Abstract
Type I interferon (IFN) induced by virus infections during pregnancy can cause placental damage, but the mechanisms and identities of IFN-stimulated genes that are involved in this damage remain under investigation. The IFN-induced transmembrane proteins (IFITMs) inhibit virus infections by preventing virus membrane fusion with cells and by inhibiting fusion of infected cells (syncytialization). Fusion of placental trophoblasts via expression of endogenous retroviral fusogens known as syncytins forms the syncytiotrophoblast, a multinucleated cell structure essential for fetal development. We found here that IFN blocks fusion of BeWo human placental trophoblasts. Stably expressed IFITM1, -2, and -3 also blocked fusion of these trophoblasts while making them more resistant to virus infections. Conversely, stable IFITM knockdowns in BeWo trophoblasts increased their spontaneous fusion and allowed fusion in the presence of IFN while also making the cells more susceptible to virus infection. We additionally found that exogenous expression of IFITMs in HEK293T cells blocked fusion with cells expressing syncytin-1 or syncytin-2, confirming the ability of IFITMs to block individual syncytin-mediated fusion. Overall, our data indicate that IFITMs inhibit trophoblast fusion and suggest that there may be a critical balance between these antifusogenic effects and the beneficial antiviral effects of IFITMs in virus infections during pregnancy.
Keywords: interferon, virus, trophoblast, membrane fusion, fusion protein, IFITM, IFITM1, IFITM2, IFITM3, influenza virus, restriction factor, Zika virus, interferon-induced transmembrane protein, syncytin, placenta, cell fusion
Introduction
Infections during pregnancy are associated with low birth weights, congenital birth defects, pregnancy complications, and miscarriages (1). Placental damage has been observed during Zika virus infections in nonhuman primates (2) and in mice during Zika virus and other flavivirus infections (3). Further, a recent landmark study demonstrated that fetal mortality during Zika virus infection is mediated by type I interferon (IFN)5 receptor signaling (4). This paradoxical effect of what are considered to be beneficial antiviral cytokines may be an evolutionary adaptation in which IFNs signal for termination of pregnancies that are unlikely to be viable due to severe or prolonged infection (4, 5). IFN signaling was shown to disrupt placental architecture in mice, leading to fetal hypoxia and demise. Placental defects caused by IFNs included disruption of the syncytiotrophoblast, a multinucleated cell structure that is critical for nutrient and gas exchange between maternal and fetal blood and that is formed by cell-to-cell fusion of trophoblasts. Importantly, continuous fusion of underlying cells with the syncytiotrophoblast is necessary for its maintenance throughout pregnancy (6, 7). Although IFNs have been known for decades to be embryotoxic molecules (8–11), the mechanism by which they induce placental damage is not fully understood.
Interestingly, placental trophoblast fusion is mediated by endogenous retroviral fusion proteins known as syncytin-1 and -2 that integrated separately into the human genome 30 and 45 million years ago, respectively, and that are uniquely expressed in this cell type (12–18). In experiments on human trophoblasts in vitro, both of the syncytins are required for maximal fusion (14–17). Although it is difficult to study the essentiality of these proteins for placentation in humans, similar proteins present in mice have been studied in vivo. Mouse syncytin-A is required for proper placenta formation, and knockout of this gene results in embryonic lethality (19). Similarly, the mouse syncytin-B is also important for development of placenta architecture, and its knockout results in fewer viable pups and low birth weights of pups that survive (20).
Our laboratory has long studied the IFN-induced transmembrane proteins (IFITMs) (IFITM1, -2, and -3), which utilize a palmitoylated amphipathic helical domain to block membrane fusion between viruses and host cells through alteration of lipid bilayers (21–27). These proteins are also capable of blocking cell-to-cell fusion (i.e. syncytia formation) of infected cells (22, 23, 28). Given that trophoblast fusion is mediated by virus-derived proteins, we hypothesized that IFN, and specifically the IFITMs, could inhibit trophoblast fusion and may thus underlie the embryotoxicity of IFN. We sought to test this hypothesis using the human BeWo trophoblast cell line, one of the most commonly used model systems for studying syncytin-mediated trophoblast fusion (15–17, 29–32). BeWo trophoblasts display minimal spontaneous fusion at baseline, but robust cell-to-cell fusion can be induced by stimulation with forskolin, a drug that increases cAMP production, resulting in syncytin up-regulation (15–17, 29–31). Here, we examine the effects of IFN and IFITMs on trophoblast fusion.
Results
Type I IFN inhibits trophoblast fusion
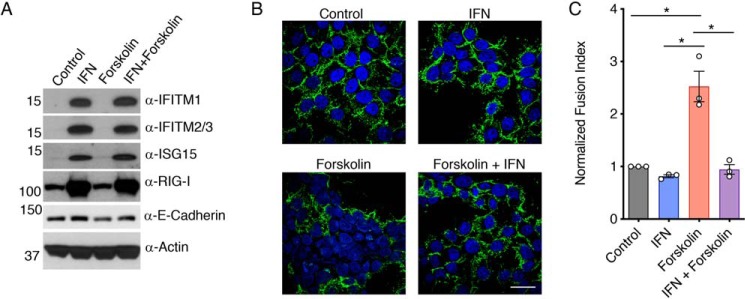
Similar to most other cell types, BeWo cells and primary trophoblasts have been reported to produce low steady-state levels of IFITM mRNAs even in the absence of IFN (33–35). However, IFITM1, -2, and -3 were difficult to detect by Western blotting in BeWo cells, indicating that baseline protein levels are low (Fig. 1A). Upon IFNβ treatment, IFITM1–3 and other IFN-stimulated genes, such as ISG15 and RIG-I, were up-regulated, demonstrating functional IFN signaling in these cells (Fig. 1A). Co-treatment with fusion-inducing forskolin had no effect on levels of the IFN-induced proteins (Fig. 1A). In these treated cells, we quantified cell-to-cell fusion by staining the cell surface with antibodies against the adhesion molecule E-cadherin, followed by counting of nuclei within syncytia versus single cells. We observed that forskolin treatment significantly increased the fusion index of the cells, as expected (Fig. 1, B and C). This fusion was significantly decreased by IFN co-treatment, demonstrating that type I IFN inhibits fusion of BeWo placental trophoblasts concomitant with induction of IFN-induced proteins, such as IFITMs (Fig. 1, A–C). E-cadherin has been known for decades to be down-regulated at the mRNA and protein level upon fusion of trophoblasts (36). We observed that indeed E-cadherin was modestly decreased in forskolin-treated cells, as measured by Western blotting, but was not decreased in forskolin/IFN co-treated cells, providing a secondary indicator of fusion inhibition by IFNβ (Fig. 1B).
