Abstract
Super-resolution microscopy techniques offer subdiffraction limited resolution that is two- to ten-fold improved compared to that offered by conventional confocal microscopy. This breakthrough in resolution for light microscopy has contributed to new findings in neuroscience and synapse biology. This review will focus on the Structured Illumination Microscopy (SIM), Stimulated emission depletion (STED) microscopy, and Stochastic optical reconstruction microscopy (STORM) / Single molecule localization microscopy (SMLM) techniques and compare them for the better understanding of their differences and their suitability for the analysis of synapse biology. In addition, we will discuss a few practical aspects of these microscopic techniques, including resolution, image acquisition speed, multicolor capability, and other advantages and disadvantages. Tips for the improvement of microscopy will be introduced; for example, information resources for recommended dyes, the limitations of multicolor analysis, and capabilities for live imaging. In addition, we will summarize how super-resolution microscopy has been used for analyses of neuromuscular junctions and synapses.
Keywords: neuromuscular junction, SIM, STED, STORM, super-resolution microscopy, synapse
Introduction
Super-resolution microscopy techniques overcome the resolution limit of light microscopy that is defined by Abbe’s diffraction limit of approximately 200 nm [2, 94]. For the recognition of this breakthrough, the 2014 Nobel Prize in Chemistry was awarded to Drs. Stefan Hell, Eric Betzig, and William E. Moerner for the development of the super-resolved fluorescence microscopy [36]. Several super-resolution microscopy techniques are commercially available, including Structured Illumination Microscopy (SIM) [45–47, 83], STimulated Emission Depletion (STED) microscopy [48, 62], and STochastic Optical Reconstruction Microscopy (STORM) / Photo Activated Localization Microscopy (PALM) [11, 50, 100, 108]. STORM and PALM are types of Single Molecule Localization Microscopy (SMLM). This review will compare the advantages and weaknesses of these super-resolution microscopy techniques and summarize how they have been applied to analyses of synapses. Neuromuscular junctions (NMJs) are large synapses formed between motor neurons and muscle fibers. Due to their large sizes and accessibility, these synapses have been studied extensively using histological and physiological methods [33, 43, 53, 72, 96, 115]. Even considering the long history of studying these synapses, recent applications of super-resolution microscopy to the examination of these synapses have revealed new findings.
Resolution
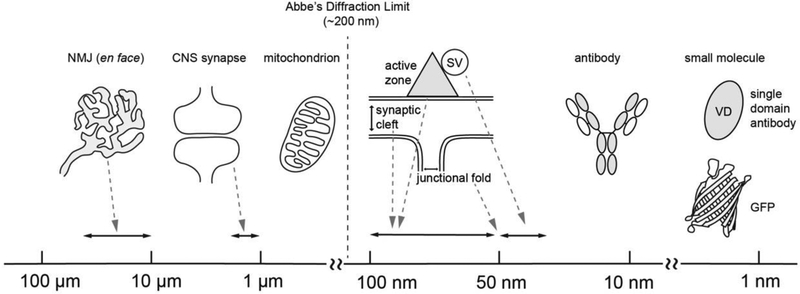
Super-resolution microscopes offer subdiffraction limited resolution, which is better than the resolution of confocal microscopes of approximately 200 nm. In neurobiology, confocal microscopes can image axons and nerves, dendritic arbor morphology, mitochondrial morphology, nuclei, synapses and NMJs. However, superresolution microscopes have been useful for imaging the detailed structures of spines, protein distribution patterns in nodes of Ranvier, nuclear pore proteins, the subsynaptic localization of pre- and postsynaptic proteins, and synaptic vesicles. An en face view of NMJs is large, with a size ranging from approximately 10 – 60 μm (human, mouse) [22, 56]; however, some of the key features of this synapse have a size that is below the diffraction limited resolution of 200 nm. For example, synaptic vesicles are 55 nm [64, 82], active zones are 50 – 100 nm [73, 82, 86], and the synaptic cleft is 50 – 100 nm [72]; in addition, the junctional fold crest width is 207 nm and the opening width is 55 nm [139] (Figure 1), the laminin protein length is 77 nm [37], and the Lrp4 protein length is approximately 60 nm [72]. Thus, the improved resolution of super-resolution microscopy is necessary for analyzing these features, and examples of such studies will be described later. In this review, we will focus on the three commercially available super-resolution microscopy techniques: SIM, STED, and STORM. The differences in their resolution will be discussed along with a brief explanation of each imaging mechanism.
Figure 1.
Abbe’s diffraction limit, the sizes of synaptic structures and tools used in super-resolution microscopy. The X-axis indicates the size in micrometers to nanometers. The sizes of the synaptic structures and objects shown in the figure are as follows: NMJs in the en face view are 10 – 60 μm (human, mouse) [22, 56], the central nervous system (CNS) synapse is 1 – 2 μm (mouse) [59], the synaptic cleft of the NMJ is 50 – 100 nm [72], the junctional fold crest width is 207 nm and the opening width is 55 nm [139], the active zone is 50 – 100 nm [73, 82, 86], the synaptic vesicle (SV) is 40 – 50 nm [72, 102], the antibody is 12 × 14 nm [68, 80], the nanobody (variable domain, VD) is 2.5 × 4 nm [60], and green fluorescent protein (GFP) is 2.4 × 4.2 nm [51].
The XY-plane resolution of these techniques is as follows: SIM, 120 nm, STED and STORM, 20 – 30 nm (Table 1). Their resolution is significantly improved compared to that of confocal microscopy, which has a resolution of approximately 200 nm in the XY plane and 500 nm along the Z-axis (for a green fluorescent dye observed with an NA=1.4 lens) [114]. The Z-axis resolution of confocal microscopy is lower than the XY-resolution because of the broad focal region, as determined by Abbe’s diffraction limited resolution theory, which was formulated by Abbe [2] and Rayleigh [94]. The Z-axis resolution of the super-resolution microscopy techniques are as follows: SIM, 300 nm, STED, 75 – 100 nm (Easy 3D, STED 3×), and STORM, 50 nm (STORM 3D). In addition to the differences in resolution, these techniques differ in the mechanisms that improve the resolution. SIM and STORM reconstruct super resolution images based on the postprocessing of the raw data micrographs. In contrast, STED microscopy generates an image with optical data only without postprocessing the raw data. More specifically, SIM generates super-resolution images using a Fourier transform that is applied to the raw data micrographs. STED microscopy generates super-resolution images by scanning the laser beam over the specimen, similar to confocal microscopy. STORM generates superresolution images by collecting thousands of micrographs and reconstructing them using Gaussian distribution approximation and assigning dots, similar to pointillism. The mechanisms of each method will be discussed next.
Table 1:
Comparison of super resolution microscopy specifications
| Parameter | SIM | STED | STORM |
|---|---|---|---|
| Resolution XY-plane | 115 – 120 nm [85, 111] | 20 – 30 nm [3, 44, 71] | 20 – 30 nm [85, 111] |
| Resolution Z-axis | 300 nm | 75 – 100 nm (Easy 3D, STED 3×) | 50 nm (STORM 3D) |
| Multi-color capability | Two channels can be acquired simultaneously and four to six colors without simultaneous acquisition [91, 140]. | Dual-color [12, 26, 86, 125], triple-color [7, 18, 110, 121, 141], and quadruple-color STED [136] have been reported. | Dual-color detection is routinely used [1, 6, 20, 25, 52, 57, 65, 66]. Triple-color STORM has been reported in combination with spectral demixing [69]. |
| Speed | Commercial model from Nikon offers a rate of 15 FPS. Zeiss Elyra 7 system offers a frame rate of 255 FPS [85, 111]. | Commercially available resonant scanners from Leica offer a rate of 50 FPS, or up to 428 FPS by restricting the region of interest [15, 16]. | Limited temporal resolution with an imaging speed of less than 1 FPS [85, 111]. |
| Live imaging | Suitable | Suitable | Not preferred |
| Post-processing | Necessary, using Fourier transform of 9 to 15 raw data micrographs [45, 47, 83]. | Not necessary, a raw data micrograph is a super resolution image [48]. | Necessary, using Gaussian fitting of tens of thousands of raw data micrographs [5, 10, 11, 50, 100, 108]. |
FPS= frames per second; SIM= structured illumination microscopy; STED= stimulated emission depletion; STORM= stochastic optical reconstruction microscopy
SIM uses a wide-field microscopy setup and rotating grid pattern illumination to generate Moiré fringes and to collect nine to 15 micrographs by changing the mask orientation, and the final super resolution image is reconstructed using a Fourier transform of the acquired micrographs [45, 47, 83]. SIM has been further developed so that this method can be applied in three dimensions to improve the resolution in the lateral and axial directions, which is called 3D-SIM [46]. The newer version of SIM uses lattice spot pattern illumination instead of grid pattern illumination to generate higher contrast images and reduce the illumination. Various SIM methods have been developed to increase spatial resolution, temporal resolution, and imaging depth by combining a custom microlens array, total internal reflection (TIRF) microscopy, laser-based point-scanning microscopy, two-photon excitation microscopy, and a lattice light sheet. These different types of SIM have been described in detail in an excellent review article [137]. These SIM techniques achieve an approximately two-fold improvement in resolution in the XY plane (lateral, 115–120 nm) and the Z-axis (axial, 300 nm) compared to confocal microscopy. SIM microscopy instruments are commercially available from various suppliers, including GE Healthcare Life Sciences, Nikon, Oxford Nanoimaging, and Zeiss.
