Abstract
Plant development is regulated by both synergistic and antagonistic interactions of different phytohormones, including a complex crosstalk between ethylene and auxin. For instance, auxin and ethylene synergistically control primary root elongation and root hair formation. However, a lack of chemical agents that specifically modulate ethylene or auxin production has precluded precise delineation of the contribution of each hormone to root development. Here, we performed a chemical genetic screen based on the recovery of root growth in ethylene-related Arabidopsis mutants with constitutive “short root” phenotypes (eto1–2 and ctr1–1). We found that ponalrestat exposure recovers root elongation in these mutants in an ethylene signal–independent manner. Genetic and pharmacological investigations revealed that ponalrestat inhibits the enzymatic activity of the flavin-containing monooxygenase YUCCA, which catalyzes the rate-limiting step of the indole-3-pyruvic acid branch of the auxin biosynthesis pathway. In summary, our findings have identified a YUCCA inhibitor that may be useful as a chemical tool to dissect the distinct steps in auxin biosynthesis and in the regulation of root development.
Keywords: chemical biology, high-throughput screening (HTS), auxin, biosynthesis, enzyme inhibitor, ethylene, flavin-containing monooxygenase, ponalrestat, root growth regulator, YUCCA
Introduction
The phytohormone auxin plays a critical role in plant growth and development processes, and indole-3-acetic acid (IAA)8 is the predominant natural auxin. Auxin regulates plant growth and development in a dose-dependent manner, which has been characterized as a bell-shaped dosage-response curve (1). For example, below the optimal IAA concentration, root growth increases with increasing IAA. Beyond the optimal concentration, however, increasing IAA inhibits root growth (2). This pattern indicates that auxin levels need to be maintained at an optimal concentration in vivo as plants develop.
In Arabidopsis, IAA is synthesized through both tryptophan (Trp)-dependent and Trp-independent pathways (3, 4). One of the major Trp-dependent auxin biosynthesis pathways is the indole-3-pyruvic acid (IPA) pathway (5). In this pathway, tryptophan is converted to IPA by “tryptophan aminotransferase of Arabidopsis1/tryptophan aminotransferase–related” (TAA1/TAR) proteins, followed by the conversion of IPA to IAA by YUCCA (YUC) enzymes, which are flavin-containing monooxygenases (FMO) (6–8). The YUC-catalyzed oxidative decarboxylation reaction has been proposed to be the rate-determining step in auxin biosynthesis (9).
Ethylene is another phytohormone involved in multiple plant developmental processes, including seed germination, seedling morphology, and root growth (10, 11). Dark-grown 3-day-old etiolated seedlings exhibit a triple response upon ethylene application: the inhibition of cell elongation in the root and hypocotyl, radial swelling of the hypocotyl, and exaggeration of the apical hook curvature (12, 13). Genetic screens for mutants based on the triple-response phenotype and molecular analyses of these mutants have uncovered a largely linear ethylene-signaling pathway (14). Perception of ethylene by its receptor leads to inactivation of “constitutive response 1” (CTR1), a kinase that plays a negative role in ethylene-signaling pathway by phosphorylating and inhibiting the positive regulator “ethylene-insensitive 2” (EIN2) (14). CTR1 inactivation releases EIN2, leading to the cleavage of the EIN2, “carboxyl end” (CEND), which translocates to the nucleus and activates the master transcription factor “ethylene-insensitive 3” (EIN3) and its homolog “EIN3-like 1” (EIL1) (14, 15).
Auxin and ethylene act synergistically to control primary root elongation and root hair formation. Multiple reports have demonstrated that the inhibition of root elongation by ethylene mainly involves its regulation of auxin biosynthesis and local auxin distribution (16–18). To identify a chemical tool that can be used to dissect the root development processes regulated by ethylene and/or auxin, we performed a high-throughput chemical genetic screen using eto1–2 and ctr1–1, both of which exhibit a short-root phenotype. Ponalrestat was identified as a stable and effective candidate compound that recovered the root elongation in eto1–2 and ctr1–1. Our subsequent analysis showed that ponalrestat promoted root elongation through the inhibition of the flavin monooxygenase YUC, which is an auxin biosynthesis enzyme. Overall, our chemical screen identified two compounds, ponalrestat and the TAA1 inhibitor Kyn (19) that affect different steps of the IPA branch of auxin biosynthesis. Several YUC inhibitors, including yucasin, PPBo, and yucasin DF (20–22), were also recently reported. Together with ponalrestat, these chemicals may be useful tools to regulate auxin biosynthesis in vivo and to elucidate the relationship of ethylene and auxin in regulating root elongation.
Results
Identification of a compound that recovers root elongation in ethylene mutants
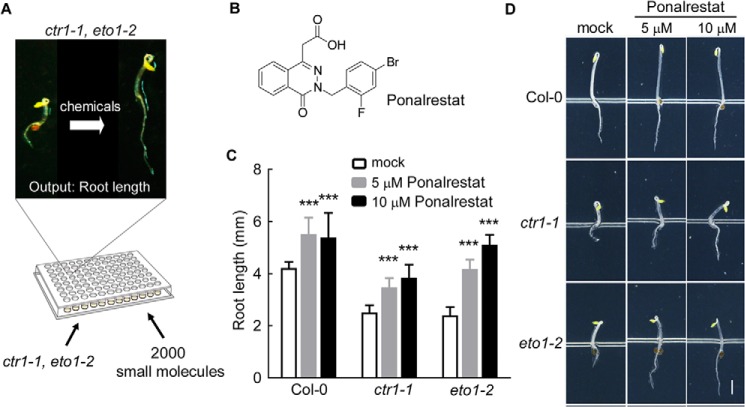
Phenotype-directed screening is a powerful strategy for bioactive compound discovery in plant studies (23–25). To identify chemical regulators of root growth and development, we performed a chemical screen using 96-well plates (Fig. 1A) along with the ethylene overproduction mutant eto1–2 (13) and constitutive triple response mutant ctr1–1 (26). When grown in the dark, these mutants exhibited the triple-response phenotype even in the absence of exogenous ethylene (13, 26). We focused on the short-root phenotype and screened for compounds that promoted root elongation. After three rounds of screening 2000 diverse compounds (SP2000), ponalrestat (Fig. 1B), an inhibitor of human aldose reductase (ALR2) (27), was found to significantly promote root elongation in eto1–2 and ctr1–1 etiolated seedlings (Fig. 1, C and D). Further analysis showed that ponalrestat promoted root elongation in both etiolated and de-etiolated seedlings (Fig. 1, C and D and Fig. S1, A and B).
Figure 1.
Chemical genetic screening for inhibitors of activated ethylene-response mutants. A, the screening strategy scheme. For the screen, 2000 small molecules from the Microsource Spectrum Chemical Library were used to identify compounds (50–100 μm) that could rescue the root elongation defects of ctr1–1 and eto1–2. B, the structure of ponalrestat. C, the root lengths of 3-day-old Col-0, eto1–2, and ctr1–1 seedlings treated with ponalrestat or DMSO as the mock control. The seeds were germinated and grown on 1/2 MS medium supplemented with ponalrestat or DMSO. Bars represent mean ± S.D. of at least 10 seedlings; a Student's t test was used to compare ponalrestat-treated and mock-treated seedlings (***, p < 0.001). The experiment was repeated for at least three times with similar results. D, representative seedlings of the genotypes in (C). Scale bar = 1 mm.
