Abstract
The internal energy deposited into analytes during the ionization process largely influences the extent of fragmentation; thus, the appearance of the resulting mass spectrum. The internal energy distributions of a series of para-substituted benzyl pyridinium cations in liquid and solid state generated by infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) were measured using the survival yield method, of which results were subsequently compared with conventional electrospray ionization (ESI). The comparable mean internal energy values (e.g., 1.8-1.9 eV at a collision energy of 15 eV) and peak widths obtained with IR-MALDESI and ESI support that IR-MALDESI is essentially a soft ionization technique where analytes do not gain considerable internal energy during the laser-induced desorption process, and/or lose energy during uptake into charged electrospray droplets. An unusual fragment ion, protonated pyridine, was only found for solid IR-MALDESI at relatively high collision energies, which is presumably resulted from direct ionization of the pre-charged analytes in form of salts. Analysis of tissue with an ice layer consistently yielded ion populations with higher internal energy than its counterpart without an ice layer, likely due to a substantially enhanced number of IR absorbers with ice. Further measurements with holo-myoglobin show that IR-MALDESI-MS retains the noncovalently bound heme-protein complexes under both native-like and denaturing conditions, while complete loss of the heme group occurred in denaturing ESI-MS, showing that the softness of IR-MALDESI is equivalent or superior to ESI for biomolecules.
Keywords: Internal Energy Deposition, IR-MALDESI, Mass Spectrometry Imaging Survival Yield Method, Thermometer Ions
Graphical abstract
Introduction
Mass spectrometry imaging (MSI) has rapidly evolved as an invaluable analytical approach in biological science over the years for its capability of simultaneously monitoring numerous biomolecules and their spatial locations. Fragmentation is not desired because it confounds the extremely complex mass spectra and places challenges to molecular weight determination and data interpretation. To address these issues, continuous efforts have focused on the development of soft ionization techniques for MSI which do not impart excessive internal energy (IE) to analytes. Infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) is a hybrid ionization technique operating at atmospheric pressure with minimum or no sample preparation, which has established its utility for visualizing the distributions of a wide variety of analytes including lipids [1, 2], metabolites [3, 4], proteins [5] and pharmaceutical drugs [6, 7] from biological tissue sections. Our previous studies have shown that spectral compositions and charge state distributions generated by IR-MALDESI are nearly identical to ESI [5], and the [M+D+]+ peak is predominant in the spectra when using a deuterated electrospray solvent system [8]. These findings indicate that the primary ionization mechanism of IR-MALDESI is the desorbed neutral species from condensed phase reach an orthogonally oriented electrospray plume which are consecutively ionized via an electrospray ionization (ESI)-like process [5, 9]. Therefore, IR-MALDESI is considered a soft ionization source as ESI, yielding and transferring molecular ions with minimum fragmentation. However, the laser irradiation process may modify the ion IE via vibrational energy-transfer from water molecules [10] and thermal activation [11], ultimately leading to mass spectra which contain fragments of labile molecules. A quantitative study of IE deposition in IR-MALDESI followed by a parallel comparison with traditional ESI is therefore necessary to accurately evaluate the softness of IR-MALDESI.
IR-MALDESI typically employs a 2940 nm laser to resonantly excite the O-H symmetric and asymmetric stretching bands of water, allowing for direct analysis of water-rich targets (e.g., biological tissues) where endogenous water serves as an energy-absorbing medium. An ice layer can also be uniformly deposited on the sample surface as a mid-IR absorbing matrix. Multifold improvements in sensitivity are commonly observed with the application of an ice layer in IR-MALDESI-MS. To better understand the role of the ice matrix in ablation dynamics, which in turn translates into greater ion signals, a series of IR-MALDESI experiments combined with shadowgraphy were conducted [12], clearly revealing remarkable distinctions in plume dynamics for fresh tissue analysis with or without an ice layer. It was observed that much finer tissue fragments and liquid droplets were ejected in the case of ablation with ice, potentially indicating a more efficient energy-transfer process. However, to what extent the ice layer affects IE deposition in IR-MALDESI remains unstudied to date.
The survival yield method based on the work of De Pauw and coworkers [13] has been extensively utilized to probe IE of ions generated by a wide range of well-recognized soft ionization techniques, e.g., ESI [14, 15], matrix-assisted laser desorption ionization (MALDI) [16, 17] and desorption electrospray ionization (DESI) [18, 19]. The basic assumption of this approach is that the ions with an IE above the critical energy (E0) undergo dissociation, while those with an IE below E0 do not. The survival yield therefore describes the fraction of precursor ions with an IE less than their E0, and can be calculated using Equation (1),
| (1) |
where I(M+) and I(F+) are the abundances of the molecular ions and the corresponding fragment ions, respectively. The function SY(E) is shown to be the integral of the IE distribution function P(E) from E = 0 to E0 [13], hence the IE distribution can be determined by taking the first derivative of SY(E). The survival yield method is applicable to a series of ions with varying E0, but identical IE distribution, e.g., thermometer ions. A set of substituted benzyl pyridinium (BP) cations are commonly used as thermometer ions because of their simple fragmentation pattern and well-characterized thermodynamics [20]. Most often, the cleavage of the C-N bond occurs between the benzyl and pyridyl groups, producing a neutral pyridine and a substituted benzyl cation for MS detection. The substitute group on the benzyl ring determines the magnitude of the E0 for the unimolecular dissociation.
In this work, we first characterized and compared the IE distribution of para-substituted BP cations generated with IR-MALDESI and ESI using the survival yield method. Different types of samples which are common targets for IR-MALDESI were investigated to reveal the differences in IE distribution related to sample physical properties. Measurements with and without the ice matrix were then performed to elucidate the effect of the ice matrix on IE deposition as a function of laser wavelengths. Myoglobin (Mb) as a noncovalent protein complex was further selected to probe the energy deposition of large and labile biomolecules. This work improves knowledge of the complex ionization mechanisms and energy transfer events underlying IR-MALDESI-MS, hence potentially enabling further enhancements in the sensitivity and specificity.
Methods
Materials
Five thermometer molecules para-methoxy-benzyl pyridinium (MeO) chloride, para-methyl-benzyl pyridinium (Me) chloride, para-chloro-benzyl pyridinium (Cl) chloride, para-cyano-benzyl pyridinium (CN) chloride, and para-nitro-benzyl pyridinium (NO2) bromide were purchased from GL Synthesis (Worcester, MA, USA) with a purity of 98% and used without further purification. Holo-myoglobin from equine skeletal muscle with a purity of 95-100% was purchased from Sigma-Aldrich (St. Louis, MO, USA). Methanol, water, formic acid and ammonium acetate in Optima grade were purchased from Fisher Scientific (Fair Lawn, NJ, USA). Nitrogen gas used for mass spectrometry and enclosure purging was obtained from Arc3 Gases (Raleigh, NC, USA).
The BP salts were first individually dissolved in 50% (v/v) aqueous methanol to prepare stock solutions at ~6 mM. Then they were mixed and further diluted with 50% (v/v) aqueous methanol to make standard mixtures at optimal concentrations for different experiments to provide molecular ion abundances between 104-105 counts, ensuring a good spectral accuracy [21] and a linear MS detector response.