STED microscopy achieves subdiffraction limited resolution by combining a confocal microscope with a depletion laser. The depletion laser has a longer wavelength than the excitation laser and illuminates the sample in a doughnut-like ring pattern and in a coaxial manner with the excitation laser. The STED depletion laser returns the excited fluorophores to the ground state by stimulated emission [48]. The emission generated from this process has the same wavelength as the STED laser and can be separated from the emission light wavelength, i.e., the fluorescent signal. Thus, the internal diameter of the doughnut-like ring pattern of the depletion laser determines the area of fluorophore excitation and therefore the resolution of an image, which is 20 – 30 nm in the XY plane [3, 44, 71]. The two lasers scan the sample in a coaxial manner and generate an image in a manner similar to confocal microscopy. Therefore, a postprocessing of the raw data is not necessary to generate super resolution images. The depletion laser illumination can be split in the axial direction to form a Z-axis doughnut ring pattern for improving the Z-axis resolution, which can reach 100 nm or below. The resolution of STED microscopy is also improved using a technique called time-gating, which restricts the time window for collecting fluorescent signals after the initiation of excitation. The combination of time-gated detection with a pulsed excitation laser contributes to improving the resolution based on the differences between the fluorescence lifetimes of the fluorophores that are depleted by the STED laser [78, 130]. The time-gating detection also contributes to decreasing the power of the STED depletion laser and maintaining super resolution. The use of fluorescent dyes with longer emission wavelengths is disadvantageous for improving resolution in confocal microscopy. However, this is not the case for STED microscopy because the resolution is defined by the size of the doughnut-like ring pattern of the depletion laser. Thus, the use of red to far-red fluorescent dyes with a pulsed 775 nm STED laser is beneficial for expanding the fluorescent dye repertoire and improving live imaging, which will be described in a later section. STED microscopy instruments are commercially available from various suppliers, including Abberior Instruments and Leica Microsystems.
STORM uses a wide-field microscopy setup, activates few fluorophores stochastically with low intensity light, collects tens of thousands of frames (micrographs), and reconstructs one super resolution image by Gaussian fitting of each fluorophore in the XY plane according to the collected images [5, 10, 11, 50, 100, 108]. The stochastic activation of fluorophores is a random event that allows each fluorophore in the XY plane to be separated temporally. This temporal separation is spread over tens of thousands of frames (micrographs) collected during one imaging session. Many frames are necessary for the Gaussian fitting of the collected signals to determine the location of single molecules; therefore, STORM is also called Single Molecule Localization Microscopy. A super resolution image is reconstructed by collecting these single molecule data in one XY plane. STORM images are generated by placing each fluorophore at a calculated location in a single plane by pointillism. Thus, the final image is composed of dots. This postprocessing calculation process improves the lateral resolution beyond the diffraction limit. The lateral resolution of STORM is up to 20 nm, which is approximately 10 times higher than that of confocal microscopy. The Z-axis resolution is improved using a different principle. Fluorescent signals that are not exactly at the focal plane are slightly deformed due to the astigmatism-induced stretch in the X or Y direction. Based on the astigmatism-induced deformation of the signal from a single fluorophore, the axial location of the single fluorophore can be calculated to estimate the distance from the focal plane and the direction in terms of the Z-axis. This method improves the axial resolution to approximately 50 nm, which is also approximately 10 times higher than that of confocal microscopy. Super-resolution microscopy techniques similar to STORM that use the stochastic activation of fluorescent signals and SMLM techniques include PALM. Single molecule localization microscopy instruments are commercially available from various suppliers, including GE Healthcare Life Sciences, Nikon, Oxford Nanoimaging Limited, and Zeiss.
Recently, a variety of other techniques have been developed to improve the resolution of imaging. Some of these techniques will be briefly introduced here. The resolution of confocal microscopy has been improved by the postprocessing optical image data using deconvolution software, which is called confocal super resolution or super resolution imaging in contrast to super resolution microscopy. This type of approach is offered by several confocal microscopy suppliers, including Leica, Nikon, and Olympus. Meanwhile, Zeiss has improved the resolution by developing an array of detectors to acquire the Airy disk pattern of a signal and use this information to reconstruct an image. These new techniques have improved the resolution by approximately two-fold compared to that of confocal microscopy, which approaches the resolution of SIM. On the other hand, lattice light sheet microscopy has been developed to image live three-dimensional structures and dynamic events at sub-second intervals [19, 74, 138]. The resolution of this technique exceeds that of 3D images generated by high numerical aperture confocal or spinning disk microscopy; however, it does not exceed the resolution of widefield SIM [19]. Another technique, expansion microscopy, has been reviewed previously [123] and has also been combined with lattice light sheet microcopy [41]. In this review, we will focus on SIM, STED, and STORM super-resolution microscopy techniques.
Number of colors
Similar to conventional fluorescence microscopy techniques, super-resolution microscopy can be performed with multiple colors. The image construction of SIM depends on the postprocessing of nine to 15 acquired images with Moiré fringes; therefore, multicolor imaging is possible if the excitation and emission wavelengths are separated between the fluorophores [91, 140]. Commercially available SIM equipment from Nikon and Zeiss acquires two channels simultaneously to meet the fast imaging needs of live imaging and four to six different colors for multicolor requirements without simultaneous acquisition.
In STED microscopy, images are generated similarly to confocal microscopy by scanning the specimen with the excitation laser and acquiring the emission wavelength of each fluorophore. The difference from confocal microscopy is the need to match the emission wavelengths of the fluorophores with the wavelength of the STED laser for the depletion of the excited fluorophores to the ground state. Specifically, the long wavelength end of the emission spectrum needs to overlap with the wavelength of the STED depletion laser. Many commercially available fluorophores have emissions at long wavelengths and can be successfully combined with STED depletion laser wavelengths (592 nm, 660 nm, 775 nm). Therefore, multicolor STED microscopy is routinely performed. For example, dual-color STED analysis has been performed to study mouse NMJs [86], synaptic protein localization in central nervous system synapses [12], neuronal and synaptic morphology of live synapses [125] and nodes of Ranvier [26]. Triple-color STED was utilized to study the spine synapses of the central nervous system [54] and other biological samples [7, 18, 110, 121, 141]. Furthermore, quadruple-color STED has been used for live imaging and for imaging of fixed samples with only one excitation laser wavelength and one depletion laser wavelength [136]. The use of one depletion laser wavelength is recommended for the alignment of images with different fluorescent wavelengths in multicolor STED microscopy. Tables with the recommended fluorescent dyes and dye combinations for multicolor analysis can be found on the microscope manufacturers’ websites [4, 71]. In addition, bright SiR dyes have been successfully used for STED live imaging [92].
STORM requires fluorophores that switch between a non-emissive state and a ground state under high intensity light and a specialized buffer environment for these switching fluorophores. The fluorophore relaxes stochastically to the ground state and is then excited to emit fluorescent light. Fluorophores with these characteristics are called photoswitchable (photoblinking) dyes (for STORM) and photoswitchable fluorescent proteins (for PALM). Tables of the available colors can be found on the microscope manufacturers’ websites [5, 10], and new dyes are being developed [23]. The direct STORM (dSTORM or activator-free STORM) method allows the use of commercially available photoswitchable dyes conjugated to secondary antibodies without the need for activator-reporter pairs of dyes, which is beneficial for multicolor analysis. For multicolor STORM, dual color detection methods have been used for many publications [1, 6, 20, 25, 52, 57, 65, 66]. Commercially available STORM systems from Nikon and Zeiss allow the detection of two different fluorescent labels. Triple-color STORM analysis has been performed by combining STORM with a spectral demixing (spectral unmixing) method to resolve fluorophores that have overlapping emission spectra [69]. However, detecting three different colors in STORM is challenging, which is explained in the later section titled “Points of consideration for Super-Resolution Microscopy analyses”.
Imaging speed and live imaging
The temporal resolution of commercially available SIM microscopes is similar to STED microscopes and better than STORM. The temporal resolution of SIM is better than that of STORM because the number of raw micrographs needed to reconstruct a super-resolution image is only nine to 15 for SIM compared to tens of thousands for STORM. In addition, SIM requires a lower light intensity for imaging compared to STED and STORM; therefore, SIM is favored for live imaging despite the limited improvement in resolution. Several SIM methods have been developed to increase the temporal resolution by combining SIM with a custom microlens array, point scanning microscopy, or a lattice light sheet [137]. A commercial SIM model from Nikon offers an imaging frame rate of 15 frames per second (FPS). The recent development of SIM using lattice light and the Zeiss Elyra 7 system improved the frame rate to a very high speed of 255 FPS. SIM live imaging has been used to analyze various nervous system structures, including growth cones [39], growth cone cytoskeletons and vesicles of cultured neurons [87], dendritic spines of cultured hippocampal neurons [58], and dendrites and dendritic spines in brains of live zebrafish larvae or live mice [127]. Furthermore, live imaging based on SIM was successfully performed in the International Space Station to analyze cultured primary human macrophages exposed to microgravity [122].
