Abstract
High CO2 retention, or hypercapnia, is associated with worse outcomes in patients with chronic pulmonary diseases. Skeletal muscle wasting is also an independent predictor of poor outcomes in patients with acute and chronic pulmonary diseases. Although previous evidence indicates that high CO2 accelerates skeletal muscle catabolism via AMPK (AMP-activated protein kinase)–FoxO3a–MuRF1 (E3-ubiquitin ligase muscle RING finger protein 1), little is known about the role of high CO2 in regulating skeletal muscle anabolism. In the present study, we investigated the potential role of high CO2 in attenuating skeletal muscle protein synthesis. We found that locomotor muscles from patients with chronic CO2 retention demonstrated depressed ribosomal gene expression in comparison with locomotor muscles from non–CO2-retaining individuals, and analysis of the muscle proteome of normo- and hypercapnic mice indicates reduction of important components of ribosomal structure and function. Indeed, mice chronically kept under a high-CO2 environment show evidence of skeletal muscle downregulation of ribosomal biogenesis and decreased protein synthesis as measured by the incorporation of puromycin into skeletal muscle. Hypercapnia did not regulate the mTOR pathway, and rapamycin-induced deactivation of mTOR did not cause a decrease in ribosomal gene expression. Loss-of-function studies in cultured myotubes showed that AMPKα2 regulates CO2-mediated reductions in ribosomal gene expression and protein synthesis. Although previous evidence has implicated TIF1A (transcription initiation factor-1α) and KDM2A (lysine-specific demethylase 2A) in AMPK-driven regulation of ribosomal gene expression, we found that these mediators were not required in the high CO2–induced depressed protein anabolism. Our research supports future studies targeting ribosomal biogenesis and protein synthesis to alleviate the effects of high CO2 on skeletal muscle turnover.
Keywords: hypercapnia, skeletal muscle, anabolism, ribosomal biogenesis
Clinical Relevance
Skeletal muscle wasting and hypercapnia both contribute to higher mortality in chronic obstructive pulmonary disease. Although accelerated CO2-induced protein degradation has been reported, no data are available on the role of high CO2 retention in protein anabolism. We demonstrate that CO2 causes AMP-activated protein kinase–mediated attenuation of protein synthesis, which occurs by downregulating ribosomal biogenesis and independently of the mTOR pathway. This research could potentially allow development of strategies to target protein anabolic suppression in chronic obstructive pulmonary disease, with effects on relevant clinical outcomes.
Skeletal muscle wasting is a frequent comorbidity in patients with acute and chronic pulmonary diseases such as chronic obstructive pulmonary disease (COPD), cystic fibrosis, and others (1–3). Muscle wasting is associated with worse clinical outcomes, including higher disability, rehospitalization rates, and mortality (1–7), and results from an imbalance between protein degradation and synthesis, which leads to net muscle loss (7, 8). Although muscle protein degradation via the ubiquitin–proteasome (9–11) and autophagy (12) pathways has been observed in COPD and other pulmonary conditions, less information is available regarding the reduced protein anabolism in the context of CO2-retaining pulmonary diseases.
Chronic CO2 retention, or hypercapnia, frequently occurs in patients with COPD and other pulmonary diseases and is associated with worse outcomes, including higher mortality (13–16). Accumulating literature indicates that CO2 regulates, in a pH-independent manner, immune response (17), wound healing (18), edema reabsorption (19), and other processes (20). Recent evidence indicates that AMP-activated protein kinase (AMPK) regulates skeletal muscle turnover (21, 22). AMPK is an important energy sensor that is activated by cellular stress such as low oxygen, high CO2, or glucose deprivation (23). The mammalian AMPK consists of a heterotrimeric complex formed by a catalytic subunit (α) and two regulatory subunits (β and γ). The phosphorylation of a conserved threonine residue (Thr-172) in the kinase domain of the catalytic subunit is needed for AMPK activation (23). We have previously reported that chronic CO2 exposure leads to skeletal muscle wasting via acceleration of protein catabolism, a process mediated by the muscle-specific MuRF1 protein (E3-ubiquitin ligase muscle RING finger protein 1) (24). Surprisingly, we have also observed an association between CO2 exposure and downregulation of ribosomal RNA expression in cultured myotubes (24). Thus, the present work was conducted to determine the role of chronic hypercapnia in the regulation of muscle protein synthesis and ribosomal biogenesis. Our central hypothesis was that high CO2–mediated AMPK phosphorylation negatively modulates protein anabolism via downregulation of ribosomal RNA. Part of this study was previously published in abstract form (25).
Methods
Human Subjects
Seven patients with or without chronic CO2 retention were enrolled in this study. Ethical approval was obtained from the Albany Medical College Committee on Research Involving Human Subjects (IRB 4619), and written informed consent was obtained from the patients at the time of study participation. Enrollment took place between December 2016 and July 2018. Patients were considered for enrollment if they were older than 18 years of age and had a history of pulmonary disease with or without chronic hypercapnia, which was based on previous CO2 or serum bicarbonate determination and confirmed with arterial blood gas sampling on the day the muscle biopsy was performed. All patients were clinically stable at the time of the study, and none of them had contraindications for the biopsy procedure, including receiving chronic anticoagulation or having a known blood-clotting abnormality. Normal international normalized ratio and platelet values were also confirmed before the procedure.
Rectus femoris cross-sectional area (CSA) and thickness measurements were performed as previously established (26). Vastus lateralis biopsy was performed as previously described (27) and using a 14-gauge automatic Bard Marquee disposable core biopsy instrument (Bard MKQ1410). In short, before the procedure, local anesthesia was provided with 2 ml of lidocaine 2%. The skin was then punctured with the insertion cannula perpendicular to the muscle until the fascia was pierced, and the needle was inserted through the cannula after removal of its trocar. A muscle sample was obtained, and the procedure was repeated three times to ensure the quality and quantity of samples. The muscle specimen was removed from the biopsy needle using a sterile scalpel, and the sample was subsequently placed in a dry tube and immediately stored at −80°C for further analyses.
Animals
Wild-type male C57BL/6 mice obtained from The Jackson Laboratory were used in this study. We accounted for different variables, determined a priori, that could confound the results: animals’ age (28, 29), sex (30), mobility (31), and food intake (32) and blood values of oxygen (33) and pH (34). Male mice 12–14 weeks old were placed in a 10% CO2 chamber (BioSpherix) for 60 days. Another group of mice was placed in hypercapnia for 3 weeks to define shorter-term effects of CO2 on protein anabolism. Age- and sex-matched (male) animals left in ambient air were used as control animals. We deliberately used mice in early adulthood to avoid the potential confounding effect of aging on the reported results (35), and to determine the lack of CO2-mediated developmental effects on skeletal integrity, we measured tibial length in the mice. We also used male mice in this study to prevent the confounding effect of hormonal cycling on the skeletal muscle anabolic response, as previously reported (36). Indeed, because we have previously observed evidence of fibers’ nuclear centralization (potentially indicating accelerated muscle turnover) in high CO2 (24), and because that process has been shown to be different between sexes (37), we reasoned that animals’ sex needed to be specifically accounted for in our experiments. Food and water were accessible ad libitum, and a 12-h/12-h light/dark cycle was maintained. Animals were weighed immediately before entering the hypercapnia chamber as well as 30 and 60 days later, and again immediately before being killed, which was accomplished via cervical dislocation. Extraction of the extensor digitorum longus (EDL) and soleus muscles was performed directly after mice were killed. The time elapsed between animals’ withdrawal from the high-CO2 environment, their killing, and muscle procurement and freezing never exceeded 4–5 minutes. All the procedures involving animals were approved by the Albany Medical College Institutional Animal Care and Use Committee (IACUC protocol 06002); animals were handled according to NIH guidelines; and all methods were performed in accordance with the relevant guidelines and regulations as stated by the Journal and public agencies.
CO2 Medium and CO2 Exposure
For the different experimental conditions, initial solutions were prepared with Dulbecco’s modified Eagle medium/F-12/Tris base/3-(N-morpholino)propanesulfonic acid (MOPS) (3:1:0.25:0.25) containing differentiation media described above. The medium buffering capacity was modified by changing its initial pH with Tris base and MOPS to obtain a pH of approximately 7.4 at carbon dioxide tension (pCO2) of 40 and 120 mm Hg. The desired CO2 and pH levels were achieved by equilibrating the medium overnight in a humidified chamber (VWR). In this chamber, cells were exposed to the desired pCO2 (5% to obtain pCO2 ∼40 mm Hg and 20% to obtain pCO2 ∼120 mm Hg) while 21% O2 balanced with N2 was maintained as previously reported (24). Before and after CO2 exposure, pH, pCO2, and oxygen tension levels in the medium were measured using an i-STAT blood gas analyzer (Abbott Point of Care). Experiments were started by replacing the culture medium with the CO2-equilibrated medium and incubating it in the chamber for the desired time.
Transfection of Myotubes with siRNA
After 3 days of differentiation, media were removed from C2C12 or primary myotubes and replaced with antibiotic-free differentiation media, and transfection was performed after 4 hours. Myotubes were transfected with mouse scrambled (control), AMPKα1, AMPKα2, and KDM2A (lysine-specific demethylase 2A) siRNA duplexes (100 pmol) obtained from Santa Cruz Biotechnology and TIF1A (transcription initiation factor-1α) siRNA (100 nM) purchased from Dharmacon by using Lipofectamine RNAiMAX (Life Technologies) according to the respective manufacturers’ recommended protocols. Cells were incubated with the RNA–Lipofectamine complexes for 30 minutes at 37°C. After this period, the transfection complex was supplemented with differentiation media up to 1.5 ml for TIF1A siRNA complexes and 3 ml for all Santa Cruz Biotechnology siRNA complexes, and experiments were performed 48 hours later.
