Abstract
Background
Breast cancer risk, development, and treatment are influenced by genetic variation in certain genes, namely those involved in cell proliferation, tumor suppression, and drug metabolism. In turn, the relevance of the aforementioned genetic variation to cancer depends on the ethnic group in question, highlighting the need for population-specific association studies. Therefore, the objective of the present study was to investigate the association between certain ESR1, ESR2, HER2, UGT1A4, and UGT2B7 single nucleotide polymorphisms and breast cancer.
Methods
Blood samples were collected from 437 Jordanian-Arab breast cancer patients and healthy volunteers and subject to genotyping using the Sequenom MassARRAY® system (iPLEX GOLD).
Results
Our findings show a significant association between breast cancer and the allelic (P = 0.02486879) and genotypic (P = 0.04793066) frequencies of the ESR1 polymorphism rs3798577, a result which was confirmed in different genetic models. No other investigated polymorphism showed a significant association with breast cancer itself in Jordanian Arabs, but the Rare Hz (GG) vs Het (AG) genetic model revealed an association of the disease with the ESR1 polymorphism rs3798577. However, several associations were found between certain polymorphisms and breast cancer’s prognostic factors.
Conclusion
This study suggests that certain polymorphisms may increase the risk of breast cancer in the Jordanian-Arab population. Future research and clinical translation could incorporate the current results in preventative breast cancer approaches tailored for Jordanian-Arab patients.
Keywords: Breast cancer, Jordanian, ESR, HER2, UGT1A4, UGT2B7
Background
Breast cancer (BC) is a complex disease that arises due to a combination of environmental and genetic factors [1]. Current approaches to understanding BC etiology focus on the identification of molecular markers that could aid in the prediction and prognosis of the disease [2, 3]. Mutations in the BRCA1 and BRCA2 genes have been well-established as risk factors for BC development, and they are responsible for approximately 90% of the disease’s genetic component [4, 5]. Moreover, certain genetic polymorphisms have been found to modulate the effects of BC chemotherapy, including the selective estrogen receptor modulator tamoxifen, which is prescribed for several BC types. Consequently, polymorphisms in genes implicated in BC pathogenesis, such as those involved in tamoxifen pharmacogenetics, such as the UGT1A4 and UGT2B7 genes, are frequent targets of BC research [6, 7].
Excessive endogenous and exogenous estrogen may cause pathological changes in many cancers cell line [8]. estrogen is a key regulator for mammary gland growth and differentiation it is also important in breast carcinoma development and progression [9]. The estrogen receptor 1 (ESR1) and estrogen receptor 2 (ESR2) genes encode for estrogen receptors alpha (ER-α) and beta (ER-β), respectively, which are activated by estrogen and interact with one another in a dimeric manner [10]. In terms of function, however, ER-α and ER-β appear to have antagonistic functions in breast tissue: ER-α stimulates cell proliferation while ER-β possesses anti-proliferative and tumor-suppressive activity [10, 11]. Thus, genetic variants in genes that encode estrogen receptors such as ESR on chromosome 6, could expose a potential risk for breast cancer. Several studies reported that about 55% of ER-positive metastatic BC patients were screened with ESR1 mutations [12–15].
The HER2 gene is a Receptor-type tyrosine kinases (RTK) which is a member of epidermal growth factor receptor (EGFR) family that encodes a 185-kDa transmembrane glycoprotein on chromosome 17 [16]. RTK are polymorphic genes that play important role in the regulation of cellular processes [17]. In addition, HER2 gene involves in human cancers including ovarian [18], bladder [19], lung [20] and stomach [21] cacinomas. In particular, HER2 overexpressed approximately in 30% of BC cases [16]. It also have been reported that overexpression of HER2 in BC substantially decrease overall survival rates and the metastatic of BC [22, 23].
Lastly, the UDP glucuronosyltransferase 1A4 (UGT1A4) and UDP glucuronosyltransferase 2B7 (UGT2B7) genes are involved in the elimination of xenobiotics such as tamoxifen, the latter of which loses its anti-estrogenic effects after being glucuronidated by UGT1A4 and UGT2B7 [24].
In fact, ESR1 polymorphisms have been found to be associated with BC susceptibility, although conflicting findings have been presented on whether such polymorphisms increase or decrease the risk of the disease [11]. Similar inconsistent reports have been found for the association between ESR2 polymorphisms and BC risk [12, 25]. However, due to the carcinogenic effects of HER2 amplification or overexpression, polymorphisms in the HER2 gene have been definitively linked with modulated BC risk [26, 27]. Likewise, polymorphisms in the UGT1A4 and UGT2B7 genes that lead to their overexpression could lead to rapid tamoxifen metabolism and lower therapeutic effect [28]. Due to the influence of interethnic genetic variation, it would not be accurate to simply extrapolate previously reported results in one population onto another, especially since cancer-related polymorphisms have been reported to have different roles in BC susceptibility and development in different populations [29]. Consequently, the aim of this study is to investigate the association of certain ESR1, ESR2, HER2, UGT1A4, and UGT2B7 single nucleotide polymorphisms (SNPs) with BC susceptibility in the Jordanian-Arab population.
Methods
Study subjects and design
Jordanian-Arab BC patients (n = 218) and healthy volunteers with patient-matched characteristics (n = 219) were enlisted from the Jordanian Royal Medical Services (JRMS) hospital. Participation in the current study entailed the withdrawal of 5 ml of blood from each subject as well as the collection of clinical, demographic, and pathologic data from patient medical records. Written informed consent was obtained from all study subjects, and ethical approval to carry out this study was obtained from Jordan University of Science and Technology’s Institutional Review Board (IRB) with an ethical approval number 14/78/2014.