STED microscopy is similar to confocal microscopy because both are scanning microscopes. A point scanning microscope allows the imaging of a smaller region of interest or a line to increase temporal resolution rather than scanning the entire field of view. STED microscopy has achieved video-rate fast live imaging; for example, 38.5 FPS was achieved for analyzing vesicle fusion and endocytosis [107], and 28 FPS was achieved for analyzing synaptic proteins in neurons [67]. A commercially available resonant scanner offers a very high scanning speed of 12,000 Hz and obtains an image of 512 lines in approximately 20 milliseconds (50 FPS), or can capture up to 428 FPS by restricting the region of interest [15, 16]. Furthermore, a custom-built STED microscope in the laboratory of Dr. Stefan W Hell reached >1,000 FPS with the use of an electro-optical deflector [103]. Leica offers two objective lens with a long working distance of 300 μm for live imaging STED microscopy: (1) a glycerol immersion lens with motorized correction collar, HC PL APO 93x/NA 1.3 Glycerol motCORR STED White lens and (2) a water immersion lens with motorized correction collar, HC PL APO 86x/NA 1.2 Water motCORR STED White lens [71]. Other examples of STED microscopy being successfully applied for live imaging include analyses of cytoskeletons in neurons [27, 28], interactions of mitochondria and endoplasmic reticulum [17], mitochondrial proteins [55], and many others [28, 54, 67, 81, 131, 132]. The use of a pulsed STED laser with a far-red 775 nm wavelength reduces phototoxicity compared to the use of a continuous wave STED lasers with shorter wavelengths. In addition, silicon-rhodamine (SiR) dyes have been used successfully for live imaging STED microscopy because these dyes have properties suitable for live imaging STED microscopy, as they are bright, photostable, without phototoxicity, and have a far-red spectrum of emission [92]. Cell-permeable SiR dyes are available commercially from Spirochrome for staining actin, tubulin, lysosome, and DNA. For live labeling of SNAP-tag or Halo-tag fusion proteins, cell permeable and fluorescently labeled SNAP substrates and Halo ligands are available from New England Biolabs and Promega [17, 38].
The STORM (PALM, SMLM) imaging speed is approximately 0.1 FPS, which is a lower temporal resolution than that of SIM and STED. The limited temporal resolution of STORM stems from the requirement for tens of thousands of frames (micrographs) to reconstruct a super resolution image [9, 57, 106, 135]. Multiple algorithms have been developed to increase the temporal resolution, including a multi-emitter fitting algorithm that identifies the locations of molecules that may have overlapping point spread functions. However, the quality of STORM image reconstruction depends significantly on the analysis software package used [101, 116, 118, 120], which will be discussed in the next section.
Points of consideration for Super-Resolution Microscopy analyses
Various super-resolution microscopy techniques are commercially available; however, each technique has advantages and disadvantages that are worth consideration prior to choosing the method used for a project. For example, if the project requires the highest resolution, STORM and STED offer the best resolution currently available as a commercially available super-resolution microscope (lateral resolution of 20 to 30 nm). The preference might be affected by the use of a reconstructed image by the former versus that of an optical image by the latter. If the project requires information of protein numbers, STORM has been demonstrated to offer quantitative information [35].
Sample preparation should follow the manufacture provided guides, which describes the required embedding/mounting condition [71, 79]. A suitable sample thickness depends on the microscopy type. SIM is suitable for tissue thickness up to 10 to 15 μm and is not suitable for thick tissue due to out of focus light interfering with the computational removal of these noise [46]. STED has been shown to image 150 μm into tissue at subdiffraction limited resolution by combining optical clearing of the specimen and the objective lens with motorized correction collar, HC PL APO 93x/NA 1.3 Glycerol motCORR STED White lens. Leica provides a protocol for this type of deep tissue imaging [70, 129]. STORM has been combined with TIRF microscopy and thus has a shallow imaging depth below one micrometer. Improved protocols have been reported to image few μm to 30 μm thick sections using STORM [42, 49].
At this level of resolution, sizes of antibodies used for immunohistochemistry become relevant. An immunoglobulin has a size of 12 to 14 nm; therefore, nanobodies or single domain antibodies are preferred due to the smaller size of 2.8 to 4.4 nm [117]. The green fluorescent protein (GFP) used for PALM is similar to nanobodies and has a size of 2.4 to 4.2 nm [51]. Also, resolution is improved by directly labeling the protein of interest using bioorthogonal click chemistry combined with small organic dyes [84].
If the study requires multicolor analysis, SIM and STED microscopy offer multicolor analysis with a larger number of colors in an image. Three-color analyses are easily achievable for STED microscopy, and four-color analyses are possible [136]. However, three-color analyses are difficult for STORM due to the specific buffer requirement for each photoswitchable fluorophore [5]. In addition to the limitation of color number, STORM requires the use of a higher concentration of primary antibodies to fulfill the requirement to saturate the signal of the specimen [5, 10]. Therefore, the staining conditions may differ from those used in the usual immunohistochemistry protocol, and the signal-to-noise ratio needs to be considered carefully.
If acquisition speed is a priority to capture fast, dynamic events by live imaging, SIM and STED offer a higher frame rate than STORM. For live imaging, the intensity of excitation light and photobleaching are significant factors for consideration, and these are common problems for any live imaging technique. SIM uses lower intensity light and is considered suitable for live imaging. Recent STED microscopes are equipped with pulsed lasers instead of continuous wave lasers and highly sensitive hybrid detectors to reduce the excitation light intensity. In addition, photobleaching becomes relevant when the project requires 3D analysis and multiple scanning, such as when generating a z-stack. Newer generation fluorophores that are bright and stable/bleach resistant have been developed recently and should be considered for improving super-resolution imaging, including Alexa Fluor & Alexa Fluor Plus (Thermo Fisher), ATTO (Sigma-Aldrich), CF (Biotium), SiR (Spirochrome, New England Biolabs), and STAR (Abberior) dyes [4, 71, 92].
The data file size of the image differs between these techniques. STED microscopy generates file sizes similar to those of confocal microscopy because STED generates an optical image raw data file as the final super resolution image. On the other hand, STORM requires tens of thousands of frames (micrographs) to reconstruct a super-resolution image. Thus, the typical data file size of STORM is much larger than that of STED and can exceed 5 gigabytes for one final super-resolution image [109]. Therefore, STORM requires significant capacity for data transfer and storage and computing power for processing the data.
Finally, the STORM image reconstruction quality depends significantly on the analysis software used, and analysis software packages were compared recently by using a common data set [101, 116]. There seems to be a considerable difference in the outcomes, which may affect the conclusion drawn from the reconstructed images. This was the case for the 3D analysis more than the 2D analysis. Validation is essential for any research technique, but it is essential for STORM to validate the use of different approaches because a reference raw data image does not exist for this technique. In contrast, STED microscopy generates optical images as raw data, which reduces concerns about postprocessing and analysis software differences. In addition, STED images can be compared against confocal microscopy images obtained using the same equipment in a sequential manner.
SIM for NMJ analysis
Three-dimensional structured illumination microscopy, 3D-SIM [46], has been used to analyze the role of the large adaptor protein Ankyrin2-L in the stabilization of Drosophila larval NMJs through the regulation of presynaptic microtubules and cell adhesion molecules [90]. 3D-SIM revealed a repetitive lattice structure with approximately 200 nm periodicity comprised of Ankyrin2-L and β-spectrin in axons but a less organized structure in presynaptic boutons. The function of Ankyrin2-L, which is to crosslink synaptic membrane proteins and spectrins to the microtubule cytoskeleton, is required to stabilize the presynaptic terminals. In addition, 3D-SIM was also used to analyze the role of the noncanonical BMP signaling pathway in the maturation of Drosophila larval NMJs [119]. Furthermore, 3D-SIM has been used to detect immunohistochemical signals and has been combined with confocal microscopy for detecting fluorescent in situ hybridization signals in Drosophila larval NMJs [124]. This publication describes the detailed protocol of the experimental procedure with the methods to check the quality of the 3D-SIM raw data and reconstruction with an open source ImageJ plugin called SIMcheck [8].
By using 3D-SIM in mouse NMJs, the distribution pattern of synaptic proteins, including the presynaptic active zone protein Piccolo and the postsynaptic proteins rapsyn, voltage-gated sodium channel, and integrin α7, was analyzed relative to that of the postsynaptic junctional folds [139]. Consistent with previous reports, these proteins were observed at the expected locations of NMJs.
STED microscopy for NMJ analysis
The first application of super-resolution microscopy for the analysis of NMJs was the imaging of the active zones of Drosophila NMJs using STED microscopy (Table 2). Active zones are synaptic vesicle release sites at the presynaptic membrane [24]. Active zones appear in electron microscopy images as T-bars in Drosophila NMJs or as small aggregates in vertebrate NMJs. In Drosophila larval NMJs, these active zones can be labeled with the nc82 antibody (DSHB), which stains the active zone-specific protein Bruchpilot. This protein was previously shown to be distributed in a punctate pattern at motor nerve terminals by confocal microscopy without any obvious structure inside each punctum. The subdiffraction-limited resolution of STED microscopy revealed a structure within each punctum for the first time and elucidated a ring-like pattern of Bruchpilot protein in each of these puncta [61]. The distribution pattern of the Bruchpilot protein was further analyzed using antibodies recognizing the N-terminus and the C-terminus (nc82) of this protein. STED microscopy revealed that the N-terminus of Bruchpilot protein resides inside the ring-like pattern of C-terminus of Bruchpilot protein, which was detected by the nc82 antibody in the en face view of the NMJs (top-down view looking at the active zones from the cytosolic side) [40]. Based on the transverse section view of the NMJ (cross-section, perpendicular view of the synapse), the N-terminus of Bruchpilot protein resided next to the voltage-gated calcium channels (VGCCs, Cacophony labeled with GFP) in the presynaptic membrane, whereas the C-terminus of Bruchpilot protein was distributed away from the presynaptic membrane. These results were consistent with the immunoelectron microscopy analysis of the T-bars at Drosophila NMJs using the same set of antibodies that recognized the N- and C-termini of Bruchpilot [40].
