Abstract
The Staphylococcus aureus Extracellular Adherence Protein (Eap) and its homologs, EapH1 and EapH2, are a family of secreted proteins that potently inhibit the neutrophil serine proteases Neutrophil Elastase (hNE), Cathepsin G, and Proteinase 3. Similarly to EapH1, inhibition of hNE by EapH2 is characterized by a rapid association rate (2.9×105 M−1s−1) coupled with a very slow dissociation rate (5.9×10−4 s−1), yielding an apparent inhibition constant of 2.11 nM. As with EapH1, inhibition of hNE by EapH2 is also time-dependent in character. A phenylalanine in EapH2 replaces the leucine in EapH1 that sits over the hNE catalytic serine and creates a potential steric clash. Indeed, the EapH1 L59F mutant is severely decreased in its ability to inhibit hNE (~9,500-fold). When compared to the EapH1:hNE co-crystal structure, a model of the EapH2:hNE complex predicts an alternative binding motif comprised of EapH2 residues 120 – 127. These putative interfacing residues were individually mutated and kinetically interrogated. The EapH2 N127A mutant resulted in the largest decrease in hNE inhibition (~200-fold) and loss of the time-dependent characteristic. Surprisingly, the time-dependent characteristic was still abolished in the EapH2 T125A mutant, even though it was less perturbed in hNE inhibition (~25-fold). T125 forms an intra-molecular hydrogen bond to the carbonyl oxygen of N127 in the EapH2 crystal structure. Given these observations, we conclude (i) that EapH2 has an altogether distinct hNE binding motif than EapH1, (ii) that N127 is the main functional determinant in EapH2, and (iii) that T125 serves an ancillary role aiding in the optimal orientation of N127.
Keywords: EapH1, EapH2, surface plasmon resonance, time-dependent inhibition, neutrophil, elastase
Introduction
Staphylococcus aureus is a Gram-positive, commensal bacterium with a human carriage rate estimated at greater than ~30 per cent. 6, 7 Infections with S. aureus are diverse in their severity, and range from superficial skin abscesses to potentially life-threatening diseases, including sepsis, endocarditis, and necrotizing pneumonia. 8–10 Upon coming into contact with the bloodstream, staphylococcal cells are rapidly opsonized by immunoglobulins and fragments of complement proteins. Opsonization by complement proteins, along with various bacterially-derived biochemical cues such as N-formylated peptides and lipoteichoic acid, provides potent chemoattractive stimuli for recruitment of neutrophils toward S. aureus cells. Once the opsonized bacteria are taken into the neutrophil through the process of receptor-mediated phagocytosis, the newly created phagosome fuses rapidly with granular compartments that are abundant in the neutrophil cytosol.11, 12 The most prominent of these are the so-called azurophilic granules, which are characterized by high concentrations of the bactericidal enzymes myeloperoxidase, 13 as well as the serine proteases Neutrophil Elastase 14, Cathepsin G 15, and Proteinase 3. 16 Hereafter, the combined activities of these anti-bacterial systems provides for efficient killing of engulfed S. aureus cells. 11, 12, 17
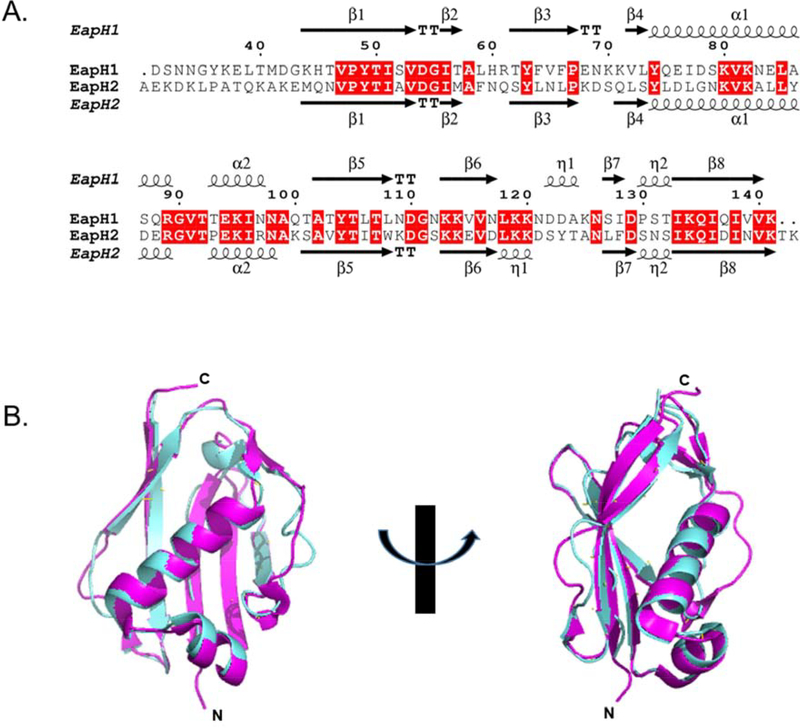
Recent work has shown that S. aureus defends itself against phagosomal destruction by secreting an array of inhibitory proteins that target the bactericidal enzymes found within neutrophils 18. One such family of these secreted inhibitors are the “Eap domain proteins” 1, which consists of the 50 – 70 kDa Extracellular Adherence Protein (i.e. Eap), as well as its two homologs EapH1 (UniprotKB: A0A0H3K0M1) and EapH2 (UniprotKB: A0A0H3JUK5). Eap domain proteins specifically target the serine proteases found in neutrophil granules. The structures of EapH11, 19, 20 and EapH2 1, 21 have been solved by X-ray crystallography and also characterized by solution-state NMR spectroscopy. Both proteins adopt a β-grasp fold consisting of an α-helix superimposed on a five-stranded mixed β-sheet. 16 EapH1 and EapH2 share an amino acid sequence similarity of 64.4 % (Fig. 1A). 4 Consistent with this, structural superimposition of EapH2 onto EapH1 yields an RMSD of 0.634 Å for all backbone atoms (Fig. 1B) and underscores the high level of physical identity between these two molecules. 5 With such an extensive level homology between EapH1 and EapH2, one would expect these two proteins to behave in a similar manner. Surprisingly, a recent study testing this hypothesis suggests that this might not be the case. 22
Figure 1. Alignment of EapH1 and EapH2.