Western Blot Analysis
C2C12 and primary myotubes were homogenized in lysis buffer (Cell Signaling Technology) with 100× Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific) and then further homogenized by sonication (Sonics and Materials, Inc.). Samples were then centrifuged at 1,600 × g for 5 minutes at 4°C. Muscle samples were homogenized on ice with cold lysis buffer in a 10-fold (wt/wt) excess of lysis buffer, pH 7.6, that contained 8.7 mM NaH2PO4, 58 mM Na2HPO4, 144 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, and 100× protease and phosphatase inhibitor cocktail as previously described using a Minilys bead homogenizer (Bertin Corp.) (24). Samples were centrifuged at 22,000 × g for 10 minutes at 4°C, and after two spins, the final supernatant was collected. Protein concentrations were determined by performing a bicinchoninic acid assay (Thermo Fisher Scientific). Proteins were separated by SDS–polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes using a wet electroblotting system (Bio-Rad Laboratories), immunoblotted, and visualized by chemiluminescence following the manufacturer’s instructions (Bio-Rad Laboratories). Used antibodies, with corresponding manufacturers and working dilutions, are presented in Table E2 in the data supplement. Membranes were stripped for reprobing with an internal lane loading control using stripping buffer, pH 2.2, that contained 100 mM glycine, 20 mM Mg(CH3COO)2 · 4H2O, and 50 mM KCl. Quantification of protein concentrations was performed by densitometric scanning with Multi Gauge V3.0 software from Fujifilm or Image Lab 6.0 software (Bio-Rad Laboratories), and image exposure and contrast were adjusted to prevent saturation and to optimize adequate comparison between different conditions. For the immunoblots presented in the main figures representing sections cropped from larger membranes, the entire image has been included in the data supplement.
Statistics
Data are expressed as the mean ± SEM. For results normalized to a reference value, we used a single-sample t test, and values represent fold change in reference to control. For comparisons performed between two groups, significance was evaluated by Student’s t test, and values represent ratios of values. For comparisons in which more than two groups were compared, ANOVA was used followed by the Dunnett test. Prism software (GraphPad Software) was used. Results were considered significant at P < 0.05. For the proteomic analysis, data were processed using MaxQuant software (version 1.6.2.3). Searches were performed against a target-decoy database (UniProt [mouse]; www.uniprot.org; October 28, 2018). Searches were conducted using a 20-ppm precursor mass tolerance and a 0.04 Da product mass tolerance. A maximum of two missed tryptic cleavages were allowed. The fixed modifications specified were carbamidomethylation of cysteine residues. The variable modifications specified were oxidation of methionine and acetylation of the N-terminus. Within MaxQuant, peptides were filtered to a 1% unique peptide false discovery rate. Characterized proteins were grouped on the basis of rules of parsimony and filtered to a 1% false discovery rate (38). Label-free quantification was performed in MaxQuant (Max Plank Institute, Germany) using MaxLFQ (39). Missing values were imputed using the Perseus tool available with MaxQuant (40). Quantitative data from each experiment were log2 transformed and mean normalized across all tissues for each given protein. Significantly changing proteins were identified using a two-sided Student’s t test in Excel (Microsoft). Gene Ontology enrichment was performed in R software (41) using Fisher’s exact test and corrected for multiple comparisons using the Hochberg correction (42). Further details on the methods can be found in the data supplement.
Results
Patients with Chronic CO2 Retention Demonstrate Depressed Ribosomal Biogenesis in Locomotor Skeletal Muscle
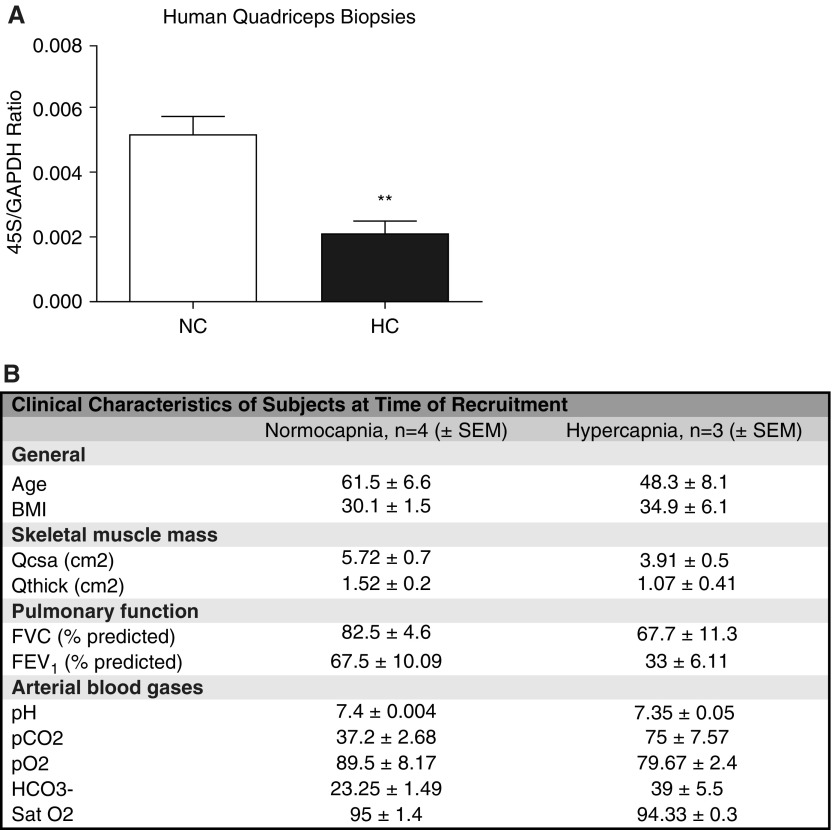
On the basis of our previous observation that high-CO2 exposure to cultured myotubes is associated with decreased expression of ribosomal RNA (24), we conducted an exploratory pilot study enrolling CO2-retaining individuals (n = 3) and nonretaining counterparts (n = 4). In these subjects, we performed percutaneous quadriceps muscle biopsies and simultaneous determination of arterial blood gases to confirm ongoing pCO2 values. Hypercapnic patients demonstrated a significant downregulation of 45S pre-RNA (pre-rRNA, a precursor of rRNA required for the assembly of the mature ribosome) compared with normocapnic control subjects (Figures 1A and 1B), which suggests, in this small cohort, that high CO2 could be associated with depressed ribosomal biogenesis.
Figure 1.
High CO2 associates with decrease of skeletal muscle ribosomal gene expression in humans. (A) Biopsies from quadriceps skeletal muscle of normocapnic (NC; n = 4) versus hypercapnic (HC; n = 3) patients were sampled, and quantitative PCR (qPCR) was performed to determine the expression level of 45S pre-RNA/GAPDH. **P < 0.01. (B) Different covariables present in both groups of patients. (No statistical analyses were performed to compare these variables, given the small number of patients in this pilot study.) BMI = body mass index; FEV1 = forced exporatory volume in 1 second; FVC = forced vital capacity; pCO2 = carbon dioxide tension; pO2 = oxygen tension; Qcsa = quadriceps cross-sectional area; Qthick = quadriceps muscle thickness; Sat O2 = arterial oxygen saturation.
High-CO2 Exposure Leads to a Proteomic Signature Suggesting Downregulation of Ribosomal Structural Components and Translation-Regulating Proteins in Murine Skeletal Muscle
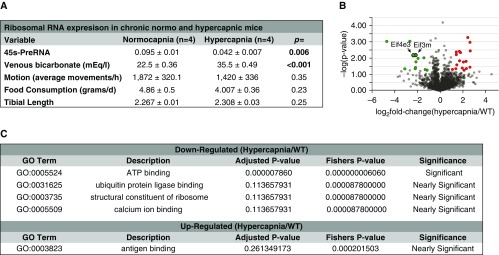
Given the presence of confounding covariables in our pilot human study (Figure 1B) that could have potentially accounted for the association of high CO2 retention with depressed ribosomal RNA expression, we conducted an animal study in which these covariables were controlled a priori (Figure 2A). Mice in normal and high CO2 environments demonstrated similar tibial lengths, suggesting that skeletal development/growth did not play a role in the effects of hypercapnia. Food intake and motion activity, as measured by aggregated vertical and horizontal motions, were not significantly different among the normocapnic and hypercapnic mice, indicating that these variables are unlikely to have played a major role in the observed effects (Figure 2A). Thus, mice were housed for 60 days in a 10% CO2 and 21% O2 (hypercapnia) environment or in room air (normocapnia), then the EDL muscle was sampled and processed for a large-scale, unbiased proteomic study. We found a significant downregulation of individual components of translation initiation, such as eukaryotic initiation factors eIF4E3 and eIF3M (P < 0.001), which recruit mRNA to the ribosome (43) (Figure 2B). Moreover, ontology enrichment analysis included “structural constituent of ribosome” among the significant or nearly significant terms generated with that dataset (Figure 2C; see the data supplement for the complete dataset). These data suggested that ribosomal biogenesis could be disrupted in skeletal muscle in the context of chronic hypercapnia and supported further studies to directly interrogate that process using validated surrogates.