Table 2:
Super resolution microscopy techniques used for NMJ analyses
| Technique | Species | Analysis |
|---|---|---|
| SIM | Drosophila | The role of Ankyrin2-L in the stabilization of larval NMJs [90]. The roles of noncanonical BMP signaling pathway for the maturation of larval NMJs [119]. |
| Combined SIM to detect immunohistochemical signals and confocal microscopy to detect fluorescent in situ hybridization signals at larval NMJs [124]. | ||
| Mouse | The distribution pattern of the presynaptic active zone protein Piccolo and the postsynaptic proteins rapsyn, voltage-gated sodium channel, integrin α7, and postsynaptic junctional folds [139]. | |
| Human | N/A | |
| STED | Drosophila | The distribution pattern of Bruchpilot protein. The N-terminus of Bruchpilot resided next to the vGcCs in the presynaptic membrane, whereas the C-terminus of Bruchpilot protein distributed away from the presynaptic membrane [40, 61]. The distribution pattern of Abp1, DCSP3, Futsch [63], Liprin-α and glutamate receptors [40], RIM-BP [73], Unc13A and Unc13B [13], and Unc18 and Syntaxin-1A [95, 128]. Active zone proteins including Bruchpilot, Rim-BP, and Unc13A show puncta number between 2 to 6 per one active zone unit structure of NMJs. [43]. |
| Mouse | The distribution patterns of Bassoon, Piccolo, and PQ-VGCC in adult and aged NMJs. This study revealed the selective degeneration of active zone proteins in aged NMJs [86]. | |
| Human | N/A | |
| STORM | Drosophila | Quantification of Bruchpilot protein number per active zone in NMJs of wild-type and Rab3 mutant larvae [35]. |
| The roles of Bruchpilot and synaptotagmin for rapid glutamate release [89]. | ||
| The roles of Bruchpilot and complexin for tethering synaptic vesicles to active zones [104]. | ||
| Mouse | The distribution pattern of acetylcholine receptor in junctional folds [139] and SNAP25 in adult NMJs [56]. | |
| Human | The distribution pattern, puncta size, density, and signal intensity of SNAP25 in adult NMJs [56]. |
Abp1= actin-binding protein 1; BMP= bone morphogenetic protein; DCSP3= Drosophila cysteine string protein 3; NMJs= neuromuscular junctions; Rab3= Ras-related protein 3; SIM= structured illumination microscopy; SNAP25= synaptosome associated protein 25; STED= stimulated emission depletion; STORM= stochastic; optical reconstruction microscopy; RIM-BP= Rab3-interacting molecule-binding protein; Unc= uncoordinated
Further analyses of Drosophila NMJs using STED microscopy revealed the nanostructure distribution patterns of synaptic proteins, including actin-binding protein 1 (Abp1), Drosophila cysteine string protein 3 (DCSP3, marker for synaptic vesicles), and Futsch (microtubule-enriched area) [63], Liprin-α and glutamate receptors [40], Rab3-interacting molecule–binding protein (RIM-BP) [73], Unc13A and Unc13B [13], and Unc18 and Syntaxin-1A [95, 128]. Based on some these of studies, a model of the molecular nanostructure of Drosophila NMJs has been proposed [43]. Interestingly, many of these active zone proteins distribute in punctate clusters and do not show diffuse distribution at Drosophila NMJs. When these active zone proteins show increases in the protein quantity, they show the increases in the number of puncta or the quantity of proteins in each puncta [14]. Active zone proteins, including Bruchpilot, Rim-BP, and Unc13A, show puncta numbers between 2 and 6 per active zone unit structure in Drosophila NMJs. Unc13A is essential for both short-term plasticity that occurs over minutes and for long-term plasticity that occurs over hours to days [14]. Importantly, Bruchpilot protein is aberrantly organized at larval NMJs in a Drosophila Amyotrophic lateral sclerosis (ALS) model, which expresses human FUS with ALS-causing mutations in motor neurons [105]. These data suggest that active zone proteins organize a dynamic structure that plays a role in synaptic plasticity and is degenerated in neurological diseases.
STED microscopy was also used to study the active zones of adult mouse NMJs. Adult NMJs rely on P/Q-type VGCCs for synaptic transmission [93, 98]. STED microscopy revealed that calcium channels cluster in puncta and are distributed at roughly evenly-spaced distances throughout the presynaptic membrane [86]. Most of these calcium channel puncta colocalized with the active zone-specific protein Bassoon, which also clustered in puncta [86]. The colocalization of P/Q-type VGCCs and Bassoon was consistent with biochemical analyses demonstrating the direct or indirect interaction of these two proteins [21, 30]. Bassoon has structural similarities with another active zone protein, Piccolo, and these two proteins are believed to colocalize at nerve terminals in the central nervous system. Unexpectedly, STED microscopy revealed that Bassoon and Piccolo reside in a side-by-side manner, in which Piccolo puncta form a “sandwich” around a Bassoon punctum [86]. Interestingly, this distribution pattern showed similarities with an active zone model based on electron microscope tomography of mouse NMJs [82]. Further study of aged NMJs using STED microscopy revealed that the levels of PQ-type VGCCs and Bassoon proteins were significantly decreased, while Piccolo protein was resistant to age-related changes and remained at a similar level as that found in NMJs in young adult mice [86]. For the first time, the selective degeneration of active zone proteins in NMJs of aged animals has been revealed owing to the subdiffraction limited resolution of STED microscopy.
STORM for NMJ analysis
Similar to STED microscopy, STORM was used initially for the analysis of Drosophila larval NMJs. Active zones of Drosophila larval NMJs were analyzed quantitatively using direct STORM (dSTORM). As a Single Molecule Localization Microscopy method, dSTORM was used to quantify endogenous proteins [35]. The number of active zone-specific Bruchpilot proteins has been estimated to be 137 proteins per active zone, and three quarters of these proteins are organized into approximately 15 different positions with seven Bruchpilot proteins per position. This quantitative dSTORM technique successfully showed an approximately 60% increase in the Bruchpilot protein number per active zone in NMJs of Rab3 mutant larvae, which were previously known to have altered synaptic transmission and enlarged Bruchpilot aggregates [35]. Furthermore, STORM was used to analyze the roles of Bruchpilot and synaptotagmin in rapid glutamate release [89] and the roles of Bruchpilot and complexin in tethering synaptic vesicles to active zones [104] in Drosophila larval NMJs. Excellent reviews have been published elsewhere describing the use of STORM for Drosophila larval NMJ studies [33, 34].
At mouse NMJs, the acetylcholine receptor distribution pattern in junctional folds was compared between SIM images, STORM images, and electron microscopy ultrastructure images. Based on the STORM images, the authors proposed a novel acetylcholine receptor distribution pattern, in which the receptor concentration is high at the shoulders of the junctional fold mouth but low in the ridges [139]. This distribution pattern is slightly different from the classical understanding, in which acetylcholine receptors have been thought to be distributed at the top and shoulders of the junctional fold ridges and not at the bottom of the folds. It would be interesting to validate this new model of the acetylcholine receptor distribution pattern using a different technique, such as immunoelectron microscopy, or using STORM with transverse section images of the NMJs because the new STORM analysis was based on en face images.
Human and mouse NMJs were compared using STORM to analyze the distribution pattern of synaptosome associated protein 25 (SNAP25), one of the SNARE proteins essential for synaptic vesicle fusion. In mouse NMJs, SNAP25 was distributed in a punctate fashion at a density (15 per μm2) [56] similar to the puncta density of the active zone proteins Bassoon and Piccolo (10 per μm2) [86]. Interestingly, human NMJs showed greater values for SNAP25 compared to mouse NMJs in terms of puncta size, puncta density per NMJ area, and the signal intensity of immunohistochemistry [56]. Based on these findings, the authors suggested that there were differences between human and mouse NMJs.
Super-Resolution Microscopy for brain synapse analyses.
Synapses of the central nervous system have been analyzed extensively using super-resolution microscopy. To mention just few examples, the first synaptic protein analyzed using STED microscopy was the presynaptic calcium sensor synaptotagmin in cultured hippocampal neurons [134]. The movement of synaptic vesicles in cultured hippocampal neurons was recorded using STED microscopy at a video rate live imaging [133]. Pre- and postsynaptic proteins in the main olfactory bulb of mice were analyzed using 3D-STORM [29]. The distribution patterns of synaptic proteins within small central nervous synapses were analyzed using STORM with fixed samples and STED live imaging [32, 54]. For a comprehensive review of the analysis of central nervous system synapses using super-resolution microscopy, many excellent reviews have been published [31, 75–77, 88, 97, 99, 112, 113, 126].
Conclusion
Super-resolution microscopy studies of Drosophila NMJs have come a long way and revealed impressive nanostructures and the molecular identity of active zones previously known as T-bars in electron micrographs. The combination of good molecular markers (for example, Bruchpilot for active zones) and rich genetic mutant resources provided a wealth of opportunity to study the molecular mechanisms of the physiological modification of Drosophila NMJs using super-resolution microscopy. Super-resolution microscopy studies of mammalian NMJs are still in their infancy. To our knowledge, these super-resolution methods have not been used for live imaging of vertebrate NMJs. However, vertebrate NMJs have been studied using electron microscope tomography in frogs and mice, which revealed the ultrastructural details of the presynaptic active zones. Super-resolution microscopy provides the opportunity to reveal the molecular identities of these modeled structures for a better understanding of vertebrate synapses. The synapse is a small structure with a complex molecular mechanism that is necessary to accomplish synaptic transmission and plasticity, and the development of super-resolution microscopy has improved the understanding of the underpinning molecular mechanisms.
Highlights.
Advantages and disadvantages of SIM, STED, and STORM super-resolution microscopy
Suitability of super-resolution microscopy types for synapse analyses
Molecular nanostructures of neuromuscular junctions revealed using super-resolution microscopy
Acknowledgement
We thank Dr. Weichun Lin for the invitation for this manuscript.
