end of RNA template
- PCA
- principal component analysis
- GO
- Gene Ontology
- WM
- whole muscle
- DMEM
- Dulbecco's modified Eagle's medium
- TA
- tibialis anterior
- HS
- horse serum.
References
- 1. Giordani L., He G. J., Negroni E., Sakai H., Law J. Y. C., Siu M. M., Wan R., Corneau A., Tajbakhsh S., Cheung T. H., and Le Grand F. (2019) High-dimensional single-cell cartography reveals novel skeletal muscle-resident cell populations. Mol. Cell 74, 609–621.e6 10.1016/j.molcel.2019.02.026 [DOI] [PubMed] [Google Scholar]
- 2. Joe A. W., Yi L., Natarajan A., Le Grand F., So L., Wang J., Rudnicki M. A., and Rossi F. M. (2010) Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat. Cell Biol. 12, 153–163 10.1038/ncb2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arnold L., Henry A., Poron F., Baba-Amer Y., van Rooijen N., Plonquet A., Gherardi R. K., and Chazaud B. (2007) Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J. Exp. Med. 204, 1057–1069 10.1084/jem.20070075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Christov C., Chrétien F., Abou-Khalil R., Bassez G., Vallet G., Authier F. J., Bassaglia Y., Shinin V., Tajbakhsh S., Chazaud B., and Gherardi R. K. (2007) Muscle satellite cells and endothelial cells: close neighbors and privileged partners. Mol. Biol. Cell 18, 1397–1409 10.1091/mbc.e06-08-0693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pette D., and Staron R. S. (1997) Mammalian skeletal muscle fiber type transitions. Int. Rev. Cytol. 170, 143–223 10.1016/S0074-7696(08)61622-8 [DOI] [PubMed] [Google Scholar]
- 6. Brooke M. H., and Kaiser K. K. (1970) Three “myosin adenosine triphosphatase” systems: the nature of their pH lability and sulfhydryl dependence. J. Histochem. Cytochem. 18, 670–672 10.1177/18.9.670 [DOI] [PubMed] [Google Scholar]
- 7. Pette D., and Staron R. S. (2000) Myosin isoforms, muscle fiber types, and transitions. Microsc. Res. Tech. 50, 500–509 [DOI] [PubMed] [Google Scholar]
- 8. Pette D., Peuker H., and Staron R. S. (1999) The impact of biochemical methods for single muscle fibre analysis. Acta Physiol. Scand. 166, 261–277 10.1046/j.1365-201x.1999.00573.x [DOI] [PubMed] [Google Scholar]
- 9. Picelli S., Faridani O. R., Björklund A. K., Winberg G., Sagasser S., and Sandberg R. (2014) Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 10.1038/nprot.2014.006 [DOI] [PubMed] [Google Scholar]
- 10. Hood D. A., and Simoneau J. A. (1989) Rapid isolation of total RNA from small mammal and human skeletal muscle. Am. J. Physiol. 256, C1092–C1096 10.1152/ajpcell.1989.256.5.C1092 [DOI] [PubMed] [Google Scholar]
- 11. Yoon S. J., Seiler S. H., Kucherlapati R., and Leinwand L. (1992) Organization of the human skeletal myosin heavy chain gene cluster. Proc. Natl. Acad. Sci. U.S.A. 89, 12078–12082 10.1073/pnas.89.24.12078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nuss J. E., Amaning J. K., Bailey C. E., DeFord J. H., Dimayuga V. L., Rabek J. P., and Papaconstantinou J. (2009) Oxidative modification and aggregation of creatine kinase from aged mouse skeletal muscle. Aging 1, 557–572 10.18632/aging.100055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nowak K. J., Ravenscroft G., and Laing N. G. (2013) Skeletal muscle α-actin diseases (actinopathies): pathology and mechanisms. Acta Neuropathol. 125, 19–32 10.1007/s00401-012-1019-z [DOI] [PubMed] [Google Scholar]
- 14. Guo Q., Minnier J., Burchard J., Chiotti K., Spellman P., and Schedin P. (2017) Physiologically activated mammary fibroblasts promote postpartum mammary cancer. JCI Insight 2, e89206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jiang D., and Rinkevich Y. (2018) Defining skin fibroblastic cell types beyond CD90. Front. Cell Dev. Biol. 6, 133 10.3389/fcell.2018.00133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ferrero E., Ferrero M. E., Pardi R., and Zocchi M. R. (1995) The platelet endothelial cell adhesion molecule-1 (PECAM1) contributes to endothelial barrier function. FEBS Lett. 374, 323–326 10.1016/0014-5793(95)01110-Z [DOI] [PubMed] [Google Scholar]
- 17. Khan S. S., Solomon M. A., and McCoy J. P. Jr. (2005) Detection of circulating endothelial cells and endothelial progenitor cells by flow cytometry. Cytometry B Clin. Cytom. 64, 1–8 [DOI] [PubMed] [Google Scholar]