Ponalrestat promotes root elongation downstream of ethylene signaling
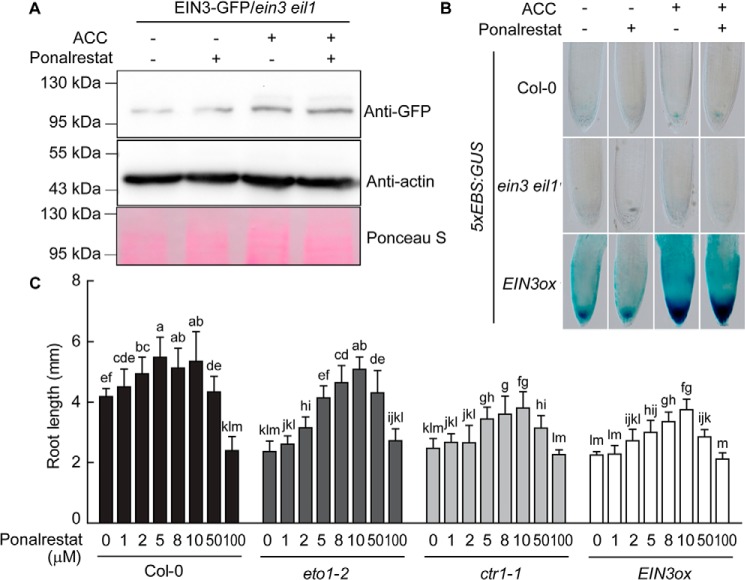
The finding that ponalrestat restored root elongation in eto1–2 and ctr1–1 raised the possibility that ponalrestat may target the ethylene-signaling pathway. To test this hypothesis, we investigated the effect of ponalrestat on the EIN3 transcription factor, a key downstream hub that mediates ethylene-responsive outputs (28). EIN3 protein can be degraded by the proteasome when it is ubiquitinated by “EIN3-binding F-BOX1” (EBF1) and EBF2 (29, 30). Ethylene treatment stabilizes EIN3 protein via EIN2, another key component of the ethylene-signaling pathway (31). Thus, EIN3 protein abundance can be used as an indicator of the impact of chemical treatment on the ethylene signal. We expressed 35S:EIN3-GFP in the ein3 eil1 genetic background, and, as expected, EIN3 protein abundance increased upon treatment with ACC, an ethylene precursor that can be converted to ethylene in vivo (Fig. 2A) (19, 31). Treatment with ponalrestat resulted in no significant inhibition of either basal or ACC-induced EIN3 protein levels (Fig. 2A). To investigate whether ponalrestat affects the transcriptional activity of EIN3, GUS driven by an EIN3-responsive promoter (5xEBS:GUS), was transformed into several genetic backgrounds (17). As shown in Fig. 2B, no significant change in GUS activity was observed in Col-0 or ein3 eil1 in response to ponalrestat treatment. GUS activity was greatly enhanced in the EIN3 overexpression line (EIN3ox) relative to that in Col-0 (WT), regardless of the presence of ponalrestat. These results indicate that ponalrestat does not inhibit EIN3 transcriptional activity.
Figure 2.
Ponalrestat rescues ethylene-induced short root growth downstream of EIN3. A, Western blotting for EIN3-GFP protein in EIN3-GFP/ein3 eil1. The seedlings were grown in the dark for 5 days on 1/2 MS medium with or without ponalrestat (5 μm) and/or ACC (10 μm). α-GFP antibody was used to detect the EIN3-GFP protein. Western blotting with actin antibody and membrane staining with Ponceau S were performed as the loading control. B, expression of 5xEBS:GUS in the roots of Col-0, ein3 eil1, and EIN3ox. The seedlings were grown in the dark for 3 days on 1/2 MS medium with or without ponalrestat (5 μm) and/or ACC (10 μm), followed by GUS staining. C, root lengths of etiolated seedlings of Col-0, eto1–2, ctr1–1, and EIN3ox grown on 1/2 MS medium supplemented with different concentrations of ponalrestat. Bars represent mean ± S.D. of at least 10 seedlings. Statistical differences between the groups were calculated with ANOVA (with Duncan's post hoc test). Bars with different letters are significantly different at p < 0.01. All the above experiments were repeated at least three times with similar results.
In accordance with the biochemical results, ponalrestat promoted root elongation in EIN3ox in the same pattern observed in Col-0, eto1–2, and ctr1–1 within a concentration range of 1 to 50 μm (Fig. 2C). In all of the analyzed genotypes, a higher concentration (100 μm) of ponalrestat failed to promote root elongation (Fig. 2C), indicating that high doses of ponalrestat may promote an unknown mechanism or have a toxic effect on root growth. Collectively, these data suggest that the ethylene-signaling pathway is not the main direct target of ponalrestat in terms of its ability to restore root elongation in the eto1–2 and ctr1–1 mutants.
Ponalrestat leads to auxin-related pleiotropic effects on root growth
To identify the mode of action of ponalrestat, we further analyzed the root responses to ponalrestat. We found that ponalrestat treatment suppressed root hair formation (Fig. 3A) and interrupted root gravitropic growth (Fig. 3, B and C), with similar changes observed in eto1–2, ctr1–1, and EIN3ox mutants (Fig. S2). Together with the dose-dependent effects of ponalrestat on root growth (Fig. 2C), these results suggest that ponalrestat may affect auxin responses. Consistent with these observations, the transcript levels of GH3.3, an auxin-induced downstream marker gene, were down-regulated upon ponalrestat application (Fig. 3D). Multiple reports have demonstrated that the inhibition of root elongation by ethylene mainly involves its regulation of the biosynthesis and local distribution of auxin (16–18). Based on these previous reports and the present findings, we hypothesize that ponalrestat may recover root growth by attenuating the auxin response in the root, which becomes more significant when the auxin response is activated by ethylene.
Figure 3.
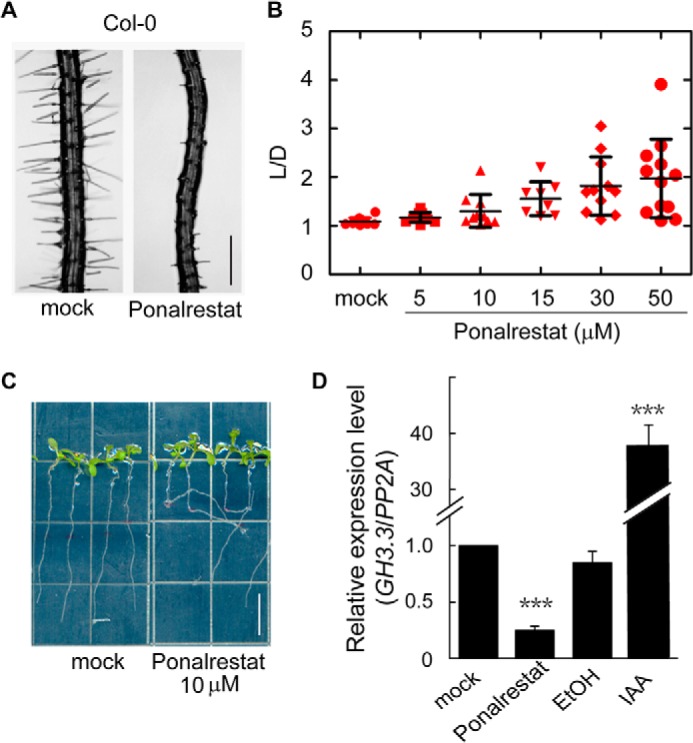
Ponalrestat treatment results in auxin-related defects for different root phenotypes. A, representative photos of root hairs of 5-day-old green seedlings of Col-0 grown on 1/2 MS medium supplemented with 5 μm ponalrestat or DMSO as the mock control. Scale bar = 200 μm. B, quantitative evaluation of root gravitropism. 10-day-old green seedlings of Col-0 were grown vertically on 1/2 MS medium supplemented with ponalrestat or DMSO as the mock control. The root gravitropic index (L/D) is the root length (L)/vertical length (D). Each red symbol represents the data for a single seedling. Bars represent mean ± S.D. of at least 10 seedlings. C, representative seedlings for the indicated groups from the analysis in (B). Scale bar = 10 mm. D, quantitative real-time PCR analysis of the auxin downstream marker gene GH3.3. 3-day-old seedlings were grown in the dark before treatment with liquid 1/2 MS supplemented with 50 μm ponalrestat (DMSO as the control) or 10 μm IAA (ethanol/EtOH as the control) for 4 h. Bars represent mean ± S.D. of three replicates; a Student's t test was used to compare ponalrestat-treated and mock-treated seedlings or IAA-treated and EtOH-treated seedlings (***, p < 0.001). All the above experiments were repeated for at least three times with similar results.