Funding: This work was supported by the National Institute of Health [1R01AG051470 for H.N.].
Abbreviations
- Abp1
actin-binding protein 1
- ALS
Amyotrophic lateral sclerosis
- BMP
bone morphogenetic protein
- CNS
central nervous system
- DCSP3
Drosophila cysteine string protein 3
- DSHB
Developmental Studies Hybridoma Bank
- FPS
frames per second
- GFP
green fluorescent protein
- NMJ
neuromuscular junctions
- Rab3
Ras-related protein 3
- SIM
structured illumination microscopy
- SiR
silicon-rhodamine
- STED
stimulated emission depletion
- STORM
stochastic optical reconstruction microscopy
- SMLM
Single molecule localization microscopy
- Lrp4
low-density lipoprotein receptor-related protein
- PALM
photoactivated localization microscopy
- RIM-BP
Rab3-interacting molecule–binding protein
- SNARE
soluble N-ethylmaleimide sensitive factor attachment protein receptor
- SNAP25
synaptosome associated protein 25
- TIRF
total internal reflection
- UNC
uncoordinated
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing financial interests: The authors declare no competing financial interests.
References
- [1].Aaron JS, Carson BD, Timlin JA, Characterization of differential Toll-like receptor responses below the optical diffraction limit, Small 8 (2012) 3041–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Abbe E, Beiträge zur Theori des Mikroskops und der mikroskopischen Wahrnehmung, Archiv fur mikroskopische Anatomy 9 (1873) 413–418. [Google Scholar]
- [3].Abberior Instruments. Abberior multicolor STED @ 775 nm, <https://www.abberiorinstruments.com/site/assets/files/1344/abberiorinstruments_sted_775nm-1.pdf> (2019).
- [4].Abberior Instruments. List of recommended dyes for STED microscopy, <https://www.abberiorinstruments.com/references/recommended-dyes/> (2019).
- [5].Allen JR, J.S. S, Schwartz SA, Davidson MW. Single-Molecule Super-Resolution Imaging, <https://www.microscopyu.com/techniques/super-resolution/single-molecule-super-resolution-imaging> (2019).
- [6].Bach JN, Giacomelli G, Bramkamp M, Sample Preparation and Choice of Fluorophores for Single and Dual Color Photo-Activated Localization Microscopy (PALM) with Bacterial Cells, Methods Mol Biol 1563 (2017) 129–141. [DOI] [PubMed] [Google Scholar]
- [7].Baharom F, Thomas OS, Lepzien R, Mellman I, Chalouni C, Smed-Sorensen A, Visualization of early influenza A virus trafficking in human dendritic cells using STED microscopy, PLoS One 12 (2017) e0177920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ball G, Demmerle J, Kaufmann R, Davis I, Dobbie IM, Schermelleh L, SIMcheck: a Toolbox for Successful Super-resolution Structured Illumination Microscopy, Scientific reports 5 (2015) 15915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Benke A, Olivier N, Gunzenhauser J, Manley S, Multicolor single molecule tracking of stochastically active synthetic dyes, Nano Lett 12 (2012) 2619–2624. [DOI] [PubMed] [Google Scholar]
- [10].Betzig E, Hess HF, Shroff H, Patterson GH, Lippincott-Schwartz J, Davidson MW. Introduction to Photoactivated Localization Microscopy, <http://zeisscampus.magnet.fsu.edu/articles/superresolution/palm/introduction.html> (2019).
- [11].Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF, Imaging intracellular fluorescent proteins at nanometer resolution, Science 313 (2006) 1642–1645. [DOI] [PubMed] [Google Scholar]
- [12].Blom H, Ronnlund D, Scott L, Westin L, Widengren J, Aperia A, Brismar H, Spatial distribution of DARPP-32 in dendritic spines, PLoS One 8 (2013) e75155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bohme MA, Beis C, Reddy-Alla S, Reynolds E, Mampell MM, Grasskamp AT, Lutzkendorf J, Bergeron DD, Driller JH, Babikir H, Gottfert F, Robinson IM, O’Kane CJ, Hell SW, Wahl MC, Stelzl U, Loll B, Walter AM, Sigrist SJ, Active zone scaffolds differentially accumulate Unc13 isoforms to tune Ca(2+) channel-vesicle coupling, Nat Neurosci 19 (2016) 1311–1320. [DOI] [PubMed] [Google Scholar]
- [14].Bohme MA, McCarthy AW, Grasskamp AT, Beuschel CB, Goel P, Jusyte M, Laber D, Huang S, Rey U, Petzoldt AG, Lehmann M, Gottfert F, Haghighi P, Hell SW, Owald D, Dickman D, Sigrist SJ, Walter AM, Rapid active zone remodeling consolidates presynaptic potentiation, Nat Commun 10 (2019) 1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Borlinghaus RT. What is a Resonant Scanner?, <https://www.leica-microsystems.com/science-lab/what-is-a-resonant-scanner/> (2019).
- [16].Borlinghaus RT. What is a Tandem Scanner?, <https://www.leica-microsystems.com/science-lab/what-is-a-tandem-scanner/> (2019).
- [17].Bottanelli F, Kromann EB, Allgeyer ES, Erdmann RS, Wood Baguley S, Sirinakis G, Schepartz A, Baddeley D, Toomre DK, Rothman JE, Bewersdorf J, Two-colour live-cell nanoscale imaging of intracellular targets, Nat Commun 7 (2016) 10778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Butkevich AN, Lukinavicius G, D’Este E, Hell SW, Cell-Permeant Large Stokes Shift Dyes for Transfection-Free Multicolor Nanoscopy, J Am Chem Soc 139 (2017) 12378–12381. [DOI] [PubMed] [Google Scholar]
- [19].Chen BC, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, Janetopoulos C, Wu XS, Hammer JA 3rd, Liu Z, English BP, Mimori-Kiyosue Y, Romero DP, Ritter AT, Lippincott-Schwartz J, Fritz-Laylin L, Mullins RD, Mitchell DM, Bembenek JN, Reymann AC, Bohme R, Grill SW, Wang JT, Seydoux G, Tulu US, Kiehart DP, Betzig E, Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution, Science 346 (2014) 1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chen C, Zong S, Wang Z, Lu J, Zhu D, Zhang Y, Cui Y, Imaging and Intracellular Tracking of Cancer-Derived Exosomes Using Single-Molecule Localization-Based Super-Resolution Microscope, ACS Appl Mater Interfaces 8 (2016) 25825–25833. [DOI] [PubMed] [Google Scholar]
- [21].Chen J, Billings SE, Nishimune H, Calcium channels link the muscle-derived synapse organizer laminin beta2 to Bassoon and CAST/Erc2 to organize presynaptic active zones, J Neurosci 31 (2011) 512–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen J, Mizushige T, Nishimune H, Active zone density is conserved during synaptic growth but impaired in aged mice, J Comp Neurol 520 (2012) 434–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Collot M, Ashokkumar P, Anton H, Boutant E, Faklaris O, Galli T, Mely Y, Danglot L, Klymchenko AS, MemBright: A Family of Fluorescent Membrane Probes for Advanced Cellular Imaging and Neuroscience, Cell Chem Biol (2019). [DOI] [PubMed] [Google Scholar]
- [24].Couteaux R, Pecot-Dechavassine M, Synaptic vesicles and pouches at the level of “active zones” of the neuromuscular junction, C R Acad Sci Hebd Seances Acad Sci D 271 (1970) 2346–2349. [PubMed] [Google Scholar]
- [25].Cremer C, Szczurek A, Schock F, Gourram A, Birk U, Super-resolution microscopy approaches to nuclear nanostructure imaging, Methods (San Diego, Calif 123 (2017) 11–32. [DOI] [PubMed] [Google Scholar]
- [26].D’Este E, Kamin D, Balzarotti F, Hell SW, Ultrastructural anatomy of nodes of Ranvier in the peripheral nervous system as revealed by STED microscopy, Proc. Natl. Acad. Sci. U.S.A. 114 (2017) E191–E199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].D’Este E, Kamin D, Gottfert F, El-Hady A, Hell SW, STED nanoscopy reveals the ubiquity of subcortical cytoskeleton periodicity in living neurons, Cell reports 10 (2015) 1246–1251. [DOI] [PubMed] [Google Scholar]
- [28].D’Este E, Kamin D, Velte C, Gottfert F, Simons M, Hell SW, Subcortical cytoskeleton periodicity throughout the nervous system, Scientific reports 6 (2016) 22741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Dani A, Huang B, Bergan J, Dulac C, Zhuang X, Superresolution Imaging of Chemical Synapses in the Brain, Neuron 68 (2010) 843–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Davydova D, Marini C, King C, Klueva J, Bischof F, Romorini S, Montenegro-Venegas C, Heine M, Schneider R, Schroder MS, Altrock WD, Henneberger C, Rusakov DA, Gundelfinger ED, Fejtova A, Bassoon specifically controls presynaptic P/Q-type Ca(2+) channels via RIM-binding protein, Neuron 82 (2014) 181–194. [DOI] [PubMed] [Google Scholar]
- [31].Delgado JY, Selvin PR, A Revised View on the Role of Surface AMPAR Mobility in Tuning Synaptic Transmission: Limitations, Tools, and Alternative Views, Front Synaptic Neurosci 10 (2018) 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Dudok B, Barna L, Ledri M, Szabo SI, Szabadits E, Pinter B, Woodhams SG, Henstridge CM, Balla GY, Nyilas R, Varga C, Lee SH, Matolcsi M, Cervenak J, Kacskovics I, Watanabe M, Sagheddu C, Melis M, Pistis M, Soltesz I, Katona I, Cell-specific STORM super-resolution imaging reveals nanoscale organization of cannabinoid signaling, Nat Neurosci 18 (2015) 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ehmann N, Owald D, Kittel RJ, Drosophila active zones: From molecules to behaviour, Neurosci Res 127 (2018) 14–24. [DOI] [PubMed] [Google Scholar]
