Abstract
Purpose
To better characterize the urinary microbiome in males and contribute to overall understanding of the urinary microbiota specifically in patients undergoing evaluation for possible bladder cancer, stratified by risk exposure to smoking.
Patients and Methods
Recruitment of 43 male patients in a sequential manner presenting for hematuria evaluation to a single institution was undertaken. Mid-stream urine specimen pellets were processed through a DNA isolation protocol before undergoing PCR amplification, purification, and 16S rRNA gene sequencing. Gene sequences were clustered into operational taxonomic units and statistical analysis was performed to determine specimen diversity and phylogenetic trends.
Results
No significant difference in microbial diversity was found between the specimens. On subgroup analysis, no significant difference was observed when stratified by either tobacco smoking history or by newly diagnosed urothelial bladder cancer. Variation in microbial diversity was seen amongst all analyzed specimens.
Conclusion
The results of our analysis of carefully selected subjects help to better characterize the urinary microbiome in males and supplements the limited available information on the interrelationship between the urinary microbiome and development of genitourinary malignancy. No significant difference was observed in our small sample size when stratified by tobacco exposure or newly diagnosed bladder cancer.
Keywords: urinary microbiome, tobacco smoking, urothelial carcinoma, hematuria
Introduction
A microbiome is a comprehensive term that represents all the components of the host, microorganism genomes, and environmental conditions within a particular habitat. In humans, body systems that have traditionally been considered sterile, such as the urinary tract, have been found to be complex microenvironments, home to unique sets of bacterial communities.1 Standard bacterial cultures used to identify urinary pathogens such as Escherichia coli and Enterococcus faecalis have traditionally isolated fast-growing organisms involved in the development of urinary tract infections. However, organisms such as Lactobacillus and Ureaplasma are slow-growing and standard methodology is not designed to grow these species.2,3 Yet, advancements in molecular target analysis technology, such as 16S rRNA gene sequencing, has allowed for the identification of a wide variety of naturally occurring species and led to the understanding of the complex composition of the human urinary tract.
Human microbiota have been shown to play an important role in homeostasis by regulating health and disease. Potential roles of the urinary microbiota include production of neurotransmitters, competition with pathogens for resources, epithelial junction maintenance, priming of host immune defenses, and creation of a barrier to uroepithelium, amongst others.4,5 With respect to the genitourinary tract, research has already shown altered microbiota in individuals with neurogenic bladder dysfunction,6 incontinence,7 and other benign conditions.8 However, despite research that has linked microbiota to the development of cancers of many non-genitourinary organs,9 there is a dearth of translational investigation regarding the relationship between the urinary microbiota and bladder cancer.
Investigation into this subject has been limited and conflicting thus far. Patients with pre-existing known bladder cancer have inconsistently been shown in studies to have alterations in urinary tract microbiota; however, the studies have been underpowered and difficult to interpret amongst confounding factors.10−12 One such confounder, tobacco smoking, has long been an established risk factor for the development of bladder cancer13 and has been shown to alter microbiomes in other non-urologic oncologic investigations.14,15 From our literature search, there has been no similar study that investigates the relationship of smoking on the urinary microbiome, or specifically in relation to the development of bladder cancer.
We sought to contribute to the overall understanding of the urinary microbiota specifically in patients undergoing evaluation for possible bladder cancer, stratified by risk exposure to smoking.
Materials and Methods
Study Design
As hematuria is the most common presenting symptom of bladder cancer and results in a urologic evaluation, these patients who presented to our institution were recruited for our study. Written informed consent in compliance with Declaration of Helsinki was obtained in a sequential manner under this Lahey Hospital & Medical Center Institutional Review Board approved study. Patients provided a mid-stream voided urine sample in the ambulatory care setting prior to diagnostic flexible cystoscopy, which is standard practice for hematuria workup. Inclusion criteria included males greater than 18 years old who presented to outpatient urology clinic for evaluation of either gross or microscopic (≥3 red blood cells per high-powered field) hematuria, according to American Urological Association guidelines. Exclusion criteria included those with urolithiasis on computed tomography scan for hematuria evaluation, probiotic use, nitrite positive urinalysis, previous antibiotic use within 6 months prior to presentation, instrumentation of lower urinary tract within past year, or a previous diagnosis of urothelial carcinoma, or history of overactive bladder or urge incontinence. Those who met the study criteria were included in the final study population. Additionally, patients who subsequently underwent cystoscopic transurethral resection of a bladder tumor were followed to determine the presence of pathologically confirmed urothelial carcinoma. Patient demographics were collected through a retrospective review.
