Abstract
Ovarian cancer (OC) is the most lethal cancer of all gynecological malignancies, while endometrial cancer (EC) is the most common one. Current strategies for OC/EC diagnosis consist of the extraction of a solid tissue from the affected area. This sample enables the study of specific biomarkers and the genetic nature of the tumor. However, the tissue extraction is risky and painful for the patient and in some cases is unavailable in inaccessible tumors. Moreover, a tissue biopsy is expensive and requires a highly skilled gynecological surgery to pinpoint accurately which cannot be applied repeatedly. New alternatives that overcome these drawbacks are rising up nowadays, such as liquid biopsy. A liquid biopsy is the analysis of biomarkers in a non-solid biological tissue, mainly blood, which has remarkable advantages over the traditional method. The most studied cancer non-invasive biomarkers are circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), and circulating free DNA (cfDNA). These circulating biomarkers play a key role in the understanding of metastasis and tumorigenesis, which could provide a better insight into the evolution of the tumor dynamics during treatment and disease progression. Liquid biopsy is an emerging non-invasive, safe and effective method with considerable potential for clinical diagnosis and treatment management in patients with OC and EC. Analysis of cfDNA and ctDNA will provide a better characterization of biomarkers and give rise to a wide range of clinical applications, such as early detection of OC/EC, the prediction of treatment responses due to the discovery of personalized tumor-related biomarkers, and therapeutic response monitoring.
Keywords: ovarian cancer, endometrial cancer, liquid biopsy, circulating cell-free DNA, circulating tumor DNA
Introduction
In recent years, the continuous development in genomic detection and analysis technology has revealed a more complicated molecular picture of cancers. Such complexity may be caused by the inter/intra-tumoral heterogeneity and clonal evolution, which brings forward a higher demand on a precise and real-time monitoring system.1
Ovarian cancer (OC), endometrial cancer (EC) and cervical cancer (CC) are the three major cancers in gynecology. Established screening projects and the prevalence of vaccination have remarkably reduced the incidence and mortality of invasive CC,2 but the situation of OC and EC is still not optimistic.
Ovarian cancer is an aggressive disease with a high mortality rate, largely due to a lack of effective screening methods or biomarkers with high sensitivity and specificity.3 A recent review on the benefits and harms of OC screening among average-risk women included 4 randomized clinical trials involving transvaginal ultrasound (TVS), cancer antigen 125 (CA125) testing, or their combination. The review illustrated no significant difference on mortality between screened women and non-screened or irregular-screened ones.4 Furthermore, although the response rate of initial treatment is satisfactory, recurrence rate in advanced-stage OC patients remain high, partly because of insufficient understanding of drug resistance mechanisms.5 As a widely used biomarker during the treatment and the follow-up, CA125 does not perform well in clinical work.6
Endometrial cancer is the most common cancer occurring in female reproductive tract.7 It can be diagnosed and treated at an early stage, due to its typical symptom of abnormal genital bleeding and the subsequent detailed examination. However, some patients present nonspecific symptoms and those with intermediate to high-risk factors are still very vulnerable to recurrence, resulting in a higher mortality rate. The sensitivity and specificity of TVS is too limited to be a screening method for EC, which can only provide a rough evaluation.8 Diagnostic dilatation and curettage is effective with high sensitivity and specificity for post-menopausal women,9 while the invasive process and possible complications may reduce the compliance of patients. More importantly, it could not be used in the treatment surveillance once the uterus is removed.
Histopathology examination has long been regarded as the gold standard for the final diagnosis and genotypic analysis of OC and EC. However, tissue biopsy is incompatible for clinical longitudinal monitoring due to its invasive nature.
Based on the above, a new strategy for these two types of gynecological malignancies is urgently needed. As a non-invasive and cost-effective method, liquid biopsy is expected to overcome the above shortcomings and has become a research hotspot.10 Circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), circulating free RNAs, tumor-derived extracellular vesicles (TEVs) and tumor-educated platelets (TEPs) are the main components released by tumors from either primary or metastatic sites that can be used in a variety of analysis.11 In 2017, Cheng et al briefly reviewed the role of circulating cell-free DNA (cfDNA) and CTC in OC,12 providing us with important information. In our review, we will focus on the potential value of cfDNA/ctDNA in OC and EC patients.
Origin and Detection of cfDNA/ctDNA
It took about 30 years from the first discovery of cfDNA in the blood of healthy people13 to the report of its relationship with cancer in 1977.14 Researchers found that higher levels of cfDNA appeared in cancer patients and those with metastatic disease compared to the healthy and non-metastatic ones, respectively. It was not until 1994 that somatic point mutations were identified in cfDNA.15 Since then, more studies have focused on this circulating molecular, especially its potential role in cancer management.
A great proportion of cfDNA comes from normal cells of the body, a small part of which is related to tumors, coming from primary tumors, metastatic sites or CTCs, and is called ctDNA. The mechanism of cfDNA being released by cells into circulation was still not fully understood. Mouliere et al16 investigated the sources of circulating DNA of three distinctive categories (normal extratumoral cells, tumor microenvironment cells and neoplastic tumor cells) in colorectal cancer patients, elucidating three sources of cfDNA: necrosis, apoptosis and active secretion. Apoptosis has been reported to be one of the sources of cfDNA, as cfDNA fragments are found to be similar in size to apoptotic DNA. Necrosis might be an important contributor to long cfDNA fragments (over 1000bp).17 Another hypothesis of cfDNA release was active secretion, with DNA included in the exosomes or DNA-lipoprotein complexes.16
Analysis of ctDNA paves us the way for a comprehensive view of the tumor genomic landscape. The ctDNA fragments (~134-144 bp) were reported to be shorter than that of cfDNA (~166bp).18,19 The proportion of ctDNA in the total cfDNA may vary greatly in accordance with clinical-pathologic features of tumor as well as the tumor microenvironment.20,21 Relatively low ctDNA levels in standard blood samples and background cfDNA unrelated to tumor cells make detection a challenge. Therefore, the standard procedure for blood collection and DNA isolation should be finalized prior to analysis.22,23
Methodologies in Exploring cfDNA/ctDNA
For blood collection, the ratio of plasma germline cfDNA is lower than that of serum; thus, it is more suitable for ctDNA isolation.24 Plasma separation should be performed within a short time since the half-time of ctDNA is only about 2 hrs in standard EDTA tubes.25 Some other special blood collection tubes with special preservatives, such as Streck, allow for delayed plasma separation.26 After centrifugation, the separated plasma should be stored at −80°C for later use.
The extraction of cfDNA/ctDNA is the key to the subsequent test. Available methods include phase isolation, silicon membrane-based spin column, and magnetic bead-based isolation.27 The extraction efficiency varies depending on the methods and the plasma volumes.28
New techniques have emerged with high sensitivity and high specificity for the analysis of cfDNA or ctDNA from a quantitative and qualitative perspective,29–31 such as polymerase chain reaction (PCR) strategies and Next-Generation Sequencing (NGS) strategies.
There is a variety of PCR-based methods, each with its characteristics, including real-time PCR, co-amplification at lower denaturation temperature-PCR, methylation-specific PCR, digital PCR or droplet digital PCR (ddPCR), “Beads, Emulsion, Amplification, Magnetics digital PCR” (BEAMing) and so on.24 Designed to detect hotspot mutations, these PCR-based methods are relatively time-saving and economical.
Next-Generation Sequencing (NGS)-based methods such as tagged-amplicon deep sequencing (TamSeq), the Safe Sequencing System (SafeSeqS), CAncer Personalized Profiling by deep Sequencing (CAPP-Seq) and targeted error correction sequencing (TEC-Seq) are available to assess genetic alterations with different detection capacities.24 Whole Genome Sequencing (WGS) and Whole Exome Sequencing (WES) technologies allow for the identification of novel alterations with no pre-existing knowledge.32,33 The ability of NGS-based methods to detect copy number variations (CNVs) is an advantage over PCR.24 Though efficient, these methods are relatively time-consuming and require professional bioinformatics analysis.