Ponalrestat targets auxin biosynthesis pathway
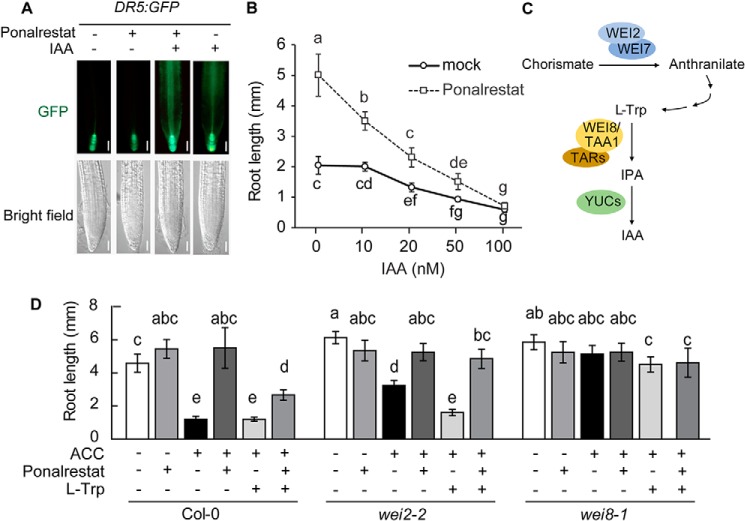
To narrow down the possible targets of ponalrestat, a DR5:GFP reporter construct was introduced into the Col-0 background to monitor the auxin signal in response to ponalrestat and IAA. As shown in Fig. 4A, IAA led to enhanced GFP fluorescence in the root, indicating an increased auxin signal. In contrast, ponalrestat suppressed the native auxin signal, as indicated by attenuated GFP fluorescence. Exogenous IAA-induced signal was not suppressed by ponalrestat (Fig. 4A). We also confirmed that ponalrestat-induced root elongation was suppressed by exogenous IAA (Fig. 4B). In another analysis, we introduced the DR5:GUS construct into ctr1–1 mutant and Col-0 backgrounds then analyzed GUS activity. Higher GUS activity was detected in ctr1–1 compared with Col-0, and ponalrestat application suppressed the relatively high GUS activity (Fig. S3). Based on these results and previous reports that ethylene promotes auxin biosynthesis (16–18), we propose that ponalrestat may affect auxin biosynthesis. To test this hypothesis, we performed genetic and pharmacological experiments with wei2–2 and wei8–1, two mutants with defects in l-tryptophan (l-Trp) and IPA biosynthesis, respectively (Fig. 4C). In accordance with the previous finding that ethylene stimulates IAA accumulation via WEI2 activation (32, 33), the root growth of wei2–2 displayed partial resistance to exogenous ACC, and l-Trp treatment recovered the sensitivity of wei2–2 to ACC (Fig. 4D). The root growth of wei8–1 showed resistance to treatment with ACC or both ACC and l-Trp (Fig. 4D), indicating that ethylene-induced root inhibition is partially dependent on a functional IAA biosynthesis pathway. Ponalrestat recovered the root elongation that was suppressed by ACC or co-treatment with ACC and l-Trp in both Col-0 and wei2–2, suggesting that ponalrestat may suppress IAA biosynthesis at a step downstream of l-Trp.
Figure 4.
Ponalrestat targets the auxin biosynthesis pathway. A, GFP fluorescence in the roots of DR5:GFP plants. 5-day-old green seedlings were treated with ponalrestat (5 μm), IAA (100 nm), both ponalrestat and IAA, or DMSO as the mock control. Scale bars = 50 μm. B, root lengths of 3-day-old eto1–2 etiolated seedlings grown on 1/2 MS medium containing ponalrestat (5 μm) or DMSO as the mock control with co-treatments of increasing concentrations of IAA. Bars represent mean ± S.D. of at least 10 seedlings. Statistical differences between the groups were calculated with ANOVA (with Tukey's HSD post hoc test). Data points with different letters are significantly different at p < 0.01. C, diagram of IPA branch of the IAA biosynthesis pathway. In this pathway, l-tryptophan (l-Trp) is sequentially derived from chorismate and anthranilate, which involves the catalytic activity of WEI2/WEI7. l-Trp is converted to IPA via aminotransferase WEI8/TAA1/TARs, then IPA is converted into the final product IAA by flavin-containing monooxygenase YUCCAs (YUCs). D, root lengths of Col-0, wei2–2, and wei8–1 seedlings with single or combinatorial treatments of 1 μm ACC, 5 μm ponalrestat, and 10 μm l-Trp. Bars represent mean ± S.D. of at least 10 seedlings. Statistical differences between the groups were calculated with ANOVA (with Tukey's HSD post hoc test). Bars with different letters are significantly different at p < 0.01. All the above experiments were repeated for at least three times with similar results.
Ponalrestat is an inhibitor of YUC monooxygenase
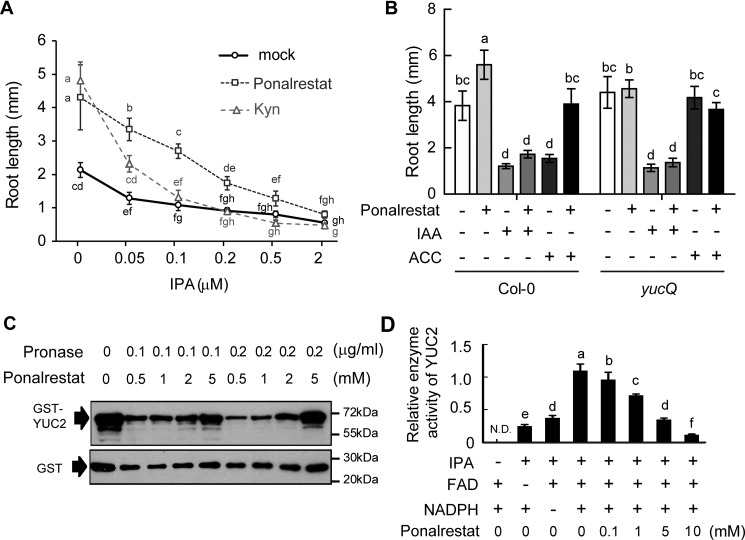
It has been reported that ethylene induces auxin biosynthesis via the TAA1-YUC branch of the auxin biosynthesis pathway (19, 34). We previously identified the small molecule Kyn (l-Kynurenine) as a competitive inhibitor of TAA1/TAR that can promote root elongation in the eto1–2 mutant (19). As shown in Fig. 5A, treatment with ponalrestat alone recovered the root growth of eto1–2 in the same manner as Kyn. However, ponalrestat but not Kyn recovered the root growth that was suppressed by 0.2 μm IPA, an intermediate metabolite that can be converted to IAA by YUC (6) (Fig. 5A). When the treatment concentrations of ponalrestat and Kyn were left unchanged, the ponalrestat-treated seedlings displayed hyposensitivity to increasing IPA concentrations compared with Kyn or mock treatment (Fig. 5A). These results are in accordance with the previous finding that Kyn targets TAA1, the upstream enzyme of IPA (19, 35), and indicate that the target of ponalrestat may be downstream of IPA, such as the YUC enzymes in the IPA pathway.