Figure 1.
IFN inhibits fusion of BeWo trophoblasts stimulated by forskolin. BeWo cells were treated for 24 h with 40 units/ml IFNβ (IFN), 50 μm forskolin, a combination of IFN and forskolin, or vehicle control. Treated cells were analyzed by Western blotting of cell lysates (A) or confocal microscopy imaging (B) after staining with anti-E-cadherin (green) or DAPI (blue). White scale bar, 10 μm. C, fusion indices were calculated and normalized to the background spontaneous fusion level observed in each experiment in vehicle control cells. Bars, averages from three identical independent experiments with individual data points shown as circles; data are representative of two additional similar experiments. Error bars, S.D. Differences between groups were evaluated by one-way ANOVA followed by Tukey's multiple-comparison test. *, p < 0.0005 for the indicated comparisons.
IFITM overexpression inhibits trophoblast fusion
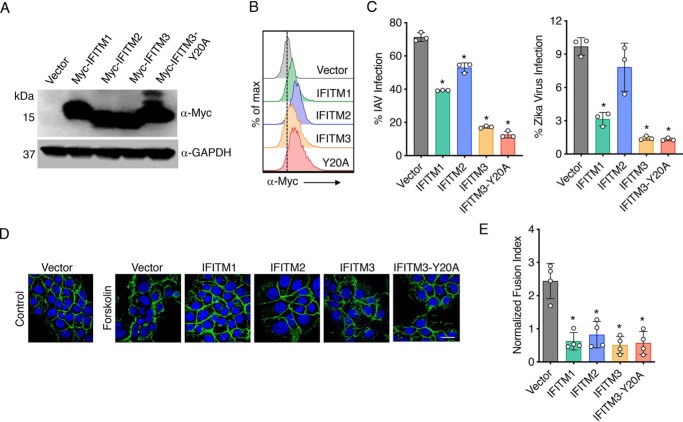
To more specifically examine the effect of the individual IFITMs on trophoblast fusion, we generated stable BeWo lines expressing IFITM1, -2, or -3 or an IFITM3 variant (Y20A) (Fig. 2, A and B). We included IFITM3-Y20A, because Tyr-20 is within a motif that facilitates endocytosis of IFITM3, and its mutation results in accumulation at the plasma membrane (37, 38), potentially providing enhanced inhibition of trophoblast fusion. To first test whether IFITMs are antivirally functional in BeWo cells, we examined infections of the stable cell lines with influenza A virus and Zika virus. We found that each of the IFITMs decreased infection with influenza virus as compared with vector control cells and that IFITM1 and -3 provided protection against Zika virus infection (Fig. 2B). We further observed that forskolin was unable to induce fusion of the IFITM-expressing BeWo lines (Fig. 2, C and D), indicating that IFITM1, -2, and -3 are each capable of inhibiting trophoblast fusion. IFITM3-Y20A was also able to inhibit trophoblast fusion, but we did not observe an enhancement of inhibition as compared with WT IFITM3. Overall, IFITM expression in trophoblasts decreases both virus infection and cell-to-cell fusion.
Figure 2.
IFITM overexpression inhibits BeWo trophoblast fusion. BeWo cells were stably transduced with lentiviruses expressing Myc-tagged IFITM constructs. A, Western blotting of cell lysates. B, flow cytometry histograms after staining with anti-Myc antibody. C, percentage of infection measured 24 h postinfection with influenza A virus (IAV) (MOI 2.5) or Zika virus (MOI 5), as measured by flow cytometry staining for viral antigens. Flow cytometry gates were set based on mock-infected controls. Bars, averages of three independent experiments with individual data points shown as circles. Error bars, S.D. Differences between groups were analyzed by one-way ANOVA followed by Dunnett's multiple-comparison test. *, p < 0.0001 compared with vector control cells. D, cell lines were treated for 48 h with 50 μm forskolin or vehicle control. Cells were then imaged by confocal microscopy after staining with anti-E-cadherin (green) or DAPI (blue). White scale bar, 10 μm. E, fusion indices from forskolin-treated cells as in D were calculated and normalized to the background spontaneous fusion levels observed in each experiment in vector control cells treated with vehicle control. Bars, averages from four independent experiments with individual data points shown as circles. Error bars, standard S.D. Differences between groups were analyzed by one-way ANOVA followed by Dunnett's multiple-comparison test. *, p < 0.0001 compared with vector control cells.