Figure 2.
Hypercapnia causes a proteomic signature suggesting depressed ribosomal function and structure. (A) Covariables chosen a priori were investigated to determine their possible confounding effect on ribosomal RNA expression and puromycin incorporation. Extensor digitorum longus muscles were sampled and submitted for a large-scale proteomic analysis. (B) Volcano plot highlighting proteins found to be downregulated (green circles) and upregulated (red circles). Eif4e3 and Eif3m are indicated by arrows and significantly downregulated (P < 0.001). (C) Ontological enrichment terms identified as significant or nearly significant in this study. n = 6 (3 normocapnia and 3 hypercapnia). Eif3m = eukaryotic translation initiation factor 3 subunit m; Eif4e3 = eukaryotic translation initiation factor 4E family member 3; GO = gene ontology; WT = wild type.
High-CO2 Exposure Leads to Depressed Ribosomal RNA Expression and Protein Synthesis in Mice
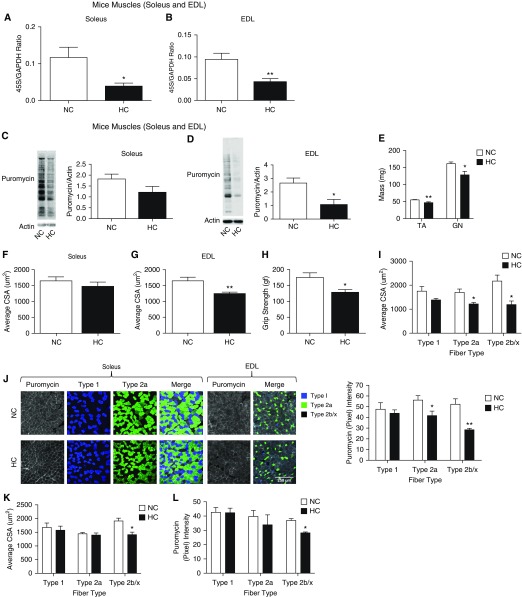
On the basis of information obtained from the previous experiments, we directly determined mouse skeletal muscle expression of 45S pre-RNA in the context of chronic high-CO2 conditions and compared that with room air counterparts. This gene expression was found to be downregulated in both soleus and EDL muscles in hypercapnic mice (Figures 3A and 3B). The housekeeping gene GAPDH, similarly to previous evidence (44, 45), was found to remain stable in high CO2 (24). To further determine whether that downregulation was associated with decreased protein synthesis, chronically hypercapnic mice received a nontoxic dose of puromycin (an aminonucleoside antibiotic with a structure that mimics tRNA, which can be immunodetected) for 30 minutes before being killed (Figure E1) (46). Immunoblot analysis of lysates from soleus and EDL muscles incubated with antipuromycin antibody demonstrated a significant decrease of puromycin incorporation in hypercapnic mice, reflecting an attenuation of protein synthesis during the time elapsed between the puromycin injection and muscle procurement (Figures 3C and 3D). Consistent with previous data, hypercapnic mice demonstrated decreased tibialis anterior and gastrocnemius muscle weight (Figure 3E). CSAs fibers’ reductions reached statistical significance only in EDL and not in soleus muscle, consistent with a more glicolytic profile of EDL (see below and Figures 3F and 3G), and in force-generation capacity as measured by the grip-strength method (Figure 3H). Interestingly, although we previously found that a 3-week high-CO2 exposure led to similar reduction of CSA among types I and II fibers (24), the present data involving longer-term hypercapnia and using a more specific identification method of fiber types were associated with more significant atrophy of type II (glycolytic) fibers, specifically types IIa and IIb/x (Figure 3I). Moreover, anabolic attenuation was not evident in the shorter-term CO2 exposure (Figures E2A and E2B), which suggests that this process is activated at a later time in the context of chronic hypercapnia.
Figure 3.
Hypercapnia causes a decrease in skeletal muscle anabolism and expression of ribosomal RNA. Age-matched male C57BL/6 mice were exposed to 10% CO2 (HC) for 60 days or maintained in room air (NC). RNA was isolated from these samples; qPCR was performed; and the amount of 45S pre-RNA/GAPDH ratio was determined in (A) soleus (n = 6) and (B) extensor digitorum longus (EDL) muscles (n = 6). After chronic high CO2 or room air exposures and 30 minutes before mice were killed, 0.040 μmol/g of puromycin was intraperitoneally injected. Then, soleus and EDL muscles were procured, samples were immunoblotted with antipuromycin antibody, and the intensity of the resultant smear was scored. (C) Soleus muscle puromycin incorporation (n = 3). (D) EDL muscle puromycin incorporation (n = 3). (E) Tibialis anterior (TA) and gastrocnemius (GN) muscles from NC and HC mice were procured and immediately weighed (n = 3). (F) Soleus muscle average fiber size in NC and HC (n = 3). (G) EDL muscle average fiber size in NC and HC (n = 3). (H) Grip strength determinations in NC and HC mice (n = 3). (I) Fiber-specific cross-sectional area (CSA) in NC and HC (n = 3). (J) Cryosectioned samples from soleus or EDL muscles were immunostained with antipuromycin antibody and specific antibodies to type I, IIa, or type IIb/x fibers. Then, the amount of fiber-incorporated puromycin in the specific fiber types of soleus and EDL muscles was scored by grayscale scoring of room air–maintained (NC) CO2 versus 10% CO2 for 60 days (HC). NC and HC mice underwent a regime of endurance exercise during 1 month. Scale bar: 250 μm. (K and L) Then, fiber-specific CSA (K) and puromycin incorporation (L) were determined and scored. *P < 0.05 and **P < 0.01.
Because we observed a more robust downregulation of ribosomal gene expression, puromycin incorporation, and atrophy in EDL than in soleus muscle (Figures 3A–3D), and because soleus muscle is richer in type I (oxidative) fibers than the type II (glycolytic)-rich EDL muscle (47), we speculated that CO2-driven anabolic attenuation could be fiber type specific. To address that hypothesis, we scored the immunofluorescence intensity of EDL and soleus muscle sections stained with both antipuromycin antibody and merged with images stained with fiber type myosin heavy chain isoform antibodies (46). We found that in hypercapnia, type II fibers demonstrate a more robust attenuation of puromycin signal than type I fibers (Figure 3J and higher magnification in Figure E3A), which is associated with the mentioned CO2-induced reduction of fiber type–specific diameter (Figure 3I). Moreover, we took advantage of the anabolic stimulation entailed by 1 month of endurance exercise in normocapnic and hypercapnic mice, which indicates that although hypercapnic (more oxidative) types I and IIa fibers regain baseline normocapnia-similar CSA and puromycin contents, more glycolytic type IIb/x fibers fail to significantly increase both variables under the same exercise protocol (Figures 3K, 3L, and E3B). These data suggest that the muscle wasting associated with chronic CO2 retention could be contributed by decreased expression of ribosomal RNA and protein synthesis and that this process is more pronounced in type II fibers than in type I fibers. Also, the magnitude of fiber-specific mass and protein puromycin incorporation is associated in the context of hypercapnia, both in a steady-state catabolic status and after endurance exercise.
Hypercapnia Leads to Depressed Ribosomal RNA Expression and Protein Synthesis In Vitro
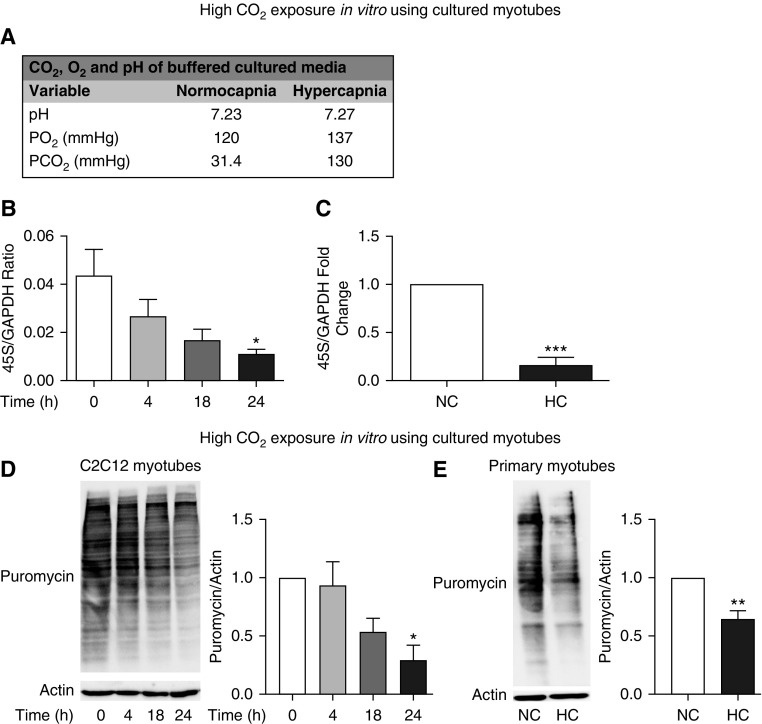
To determine whether in vitro hypercapnic conditions recapitulate the observed in vivo decrease in ribosomal RNA transcription and protein synthesis, we established two cell culture models: C2C12 cells, which are nontransformed mouse skeletal muscle myotubes (48); and primary myotubes, obtained from in vitro differentiation of epitope-specific isolated myoblasts (49). In both cells, exposure to a culture medium buffered to maintain normal pH and oxygen but high CO2 (Figure 4A) resulted in a significant reduction of ribosomal RNA expression (Figures 4B and 4C) and puromycin incorporation (Figures 4D and 4E). Importantly, these cells demonstrate morphological and molecular characteristics of skeletal muscle that are not affected by exposure to puromycin and high CO2 (Figures E4A–E4C). These data indicate that an in vitro model using cultured myotubes recapitulates the effects of high CO2 on ribosomal gene expression and protein synthesis as reflected by the puromycin incorporation and can be used as a system to perform mechanistic studies.