- [34].Ehmann N, Sauer M, Kittel RJ, Super-resolution microscopy of the synaptic active zone, Front Cell Neurosci 9 (2015) 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ehmann N, van de Linde S, Alon A, Ljaschenko D, Keung XZ, Holm T, Rings A, DiAntonio A, Hallermann S, Ashery U, Heckmann M, Sauer M, Kittel RJ, Quantitative super-resolution imaging of Bruchpilot distinguishes active zone states, Nat Commun 5 (2014) 4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ehrenberg M. The Nobel Prize in Chemistry 2014 “for the development of super-resolved fluorescence microscopy”, <https://www.nobelprize.org/prizes/chemistry/2014/press-release/> (2014). [DOI] [PubMed]
- [37].Engel J, Odermatt E, Engel A, Madri JA, Furthmayr H, Rohde H, Timpl R, Shapes, domain organizations and flexibility of laminin and fibronectin, two multifunctional proteins of the extracellular matrix, Journal of molecular biology 150 (1981) 97–120. [DOI] [PubMed] [Google Scholar]
- [38].Erdmann RS, Baguley SW, Richens JH, Wissner RF, Xi Z, Allgeyer ES, Zhong S, Thompson AD, Lowe N, Butler R, Bewersdorf J, Rothman JE, St Johnston D, Schepartz A, Toomre D, Labeling Strategies Matter for Super-Resolution Microscopy: A Comparison between HaloTags and SNAP-tags, Cell Chem Biol 26 (2019) 584–592 e586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Fiolka R, Shao L, Rego EH, Davidson MW, Gustafsson MG, Time-lapse two-color 3D imaging of live cells with doubled resolution using structured illumination, Proc. Natl. Acad. Sci. U.S.A. 109 (2012) 5311–5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Fouquet W, Owald D, Wichmann C, Mertel S, Depner H, Dyba M, Hallermann S, Kittel RJ, Eimer S, Sigrist SJ, Maturation of active zone assembly by Drosophila Bruchpilot, J Cell Biol 186 (2009) 129–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Gao R, Asano SM, Upadhyayula S, Pisarev I, Milkie DE, Liu TL, Singh V, Graves A, Huynh GH, Zhao Y, Bogovic J, Colonell J, Ott CM, Zugates C, Tappan S, Rodriguez A, Mosaliganti KR, Sheu SH, Pasolli HA, Pang S, Xu CS, Megason SG, Hess H, Lippincott-Schwartz J, Hantman A, Rubin GM, Kirchhausen T, Saalfeld S, Aso Y, Boyden ES, Betzig E, Cortical column and whole-brain imaging with molecular contrast and nanoscale resolution, Science 363 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].German CL, Gudheti MV, Fleckenstein AE, Jorgensen EM, Brain Slice Staining and Preparation for Three-Dimensional Super-Resolution Microscopy, Methods Mol Biol 1663 (2017) 153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ghelani T, Sigrist SJ, Coupling the Structural and Functional Assembly of Synaptic Release Sites, Front Neuroanat 12 (2018) 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gottfert F, Wurm CA, Mueller V, Berning S, Cordes VC, Honigmann A, Hell SW, Coaligned dual-channel STED nanoscopy and molecular diffusion analysis at 20 nm resolution, Biophysical journal 105 (2013) L01–03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Gustafsson MG, Agard DA, Sedat JW, I5M: 3D widefield light microscopy with better than 100 nm axial resolution, J Microsc 195 (1999) 10–16. [DOI] [PubMed] [Google Scholar]
- [46].Gustafsson MG, Shao L, Carlton PM, Wang CJ, Golubovskaya IN, Cande WZ, Agard DA, Sedat JW, Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination, Biophysical journal 94 (2008) 4957–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Heintzmann R, Cremer CG, Laterally modulated excitation microscopy: improvement of resolution by using a diffraction grating, Proceedings of the SPIE 3568 (1999) 185–196. [Google Scholar]
- [48].Hell SW, Wichmann J, Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy, Opt Lett 19 (1994) 780–782. [DOI] [PubMed] [Google Scholar]
- [49].Herrmannsdorfer F, Flottmann B, Nanguneri S, Venkataramani V, Horstmann H, Kuner T, Heilemann M, 3D d STORM Imaging of Fixed Brain Tissue, Methods Mol Biol 1538 (2017) 169–184. [DOI] [PubMed] [Google Scholar]
- [50].Hess ST, Girirajan TP, Mason MD, Ultra-high resolution imaging by fluorescence photoactivation localization microscopy, Biophysical journal 91 (2006) 4258–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hink MA, Griep RA, Borst JW, van Hoek A, Eppink MH, Schots A, Visser AJ, Structural dynamics of green fluorescent protein alone and fused with a single chain Fv protein, J Biol Chem 275 (2000) 17556–17560. [DOI] [PubMed] [Google Scholar]
- [52].Hoess P, Mund M, Reitberger M, Ries J, Dual-Color and 3D Super-Resolution Microscopy of Multiprotein Assemblies, Methods Mol Biol 1764 (2018) 237–251. [DOI] [PubMed] [Google Scholar]
- [53].Homan AE, Meriney SD, Active zone structure-function relationships at the neuromuscular junction, Synapse 72 (2018) e22057. [DOI] [PubMed] [Google Scholar]
- [54].Hruska M, Henderson N, Le Marchand SJ, Jafri H, Dalva MB, Synaptic nanomodules underlie the organization and plasticity of spine synapses, Nat Neurosci 21 (2018) 671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ishigaki M, Iketani M, Sugaya M, Takahashi M, Tanaka M, Hattori S, Ohsawa I, STED superresolution imaging of mitochondria labeled with TMRM in living cells, Mitochondrion 28 (2016) 79–87. [DOI] [PubMed] [Google Scholar]
- [56].Jones RA, Harrison C, Eaton SL, Llavero Hurtado M, Graham LC, Alkhammash L, Oladiran OA, Gale A, Lamont DJ, Simpson H, Simmen MW, Soeller C, Wishart TM, Gillingwater TH, Cellular and Molecular Anatomy of the Human Neuromuscular Junction, Cell reports 21 (2017) 2348–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Jones SA, Shim SH, He J, Zhuang X, Fast, three-dimensional super-resolution imaging of live cells, Nat Methods 8 (2011) 499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Kashiwagi Y, Higashi T, Obashi K, Sato Y, Komiyama NH, Grant SGN, Okabe S, Computational geometry analysis of dendritic spines by structured illumination microscopy, Nat Commun 10 (2019) 1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Kasthuri N, Hayworth KJ, Berger DR, Schalek RL, Conchello JA, Knowles-Barley S, Lee D, Vazquez-Reina A, Kaynig V, Jones TR, Roberts M, Morgan JL, Tapia JC, Seung HS, Roncal WG, Vogelstein JT, Burns R, Sussman DL, Priebe CE, Pfister H, Lichtman JW, Saturated Reconstruction of a Volume of Neocortex, Cell 162 (2015) 648–661. [DOI] [PubMed] [Google Scholar]
- [60].Khodabakhsh F, Behdani M, Rami A, Kazemi-Lomedasht F, Single-Domain Antibodies or Nanobodies: A Class of Next-Generation Antibodies, Int Rev Immunol 37 (2018) 316–322. [DOI] [PubMed] [Google Scholar]
- [61].Kittel RJ, Wichmann C, Rasse TM, Fouquet W, Schmidt M, Schmid A, Wagh DA, Pawlu C, Kellner RR, Willig KI, Hell SW, Buchner E, Heckmann M, Sigrist SJ, Bruchpilot Promotes Active Zone Assembly, Ca2+ Channel Clustering, and Vesicle Release, Science 312 (2006) 1051–1054. [DOI] [PubMed] [Google Scholar]
- [62].Klar TA, Jakobs S, Dyba M, Egner A, Hell SW, Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission, Proc. Natl. Acad. Sci 97 (2000) 8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Koch N, Kobler O, Thomas U, Qualmann B, Kessels MM, Terminal axonal arborization and synaptic bouton formation critically rely on abp1 and the arp2/3 complex, PLoS One 9 (2014) e97692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Kriebel ME, Hanna R, Muniak C, Synaptic vesicle diameters and synaptic cleft widths at the mouse diaphragm in neonates and adults, Brain Res 392 (1986) 19–29. [DOI] [PubMed] [Google Scholar]
- [65].Lagache T, Grassart A, Dallongeville S, Faklaris O, Sauvonnet N, Dufour A, Danglot L, Olivo-Marin JC, Mapping molecular assemblies with fluorescence microscopy and object-based spatial statistics, Nat Commun 9 (2018) 698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Lampe A, Haucke V, Sigrist SJ, Heilemann M, Schmoranzer J, Multi-colour direct STORM with red emitting carbocyanines, Biol Cell 104 (2012) 229–237. [DOI] [PubMed] [Google Scholar]
- [67].Lauterbach MA, Keller J, Schonle A, Kamin D, Westphal V, Rizzoli SO, Hell SW, Comparing video-rate STED nanoscopy and confocal microscopy of living neurons, J Biophotonics 3 (2010) 417–424. [DOI] [PubMed] [Google Scholar]
- [68].Lee JF, Stovall GM, Ellington AD, Aptamer therapeutics advance, Curr Opin Chem Biol 10 (2006) 282–289. [DOI] [PubMed] [Google Scholar]
- [69].Lehmann M, Lichtner G, Klenz H, Schmoranzer J, Novel organic dyes for multicolor localization-based super-resolution microscopy, J Biophotonics 9 (2016) 161–170. [DOI] [PubMed] [Google Scholar]
- [70].Leica Microsystems. 3D STED Deep Nanoscopy, <https://www.leica-microsystems.com/applications/life-science/3d-sted-nanoscopy/> (2019).