- 18. Tomaru T., Steger D. J., Lefterova M. I., Schupp M., and Lazar M. A. (2009) Adipocyte-specific expression of murine resistin is mediated by synergism between peroxisome proliferator-activated receptor gamma and CCAAT/enhancer-binding proteins. J. Biol. Chem. 284, 6116–6125 10.1074/jbc.M808407200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. AbuSamra D. B., Aleisa F. A., Al-Amoodi A. S., Jalal Ahmed H. M., Chin C. J., Abuelela A. F., Bergam P., Sougrat R., and Merzaban J. S. (2017) Not just a marker: CD34 on human hematopoietic stem/progenitor cells dominates vascular selectin binding along with CD44. Blood Adv. 1, 2799–2816 10.1182/bloodadvances.2017004317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Waddell L. A., Lefevre L., Bush S. J., Raper A., Young R., Lisowski Z. M., McCulloch M. E. B., Muriuki C., Sauter K. A., Clark E. L., Irvine K. M., Pridans C., Hope J. C., and Hume D. A. (2018) ADGRE1 (EMR1, F4/80) Is a rapidly-evolving gene expressed in mammalian monocyte-macrophages. Front. Immunol. 9, 2246 10.3389/fimmu.2018.02246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sandonà D., Desaphy J. F., Camerino G. M., Bianchini E., Ciciliot S., Danieli-Betto D., Dobrowolny G., Furlan S., Germinario E., Goto K., Gutsmann M., Kawano F., Nakai N., Ohira T., Ohno Y., et al. (2012) Adaptation of mouse skeletal muscle to long-term microgravity in the MDS mission. PLoS One 7, e33232 10.1371/journal.pone.0033232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sheng J. J., and Jin J. P. (2016) TNNI1, TNNI2 and TNNI3: evolution, regulation, and protein structure-function relationships. Gene 576, 385–394 10.1016/j.gene.2015.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wei B., and Jin J. P. (2016) TNNT1, TNNT2, and TNNT3: isoform genes, regulation, and structure-function relationships. Gene 582, 1–13 10.1016/j.gene.2016.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hwang A. B., and Brack A. S. (2018) Muscle stem cells and aging. Curr. Top. Dev. Biol. 126, 299–322 10.1016/bs.ctdb.2017.08.008 [DOI] [PubMed] [Google Scholar]
- 25. Thorley M., Malatras A., Duddy W., Le Gall L., Mouly V., Butler Browne G., and Duguez S. (2015) Changes in communication between muscle stem cells and their environment with aging. J. Neuromuscul. Dis. 2, 205–217 10.3233/JND-150097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cui C. Y., Driscoll R. K., Piao Y., Chia C. W., Gorospe M., and Ferrucci L. (2019) Skewed macrophage polarization in aging skeletal muscle. Aging Cell 18, e13032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bird L. (2016) Regulatory T cells: ageing muscles lose T Reg-eneration. Nat. Rev. Immunol. 16, 204 10.1038/nri.2016.30 [DOI] [PubMed] [Google Scholar]
- 28. Ancel S., Mashinchian O., and Feige J. N. (2019) Adipogenic progenitors keep muscle stem cells young. Aging 11, 7331–7333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chakkalakal J. V., Jones K. M., Basson M. A., and Brack A. S. (2012) The aged niche disrupts muscle stem cell quiescence. Nature 490, 355–360 10.1038/nature11438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alway S. E., Myers M. J., and Mohamed J. S. (2014) Regulation of satellite cell function in sarcopenia. Front. Aging Neurosci. 6, 246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ravenscroft G., Zaharieva I. T., Bortolotti C. A., Lambrughi M., Pignataro M., Borsari M., Sewry C. A., Phadke R., Haliloglu G., Ong R., Goullée H., Whyte T., Consortium U. K., Manzur A., Talim B., et al. (2018) Bi-allelic mutations in MYL1 cause a severe congenital myopathy. Hum. Mol. Genet. 27, 4263–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Laing N. G., Dye D. E., Wallgren-Pettersson C., Richard G., Monnier N., Lillis S., Winder T. L., Lochmüller H., Graziano C., Mitrani-Rosenbaum S., Twomey D., Sparrow J. C., Beggs A. H., and Nowak K. J. (2009) Mutations and polymorphisms of the skeletal muscle α-actin gene (ACTA1). Hum. Mutat. 30, 1267–1277 10.1002/humu.21059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yin J., Yang L., Xie Y., Liu Y., Li S., Yang W., Xu B., Ji H., Ding L., Wang K., Li G., Chen L., and Hu P. (2018) Dkk3 dependent transcriptional regulation controls age related skeletal muscle atrophy. Nat. Commun. 9, 1752 10.1038/s41467-018-04038-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Deponti D., François S., Baesso S., Sciorati C., Innocenzi A., Broccoli V., Muscatelli F., Meneveri R., Clementi E., Cossu G., and Brunelli S. (2007) Necdin mediates skeletal muscle regeneration by promoting myoblast survival and differentiation. J. Cell Biol. 179, 305–319 10.1083/jcb.200701027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sciorati C., Touvier T., Buono R., Pessina P., François S., Perrotta C., Meneveri R., Clementi E., and Brunelli S. (2009) Necdin is expressed in cachectic skeletal muscle to protect fibers from tumor-induced wasting. J. Cell Sci. 122, 1119–1125 10.1242/jcs.041665 [DOI] [PubMed] [Google Scholar]