(A) Primary sequence alignment generated with the EMBL-EBI Clustal Omega suite. 4 Conserved residues shaded in red. Residue numbering is based on the EapH1 sequence. (B) Superimposition of EapH1 (PDB: 1YN4 1,cyan) and EapH2 (PDB: 1YN5 1, magenta) generated with PyMol. 5
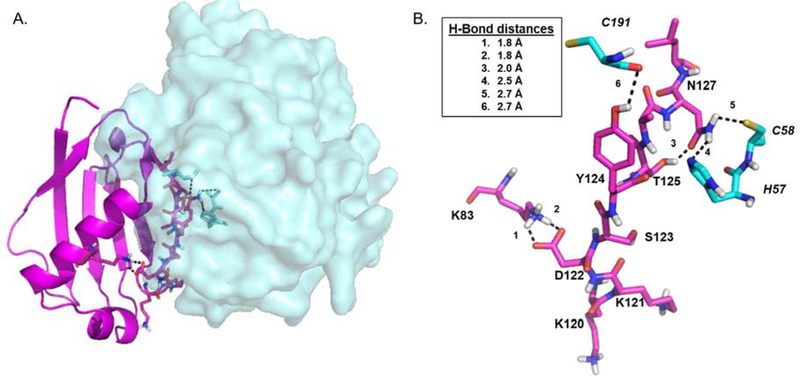
Using the EapH1:hNE co-crystal structure as a guide 2, conservative mutations of the EapH1 residues found at the NE interface (R89, E94, K95) were prepared and rigorously evaluated. From this, EapH1 R89 was identified as the major contributor to the time-dependent inhibition of human neutrophil elastase (hNE, UniProtKB: P08246). 3 These residues are conserved in EapH2 (Fig. 1A). However, when they were collectively mutated to alanine, only a minimal effect on the binding of EapH2 to hNE was observed (~2-fold). 22 These data were the first to suggest that EapH2 recognizes hNE through an alternative binding mode than EapH1. To provide a basis for exploring this hypothesis, an EapH2:hNE model was created via protein docking (Fig. 2). 22 This model suggested a different hNE binding region on EapH2 consisting of residues 120 – 127 when compared to EapH1 (Fig. S1). Indeed, the affinity for hNE was dramatically reduced (~13,000-fold) when these residues were collectively mutated to alanine. 22
Figure 2. Energy minimized model of S. aureus EapH2:hNE.
(A) overall view of EapH2 (magenta, depicted as cartoon):hNE (cyan, depicted as space-fill). (B) Stick representation of EapH2 interface residues (magenta, normal font) and hNE residues (cyan, italic font). Potentially important Hydrogen bonds shown as black dashes. Inset: Hydrogen bond distances.
In this study, we explored the prediction and nature of the alternative hNE binding motif of EapH2 in comparison to EapH1. Considering the EapH1 residue responsible for the potent inhibition of hNE (R89) is conserved in EapH2, the corresponding residue (R90, Fig. 1A) was conservatively mutated and kinetically evaluated. The EapH2 R90M mutant inhibited hNE similar to the WT protein. In the co-crystal structure of EapH1:hNE (PDB: 4NZL 2), EapH1 L59 sits over the hNE active site serine. In EapH2, the leucine is replaced with a phenylalanine (Fig. 1A). The EapH1 L59F mutant was therefore created and the effect on hNE inhibition was determined. The EapH1 L59F mutant displayed a significant inflation in the apparent inhibition constant and loss of time-dependent characteristic. Finally, the putative “new” binding region in EapH2 was interrogated by individually mutating the putative interfacing residues (K120 – N127) to evaluate their contribution to hNE inhibition. Of these residues, two mutants (T125A and N127A) lost the time-dependent inhibition characteristic with N127A being the most perturbed. Collectively, these experiments confirm an alternative hNE-binding mode for EapH2 with N127 as the major contributor to hNE inhibition as well as being the source of the time-dependent characteristic. We further propose that the role of EapH2 T125 is to optimally orient N127 for interaction with hNE.
Materials and Methods
Materials
Kanamycin monosulfate, isopropyl-β-D-thiogalactopyranoside (IPTG), tris(hydroxymethyl)aminomethane (TRIS) and Nickel-IDA agarose were purchased from Gold Biotechnology. Methoxysuccinyl-Ala-Ala-Pro-Val-P-nitroanilide (MSA2PV-pNA), Triton X-100, dimethylsulfoxide (DMSO), succinic acid and Luria Broth powder were purchased from Sigma-Aldrich. Nickel (II) Chloride-hexahydrate was purchased from Acros Organics. Imidazole was purchased from Alfa Aesar. 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) was purchased from MP Biomedical, LLC. Human neutrophil elastase was purchased from Elastin Products Company. Mutagenic oligonucleotides were purchased from Integrated DNA Technologies. Phusion High-Fidelity DNA Polymerase, dNTPs and Dpn I were purchased from New England Biolabs. Plasmid DNA was purified using the Wizard Plus SV Miniprep DNA Purification System (Promega).
Mutagenesis, protein expression and purification
Mutagenesis was accomplished using a full-circle site-directed PCR strategy. The integrity of the coding sequence and the presence of the mutation was verified by DNA sequencing (GENEWIZ). Due to the presence of repeated primer insertions during the PCR, three of the EapH2 mutations (K120M, D122A, and T125A) were prepared from synthetic gene fragments specific for each set of mutations and cloned into the pT7HMT expression plasmid (GenScript USA Inc.). 23
The wild-type and mutant EapH2 proteins were expressed in E. coli BL21 (λDE3) and purified as previously described. 3 The concentrations of wild-type/mutant EapH1 and EapH2 proteins were determined spectrophotometrically (DS-11 Spectrophotometer, DeNovix) using an extinction coefficient calculated from the deduced protein composition (EapH1: ε280 = 7.45 mM−1 cm−1, EapH2: ε280 = 14.44 mM−1 cm−1). Extinction coefficients were determined using the ExPASy suite ProtParam. 24
Preparation of human neutrophil elastase and substrate
Lyophilized human neutrophil elastase (1 mg) was dissolved in 200 μL 20 mM Sodium Acetate/50 % (v/v) glycerol (pH: 5.0, RT). The concentration of the dissolved elastase was determined using the deduced protein composition (ε280 = 20.23 mM−1 cm−1). The enzyme (10 μL aliquots) was stored at −20 °C until use. The substrate, Methoxysuccinyl-Ala-Ala-Pro-Val-p-nitroanalide (MSA2PV-pNA), was initially dissolved in 100% (v/v) DMSO and then diluted to a working concentration with 50 mM HEPES/140 mM NaCl/0.05 % (v/v) Triton X-100 (pH: 7.4, RT) and quantified by end-point assays (ε400 = 12.3 mM−1 cm−1).
Steady-state kinetic analysis of human neutrophil elastase
The steady-state kinetic constants for human neutrophil elastase have been previously determined.3 Briefly, initial velocities of the elastase (4 nM) reaction were measured spectrophotometrically at 400 nm in a plate-based assay using a Versamax tunable microplate reader (Molecular Devices). These data (n = 9) were fit to the Michaelis-Menten equation to determine the kcat and Km (SigmaPlot 10.0/Enzyme Kinetics Module 1.3, Systat Software, Inc.). The Km determined from the plate assay was used in the script for progress curve analysis. The reaction buffer contained 50 mM HEPES/140 mM NaCl/0.05 % (v/v) Triton X-100 (pH: 7.4, RT).
Reversible behavior of human neutrophil elastase inhibition by EapH2
To determine the reversible nature of EapH2 inhibition of hNE, 112 nM hNE was pre-incubated with 492 nM EapH2 then diluted to a final concentration of 10.6 nM hNE, 46 nM EapH2 with 50 mM HEPES/140 mM NaCl/0.05 % (v/v) Triton X-100 (pH: 7.4, RT) containing 504 μM MSA2PV-pNA. The reaction was followed at 400 nm for 80 minutes. As a control, the reaction was separately followed without the pre-incubation of enzyme and inhibitor. The concentrations of enzyme and inhibitor were the same as the pre-incubation concentrations post-dilution.