Figure 5.
Ponalrestat is an inhibitor of monooxygenase YUCs. A, root lengths of 3-day-old eto1–2 etiolated seedlings grown on 1/2 MS medium containing ponalrestat (5 μm), Kyn (1.5 μm), or DMSO as the mock control with co-treatments of increasing concentrations of IPA. Bars represent mean ± S.D. of at least 10 seedlings. Statistical differences between the groups were calculated with ANOVA (with Tukey's HSD post hoc test). Data points with different letters are significantly different at p < 0.01. B, root lengths of Col-0 or yucQ with single or combinatorial treatments of 1 μm ACC, 5 μm ponalrestat, and 100 nm IAA. Bars represent mean ± S.D. of at least 10 seedlings. Statistical differences between the groups were calculated with ANOVA (with Tukey's HSD post hoc test). Data points with different letters are significantly different at p < 0.01. C, DARTS assay. Recombinant GST-YUC2 was pretreated with increasing concentration of ponalrestat (0.5 mm to 5 mm, along with a control without ponalrestat pretreatment) then digested by Pronase protease. Protein levels were detected using GST antibody. As a control, GST tag was subjected to the same digestion treatment. D, in vitro assay of GST-YUC2 enzymatic activity. Purified GST-YUC2 recombinant protein was used for the enzyme activity assay. FAD and NADPH were added to the reaction buffer as cofactors. IPA was fed to YUC2 in the presence of increasing concentration of ponalrestat (0.1 mm, 1 mm, 5 mm, and 10 mm). The IAA product was detected by LC/MS, and the relative enzyme activity was defined as the level of product. Bars represent mean ± S.D. of three replicates experiments. Statistical differences between the groups were calculated with ANOVA (with Tukey's HSD post hoc test). Bars with different letters are significantly different at p < 0.01. All the above experiments were repeated for at least three times with similar results.
Consistent with this hypothesis and our previous results, the YUC quintuple (yucQ) mutant, with mutations in YUC3, YUC5, YUC7, YUC8, and YUC9 (36), lost sensitivity to both the negative effects of ACC and the positive effects of ponalrestat in root elongation (Fig. 5B). Moreover, ponalrestat suppressed the leaf epinastic curvature of yuc1D, a gain-of-function YUC1 mutant with excessive IAA biosynthesis (Fig. S4). These results indicate that YUC enzymes may be the targets of ponalrestat.
To further investigate whether ponalrestat directly acts through the auxin biosynthesis enzyme YUC, we performed several in vitro biochemical experiments. Drug affinity responsive target stability (DARTS) is a widely used strategy for validating protein-ligand interaction. This approach is based on the ability of many ligands to stabilize and thereby protect their target proteins from protease digestion (37, 38). In vitro–purified GST-YUC2 fusion protein was incubated with ponalrestat for 2 h before treatment with a Pronase, protease mixture, according to the methodology described by Lomenick et al. (38). Western blotting showed that ponalrestat protected GST-YUC2 protein from Pronase degradation in a concentration-dependent manner (Fig. 5C), indicating a direct interaction between ponalrestat and YUC2. We subsequently performed an enzymatic assay with GST-YUC2 according to a previously reported procedure (5). In the optimized reaction buffer system, IPA was converted into IAA with the catalysis of GST-YUC2 (Fig. 5D). Cofactors FAD and NADPH were essential for the enzyme activity as the removal of either cofactor largely decreased the enzyme activity levels (Fig. 5D). Using this enzymatic assay, we found that IAA production was reduced by ponalrestat treatment in a concentration-dependent manner, suggesting that ponalrestat has an inhibitory effect on YUC enzyme activity (Fig. 5D).
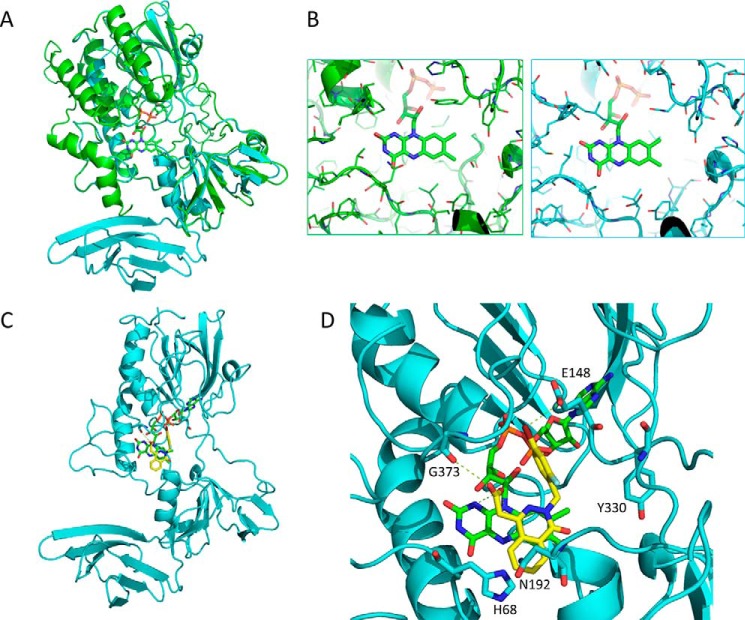
YUC2 homology modeling and ponalrestat docking simulation
For further insight into the mode of action of ponalrestat, we performed homology modeling of YUC2 based on the structure of its ortholog FMO in yeast (2GVC). Structure alignment (Fig. 6A) showed that YUC2 has a core FMO-like domain and a β-barrel structure in the C terminus. The β-barrel contains two layers of β sheets, and each layer includes three anti-parallel β strands. Structural analysis of FMO in complex with MET and NAD indicated a substrate-binding site in the center of the active site pocket right next to the co-factor FAD (Fig. S5) (39). The postulated pocket of YUC2 and that of FMO showed high similarity (Fig. 6A), suggesting conserved enzyme activity. However, there were some different residues in the pocket which potentially allow for the selection of specific substrates (Fig. 6B). Molecular docking simulation of ponalrestat to YUC2 revealed that ponalrestat could fit into the active site pocket of YUC2 and may act as a substrate antagonist (Fig. 6C). Several hydrogen bonds between ponalrestat and residues around the YUC2 active site were also predicted (Fig. 6D). The bromo group on the phenyl ring and the carboxylic acid group of ponalrestat were predicted to interact with Glu-148 and Gly-373, respectively. These residues are highly conserved in YUC homologs (Fig. S6), suggesting that ponalrestat may affect multiple YUC family members.
Figure 6.
Homology modeling of YUC2 and molecular docking of ponalrestat. A, structure of YUC2 (in cyan) based on homology modeling with FMO (in green). B, structural details of the binding pocket of FMO (in green) and YUC2 (in cyan) in complex with FAD. C, a complex structure of YUC2 and ponalrestat based on a docking simulation. D, structural details of the binding pocket of YUC2 in complex with ponalrestat and FAD. Dashed lines indicate hydrogen bonds.