DNA extraction and genotyping
Genomic DNA was extracted from each blood sample using the Wizard® Genomic DNA Purification Kit (Promega Corporation, USA) according to the manufacturer’s instructions. The quality and quantity of the purified DNA was ascertained via agarose gel electrophoresis and the Nano-Drop ND-1000 UV-Vis Spectrophotometer (BioDrop, UK), respectively. DNA samples were then diluted with nuclease-free water in order to achieve a final concentration of 20 ng/μl and a final volume ranging between 50 and 500 μl. Afterwards, samples were shipped on ice to Melbourne node of the Australian Genome Research Facility (AGRF) for custom genotyping on the Sequenom MassARRAY® system (iPLEX GOLD) (Sequenom, USA).
Data analysis
Both the Hardy-Weinberg equilibrium (p2 + 2pq + q2 = 1) (http://www.oege.org/software/hwe-mr-calc.html) and the χ2 test were employed to assess the genotypic and allelic frequencies [30]. The genetic association, different genetic models and phenotype-genotype analyses were conducted using the Statistical Package for the Social Sciences (SPSS), version 25.0 (SPSS, Inc., Chicago, IL). For the present study, statistical significance was set at p-value < 0.05.
Correction for multiple testing
According to Li and Ji (2005) a method was used to estimate the effective number of SNPs (Nem) that employs a modification of an earlier approach by Nyholt (2004) [31, 32]. Modified Bonferroni procedure was applied to determine a target alpha level (0.05/ Nem) that would maintain an overall significance level of 0.05 or less.
Results
Candidate SNPs and their minor allelic frequencies
Table 1 lists the ESR1, ESR2, HER2, UGT1A4, and UGT2B7 SNPs investigated by the current study, in addition to the minor alleles of the variants and their frequencies. Genetic variants were selected based on their clinical and pathological significant in addition they were chosen from published polymorphisms associated with BC.
Table 1.
Minor allele frequencies of gene polymorphisms in breast cancer patients and healthy controls
| Gene | SNP ID | Cases (n = 218) | Controls (n = 219) | ||||
|---|---|---|---|---|---|---|---|
| MAa | MAFb | HWEc p-value |
MAa | MAFb | HWEc p-value |
||
| ESR1 | rs3020410 | A | 0.1 | 0.44 | A | 0.08 | 0.63 |
| rs3798577 | C | 0.41 | 0.33 | C | 0.48 | 0.68 | |
| rs2234693 | T | 0.49 | 0.34 | T | 0.49 | 0.03 | |
| rs9340799 | G | 0.47 | 0.5 | G | 0.46 | 0.02 | |
| ESR2 | rs1256049 | T | 0.02 | 1 | T | 0.02 | 1 |
| HER2 | rs1058808 | C | 0.32 | 0.76 | C | 0.32 | 0.21 |
| UGT1A4 | rs12468274 | C | 0.08 | 0.37 | C | 0.07 | 0.61 |
| rs2011425 | G | 0.09 | 0.23 | G | 0.09 | 0.38 | |
| rs6755571 | A | 0.06 | 0.54 | A | 0.05 | 0.11 | |
| UGT2B7 | rs28365062 | G | 0.16 | 0.2 | G | 0.17 | 0.47 |
| rs4348159 | T | 0.16 | 0.13 | T | 0.17 | 0.13 | |
aMA: minor allele. bMAF: minor allele frequency. cHWE: Hardy—Weinberg equilibrium. N/A not applicable
Association between BC and ESR1, ESR2, HER2, UGT1A4, and UGT2B7 SNPs
Table 2 summarizes the findings of the present study with regard to genetic association with BC. A correlation was found between BC and the allelic (P = 0.02) and genotypic (P = 0.04) frequencies of the ESR1 polymorphism rs3798577. Regarding this, the distribution of the variant allele of the aforementioned SNP (C) within cases were slightly higher than it among control 48 and 41% respectively. Suggesting that the C allele of ESR1 gene variant ‘rs3798577’ may be considered as BC risk factor.
Table 2.
Association of the investigated ESR1, ESR2, HER2, UGT1A4, and UGT2B7 SNPs and breast cancer (BC)