Tissue
Rat liver tissue was obtained from the North Carolina State University Department of Biological Sciences, frozen in isopentane/dry ice and stored at −80 °C until sectioning. The animal was managed in accordance with the Institute for Laboratory Animal Research Guide, and all husbandry practices were approved by North Carolina State University Institutional Animal Care and Use Committee (IACUC).
Tissue sections of 10 μm thickness were cut using a Leica CM1950 cryostat (Buffalo Grove, IL, USA) at a temperature of −15 °C. The sections were then thaw mounted onto standard glass microscope slides with and without BP salts applied and analyzed immediately thereafter.
IR-MALDESI-MS Analysis
The in-house built IR-MALDESI source which has been detailed in previous publications [12, 22] was used for all experiments. A tunable-wavelength (2700 - 3100 nm) IR laser operating at 20 Hz with a pulse width of 5 - 7 ns (IR-Opolette 2731, Opotek, Carlsbad, CA, USA) is employed and focused to a spot diameter of approximately 150 pm on the sample surface to resonantly excite water, facilitating the desorption of materials from the sample. The sample stage is mounted on a water-cooled Peltier stage to achieve −10 °C sample temperature when the ice matrix was used. The incident wavelength was set to 2940 nm to match the O-H stretching band of water unless otherwise noted. The laser energy was measured to be 1.3 mJ/pulse at the exit of the optical path. For all IR-MALDESI experiments except one noted as 1 pulse, each voxel was subjected to two laser pulses to completely ablate probed sample materials. For the measurements of thermometer ions, electrospray solvent of 50% (v/v) aqueous methanol modified by 0.2% formic acid was delivered by a Fusion 101 syringe pump (Thermo Fisher Scientific, Bremen, Germany) at 2 μL·min−1 flow rate. A Q-Exactive-Plus Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) was coupled to the IR-MALDESI source. The mass spectrometer was operated in positive ionization mode with a spray voltage of 3.8 kV and a capillary temperature of 315 °C. The automatic gain control function (AGC) was disabled and the injection time (IT) was fixed to 75 or 25 ms for IR-MALDESI experiments with 2 laser pulses or 1 laser pulse, respectively, to coordinate the laser desorption and ion acquisition events. All BP data sets were collected at m/z 75-250 with a resolving power of 140,000 (FWHM, m/z = 200), while Mb data sets were acquired at m/z 500-2000 or 700-2800 depending on the charge states.
To measure the IE deposition in liquid IR-MALDESI, 10 μL fresh BP solution at 4 μM was dropped on a standard glass slide and analyzed by IR-MALDESI-MS at each collision energy. Solid samples for IR-MALDESI were prepared by spraying solutions of BP salts evenly on glass slides at 0.1 and 1 μg·cm−2 using a HTX TM Sprayer (HTX Technologies, Chapel Hill, NC, USA) for sprayed slide samples and tissue section samples, respectively. The TM Sprayer parameters were optimized earlier [7] and are detailed in Table S1. For experiments evaluating the ice matrix, 100 μL of BP solution at ~1.5 mM was deposited on top of a tissue section and air-dried. 1 mM Mb was prepared in water, then diluted to 20 μM with different solvents to produce working solutions, of which 10 μL was deposited on a glass slide for liquid IR-MALDESI-MS. For all measurements, a region-of-interest of 6-by-6 was sampled at each collision energy to provide 36 replicates for statistical analysis.
ESI-MS Analysis
Mixed BP salts or Mb doped in electrospray solution at 0.25 μM were directly infused into the mass spectrometer by an ESI source at 2 μL·min−1 flow rate. AGC was performed in prescan mode with AGC target of 3 × 106 for BP samples and 1 × 106 for Mb samples. Other experimental settings were kept constant for IR-MALDESI and ESI experiments to offer meaningful comparisons. For MS/MS acquisition, BP molecular ions were selected with a 4m/z isolation window then fragmented in the HCD cell at a normalized collision energy (NCE) of 10% for MeO, Me and Cl or 30% for CN and NO2.
Data Processing
Raw data files (.raw) were converted into the mzML format using MSConvert from the ProteoWizard toolkit [23], followed by conversion into an imzML image file by imzMLConverter [24]. These files were subsequently loaded in MatLab (R2018a; MathWorks, Natick, MA, USA) environment using MSiReader [25, 26]. Then a list of M+ and F+ m/z was created and the abundances were determined by peak intensity then exported using the built-in MSiExport tool. The sigmoidal curve fitting was carried out in Origin 2019 (OriginLab, Northampton, MA, USA).
Results and Discussion
Calculation of Survival Yield
To determine the m/z of M+ and F+ used for the calculation of survival yields, BP salt solutions were directly sprayed into the mass spectrometer with ESI to perform MS/MS analysis. MS/MS spectra (Figure S1) show that as reported in literature [13], the predominant fragmentation pathway is the loss of pyridine ([M+ - Py]+), yielding para-substituted benzyl ions (Scheme 1). The M+ and F+ m/z values along with the E0 using AM1 calculations for each BP are shown in Table 1, while readers are referred to prior literature for E0 values using ab initio computations [13, 17, 27]. Potential differences between the S-lens and the injection flatapole ranging from 0 to 35 eV were implemented to accelerate ions moving towards the mass analyzer. Subsequent inelastic collisions of ions with inert gas molecules cause a portion of their kinetic energy to be converted to IE, leading to in source-collision induced dissociation (IS-CID) when the reaction time is sufficient. The mass spectra acquired using ESI and IR-MALDESI at IS-CID of 0, 10 and 20 eV are shown in Figure 1, showing that the relative abundances of [M+ - Py]+ increase with collision energy, which is in accordance with the increases in IE of BP cations. The sum of the absolute abundances of M+ and F+ with ESI direct infusion were fairly constant at IS-CID energies of 0 to 20 eV, showing %RSDs less than 15. The amounts of M+ + F+ increased observably with the IS-CID > 20 eV, which might be a result of more efficient desolvation and/or ion transmission. However, the Method presented here should still be robust because the comparisons were done at the same collision energy.
Scheme 1.
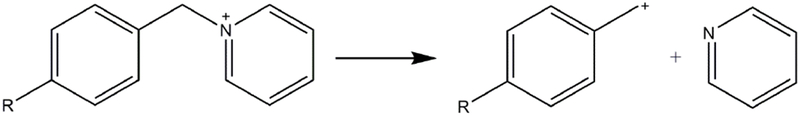
The fragmentation pattern of p-substituted benzyl pyridinium cation into a substituted benzyl cation and a neutral pyridine.
Table 1.
Molecular (M+), fragmental (M+ - Py) m/z and critical energy (E0) of BP cations. E0 values using AM1 calculations were taken from literature [14].
| Substituted BP | Abbreviation | m/z (M+) | m/z (M+ - Py) | E0 (eV) |
|---|---|---|---|---|
| para-methoxy- | MeO | 200.1069 | 121.0647 | 1.51 |
| para-methyl- | Me | 184.1120 | 105.0698 | 1.77 |
| para-chloro- | Cl | 204.0574 | 125.0152 | 1.90 |
| para-cyano- | CN | 195.0916 | 116.0494 | 2.10 |
| para-nitro- | NO2 | 215.0815 | 136.0393 | 2.35 |
Figure 1.