Structure-activity relationship (SAR) analysis of ponalrestat
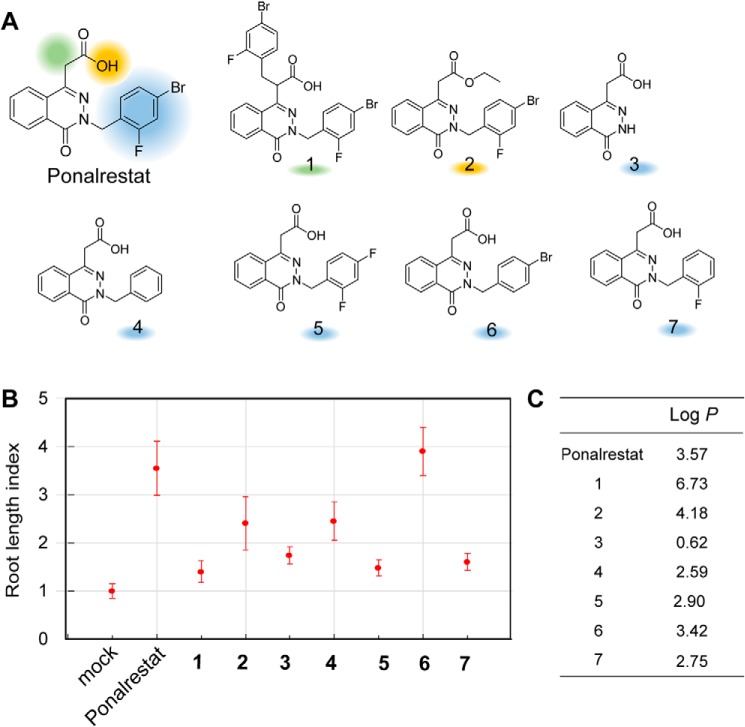
We synthesized several ponalrestat derivatives based on its backbone structure and estimated their biological activity using the root length index (Fig. 7A), i.e. the relative root length of derivative-treated seedlings divided by the average root length of mock-treated seedlings. Higher index values indicate higher ponalrestat-like activity.
Figure 7.
Structure-activity relationship analysis for ponalrestat. A, chemical structures of ponalrestat and its derivatives. B, estimation of chemical activities with root length index. Col-0 seeds were germinated and grown on 1/2 MS supplemented with or without 10 μm ponalrestat derivatives. The root lengths of 3-day-old seedlings were measured. The root length index is defined as the average root length of derivative-treated seedlings divided by the average root length of mock-treated seedlings. Bars represent mean ± S.D. of at least 10 seedlings. C, log P data for ponalrestat and derivatives calculated with ChemDraw.
As shown in Fig. 7B, the root length index value of ponalrestat is 3.5. Introducing an aromatic ring on the carboxylic acid chain (compound 1) and esterification on the carboxylic acid (compound 2) to ponalrestat dramatically impaired its root growth–promoting activity (root length index values dropped to 1.5 and 2.5, respectively), suggesting that modifications on the carboxylic acid chain are not suitable for enhancing ponalrestat activity. This observation is consistent with the docking simulation result that the carboxylic acid is important for interaction with Gly-373 and FAD (Fig. 6D). Deletion of the halogen aromatic ring (compound 3) led to decreasing activity. In contrast, introducing a benzyl to compound 3 (compound 4) recovered the activity to some extent (root length index value was increased from 1.8 to 2.5), but it was still weaker than the activity of ponalrestat, which possesses halogens on the benzene ring. Therefore, we focused on modifying the benzene ring with different combinations of halogens (compounds 5–7). Compared with ponalrestat, the low activities of compound 5 (in which the bromo group was replaced with a fluoro group) and compound 7 (with deletion of the bromo group) suggest that the para-bromo group rather than ortho-fluoro on the benzene ring is important for the bioactivity. In accordance with this hypothesis, deletion of para-fluoro group (compound 6) resulted in bioactivity that was almost the same as that of ponalrestat.
Log P, a measure of the partition coefficient of a molecule between a hydrocarbon solvent and water, is a key factor in biological SARs (40). According to Lipinski's “Rule of Five” for drug discovery and Briggs' “Rule of Three” for agrochemicals, the log P of molecules should be less than five or three for their bioavailability (41). Changes in the log P value were observed for all of the analyzed ponalrestat derivatives; in particular, compound 1 had a high log P value of 6.73 (Fig. 7C). Thus, the different bioactivities of the ponalrestat derivatives may be attributable to both their affinities for YUC proteins and their log P values. In summary, our SAR studies provide information for further chemical design of ponalrestat analogs.
Discussion
Chemical genetic screening is a powerful tool for identifying chemical regulators of plant growth and development. There are basically two strategies for high-throughput screening: Phenotype-directed screening and target-directed screening (24, 25). In the present study, we performed a phenotype-directed screen using ethylene signaling-activated mutants and found that ponalrestat, an inhibitor of human aldose reductase, could rescue the short root phenotype induced by the ethylene signal (Fig. 1 and Fig. S1). However, the ability of ponalrestat to recover root elongation was independent of the ethylene signal, based on the findings that ponalrestat did not significantly affect the protein level or transcriptional activity of EIN3, a master transcription factor of ethylene-signaling pathway (Fig. 2, A and B). Moreover, ponalrestat was still functional in the EIN3 overexpression line (Fig. 2C). Genetic and biochemical analysis indicated that ponalrestat acts on the IAA biosynthesis enzyme YUC by directly binding to it and inhibiting its catalysis of IPA to IAA (Figs. 4 and 5). Together with previous reports that ethylene stimulates IAA biosynthesis (16–18), our results suggest that ponalrestat recovers root elongation in eto1–1 and ctr1–1 by inhibiting endogenous IAA biosynthesis and that its target is the YUC enzyme.
We noticed that ponalrestat showed a high IC50 on YUC2 enzyme activity (Fig. 5D). One possible reason may result from the different affinity of ponalrestat for YUC family. The microarray data show that YUC3, YUC5, YUC7, YUC8, and YUC9 are highly expressed in Arabidopsis root, whereas the other YUCs including YUC2 have low or no expression in the root (36). The biological activity of ponalrestat in root may be contributed by its inhibition on YUCs with high expression levels in root rather than YUC2 we tested here. It is intriguing to perform a comparison for the affinities of ponalrestat for different YUCs based on biochemical experiment or computational modeling in future study. Another possibility for the high IC50 of ponalrestat on YUC2 enzyme activity is that ponalrestat might not be the optimal inhibitor of YUC. It could be modified or metabolized into a different compound in plants that is either more effective for transport or more potent to inhibit YUC enzymes.
In light of the complexity and importance of IAA biosynthesis in Arabidopsis, there have been many efforts to identify potential inhibitors of the key pathway components. In the IPA branch of IAA biosynthesis, Kyn and AOPP were previously identified as competitive inhibitors of TAA1 (19, 42). Yucasin and PPBo were identified as YUC inhibitors in separate studies (20, 21). Yucasin was identified in a screen using a maize coleoptile system, whereas PPBo was identified using a direct YUC enzyme assay system. In our study, the identification of ponalrestat was identified based on the root length phenotype. Although these different compounds all target the YUC enzyme, their effects on plant development are not identical. One reason may be that their structural differences lead to specificity for different YUC enzymes with different spatiotemporal expression patterns. Yucasin has a 1,2,4-triazole-3(4H)-thione moiety with a substructure similar to that of methimazole, an artificial substrate that blocks yeast FMO function (39). This moiety is crucial for the inhibitory effect of yucasin on YUC, which is an FMO enzyme (20). PPBo is an aromatic boronic acid that is extensively used in organic chemistry as building block (43). The structure of ponalrestat is clearly distinct from that of yucasin and aromatic boronic acids, and it is an aldose reductase inhibitor with potential application for the treatment of diabetes in humans. Our findings uncover the effects of ponalrestat in plants and provide a new chemical scaffold for developing YUC inhibitors.
