Abstract
Background
Overcoming β-lactam resistance in pathogens such as Pseudomonas aeruginosa is a major clinical challenge. Rapid molecular diagnostics (RMDs) have the potential to inform selection of empiric therapy in patients infected by P. aeruginosa.
Methods
In this study, we used a heterogeneous collection of 197 P. aeruginosa that included multidrug-resistant isolates to determine whether 2 representative RMDs (Acuitas Resistome test and VERIGENE gram-negative blood culture test) could identify susceptibility to 2 newer β-lactam/β-lactamase inhibitor (BL-BLI) combinations, ceftazidime/avibactam (CZA) and ceftolozane/tazobactam (TOL/TAZO).
Results
We found that the studied RMD platforms were able to correctly identify BL-BLI susceptibility (susceptibility sensitivity, 100%; 95% confidence interval [CI], 97%, 100%) for both BLs-BLIs. However, their ability to detect resistance to these BLs-BLIs was lower (resistance sensitivity, 66%; 95% CI, 52%, 78% for TOL/TAZO and 33%; 95% CI, 20%, 49% for CZA).
Conclusions
The diagnostic platforms studied showed the most potential in scenarios where a resistance gene was detected or in scenarios where a resistance gene was not detected and the prevalence of resistance to TOL/TAZO or CZA is known to be low. Clinicians need to be mindful of the benefits and risks that result from empiric treatment decisions that are based on resistance gene detection in P. aeruginosa, acknowledging that such decisions are impacted by the prevalence of resistance, which varies temporally and geographically.
Keywords: Pseudomonas aeruginosa, ceftolozane/tazobactam, ceftazidime/avibactam, antimicrobial resistance
A collection of 197 Pseudomonas aeruginosa was used to determine whether 2 rapid molecular diagnostics could predict resistance to ceftazidime/avibactam and ceftolozane/tazobactam. The greatest potential was observed when a defined resistance gene was detected or when not detected and resistance prevalence was low.
Pseudomonas aeruginosa is a frequently observed and notorious healthcare-associated pathogen that causes severe infections in immunocompromised hosts, patients with cystic fibrosis pulmonary disease, and those with surgical site and burn infections. In US hospitals, P. aeruginosa is also a common cause of hospital-acquired infections, including ventilator-associated pneumonia and urinary tract and bloodstream infections. More than 50000 healthcare-associated P. aeruginosa infections occur in the United States each year. Rates of resistance to carbapenems, piperacillin/tazobactam, and the expanded-spectrum cephalosporins (ceftazidime and cefepime) are steadily increasing in P. aeruginosa. Monitoring agencies are also reporting a significant increase in multidrug resistance in isolates recovered from intensive care units [1]. Despite decades of advancements in therapy and infection control, the Centers for Disease Control and Prevention have designated P. aeruginosa as “a serious threat”.
Overcoming β-lactam resistance in P. aeruginosa is a major clinical challenge as diverse mechanisms, including many different β-lactamases encoded by bla genes, porin changes, efflux pump overexpression, and changes in penicillin-binding proteins (PBPs), are responsible for resistance [2]. The most problematic resistance phenotypes for clinicians are resistance to imipenem, meropenem, piperacillin/tazobactam, ceftazidime, and cefepime. Losing the ability to empirically or definitely administer these “last-line” drugs creates a distressing clinical situation as alternatives (eg, polymyxins) may be toxic, difficult to monitor, and have challenging pharmacokinetic properties.
Fortunately, novel therapeutics that have promise in overcoming β-lactam resistance in P. aeruginosa are becoming available. Two recently available agents are ceftazidime/avibactam (CZA) and ceftolozane/tazobactam (TOL/TAZO). Both of these new β-lactam/β-lactamase inhibitor (BL-BLI) combinations possess microbiological and pharmacological properties that have the potential to overcome many of the common resistance mechanisms in P. aeruginosa. CZA contains the BLI, avibactam, which readily inactivates the chromosomal AmpC of P. aeruginosa (PDC β-lactamase) and other class A β-lactamases such as KPC [3]. TOL/TAZO has a BLI, tazobactam, which has modest AmpC inhibitory activity but is paired with ceftolozane, an advanced-generation cephalosporin that is poorly hydrolyzed by the AmpC of P. aeruginosa [4, 5]. Additionally, TOL/TAZO does not require the OprD porin channel to be intact for penetration into the periplasmic space to reach its target, the PBPs [6], and is also not affected by drug efflux systems such as MexAB-OprM, MexCD-OprJ, and MexXY-OprM [7].