Application of cfDNA/ctDNA in Ovarian and Endometrial Cancer
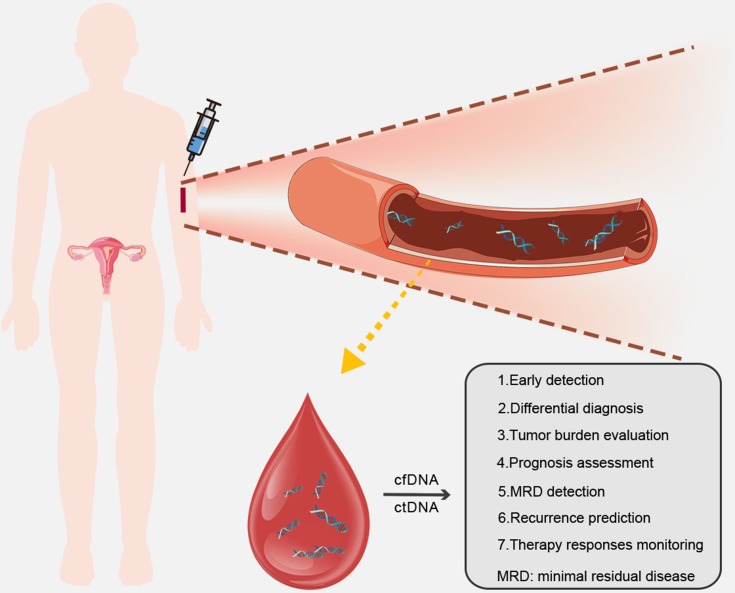
As a promising effective tool in oncology, cfDNA/ctDNA is of essential value in clinical management of ovarian and endometrial cancer patients from the diagnostic, predictive and prognostic aspects (Table 1; Figure 1).
Table 1.
Studies with Analysis of cfDNA/ctDNA in OC or EC Patients
| References | Country | Cancer Type | Sources | Cancer/Control | Abnormalities | Methodology | Clinical Relevance |
|---|---|---|---|---|---|---|---|
| Shao et al34 | China | OC | Serum | 36/41 | Level | bDNA technique | Diagnosis |
| Kamat et al35 | USA | OC | Plasma | 164/124 | Level | RT-PCR | Diagnosis |
| Kamat et al36 | USA | OC | Plasma | 19/12 | Level | RT-PCR | Diagnosis |
| Zachariah et al37 | Switzerland | OC | Serum/plasma | 21/83 | Level | RT-PCR | Diagnosis |
| Dobrzycka et al38 | Poland | OC | Plasma | 126/NA | Level/KRAS mutation | PCR-RFLP | Diagnosis/prognosis |
| Cicchillitti et al39 | Italy | EC | seRum | 59/NA | Level | RT-PCR | Diagnosis |
| Dobrzycka et al40 | Poland | EC | Plasma | 109/NA | Level | PCR-RFLP | Diagnosis |
| Vizza et al41 | Italy | EC | Serum | 60/NA | Level | RT-PCR | Diagnosis |
| Tanaka et al42 | Japan | EC | Plasma | 53/24 | Level | RT-PCR | Diagnosis/prognosis |
| Hickey et al43 | United Kingdom | OC | Serum | 20/NA | Mutations, LOH and MI | PCR | Diagnosis |
| Otsuka et al46 | Japan | OC | Plasma | 27/NA | p53 mutation | PCR | Diagnosis/prognosis |
| Park et al47 | Korea | OC | Plasma | 4/NA | TP53 mutation | Digital PCR | Diagnosis |
| Cohen et al49 | Australia | OC | Plasma | 32/32 | Chromosomal instability | Whole genome NIPT platform | Diagnosis |
| Vanderstichele et al50 | Belgium | OC | Plasma | 57/11 | Chromosomal instability | NGS | Diagnosis |
| Phallen et al51 | USA | OC | Plasma | 42/NA | Somatic mutations | TEC-Seq | Diagnosis |
| Farkkila et al52 | Finland | OC | Plasma | 35/NA | FOXL2 mutation | ddPCR | Diagnosis/prognosis |
| Sun et al53 | China | EC | PBLs | 139/139 | mtDNA copy number value | RT-PCR | Diagnosis |
| Giannopoulou et al56 | Greece | OC | Plasma | 59/NA | RASSF1A methylation | MSP technique | Diagnosis |
| Zhang et al57 | China | OC | Serum | 87/115 | (APC, RASSF1A, CDH1, RUNX3, TFPI2, SFRP5, OPCML) methylation | MSP technique | Diagnosis |
| Liggett et al58 | USA | OC | Plasma | 30/60 | (RASSF1A, CALCA, EP300, BRCA1, CDKN1C, PGR-PROX) methylation | MethDet 56 | Diagnosis |
| Melnikov et al59 | USA | OC | Plasma | 33/33 | (BRCA1, HIC1, PAX5, PGR, THBS1) methylation | MethDet 56 | Diagnosis |
| Widschwendter et al60 | UK | OC | Serum | 25/598 | (COL23A1, C2CD4D, WNT6) methylation | NGS | Diagnosis/prognosis |
| Li et al61 | China | OC | Peripheral blood | 206/205 | Methylation at multiple sites | NGS | Diagnosis |
| Wu et al62 | China | OC | Plasma | 47/24 | RASSF2A methylation | MSP technique | Diagnosis |
| Ibanez et al63 | USA | OC | Serum/plasma | 50/NA | (BRCA1, RASSF1A) methylation | MSP technique | Diagnosis |
| Margolin et al67 | USA | EC | A computational simulation | 42/8 | ZNF154 methylation | NGS | Diagnosis |
| Wimberger et al71 | Germany | OC | plasma | 62/28 | Level | RT-PCR | prognosis |
| Perkins et al72 | United Kingdom | OC | Plasma | 105/20 | Level/mutations in 19 genes | NGS | Prognosis |
| Steffensen et al73 | Denmark | OC | Plasma | 144/NA | Level | RT-PCR | Prognosis |
| No et al74 | Korea | OC | Plasma | 36/16 | (B2M, RAB25, CLDN4, ABCF2) mutation |
RT-PCR | Prognosis |
| Kuhlmann et al75 | Germany | OC | Serum | 63/20 | Level/LOH | Fluorescence-labeled PCR | Prognosis |
| Swisher et al48 | USA | OC | Plasma/serum | 137/NA | p53 mutation | DNA sequencing | Prognosis |
| Giannopoulou et al76 | Greece | OC | Plasma | 129/NA | ESR1 methylation | MSP technique | Prognosis |
| Bolivar et al70 | USA | EC | Plasma | 48/NA | (CTNNB1, KRAS, PTEN, PIK3CA) mutation | NGS | Prognosis |
| Harris et al77 | USA | OC | Plasma | 8/NA | Chromosomal junctions | Quantitative PCR | Prognosis |
| Pereira et al78 | USA | OC and EC | Serum | 22OC,17EC/NA | Level | Digital PCR | Prognosis |
| Du et al79 | China | OC | Plasma | 21/NA | Mutations/CNV | NGS | Prognosis |
| Martignetti et al80 | USA | OC | Plasma/serum | 1/NA | FGFR fusions | NGS/RT-PCR | Prognosis |
| Choudhuri et al82 | India | OC | Plasma | 100/NA | Level | RT-PCR | Treatment response |
| Capizzi et al83 | Italy | OC | Plasma | 22/NA | Level | RT-PCR | Treatment response |
| Arend et al84 | USA | OC | plasma | 14/NA | level | NGS | Treatment response |
| Kamat et al69 | USA | OC in mice | Plasma | – | Level | RT-PCR | Treatment response |
| Parkinson et al68 | United Kingdom | OC | Plasma | 40/NA | TP53 mutation | Digital PCR | Treatment response |
| Gifford et al85 | United Kingdom | OC | Plasma | 138/NA | hMLH1 methylation | Microsatellite PCR | Treatment response |
| Flanagan et al86 | United Kingdom | OC | Peripheral blood | 247/NA | Methylation at CpG sites | NGS | Treatment response |
| Matulonis et al90 | USA | OC | Plasma | 67/NA | Mutations in components/modulators of the PI3K pathway | NGS/Sanger sequencing | Treatment response |
| Weigelt et al87 | USA | OC | Plasma | 19/NA | BRCA reversion mutation | NGS | Treatment response |
| Christie et al88 | Australia | OC | Plasma | 30/NA | BRCA reversion mutation | NGS | Treatment response |
| Lin et al89 | USA | OC | Plasma | 209/NA | BRCA reversion mutation | NGS | Treatment response |
Abbreviations: OC, ovarian cancer; EC, endometrial cancer; bDNA, branched DNA; PCR-RFLP, PCR-restriction fragment length polymorphism; LOH, loss of heterozygosity; MI, microsatellite instability; NGS, next-generation sequencing; mtDNA, mitochondrial DNA; ddPCR, digital droplet PCR; PBLs, peripheral blood leukocytes; MSP, methylation-specific PCR; CNV, copy number variation; NK, do not know.
Figure 1.
Applications of cfDNA/ctDNA in ovarian or endometrial cancer patients.