Figure 4.
Cultured myotubes demonstrate hypercapnia-driven decrease in protein synthesis and ribosomal gene expression. (A) Values of pH, pCO2, and pO2 as measured in buffered media under NC and high-CO2 (HC) conditions. (B and C) C2C12 myotubes exposed to 20% CO2 for 4, 18, and 24 hours as compared with cells maintained in 5% CO2 (0 h) were sampled, and 45S-preRNA/GAPDH expression was determined with qPCR (n = 3). (C) Primary myotubes exposed to 20% CO2 for 24 hours as compared with cells maintained in 5% CO2 (0 h) were sampled, and expression of 45S-preRNA/GAPDH expression was determined with qPCR (n = 3). (D) Representative immunoblot of C2C12 myotubes exposed to 20% CO2 for 4, 18, and 24 hours as compared with cells maintained in 5% CO2 (0 h). All cells were kept on a puromycin-containing medium, and then lysates were immunoblotted with antipuromycin antibody, and the intensity was interpreted as the index of puromycin incorporation over the analyzed period. Membranes were then stripped and probed with antiactin antibody, used as a lane loading control (n = 3). (E) Representative immunoblot of primary myotubes exposed to 20% CO2 for 24 hours as compared with cells maintained in 5% CO2 (0 h). Cells were kept on a puromycin-containing medium, and then lysates were probed with antipuromycin antibody, and the intensity was interpreted as the index of puromycin incorporation over the analyzed period. Membranes were then stripped and probed with antiactin antibody, used as a lane loading control (n = 3). *P < 0.05, **P < 0.01, and ***P < 0.001.
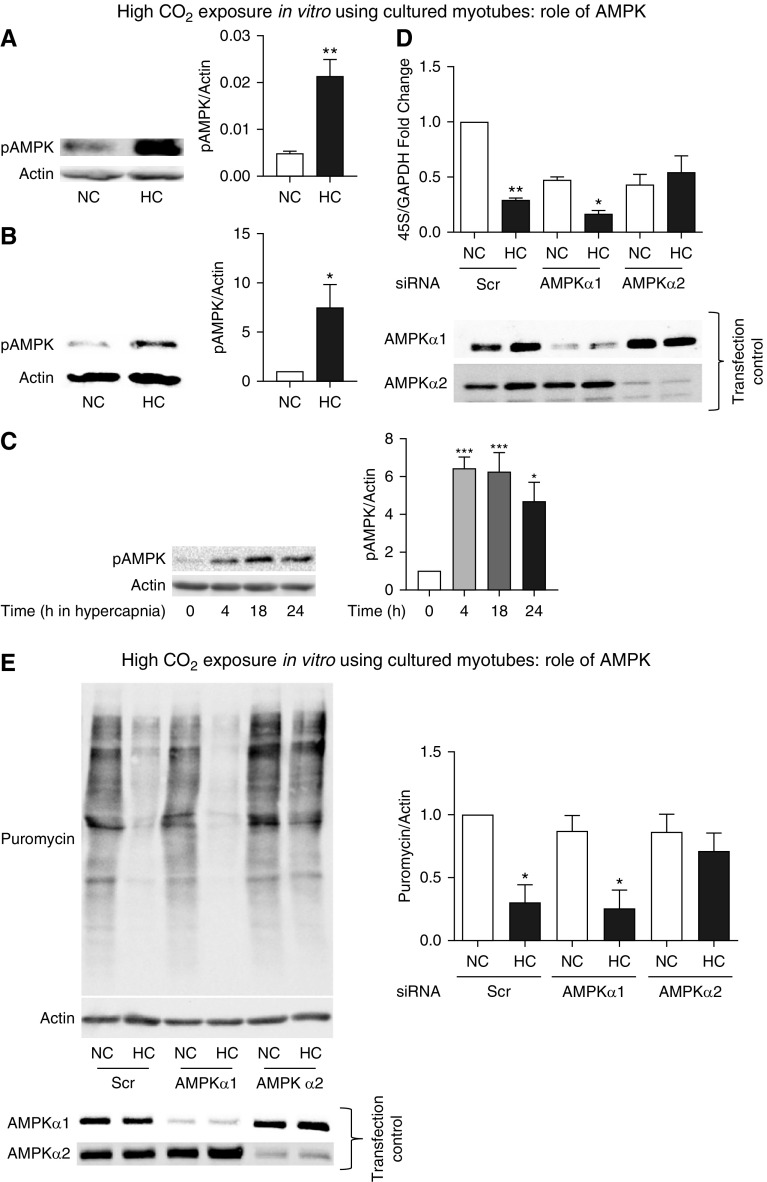
High-CO2 Effect on rRNA Expression and Protein Synthesis Depends on AMPKα2
Exposure to high CO2 has previously been shown to cause AMPK phosphorylation in different cell lines, including fibroblasts, alveolar epithelial cells, and skeletal muscle myotubes (19, 24). We confirmed this process in chronically hypercapnic mice (Figure 5A) and in cultured myotubes (Figures 5B and 5C). We then investigated the possible role of AMPK in regulating ribosomal RNA and protein synthesis by transfecting C2C12 cells with scrambled or AMPK isoform–specific siRNAs. We found that knockdown of AMPKα2, but not AMPKα1, attenuated the high CO2–mediated reduction in 45S pre-RNA expression (Figure 5D) and puromycin incorporation (Figure 5E). Because AMPK regulation by the mTOR pathway has been reported to modulate ribosomal biogenesis and protein synthesis (50, 51), we investigated the potential role of that signaling in the context of high-CO2 exposure. Although myotubes treated with rapamycin demonstrated a robust dephosphorylation of mTOR, no such effect was observed in the context of CO2 stimulation (Figure E5A). Moreover, although high CO2 causes robust and significant downregulation of 45S pre-RNA expression and puromycin incorporation (Figures 4B–4E), rapamycin exerts no significant effect in 45S pre-RNA (Figure E5B); yet, it causes a significant reduction of puromycin incorporation (Figure E5C). These mTOR pathway effects demonstrated in cultured myotubes are similarly observed in vivo (Figures E5D–E5F). Taken together, our data suggest that CO2 leads to AMPK activation that negatively regulates ribosomal RNA expression and protein synthesis specifically via AMPKα2 in cultured myotubes, effects that are not mimicked by rapamycin-induced deactivation of the mTOR pathway.
Figure 5.
AMP-activated protein kinase (AMPK) regulates protein anabolism during exposure to high CO2 in cultured myotubes. (A) AMPK phosphorylation (pAMPK) in skeletal muscle from mice exposed to high CO2 (HC) versus room air (NC) for 60 days. Actin was used as a lane loading control (n = 3). (B) pAMPK in primary myotubes exposed to 40 mm Hg (NC) or 120 mm Hg (HC) CO2 for 24 hours. Actin was used as a lane loading control (n = 3). (C) pAMPK in C2C12 myotubes exposed to 40 mm Hg (NC) or 120 mm Hg (HC) CO2 for different durations. Actin was used as a lane loading control. (D) Myotubes were transfected with scrambled (Scr), AMPKα1, or AMPKα2 siRNA and then exposed to normal (NC) or high CO2 (HC). Representative figures depict results of qPCR with primers to amplify 45S pre-RNA/GAPDH. Cell samples were processed for Western blotting and probed with specific antibodies to indicate AMPKα1 or AMPKα2 siRNA expression as a control of transfection and silencing efficiency in each condition (n = 3). (E) Conditions similar to D with myotubes exposed to NC and HC for 24 hours in the presence of puromycin-containing medium. Samples were then lysed and immunoblotted with antipuromycin antibody, and the intensity of the smears was scored as shown. Membrane was then stripped and probed with actin as a lane loading control. Transfection control was carried out by Western blotting and probing with specific antibodies to indicate specific silencing of AMPKα1 or AMPKα2 siRNA (n = 3). *P < 0.05, **P < 0.01, and ***P < 0.001.