| Gene | SNP ID | Allelic and Genotypic Frequencies in Cases and Controls | ||||
|---|---|---|---|---|---|---|
| Allele/Genotype | Cases (n = 218) |
Controls (n = 219) |
P-value | Chi-square | ||
| ESR1 | rs2234693 | C | 222(0.51) | 221(0.51) | 0.943 | 0.005 |
| T | 216(0.49) | 213(0.49) | ||||
| CC | 60 (27.4) | 48 (22.1) | 0.069 | 5.328 | ||
| TC | 102 (46.6) | 125 (57.6) | ||||
| TT | 57 (26) | 44 (20.3) | ||||
| rs9340799 | A | 231(0.53) | 234(0.54) | 0.782 | 0.076 | |
| G | 205(0.47) | 200 (0.46) | ||||
| AA | 64 (29.4) | 54 (24.9) | 0.067 | 5.383 | ||
| AG | 103 (47.2) | 126 (58.1) | ||||
| GG | 51 (23.4) | 37 (17.1) | ||||
| rs3020410 | C | 399(0.9) | 399(0.92) | 0.387 | 0.748 | |
| A | 43(0.1) | 35(0.08) | ||||
| CC | 181 (81.9) | 184 (84.8) | 0.698 | 0.718 | ||
| CA | 37 (16.7) | 31 (14.3) | ||||
| AA | 3 (1.4) | 2 (0.9) | ||||
| rs3798577 | T | 258(0.59) | 224(0.52) | 0.024 | 5.033 | |
| C | 178(0.41) | 210(0.48) | ||||
| TT | 80 (36.7) | 56 (25.8) | 0.047 | 6.076 | ||
| TC | 98 (45) | 112 (51.6) | ||||
| CC | 40 (18.4) | 49 (22.6) | ||||
| ESR2 | rs1256049 | C | 434(0.98) | 425(0.98) | 0.777 | 0.08 |
| T | 8(0.02) | 9(0.02) | ||||
| CC | 213 (96.4) | 208 (95.8) | 0.774 | 0.082 | ||
| CT | 8 (3.6) | 9 (4.2) | ||||
| HER2 | rs1058808 | G | 300(0.68) | 296(0.68) | N/A | N/A |
| C | 140(0.32) | 138(0.32) | ||||
| GG | 101 (45.9) | 105 (48.4) | 0.503 | 1.372 | ||
| GC | 98 (44.5) | 86 (39.6) | ||||
| CC | 21 (9.6) | 26 (12) | ||||
| UGT1A4 | rs12468274 | T | 400(0.92) | 402(0.93) | 0.627 | 0.236 |
| C | 36 (0.08) | 32(0.07) | ||||
| TT | 182 (83.5) | 185 (85.2) | 0.611 | 0.258 | ||
| CT | 36 (16.5) | 32 (14.8) | ||||
| rs2011425 | T | 399(0.91) | 392(0.91) | 0.974 | 0.001 | |
| G | 39(0.09) | 38 (0.09) | ||||
| TT | 180 (82.2) | 177 (82.3) | 0.974 | 0.001 | ||
| TG | 39 (17.8) | 38 (17.7) | ||||
| rs6755571 | C | 416(0.94) | 413(0.95) | 0.694 | 0.154 | |
| A | 26(0.06) | 23(0.05) | ||||
| CC | 196 (88.7) | 197 (90.4) | 0.638 | 0.897 | ||
| CA | 24 (10.9) | 19 (8.7) | ||||
| AA | 1 (0.4) | 2 (0.9) | ||||
| UGT2B7 | rs28365062 | A | 371(0.84) | 362(0.83) | 0.605 | 0.267 |
| G | 69 (0.16) | 74(0.17) | ||||
| AA | 159 (72.3) | 152 (69.7) | 0.829 | 0.374 | ||
| GA | 53 (24.1) | 58 (26.6) | ||||
| GG | 8 (3.6) | 8 (3.7) | ||||
| rs4348159 | C | 369(0.84) | 361(0.83) | 0.785 | 0.074 | |
| T | 71(0.16) | 73(0.17) | ||||
| CC | 158 (71.8%) | 152 (70) | 0.860 | 0.3 | ||
| TC | 53 (24.1%) | 57 (26.3) | ||||
| TT | 9 (4.1%) | 8 (3.7) | ||||
P-Value < 0.05 was considered as significant
Fig. 1 illustrates the scatter pattern of genotypic distribution for the rs3798577 polymorphism. However, the other investigated ESR1 and ESR2 SNPs did not show any significant relationship with BC. Incorporating different genetic models into the association analysis revealed a significant association between BC and the ESR1 polymorphism rs9340799 for the Rare Hz (GG) vs Het (AG) genetic model (χ2 = 4.29). Moreover, a correlation was found between BC and the ESR1 polymorphism rs3798577 for both the Het (CT) vs Common Hz (TT) (χ2 = 4.88) and the Rare Hz (CC) vs Common Hz (TT) (χ2 = 4.16) genetic models (Table 3). On the other hand, no significant association was found between the investigated HER2, UGT1A4, and UGT2B7 polymorphisms and BC in the Jordanian-Arab population sample (Tables 2 and 3).
Fig. 1.
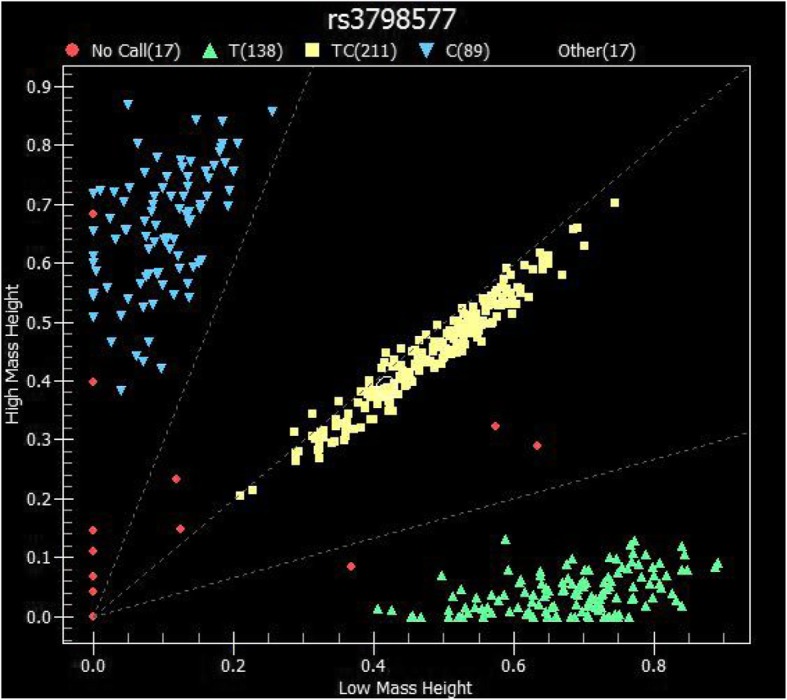
Scatter plot representing Sequenom data for the rs3798577 SNP of the ESR1 gene. Each dot refers to a single sample, and each color indicates a different genotype
Table 3.