Early Detection and Differential Diagnosis
As previously mentioned, the diagnostic value of blood markers and imaging examinations for early detection and differential diagnosis of ovarian and endometrial tumors is limited. Plenty of studies have been carried out to evaluate the competency of cfDNA/ctDNA in cancer screening.
Quantification of cfDNA/ctDNA
Ovarian Cancer
Changes in levels of cfDNA/ctDNA can be a hint for the existence of malignance, which could be more accurate than CA125 or HE4 in some OC studies. Shao et al reported a significant increment of cfDNA levels in the OC group compared with the control group (P<0.01). They also noticed a higher sensitivity and specificity (88.9% and 89.5%) of cfDNA than CA125 (75% and 52.6%) and HE4 (80.6% and 68.4%) in OC detection. Sensitivity and specificity were promoted (91.67% and 84.21%) when the above biomarkers were combined.34 Similarly, a cohort study of OC achieved a sensitivity and specificity of 87–91.5% and 85-87%, respectively. By comparison, they found a lower false-positive rate of cfDNA level than CA125.35 Another small sample study focusing on three endogenous loci (GAPDH, β-actin and β-globin) identified significantly higher cfDNA levels among OC patients compared to controls, with the greatest difference occurring at β-actin locus.36 Quantitative alterations of circulating cell-free nuclear DNA and circulating cell-free mitochondrial DNA were evaluated to confirm their values in diagnosing OC with moderate sensitivity and specificity.37 Dobrzycka et al found that the detection rate of ovarian serous carcinoma was higher than other types, suggesting the role of cfDNA in differential diagnosis.38
Endometrial Cancer
The role of cfDNA levels for EC screening and classification appears to be controversial. Significantly elevated total cfDNA levels were detected in EC group compared with the controls, with higher levels in high-grade groups (G2 and G3).39 Another study reported a dramatic difference between the mean level of cfDNA in patients with type I and type II EC.40 Vizza et al observed a remarkably increased total cfDNA content in high-grade EC patients. They adopted a special calculation method applying Alu-quantitative real-time PCR (qPCR) technology to investigate the role of cfDNA. The serum DNA integrity index, defined as qPCR-Alu247 value/qPCR-Alu115 value of each sample, was significantly reduced in high-grade EC patients with hypertension and obesity. Unfortunately, this index presented a low predictive accuracy to differentiate high grade from low-grade EC patients.41 The study carried out by Tanaka et al showed no significant difference of cfDNA levels between EC and the controls, partly due to the small sample size or differences in methods.42
Genomic Alterations of cfDNA/ctDNA
Ovarian Cancer
Detection of molecular alterations in cfDNA/ctDNA dates back to 1999, when Hickey et al used PCR to detect loss of heterozygosity (LOH) and microsatellite instability in 20 OC patients.43 TP53 mutations were reported as the most common in OC, accounting for approximately 96% of the somatic mutations.44,45 Articles targeting TP53 mutations in cfDNA/ctDNA with different detection techniques have been published, implying their roles in OC management.46–48 Researches centered on other genetic alterations or gene panels with several selected genes of cfDNA/ctDNA were also carried out. Cohen et al conducted a proof-of-concept study for subchromosomal with a low coverage sequencing approach called non-invasive prenatal testing platform, observing a relatively low sensitivity of 40.6%, but a high specificity of 93.8%.49 A recent study focused on the chromosomal instability of cfDNA included 68 patients presenting adnexal masses. In terms of the area under the curve (AUC), a much higher sensitivity over CA125 appeared, when the specificity was set to 99.6%.50 An ultrasensitive approach called TEC-Seq was applied to detect ctDNA alterations in different kinds of tumors based on a well-designed combination of genes. Of the 42 enrolled OC patients, 71% were found with ctDNA alterations, and the fraction turned into 68% when researchers focused on early-stage patients.51 Farkkila et al developed a study to detect the FOXL2 mutation in ctDNA of adult granulosa cell tumor (AGCT) patients with ddPCR assay, revealing a sensitivity and specificity of 23% and 90%, respectively.52
Endometrial Cancer
With regard to EC patients, Sun et al conducted a special research on mitochondrial DNA (mtDNA) copy number detection in peripheral blood leukocytes (PBLs) rather than serum or plasma.53 Alterations in mtDNA may lead to mitochondrial dysfunction, thus contributing to tumorigenesis.54 They reported that low mtDNA copy number indicated a more than five-fold increase in the risk of EC.53
Epigenetic Changes of cfDNA/ctDNA
Alteration of DNA methylation has been proven to be an early event of tumorigenesis, making the analysis of circulating DNA methylation patterns a potential method to detect OC and EC.55
Ovarian Cancer
Published studies applied similar research methods for OC, while focused on different genes.56–62 Ibanez et al developed a promoter hypermethylation analysis of 6-gene panel in the serum of OC patients (ranging from early to advanced-stage patients), observing a sensitivity of 82% and a specificity of 100%.63 A 7-gene panel study showed higher sensitivity and specificity (85.3% and 90.5%, respectively) of cfDNA than CA125 (56.1% and 64.15%, respectively) in stage I OC.57 There are similar studies published with different results of sensitivities and specificities, which may attribute to different markers they adopted.58,59 Although Widschwendter’s group observed sensitivities of 41.4% and 82.8%, and specificities of 90.7% and 87.1%, respectively, for a three-DNA-methylation-serum-marker panel and CA125, they noticed that cfDNA can detect OC much earlier than the exact diagnosis.60 In addition, Li et al obtained a prediction accuracy of 77.3% in the included OC population for the six validated CPG sites.61 It is worth noticing that four of the studies mentioned above selected RASSF1A gene as one of the biomarkers.56,57,59,63 Wu et al evaluated the methylation status of RASSF2A, observing an aberrant rate of 51.1% in tissues and 36.2% in corresponding plasma samples.62 The RASSF gene family has been reported to play vital roles in tumorigenesis of various malignancies.64–66 Epigenetic alterations of RASSF gene family, especially RASSF1A and RASSF2A may be promising as a marker that warrants further investigations.
Endometrial Cancer
Margolin et al developed a study to detect the hypermethylation of the ZNF154 CpG island in five types of tumor, including 42 EC patients. Apart from the tissue samples, a computational simulation of ctDNA (1% tumor DNA in 99% normal DNA) was also applied in their study, and an AUC of 0.79 was proved to show the best performance in endometrial tumors.67
Tumor Burden Evaluation and Prognosis Assessment
Analysis of cfDNA/ctDNA allows doctors to gain insight into tumor burden, thus providing references for subsequent treatment. Information on survival outcomes predicted by cfDNA/ctDNA could assist us in developing therapeutic projects and personal follow-up plans for cancer patients.
Tumor Burden Evaluation
Ovarian Cancer
A significant discrepancy of cfDNA levels between stage I-II and stage III-IV (P<0.01) was reported based on an analysis of 36 OC cases,34 and a strong relationship between ctDNA level and tumor volume was also documented in a group of high-grade serous ovarian carcinoma patients,68 suggesting the potential value of cfDNA/ctDNA in evaluating tumor burden. A study in mice confirmed that elevated ctDNA levels indicated elevated tumor burden.69 In addition to quantification, specific genetic mutations can also reflect tumor burden. Larger median tumor size was observed in AGCT patients with FOXL2 ctDNA mutations by a research group.52
Endometrial Cancer
Relative researches about EC are rarely seen, but one has found that the appearance of plasma DNA mutations was remarkably correlated with primary tumor size.70
Prognosis Assessment
Ovarian Cancer
Several studies drew similar conclusions on the prognostic role of cfDNA levels, that is, an increased cfDNA levels indicate a significant reduction in overall survival (OS) in OC patients.35,38,71–73 From the perspective of genetic changes, certain genes were verified to be useful to assess prognosis. No et al reported that RAB25 levels in serum cfDNA were remarkably related to disease-free survival (HR=18.2, 95% CI=2.0–170.0) and OS (HR=33.6, 95% CI=1.8–634.8) of advanced-stage OC patients.74 LOH proximal to M6P/IGF2R locus (D6S1581) was found to be associated with OS (P=0.030) in another study.75 The level of cfDNA72 or presence of specific ctDNA mutation48 was even identified as an independent predictor of OS for OC patients. In addition, the presence of ESR1 methylation in primary tumor samples was significantly correlated with better survival outcomes, though the correlation was not significant in plasma samples.76
Endometrial Cancer
Plasma DNA mutations in EC patients were found to be significantly related to deep myometrial invasion and lymphatic/vascular invasion, suggesting a possibly poorer survival outcome than those with only wildtype DNA.70
Minimal Residual Disease (MRD) Detection and Recurrence Prediction
Postoperative residual disease may occur in advanced OC and EC patients even under standard surgical procedures. As a potential source of relapse, it poses a serious threat to the patient’s survival outcome. As aforementioned, despite emerging new treatment strategies, recurrences of OC and EC remain as urgent problems to be resolved. The evaluation of residual tumor burden and recurrence is thus important, and cfDNA/ctDNA may offer assistance.
