Abstract
Influenza A virus (IAV) effectively manipulates host machinery to replicate. There is a growing evidence that an optimal acetylation environment in the host cell is favorable to IAV proliferation and vice versa. The histone deacetylases (HDACs), a family of 18 host enzymes classified into four classes, are central to negatively regulating the acetylation level, hence the HDACs would not be favorable to IAV. Indeed, by using the RNAi and overexpression strategies, we found that human HDAC4, a class II member, possesses anti-IAV properties and is a component of host innate antiviral response. We discovered that IAV multiplication was augmented in HDAC4-depleted cells and abated in HDAC4-supplemented cells. Likewise, the expression of IFITM3, ISG15, and viperin, some of the critical markers of host anti-IAV response was abated in HDAC4-depleted cells and augmented in HDAC4-supplemented cells. In turn, IAV strongly antagonizes the HDAC4, by down-regulating its expression both at the mRNA level via viral RNA endonuclease PA-X and at the polypeptide level by inducing its cleavage via host caspase 3 in infected cells. Such HDAC4 polypeptide cleavage resulted in a ∼30 kDa fragment that is also observed in some heterologous systems and may have a significant role in IAV replication.
Keywords: influenza virus, histone deacetylase 4 (HDAC4), innate immunity, host-pathogen interaction, signal transducers and activators of transcription 1 (STAT1), host defense, IFITM3, ISG15, PA, PA-X, viperin
Introduction
Influenza virus is an ever-evolving human pathogen, and continues to be a concern for global public as well as animal health. Influenza virus impacts the global human population in several ways, namely by causing 1) regular seasonal epidemics, which alternate in Northern and Southern hemispheres; 2) intermittent unpredictable pandemics; and 3) zoonotic outbreaks, which have become more frequent lately. All of these events result in significant morbidity and mortality as well as productivity and economic losses worldwide (1–5). The zoonotic outbreaks of newly-emerged avian influenza A virus (IAV)2 subtypes in humans result in an unusually high mortality rate (∼35–50%). Furthermore, they pose the threat of the emergence of a highly virulent pandemic IAV. These events are aided by the segmented nature of influenza virus RNA genome and constant circulation of influenza viruses in various clinical and reservoir hosts globally (6). Consequently, the individual infections and co-infections of each host with influenza virus give rise to genetically diverse influenza progeny populations due to de novo mutations and genetic reassortments. Such rapid evolution of influenza viruses has precluded the development of a universal influenza virus vaccine and makes the annually formulated influenza vaccines only variably effective (7). This phenomenon also aided the rapid emergence of drug resistance. Consequently, half of the available anti-influenza virus drugs, i.e. adamantanes, have become practically obsolete and the other half, i.e. neuraminidase, inhibitors prone to be ineffective over time (8). All these influenza virus characteristics combined also mean that it will be practically impossible to eradicate, particularly the type A influenza viruses from nature. Therefore, there is an undeniable need to identify the missing links, both host and viral, that are critical for influenza virus multiplication and pathogenesis to aid the development of alternative, effective and long-lasting anti-influenza virus strategies. One such strategy is to identify and target the naturally occurring antiviral defenses already present in the host cell. These defenses play a pivotal role as part of the host antiviral response by targeting nearly every stage of virus life cycle. Influenza virus is potentially less likely to evolve rapidly against the host-directed therapies.
We have identified and characterized a role of multiple human histone deacetylases (HDACs) in IAV infection (9–12). The HDACs are a family of enzymes that were originally described to catalyze the deacetylation of acetylated histones (13). Now, a variety of nonhistone proteins, both cytoplasmic and nuclear are known to be the HDAC substrates (13). HDACs work in equilibrium with histone acetyltransferases to control the level of protein acetylation, a post-translational modification, and influence diverse biological processes like gene expression (14), protein trafficking (15), and the innate immune response (16). Consequently, the imbalance in protein acetylation due to aberrant function of HDACs or histone acetyltransferases contributes to multiple human diseases, such as cancer (17), neurodegeneration (18), and infection (19, 20). Since our first observation in 2009 (21), an important role of host acetylation machinery in IAV infection is also emerging (22–25).
So far, 18 HDACs have been identified in the human HDAC family and classified into four main classes based on sequence similarities to yeast proteins (13). The class I comprises four members, HDAC1, -2, -3, and -8. The class II has been subclassified into class IIa and class IIb with four members (HDAC4, -5, -7, -9) and two members (HDAC6 and -10), respectively. The class III possesses seven members, which are commonly known as sirtuins (SIRT 1–7). Finally, class IV is comprised of only one member, HDAC11. We and others have demonstrated that at least one member of each class/subclass (i.e. HDAC1, -2, -6, -11, and SIRT 1–7) possesses anti-IAV properties (9–12, 26). We have also demonstrated that, in turn, IAV antagonizes the antiviral function of identified HDACs by employing various strategies, such as down-regulation of their expression at mRNA level (e.g. HDAC11 (12)) and facilitating their degradation by host proteasome machinery (e.g. HDAC1 and HDAC2, Ref. 10, 11). However, a role of class IIa members in IAV infection has not been investigated. Here we demonstrate that HDAC4 (a class IIa member) possesses anti-IAV properties and, in turn, IAV antagonizes the antiviral function of HDAC4, rather strongly, by down-regulating its mRNA level via viral protein PA-X as well as facilitating the proteolytic degradation of its polypeptide by host caspases.
Results
Human HDAC4 possesses the anti-IAV properties
To investigate a role for HDAC4 in IAV infection, we employed the HDAC4 depletion and overexpression strategies. For depletion of HDAC4 expression, we employed RNAi and obtained a predesigned small interfering RNA (siRNA) oligonucleotide targeting human HDAC4. The A549 cells were transfected with different nanomolar (nm) quantities of HDAC4-targeting siRNA and a nontargeting control siRNA. After 72 h, the depletion of HDAC4 polypeptide was analyzed and quantified by Western blotting. To quantify the depletion efficiency, the intensity of HDAC4 and protein-disulfide isomerase (PDI, as loading control) polypeptide bands on blots was quantified using Image Studio Lite software (version 5.0, LI-COR). Then, the amount of HDAC4 was normalized with the corresponding PDI amount. Finally, the normalized amount of HDAC4 polypeptide in control siRNA-transfected cells was considered 100% to compare its level in HDAC4 siRNA-transfected cells. We found that 1 nm quantity of HDAC4 siRNA was sufficient to deplete HDAC4 polypeptide expression by >98% (Fig. 1, A, UNI/HD4 lane, and B), without significantly influencing the cell viability (Fig. 1C). For further experiments, 1 nm concentration of HDAC4, as well as control siRNA was used to deplete HDAC4 expression. Next, A549 cells, transfected with HDAC4 siRNA or control siRNA were either infected with influenza virus A/PR/8/34(H1N1) strain (henceforth referred to as PR8) or influenza A/California/07/2009(H1N1) strain (henceforth referred to as CA09) at a multiplicity of infection (m.o.i.) of 1.0. After 24 h, the culture medium and the infected cells were harvested separately. The culture medium was either titrated by microplaque assay on MDCK cells or analyzed by Western blotting detection of viral hemagglutinin (HA) and nucleoprotein (NP). The former was employed to quantify the amounts of released infectious viral progeny, whereas the latter was employed to measure the released total viral progeny. On the other hand, the infected cells were processed either for Western blot to detect the intracellular level of HDAC4, viral NP, or PDI polypeptides or for quantitative real-time PCR (qPCR) to measure the intracellular level of viral NP and M mRNAs. By employing these methods, we found that the growth of IAV was higher in HDAC4-depleted cells compared with control cells (Fig. 1, D–H), indicating an antiviral function of host HDAC4. Specifically, there was a significant 2.6- (p = 0.0123) and 2.7-fold (p = 0.0088) increase in the release of PR8 and CA09 infectious progeny, respectively, from HDAC4-depleted cells than the control cells (Fig. 1D). Similar results were obtained when PR8 progeny release was measured from the infected cells treated with HDAC inhibitors, trichostatin A and SAHA (Fig. S1). Furthermore, the ratio of viral NP and HA levels detected by Western blotting (Fig. 1E) in culture medium harvested from the control cells and HDAC4-depleted cells was 1:2.5 (n = 3, p = 0.006) and 1:1.7 (n = 3, p = 0.001), respectively. Furthermore, there was a significant 1.9- (p = 0.0087) and 3.8-fold (p = 0.0158) increase in the intracellular level of PR8 NP and M mRNAs, respectively, in HDAC4-depleted cells compared with control cells (Fig. 1F). Likewise, the intracellular level of CA09 NP and M mRNAs was also increased by significant 1.8- (p = 0.0004) and 3.0-fold (p = 0.048), respectively, in HDAC4-depleted cells compared with control cells (Fig. 1F). Finally, there was a significant 1.9-fold (p = 0.027) increase in the intracellular NP polypeptide level in HDAC4-depleted cells compared with control cells (Fig. 1G). The immunofluorescent staining of viral NP and host cell nuclei after 12 h of infection followed by cell counting was used to ascertain that a similar percentage of HDAC4 siRNA-transfected cells (71%, n = 4, p = <0.0001) and control siRNA-transfected cells (69%, n = 4, p = <0.0001) were infected with PR8. The depletion of HDAC4 at the polypeptide level in infected cells was confirmed by Western blotting (Fig. 1A, INF/HD4 lane). However, we noticed that, after 24 h of infection, the level of HDAC4 polypeptide in infected cells (Fig. 1A, INF/CT lane) was much lower than its level in uninfected cells (Fig. 1A, UNI/CT lane) (see Fig. S1). This indicated that, like HDAC1 (10) and HDAC2 (11), IAV is also inducing the down-regulation of HDAC4 polypeptide in infected cells to antagonize its antiviral function. Such HDAC4 polypeptide down-regulation is potentially the reason we observed a modest increase in IAV growth characteristics in HDAC4-depleted infected cells when compared with control infected cells above. IAV on its own was able to deplete the HDAC4 level in control cells hence skewing the real comparison with HDAC4-depleted cells. Before further investigating the HDAC4 down-regulation and its mechanism in response to IAV infection, we analyzed the IAV growth kinetics in HDAC4-depleted cells infected with a lower m.o.i. to minimize the extent of HDAC4 antagonism in control cells and aid the comparison. For this, A549 cells, transfected with HDAC4 or control siRNA as above, were infected with PR8 at a m.o.i. of 0.1. Subsequently, the release of viral progeny in culture medium was quantified by microplaque assay after 6, 12, and 24 h of infection. Consistent with above data, IAV exhibited faster growth kinetics in HDAC4-depleted cells compared with control cells (Fig. 1H). But, here the magnitude of the amount of released viral progeny from HDAC4-depleted cells was significantly increased to over 4.4-fold (p = <0.0001) than control cells after 24 h infection (Fig. 1H).
Figure 1.