IFITMs inhibit HEK293T cell fusion mediated by exogenous syncytin expression
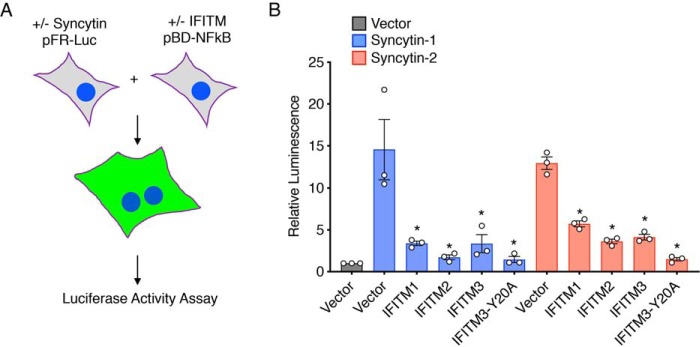
To determine whether IFITMs are specifically capable of inhibiting fusion mediated individually by syncytin-1 or -2, we employed a cell-to-cell fusion assay utilizing HEK293T cells, which naturally lack expression of endogenous fusion proteins and endogenous IFITMs, and that we previously validated for studying effects of IFITMs on cell-to-cell fusion mediated by viral fusion proteins (23, 28, 39). In short, one population of cells was transfected with vector control or syncytin-1 or -2, whereas target cells were transfected with individual IFITM expression plasmids or vector control. Additionally, the two groups of cells were co-transfected with distinct plasmids that produce luciferase only when the plasmids are together in the same cell (40) (Fig. 3A). Thus, after mixing the two cell populations, luciferase activity serves as a quantitative readout of cell-to-cell fusion between the two populations (23, 28, 40). We first established that both syncytin-1 and -2 were able to induce fusion of HEK293T cells (Fig. 3B). Fusion mediated by either syncytin was significantly inhibited by expression of IFITMs in target cells, further establishing that IFITMs are capable of inhibiting cell-to-cell fusion mediated by these essential trophoblast fusogens (Fig. 3B). Further, these data demonstrate that inhibition of syncytin-mediated cell fusion by IFITMs does not require intracellular interaction between syncytins and IFITMs, as these proteins were expressed in distinct cell populations in these experiments.
Figure 3.
IFITM inhibition of syncytin-mediated fusion of HEK293T cells. A, schematic representation of the HEK293T cell fusion assay. One population of cells was transfected with syncytin-1 or -2 constructs or vector control plus pFR-Luc plasmid. A second population of cells was transfected with IFITM constructs or vector control plus pBD-NF-κB. Cells were then mixed and allowed to fuse for 24 h. Luciferase activity in cell lysates was then measured as a quantitative readout of cell fusion. B, luciferase activity was measured from cells prepared as described in A. Bars, average results from three independent experiments with individual data points shown as circles. Error bars, S.D. Differences between groups were analyzed by one-way ANOVA followed by Tukey's multiple-comparison test. *, p < 0.01 compared with the respective syncytin-transfected vector control cells.
Endogenous IFITMs inhibit trophoblast fusion
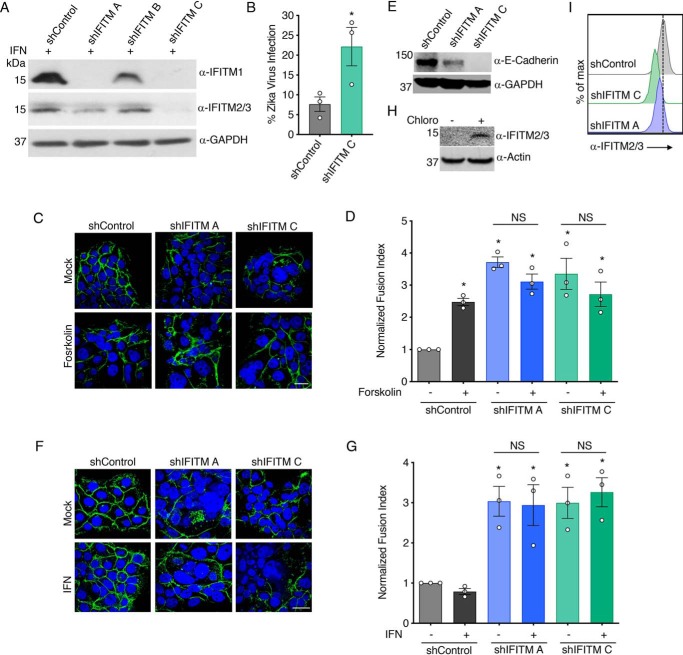
To determine whether endogenous levels of IFITMs are capable of inhibiting trophoblast fusion, we generated stable BeWo cell lines expressing shRNAs targeting the IFITMs. We generated two cell lines (labeled shIFITM A and C) with significant knockdowns of IFITM1–3, resulting in low IFITM levels as compared with control cells with IFN treatment (Fig. 4A), and confirmed that IFITM knockdown results in increased susceptibility to virus infection (Fig. 4B). Surprisingly, we found that these cells were difficult to maintain when using standard BeWo passaging methods but that the cells could be readily expanded when passaged frequently at high dilution. We determined that this was likely due to increased spontaneous fusion of these cells and thus their terminal differentiation when grown at high density. We quantified fusion of these cells when plated at high density and found that increased spontaneous fusion was indeed occurring for both of the IFITM knockdown cell lines as compared with control cells, and this fusion was not increased further by forskolin treatment (Fig. 4, C and D). Strikingly, E-cadherin was down-regulated under these conditions more than we have observed with forskolin treatments, providing a secondary confirmation of the robust spontaneous fusion of these IFITM knockdown BeWo cells (Fig. 4E). Additionally, we observed that IFN was unable to inhibit the spontaneous fusion of these cells, identifying that inhibition of trophoblast fusion by IFN requires the IFITMs (Fig. 4, F and G). These results suggest that BeWo cells are capable of fusing without forskolin treatment but that this is prevented by a low baseline level of IFITMs. Indeed, we were able to detect basal IFITM2/3 in WT BeWo cells when we prevented IFITM turnover in lysosomes by treatment of cells with chloroquine (41) (Fig. 4H). We further confirmed baseline expression of IFITM2/3 in BeWo cells as well as their knockdown in the stable shIFITM lines using flow cytometry analysis of the cells with IFITM2/3 antibody staining (Fig. 4I). Overall, we have employed gain- and loss-of-function experiments to identify that IFITMs inhibit virus infection of trophoblasts while also inhibiting critical cell-to-cell fusion required for syncytiotrophoblast formation.