Genetic association analysis for the ESR1, HER2, UGT1A4, and UGT2B7 SNPs using different genetic models
| Gene | SNP ID | Category Test | Odds Ratio | 95% CI | Chi square* |
|---|---|---|---|---|---|
| ESR1 | rs2234693 | Het (GT) vs Common Hz (GG) | 0.65 | 0.41–1.04 | 3.31 |
| Rare Hz (TT) vs Het (GT) | 1.59 | 0.99–2.55 | 3.7 | ||
| Rare Hz (TT) vs Common Hz (GG) | 1.04 | 0.6–1.79 | 0.02 | ||
| rs9340799 | Het (AG) vs Common Hz (AA) | 0.69 | 0.44–1.08 | 2.67 | |
| Rare Hz (GG) vs Het (AG) | 1.69 | 1.03–2.77 | 4.29 | ||
| Rare Hz (GG) vs Common Hz (AA) | 1.16 | 0.67–2.03 | 0.28 | ||
| rs3020410 | Het (CT) vs Common Hz (CC) | 1.21 | 0.72–2.04 | 0.53 | |
| Rare Hz (TT) vs Het (AG) | 1.26 | 0.2–8.01 | 0.06 | ||
| Rare Hz (TT) vs Common Hz (CC) | 1.52 | 0.25–9.23 | 0.21 | ||
| rs3798577 | Het (GT) vs Common Hz (GG) | 0.61 | 0.4–0.95 | 4.88 | |
| Rare Hz (TT) vs Het (GT) | 0.93 | 0.57–1.53 | 0.07 | ||
| Rare Hz (TT) vs Common Hz (GG) | 0.57 | 0.33–0.98 | 4.16 | ||
| HER2 | rs1058808 | Het (GA) vs Common Hz (GG) | 1.18 | 0.8–1.76 | 0.7 |
| Rare Hz (AA) vs Het (GA) | 0.71 | 0.37–1.35 | 1.1 | ||
| Rare Hz (AA) vs Common Hz (GG) | 0.84 | 0.44–1.59 | 0.29 | ||
| UGT1A4 | rs6755571 | Het (GA) vs Common Hz (AA) | 1.27 | 0.67–2.39 | 0.55 |
| Rare Hz (GG) vs Het (GA) | 0.4 | 0.03–4.7 | 0.57 | ||
| Rare Hz (GG) vs Common Hz (AA) | 0.5 | 0.05–5.59 | 0.33 | ||
| UGT2B7 | rs28365062 | Het (CT) vs Common Hz (CC) | 0.87 | 0.57–1.35 | 0.37 |
| Rare Hz (TT) vs Het (CT) | 1.09 | 0.38–3.12 | 0.03 | ||
| Rare Hz (TT) vs Common Hz (CC) | 0.96 | 0.35–2.61 | 0.01 | ||
| rs4348159 | Het (CT) vs Common Hz (CC) | 0.89 | 0.58–1.38 | 0.25 | |
| Rare Hz (TT) vs Het (CT) | 1.21 | 0.43–3.37 | 0.13 | ||
| Rare Hz (TT) vs Common Hz (CC) | 1.08 | 0.41–2.88 | 0.03 |
* For significant association χ2 should be > 3.84 with P < 0.025
CI indicates confidence interval
Association of the Clinical and Pathological Factors of BC with ESR1, ESR2, HER2, UGT1A4, and UGT2B7 SNPs
In the present study, a group of known clinical and pathological BC factors were investigated for their association with the ESR1 and ESR2 SNPs (Table 4). The ESR1 SNPs rs3798577 (CC vs CT vs TT) and rs9340799 (AA vs AG vs GG) were associated with family history of BC (P = 0.032) and body mass index (P = 0.007), respectively. While the ESR1 SNP rs3020410 (CC vs CA vs AA) was correlated with both estrogen receptor status (P = 0.012) and tumor size (P = 0.032). The ESR2 polymorphism rs1256049 (CC vs CT) exhibited an association with age at BC diagnosis (P = 0.019).
Table 4.
Association between different ESR1 and ESR2 SNP genotypes and the Clinico-pathological attributes of breast cancer (BC)
| Clinical attributes of BC |
ESR1 | ESR2 | |||
|---|---|---|---|---|---|
| rs3020410 CC vs CA vs AA |
rs3798577 CC vs CT vs TT |
rs2234693 CC vs CT vs TT |
rs9340799 AA vs AG vs GG |
rs1256049 CC vs CT |
|
| Age at BC diagnosis b | 0.632 | 0.528 | 0.179 | 0.190 | 0.019 |
| Age at first pregnancy b | 0.904 | 0.295 | 0.128 | 0.318 | 0.634 |
| Age at menarche b | 0.741 | 0.866 | 0.154 | 0.138 | 0.570 |
| Age at menopause b | 0.965 | 0.077 | 0.627 | 0.664 | 0.533 |
| Allergy a | 0.300 | 0.893 | 0.886 | 0.749 | 0.625 |
| Body mass index b | 0.627 | 0.209 | 0.126 | 0.007 | 0.983 |
| Breastfeeding status a | 0.206 | 0.497 | 0.895 | 0.540 | 0.448 |
| Co-morbidity a | 0.914 | 0.719 | 0.485 | 0.615 | 0.868 |
| Family history a | 0.450 | 0.032 | 0.674 | 0.706 | 0.497 |
| Smoking a | 0.067 | 0.722 | 0.868 | 0.575 | 0.415 |
| Pathological attributes of breast cancer (BC) | |||||
| Axillary lymph nodes a | 0.434 | 0.314 | 0.078 | 0.266 | 0.805 |
| Estrogen receptor status a | 0.012 | 0.398 | 0.803 | 0.517 | 0.569 |
| HER2 a | 0.561 | 0.642 | 0.152 | 0.420 | 0.492 |
| Histology classification a | 0.702 | 0.610 | 0.818 | 0.898 | 0.806 |
| Lymph node involvement a | 0.772 | 0.362 | 0.318 | 0.255 | 0.534 |
| Progesterone receptor status a | 0.966 | 0.756 | 0.536 | 0.495 | 0.736 |
| Tumor differentiation a | 0.970 | 0.399 | 0.596 | 0.849 | 0.056 |
| Tumor size b | 0.032 | 0.177 | 0.637 | 0.619 | 0.536 |
| Tumor stage a | 0.793 | 0.158 | 0.199 | 0.155 | 0.614 |
aPearson’s chi-squared test was used to determine genotype-phenotype association
bAnalysis of variance (ANOVA) was used to determine genotype-phenotype association
The association between the HER2, UGT1A4, and UGT2B7 SNPs and the clinical and pathological BC factors was also examined (Table 5). The HER2 rs1058808 (GG vs GC vs CC) SNP was associated with both progesterone receptor status (P = 0.01) and tumor size (P = 0.013). Regarding UGT1A4, its rs12468274 (TT vs CT) and rs2011425 SNPs were correlated with allergy (P = 0.001) and tumor size (P = 0.002). However, no such significant association was found between the investigated UGT2B7 SNPs and the clinical or pathological features of BC.