Ovarian Cancer
Wimberger et al demonstrated that serum DNA levels of OC patients were significantly correlated with postoperative residual tumor load of >1 cm (p=0.0001) and a higher risk of relapse (p=0.002).71 Similar relationship between preoperative cfDNA levels and residual tumor load (p=0.017) was mentioned in another study.75 A research focusing on selected chromosomal junctions reported presence of ctDNA after surgery in patients with detectable disease, while no ctDNA in those without the disease.77
ctDNA may provide opportunities of timely treatment of recurrent lesions that were not detectable through imaging. One research revealed that ctDNA could make a diagnosis for relapsed OC cases 7 months earlier than CT scanning.78 In patients with recurrent OC, the coincidence rates of TP53 and BRCA1 in cfDNA and tumor tissue DNA were 76.2% and 95.24%, respectively, which means that cfDNA/ctDNA could assist the monitoring of disease progression.79 Otsuka et al noticed in a follow-up survey that one patient with re-emerging p53 mutation after surgery died shortly after, while the other one with no p53 mutation survived.46 An American study revealed a more sensitive and specific biomarker, the fibroblast growth factor receptor 2 (FGFR2) fusion ctDNA biomarker, to detect OC. In a series of 28 measurements during a 4-year follow-up of a specific patient, FGFR2 fusion better reflected the evolvement of the disease than CA125, especially the tumor recurrences.80
Endometrial Cancer
Levels of cfDNA were evaluated in a cohort of 25 EC patients before and after operation, and 3 cases whose postoperative cfDNA levels did not decrease as expected relapsed during the follow up.41
Therapy Responses Monitoring
As the molecular mechanisms of tumors continue to unlock, different types of targeted drugs have been validated in clinical trials. Treatment resistance occurs commonly, possibly because of the genomic heterogeneity or tumor subclonal evolution under selection pressure.81 Real-time monitoring of therapeutic response is pivotal as it helps to understand the dynamic development of the disease. Detection of cfDNA/ctDNA may achieve the goal of screening out potential drug-resistant patients and instituting candidate-tailored therapeutic project to reduce the recurrence rate and improving survival rate.
Ovarian Cancer
Several studies, including a study of the OC mouse model, reported a decrease in cfDNA levels after ovarian cancer treatment, and some studies have shown that it is associated with post-therapy survival.69,82,83 On the other hand, significant changes in genetic variants of cfDNA were noticed after chemotherapy.84 In relapsed patients, a >60% reduction in TP53 mutant allele fraction was reported as an independent predictor of time to progression after one cycle of chemotherapy.68 In one study, methylation changes of the hMLH1 gene in plasma DNA were detected (before chemotherapy and at relapse) in 138 patients with stage IC–IV OC who experienced a relapse. A significant (P<0.001) increase of the positive rate (from 12% to 33%) in hMLH1 methylation was observed, together with a poor OS. This study provided evidence to support the idea that the loss of DNA mismatch repair (MMR) might involve in the process of acquired drug resistance.85 Another study also documented changes in plasma DNA methylation in patients with OC who relapsed after platinum-based chemotherapy.86 The identification of chemotherapy responders and non-responders was essential for the subsequent treatment plan, and the analysis of DNA-methylation-serum-marker panel will be useful.58
Recently, a couple of studies have been developed to explore the acquisition of BRCA reversion mutations and their roles in drug resistance by analyzing cfDNA/ctDNA in OC patients who received platinum-based chemotherapies or PARP inhibitors.87–89 Detection of BRCA reversion mutations in cfDNA/ctDNA has been proven to be useful to predict possible drug resistance and guide treatment strategies.
Endometrial Cancer
Matulonis et al conducted a study to assess treatment response of PI3K inhibitor pilaralisib in advanced or recurrent EC cases. Different degrees of consistency were noticed between ctDNA and paired tumor tissue on the status of PIK3CA, KRAS, and BRAF genes.90
Possible Applications of cfDNA/ctDNA in Specific Situations
Utility of cfDNA/ctDNA in Fertility-Sparing Treatment
Advanced techniques, including detection techniques of cfDNA/ctDNA, have improved the detection rate of early-stage disease. Choices of fertility-sparing approaches for gynecological cancer patients are receiving greater attention, since an increasing number of patients are diagnosed at productive age and desire to have children. Gynecological cancers have a negative impact on the sexuality and fertility of patients, increasing their stress level and reducing personal identity, which may pose a threat to the quality of their lives.91
Most OC cases are diagnosed at an advanced stage, for whom cytoreduction remains optimal. However, fertility-sparing surgery is still recommended for selected patients by several society recommendations, with the premise of a comprehensive surgical staging.92,93
As for EC patients, total hysterectomy and bilateral salpingo-oophorectomy with or without surgical staging are regarded as the standard treatment. Fertility-sparing surgery is a choice for reproductive-aged patients with stage IA type I and G2 EC.94 Furthermore, previous studies have evaluated the accuracy of preoperative methods, such as magnetic resonance imaging (MRI), TVS and hysteroscopically directed biopsies, for predicting the nodal-spread risk of EC.95,96 They concluded that these preoperative mapping methods worked well with high accuracy, avoiding over- or under-treatment of EC patients, while more evidence in this field is still needed. Conservative treatment such as progestational agents has attracted widespread attention among premenopausal women who are eager to give birth and have achieved favorable outcomes in early-stage patients.97–100
Given the risk of recurrences, monitoring must be strengthened for patients receiving conservative treatment. Non-invasive detection of cfDNA/ctDNA provides a more convenient tool for tracking disease progression, which helps to assess treatment response. Nevertheless, no relative data have been published by now, and the application of cfDNA/ctDNA analysis in this field needs further investigations.
Utility of cfDNA/ctDNA in the Management of Elderly Patients
The incidence of cancer in elderly population is much higher than the young group. However, the management of elderly patients remains a challenge. In the clinical setting, treatment patterns of elderly patients with OC and EC often differ from younger patients, and are less aggressive, due to their seemingly fragile bodies.101,102 Studies supported that age itself was not a prognostic factor for survival outcome, and elderly patients could also receive standard treatments.102 The management of elderly OC or EC patients should be personalized according to the performance status of the patients, the extent of the disease and their life goals.102 The detection of cfDNA/ctDNA may act as an assistant examination during the whole course of the management, given its non-invasive nature.
Detection of cfDNA/ctDNA in Non-Blood Fluids
In addition to blood, cfDNA/ctDNA is also detectable in other body fluids, among which urine, peritoneal fluid and uterine lavage fluid were reported to be utilized in the management of OC or EC patients (Table 2). Compared to bloodstream, the concentration of ctDNA shed in non-blood fluids can be higher in some certain cancer types.103 However, a lack of relative researches and protocols, as well as rare experience in the exploration of these tests remain problems for non-blood-based liquid biopsy.
Table 2.
Studies with Analysis of cfDNA/ctDNA in Other Body Fluids of OC or EC Patients
| References | Country | Cancer Type | Fluid Type | Number of Samples | Abnormalities | Methodology | Clinical Relevance |
|---|---|---|---|---|---|---|---|
| Hickey et al43 | United Kingdom | OC | Peritoneal fluid | 20 | Mutations, LOH and MI | PCR | Diagnosis |
| Swisher et al48 | USA | OC | Peritoneal fluid | 30 | p53 mutation | DNA sequencing | Prognosis |
| Krimmel et al104 | USA | OC | Peritoneal fluid | 37 | TP53 mutation | Duplex sequencing | Diagnosis |
| Parrella et al105 | Italy | OC | Peritoneal fluid | 15 | (p53,KRAS) mutation/LOH | DNA sequencing | Diagnosis |
| Barquin et al106 | Spain | OC | Peritoneal fluid | 10 | BRCA mutation | DNA sequencing | Prognosis |
| Muller et al107 | Austria | OC | Peritoneal fluid | 61 | methylation of 15 selected genes | MethyLight | Prognosis |
| Ibanez et al63 | USA | OC | Peritoneal fluid | 42 | (BRCA1, RASSF1A) methylation | MSP technique | Diagnosis |
| Du et al79 | China | OC | Urine and ascites | 21 and 13 | Mutations/CNV | NGS | Prognosis |
| Nair et al108 | USA | EC | Uterine lavage fluid | 107 | Mutations of selected genes | NGS | Diagnosis |
| Maritschnegg et al109 | Austria | OC and EC | Uterine lavage fluid | 30 and 5 | Somatic mutations | NGS | Diagnosis |
| Kinde et al110 | USA | OC and EC | Liquid-based Pap smear | 22and 24 | Somatic mutations | NGS | Diagnosis |
| Wang et al111 | USA | OC and EC | Pap brush samples | 245 and 382 | Somatic mutation | Safe-sequencing system | Diagnosis |
Abbreviations: OC, ovarian cancer; EC, endometrial cancer; LOH, loss of heterozygosity; MI, microsatellite instability; MSP, methylation-specific PCR; CNV, copy number variation; NGS, next-generation sequencing; Pap, Papanicolaou.