Clinical trials are underway to test these agents in the treatment of P. aeruginosa infection. As these trials are being conducted, the biomedical community is also investigating whether rapid molecular diagnostics (RMDs) can help clinicians chose appropriate empiric therapy and practice antimicrobial stewardship [8]. PRIMERS (Platforms for Rapid Identification of MDR-gram negative bacteria and Evaluation of Resistance Studies) is a series of studies launched by the Antibacterial Resistance Leadership Group, which is dedicated to the evaluation of RMDs. In PRIMERS I and II, 2 RMD platforms were used to assess extended-spectrum cephalosporin and carbapenem susceptibility in Escherichia coli and Klebsiella pneumoniae [9]. PRIMERS III established that RMDs can discriminate between carbapenem resistance and susceptibility in Acinetobacter species [10].
Our objective in this study was to determine whether representative RMDs can accurately detect resistance to 2 novel BLs-BLIs, CZA and TOL/TAZO, and potentially predict their activity against multidrug-resistant P. aeruginosa. We reasoned that finding β-lactamase gene “targets” (bla genes) may facilitate appropriate clinical decision-making regarding β-lactam selection and antibiotic stewardship. We selected agents for use (CZA and TOL/TAZO) against P. aeruginosa that are known to be active in microbiological survey studies. We adopted this challenge as it captures the current dilemma faced by both clinicians and industry trying to evaluate novel therapeutic agents using established and developing RMD platforms.
METHODS
A collection of 197 well-characterized clinical P. aeruginosa isolates was tested. These isolates came primarily from northeast Ohio and the mid-Atlantic states and were stored in the investigators’ laboratories (R. A. B. and B. N. K.) [11]. Approximately half were previously determined by phenotypic testing from clinical laboratories in these geographic areas to be resistant to carbapenems and/or expanded-spectrum cephalosporins.
Antimicrobial susceptibility testing (AST) was performed using broth microdilution as previously described, with results interpreted according to Clinical and Laboratory Standards Institute and US Food and Drug Administration guidelines [10, 12]. American Type Culture Collection control strains P. aeruginosa 27853 and E. coli 25922 were used as quality control strains.
Two RMD platforms, the Acuitas Resistome test (OpGen Inc., Gaithersburg, MD) and VERIGENE gram-negative blood culture test (VERIGENE BC-GN, Luminex Corporation, Austin, TX), were used to identify genes that potentially confer resistance to TOL/TAZO or CZA. The OpGen Clinical Services Laboratory tested colony isolates using the Acuitas Resistome test. This platform is a microfluidic polymerase chain reaction (PCR) array that analyzes gram-negative bacilli for 46 antibiotic-resistant gene families across several hundred variants, including genes for carbapenemases, extended-spectrum β-lactamases, and AmpC β-lactamases.
Nucleic acids were extracted from 500 µL of colonies grown to a McFarland standard of 0.5, using the Roche MagNA Pure 96 DNA and Viral NA large volume kit on the MagNA Pure 96 system. PCR was performed using primers and fluorescent reporter probes (Applied Biosystems Custom TaqMan MGB probes with 5’-FAM and a 3’ non-fluorescent quencher). All PCRs used dUTP instead of TTP along with uracil-DNA glycosylase to guard against accidental amplicon contamination. An internal amplification control (gBlocks Gene Fragment from Integrated DNA Technologies) was prepared in 1 µg/mL of calf thymus DNA in Tris-EDTA, pH 8, and added to all samples to monitor potential PCR inhibition. gBlocks covering all target amplicon sequences were used as positive PCR control samples.
PCR was performed with Fluidigm’s BioMark HD system using 96.96 Dynamic Array IFC arrays, a microfluidic system capable of analyzing 96 samples with 96 separate PCR assays. Each PCR contained 3 nL of extracted DNA plus 610 nmol/L of each PCR primer, 340 nmol/L fluorescent reporter probe, and 0.91X ThermoFisher TaqPath qPCR MasterMix. PCR was performed with the following cycling program: 2 minutes at 50°C, 10 minutes at 95°C, followed by 40 cycles of 15 seconds at 95°C and 1 minute at 60°C.
The VERIGENE BC-GN assay is a nonamplified gene-array detection system that identifies genus, species, and genetic resistance determinants for a panel of gram-negative bacteria directly from bacterial suspensions in positive blood culture bottles within 2 hours. Bacteria identified are E. coli, K. pneumoniae, Klebsiella oxytoca, P. aeruginosa, Acinetobacter spp., Citrobacter spp., Enterobacter spp., and Proteus spp. Resistance determinants detected include blaCTX-M, blaKPC, blaOXA, blaNDM, blaVIM, and blaIMP. The VERIGENE BC-GN assay was used to test bacterial suspensions of isolates grown on sheep blood agar plates and suspended in saline at a density matching a 0.5 McFarland standard. These suspensions were tested using GN-BC cartridges according to the manufacturer’s instructions for positive blood culture bottles containing gram-negative bacteria.