Table 5.
Association between different HER2, UGT1A4, and UGT2B7 SNP genotypes and the Clinico-pathological attributes of breast cancer (BC)
| Clinical attributes of BC |
HER2 | UGT1A4 | UGT2B7 | |||
|---|---|---|---|---|---|---|
| rs1058808 GG vs GC vs CC |
rs12468274 TT vs CT |
rs2011425 TT vs TG |
rs6755571 CC vs CA vs AA |
rs28365062 AA vs AG vs GG |
rs4348159 CC vs CT vs TT |
|
| Age at BC diagnosis b | 0.457 | 0.443 | 0.677 | 0.958 | 0.249 | 0.242 |
| Age at first pregnancy b | 0.712 | 0.363 | 0.280 | 0.593 | 0.416 | 0.258 |
| Age at menarche b | 0.352 | 0.733 | 0.632 | 0.610 | 0.303 | 0.301 |
| Age at menopause b | 0.369 | 0.198 | 0.257 | 0.802 | 0.817 | 0.477 |
| Allergy a | 0.393 | 0.001 | 0.901 | 0.820 | 0.296 | 0.363 |
| Body mass index b | 0.373 | 0.264 | 0.177 | 0.729 | 0.806 | 0.796 |
| Breastfeeding status a | 0.107 | 0.424 | 0.556 | 0.058 | 0.839 | 0.726 |
| Co-morbidity a | 0.137a | 0.2802 | 0.884 | 0.936 | 0.895 | 0.889 |
| Family history a | 0.46 | 0.882 | 0.337 | 0.221 | 0.418 | 0.686 |
| Smoking a | 0.275 | 0.380 | 0.150 | 0.273 | 0.667 | 0.403 |
| Pathological attributes of BC | ||||||
| Axillary lymph nodesa | 0.645 | 0.994 | 0.607 | 0.447 | 0.967 | 0.451 |
| Estrogen receptora | 0.051 | 0.555 | 0.583 | 0.705 | 0.798 | 0.121 |
| HER2a | 0.054 | 0.223 | 0.295 | 0.968 | 0.223 | 0.567 |
| Histology classification a | 0.786 | 0.916 | 0.201 | 0.535 | 0. 820 | 0.927 |
| IHC profilea | 0.252 | 0.472 | 0.409 | 0.918 | 0.472 | 0.826 |
| Lymph node involvement a | 0.875 | 0.368 | 0.658 | 0.386 | 0.769 | 0.317 |
| Progesterone receptor a | 0.010 | 0. 770 | 0.109 | 0.422 | 0.919 | 0.496 |
| Tumor differentiationa | 0.288 | 0.426 | 0.690 | 0.373 | 0.373 | 0.855 |
| Tumor size b | 0.013 | 0.323 | 0.002 | 0.232 | 0.359 | 0.941 |
| Tumor stage a | 0.580 | 0.712 | 0.347 | 0.322 | 0.675 | 0.788 |
aPearson’s chi-squared test was used to determine genotype-phenotype association
bAnalysis of variance (ANOVA) test was used to determine genotype-phenotype association
Haplotype analysis
The ESR1, ESR2, and UGT1A4 SNPs were subject to haplotype analysis. Our results revealed two separate blocks: ESR (rs3020410, rs3798577, rs1256049, rs2234693, and rs9340799) and UGT1A4 (rs12468274, rs2011425, and rs6755571). Table 6 shows the frequency ratios for cases and controls as well as the p-values for each block, and no association was deduced between the aforementioned haplotypes and BC risk in the present study.
Table 6.
Haplotypic analysis of ESR1, ESR2, and UGT1A4 polymorphisms
| Haplotype | Frequency of block | Frequency ratio (case:control) (%) |
Odds ratio (95% CI) |
P-value |
|---|---|---|---|---|
| ESR1 and ESR2 Block (rs3020410, rs3798577, rs1256049, rs2234693, and rs9340799) | ||||
| CTCCG | 0.2417 | 0.2761: 0.232 | 1:00 | N.A |
| CTCTA | 0.2358 | 0.2172: 0.2345 | 0.90 (0.56–1.45) | 0.66 |
| CCCCG | 0.1957 | 0.2025: 0.1681 | 0.61 (0.35–1.04) | 0.071 |
| CCCTA | 0.1702 | 0.2008: 0.1538 | 0.65 (0.42–1.02) | 0.061 |
| ACCTA | 0.0355 | 0.0266: 0.0391 | 0.73 (0.27–1.94) | 0.53 |
| ATCTA | 0.0355 | 0.0414: 0.0382 | 1.16 (0.46–2.89) | 0.76 |
| CCCCA | 0.0277 | 0.0291: 0.0274 | 0.80 (0.31–2.04) | 0.64 |
| CTCCA | 0.0186 | 0.0168: 0.0194 | 0.81 (0.22–2.92) | 0.74 |
| Global haplotype association p-value: 0.47 | ||||
| UGT1A4 Block (rs12468274, rs2011425, and rs6755571) | ||||
| TTC | 0.8552 | 0.8557:0.8548 | 1.00 | N.A |
| CGC | 0.0771 | 0.0726: 0.0816 | 1.12 (0.65–1.92) | 0.69 |
| TTA | 0.0526 | 0.0501:0.0551 | 1.07 (0.59–1.95) | 0.82 |
| TGC | 0.0118 | 0.019: 0.0048 | 0.25 (0.05–1.21) | 0.086 |
| Global haplotype association p-value: 0.39 | ||||
Discussion
Studies focusing on breast cancer (BC) genetics are increasingly shedding light on the etiology, progression, and treatment of the disease [33, 34]. However, the presence of genetic differences at the ethnic level mandates that cancer-related polymorphisms reported in one group be similarly investigated for any such association in other groups [35, 36]. This rings true for Arab populations especially, which are neither homogenous in their cancer distribution nor identical in their cancer genetic profiles [37]. Therefore, the aim of the present study was to investigate the association of specific ESR1, ESR2, HER2, UGT1A4, and UGT2B7 SNPs with BC in Jordanian-Arabs.