Several studies focused on tumor-specific genetic alterations of DNA extracted from peritoneal fluid samples of OC patients, receiving relatively high sensitivities in detection.43,48,104–106 In 2004, methylation of peritoneal DNA was first reported as an independent factor in OC survival.107 Shortly after, hypermethylation was detected in peritoneal fluid DNA with relatively high sensitivity and specificity, including 3 cytologically negative patients.63 Du and his colleagues included urine samples into their study for genetic mutation analysis, identifying a detection rate of 86%.79 Nair et al conducted a study to detect endometrial driver mutations in uterine lavage in seven EC patients, of whom six were at early stage.108 In another similar study, specific mutations were identified in 80% (24/30) OC and 100% (5/5) EC patients.109 It has been confirmed that tumor DNA is detectable in detached cells from ovarian and endometrial malignancies collected at the cervix.110 Recently, genetic analysis of Papanicolaou (Pap) test using liquid-based methods rather than traditional Pap smear revealed potential advantages in early detection of OC and EC patients.110,111
Conclusion
On June 1, 2016, cobas EGFR Mutation Test v2 was approved by the U. S. Food and Drug Administration (FDA) to identify patients with metastatic non-small cell lung cancer (NSCLC) appropriate for treatment with Tarceva® (erlotinib).112 This test, focusing on the mutation of the epidermal growth factor receptor (EGFR) gene in the cfDNA isolated from the blood samples of patients, is the first cfDNA/ctDNA test officially applied to clinical work.112 Currently, no cfDNA/ctDNA-related test has been approved by FDA in the field of OC and EC. Encouragingly, as mentioned above, published studies supported the feasibility of cfDNA/ctDNA’s application in the interrogation of tumor genome profiles and real-time tracing of the tumor conditions in OC and EC patients. The analysis of levels, genetic changes and epigenetic alterations of cfDNA/ctDNA hold tremendous promise in a wide range of applications: cancer screening, tumor burden evaluation, prognosis assessment, MRD detection, recurrence surveillance and treatment monitoring. In addition, a large number of clinical trials exploring the role of cfDNA/ctDNA in the management of OC and EC are underway (for example, NCT03691012), which may provide us with strong evidence.
It should be noted that well-defined preanalytical, analytical and postanalytical protocols are the prerequisites to obtain convincing results from large-scale clinical trials. Efforts are still needed to overcome challenges in detection techniques such as low amounts of cfDNA/ctDNA and high background signals. Combination of this new liquid biopsy method with other traditional methods may yield more satisfactory results. Accumulated evidence for survival benefits is required to integrate this novel diagnostic approach into the clinical scenario in the foreseeable future.
Acknowledgments
This study was funded by CAMS Youth Talent Award Project (No. 2018RC320006) and CAMS Innovation Fund for Medical Sciences (CIFMS; No. 2016-I2M-1-002).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017;168(4):613–628. doi: 10.1016/j.cell.2017.01.018 [DOI] [PubMed] [Google Scholar]
- 2.Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical cancer. Lancet. 2019;393(10167):169–182. doi: 10.1016/S0140-6736(18)32470-X [DOI] [PubMed] [Google Scholar]
- 3.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 4.Henderson JT, Webber EM, Sawaya GF. Screening for ovarian cancer: updated evidence report and systematic review for the US preventive services task force. JAMA. 2018;319(6):595–606. doi: 10.1001/jama.2017.21421 [DOI] [PubMed] [Google Scholar]
- 5.Parmar MK, Ledermann JA, Colombo N, et al. Paclitaxel plus platinum-based chemotherapy versus conventional platinum-based chemotherapy in women with relapsed ovarian cancer: the ICON4/AGO-OVAR-2.2 trial. Lancet. 2003;361(9375):2099–2106. [DOI] [PubMed] [Google Scholar]
- 6.Bast RC Jr. CA 125 and the detection of recurrent ovarian cancer: a reasonably accurate biomarker for a difficult disease. Cancer. 2010;116(12):2850–2853. doi: 10.1002/cncr.v116:12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 8.Yasa C, Dural O, Bastu E, Ugurlucan FG, Nehir A, Iyibozkurt AC. Evaluation of the diagnostic role of transvaginal ultrasound measurements of endometrial thickness to detect endometrial malignancy in asymptomatic postmenopausal women. Arch Gynecol Obstet. 2016;294(2):311–316. doi: 10.1007/s00404-016-4054-5 [DOI] [PubMed] [Google Scholar]
- 9.Dijkhuizen FP, Mol BW, Brolmann HA, Heintz AP. The accuracy of endometrial sampling in the diagnosis of patients with endometrial carcinoma and hyperplasia: a meta-analysis. Cancer. 2000;89(8):1765–1772. doi: [DOI] [PubMed] [Google Scholar]
- 10.Lianidou E, Pantel K. Liquid biopsies. Genes Chromosomes Cancer. 2019;58(4):219–232. doi: 10.1002/gcc.v58.4 [DOI] [PubMed] [Google Scholar]
- 11.Jia S, Zhang R, Li Z, Li J. Clinical and biological significance of circulating tumor cells, circulating tumor DNA, and exosomes as biomarkers in colorectal cancer. Oncotarget. 2017;8(33):55632–55645. doi: 10.18632/oncotarget.v8i33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng X, Zhang L, Chen Y, Qing C. Circulating cell-free DNA and circulating tumor cells, the “liquid biopsies” in ovarian cancer. J Ovarian Res. 2017;10(1):75. doi: 10.1186/s13048-017-0369-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mandel P, Metais P. Les acides nucléiques du plasma sanguin chez l’homme. C R Seances Soc Biol Fil. 1948;142(3–4):241–243. [PubMed] [Google Scholar]
- 14.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37(3):646–650. [PubMed] [Google Scholar]
- 15.Vasioukhin V, Anker P, Maurice P, Lyautey J, Lederrey C, Stroun M. Point mutations of the N-ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemia. Br J Haematol. 1994;86(4):774–779. doi: 10.1111/j.1365-2141.1994.tb04828.x [DOI] [PubMed] [Google Scholar]
- 16.Mouliere F, Thierry AR. The importance of examining the proportion of circulating DNA originating from tumor, microenvironment and normal cells in colorectal cancer patients. Expert Opin Biol Ther. 2012;12(Suppl 1):S209–S215. doi: 10.1517/14712598.2012.688023 [DOI] [PubMed] [Google Scholar]
- 17.Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61(4):1659–1665. [PubMed] [Google Scholar]
- 18.Jiang P, Chan CW, Chan KC, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci U S A. 2015;112(11):E1317–E1325. doi: 10.1073/pnas.1500076112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Underhill HR, Kitzman JO, Hellwig S, et al. Fragment length of circulating tumor DNA. PLoS Genet. 2016;12(7):e1006162. doi: 10.1371/journal.pgen.1006162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra224. doi: 10.1126/scitranslmed.3007094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tug S, Helmig S, Deichmann ER, et al. Exercise-induced increases in cell free DNA in human plasma originate predominantly from cells of the haematopoietic lineage. Exerc Immunol Rev. 2015;21:164–173. [PubMed] [Google Scholar]
- 22.Pandoh PK, Corbett RD, McDonald H, et al. A high-throughput protocol for isolating cell-free circulating tumor DNA from peripheral blood. Biotechniques. 2019;66(2):85–92. doi: 10.2144/btn-2018-0148 [DOI] [PubMed] [Google Scholar]
- 23.Fettke H, Kwan EM, Azad AA. Cell-free DNA in cancer: current insights. Cell Oncol (Dordr). 2019;42(1):13–28. doi: 10.1007/s13402-018-0413-5 [DOI] [PubMed] [Google Scholar]