Steady-state inhibition kinetics of human neutrophil elastase by wild-type/mutant EapH2 proteins
Progress curves were collected for 30 minutes using a VersaMax tunable microplate reader (Molecular Devices). A total of 57 progress curves were collected for each EapH1/H2 inhibitor (three hNE concentrations (~4, 8, 12 nM), three substrate concentrations (~Km, 2 Km, 3 Km) per hNE concentration, five inhibitor concentrations per substrate concentration). To obtain a more accurate estimate of the substrate on-rate, two hNE concentrations with 6 substrate concentrations per hNE concentration were included in the global fitting. These data were fit to a rapid-equilibrium competitive inhibition mechanism (Scheme 1) using Dynafit (Biokin, Ltd). 25 A detailed description of the Dynafit script can be found in the second supplemental figure in Herdendorf and Geisbrecht (Biochemistry (57), 2018). 3
To decrease the search space for fitting the progress curves, the substrate off-rate (k2) was linked to the on-rate by the estimated Michaelis constant (Km) previously determined from steady-state kinetic analysis using the plate-based assay.3 The inhibition constant was calculated from the on- and off-rate estimates determined from the fits. The values reported are the average of at least three different sets of data, while the errors are the standard deviation.
The inhibition constants for the L59F EapH1 mutant and the EapH2 mutants (T125A and N127A) were determined using a classical 5 5 assay. The initial velocity of hNE (~4 nM) was determined at five substrate concentrations (~ 0.5 Km − 4 Km) using five inhibitor concentrations (0 − ~5 Ki) per substrate concentration. The data (n = 3) were globally fit to a competitive inhibition model using SigmaPlot 10.0/Enzyme Kinetics Module 1.3 (Systat Software, Inc). The Ki estimates reported are an average of three individual data sets. Errors reported are the standard deviation.
On- and off-rate estimates of the L59F EapH1 and the T125A and N127A EapH2 mutants
The association/dissociation rates for the L59F EapH1 and the T125A, N127A EapH2 mutants could not be determined by progress curve analysis. Therefore, Surface Plasmon Resonance (SPR) was utilized as an alternative approach to obtain these values. hNE, previously modified to contain an N-linked glycan ethylenediamine moiety 22, was immobilized on CMD50M and CMD200M sensor chips (Xantec Bioanalytics; Dusseldorf, Germany) via EDC/NHS coupling chemistry to ~2000 RU (CMD50M) or ~1000 RU (CMD200M). A reference flow cell was created by flowing the EDC/NHS solution over the surface followed by quenching the activated carboxymethyl-dextran with 1 M ethanolamine (pH: 9.0, RT).
All injections were carried out using a running buffer of 50 mM HEPES/140 mM NaCl/0.05 % (v/v) Tween-20 (pH: 7.4, RT). The parameters for binding of wild-type EapH2 to hNE were estimated by both single-cycle kinetics ([EapH2] (nM): 6.25, 12.5, 25, 50, 100) and conventional dose-response kinetics ([EapH2] (nM): 3.91, 7.8125, 15.625, 31.25, 62.5, 125, 250). The analyte contact time for the single-cycle approach was 120 s with a final dissociation phase of 3600 s at a flow rate of 60 μL/min; similarly, the analyte contact time for the conventional dose-response runs was 120 s followed by a dissociation phase of 3600 s at a flow rate of 60 μL/min for each analyte concentration. The microscopic rate constants for EapH1 L59F and the EapH2 mutants (T125A, N127A) were estimated by conventional dose-response kinetics ([EapH1/2] (nM): 62.5, 125, 250, 500, 1000, 2000, 4000), with the exception of T125A ([EapH2] (nM): 15.625, 31.25, 62.5, 125, 250, 500, 1000). The EapH2 T125A mutant was also analyzed using single cycle kinetics ([EapH2] (nM): 15.625, 31.25, 62.5, 125, 250). The analyte contact time was 120 s with a dissociation time of 240 s at a flow rate of 60 μL/min. The chip surface was regenerated after each injection with 10 mM glycine/1.5 M NaCl (pH: 2.2, RT) for 30 s at a flow rate of 20 μL/min. All reference-subtracted sensorgrams were fit to a 1:1 Langmuir binding model using the BiaCore T-200 Evaluation Software (GE Life Sciences). The microscopic rate estimates reported are the average of at least six individual sensorgram fits. The errors on the microscopic rate constants are the standard deviation. The dissociation constant was estimated from the averaged microscopic rate constants. The Kd errors were propagated from the on- and off-rate standard deviations.
Circular Dichroism (CD) Spectroscopy
CD studies on the WT/mutant EapH1/H2 proteins were performed on a Jasco J-815 CD spectrometer in a 1-mm path length cell using a protein concentration of 15 μM. The buffer consisted of 15 mM HEPES, 140 mM NaCl, (pH 7.4). Spectra were collected from 200 to 260 nm in increments of 1 nm. Each spectrum was buffer-corrected, and the mutant spectra were normalized to the ellipticity of the WT protein at 212 nm to correct for minor differences in protein concentration (Fig. S2).
Sequence alignment and structure generation
Protein sequence alignment was generated using ESPript3 internet suite.26 The sequence alignment file utilized by ESPript3 was generated using the EMBL-EBI Clustal Omega suite. 4 The EapH2:hNE model was created using the ClusPro 2.0: protein-protein docking server. 22, 27 Briefly, the structures of EapH2 (PDB: 1YN51)and hNE (PDB:1HNE 28) were used as the inputs to the server with no experimentally derived constraints on the modeling calculations. 22 Both the buried surface area calculations and hydrogen bond identification were performed by the EBI-PISA server. 22, 29 All structural representations were rendered from the corresponding PDB files using PyMol (Shrodinger, LLC).5
Results
Rationale behind residue selection for mutagenesis and kinetic evaluation
We designed a panel of site-directed point mutants to probe the structure/function relationships of EapH2 and to provide a basis for its comparison to the better understood EapH1. These mutants could be considered as falling into either of two different categories. Mutants in the first category aimed to identify global differences in the hNE binding sites of EapH1 and EapH2, while mutants in the second category aimed to define the contributions of individual residues to EapH2 function. The specific nature of these mutations are described below.
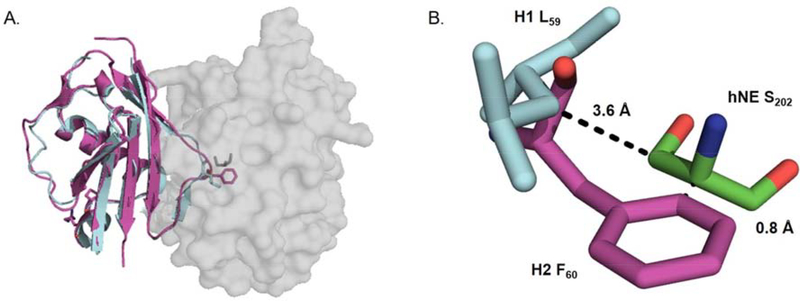
Whereas mutation of three EapH1 residues to alanine (R89, E94, K95) essentially eliminated binding to hNE22, mutation of corresponding residues in EapH2 (R90, E95, K96) produced only a mild effect (~ 2-fold) 22. Since we have recently shown that EapH1 R89 is responsible for its potent inhibition of hNE 3, We opted to mutate the corresponding EapH2 residue (i.e. R90) to methionine to further investigate the role of this position in EapH2 function. Separately, examination of EapH2 (PDB:1YN51) superimposed upon the EapH1:hNE co-crystal structure (PDB:4NZL 2) revealed that the position occupied by EapH1 residue L59 is replaced by a phenylalanine in EapH2 (Fig. 3). Such a change could be considered a relatively conservative mutation in the absence of further information. In that regard, while the EapH1 L59 sidechain lies some 3.6 Å from the hNE active-site serine (S202) in the EapH1:hNE co-crystal structure, the sidechain of EapH2 F60 appears likely to result in a steric clash if the superimposition model were correct (0.8 Å) (Fig. 3B). To examine this possibility at the level of protein function, we mutated EapH1 L59 to phenylalanine and kinetically evaluated the inhibitory activity of this variant.