Figure 4.
Knockdown of IFITMs promotes fusion of BeWo trophoblasts. BeWo cells were stably transduced with lentiviruses expressing control shRNA (shControl) or different shRNAs targeting IFITMs (shIFITM A, B, or C). A, Western blotting of cell lysates after 18-h treatment with 40 units/ml IFNβ demonstrates IFITM knockdown in specific cell lines. B, percentage of infection of shControl cells and shIFITM C cells 24 h postinfection with Zika virus (MOI 5), as measured by flow cytometry staining for viral antigen. Flow cytometry gates were set based on mock-infected controls. Bars, averages of three independent experiments with individual data points shown as circles. Error bars, S.D. *, p < 0.05 as compared with shControl cells by paired t test. C, the indicated cell lines were treated for 48 h with 50 μm forskolin or vehicle control. Cells were then imaged by confocal microscopy after staining with anti-E-cadherin (green) or DAPI (blue). White scale bar, 10 μm. D, fusion indices from cells as in C were calculated and normalized to the background spontaneous fusion levels observed in each experiment in shControl cells treated with vehicle control. Bars, averages from three independent experiments with individual data points shown as circles. Error bars, S.D. Differences between groups were evaluated by one-way ANOVA followed by Tukey's multiple-comparison test. *, p < 0.02 compared with shControl without forskolin treatment. NS, selected comparisons of interest that are not significantly different as judged by a p value > 0.05. E, Western blotting of cell lysates demonstrates down-regulation of E-cadherin in IFITM knockdown lines. F, the indicated cell lines were treated for 48 h with 40 units/ml IFNβ or vehicle control. Cells were then imaged by confocal microscopy after staining with anti-E-cadherin (green) or DAPI (blue). White scale bar, 10 μm. G, fusion indices from cells as in F were calculated and normalized to the background spontaneous fusion levels observed in each experiment in shControl cells treated with vehicle control. Bars, averages from three independent experiments with individual data points shown as circles. Error bars, S.D. Differences between groups were evaluated by one-way ANOVA followed by Tukey's multiple-comparison test. *, p < 0.01 compared with shControl without IFNβ treatment. NS, selected comparisons of interest that are not significantly different as judged by a p value > 0.05. H, Western blotting of WT BeWo cells after 24-h treatment with 40 μm chloroquine (chloro) or vehicle control (mock) demonstrates accumulation of baseline IFITM2/3 when lysosomal degradation is inhibited. I, flow cytometry histograms for untreated/unstimulated cell lines after staining with anti-IFITM2/3 antibody confirm that low baseline levels of IFITMs are reduced by shRNAs targeting IFITMs.
Discussion
The results presented here have significant implications for our understanding of the embryotoxic effects of IFNs in infections during pregnancy because they suggest that IFITMs are a critical contributor to this toxicity through effects on placenta syncytiotrophoblast development. The IFITMs may provide antiviral protection during moderate pregnancy-associated infections, including protection against direct viral infections of trophoblasts or the syncytiotrophoblast. Indeed, we have shown for the first time that IFITMs are able to inhibit virus infections of trophoblasts (Figs. 2C and 4B). However, we also observed that the IFITMs inhibit trophoblast cell-to-cell fusion (Figs. 2 and 4) and, specifically, that the IFITMs can block fusion mediated by the trophoblast syncytin-1 and syncytin-2 proteins (Fig. 3). We posit that IFITM1, -2, and -3 may disrupt the formation/maintenance of the syncytiotrophoblast during severe or lengthy infections, resulting in fetal demise. These observations may explain the need for tight regulation of baseline cellular levels of IFITM3 by posttranslational modifications, including phosphorylation and ubiquitination, in the absence of infection (25, 37, 41). Our data using unique tools and readouts also independently corroborate a groundbreaking recent study showing that IFITMs are required for placental malformations and fetal resorption induced by the viral mimic poly(I:C) in mice (32) and should allow the field to confidently build upon these results regarding the negative effects of IFITMs on the placenta.
Through the course of our work, we have generated interesting new reagents that should be of value to the placenta research community. BeWo trophoblast lines overexpressing IFITMs fail to fuse upon forskolin treatment (Fig. 2) and thus provide a new tool to study unfused trophoblasts as well as to parse effects of forskolin treatment versus the fusion process itself in regulating trophoblast gene expression programs. Moreover, through stable IFITM knockdown, we have generated BeWo lines that show dramatically increased spontaneous fusion that will allow studies of trophoblast fusion in this system without confounding effects of forskolin treatment (Fig. 4). These are among the first reported BeWo lines capable of robust fusion that do not overexpress fusion-promoting proteins. Additionally, results from IFITM knockdown cells may clarify why BeWo cells are poorly fusogenic at baseline despite their moderate expression of syncytins. We also speculate based on these results that expression of IFITMs may also be involved in preventing inappropriate cell-to-cell fusion in the placenta and maternal environment.