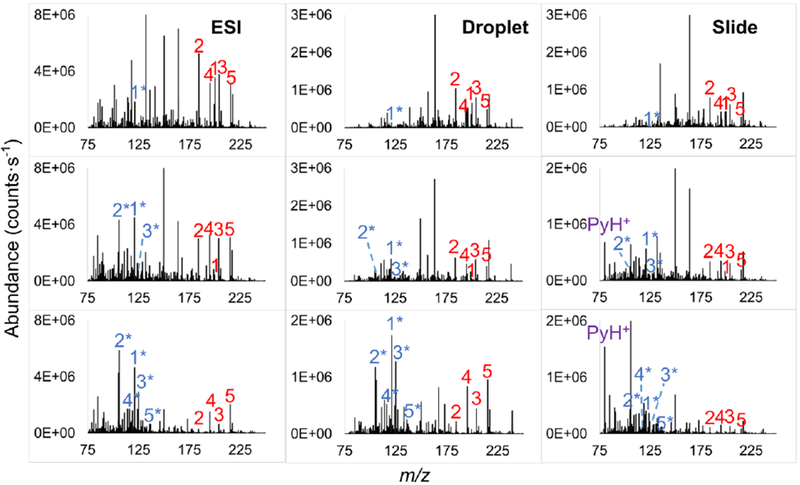
Representative mass spectra acquired with direct infusion ESI, and IR-MALDESI of solution droplets and sprayed glass slides at IS-CID of 0 (top), 10 (middle) and 20 eV (bottom). The precursor ions of MeO, Me, Cl, CN and NO2 are numbered consecutively from 1 to 5 in red and the corresponding benzyl fragment ions are numbered in blue with asterisks.
Survival yields were calculated for the five BP cations using Equation (1) with the abundances of intact benzyl pyridinium cations as M+ and the corresponding benzyl cations as F+. Then the calculated survival yields were plotted as a function of E0. Two limit points (E0 = 0, survival yield = 0 and E0 = 3.5, survival yield = 100%) were added to the plots to indicate the boundaries of the IE distribution, corresponding to complete and no dissociation, respectively. A Boltzmann-type sigmoidal curve Equation (2) was then fitted to the data points,
| (2) |
where E is the center of IE, x is E0, and A1 and A2 are initial and final survival yield values, respectively. Good fitting results with correlation coefficients R2 > 0.9 were achieved with this curve function.
Comparison of IE Deposition in ESI and IR-MALDESI of Liquid Samples
The survival yields of five BP molecular ions from solutions were determined with ESI-MS or IR-MALDESI-MS and plotted as a function of IS-CID energy to reveal the survival yield breakdown curves (Figure 2a and 2b). The survival yields rank accordingly with E0 as expected, meaning high E0 BP cations dissociate less than the low E0 ones. Noticeably, breakdown curves from both techniques show remarkably similar descending trends. Survival yields at IS-CID energies of 10-25 eV were chosen to fit the sigmoidal curves because at this range the survival yield values spread widely between 0 and 100%, corresponding to no and complete dissociation, respectively. As mentioned before, the IE distributions were then obtained by taking the first derivative of the survival yield sigmoidal curves, and the mean values with full-width at half-maximum (FWHM) reported directly from IE distributions. The summary of fitted values is available in Table 2. It should be stressed that the determined IEs are not absolute values since the kinetic shifts were not accounted for in our work. Actually, energies higher than the E0 are needed for an appreciable number of fragment ions to be detected, therefore the mean IE values determined by the survival yield method are underestimated [28]. But the IE estimations we present here are sufficient for the comparison of IR-MALDESI and ESI because all relevant MS parameters were kept constant throughout the entire experiment.
Figure 2.
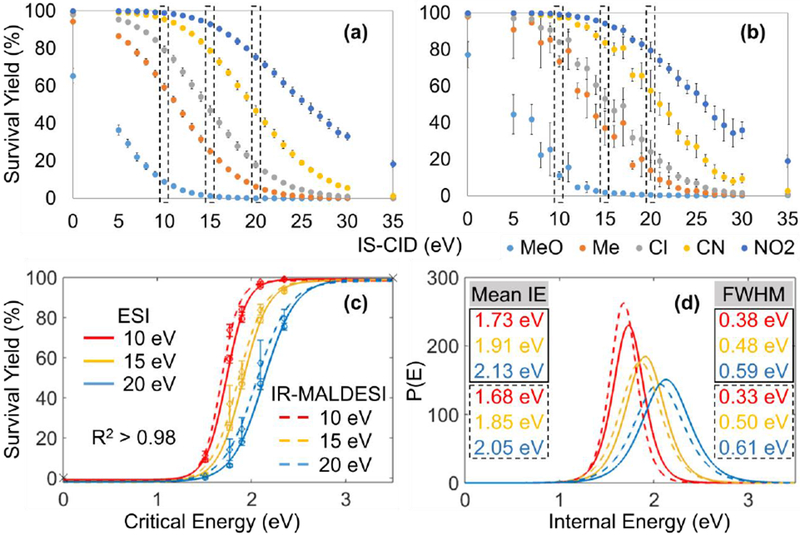
Survival yields of five BP cations versus IS-CID energies for (a) ESI and (b) IR-MALDESI of sample solutions. The comparison of (c) Boltzmann’s sigmoidal curves and (d) IE distributions between ESI and IR-MALDESI were made at IS-CID of 10, 15 and 20 eV. The mean IE and FWHM values for ESI and IR-MALDESI are solid- and dashed-outlined, respectively. Error bars represent one standard deviation of 36 replicate measurements.
Table 2.
Mean IE values (eV) with 95% confidence intervals (CI) and full-width at half maximum (FWHM, eV) of BP cations for ESI and IR-MALDESI determined at IS-CID of 10-25 eV.
| Source | ESI | IR-MALDESI | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample Type | Solution | Droplet | Sprayed Slide | ||||||
| IS-CID (eV) | R2 | Mean IE [CI] | FWHM | R2 | Mean IE [CI] | FWHM | R2 | Mean IE [CI] | FWHM |
| 10 | 0.998 | 1.73 [1.73, 1.73] | 0.38 | 0.995 | 1.68 [1.68, 1.69] | 0.33 | 0.988 | 1.74 [1.73, 1.75] | 0.39 |
| 15 | 0.998 | 1.91 [1.91, 1.91] | 0.48 | 0.984 | 1.85 [1.85, 1.87] | 0.50 | 0.983 | 1.86 [1.85, 1.87] | 0.44 |
| 20 | 0.996 | 2.13 [2.13, 2.13] | 0.59 | 0.987 | 2.05 [2.04, 2.06] | 0.61 | 0.971 | 1.97 [1.96, 1.99] | 0.59 |
| 25 | 0.999 | 2.33 [2.33, 2.33] | 0.56 | 0.990 | 2.29 [2.28, 2.30] | 0.64 | 0.962 | 2.11 [2.09, 2.13] | 0.74 |
| Source | IR-MALDESI | |||||
|---|---|---|---|---|---|---|
| Sample Type | Sprayed Slide (1 Laser Pulse) | Sprayed Slide underneath A Tissue Section | ||||
| IS-CID (eV) | R2 | Mean IE [CI] | FWHM | R2 | Mean IE [CI] | FWHM |
| 10 | 0.998 | 1.74 [1.74, 1.75] | 0.39 | 0.994 | 1.70 [1.69, 1.70] | 0.27 |
| 15 | 0.996 | 1.88 [1.87, 1.88] | 0.47 | 0.986 | 1.76 [1.75, 1.77] | 0.33 |
| 20 | 0.991 | 2.05 [2.04, 2.06] | 0.59 | 0.981 | 1.85 [1.84, 1.86] | 0.39 |
| 25 | 0.983 | 2.22 [2.20, 2.23] | 0.73 | 0.953 | 1.93 [1.91, 1.95] | 0.68 |
R2 stands for the regression coefficient of the sigmoidal fit.
