Abstract
Background
Erwinia chrysanthemi (Ec) is a destructive pathogen which causes soft-rot diseases in diverse plant species including orchids. We investigated whether colonization of Oncidium roots by the endophytic fungus Piriformospora indica (Pi) restricts Ec-induced disease development in leaves, and whether this might be related to the regulation of nucleotide binding site-leucine rich repeat (NBS-LRR) Resistance (R) genes.
Results
Root colonization of Oncidium stackings by Pi restricts progression of Ec-induced disease development in the leaves. Since Pi does not inhibit Ec growth on agar plates, we tested whether NBS-LRR R gene transcripts and the levels of their potential target miRNAs in Oncidium leaves might be regulated by Pi. Using bioinformatic tools, we first identified NBS-LRR R gene sequences from Oncidium, which are predicted to be targets of miRNAs. Among them, the expression of two R genes was repressed and the accumulation of several regulatory miRNA stimulated by Ec in the leaves of Oncidium plants. This correlated with the progression of disease development, jasmonic and salicylic acid accumulation, ethylene synthesis and H2O2 production after Ec infection of Oncidium leaves. Interestingly, root colonization by Pi restricted disease development in the leaves, and this was accompanied by higher expression levels of several defense-related R genes and lower expression level of their target miRNA.
Conclusion
Based on these data we propose that Pi controls the levels of NBS-LRR R mRNAs and their target miRNAs in leaves. This regulatory circuit correlates with the protection of Oncidium plants against Ec infection, and molecular and biochemical investigations will demonstrate in the future whether, and if so, to what extent these two observations are related to each other.
Keywords: Oncidium, Piriformospora indica, Erwinia chrysanthemi, microRNA, Resistance gene
Background
Orchids such as Oncidium, Phalaenopsis and Dendrobium have high commercial value and are becoming globally important for the agro-industry. However, modern commercial orchid cultivars are selected by artificial pollination and multiplied by micro-propagation; as a consequence, the low gene diversity and large scale cultivation make them susceptible to pathogen infection causing great economic loss. For example, Erwinia chrysanthemi (Ec, also known as Dickeya dadantii or Pectobacterium chrysanthemi) is one of the soft rot pathogens [1, 2] which causes economic losses in a wide variety of crops and orchids, including Oncidium (cultivar Onc. ‘Gower Ramsey’, the most often commercialized cultivar in Taiwan, South East Asia and China) [3, 4]. Up to now, no resistance (R) genes against soft rot disease have been reported in orchids. Moreover, most orchids have long vegetative growth, and the long breeding cycle prevents an improvement of single characteristics via hybridization. Identification and characterization of R genes from the existing orchid germplasm resources would be helpful in breeding high-resistance orchid cultivars and in genetic engineering programs.
Piriformospora indica (Pi), originally isolated from the woody shrubs rhizosphere in an Indian desert, is a root-colonizing endophytic fungus with a broad range of host plants. It confers diverse beneficial effects on host plants by improving nutrition uptake [5, 6], promoting biomass production [7–9], stimulating the accumulation of secondary metabolites [10, 11], and strengthening resistance against biotic and abiotic stresses [12–16].
The nucleotide binding site-leucine rich repeat (NBS-LRR) resistance (R) proteins function as molecular switches. They are characterized by highly conserved motifs, including the P-Loop/Kinase-1a [GGV(I/M)GKTT], Kinase-2 [LVDDVW(D)], Kinase-3a (GSRIIITTRD) and GLPL [GL(F)PL(F)AL] motifs in all plant species [17]. R proteins recognize directly or indirectly pathogens’ effectors and trigger or suppress downstream defense responses in plants. Thus far, 149 and 480 genes for NBS-LRR R and LRR domain proteins were identified in Arabidopsis and rice, respectively [18, 19]. They are classified into two major groups: Toll/Interleukin-1 receptors (TIR) and non-TIR-NBS-LRR proteins, based on the N-terminal TIR or curly coiled-coil structure [20].
Micro-RNAs (miRNAs), small non-coding RNAs, silence gene expression at transcriptional and post-transcriptional levels, and participate in numerous processes including plant defense [21–23]. For example, miR393 is a pathogen-associated molecular pattern-responsive miRNA which contributes to disease resistance, and pathogen effectors can suppress miR393 accumulation to facilitate disease development [24, 25]. Additionally, miRNA-mediated R gene silencing plays an important role in the development of plant-microbial symbiosis and systemic resistance [26–28]. In Medicago truncatula, the expression levels of miRNAs such as miR399k*, miR1507, miR1510a*, miR2678 and miR5213, which have been verified to regulate NBS-LRR R genes, were suppressed during the mycorrhiza formation; moreover, miR5213 is only found in mycorrhizal plants [29].
In a previous study, we reported that colonization of Oncidium roots by Pi regulates a group of miRNAs and related target R genes [30]. 1083 miRNAs belonging to 56 families were detected in a transcriptomic library from Pi-colonized roots, but not from uncolonized roots. Furthermore, the expression patterns of miRNAs and their target genes during the symbiotic process showed significant changes during root colonisation. Especially, the miRNAs involved in auxin signaling functions and root development responded to Pi colonization. The work suggested that Pi promoted plant growth through regulating the expression level of miRNAs and their target genes. In the present work, we focus on the mechanism of pathogen resistance induced by Pi-colonization. The expression pattern of miRNAs and their target transcripts for NBS-LRR R proteins was investigated in Ec-infected leaves of Pi-colonized and uncolonized Oncidium. Our work revealed that root colonization by Pi activates the expression of NBS-LRR R genes in the leaves which correlated with an increase resistance against Ec infection. Furthermore, when a leaf is infected by Ec, Pi can suppress the accumulation of Ec-induced miRNAs in the leaves which results in high expression of their target R genes. The counteracting mechanism between Pi and Ec is discussed in the context of root-to-shoot signaling.
Results
Mining for NBS-LRR-type R genes in Oncidium
NBS-LRR R proteins play an important role in plant resistance against abiotic and biotic stress and act as a molecular switch to regulate defense in plant-microorganism interactions [31, 32]. In a previous study, we identified miRNAs which were significantly up-regulated in Oncidium roots in response to Pi colonization [30]. Closer inspection of these miRNAs uncovered that many of them are predicted to target R gene transcripts. This prompted us to investigate the regulation of R genes/transcripts and their potential target miRNAs in the tripartite interaction between Oncidium, Ec and Pi in greater details.
To identify R genes in Oncidium, we used the previously described transcriptomic datasets (accession: PRJNA428913, [30]). 24,616 deduced amino acid sequences from the transcripts were scanned for pfam NB-ARC HMM profiles (pfam: PF00931) by an E-value cut-off of < 1− 40 with the HMMER software. We identified 99 candidate R gene sequences using an E-value cut-off of < 1− 2. The proteins deduced from these sequences contained between 62 and 610 amino acids (average length: 229 amino acids) (Additional file 3: Table S1). Similar methods were used for the identification of R genes in Phalaenopsis equestris [33] and Dendrobium officinale [34] and resulted in 63 and 94 candidate sequences with the respective whole genomic sequence information.
Subsequently, the candidate R protein sequences from Oncidium were used for BLAST searches against the NCBI nr database using BLASTP. Ninety-six of the 99 candidates matched to known disease R proteins with 50.2 to 88.0% similarity (E-values: between 0 and 1.56− 11). Among them, 57 and 36 proteins were highly similar to R proteins from D. officinale and P. equestri, respectively (Additional file 1: Figure S1).
NBS-LRR R proteins are characterized by their amino-terminal TIR domains or CC motifs, and a carboxyl-terminal LRR domain. The 99 candidate sequences were further analyzed with the InterProScan program on the BLAST2GO software. We identified 87 sequences with NB-ARC domains, 16 with CC motifs, 36 with LRR domains, and none with a TIR domain (Additional file 3: Table S1). TIR domains were also not detected in the predicted R protein sequences of D. officinale and P. equestri.
The NB-ARC motif-containing R proteins identified by the MEME analyses were identical to the Oncidium R proteins found with the InterProScan program. Eight types of major motifs, including the P-loop, RNBS-A-non-TIR motif, kinase-2, RNBS-B, GLPL, RNBS-C, RNBS-D and an MHD-like motif, were identified in Oncidium R proteins (Table 1). Among them, the P-loop, RNBS-A, RNBS-B and RNBS-C motifs showed the highest conservation. However, the GLPL was replaced by the GC/SPLAA motif in Oncidium. The same replacement was also found in P. equestri, but not in D. officinale where the original sequence GLPLAL/I was conserved. Furthermore, the MHDL motif was replaced by a MHD-like motif in Oncidium, which is also found in apple [35] and Phalaenopsis [33]. Finally, a highly conserved FxKxDLVRMW motif, located ~ 40 amino acids N-terminal to the MHD-like motif sequence, is also presented in P. equestri and D. officinale, but not found in other species including Arabidopsis [17], Japonica rice [18], Populus trichocarpa [36], soybean [37], Solanum tuberosum [38] and Lotus japonicas [39].
Table 1.