Sample Collection and DNA Isolation
Mid-stream urine samples were collected from patients who met study criteria and stored for up to one hour at 2–8°C before frozen to −80°C. Prior to analysis, samples were then thawed at 2–8°C and centrifuged at 4°C for 15 mins to form a pellet. The pellet was used for bacterial genomic DNA extraction using the PowerMag Soil DNA Isolation Kit (Catalog No. 27100; MO BIO Laboratories, Qiagen Inc., Carlsbad, CA). Negative extraction controls were used to reduce the influence of potential contamination on our analysis.
PCR Amplification, Purification, and Sequencing
The 16S rRNA gene sequencing methods have been adapted from methods developed for the NIH-Human Microbiome Project and were conducted by Diversigen, Inc (Houston, TX).16,17 The 16S rRNA hypervariable region V4 is amplified from the extracted community DNA by PCR and sequenced in the MiSeq platform (Illumina) using the 2x250 bp paired-end protocol. This generates paired end reads that overlap almost completely. The primers used for amplification contain adapters for MiSeq sequencing and single-end barcodes allowing pooling and direct sequencing of PCR products.
DNA from extracted samples is amplified using Invitrogen’s AccuPrime High Fidelity kit (Catalog No. 12346094). Each PCR reaction is prepared by combining 16 μL of the master mix (13.85 μL water + 2 μL 10 × reaction buffer + 1.5 μL Taq DNA polymerase), 2μl template DNA, 2 μL forward (515F) and reverse (806R) primers. PCR primers used for amplification incorporate adapters enabling DNA sequencing of the amplified product using an Illumina MiSeq.18 Primer sequences are shown below:
515F:
5ʹ AATGATACGGCGACCACCGAGATCTACACTATGGTAATTGTGTGCCAGCMGCCGCGGTAA 3ʹ
806R:
5ʹ CAAGCAGAAGACGGCATACGAGATTCCCTTGTCTCCAGTCAGTCAGCCGGACTACHVGGGTWTCTAAT ‘3
The DNA samples are amplified using the following thermocycler conditions: Initial denaturation at 95°C for 2 mins followed by 33 amplification cycles of 20 s at 95°C, 45 s at 50°C, 90 s at 72°C followed by a final extension at 72°C for 10 mins. The PCR product is purified using the QIAquick PCR purification kit (Catalogue# 28104) and the yield is quantified using Invitrogen’s Quant-iT Picogreen dsDNA assay kit (Catalogue # P7589). Amplicons are pooled, barcoded, and cleaned using the Invitrogen charge switch kit for 16S rRNA sequencing.
Compositional Analysis
The 16S rRNA gene analysis pipeline utilizes a combination of public tools and custom analytic packages developed at Diversigen to provide summary statistics and quality control measurements for each sequencing run, as well as multi-run reports and data-merging capabilities for validating built-in controls and characterizing microbial communities across large numbers of samples or sample groups.
The 16S pipeline incorporates phylogenetic and alignment-based approaches to maximize data resolution. The read pairs are demultiplexed based on their unique molecular barcodes, denoised and merged using DADA2,19 and subject to chimera removal using VSEARCH.20 16S rRNA gene sequences are clustered into Operational Taxonomic Units (OTUs) at a similarity cutoff value of 97%. Taxonomic identities are assigned to each out using the scikit-learn classifier and an optimized, variable region-specific version of the SILVA Database.21 Custom scripts construct a rarefied OTU table from the output files generated in the previous two steps for downstream analyses of alpha-diversity, beta-diversity, and phylogenetic trends.22 Alpha-diversity is used to describe the bacteria within the sample with regard to the number of species in the community and how close in numbers each species are to given a representation of species diversity. On the other hand, beta-diversity is a comparison of the resemblance of the bacterial communities between the different samples. Downstream statistical analysis and data visualization performed in R and SPSS. The continuous variables were compared between groups using a two-tailed independent sample Mann–Whitney U-test or the Kruskal–Wallis test, as appropriate. Categorical variables were analysed using Chi-squared tests or Fisher’s Exact Test, as appropriate. A P-value less than 0.05 was considered statistically significant, and adjusted for multiple comparisons using the Benjamini-Hochberg procedure.