As shown in Table 2, the increase in potential differences shift the mean IE toward higher values with broadening distribution widths, which is in agreement with other studies [29, 30]. IR-MALDESI of BP solutions resulted in IE distributions basically equivalent to ESI. For instance, at IS-CID of 15 eV the mean IE values were 1.91 eV for ESI and 1.85 eV for IR-MALDESI with similar peak widths (Figure 2c and 2d), corroborating the view that the ions produced by IR-MALDESI did not gain considerably excess energy. The result can be attributed to either the energy dissipation from excited molecules to surrounding solvent molecules, or the complete collisional cooling of analytes during capture by charged electrospray droplets, or a combination of these effects.
Comparison of IE Deposition in ESI and IR-MALDESI of Solid Samples
Our earlier investigation of plume ablation dynamics has manifested substantial variations for liquid droplets and solid samples [12], implying potential differences in IE deposition associated with sample physical properties. In order to explore such effects, the IR-MALDESI measurements were repeated for solid samples including glass microscope slides sprayed with BP salts and sprayed slides underneath tissue sections. Unlike solution samples, the sprayed slides are dry and thus transparent to the incident laser at 2940 nm, so a thin ice layer was formed on top of each sample to absorb IR so to assist analyte desorption.
The mass spectra of solid samples show a prevalent peak at m/z 80.0502 when applying relatively high collision energies (Figure 1). Its absence from solvent-coated glass slides and its occurrence depending on the critical energies of BP substituents (Figure S2) demonstrated that this peak originated from BP salts, which was then identified as protonated pyridine (PyH+). The observation is consistent with our hypothesis of different ablation mechanisms for liquid- and solid-phase samples. Few studies on IE deposition have reported the pyridine feature. One example by Flanigan et al. [31] observed PyH+ in dried BP salts with a 800 nm laser electrospray mass spectrometry (LEMS). Two-photon resonant-excitation of BP salts was proposed to cause their dissociation before entering the mass spectrometer, enabling the neutral pyridine to be ionized by nanospray. Gabelica et al. [17] found PyH+ with a vacuum-MALDI time-of-flight MS, and attributed this to a hydrogen radical reaction in the dense plume happening on a nanosecond timescale after laser impact. Although these hypotheses worked well in those studies, they appear less likely to explain PyH+ in IR-MALDESI-MS. In the case of IR-MALDESI, PyH+ is produced mainly inside the mass spectrometer since it can be barely found at no or a low collision energy (Figure 1 and Figure S2). The radical theory is also less suitable in our case considering the instrumental configuration of IR-MALDESI, where the ablated sample plume and the MS inlet capillary are perpendicular, making hydrogen radicals, if they exist, unlikely to travel into the mass spectrometer.
To determine how PyH+ is produced in IR-MALDESI, a series of control IR-MALDESI experiments with the electrospray flow rate off were carried out on sprayed slides, showing dramatically enhanced abundances of PyH+ than typical experiments with electrospray on (Figure S3). In this regard, we postulate that the desorbed BP salts experience an ionization mechanism of both electrospray post-ionization and direct ionization. Under the typical IR-MALDESI conditions, a fraction of the BP salts go through electrospray post-ionization which are cooled down by the electrospray droplets through vibrational relaxation within 10−14 - 10−12 seconds [32], only decomposing into substituted benzyl ions. Nonetheless, the other BP salts which form ions through direct ionization by charge splitting do not partition into ESI droplets and thereby enter the mass spectrometer retaining the excess IE. In this case, the excited BP cations have two competitive fragmentation channels producing either benzyl ions or protonated pyridines. Given the experimental observations that when the electrospray is off, PyH+ was always more abundant than the total benzyl fragment ions (Figure S4), and the survival yields calculated only with benzyl ions displayed significantly slower declines as collision energy increases (Figure S5), we conclude that the PyH+ fragmentation pathway proceeds at faster reaction rates than the loss of neutral pyridine, and becomes dominantly favorable at sufficiently high collision energies.
Although the pyridine feature is observed, it was excluded from the calculation of survival yields for two reasons. Foremost, our previous work demonstrated that the majority of the laser-ablated materials from biological samples are neutral and mainly ionized by electrospray [8], so the direct ionization pathway which is specific to preformed ions falls outside the range of our current focus. Moreover, including PyH+ requires injecting each BP salt separately because it is a common fragment for all benzyl-pyridinium structured ions, which not only is time-consuming, but also introduces repeatability issues [33]. Table S2 shows the similar IE distributions of BP cations with and without electrospray at IS-CID ≤ 20 eV, suggesting that the direct ionization pathway has negligible effects on the measurements of IE distributions of ESI-formed ions at this energy range. Therefore, comparing survival yields and IE distributions without including PyH+ as a fragment ion at a moderate IS-CID energy (e.g., 10-20 eV) is appropriate for our current study.
Figure 3a displays the survival yields of five BP cations versus IS-CID energies for sprayed slides. The results clearly show that only a minor fraction of para-MeO substituents dissociate at 0 eV collision energy, and the survival yields are not statistically different between IR-MALDESI and ESI at a significance level of 0.05. It is therefore reasonable to believe that IR-MALDESI mainly gives precursor ions under routine experimental conditions (i.e., collision energy = 0). A representative comparison of sigmoidal curves and IE depositions at 15 eV between ESI and IR-MALDESI is displayed in Figure 3c and 3d, and more results can be found in Table 2. The strongly similar mean IE values (e.g., 1.86 eV for IR-MALDESI and 1.91 eV for ESI at IS-CID of 15 eV) and distribution peak widths indicate IR-MALDESI of sprayed slides deposits IE into analytes comparable to ESI. No significant difference in IE distribution was found between experiments of sprayed glass slides with 1 and 2 laser pulses on each location (Table 2), suggesting the influence of cumulated laser exposure on the ion IE is negligible. Relatively smaller mean IE values and narrower distributions were observed for more complex matrices such as animal tissues, e.g. 1.76 and 0.33 eV for mean IE and FWHM at IS-CID of 15 eV, respectively, and the differences became more apparent at high IS-CID energies (Figure 3b, 3d and Table 2). The lower degree of fragmentation may be due to the significantly higher sample density of animal tissues so that less energy is imparted into each molecule, preserving more M+.
Figure 3.