The motifs of R protein sequences identified by MEME
| No. | Best match | NBS motif | E-value |
|---|---|---|---|
| 1 | FCxxFxQDHxFDKDDLVRMW | 9.1− 310 | |
| 2 | LsVVGH/MGGMKxTLLQHVY | P-loop | 1.0− 295 |
| 3 | MVxKLxGC/SPLAAKVIGGILN | GLPL | 7.8− 266 |
| 4 | SYxxLPxxLxxCFxFCxxFP | RNBS-D | 8.3−250 |
| 5 | FxVK/QxW/FV/ACVSxNFxAxxVIX | RNBS-A-non-TIR | 1.4−266 |
| 6 | xYKMHDLLHELAQS/EVSxxEx | MHD-like | 3.1− 256 |
| 7 | VLAPLxxGSS/LGSKxLITTRx | RNBS-B | 2.2−259 |
| 8 | DxGRcYFN/DILVxxSFFDEFx | 9.9− 245 | |
| 9 | D/ExCLxLF/LxxH/YAFA/FGVENPDD | RNBS-C | 2.2− 243 |
| 10 | KRFLL/IVxDDI/VWExDExxWxN | Kinase-2 | 5.2−234 |
*If the bit value of the amino acid at this position is < 1, it is replaced by an x; conserved amino acid sequences are shown in bold letters
Phylogenetic analysis of NB-ARC domain-containing R proteins from Oncidium
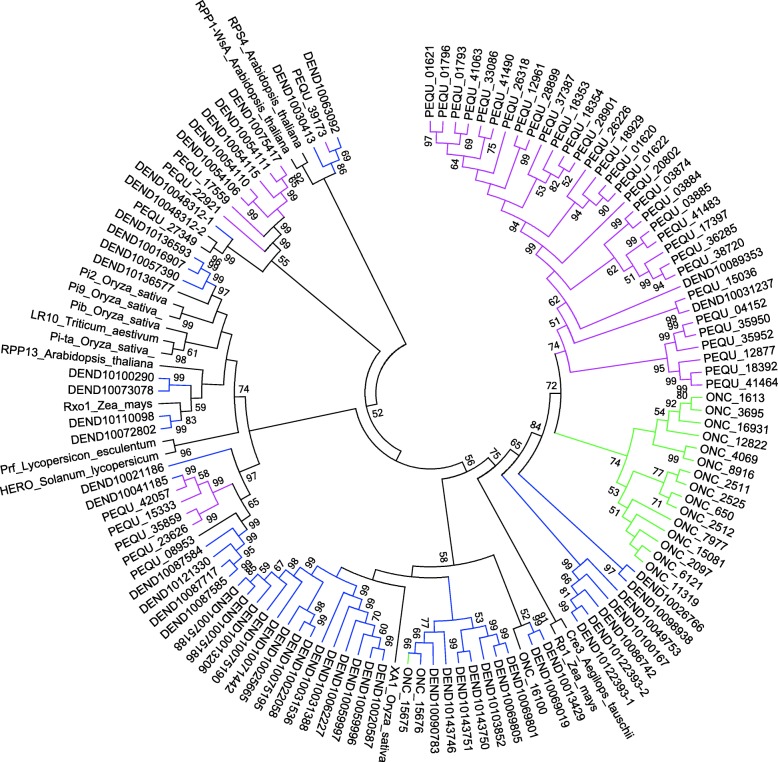
To study the evolutionary relationships of Oncidium R proteins, a Neighbor-Joining phylogenetic tree was built based on the conserved NB-ARC domain (from P-loop to MHD-like motif) by using the MEGA6.06 software. Eighteen of the 99 Oncidium R protein sequences contain the complete NB-ARC domain. Together with 15 well-known R protein sequences from other species, they were used for the phylogenetic analysis (Fig. 1). As expected, two well-known TIR type R proteins were grouped into the TNL clade, while the Oncidium sequences were grouped into the non-TNL clade together with the well characterized R proteins from the other species. Furthermore, 15 of the 18 Oncidium R proteins form an independent clade and show only a distant relationship to the known XA1 (rice), Cre3 (wheat) and Rp1 (maize) R proteins. The result suggests that the diversification of R genes in Oncidium and other orchids has a unique evolutionary history.
Fig. 1.
Phylogenetic analysis of R proteins of Oncidium, Phalaenopsis equestris and Dendrobium officinale orchids. Bootstrap Neighbor-Joint tree was constructed for the R proteins from Oncidium (ONC, green), Phalaenopsis equestris (PEQU, red) and Dendrobium officinale (DEND, blue) using MEGA6.01 and the respective NBARC domains (from P-loop to MHD-like domain) (Fig. 2). The sequences were compared to 15 known R protein sequences: TNL: RPP-1 (AAC72977), RPS4 (BAB11393); NL: Pi9 (ABB88855), Pi2 (ABC94599), Pib (BAA76281); XNL: Prf (U65391); CNL: Rp1 (AAP81262), RXO1 (AY935244), Xa1 (BAA25068), Pita (AAK00132), Cre3 (AAC05834), Lr10 (aaq01784), RPM1 (NP187360), RPP13 (AF209732) and HERO (CAD29728)
Prediction of miRNA target sequences in Oncidium R gene mRNAs
The putative 99 R gene sequences were used to identify target sequences for miRNAs which had been identified previously in Pi-colonized Oncidium (Accession: SRP031471, as described in [30]), by using the psRNAtarget search program with E value < 3. We performed a BLAST search against the miRBase1.9 (plant section) which contained 4562 miRNA sequences. As shown in the Table 2 and Additional file 5: Table S3, 43 of the 99 R gene sequences were positively predicted to be targeted by 46 miRNAs. Among them, miR1507, miR1510a*, miR2118 and miR482/472 were commonly reported in controlling NBS-LRR R gene expression in M. truncatula [29], cotton [40] and potato [41, 42]. Furthermore, several of the identified miRNAs are predicted to target multiple R messages, such as miR1514 (16 R mRNAs), miR1510a* (14 R mRNAs), miR5246 (13 R mRNAs) and miR5654 (12 R mRNAs). Therefore, it appears that the mRNA levels of many R genes might be regulated by more than one miRNA species.
Table 2.
Predicted R genes and the targeting miRNAs
| Target | Counts (RPKM) | Annotation | Predicted regulator miRNA | |
|---|---|---|---|---|
| CK | P | |||
| Onc 235 | 7.9 | 15.4 | uncharacterized protein LOC100279616 | miR2088, miR5654 |
| Onc 649 | 12,.0 | 14.0 | OsJNBa0083D01.14 | miR1514 |
| Onc 650 | 7.8 | 3.0 | hypothetical protein OsI_07084 | miR1510a*, miR1514 |
| Onc 651 | 0 | 14.7 | putative disease resistance protein I2 | miR1514 |
| Onc 1207 | 59.9 | 93.6 | hypothetical protein OsI_15587 | miR1510a* |
| Onc 1471 | 60.9 | 81.6 | putative disease resistance protein RGA4 | miR1514, miR5654 |
| Onc 1537 | 86.2 | 85.7 | hypothetical protein OsJ_14506 | miR1514, miR2088, miR5654 |
| Onc 1613 | 13.2 | 27.5 | disease resistance protein I2 | miR1510, miR529g |
| Onc 1615 | 28.1 | 15.7 | NBS-containing resistance-like protein | miR5246 |
| Onc 1618 | 0.0 | 10.7 | hypothetical protein OsI_07084 | miR1510a* |
| Onc 1724 | 89.8 | 99.4 | hypothetical protein SORBIDRAFT_08g020630 | miR1514 |
| Onc 2107 | 8.5 | 15.0 | putative disease resistance protein | miR156k, miR2088, miR5654 |
| Onc 2524 | 18.5 | 27.2 | putative disease resistance protein RGA2 | miR2088 |
| Onc 2555 | 24.6 | 60.0 | putative disease resistance protein RGA4 | miR1514 |
| Onc 3695 | 11.7 | 22.9 | CC-NBS-LRR R protein | miR1510a*, miR529g |
| Onc 4069 | 36.1 | 32.0 | hypothetical protein OsI_15587 | miR1510a*, miR1514, miR2088, miR5246, miR5654 |
| Onc 4126 | 0.0 | 17.4 | CC-NBS-LRR R protein | miR1510a* |
| Onc 4434 | 16.5 | 10.8 | hypothetical protein VITISV_025836 | miR2088, miR5654 |
| Onc 4722 | 8.6 | 20.2 | hypothetical protein OsI_07084 | miR1510a*, miR894 |
| Onc 4724 | 41.8 | 107.4 | NB-ARC domain-containing protein | miR1510a*, miR857 |
| Onc 5026 | 0.0 | 15.7 | disease resistance protein I2 | miR166c |
| Onc 5046 | 26.3 | 70.0 | uncharacterized protein LOC100279616 | miR5654 |
| Onc 5277 | 25.9 | 52.6 | hypothetical protein OsI_07084 | miR1510, miR5246 |
| Onc 5425 | 41.1 | 48.2 | putative disease resistance protein | miR1514, miR5654 |
| Onc 5583 | 119.0 | 203.5 | hypothetical protein VITISV_018147 | miR482 |
| Onc 6091 | 5.2 | 17.7 | putative disease resistance protein | miR156k, miR2088, miR5654 |
| Onc 6121 | 3.2 | 7.5 | hypothetical protein OsI_15587 | miR1514, miR5246 |
| Onc 7005 | 58.9 | 62.4 | putative disease resistance RPP13 protein | miR1514 |
| Onc 7221 | 20.1 | 50.3 | hypothetical protein OsI_07084 | miR1510a* |
| Onc 7977 | 32.2 | 25.1 | putative disease resistance protein | miR1510a*, miR5246 |
| Onc 8764 | 0.0 | 19.9 | OsJNBa0083D01.14 | miR5654 |
| Onc 8916 | 23.4 | 40.2 | hypothetical protein OsI_07084 | miR156k, miR5246 |
| Onc 9873 | 0.0 | 15.9 | NB-ARC domain containing protein | miR1510a*, miR5246 |
| Onc 11,319 | 9.1 | 10.6 | putative disease resistance protein RGA3-like | miR1514, miR5246 |
| Onc 12,822 | 31.6 | 53.9 | hypothetical protein OsI_15587 | miR1510a*, miR156k |
| Onc 13,214 | 0.0 | 11.9 | NBS-LRR protein | miR1514, miR529g |
| Onc 15,037 | 0.0 | 9.0 | NB-ARC domain containing protein | miR1510a* |
| Onc 15,081 | 26.7 | 20.8 | putative disease resistance RPP13 protein | miR5246 |
| Onc 16,931 | 7.8 | 21.2 | hypothetical protein OsI_15587 | miR1510a*, miR1514, miR166c, miR2088, miR5246, miR529g |
| Onc 19,773 | 6.3 | 0.0 | putative disease resistance protein At3g14460 | miR2088, miR5654 |
| Onc 19,900 | 0.0 | 3.4 | OSIGBa0148A10.13 | miR5654 |
| Onc 20,607 | 0.0 | 6.5 | hypothetical protein OsJ_14506 | miR838 |
| Onc 28,117 | 9.1 | 3.6 | NBS-LRR R protein | miR2118 |
Count normalized by RPKM (reads per kilobase million). CK, control RNA from uncolonized Oncidium roots; P, RNA from Pi-colonized Oncidium roots
Pi protects Oncidium against Ec infection, but does not inhibit Ec growth on agar plates
Further on, we wanted to assess how the expression of the identified R genes is related to Oncidium defence against Ec, and also if it could be modified by colonization with Pi. First of all, orchid cuttings were inoculated with Pi for 2 weeks. Microscopic observation demonstrates that Pi mycelia and spores were present in the cortex and velamen of Oncidium roots (Additional file 2: Figure S2A and B), indicating successful colonization. Subsequently, the second leaf of Pi-colonized and uncolonized control cuttings was infected with Ec bacteria, as described previously [3]. The treated plants were monitored on the 1st, 3rd, 7th and 21th day after infection (dai) by visible examination and counting the cell number of the pathogenic bacteria. At 1st dai, Ec caused obvious necrosis on the inoculated leaves of both Pi-pretreated and control cuttings. Until the 3rd dai, control plants not pre-treated with Pi showed pathogen-induced disease symptoms in the infected and the neighboring uninfected leaf, as well as in the connecting stems. However, the disease symptoms in the Pi-colonized cuttings were restricted to the infected leaf. Until the 7th dai, the control without Pi displayed necrosis in the whole plant, i.e. in all leaves, stems and roots. Moreover, Ec grew widespread on the MS medium and started to infect the neighboring healthy plants via the roots. In contrast, bacterial growth in Pi-colonized plants was mainly detectable on the inoculated leaf, and little infection was visible in neighboring leaves. No Ec bacteria could be observed on the MS medium. Even at the 21th dai, the Pi–colonized plants continued to grow (Additional file 2: Figure S2D), while the control plants were dead. These results indicate that Pi confers resistance against Ec infection and inhibits growth and propagation of the bacterium.