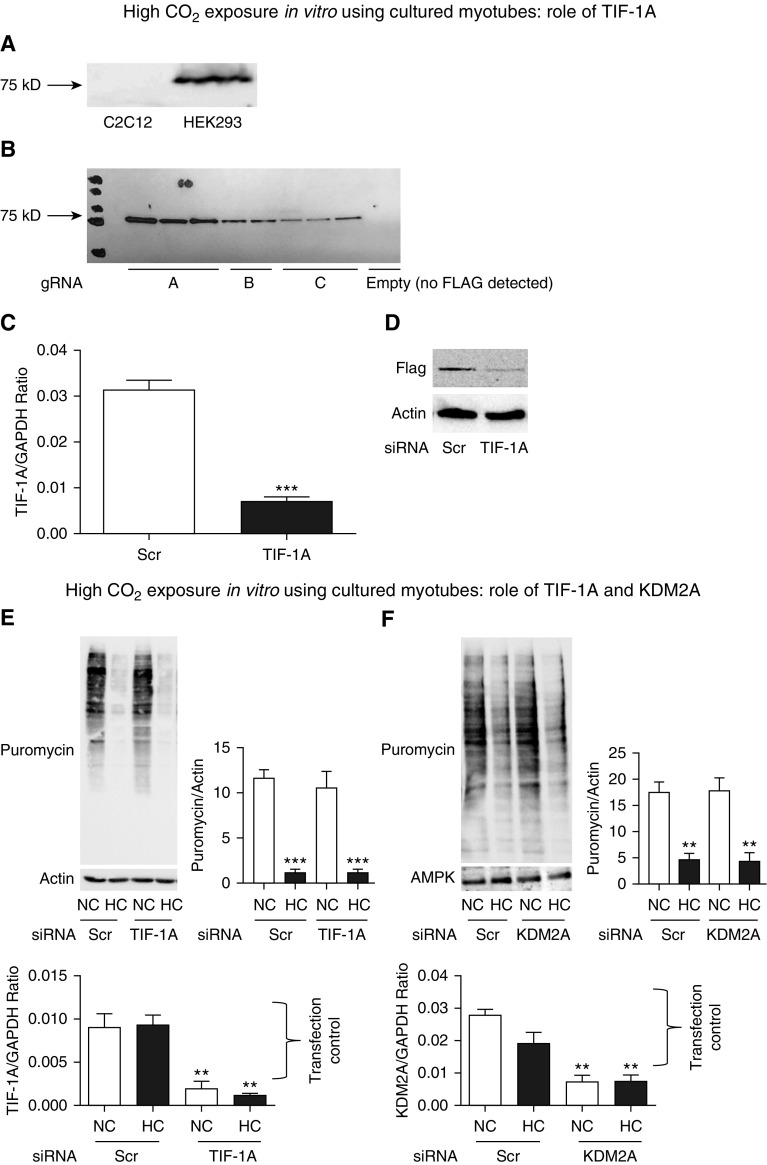
CO2-mediated AMPK Regulation of Protein Synthesis Is Independent of TIF1A or KDM2A
Because AMPK has previously been shown to phosphorylate the transcription factor TIF1A, leading to inhibition of ribosomal RNA expression (52, 53), we hypothesized that this mechanism could be relevant in the context of high CO2. Using different commercially available antibodies, we were unable to reliably detect a TIF1A product of approximately 75 kD in lysates for C2C12 cells or primary myotubes, whereas a robust band of that molecular weight was detected in lysates from human embryonic kidney cells (HEK293) (Figure 6A, showing one antibody as an example). These antibodies are reported to detect TIF1A in mouse cells (54), which made it unlikely that the lack of a visible C2C12 or primary myotube product was due to a species-related reactivity. Surprisingly, both C2C12 cells and primary myotubes expressed a 50 kD product that was detectable by various TIF1A antibodies and regulated during the transition from myoblast into myotube form (Figures E6A–E6D). Thus, we directly evaluated TIF1A expression in C2C12 myotubes using a Crispr/Cas9-mediated addition of a 3×FLAG tag to exon 18 of the TIF1A gene. Using this strategy, we were able to detect a band at 75 kD (Figure 6B). Silencing TIF1A with specific siRNA led to downregulation of both the RNA expression and FLAG-tagged protein product, confirming TIF1A expression (Figures 6C and 6D) in the cultured skeletal muscle cells, despite our inability to detect the endogenous wild-type form by Western blotting with commercial antibodies. To screen for a possible role of TIF1A in the CO2-mediated downregulation of muscle anabolism, we incubated C2C12 cells previously transfected with scrambled or TIF1A-specific siRNA with normal or high CO2. Surprisingly, silencing of TIF1A did not prevent the decrease in puromycin incorporation into myotubes in response to hypercapnia (Figure 6E).
Figure 6.
CO2-driven skeletal muscle anabolic attenuation is independent of TIF1A and KDM2A. (A) Example of commercially available anti–TIF1A antibody that does not detect an approximately 75 kD band in lysates of C2C12 cells, whereas it does in HEK293 cells. (B) Cas9-expressing C2C12 cells were transfected with a donor clone holding the sequence of 3×FLAG and with three different guide RNAs (gRNA; A, B, and C) or no guide RNA (last lane with no FLAG detection) to localize the donor clone to exon 18 of the TIF1A gene. Cells were then lysed and immunoblotted with anti-FLAG antibody, demonstrating a band of approximately 75 kD (expected weight of TIF1A). (C) C2C12 cells from (B) (clone A) were transfected with Scr or specific TIF1A siRNA and then sampled for qPCR using TIF1A-amplifying primers, which demonstrate a decrease in the gene product in the siRNA-transfected group. GAPDH was used as a housekeeping control (n = 3). (D) The same C2C12 cells as in (C) were processed for Western blotting and probed with anti-FLAG antibody, demonstrating a significant decrease in the FLAG band intensity in the silenced group. Actin was used as a lane loading control (n = 3). (E) Myotubes were transfected with Scr or TIF1A siRNA and then exposed to NC or HC for 24 hours in the presence of puromycin-containing medium. Samples were then lysed and immunoblotted with antipuromycin antibody, and the intensity of the smears was interpreted as the amount of protein synthesis occurring in 24 hours. Actin was used as a lane loading control. Representative graphs depict results of qPCR with primers to amplify TIF1A and GAPDH used as a housekeeping control, demonstrating the siRNA transfection and silencing efficiency (n = 3). (F) Myotubes were transfected with Scr or KDM2A siRNA and then exposed to NC or HC for 24 hours in the presence of puromycin-containing medium. Samples were then lysed and immunoblotted with antipuromycin antibody, and the intensity of the smears was interpreted as the amount of protein synthesis occurring in 24 hours. Representative graphs depict the results of qPCR with primers to amplify KDM2A and GAPDH as a housekeeping control, demonstrating the siRNA transfection and silencing efficiency (n = 3). **P < 0.01 and ***P < 0.001. C2C12 = immortalized mouse myoblast cell line; Cas9 = CRISPR associated protein 9; FLAG = octapeptide sequence added to the TIF1A gene is DYKDDDDK; HEK293 = human embryonic kidney 293; KDM2A = lysine demethylase 2A; T1F1A = transcription intermediary factor 1-α.
Previous research has also shown that AMPK regulates ribosomal biogenesis via KDM2A-mediated H3K36me2 demethylation in the rRNA gene, and thus we entertained the possibility that this process could also be relevant in hypercapnia. We therefore transfected myotubes with scrambled and specific KDM2A siRNA, and we found that, similarly to TIF1A knockout, it failed to protect hypercapnia-exposed cells from depressed protein synthesis (Figure 6F). Consistent with previous literature, silencing of TIF1A (52) or KDM2A (55) had no significant effect on 45S pre-RNA expression in baseline conditions (Figures E6F and E6G). Taken together, our data indicate that an unidentified mechanism involving neither TIF1A nor KDM2A mediates the CO2-triggered downregulation of protein synthesis in skeletal muscle.
Discussion
Skeletal muscle dysfunction is a major comorbidity in patients with acute and chronic pulmonary diseases, including COPD and others (1, 2), and is independently associated with mortality in these populations (3–5, 16, 56). Moreover, recovery of skeletal muscle mass and function in the context of pulmonary rehabilitation has demonstrated benefits in important outcomes, including hospital readmissions, rate of exacerbations, and others (7, 57–59). Muscle wasting is associated with an imbalance between protein synthesis and degradation, and we are reporting that in the present study, chronic CO2 retention or hypercapnia, which is prevalent in patients with COPD, was associated with a dysfunctional protein anabolism contributed by downregulated ribosomal biogenesis. Specifically, we conducted a small exploratory pilot study that suggested a potential association between chronic hypercapnia and decreased skeletal muscle ribosomal RNA expression in humans. We also performed an unbiased analysis of the skeletal muscle proteome from animals housed in normal and high CO2 environments, which also suggested an association between hypercapnia and depressed ribosome-mediated anabolism. The process was then directly confirmed in high CO2–exposed mice and in cultured cells using a validated surrogate of ribosomal biogenesis. Importantly, animal and cellular data also indicate a hypercapnia-induced depressed muscle puromycin incorporation, suggesting attenuated protein synthesis in that setting (46), which could depend on ribosomal biogenesis. Rapamycin-induced deactivation of the mTOR pathway was unable to recapitulate the hypercapnia effects on ribosomal gene expression, and hypercapnia exposure did not regulate mTOR phosphorylation status, indicating that although CO2 stimulation and rapamycin-driven deactivation of mTOR are both able to attenuate muscle protein anabolism, they operate through distinct signaling pathways. Common confounders that could potentially lead to muscle wasting, such as lower food intake, motion activity, or skeletal development, are not significantly different between animals in normocapnic and high-CO2 environments, indicating that they are unlikely to have played a major role in the current model and facilitating the use of this setting to interrogate the association between hypercapnia and attenuated anabolism.
Although deceleration of protein synthesis does not necessarily require a decrease in the ribosomal mass (45), lower transcription of the 45S pre-rRNA has been described to downregulate cellular anabolism in different settings, such as skeletal muscle atrophy and others (54, 60–62). Moreover, dysfunctional ribosomal biogenesis has been described in age-related sarcopenia (45).