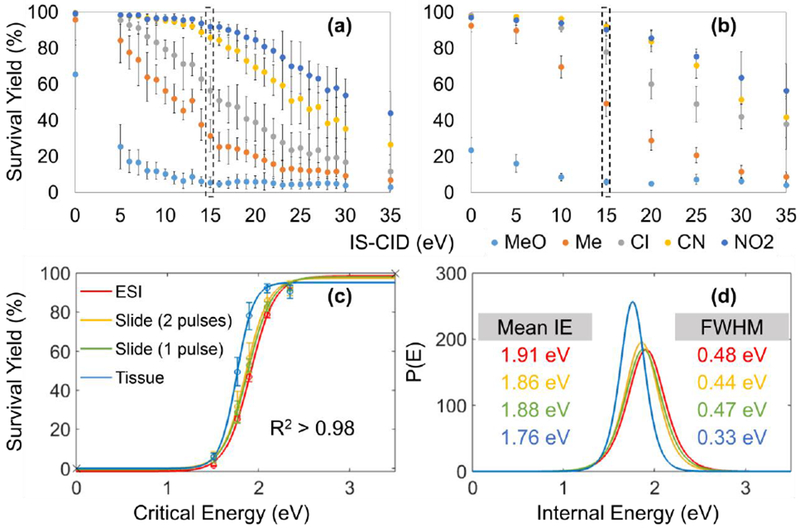
Survival yields of five BP cations versus IS-CID energies for IR-MALDESI analysis of (a) sprayed slides and (b) tissue sections on sprayed slides. The comparison of (c) Boltzmann’s sigmoidal curves and (d) IE distributions between ESI and IR-MALDESI was made at 15 eV IS-CID.
The Effect of an Ice Matrix on IE Deposition in IR-MALDESI
To assess the influence of an ice matrix on ion IE input, BP salts air-dried on the surface of rat liver sections were analyzed by IR-MALDESI-MS both with and without an ice matrix. The wavelength dependence was also evaluated by repeating the measurements with two other mid IR-wavelengths (2700 and 3100 nm). The comparison of M+ survival yields with and without an ice matrix is displayed in Figure 4. The representative survival yield sigmoidal curves and IE distributions at IS-CID of 20 eV are shown in Figure 5, and results at other collision energies are summarized in Table 3. As shown in these results, ions produced with an ice matrix consistently exhibit lower survival yields and as a consequence higher mean IE values than their counterparts without an ice matrix. The differences in mean IE values approximately range from approximately 0.1 to 0.3 eV and become more pronounced at higher collision energies. Different laser wavelengths show inconsequential differences in IE distribution (Table S3), which is in accordance with our previous findings [34].
Figure 4.
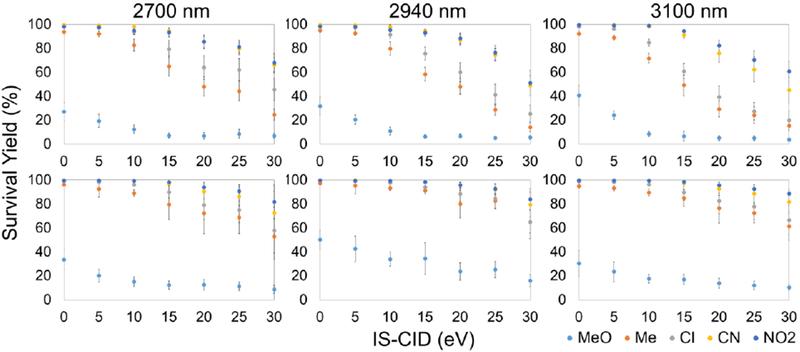
Survival yields of BP cations from tissue sections are plotted as a function of IS-CID energies for IR-MALDESI analysis either with (top) and without an ice matrix (bottom) at laser wavelengths of 2700, 2940 and 3100 nm.
Figure 5.
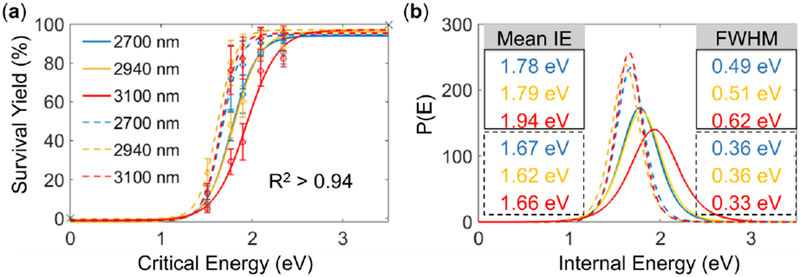
(a) Sigmoidal curves and (b) IE distributions for IR-MALDESI analysis either with (solid lines) or without the ice matrix (dashed lines) at IS-CID of 20 eV. The mean IE and FWHM values for IR-MALDESI with and without the ice matrix are solid- and dashed-outlined, respectively.
Table 3.
Internal energy distribution of BP Cations for IR-MALDESI with and without the ice matrix at an incident wavelength of 2940 nm.
| With Ice | Without Ice | |||||
|---|---|---|---|---|---|---|
| IS-CID (eV) | R2 | Mean IE [CI] | FWHM | R2 | Mean IE [CI] | FWHM |
| 10 | 0.994 | 1.66 [1.66, 1.67] | 0.27 | 0.996 | 1.56 [1.56, 1.57] | 0.28 |
| 15 | 0.988 | 1.74 [1.73, 1.74] | 0.37 | 0.993 | 1.57 [1.56, 1.57] | 0.30 |
| 20 | 0.977 | 1.79 [1.78, 1.80] | 0.51 | 0.975 | 1.62 [1.61, 1.63] | 0.36 |
| 25 | 0.964 | 1.93 [1.91, 1.95] | 0.67 | 0.972 | 1.61 [1.59, 1.62] | 0.35 |
R2 stands for the regression coefficient of the sigmoidal fit. CI represents 95% confidence interval on the mean.
Overall, the results suggest that performing tissue imaging with an externally deposited layer of ice induces more laser energy into the system. This observation can be plausibly explained by a considerably larger number of IR absorbers when the ice layer is formed absorbing more laser energy, which is consistent with previous IR-LDI work showing the significant effect of water content on ion yields [35]. Albeit it is ideal to have IE input as low as possible for biological samples to reduce the complexity of the mass spectra, the use of an ice matrix will still be suggested in most cases to: (1) avoid signal variation originating from varying contents of endogenous water in heterogeneous samples [36]; (2) prevent sublimation of water [37, 38]; (3) preserve sample structures and eliminate molecular degradation throughout the imaging process [34, 39]; and (4) enhance detection sensitivity [6]. Besides, the effect of increased fragmentation can be less detectable for relatively large biomolecules that undergo collisional cooling during integration into the electrospray droplets. However, performing measurements in a relatively small region without applying an ice matrix may benefit the detection of labile analytes by reducing the dissociation rate without introducing consequential biases.
Analysis of noncovalent protein complex
The softness of IR-MALDESI was further probed by performing measurements on heme-myoglobin complex (holo-Mb, MW ~17.6 kDa). In its native state, a heme prosthetic group is noncovalently bound to the polypeptide chain(s) [40] with a binding energy reported to be 0.7 to 1.0 eV [41]. The Mb which loses the heme moiety is referred to as apo-myoglobin (apo-Mb, MW ~16.9 kDa). Previous measurements have shown both intact holo-Mb and apo-Mb features using laser desorption-based mass spectrometry including IR-MALDI [37, 42], LEMS [43, 44] and electrospray-assisted laser desorption/ionization (ELDI) [45]. In this work, Mb working solutions were prepared with either a buffer solution of 70:30 (v/v) 5 mM ammonium acetate/methanol at pH 5.5, or an unbuffered solution of 1 mM formic acid in 50:50 (v/v) water/methanol which is typically used in IR-MALDESI-MS. The electrospray solvent employed in IR-MALDESI experiments was matched with the sample dissolving solvent to achieve more valid comparisons.