Better performance of the Pi-colonized plants after Ec infection might be caused by a direct inhibition of Ec growth, or by stimulating plant immunity. To investigate the Pi-mediated mechanism, both microbes were co-cultured on an agar plate. As shown in Additional file 1: Figure S1E, there is no inhibition zone between Pi mycelium and the Ec colony. On the contrary, Ec actively inhibits the mycelial growth of Pi hyphae, as visible by comparison of the growth of Pi on plates without the bacterium (Additional file 2: Figure S2E). This suggests that better performance of Pi-colonized Oncidium plants after Ec infection is probably not caused by direct inhibition of the bacterial growth in the plant. Rather, the acquired resistance might be caused by Pi-stimulated defense in the host. This differs from a previously report in which Pi protected Arabidopsis seedlings from Verticillium dahliae infection by inhibiting V. dahliae growth both on plates and in the plant [14].
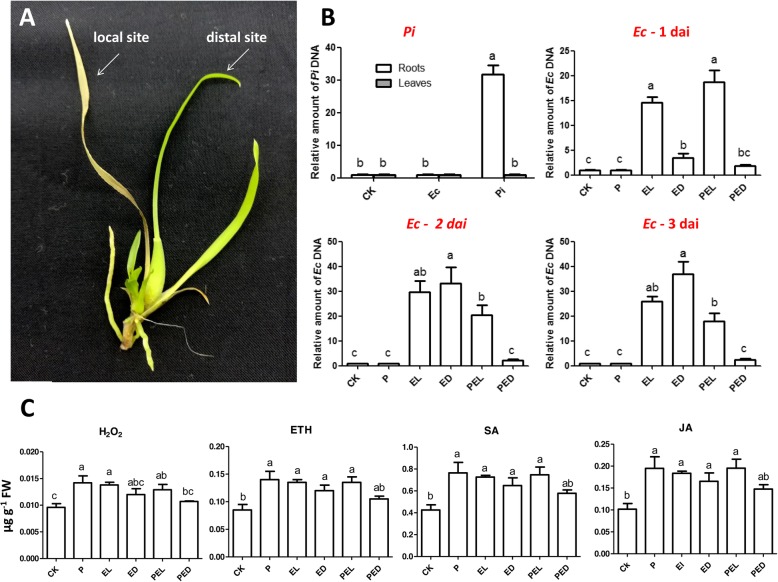
To investigate how Pi protects Oncidium plants against Ec-induced disease symptom development, we determined growth and propagation of the pathogen in the plant by real-time quantitative PCR (qPCR). Ec-inoculated leaves and the non-infected neighboring leaves (Fig. 2a) were separately harvested. We used 16S rDNA to detect the propagation of Ec in the different leaves and compared the results of Pi-colonized and un-colonized Oncidium plants. As shown in Fig. 2b, in the absence of Pi, Ec can be detected in the local infected leaf (EL) and the distal leaf (ED) 2th dai, confirming that the pathogen is highly infectious to Oncidium. In contrast, Ec could only be detected in the local infected leaf of Pi-colonized plants (PEL, Fig. 2b), and was not detectable in the distal leaf of the same plant (PED, Fig. 2b). We also examined the salicylic acid (SA), jasmonic acid (JA) and ethylene (ETH) levels and compared the hormone levels with those for H2O2 24 h after Ec-infection in Pi-colonized and uncolonized plants. The results of the ELISA assays indicate that the hormone and H2O2 levels increased 1.5–2.0-fold in the Ec-infected and none-infected leaves of Pi-colonized and uncolonized plants (Fig. 2c), but the effects were always lower in the distal leaf of Pi-colonized plants although not always significantly (PED, Fig. 2c). These results indicate that the microbes control hormone and H2O2 responses in the leaves. Apparently, in tissues where Pi restricts propagation of the pathogen and disease development, the phytohormone and H2O2 levels are lower (Fig. 2c, PED).
Fig. 2.
Detection of the pathogen in leaf tissues in Pi-colonized/−uncolonized Oncidium. a E. chrysenthemi (Ec) was locally inoculated on the second leaf of Pi-colonized/−uncolonized cuttings, respectively. Local and distal leaves were collected separately. b Ec DNA levels in leaves were detected by qPCR of 16S rDNA 1, 2 and 3 days after infection, Pi DNA in leaves and roots were detected with EF-hand DNA primer pair 10 days after inoculation, data represent the means ± SE of 3 replicates and were normalized to the plant ACTIN DNA level, values with the same letter were not significantly different (p < 0.05). c Levels of endogenous salicylic acid, jasmonic acid, ethylene and H2O2 24 h after infection of the leaf with Ec. Data represent the means ± SE of 3 replicates, values with the same letter were not significantly different (p < 0.05). PI: qPCR for Pi and Ec DNA in roots/leaves of Pi-colonized cuttings. CK: uncolonized plants. EC1d, EC2d and EC3d indicates the detection of the presence Pi and Ec in Pi-colonized/−uncolonized plants 1, 2, or 3 days after Ec infection, relative values normalized to the plant ACTIN DNA level. CK: control plant. P: Pi-colonized plants; (P)EL: local infected leaf of Pi-uncolonized (EL) or -colonized (PEL) plants inoculated with Ec. (P)ED: distal leaves of Pi-uncolonized (ED) or –colonized (PED) plants inoculated with Ec
Colonization of Oncidium roots by Pi affects the expression of R genes and the accumulation of their target miRNA levels in leaves
Analysis of previously performed expression profiles [30] demonstrated that most of the 43 R mRNA levels predicted to be targeted by miRNAs responded to Pi colonization in Oncidium roots (Table 2 and Additional file 4: Table S2). Transcripts for 24 R genes were up-regulated and for 8 R genes down-regulated by the fungus. Transcripts for 10 R genes could only be detected in Pi-colonized plants. One R gene was only expressed in uncolonized roots. However, it is worth noting that almost all miRNA levels which are predicted to target the messages of R genes were present in low abundance in our high-throughput sequencing data (Additional file 4: Table S2).
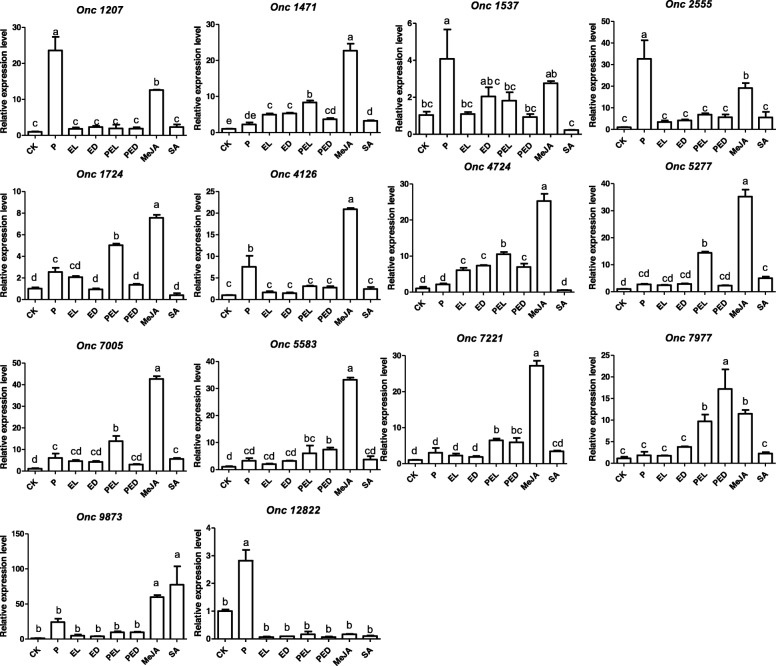
As shown in the Additional file 1: Figure S1 and Additional file 2: Figure S2, the Pi-colonized Oncidium showed increased resistance against Ec infection in the leaf tissues. This suggests that defense information is transmitted from roots to leaves. To investigate the expression levels of R genes and the accumulation of miRNAs in response to either Pi colonization or Ec infection or both, qPCR was performed with RNA preparations from leaves for all 43 R genes predicted to be targeted by miRNAs (Table 2 and Additional file 4: Table S2). As shown in Fig. 3, 7 R genes (Onc1207, Onc1537, Onc1724, Onc2555, Onc4126, Onc12822 and Onc7005) were significantly up-regulated in the leaves of Oncidium plants which were colonized by Pi (Fig. 3, P). This is consistent with our previous results from the high-throughput sequence data and suggests that signals transmitted from the Pi-colonized roots up-regulate these R genes in the leaves, although they were not yet exposed to any threat.
Fig. 3.
Expression of R genes after Ec infection of leaves of Pi-colonized or -uncolonized Oncidium. Expression levels of R genes 24 h after Ec infection of Pi-colonized (2 weeks) or –uncolonized Oncidium plants. 24 h after infection, the leaves were harvested for qRT-PCR analyses. CK: control plant without Pi colonization and Ec infection. P: Pi-colonized plants; (P)EL: local infected leaf of Pi-uncolonized (EL) or -colonized (PEL) plants. (P)ED: distal leaves of Pi-uncolonized (ED) or –colonized (PED) plants. SA: leaves treated with 1.0 mM salicylic acid for 24 h. MeJA: leaves treated with 0.1 mM methyl jasmonic acid for 24 h. Data represent the means ± SE of 3 replicates and were normalized to the Oncidium ACTIN mRNA level, values with the same letter were not significantly different (p < 0.05)
Twenty-four h after leaf infection by Ec, Onc1471 and Onc4724 were induced in the infected leaf (Fig. 3, EL, PEL) and the neighboring uninfected leaf (Fig. 3, ED and PED) of both colonized and uncolonized plants (Fig. 3, CK and P). However, the transcript levels for the R genes Onc1471, Onc1724, Onc4724, Onc5277, Onc7005, Onc7221 and Onc 7977 were higher in Ec-infected leaves of Pi-colonized plants (Fig. 3, PEL) than in the leaves of Pi-uncolonized plants (Fig. 3, EL). Furthermore, the expression of the R genes Onc5583, Onc7221 and Onc7977 in none-infected neighboring leaves were higher in Pi-colonized Oncidium than in the Pi-uncolonized control plants (Fig. 3, PED and ED). These data suggest that the R transcript levels in the leaves respond to Pi colonization of the roots.
Defense hormones in the resistance response
Stein et al. [43] demonstrated that JA signalling and the cytoplasmic, but not nuclear localization of NPR1 is required for Pi-induced resistance against powdery mildrew infection. In order to test whether defence hormones are involved in the Oncidium resistance response against Ec, leaves were treated with 1 mM SA or 0.1 mM MeJA. Interestingly, all R genes (except Onc 12,822) are significantly up-regulated after treatment with 0.1 mM MeJA (Fig. 3, MeJA), but only 4 R genes (Onc1471, Onc5277, Onc7055 and Onc9873) were up-regulated after treatment with 1 mM SA (Fig. 3, SA).