Our findings show that the ESR1 polymorphism rs3798577 was significantly associated with BC and history of BC in the Jordanian-Arab population, and it was similarly found to confer higher BC risk in the Tunisian-Arab population [38]. rs3978577 polymorphism is located in the 3′ UTR of ER-α, and it has been suggested to increased the overall risk of BC [25]. Moreover, it has been revealed that T allele of ESR1 rs3798577 serve as binding site for forkhead box transcription factor (FOXP1). FOXP1 is involved in proliferation, differentiation in addition to malignant transformation. Fox et al. (2004) indicated that FOXP1 might act as coregulator of ESR1 Expression [39]. While C allele may serve as Sex determining region Y-box 5 (SOX5) binding site which is a transcription factor that binds to ESR1 promoter and play role in embryonic development and determination of the cell fate [40].
In contrast, Ghali et al. (2018) found that the ESR1 rs2234693 and the ESR2 rs1256049 SNPs were positively and negatively associated with BC in Tunisian Arabs, respectively, while our results only showed an association between rs1256049 and age at BC diagnosis in Jordanian Arabs [38]. In contrast with our results, the ESR1 rs2234693 SNP was significantly associated with BC in a meta-analysis covering 44 case-control studies, and different levels of association between the ESR2 rs1256049 SNP and BC were reported in non-Arab populations [10, 11, 41]. Lastly, no significant association with BC was found for the ESR1 SNPs rs3020410 and rs9340799 in Jordanian Arabs. However, our results show an association between these SNPs and certain BC prognostic factors: rs9340799 was associated with body mass index while rs3020410 was linked to both estrogen receptor status and tumour size in Jordanian Arabs. In older Caucasian females, the rs9340799 SNP protected against BC, while the C allele of the rs3020410 SNP was associated with increased relapse risk [42, 43].
With regard to the HER2 gene, it has been well-documented that its overexpression or its amplification can negatively affect BC survival, chemotherapy, and remission [44]. In the present study, no significant association was found between the HER2 rs1058808 SNP and BC in Jordanian Arabs, but it was significantly associated with progesterone receptor status and tumor size. Conflictingly, this SNP was significantly associated with HER2 protein expression in Han Chinese BC patients, while another study found no BC association of rs1058808 in the same ethnic group [26, 45]. Moreover, no significant BC association was found for rs1058808 in Mexican and Vietnamese BC patients [46].
In terms of BC pharmacogenetics, the UGT genes play an important role in the metabolism of tamoxifen, a first-therapy for several types of BC [24]. Concerning UGT1A4 and UGT2B7, our results showed no significant association between the investigated SNPs and BC in Jordanian Arabs. However, the UGT1A4 rs12468274 and rs2011425 SNPs were found to be associated with allergy and tumor size, respectively. In Spanish Caucasians, the homozygous mutant form of the rs2011425 SNP was associated with lower concentrations of active tamoxifen metabolites [24].
Conclusions
In conclusion, it can be seen that the influence of certain ESR1, ESR2, HER2, UGT1A4, and UGT2B7 SNPs on BC in Jordanian Arabs differs from that in other populations. The findings of the present study identified the ESR1 SNP rs3798577 as being significantly associated with BC, which could potentially be taken into consideration in preventative approaches to BC in the Jordanian population. Further characterization of the role of such variants in specific populations will definitely aid in understanding BC etiology, progression, and treatment.
Acknowledgements
The authors thank the Jordanian Royal Medical Services (JRMS), Amman, Jordan, for approving this study in the first instance and making the clinical data and samples available for the study.
Abbreviations
- AGRF
Australian genome research facility
- BC
Breast cancer
- χ2
Chi squared value
- DNA
Deoxyribonucleic acid
- ESR
Etrogen receptor
- HER2
Human epidermal growth factor receptor 2 marker
- Het
Heterozygote
- HWE
Hardy-Weinberg equilibrium
- Hz
Homozygote (Hz)
- IRB
Institutional review board
- JRMS
Jordanian Royal medical services
- PR
Progesterone receptor
- SNPs
Single nucleotide polymorphisms
- SPSS
Statistical package for the social sciences (SPSS)
- UGTs
UDP glucuronosyltransferases
Authors’ contributions
LNA-E designed the method study and supervised the study. LNA-E, DMR and MAA lead the implementation of the method, performed the data analysis and drafted the manuscript. LNA-E, DMR, MAA and RHK helped with the interpretation, and description of the results. All authors read and approved the final manuscript.