With the use of the buffer solution, the dominant species present in ESI and IR-MALDESI spectra correspond to holo-Mb with highly similar charge state distributions, displaying the average charge state (ACS) of 8.2 for ESI and 8.1 for IR-MALDESI (Figure 6a and 6b). This reveals the similarity in ionization mechanisms of those two ionization techniques. Apo-Mb peaks with low abundances in higher charge states +7 to +18 were observed in the IR-MALDESI spectrum as well, which might be a result of energy-induced partial cleavage of the noncovalent bond during the desorption process. The survival yield of noncovalent protein complex which is defined as Σ Iholo/(Σ Iholo + Σ Iapo) achieves ~70%, where Iholo and Iapo represent the abundances of central isotopic holo- and apo-Mb ions, respectively, implying the laser-induced desorption is gentle and does not deposit considerable energy into analytes. Sodiated Mb ions were present in both mass spectra (inserts in Figure 6a and 6b), yielding a tailing of the signals on the higher m/z side.
Figure 6.
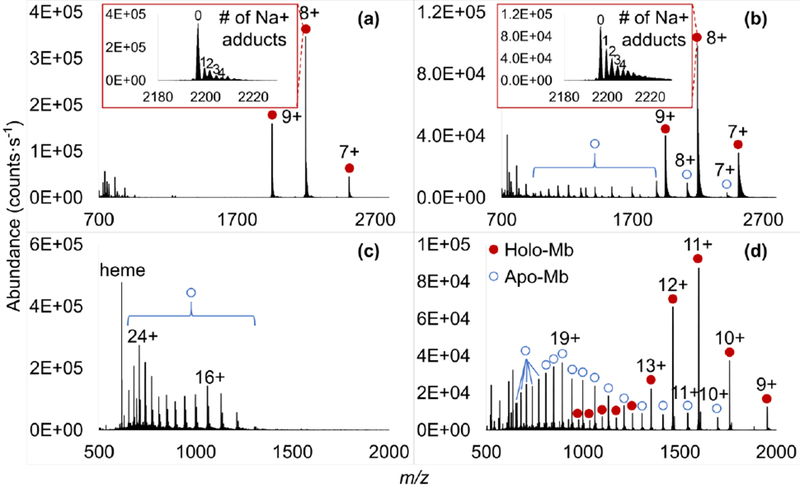
ESI and IR-MALDESI mass spectra of myoglobin in 70:30 (v/v) 5 mM ammonium acetate/ methanol (a and b), and 1 mM formic acid in 50:50 (v/v) water/methanol (c and d), respectively. Inserts: expanded spectra of m/z 2180-2230 show sodiated holo-Mb ions in charge state 8+.
Under the unbuffered condition, direct infusion ESI only generated ions corresponding to apo-Mb in high charge states +14 to +26, and a strong signal at m/z 616.1766 corresponding to Fe (III)-heme [40] was readily observed (Figure 6c). The result is in agreement with the known instability of Mb in denaturing ESI-MS [46–48]. Interestingly, the IR-MALDESI spectrum contains abundant holo-Mb peaks in charge states +9 to +20 with +11 the most abundant, together with some apo-Mb species in charge states +9 to +26, obtaining a survival yield of noncovalent protein complex of ~45% (Figure 6d). The significant differences in detected species between these two techniques could be attributed to electrochemical oxidation reactions such as 2H2O → O2 + 4H+ + 4e− [49] occurring in the electrospray emitter which decreases the pH of unbuffered solution dramatically, in some cases down to pH 1.4 [50]. Myoglobin for direct infusion ESI stayed in the acidic condition for a longer time, inducing denaturation of the protein thus the expulsion of the heme group along with more complete protonation [51]. The denaturing solvent likewise shifted holo-Mb ions in IR-MALDESI toward higher charge states with ACS of 12.0. Comparative IR-MALDESI experiments were further carried out on Mb dissolved in 50:50 water/methanol or pure water while the electrospray solvent was kept as 1 mM formic acid in 50:50 water/methanol. It can be seen in Figure S6 that IR-MALDESI from liquid samples with reduced severity of denaturing conditions retained up to 90% intact protein complexes, suggesting that the dissociation of holo-Mb into apo-Mb and heme group in IR-MALDESI primarily originated from the denaturing solvent instead of the desorption process.
As a summary of our discussion, the key energy-transfer events occurring during the laser-induced desorption and following ionization processes in IR-MALDESI are illustrated in Figure 7. IR-MALDESI is overall a multistep technique with overwhelmingly complex physical and chemical events happening during the formation of ions and contributing to ion IE content in different ways. Analyte activation can occur due to laser irradiation, whereas processes such as plume expansions [20, 52], collisional cooling at atmospheric pressure condition [53, 54] and integration into electrospray droplets [34] can lead to deactivation.
Figure 7.
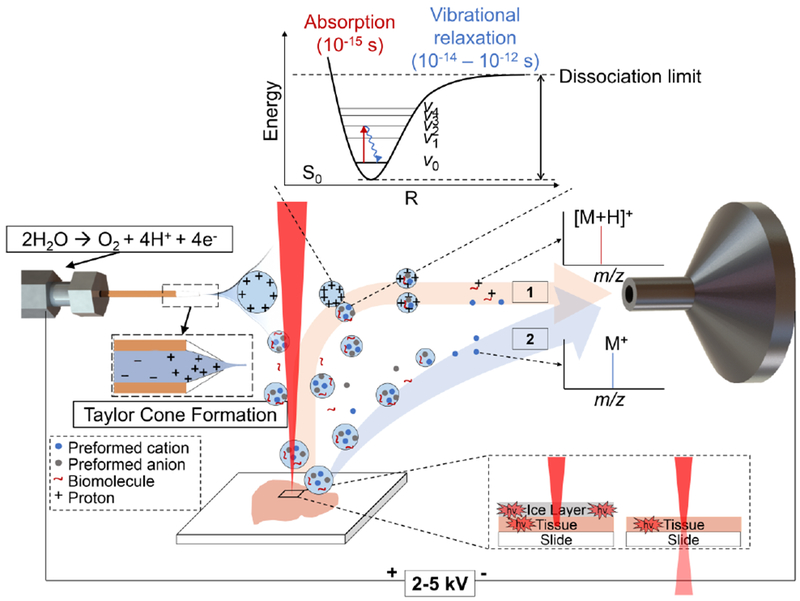
The diagram graphically exhibits the key energy transfer and ionization processes underlying positive mode-IR-MALDESI proposed at the current stage. The process starts with the IR-laser absorption solely by inherent water molecules in biological samples, or combinedly by inherent water and exogenous ice with higher energy intake. Then the surface evaporation and phase explosions result in a primary material expulsion, followed by a recoil-induced secondary material ejection [55, 56]. The ejected biomolecules in droplets or particulates are subsequently captured by an orthogonally oriented electrospray plume and lose their internal energy on the picosecond timescale via vibrational relaxation, producing analyte-containing charged droplets. Gas-phase protonated ions are eventually formed following an ESI-like mechanism (denoted by 1 in the diagram) with internal energy indistinguishable from ions generated by conventional ESI. Pre-charged species in the form of salts proceed through either electrospray post-ionization or direct ionization by disintegrating their counter ions (denoted by 2 in the diagram). These directly-formed ions preserve the internal energy input during laser-induced desorption process and thus may dissociate more drastically and in uncommon ways.