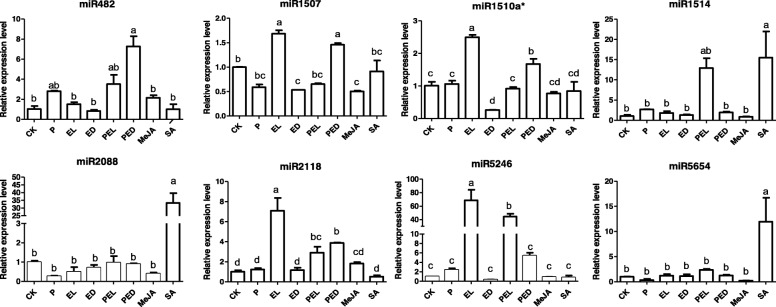
Next, we examined the levels of the identified miRNAs in response to Pi-colonization in Oncidium leaves. In contrast to the responses of the R mRNA levels, 7 miRNA levels did not change in the leaves when the roots were colonized by Pi. The only exception is the miR482 level which was always higher in the leaves of Pi-colonised plants (Fig. 4, P). Ec stimulated the miR1507, miR1510a*, miR2118 and miR5246 levels in the infected leaves, and the effect was no longer detectable in the neighboring, none-infected leaves (Fig. 4, EL and ED). Up-regulation of these miRNA levels in the Ec-infected leaves was reduced when the roots were colonized by Pi (Fig. 4, EL and PED). Thus, the beneficial fungus influences the Ec-induced miRNA levels in the leaves.
Fig. 4.
Expression of miRNAs after leaf infection of Pi-colonized or –uncolonized Oncidium with Ec. miRNA levels in Pi-colonized (2 weeks) or –uncolonized Oncidium. 24 h after Ec infection, the leaves were harvested for qRT-PCR analyses. CK: control plant without Pi colonization and Ec infection. P: Pi-colonized plants; (P)EL: local infected leaf of Pi-uncolonized (EL) or -colonized (PEL) plants inoculated with Ec for 24 h. (P)ED: distal leaves of Pi-uncolonized (ED) or –colonized (PED) plants inoculated with Ec for 24 h. SA: leaves treated with 1.0 mM salicylic acid for 24 h. MeJA: leaves treated with 0.1 mM methyl jasmonic acid for 24 h. Data represent the means ± SE of 3 replicates and were normalized to the U6 snRNA level, values with the same letter were not significantly different (p < 0.05)
Moreover, quite different from the R mRNAs, none of the miRNAs responded to exogenous application of MeJA (Fig. 4, MeJA), similar to the results obtained by Pi colonization. However, the miR1514, miR2088 and miR5654 levels were significantly up-regulated after SA treatment (Fig. 4, SA).
Discussion
Pi protects Oncidium against Ec infection
As reported previously for several pathosystems [44, 45], colonization of the roots by Pi confers resistance against leaf pathogens [13, 15, 16, 46–48]. We demonstrate that the severe disease symptom development induced by Ec in Oncidium leaves is partially restricted when the roots are colonized by Pi. Ec infection stimulates the accumulation of the defense hormones SA, JA and ETH in the leaves (Fig. 2c). The elevated hormone levels presumably participate in activating R gene expression which might participate in restricting disease development and propagation of the pathogen. Our data suggest that the regulation of NBS-LRR R genes and their related miRNA levels in the leaves could play a role in Pi-induced resistance against Ec infection, however, we did not provide any direct evidence for a link between the regulation of the R mRNA / miRNA levels and the disease symptom development in this study. Since Ec produce quite different virulence factors, proteins and metabolites (cf. below) which cause the disease symptoms in the infected plants, it is likely that additional plant defense compounds, mechanisms and strategies are involved in the host response. The R genes and miRNAs investigate here were identified in transcriptomic datasets generated from Pi-colonized Oncidium roots [30]. If they participate in the disease resistance phenotype, a possible and testable scenario could be that Ec counteracts the plant defense response by promoting the accumulation of miRNAs against the R messages (Fig. 5). Signals from the Pi-colonized roots might restrict miRNA accumulation in the leaves and thus support the plant defense against Ec infection (Figs. 3 and 5). The beneficial effect of Pi can be clearly seen by the restriction of Ec propagation in the non-infected neighboring Oncidium leaves. However, since the levels of all three defense-related phytohormones increased in response to Ec infection, we did not observe specific phytohormone effects, which allow allocation of the plant response to the JA/ETH-based defense against necrotrophs or the SA-based defense against biotrophs. This might be due to the massive destruction of the Ec-infected tissue which is associated with cell death processes and a collapse of a coordinated activation of the host defense system against the propagating pathogen. Ec is an opportunistic necrotrophic pathogen that does not appear to invade host cells internally in the pathogenic phase [49]. The bacteria remain in the intercellular spaces of infected plant tissue and use several secretion systems to inject virulence factors into host cells. Well-studied virulence determinants are also extracellular enzymes such as pectate lyase, pectinase, and cellulase; siderophore-dependent iron uptake systems, as well as the sap and msrA genes [49–55]. In addition to causing local disease, the bacteria enter vascular elements of infected plants, thereby moving rapidly through the host [50, 56–59]. This is consistent with the observed rapid collapse of the host defense system and highlights the importance for the search for strategies to restrict Ec propagation in infected plants. We propose that signals derived from Pi-colonized roots might be an interesting tool to control Ec-induced soft rod, wilts and blight diseases [49].
Fig. 5.
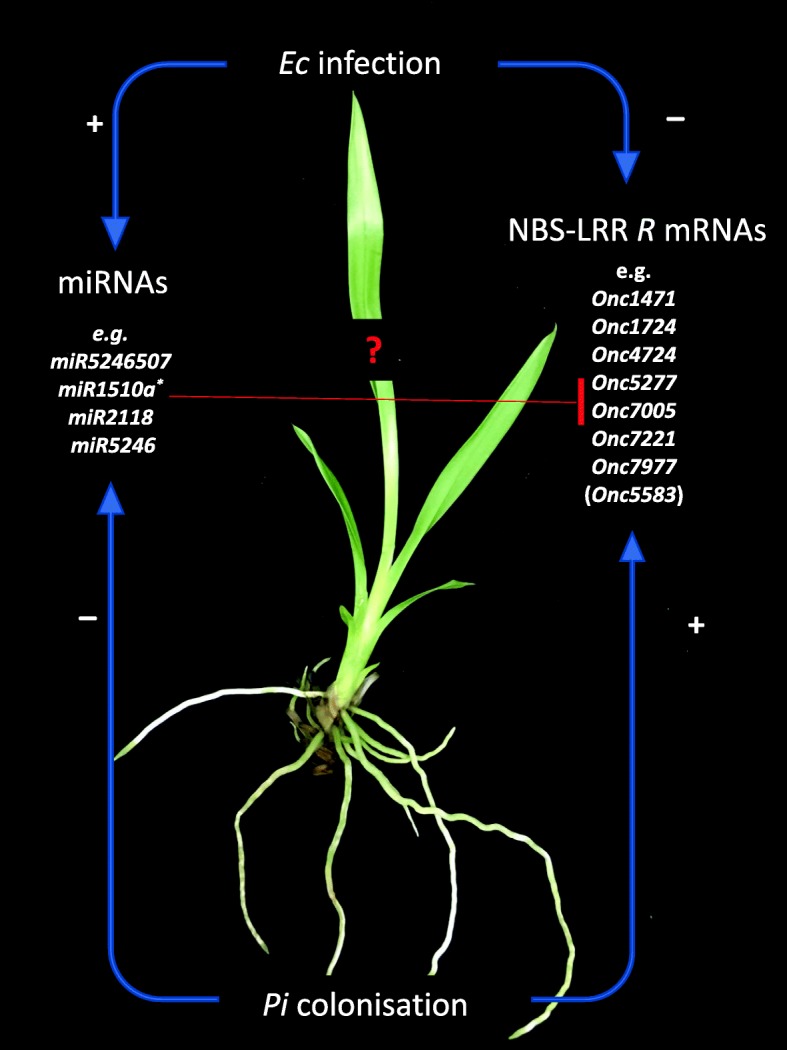
A model describing the regulation of miRNA and NB-LRR R mRNA levels in Oncidium leaves after Ec infection and root colonization by Pi
Pi suppressed the accumulation of Ec-induced miRNAs
The involvement of miRNA in the regulation of host immune responses following fungal exposure has been described in many systems [60]. Interestingly, in human and animals, pathogen-induced changes in expression profiles have identified the same critical miRNAs which are also involved in inflammation and allergy responses [60], suggesting a broad conservation in the mechanisms. In plants, miRNAs play important roles in immune responses and defense gene activation [61–66]. Together with siRNAs (small interfering RNAs), they guide sequence-specific silencing of genes, and recognize repetitive DNA and virus nucleic acids through base complementarily [67]. In our study, we discovered miRNAs from transcriptomic datasets, which were predicted to silence R genes encoding NBS-LRR R proteins (Fig. 4). We showed that the miR1507, miR1510a*, miR2118 and miR5246 levels were induced after Ec infection (Fig. 4, EL), and this stimulation was either completely or partially prevented when the roots were colonized by Pi (Fig. 4, PEL). More importantly, the transcript levels for the R genes Onc1471, Onc1724, Onc4724, Onc5277, Onc7005, Onc7221 and Onc 7977 were higher in Ec-infected local leaves of Pi-colonized plants (Fig. 3, PEL) than in the leaves of plants without Pi colonization (Fig. 3, EL). This suggests that Pi in and around the roots repress miRNAs accumulation in the leaves to maintain relative high R genes levels. Notably, similar results also have been reported for fusiform rust gall development in Pinus taeda, which is controlled by the inhibition of miRNA biosynthesis for target R mRNAs [68]. Besides, during early stage of nodulation, miRNAs as miR482, miR1507, miR2118 accumulate to avoid plant immunity responses against the colonizing microbes [26, 29, 69]. Although there is little known about miRNAs and R genes involved in resistance effects conferred by Pi, our data demonstrate that they might be important regulatory components for disease control. The control of R mRNAs by the miRNAs is based on bioinformatic predictions of target sequences and similarities described for other pathosystems and requires experimental verifications. However, for some miRNAs, the molecular mechanisms have been investigated. During symbiosis development, miR1510a*-mediated cleavage was confirmed by degradome analyses and miR1507, miR5213 and miR2118 are predicted to target R genes [29]. The participation of miR482, miR1705 and miR2118 in the regulation of NB-LRR R transcripts was confirmed by 5′-RACE [41, 42, 70]. miR1514, which was reported to target R genes in Oncidium ([30] and ref. therein), was shown to target NAC-transcription factor NAM messages by degradome analyses [71]. miR5654 targets transcripts of a MYB transcription factor [72], but this has not been confirmed experimentally. No experimental evidence for the mechanism has been shown yet for miR5246 and miR2088. Furthermore, some miRNAs might also be involved in other responses, such as miR482, which is not stimulated by Ec, but by Pi, or miR2088 and miR5654, which do not respond significantly to both microbes (Fig. 4). Furthermore, the miR1507, miR1510a*, miR2118 and miR5246 levels were repressed by Pi in the infected leaves, but not in the neighboring leaves. A possible explanation could be that the slower progression of the Ec-induced disease development in plants with Pi-colonized roots did not yet result in the activation of the R/miRNA-based defense mechanism, because the pathogen titer in the distal leaves of the Pi-pretreated plants is too low. Finally, since miRNAs target multiple R messages, such as miR1514 (16 R mRNAs), miR1510a* (14 R mRNAs), miR5246 (13 R mRNAs) and miR5654 (12 R mRNAs), and a particular R mRNA can be targeted by different miRNA species, the results shown in the Figs. 3 and 4 provide only a basis for future investigations.