The knockdown of HDAC4 expression promotes IAV infection. A–F, A549 cells were transfected with 1 nm control siRNA or HDAC4 siRNA for 72 h. The cells were then infected with IAV PR8 or CA09 strains at a m.o.i. of 1.0. After 24 h, the cells and the medium was harvested separately. A, total lysates of uninfected (UNI) and PR8-infected (INF) cells transfected with control siRNA (CT) or HDAC4 siRNA (HD4) were prepared, and the HDAC4 (140 kDa), PDI (57 kDa), and viral NP (56 kDa) polypeptides were detected by Western blotting. Note: the HDAC4 blot was reused to probe for PDI and NP, and the MW, UNI/CT, and UNI/HD4 lanes were combined with INF/CT and INF/HD4 lanes after cropping out the less relevant middle lanes of same blot. B, the HDAC4 and PDI bands in panel A were quantified using the Image Studio Lite software (LI-COR). The level of HDAC4 polypeptide in each sample was then normalized with corresponding PDI levels. Finally, the normalized level of HDAC4 polypeptide in control siRNA-transfected cells was considered 100% to compare its level in HDAC4 siRNA-transfected cells. C, A549 cells were transfected with either Lipofectamine (LF) alone or in complex with control siRNA or HDAC4 siRNA. After 72 h, the viability of cells was determined by MTT assay. Then, the viability of Lipofectamine only transfected cells was considered 100% to compare the viability of the cells transfected with control siRNA and HDAC4 siRNA. D, the culture medium from the cells transfected with control siRNA or HDAC4 siRNA and infected with IAV PR8 or CA09 strains was titrated on MDCK cells by microplaque assay. Then, the amount of viral progeny released from control siRNA-transfected cells was considered 1-fold to compare the amount of viral progeny released from HDAC4 siRNA-transfected cells. E, the total culture medium from the cells transfected with control siRNA or HDAC4 siRNA and infected with PR8 was concentrated by TCA precipitation, and the levels of viral HA (68 kDa) and viral NP were detected by Western blotting. Note: the HA blot was reused to probe for NP, and the MW lane was combined with CT and HD4 lanes after cropping out the less relevant middle lanes of same blot. F, total RNA from the cells transfected with control siRNA or HDAC4 siRNA and infected with PR8 or CA09 was isolated, and the levels of viral NP, viral M, and actin mRNAs were detected by qPCR. Then, the levels of NP and M mRNAs were normalized with the level of corresponding actin mRNA. Finally, the normalized levels of each viral mRNA in control siRNA-transfected cells were considered 1-fold to compare their levels in HDAC4 siRNA-transfected cells. G, the NP polypeptide level in infected cells in panel A was quantified and normalized as in panel B. Then, the normalized level of NP in control siRNA-transfected cells was considered 1-fold to compare its level in HDAC4 siRNA-transfected cells. H, A549 cells, transfected with control siRNA or HDAC4 siRNA for 72 h, were infected with PR8 at a m.o.i. of 0.1. After 6, 12, and 24 h of infection, the culture medium was titrated by microplaque assay to determine the titers of released viral progeny. Error bars represent the mean ± S.E. of three independent experiments (B, D, F, and G), biological replicates (H), or technical replicates (C). The asterisks represent p values mentioned in the text calculated by unpaired t test (B, F, and G) or ANOVA (C, D, and H), and indicate the significant differences in means. MW, molecular weight; CT, control siRNA; HD4, HDAC4 siRNA.
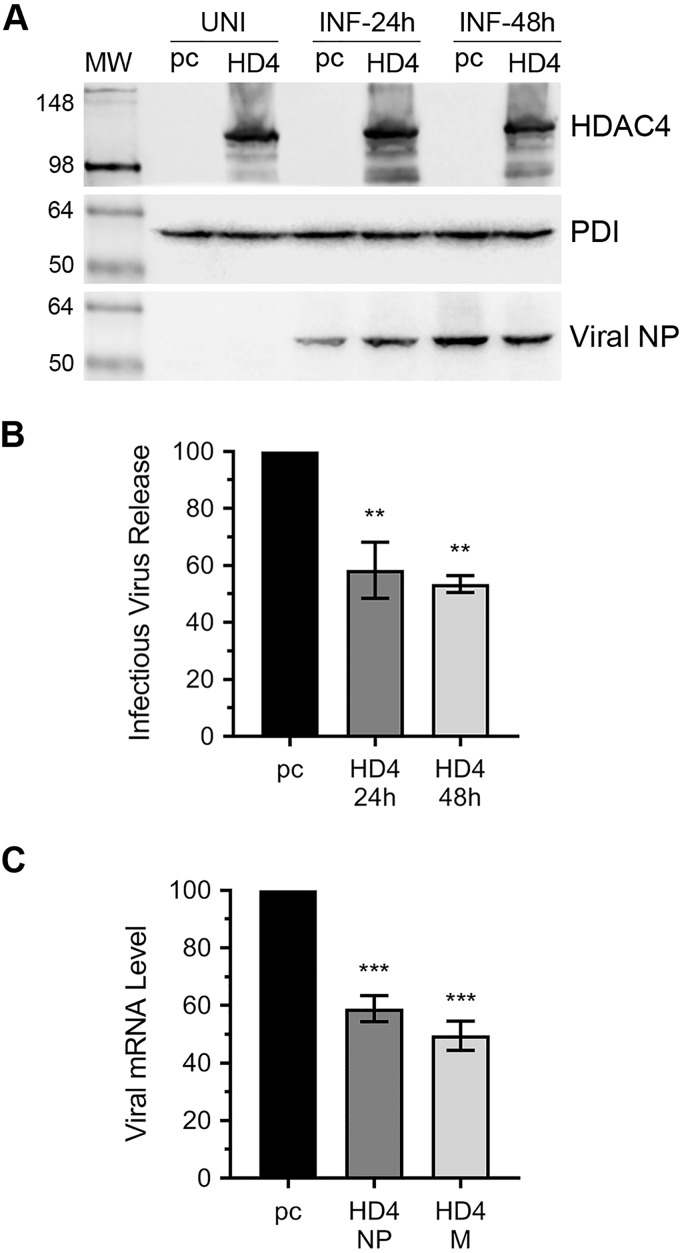
To further validate that host HDAC4 possesses anti-IAV properties, we analyzed IAV growth characteristics in A549 cells overexpressing human HDAC4 from a plasmid. However, like endogenously-expressed HDAC4 polypeptide, the level of plasmid-expressed HDAC4 polypeptide was also significantly decreased in A549 (and MDCK) cells after IAV infection. Therefore, we switched to HEK-293T cells, and transfected them with HDAC4 plasmid or an empty plasmid and subsequently infected them with PR8 at a m.o.i. of 1.0. After 24 and 48 h of infection, the culture medium was titrated by microplaque assay to quantify released infectious PR8 progeny and the cell lysates were analyzed by Western blotting to confirm HDAC4 overexpression. In HEK-293T cells, the level of ectopically-expressed HDAC4 polypeptide under the uninfected and infected conditions were comparable although there was sign of its degradation under the latter (Fig. 2A). Consistent with the results obtained with HDAC4 depletion, overexpression of HDAC4 inhibited the IAV infection. Specifically, compared with control cells, there was a significant 41.6 (p = 0.004) and 46.5% (p = 0.002) decrease in the release of PR8 progeny from HDAC4-overexpressing cells after 24 and 48 h of infection, respectively (Fig. 2B). Furthermore, compared with control cells, there was a significant 41 (p = 0.0006) and 50.4% (p = 0.0002) decrease in the intracellular level of viral NP and M mRNAs, respectively, in HDAC4-overexpressing cells after 24 h of infection (Fig. 2C).
Figure 2.
The overexpression of HDAC4 inhibits IAV infection. A–C, HEK-293T cells were transfected with either empty plasmid pcDNA3 (pc) or HDAC4 (HD4) plasmid for 24 h. The cells were then either harvested as uninfected (UNI) sample or infected (INF) with PR8 at a m.o.i. of 1.0. After 24 or 48 h of infection, the culture medium and the cells were harvested separately. A, total cell lysates were prepared, and the HDAC4, PDI, and viral NP polypeptides were detected by Western blotting. Note: the HDAC4 blot was reused to probe for PDI and NP. B, the culture medium was titrated on MDCK cells by microplaque assay. Then, the amount of viral progeny released from pcDNA3-transfected cells (pc) was considered 100% to compare the amount of viral progeny released from HDAC4-transfected cells (HD4) after 24 and 48 h of infection. C, total RNA from the cells infected for 24 h as above was isolated, and the viral NP, viral M, and actin mRNA levels were detected by qPCR. Then, the level of NP and M mRNAs were normalized with the level of corresponding actin mRNA. Finally, the normalized levels of each viral mRNA in pcDNA3-transfected cells (pc) were considered 100% to compare their levels in the HDAC4-transfected cells (HD4). Error bars represent the mean ± S.E. of three independent experiments (B) or biological replicates (C). The asterisks represent p values mentioned in the text calculated by ANOVA, and indicate significant differences in means. MW, molecular weight.
IAV down-regulates the HDAC4 polypeptide level in mammalian cells
As mentioned above, the levels of both endogenously- and ectopically-expressed HDAC4 polypeptides were down-regulated in A549 cells in response to IAV infection. We next endeavored to understand the kinetics as well as the mechanism of this phenomenon. To understand the former, A549 or MDCK cells were infected with PR8 at a m.o.i. of 0.5 or 5.0. After 24 h, the HDAC4 polypeptide levels in total cell lysates were analyzed by Western blotting. We found that, indeed, IAV down-regulated the HDAC4 polypeptide level in both MDCK (Fig. 3A) and A549 (Fig. 3B) cells in a dose-dependent manner. To quantify the extent of HDAC4 polypeptide down-regulation, the intensity of HDAC4 and actin (loading control) bands was quantified as above. Then, the amount of HDAC4 was normalized with a corresponding actin amount. Finally, the normalized amount of HDAC4 in uninfected cells was considered 100% to compare its amount in infected cells. We found that, in MDCK cells, there was a significant dose-dependent 53.8 (p = 0.03) and 70.5% (p = 0.02) decrease in HDAC4 polypeptide level in response to infection with PR8 at m.o.i. of 0.5 and 5.0, respectively (Fig. 3F). Likewise, in A549 cells, there was a significant dose-dependent 40 (p = 0.04) and 63.7% (p = 0.03) decrease in HDAC4 polypeptide level in response to infection with PR8 at m.o.i. of 0.5 and 5.0, respectively (Fig. 3F). Next, to determine whether the down-regulation of HDAC4 polypeptide in response to IAV infection was strain-independent, MDCK cells were infected with IAV CA09 or WSN strains at a m.o.i. of 0.5 or 5.0. Subsequently, the level of HDAC4 polypeptide was detected and quantified as above. Consistent with above results, both CA09 (Fig. 3C) and WSN (Fig. 3D) strains down-regulated the HDAC4 polypeptide level in a dose-dependent manner. Specifically, there was 55.3 (n = 1) and 86.3% (n = 1) reduction in the HDAC4 polypeptide level in response to infection with CA09 at m.o.i. of 0.5 and 5.0, respectively. Similarly, the level of HDAC4 polypeptide was reduced by 56.7 (n = 1) and 78.8% (n = 1) in the cells infected with WSN at m.o.i. of 0.5 and 5.0, respectively. Finally, by using an UV-irradiated PR8 inoculum, we confirmed that a replication-competent IAV was required to down-regulate the HDAC4 polypeptide level in infected cells (Fig. 3E). Next, to determine the kinetics of HDAC4 polypeptide down-regulation, MDCK cells were infected with PR8 at a m.o.i. of 0.5, and subsequently harvested after 2, 6, 12, and 24 h after infection. The level of HDAC4 polypeptide at each time point was then analyzed and quantified as above. Here, the level of HDAC4 polypeptide decreased with the progress of infection (Fig. 3G), and it was at its minimum, a significantly low 16.8% (p = 0.006) compared with control (Fig. 3H), after 24 h infection. In addition, concurrent to the disappearance of full-length HDAC4 polypeptide, we also observed the appearance of at least two smaller polypeptide fragments, one resolving around the 98-kDa mark and the other resolving around the 36-kDa mark on the HDAC4 blot, between 12 and 24 h infection (Fig. 3G, arrows). Incidentally, the appearance of these two HDAC4 polypeptide fragments coincided with already known (9) caspase-mediated cleavage of viral NP (Fig. 3G) as well as the proteolytic activation of caspase 3 in IAV-infected cells (Fig. 3I). This suggested that the HDAC4 polypeptide is undergoing a caspase-mediated proteolytic degradation in IAV-infected cells.