- [71].Leica Microsystems. Quick Guide to STED Sample Preparation, <https://www.leica-microsystems.com/science-lab/quick-guide-to-sted-sample-preparation/> (2019).
- [72].Li L, Xiong WC, Mei L, Neuromuscular Junction Formation, Aging, and Disorders, Annual review of physiology 80 (2018) 159–188. [DOI] [PubMed] [Google Scholar]
- [73].Liu KS, Siebert M, Mertel S, Knoche E, Wegener S, Wichmann C, Matkovic T, Muhammad K, Depner H, Mettke C, Buckers J, Hell SW, Muller M, Davis GW, Schmitz D, Sigrist SJ, RIM-binding protein, a central part of the active zone, is essential for neurotransmitter release, Science 334 (2011) 1565–1569. [DOI] [PubMed] [Google Scholar]
- [74].Liu TL, Upadhyayula S, Milkie DE, Singh V, Wang K, Swinburne IA, Mosaliganti KR, Collins ZM, Hiscock TW, Shea J, Kohrman AQ, Medwig TN, Dambournet D, Forster R, Cunniff B, Ruan Y, Yashiro H, Scholpp S, Meyerowitz EM, Hockemeyer D, Drubin DG, Martin BL, Matus DQ, Koyama M, Megason SG, Kirchhausen T, Betzig E, Observing the cell in its native state: Imaging subcellular dynamics in multicellular organisms, Science 360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].MacGillavry HD, Kerr JM, Blanpied TA, Lateral organization of the postsynaptic density, Molecular and cellular neurosciences 48 (2011) 321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Maidorn M, Rizzoli SO, Opazo F, Tools and limitations to study the molecular composition of synapses by fluorescence microscopy, Biochem J 473 (2016) 3385–3399. [DOI] [PubMed] [Google Scholar]
- [77].Micheva KD, Bruchez MP, The gain in brain: novel imaging techniques and multiplexed proteomic imaging of brain tissue ultrastructure, Curr Opin Neurobiol 22 (2012) 94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Moffitt JR, Osseforth C, Michaelis J, Time-gating improves the spatial resolution of STED microscopy, Opt Express 19 (2011) 4242–4254. [DOI] [PubMed] [Google Scholar]
- [79].Münter S, Niyaz Y, Carl Zeiss Microscopy GmbH. Sample Preparation for Superresolution Microscopy – a Quick Guide, ZEISS ELYRA, <https://applications.zeiss.com/C125792900358A3F/0/5BCE097470446D4EC1257C3C005C545F/$FILE/EN_41_011_065_ELYRA_Sample-prep-QuickGuide.pdf> (2013).
- [80].Murphy RM, Slayter H, Schurtenberger P, Chamberlin RA, Colton CK, Yarmush ML, Size and structure of antigen-antibody complexes. Electron microscopy and light scattering studies, Biophysical journal 54 (1988) 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Nagerl UV, Willig KI, Hein B, Hell SW, Bonhoeffer T, Live-cell imaging of dendritic spines by STED microscopy, Proc. Natl. Acad. Sci. U.S.A. 105 (2008) 18982–18987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Nagwaney S, Harlow ML, Jung JH, Szule JA, Ress D, Xu J, Marshall RM, McMahan UJ, Macromolecular Connections of Active Zone Material to Docked Synaptic Vesicles and Presynaptic Membrane at Neuromuscular Junctions of Mouse, J Comp Neurol 513 (2009) 457–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Neil MA, Juskaitis R, Wilson T, Method of obtaining optical sectioning by using structured light in a conventional microscope, Opt Lett 22 (1997) 1905–1907. [DOI] [PubMed] [Google Scholar]
- [84].Neubert F, Beliu G, Terpitz U, Werner C, Geis C, Sauer M, Doose S, Bioorthogonal Click Chemistry Enables Site-specific Fluorescence Labeling of Functional NMDA Receptors for Super-Resolution Imaging, Angew Chem Int Ed Engl 57 (2018) 16364–16369. [DOI] [PubMed] [Google Scholar]
- [85].Nikon Instrumetns Inc. Super Resolution Microscopes, <https://d33b8×22mym97j.cloudfront.net/phase4/literature/Brochures/2ce-scjk-1-2.pdf?mtime=20190408074637> (2018).
- [86].Nishimune H, Badawi Y, Mori S, Shigemoto K, Dual-color STED microscopy reveals a sandwich structure of Bassoon and Piccolo in active zones of adult and aged mice, Scientific reports 6 (2016) 27935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Nozumi M, Nakatsu F, Katoh K, Igarashi M, Coordinated Movement of Vesicles and Actin Bundles during Nerve Growth Revealed by Superresolution Microscopy, Cell reports 18 (2017) 2203–2216. [DOI] [PubMed] [Google Scholar]
- [88].Okabe S, Molecular dynamics of the excitatory synapse, Adv Exp Med Biol 970 (2012) 131–152. [DOI] [PubMed] [Google Scholar]
- [89].Paul MM, Pauli M, Ehmann N, Hallermann S, Sauer M, Kittel RJ, Heckmann M, Bruchpilot and Synaptotagmin collaborate to drive rapid glutamate release and active zone differentiation, Front Cell Neurosci 9 (2015) 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Pielage J, Cheng L, Fetter RD, Carlton PM, Sedat JW, Davis GW, A presynaptic giant ankyrin stabilizes the NMJ through regulation of presynaptic microtubules and transsynaptic cell adhesion, Neuron 58 (2008) 195–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Pinnington SJL, Marshall JF, Wheeler AP, Correlative 3D Structured Illumination Microscopy and Single-Molecule Localization Microscopy for Imaging Cancer Invasion, Methods Mol Biol 1764 (2018) 253–265. [DOI] [PubMed] [Google Scholar]
- [92].Pitsch S, Köster I. A Bright Dye for Live-Cell STED Microscopy, <https://www.leica-microsystems.com/science-lab/a-bright-dye-for-live-cell-sted-microscopy/> (2015).
- [93].Protti DA, Reisin R, Mackinley TA, Uchitel OD, Calcium channel blockers and transmitter release at the normal human neuromuscular junction, Neurology 46 (1996) 1391–1396. [DOI] [PubMed] [Google Scholar]
- [94].Rayleigh L, On the theory of optical images, with special reference to microscopy, Philosophical Magazine 42 (1896) 167–195. [Google Scholar]
- [95].Reddy-Alla S, Bohme MA, Reynolds E, Beis C, Grasskamp AT, Mampell MM, Maglione M, Jusyte M, Rey U, Babikir H, McCarthy AW, Quentin C, Matkovic T, Bergeron DD, Mushtaq Z, Gottfert F, Owald D, Mielke T, Hell SW, Sigrist SJ, Walter AM, Stable Positioning of Unc13 Restricts Synaptic Vesicle Fusion to Defined Release Sites to Promote Synchronous Neurotransmission, Neuron 95 (2017) 1350–1364 e1312. [DOI] [PubMed] [Google Scholar]
- [96].Ribchester RR, Mammalian neuromuscular junctions: modern tools to monitor synaptic form and function, Current Opinion in Pharmacology 9 (2009) 297–305. [DOI] [PubMed] [Google Scholar]
- [97].Robinson CM, Patel MR, Webb DJ, Super resolution microscopy is poised to reveal new insights into the formation and maturation of dendritic spines, F1000Res 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Rosato Siri MD, Uchitel OD, Calcium channels coupled to neurotransmitter release at neonatal rat neuromuscular junctions, J Physiol 514 (Pt 2) (1999) 533–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Rossy J, Pageon SV, Davis DM, Gaus K, Super-resolution microscopy of the immunological synapse, Curr Opin Immunol 25 (2013) 307–312. [DOI] [PubMed] [Google Scholar]
- [100].Rust MJ, Bates M, Zhuang X, Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM), Nat Methods 3 (2006) 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Sage D, Pham TA, Babcock H, Lukes T, Pengo T, Chao J, Velmurugan R, Herbert A, Agrawal A, Colabrese S, Wheeler A, Archetti A, Rieger B, Ober R, Hagen GM, Sibarita JB, Ries J, Henriques R, Unser M, Holden S, Super-resolution fight club: assessment of 2D and 3D single-molecule localization microscopy software, Nat Methods 16 (2019) 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Sanes JR, Lichtman JW, Development of the vertebrate neuromuscular junction, Annual review of neuroscience 22 (1999) 389–442. [DOI] [PubMed] [Google Scholar]
- [103].Schneider J, Zahn J, Maglione M, Sigrist SJ, Marquard J, Chojnacki J, Krausslich HG, Sahl SJ, Engelhardt J, Hell SW, Ultrafast, temporally stochastic STED nanoscopy of millisecond dynamics, Nat Methods 12 (2015) 827–830. [DOI] [PubMed] [Google Scholar]
- [104].Scholz N, Ehmann N, Sachidanandan D, Imig C, Cooper BH, Jahn O, Reim K, Brose N, Meyer J, Lamberty M, Altrichter S, Bormann A, Hallermann S, Pauli M, Heckmann M, Stigloher C, Langenhan T, Kittel RJ, Complexin cooperates with Bruchpilot to tether synaptic vesicles to the active zone cytomatrix, J Cell Biol 218 (2019) 1011–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Shahidullah M, Le Marchand SJ, Fei H, Zhang J, Pandey UB, Dalva MB, Pasinelli P, Levitan IB, Defects in synapse structure and function precede motor neuron degeneration in Drosophila models of FUS-related ALS, J Neurosci 33 (2013) 19590–19598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Shim SH, Xia C, Zhong G, Babcock HP, Vaughan JC, Huang B, Wang X, Xu C, Bi GQ, Zhuang X, Super-resolution fluorescence imaging of organelles in live cells with photoswitchable membrane probes, Proc. Natl. Acad. Sci. U.S.A. 109 (2012) 13978–13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Shin W, Ge L, Arpino G, Villarreal SA, Hamid E, Liu H, Zhao WD, Wen PJ, Chiang HC, Wu LG, Visualization of Membrane Pore in Live Cells Reveals a Dynamic-Pore Theory Governing Fusion and Endocytosis, Cell 173 (2018) 934–945 e912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Shroff H, Galbraith CG, Galbraith JA, Betzig E, Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics, Nat Methods 5 (2008) 417–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Shroff H, Hess HF, Betzig E, Patterson GH, Lippincott-Schwartz J, Davidson MW. Practical Aspects of PALM Imaging, <http://zeisscampus.magnet.fsu.edu/articles/superresolution/palm/practicalaspects.html> (2019).
