Abstract
Background
Human immunodeficiency virus type 1 (HIV-1) populations are detected in cerebrospinal fluid (CSF) of some people on suppressive antiretroviral therapy (ART). Detailed analysis of these populations may reveal whether they are produced by central nervous system (CNS) reservoirs.
Methods
We performed a study of 101 asymptomatic participants on stable ART. HIV-1 RNA concentrations were cross-sectionally measured in CSF and plasma. In participants with CSF HIV-1 RNA concentrations sufficient for analysis, viral populations were genetically and phenotypically characterized over multiple time points.
Results
For 6% of participants (6 of 101), the concentration of HIV-1 RNA in their CSF was ≥0.5 log copies/mL above that of plasma (ie, CSF escape). We generated viral envelope sequences from CSF of 3 participants. One had a persistent CSF escape population that was macrophage-tropic, partially drug resistant, genetically diverse, and closely related to a minor macrophage-tropic lineage present in the blood prior to viral suppression and enriched for after ART. Two participants (1 suppressed and 1 not) had transient CSF escape populations that were R5 T cell-tropic with little genetic diversity.
Conclusions
Extensive analysis of viral populations in 1 participant revealed that CSF escape was from a persistently replicating population, likely in macrophages/microglia, present in the CNS over 3 years of ART. CSF escape in 2 other participants was likely produced by trafficking and transient expansion of infected T cells in the CNS. Our results show that CNS reservoirs can persist during ART and that CSF escape is not exclusively produced by replicating CNS reservoirs.
Keywords: CNS, CSF escape, HIV reservoirs, drug resistance, persistence
Analyses of human immunodeficiency virus type-1 in the cerebrospinal fluid of antiretroviral therapy (ART)-treated, virologically-suppressed people suggest these populations are produced by replicating central nervous system (CNS) reservoirs present during long-term ART or infected T cells trafficking into the CNS.
Antiretroviral therapy (ART) typically reduces plasma viral loads to levels that are undetectable by standard assays though often still detectable by ultrasensitive methods [1–3]. Sequence analyses of these low-level populations in the plasma [4, 5] and proviral DNA isolated from the blood [6] and lymph nodes [7] have not shown evidence of viral evolution during ART. Thus, our current understanding is that persistent viral reservoirs in the periphery during suppressive ART are not maintained by ongoing replication and that viral production during this time is likely due to reactivation of latently infected cells. Human immunodeficiency virus type 1 (HIV-1) RNA in the cerebrospinal fluid (CSF) of patients on suppressive ART may be due to reactivation from persistent reservoirs in the central nervous system (CNS) compartment or from cells that enter the CNS from the periphery. Ongoing replication of HIV in the CNS may occur in the context of subtherapeutic drug concentration in the CNS (reviewed by Yilmaz et al [8]) and/or poor immune control due to small numbers of CD8+ T cells in that compartment [9].
Analysis of DNA from CNS cells of infected people [10] and simian immunodeficiency virus (SIV)—or simian human immunodeficiency virus—infected macaques [11, 12] has shown viral DNA in the brain after extensive ART treatment. However, whether these proviral genomes are intact and therefore capable of generating rebound virus if ART were stopped is unknown. Recently, a study of macaques infected with a highly neuropathogenic SIV showed that macrophages isolated from the brain produced SIV in a quantitative viral outgrowth assay (QVOA) [13]. While the study clearly indicated that viral reservoirs can persist in the CNS during ART, CNS reservoirs in that model may not be representative of reservoirs in humans.
An alternative approach to studying CNS reservoirs is to examine viral populations in the CSF of ART-treated people. In this study, we identified people with viral suppression systemically but who had at least 0.5 log higher levels of HIV-1 RNA in their CSF than in their plasma—a condition termed CSF escape [14, 15]. We characterized viral populations in 3 participants who had CSF escape and concentrations of CSF RNA levels that were high enough to facilitate genetic analyses of the virus, allowing us to infer the source of CSF escape.
METHODS
Study Design
Participants were enrolled as part of the Tropism of HIV-1, Inflammation and NeuroCognition (THINC) Study. Participants were HIV-positive men and women aged ≥18 years on stable ART with a plasma viral load of <50 copies/mL for at least 12 months prior to enrollment with blips up to 200 copies/mL allowed. Participants had no change to their ART regimen for at least 3 months prior to enrollment. Individuals with active psychiatric illness or active brain infections were excluded. A total of 101 participants met these criteria and were enrolled for a cross-sectional study visit at 1 of 3 study sites: University of North Carolina at Chapel Hill (UNC), University of California at San Francisco, or Yale University (New Haven, CT). The study visit consisted of a blood draw, a lumbar puncture with CSF collection, and a neurocognitive assessment. Local institutional review boards at all 3 sites approved the study, and participants gave informed consent prior to study enrollment.
Sample Collection and Viral Load Analyses
Whole blood was collected from participants in EDTA. Blood plasma was separated from the whole blood pellet and stored at −80°C. Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll purification and viably stored. CSF was collected into uncoated tubes and immediately placed on ice. The CSF was then centrifuged to pellet cells. The supernatant was aliquoted into cryovials and stored at −80°C.