Conclusions
The survival yield measurements show that IR-MALDESI produces para-substituted BP cations with IE distributions comparable to those obtained with a conventional ESI. The results with Mb demonstrate that IR-MALDESI not only generates ESI-like intact multiply charged protein ions under native-like conditions, but also maintains the native structure of the weakly-bound noncovalent complexes under harsh solvent conditions, offering superior performance over ESI-MS which only reflects dissociated species in this case. These findings provide reasonable evidences that an ionization mechanism in analogy to ESI occurs at least in the later stages, and analytes do not experience significant energy gain in IR-MALDESI source so that little or no dissociation can be expected. The uniformly deposited ice layer is a harder energy-absorbing matrix than the endogenous water in animal tissues, leading to higher IE build-up and consequently inducing more fragmentation. A larger amount of IR absorbers with the use of ice matrix giving rise to a stronger energy absorption may explain this observation. A trade-off for labile species between a “softer” analysis and sample preservation thus should be fully considered when choosing the matrix.
Supplementary Material
Acknowledgements
All mass spectrometry measurements were carried out in the Molecular Education, Technology, and Research Innovation Center (METRIC) at North Carolina State University. The authors gratefully acknowledge the financial support received from the National Institutes of Health (R01GM087964) and North Carolina State University.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Disclosures
The authors declare no competing financial interests.
References
- 1.Loziuk P, Meier F, Johnson C, Ghashghaei HT, Muddiman DC: TransOmic analysis of forebrain sections in Sp2 conditional knockout embryonic mice using IR-MALDESI imaging of lipids and LC-MS/MS label-free proteomics. Anal. Bioanal. Chem 408, 3453–3474 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meier F, Garrard KP, Muddiman DC: Silver dopants for targeted and untargeted direct analysis of unsaturated lipids via infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI). Rapid Commun. Mass Spectrom 28, 2461–2470 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nazari M, Muddiman DC: Polarity switching mass spectrometry imaging of healthy and cancerous hen ovarian tissue sections by infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI). Analyst. 141, 595–605 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nazari M, Bokhart MT, Loziuk PL, Muddiman D: Quantitative mass spectrometry imaging of glutathione in healthy and cancerous hen ovarian tissue sections by infrared matrix- assisted laser desorption electrospray ionization (IR-MALDESI). Analyst. 143, 654–661 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sampson JS, Hawkridge AM, Muddiman DC: Generation and detection of multiply-charged peptides and proteins by matrix-assisted laser desorption electrospray ionization (MALDESI) Fourier transform ion cyclotron resonance mass spectrometry. J. Am. Soc. Mass Spectrom 17, 1712–1716 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Barry JA, Robichaud G, Bokhart MT, Thompson C, Sykes C, Kashuba ADM, Muddiman DC: Mapping antiretroviral drugs in tissue by IR-MALDESI MSI coupled to the Q Exactive and comparison with LC-MS/MS SRM assay. J. Am. Soc. Mass Spectrom 25, 2038–2047 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bokhart MT, Rosen E, Thompson C, Sykes C, Kashuba ADM, Muddiman DC: Quantitative mass spectrometry imaging of emtricitabine in cervical tissue model using infrared matrix-assisted laser desorption electrospray ionization. Anal. Bioanal. Chem 407, 2073–2084 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixon RB, Muddiman DC: Study of the ionization mechanism in hybrid laser based desorption techniques. Analyst. 135, 880–882 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Sampson JS, Hawkridge AM, Muddiman DC: Development and characterization of an ionization technique for analysis of biological macromolecules: liquid matrix-assisted laser desorption electrospray ionization. Anal. Chem 80, 6773–6778 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schulz E, Karas M, Rosu F, Gabelica V: Influence of the matrix on analyte fragmentation in atmospheric pressure MALDI. J. Am. Soc. Mass Spectrom 17, 1005–1013 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Moskovets E: Ghost peaks observed after atmospheric pressure matrix-assisted laser desorption/ionization experiments may disclose new ionization mechanism of matrix-assisted hypersonic velocity impact ionization. Rapid Commun. Mass Spectrom 29, 1501–1512 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robichaud G, Barry JA, Muddiman DC: IR-MALDESI mass spectrometry imaging of biological tissue sections using ice as a matrix. J. Am. Soc. Mass Spectrom 25, 319–328 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gabellca V, De Pauw E: Internal energy and fragmentation of ions produced in electrospray sources. Mass Spectrom. Rev 24, 566–587 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Gabelica V, De Pauw E, Karas M: Influence of the capillary temperature and the source pressure on the internal energy distribution of electrosprayed ions. Int. J. Mass Spectrom 231, 189–195 (2004). [Google Scholar]
- 15.Harris GA, Hostetler DM, Hampton CY, Fernández FM: Comparison of the internal energy deposition of direct analysis in real time and electrospray ionization time-of-flight mass spectrometry. J. Am. Soc. Mass Spectrom 21, 855–863 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Luo G, Marginean I, Vertes A: Internal energy of ions generated by matrix-assisted laser desorption/ionization. Anal. Chem 74, 6185–6190 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Gabelica V, Schulz E, Karas M: Internal energy build-up in matrix-assisted laser desorption/ionization. J. Mass Spectrom 39, 579–593 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Nefliu M, Smith JN, Venter A, Cooks RG: Internal energy distributions in desorption electrospray ionization (DESI). J. Am. Soc. Mass Spectrom 19, 420–427 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Badu-Tawiah A, Bland C, Campbell DI, Cooks RG: Non-aqueous spray solvents and solubility effects in desorption electrospray ionization. J. Am. Soc. Mass Spectrom 21, 572–579 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Greisch JF, Gabelica V, Remacle F, De Pauw E: Thermometer ions for matrix-enhanced laser desorption/ionization internal energy calibration. Rapid Commun. Mass Spectrom 17, 1847–1854 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Khodjaniyazova S, Nazari M, Garrard KP, Matos MPV, Jackson GP, Muddiman DC: Characterization of the spectral accuracy of an orbitrap mass analyzer using isotope ratio mass spectrometry. Anal. Chem 90, 1897–1906 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Robichaud G, Barry JA, Garrard KP, Muddiman DC: Infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI) imaging source coupled to a FT-ICR mass spectrometer. J. Am. Soc. Mass Spectrom 24, 92–100 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kessner D, Chambers M, Burke R, Agus D, Mallick P: ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics. 24, 2534–2536 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Race AM, Styles IB, Bunch J: Inclusive sharing of mass spectrometry imaging data requires a converter for all. J. Proteomics 75, 5111–5112 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Robichaud