Figure 3. Crystal structure of S. aureus EapH2 (PDB: 1YN5 1) aligned upon the EapH1:hNE crystal structure (PDB: 4NZL 2).
(A) EapH1 (cyan/H2 (magenta) depicted as cartoon, hNE depicted as space-fill model. (B) close up view of EapH1 L59/EapH2 F60 in relation to hNE S202.
In a previous study, Stapels et al. simultaneously mutated EapH2 residues 120 – 129 to alanine and determined that this change resulted in a large inflation of the Kd for the EapH2:hNE complex (~13,000- fold). 22 Here, we opted to individually and conservatively mutate each of these positions to discern the effects on hNE inhibition. Positions occupied by residues with large sidechains in wild-type EapH2 (e.g. arginine and lysine) were replaced by a reasonably isosteric but uncharged residue in methionine, while positions occupied by small polar sidechains were replaced by the small uncharged residue alanine. Using this approach, we prepared a library of eight EapH2 point mutants for structure/function investigation.
EapH2 is a reversible, time-dependent inhibitor of human neutrophil elastase
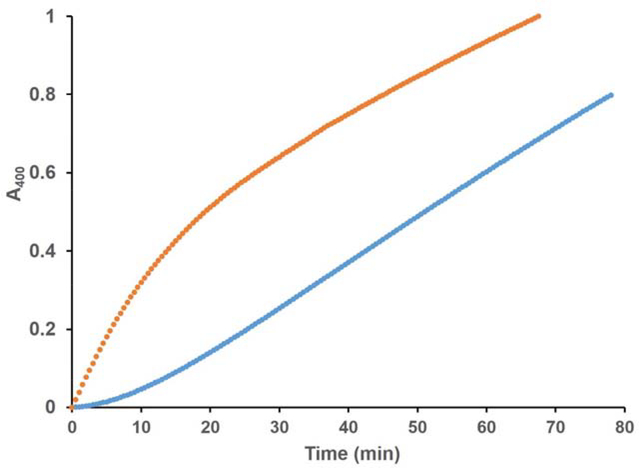
We have previously shown that S. aureus EapH1 is a reversible, time-dependent inhibitor of hNE. 3 To determine if S. aureus EapH2 also displayed this characteristic, we performed traditional steady-state kinetics by exposing the enzyme to the substrate and inhibitor simultaneously. The observed progress curve of the hNE reaction was non-linear under these conditions (Fig. 4). The velocity of the initial phase was approximately equal to the rate of the uninhibited reaction and decreased as a function of time to a steady-state inhibited rate, consistent with time-dependent inhibition. 30 To determine whether this inhibition was reversible, we first pre-incubated with hNE EapH2 and then diluted the reaction with a saturating concentration of substrate. A pronounced lag followed by a steady-state phase that corresponded to the rate of the reaction determined at the diluted EapH2:hNE concentrations was observed (Fig. 4). Together, these data demonstrate that EapH2 is a reversible, time-dependent inhibitor of hNE.
Figure 4. EapH2 is a Time-Dependent, Reversible Inhibitors of Human Neutrophil Elastase.
( ) 504 μM MSA2PV-pNA incubated with 46 nM EapH2, reaction started with 10.6 nM elastase; (
) 504 μM MSA2PV-pNA incubated with 46 nM EapH2, reaction started with 10.6 nM elastase; ( ) 492 nM EapH2 pre-incubated with 112 nM elastase diluted to 46 nM EapH2, 10.6 nM elastase with buffer containing 504 μM MSA2PV-pNA.
) 492 nM EapH2 pre-incubated with 112 nM elastase diluted to 46 nM EapH2, 10.6 nM elastase with buffer containing 504 μM MSA2PV-pNA.
Steady-state inhibition kinetics of human neutrophil elastase by wild-type/mutant EapH1/H2 proteins
As with EapH1, the impact of wild-type EapH2 on hNE activity could not be studied by initial velocity analysis since its inhibitory effect is time-dependent. 3 We therefore obtained a series of hNE reaction progress curves across a range of enzyme, substrate, and inhibitor concentrations and globally fit these to quantify inhibition of hNE activity by wild-type and mutant EapH2. In this regard, we previously reported the on- (k1) and off-rates (k2), as well as the turnover number (k3) for the synthetic substrate, MSA2PV-pNA, for hNE. 3 Moreover, as we have previously noted, “the microscopic rate constants relating to the hNE mechanism should be consistent between wild-type and mutant EapH1/H2 proteins, while those constants relating to the EapH1/H2 inhibitors may vary between the wild-type and mutant proteins.” 3 The kcat values (Table 1, k3) for the turnover of substrate by hNE estimated during inhibition with wild-type and mutant EapH1/H2 were consistent throughout and in good agreement to those determined by steady-state kinetics 3 and the manufacturer’s specifications (5.8 – 8.4 s−1, Table 1). The hNE substrate on-rates (Table 1, k1) were likewise consistent between the wild-type and mutant EapH1/H2 proteins (1.9 105 – 6.9 105 M−1 s−1, Table 1). We subsequently determined the estimated on-rate (k4) for wild-type EapH2 to be 2.9 105 ± 4.3 104 M−1s−1, while the estimated off-rate (k5) for the wild-type protein was 5.9 10−4 ± 2.1 10−5 s−1. Using these microscopic rate constants, we arrived at a value of 2.11± 0.32 nM for the macroscopic inhibition constant (Ki) for wild-type EapH2.
Table 1.