The absence or presence of the above-listed bla genes was queried against minimum inhibitory concentration (MIC) values determined by standard broth microdilution. For TOL/TAZO predictive analysis, blaKPC, blaNDM, blaVIM, and blaIMP were queried; only blaNDM,blaVIM, and blaIMP were included in the CZA predictive analysis.
Analytical methods using susceptibility and resistance sensitivities were estimated with 95% confidence intervals (CIs). Predictive values for susceptibility (SPVs) and resistance (RPVs) were also estimated as a function of susceptibility prevalence using an adjusted logit transformation and previously described methods [9, 10, 13]. BED-FRAME analyses [14], using the average weighted accuracy (AWA, accuracy adjusted for the relative importance of diagnostic errors over the relevant range of P. aeruginosa resistance [5%–20%]), were used to evaluate the global utility of these RMD strategies relative to the best random test (BRT), the random test that predicts resistance with a fixed probability resulting in the largest AWA.
RESULTS
The AST determinations and MIC distributions for isolates included in PRIMERS IV are presented in Figure 1 and Tables 1–3. Table 1 summarizes the susceptibility and resistance phenotypes of this P. aeruginosa collection against 17 clinically important anti-pseudomonal agents. To assist with the analysis of AST data, we categorized isolates into 4 groups: carbapenem susceptible, carbapenem resistant without blaKPC or blaVIM, carbapenem resistant with blaKPC, and carbapenem resistant with blaVIM. The carbapenem-resistant group without blaKPC or blaVIM likely represents the most common genotype of carbapenem-resistant P. aeruginosa in the United States as the prevalence of blaKPC and blaVIM in this pathogen is still limited. Alteration of OprD and/or upregulation of efflux systems are most likely the molecular basis of this phenotype in the United States. We note that in this group, 25.3% and 22.8% of isolates are resistant to CZA and TOL/TAZO, respectively, and those isolates are significantly resistant to other commonly used agents. In the group of carbapenem resistant with blaVIM or blaKPC, all were resistant to TOL/TAZO.
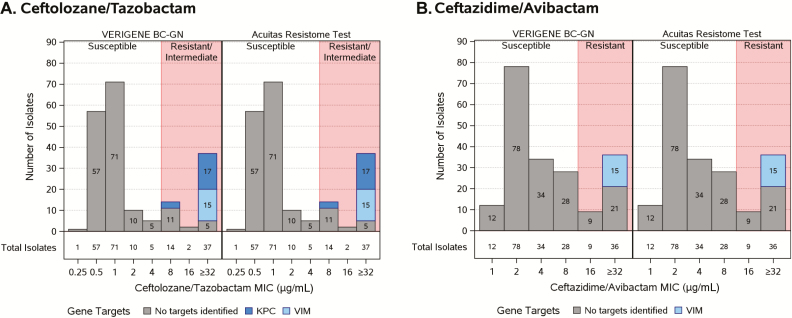
Figure 1.
Isolates identified by minimum inhibitory concentration susceptibility status, platform, and gene targets. For ceftolozane/tazobactam the gene targets were blaNDM, -VIM, -IMP, and -KPC. For ceftazidime/avibactam the gene targets were blaNDM, -VIM, and -IMP. All gene targets were examined; only genes identified are presented. Abbreviations: BC-GN, gram-negative blood culture test; MIC, minimum inhibitory concentration.
Table 1.
Antimicrobial Susceptibility Testing Results: Percent of Isolates Susceptible to Antimicrobial Agents
| Agent | Percent of Isolates Susceptible to Antimicrobial Agents | ||||
|---|---|---|---|---|---|
| Carbapenem Susceptible (to All Tested Carbapenems) (N = 83) |
Carbapenem Resistanta Without KPC or VIM (N = 79)b |
Carbapenem Resistant With KPC (N = 20) |
Carbapenem Resistant With VIM (N = 15) |
All Isolates (N = 197) |
|
| Imipenem | 100 | 2.5 | 0.0 | 0.0 | 43.1 |
| Meropenem | 100 | 24.1 | 0.0 | 13.3 | 52.8 |
| Doripenem | 100 | 29.1 | 0.0 | 6.7 | 54.3 |
| Ticarcillin-clavulanate | 21.7 | 5.1 | 0.0 | 0.0 | 11.2 |
| Piperacillin-tazobactam | 94.0 | 34.2 | 0.0 | 0.0 | 53.3 |
| Aztreonam | 94.0 | 30.4 | 0.0 | 40.0 | 54.8 |
| Cefepime | 100 | 58.2 | 0.0 | 0.0 | 65.5 |
| Ceftazidime | 98.8 | 49.4 | 0.0 | 0.0 | 61.4 |
| Ceftazidime-avibactam | 100 | 74.7 | 50.0 | 0.0 | 77.2 |
| Ceftolozane-tazobactam | 100 | 77.2 | 0.0 | 0.0 | 73.1 |
| Gentamicin | 98.8 | 78.5 | 30.0 | 6.7 | 76.6 |
| Tobramycin | 100 | 82.3 | 30.0 | 0.0 | 78.2 |
| Amikacin | 100 | 96.2 | 60.0 | 6.7 | 87.3 |
| Ciprofloxacin | 97.6 | 36.7 | 15.0 | 0.0 | 57.4 |
| Levofloxacin | 97.6 | 34.2 | 15.0 | 0.0 | 56.3 |
| Colistin | 100 | 94.9 | 100.0 | 100.0 | 98.0 |
| Polymixin B | 100 | 96.2 | 100.0 | 100.0 | 98.5 |
aFor purposes of this study, an intermediate phenotype was considered as resistant.