HIV-1 RNA levels were measured in blood plasma and CSF samples using assays with a lower limit of quantification of 40 (Abbott RealTime HIV-1 RNA assay) or 20 copies/mL (COBAS Ampliprep/COBAS Taqman version 2.0). Participants were defined as having CSF escape when viral load measured by standard assays indicated that their CSF viral load was >40 copies/mL and their plasma viral load was <40 copies/mL or their CSF viral load was ≥0.5 log plasma viral load.
QVOA Procedure
Resting CD4+ T cells were isolated from PBMCs using negative selection for CD4+ cells with depletion of CD25 high cells (custom kit, Stem Cell). Resting cells were cultured at a density of 100 000 cells/well in previously described outgrowth conditions [16]. On days 15 and 19, p24 concentration was measured, and positive supernatants were stored at −80°C.
Deep Sequencing
Illumina MiSeq 300 base paired-end multiplex library preparation was performed using our Primer ID approach that avoids resampling and corrects for mutations generated during polymerase chain reaction assay and sequencing [17]. Briefly, viral RNA was extracted from blood plasma and CSF, and cDNA was generated using a pool of 4 primers for the V1–V3 region of env and 3 regions of pol (partial RT, IN, and PR). Each cDNA primer included a random 11 base tag (Primer ID). cDNAs were amplified and sequenced using the Illumina MiSeq platform. A template consensus sequence (TCS) was generated for each Primer ID. TCSs were aligned (multiple sequence comparison by log-expectation [MUSCLE]), and neighbor-joining phylogenetic trees were generated.
Phenotyping
Single-genome amplification was performed as previously described [18] and full-length env genes were cloned into the pcDNA3.1D/V5-His-TOPO expression vector (Invitrogen) using the pcDNA 3.1 directional TOPO expression kit (Invitrogen). We used our established protocol [19, 20] to assess the ability of cloned env genes to facilitate entry of a pseudotyped reporter virus into cells expressing a low density of CD4 (a marker for macrophage tropism). Macrophage- and R5 T cell-tropic HIV-1 controls have been previously described [19, 21, 22].
PK Analyses
Antiretroviral concentrations in the plasma and the CSF were analyzed using liquid chromatography–tandem mass spectroscopy assays available at the UNC Center for AIDS Research Clinical Pharmacology and the Analytical Chemistry Laboratory at UNC [23].
Statistical Analyses
All statistical analyses were performed using R statistical software (version 3.3.3).
RESULTS
CSF Escape in the THINC Study
A total of 101 ART-treated, neurologically asymptomatic participants were enrolled in the THINC study from 2011 to 2017. The cohort was 86% male, 44% black or African American, 52% white, and 4% other. The median age was 50 (interquartile range, 42–55). Six participants (6%) had CSF escape, and the remaining participants had CSF and plasma viral loads below the limit of detection (Table 1). The median nadir CD4+ T-cell count of the cohort was 133 cells/mm3. Participants with CSF escape had lower CD4+ T-cell counts at study entry (Wilcoxon rank sum, P = .03). Participants with CSF escape had higher CSF white blood cell (WBC) counts, lower nadir CD4 T-cell counts, and shorter time on therapy, but these differences were not significant (P > .05). In this, and a previous study [14], it was found that some individuals with asymptomatic CSF escape had very low nadir CD4+ T-cell counts, but neither study found a statistically significant association between asymptomatic CSF escape and nadir CD4+ T-cell counts. In contrast, symptomatic CSF escape has been associated with low nadir CD4 T-cell counts [15, 24, 25].
Table 1.
Background and Clinical Characteristics
| Variable | 503 | 1018 | 3017 | 340 | 3025 | 3026 | Escape (Median)a | Suppressed (Median, Interquartile Range) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gender | Female | Male | Male | Male | Male | Male | … | … | ||||
| Race | Black or African American | Black or African American | Black or African American | American Indian/Alaska Native | White | White | … | … | ||||
| T1 | T2 | T1 | T2 | T1 | T2 | T3 | … | … | ||||
| Age (years) | 52 | 53 | 48 | 48 | 48 | 39 | 39 | 42 | 42 | 47 | 48 | 50 (42–56) |
| CSF VL (copies/mL) | 117 | 89 | 47 | 209 | 54 | 1295 | <40 | 265 | 356 | <40 | 117 | <40b |
| Plasma VL (copies/mL) | <40 | <40 | <40 | 62 | <40 | <40 | <40 | <40 | <40 | <40 | <40 | <40b |
| CD4 (cells/mm3) | 430 | 567 | 325 | 327 | 210 | 261 | 200 | 309 | 503 | 326 | 539 (389–693)b | |
| Nadir CD4 (cells/mm3) | 298 | 296 | 150 | 103 | 12 | 10 | 127 | 133 (34–253)c | ||||
| CSF white blood cell count (cells/mm3) | 3 | 134 | 1 | 1 | 87 | 4 | 2 | 1 | 2 | 2 | 1 (1–3)b | |
| Months on ART | 12 | 13 | 32 | 83 | 85 | 19 | 28 | 23 | 31 | 83 | 23 | 62 (19–123)d |
| ART regimen | EFV, FTC, TDF | EFV, FTC, TDF | EFV, FTC, TDF | DRV/r, TDF, FTC | DRV/r, TDF, FTC | DRV/r, TDF, FTC | DRV/r, TDF, FTC | DRV/r, TDF, FTC | DRV/r, TDF, FTC | BIC, FTC, TAF | … | … |
| ART regimen resistance mutations | N/A | N/A | N/A | N/A | N/A | None | N/A | M184V | M184V | N/A | … | … |
Abbreviations: ART, antiretroviral therapy; BIC, bictegravir; CSF, cerebrospinal fluid; DRV/r, ritonavir-boosted darunavir; EFV, efavirenz; FTC, emtricitabine; N/A, not measured; T1, time point 1; T2, time point 2; T3, time point 3; TDF, tenofovir disoproxil fumarate; VL, viral load.
aFor participants with multiple escape time points, an average value was generated and used to calculate the median.
bA total of 95 participants.
cA total of 94 participants.
dA total of 92 participants.