Ribosomal biogenesis involves the generation and processing of the four rRNAs and more than 80 ribosomal proteins that form the mature 80S eukaryotic ribosome (63). Three classes of RNA polymerases participate in that process, which also requires the synthesis of an array of proteins related to processing, assembly, and nuclear import/export of ribosomes (64). Synthesis of rRNA is a major rate-limiting step in ribosomal biogenesis, with rRNA comprising 85% of total cellular RNA (45). Specifically, of the four rRNAs, three (28S, 18S, and 5.8S rRNAs) are transcribed from a single gene (ribosomal DNA [rDNA]) that exists in hundreds of tandem repeats throughout the genome (65). Transcription of rDNA via RNA polymerase I (Pol I) leads to the generation of a precursor rRNA, 45S pre-rRNA, which is processed to form the 28S, 18S, and 5.8S rRNAs. Evidence indicates that TIF1A regulates Pol I activation in different cellular models (65).
Previously, using 3-week hypercapnia stimulation, we found that CO2-triggered muscle catabolism is not associated with different effects in type I versus type II fibers (24). In this study, using more specific fiber type identification methods, we found that 8-week hypercapnia-induced anabolic attenuation and reduction of CSA are both more pronounced in the glycolytic type IIa fibers and even more in type IIb/x than in the oxidative type I fibers, suggesting that less oxidative myofibers are more vulnerable to this wasting signal. Indeed, the association between fiber-specific reduction of anabolism and fiber mass persists after 1 month of endurance exercise, which leads to regaining of puromycin incorporation and CSA in the more oxidative type I and type IIa fibers, whereas the more glycolytic type IIb/x fibers remain relatively atrophic and puromycin poor. The timing of CO2-induced altered muscle turnover indicates that attenuation of puromycin incorporation seen at 8 weeks is not observed at earlier time points. Similar observations regarding higher type II fiber lability have been made in different human and animal models, including COPD (66). The causes of fiber-specific anabolic attenuation remain to be determined and are beyond the scope of this report. Possible mechanisms regulating higher vulnerability of less oxidative fibers involve previously reported CO2-induced microRNA-183–mediated mitochondrial dysfunction, which could have more impact on cells with lower oxidative capacity (18).
Previous work by different groups has implicated AMPK in the regulation of muscle wasting (12, 21, 24), and we have reported that high CO2–induced muscle atrophy is mediated by AMPKα2 (24). In this work, we exposed cultured myotubes to high CO2 and found an association between the time-dependent, CO2-induced AMPK phosphorylation, downregulation of 45S pre-RNA, and reduction of puromycin incorporation. The silencing of AMPKα2, but not AMPKα1, attenuated the effects of high CO2 on 45S pre-RNA and puromycin. AMPKα2 is the major catalytic subunit expressed in skeletal muscle and has also been shown to mediate activation of proteasomal degradation and muscle atrophy induced by high CO2 (24).
The precise signaling mechanism leading to CO2-induced, AMPKα2-dependent downregulation of 45S pre-RNA and protein synthesis remains undefined. We investigated previously reported pathways that were found not to be relevant in this model. The transcription factor TIF1A was reported to be phosphorylated and inhibited by AMPK, leading to decreased assembly of a functional transcription initiation complex. Interestingly, although TIF1A molecular weight is approximately 75 kD, we found that various commercially available antibodies raised to the N- and C-terminal areas of the protein detect, in myotubes, an atypical band of approximately 50 kD that is regulated during the transition from myoblasts into myotubes and is specifically pulled down upon IP with these antibodies. To determine whether this band represented TIF1A or a nonspecific binding to commercial antibodies, we used Crispr/Cas9 technology to introduce a 3×FLAG tag to the TIF1A gene and confirmed the expression of a product with expected molecular weight (∼75 kD, not 50 kD) in C2C12 cells. Further confirmation of the product’s identity was obtained by consistent siRNA-mediated silencing in both C2C12 (3×FLAG and RNA) and primary myotubes (RNA). Although AMPK regulation of TIF1A was originally investigated in nonmuscle cells and upon glucose deprivation (52), and recently in myocardial cells (53), our data suggest that CO2-driven downregulation of skeletal muscle protein synthesis is not mediated by TIF1A.
Another reported mechanism of AMPK-driven depressed ribosomal biogenesis is KDM2A-mediated H3K36me2 demethylation in the rRNA gene. However, silencing KDM2A with specific siRNA did not attenuate the effect of high CO2 on protein synthesis. Thus, it appears that some unidentified molecular link between AMPK and ribosomal biogenesis participates and regulates the skeletal muscle anabolic suppression observed in the context of chronic hypercapnia. Identification of this mechanism potentially involving epigenetic changes leading to modulation of DNA methylation, histone acetylation and methylation (67), and regulation of noncoding RNAs such as microRNAs (68, 69) will be a focus of future studies.
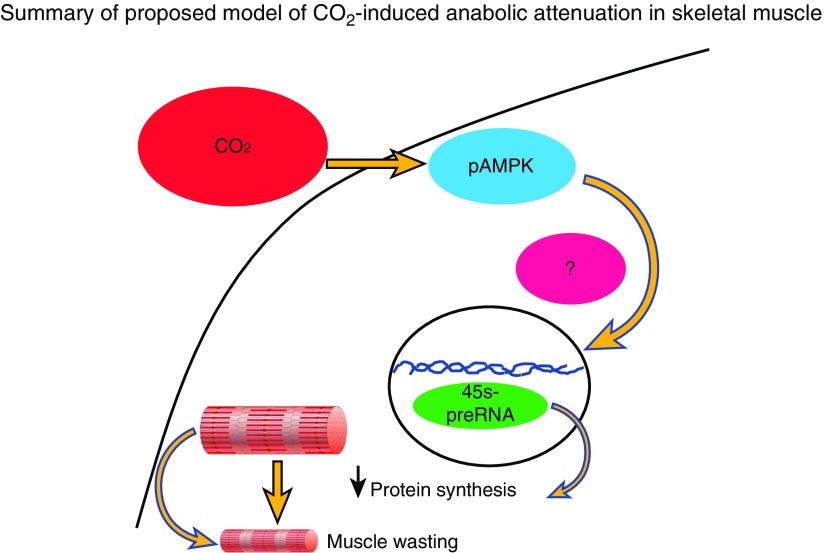
The main strengths of this study include being, to our knowledge, the first to establish a clear link between chronic CO2 retention, ribosomal biogenesis, and protein synthesis in humans and mice, which is not mimicked by pharmacological deactivation of mTOR pathway and depends, at least in cultured myotubes, on AMPKα2. Given the relevance of hypercapnia and muscle integrity to pulmonary diseases outcomes, this process could be a potential target to alleviate an important comorbidity. Although the endurance exercise protocol we used allowed us to investigate the fiber-specific association between fiber anabolism and mass, the present data could stimulate future studies focused on the effects of chronic hypercapnia on skeletal muscle response to exercise, which could add potentially important insight related to rehabilitation strategies commonly used in patients with chronic pulmonary diseases. Also, and given the relevance of ribosomal function on skeletal muscle turnover, we present a highly validated instrument to investigate the role of TIF1A that is particularly useful, considering the limited value of available antibodies in that setting. Our study has some limitations, including the lack of a mechanism that links AMPK to ribosomal biogenesis, the small size of our human cohort, and the lack of a transgenic AMPK animal that validates our findings in vivo. Another limitation is that although the association between fiber-specific puromycin incorporation and CSA, both in steady-state hypercapnia and after endurance exercise, suggests a contribution of CO2-induced anabolic attenuation to muscle wasting, owing to the overlap between protein degradation and synthesis, we are unable to determine the magnitude contributed by each individual process to the fiber phenotype. Future studies could provide more quantitative data. In summary, we report that hypercapnia contributes to skeletal muscle wasting by causing anabolic suppression via AMPKα2-dependent depressed ribosomal biogenesis and protein synthesis (Figure 7), a pathway that seems to be of pathophysiological significance in patients with chronic CO2-retaining pulmonary diseases.
Figure 7.
A proposed schematic model showing that hypercapnia triggers skeletal muscle anabolic attenuation though a pathway that involves the activation of AMPKα2, downregulation of ribosomal RNA expression, and protein synthesis. We were unable to identify the mechanism that links AMPKα2 with ribosomal gene expression in this setting.
Supplementary Material
Acknowledgments
Acknowledgment
The authors appreciate the help provided by Nina Martino with the molecular biology assays, Darren Lydon for his assistance in the blood gases determination, and Leahruth Saavedra and Indrawattie Sukhu with the patients’ enrollment and muscle biopsy performances. The authors acknowledge the National Center for Quantitative Biology of Complex Systems (NCQBCS; P41 GM108538) at the University of Wisconsin, Madison, for graciously providing the liquid chromatography–tandem mass spectrometry instrument time required to analyze our proteomic samples. The authors especially thank Alexander S. Hebert and Joshua J. Coon for facilitating sample transport and data acquisition at the NCQBCS.
Footnotes
Supported in part by the National Heart, Lung, and Blood Institute (NHLBI) of the National Institutes of Health (NIH) under award number NIH/NHLBI K01-HL130704 (A.J.), the Collins Family Foundation Endowment (A.J.), American Heart Association grant 18TPA34170561 and NIH/National Institute of General Medical Science grant 1R01GM124133 (A.P.A.), and NIH/NHLBI grant 5R01HL049426 (H.A.S.).