bIncludes 54 isolates resistant to all 3 carbapenems, 19 resistant to imipenem only, 1 resistant to meropenem only, 4 resistant to meropenem and imipenem only, and 1 resistant to doripenem and meropenem only, N = 79.
Table 3.
Summary of Platform Results by Minimum Inhibitory Concentration Susceptibility Status
| Platform | Ceftolozane/Tazobactam | Ceftazidime/Avibactam | |||||
|---|---|---|---|---|---|---|---|
| S (N = 144) | R (N = 53) | Total (N = 197) | S (N = 152) | R (N = 45) | Total (N = 197) | ||
| VERIGENE gram-negative blood culture test | Negative | 144 (100%) | 18 (34%) | 162 (82%) | 152 (100%) | 30 (67%) | 182 (92%) |
| Positive | 0 (0%) | 35 (66%) | 35 (18%) | 0 (0%) | 15 (33%) | 15 (8%) | |
| Acuitas Resistome test | Negative | 144 (100%) | 18 (34%) | 162 (82%) | 152 (100%) | 30 (67%) | 182 (92%) |
| Positive | 0 (0%) | 35 (66%) | 35 (18%) | 0 (0%) | 15 (33%) | 15 (8%) | |
All carbapenemase genes were queried for the ceftolozane/tazobactam predictive analysis; only the blaNDM, -VIM, -IMP genes were included in the ceftazidime/avibactam predictive analysis. For the purposes of this study, an intermediate phenotype was also considered as R.
Abbreviations: R, resistant; S, susceptible.
The genotype profile is presented in Figure 1 and Table 4. In this collection, 27% (n = 53) of the isolates chosen were resistant to TOL/TAZO and 23% (n = 45) were resistant to CZA by MIC determinations (Figure 1, Table 2). These susceptibility data served as the reference standard to compare the ability of RMDs to inform therapy. In Figure 1A, the relevant targets identified include blaVIM and blaKPC as determinants of TOL/TAZO resistance. In Figure 1B, the only resistance determinant is blaVIM as avibactam inhibits most KPC variants, and blaIMP and blaNDM were not detected in any of the isolates (a limitation of this study). Although blaIMP and blaNDM are rare causes of carbapenem resistance in the United States, these determinants may be more relevant in other parts of the world.
Table 4.
Genotypic Profile of 197 Isolates Studied via Acuitas Resistome Test and VERIGENE Gram-negative Blood Culture Test Platforms
| No. of isolates | IMP | KPC | NDM | VIM |
|---|---|---|---|---|
| 162/162 | – | – | – | – |
| 15/15 | – | – | – | + |
| 20/20 | – | + | – | – |
Isolate count reflects number of isolates for Acuitas Resistome test/VERIGENE gram-negative blood culture test platforms, respectively.
Table 2.
Phenotypic Profile of 197 Isolates Studied
| Susceptibility Status by Drug | ||
|---|---|---|
| No. of Isolates | Ceftolozane/Tazobactam | Ceftazidime/Avibactam |
| 138 | S | S |
| 6 | S | R |
| 14 | R | S |
| 39 | R | R |
| Total | 144 S, 53 R | 152 S, 45 R |
US Food and Drug Administration breakpoints were used for determining ceftolozane/tazobactam (S ≤ 4, I = 8, R > 8) and ceftazidime/avibactam (S ≤ 8, R > 8) susceptibility status. For the purposes of this study, an intermediate phenotype was also considered as R.
Abbreviations: R, resistant; S, susceptible.