Interestingly, our screening system based on the recovery of root elongation in mutants identified both Kyn and ponalrestat as inhibitors, and these two compounds have different targets in the IPA branch of auxin biosynthesis. In future studies, combinations of Kyn and ponalrestat at different concentrations could lead to wide-ranging effects, possibly allowing for precise regulation of root growth and fine-tuned investigation of the relationship between endogenous IAA concentrations and the output root length.
Materials and methods
Plant materials and growth conditions
Unless otherwise indicated, all Arabidopsis mutants and transgenic lines in this study, are in the Col-0 background. eto1–2 (13), ctr1–1 (26), EIN3ox (29), EIN3:GFP/ein3 eil1 (29), 5xEBS:GUS (17), and DR5:GFP (44) were described previously. wei2–2 (33) and wei8–1 (34), were gifts from Dr. Jose Alonso at North Carolina State University. yucQ (36) and yuc1D (45) were gifts from Dr. Yunde Zhao at the University of California San Diego.
All the seeds were stepwise surface-sterilized with 75% ethanol and 100% ethanol then sown on 1/2 Murashige and Skoog (MS) basic medium with 1% sucrose. All plated seeds were imbibed in a 4 °C refrigerator for 3 days to control the germination rate. For etiolated seedlings, seeds were wrapped in aluminum foil and incubated in the dark at 22 °C for 3 days. For green seedlings, seeds were maintained in a greenhouse at 22 °C under a 16 h/8 h light/dark cycle for 5 days. For the transient treatment experiments, 3-day-old seedlings were transferred into liquid MS medium supplemented with the indicated compounds. All the plant experiments were repeated at least three times with similar results.
Chemical genetic screening
The chemical genetic screening was performed at the University of California, Riverside with a selection of 2000 diverse compounds (Microsource Spectrum SP2000). The average stock concentration was 5 mg/ml in DMSO. The library was screened at a concentration of 50–100 μm in liquid MS medium. Seeds were surface sterilized and distributed by pipette into 96-well plates containing liquid 1/2 MS (0.1% agar). After 3 days at 4 °C, automated robot (Beckman Coulter Biomek FX) added the compounds from 384-well plates into the 96-well plates. DMSO served as the mock control. Seeds were germinated and grown in the dark for 3 days. The root lengths of the seedlings in each well were analyzed to identify longer roots compared with that of the mock-treated control.
Chemical preparation
Ponalrestat and its derivatives were synthesized by the Yang Lab at Peking University according to the procedures reported by Sriram et al. (46). These compounds were confirmed by hydrogen NMR (File S1). Other compounds used in this study were purchased from Sigma-Aldrich. All the compound stocks were dissolved in DMSO except for IAA which was dissolved in ethanol.
GUS staining
Seedlings were grown on the indicated media for 3 days in the dark or 5 days under light, then collected and washed with PBS buffer (100 mm Na2HPO4·12H2O, and NaH2PO4·2H2O) three times. After treatment with the staining buffer (100 mm PBS buffer, 10 mm Na2EDTA, 0.5 mm K4 [Fe(CN)6])·3H2O, 0.5 mm K3 [Fe(CN)6], 0.1% Triton X-100, and 1 mg/ml X-Gluc), 70% ethanol was used to terminate the staining reaction. The seedlings were mounted on slides with Hoyer's solution (chloral hydrate:water:glycerol = 8:3:1) and examined by differential interference contrast microscopy.
EIN3 protein level quantification
5-day-old EIN3-GFP/ein 3eil1 seedlings grown in the dark were frozen in liquid nitrogen and ground into powder. Total proteins were extracted from 20-mg seedlings treated by the mock control or chemicals with 60 μl extraction buffer (15 mm Tris-HCl, pH 7.5, 0.3 m urea, 7% SDS, 0.35% BPB, 35% glycerol, 50 mm DTT) and boiled for 5 min followed by centrifugation at 13,000 rpm at 4 °C. 15-μl supernatants (total proteins from 5 mg ground powder) were loaded into 7.5% SDS-PAGE gel. After separation SDS-PAGE, the proteins were transferred to nitrocellulose membrane followed by staining with Ponceau S. Western blotting was performed following standard procedures using an α-GFP antibody (1:5000 dilution) and anti-Rabbit HRP (10,000 dilution). Immobilon Western HRP Substrate and Tanon 5200 Chemi-Image System were used for chemiluminescent detection. A second-round Western blotting was performed for actin as a loading reference with anti-actin and anti-mouse.
RNA extraction and real-time quantitative RT-PCR analysis of gene expression
Total RNA was prepared using TRIzol reagent (Invitrogen), and 2 μg RNA was added to a 20 μl reverse transcription reaction containing M-MLV reverse transcriptase (Promega). The cDNA product was then used for a PCR amplification reaction to test the quality of the cDNA. Real-time PCR was performed using SYBR Green Mix (Takara). The primers for the auxin downstream marker gene GH3.3 were (5′–3′) forward, ACAATTCCGCTCCACAGTTC and reverse, ACGAGTTCCTTGCTCTCCAA. The final expression level was normalized to the PP2A reference control. The present data are mean values of three biological replicates with standard deviation calculated by Roche 480.
Confocal laser microscopy
All seedlings were grown on 1/2 MS medium in the dark for 3 days then transferred into liquid MS medium supplemented with the indicated compounds for 4 h prior to mounting on glass slides. A Zeiss LSM-710 microscope with 20× and 40× objectives was used to detect GFP fluorescence. The excitation wavelength was 488 nm, and a band-pass filter of 510 to 525 nm was used for emission.
In vitro expression of YUCs and enzyme activity assays
The YUC2 coding sequence was amplified from cDNA by PCR (forward, 3′–5′ AACGGATCCATGGAGTTTGTTACAG and reverse, 5′–3′, TAAGTCGACTTAACAATGTTGAGGACGAG) then inserted into expression vector pGEX-6p-1. Escherichia coli BL21 strain was used for expression of GST-YUC2. The cells were cultured in LB media (A600 = 0.65) and induced with 100 μm Isopropyl β-D-1-thiogalactopyranoside at 37 °C for 2 h. Cells were harvested and suspended with lysis buffer (50 mm pH 7.5 Tris-HCl, 1 mm EDTA, 100 mm NaCl, and 20% glycerol) and sonicated. After centrifugation at 10,000 × g, the supernatants were purified using the GE AKTA PURE system with GSH columns according to the manufacturer's instructions. 1× PBS buffer (pH 7.4) was used as wash buffer and 50 mm Tris-HCl (pH 8.0) containing 10 mm reduced GSH was used as elution buffer. Purified proteins were concentrated using Amicon Ultra-4 10K (Millipore). The protein concentrations were calculated using NanoDrop (extinction coefficient (ϵ) = 94.62 × 1000). The purified and concentrated proteins were aliquoted then immediately frozen in liquid nitrogen and stored at −80 °C.
The enzyme activity assay for GST-YUC2 was performed as described in literature (5). The protein concentration of GST-YUC2 was determined using Quick StartTM Bradford Protein Assay (Bio-Rad), a procedure based on the classic method of Bradford. The reaction (100 μl standard reaction buffer containing 20 μg GST-YUC2 protein, 100 μm substrate IPA, 40 μm cofactor FAD, 1 mm cofactor NADPH in PBS buffer, pH 7.4) was incubated at 30 °C for 30 min then terminated with 100 μl acetonitrile. The final products were extracted with ethyl acetate and dissolved in methanol followed by analysis with an HPLC coupled with electrospray ionization MS (HPLC/ESI-MS). Briefly, the samples were injected into a C18 column (1.9 μm, 2.1 × 100 mm; Thermo Fisher) connected with UltiMate 3000 HPLC system (Thermo Fisher) followed by a gradient elution (solvent A: 0.1% acetate, 5% methanol in water; solvent B: 0.1% acetate in methanol) at a flow rate of 0.3 ml/min. The gradient profile was applied as follows: 0–65% solvent B (0–5 min), 65–100% solvent B (5–6 min), 100–0% solvent B (6.1–8 min). The effluent was introduced into the MS, Q Exactive (Thermo Fisher) for analysis. All the data were quantified by Excalibur software.