Author Contributions: T.C.K., J.B., D.V.S., and A.J. designed and performed experiments. J.B. and C.E.V. performed proteomic analyses. A.C. and A.J. performed human biopsies. A.P.A., R.G., H.A.S., and A.J. designed the experiments and wrote the current manuscript.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2019-0061OC on July 2, 2019
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Jaitovich A, Barreiro E. Skeletal muscle dysfunction in chronic obstructive pulmonary disease: what we know and can do for our patients. Am J Respir Crit Care Med. 2018;198:175–186. doi: 10.1164/rccm.201710-2140CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaitovich A, Khan MMHS, Itty R, Chieng HC, Dumas CL, Nadendla P, et al. ICU admission muscle and fat mass, survival, and disability at discharge: a prospective cohort study. Chest. 2019;155:322–330. doi: 10.1016/j.chest.2018.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma R, Florea VG, Bolger AP, Doehner W, Florea ND, Coats AJ, et al. Wasting as an independent predictor of mortality in patients with cystic fibrosis. Thorax. 2001;56:746–750. doi: 10.1136/thorax.56.10.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marquis K, Debigaré R, Lacasse Y, LeBlanc P, Jobin J, Carrier G, et al. Midthigh muscle cross-sectional area is a better predictor of mortality than body mass index in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2002;166:809–813. doi: 10.1164/rccm.2107031. [DOI] [PubMed] [Google Scholar]
- 5.Shrikrishna D, Patel M, Tanner RJ, Seymour JM, Connolly BA, Puthucheary ZA, et al. Quadriceps wasting and physical inactivity in patients with COPD. Eur Respir J. 2012;40:1115–1122. doi: 10.1183/09031936.00170111. [DOI] [PubMed] [Google Scholar]
- 6.Puthucheary ZA, Rawal J, McPhail M, Connolly B, Ratnayake G, Chan P, et al. Acute skeletal muscle wasting in critical illness. JAMA. 2013;310:1591–1600. doi: 10.1001/jama.2013.278481. [DOI] [PubMed] [Google Scholar]
- 7.Barreiro E, Jaitovich A. Skeletal muscle dysfunction in COPD: relevance of nutritional support and pulmonary rehabilitation. J Thorac Dis. 2018;10(Suppl 12):S1330–S1331. doi: 10.21037/jtd.2018.05.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodine SC, Baehr LM. Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1. Am J Physiol Endocrinol Metab. 2014;307:E469–E484. doi: 10.1152/ajpendo.00204.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doucet M, Russell AP, Léger B, Debigaré R, Joanisse DR, Caron MA, et al. Muscle atrophy and hypertrophy signaling in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2007;176:261–269. doi: 10.1164/rccm.200605-704OC. [DOI] [PubMed] [Google Scholar]
- 10.Crul T, Testelmans D, Spruit MA, Troosters T, Gosselink R, Geeraerts I, et al. Gene expression profiling in vastus lateralis muscle during an acute exacerbation of COPD. Cell Physiol Biochem. 2010;25:491–500. doi: 10.1159/000303054. [DOI] [PubMed] [Google Scholar]
- 11.Plant PJ, Brooks D, Faughnan M, Bayley T, Bain J, Singer L, et al. Cellular markers of muscle atrophy in chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2010;42:461–471. doi: 10.1165/rcmb.2008-0382OC. [DOI] [PubMed] [Google Scholar]
- 12.Guo Y, Gosker HR, Schols AM, Kapchinsky S, Bourbeau J, Sandri M, et al. Autophagy in locomotor muscles of patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2013;188:1313–1320. doi: 10.1164/rccm.201304-0732OC. [DOI] [PubMed] [Google Scholar]
- 13.Belkin RA, Henig NR, Singer LG, Chaparro C, Rubenstein RC, Xie SX, et al. Risk factors for death of patients with cystic fibrosis awaiting lung transplantation. Am J Respir Crit Care Med. 2006;173:659–666. doi: 10.1164/rccm.200410-1369OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinberger SE, Schwartzstein RM, Weiss JW. Hypercapnia. N Engl J Med. 1989;321:1223–1231. doi: 10.1056/NEJM198911023211804. [DOI] [PubMed] [Google Scholar]
- 15.Brat K, Plutinsky M, Hejduk K, Svoboda M, Popelkova P, Zatloukal J, et al. Respiratory parameters predict poor outcome in COPD patients, category GOLD 2017 B. Int J Chron Obstruct Pulmon Dis. 2018;13:1037–1052. doi: 10.2147/COPD.S147262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nin N, Muriel A, Peñuelas O, Brochard L, Lorente JA, Ferguson ND, et al. VENTILA Group. Severe hypercapnia and outcome of mechanically ventilated patients with moderate or severe acute respiratory distress syndrome. Intensive Care Med. 2017;43:200–208. doi: 10.1007/s00134-016-4611-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu Z, Casalino-Matsuda SM, Nair A, Buchbinder A, Budinger GRS, Sporn PHS, et al. A role for heat shock factor 1 in hypercapnia-induced inhibition of inflammatory cytokine expression. FASEB J. 2018;32:3614–3622. doi: 10.1096/fj.201701164R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vohwinkel CU, Lecuona E, Sun H, Sommer N, Vadász I, Chandel NS, et al. Elevated CO2 levels cause mitochondrial dysfunction and impair cell proliferation. J Biol Chem. 2011;286:37067–37076. doi: 10.1074/jbc.M111.290056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vadász I, Dada LA, Briva A, Trejo HE, Welch LC, Chen J, et al. AMP-activated protein kinase regulates CO2-induced alveolar epithelial dysfunction in rats and human cells by promoting Na,K-ATPase endocytosis. J Clin Invest. 2008;118:752–762. doi: 10.1172/JCI29723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vadász I, Hubmayr RD, Nin N, Sporn PH, Sznajder JI. Hypercapnia: a nonpermissive environment for the lung. Am J Respir Cell Mol Biol. 2012;46:417–421. doi: 10.1165/rcmb.2011-0395PS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakashima K, Yakabe Y. AMPK activation stimulates myofibrillar protein degradation and expression of atrophy-related ubiquitin ligases by increasing FOXO transcription factors in C2C12 myotubes. Biosci Biotechnol Biochem. 2007;71:1650–1656. doi: 10.1271/bbb.70057. Published erratum appears in Biosci Biotechnol Biochem 72:2008E2. [DOI] [PubMed] [Google Scholar]
- 22.Nystrom GJ, Lang CH. Sepsis and AMPK activation by AICAR differentially regulate FoxO-1, -3 and -4 mRNA in striated muscle. Int J Clin Exp Med. 2008;1:50–63. [PMC free article] [PubMed] [Google Scholar]
- 23.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaitovich A, Angulo M, Lecuona E, Dada LA, Welch LC, Cheng Y, et al. High CO2 levels cause skeletal muscle atrophy via AMP-activated kinase (AMPK), FoxO3a protein, and muscle-specific Ring finger protein 1 (MuRF1) J Biol Chem. 2015;290:9183–9194. doi: 10.1074/jbc.M114.625715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaitovich AA, Korponay T, Balnis J, Nguyen T, Singer H. Hypercapnia suppresses protein anabolism in skeletal muscle via AMPK-driven downregulation of ribosomal biogenesis [abstract] Am J Respir Crit Care Med. 2018;197:A7141. [Google Scholar]
- 26.Greening NJ, Harvey-Dunstan TC, Chaplin EJ, Vincent EE, Morgan MD, Singh SJ, et al. Bedside assessment of quadriceps muscle by ultrasound after admission for acute exacerbations of chronic respiratory disease. Am J Respir Crit Care Med. 2015;192:810–816. doi: 10.1164/rccm.201503-0535OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayot M, Michaud A, Koechlin C, Caron MA, Leblanc P, Préfaut C, et al. Skeletal muscle microbiopsy: a validation study of a minimally invasive technique. Eur Respir J. 2005;25:431–440. doi: 10.1183/09031936.05.00053404. [DOI] [PubMed] [Google Scholar]
- 28.Thornton AM, Zhao X, Weisleder N, Brotto LS, Bougoin S, Nosek TM, et al. Store-operated Ca2+ entry (SOCE) contributes to normal skeletal muscle contractility in young but not in aged skeletal muscle. Aging (Albany NY) 2011;3:621–634. doi: 10.18632/aging.100335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernet JD, Doles JD, Hall JK, Kelly Tanaka K, Carter TA, Olwin BB. p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nat Med. 2014;20:265–271. doi: 10.1038/nm.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scalzo RL, Peltonen GL, Binns SE, Shankaran M, Giordano GR, Hartley DA, et al. Greater muscle protein synthesis and mitochondrial biogenesis in males compared with females during sprint interval training. FASEB J. 2014;28:2705–2714. doi: 10.1096/fj.13-246595. [DOI] [PubMed] [Google Scholar]
- 31.Fox DK, Ebert SM, Bongers KS, Dyle MC, Bullard SA, Dierdorff JM, et al. p53 and ATF4 mediate distinct and additive pathways to skeletal muscle atrophy during limb immobilization. Am J Physiol Endocrinol Metab. 2014;307:E245–E261. doi: 10.1152/ajpendo.00010.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ebert SM, Monteys AM, Fox DK, Bongers KS, Shields BE, Malmberg SE, et al. The transcription factor ATF4 promotes skeletal myofiber atrophy during fasting. Mol Endocrinol. 2010;24:790–799. doi: 10.1210/me.2009-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Theije CC, Langen RC, Lamers WH, Schols AM, Köhler SE. Distinct responses of protein turnover regulatory pathways in hypoxia- and semistarvation-induced muscle atrophy. Am J Physiol Lung Cell Mol Physiol. 2013;305:L82–L91. doi: 10.1152/ajplung.00354.2012. [DOI] [PubMed] [Google Scholar]
- 34.Mutsvangwa T, Gilmore J, Squires JE, Lindinger MI, McBride BW. Chronic metabolic acidosis increases mRNA levels for components of the ubiquitin-mediated proteolytic pathway in skeletal muscle of dairy cows. J Nutr. 2004;134:558–561. doi: 10.1093/jn/134.3.558. [DOI] [PubMed] [Google Scholar]
- 35.Sousa-Victor P, Gutarra S, García-Prat L, Rodriguez-Ubreva J, Ortet L, Ruiz-Bonilla V, et al. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature. 2014;506:316–321. doi: 10.1038/nature13013. [DOI] [PubMed] [Google Scholar]
- 36.Toledo-Arruda AC, Vieira RP, Guarnier FA, Suehiro CL, Caleman-Neto A, Olivo CR, et al. Time-course effects of aerobic physical training in the prevention of cigarette smoke-induced COPD. J Appl Physiol (1985) 2017;123:674–683. doi: 10.1152/japplphysiol.00819.2016. [DOI] [PubMed] [Google Scholar]
- 37.Deasy BM, Lu A, Tebbets JC, Feduska JM, Schugar RC, Pollett JB, et al. A role for cell sex in stem cell-mediated skeletal muscle regeneration: female cells have higher muscle regeneration efficiency. J Cell Biol. 2007;177:73–86. doi: 10.1083/jcb.200612094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nesvizhskii AI, Aebersold R. Interpretation of shotgun proteomic data: the protein inference problem. Mol Cell Proteomics. 2005;4:1419–1440. doi: 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 41.R Studio Team RStudio: Integrated Development for R. RStudio, Inc. Boston, MA: R Studio. 2015 [accessed 2019 Feb 18]. Available from: http://www.rstudio.com/.