- [110].Sidenstein SC, D’Este E, Bohm MJ, Danzl JG, Belov VN, Hell SW, Multicolour Multilevel STED nanoscopy of Actin/Spectrin Organization at Synapses, Scientific reports 6 (2016) 26725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Siebenmorgen J, Novikau Y, Wolleschensky R, Weisshart K, Kleppe I. Introducing Lattice SIM for ZEISS Elyra 7 Structured Illumination Microscopy with a 3D Lattice for Live Cell Imaging, <https://applications.zeiss.com/C125792900358A3F/0/31094A41795C8E17C125835A005391AF/$FILE/EN_41_013_188-WP_Introducing_Lattice_SIM_for_Elyra-7.pdf> (2018).
- [112].Sigal YM, Zhou R, Zhuang X, Visualizing and discovering cellular structures with super-resolution microscopy, Science 361 (2018) 880–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Sigrist SJ, Sabatini BL, Optical super-resolution microscopy in neurobiology, Curr Opin Neurobiol 22 (2012) 86–93. [DOI] [PubMed] [Google Scholar]
- [114].Silfies JS, Schwartz SA, Davidson MW. The Diffraction Barrier in Optical Microscopy, <https://www.microscopyu.com/techniques/super-resolution/the-diffraction-barrier-in-optical-microscopy> (2019).
- [115].Slater CR, The Structure of Human Neuromuscular Junctions: Some Unanswered Molecular Questions, International journal of molecular sciences 18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Small A, Stahlheber S, Fluorophore localization algorithms for super-resolution microscopy, Nat Methods 11 (2014) 267–279. [DOI] [PubMed] [Google Scholar]
- [117].Stijlemans B, Conrath K, Cortez-Retamozo V, Van Xong H, Wyns L, Senter P, Revets H, De Baetselier P, Muyldermans S, Magez S, Efficient targeting of conserved cryptic epitopes of infectious agents by single domain antibodies. African trypanosomes as paradigm, J Biol Chem 279 (2004) 1256–1261. [DOI] [PubMed] [Google Scholar]
- [118].Stracy M, Kapanidis AN, Single-molecule and super-resolution imaging of transcription in living bacteria, Methods (San Diego, Calif 120 (2017) 103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Sulkowski MJ, Han TH, Ott C, Wang Q, Verheyen EM, Lippincott-Schwartz J, Serpe M, A Novel, Noncanonical BMP Pathway Modulates Synapse Maturation at the Drosophila Neuromuscular Junction, PLoS Genet 12 (2016) e1005810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Takeshima T, Takahashi T, Yamashita J, Okada Y, Watanabe S, A multi-emitter fitting algorithm for potential live cell super-resolution imaging over a wide range of molecular densities, J Microsc 271 (2018) 266–281. [DOI] [PubMed] [Google Scholar]
- [121].Terriac E, Coceano G, Mavajian Z, Hageman TA, Christ AF, Testa I, Lautenschlager F, Gad AK, Vimentin Levels and Serine 71 Phosphorylation in the Control of Cell-Matrix Adhesions, Migration Speed, and Shape of Transformed Human Fibroblasts, Cells 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Thiel CS, Tauber S, Seebacher C, Schropp M, Uhl R, Lauber B, Polzer J, Neelam S, Zhang Y, Ullrich O, Real-Time 3D High-Resolution Microscopy of Human Cells on the International Space Station, International journal of molecular sciences 20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Tillberg PW, Chen F, Piatkevich KD, Zhao Y, Yu CC, English BP, Gao L, Martorell A, Suk HJ, Yoshida F, DeGennaro EM, Roossien DH, Gong G, Seneviratne U, Tannenbaum SR, Desimone R, Cai D, Boyden ES, Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies, Nat Biotechnol 34 (2016) 987–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Titlow JS, Yang L, Parton RM, Palanca A, Davis I, Super-Resolution Single Molecule FISH at the Drosophila Neuromuscular Junction, Methods Mol Biol 1649 (2018) 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Tønnesen J, Nadrigny F, Willig KI, Wedlich-Soldner R, Nagerl UV, Two-color STED microscopy of living synapses using a single laser-beam pair, Biophysical journal 101 (2011) 2545–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Tonnesen J, Nagerl UV, Superresolution imaging for neuroscience, Exp Neurol 242 (2013) 33–40. [DOI] [PubMed] [Google Scholar]
- [127].Turcotte R, Liang Y, Tanimoto M, Zhang Q, Li Z, Koyama M, Betzig E, Ji N, Dynamic superresolution structured illumination imaging in the living brain, Proc. Natl. Acad. Sci. U.S.A. 116 (2019) 9586–9591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Ullrich A, Bohme MA, Schoneberg J, Depner H, Sigrist SJ, Noe F, Dynamical Organization of Syntaxin-1A at the Presynaptic Active Zone, PLoS computational biology 11 (2015) e1004407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Unnersjo-Jess D, Scott L, Blom H, Brismar H, Super-resolution stimulated emission depletion imaging of slit diaphragm proteins in optically cleared kidney tissue, Kidney Int 89 (2016) 243–247. [DOI] [PubMed] [Google Scholar]
- [130].Vicidomini G, Moneron G, Han KY, Westphal V, Ta H, Reuss M, Engelhardt J, Eggeling C, Hell SW, Sharper low-power STED nanoscopy by time gating, Nat Methods 8 (2011) 571–573. [DOI] [PubMed] [Google Scholar]
- [131].Wegner W, Ilgen P, Gregor C, van Dort J, Mott AC, Steffens H, Willig KI, In vivo mouse and live cell STED microscopy of neuronal actin plasticity using far-red emitting fluorescent proteins, Scientific reports 7 (2017) 11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Wegner W, Mott AC, Grant SGN, Steffens H, Willig KI, In vivo STED microscopy visualizes PSD95 sub-structures and morphological changes over several hours in the mouse visual cortex, Scientific reports 8 (2018) 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Westphal V, Rizzoli SO, Lauterbach MA, Kamin D, Jahn R, Hell SW, Video-rate far-field optical nanoscopy dissects synaptic vesicle movement, Science 320 (2008) 246–249. [DOI] [PubMed] [Google Scholar]
- [134].Willig KI, Rizzoli SO, Westphal V, Jahn R, Hell SW, STED microscopy reveals that synaptotagmin remains clustered after synaptic vesicle exocytosis, Nature 440 (2006) 935–939. [DOI] [PubMed] [Google Scholar]
- [135].Wilmes S, Staufenbiel M, Lisse D, Richter CP, Beutel O, Busch KB, Hess ST, Piehler J, Triple-color super-resolution imaging of live cells: resolving submicroscopic receptor organization in the plasma membrane, Angew Chem Int Ed Engl 51 (2012) 4868–4871. [DOI] [PubMed] [Google Scholar]
- [136].Winter FR, Loidolt M, Westphal V, Butkevich AN, Gregor C, Sahl SJ, Hell SW, Multicolour nanoscopy of fixed and living cells with a single STED beam and hyperspectral detection, Scientific reports 7 (2017) 46492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Wu Y, Shroff H, Faster, sharper, and deeper: structured illumination microscopy for biological imaging, Nat Methods 15 (2018) 1011–1019. [DOI] [PubMed] [Google Scholar]
- [138].Yamashita N, Morita M, Legant WR, Chen BC, Betzig E, Yokota H, Mimori-Kiyosue Y, Three-dimensional tracking of plus-tips by lattice light-sheet microscopy permits the quantification of microtubule growth trajectories within the mitotic apparatus, J Biomed Opt 20 (2015) 101206. [DOI] [PubMed] [Google Scholar]
- [139].York AL, Zheng JQ, Super-Resolution Microscopy Reveals a Nanoscale Organization of Acetylcholine Receptors for Trans-Synaptic Alignment at Neuromuscular Synapses, eNeuro 4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Young LJ, Strohl F, Kaminski CF, A Guide to Structured Illumination TIRF Microscopy at High Speed with Multiple Colors, J Vis Exp (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Zou W, Wang Z, Xiong M, Chen AY, Xu P, Ganaie SS, Badawi Y, Kleiboeker S, Nishimune H, Ye SQ, Qiu J, Human Parvovirus B19 Utilizes Cellular DNA Replication Machinery for Viral DNA Replication, Journal of virology 92 (2018) e01881–01817. [DOI] [PMC free article] [PubMed] [Google Scholar]