Results
Urine samples were obtained from 43 consecutively enrolled patients that met the study criteria. Two of these samples were later excluded because of too few sequencing reads. Of the 41 samples retained in the analysis (Table 1), 24 were from patients that reported not having a history of smoking while 17 were from current or former smokers (4 and 13, respectively). Not surprisingly, there was a significantly higher proportion of patients subsequently diagnosed with bladder cancer in smokers vs non-smokers (35% vs 8%, respectively). Although age was not significantly different between the groups (p=0.190), there was a greater range of age in the non-smoking group, because three of these patients were under the age of 50. Consistent with previous studies that demonstrated higher prevalence rates of microscopic hematuria in males over the age of 60 with a history of smoking, those with a smoking history in our population had a higher proportion of patients who presented with microscopic compared to gross hematuria (88% vs 63%).
Table 1.
Clinical and Demographic Characteristics for the Patients Providing Samples for This Study
| Characteristic | Never Smoker | Ever Smoker | P-value |
|---|---|---|---|
| Number of samples | 24 | 17 | |
| Age, year, median (range) | 61 (22–84) | 67 (51–87) | 0.190 |
| BMI, kg/m2, median (range) | 27.9 (20–36.7) | 27 (22.3–32.6) | 0.705 |
| Anticoagulation/Antiplatelet, n (%) | 12 (50) | 9 (53) | 0.853 |
| Bladder cancer, n (%) | 2 (8) | 6 (35) | 0.049 |
| Diabetic, n (%) | 4 (17) | 0(0) | 0.128 |
| Hematuria_G | 16 (67) | 8 (47) | 0.209 |
| Hematuria_M | 15 (63) | 15(88) | 0.085 |