DARTS Assay
The assay was modified from previously described protocols (37, 38). The purified protein (as described above under “In vitro expression of YUCs and enzyme activity assays”) was adjusted to an appropriate concentration with 50 mm Tris-HCl buffer (pH 7.5). The protein was pre-incubated with compounds at 4 °C for 2 h. The reaction was initiated by adding 2 μl 1:100 or 1:500 Pronase solution to one aliquot of compound-treated sample or DMSO as the control. After incubation at room temperature for 30 min, the reaction was stopped. A Western blotting was performed to detect GST-YUC2 protein with anti-GST antibody.
Computational docking and molecular modeling
The structure of YUC was modeled using SWISS-MODEL (47) based on the structure of FMO (PDB ID 2GVC). The alignment of MET and NADPH was based on the structures of FMO-MET (PDB ID 2GVC) and FMO-NADPH (PDB ID 2GV8). Ponalrestat was docked into YUC2 using PatchDock docking software (48). The structure alignment was performed using the program PyMOL.
Author contributions
Y. Z. and K. J. formal analysis; Y. Z., H. L., Y. W., and K. J. investigation; Y. Z. and K. J. writing-original draft; H. L., Q. S., J. W., Y. W., W. S., and Z. Y. methodology; Y. X., W. H., K. J., and H. G. writing-review and editing; K. J. and H. G. supervision; H. G. conceptualization; H. G. funding acquisition; H. G. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Natasha Raikhel, the Center for Plant Cell Biology at the University of California, for support on the chemical screen and Dr. Yunde Zhao, Division of Biological Sciences at the University of California, for providing the yucQ mutant.
This work was supported by the National Natural Science Foundation of China Grants 31570286 and 91740203 (to H. G.) and Grant 21907049 (to K. J.), the Guangdong Innovative and Entrepreneurial Research Team Program Grant 2016ZT06S172 (to K. J.), and the Shenzhen Sci-Tech Fund Grant KYTDPT20181011104005 (to K. J.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S6 and File S1.
- IAA
- indole-3-acetic acid
- Trp
- tryptophan
- IPA
- indole-3-pyruvic acid
- YUC
- YUCCA
- FMO
- flavin-containing monooxygenase
- Kyn
- l-kynurenine
- DARTS
- drug affinity responsive target stability
- SAR
- structure-activity relationship
- MS
- Murashige and Skoog
- ANOVA
- analysis of variance.
References
- 1. Thimann K. (1938) Hormones and the analysis of growth. Plant Physiol. 13, 437–449 10.1104/pp.13.3.437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Evans M. L., Ishikawa H., and Estelle M. A. (1994) Responses of Arabidopsis roots to auxin studied with high temporal resolution: Comparison of wild type and auxin-response mutants. Planta 194, 215–222 10.1007/BF00196390 [DOI] [Google Scholar]
- 3. Korasick D. A., Enders T. A., and Strader L. C. (2013) Auxin biosynthesis and storage forms. J. Exp. Bot. 64, 2541–2555 10.1093/jxb/ert080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Woodward A. W., and Bartel B. (2005) Auxin: Regulation, action, and interaction. Ann. Bot. 95, 707–735 10.1093/aob/mci083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhao Y. (2012) Auxin biosynthesis: A simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant. 5, 334–338 10.1093/mp/ssr104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mashiguchi K., Tanaka K., Sakai T., Sugawara S., Kawaide H., Natsume M., Hanada A., Yaeno T., Shirasu K., Yao H., McSteen P., Zhao Y., Hayashi K., Kamiya. Y., and Kasahara H. (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 18512–18517 10.1073/pnas.1108434108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stepanova A. N., Yun J., Robles L. M., Novak O., He W., Guo H., Ljung K., and Alonso J. M. (2011) The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23, 3961–3973 10.1105/tpc.111.088047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Won C., Shen X., Mashiguchi K., Zheng Z., Dai X., Cheng Y., Kasahara H., Kamiya Y., Chory J., and Zhao Y. (2011) Conversion of tryptophan to indole-3-acetic acid by tryptophan aminotransferases of Arabidopsis and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 18518–18523 10.1073/pnas.1108436108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dai X., Mashiguchi K., Chen Q., Kasahara H., Kamiya Y., Ojha S., DuBois J., Ballou D., and Zhao Y. (2013) The biochemical mechanism of auxin biosynthesis by an Arabidopsis YUCCA flavin-containing monooxygenase. J. Biol. Chem. 288, 1448–1457 10.1074/jbc.M112.424077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Abeles F., Morgan P., and Saltveit M. Jr. (1992) Ethylene in Plant Biology, 2nd ed Academic Press, Cambridge, MA: 10.1016/C2009-0-03226-7 [DOI] [Google Scholar]
- 11. Zhao Q., and Guo H. W. (2011) Paradigms and paradox in the ethylene signaling pathway and interaction network. Mol. Plant 4, 626–634 10.1093/mp/ssr042 [DOI] [PubMed] [Google Scholar]
- 12. Bleecker A. B., Estelle M. A., Somerville C., and Kende H. (1988) Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science 241, 1086–1089 10.1126/science.241.4869.1086 [DOI] [PubMed] [Google Scholar]
- 13. Guzmán P., and Ecker J. R. (1990) Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2, 513–523 10.1105/tpc.2.6.513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guo H., and Ecker J. R. (2004) The ethylene signaling pathway: new insights. Curr. Opin. Plant Biol. 7, 40–49 10.1016/j.pbi.2003.11.011 [DOI] [PubMed] [Google Scholar]
- 15. Ju C., and Chang C. (2015) Mechanistic insights in ethylene perception and signal transduction. Plant Physiol. 169, 85–95 10.1104/pp.15.00845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rzicka K., Ljung K., Vanneste S., Podhorská R., Beeckman T., Friml J., and Benková E. (2007) Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19, 2197–2212 10.1105/tpc.107.052126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stepanova A. N., Yun J., Likhacheva A. V., and Alonso J. M. (2007) Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19, 2169–2185 10.1105/tpc.107.052068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Swarup R., Perry P., Hagenbeek D., Van Der Straeten D., Beemster G. T. S., Sandberg G., Bhalerao R., Ljung K., and Bennett M. J. (2007) Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19, 2186–2196 10.1105/tpc.107.052100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. He W., Brumos J., Li H., Ji Y., Ke M., Gong X., Zeng Q., Li W., Zhang X., An F., Wen X., Li P., Chu J., Sun X., Yan C., et al. (2011) A small-molecule screen identifies l-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 23, 3944–3960 10.1105/tpc.111.089029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nishimura T., Hayashi K. I., Suzuki H., Gyohda A., Takaoka C., Sakaguchi Y., Matsumoto S., Kasahara H., Sakai T., Kato J. I., Kamiya Y., and Koshiba T. (2014) Yucasin is a potent inhibitor of YUCCA, a key enzyme in auxin biosynthesis. Plant J. 77, 352–366 10.1111/tpj.12399 [DOI] [PubMed] [Google Scholar]