Among the 6 participants with CSF escape, the 3 with the highest CSF viral loads (participants 340, 3025, 3026) also had the lowest nadir CD4+ T-cell counts off ART but otherwise were not distinguishable from the other 3 CSF escape participants (Table 1). We further characterized the CSF virus from these 3 participants and collected follow-up CSF samples to evaluate the stability of the viral CSF escape populations. Since interpretation of the nature of the CSF escape virus varied between the cases, each is presented separately below.
Persistent CSF Escape in a Participant With Extensive Longitudinal Sampling
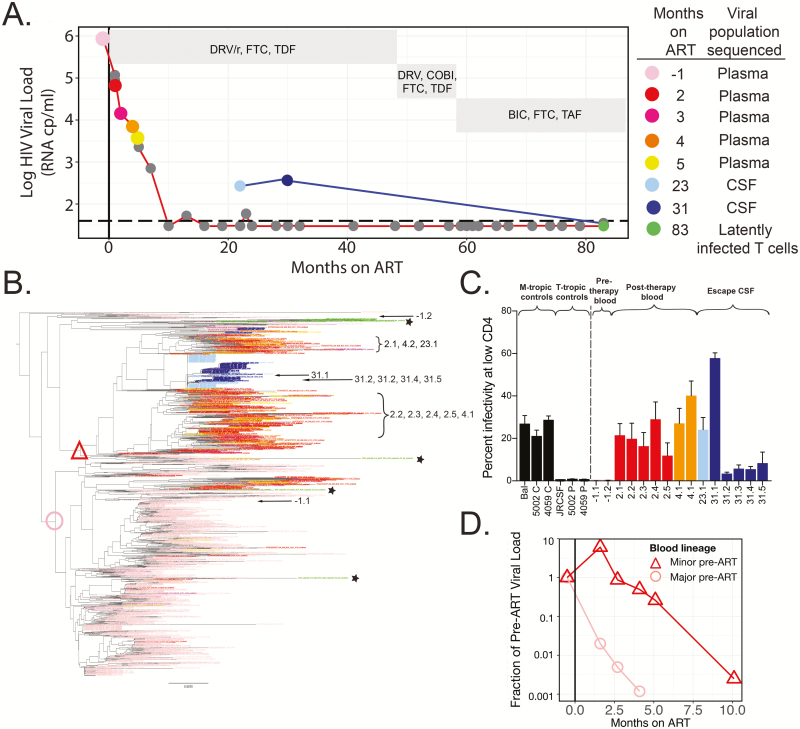
Participant 3026 initiated ART approximately 40 months after diagnosis (CD4 nadir = 10 cells/mm3). We sequenced the plasma virus population 2 weeks prior to ART initiation and at 4 time points post-ART initiation when plasma HIV RNA remained detectable (Figure 1A). In addition, this participant received 3 lumbar punctures while on ART, and we sequenced the CSF population at the 2 time points with CSF escape (ie, T1 and T2, 23 and 31 months post-ART). At the time of the third lumbar puncture (ie, T3, 83 months post-ART), the CSF viral load was undetectable after an earlier change in the drug regimen (see Figure 1A for timeline). At time point 3, resting CD4+ T cells were isolated from the blood and analyzed by QVOA, which gave a latent reservoir size of 2.7 infectious units per million resting CD4+ T cells.
Figure 1.