Figure 3.
IAV down-regulates the level of HDAC4 polypeptide in epithelial cells. MDCK (A and C) or A549 (B and D) cells were infected with IAV PR8 (A and B), CA09 (C), or WSN (D) strains at a m.o.i. of 0.5 or 5.0. After 24 h, the uninfected (UNI) and infected (0.5, 5.0) cells were harvested, total cell lysates were prepared, and the HDAC4, actin (42 kDa), and viral NP polypeptides were detected by Western blotting. E, MDCK cells were infected with live (INF) or UV-irradiated (INFUV) PR8 at an m.o.i. of 0.5. After 24 h, total cell lysates were prepared, and the HDAC4, actin, and NP polypeptides were detected by Western blotting. Note: the HDAC4 blots were reused to probe for actin. F, the level of HDAC4 polypeptide in panels A and B blots were quantified and normalized as described in the legend to Fig. 1B. Then, the normalized level of HDAC4 polypeptide in uninfected cells (UNI) was considered 100% to compare its level in cells infected (INF) with 0.5 and 5.0 m.o.i. G, MDCK cells were infected with PR8 at a m.o.i. of 0.5, and harvested after 2, 6, 12, and 24 h of infection. The total cell lysates were resolved on 7–12% gradient SDS-PAGE, and the HDAC4, PDI, and NP polypeptides were detected by Western blotting. Arrows indicate the HDAC4 polypeptide cleavage products. Note: the HDAC4 blot was reused to probe for PDI, and the MW lane was combined with the rest of the lanes after cropping out the inconsistent second lane of same blot. H, the level of full-length HDAC4 polypeptide in panel G was quantified and normalized as described in the legend to Fig. 1B. Then, the normalized level of HDAC4 polypeptide at 2 h infection was considered 100% to compare its levels at 6, 12, and 24 h infection. I, MDCK cells were infected with PR8 and harvested at the times indicated in panel G. Then, the full-length caspase 3 (Cas3-FL) and cleaved caspase 3 (Cas3-CL) polypeptides were detected in total cell lysates by Western blotting. Note: the MW lane was combined with rest of the lanes after cropping out the less relevant middle lanes of same blot. Error bars represent the mean ± S.E. of three independent experiments. The asterisks represent p values mentioned in the text calculated by unpaired t test (F) or ANOVA (H), and indicate the significant differences in means. MW, molecular weight.
The caspase 3 cleaves HDAC4 polypeptide in infected cells
Next, we endeavored to further understand the mode of proteolytic degradation of HDAC4 polypeptide in IAV-infected cells. Primarily, lysosome-mediated and proteasome-mediated pathways facilitate the degradation of polypeptides in mammalian cells. To identify the pathway leading to the degradation of HDAC4 polypeptide in IAV-infected cells, we perturbed these two pathways. For this, MDCK cells were infected with PR8 as above and subsequently treated with lysosome inhibitor, NH4Cl, or proteasome inhibitor, MG132. In addition, one set of infected cells was also treated with a caspase 3 inhibitor (Cas3-I). After 24 h, the HDAC4 polypeptide levels were analyzed and quantified as described above. We found that the treatments with NH4Cl and Cas3-I rescued the HDAC4 polypeptide level in infected cells. Specifically, compared with 81% (p = <0.0001) reduction in the HDAC4 polypeptide level in mock-treated infected cells, there was only 54 and 46% reduction in HDAC4 polypeptide levels in NH4Cl–treated and Cas3-I–treated infected cells, respectively (Fig. 4B). In other words, the level of HDAC4 polypeptide in infected cells recovered from 19 to 46% (statistically nonsignificant) and a significant 54% (p = 0.02) after the treatment with NH4Cl and Cas3-I, respectively (Fig. 4B). Such recovery in the HDAC4 polypeptide level in Cas3-I–treated infected cells also coincided with the inhibition of proteolytic activation of caspase 3 in infected cells (Fig. 4A, last panel, Cas3-I/INF lane). Furthermore, the proteolytic activation of caspase 3 was also slightly reduced in NH4Cl–treated infected cells (Fig. 4A, last panel, compare Mock/INF and NH4Cl/INF lanes). This indicated that the proteolytic activation of caspase 3 was associated with lysosomal activity. On the contrary, treatment with MG132 worsened the degradation of HDAC4 polypeptide in infected cells (Fig. 4A, MG132/INF lane). Specifically, compared with 81% (p = <0.0001) reduction in the HDAC4 polypeptide level in mock-treated infected cells, there was a significant 94.4% (p = 0.0039) reduction in the HDAC4 polypeptide level in MG132–treated infected cells (Fig. 4B). The MG132 treatment even induced the HDAC4 polypeptide degradation in uninfected cells (Fig. 4A, MG132/UNI lane). This effect of MG132 treatment on the HDAC4 polypeptide level, both in uninfected and infected cells, further indicated that the HDAC4 polypeptide is degraded by caspases. It is known that MG132 treatment induces the apoptosis, hence the activation of caspases in mammalian cells (27). Consistent with this, we observed a rather profound proteolytic activation of caspase 3 both in uninfected and infected cells treated with MG132 (Fig. 4A, last panel, MG132/UNI and MG132/INF lanes). Interestingly, MG132 treatment of infected cells also increased the Western blot detection limit and revealed that the HDAC4 polypeptide was actually being cleaved into two fragments, resolving around the 36-kDa marker (indicated by arrows a and b in Fig. 4A) instead of one observed in mock–treated infected cells. To further confirm the role of caspase 3 in HDAC4 polypeptide degradation in response to IAV infection, we knocked down the expression of caspase 3 in A549 cells by RNAi. Then, A549 cells were infected with PR8 and the level of HDAC4 polypeptide was analyzed and quantified by Western blotting as described above. Consistent with the above observations, the knockdown of caspase 3 expression (>90%, confirmed by qPCR, Fig. 4E) rescued the level of HDAC4 polypeptide in infected cells (Fig. 4C). Specifically, compared with 77% (p = <0.0001) reduction in the HDAC4 polypeptide level in infected cells transfected with control siRNA, there was only 28% (p = 0.004) reduction in the HDAC4 polypeptide level in infected cells transfected with caspase 3 siRNA (Fig. 4D). In other words, the level of HDAC4 polypeptide in infected cells recovered from 22.7% to a significant 71.7% (p = 0.0002) after the depletion of caspase 3 expression (Fig. 4D).
Figure 4.
The caspase 3 cleaves HDAC4 polypeptide in infected cells. A, MDCK cells were infected with PR8 at a m.o.i. of 0.5 and, after removing the virus inoculum treated with NH4Cl (10 mm), MG132 (20 μm), or caspase 3 inhibitor (Cas3-I; 40 μm). After 24 h, the uninfected (UNI) and infected (INF) cells were harvested, total cell lysates were prepared, and the HDAC4, PDI, viral NP, and full-length caspase 3 (Cas3-FL) and cleaved caspase 3 (Cas3-CL) polypeptides were detected by Western blotting. Note: the HDAC4 blot was reused to probe for PDI and NP. Arrows indicate the HDAC4 polypeptide cleavage products. B, the full-length HDAC4 polypeptide level in panel A was quantified and normalized as described in the legend to Fig. 1B. Then, the normalized level of HDAC4 polypeptide in mock-treated uninfected (UNI) cells was considered 100% to compare its levels in mock-, NH4Cl-, MG132-, and Cas3-I-treated infected (INF) cells. C, A549 cells were transfected in duplicate with 10 nm control siRNA (CT) or caspase 3 siRNA (Cas3) for 72 h. One set of cells was infected with PR8 at a m.o.i. of 1.0. After 24 h, cells were harvested and the HDAC4, NP, and actin polypeptides were detected in total cell lysates by Western blotting. Note: the HDAC4 blot was reused to probe for actin and NP. Arrows indicate the HDAC4 polypeptide cleavage products in the higher exposure of the dotted rectangle. D, the full-length HDAC4 polypeptide level in panel C was quantified and normalized as described in the legend to Fig. 1B. Then, the normalized level of HDAC4 polypeptide in uninfected/control siRNA-transfected cells (UNI/CT) was considered 100% to compare its levels in infected/control siRNA-transfected cells (INF/CT) and infected/caspase 3 siRNA-transfected cells (INF/Cas3). E, the second set of cells from panel C was processed to detect the levels of caspase 3 and actin mRNAs by qPCR. Then, the levels of caspase 3 mRNA in control siRNA-transfected cells and caspase 3 siRNA-transfected cells were normalized with corresponding actin mRNA levels. Finally, the normalized level of caspase 3 mRNA in control siRNA-transfected cells (CT) was considered 100% to compare its level in caspase 3 siRNA-transfected cells (Cas3). Error bars represent the mean ± S.E. of three independent experiments. The asterisks represent p values mentioned in the text calculated by ANOVA (B and D) or unpaired t test (E), and indicate the significant differences in means. MW, molecular weight.