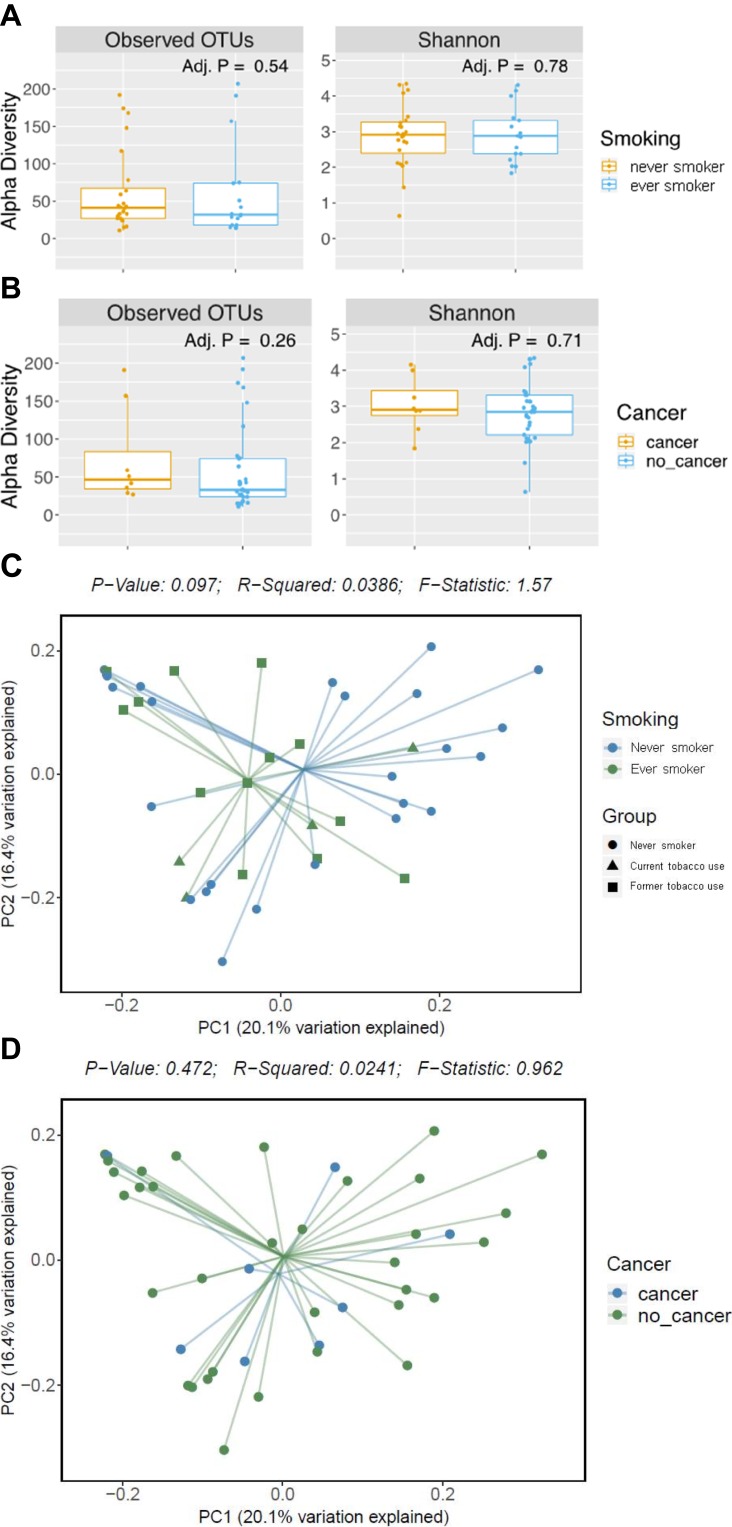
Two subgroups of particular interest were those with a smoking history and those found to have a pathologically proven new diagnosis of urothelial carcinoma. To determine a potential relationship, the subgroups were analyzed with a weighted UniFrac statistical method, which takes OTU presence and abundance into account and thereby helps to represent the community in the sample. When stratified by smoking, there was no significant difference in alpha or beta-diversity detected after false discovery rate (FDR) corrections between the smoking status. Specifically, there was no detected significant beta-diversity of those with any prior or current smoking history and those without a smoking history when analyzed with weighted UniFrac methodology (p=0.097). Subgroup analysis of those with bladder cancer did not reveal any significant difference in the alpha- or beta-diversity. Weighted UniFrac analysis of the beta-diversity between those with (n = 8) and without (n= 35) cancer did not reveal any significant difference in bacterial presence or abundance (p = 0.472) with principal coordinate analysis showing no significant clustering between either group (Figure 1).
Figure 1.
Alpha- and beta-diversity measurements for the groups in this study. No significant differences were observed in alpha-diversity measurements between never and ever smokers (A) nor patients with or without bladder cancer (B). No significant differences were observed in weighted UniFrac beta-diversity measurement between never and ever smokers (C) nor patients with or without bladder cancer (D).