Persistent asymptomatic CSF escape variants are macrophage-tropic, produced by a population of infected cells in the central nervous system during ART, and genetically similar to macrophage-tropic variants detected in the plasma after ART initiation (participant 3026). A, HIV-1 viral loads in the plasma (values connected by a red line) and CSF (values connected by a blue line) were measured at multiple time points for participant 3026. HIV-1 was suppressed in the blood plasma after 10 months of ART, and CSF escape was observed after 23 and 31 months of ART but undetectable after 83 months of ART. Nongray dots indicate time points at which viral RNA was extracted from plasma and/or CSF and a MiSeq with Primer ID approach was used to generate partial env sequences or an outgrowth analysis was performed. ART regimens are shown in gray boxes. B, A neighbor-joining phylogenetic tree was constructed to compare partial env sequences from the plasma and CSF at multiple time points. Prior to ART, 92% of virus in the plasma formed a single major lineage (node marked with a pink circle), 4% of the plasma population was found in a separate, minor lineage (node marked with a red triangle), and the remaining virus was found in 4 small lineages representing approximately 4% of the pre-ART plasma population. After 2 months of ART, the initially rare lineage represented 82% of variants in the plasma. CSF escape variants were largely monophyletic and most closely related to the lineage that was rare pre-ART. Single-genome amplification was used to amplify 15 full-length env genes from the CSF and plasma at multiple time points (amplicon names indicate months on ART and are designated on the tree) and amplicons were cloned into expression vectors for entry phenotype analyses. C, Cloned HIV-1 envs were used to produce Env-pseudotyped reporter viruses and perform single-cycle infection of CD4lowCCR5high Affinofile cells to determine whether clones were macrophage-tropic. The entry phenotypes of previously characterized macrophage-tropic and T cell-tropic controls are shown. Two clones from the blood population pre-ART, one from the majority lineage pre-ART (pink circle) and another from elsewhere in the tree, were found to be T cell-tropic due to their inability to efficiently enter cells expressing a low density of CD4. In contrast, envs cloned from the initially rare lineage (red triangle) and CSF escape variants were all macrophage-tropic. D, Sequence and viral load data were used to estimate the viral load of the lineages at each time point and to plot the viral load of each lineage as a fraction of its pre-ART viral load. Prior to ART (to the left of the vertical line), each lineage starts at 1, but by 5 months post-ART, the initially common lineage (pink circle) had decayed 3 log10 from its pre-ART frequency, while the initially rare lineage (red triangle) only decayed 0.7 log10. This difference was not simply due to differences in the post-ART decay rate but also due to expansion of this population near the time of ART initiation. Abbreviations: ART, antiretroviral therapy; BIC, bictegravir; COBI, cobicistat; CSF, cerebrospinal fluid; DRV/r, ritonavir-boosted darunavir; FTC, emtricitabine; HIV, human immunodeficiency virus; TAF, tenofovir alafenamide; TDF, tenofovir disoproxil fumarate.
The origin of CSF escape was explored using genetic and phenotypic analyses. Deep sequencing revealed that populations present at the 2 time points were genetically distinct but highly related (Figure 1B; T1 in light blue and T2 in dark blue). In addition, CSF escape viruses at T1 and T2 had an M184V mutation, conferring resistance to emtricitabine (FTC); at T1, the CSF escape population also had a T215I mutation (a partial reversion of an azidothymidine [AZT] resistance marker) that was no longer detected by T2. Given that partial reversions at position 215 are transmitted in the absence of AZT and have at most a modest impact on viral fitness [26], the reason for the loss of the T215I allele from this CSF escape population is difficult to interpret. It may have been due to selection at linked sites, genetic drift, or the result of selective pressure to the wild-type threonine allele. Alternatively, in the absence of viral replication, T215I could have been lost from the CSF escape population if the population of cells producing virus at T1 was different from the population of cells producing virus at T2. While we cannot rule out the latter possibility, we view it as unlikely.
Analyses of full-length env genes from CSF escape populations revealed that they had a moderate to extremely high ability to use low levels of CD4 for entry (Figure 1C). This is a marker for macrophage tropism and indicative of viral adaptation to the CNS compartment, again consistent with a CNS source of this virus. Changes in viral diversity and drug resistance along with the observed macrophage-tropism are most consistent with a partially resistant, replicating population in CNS macrophage/microglia present 2 years after the initiation of ART. The ability of HIV-1 to establish macrophage-tropic populations in the CNS has been previously illustrated in studies of viral populations in CSF collected from untreated people, often with HIV-associated dementia [22, 27–32]. This raises the possibility that once established, a macrophage-tropic population may persist in the CNS of some ART-treated people and possibly give rise to CSF escape virus.
In an effort to understand the origin of the persistently replicating virus in the CNS, we examined virus in the blood plasma just before therapy (Figure 1A; light pink symbol) and during the initial slow decay period on therapy (Figure 1A; red, dark pink, orange, yellow symbols). Using deep sequencing, we found a dramatic shift in the composition of the viral population in the blood plasma with the initiation of therapy, with the major lineage found in the plasma 2 weeks prior to ART largely absent from the plasma after 2 months on ART (Figure 1B; same color scheme as Figure 1A). Conversely, after 2 months on ART, the plasma was dominated by what had initially been a very minor population prior to ART (designated at its internal node in the tree with a red triangle) and one that was closely related to the replicating CSF escape population found nearly 2 years later. Figure 1D illustrates that 2 months after ART initiation, this initially rare blood plasma population before ART became the dominant population. This suggests that the initially minor lineage was expanding when ART was initiated or that infected cells continued to produce these variants as the overall viral load dropped with ART. As with the CSF escape population, this minor lineage in the blood was also macrophage-tropic, while the major population in the blood required high levels of CD4 for entry and was thus R5 T cell-tropic. This macrophage-tropic lineage in the blood was drug sensitive, which is more consistent with viral production from long-lived cells than persistent viral replication. Finally, we sequenced 15 outgrowth viruses from the QVOA (Figure 1B; green sequences designated with asterisks) and found that they clustered with R5 T cell-tropic variants and were not found in the lineage containing macrophage-tropic variants (red triangle). Given that QVOA viruses were both cultured from CD4+ T cells and most closely related to T cell-tropic viruses in the phylogenetic tree, we conclude that QVOA viruses are most likely R5 T cell-tropic.