The IAV protein PA-X is involved in down-regulation of HDAC4 in infected cells
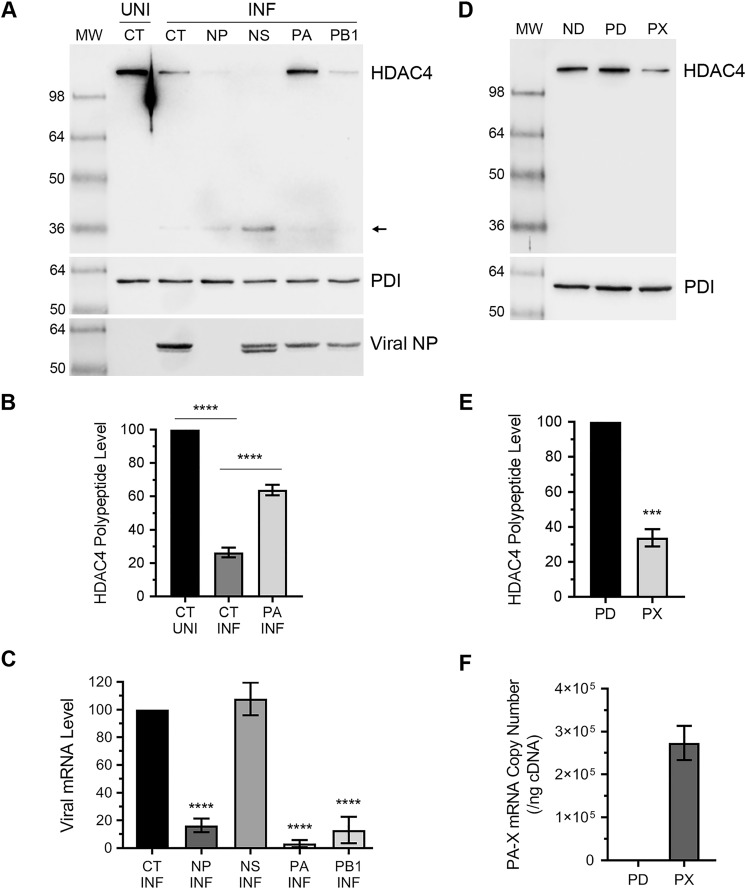
To further understand the mechanism of HDAC4 antagonism, we next endeavored to identify the viral gene(s) IAV is employing to down-regulate the HDAC4 expression in infected cells. To accomplish this, we employed RNAi to knockdown the expression of 6 internal IAV genes: M, NP, NS, PA, PB1, and PB2, by using the previously described siRNAs (28). However, the M and NS siRNAs did not work in our hands and potency of PB2 siRNA could not be confirmed. Nevertheless, we delivered control, NP, NS (as negative control), PA, and PB1 siRNAs to A549 cells and subsequently infected them with PR8 at a m.o.i. of 1.0. Then, the levels of HDAC4 polypeptide in each sample were analyzed and quantified by Western blotting as described above. Of the viral genes that were targeted, only the knockdown of PA gene expression rescued the level of HDAC4 polypeptide in infected cells (Fig. 5A). Specifically, compared with 73.6% (p = <0.0001) reduction in the HDAC4 polypeptide level in infected cells transfected with control siRNA, there was only 36% (p = 0.0001) reduction in the HDAC4 polypeptide level in infected cells transfected with PA siRNA (Fig. 5B). In other words, the level of HDAC4 polypeptide in infected cells recovered from 26.4% to a significant 63.8% (p = <0.0001) after the knockdown of PA expression (Fig. 5B). The knockdown in NP, PA, and PB1 mRNA expression by a significant 83.7, 96.7, and 87% (p = <0.0001), respectively, was confirmed by qPCR (Fig. 5C). The depletion of NP polypeptide was also confirmed by Western blotting (Fig. 5A). Although knockdown of PA and PB1 expression at the mRNA level above reduced the viral gene expression (as indicated by reduced NP polypeptide level detected in Fig. 5A) and the yield of released viral progeny (not shown), it did not fatally affect the viral growth. Therefore, this and nonworking NS siRNA helped to conclude that the rescue of HDAC4 polypeptide in PA mRNA knockdown cells was specific. Next, we wondered whether IAV PA-X was involved in the down-regulation of HDAC4 expression. The PA-X is a second product of PA mRNA resulting from the ribosomal frameshifting during translation (29), and is implicated in shutoff of host gene expression (30) and consequently dampening of host innate anti-IAV response. We expressed PA-X in A549 cells from a plasmid (31) and subsequently analyzed and quantified the HDAC4 polypeptide level by Western blotting as described above. Indeed, like IAV infection, the expression of PA-X alone down-regulated the HDAC4 polypeptide level in A549 cells (Fig. 5D). Specifically, when compared with the cells transfected with empty plasmid, there was a significant 66% (p = 0.0002) reduction in the HDAC4 polypeptide level in cells transfected with PA-X plasmid (Fig. 5E). The expression of PA-X in A549 cells was confirmed by absolute qPCR (Fig. 5F).
Figure 5.
The IAV protein PA-X is involved in down-regulation of HDAC4 polypeptide. A–C, A549 cells were transfected in duplicate with 50 nm control siRNA (CT) or viral NP, NS, PA, or PB1 siRNA for 24 h. Cells were then infected with PR8 at a m.o.i. of 1.0 for further 24 h. A, one set of cells was harvested and the HDAC4, PDI, and NP polypeptides were detected in total uninfected (UNI) and infected (INF) cell lysates by Western blotting. Note: the HDAC4 blot was reused to probe for PDI and NP. B, the full-length HDAC4 polypeptide levels in UNI/CT, INF/CT, and INF/PA lanes of panel A were quantified and normalized as described in the legend to Fig. 1B. Then, the normalized level of the HDAC4 polypeptide in uninfected/control siRNA-transfected (UNI/CT) cells was considered 100% to compare its levels in infected/control siRNA-transfected (INF/CT) cells and the PA siRNA-transfected/infected (INF/PA) cells. C, the second set of cells was processed to detect the levels of NP, NS, PA, PB1, and actin mRNAs by qPCR. Then, the levels of viral gene mRNAs in control siRNA-transfected and viral gene siRNA-transfected cells were normalized with corresponding actin mRNA levels. Finally, the normalized levels of viral mRNAs in control siRNA-transfected cells were considered 100% to compare their levels in viral gene siRNA-transfected cells. D–F, A549 cells were transfected in duplicate with no DNA (ND), empty plasmid pcDNA3 (PD), or PA-X plasmid (PX) for 48 h. D, one set of cells were harvested and the HDAC4 and PDI polypeptides were detected in total cell lysates by Western blotting. Note: the HDAC4 blot was reused to probe for PDI. E, the full-length HDAC4 polypeptide level in panel D was quantified and normalized as described in the legend to Fig. 1B. Then, the normalized level of HDAC4 polypeptide in pcDNA3-transfected cells (PD) was considered 100% to compare its level in the PA-X plasmid-transfected cells (PX). F, the second set of cells was processed to determine the PA-X mRNA copy number by absolute qPCR using the PA-X plasmid as a calibrator. For this, PA-X cDNA from PA-X plasmid-transfected cells and 10-fold serial dilutions of PA-X plasmid was amplified by qPCR. In parallel, the number of PA-X cDNA copies in 10-fold serial dilutions were calculated using the known plasmid DNA concentration and plasmid molecular weight. Then, a standard curve of cycle threshold (CT) value from qPCR versus cDNA copy numbers was generated and was used as a reference to calculate the PA-X mRNA copy numbers in PA-X plasmid-transfected cells. Error bars represent the mean ± S.E. of three independent experiments. The asterisks represent p values mentioned in the text calculated by ANOVA (B and C) or unpaired t test (E), and indicate the significant differences in means. MW, molecular weight.
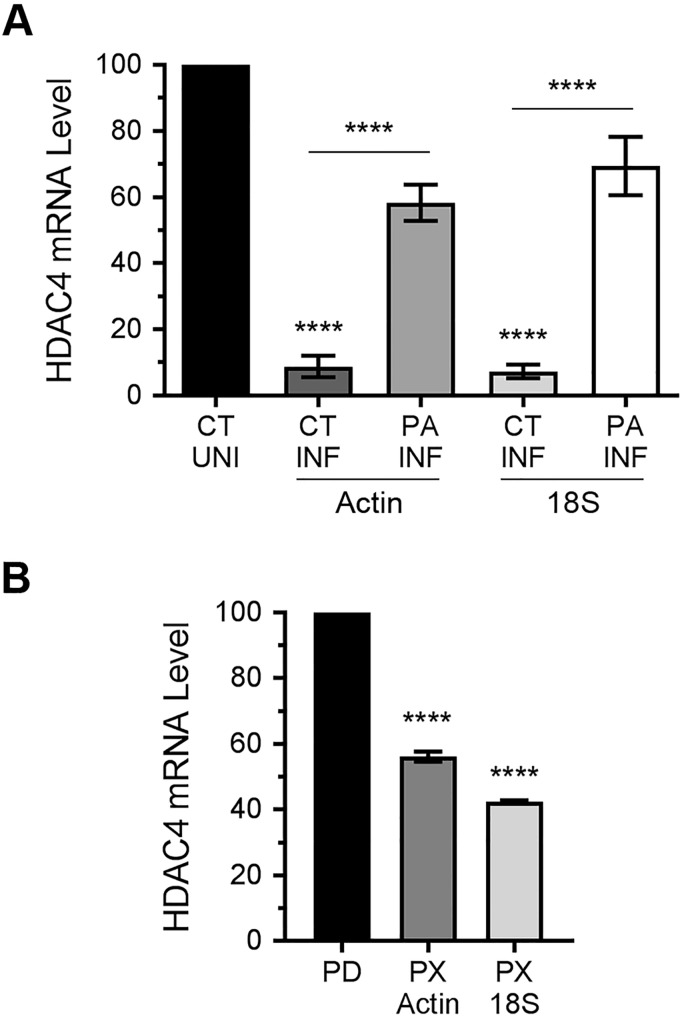
Interestingly, like in infected cells, we could not detect a cleavage product of HDAC4 polypeptide in the cells expressing PA-X alone (compare HDAC4 blots in Fig. 5, A and D). Furthermore, PA-X–mediated down-regulation of full-length HDAC4 polypeptide could not be rescued by Cas3-I treatment (Fig. S2). This indicated that, in addition to down-regulating the HDAC4 expression at the polypeptide level via caspase 3, IAV is also down-regulating HDAC4 expression at the mRNA level; and IAV is accomplishing this by employing PA-X, a RNA endonuclease (29). To investigate this further, A549 cells were transfected with control siRNA or PA-targeting siRNA, and subsequently infected with PR8 at a m.o.i. of 1.0. After 24 h infection, cells were processed to measure the levels of HDAC4 mRNA and actin mRNA and 18S rRNA (18S RNA) as reference by qPCR. Indeed, we discovered that the level of HDAC4 mRNA in A549 cells was reduced in response to IAV infection (Fig. 6A). When normalized to the actin mRNA level and compared with uninfected cells, there was a significant 91% (p = 0.0001) reduction in the HDAC4 mRNA level in infected cells (Fig. 6A). Similarly, when normalized to the 18S RNA level and compared with uninfected cells, there was a significant 92% (p = 0.0001) decrease in the HDAC4 mRNA level in infected cells (Fig. 6A). However, in the infected cells that were transfected with PA siRNA, such reduction in the HDAC4 mRNA level was only 41.6 (p = 0.0005) and 30.6% (p = 0.004) when normalized to actin mRNA and 18S RNA levels, respectively (Fig. 6A). In other words, the level of HDAC4 mRNA in infected cells recovered from 8.7 and 7.2% to a significant 58.3 (p = <0.0001) and 69.4% (p = <0.0001) when normalized to actin mRNA and 18S RNA levels, respectively, after the knockdown of PA mRNA expression (Fig. 6A). Finally, to confirm that PA-X was involved in the down-regulation of HDAC4 mRNA, we expressed PA-X in A549 cells from the plasmid and subsequently analyzed and quantified the HDAC4 mRNA level by qPCR as above. Indeed, like IAV infection, the expression of PA-X alone down-regulated the HDAC4 mRNA level in A549 cells (Fig. 6B). Compared with the cells transfected with empty plasmid, there was a significant 43.8 (p = 0.0001) and 57.6% (p = 0.0001) decrease in the HDAC4 mRNA level in the cells transfected with PA-X plasmid when normalized to actin mRNA and 18S RNA levels, respectively (Fig. 6B).
Figure 6.