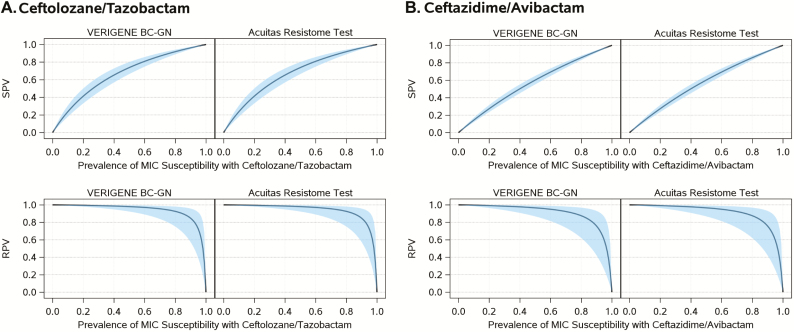
For the CZA and TOL/TAZO treatment combination, estimates of susceptibility and resistance sensitivities and of SPV and RPV were the same for both platforms. The susceptibility sensitivity was 100% for both BLs-BLIs (95% CIs, 97%, 100% and 98%, 100%; Figure 2A). In contrast, resistance sensitivity estimates were lower: 66% (95% CI, 52%, 78%) for TOL/TAZO and 33% (95% CI, 20%–49%) for CZA (Figure 2B). The framework in which PRIMERS I–IV was designed allows us to interpret these data in a “real world” setting. Assuming 95% prevalence of susceptibility for both combinations, for example, obtained from local and current antibiograms (Figures 2C and 2D), our analysis revealed that the SPVs would be ≥97% across both platforms for both combinations (95% CIs, 97%, 99% and 96%, 97%). In contrast, the RPVs would be 72% (95% CI, 39%, 92%) and 60% (95% CI, 26%, 86%) for TOL/TAZO and CZA, respectively. In areas where, or time periods when, resistance to TOL/TAZO and CZA are higher (eg, a population of P. aeruginosa isolates with 80% susceptibility to TOL/TAZO and CZA; Figures 2E and 2F), we would expect SPVs of 92% (95% CI, 89%, 94%) for TOL/TAZO and 86% (95% CI, 83%, 88%) for CZA and RPVs of 93% (95% CI, 75%, 98%) with TOL/TAZO and 88% (95% CI, 62%, 97%) with CZA.
Figure 2.
Estimates of sensitivities and predictive values with 95% confidence intervals by drug and platform. Prevalence refers to either 95% or 80% prevalence of susceptibility, as indicated. Abbreviation: BC-GN, gram-negative blood culture test.
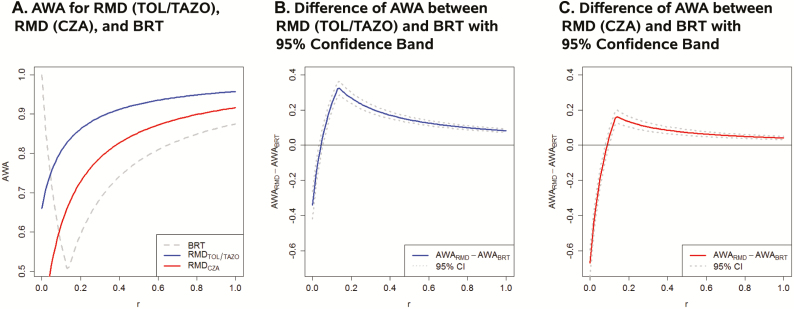
To assess the global utility of these strategies for using RMDs, we estimated the AWA over the relevant range of P. aeruginosa resistance (5%–20%) for each BL-BLI combination (Figure 3). The AWA for the Acuitas Resistome test and VERIGENE BC-GN platforms are equivalent given the same resistance and susceptibility sensitivities. The AWA for these platforms for TOL/TAZO and CZA are presented with that of the BRT as a function of the relative importance (r) of a false resistance determination relative to a false susceptibility determination (Figure 3A). The 95% confidence bands for the differences between the AWA for the platforms with the BRT as a function of r are provided for TOL/TAZO and CZA (Figure 3B and 3C). The platforms are observed to be statistically superior to the BRT when the relative importance of a false resistance is more than 4% of a false susceptible for TOL/TAZO and more than 9% of a false susceptible for CZA.
Figure 3.