Episodic CSF Escape
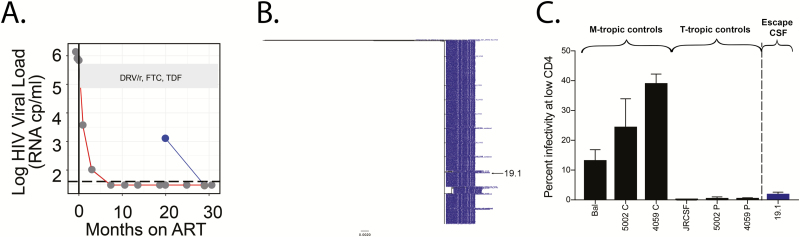
Participant 3025 initiated ART approximately 84 months after diagnosis (CD4 nadir = 10 cells/mm3). This participant received 2 lumbar punctures as part of the THINC study (at 19 and 28 months after ART initiation). There was CSF escape virus at the first time point, with elevated CSF WBC, but virus was undetectable at the time of the second lumbar puncture (Figure 2A and Table 1). At the CSF escape time point, the viral population was extremely homogeneous (Figure 2B), drug sensitive, and, based on the V3 sequence, capable of using either CXCR4 or CCR5 for entry. We cloned a single env gene from this population and found that it was inefficient at facilitating entry into cells expressing a low density of CD4 and therefore T cell-tropic (Figure 2C). Because the population was homogeneous, this env gene represented the phenotype of the entire CSF escape population. The lack of sequence diversity, the transient nature of the virus in the CSF, and the lack of resistance mutations suggest this viral population was not the product of ongoing viral replication. The high CSF viral load makes it unlikely that this variant was produced by a single cell but instead was released from a population of infected, clonally expanded T cells that reactivated in the CNS.
Figure 2.
Episodic asymptomatic CSF escape variants are most likely produced by clonally expanded CD4+ T cells present in the CNS during ART (participant 3025). A, HIV-1 viral loads in the plasma (values connected by a red line) and CSF (values connected by a blue line) were measured at multiple time points for participant 3025. CSF escape was observed after 19 months of ART but was undetectable at 27 months post-ART. A MiSeq with Primer ID approach was used to generate partial env sequences from the CSF escape population (blue dot). ART regimen is shown in the gray box. B, A neighbor-joining phylogenetic tree of 733 partial env sequences illustrates that the CSF escape population was nearly homogeneous. C, A full-length HIV-1 env cloned from this homogeneous population was pseudotyped and shown to be T cell-tropic in a CD4lowCCR5high Affinofile entry assay. Abbreviations: ART, antiretroviral therapy; CSF, cerebrospinal fluid; DRV/r, ritonavir-boosted darunavir; FTC, emtricitabine; HIV, human immunodeficiency virus; TDF, tenofovir disoproxil fumarate.
CSF Escape in a Poorly Suppressed Participant
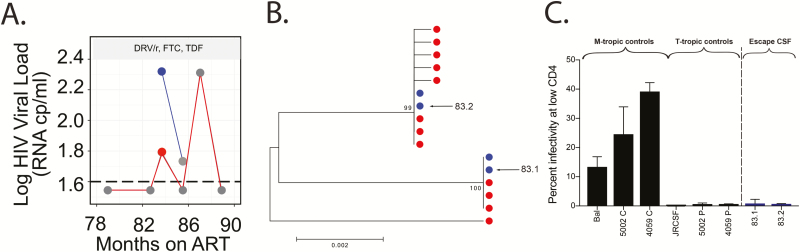
Participant 340 initiated ART approximately 14 years after diagnosis, at which time his CD4+ T-cell count was 103 cells/mm3. CSF escape was identified 84 months after ART initiation at a time when low-level virus was detected in the blood (Figure 3A and Table 1). The small number of sequences sampled from the CSF and plasma at the first time point were relatively homogeneous and largely mixed between the 2 compartments (Figure 3B). Entry analysis and V3 sequencing revealed that they were R5 T cell-tropic (Figure 3C). The fact that the CSF and plasma contained identical variants is most easily explained by virus production from clonally expanded, infected T cells trafficking into the CNS.
Figure 3.
The source of asymptomatic CSF escape in a poorly suppressed participant is unclear (participant 340). A, HIV-1 viral loads in the plasma (values connected by a red line) and CSF (values connected by a blue line) were measured at multiple time points for participant 340. Nongray dots indicate the time point at which viral RNA was extracted from plasma and/or CSF, and single-genome amplification (SGA) was used to generate full-length env genes. Antiretroviral therapy regimen is shown in the gray box. B, A neighbor-joining phylogenetic tree of full-length env sequences illustrates that identical sequences are found in the CSF and blood plasma. C, Full-length HIV-1 envs cloned from the CSF population were pseudotyped and shown to be T cell-tropic based on an inability to efficiently enter Affinofile cells expressing a low surface density of CD4. Abbreviations: ART, antiretroviral therapy; CSF, cerebrospinal fluid; DRV/r, ritonavir-boosted darunavir; FTC, emtricitabine; HIV, human immunodeficiency virus; TDF, tenofovir disoproxil fumarate.