Root-to-shoot information transfer
Better protection of the leaves against Ec infection by Pi requires root-to-shoot information transfer. Induced systemic resistance (ISR) is well established [73] and Stein et al. [43] demonstrated for Pi that JA signalling and the cytoplasmic, but not nuclear localization of NPR1 is required for resistance against powdery mildew G. orontii infection. The JA-insensitive mutants jasmonate-resistant 1 (jar1–1) [74] and jasmonate-insensitive 1 (jin 1) [75] as well as the null mutant npr1–1 [nonexpressor of pathogenesis-related (PR) genes 1, also known as NIM1] [76] are compromised in Pi-mediated resistance [43]. The ISR is independent of SA and SA signalling [43, 73], since NahG plants expressing a bacterial salicylate-hydroxylase [77] and the npr1–3 mutant, lacking the nuclear-localisation signal, were not affected in Pi-mediated resistance to G. orontii [73]. Whether the protective function of Pi against Ec infection in Oncidium leaves is mediated by a JA-dependent ISR, requires studies with plant hormone mutants, which are not available at present for Oncidium. Numerous other mechanisms are also possible. Symbiosis-specific compounds from Pi might travel from the roots to the leaves, the beneficial fungus might influence the metabolomic stage, or the transport efficiency of defense relevant compounds, to mention a few.
The increase in ETH emission after Ec infection demonstrates that this phytohormone is also involved in the defense response. ETH is involved in ISR conferred by Pseudomonas fluorescens WCS417r, [78, 79], and also Nie et al. [80] demonstrated that ISR against Botrytis cinerea by Bacillus cereus AR156 is mediated through a JA/ETH- and NPR1-dependent signaling pathway in Arabidopsis. However, ISR mediated by P. fluorescens CHA0r against Peronospora parasitica is independent of the ETH receptor ETR1 and the downstream signaling component EIN2 [81–83]. It appears that also Pi-mediated resistance does not require ETH signaling ([84], and ref. therein). Altered phytohomone levels in the leaves were proposed to suppress host immunity or to prime the aerial parts for better resistance against pathogen attack (cf. [43, 44, 46] and ref. therein, [85]). Stimulation of JA and JA-responsive genes by Alternaria brassiacae infection was strongly inhibited when the plants are colonized by Pi [86, 87], and the involvement of jasmonate signalling has been well confirmed for Pi-mediated ISR responses [43, 46, 88]. A similar ISR mechanism has been shown for a non-pathogenic Rhizobium radiobacter strain which forms a symbiotic interaction with Pi [89], and the authors proposed that the beneficial activity assigned to Pi may be at least partly allocated to its symbiotic bacterium. Systemic signals also stimulate defense-related responses in distal, not Pi-colonized root areas which inhibit secondary colonisation of the roots by the fungus [90]. Kinetical studies after infection of Oncidium with Ec in the presence or absence of Pi will shine light on the role of ETH in this system.
ISR is characterized by a weak or not detectable systemic regulation of defense-related transcripts in the absence of a challenging pathogen [91, 92] and only after a pathogen attack a stronger defense response was observed [cf. 43, 73]. A similar response was described for rhizobacteria-induced ISR in Arabidopsis [93]. Since R genes which respond to Pi also respond to MeJA application, it is tempting to speculate that the information flow from the roots to the shoots is based on a JA-dependent information flow. However, this requires more detailed kinetic analyses and a better understanding of the connection between R mRNA/miRNA and phytohormone levels (cf. Discussion in [94]).
Besides the involvement of phytohormones in Pi-induced systemic resistance responses, Felle et al. [95] showed that the beneficial fungus induces fast root-surface pH signaling which primes systemic alkalization of the leaf apoplast upon powdery mildew infection. Rapid propagation of information within the plant body has also been associated with combined electric, Ca2+ and ROS waves [96], and an Arabidopsis mutant which is unable to respond to Pi and fails to confer resistance to the pathogen in systemic tissue is impaired in all three responses [16], and ref. therein).
Pi has also been reported to directly inhibit growth of pathogens, such as of Verticillium dahliae on agar plates and in colonized Arabidopsis roots [14]. Since growth of Ec was not inhibited by Pi on agar plates, the protection of colonized Oncidium orchids against Ec propagation in the leaves is likely caused by a Pi-induced root response, rather than a direct inhibition of the propagation of the pathogenic bacterium.
Ec has a wide host range, and rapidly kills host tissues mainly by type II secreted macerating isoenzymes. Virulence effector proteins secreted by the type III secretion system may be less important for the disease development [97, 98]. During Ec infection in plants, accumulation of reactive oxygen species [99, 100] and phenolic compounds [101], as well as the expression of JA-, SA-, abscisic acid- and ETH-responsive defense genes have been observed [100]. In Phaleanopsis [4], besides WRKY and MYB genes, also NBS-LRR R genes responded to Ec. To date, no monogenic resistance mechanism has been described for orchids [100]. We propose that NBS-LRR R genes are targeted and enhanced to express by the signals from Pi-colonized roots to restrict Ec growth in the leaves. In parallel, repression of their target miRNAs occurs.
Conclusion
These results indicated that Pi and MeJA promotes R gene expression in both local and distal leaves of Oncidium, while Ec and SA triggers the accumulation of their target miRNAs. It appears that Ec prevents R mRNA accumulation by stimulating the accumulation of their miRNAs in Oncidium, and Pi counteracts this effect (Fig. 5). How these regulatory processes are related to the protection of the plants against the bacterial infection, remains to be investigated.
Methods
Growth of the plant and microbes, co-cultivation and infection procedure
Oncidium (cultivar Onc. ‘Gower Ramsey’), a commercialized hybrid orchid, was originally obtained from the flower market in Fouzhou, China, the morphology and biology characteristics were identified [102]. The flower stalk buds were propagated at the Institute of Horticultural Biotechnology (Fujian Agriculture and Forestry University, Fuzhou, Fujian, China) and at the Sanming Academy of Agricultural Sciences (Sanming, Fujian, China). The plant material is commercially available at the two research institutions. The regenerated cuttings were propagated in sterile tissue culture on MS medium supplemented with benzyl adenine (2.0 mg/l), 2% sucrose and 0.6% agar at pH 5.8. The Ec bacteria were isolated from Oncidium seedling and conserved at the Sanming Academy of Agricultural Sciences (Sanming, Fujian, China), and the Pi fungal strain which was used for these studies is available from the Matthias-Schleiden-Institute, Plant Physiology (Friedrich Schiller University, Germany).
Co-cultivation of Oncidium with Pi in flasks were conducted as described previously [30]. Briefly, cuttings of about 6 cm height were transferred to fresh ½-strength Murashige-Skoog (MS) medium. After 10 day of acclimation, one agar block with Pi mycelium (or without, mock treatment) of 5 mm diameter was placed at a distance of 1 cm from the adventitious roots. The plants were cultured at a 16 h light/8 h dark photoperiod (100 μmol m− 2 s− 1) at 25 °C.
For pathogen resistance analysis, Ec was isolated and inoculated to Oncidium as described [3]. Briefly, the second leaves of Oncidium cuttings were punctured with a sterile tip, containing 2 μl of an Ec bacteria solution (OD600 = 1.0) in LB liquid medium, or LB medium alone (control). The plants were cultured on ½-strength MS medium at 25 °C in the flasks. The amounts of Pi and Ec DNA relative to the plant ACTIN DNA were detected by qPCR, primers were designed according to Pi EF-hand DNA (accession: FJ944820) and Ec 16S rDNA (assesion: KY020447) [3], respectively. Each PCR reaction was repeated three times with 3 independent biological samples. The primers for the target and reference genes are shown in Additional file 5: Table S3.
The levels of SA, JA, ETH and H2O2 in the leaves of plants co-cultivated with or without Pi were examined 24 h after Ec infection using the respective ELISA kits (ChunDu, China). Samples were extracted with phosphate buffer (100 mM, pH 7.2) in liquid nitrogen and measured with the Tecan M200 PRO plate reader (Switzerland) as described previously [103].
Root dissection
Root samples were fixed as described previously [30]. Thin sections were cut by free hand and stained with lactophenol cotton blue solution [9] or acridine orange [104]. Sections were analyzed with an Olympus BX53 microscope system (Japan), fluorescence images were excited at 485 nm and detected at 540 nm before photography.
Prediction of R genes from orchids and transcriptome analyses
The assembled transcriptome dataset from Oncidium Gower Ramsey was downloaded from the NCBI database (PRJNA428913). The D. officinale assembly and annotated genome V2.0 and the P. equestris assembly and annotated genome V5.0 were downloaded from the NCBI database (http://202.203.187.112/herbalplant; PRJNA192198) [33, 34]. R genes were predicted using HMMER v3 (http://pfam.xfam.org/) as described [105]. The NBS HMM file (PF00931) was downloaded from pfam (http://pfam.xfam.org/). R protein sequences from different orchid species were obtained using the raw NBS HMM with an E-value < 1− 40 after manual verification of the existence of NBS domains. Different orchid-specific NBS HMM files were generated, and these new orchid-specific HMM files were used to screen all proteins with E-value < 1− 2.
R genes were further analyzed based on manual verification of the existence of NBS domains. The annotation and identification of conserved domains were performed on BLAST2GO, sequence analyses were conducted with BLASTP against the NCBI nr database, and the conserved domains and the CC motif were analyzed using the InterProScan program. Additional motif analyses were conducted using MEME (meme-suite.org/tools/meme), the maximum number of motifs was set as 10, the minimum motif width as 6, the maximum motif width as 20, and the maximum sites per motif as 20.
Alignment and phylogenetic analysis
The alignment and phylogenetic analysis were conducted as described [106]. Briefly, multiple alignment of the conserved NBS domain sequences (from P-loop to MDH-like motif) of the Oncidium, Denddrobium and Phaleanopsis R proteins were performed using ClustalW, a neighbor-joint phylogenetic tree was build using MEGA6.06, and 15 well-known R protein sequences from other species were also included. Bootstrap analysis was set with 1000 replicates to assess the stability of internal nodes. R protein sequences with < 10% intactness of the NBS domain were manually removed.
Prediction of regulatory miRNAs for R genes in Oncidium
The Oncidium R gene sequences were submitted to psRNATarget (plantgrn.noble.org/psRNATarget/) and aligned to Oncidium miRNA sequences (accession: SRP031471) as described [30].
qPCR expression analysis of R genes and their regulatory miRNAs in Oncidium
The leaves (Ec-infected or mock-treated leaves, or neighboring not infected leaves) of Oncidium plants pre-treated with Pi or mock-treated were collected for RNA isolation using isopropanol and LiCl methods as described [36]. Leaves treated with 1 mM SA and 0.1 mM methyl-JA (MeJA) for 24 h were also collected for RNA analyses. cDNA was synthesized using the PrimeScript™ RT reagent Kit with gDNA Eraser (RR047A; TAKARA) for R genes and miRcut (KR201; TIANGEN) for miRNAs, respectively. Expression analyses were performed using the ABI Q3 Real-Time PCR System with the SYBR Advantage qPCR Premix kit (639,676; Clontech). The qPCR reaction was performed in a total volume of 20 μl. Each reaction was repeated three times. The primers for the target and reference genes are shown in Additional file 5: Table S3.