Funding
This study was funded by the Deanship of Research (RN: 20140204), Jordan University of Science and Technology. The Deanship of Research has no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The datasets generated and/or analysed over the course of the study are not publicly available but are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Institutional Review Board (IRB) at Jordan University of Science and Technology with ethical code number (14/78/2014). Written informed consent was obtained from all individual participants included in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hankinson SE, Colditz GA, Willett WC. Towards an integrated model for breast cancer etiology: the lifelong interplay of genes, lifestyle, and hormones. Breast Cancer Res. 2004;6:213. doi: 10.1186/bcr921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nowsheen S, Aziz K, Panayiotidis MI, Georgakilas AG. Molecular markers for cancer prognosis and treatment: Have we struck gold? Cancer Lett. 2012;327:142–152. doi: 10.1016/J.CANLET.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 3.AL-Eitan L, Rababa’h D, Alghamdi M, Khasawneh R. Correlation between candidate single nucleotide variants and several Clinicopathological risk factors related to breast Cancer in Jordanian women: a genotype-phenotype study. J Cancer. 2019;10(19):4647–4654. doi: 10.7150/jca.33857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LN AL-E, Jamous RI, Khasawneh RH. Candidate gene analysis of breast Cancer in the Jordanian population of Arab descent: a case-control study. Cancer Investig. 2017;35:256–270. doi: 10.1080/07357907.2017.1289217. [DOI] [PubMed] [Google Scholar]
- 5.Gage M, Wattendorf D, Henry LR. Translational advances regarding hereditary breast cancer syndromes. J Surg Oncol. 2012;105:444–451. doi: 10.1002/jso.21856. [DOI] [PubMed] [Google Scholar]
- 6.Lu H, Chen D, Hu LP, et al. Estrogen receptor alpha gene polymorphisms and breast cancer risk: a case-control study with meta-analysis combined. Asian Pac J Cancer Prev. 2014;14:6743–6749. doi: 10.7314/APJCP.2013.14.11.6743. [DOI] [PubMed] [Google Scholar]
- 7.Lazarus P, Sun D. Potential role of UGT pharmacogenetics in cancer treatment and prevention: focus on tamoxifen and aromatase inhibitors. Drug Metab Rev. 2010;42:182–194. doi: 10.3109/03602530903208652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krishna BM, Chaudhary S, Panda AK, et al. Her2 Ile655Val polymorphism and its association with breast cancer risk: an updated meta-analysis of case-control studies. Sci Rep. 2018;8:7427. doi: 10.1038/s41598-018-25769-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Medina D, Sivaraman L, Hilsenbeck SG, et al. Mechanisms of hormonal prevention of breast cancer. Ann N Y Acad Sci. 2001;952:23–35. doi: 10.1111/j.1749-6632.2001.tb02725.x. [DOI] [PubMed] [Google Scholar]
- 10.Yu K-D, Rao N-Y, Chen A-X, et al. A systematic review of the relationship between polymorphic sites in the estrogen receptor-beta (ESR2) gene and breast cancer risk. Breast Cancer Res Treat. 2011;126:37–45. doi: 10.1007/s10549-010-0891-2. [DOI] [PubMed] [Google Scholar]
- 11.Maguire P, Margolin S, Skoglund J, et al. Estrogen receptor Beta (ESR2) polymorphisms in familial and sporadic breast Cancer. Breast Cancer Res Treat. 2005;94:145–152. doi: 10.1007/s10549-005-7697-7. [DOI] [PubMed] [Google Scholar]
- 12.Jeselsohn R, Yelensky R, Buchwalter G, et al. Emergence of constitutively active estrogen receptor-α mutations in pretreated advanced estrogen receptor positive breast cancer. Clin Cancer Res. 2014;20(7):1757–1767. doi: 10.1158/1078-0432.CCR-13-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merenbakh-Lamin K, Ben-Baruch N, Yeheskel A, et al. D538G mutation in estrogen receptor-α: a novel mechanism for acquired endocrine resistance in breast Cancer. Cancer Res. 2013;73(23):6856–6864. doi: 10.1158/0008-5472.CAN-13-1197. [DOI] [PubMed] [Google Scholar]
- 14.Robinson DR, Wu Y-M, Vats P, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013;45(12):1446–1451. doi: 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toy W, Shen Y, Won H, et al. ESR1 ligand binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45(12):1439–1445. doi: 10.1038/ng.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan M, Yu D. Molecular mechanisms of ErbB2-mediated breast Cancer Chemoresistance. In: Madame curie bioscience database [internet] Austin (TX): Landes Bioscience; 2000. [DOI] [PubMed] [Google Scholar]
- 17.Tein RA, Staros JV. Evolutionary analysis of the ErbB receptor and ligand families. J Mol Evol. 2000;50(5):397–412. doi: 10.1007/s002390010043. [DOI] [PubMed] [Google Scholar]
- 18.Slamon DJ, Godolphin W, Jones LA, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244(4905):707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 19.Sauter G, Moch H, Moore D, et al. Heterogeneity of erbB-2 gene amplification in bladder cancer. Cancer Res. 1993;53(10 Suppl):2199–2203. [PubMed] [Google Scholar]
- 20.Tateishi M, Ishida T, Mitsudomi T, et al. Prognostic value of c-erbB-2 protein expression in human lung adenocarcinoma and squamous cell carcinoma. Eur J Cancer. 1991;27(11):1372–1375. doi: 10.1016/0277-5379(91)90012-3. [DOI] [PubMed] [Google Scholar]
- 21.Lemoine NR, Jain S, Silvestre F, et al. Amplification and overexpression of the EGF receptor and c-erbB-2 proto-oncogenes in human stomach cancer. Br J Cancer. 1991;64(1):79–83. doi: 10.1038/bjc.1991.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan M, Yao J, Yu D. Overexpression of the c-erbB-2 gene enhanced intrinsic metastatic potential in human breast cancer cells without increasing their transformation abilities. Cancer Res. 1997;57:1199–1205. [PubMed] [Google Scholar]