A, Estimated platform average weighted accuracy (AWA) for ceftolozane/tazobactam (TOL/TAZO) and ceftazidime/avibactam (CZA) as a function of the relative importance (r) of a false resistance determination relative to a false susceptibility determination over the relevant prevalence range for Pseudomonas aeruginosa resistance (5%–20%). The AWA for the Acuitas Resistome test and VERIGENE gram-negative blood culture test platforms are equivalent given the same resistance and susceptibility sensitivities. The AWA for the best random test (BRT) is also presented. B, Estimated difference and associated 95% confidence bands in platform AWA for TOL/TAZO and the BRT as a function of the relative importance (r) of a false resistance determination relative to a false susceptibility determination over the relevant prevalence range for P. aeruginosa resistance (5%–20%). The platforms are statistically superior to the BRT when the relative importance of a false resistance is more than 4% of a false susceptibility determination for TOL/TAZO. C, Estimated difference and associated 95% confidence bands in platform AWA for CZA and the BRT as a function of the relative importance (r) of a false resistance determination relative to a false susceptibility determination over the relevant prevalence range for P. aeruginosa resistance (5%–20%). The platforms are statistically superior to the BRT when the relative importance of a false resistance is more than 9% of a false susceptibility determination for CZA. Abbreviations: AWA, average weighted accuracy; BRT, best random test; CZA, ceftazidime/avibactam; RMD, rapid molecular diagnostic; TAZO, tazobactam; TOL, ceftolozane.
Determination of the relative importance of diagnostic errors (r) is also an important area for development. Data-driven perspectives on the relative importance can be obtained from a survey of experts or via cost-effectiveness analyses of false-resistance vs false-susceptibility determinations. Acknowledging that perspectives may vary is important. Thus, we have conducted sensitivity analyses displaying how the diagnostic yield from using RMDs compares with alternatives, when varying the relative importance (Figure 3).
In this study it is important to note that when a resistance gene was found, the isolate was always resistant to TOL/TAZO and CZA (Table 3, Figure 4). Simply put, if a clinician finds 1 of the queried carbapenemase genes in these platforms (with the exception of blaKPC for CZA), they should avoid the use of TOL/TAZO or CZA and consider the use of alternative anti-Pseudomonas therapy. However, there were resistant isolates in this study that did not possess the targeted bla genes. This can be an unsettling situation particularly when one considers strains of P. aeruginosa that have upregulated efflux pumps, porin mutations, or hyperexpressed chromosomal β-lactamases. Additional work is needed to incorporate detection of these more complex resistance mechanisms into RMDs.
Figure 4.
Estimates of susceptibility and resistance predictive values for range of minimum inhibitory concentration susceptibility prevalence with 95% confidence interval bands by platform and drug. Abbreviations: BC-GN, gram-negative blood culture test; MIC, minimum inhibitory concentration; RPV, predictive value for resistance; SPV, predictive value for susceptibility.
The goals of PRIMERS I–IV were to develop analytical strategies that integrate detailed microbiologic data and genetic analyses in order to determine if RMD platforms can accurately identify resistance and susceptible phenotypes. In this context, PRIMERS IV was designed to obtain precise estimates of resistance sensitivities, susceptibility sensitivities, SPVs, RPVs, and AWAs to CZA and TOL/TAZO in order to determine if RMDs can assist with the decision to use each of these agents based upon the detection of certain resistance genes in P. aeruginosa. Both susceptible and resistant isolates were selected to reflect “real-world” clinical practice.
This study shows that the decision to use CZA or TOL/TAZO as empiric therapy against P. aeruginosa can be informed by results of RMDs, but that the complexity of nontested determinants of β-lactam resistance (eg, OprD changes and efflux) limit interpretations. However, these results could potentially be used for registration trial design using RMDs to tailor enrollment into a study evaluating CZA or TOL/TAZO.
Illustrative Case
To highlight the utility of this strategy, we present the following hypothetical clinical case study. A patient presents with a syndrome suggestive of healthcare-associated pneumonia while recovering in a surgical intensive care unit (SICU). A respiratory sample is obtained and sent to the clinical microbiology laboratory. Gram-negative bacilli and numerous white blood cells are noted on Gram stain. A 2-year retrospective microbiological survey (antibiogram) of P. aeruginosa isolates reveals that in this SICU, 20% of all P. aeruginosa are resistant to CZA and TOL/TAZO (80% susceptible). Processing the sample on an RMD platform identifies P. aeruginosa and does not identify blaKPC or blaVIM. Using the paradigms that we developed, there is a 92% (95% CI, 89%, 94%; Figures 2 and 4) chance of this isolate being susceptible to TOL/TAZO. However, if the RMD does detect 1 of the queried carbapenemase genes for each specific BL-BLI combination, a physician would not want to use that BL-BLI combination and should instead use an alternative anti-Pseudomonas therapy.
In conclusion, clinicians need to be mindful of the benefits and risks that result from treatment decisions that are based upon resistance gene detection in P. aeruginosa, acknowledging that such decisions are impacted by the prevalence and mechanisms of resistance, which can vary temporally and geographically. An important question emerges here: would one feel comfortable using empiric therapy when there is approximately a 10% or greater chance of choosing inadequate therapy?