Supplementary information
Additional file 1: Figure S1. Oncidium R genes Blast2GO results; Multi-alignment of Oncidium R gene protein sequences. (PPTX 202 kb)
Additional file 2: Figure S2. P. indica colonization of Oncidium roots confers resistance against E. chrysanthemi. (PPTX 1961 kb)
Additional file 3: Table S1. Oncidium R genes Blast2GO results.
Additional file 4: Table S2. R gene sequences and their regulatory miRNAs.
Additional file 5: Table S3. The primer sequence information.
Acknowledgements
The entire research team in Sanming Academy of Agricultural Sciences, Fujian, China acknowledges support by the research teams from National Taiwan University, Taipei, Taiwan, as well as the Friedrich-Schiller University Jena, Germany.
Abbreviations
- Ec
Erwinia chrysanthem
- ETH
Ethylene
- ISR
Induced systemic resistance
- JA
Jasmonic acid
- MeJA
Methyl-jasmonate
- Pi
Piriformospora indica
- RT-PCR
Reverse transcription polymerase chain reaction
- SA
Salicylic acid
Authors’ contributions
Original idea was conceived by WY, all authors designed the experiments. WY and JJ performed the experiments, K-WY, ZL, XX and RO monitored the experiment and discussed the results and strategies. WY, K-WY, ZL and RO wrote and corrected manuscript and financed the project. All authors read, edited and approved the final manuscript.
Funding
National Natural Science Foundation of China (31761130073 and 31422047), Ministry of Science and Technology (KXGH17005), Taiwan supported this work. RO was supported by CRC1127 and DAAD. The funding bodies were not involved in the design of the study, analysis, and interpretation of data in the manuscript.
Availability of data and materials
All data generated or analyzed during this study will be freely available upon request to corresponding author: Wei Ye [E-mail:yewei922@qq.com] for scientific use.
Ethics approval and consent to participate
This study does not contain any research requiring ethical consent or approval.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Wei Ye, Email: yewei922@qq.com.
Jinlan Jiang, Email: 420465520@qq.com.
Yuling Lin, Email: 115635542@qq.com.
Kai-Wun Yeh, Email: ykwbppp@ntu.edu.tw.
Zhongxiong Lai, Email: laizx_01@163.com.
Xuming Xu, Email: 695299859@qq.com.
Ralf Oelmüller, Email: b7oera@uni-jena.de.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12870-019-2105-3.
References
- 1.Samson R, Legendre JB, Christen R, Fischer-Le Saux M, Achouak W, et al. Transfer of Pectobacterium chrysanthemi and Brenneria paradisiaca to the genus Dickeya gen. Nov. as Dickeya chrysanthemi comb. nov. and Dickeya paradisiaca comb. nov. and delineation of four novel species, Dickeya dadantii sp. nov., Dickeya dianthicola sp. nov., Dickeya dieffenbachiae sp. nov. and Dickeya zeae sp. nov. Int J Syst Evol Microbiol. 2005;55:1415–1427. doi: 10.1099/ijs.0.02791-0. [DOI] [PubMed] [Google Scholar]
- 2.Toth IK, Bell KS, Holeva MC, Birch PR. Soft rot erwiniae: from genes to genomes. Mol Plant Pathol. 2003;4:17–30. doi: 10.1046/j.1364-3703.2003.00149.x. [DOI] [PubMed] [Google Scholar]
- 3.Yan X, Ye W, Li Y, Jiang J, Cao Y, et al. Isolation, identification and pahtogenicity analysis of soft rot pathogen from Oncidium 'Gower Ramsey'. Subtropical Plant Sci. 2017;46:201–208. [Google Scholar]
- 4.Fu SF, Tsai TM, Chen YR, Liu CP, Haiso LJ, et al. Characterization of the early response of the orchid, Phalaenopsis amabilis, to Erwinia chrysanthemi infection using expression profiling. Physiol Plant. 2012;145:406–425. doi: 10.1111/j.1399-3054.2012.01582.x. [DOI] [PubMed] [Google Scholar]
- 5.Sherameti I, Shahollari B, Venus Y, Altschmied L, Varma A, et al. The endophytic fungus Piriformospora indica stimulates the expression of nitrate reductase and the starch-degrading enzyme glucan-water dikinase in tobacco and Arabidopsis roots through a homeodomain transcription factor that binds to a conserved motif in their promoters. J Biol Chem. 2005;280:26241–26247. doi: 10.1074/jbc.M500447200. [DOI] [PubMed] [Google Scholar]
- 6.Yadav V, Kumar M, Deep DK, Kumar H, Sharma R, et al. A phosphate transporter from the root endophytic fungus Piriformospora indica plays a role in phosphate transport to the host plant. J Biol Chem. 2010;285:26532–26544. doi: 10.1074/jbc.M110.111021. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Peśkan-Berghöfer T, Shahollari B, Giong PH, Hehl S, Markert C, et al. Association of Piriformospora indica with Arabidopsis thaliana roots represents a novel system to study beneficial plant-microbe interactions and involves early plant protein modifications in the endoplasmic reticulum and at the plasma membrane. Physiol Plant. 2004;122:465–477. doi: 10.1111/j.1399-3054.2004.00424.x. [DOI] [Google Scholar]
- 8.Shahollari B, Varma A, Oelmüller R. Expression of a receptor kinase in Arabidopsis roots is stimulated by the basidiomycete Piriformospora indica and the protein accumulates in triton X-100 insoluble plasma membrane microdomains. J Plant Physiol. 2005;162:945–958. doi: 10.1016/j.jplph.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Lee Y-C, Johnson JM, Chien C-T, Yeh K-W. Growth promotion of Chinese cabbage and Arabidopsis by Piriformospora indica is not stimulated by mycelium-synthesized Auxin. MPMI. 2011;20:421–431. doi: 10.1094/MPMI-05-10-0110. [DOI] [PubMed] [Google Scholar]
- 10.Dolatabadi HK, Goltapeh EM, Jaimand K, Rohani N, Varma A. Effects of Piriformospora indica and Sebacina vermifera on growth and yield of essential oil in fennel (Foeniculum vulgare) under greenhouse conditions. J Basic Microbiol. 2011;51:33–39. doi: 10.1002/jobm.201000214. [DOI] [PubMed] [Google Scholar]
- 11.Hua MD, Senthil Kumar R, Shyur LF, Cheng YB, Tian Z, et al. Metabolomic compounds identified in Piriformospora indica-colonized Chinese cabbage roots delineate symbiotic functions of the interaction. Sci Rep. 2017;7:9291. doi: 10.1038/s41598-017-08715-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varma A, Savita V. Sudha, Sahay N, Butehorn B, et al. Piriformospora indica, a cultivable plant-growth-promoting root endophyte. Appl Environ Microbiol. 1999;65:2741–2744. doi: 10.1128/AEM.65.6.2741-2744.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waller F, Achatz B, Baltruschat H, Fodor J, Becker K, et al. The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. Proc Natl Acad Sci U S A. 2005;102:13386–13391. doi: 10.1073/pnas.0504423102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun C, Shao Y, Vahabi K, Lu J, Bhattacharya S, Dong S, Yeh K-W, Sherameti I, Lou B, Baldwin IT, et al. The beneficial fungus Piriformospora indica protects Arabidopsis from Verticillium dahliaeinfection by downregulation plant defense responses. BMC Plant Biol. 2014;14(1):268. [DOI] [PMC free article] [PubMed]
- 15.Matsuo M, Johnson JM, Hieno A, Tokizawa M, Nomoto M, et al. High REDOX RESPONSIVE TRANSCRIPTION FACTOR1 levels result in accumulation of reactive oxygen species in Arabidopsis thaliana shoots and roots. Mol Plant. 2015;8:1253–1273. doi: 10.1016/j.molp.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 16.Johnson JM, Thürich J, Petutschnig EK, Altschmied L, Meichsner D, et al. A poly(a) ribonuclease controls the cellotriose-based interaction between Piriformospora indica and its host Arabidopsis. Plant Physiol. 2018;176:2496–2514. doi: 10.1104/pp.17.01423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou T, Wang Y, Chen JQ, Araki H, Jing Z, et al. Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol Gen Genomics. 2004;271:402–415. doi: 10.1007/s00438-004-0990-z. [DOI] [PubMed] [Google Scholar]
- 19.EAvd B, JDG J. The NB-ARC domain:a novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr Biol. 1998;8:226–228. doi: 10.1016/S0960-9822(98)70145-9. [DOI] [PubMed] [Google Scholar]
- 20.Pan Q, Wendel J, Fluhr R. Divergent evolution of plant NBS-LRR resistance gene homologues in dicot and cereal genomes. J Mol Evol. 2000;50:203–213. doi: 10.1007/s002399910023. [DOI] [PubMed] [Google Scholar]
- 21.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 22.Hajdarpasic A, Ruggenthaler P. Analysis of miRNA expression under stress in Arabidopsis thaliana. Bosn J Basic Med Sci. 2012;12:169–176. doi: 10.17305/bjbms.2012.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Zhang Q, Zhang J, Wu L, Qi Y, et al. Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol. 2010;152:2222–2231. doi: 10.1104/pp.109.151803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 25.Navarro L, Jay F, Nomura K, He SY, Voinnet O. Suppression of the microRNA pathway by bacterial effector proteins. Science. 2008;321:964–967. doi: 10.1126/science.1159505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Deng Y, Wu T, Subramanian S, Yu O. Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiol. 2010;153:1759–1770. doi: 10.1104/pp.110.156950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jue D, Sang X, Liu L, Shu B, Wang Y, et al. Identification of WRKY gene family from Dimocarpus longan and its expression analysis during flower induction and abiotic stress responses. Int J Mol Sci. 2018;19(8):E2169. doi: 10.3390/ijms19082169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang L, Mu X, Liu C, Cai J, Shi K, et al. Overexpression of potato miR482e enhanced plant sensitivity to Verticillium dahliae infection. J Integr Plant Biol. 2015;57:1078–1088. doi: 10.1111/jipb.12348. [DOI] [PubMed] [Google Scholar]
- 29.Devers EA, Branscheid A, May P, Krajinski F. Stars and symbiosis: microRNA- and microRNA*-mediated transcript cleavage involved in arbuscular mycorrhizal symbiosis. Plant Physiol. 2011;156:1990–2010. doi: 10.1104/pp.111.172627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye W, Shen C-H, Lin Y, Chen P-J, Xu X, et al. Growth promotion-related miRNAs in oncidium orchid roots colonized by the endophytic fungus Piriformospora indica. PLoS One. 2014;9:e84920. doi: 10.1371/journal.pone.0084920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, et al. Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 1999;20:317–332. doi: 10.1046/j.1365-313X.1999.t01-1-00606.x. [DOI] [PubMed] [Google Scholar]