- 42.Hochberg Y. A sharper Bonferroni procedure for multiple tests of significance. Biometrika. 1988;75:800–802.42. [Google Scholar]
- 43.Joshi B, Cameron A, Jagus R. Characterization of mammalian eIF4E-family members. Eur J Biochem. 2004;271:2189–2203. doi: 10.1111/j.1432-1033.2004.04149.x. [DOI] [PubMed] [Google Scholar]
- 44.Nader GA, von Walden F, Liu C, Lindvall J, Gutmann L, Pistilli EE, et al. Resistance exercise training modulates acute gene expression during human skeletal muscle hypertrophy. J Appl Physiol (1985) 2014;116:693–702. doi: 10.1152/japplphysiol.01366.2013. [DOI] [PubMed] [Google Scholar]
- 45.Stec MJ, Mayhew DL, Bamman MM. The effects of age and resistance loading on skeletal muscle ribosome biogenesis. J Appl Physiol (1985) 2015;119:851–857. doi: 10.1152/japplphysiol.00489.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goodman CA, Mabrey DM, Frey JW, Miu MH, Schmidt EK, Pierre P, et al. Novel insights into the regulation of skeletal muscle protein synthesis as revealed by a new nonradioactive in vivo technique. FASEB J. 2011;25:1028–1039. doi: 10.1096/fj.10-168799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev. 2011;91:1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- 48.Silberstein L, Webster SG, Travis M, Blau HM. Developmental progression of myosin gene expression in cultured muscle cells. Cell. 1986;46:1075–1081. doi: 10.1016/0092-8674(86)90707-5. [DOI] [PubMed] [Google Scholar]
- 49.Sincennes MC, Wang YX, Rudnicki MA. Primary mouse myoblast purification using magnetic cell separation. Methods Mol Biol. 2017;1556:41–50. doi: 10.1007/978-1-4939-6771-1_3. [DOI] [PubMed] [Google Scholar]
- 50.Nader GA, McLoughlin TJ, Esser KA. mTOR function in skeletal muscle hypertrophy: increased ribosomal RNA via cell cycle regulators. Am J Physiol Cell Physiol. 2005;289:C1457–C1465. doi: 10.1152/ajpcell.00165.2005. [DOI] [PubMed] [Google Scholar]
- 51.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoppe S, Bierhoff H, Cado I, Weber A, Tiebe M, Grummt I, et al. AMP-activated protein kinase adapts rRNA synthesis to cellular energy supply. Proc Natl Acad Sci USA. 2009;106:17781–17786. doi: 10.1073/pnas.0909873106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cao Y, Bojjireddy N, Kim M, Li T, Zhai P, Nagarajan N, et al. Activation of γ2-AMPK suppresses ribosome biogenesis and protects against myocardial ischemia/reperfusion injury. Circ Res. 2017;121:1182–1191. doi: 10.1161/CIRCRESAHA.117.311159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen XT, Mitchell BS. Akt activation enhances ribosomal RNA synthesis through casein kinase II and TIF-IA. Proc Natl Acad Sci USA. 2013;110:20681–20686. doi: 10.1073/pnas.1313097110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tanaka Y, Yano H, Ogasawara S, Yoshioka S, Imamura H, Okamoto K, et al. Mild glucose starvation induces KDM2A-mediated H3K36me2 demethylation through AMPK to reduce rRNA transcription and cell proliferation. Mol Cell Biol. 2015;35:4170–4184. doi: 10.1128/MCB.00579-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swallow EB, Reyes D, Hopkinson NS, Man WD, Porcher R, Cetti EJ, et al. Quadriceps strength predicts mortality in patients with moderate to severe chronic obstructive pulmonary disease. Thorax. 2007;62:115–120. doi: 10.1136/thx.2006.062026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rochester CL, Vogiatzis I, Holland AE, Lareau SC, Marciniuk DD, Puhan MA, et al. ATS/ERS Task Force on Policy in Pulmonary Rehabilitation. An official American Thoracic Society/European Respiratory Society Policy Statement: enhancing implementation, use, and delivery of pulmonary rehabilitation. Am J Respir Crit Care Med. 2015;192:1373–1386. doi: 10.1164/rccm.201510-1966ST. [DOI] [PubMed] [Google Scholar]
- 58.Puhan MA, Gimeno-Santos E, Scharplatz M, Troosters T, Walters EH, Steurer J. Pulmonary rehabilitation following exacerbations of chronic obstructive pulmonary disease. Cochrane Database Syst Rev. 2011;(10):CD005305. doi: 10.1002/14651858.CD005305.pub3. [DOI] [PubMed] [Google Scholar]
- 59.Puhan MA, Lareau SC. Evidence-based outcomes from pulmonary rehabilitation in the chronic obstructive pulmonary disease patient. Clin Chest Med. 2014;35:295–301. doi: 10.1016/j.ccm.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 60.White RJ. RNA polymerases I and III, growth control and cancer. Nat Rev Mol Cell Biol. 2005;6:69–78. doi: 10.1038/nrm1551. [DOI] [PubMed] [Google Scholar]
- 61.Nader GA. Ribosomes ‘muscle up’ postnatal muscle growth. J Physiol. 2014;592:5143. doi: 10.1113/jphysiol.2014.284828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.von Walden F, Liu C, Aurigemma N, Nader GA. mTOR signaling regulates myotube hypertrophy by modulating protein synthesis, rDNA transcription, and chromatin remodeling. Am J Physiol Cell Physiol. 2016;311:C663–C672. doi: 10.1152/ajpcell.00144.2016. [DOI] [PubMed] [Google Scholar]
- 63.Chaillou T, Kirby TJ, McCarthy JJ. Ribosome biogenesis: emerging evidence for a central role in the regulation of skeletal muscle mass. J Cell Physiol. 2014;229:1584–1594. doi: 10.1002/jcp.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384–6391. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- 65.Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu Rev Pharmacol Toxicol. 2010;50:131–156. doi: 10.1146/annurev.pharmtox.010909.105844. [DOI] [PubMed] [Google Scholar]
- 66.Ciciliot S, Rossi AC, Dyar KA, Blaauw B, Schiaffino S. Muscle type and fiber type specificity in muscle wasting. Int J Biochem Cell Biol. 2013;45:2191–2199. doi: 10.1016/j.biocel.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 67.Salminen A, Kauppinen A, Kaarniranta K. AMPK/Snf1 signaling regulates histone acetylation: impact on gene expression and epigenetic functions. Cell Signal. 2016;28:887–895. doi: 10.1016/j.cellsig.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 68.Barreiro E, Sznajder JI, Nader GA, Budinger GR. Muscle dysfunction in patients with lung diseases: a growing epidemic. Am J Respir Crit Care Med. 2015;191:616–619. doi: 10.1164/rccm.201412-2189OE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barreiro E, Sznajder JI. Epigenetic regulation of muscle phenotype and adaptation: a potential role in COPD muscle dysfunction. J Appl Physiol (1985) 2013;114:1263–1272. doi: 10.1152/japplphysiol.01027.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.