Kinetic Rate Constant Estimates determined by Progress Curve Analysis a
| Inhibitor | k1 (M−1s−1) | SD | k3 (s−1) | SD | k4 (M−1s−1) | SD | k5 (s−1) | SD | Ki (nM) | SD |
|---|---|---|---|---|---|---|---|---|---|---|
| EapH1 WTb | 6.0×105 | 2.2×105 | 7.4 | 0.8 | 2.0×106 | 1.4×105 | 4.3×10−5 | 1.3×10−5 | 0.021 | 0.005 |
| EapH2 WT | 3.1×105 | 4.5×104 | 7.8 | 0.2 | 2.9×105 | 4.3×104 | 5.9×10−4 | 2.1×10−5 | 2.11 | 0.32 |
| H1 R89Mb,c | - | - | 7.0 | 0.1 | - | - | 321 | 12 | ||
| H2 R90M | 2.4×105 | 1.3×105 | 7.2 | 1.7 | 2.8×105 | 1.3×105 | 5.5×10−4 | 4.1×10−5 | 2.3 | 1 |
| H1 L59Fc | - | - | 6.3 | 0.3 | - | - | 200 | 5 | ||
| H2 K120M | 3.2×105 | 5.5×104 | 8.3 | 1.1 | 2.6×105 | 1.1×105 | 8.3×10−4 | 1.8×10−5 | 3.8 | 2.0 |
| H2 K121M | 4.3×105 | 2.3×105 | 7.0 | 0.7 | 3.3×105 | 6.6×104 | 9.1×10−4 | 6.9×10−5 | 2.9 | 0.7 |
| H2 D122N | 3.5×105 | 4.6×104 | 7.9 | 1.5 | 2.6×105 | 5.8×104 | 3.7×10−4 | 1.6×10−5 | 1.5 | 0.4 |
| H2 D122A | 6.9×105 | 5.0×105 | 8.3 | 2.3 | 1.2×105 | 1.5×104 | 3.1×10−3 | 1.2×10−4 | 26 | 3 |
| H2 S123A | 1.9×105 | 5.4×104 | 6.6 | 0.5 | 1.6×105 | 7.8×104 | 4.1×10−4 | 2.6×10−5 | 3.2 | 2.1 |
| H2 Y124F | 5.5×105 | 1.3×105 | 8.4 | 0.2 | 1.2×105 | 3.8×104 | 2.2×10−3 | 5.1×10−5 | 18.6 | 4.9 |
| H2 T125Ac | - | - | 6.2 | 0.2 | - | - | 53 | 4 | ||
| H1 N126A | 2.4×105 | 6.4×104 | 6.3 | 0.5 | 9.5×105 | 5.0×105 | 1.0×10−4 | 2.5×10−5 | 0.166 | 0.163 |
| H2 N127Ac | - | - | 5.8 | 0.5 | - | - | 397 | 66 | ||
| (-)b,d | 4.0×105 | 2.2×105 | 7.4 | 0.8 | - | - | - |
Microscopic rate constant estimates are the average of at least three individual sets of 57 progress curves (three [elastase], three [substrate] per [elastase], and five [inhibitor] per [substrate].
values previously determined by Herdendorf and Geisbrecht.3
Due to the loss of the time-dependent inhibition characteristic, these mutants were analyzed by a classical 5×5 inhibition assay. The data (n = 3) were globally fit to a competitive inhibition model using SigmaPlot 10.0/Enzyme Kinetics Module 1.3 (Systat Software, Inc). The Ki estimates reported are an average of three individual data sets. Errors reported are the standard deviation. k4 and k5 were estimated by following the EapH2 mutants binding to immobilized elastase using surface plasmon resonance (See Table 2).
Microscopic rate constant estimates are the average of 10 individual sets of 12 progress curves (two [elastase], six [substrate] per [elastase].
Evaluation of the EapH1 L59F and EapH2 R90M mutants suggests a difference in hNE binding
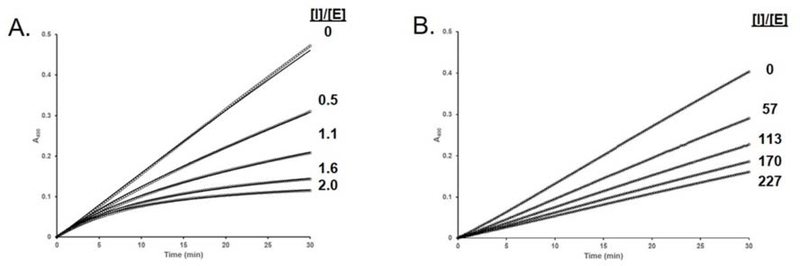
As stated above, the first class of mutants we investigated were designed to test for global differences in the hNE binding sites of EapH1 and EapH2. Whereas the R89M mutation had a profound effect on hNE inhibition by EapH1 (~15,000-fold) 3, the corresponding R90M mutation in EapH2 had no discernable effect on hNE inhibition by this protein (Table 1). This result strongly suggested that the critical role played by EapH1 R89 must instead be accounted for by another position in EapH2. Separately, the L59F mutant of EapH1 was significantly perturbed in terms of hNE inhibition (~ 9,500-fold, Table 1). The time-dependent characteristic was also abolished (Fig. 5). This result argued that the phenylalanine in this position of EapH2 was incompatible with forming a structurally-equivalent complex to that seen in the EapH1:hNE co-crystal structure (PDB:4NZL 2). Together, these observations provided further evidence that EapH2 must recognize hNE through a distinct binding mode when compared to EapH1.
Figure 5. Representative Fitted Inhibition Progress Curves.
A. WT EapH1 inhibition: assay contained 3.9 nM Elastase, 147 μM MSA2PV-pNA.3 B. L59F EapH1 inhibition: assay contained 4.5 nM Elastase, 134 μM MSA2PV-pNA.
N127 is a major contributor and T125 is a minor contributor to EapH2 inhibition of hNE
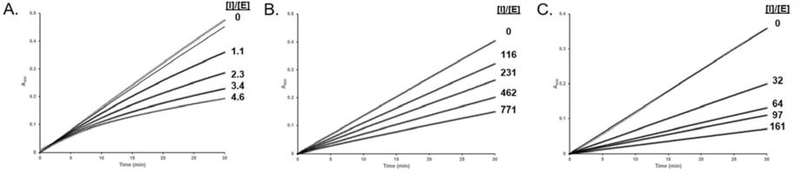
Using the EapH2:hNE model and previous alanine scanning mutagenesis data as a guide 22, we prepared a panel of site-directed single mutants of EapH2 affecting positions K120 – N127. We then kinetically characterized these EapH2 variants. Four of these mutants (K120M, K121M, D122N, and S123A) were similar to wild-type in terms of the on- and off- rates and subsequent Ki estimates (Table 1). Two of the mutants (D122A and Y124F) were mildly perturbed (~ 10-fold). These effects were due to a 10-fold increase in the off-rate of the inhibitor when compared to the wild-type protein (Table 1). While the T125A mutation was slightly more perturbed (~25-fold) in its ability to inhibit hNE (Table 1), the time-dependent aspect of hNE inhibition was abolished in this mutant (Fig. 6, panel C). Mutation of N127 to alanine resulted in the largest diminution of hNE inhibition (~188-fold, Table 1) and, similarly to T125A, this mutation also abolished the time-dependent characteristic (Fig. 6, panel B). Considering the large impact that the N127A mutation had on hNE inhibition, we mutated the corresponding residue in EapH1 (N126) to an alanine and evaluated this variant kinetically. Although this mutation did exhibit an effect on the inhibition of hNE when compared to wild-type EapH1 (~ 8-fold, Table 1), it was much less pronounced than that of EapH2 N127A and the time-dependent characteristic was also maintained. Finally, while the EapH2:hNE model suggests the existence of an inter-molecular hydrogen bond between Y124 and C191 of hNE (Fig. 2B), the EapH2 Y124F mutation had a lesser effect on hNE inhibition (~10-fold), when compared to the T125A and N127A mutations.
Figure 6. Representative Fitted Inhibition Progress Curves.
A. WT EapH2 inhibition: assay contained 4.2 nM elastase, 152 mM MSA2PV-pNA. B. N127A EapH2 inhibition: 4.2 nM elastase, 128 mM MSA2PV-pNA. C. T125A EapH2 inhibition: 4.0 nM elastase, 115 mM MSA2PV-pNA.