- 32.Chini A, Grant JJ, Seki M, Shinozaki K, Loake GJ. Drought tolerance established by enhanced expression of the CC-NBS-LRR gene, ADR1, requires salicylic acid, EDS1 and ABI1. Plant J. 2004;38:810–822. doi: 10.1111/j.1365-313X.2004.02086.x. [DOI] [PubMed] [Google Scholar]
- 33.Cai J, Liu X, Vanneste K, Proost S, Tsai W-C, et al. The genome sequence of the orchid Phalaenopsis equestris. Nat Genet. 2015;47:65–72. doi: 10.1038/ng.3149. [DOI] [PubMed] [Google Scholar]
- 34.Yan L, Wang X, Liu H, Tian Y, Lian J, et al. The genome of Dendrobium officinale illuminates the biology of the important traditional Chinese orchid herb. Mol Plant. 2015;8:922–934. doi: 10.1016/j.molp.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 35.Arya P, Kumar G, Acharya V, Singh AK. Genome-wide identification and expression analysis of NBS-encoding genes in Malus x domestica and expansion of NBS genes family in Rosaceae. PLoS One. 2014;9:e107987. doi: 10.1371/journal.pone.0107987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kohler A, Rinaldi C, Duplessis S, Baucher M, Geelen D, et al. Genome-wide identification of NBS resistance genes in Populus trichocarpa. Plant Mol Biol. 2008;66:619–636. doi: 10.1007/s11103-008-9293-9. [DOI] [PubMed] [Google Scholar]
- 37.Kang YJ, Kim KH, Shim S, Yoon MY, Sun S, et al. Genome-wide mapping of NBS-LRR genes and their association with disease resistance in soybean. BMC Plant Biol. 2012;12:139. doi: 10.1186/1471-2229-12-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lozano R, Ponce O, Ramirez M, Mostajo N, Orjeda G. Genome-wide identification and mapping of NBS-encoding resistance genes in Solanum tuberosum group phureja. PLoS One. 2012;7:e34775. doi: 10.1371/journal.pone.0034775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song H, Wang PF, Li TT, Xia H, Zhao SZ, et al. Genome-wide identification and evolutionary analysis of nucleotide-binding site-encoding resistance genes in Lotus japonicus (Fabaceae) Genet Mol Res. 2015;14:16024–16040. doi: 10.4238/2015.December.7.16. [DOI] [PubMed] [Google Scholar]
- 40.Zhu Q, Fan L, Liu Y, Xu H, Llewellyn D, et al. miR482 Regulation of NBS-LRR Defense Genes during Fungal Pathogen Infection in Cotton. PLoS One. 2013;8:e84390. doi: 10.1371/journal.pone.0084390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhai J, Jeong DH, De Paoli E, Park S, Rosen BD, et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011;25:2540–2553. doi: 10.1101/gad.177527.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shivaprasad PV, Chen HM, Patel K, Bond DM, Santos BA, et al. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell. 2012;24:859–874. doi: 10.1105/tpc.111.095380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stein E, Molitor A, Kogel KH, Waller F. Systemic resistance in Arabidopsis conferred by the mycorrhizal fungus Piriformospora indica requires jasmonic acid signaling and the cytoplasmic function of NPR1. Plant Cell Physiol. 2008;49:1747–1751. doi: 10.1093/pcp/pcn147. [DOI] [PubMed] [Google Scholar]
- 44.Fu ZQ, Dong X. Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol. 2013;64:839–863. doi: 10.1146/annurev-arplant-042811-105606. [DOI] [PubMed] [Google Scholar]
- 45.Choudhary DK, Johri BN. Interactions of Bacillus spp. and plants -with special reference to induced systemic resistance (ISR) Microbiol Res. 2009;164:493–513. doi: 10.1016/j.micres.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 46.Waller F, Mukherjee K, Deshmukh SD, Achatz B, Sharma M, et al. Systemic and local modulation of plant responses by Piriformospora indica and related Sebacinales species. J Plant Physiol. 2008;165:60–70. doi: 10.1016/j.jplph.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 47.Schäfer P, Khatabi B, Kogel KH. Root cell death and systemic effects of Piriformospora indica: a study on mutualism. FEMS Microbiol Lett. 2007;275:1–7. doi: 10.1111/j.1574-6968.2007.00848.x. [DOI] [PubMed] [Google Scholar]
- 48.Schäfer P, Pfiffi S, Voll LM, Zajic D, Chandler PM, et al. Manipulation of plant innate immunity and gibberellin as factor of compatibility in the mutualistic association of barley roots with Piriformospora indica. Plant J. 2009;59:461–474. doi: 10.1111/j.1365-313X.2009.03887.x. [DOI] [PubMed] [Google Scholar]
- 49.Yang S, Perna NT, Cooksey DA, Okinaka Y, Lindow SE, Ibekwe AM, Keen NT, Yang C-H. Genome-wide identification of plant-Upregulated genes of Erwinia chrysanthemi 3937 using a GFP-based IVET leaf Array. Mol Plant-Microb Interact. 2004;17:999–1008. doi: 10.1094/MPMI.2004.17.9.999. [DOI] [PubMed] [Google Scholar]
- 50.Expert D. Withholding and exchanging iron: interactions between Erwinia spp. and their plant hosts. Annu. Rev. Phytopathol. 1999;37:307–334. doi: 10.1146/annurev.phyto.37.1.307. [DOI] [PubMed] [Google Scholar]
- 51.Franza T, Expert D. The virulence-associated chrysobactin iron uptake system of Erwinia chrysanthemi 3937 involves an operon encoding transport and biosynthetic functions. J Bacteriol. 1991;173:6874–6881. doi: 10.1128/jb.173.21.6874-6881.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Franza T, Sauvage C, Expert D. Iron regulation and pathogenicity in Erwinia chrysanthemi 3937: role of the Fur repressor protein. Mol Plant-Microbe Interact. 1999;12:119–128. doi: 10.1094/MPMI.1999.12.2.119. [DOI] [PubMed] [Google Scholar]
- 53.Hassouni ME, Chambost JP, Expert D, Van Gijsegem F, Barras F. The minimal gene set member msrA, encoding peptide methionine sulfoxide reductase, is a virulence determinant of the plant pathogen Erwinia chrysanthemi. Proc Natl Acad Sci U S A. 1999;96:887–892. doi: 10.1073/pnas.96.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.López-Solanilla E, Llama-Palacios A, Collmer A, García-Olmedo F, Rodríguez-Palenzuela P. Relative effects on virulence of mutations in the sap, pel, and hrp loci of Erwinia chrysanthemi. Mol Plant-Microbe Interact. 2001;14:386–393. doi: 10.1094/MPMI.2001.14.3.386. [DOI] [PubMed] [Google Scholar]
- 55.Tardy F, Nasser W, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. Comparative analysis of the five major Erwinia chrysanthemi pectate lyases: enzyme characteristics and potential inhibitors. J Bacteriol. 1997;179:2503–2511. doi: 10.1128/jb.179.8.2503-2511.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chatterjee AK, Dumenyo CK, Liu Y, Chatterjee A. Erwinia: Genetics of pathogenicity factors. In: Lederberg J, editor. Encyclopedia of Microbiology. 2. New York: Academic Press; 2000. pp. 236–259. [Google Scholar]
- 57.Collmer A. Keen NT the role of pectic enzymes in plant pathogenesis. Annu Rev Phytopathol. 1986;24:383–409. doi: 10.1146/annurev.py.24.090186.002123. [DOI] [Google Scholar]
- 58.Hugouvieux-Cotte-Pattat N, Condemine G, Nasser W, Reverchon S. Regulation of pectinolysis in Erwinia. Annu Rev Microbiol. 1996;50:213–257. doi: 10.1146/annurev.micro.50.1.213. [DOI] [PubMed] [Google Scholar]
- 59.Perombelon MCM, Kelman A. Ecology of the soft rot erwinias. Annu. Rev. Phytopathol. 1980;18:361–87.49. doi: 10.1146/annurev.py.18.090180.002045. [DOI] [Google Scholar]
- 60.Croston TL, Lemons AR, Beezhold DH, Green BJ. MicroRNA regulation of host immune responses following fungal exposure. Front Immunol. 2018;9:170. doi: 10.3389/fimmu.2018.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Islas-Flores T, Rahman A, Ullah H, Villanueva MA. The receptor for activated C kinase in plant signaling: tale of a promiscuous little molecule. Front Plant Sci. 2015;6:1090. doi: 10.3389/fpls.2015.01090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Claycomb JM. Ancient endo-siRNA pathways reveal new tricks. Curr Biol. 2014;24:R703–R715. doi: 10.1016/j.cub.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 63.Seo JK, Wu J, Lii Y, Li Y, Jin H. Contribution of small RNA pathway components in plant immunity. Mol Plant-Microbe Interact. 2013;26:617–625. doi: 10.1094/MPMI-10-12-0255-IA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Staiger D, Korneli C, Lummer M, Navarro L. Emerging role for RNA-based regulation in plant immunity. New Phytol. 2013;197:394–404. doi: 10.1111/nph.12022. [DOI] [PubMed] [Google Scholar]
- 65.Naqvi AR, Sarwat M, Hasan S, Roychodhury N. Biogenesis, functions and fate of plant microRNAs. J Cell Physiol. 2012;227:3163–3168. doi: 10.1002/jcp.24052. [DOI] [PubMed] [Google Scholar]
- 66.Hohn T, Vazquez F. RNA silencing pathways of plants: silencing and its suppression by plant DNA viruses. Biochim Biophys Acta. 1809;2011:588–600. doi: 10.1016/j.bbagrm.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 67.Li Z, Rana TM. Molecular mechanisms of RNA-triggered gene silencing machineries. Acc Chem Res. 2012;45:1122–1131. doi: 10.1021/ar200253u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu S, Sun YH, Amerson H, Chiang VL. MicroRNAs in loblolly pine (Pinus taeda L.) and their association with fusiform rust gall development. Plant J. 2007;51:1077–1098. doi: 10.1111/j.1365-313X.2007.03208.x. [DOI] [PubMed] [Google Scholar]
- 69.Bazin J, Bustos-Sanmamed P, Hartmann C, Lelandais-Briere C, Crespi M. Complexity of miRNA-dependent regulation in root symbiosis. Philos Trans R Soc B Biol Sci. 2012;367:1570–1579. doi: 10.1098/rstb.2011.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li F, Pignatta D, Bendix C, Brunkard JO, Cohn MM, Tung J, Sun H, Kumar P, Baker B. MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci U S A. 2012;109(5):1790–1795. doi: 10.1073/pnas.1118282109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu Z, Jiang Q, Ni Z, Chen R, Xu S, Zhang H. Analyses of a Glycine max degradome library identify microRNA targets and microRNAs that trigger secondary siRNA biogenesis. J Integr Plant Biol. 2013;55(2):160–176. doi: 10.1111/jipb.12002. [DOI] [PubMed] [Google Scholar]