- 24.Franczak C, Filhine-Tresarrieu P, Gilson P, Merlin JL, Au L, Harle A. Technical considerations for circulating tumor DNA detection in oncology. Expert Rev Mol Diagn. 2019;19(2):121–135. doi: 10.1080/14737159.2019.1568873 [DOI] [PubMed] [Google Scholar]
- 25.Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–990. doi: 10.1038/nm.1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medina Diaz I, Nocon A, Mehnert DH, Fredebohm J, Diehl F, Holtrup F. Performance of streck cfDNA blood collection tubes for liquid biopsy testing. PLoS One. 2016;11(11):e0166354. doi: 10.1371/journal.pone.0166354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu JL, Liang ZY. Circulating free DNA in the era of precision oncology: pre- and post-analytical concerns. Chronic Dis Transl Med. 2016;2(4):223–230. doi: 10.1016/j.cdtm.2016.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haselmann V, Ahmad-Nejad P, Geilenkeuser WJ, et al. Results of the first external quality assessment scheme (EQA) for isolation and analysis of circulating tumour DNA (ctDNA). Clin Chem Lab Med. 2018;56(2):220–228. doi: 10.1515/cclm-2017-0283 [DOI] [PubMed] [Google Scholar]
- 29.Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4(136):136ra168. doi: 10.1126/scitranslmed.3003726 [DOI] [PubMed] [Google Scholar]
- 30.Buono G, Gerratana L, Bulfoni M, et al. Circulating tumor DNA analysis in breast cancer: is it ready for prime-time? Cancer Treat Rev. 2019;73:73–83. doi: 10.1016/j.ctrv.2019.01.004 [DOI] [PubMed] [Google Scholar]
- 31.Lise BA, Ostrup O. Toward liquid biopsies in cancer treatment: application of circulating tumor DNA. APMIS. 2019;127:329–336. [DOI] [PubMed] [Google Scholar]
- 32.Butler TM, Johnson-Camacho K, Peto M, et al. Exome sequencing of cell-free DNA from metastatic cancer patients identifies clinically actionable mutations distinct from primary disease. PLoS One. 2015;10(8):e0136407. doi: 10.1371/journal.pone.0136407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mouliere F, Chandrananda D, Piskorz AM, et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med. 2018;10(466):eaat4921. doi: 10.1126/scitranslmed.aat4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shao X, He Y, Ji M, et al. Quantitative analysis of cell-free DNA in ovarian cancer. Oncol Lett. 2015;10(6):3478–3482. doi: 10.3892/ol.2015.3771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamat AA, Baldwin M, Urbauer D, et al. Plasma cell-free DNA in ovarian cancer: an independent prognostic biomarker. Cancer. 2010;116(8):1918–1925. doi: 10.1002/cncr.v116:8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamat AA, Sood AK, Dang D, Gershenson DM, Simpson JL, Bischoff FZ. Quantification of total plasma cell-free DNA in ovarian cancer using real-time PCR. Ann N Y Acad Sci. 2006;1075:230–234. doi: 10.1196/annals.1368.031 [DOI] [PubMed] [Google Scholar]
- 37.Zachariah RR, Schmid S, Buerki N, Radpour R, Holzgreve W, Zhong X. Levels of circulating cell-free nuclear and mitochondrial DNA in benign and malignant ovarian tumors. Obstet Gynecol. 2008;112(4):843–850. doi: 10.1097/AOG.0b013e3181867bc0 [DOI] [PubMed] [Google Scholar]
- 38.Dobrzycka B, Terlikowski SJ, Kinalski M, Kowalczuk O, Niklinska W, Chyczewski L. Circulating free DNA and p53 antibodies in plasma of patients with ovarian epithelial cancers. Ann Oncol. 2011;22(5):1133–1140. doi: 10.1093/annonc/mdq584 [DOI] [PubMed] [Google Scholar]
- 39.Cicchillitti L, Corrado G, De Angeli M, et al. Circulating cell-free DNA content as blood based biomarker in endometrial cancer. Oncotarget. 2017;8(70):115230–115243. doi: 10.18632/oncotarget.v8i70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobrzycka B, Terlikowski SJ, Mazurek A, et al. Circulating free DNA, p53 antibody and mutations of KRAS gene in endometrial cancer. Int J Cancer. 2010;127(3):612–621. doi: 10.1002/ijc.v127:3 [DOI] [PubMed] [Google Scholar]
- 41.Vizza E, Corrado G, De Angeli M, et al. Serum DNA integrity index as a potential molecular biomarker in endometrial cancer. J Exp Clin Cancer Res. 2018;37(1):16. doi: 10.1186/s13046-018-0688-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka H, Tsuda H, Nishimura S, et al. Role of circulating free alu DNA in endometrial cancer. Int J Gynecol Cancer. 2012;22(1):82–86. doi: 10.1097/IGC.0b013e3182328c94 [DOI] [PubMed] [Google Scholar]
- 43.Hickey KP, Boyle KP, Jepps HM, Andrew AC, Buxton EJ, Burns PA. Molecular detection of tumour DNA in serum and peritoneal fluid from ovarian cancer patients. Br J Cancer. 1999;80(11):1803–1808. doi: 10.1038/sj.bjc.6690601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rivlin N, Brosh R, Oren M, Rotter V. Mutations in the p53 tumor suppressor gene: important milestones at the various steps of tumorigenesis. Genes Cancer. 2011;2(4):466–474. doi: 10.1177/1947601911408889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Otsuka J, Okuda T, Sekizawa A, et al. Detection of p53 mutations in the plasma DNA of patients with ovarian cancer. Int J Gynecol Cancer. 2004;14(3):459–464. doi: 10.1111/ijg.2004.14.issue-3 [DOI] [PubMed] [Google Scholar]
- 47.Park YR, Kim YM, Lee SW, et al. Optimization to detect TP53 mutations in circulating cell-free tumor DNA from patients with serous epithelial ovarian cancer. Obstet Gynecol Sci. 2018;61(3):328–336. doi: 10.5468/ogs.2018.61.3.328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swisher EM, Wollan M, Mahtani SM, et al. Tumor-specific p53 sequences in blood and peritoneal fluid of women with epithelial ovarian cancer. Am J Obstet Gynecol. 2005;193(3 Pt 1):662–667. doi: 10.1016/j.ajog.2005.01.054 [DOI] [PubMed] [Google Scholar]
- 49.Cohen PA, Flowers N, Tong S, Hannan N, Pertile MD, Hui L. Abnormal plasma DNA profiles in early ovarian cancer using a non-invasive prenatal testing platform: implications for cancer screening. BMC Med. 2016;14(1):126. doi: 10.1186/s12916-016-0667-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vanderstichele A, Busschaert P, Smeets D, et al. Chromosomal instability in cell-free DNA as a highly specific biomarker for detection of ovarian cancer in women with adnexal masses. Clin Cancer Res. 2017;23(9):2223–2231. doi: 10.1158/1078-0432.CCR-16-1078 [DOI] [PubMed] [Google Scholar]
- 51.Phallen J, Sausen M, Adleff V, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. 2017;9(403). doi: 10.1126/scitranslmed.aan2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Farkkila A, McConechy MK, Yang W, et al. FOXL2 402C>G mutation can be identified in the circulating tumor DNA of patients with adult-type granulosa cell tumor. J Mol Diagn. 2017;19(1):126–136. doi: 10.1016/j.jmoldx.2016.08.005 [DOI] [PubMed] [Google Scholar]
- 53.Sun Y, Zhang L, Ho SS, Wu X, Gu J. Lower mitochondrial DNA copy number in peripheral blood leukocytes increases the risk of endometrial cancer. Mol Carcinog. 2016;55(6):1111–1117. doi: 10.1002/mc.v55.6 [DOI] [PubMed] [Google Scholar]
- 54.Clay Montier LL, Deng JJ, Bai Y. Number matters: control of mammalian mitochondrial DNA copy number. J Genet Genomics. 2009;36(3):125–131. doi: 10.1016/S1673-8527(08)60099-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li L, Li C, Mao H, et al. Epigenetic inactivation of the CpG demethylase TET1 as a DNA methylation feedback loop in human cancers. Sci Rep. 2016;6:26591. doi: 10.1038/srep26591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giannopoulou L, Chebouti I, Pavlakis K, Kasimir-Bauer S, Lianidou ES. RASSF1A promoter methylation in high-grade serous ovarian cancer: a direct comparison study in primary tumors, adjacent morphologically tumor cell-free tissues and paired circulating tumor DNA. Oncotarget. 2017;8(13):21429–21443. doi: 10.18632/oncotarget.v8i13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Q, Hu G, Yang Q, et al. A multiplex methylation-specific PCR assay for the detection of early-stage ovarian cancer using cell-free serum DNA. Gynecol Oncol. 2013;130(1):132–139. doi: 10.1016/j.ygyno.2013.04.048 [DOI] [PubMed] [Google Scholar]