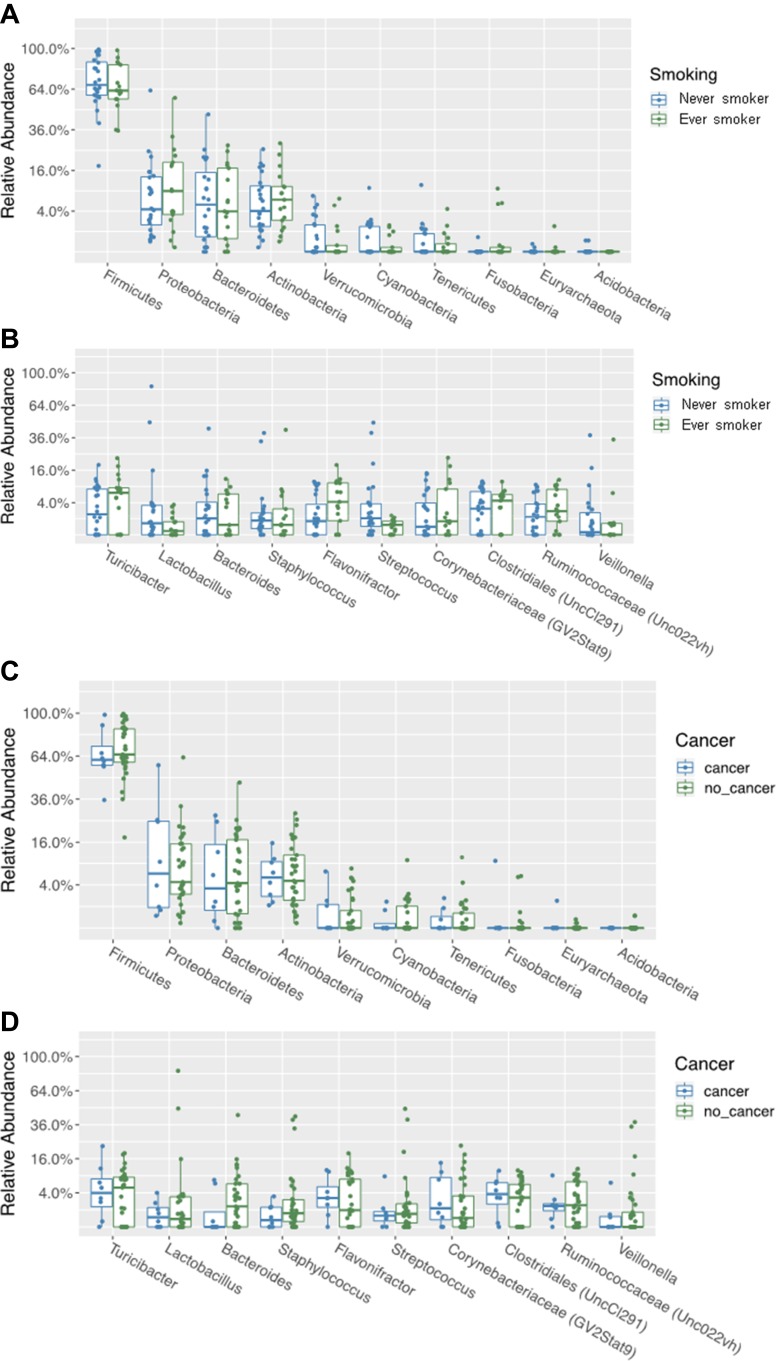
An evaluation of the relative abundance of specific phyla for our samples overall revealed Firmicutes, Proteobacteria, and Bacteroides as the most abundant. Focusing on the genera for overall samples demonstrated Turicibacter, Lactobacillus, and Bacteroides were most common. On subgroup analysis, there were no significant differences either based on tobacco smoking history or cancer diagnosis after false discovery rate corrections (Figure 2). There was wide variability in the inter-sample species variability, as is reflexed in the insignificant beta-diversity analysis.
Figure 2.
Relative abundance of phyla (A) and genera (B) between those with and without smoking history. Relative abundance of phyla (C) and genera (D) between those with and without bladder cancer.
Discussion
Our investigation of the urinary microbiome utilized 16S rRNA genomic sequencing to characterize the microbiome of patients presenting with hematuria for urologic evaluation. We were unable to demonstrate a significant difference in our sample of chronologically sequential specimens between patients who had a history of smoking or in those who were newly diagnosed with bladder urothelial carcinoma. Our cohort consisted entirely of males by design, but were also primarily in the seventh decade of life and overweight. We chose to exclusively study male patients as the female urinary microbiome has been found to be inherently different,2 likely due to hormonal and anatomic influences. With regard to our cohort mean age, our findings showed a lack of observed difference between those with similar mean age. Similar to Dong et al, we found a high relative abundance of Lactobacillus;23 however, we differ from previous reports in finding a high abundance of Turicibacter and Bacteroides.