- 72.Shen D, Suhrkamp I, Wang Y, Liu S, Menkhaus J, Verreet JA, Fan L, Cai D. Identification and characterization of microRNAs in oilseed rape (Brassica napus) responsive to infection with the pathogenic fungus Verticillium longisporum using Brassica AA (Brassica rapa) and CC (Brassica oleracea) as reference genomes. New Phytol. 2014;204(3):577–594. doi: 10.1111/nph.12934. [DOI] [PubMed] [Google Scholar]
- 73.Molitor A, Kogel KH. Induced resistance triggered by Piriformospora indica. Plant Signal Behav. 2009;4:215–216. doi: 10.4161/psb.4.3.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Staswick PE, Tiryaki I. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell. 2004;16:2117–2127. doi: 10.1105/tpc.104.023549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell. 2004;16:1938–1950. doi: 10.1105/tpc.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cao H, Bowling SA, Gordon AS, Dong X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell. 1994;6:1583–1592. doi: 10.2307/3869945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lawton K, Weymann K, Friedrich L, Vernooij B, Uknes S, Ryals J. Systemic acquired resistance in Arabidopsis requires salicylic acid but not ethylene. Mol Plant-Microbe Interact. 1995;8:863–870. doi: 10.1094/MPMI-8-0863. [DOI] [PubMed] [Google Scholar]
- 78.Pieterse CM, Van Wees SC, Van Pelt JA, Knoester M, Laan R, Gerrits H, et al. A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell. 1998;10:1571–1580. doi: 10.1105/tpc.10.9.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Knoester M, Pieterse CMJ, Bol JF, Van Loon LC. Systemic resistance in Arabidopsis induced by rhizobacteria requires ethylene-dependent signaling at the site of application. Mol Plant-Microbe Interact. 1999;12:720–727. doi: 10.1094/MPMI.1999.12.8.720. [DOI] [PubMed] [Google Scholar]
- 80.Nie P, Li X, Wang S, Guo J, Zhao H, et al. Induced systemic resistance against Botrytis cinerea by Bacillus cereus AR156 through a JA/ET- and NPR1-dependent signaling pathway activates PAMP-triggered immunity in Arabidopsis. Front Plant Sci. 2017;8:238. doi: 10.3389/fpls.2017.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Iavicoli A, Boutet E, Buchala A, Metraux JP. Induced systemic resistance in Arabidopsis thaliana in response to root inoculation with Pseudomonas fluorescens CHA0. Mol Plant-Microbe Interact. 2003;16:851–858. doi: 10.1094/MPMI.2003.16.10.851. [DOI] [PubMed] [Google Scholar]
- 82.Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- 83.Bleecker AB, Estelle MA, Somerville C, Kende H. Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science. 1988;241:1086–1089. doi: 10.1126/science.241.4869.1086. [DOI] [PubMed] [Google Scholar]
- 84.Jacobs S, Zechmann B, Molitor A, Trujillo M, Petutschnig E, et al. Broad-spectrum suppression of innate immunity is required for colonization of Arabidopsis roots by the fungus Piriformospora indica. Plant Physiol. 2011;156:726–740. doi: 10.1104/pp.111.176446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Molitor A, Zajic D, Voll LM, Pons-K Hnemann J, Samans B, Kogel KH, Waller F. Barley leaf transcriptome and metabolite analysis reveals new aspects of compatibility and Piriformospora indica-mediated systemic induced resistance to powdery mildew. Mol Plant-Microbe Interact. 2011;24:1427–1439. doi: 10.1094/MPMI-06-11-0177. [DOI] [PubMed] [Google Scholar]
- 86.Vahabi K, Camehl I, Sherameti I, Oelmüller R. Growth of Arabidopsis seedlings on high fungal doses of Piriformospora indica has little effect on plant performance, stress, and defense gene expression in spite of elevated jasmonic acid and jasmonic acid-isoleucine levels in the roots. Plant Signal Behav. 2013;8:e26301. doi: 10.4161/psb.26301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vahabi K, Dorcheh SK, Monajembashi S, Westermann M, Reichelt M, et al. Stress promotes Arabidopsis - Piriformospora indica interaction. Plant Signal Behav. 2016;11:e1136763. doi: 10.1080/15592324.2015.1136763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xu L, Wu C, Oelmüller R, Zhang W. Role of Phytohormones in Piriformospora indica-induced growth promotion and stress tolerance in plants: more questions than answers. Front Microbiol. 2018;9:1646. doi: 10.3389/fmicb.2018.01646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Glaeser SP, Imani J, Alabid I, Guo H, Kumar N, Kämpfer P, Hardt M, Blom J, Goesmann A, Rothballer M, Hartmann A, Kogel KH. Non-pathogenic rhizobium radiobacter F4 deploys plant beneficial activity independent of its host Piriformospora indica. ISME J. 2016;10:871–884. doi: 10.1038/ismej.2015.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pedrotti L, Mueller MJ, Waller F. Piriformospora indica root colonization triggers local and systemic root responses and inhibits secondary colonization of distal roots. PLoS One. 2013;8:e69352. doi: 10.1371/journal.pone.0069352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang YQ, Ohara Y, Nakayashiki H, Tosa Y, Mayama S. Microarray analysis of the gene expression profile induced by the endophytic plant growth-promoting rhizobacteria Pseudomonas fluorescens FPT9601-T5 in Arbidopsis. Mol Plant-Microbe Interact. 2005;18:385–396. doi: 10.1094/MPMI-18-0385. [DOI] [PubMed] [Google Scholar]
- 92.Verhagen BWM, Glazebrook J, Zhu T, Chang HS, Van Loon LC, Pieterse CMJ. The transcriptome of rhizobacteria induced systemic resistance in Arabidopsis. Mol Plant-Microbe Interact. 2004;17:895–908. doi: 10.1094/MPMI.2004.17.8.895. [DOI] [PubMed] [Google Scholar]
- 93.Van Wees SCM, Luijendijk M, Smoorenburg I, Van Loon LC, Pieterse CMJ. Rhizobacteria-mediated induced systemic resistance (ISR) in Arabidopsis is not associated with a direct effect of expression of known defense-related genes but stimulates the expression of the jasmonate-inducible gene Atvsp upon challenge. Plant Mol Biol. 1999;41:537–549. doi: 10.1023/A:1006319216982. [DOI] [PubMed] [Google Scholar]
- 94.Lucas SJ, Baştaş K, Budak H. Exploring the interaction between small RNAs and R genes during Brachypodium response to Fusarium culmorum infection. Gene. 2014;536:254–264. doi: 10.1016/j.gene.2013.12.025. [DOI] [PubMed] [Google Scholar]
- 95.Felle HH, Waller F, Molitor A, Kogel KH. The mycorrhiza fungus Piriformospora indica induces fast root-surface pH signaling and primes systemic alkalinization of the leaf apoplast upon powdery mildew infection. Mol Plant-Microbe Interact. 2009;22:1179–1185. doi: 10.1094/MPMI-22-9-1179. [DOI] [PubMed] [Google Scholar]
- 96.Gilroy S, Białasek M, Suzuki N, Górecka M, Devireddy AR, Karpiński S, Mittler R. ROS, calcium, and electric signals: key mediators of rapid systemic signaling in plants. Plant Physiol. 2016;171:1606–1615. doi: 10.1104/pp.16.00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bauer DW, Bogdanove AJ, Beer SV, Collmer A. Erwinia chrysanthemi hrp genes and their involvement in soft rot pathogenesis and elicitation of the hypersensitive response. Mol Plant-Microbe Interact. 1994;7:573–581. doi: 10.1094/MPMI-7-0573. [DOI] [PubMed] [Google Scholar]
- 98.Rojas CM, Ham JH, Schechter LM, Kim JF, Beer SV, et al. The Erwinia chrysanthemi EC16 hrp/hrc gene cluster encodes an active Hrp type III secretion system that is flanked by virulence genes functionally unrelated to the Hrp system. Mol Plant-Microbe Interact. 2004;17:644–653. doi: 10.1094/MPMI.2004.17.6.644. [DOI] [PubMed] [Google Scholar]
- 99.Fagard M, Dellagi A, Roux C, Perino C, Rigault M, et al. Arabidopsis thaliana expresses multiple lines of defense to counterattack Erwinia chrysanthemi. Mol Plant-Microbe Interact. 2007;20:794–805. doi: 10.1094/MPMI-20-7-0794. [DOI] [PubMed] [Google Scholar]
- 100.Kraepiel Y, Pedron J, Patrit O, Simond-Cote E, Hermand V, et al. Analysis of the plant bos1 mutant highlights necrosis as an efficient defence mechanism during D. dadantii/Arabidospis thaliana interaction. PLoS One. 2011;6:e18991. doi: 10.1371/journal.pone.0018991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Perez-Bueno ML, Granum E, Pineda M, Flors V, Rodriguez-Palenzuela P, et al. Temporal and spatial resolution of activated plant defense responses in leaves of Nicotiana benthamiana infected with Dickeya dadantii. Front Plant Sci. 2015;6:1209. doi: 10.3389/fpls.2015.01209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li Y, Ye W, Jiang J. Observation on biological characteristics of 24 germplasm resources of Oncidium. Fujian Agric Sci Technol. 2013;Z1:99–101. [Google Scholar]
- 103.Chen SP, Lin IW, Chen X, Huang YH, Chang SC, et al. Sweet potato NAC transcription factor, IbNAC1, upregulates sporamin gene expression by binding the SWRE motif against mechanical wounding and herbivore attack. Plant J. 2016;86:234–248. doi: 10.1111/tpj.13171. [DOI] [PubMed] [Google Scholar]
- 104.Senthilkumar S, Krishnamurthy KV, Britto SJ, Arockiasamy DI. Visualization of orchid mycorrhizal fungal structures with fluorescence dye using epifluorescence microscopy. Curr Sci. 2000;79:1527–1528. [Google Scholar]
- 105.Lozano R, Hamblin MT, Prochnik S, Jannink JL. Identification and distribution of the NBS-LRR gene family in the cassava genome. BMC Genomics. 2015;16:360. doi: 10.1186/s12864-015-1554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.An FM, Hsiao SR, Chan MT. Sequencing-based approaches reveal low ambient temperature-responsive and tissue-specific microRNAs in Phalaenopsis orchid. PLoS One. 2011;6:e18937. doi: 10.1371/journal.pone.0018937. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Oncidium R genes Blast2GO results; Multi-alignment of Oncidium R gene protein sequences. (PPTX 202 kb)
Additional file 2: Figure S2. P. indica colonization of Oncidium roots confers resistance against E. chrysanthemi. (PPTX 1961 kb)
Additional file 3: Table S1. Oncidium R genes Blast2GO results.
Additional file 4: Table S2. R gene sequences and their regulatory miRNAs.
Additional file 5: Table S3. The primer sequence information.
Data Availability Statement
All data generated or analyzed during this study will be freely available upon request to corresponding author: Wei Ye [E-mail:yewei922@qq.com] for scientific use.