IAV protein PA-X down-regulates HDAC4 at mRNA level. A, A549 cells were transfected with the control siRNA or viral PA siRNA and subsequently infected with PR8 at a m.o.i. of 1.0 for 24 h. Then, the levels of HDAC4 mRNA, actin mRNA, and 18S RNA were detected by qPCR. The levels of HDAC4 mRNA in all samples were normalized with corresponding actin mRNA or 18S RNA levels. Finally, the normalized level of HDAC4 mRNA in control siRNA-transfected/uninfected (CT/UNI) cells were considered 100% to compare its levels in the control siRNA-transfected/infected (CT/INF) cells and PA siRNA-transfected/infected (PA/INF) cells. B, A549 cells were transfected with empty plasmid pcDNA3 (PD) or PA-X plasmid (PX) as above, and the levels of HDAC4 mRNA, actin mRNA, and 18S RNA were detected by qPCR. Then, the levels of HDAC4 mRNA in all samples were normalized with corresponding actin mRNA or 18S RNA levels. Finally, the normalized level of HDAC4 mRNA in pcDNA3-transfected cells (PD) were considered 100% to compare its level in the PA-X plasmid-transfected cells (PX). Error bars represent the mean ± S.E. of three independent experiments. The asterisks represent p values mentioned in the text calculated by ANOVA, and indicate the significant differences in means. MW, molecular weight.
HDAC4 is involved in IAV-induced host innate antiviral response
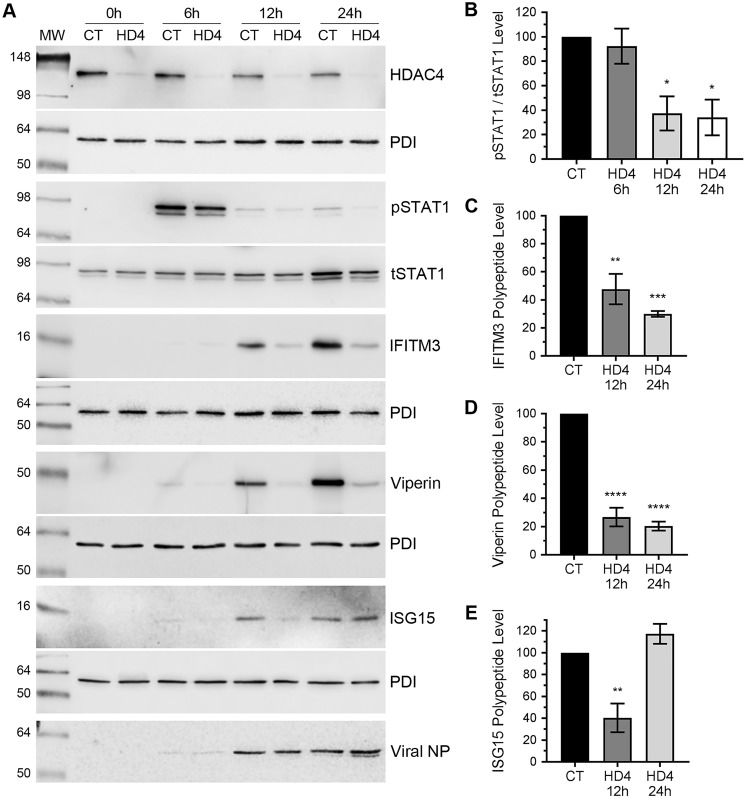
The data presented above demonstrate anti-IAV properties of HDAC4 and, in turn, a strong antagonism of HDAC4, both at mRNA and polypeptide levels, by IAV. Next, we investigated the anti-IAV mechanism of HDAC4. Previously, we have demonstrated that HDAC1, HDAC2, and HDAC11 exert their anti-IAV function through their involvement in IAV-induced host innate antiviral response (10–12). Hence, to get insight into the anti-IAV mechanism of HDAC4, we examined the role of HDAC4 in IAV-induced signaling of signal transducer and activator of transcription 1 (STAT1), one of the main inducers of innate antiviral cascade and subsequent expression of interferon-stimulated genes (ISGs) in IAV-infected cells (32–34). We compared the kinetics of STAT1 phosphorylation and subsequently ISG expression in IAV-infected cells with the normal or depleted expression of HDAC4. For this, A549 cells, transfected with the control siRNA or HDAC4 siRNA, were infected with PR8 at a m.o.i. of 1.0. After 0, 6, 12, and 24 h of infection, cells were processed, and the level of phosphorylated STAT1 (pSTAT1) were analyzed and quantified by Western blotting as described above. We found that there was a noticeable decrease in the pSTAT1 level in HDAC4-depleted cells, particularly after 12 and 24 h of infection (Fig. 7A). Specifically, compared with normal cells, the level of pSTAT1 in HDAC4-depleted cells was reduced by a significant 62.6 (p = 0.018) and 66% (p = 0.013) after 12 and 24 h of infection, respectively (Fig. 7B). This indicated a reduced induction of host innate antiviral response against IAV in the absence of HDAC4. To investigate this further, we compared the expression kinetics of ISGs: interferon-induced transmembrane protein 3 (IFITM3), ISG15, and viperin (also known as RSAD2) polypeptides in control and HDAC4-depleted cells in response to IAV infection. These three ISGs have been directly implicated in the inhibition of IAV infection (35–37). To do this, A549 cells were subjected to the same conditions as used to detect the pSTAT1 level above. Under such conditions, the IAV-induced expression of IFITM3, ISG15, and viperin polypeptides in control cells was first detected after 6 h of infection that reached to a significant level after 24 h of infection (Fig. 7A). However, in HDAC4-depleted cells, the IAV-induced expression of all three polypeptides was reduced at the corresponding time points (Fig. 7A). Particularly, their level in HDAC4-depleted cells were barely detectable after 12 h of infection. We quantified these levels and found that, after 12 h of infection, there was a significant 52 (p = 0.002), 73 (p = 0.0001), and 59.6% (p = 0.006) reduction in IAV-induced expression of IFITM3 (Fig. 7C), viperin (Fig. 7D), and ISG15 (Fig. 7E) polypeptides, respectively, in HDAC4-depleted cells. Similarly, after 24 h of infection, there was a significant 70 (p = 0.0005) and 79.6% (p = 0.0001) reduction in IAV-induced expression of IFITM3 (Fig. 7C) and viperin (Fig. 7D) polypeptides, respectively, in HDAC4-depleted cells. Intriguingly, after 24 of infection, the level of ISG15 polypeptide in HDAC4-depleted cells recovered to its level as in control cells (Fig. 7E).
Figure 7.
The IAV-induced phosphorylation of STAT1 and the expression of ISGs: IFITM3, ISG15, and viperin are reduced in HDAC4-depleted cells. A–E, A549 cells were transfected with 1 nm of either control siRNA (CT) or HDAC4 (HD4) siRNA for 72 h. Cells were then infected with PR8 at a m.o.i. of 1.0. A, after 0, 6, 12, and 24 h of infection, cells were harvested and the total cell lysates were prepared. Then, the pSTAT1 (91/84 kDa), tSTAT1 (91/84 kDa), IFITM3 (15 kDa), ISG15 (15 kDa), viperin (42 kDa), PDI, and viral NP polypeptides were detected by Western blotting. Note: the HDAC4, IFITM3, viperin, and ISG15 blots were reused to probe for corresponding PDI, the pSTAT1 blot was reused to probe for tSTAT1 and viperin blot was reused to probe for NP. B–E, the levels of pSTAT1 and IFITM3, ISG15, and viperin in panel A were quantified and normalized with tSTAT1 and PDI, respectively, as described in the legend to Fig. 1B. Then, the normalized levels of pSTAT1 (B), IFITM3 (C), ISG15 (D), and viperin (E) at each time point in control siRNA-transfected cells were considered 100% to compare their levels in HDAC4 siRNA-transfected cells at the respective time points. Error bars represent the mean ± S.E. of three independent experiments. The asterisks represent p values mentioned in the text calculated by ANOVA, and indicate the significant differences in means. MW, molecular weight.
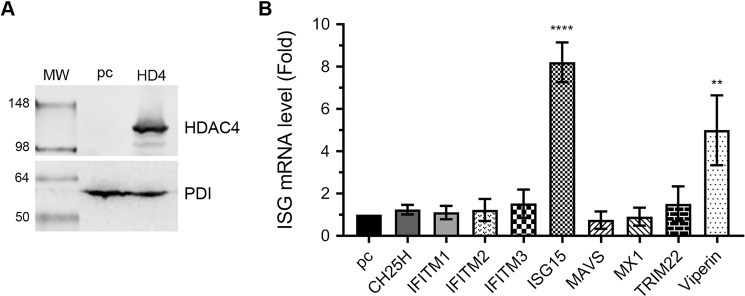
Next, we compared the IAV-induced expression of ISGs in normal and HDAC4-overexpressing cells to conversely examine the involvement of HDAC4 in host innate anti-IAV response. In this experiment, besides IFITM3, ISG15, and viperin, we also examined additional ISGs: cholesterol-25-hydroxylase (CH25H), IFITM1 and -2, myxovirus-resistance protein-1 (MX-1), and tripartite motif protein 22 (TRIM22). These ISGs also have been implicated in IAV infection (32, 33). For this, HEK-293T cells were transfected with either empty plasmid or HDAC4-expressing plasmid. After 24 h, one set of cells was processed to confirm the HDAC4 overexpression by Western blotting (Fig. 8A), whereas the other set of cells was infected with PR8 at a m.o.i. of 1.0. After 6 h of infection, cells were processed, and the levels of mRNA of above ISGs and actin and 18S RNA were detected by qPCR. Consistent with the above findings, when normalized to the actin mRNA level and compared with control cells, there was a significant 8.2- (p = <0.0001) and 5.0-fold (p = 0.007) increase in the mRNA levels of ISG15 and viperin, respectively, in HDAC4-overexpressing cells (Fig. 8B). Similar results were obtained when ISG15 and viperin mRNA levels were normalized to the 18S RNA level (data not shown). Intriguingly, the mRNA level of IFITM3 increased only by 1.5-fold (statistically nonsignificant) in HDAC4-overexpressing cells (Fig. 8B). No noticeable change was observed in the mRNA levels of other ISGs (except TRIM22, by 1.5-fold) in HDAC4-overexpressing cells (Fig. 8B).
Figure 8.
The IAV-induced expression of ISG15 and viperin is augmented in HDAC4-supplemented cells. HEK-293T cells were transfected in duplicate with either empty plasmid pcDNA3 (pc) or HDAC4 (HD4) plasmid for 24 h. A, one set of cells was harvested, total cell lysates were prepared, and the HDAC4 and PDI polypeptides were detected by Western blotting. Note: the HDAC4 blot was reused to probe for PDI. B, the other set of cells was infected with PR8 at a m.o.i. of 1.0 for 6 h. Then, the mRNA levels of the indicated ISGs and actin were detected by qPCR. The levels of each ISGs in both samples were normalized with corresponding actin mRNA levels. Finally, the normalized mRNA levels of each ISG in pcDNA3-transfected cells were considered 1-fold to compare their levels in HDAC4-transfected cells. Error bars represent the mean ± S.E. of three biological replicates. The asterisks represent p values mentioned in the text calculated by ANOVA, and indicate significant differences in means. MW, molecular weight.