We employed more stringent exclusion criteria than previous investigations of the UM by other authors with the goal of helping eliminate confounding factors affecting the UM. We excluded patients that had radiographic evidence of urolithiasis due to the emerging research that the urinary and intestinal microbiomes are associated with urinary stone formation, indicating that unique groups of bacteria are likely present in those with urinary stone disease and could potentially impact our findings.24 Patients with UTIs identified through standard laboratory means within the past 6 months were excluded from this study to reflect the numerous publications that demonstrate the incidence of UTIs is associated with changes in microbial diversity.5,7 Those patients with documented overactive bladder or urinary incontinence were also excluded given recent findings that demonstrate a variation of UM species in those with these conditions.25,26 Probiotic use is thought to alter urogenital microbiota in small clinical studies and molecular biology investigations.27 Additionally, the administration of Lactobacillus casei has been shown to alter the recurrence of superficial bladder cancer,28 so patients who reported taking an oral probiotic were not incorporated into our study. Lastly, previous studies that have looked at microbiota and bladder cancer have used patients with pre-existing urothelial carcinoma and it is therefore unclear whether any observed changes in the microbiome are secondary to the malignancy or are contributing to the development of the disease process. Through careful patient selection, our work hopes to further contribute to the growing body of evidence that seeks to understand this relationship.
To our knowledge, a focus on the effect of smoking on the human UM has not been studied before. Other authors have attempted to study the effect of smoking on bladder cancer protein expression29 and gene mutations;30,31 however, the importance of these molecular alterations in patients with bladder cancer with a smoking history is unclear. Since bacteria and microbial processes are known to interact with environmental exposures that can lead to human disease and immunological function,9 we hypothesized there would be a difference in the bacterial community in those with a smoking history. However, our study found no significant differences in terms of alpha diversity, beta diversity, or taxonomy after FDR corrections. This warrants further attention with a larger study population to not only validate these findings, but seek to better understand the potential role of metabolic byproducts of tobacco consumption on the pathophysiology of bladder cancer.
Our subgroup analysis of the small number of patients with biopsy-proven urothelial carcinoma in our study additionally found no significant difference in the microbial communities between those with cancer and those without. A recent study by Wu et al in China compared a small group of patients with variable grades of bladder urothelial cancer against controls evaluated for non-neoplastic processes and found both increased bacterial richness and a dysbiosis of urinary microbial communities in the cancer group.32 Our study was unable to conclude this difference. Conversely, Bucevic Popovic et al in Croatia similarly studied the urinary microbiome with regard to bladder cancer and found no significant difference in those with and without cancer.11 It is unclear what role ethnicity, geographics, diet, and other patient characteristics plays in the urinary microbiome that may lead to our conflicting results in the context of previously published literature. Additionally unclear is at what point a potential dysbiosis of the urinary microbiome might play on malignant pathophysiology. This question thus far seems not to be addressed in the available literature and we hoped our study can contribute to this discovery. Based on our admittedly small subgroup, we would think that it is actually the neoplastic process that causes a shift in microbial diversity rather than the shift being the instigating feature.
There are several limitations to our study. First, this is a small sample size and as a result the findings are observational and often do not meet strict scientific statistical significance. We hope to augment our cohort with a larger sample size and prolonged follow up to both strengthen our findings and determine if any of the initially negative evaluations eventually develop a urothelial malignancy. Additionally, we used mid-stream urine samples to collect our specimens, which introduces the possibility of distal urethra and urogenital skin contamination.8,12 We chose this method rather than more invasive approaches such as catheterization or suprapubic aspiration due to its practicality and potential to be used as a non-invasive diagnostic tool. Interpretation of our results must also be undertaken cautiously, as we are sampling from the unique geographic and demographic cohort that our institution serves, and there are likely differences in microbiota across different diets, geographies, and other yet to be identified influences. There we also did not use duplicate or triplicate samples to confirm the lack of sample contamination. Finally, our study focused on bacterial communities and did not specifically address the urinary mycobiome or virome that, although less abundant than bacteria, may play a role in the microbial balance.