- 36. Martinet C., Monnier P., Louault Y., Benard M., Gabory A., and Dandolo L. (2016) H19 controls reactivation of the imprinted gene network during muscle regeneration. Development 143, 962–971 10.1242/dev.131771 [DOI] [PubMed] [Google Scholar]
- 37. Geng T., Liu Y., Xu Y., Jiang Y., Zhang N., Wang Z., Carmichael G. G., Taylor H. S., Li D., and Huang Y. (2018) H19 lncRNA promotes skeletal muscle insulin sensitivity in part by targeting AMPK. Diabetes 67, 2183–2198 10.2337/db18-0370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Barzilai N., Huffman D. M., Muzumdar R. H., and Bartke A. (2012) The critical role of metabolic pathways in aging. Diabetes 61, 1315–1322 10.2337/db11-1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fernández-Verdejo R., Vanwynsberghe A. M., Essaghir A., Demoulin J. B., Hai T., Deldicque L., and Francaux M. (2017) Activating transcription factor 3 attenuates chemokine and cytokine expression in mouse skeletal muscle after exercise and facilitates molecular adaptation to endurance training. FASEB J. 31, 840–851 10.1096/fj.201600987R [DOI] [PubMed] [Google Scholar]
- 40. Fernández-Verdejo R., Vanwynsberghe A. M., Hai T., Deldicque L., and Francaux M. (2017) Activating transcription factor 3 regulates chemokine expression in contracting C2C12 myotubes and in mouse skeletal muscle after eccentric exercise. Biochem. Biophys. Res. Commun. 492, 249–254 10.1016/j.bbrc.2017.08.059 [DOI] [PubMed] [Google Scholar]
- 41. Peake J., Della Gatta P., and Cameron-Smith D. (2010) Aging and its effects on inflammation in skeletal muscle at rest and following exercise-induced muscle injury. Am. J. Physiol. Regul. Integr. Comp. Physiol. 298, R1485–R1495 10.1152/ajpregu.00467.2009 [DOI] [PubMed] [Google Scholar]
- 42. Stamler J. S., and Meissner G. (2001) Physiology of nitric oxide in skeletal muscle. Physiol. Rev. 81, 209–237 10.1152/physrev.2001.81.1.209 [DOI] [PubMed] [Google Scholar]
- 43. Urciuolo A., Quarta M., Morbidoni V., Gattazzo F., Molon S., Grumati P., Montemurro F., Tedesco F. S., Blaauw B., Cossu G., Vozzi G., Rando T. A., and Bonaldo P. (2013) Collagen VI regulates satellite cell self-renewal and muscle regeneration. Nat. Commun. 4, 1964 10.1038/ncomms2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Baghdadi M. B., Castel D., Machado L., Fukada S. I., Birk D. E., Relaix F., Tajbakhsh S., and Mourikis P. (2018) Reciprocal signalling by Notch-Collagen V-CALCR retains muscle stem cells in their niche. Nature 557, 714–718 10.1038/s41586-018-0144-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim D., Langmead B., and Salzberg S. L. (2015) HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liao Y., Smyth G. K., and Shi W. (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 47. Anders S., Pyl P. T., and Huber W. (2015) HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Frankish A., Diekhans M., Ferreira A. M., Johnson R., Jungreis I., Loveland J., Mudge J. M., Sisu C., Wright J., Armstrong J., Barnes I., Berry A., Bignell A., Carbonell Sala S., et al. (2019) GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47, D766–D773 10.1093/nar/gky955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Love M. I., Huber W., and Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ignatiadis N., Klaus B., Zaugg J. B., and Huber W. (2016) Data-driven hypothesis weighting increases detection power in genome-scale multiple testing. Nat. Methods 13, 577–580 10.1038/nmeth.3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., Haw R., Jassal B., Korninger F., May B., Milacic M., Roca C. D., Rothfels K., Sevilla C., Shamovsky V., et al. (2018) The Reactome Pathway Knowledgebase. Nucleic Acids Res. 46, D649–D655 10.1093/nar/gkx1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.