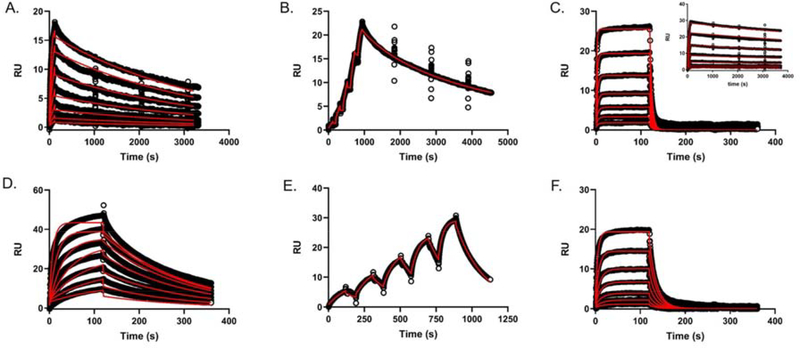
Two EapH2 mutations (T125A and N127A) and one EapH1 mutation (L59F) had the effect of losing time-dependent characteristic inhibition of hNE. We therefore carried out a classical 5 5 assay to determine the inhibition constants of these mutants. The Ki values for EapH2 T125A, N127A, and EapH1 L59F were estimated to be 53 nM, 397 nM, and 200 nM, respectively (Fig. 7, Table 1). To further determine estimates for the individual microscopic rate constants, we utilized surface plasmon resonance to characterize the interaction between various EapH1/H2 proteins and immobilized hNE.22 Wild-type EapH2 was flowed over the hNE sensor surface using both single cycle and dose response kinetic methods to establish the reproducibility of previous results. The single-cycle derived on- and off-rates (Table 2; 2.3 105 M−1s−1, 3.69 10−4 s−1, respectively) were comparable to values previously determined (3.75 105 M−1s−1, 4.47 10−4 s−1). 22 A more extensive dose-response kinetic analysis (i.e. long duration dissociation phase (3600 s) following each concentration injection) of wild-type EapH2 binding to hNE resulted in essentially equivalent values with an estimated a Kd of 2.16 nM (Fig. 8A, Table 2) and on- and off-rates of 1.28 105 M−1s−1 and 2.76 10−4 s−1, respectively. The on-rate for the EapH1 L59F mutant remained largely unperturbed (~ 2-fold, Table 2); however, the off-rate was significantly increased (~3,000-fold, Table 2, Fig. 8C). The EapH2 mutations also had a minimal effect on the on-rates (Table 2, T125A (~1.6-fold), N127A (~3.3-fold)). The off-rates were significantly more affected (T125A (~15-fold), N127A (~200-fold), Table 2, Fig. 8D–F). Considering the T125A off-rate most closely approximated the wild-type off-rate, single-cycle kinetics were also performed. The microscopic rate constants estimated by single-cycle agreed with those determined by the dose-response kinetics (Table 2). Together, these results are consistent with the interpretation that loss of function for these EapH2 mutants is driven primarily by increasing the off-rates of the interaction with hNE.
Figure 7. Competitive Inhibition of hNE by EapH1/H2 Mutants.
(A) EapH2 T125A, (B) EapH2 N127A, (C) EapH1 L59F. All assays were done with ~ 4nM hNE in 50 mM HEPES/140 mM NaCl/0.05 %Triton X-100 (pH: 7.4, RT). The representative inhibitor concentrations are inset within the legend box at the right side of each panel.
Table 2.
Kinetic Rate Constant Estimates determined by Surface Plasmon Resonance a
| Protein | kon (M−1s−1) | SD | koff (s−1) | SD | Kd (nM) | errorb | Ki (nM)c | Fold differencef |
|---|---|---|---|---|---|---|---|---|
| H1 WTd | 2.23×105 | 5.0×103 | 6.03×10−5 | 3.1×10−6 | 0.271 | 0.015 | 0.021 | 12.9 |
| H1 WTd,e | 4.85×105 | 2.06×105 | 7.10×10−5 | 5.6×10−6 | 0.146 | 0.063 | 0.021 | 7.0 |
| H2 WT | 1.28×105 | 1.54×104 | 2.76×10−4 | 2.32×10−5 | 2.16 | 0.32 | 2.11 | 1.02 |
| H2 WTe | 2.3×105 | 2.39×104 | 3.69×10−4 | 6.45×10−5 | 1.61 | 0.32 | 2.11 | 0.8 |
| H1 L59F | 1.12×105 | 7.00×103 | 0.2020 | 0.0351 | 2050 | 478 | 200 | 10 |
| H2 T125A | 7.21×104 | 4.61×103 | 5.3×10−3 | 3.0×10−4 | 73.5 | 6.3 | 53 | 1.4 |
| H2 T125Ae | 1.33×105 | 2.21×104 | 4.8×10−3 | 6.8×10−4 | 36.5 | 8.0 | 53 | 0.7 |
| H2 N127A | 3.48×104 | 6.81×102 | 0.0591 | 0.0084 | 1710 | 773 | 397 | 4.3 |
Microscopic rate constant estimates are the average of three individual sensorgrams.
The dissociation constant error was propagated from the on- and off-rate standard deviations.
The inhibition constants determined by progress curve analysis (WT) or 5×5 assay (EapH1 L59F, EapH2 T125A and N127A; Fig. 7).
Values previously determined by Herdendorf and Geisbrecht. 3.
Microscopic rate constant estimates determined by single-cycle binding kinetics.
The fold difference depicts the difference between the dissociation constant determined by SPR and the inhibition constant determined by kinetic analysis.
Figure 8. Surface Plasmon Resonance of Wild-Type and Mutant EapH1/H2 binding to immobilized hNE.
(A) EapH2 WT (dose-response kinetics with elongated dissociation phase, [EapH2] ranging from ~1.8x – 118x the EapH2 Ki), (B) EapH2 WT (single-cycle kinetics, [EapH2] ranging from ~3x – 47x the EapH2 Ki), (C) EapH1 L59F ([EapH1] ranging from` 0.3x – 20x the L59F Ki); inset: WT EapH1 dose-response sensorgram 3, (D) EapH2 T125A, ([EapH2] ranging from ~0.3x – 19x the T125A Ki), (E) EapH2 T125A, (single-cycle kinetics, [EapH2] ranging from ~0.3x – 5x the T125A Ki), (F) EapH2 N127A ([EapH2] ranging from ~ 0.2x – 10x the N127A Ki). All sensorgram series were evaluated with a Langmuir kinetic model as described in the materials and methods section.
Discussion
EapH1 and EapH2 were originally identified as hypothetical secreted proteins that shared a high level of sequence similarity to one another 1. While determination of their three-dimensional structures allowed for a physical description of the so-called “EAP domain” 1, nearly a decade passed before Stapels and coworkers characterized these proteins as high-affinity inhibitors of neutrophil serine proteases 2. Yet even though EapH1 and EapH2 both appear to reversibly inhibit the activity of hNE 3, evidence suggesting that EapH1 and EapH2 achieve this outcome through different means has begun to emerge. In this regard, EapH2 has displayed consistently weaker inhibitory potency than EapH1 against hNE, Cathepsin G, and Proteinase 3 2, 22. Insofar as the case of hNE inhibition is concerned, the difference in Ki is approximately 100-fold for EapH2 when compared to EapH1 and arises from a 10-fold decrease in inhibitor on-rate (2.9 105 M−1s−1) coupled to a 10-fold increase in the off-rate (5.9 10−4 s−1, Table 1). Given that the EapH2 structure superimposes on that of EapH1 with an RMSD of 0.634 Å for all backbone atoms, it seemed reasonable that sequence-level differences between EapH2 and EapH1 were responsible for the differences in potency observed. Ultimately, this led to the hypothesis that EapH1 and EapH2 recognize their target proteases through fundamentally different binding modes.