Discussion
Acetylation is one of the most ubiquitous post-translation modifications of proteins as it is now known to exist in all three domains of life and control diverse cellular processes (38, 39). Unsurprisingly, a clear role of acetylation in the infection of viruses that cause both acute and persistent infections is also emerging (40). Hence, HDACs, the enzymes central to regulating the acetylation are bound to play a crucial role, either proviral or antiviral, in the virus infections. So far, we and others have demonstrated that at least one member of each class/subclass of HDAC family possesses an anti-IAV property (9–12, 26). On a quest to investigate the interplay of individual HDAC with IAV, we have demonstrated herein the anti-IAV properties of class IIa HDAC, HDAC4, and a rather remarkably strong antagonism of HDAC4 by the IAV. We were able to determine that, compared with control cells, IAV replicated more efficiently in HDAC4-depleted cells and less efficiently in HDAC4-supplemented cells. We analyzed several parameters of IAV multiplication, viz. extracellular (total and infectious) virion release, intracellular viral mRNA and protein levels, and utilized two different IAV H1N1 strains to show that the IAV growth was 2–3–fold higher in HDAC4-depleted cells when infected at m.o.i. of 1.0. One could argue that although statistically significant, this is a modest increase in virus growth. However, one obvious and simple explanation for this could be the profound down-regulation of HDAC4 expression in infected cells. Before the HDAC4 was able to effectively assert its antiviral effect, its expression in infected control cells came down to a level that was only subtly higher to its level in infected cells depleted with HDAC4 (Fig. 1A, compare INF/CT and INF/HD4 lanes). Hence, the resulting difference observed on virus growth could not be as profound. This argument is well-supported by 4.4-fold more virus released from HDAC4-depleted cells when they were infected with a low m.o.i. of 0.1 (Fig. 1H). Furthermore, this argument also finds support from a recent finding where the ectopic expression of HDAC4 protected human cells from vesicular stomatitis virus-induced death, but only when infection was at a lower m.o.i. of 0.1 (41).
IAV antagonized the HDAC4 by down-regulating its expression at both mRNA and polypeptide levels. Although the degradation of HDAC4 in IAV-infected cells is consistent with the degradation of HDAC1 and HDAC2, the recently identified anti-IAV HDACs, in IAV-infected cells (10, 11) their mechanisms of degradation are distinct. Although the HDAC4 is degraded by caspases as demonstrated herein, HDAC1 and HDAC2 are degraded by proteasome as both polypeptides are known to undergo ubiquitination (10, 11). This indicates that IAV is antagonizing various HDACs based upon the inherent characteristics of their polypeptides. Evidently, an earlier report revealed that HDAC4 is highly unstable in several mammalian cell lines, with its mRNA half-life less than 4 h and polypeptide half-life less than 8 h (42). Furthermore, the same report showed that the HDAC4 polypeptide degradation could be further induced by nonviral induction of apoptosis in those cell lines. Hence, IAV, which is known to induce apoptosis in mammalian cells, was able to create a favorable environment for the degradation of the HDAC4 polypeptide in infected cells. This is evident from the degradation kinetics of HDAC4 polypeptide coinciding with the proteolytic activation of caspase 3, one of the executor proteases of apoptotic pathway, in infected cells. Such HDAC4 degradation and caspase 3 activation, and the infection-mediated cell death (Fig. S3), were significantly reversed by a caspase 3 inhibitor. Furthermore, the HDAC4 degradation was also significantly reversed by RNAi-mediated depletion of caspase 3 in infected cells. However, unlike HDAC4 polypeptide cleavage product “a,” neither the inhibition nor the depletion of caspase 3 made the HDAC4 polypeptide cleavage product “b” completely disappear (Fig. 4, A and C). Instead, after these manipulations, an additional HDAC4 polypeptide fragment (indicated by arrow “c”) lagging fragment b appeared in infected cells (Fig. 4, A and C). This indicated that either different caspases or other host proteases, independently or in conjunction with caspases, are also causing the HDAC4 polypeptide degradation in IAV-infected cells. Among other host proteases, the obvious candidates are lysosomal proteases, cathepsins, because here a lysosomal inhibitor did rescue, albeit partially, the IAV-induced HDAC4 polypeptide degradation (Fig. 4, A and B). Consistent with this observation, the HDAC4 polypeptide has recently been shown to undergo cleavage by cathepsin H in a heterologous system (43). Interestingly, the nature of the degradation of HDAC4 polypeptide in IAV-infected cells discovered here is very similar to the lysosome-associated caspase-mediated degradation of host cortactin polypeptide in IAV-infected cells reported by us recently (44). Therefore, we believe that the lysosomes are closely associated with the apoptotic pathway in IAV-infected cells. Evidently, lysosomes have been described to be involved in the induction of both intrinsic and extrinsic apoptotic pathways in heterologous systems (45–48). A further detailed investigation is needed to determine the precise role of lysosomal proteases and their linkages with intrinsic or extrinsic apoptotic pathways in the degradation of HDAC4 polypeptide in IAV-infected cells.
In addition to polypeptide, the HDAC4 mRNA level was also significantly down-regulated in IAV-infected cells. Using RNAi and overexpression strategies, we identified that IAV is employing its protein PA-X to accomplish this. PA-X is a newly identified IAV protein, and is highly conserved in IAV genome, including PR8 and CA09 strains used here (29, 49). PA-X is a RNA endonuclease and selectively degrade the host mRNAs (30), which results in shutoff of host gene expression and allows IAV to escape host innate antiviral response (29, 30, 49). Potentially, the combined HDAC4 characteristics, anti-IAV activity through involvement in host antiviral response and reduced mRNA half-life, made the HDAC4 mRNA an ideal target for PA-X endonuclease-mediated degradation. The selectivity of PA-targeting siRNA rescuing the HDAC4 expression in infected cells and complementarity of PA-X expression alone reducing the HDAC4 expression in cells confirmed the antagonism of HDAC4 by IAV via PA-X.
Like HDAC1, HDAC2, and HDAC11 that exhibit anti-IAV function, HDAC4 is part of the host innate anti-IAV response. In the absence of HDAC4 expression, the extent of IAV-induced phosphorylation of STAT1 and subsequent signaling cascade was reduced in A549 cells. Consequently, the IAV-induced expression of ISGs, IFITM3, ISG15, and viperin, which have been previously implicated to control IAV infection, was down-regulated in HDAC4-depleted cells. Conversely, the IAV-induced expression of these ISGs was up-regulated in HDAC4-supplemented cells. Nevertheless, such involvement of HDAC4 in the IAV-induced host innate antiviral response as well as in IAV-induced apoptosis in host cells needs to be investigated in more detail. Furthermore, it remains to be determined where and how many times on HDAC4 polypeptide the proteolytic cleavage occurs. One likely cleavage position is the aspartic acid residue at position 289, which has been described previously to be targeted by caspase 3 in heterologous systems (42, 50, 51). The cleavage at this position generates an N-terminal fragment that is quite similar to the size of fragments resolving around the 36-kDa mark detected here by HDAC4 antibody recognizing an N-terminal epitope (42, 50, 51) (Fig. 3G). It is also critical to understand whether proteolytic cleavage of the HDAC4 polypeptide during IAV infection occurs just to antagonize its anti-IAV function or the HDAC4 cleavage products, particularly the above-mentioned N-terminal fragment, also play some significant roles in IAV replication. It has been reported that this N-terminal fragment of the HDAC4 polypeptide is quite stable and translocate to the nucleus, where it binds to the MEF2 (myocyte enhancer factor) transcription factor and represses its activity (52). Furthermore, this HDAC4 N-terminal fragment also induces the intrinsic apoptotic pathway in mammalian cells under heterologous conditions (50, 51). Evidently, both the repressed host transcription and the apoptosis are favorable environments for IAV to proliferate (53). Further investigations will delineate that IAV manipulates host HDAC4 not just to antagonize its antiviral function but also as a means to promote its own replication. In addition, it will also be useful to investigate the role of HDAC4 in IAV-induced respiratory tract tissue damage during flu.
In summary, we have provided here evidence that a class IIa HDAC (HDAC4) joins the list of HDACs to exhibit anti-IAV properties. This report also reinforces the notion that the HDACs are a family of anti-IAV factors. Next, the understanding of the intermolecular interactions between various HDACs, their host partners, and viral proteins during IAV infection will help understand the mechanisms of their antiviral function as well as their antagonism. Consequently, the resulting knowledge will help to assess the potential of HDACs as the targets for host-directed anti-influenza virus therapies.
Experimental procedures
Cells, viruses, and plasmids
A549, MDCK, and HEK293 cells were grown and maintained in complete minimum essential medium (MEM) supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, and 1% l-glutamine (Life Technologies) at 37 °C and under 5% CO2 atmosphere. Influenza virus strains A/Puerto Rico/8/1934(H1N1), A/California/07/2009/(H1N1), and A/WSN/1933(H1N1) (WSN) were propagated in 10-day-old embryonated chicken eggs, and titrated on MDCK cell monolayers. Influenza A virus PA-X (WSN) plasmid (gifted by Yoshihiro Kawaoka, The University of Tokyo, Japan) and human HDAC4 plasmid (gifted by Eric Verdin, Addgene plasmid number 13821) were amplified in Escherichia coli DH5α strain and extracted using a plasmid purification kit (Qiagen).
Infection
The virus inoculum was prepared in serum-free MEM and added to the cell monolayers, which had been washed twice with serum-free MEM. For infection of MDCK cells, 1 μg/ml of TPCK (tosylphenylalanyl chloromethyl ketone)-trypsin; Sigma-Aldrich) was added to the virus inoculum. After 1 h of incubation at 35 °C, the inoculum was removed and cells were washed once with serum-free MEM. Fresh serum-free MEM was added and the cells were incubated back at 35 °C. In some experiments, serum-free MEM was supplemented with trichostatin A (Sigma-Aldrich), SAHA (Sigma-Aldrich), NH4Cl (Sigma-Aldrich), MG132 (Calbiochem), or caspase 3 inhibitor: Z-DEVD-fmk (Calbiochem). In some experiments, the virus inoculum was irradiated with a 30 W ultraviolet (UV) bulb for 30 min before adding to the cells and proceeding with the infection.
Quantitative real-time PCR
Total RNA from the cells was isolated using a Nucleospin RNA isolation kit (Macherey-Nagel) and the cDNA was synthesized using PrimeScript RT reagent kit (Takara) by following the manufacture's protocol. Quantitative real-time PCR was performed on ViiA 6 Real-time PCR system (Applied Biosystems) using SYBR Green Select Master Mix (Life Technologies) and predesigned or custom-designed primers. For HDAC4, caspase 3, and interferon-stimulated genes, the predesigned KiCqStart primers were obtained from Sigma-Aldrich. The custom designed primers were: 18S rRNA, forward, 5′-ATCGGGGATTGCAATTATTC-3′, reverse, 5′-TCACTAAACCATCCAATCG-3′; β-actin, forward, 5′-GACGACATGGAGAAAATCTG-3′, reverse, 5′-ATGATCTGGGTCATCTTCTC-3′; IAV M gene, forward, 5′-TTGCTGATTCACAGCATCGG-3′, reverse, 5′-CAGATGGCTGGATCGAGTGA-3′; IAV NP gene, forward, 5′-AGAAATAAGGAGAGTTTGGCGCCTAGC-3′, reverse, 5′-CATTCCGGTGCGAACAAGCGC-3′; IAV PA, forward, 5′-GCTTCTTATCGTTCAGGCTCTTAGG-3′, reverse, 5′-CCGAGAAGCATTAAGCAAAACCCAG-3′; IAV PA-X gene, forward, 5′-GCGACAATGCTTCAATCCGA-3′, reverse, 5′-TTGACTCGCCTTGCTCATTG-3′. The level of β-actin mRNA and/or 18S rRNA were used as reference for normalizing the levels of target gene mRNAs. The fold or percentage change in the mRNA levels of target gene were calculated using the 2−ΔΔCT method.