A better understanding of the male microbiome presenting for potential bladder cancer evaluation and the potential existence of differential microbiomes carries multiple clinical implications. First, there is great potential to augment current diagnostic tools both in the smoking and non-smoking population by comparing microbiomes. Diagnostic techniques for bladder cancer rely on surveillance and repeat biopsies, but predictive models and tests are currently either poor or nonexistent. A relatively non-invasive test such as a urine sample, even as an adjunct to predictive models, would represent a significant contribution to the diagnostic model. In particular, stratification by smoking history and by development of bladder cancer would allow for applications to distinct clinical scenarios. Second, there is the future possibility of prophylaxis by adjusting the microbiome of the genitourinary tract to influence outcomes that could range from disease progression to recurrence. By applying knowledge of the microbiomes in patients who do not develop bladder cancer, we may at some point be able to adjust the bladder microbiome to decrease the risk of bladder cancer development, perhaps by probiotic inoculation. Future studies can utilize the results of this initial study to investigate such therapeutic potential.
Conclusion
This study contributes to the understanding of the male urinary microbiome and its potential relationship to the development of genitourinary malignancy. This single institutional study is the first to specifically address the potential interrelationship between tobacco exposure and development of urothelial carcinoma in the cotext of microbial changes. Although no significant differences were found amongst this small sample size, future studies can utilize the results of this initial study to investigate potential diagnostic and therapeutic interventions.
Acknowledgments
Specimen analysis and statistical methodology was performed in conjunction with Duy Dinh, PhD at Diversigen, Inc.
Disclosure
Matthew J Moynihan reports grants from Lahey Hospital & Medical Center, during the conduct of the study. The authors report no other conflicts of interest in this work.
This work was supported in part by an institutional grant from the R.E. Wise Research and Education Institute. Specimen analysis and statistical methodology were performed in conjunction with Diversigen, Inc.
References
- 1.Thomas-White K, Brady M, Wolfe AJ, Mueller ER. The bladder is not sterile: history and current discoveries on the urinary microbiome. Curr Bladder Dysfunct Rep. 2016;11(1):18–24. doi: 10.1007/s11884-016-0345-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hilt EE, McKinley K, Pearce MM, et al. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol. 2014;52:871–876. doi: 10.1128/JCM.02876-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.price TK, Dune T, Hilt EE, et al. The clinical urine culture: enhanced techniques improve detection of clinical relevant microorganisms. J Clin Microbiol. 2016;54(5):1216–1222. doi: 10.1128/JCM.00044-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whiteside SA, Razvi H, Dave S, Reid G, Burton JP. The microbiome of the urinary tract – a role beyond infection. Nat Rev Urol. 2015;12:81–90. doi: 10.1038/nrurol.2014.361 [DOI] [PubMed] [Google Scholar]
- 5.Nienhouse V, Gao X, Dong Q, et al. Interplay between bladder microbiota and urinary antimicrobial peptides: mechanisms for human urinary tract infection risk and symptom severity. PLoS One. 2014;9(12):e114185. doi: 10.1371/journal.pone.0114185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fouts DE, Pieper R, Szpakowski S, et al. Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. J Transl Med. 2012;28(10):174. doi: 10.1186/1479-5876-10-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearce MM, Hilt EE, Rosenfeld AB, et al. The female urinary microbiome: a comparison of women with and without urgency urinary incontinence. MBio. 2014;5(4):e01283–14. doi: 10.1128/mBio.01283-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bajic P, Van Kuiken ME, Burge BK, et al. Male bladder microbiome relates to lower urinary tract symptoms. Eur Urol Focus. 2018;21. [DOI] [PubMed] [Google Scholar]