To address this possibility, Stapels and coworkers utilized alanine scanning mutagenesis to better define the hNE binding sites on EapH1 and EapH2 at a functional level 22. They found that while simultaneous mutation of EapH1 residues R89, E94, and E95 dramatically decreased the affinity for hNE, corresponding loss of EapH2 residues R90, E95, and E96 had only a very modest effect (~2-fold) 22. We recently expanded upon the work of Stapels et al. by identifying EapH1 R89 as the principal residue required for high-affinity binding to and time-dependent inhibition of hNE activity 3. Herein we prepared the corresponding R90 mutant in EapH2 and evaluated the properties of this protein. Consistent with previous work22, we found that the R90M mutation had no effect on EapH2 function (Table 1). This observation supports the hypothesis that EapH1 and EapH2 recognize hNE through distinct binding modes. Still further support for this idea arises from analysis of the sidechain found in the corresponding positions of L59 in EapH1 and F60 in EapH2 (Fig. 1A). In the EapH1:hNE co-crystal structure (PDB: 4NZL 2), L59 “caps” the approach channel to the hNE active site serine (i.e. S202). However, when EapH2 was superimposed onto the EapH1:hNE co-crystal structure5, we observed the potential for an obvious steric clash between EapH2 F60 and the hNE active site serine if EapH2 were to bind hNE identically to EapH1 (Fig. 3B). Upon evaluating the L59F mutant of EapH1, we found that both the affinity for and inhibition of hNE were dramatically reduced (~7,500-fold (Table 2) and ~9,500-fold (Table 1), respectively). This mutation also abolished the time-dependent inhibitory characteristic of EapH2 (Fig. 5). Together, these data argue that the sequence of EapH2 is incompatible with hNE binding as seen in the EapH1:hNE co-crystal structure. We therefore conclude that EapH1 and EapH2 must recognize hNE through fundamentally different binding modes. Whether this is also the case for EapH1 and EapH2 binding to Cathepsin G or Proteinase 3 in an intriguing question, and remains a topic for future investigation.
Since a co-crystal structure of EapH2:hNE has not been determined, nearly all physical understanding of this complex has been obtained from molecular modeling predictions. 22, 27 Interestingly, inspection of an EapH2:hNE model implicates a different region of the EapH2 protein (i.e. K120 – N127) in mediating hNE binding (Fig. 2). Staples and co-workers simultaneously mutated these residues to alanine and found that this mutant protein was severely perturbed in hNE binding (~ 13,000-fold). 22 To further expand upon that work, we mutated each residue in this region of EapH2 to determine its individual contribution to binding and inhibition of hNE. Residues K120 and K121 were conservatively mutated to methionine, but these changes had no effect on inhibition of hNE (Table 1). The conservative mutation of D122N also had no effect on inhibition of hNE. However, since D122 forms intra-molecular hydrogen bonds to K83 (Fig. 2B), it is possible that this mutation maintained these interactions and led to no consequence. To further probe the importance of these interactions, we created a D122A protein to remove the possibility of any hydrogen bond formation. Significantly, we found that the D122A mutant displayed a 10-fold (Table 1) weaker ability to inhibit hNE. Assuming that the published EapH2:hNE model is accurate 22, this effect suggests that the role of D122 is to aid in organizing the EapH2 loop found at the interface with hNE. Whereas the S123A mutation was essentially wild-type in its ability to inhibit hNE, the EapH2:hNE model suggests an inter-molecular interaction between EapH2 Y124 and hNE C191 (Fig. 2B). Despite this, we found that the Y124F mutant had only a relatively minor impact on function (~10-fold, Table 1) when compared to other residues that form intermolecular contacts with hNE, most notably EapH1 R89 (~15,000-fold). 3
Of all the point mutants we prepared in EapH2, we found T125A and N127A to be the most insightful vis-à-vis EapH2 function. Even though the decrease in apparent inhibition constant of the T125A mutation was of similar magnitude to that of Y124F (~25-fold, Table 1), we also determined that loss of this threonine abolished the time-dependent inhibition characteristic (Fig. 6). Based on the EapH2 crystal structure (PDB: 1YN5 1), T125 forms an intra-molecular hydrogen bond to the carbonyl oxygen of N127 (Fig. 2B). This observation suggests the role of T125 is to orient N127 for optimal interaction with hNE. A role in orienting N127 is relevant because we found that the N127A mutation resulted in the largest decrease in inhibition of hNE (~200-fold, Table 1) as well as loss of the time-dependent characteristic. The EapH2:hNE model suggests that N127 participates in two inter-molecular hydrogen bonds, one to the sulfur of C58 and a second to the ε2 nitrogen of hNE H57 (Fig. 2B). We recently demonstrated that one EapH1 residue (R89) is responsible for the potent, time-dependent inhibition of hNE by forming two intermolecular hydrogen bonds with the protease 3. Our data concerning EapH2 N127 therefore invokes many parallels with EapH1 R89. It is interesting to note that this EapH2 asparagine (N127) is conserved in EapH1 (N126, Fig. 1A). However, the EapH1 N126A mutation had only a minimal effect on the inhibition of hNE (~8-fold). Together, these results not only established N127 as the crucial residue for EapH2 function, they also further support the idea that EapH1 and EapH2 must recognize hNE through alternative binding modes.
Conclusion
Although the hNE binding mode of EapH1 has been defined by a co-crystal structure 2 and mutagenesis studies 3, 22, the nature of the EapH2:hNE interaction has remained poorly understood. In this study, we used a model of the EapH2:hNE complex 22 to guide investigation of this interaction at the individual residue level. Of the EapH2 residues (120 – 127) implicated by this model, only the N127A mutant was substantially perturbed in its ability to inhibit hNE (~200-fold). This mutation was also noteworthy because it abolished the time-dependent inhibition characteristic of EapH2. Our results suggest that N127, which potentially forms two inter-molecular hydrogen bonds with hNE, acts much like R89 in EapH1. 3 Although EapH2 T125A was less perturbed in its ability to inhibit hNE (~25-fold) than N127A, this mutant protein was likewise no longer a time-dependent inhibitor of hNE. EapH2 T125 forms an intra-molecular hydrogen bond to the carbonyl oxygen of N127 1, which suggests that T125 acts to optimally orient N127 for interaction with hNE residues H57 and C58 22. Together, these data demonstrate that EapH2 has a different hNE binding motif than EapH1 and that EapH2 N127 serves a critical role.
Supplementary Material
Acknowledgements
We would like to thank Dr. D.J. Black, Principal Scientist, BioLogic USA for useful discussions during this work.
Funding Source
Funding was provided by a grant from the National Institute of General Medical Sciences (R01-GM121511) to B.V.G.
Footnotes
Conflict of interest: The authors declare that they have no conflicts of interest with the contents of this article.