From a therapeutic perspective, the antiviral activity of IFITMs is counteracted by the clinically approved drug amphotericin B (28, 42), suggesting that this drug may also hold promise for countering the negative effects of IFITMs on the placenta. Likewise, short treatments of cells with rapamycin have been shown to result in degradation of IFITMs via the autophagy pathway (43), and resveratrol trimers have shown a similar effect in decreasing cellular IFITM levels (44). Thus, the identification of the IFITMs as a missing link between IFN and placental disruption may provide specific molecular targets for mitigating negative effects of IFNs during pregnancy-associated infections.
Experimental procedures
Cell culture and virus infections
An initial stock of BeWo cells was kindly provided by Dr. Stephanie Seveau (Ohio State University). We confirmed the identity of these cells by confirming their expression of syncytins and by confirming their fusion in the presence of forskolin. BeWo cells were cultured in Ham's F-12K medium supplemented with 10% EquaFETAL bovine serum (Atlas Biologicals). HEK293T and Vero cells were purchased from the ATCC and were grown in Dulbecco's modified Eagle's medium supplemented with 10% EquaFETAL bovine serum. All cells were cultured in a humidified incubator at 37 °C with 5% CO2 and were confirmed to be free of mycoplasma contamination by spot checking with the Lonza MycoAlert mycoplasma detection kit. In some experiments, BeWo cells were treated with IFNβ (EMD Millipore) at a concentration of 40 units/ml. For infections, influenza virus A/PR/8/34 (H1N1) (provided by Dr. Thomas Moran of the Icahn School of Medicine at Mt. Sinai) was propagated in embryonated chicken eggs and titered as described previously (45), and Zika virus strain PRVABC59 (National Institutes of Health BEI Resources) was propagated and titered in Vero cells. For detection of influenza virus–infected cells, anti-nucleoprotein antibody (National Institutes of Health BEI Resources, NR-19868) staining was used. For detection of Zika virus infection, anti-Zika E antibody (Kerafast, EVU302) was used. Percentage infection was measured by flow cytometry with mock-infected cells serving as controls for gating. Samples were analyzed using FlowJo software.
Generation of stable BeWo cell lines
For stable expression of IFITM proteins, IFITM coding sequences were cloned into the pLenti-puro vector as described previously (28). Lentiviruses were generated in HEK293T cells and were concentrated using Lenti-X concentrator reagent (Takara Bio) before transduction of BeWo cells. Stable IFITM-expressing BeWo lines were selected and subsequently maintained with 1 μg/ml puromycin in the medium. IFITM knockdown lines were generated and maintained similarly using lentiviral shRNA constructs purchased from Sigma as described previously (46).
BeWo cell fusion assays
For analysis of cell fusion, BeWo cells were plated to achieve ∼75% confluence on glass slides. To induce fusion, 50 μm forskolin (Sigma) was added to culture medium for 48–72 h. For imaging, cells were fixed with 4% paraformaldehyde, permeabilized with PBS/0.1% Triton X-100, and blocked with PBS/2% EquaFETAL bovine serum. Cells were then stained with anti-E-cadherin antibody (Abcam, ab1416) at 1:100 in PBS/0.1% Triton X-100, followed by staining with Alexa Fluor 488–labeled goat anti-mouse secondary antibody (Life Technologies, Inc., A11029) at 1:1000 in PBS/0.1% Triton X-100. Slides were mounted using Prolong Gold Antifade Mountant with DAPI. Cells were imaged on an Olympus FluoView confocal microscope. For each sample, 10 randomly chosen images were collected. In each field, nuclei in mono- versus multinucleated cells (three or more nuclei within a continuous E-cadherin–stained cell border) were counted. E-cadherin staining was digitally enhanced using ImageJ software (National Institutes of Health) to assist with identification of fused and unfused cells. The percentage of fused cells in each field was calculated as nuclei in multinucleated cells divided by the total number of nuclei in the field. Values for each of the 10 fields were averaged to determine the percentage of fusion for each sample. Generally, more than 400 nuclei were counted for each sample. The percentage of fusion value for the negative control in each experiment (WT BeWo or vector-transduced BeWo without forskolin) was set to 1, and all other samples were normalized to this value. The percentage of spontaneous fusion for the negative controls varied from roughly 8 to 20% in different experiments and likely represented cells that were fused prior to plating and cells that spontaneously fused during the experiment, and it may have potentially also included a small number of false positive multinucleated cells. Fusion induced by forskolin or knockdown of the IFITMs was generally 2–3-fold above the negative control and represented values of roughly 20–60% fusion in different experiments.
HEK293T cell fusion assays
IFITM constructs cloned into the pCMV-HA vector (Clontech) were described previously (24, 37), and Myc-FLAG-tagged syncytin-1 and -2 expression constructs in the pCMV6 vector were purchased from Origene (RC213951 and RC209875). pFR-Luc and pBD-NF-κB (Agilent) were co-transfected with plasmids as indicated in Fig. 3A. Cell-to-cell fusion assays were performed as outlined and validated previously (23, 28, 40). In short, transfections were performed overnight using LipoJet transfection reagent (Signagen). Cell populations were then mixed, replated, and incubated at 37 °C in standard culture medium for 24 h. Luciferase activity was measured using the Promega Dual-Luciferase Reporter Assay System.