Analysis of Drug Concentrations
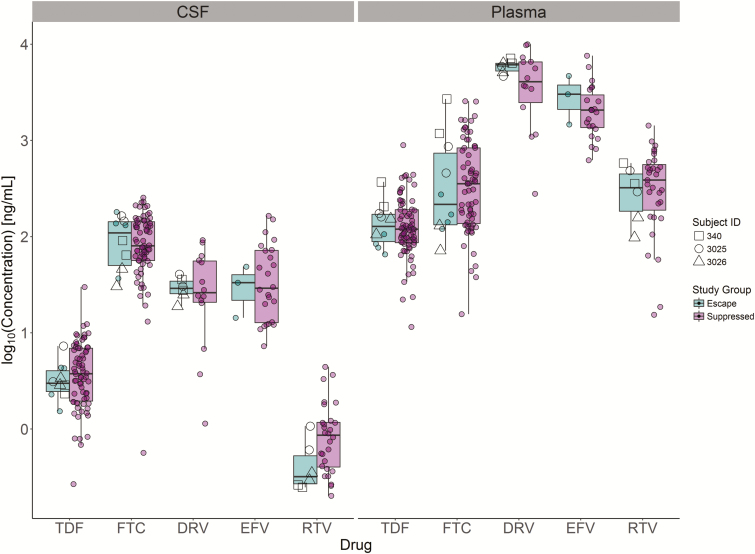
Analysis of 5 drugs common in the THINC cohort revealed that antiviral drug concentrations were not lower in the CSF of the 6 individuals with CSF escape (Figure 4; Kruskal-Wallis rank sum test, P > .05). This suggests that the detection of CSF escape virus in this study was not the result of especially low drug exposure in a subset of participants. It is, however, worth noting that the 3 participants with the highest CSF escape viral loads (participants 340, 3025, and 3026) were all on boosted protease inhibitor–based regimens.
Figure 4.
Individuals with CSF escape had drug concentrations similar to those of well-suppressed individuals. CSF and plasma collected from the THINC (Tropism of HIV-1, Inflammation and NeuroCognition) cohort were analyzed for the concentration of 5 of the most common drugs used in the cohort. As expected, CSF drug concentrations were lower than that of plasma. On average, drug concentrations were not lower in the individuals with CSF escape (green) relative to individuals who were virologically suppressed (purple). Abbreviations: CSF, cerebrospinal fluid; DRV, darunavir; EFV, efavirenz; FTC, emtricitabine; RTV, ritonavir; TDF, tenofovir disoproxil fumarate.
DISCUSSION
We observed that 6% of individuals in this cohort had asymptomatic CSF escape. This is similar to a previous cross-sectional analysis of neurologically asymptomatic people that found that 10% of individuals receiving combination ART had asymptomatic CSF escape [14]. We performed extensive genetic and phenotypic analyses of CSF viral populations in 3 people with CSF viral escape to determine whether these populations were the result of a persistent viral reservoir in the CNS. To our knowledge, this and our previous study of a single participant with symptomatic CSF escape [33] are the only studies that have phylogenetically examined CSF escape populations, and this is the first study to assess entry phenotypes of CSF escape virus. Combined, these approaches give new insight into the origin of CSF escape virus.
One participant (3026) had strong evidence of CSF escape produced by persistent viral replication in the CNS (Figure 1). The CSF escape population in this participant was observed 8 months apart (23 and 31 months post-ART). At both times, the blood plasma viral load was <40 copies/mL, indicating no detectable systemic virus production/replication. This participant did, however, have plasma blips near the time of escape, which is consistent with previous studies showing that low-level plasma viremia is associated with CSF escape [14, 25]. Analysis of viral entry phenotype revealed that the CSF escape virus was macrophage-tropic. This rare entry phenotype has primarily been observed in virus populations isolated from the CNS (reviewed by Joseph and Swanstrom [34]). Together, these observations indicate that this participant had a population of HIV-infected cells that persisted in the CNS after 31 months of ART and that the cells supporting viral replication were most likely macrophage/microglia. The observed changes in env genetic diversity and drug resistance between the first and second CSF escape time points further indicate that the escape population not only persisted during this period but was replicating and evolving. While most evidence suggests that ART suppresses viral replication of drug-sensitive variants [6, 35], the CSF escape variants had an M184V mutation that made them highly resistant to FTC that was part of their ART regimen. Given that his CSF contained a low concentration of the other drugs in his regimen (ritonavir-boosted darunavir and tenofovir disoproxil fumarate) relative to concentrations in the blood plasma (Figure 4), resistance to FTC may have been sufficient to allow replication in the CNS.
We observed that as the plasma viral load declined after ART initiation, the plasma virus population shifted from the T cell-tropic population that dominated the plasma pre-ART to a macrophage-tropic lineage that was a minor lineage pre-ART and was closely related to the CSF escape populations. The simplest explanation is that the macrophage-tropic population detected in the plasma post-ART is ancestral to the CSF escape population and was replicating in macrophage/microglia in the CNS (and possibly infected macrophages in other tissues) before ART initiation. After ART initiation, these long-lived cells continued producing virus that could be detected in the plasma before ultimately decaying to levels that could no longer be detected in the plasma. During this post-ART period, viral production in the CNS (with its low drug exposure) allowed the evolution of FTC resistance.