- 23.Moody SE, Sarkisian CJ, Hahn KT, et al. Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer Cell. 2002;2(6):451–461. doi: 10.1016/S1535-6108(02)00212-X. [DOI] [PubMed] [Google Scholar]
- 24.Romero-Lorca A, Novillo A, Gaibar M, et al. Impacts of the Glucuronidase genotypes UGT1A4, UGT2B7, UGT2B15 and UGT2B17 on Tamoxifen metabolism in breast Cancer patients. PLoS One. 2015;10:e0132269. doi: 10.1371/journal.pone.0132269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li N, Dong J, Hu Z, et al. Potentially functional polymorphisms in ESR1 and breast cancer risk: a meta-analysis. Breast Cancer Res Treat. 2010;121:177–184. doi: 10.1007/s10549-009-0532-9. [DOI] [PubMed] [Google Scholar]
- 26.Su Y, Jiang Y, Sun S, et al. Effects of HER2 genetic polymorphisms on its protein expression in breast cancer. Cancer Epidemiol. 2015;39:1123–1127. doi: 10.1016/J.CANEP.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 27.De Almeida FC, Banin Hirata BK, Ariza CB, et al. HER2 Ile655Val polymorphism is negatively associated with breast cancer susceptibility. J Clin Lab Anal. 2018;32:e22406. doi: 10.1002/jcla.22406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sutiman N, Lim JSL, Muerdter TE, et al. Pharmacogenetics of UGT1A4, UGT2B7 and UGT2B15 and their influence on Tamoxifen disposition in Asian breast Cancer patients. Clin Pharmacokinet. 2016;55:1239–1250. doi: 10.1007/s40262-016-0402-7. [DOI] [PubMed] [Google Scholar]
- 29.Jing L, Su L, Ring BZ. Ethnic background and genetic variation in the evaluation of cancer risk: a systematic review. PLoS One. 2014;9:e97522. doi: 10.1371/journal.pone.0097522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Preacher KJ. Calculation for the chi-square test: an interactive calculation tool for chi-square tests of goodness of fit and independence. 2001. [Google Scholar]
- 31.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 32.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.AL-Eitan L, Rababa’h D, Alghamdi M, et al. Association of GSTM1, GSTT1 and GSTP1 polymorphisms with breast Cancer among Jordanian women. OncoTargets and Therapy. 2019;12:7757–7765. doi: 10.2147/OTT.S207255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.AL-Eitan Laith N., Rababa’h Doaa M., Alghamdi Mansour A., Khasawneh Rame H. Role of Four ABC Transporter Genes in Pharmacogenetic Susceptibility to Breast Cancer in Jordanian Patients. Journal of Oncology. 2019;2019:1–8. doi: 10.1155/2019/6425708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaukat U, Ismail M, Mehmood N. Epidemiology, major risk factors and genetic predisposition for breast cancer in the Pakistani population. Asian Pac J Cancer Prev. 2013;14:5625–5629. doi: 10.7314/APJCP.2013.14.10.5625. [DOI] [PubMed] [Google Scholar]
- 36.AL-Eitan L, Rababa’h D, Alghamdi M, et al. Association of CYP gene polymorphisms with breast cancer risk and prognostic factors in the Jordanian population.L. BMC Med Genet. 2019;20:148. doi: 10.1186/s12881-019-0884-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tadmouri Ghazi O., Sastry Konduru S., Chouchane Lotfi. Arab gene geography: From population diversities to personalized medical genomics. Global Cardiology Science and Practice. 2014;2014(4):54. doi: 10.5339/gcsp.2014.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghali RM, Al-Mutawa MA, Al-Ansari AK, et al. Differential association of ESR1 and ESR2 gene variants with the risk of breast cancer and associated features: a case-control study. Gene. 2018;651:194–199. doi: 10.1016/J.GENE.2018.02.011. [DOI] [PubMed] [Google Scholar]
- 39.Fox SB, Brown P, Han C, et al. Expression of the forkhead transcription factor FOXP1 is associated with estrogen receptor alpha and improved survival in primary human breast carcinomas. Clin Cancer Res. 2004;10(10):3521–3527. doi: 10.1158/1078-0432.CCR-03-0461. [DOI] [PubMed] [Google Scholar]
- 40.Sa-Nguanraksa D, Suntiparpluacha M, Kulprom A, et al. Association of Estrogen Receptor Alpha and Interleukin 6 Polymorphisms with Lymphovascular Invasion, Extranodal Extension, and Lower Disease-Free Survival in Thai Breast Cancer Patients. APJCP. 2016;17(6):2935–2940. [PubMed] [Google Scholar]
- 41.Hu X, Jiang L, Tang C, et al. Association of three single nucleotide polymorphisms of ESR1with breast cancer susceptibility: a meta-analysis. J Biomed Res. 2017;31:213–225. doi: 10.7555/JBR.31.20160087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Higuchi R, Modugno F, et al. Estrogen receptor alpha haplotypes and breast cancer risk in older Caucasian women. Breast Cancer Res Treat. 2007;106:273–280. doi: 10.1007/s10549-007-9497-8. [DOI] [PubMed] [Google Scholar]
- 43.Tapper W, Hammond V, Gerty S, et al. The influence of genetic variation in 30 selected genes on the clinical characteristics of early onset breast cancer. Breast Cancer Res. 2008;10:R108. doi: 10.1186/bcr2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arteaga CL, Sliwkowski MX, Osborne CK, et al. Treatment of HER2-positive breast cancer: current status and future perspectives. Nat Rev Clin Oncol. 2012;9:16–32. doi: 10.1038/nrclinonc.2011.177. [DOI] [PubMed] [Google Scholar]
- 45.Breyer JP, Sanders ME, Airey DC, et al. Heritable variation of ERBB2 and breast cancer risk. Cancer Epidemiol Biomark Prev. 2009;18:1252–1258. doi: 10.1158/1055-9965.EPI-08-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Wang X, Vural S, et al. Exome analysis reveals differentially mutated gene signatures of stage, Grade and Subtype in Breast Cancers. PLoS One. 2015;10:e0119383. doi: 10.1371/journal.pone.0119383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analysed over the course of the study are not publicly available but are available from the corresponding author on reasonable request.