Western blotting
For Western blotting, cells were lysed in 1% SDS buffer (1% SDS, 150 mm NaCl, 50 mm triethanolamine, pH 7.4), supplemented with cOmplete EDTA-free protease inhibitor mixture (Sigma) at 5 times the recommended concentration. Western antibodies used in this study include those specific for the following proteins: E-cadherin (Abcam ab1416), ISG15 (Cell Signaling, 2743S), RIG-I (Cell Signaling, 3743S), IFITM1 (Cell Signaling, 13126S), IFITM3 (ProteinTech, 11714-1-AP, which detects both IFITM2 and IFITM3), and GAPDH (Life Technologies, 39-8600).
Author contributions
A. Z., J. J. K., and J. S. Y. conceptualization; A. Z., L. Z., M. C., and J. S. Y. data curation; A. Z., J. J. K., and J. S. Y. formal analysis; A. Z., J. J. K., and J. S. Y. funding acquisition; A. Z., L. Z., T. M. M., A. D. K., M. C., and J. S. Y. investigation; A. Z., L. Z., T. M. M., A. D. K., S.-L. L., and J. S. Y. methodology; A. Z., A. D. K., J. J. K., S.-L. L., and J. S. Y. writing-review and editing; J. S. Y. supervision; J. S. Y. writing-original draft; J. S. Y. project administration.
Acknowledgment
We thank Dr. Stephanie Seveau for supplying an initial stock of BeWo cells and providing technical advice for fusion assays.
This work was supported by National Institutes of Health Grants AI130110 and AI142256 (to J. S. Y.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- IFN
- interferon
- IFITM
- IFN-induced transmembrane protein
- DAPI
- 4′,6-diamidino-2-phenylindole
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- ANOVA
- analysis of variance
- MOI
- multiplicity of infection.
References
- 1. Coyne C. B., and Lazear H. M. (2016) Zika virus: reigniting the TORCH. Nat. Rev. Microbiol. 14, 707–715 10.1038/nrmicro.2016.125 [DOI] [PubMed] [Google Scholar]
- 2. Hirsch A. J., Roberts V. H. J., Grigsby P. L., Haese N., Schabel M. C., Wang X., Lo J. O., Liu Z., Kroenke C. D., Smith J. L., Kelleher M., Broeckel R., Kreklywich C. N., Parkins C. J., Denton M., et al. (2018) Zika virus infection in pregnant rhesus macaques causes placental dysfunction and immunopathology. Nat. Commun. 9, 263 10.1038/s41467-017-02499-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Platt D. J., Smith A. M., Arora N., Diamond M. S., Coyne C. B., and Miner J. J. (2018) Zika virus-related neurotropic flaviviruses infect human placental explants and cause fetal demise in mice. Sci. Transl. Med. 10, eaao7090 10.1126/scitranslmed.aao7090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yockey L. J., Jurado K. A., Arora N., Millet A., Rakib T., Milano K. M., Hastings A. K., Fikrig E., Kong Y., Horvath T. L., Weatherbee S., Kliman H. J., Coyne C. B., and Iwasaki A. (2018) Type I interferons instigate fetal demise after Zika virus infection. Sci. Immunol. 3, eaao1680 10.1126/sciimmunol.aao1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Casazza R. L., and Lazear H. M. (2018) Antiviral immunity backfires: pathogenic effects of type I interferon signaling in fetal development. Sci. Immunol. 3, eaar3446 10.1126/sciimmunol.aar3446 [DOI] [PubMed] [Google Scholar]
- 6. Gauster M., Moser G., Orendi K., and Huppertz B. (2009) Factors involved in regulating trophoblast fusion: potential role in the development of preeclampsia. Placenta 30, S49–S54 10.1016/j.placenta.2008.10.011 [DOI] [PubMed] [Google Scholar]
- 7. Gauster M., and Huppertz B. (2010) The paradox of caspase 8 in human villous trophoblast fusion. Placenta 31, 82–88 10.1016/j.placenta.2009.12.007 [DOI] [PubMed] [Google Scholar]
- 8. Gresser I., Morel-Maroger L., Rivière Y., Guillon J. C., Tovey M. G., Woodrow D., Sloper J. C., and Moss J. (1980) Interferon-induced disease in mice and rats. Ann. N.Y. Acad. Sci. 350, 12–20 10.1111/j.1749-6632.1980.tb20602.x [DOI] [PubMed] [Google Scholar]
- 9. Cameo M., Fontana V., Cameo P., Vauthay L. G., Kaplan J., and Tesone M. (1999) Similar embryotoxic effects of sera from infertile patients and exogenous interferon-γ on long-term in-vitro development of mouse embryos. Hum. Reprod. 14, 959–963 10.1093/humrep/14.4.959 [DOI] [PubMed] [Google Scholar]
- 10. Ihara T., Oneda S., Yamamoto T., Boudrel L., Lau D., Miller D., and Nagata R. (1999) An embryotoxic/teratogenic potential and abortifacient effect study of interferon alfacon-1 (Infergen®) via subcutaneous administration to rhesus monkeys. Congenital Anomalies 39, 223–242 10.1111/j.1741-4520.1999.tb00562.x [DOI] [Google Scholar]
- 11. Zusman I., Engelhard D., Yaffe P., Ron A., Panet A., and Ornoy A. (1984) Effects of interferon and encephalomyocarditis virus on in vitro development of preimplantation mouse embryos with and without the zona pellucida. Teratology 29, 405–409 10.1002/tera.1420290311 [DOI] [PubMed] [Google Scholar]