We also examined CSF escape populations from 2 additional participants with asymptomatic CSF escape (Figures 2 and 3). In both cases, the viral CSF escape population was relatively homogeneous and T cell-tropic. In addition, the 1 participant that we were able to test for drug resistance (participant 3025) had virus that was drug sensitive. Both examples of CSF escape are most likely due to virus production from clonally expanded T cells that traffic into the CNS, not virus production from long-lived reservoirs in the CNS. In contrast, it is highly unlikely that the persistent CSF escape population in participant 3026 was produced by cells migrating into the CNS from the periphery. Since the vast majority of variants in the periphery pre-ART are T cell-tropic, production of a diverse macrophage-tropic CSF escape population would require that many cells migrate into the CNS during ART and release an extremely rare type of HIV-1 without releasing the typical T cell-tropic HIV-1.
Our in-depth analysis of 3 HIV-infected participants illustrates that asymptomatic CSF viral escape can be generated by at least 2 mechanisms and that a subset of people may have CSF escape populations produced by an actively replicating CNS reservoir. We were only able to identify CNS reservoirs that are being expressed at the time of CSF sampling and accumulate in the CSF at a sufficiently high concentration to allow detection. Despite these limitations, this study provides the first genotypic and phenotypic characterization of a replication-competent viral reservoir in the CNS of an ART-suppressed person.
Notes
Financial support. This work was supported by National Institutes of Health (NIH) grants (P01 MH094177 to R. S. and R01 NS094067 to R. W. P.). The work was also supported by the University of North Carolina at Chapel Hill (UNC) Center for AIDS Research (NIH award P30 AI050410) and the UNC Lineberger Comprehensive Cancer Center (NIH award P30 CA16068). N. M. B. received travel support to present study data at a conference. N. A.’s institution received grant funding from the National Institute of Allergy and Infectious Diseases.
Potential conflicts of interest. The UNC is pursuing intellectual property protection for Primer ID (Cellular Research, Inc.), and R. S. is listed as a coinventor and has received nominal royalties from Cellular Research, Inc. J. J. E. received payment for consultancy to Merck, Gilead Sciences, Janssen, and ViiV Healthcare; J. J. E.’s institution has received grants/grants pending from ViiV Healthcare, Janssen, and Gilead Sciences outside the submitted work. M. G. has received grants from Gilead, has served on scientific advisory boards for Merck Sharp & Dohme, Gilead, and GlaxoSmithKline/ViiV, and has received personal fees from Gilead, Janssen, and Bristol-Myers Squibb outside the submitted work. S. S. directed a clinical trial for which ViiV Healthcare provided study medication, and S. S.’s institution has received grants/grants pending from National Institute of Mental Health and National Institute of Neurological Disorders and Stroke at NIH. All other authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Dinoso JB, Kim SY, Wiegand AM, et al. Treatment intensification does not reduce residual HIV-1 viremia in patients on highly active antiretroviral therapy. Proc Natl Acad Sci U S A 2009; 106:9403–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Doyle T, Smith C, Vitiello P, et al. Plasma HIV-1 RNA detection below 50 copies/ml and risk of virologic rebound in patients receiving highly active antiretroviral therapy. Clin Infect Dis 2012; 54:724–32. [DOI] [PubMed] [Google Scholar]
- 3. Maldarelli F, Palmer S, King MS, et al. ART suppresses plasma HIV-1 RNA to a stable set point predicted by pretherapy viremia. PLoS Pathog 2007; 3:e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hermankova M, Ray SC, Ruff C, et al. HIV-1 drug resistance profiles in children and adults with viral load of <50 copies/ml receiving combination therapy. JAMA 2001; 286:196–207. [DOI] [PubMed] [Google Scholar]
- 5. Kieffer TL, Finucane MM, Nettles RE, et al. Genotypic analysis of HIV-1 drug resistance at the limit of detection: virus production without evolution in treated adults with undetectable HIV loads. J Infect Dis 2004; 189:1452–65. [DOI] [PubMed] [Google Scholar]
- 6. Van Zyl GU, Katusiime MG, Wiegand A, et al. No evidence of HIV replication in children on antiretroviral therapy. J Clin Invest 2017; 127:3827–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McManus WR, Bale MJ, Spindler J, et al. No evidence for ongoing HIV replication in lymph nodes during suppressive ART. Boston: CROI, 2018. [Google Scholar]
- 8. Yilmaz A, Price RW, Gisslén M. Antiretroviral drug treatment of CNS HIV-1 infection. J Antimicrob Chemother 2012; 67:299–311. [DOI] [PubMed] [Google Scholar]
- 9. Ousman SS, Kubes P. Immune surveillance in the central nervous system. Nat Neurosci 2012; 15:1096–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lamers SL, Rose R, Maidji E, et al. HIV DNA is frequently present within pathologic tissues evaluated at autopsy from combined antiretroviral therapy-treated patients with undetectable viral loads. J Virol 2016; 90:8968–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clements JE, Gama L, Graham DR, Mankowski JL, Zink MC. A simian immunodeficiency virus macaque model of highly active antiretroviral treatment: viral latency in the periphery and the central nervous system. Curr Opin HIV AIDS 2011; 6:37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Estes JD, Kityo C, Ssali F, et al. Defining total-body AIDS-virus burden with implications for curative strategies. Nat Med 2017; 23:1271–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Avalos CR, Abreu CM, Queen SE, et al. Brain macrophages in simian immunodeficiency virus-infected, antiretroviral-suppressed macaques: a functional latent reservoir. mBio 2017; 8:doi: 10.1128/mBio.01186-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Edén A, Fuchs D, Hagberg L, et al. HIV-1 viral escape in cerebrospinal fluid of subjects on suppressive antiretroviral treatment. J Infect Dis 2010; 202:1819–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ferretti F, Gisslen M, Cinque P, Price RW. Cerebrospinal fluid HIV escape from antiretroviral therapy. Curr HIV/AIDS Rep 2015; 12:280–8. [DOI] [PubMed] [Google Scholar]