- 58.Liggett TE, Melnikov A, Yi Q, et al. Distinctive DNA methylation patterns of cell-free plasma DNA in women with malignant ovarian tumors. Gynecol Oncol. 2011;120(1):113–120. doi: 10.1016/j.ygyno.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Melnikov A, Scholtens D, Godwin A, Levenson V. Differential methylation profile of ovarian cancer in tissues and plasma. J Mol Diagn. 2009;11(1):60–65. doi: 10.2353/jmoldx.2009.080072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Widschwendter M, Zikan M, Wahl B, et al. The potential of circulating tumor DNA methylation analysis for the early detection and management of ovarian cancer. Genome Med. 2017;9(1):116. doi: 10.1186/s13073-017-0500-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li L, Zheng H, Huang Y, et al. DNA methylation signatures and coagulation factors in the peripheral blood leucocytes of epithelial ovarian cancer. Carcinogenesis. 2017;38(8):797–805. doi: 10.1093/carcin/bgx057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu Y, Zhang X, Lin L, Ma XP, Ma YC, Liu PS. Aberrant methylation of RASSF2A in tumors and plasma of patients with epithelial ovarian cancer. Asian Pac J Cancer Prev. 2014;15(3):1171–1176. doi: 10.7314/APJCP.2014.15.3.1171 [DOI] [PubMed] [Google Scholar]
- 63.Ibanez de Caceres I, Battagli C, Esteller M, et al. Tumor cell-specific BRCA1 and RASSF1A hypermethylation in serum, plasma, and peritoneal fluid from ovarian cancer patients. Cancer Res. 2004;64(18):6476–6481. doi: 10.1158/0008-5472.CAN-04-1529 [DOI] [PubMed] [Google Scholar]
- 64.Iwasa H, Hossain S, Hata Y. Tumor suppressor C-RASSF proteins. Cell Mol Life Sci. 2018;75(10):1773–1787. doi: 10.1007/s00018-018-2756-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fernandes MS, Carneiro F, Oliveira C, Seruca R. Colorectal cancer and RASSF family – a special emphasis on RASSF1A. Int J Cancer. 2013;132(2):251–258. doi: 10.1002/ijc.27696 [DOI] [PubMed] [Google Scholar]
- 66.Schmidt ML, Hobbing KR, Donninger H, Clark GJ. RASSF1A deficiency enhances RAS-driven lung tumorigenesis. Cancer Res. 2018;78(10):2614–2623. doi: 10.1158/0008-5472.CAN-17-2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Margolin G, Petrykowska HM, Jameel N, Bell DW, Young AC, Elnitski L. Robust detection of DNA hypermethylation of ZNF154 as a pan-cancer locus with in silico modeling for blood-based diagnostic development. J Mol Diagn. 2016;18(2):283–298. doi: 10.1016/j.jmoldx.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parkinson CA, Gale D, Piskorz AM, et al. Exploratory analysis of TP53 mutations in circulating tumour DNA as biomarkers of treatment response for patients with relapsed high-grade serous ovarian carcinoma: a retrospective study. PLoS Med. 2016;13(12):e1002198. doi: 10.1371/journal.pmed.1002198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kamat AA, Bischoff FZ, Dang D, et al. Circulating cell-free DNA: a novel biomarker for response to therapy in ovarian carcinoma. Cancer Biol Ther. 2006;5(10):1369–1374. doi: 10.4161/cbt.5.10.3240 [DOI] [PubMed] [Google Scholar]
- 70.Bolivar AM, Luthra R, Mehrotra M, et al. Targeted next-generation sequencing of endometrial cancer and matched circulating tumor DNA: identification of plasma-based, tumor-associated mutations in early stage patients. Modern Pathol. 2019;32(3):405–414. doi: 10.1038/s41379-018-0158-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wimberger P, Roth C, Pantel K, Kasimir-Bauer S, Kimmig R, Schwarzenbach H. Impact of platinum-based chemotherapy on circulating nucleic acid levels, protease activities in blood and disseminated tumor cells in bone marrow of ovarian cancer patients. Int J Cancer. 2011;128(11):2572–2580. doi: 10.1002/ijc.v128.11 [DOI] [PubMed] [Google Scholar]
- 72.Perkins G, Yap TA, Pope L, et al. Multi-purpose utility of circulating plasma DNA testing in patients with advanced cancers. PLoS One. 2012;7(11):e47020. doi: 10.1371/journal.pone.0047020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Steffensen KD, Madsen CV, Andersen RF, Waldstrom M, Adimi P, Jakobsen A. Prognostic importance of cell-free DNA in chemotherapy resistant ovarian cancer treated with bevacizumab. Eur J Cancer. 2014;50(15):2611–2618. doi: 10.1016/j.ejca.2014.06.022 [DOI] [PubMed] [Google Scholar]
- 74.No JH, Kim K, Park KH, Kim YB. Cell-free DNA level as a prognostic biomarker for epithelial ovarian cancer. Anticancer Res. 2012;32(8):3467–3471. [PubMed] [Google Scholar]
- 75.Kuhlmann JD, Schwarzenbach H, Wimberger P, Poetsch M, Kimmig R, Kasimir-Bauer S. LOH at 6q and 10q in fractionated circulating DNA of ovarian cancer patients is predictive for tumor cell spread and overall survival. BMC Cancer. 2012;12:325. doi: 10.1186/1471-2407-12-325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Giannopoulou L, Mastoraki S, Buderath P, et al. ESR1 methylation in primary tumors and paired circulating tumor DNA of patients with high-grade serous ovarian cancer. Gynecol Oncol. 2018;150(2):355–360. doi: 10.1016/j.ygyno.2018.05.026 [DOI] [PubMed] [Google Scholar]
- 77.Harris FR, Kovtun IV, Smadbeck J, et al. Quantification of somatic chromosomal rearrangements in circulating cell-free DNA from ovarian cancers. Sci Rep. 2016;6:29831. doi: 10.1038/srep29831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pereira E, Camacho-Vanegas O, Anand S, et al. Personalized circulating tumor DNA biomarkers dynamically predict treatment response and survival in gynecologic cancers. PLoS One. 2015;10(12):e0145754. doi: 10.1371/journal.pone.0145754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Du ZH, Bi FF, Wang L, Yang Q. Next-generation sequencing unravels extensive genetic alteration in recurrent ovarian cancer and unique genetic changes in drug-resistant recurrent ovarian cancer. Mol Genet Genomic Med. 2018. doi: 10.1002/mgg3.414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martignetti JA, Camacho-Vanegas O, Priedigkeit N, et al. Personalized ovarian cancer disease surveillance and detection of candidate therapeutic drug target in circulating tumor DNA. Neoplasia. 2014;16(1):97–103. doi: 10.1593/neo.131900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rojas V, Hirshfield KM, Ganesan S, Rodriguez-Rodriguez L. Molecular characterization of epithelial ovarian cancer: implications for diagnosis and treatment. Int J Mol Sci. 2016;17(12):2113. doi: 10.3390/ijms17122113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Choudhuri S, Sharma C, Banerjee A, Kumar S, Kumar L, Singh N. A repertoire of biomarkers helps in detection and assessment of therapeutic response in epithelial ovarian cancer. Mol Cell Biochem. 2014;386(1–2):259–269. doi: 10.1007/s11010-013-1863-8 [DOI] [PubMed] [Google Scholar]
- 83.Capizzi E, Gabusi E, Grigioni AD, et al. Quantification of free plasma DNA before and after chemotherapy in patients with advanced epithelial ovarian cancer. Diagn Mol Pathol. 2008;17(1):34–38. doi: 10.1097/PDM.0b013e3181359e1f [DOI] [PubMed] [Google Scholar]