We stress that the utility of the RMDs for guidance in the choice of antibiotic therapy for P. aeruginosa should be interpreted in the context of how well physicians choose empiric therapy when faced with a challenging clinical situation. Studies are needed to compare the clinical and antibiotic use outcomes that result from empirical treatment selection guided by RMDs vs empirical treatment selection guided by clinician decision alone. Biomarker trial designs, for example, biomarker stratified designs, enrichment designs, and biomarker strategy designs, can be used to address these questions [15]. For example, a randomized comparison of 2 strategies—at the initial stage an RMD is used to guide the empiric therapy decision followed by obtaining MICs and adjusting accordingly or at the initial stage using the clinician’s best decision, followed by obtaining MICs and adjusting accordingly—may inform us regarding the pragmatic utility of the use of RMDs to direct therapy. Thus, a thorough assessment of the clinical utility of RMDs needs to be measured not only by a comparison of their diagnostic accuracy statistics to that of clinician choice at the time of empiric antibiotic treatment but also by a comparison of the outcomes that result from these treatment decisions within the context of clinical practice.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health (NIH) or the Department of Veterans Affairs.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID) of the NIH (award UM1AI104681). Additional NIH support to R. A. B. includes R01AI100560, R01AI063517, and R01AI072219. This study was also supported in part by funds and/or facilities provided by the Department of Veterans Affairs (award 1I01BX001974 to R. A. B.), the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development, and the Geriatric Research Education and Clinical Center VISN 10, and by funds provided by Merck (Merck Investigator Studies Program 53818 to R. A. B.). S. R. E. and R. A. B. had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Potential conflicts of interest B. N. K. consults for Pfizer and Abbott. R. A. B. reports grants from Duke Clinical Research Institute during the conduct of the study and grants from Wockhardt, Merck, Entasis, Roche, GlaxoSmithKline (GSK), Allecra, and Shionogi outside the submitted work. R. P. reports grants from CD Diagnostics, BioFire, Curetis, Merck, Hutchison Biofilm Medical Solutions, Accelerate Diagnostics, Allergan, and the Medicines Company. R. P. is a consultant to Curetis; monies are paid to Mayo Clinic. In addition, R. P. reports grants from CD Diagnostics, BioFire, Curetis, Merck, Hutchison Biofilm Medical Solutions, Accelerate Diagnostics, Allergan, and the Medicines Company. R. P. is or has been a consultant to Curetis, Qvella, Specific Technologies, Selux Dx, GenMark Diagnostics, PathoQuest, and Genentech; monies are paid to Mayo Clinic. R. P. has a patent on Bordetella pertussis/parapertussis polymerase chain reaction issued, a patent on a device/method for sonication with royalties paid by Samsung to Mayo Clinic, and a patent on an anti-biofilm substance issued. R. P. has served on an Actelion data monitoring board; receives travel reimbursement from American Society for Microbiology (ASM) and the Infectious Diseases Society of America (IDSA); has received an editor’s stipend from ASM and IDSA; and has received honoraria from the National Board of Medical Examiners, Up-to-Date, and the Infectious Diseases Board Review Course. M. F. and K. M. P. are salaried employees of OpGen. M. F. and K. M. P. have patents US 20170253917, EP 3149639 A1, and EP 3117010 A2 pending. F. P. reports grants from Duke Clinical Research Institute during the conduct of the study and grants from Pfizer and Merck outside the submitted work. H. F. C. reports personal fees from Allergan outside the submitted work and personal fees from Allergan outside the submitted work. K. M. H., A. M. H., and T. N. D. report grants from Duke Clinical Research Institute during the conduct of the study. M. R. J. reports grants from ALRG during the conduct of the study. V. G. F. served as chair of the V710 Scientific Advisory Committee (Merck); has received grant support from Cerexa/Actavis/Allergan, Pfizer, Advanced Liquid Logics, NIH, MedImmune, Basilea Pharmaceutica, Karius, ContraFect, Regeneron Pharmaceuticals, and Genentech; has NIH Small Business Technology Transfer/Small Business Innovation Research grants pending with Affinergy, Locus, and Medical Surface, Inc; has been a consultant for Achaogen, AmpliPhi Biosciences, Astellas Pharma, Arsanis, Affinergy, Basilea Pharmaceutica, Bayer, Cerexa Inc., ContraFect, Cubist, Debiopharm, Durata Therapeutics, Grifols, Genentech, MedImmune, Merck, the Medicines