- 12. Blaise S., de Parseval N., Bénit L., and Heidmann T. (2003) Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl. Acad. Sci. U.S.A. 100, 13013–13018 10.1073/pnas.2132646100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blond J. L., Lavillette D., Cheynet V., Bouton O., Oriol G., Chapel-Fernandes S., Mandrand B., Mallet F., and Cosset F. L. (2000) An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 74, 3321–3329 10.1128/JVI.74.7.3321-3329.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Frendo J. L., Olivier D., Cheynet V., Blond J. L., Bouton O., Vidaud M., Rabreau M., Evain-Brion D., and Mallet F. (2003) Direct involvement of HERV-W Env glycoprotein in human trophoblast cell fusion and differentiation. Mol. Cell. Biol. 23, 3566–3574 10.1128/MCB.23.10.3566-3574.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mi S., Lee X., Li X., Veldman G. M., Finnerty H., Racie L., LaVallie E., Tang X. Y., Edouard P., Howes S., Keith J. C. Jr., and McCoy J. M. (2000) Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403, 785–789 10.1038/35001608 [DOI] [PubMed] [Google Scholar]
- 16. Vargas A., Moreau J., Landry S., LeBellego F., Toufaily C., Rassart E., Lafond J., and Barbeau B. (2009) Syncytin-2 plays an important role in the fusion of human trophoblast cells. J. Mol. Biol. 392, 301–318 10.1016/j.jmb.2009.07.025 [DOI] [PubMed] [Google Scholar]
- 17. Chen C. P., Chen L. F., Yang S. R., Chen C. Y., Ko C. C., Chang G. D., and Chen H. (2008) Functional characterization of the human placental fusogenic membrane protein syncytin 2. Biol. Reprod. 79, 815–823 10.1095/biolreprod.108.069765 [DOI] [PubMed] [Google Scholar]
- 18. Lavialle C., Cornelis G., Dupressoir A., Esnault C., Heidmann O., Vernochet C., and Heidmann T. (2013) Paleovirology of “syncytins”, retroviral env genes exapted for a role in placentation. Philos Trans. R. Soc. Lond. B Biol. Sci. 368, 20120507 10.1098/rstb.2012.0507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dupressoir A., Vernochet C., Bawa O., Harper F., Pierron G., Opolon P., and Heidmann T. (2009) Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc. Natl. Acad. Sci. U.S.A. 106, 12127–12132 10.1073/pnas.0902925106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dupressoir A., Vernochet C., Harper F., Guégan J., Dessen P., Pierron G., and Heidmann T. (2011) A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc. Natl. Acad. Sci. U.S.A. 108, E1164–E1173 10.1073/pnas.1112304108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Desai T. M., Marin M., Chin C. R., Savidis G., Brass A. L., and Melikyan G. B. (2014) IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLoS Pathog. 10, e1004048 10.1371/journal.ppat.1004048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li K., Markosyan R. M., Zheng Y. M., Golfetto O., Bungart B., Li M., Ding S., He Y., Liang C., Lee J. C., Gratton E., Cohen F. S., and Liu S. L. (2013) IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 9, e1003124 10.1371/journal.ppat.1003124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chesarino N. M., Compton A. A., McMichael T. M., Kenney A. D., Zhang L., Soewarna V., Davis M., Schwartz O., and Yount J. S. (2017) IFITM3 requires an amphipathic helix for antiviral activity. EMBO Rep. 18, 1740–1751 10.15252/embr.201744100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yount J. S., Moltedo B., Yang Y. Y., Charron G., Moran T. M., López C. B., and Hang H. C. (2010) Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat. Chem. Biol. 6, 610–614 10.1038/nchembio.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yount J. S., Karssemeijer R. A., and Hang H. C. (2012) S-Palmitoylation and ubiquitination differentially regulate interferon-induced transmembrane protein 3 (IFITM3)-mediated resistance to influenza virus. J. Biol. Chem. 287, 19631–19641 10.1074/jbc.M112.362095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Percher A., Ramakrishnan S., Thinon E., Yuan X., Yount J. S., and Hang H. C. (2016) Mass-tag labeling reveals site-specific and endogenous levels of protein S-fatty acylation. Proc. Natl. Acad. Sci. U.S.A. 113, 4302–4307 10.1073/pnas.1602244113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McMichael T. M., Zhang L., Chemudupati M., Hach J. C., Kenney A. D., Hang H. C., and Yount J. S. (2017) The palmitoyltransferase ZDHHC20 enhances interferon-induced transmembrane protein 3 (IFITM3) palmitoylation and antiviral activity. J. Biol. Chem. 292, 21517–21526 10.1074/jbc.M117.800482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McMichael T. M., Zhang Y., Kenney A. D., Zhang L., Zani A., Lu M., Chemudupati M., Li J., and Yount J. S. (2018) IFITM3 restricts human metapneumovirus infection. J. Infect. Dis. 218, 1582–1591 10.1093/infdis/jiy361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kudo Y., and Boyd C. A. (2002) Changes in expression and function of syncytin and its receptor, amino acid transport system B(0) (ASCT2), in human placental choriocarcinoma BeWo cells during syncytialization. Placenta 23, 536–541 10.1053/plac.2002.0839 [DOI] [PubMed] [Google Scholar]
- 30. Borges M., Bose P., Frank H. G., Kaufmann P., and Pötgens A. J. (2003) A two-colour fluorescence assay for the measurement of syncytial fusion between trophoblast-derived cell lines. Placenta 24, 959–964 10.1016/S0143-4004(03)00173-5 [DOI] [PubMed] [Google Scholar]