- 84.Arend RC, Londono AI, Montgomery AM, et al. Molecular response to neoadjuvant chemotherapy in high-grade serous ovarian carcinoma. Mol Cancer Res. 2018;16(5):813–824. doi: 10.1158/1541-7786.MCR-17-0594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gifford G, Paul J, Vasey PA, Kaye SB, Brown R. The acquisition of hMLH1 methylation in plasma DNA after chemotherapy predicts poor survival for ovarian cancer patients. Clin Cancer Res. 2004;10(13):4420–4426. doi: 10.1158/1078-0432.CCR-03-0732 [DOI] [PubMed] [Google Scholar]
- 86.Flanagan JM, Wilson A, Koo C, et al. Platinum-based chemotherapy induces methylation changes in blood DNA associated with overall survival in patients with ovarian cancer. Clin Cancer Res. 2017;23(9):2213–2222. doi: 10.1158/1078-0432.CCR-16-1754 [DOI] [PubMed] [Google Scholar]
- 87.Weigelt B, Comino-Mendez I, de Bruijn I, et al. Diverse BRCA1 and BRCA2 reversion mutations in circulating cell-free DNA of therapy-resistant breast or ovarian cancer. Clin Cancer Res. 2017;23(21):6708–6720. doi: 10.1158/1078-0432.CCR-17-0544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Christie EL, Fereday S, Doig K, Pattnaik S, Dawson SJ, Bowtell DDL. Reversion of BRCA1/2 germline mutations detected in circulating tumor DNA from patients with high-grade serous ovarian cancer. J Clin Oncol. 2017;35(12):1274–1280. doi: 10.1200/JCO.2016.70.4627 [DOI] [PubMed] [Google Scholar]
- 89.Lin KK, Harrell MI, Oza AM, et al. BRCA reversion mutations in circulating tumor DNA predict primary and acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov. 2019;9(2):210–219. doi: 10.1158/2159-8290.CD-18-0715 [DOI] [PubMed] [Google Scholar]
- 90.Matulonis U, Vergote I, Backes F, et al. Phase II study of the PI3K inhibitor pilaralisib (SAR245408; XL147) in patients with advanced or recurrent endometrial carcinoma. Gynecol Oncol. 2015;136(2):246–253. doi: 10.1016/j.ygyno.2014.12.019 [DOI] [PubMed] [Google Scholar]
- 91.La Rosa VL, Shah M, Kahramanoglu I, et al. Quality of life and fertility preservation counseling for women with gynecological cancer: an integrated psychological and clinical perspective. J Psychosom Obstet Gynaecol. 2019;1–7. doi: 10.1080/0167482X.2019.1648424 [DOI] [PubMed] [Google Scholar]
- 92.Calle EE, Kaaks R. Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nat Rev Cancer. 2004;4(8):579–591. doi: 10.1038/nrc1408 [DOI] [PubMed] [Google Scholar]
- 93.Ledermann JA, Raja FA, Fotopoulou C, et al. Newly diagnosed and relapsed epithelial ovarian carcinoma: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2013;24(Suppl 6):vi24–vi32. doi: 10.1093/annonc/mdt333 [DOI] [PubMed] [Google Scholar]
- 94.Vitale SG, Rossetti D, Tropea A, Biondi A, Lagana AS. Fertility sparing surgery for stage IA type I and G2 endometrial cancer in reproductive-aged patients: evidence-based approach and future perspectives. Updates Surg. 2017;69(1):29–34. doi: 10.1007/s13304-017-0419-y [DOI] [PubMed] [Google Scholar]
- 95.Cignini P, Vitale SG, Lagana AS, Biondi A, La Rosa VL, Cutillo G. Preoperative work-up for definition of lymph node risk involvement in early stage endometrial cancer: 5-year follow-up. Updates Surg. 2017;69(1):75–82. doi: 10.1007/s13304-017-0418-z [DOI] [PubMed] [Google Scholar]
- 96.Ortoft G, Dueholm M, Mathiesen O, et al. Preoperative staging of endometrial cancer using TVS, MRI, and hysteroscopy. Acta Obstet Gynecol Scand. 2013;92(5):536–545. doi: 10.1111/aogs.2013.92.issue-5 [DOI] [PubMed] [Google Scholar]
- 97.Pronin SM, Novikova OV, Andreeva JY, Novikova EG. Fertility-sparing treatment of early endometrial cancer and complex atypical hyperplasia in young women of childbearing potential. Int J Gynecol Cancer. 2015;25(6):1010–1014. doi: 10.1097/IGC.0000000000000467 [DOI] [PubMed] [Google Scholar]
- 98.Pal N, Broaddus RR, Urbauer DL, et al. Treatment of low-risk endometrial cancer and complex atypical hyperplasia with the levonorgestrel-releasing intrauterine device. Obstet Gynecol. 2018;131(1):109–116. doi: 10.1097/AOG.0000000000002390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Janda M, McGrath S, Obermair A. Challenges and controversies in the conservative management of uterine and ovarian cancer. Best Pract Res Clin Obstet Gynaecol. 2019;55:93–108. doi: 10.1016/j.bpobgyn.2018.08.004 [DOI] [PubMed] [Google Scholar]
- 100.Greenwald ZR, Huang LN, Wissing MD, Franco EL, Gotlieb WH. Does hormonal therapy for fertility preservation affect the survival of young women with early-stage endometrial cancer? Cancer. 2017;123(9):1545–1554. doi: 10.1002/cncr.v123.9 [DOI] [PubMed] [Google Scholar]
- 101.Schuurman MS, Kruitwagen R, Portielje JEA, Roes EM, Lemmens V, van der Aa MA. Treatment and outcome of elderly patients with advanced stage ovarian cancer: a nationwide analysis. Gynecol Oncol. 2018;149(2):270–274. doi: 10.1016/j.ygyno.2018.02.017 [DOI] [PubMed] [Google Scholar]
- 102.Vitale SG, Capriglione S, Zito G, et al. Management of endometrial, ovarian and cervical cancer in the elderly: current approach to a challenging condition. Arch Gynecol Obstet. 2019;299(2):299–315. doi: 10.1007/s00404-018-5006-z [DOI] [PubMed] [Google Scholar]
- 103.Ponti G, Manfredini M, Tomasi A. Non-blood sources of cell-free DNA for cancer molecular profiling in clinical pathology and oncology. Crit Rev Oncol Hematol. 2019;141:36–42. doi: 10.1016/j.critrevonc.2019.06.005 [DOI] [PubMed] [Google Scholar]
- 104.Krimmel JD, Schmitt MW, Harrell MI, et al. Ultra-deep sequencing detects ovarian cancer cells in peritoneal fluid and reveals somatic TP53 mutations in noncancerous tissues. Proc Natl Acad Sci U S A. 2016;113(21):6005–6010. doi: 10.1073/pnas.1601311113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Parrella P, Zangen R, Sidransky D, Nicol T. Molecular analysis of peritoneal fluid in ovarian cancer patients. Modern Pathol. 2003;16(7):636–640. doi: 10.1097/01.MP.0000076979.28106.ED [DOI] [PubMed] [Google Scholar]
- 106.Barquin M, Maximiano C, Perez-Barrios C, et al. Peritoneal washing is an adequate source for somatic BRCA1/2 mutation testing in ovarian malignancies. Pathol Res Pract. 2019;215(2):392–394. doi: 10.1016/j.prp.2018.10.028 [DOI] [PubMed] [Google Scholar]
- 107.Muller HM, Millinger S, Fiegl H, et al. Analysis of methylated genes in peritoneal fluids of ovarian cancer patients: a new prognostic tool. Clin Chem. 2004;50(11):2171–2173. [DOI] [PubMed] [Google Scholar]
- 108.Nair N, Camacho-Vanegas O, Rykunov D, et al. Genomic analysis of uterine lavage fluid detects early endometrial cancers and reveals a prevalent landscape of driver mutations in women without histopathologic evidence of cancer: a prospective cross-sectional study. PLoS Med. 2016;13(12):e1002206. doi: 10.1371/journal.pmed.1002206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Maritschnegg E, Wang Y, Pecha N, et al. Lavage of the uterine cavity for molecular detection of mullerian duct carcinomas: a proof-of-concept study. J Clin Oncol. 2015;33(36):4293–4300. doi: 10.1200/JCO.2015.61.3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kinde I, Bettegowda C, Wang Y, et al. Evaluation of DNA from the Papanicolaou test to detect ovarian and endometrial cancers. Sci Transl Med. 2013;5(167):167ra164. doi: 10.1126/scitranslmed.3004952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang Y, Li L, Douville C, et al. Evaluation of liquid from the Papanicolaou test and other liquid biopsies for the detection of endometrial and ovarian cancers. Sci Transl Med. 2018;10(433). doi: 10.1126/scitranslmed.aap8793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kwapisz D. The first liquid biopsy test approved. Is it a new era of mutation testing for non-small cell lung cancer? Ann Transl Med. 2017;5(3):46. doi: 10.21037/atm [DOI] [PMC free article] [PubMed] [Google Scholar]