- 16. Crooks AM, Bateson R, Cope AB, et al. Precise quantitation of the latent HIV-1 reservoir: implications for eradication strategies. J Infect Dis 2015; 212:1361–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou S, Jones C, Mieczkowski P, Swanstrom R. Primer ID validates template sampling depth and greatly reduces the error rate of next-generation sequencing of HIV-1 genomic RNA populations. J Virol 2015; 89:8540–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ping LH, Joseph SB, Anderson JA, et al. ; CAPRISA Acute Infection Study and the Center for HIV-AIDS Vaccine Immunology Consortium. Comparison of viral Env proteins from acute and chronic infections with subtype C human immunodeficiency virus type 1 identifies differences in glycosylation and CCR5 utilization and suggests a new strategy for immunogen design. J Virol 2013; 87:7218–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Joseph SB, Arrildt KT, Swanstrom AE, et al. Quantification of entry phenotypes of macrophage-tropic HIV-1 across a wide range of CD4 densities. J Virol 2014; 88:1858–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Joseph SB, Lee B, Swanstrom R. Affinofile assay for identifying macrophage-tropic HIV-1. Bio-Protocol 2014; 4:e1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Arrildt KT, LaBranche CC, Joseph SB, et al. Phenotypic correlates of HIV-1 macrophage tropism. J Virol 2015; 89:11294–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schnell G, Joseph S, Spudich S, Price RW, Swanstrom R. HIV-1 replication in the central nervous system occurs in two distinct cell types. PLoS Pathog 2011; 7:e1002286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rezk NL, Tidwell RR, Kashuba AD. High-performance liquid chromatography assay for the quantification of HIV protease inhibitors and non-nucleoside reverse transcriptase inhibitors in human plasma. J Chromatogr B Analyt Technol Biomed Life Sci 2004; 805:241–7. [DOI] [PubMed] [Google Scholar]
- 24. Peluso MJ, Ferretti F, Peterson J, et al. Cerebrospinal fluid HIV escape associated with progressive neurologic dysfunction in patients on antiretroviral therapy with well controlled plasma viral load. AIDS 2012; 26:1765–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nightingale S, Geretti AM, Beloukas A, et al. Discordant CSF/plasma HIV-1 RNA in patients with unexplained low-level viraemia. J Neurovirol 2016; 22:852–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Garcia-Lerma JG, Nidtha S, Blumoff K, Weinstock H, Heneine W. Increased ability for selection of zidovudine resistance in a distinct class of wild-type HIV-1 from drug-naive persons. Proc Natl Acad Sci U S A 2001; 98:13907–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Duenas-Decamp MJ, Peters PJ, Burton D, Clapham PR. Determinants flanking the CD4 binding loop modulate macrophage tropism of human immunodeficiency virus type 1 R5 envelopes. J Virol 2009; 83:2575–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dunfee RL, Thomas ER, Gorry PR, et al. The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia. Proc Natl Acad Sci U S A 2006; 103:15160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gorry PR, Taylor J, Holm GH, et al. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J Virol 2002; 76:6277–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martín-García J, Cao W, Varela-Rohena A, Plassmeyer ML, González-Scarano F. HIV-1 tropism for the central nervous system: brain-derived envelope glycoproteins with lower CD4 dependence and reduced sensitivity to a fusion inhibitor. Virology 2006; 346:169–79. [DOI] [PubMed] [Google Scholar]
- 31. Peters PJ, Bhattacharya J, Hibbitts S, et al. Biological analysis of human immunodeficiency virus type 1 R5 envelopes amplified from brain and lymph node tissues of AIDS patients with neuropathology reveals two distinct tropism phenotypes and identifies envelopes in the brain that confer an enhanced tropism and fusigenicity for macrophages. J Virol 2004; 78:6915–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peters PJ, Sullivan WM, Duenas-Decamp MJ, et al. Non-macrophage-tropic human immunodeficiency virus type 1 R5 envelopes predominate in blood, lymph nodes, and semen: implications for transmission and pathogenesis. J Virol 2006; 80:6324–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Trunfio M, Joseph SB, Ghisetti V, et al. Symptomatic cerebrospinal fluid HIV-1 escape with no resistance-associated mutations following low-level plasma viremia. J Neurovirol 2018; 24:132–6. [DOI] [PubMed] [Google Scholar]
- 34. Joseph SB, Swanstrom R. The evolution of HIV-1 entry phenotypes as a guide to changing target cells. J Leukoc Biol 2018; 103:421–31. [DOI] [PubMed] [Google Scholar]
- 35. Kearney MF, Spindler J, Shao W, et al. Lack of detectable HIV-1 molecular evolution during suppressive antiretroviral therapy. PLoS Pathog 2014; 10:e1004010. [DOI] [PMC free article] [PubMed] [Google Scholar]