- 31. Knerr I., Schubert S. W., Wich C., Amann K., Aigner T., Vogler T., Jung R., Dötsch J., Rascher W., and Hashemolhosseini S. (2005) Stimulation of GCMa and syncytin via cAMP mediated PKA signaling in human trophoblastic cells under normoxic and hypoxic conditions. FEBS Lett. 579, 3991–3998 10.1016/j.febslet.2005.06.029 [DOI] [PubMed] [Google Scholar]
- 32. Buchrieser J., Degrelle S. A., Couderc T., Nevers Q., Disson O., Manet C., Donahue D. A., Porrot F., Hillion K. H., Perthame E., Arroyo M. V., Souquere S., Ruigrok K., Dupressoir A., Heidmann T., Montagutelli X., Fournier T., Lecuit M., and Schwartz O. (2019) IFITM proteins inhibit placental syncytiotrophoblast formation and promote fetal demise. Science 365, 176–180 10.1126/science.aaw7733 [DOI] [PubMed] [Google Scholar]
- 33. Bayer A., Lennemann N. J., Ouyang Y., Bramley J. C., Morosky S., Marques E. T. Jr., Cherry S., Sadovsky Y., and Coyne C. B. (2016) Type III interferons produced by human placental trophoblasts confer protection against Zika virus infection. Cell Host Microbe 19, 705–712 10.1016/j.chom.2016.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Corry J., Arora N., Good C. A., Sadovsky Y., and Coyne C. B. (2017) Organotypic models of type III interferon-mediated protection from Zika virus infections at the maternal-fetal interface. Proc. Natl. Acad. Sci. U.S.A. 114, 9433–9438 10.1073/pnas.1707513114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thul P. J., Åkesson L., Wiking M., Mahdessian D., Geladaki A., Ait Blal H., Alm T., Asplund A., Bjork L., Breckels L. M., Bäckstrom A., Danielsson F., Fagerberg L., Fall J., Gatto L., et al. (2017) A subcellular map of the human proteome. Science 356, eaal3321 10.1126/science.aal3321 [DOI] [PubMed] [Google Scholar]
- 36. Coutifaris C., Kao L. C., Sehdev H. M., Chin U., Babalola G. O., Blaschuk O. W., and Strauss J. F. 3rd (1991) E-cadherin expression during the differentiation of human trophoblasts. Development 113, 767–777 [DOI] [PubMed] [Google Scholar]
- 37. Chesarino N. M., McMichael T. M., Hach J. C., and Yount J. S. (2014) Phosphorylation of the antiviral protein interferon-inducible transmembrane protein 3 (IFITM3) dually regulates its endocytosis and ubiquitination. J. Biol. Chem. 289, 11986–11992 10.1074/jbc.M114.557694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jia R., Pan Q., Ding S., Rong L., Liu S. L., Geng Y., Qiao W., and Liang C. (2012) The N-terminal region of IFITM3 modulates its antiviral activity by regulating IFITM3 cellular localization. J. Virol. 86, 13697–13707 10.1128/JVI.01828-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Melvin W. J., McMichael T. M., Chesarino N. M., Hach J. C., and Yount J. S. (2015) IFITMs from mycobacteria confer resistance to influenza virus when expressed in human cells. Viruses 7, 3035–3052 10.3390/v7062759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Battles M. B., Langedijk J. P., Furmanova-Hollenstein P., Chaiwatpongsakorn S., Costello H. M., Kwanten L., Vranckx L., Vink P., Jaensch S., Jonckers T. H., Koul A., Arnoult E., Peeples M. E., Roymans D., and McLellan J. S. (2016) Molecular mechanism of respiratory syncytial virus fusion inhibitors. Nat. Chem. Biol. 12, 87–93 10.1038/nchembio.1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chesarino N. M., McMichael T. M., and Yount J. S. (2015) E3 ubiquitin ligase NEDD4 promotes influenza virus infection by decreasing levels of the antiviral protein IFITM3. PLoS Pathog. 11, e1005095 10.1371/journal.ppat.1005095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lin T. Y., Chin C. R., Everitt A. R., Clare S., Perreira J. M., Savidis G., Aker A. M., John S. P., Sarlah D., Carreira E. M., Elledge S. J., Kellam P., and Brass A. L. (2013) Amphotericin B increases influenza A virus infection by preventing IFITM3-mediated restriction. Cell Rep. 5, 895–908 10.1016/j.celrep.2013.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shi G., Ozog S., Torbett B. E., and Compton A. A. (2018) mTOR inhibitors lower an intrinsic barrier to virus infection mediated by IFITM3. Proc. Natl. Acad. Sci. U.S.A. 115, E10069–E10078 10.1073/pnas.1811892115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ozog S., Timberlake N. D., Hermann K., Garijo O., Haworth K. G., Shi G., Glinkerman C. M., Schefter L. E., D'Souza S., Simpson E., Sghia-Hughes G., Carillo R. R., Boger D. L., Kiem H. P., Slukvin I., et al. (2019) Resveratrol trimer enhances gene delivery to hematopoietic stem cells by reducing antiviral restriction at endosomes. Blood 134, 1298–1311 10.1182/blood.2019000040, 10.1182/blood-2019-125931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moltedo B., Li W., Yount J. S., and Moran T. M. (2011) Unique type I interferon responses determine the functional fate of migratory lung dendritic cells during influenza virus infection. PLoS Pathog. 7, e1002345 10.1371/journal.ppat.1002345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu J., Li M., Wilkins J., Ding S., Swartz T. H., Esposito A. M., Zheng Y. M., Freed E. O., Liang C., Chen B. K., and Liu S. L. (2015) IFITM proteins restrict HIV-1 infection by antagonizing the envelope glycoprotein. Cell Rep. 13, 145–156 10.1016/j.celrep.2015.08.055 [DOI] [PMC free article] [PubMed] [Google Scholar]