Western blotting
Cells were lysed in lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.5% SDS, 0.5% sodium deoxycholate, 1% Triton X-100, and 1× protease inhibitor mixture (Roche)) to prepare the total cell lysates. The amount of protein was quantified by a BCA kit (Thermo). Then, equal amounts of protein were resolved on an appropriate percentage of Tris glycine SDS-PAGE along with SeeBluePlus 2 pre-stained protein standards (Life Technologies), and transferred on to Protran® Premium nitrocellulose membrane (GE Healthcare). Subsequently, membranes were probed with the rabbit anti-HDAC4 (1:1,000; D15C3, Cell Signaling), rabbit anti-β-actin (1:10,000; ab8227, Abcam), goat anti-NP (1:10,000; G150, kindly provided by Richard Webby, St. Jude Children's Research Hospital), mouse anti-NP (1:10,000; NR-19868, obtained through BEI Resources, NIAID, NIH), goat anti-HA (1:1,000; G57, kindly provided by Richard Webby), rabbit anti-PDI (1:10,000; P7496, Sigma-Aldrich), rabbit anti-caspase 3 (1:1,000; 8G10, Cell Signaling), mouse anti-pSTAT1 (1:1,000; pY701, BD Bioscience), mouse anti-STAT1 (1:1,000; 610185, BD Bioscience), rabbit anti-viperin (1:1,000; D5T2X, Cell Signaling), rabbit anti-IFITM3 (1:1000; ab15592, Abcam), or rabbit anti-ISG15 (1:1,000; F-9, Cell Signaling) followed by horseradish peroxidase-conjugated anti-rabbit, anti-mouse, or anti-goat IgG antibody (1:2,000 or 1:5,000, Life Technologies). Finally, the protein bands were visualized using ECL or ECL-Prime Western blotting Systems (GE Healthcare) and images were acquired on Odyssey Fc imaging system utilizing Image Studio software version 5.0 (Li-COR). Images were then exported as TIFF files, minimally adjusted for brightness and contrast, and composed in figures in Adobe Photoshop CC 2015.
RNAi
The predesigned siRNA targeting the human HDAC4 gene (ID: SASI_Hs02_00340778; 5′-CAGCCAAGCUUCUGCAGCA-3′) and caspase 3 gene (ID: SASI_Hs01_00139105; 5′-CAGCUUUCAUGAUUAGCAA-3′), the siRNAs targeting the individual influenza gene segments (sequence mentioned in Ref. 28), and a nontargeting siRNA (MISSION universal negative control #1) were obtained from Sigma-Aldrich. First, different nanomolar quantities of siRNAs and 2 μl of Lipofectamine RNAiMAX reagent (Life Technologies) were diluted separately in OptiMEM I reduced serum medium (Life Technologies). Then both solutions were mixed together, and incubated for 30 min at room temperature for siRNA-RNAiMAX complex to form. This complex was then mixed with the cell suspensions, and cells were seeded in a cell culture plate. The cells were then incubated at 37 °C under 5% CO2 atmosphere for 24–72 h before infection or further processing.
Overexpression
Cells were grown to 80–90% confluence in a cell culture plate, and transfected with the plasmids using Lipofectamine 2000 reagent (Life Technologies). First, 1 μg of plasmid DNA and 3 μl of Lipofectamine 2000 were diluted separately in OptiMEM. Then both solutions were mixed together, and incubated for 30 min at room temperature for a DNA-Lipofectamine complex to form. The DNA-Lipofectamine complex was then added to the cells, and cells were incubated at 37 °C for 24–48 h before infection or further processing.
Cell viability assay
The viability of cells was evaluated by MTT assay. The MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) reagent (Sigma-Aldrich) was added to the cells, which were then incubated at 37 °C under 5% CO2 atmosphere for 1 h. Subsequently, 1 milliliter of dimethyl sulfoxide (DMSO; Calbiochem) was added to the cells and cells were rocked at room temperature for 15 min. Finally, the absorbance was measured at 570 nanometer wavelength using a 680 microplate reader (Bio-Rad).
Virus release assay
The culture medium from infected cells was collected and cleared off of cell debris by centrifugation at 17,000 × g for 1 min. Such cleared medium was then divided into two parts. One part was subjected to the protein precipitation by trichloroacetic acid (TCA) (Calbiochem) and the other part was mixed with 0.3% BSA (Sigma-Aldrich) and titrated by microplaque assay. For protein precipitation, the medium was mixed the ice-cold TCA to a final concentration of 20% and incubated on ice for 30 min. The mixture was then centrifuged at 20,000 × g and 4 °C for 30 min. The supernatant was removed carefully and the pellet was washed twice with ice-cold acetone by centrifugation as above. The pellet was then air dried and directly suspended in SDS-PAGE sample buffer (50 mm Tris-HCl, pH 6.8, 2% SDS, 30% glycerol, 5% 2-mercaptoethanol, and 0.04% bromphenol blue) and proteins were resolved on SDS-PAGE. The viral HA and NP was then detected by Western blotting. For microplaque assay, the confluent monolayers of MDCK cells were infected with 10-fold serial dilutions of the culture medium as described above. The inoculum was then removed, and the cells were overlaid with serum-free MEM containing 0.8% Avicel (RC-185; FMC Biopolymer) and 1 μg/ml of TPCK-treated trypsin (Sigma-Aldrich). After 20–24 h incubation at 37 °C under 5% CO2 atmosphere, the MEM overlay was removed and the cells were fixed with 4% formalin (Sigma-Aldrich) and permeabilized with 0.5% Triton X-100 prepared in 20 mm glycine. Cells were then stained with mouse anti-NP (1:1,000) followed by horseradish peroxidase-conjugated anti-mouse IgG antibody (1:1,000). The plaques were visualized by using TrueBlue substrate (KPL Biosciences).
Statistical analysis
The statistical analyses were performed using Prism 6 or 8 (GraphPad). The p values were calculated using unpaired t tests for pairwise data comparisons and one-way analysis of variance (ANOVA) or two-way ANOVA for multiple data set comparisons. A p value of ≤0.05 was considered significant.
Author contributions
H. D. G. writing-original draft; M. H. conceptualization; M. H. resources; M. H. supervision; M. H. funding acquisition; M. H. project administration; M. H. writing-review and editing; H. D. G. designed the study, carried out the experiments, interpreted the results and wrote the manuscript; M. H. designed the study, interpreted the results and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Sonya Mros for kindly gifting chicken eggs and Dr. Prashanth Nagesh for technical troubleshooting.
This work was supported by the H. S. and J. C. Anderson Charitable Trust, a University of Otago Ph.D. scholarship (to H. D. G.), and the Department of Microbiology and Immunology and the School of Biomedical Sciences (to M. H.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3.
- IAV
- influenza A virus
- HDAC
- histone deacetylase
- PDI
- protein-disulfide isomerase
- m.o.i.
- multiplicity of infection
- MDCK
- Madin-Darby canine kidney cells
- HA
- hemagglutinin
- NP
- nucleoprotein
- qPCR
- quantitative real-time PCR
- STAT
- signal transducer and activator of transcription
- ISG
- interferon-stimulated gene
- IFITM3
- interferon-induced transmembrane protein 3
- MEM
- minimum essential medium
- TPCK
- tosylphenylalanyl chloromethyl ketone
- Z
- benzyloxycarbonyl
- fmk
- fluoromethyl ketone
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- ANOVA
- analysis of variance
- TCA
- trichloroacetic acid.