G, Garrard KP, Barry JA, Muddiman DC: MSiReader: An open-source interface to view and analyze high resolving power MS imaging files on matlab platform. J. Am. Soc. Mass Spectrom 24, 718–721 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bokhart MT, Nazari M, Garrard KP, Muddiman DC: MSiReader v1.0: Evolving open-source mass spectrometry imaging software for targeted and untargeted analyses. J. Am. Soc. Mass Spectrom 29, 8–16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brendle K, Kordel M, Schneider E, Wagner D, Bräse S, Weis P, Kappes MM: Collision induced dissociation of benzylpyridinium-substituted porphyrins: Towards a thermometer scale for multiply charged ions? J. Am. Soc. Mass Spectrom 29, 382–392 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Hampton CY, Silvestri CJ, Forbes TP, Varady MJ, Meacham JM, Fedorov AG, Degertekin FL, Fernández FM: Comparison of the internal energy deposition of Venturi-assisted electrospray ionization and a Venturi-assisted array of micromachined ultrasonic electrosprays (AMUSE). J. Am. Soc. Mass Spectrom 19, 1320–1329 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nemes P, Huang H, Vertes A: Internal energy deposition and ion fragmentation in atmospheric-pressure mid-infrared laser ablation electrospray ionization. Phys. Chem. Chem. Phys 14, 2501–2507 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Collette C, De Pauw E: Calibration of the internal energy distribution of ions produced by electrospray. Rapid Commun. Mass Spectrom 12, 165–170 (1998). [Google Scholar]
- 31.Flanigan PM, Shi F, Archer JJ, Levis RJ: Internal energy deposition for low energy, femtosecond laser vaporization and nanospray post-ionization mass spectrometry using thermometer ions. J. Am. Soc. Mass Spectrom 26, 716–724 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Omary MA, Patterson HH: Luminescence, Theory. Encycl. Spectrosc. Spectrom 636–653 (2017). [Google Scholar]
- 33.Tu A, Muddiman DC: Systematic evaluation of repeatability of IR-MALDESI-MS and normalization strategies for correcting the analytical variation and improving image quality. Anal. Bioanal. Chem 10.1007/s00216-019-01953-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosen EP, Bokhart MT, Ghashghaei HT, Muddiman DC: Influence of desorption conditions on analyte sensitivity and internal energy in discrete tissue or whole body imaging by IR-MALDESI. J. Am. Soc. Mass Spectrom 26, 899–910 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dreisewerd K, Draude F, Kruppe S, Rohlfing A, Berkenkamp S, Pohlentz G: Molecular analysis of native tissue and whole oils by infrared laser mass spectrometry. Anal. Chem 79, 4514–4520 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Nazari M, Bokhart MT, Muddiman DC: Whole-body mass spectrometry imaging by infrared matrix-assisted laser desorption electrospray ionization (IR-MALDESI). J. Vis. Exp 109, e53942 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pirkl A, Soltwisch J, Draude F, Dreisewerd K: Infrared matrix-assisted laser desorption/ionization orthogonal-time-of-flight mass spectrometry employing a cooling stage and water ice as a matrix. Anal. Chem 84, 5669–5676 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Von Seggern CE, Gardner BD, Cotter RJ: Infrared atmospheric pressure MALDI ion trap mass spectrometry of frozen samples using a peltier-cooled sample stage. Anal. Chem. 76, 5887–5893 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Goodwin RJA, Iverson SL, Andren PE: The significance of ambient-temperature on pharmaceutical and endogenous compound abundance and distribution in tissues sections when analyzed by matrix-assisted laser desorption/ionization mass spectrometry imaging. Rapid Commun. Mass Spectrom 26, 494–498 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Li Y-T, Hsieh Y-L, Henion JD, Ganem B: Studies on heme binding in myoglobin, hemoglobin, and cytochrome c by ion spray mass spectrometry. J. Am. Soc. Mass Spectrom 4, 631–637 (1993). [DOI] [PubMed] [Google Scholar]
- 41.Chen Y-L, Campbell JM, Collings BA, Konermann L, Douglas DJ: Stability of a highly charged noncovalent complex in the gas phase: Holomyoglobin. Rapid Commun. Mass Spectrom 12, 1003–1010 (1998). [DOI] [PubMed] [Google Scholar]
- 42.Zhang W, Niu S, Chait BT: Exploring infrared wavelength matrix- assisted laser desorption/ionization of proteins with delayed-extraction time-of-flight mass spectrometry. 305, (1998). [DOI] [PubMed] [Google Scholar]
- 43.Karki S, Sistani H, Archer JJ, Shi F, Levis RJ: Isolating Protein Charge State Reduction in Electrospray Droplets Using Femtosecond Laser Vaporization. J. Am. Soc. Mass Spectrom 28, 470–478 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Karki S, Flanigan PM, Perez JJ, Archer JJ, Levis RJ: Increasing protein charge state when using laser electrospray mass spectrometry. J. Am. Soc. Mass Spectrom 26, 706–715 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Shiea J, Yuan CH, Huang MZ, Cheng SC, Ma YL, Tseng WL, Chang HC, Hung WC: Detection of native protein ions in aqueous solution under ambient conditions by electrospray laser desorption/ionization mass spectrometry. Anal. Chem 80, 4845–4852 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Loo JA, Giordani AB, Muenster H: Observation of intact (heme- bound) myoglobin by electrospray ionization on a double- focusing mass spectrometer. Rapid Commun. Mass Spectrom 7, 186–189 (1993). [Google Scholar]
- 47.Brenner-Weiss G, Kirschhöfer F, Kühl B, Nusser M, Obst U: Analysis of non-covalent protein complexes by capillary electrophoresis-time-of-flight mass spectrometry. J. Chromatogr. A 1009, 147–153 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Mortensen DN, Williams ER: Investigating protein folding and unfolding in electrospray nanodrops upon rapid mixing using theta-glass emitters. Anal. Chem 87, 1281–1287 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Berkel GJ, Kertesz V: Using the electrochemistry of the electrospray ion source. Anal. Chem 79, 5510–5520 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Konermann L: Addressing a common misconception: Ammonium acetate as neutral pH “buffer” for native electrospray mass spectrometry. J. Am. Soc. Mass Spectrom 28, 1827–1835 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Katta V, Chait BT: Observation of the heme-globin complex in native myoglobin by electrospray-ionization mass spectrometry. J. Am. Chem. Soc 113, 8534–8535 (1991). [Google Scholar]
- 52.Bae YJ, Moon JH, Kim MS: Expansion cooling in the matrix plume is under-recognized in MALDI mass spectrometry. J. Am. Soc. Mass Spectrom 22, 1070–1078 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Van Berkel GJ, Pasilis SP, Ovchinnikova O: Established and emerging atmospheric pressure surface sampling/ionization techniques for mass spectrometry. J. Mass Spectrom 43, 1161–1180 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Li G, Cao Q, Liu Y, DeLaney K, Tian Z, Moskovets E, Li L: Characterizing and alleviating ion suppression effects in atmospheric pressure matrix-assisted laser desorption/ionization. 33, 327–335 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Z, Vertes A: Early plume expansion in atmospheric pressure midinfrared laser ablation of water-rich targets. Phys. Rev. E Stat. Nonlin. Soft Matter Phys 77, 36316 (2008). [DOI] [PubMed] [Google Scholar]
- 56.Apitz I, Vogel A: Material ejection in nanosecond Er:YAG laser ablation of water, liver, and skin. Appl. Phys. A 81, 329–338 (2005). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