UniProt Accession ID for Proteins
EapH1, UniprotKB: A0A0H3K0M1
EapH2, UniprotKB: A0A0H3JUK5
Human Neutrophil Elastase, UniProtKB: P08246
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Geisbrecht BV, Hamaoka BY, Perman B, Zemla A, and Leahy DJ (2005) The crystal structures of EAP domains from Staphylococcus aureus reveal an unexpected homology to bacterial superantigens, J Biol Chem 280, 17243–17250. [DOI] [PubMed] [Google Scholar]
- [2].Stapels DA, Ramyar KX, Bischoff M, von Kockritz-Blickwede M, Milder FJ, Ruyken M, Eisenbeis J, McWhorter WJ, Herrmann M, van Kessel KP, Geisbrecht BV, and Rooijakkers SH (2014) Staphylococcus aureus secretes a unique class of neutrophil serine protease inhibitors, Proc Natl Acad Sci U S A 111, 13187–13192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Herdendorf TJ, and Geisbrecht BV (2018) Investigation of Human Neutrophil Elastase Inhibition by Staphylococcus aureus EapH1: The Key Role Played by Arginine 89, Biochemistry 57, 6888–6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, and Lopez R (2019) The EMBL-EBI search and sequence analysis tools APIs in 2019, Nucleic acids research 47, W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Schrodinger LLC. (2015) The PyMOL Molecular Graphics System, Version 1.8 [Google Scholar]
- [6].Gorwitz RJ, Kruszon-Moran D, McAllister SK, McQuillan G, McDougal LK, Fosheim GE, Jensen BJ, Killgore G, Tenover FC, and Kuehnert MJ (2008) Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001–2004, J Infect Dis 197, 1226–1234. [DOI] [PubMed] [Google Scholar]
- [7].Krismer B, Weidenmaier C, Zipperer A, and Peschel A (2017) The commensal lifestyle of Staphylococcus aureus and its interactions with the nasal microbiota, Nat Rev Microbiol 15, 675–687. [DOI] [PubMed] [Google Scholar]
- [8].Fernandez Guerrero ML, Gonzalez Lopez JJ, Goyenechea A, Fraile J, and de Gorgolas M (2009) Endocarditis caused by Staphylococcus aureus: A reappraisal of the epidemiologic, clinical, and pathologic manifestations with analysis of factors determining outcome, Medicine (Baltimore) 88, 1–22. [DOI] [PubMed] [Google Scholar]
- [9].Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK, and Active Bacterial Core surveillance, M. I. (2007) Invasive methicillin-resistant Staphylococcus aureus infections in the United States, JAMA 298, 1763–1771. [DOI] [PubMed] [Google Scholar]
- [10].Kobayashi SD, Malachowa N, and DeLeo FR (2015) Pathogenesis of Staphylococcus aureus abscesses, Am J Pathol 185, 1518–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Amulic B, Cazalet C, Hayes GL, Metzler KD, and Zychlinsky A (2012) Neutrophil function: from mechanisms to disease, Annu Rev Immunol 30, 459–489. [DOI] [PubMed] [Google Scholar]
- [12].Nauseef WM (2007) How Human Neutrophils Kills and Degrade Microbes: an Integrated View, Immunol. Rev. 219, 88–102. [DOI] [PubMed] [Google Scholar]
- [13].Borregaard N, and Cowland JB (1997) Granules of the human neutrophilic polymorphonuclear leukocyte, Blood 89, 3503–3521. [PubMed] [Google Scholar]
- [14].Ohlsson K, and Olsson I (1974) The neutral proteases of human granulocytes. Isolation and partial characterization of granulocyte elastases, Eur J Biochem 42, 519–527. [DOI] [PubMed] [Google Scholar]
- [15].Odeberg H, and Olsson I (1975) Antibacterial activity of cationic proteins from human granulocytes, J Clin Invest 56, 1118–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kao RC, Wehner NG, Skubitz KM, Gray BH, and Hoidal JR (1988) Proteinase 3. A distinct human polymorphonuclear leukocyte proteinase that produces emphysema in hamsters, J Clin Invest 82, 1963–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Odeberg H, and Olsson I (1976) Microbicidal Mechanisms of Human Granulocytes: Synergisitc Effects of Granulocyte Elastase and Myeloperoxidase or Chymotrypsin-Like Cationic Protein, Infect. Immun. 14, 1276–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].de Jong NWM, van Kessel KPM, and van Strijp JAG (2019) Immune Evasion by Staphylococcus aureus Microbiol. Spectr. 7, GPP3–0061-2019. [DOI] [PubMed] [Google Scholar]
- [19].Herrera AI, Ploscariu NT, Geisbrecht BV, and Prakash O (2018) (1)H, (15)N, and (13)C resonance assignments of the third domain from the S. aureus innate immune evasion protein Eap, Biomol NMR Assign 12, 175–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Woehl JL, Takahashi D, Herrera AI, Geisbrecht BV, and Prakash O (2016) (1)H, (15)N, and (13)C resonance assignments of Staphylococcus aureus extracellular adherence protein domain 4, Biomol NMR Assign 10, 301–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Herrera AI, Dubey A, Geisbrecht BV, Arthanari H, and Prakash O (2019) Backbone Resonance Assignments of Innate Immune Evasion Protein EapH2 from the S. aureus, Biomol. NMR Assign. 13, 219–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Stapels DAC, Woehl JL, Milder FJ, Tromp AT, van Batenburg AA, de Graaf WC, Broll SC, White NM, Rooijakkers SHM, and Geisbrecht BV (2018) Evidence for multiple modes of neutrophil serine protease recognition by the EAP family of Staphylococcal innate immune evasion proteins, Protein Sci 27, 509–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Geisbrecht BV, Bouyain S, and Pop M (2006) An optimized system for expression and purification of secreted bacterial proteins, Protein Expr Purif 46, 23–32. [DOI] [PubMed] [Google Scholar]
- [24].Walker JM (2005) The proteomics protocols handbook, Humana Press, Totowa, N.J. [Google Scholar]
- [25].Kuzmic P (1996) Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase, Anal Biochem 237, 260–273. [DOI] [PubMed] [Google Scholar]
- [26].Robert X, and Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server, Nucleic Acids Res 42, W320–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, Beglov D, and Vajda S (2017) The ClusPro web server for protein-protein docking, Nat Protoc 12, 255–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Navia MA, McKeever BM, Springer JP, Lin TY, Williams HR, Fluder EM, Dorn CP, and Hoogsteen K (1989) Structure of human neutrophil elastase in complex with a peptide chloromethyl ketone inhibitor at 1.84-A resolution, Proc Natl Acad Sci U S A 86, 7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Krissinel E, and Henrick K (2007) Inference of macromolecular assemblies from crystalline state, J Mol Biol 372, 774–797. [DOI] [PubMed] [Google Scholar]
- [30].Strelow J, Dewe W, Iversen PW, Brooks HB, Radding JA, McGee J, and Weidner J (2004) Mechanism of Action Assays for Enzymes, In Assay Guidance Manual (Sittampalam GS, Coussens NP, Brimacombe K, Grossman A, Arkin M, Auld D, Austin C, Baell J, Bejcek B, Caaveiro JMM, Chung TDY, Dahlin JL, Devanaryan V, Foley TL, Glicksman M, Hall MD, Haas JV, Inglese J, Iversen PW, Kahl SD, Kales SC, Lal-Nag M, Li Z, McGee J, McManus O, Riss T, Trask OJ Jr., Weidner JR, Wildey MJ, Xia M, and Xu X, Eds.), Bethesda (MD). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.