References
- 1. Husain M. (2014) Avian influenza A (H7N9) virus infection in humans: epidemiology, evolution, and pathogenesis. Infect. Genet. Evol. 28, 304–312 10.1016/j.meegid.2014.10.016 [DOI] [PubMed] [Google Scholar]
- 2. Iuliano A. D., Roguski K. M., Chang H. H., Muscatello D. J., Palekar R., Tempia S., Cohen C., Gran J. M., Schanzer D., Cowling B. J., Wu P., Kyncl J., Ang L. W., Park M., Redlberger-Fritz M., et al. (2018) Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet 391, 1285–1300 10.1016/S0140-6736(17)33293-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Petrie J. G., Cheng C., Malosh R. E., VanWormer J. J., Flannery B., Zimmerman R. K., Gaglani M., Jackson M. L., King J. P., Nowalk M. P., Benoit J., Robertson A., Thaker S. N., Monto A. S., and Ohmit S. E. (2016) Illness severity and work productivity loss among working adults with medically attended acute respiratory illnesses: US influenza vaccine effectiveness network 2012–2013. Clin. Infect. Dis. 62, 448–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ortiz J. R., Neuzil K. M., Shay D. K., Rue T. C., Neradilek M. B., Zhou H., Seymour C. W., Hooper L. G., Cheng P. Y., Goss C. H., and Cooke C. R. (2014) The burden of influenza-associated critical illness hospitalizations. Crit. Care Med. 42, 2325–2332 10.1097/CCM.0000000000000545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Webster R. G., and Govorkova E. A. (2014) Continuing challenges in influenza. Ann. N.Y. Acad. Sci. 1323, 115–139 10.1111/nyas.12462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xue K. S., Moncla L. H., Bedford T., and Bloom J. D. (2018) Within-host evolution of human influenza virus. Trends Microbiol. 26, 781–793 10.1016/j.tim.2018.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim H., Webster R. G., and Webby R. J. (2018) Influenza virus: dealing with a drifting and shifting pathogen. Viral Immunol. 31, 174–183 10.1089/vim.2017.0141 [DOI] [PubMed] [Google Scholar]
- 8. Hussain M., Galvin H. D., Haw T. Y., Nutsford A. N., and Husain M. (2017) Drug resistance in influenza A virus: the epidemiology and management. Infect. Drug Resist. 10, 121–134 10.2147/IDR.S105473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Husain M., and Cheung C. (2014) Histone deacetylase 6 inhibits influenza A virus release by downregulating the trafficking of viral components to the plasma membrane via its substrate acetylated microtubules. J. Virol. 81, 11229–11239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nagesh P. T., and Husain M. (2016) Influenza A virus dysregulates host histone deacetylase 1 that inhibits viral infection in lung epithelial cells. J. Virol. 90, 4614–4625 10.1128/JVI.00126-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nagesh P. T., Hussain M., Galvin H. D., and Husain M. (2017) Histone deacetylase 2 is a component of influenza A virus-induced host antiviral response. Front. Microbiol. 8, 1315 10.3389/fmicb.2017.01315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nutsford A. N., Galvin H. D., Ahmed F., and Husain M. (2019) The Class IV human deacetylase, HDAC11, exhibits anti-influenza A virus properties via its involvement in host innate antiviral response. Cell Microbiol. 21, e12989 10.1111/cmi.12989 [DOI] [PubMed] [Google Scholar]
- 13. Seto E., and Yoshida M. (2014) Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 6, a018713 10.1101/cshperspect.a018713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sterner D. E., and Berger S. L. (2000) Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64, 435–459 10.1128/MMBR.64.2.435-459.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li L., and Yang X. J. (2015) Tubulin acetylation: responsible enzymes, biological functions and human diseases. Cell. Mol. Life Sci. 72, 4237–4255 10.1007/s00018-015-2000-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou Y., He C., Wang L., and Ge B. (2017) Post-translational regulation of antiviral innate signaling. Eur. J. Immunol. 47, 1414–1426 10.1002/eji.201746959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu N., Li S., Wu N., and Cho K. S. (2017) Acetylation and deacetylation in cancer stem-like cells. Oncotarget 8, 89315–89325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cook C., Stankowski J. N., Carlomagno Y., Stetler C., and Petrucelli L. (2014) Acetylation: a new key to unlock tau's role in neurodegeneration. Alzheimers Res. Ther. 6, 29 10.1186/alzrt259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Song G., and Walley J. W. (2016) Dynamic protein acetylation in plant-pathogen interactions. Front Plant Sci. 7, 421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jeng M. Y., Ali I., and Ott M. (2015) Manipulation of the host protein acetylation network by human immunodeficiency virus type 1. Crit. Rev. Biochem. Mol. Biol. 50, 314–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Husain M., and Harrod K. S. (2009) Influenza A virus-induced caspase-3 cleaves the histone deacetylase 6 in infected epithelial cells. FEBS Lett. 583, 2517–2520 10.1016/j.febslet.2009.07.005 [DOI] [PubMed] [Google Scholar]
- 22. Giese S., Ciminski K., Bolte H., Moreira É A., Lakdawala. S., Hu Z., David Q., Kolesnikova L., Götz V., Zhao Y., Dengjel J., Chin Y. E., Xu K., and Schwemmle M. (2017) Role of influenza A virus NP acetylation on viral growth and replication. Nat. Commun. 8, 1259 10.1038/s41467-017-01112-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hatakeyama D., Shoji M., Yamayoshi S., Yoh R., Ohmi N., Takenaka S., Saitoh A., Arakaki Y., Masuda A., Komatsu T., Nagano R., Nakano M., Noda T., Kawaoka Y., and Kuzuhara T. (2018) Influenza A virus nucleoprotein is acetylated by histone acetyltransferases PCAF and GCN5. J. Biol. Chem. 293, 7126–7138 10.1074/jbc.RA117.001683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oishi K., Yamayoshi S., Kozuka-Hata H., Oyama M., and Kawaoka Y. (2018) N-terminal acetylation by NatB is required for the shutoff activity of influenza A virus PA-X. Cell Rep. 24, 851–860 10.1016/j.celrep.2018.06.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen H., Qian Y., Chen X., Ruan Z., Ye Y., Chen H., Babiuk L. A., Jung Y. S., and Dai J. (2019) HDAC6 restricts influenza A virus by deacetylation of the RNA polymerase PA subunit. J. Virol. 93, e01896–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koyuncu E., Budayeva H. G., Miteva Y. V., Ricci D. P., Silhavy T. J., Shenk T., and Cristea I. M. (2014) Sirtuins are evolutionarily conserved viral restriction factors. MBio. 5, e02249–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guo N., and Peng Z. (2013) MG132, a proteasome inhibitor, induces apoptosis in tumor cells. Asia Pac. J. Clin. Oncol. 9, 6–11 10.1111/j.1743-7563.2012.01535.x [DOI] [PubMed] [Google Scholar]
- 28. Ge Q., McManus M. T., Nguyen T., Shen C. H., Sharp P. A., Eisen H. N., and Chen J. (2003) RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc. Natl. Acad. Sci. U.S.A. 100, 2718–2723 10.1073/pnas.0437841100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jagger B. W., Wise H. M., Kash J. C., Walters K. A., Wills N. M., Xiao Y. L., Dunfee R. L., Schwartzman L. M., Ozinsky A., Bell G. L., Dalton R. M., Lo A., Efstathiou S., Atkins J. F., Firth A. E., Taubenberger J. K., and Digard P. (2012) An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337, 199–204 10.1126/science.1222213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Khaperskyy D. A., Schmaling S., Larkins-Ford J., McCormick C., and Gaglia M. M. (2016) Selective degradation of host RNA polymerase II transcripts by influenza A virus PA-X host shutoff protein. PLoS Pathog. 12, e1005427 10.1371/journal.ppat.1005427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oishi K., Yamayoshi S., and Kawaoka Y. (2018) Identification of novel amino acid residues of influenza virus PA-X that are important for PA-X shutoff activity by using yeast. Virology 516, 71–75 10.1016/j.virol.2018.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen X., Liu S., Goraya M. U., Maarouf M., Huang S., and Chen J.-L. (2018) Host immune response to influenza A virus infection. Front. Immunol. 9, 320 10.3389/fimmu.2018.00320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Iwasaki A., and Pillai P. S. (2014) Innate immunity to influenza virus infection. Nat. Rev. Immunol. 14, 315–328 10.1038/nri3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schneider W. M., Chevillotte M. D., and Rice C. M. (2014) Interferon-stimulated genes: a complex web of host defenses. Ann. Rev. Immunol. 32, 513–545 10.1146/annurev-immunol-032713-120231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brass A. L., Huang I. C., Benita Y., John S. P., Krishnan M. N., Feeley E. M., Ryan B. J., Weyer J. L., van der Weyden L., Fikrig E., Adams D. J., Xavier R. J., Farzan M., and Elledge S. J. (2009) The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139, 1243–1254 10.1016/j.cell.2009.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lenschow D. J., Lai C., Frias-Staheli N., Giannakopoulos N. V., Lutz A., Wolff T., Osiak A., Levine B., Schmidt R. E., García-Sastre A., Leib D. A., Pekosz A., Knobeloch K. P., Horak I., and Virgin H. W. 4th (2007) IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc. Natl. Acad. Sci. U.S.A. 104, 1371–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang X., Hinson E. R., and Cresswell P. (2007) The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2, 96–105 10.1016/j.chom.2007.06.009 [DOI] [PubMed] [Google Scholar]
- 38. Verdin E., and Ott M. (2015) 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 16, 258–264 10.1038/nrm3931 [DOI] [PubMed] [Google Scholar]
- 39. Soppa J. (2010) Protein acetylation in archaea, bacteria, and eukaryotes. Archaea 2010, pii: 820681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Conrad R. J., and Ott M. (2016) Therapeutics targeting protein acetylation perturb latency of human viruses. ACS Chem. Biol. 11, 669–680 10.1021/acschembio.5b00999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moon J., Kaowinn S., Cho I. R., Min D. S., Myung H., Oh S., Kaewpiboon C., Kraemer O. H., and Chung Y. H. (2016) Hepatitis C virus core protein enhances hepatocellular carcinoma cells to be susceptible to oncolytic vesicular stomatitis virus through down-regulation of HDAC4. Biochem. Biophys. Res. Commun. 474, 428–434 10.1016/j.bbrc.2016.05.005 [DOI] [PubMed] [Google Scholar]
- 42. Liu F., Dowling M., Yang X. J., and Kao G. D. (2004) Caspase-mediated specific cleavage of human histone deacetylase 4. J. Biol. Chem. 279, 34537–34546 10.1074/jbc.M402475200 [DOI] [PubMed] [Google Scholar]
- 43. Yang Z., Liu Y., Qin L., Wu P., Xia Z., Luo M., Zeng Y., Tsukamoto H., Ju Z., Su D., Kang H., Xiao Z., Zheng S., Duan Z., Hu R., Wang Q., Pandol S. J., and Han Y. P. (2017) Cathepsin H-mediated degradation of HDAC4 for matrix metalloproteinase expression in hepatic stellate cells: implications of epigenetic suppression of matrix metalloproteinases in fibrosis through stabilization of class IIa histone deacetylases. Am. J. Pathol. 187, 781–797 10.1016/j.ajpath.2016.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen D. Y., and Husain M. (2016) Caspase-mediated degradation of host cortactin that promotes influenza A virus infection in epithelial cells. Virology 497, 146–156 10.1016/j.virol.2016.07.017 [DOI] [PubMed] [Google Scholar]
- 45. Ferri K. F., and Kroemer G. (2001) Organelle-specific initiation of cell death pathways. Nat. Cell Biol. 3, E255–E263 10.1038/ncb1101-e255 [DOI] [PubMed] [Google Scholar]
- 46. Foghsgaard L., Wissing D., Mauch D., Lademann U., Bastholm L., Boes M., Elling F., Leist M., and Jäättelä M. (2001) Cathepsin B acts as a dominant execution protease in tumor cell apoptosis induced by tumor necrosis factor. J. Cell Biol. 153, 999–1010 10.1083/jcb.153.5.999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Guicciardi M. E., Deussing J., Miyoshi H., Bronk S. F., Svingen P. A., Peters C., Kaufmann S. H., and Gores G. J. (2000) Cathepsin B contributes to TNF-α-mediated hepatocyte apoptosis by promoting mitochondrial release of cytochrome c. J. Clin. Invest. 106, 1127–1137 10.1172/JCI9914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Roberts L. R., Kurosawa H., Bronk S. F., Fesmier P. J., Agellon L. B., Leung W. Y., Mao F., and Gores G. J. (1997) Cathepsin B contributes to bile salt-induced apoptosis of rat hepatocytes. Gastroenterology 113, 1714–1726 10.1053/gast.1997.v113.pm9352877 [DOI] [PubMed] [Google Scholar]
- 49. Hayashi T., MacDonald L. A., and Takimoto T. (2015) Influenza A virus protein PA-X contributes to viral growth and suppression of the host antiviral and immune responses. J. Virol. 89, 6442–6452 10.1128/JVI.00319-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Paroni G., Mizzau M., Henderson C., Del Sal G., Schneider C., and Brancolini C. (2004) Caspase-dependent regulation of histone deacetylase 4 nuclear-cytoplasmic shuttling promotes apoptosis. Mol. Biol. Cell. 15, 2804–2818 10.1091/mbc.e03-08-0624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Paroni G., Fontanini A., Cernotta N., Foti C., Gupta M. P., Yang X. J., Fasino D., and Brancolini C. (2007) Dephosphorylation and caspase processing generate distinct nuclear pools of histone deacetylase 4. Mol. Cell. Biol. 27, 6718–6732 10.1128/MCB.00853-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Backs J., Worst B. C., Lehmann L. H., Patrick D. M., Jebessa Z., Kreusser M. M., Sun Q., Chen L., Heft C., Katus H. A., and Olson E. N. (2011) Selective repression of MEF2 activity by PKA-dependent proteolysis of HDAC4. J. Cell Biol. 195, 403–415 10.1083/jcb.201105063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wurzer W. J., Planz O., Ehrhardt C., Giner M., Silberzahn T., Pleschka S., and Ludwig S. (2003) Caspase 3 activation is essential for efficient influenza virus propagation. EMBO J. 22, 2717–2728 10.1093/emboj/cdg279 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.