- 21. Kakei Y., Yamazaki C., Suzuki M., Nakamura A., Sato A., Ishida Y., Kikuchi R., Higashi S., Kokudo Y., Ishii T., Soeno K., and Shimada Y. (2015) Small-molecule auxin inhibitors that target YUCCA are powerful tools for studying auxin function. Plant J. 84, 827–837 10.1111/tpj.13032 [DOI] [PubMed] [Google Scholar]
- 22. Tsugafune S., Mashiguchi K., Fukui K., Takebayashi Y., Nishimura T., Sakai T., Shimada Y., Kasahara H., Koshiba T., and Hayashi K. I. (2017) Yucasin DF, a potent and persistent inhibitor of auxin biosynthesis in plants. Sci. Rep. 7, 13992 10.1038/s41598-017-14332-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hicks G. R., and Raikhel N. V (2014) Plant chemical biology: Are we meeting the promise? Front. Plant Sci. 5, 455 10.3389/fpls.2014.00455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dejonghe W., and Russinova E. (2017) Plant chemical genetics: From phenotype-based screens to synthetic biology. Plant Physiol. 174, 5–20 10.1104/pp.16.01805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jiang K., and Asami Tadao (2018) Chemical regulators of plant hormones and their applications in basic research and agriculture. Biosci. Biotechnol. Biochem. 82, 1265–1300 10.1080/09168451.2018.1462693 [DOI] [PubMed] [Google Scholar]
- 26. Kieber J. J., Rothenberg M., Roman G., Feldmann K. A., and Ecker J. R. (1993) CTR1, a negative regulator of the ethylene pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72, 427–441 10.1016/0092-8674(93)90119-B [DOI] [PubMed] [Google Scholar]
- 27. Chung S., and LaMendola J. (1989) Cloning and sequence determination of human placental aldose reductase gene. J. Biol. Chem. 264, 14775–14777 [PubMed] [Google Scholar]
- 28. Chao Q., Rothenberg M., Solano R., Roman G., Terzaghi W., and Ecker J. R. (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell. 89, 1133–1144 10.1016/S0092-8674(00)80300-1 [DOI] [PubMed] [Google Scholar]
- 29. Guo H., and Ecker J. R. (2003) Plant responses to ethylene gas are mediated by SCFEBF1/EBF2-dependent proteolysis of EIN3 transcription factor. Cell 115, 667–677 10.1016/S0092-8674(03)00969-3 [DOI] [PubMed] [Google Scholar]
- 30. Potuschak T., Lechner E., Parmentier Y., Yanagisawa S., Grava S., Koncz C., and Genschik P. (2003) EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115, 679–689 10.1016/S0092-8674(03)00968-1 [DOI] [PubMed] [Google Scholar]
- 31. An F., Zhao Q., Ji Y., Li W., Jiang Z., Yu X., Zhang C., Han Y., He W., Liu Y., Zhang S., Ecker J. R., and Guo H. (2010) Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22, 2384–2401 10.1105/tpc.110.076588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mao J. L., Miao Z. Q., Wang Z., Yu L. H., Cai X. T., and Xiang C. (2016) Arabidopsis ERF1 mediates cross-talk between ethylene and auxin biosynthesis during primary root elongation by regulating ASA1 expression. PLoS Genet. 12, e1005760 10.1371/journal.pgen.1005760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stepanova A. N., Hoyt J., Hamilton A., and Alonso J. M. (2005) A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17, 2230–2242 10.1105/tpc.105.033365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stepanova A. N., Robertson-Hoyt J., Yun J., Benavente L. M., Xie D. Y., Doležal K., Schlereth A., Jürgens G., and Alonso J. M. (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133, 177–191 10.1016/j.cell.2008.01.047 [DOI] [PubMed] [Google Scholar]
- 35. Alonso J. M., Stepanova A. N., Solano R., Wisman E., Ferrari S., Ausubel F. M., and Ecker J. R. (2003) Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 100, 2992–2997 10.1073/pnas.0438070100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen Q., Dai X., De-Paoli H., Cheng Y., Takebayashi Y., Kasahara H., Kamiya Y., and Zhao Y. (2014) Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol. 55, 1072–1079 10.1093/pcp/pcu039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lomenick B., Hao R., Jonai N., Chin R. M., Aghajan M., Warburton S., Wang J., Wu R. P., Gomez F., Loo J. A., Wohlschlegel J. A., Vondriska T. M., Pelletier J., Herschman H. R., Clardy J., Clarke C. F., and Huang J. (2009) Target identification using drug affinity responsive target stability (DARTS). Proc. Natl. Acad. Sci. U.S.A. 106, 21984–21989 10.1073/pnas.0910040106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lomenick B., Jung G., Wohlschlegel J. A., and Huang J. (2011) Target identification using drug affinity responsive target stability (DARTS). Curr. Protoc. Chem. Biol. 3, 163–180 10.1002/9780470559277.ch110180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Eswaramoorthy S., Bonanno J. B., Burley S. K., and Swaminathan S. (2006) Mechanism of action of a flavin-containing monooxygenase. Proc. Natl. Acad. Sci. U.S.A. 103, 9832–9837 10.1073/pnas.0602398103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Martin Y. C. (2018) How medicinal chemists learned about log P. J. Comput. Aided Mol. Des. 32, 809–819 10.1007/s10822-018-0127-9 [DOI] [PubMed] [Google Scholar]
- 41. Lindell S. D., Pattenden L. C., and Shannon J. (2009) Combinatorial chemistry in the agrosciences. Bioorg. Med. Chem. 17, 4035–4046 10.1016/j.bmc.2009.03.027 [DOI] [PubMed] [Google Scholar]
- 42. Narukawa-Nara M., Nakamura A., Kikuzato K., Kakei Y., Sato A., Mitani Y., Yamasaki-Kokudo Y., Ishii T., Hayashi K. I., Asami T., Ogura T., Yoshida S., Fujioka S., Kamakura T., Kawatsu T., Tachikawa M., Soeno K., and Shimada Y. (2016) Aminooxy-naphthylpropionic acid and its derivatives are inhibitors of auxin biosynthesis targeting l-tryptophan aminotransferase: structure-activity relationships. Plant J. 87, 245–257 10.1111/tpj.13197 [DOI] [PubMed] [Google Scholar]
- 43. Kubo Y., Nishiyabu R., and James T. D. (2015) Hierachical supramolecules and organization using boronic acid building blocks. Chem. Commun. 51, 2005–2020 10.1039/C4CC07712A [DOI] [PubMed] [Google Scholar]
- 44. Ottenschläger I., Wolff P., Wolverton C., Bhalerao R. P., Sandberg G., Ishikawa H., Evans M., and Palme K. (2003) Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc. Natl. Acad. Sci. U.S.A. 100, 2987–2991 10.1073/pnas.0437936100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhao Y., Christensen S. K., Fankhauser C., Cashman J. R., Cohen J. D., Weigel D., and Chory J. (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291, 306–309 10.1126/science.291.5502.306 [DOI] [PubMed] [Google Scholar]
- 46. Sriram D., Yogeeswari P., Senthilkumar P., Sangaraju D., Nelli R., Banerjee D., Bhat P., and Manjashetty T. H. (2010) Synthesis and antimycobacterial evaluation of novel phthalazin-4-ylacetamides against log- and starved phase cultures. Chem. Biol. Drug Des. 75, 381–391 10.1111/j.1747-0285.2010.00947.x [DOI] [PubMed] [Google Scholar]
- 47. Arnold K., Bordoli L., Kopp J., and Schwede T. (2006) The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 22, 195–201 10.1093/bioinformatics/bti770 [DOI] [PubMed] [Google Scholar]
- 48. Mashiach E., Schneidman-Duhovny D., Peri A., Shavit Y., Nussinov R., and Wolfson H. J. (2010) An integrated suite of fast docking algorithms. Proteins 78, 3197–3204 10.1002/prot.22790 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.