- 9.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–812. doi: 10.1038/nrc3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu W, Yang L, Lee P, et al. Mini-review: perspective of the microbiome in the pathogenesis of urothelial carcinoma. Am J Clin Exp Urol. 2014;2(1):57–61. [PMC free article] [PubMed] [Google Scholar]
- 11.Bucevic Popovic V, Situm M, Chow CET, Chan LS, Roje B, Terzic J. The urinary microbiome associated with bladder cancer. Sci Rep 2017;8(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bajic P, Wofe AJ, Gupta GN. The urinary microbiome: implications in bladder cancer pathogenesis and therapeutics. Urology. 2019;126:10–15. doi: 10.1016/j.urology.2018.12.034 [DOI] [PubMed] [Google Scholar]
- 13.Freedman ND, Silverman DT, Hollenbeck AR, Schatzkin A, Abnet CC. Association between smoking and risk of bladder cancer among men and women. JAMA. 2011;306(7):737–745. doi: 10.1001/jama.2011.1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu G, Phillips S, Gail MH, et al. The effect of cigarette smoking on the oral and nasal microbiota. Microbiome. 2017;5:3. doi: 10.1186/s40168-016-0226-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biedermann L, Zeitz J, Mwinyi J, et al. Smoking cessation induces profound changes in the composition of the intestinal microbiota in humans. PLoS One. 2013;8(3):e59260. doi: 10.1371/journal.pone.0059260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Human Microbiome Project, C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Human Microbiome Project, C. A framework for human microbiome research. Nature. 2012;486(7402):215–221. doi: 10.1038/nature11209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high-throughput microbial community analysis on the illumina HiSeq and MiSeq platforms. ISME J. 2012;6(8):1621–1624. doi: 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Callahan BJ, McMurdie PJ, Rosen MJ, et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rognes T, Flouri T, Nichols B, et al. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41(Database issue):D590–D596. doi: 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong Q, Nelson DE, Toh E, Diao L, Gao X, Fortenberry JD, Van Der Pol B. The microbial communities in male first catch urine are highly similar to those in paired urethral swab specimens. PLoS One. 2011;6(5):e19709. doi: 10.1371/journal.pone.0019709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mehta M, Goldfarb DS, Nazzal L. The role of the microbiome in kidney stone formation. Int J Surg. 2016;36(Pt D):607–612. doi: 10.1016/j.ijsu.2016.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karstens L, Asquith M, Davin S, et al. Does the urinary microbiome play a role in urgency urinary incontinence and its severity? Front Cell Infect Microbiol. 2016;27(6):78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearce MM, Zilliox MJ, Rosenfeld AB, et al. Pelvic floor disorders network. The female urinary microbiome in urgency urinary incontinence. Am J Obstet Gynecol. 2015;213(3):347. E1–E11. doi: 10.1016/j.ajog.2015.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amdekar S, Singh V, Singh DD. Probiotic therapy: immunomodulating approach toward urinary tract infection. Curr Microbiol. 2011;63(5):484–490. doi: 10.1007/s00284-011-0006-2 [DOI] [PubMed] [Google Scholar]
- 28.Aso Y, Akaza H, Kotake T, Tsukamoto T, Imai K, Naito S. Preventive effect of a Lactobacillus casei preparation on the recurrence of superficial bladder cancer in a double-blind trial. The BLP Study Group. Eur Urol. 1995;27(2):104–109. doi: 10.1159/000475138 [DOI] [PubMed] [Google Scholar]
- 29.Joshi M, Millis SZ, Arguello D, et al. Molecular characterization of bladder cancer in smokers versus nonsmokers. Eur Urol Focus. 2016;4(1). Pii: S2405-4569(16)30071–2. [DOI] [PubMed] [Google Scholar]
- 30.Stern MC, Johnson LR, Bell DA, Taylor JA, Codon XPD. 751 polymorphism, metabolism genes, smoking, and bladder cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11(10 Pt 1):1004–1011. [PubMed] [Google Scholar]
- 31.Neumann AS, Sturgis EM, Wei Q. Nucleotide excision repair as a marker for susceptibility to tobacco-related cancers: a review of molecular epidemiological studies. Mol Carcinog. 2005;42:65–92. doi: 10.1002/(ISSN)1098-2744 [DOI] [PubMed] [Google Scholar]
- 32.Wu P, Zhang G, Zhao J, et al. Profiling the urinary microbiota in male patients with bladder cancer in China. Front Cell Infect Microbiol. 2018;31(8):167. doi: 10.3389/fcimb.2018.00167 [DOI] [PMC free article] [PubMed] [Google Scholar]