Company, Pfizer, Novartis, NovaDigm Therapeutics Inc., Theravance Biopharma, Inc., and XBiotech; has received honoraria from Theravance Biopharma, Inc. and Green Cross; and has a patent pending in sepsis diagnostics. T. T. T. T. reports grants from NIAID during the conduct of the study. B. M. W. reports grants from Duke Clinical Research Institute during the conduct of the study. S. R. E. reports grants from NIAID/NIH during the conduct of the study; personal fees from Takeda/Millennium, Pfizer, Roche, Novartis, Achaogen, Huntington’s Study Group, Auspex, Alcon, Merck, Chelsea, Mannkind, QRx Pharma, ACTTION, Genentech, Affymax, FzioMed, Amgen, GSK, Boehringer-Ingelheim, American Statistical Association, the US Food and Drug Administration, Osaka University, City of Hope, National Cerebral and Cardiovascular Center of Japan, NIH, Muscle Study Group, Society for Clinical Trials, Drug Information Association, University of Rhode Island, New Jersey Medical School/Rutgers, PPRECISE, Statistical Communications in Infectious Diseases, Cubist, AstraZeneca, Teva, Repros, Austrian Breast & Colorectal Cancer Study Group/Breast International Group, the Alliance Foundation Trials, Zeiss, Dexcom, American Society for Microbiology, Taylor and Francis, Claret Medical, Vir, Arrevus, Five Prime, Shire, Alexion, Gilead, Spark, Clinical Trials Transformation Initiative, Nuvelution, and Tracon outside the submitted work. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. MacVane SH. Antimicrobial resistance in the intensive care unit: a focus on gram-negative bacterial infections. J Intensive Care Med 2017; 32:25–37. [DOI] [PubMed] [Google Scholar]
- 2. Wolter DJ, Lister PD. Mechanisms of β-lactam resistance among Pseudomonas aeruginosa. Curr Pharm Des 2013; 19:209–22. [PubMed] [Google Scholar]
- 3. Papp-Wallace KM, Bonomo RA. New β-lactamase inhibitors in the clinic. Infect Dis Clin North Am 2016; 30:441–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cabot G, Bruchmann S, Mulet X, et al. . Pseudomonas aeruginosa ceftolozane-tazobactam resistance development requires multiple mutations leading to overexpression and structural modification of AmpC. Antimicrob Agents Chemother 2014; 58:3091–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Takeda S, Ishii Y, Hatano K, Tateda K, Yamaguchi K. Stability of FR264205 against AmpC beta-lactamase of Pseudomonas aeruginosa. Int J Antimicrob Agents 2007; 30:443–5. [DOI] [PubMed] [Google Scholar]
- 6. van Duin D, Bonomo RA. Ceftazidime/avibactam and ceftolozane/tazobactam: second-generation β-lactam/β-lactamase inhibitor combinations. Clin Infect Dis 2016; 63:234–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Livermore DM. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare?Clin Infect Dis 2002; 34:634–40. [DOI] [PubMed] [Google Scholar]
- 8. Satlin MJ, Chen L, Patel G, et al. . Multicenter clinical and molecular epidemiological analysis of bacteremia due to carbapenem-resistant Enterobacteriaceae (CRE) in the CRE epicenter of the United States. Antimicrob Agents Chemother 2017; 61:e02349-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Evans SR, Hujer AM, Jiang H, et al. ; Antibacterial Resistance Leadership Group Rapid molecular diagnostics, antibiotic treatment decisions, and developing approaches to inform empiric therapy: PRIMERS I and II. Clin Infect Dis 2016; 62:181–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Evans SR, Hujer AM, Jiang H, et al. . Informing antibiotic treatment decisions: evaluating rapid molecular diagnostics to identify susceptibility and resistance to carbapenems against Acinetobacter spp. in PRIMERS III. J Clin Microbiol 2017; 55:134–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Perez F, Hujer AM, Marshall SH, et al. . Extensively drug-resistant Pseudomonas aeruginosa isolates containing blaVIM-2 and elements of Salmonella genomic island 2: a new genetic resistance determinant in northeast Ohio. Antimicrob Agents Chemother 2014; 58:5929–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. CLSI Performance Standards for Antimicrobial Susceptibility Testing. 28th Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute, 2017. [Google Scholar]
- 13. Mercaldo ND, Lau KF, Zhou XH. Confidence intervals for predictive values with an emphasis to case-control studies. Stat Med 2007; 26:2170–83. [DOI] [PubMed] [Google Scholar]
- 14. Evans SR, Pennello G, Pantoja-Galicia N, et al. ; Antibacterial Resistance Leadership Group Benefit-risk evaluation for diagnostics: a framework (BED-FRAME). Clin Infect Dis 2016; 63:812–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Evans S, Ting N.. Fundamental Concepts for New Clinical Trialists. Boca Raton, FL: Chapman & Hall/CRC Press Biostatistics Series, 2015. [Google Scholar]